Introduction
Sepsis is an emergency systemic illness that is due
to the combined effects of the under- and overactivity of the
immune systems of patients. Initial symptoms of sepsis include
multiple organ impairment and can possibly result in severe
secondary infections. The immune systems of patients react to
infection as a response to high febricity, decreased blood pressure
and organ dysfunction, which was defined in the 1970s-1980s
(1). In the mid-last century, a
previous study on microbiology and immunology showed that several
hallmarks of infectious diseases were not only generally caused by
the body's own immune response but also by invading pathogens
(2). Previously, life-threatening
sepsis was shown to result in organ dysfunction; the definition of
the new Sepsis-3 indicates a 60% possibility of mortality within 28
days (3-6).
Bacterial endotoxins, including lipopolysaccharide
(LPS), are potential inducers of inflammation. These endotoxins
induce a complex pathogen-host interaction that enables the host to
respond to infection and results in hyperinflammation. LPS affects
innate and acquired immune responses in several systems, such as
the nervous, respiratory, circulatory, endocrine and metabolic
systems (7). Inflammation of the
vessels and endocardium, and persistent activation of inflammatory
signals are the initial signs of cardiac dysfunction in sepsis
(8). The role and molecular
mechanism of inflammatory signal transduction in the myocardial
injury of sepsis are important to elucidate the molecular mechanism
of this disease.
Molecular research has further hinted at the
underlying mechanisms of sepsis. A genome-wide association (GWA)
study and high-throughput omic technologies have facilitated
studies on the diagnosis of sepsis by screening sepsis-specific
molecular indicators as biomarkers. Common sepsis biomarkers
include high-mobility group box 1, C-reactive protein, tumor
necrosis factor (TNF)-α, interleukin (IL)-1, IL-6, IL-10 and
macrophage migration inhibitory factor (9-16).
GWA revealed that the FER tyrosine kinase (FER) gene serves
an important role in intercellular signaling in patients with
sepsis (17), as well as vacuolar
protein sorting 13 homolog A and cysteine rich secretory protein
LCCL domain containing 2, which may be involved in survival after
28 days (18). Methylthioadenosine
is a sepsis biomarker that is involved in the inflammatory response
and is related to high rates of fever-induced host cell death
(19). Several previous studies have
shown that sepsis is associated with the inflammatory response in
the heart (8,20,21), but
the detailed molecular mechanism remains unclear. The Wnt and
inflammatory response pathways were associated with T cell-specific
transcription factor (TCF)/lymphoid enhancer-binding factor (LEF)
(22,23). The typical Wnt signaling pathway,
also known as the β-catenin pathway or β-catenin/TCF pathway
(24), affects a wide array of
signaling channels (25-27).
A major characteristic of the WNT/β-catenin pathway includes the
stabilization of cytosolic β-catenin. Before stimulation, β-catenin
is phosphorylated by a destruction complex that contains glycogen
synthase kinase-3β (GSK3β); phosphorylated β-catenin becomes
ubiquitinated and is degraded by the proteasome (23,28). The
stimulation of Wnt signaling reduces GSK3β activity, thereby
resulting in the accumulation of β-catenin in the nucleus, which
results in the transcription of Wnt target genes by TCF/LEF to
regulate biological processes, such as cell proliferation,
differentiation and survival (23).
Furthermore, Wnt signaling has been reported to be associated with
inflammatory response signaling to control disease (28), but whether Wnt signaling is involved
in sepsis development has not yet been investigated.
In the present study transcriptome changes were
analyzed using heart tissues from LPS-injected mice as a sepsis
model. Furthermore, the differentially expressed genes were
classified by Gene Ontology (GO) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) terms. Among the pathways enriched, Wnt
signaling was further examined for its roles in the regulation of
the inflammatory responsive pathway and cardiac tissue. The present
results might provide a theoretical basis for improving the
clinical treatment protocol of sepsis in the future.
Materials and methods
Ethics statement
Animals were obtained from The Laboratory Animal
Center of Fuzhou Wushi Animal Center. All animal studies were
performed according to the National Institutes of Health Guide
Concerning the Care and Use of Laboratory Animals (29) with the approval of the Animal
Experimentation Ethics Committee of The Affiliated Hospital of
Putian University.
Animal model treatments
A total of 90 female C57BL/6 mice (age, 8 weeks;
weight, 220-250 g) were purchased from The Nanjing Animal Center.
All mice were housed in cages in an SPF animal room with a
temperature of 20±2˚C and a humidity of 55±5% under a 12-h
light/dark cycle. All rats had free access to food and water. The
animals received LPS in sterile PBS by intraperitoneal injection.
In total, 50 of the 90 mice were divided into the following five
groups (n=10): Control group (intraperitoneal injection with 0.9%
saline solution) and four LPS treatment groups (intraperitoneal
injection with LPS at 50 mg/kg body weight for 1, 2, 3 and 4
days).
The remaining 40 mice were used to investigate
whether the Wnt signaling is involved in LPS-induced heart damage.
In the treatment group, mice weighing 220-250 g were injected with
LPS (50 mg/kg body weight), LPS + Wnt3a (100 ng/kg body weight) or
IWR (100 ng/kg body weight). Mice in the control group were
administered an equivalent volume of vehicle (DMSO) for 1 h before
LPS injection. Mice with matched ages, sexes and weights were
randomly divided into the normal control group (10 mice), the LPS
model group (10 mice), LPS+Wnt3a group (10 mice) and IWR-injected
group (10 mice). LPS from Escherichia coli serotypes
0111:B4, Wnt3a and IWR were all purchased from Sigma-Aldrich; Merck
KGaA. The mice were sacrificed by an intraperitoneal injection of
sodium pentobarbital (150 mg/kg) following the AVMA Guidelines for
the Euthanasia of Animals (30).
Death was then confirmed by a certain set of criteria: Respiratory
arrest, cardiac arrest, dilation of the pupils and disappearance of
nerve reflex.
Histopathological observation of
damaged myocardial tissue
The heart samples were taken immediately after 4
days of DMSO, LPS, Wnt3a or IWR injection, and were subsequently
fixed in 10% formaldehyde solution for 20 min at room temperature.
The tissue was dehydrated, and paraffin embedded, and pathological
sections were cut at 4-µM thickness. Hematoxylin and eosin (HE)
staining for 20 min at room temperature was performed to analyze
the pathological events of the myocardium under a light microscope
at x200 magnification to examine heart muscle arrangement.
RNA isolation and cDNA reverse
transcription
Ground heart tissues (30 min post stimulation) were
preserved in TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific Inc.) at -80˚C until use. Total RNA isolation was
performed by following the TRIzol® manufacturer's
protocol with an RNeasy Mini kit (Qiagen, Inc.) for RNA
purification. DNase I. SuperScript III First-Strand Synthesis
SuperMix (Thermo Fisher Scientific, Inc.) was used for reverse
transcription-quantitative PCR (RT-qPCR) to produce cDNA from 2 µg
total RNA. The reaction protocol consisted of 25˚C for 5 min, 42˚C
for 60 min and 70˚C for 15 min.
RT-qPCR
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. Subsequently, the selected results
from RNA-sequencing (RNA-seq) were verified by RT-qPCR analysis.
cDNA was produced using a Prime Script™ RT-qPCR kit
(Takara Bio, Inc.) according to the manufacturer's protocol. qPCR
was performed using SYBR® Premix Ex Taq™ (Takara Bio,
Inc.) on a 7900HT fast RT-qPCR instrument (Applied Biosystems;
Thermo Fisher Scientific, Inc.) with technical triplicates. The
gene-specific primers used for RT-qPCR are listed in Table I. Thermocycling conditions were as
follows: Initial denaturation at 95˚C for 5 min, followed with 35
cycles at 95˚C for 30 sec and 60˚C for 30 sec. The relative gene
expression levels were determined using the
2-ΔΔCq method (31).
 | Table IPrimer sequences for reverse
transcription PCR. |
Table I
Primer sequences for reverse
transcription PCR.
| Gene | Forward Primer
sequences, 5'-3' | Reverse Primer
sequences, 5'-3' |
|---|
| FZD8 |
TCTACAACCGCGTCAAGACC |
GGCCGTTCCGGGTACTTAAA |
| PAI-1 |
AGGCTGGTCCTCGTTAATGC |
CACAGAGAGCTGAGCCAACA |
| IL-8 |
TGTGTGCATAGCCATGTGGT |
GAACAGCATGATGAGCAGCG |
| FGF21 |
AGCATACCCCATCCCTGACT |
TCCTCCCTGATCTCCAGGTG |
| GAPDH |
AGGCCGGTGAGTATGC |
TGCCTGCACCACCTTC |
Sequencing analysis
The RNA was extracted using RNAiso Reagent (Takara
Biotechnology Co., Ltd.) according to the manufacturer's
instructions. The concentration and purity of RNA were determined
by Agilent 2100 Bioanaylzer (Agilent Technologies, Inc.). All the
samples had RNA integrity numbers of >8.5 and concentration
>200 ng/µl. Sequencing libraries were generated using TIANSeq
Fast RNA Library Kit for Illumina® (cat. no. NR102;
Tiangen Biotech Co., Ltd.) following the manufacturer's protocols.
After completing the library, paired-end (150 bp) sequencing of the
cDNA libraries (10 nM) was performed on the GAIIx instrument
(Illumina, Inc.) using the reagents provided in the TruSeq PE
Cluster Kit v5-CS-GA (cat. no. PE-203-5001; Illumina, Inc.). A
standard analysis protocol was performed (32), and sequencing data were aligned to
the hg19 human genome using TopHat version 2.06(33) and Bowtie2 2.0.0(34) and mapped to Ensembl transcripts
(http://www.ensembl.org). NovelBio Bio-Pharm
Technology Co., Ltd. (http://www.novelbio.com/) performed the library
construction and sequence using LPS-treated and untreated mouse
heart samples.
Analysis of GO categories and KEGG
analysis
Differentially expressed genes were classified into
diverse biological processes according to the GO terms (35). GO categories were assessed using a
one-tailed Fisher's exact test with a P-value, and corrected for
using the false discovery rate (FDR) (36). P<0.05 was considered as
significant. GO enrichment analysis was also performed using a
Fisher's exact test using 2x2 contingency tables (37). As the enrichment increases, the
corresponding function becomes more specific. Pathway analysis
focuses on the significance of KEGG (http://www.kegg.jp/) (38), Biocarta (http://biocarta.com) (39) and Reatome (http://www.reactome.org) (40). Benjamini-Hochberg multiple testing
was used further to correct the Fisher's exact test results, and
the threshold of significance was defined as P-value<0.05 and
FDR<0.05(41).
Western blot analysis
RIPA lysis buffer (Takara Bio, Inc.) supplemented
with complete protease inhibitor cocktail (Roche Diagnostics) was
used to lyse the cells. The mouse heart tissues were ground in
liquid nitrogen, and protein was extracted by adding cold RIPA
lysis buffer. A Bradford reagent protein assay kit (Bio-Rad
Laboratories, Inc.) was used to determine protein concentrations
using bovine serum albumin as the control. Subsequently, a total of
30 µg sample protein was loaded per lane and separated using
SDS-PAGE on a 10-20% gel, then subsequently transferred PVDF
membranes. The membranes were blocked with buffer (5% skimmed milk
in 20 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20) for 1 h at room
temperature and incubated with primary antibodies at 4˚C overnight;
The primary antibodies used were anti-IL-6 antibody (1:1,000
dilution; cat. no. ab7737; Abcam), anti-TNF-α antibody (1:1,000
dilution; cat. no. ab11564; Abcam), anti-PAI-1 (1:1,000 dilution;
cat. no. ab222754; Abcam), anti-IκBα antibody (1:1,000 dilution;
cat. no. sc-371; Santa Cruz Biotechnology, Inc.), anti-p-IκBα
antibody (1:200 dilution; cat. no. sc-8404; Santa Cruz
Biotechnology, Inc.) and anti-GAPDH (1:5,000 dilution; cat. no.
ab9485; Abcam). After two washes with 1x TBS, the membranes were
incubated with a horseradish peroxidase-conjugated secondary
antibody (1:1,000, cat. no. 7074; Cell Signaling Technology, Inc.)
for 2 h at room temperature, and the protein levels were detected
using ECL western blotting reagents (cat. no. RPN2135, GE
Healthcare) and Image lab v3.0 software (Bio-Rad Laboratories,
Inc.).
Statistical analysis
Data are presented as the mean ± the standard error
of the mean. GraphPad Prism version 5 (GraphPad Software, Inc.) was
used to perform the statistical analysis. Differences between two
group were compared using a Student's t-test, and the differences
among multiple groups were analyzed using a one-way ANOVA. All
experiments were repeated three times. P<0.05 was considered to
indicate a statistically significant difference.
Results
LPS injection induces inflammation and
cardiac damage
To understand the molecular mechanism of sepsis, LPS
was injected into mice to generate a sepsis model. Furthermore, an
RNA-Seq-based transcriptome study was performed to understand the
molecular basis of this illness. Before performing RNA-Seq, the
effects of LPS on heart tissue damage were examined in mice at
different time points. After 1, 2, 3 and 4 days of LPS injection,
the heart tissue was sampled, and the inflammatory response was
examined by western blot analysis. The results showed that PAI-1
and TNF-α levels were induced by LPS injection in heart tissue, and
the highest levels were detected in the 4-day sample; therefore,
the subsequent experiments were performed using 4-day LPS-injected
mouse hearts (Fig. 1A). After 4 days
of LPS injection, the heart tissue began to show an abnormal
structure with aberrant cardiac muscle arrangement.
Histopathological analysis was performed using HE staining. The
results showed major cardiac disorder in LSP-injected hearts, and
the symptoms included increased disorganization and discontinuation
of the myocardial structure (Fig.
1B). In addition, the levels of inflammatory responsive
proteins were assessed using western blot analysis. The three
inflammation-responsive markers tested, IL-6, IκBα and p-IκB,
exhibited higher levels in LPS-injected mouse hearts compared with
the control group (without injection) (Fig. 1C). These results suggest that LPS
injection induces inflammatory responses and cardiac damage in
mice.
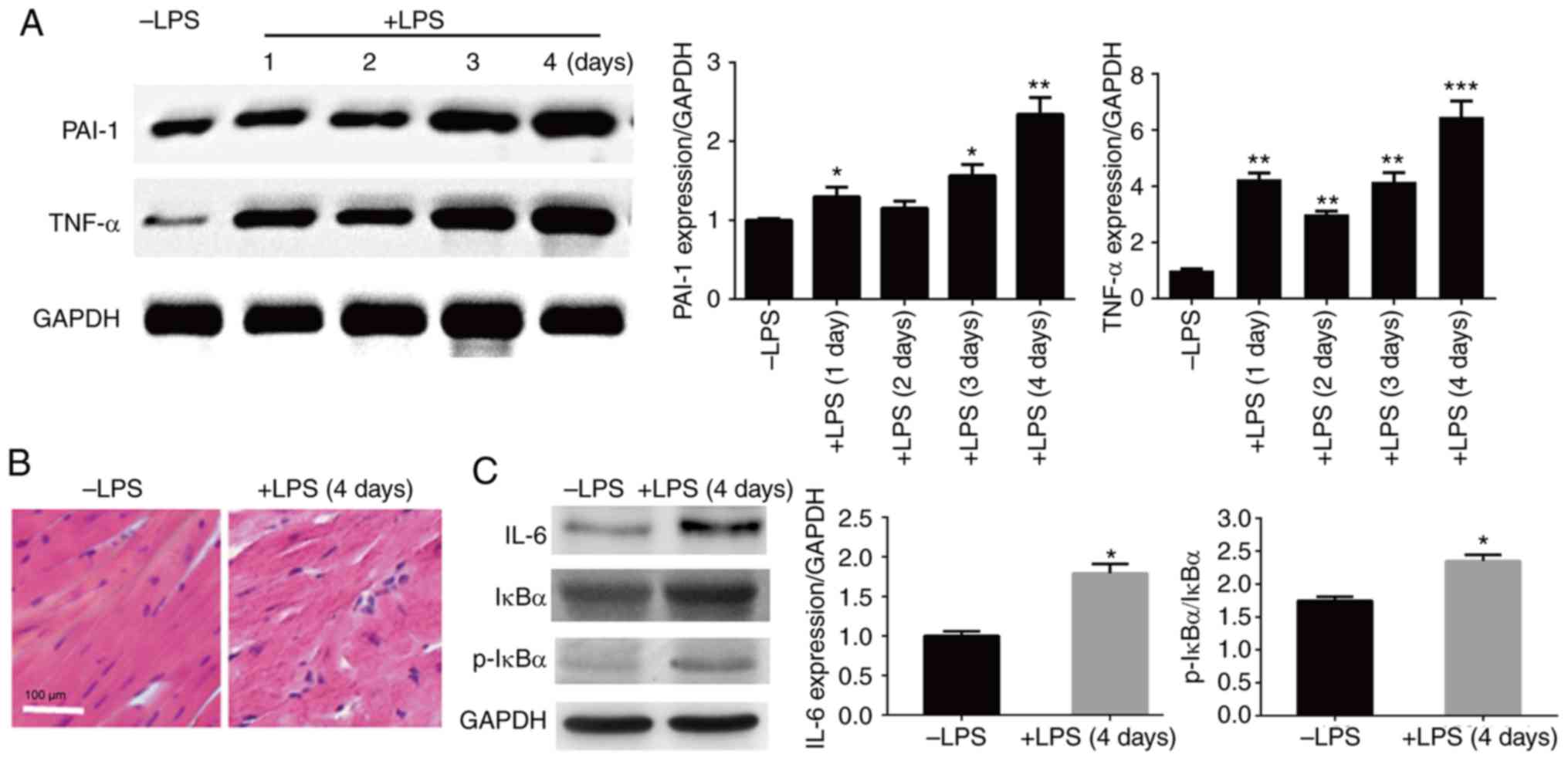 | Figure 1Effects of LPS on heart tissue
structure and inflammation response marker levels. (A) Western blot
analysis was performed to evaluate the levels of PAI-1 and TNF-α in
the heart tissues after 1, 2, 3 and 4 days of LPS injection. GAPDH
was used as a loading control. (B) Histopathology of heart tissues
with or without LPS injection for 4 days was assessed by staining
tissues with hematoxylin and eosin under a light microscope. scale
bar, 100 µm. (C) Western blot analysis was performed to evaluate
the levels of IL-6, IκBα and p-IκBα in the heart tissues with or
without LPS injection for 4 days. GAPDH was used as a loading
control. *P<0.05, **P<0.01,
***P<0.001 vs. the control group. LPS,
lipopolysaccharide; PAI-1, plasminogen activator inhibitor 1;
TNF-α, tumor necrosis factor-α; HE, Hematoxylin eosin; IL-6,
interleukin 6; IκBα, inhibitor of nuclear factor κB kinase α; p,
phosphorylated. |
LPS treatment alters the expression of
several genes
To further investigate the molecular mechanism
underlying sepsis illness, RNA-Seq experiments were carried out
using mouse heart tissues with or without LPS injection for 4 days.
The RNA-Seq results demonstrated that 5,094 genes were
significantly altered by LPS (>two-fold changes; P<0.05).
Among them, 3,326 and 1,769 genes were upregulated and
downregulated, respectively, by LPS injection in mouse hearts
(Fig. 2A). The following genes were
assessed by RT-qPCR to confirm the RNA-Seq data: Frizzled class
receptor 8 (FZD8), IL-8, PAI1 and fibroblast
growth factor 21 (FGF21). The inflammatory response genes
(IL-8 and PAI-1) and FGF21 were upregulated,
whereas a Wnt signaling gene (FZD8) decreased after LPS
stimulation, and the RT-qPCR results were similar to the RNA-Seq
data (Fig. 2B).
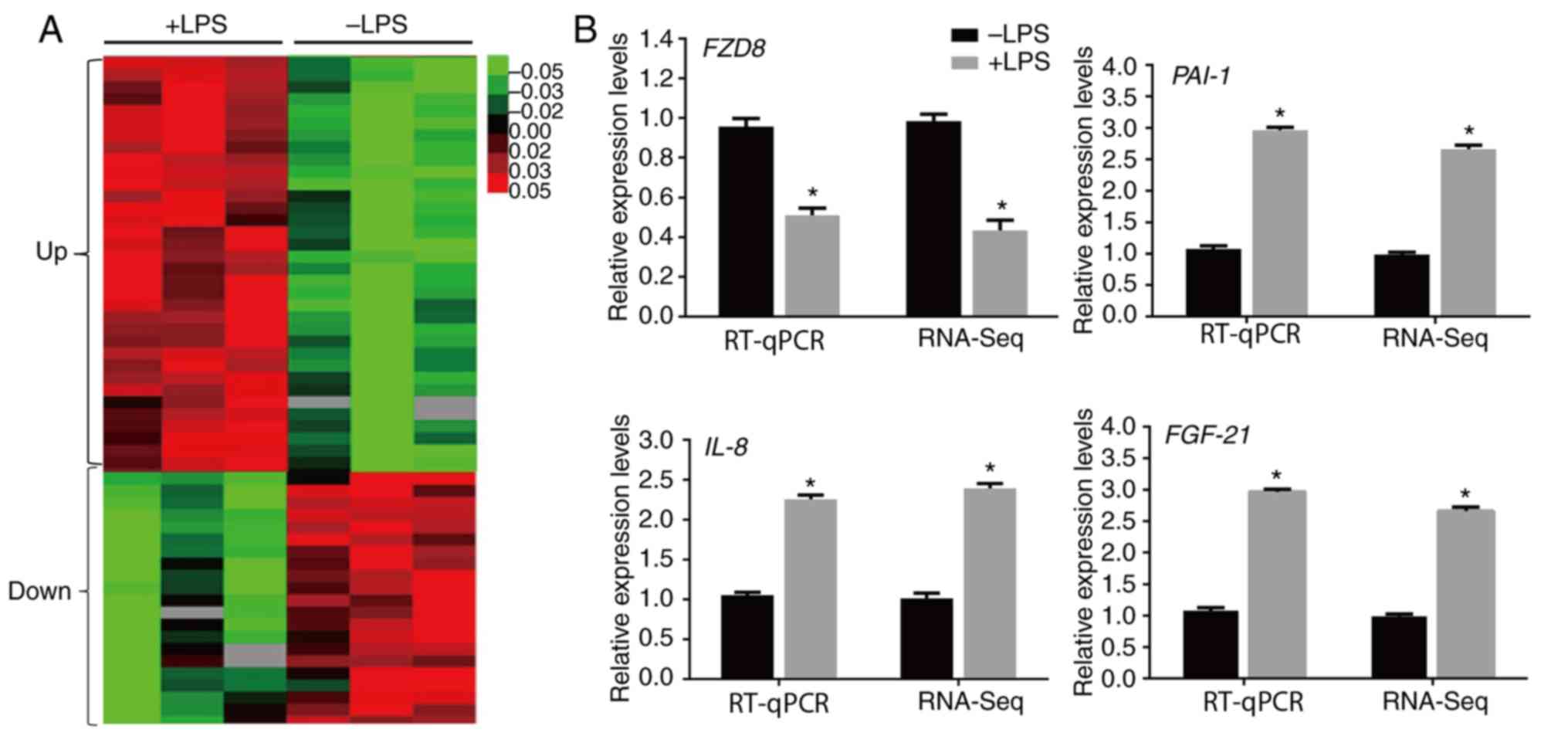 | Figure 2LPS-regulated transcriptome profile
in heart. (A) Heat map representing the genes with expression
levels that were altered after 4 days of LPS treatment in the mouse
heart. Gene expression is shown with a pseudocolor scale, where red
denotes higher gene expression levels and green denotes lower gene
expression levels (P<0.05). (B) RT-qPCR was performed to verify
the expression levels of FZD8, IL-8, PAI-1 and
FGF-21, and the data were compared with the RNA-sequencing
data. GAPDH was used as the internal control. *P<0.05
vs. the control group. LPS, lipopolysaccharide; RT-qPCR, reverse
transcription-quantitative PCR; FZD8, frizzled class
receptor 8; IL-8, interleukin 8; PAI-1, plasminogen
activator inhibitor 1; FGF21, fibroblast growth factor
21. |
Differentially expressed genes are
classified into diverse biological categories
In total, 5,094 differentially expressed genes
regulated by LPS were further classified into GO and KEGG terms. GO
analysis demonstrated that 41 GO terms were highly enriched
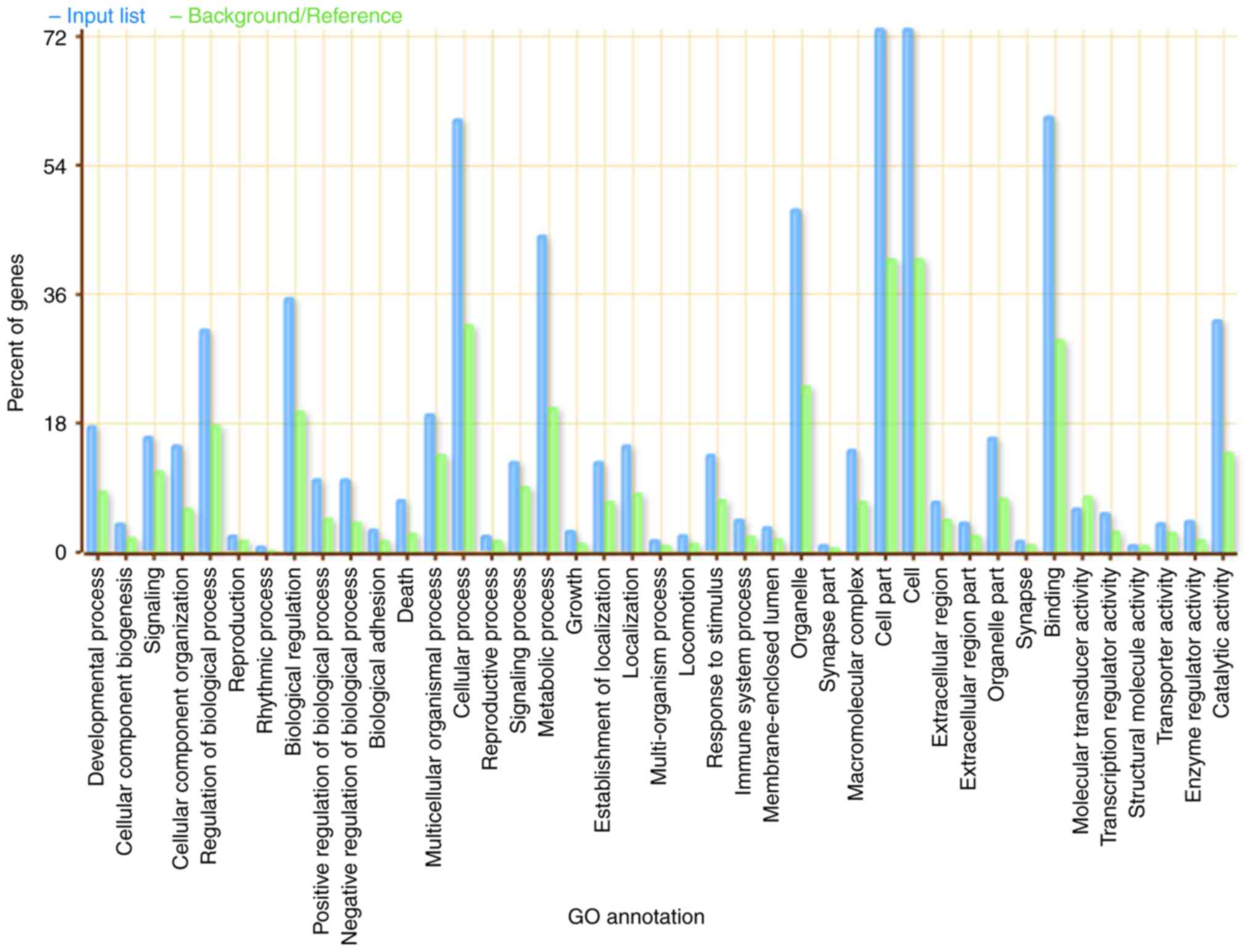
(P<0.01; Fig. 3). These genes are
involved in multiple biological processes, including ‘developmental
process’, ‘cell part’, ‘binding’ and ‘cellular process’ (Fig. 3). Several genes were enriched in the
‘cell’, ‘cell part’ and ‘cellular process’ biological categories.
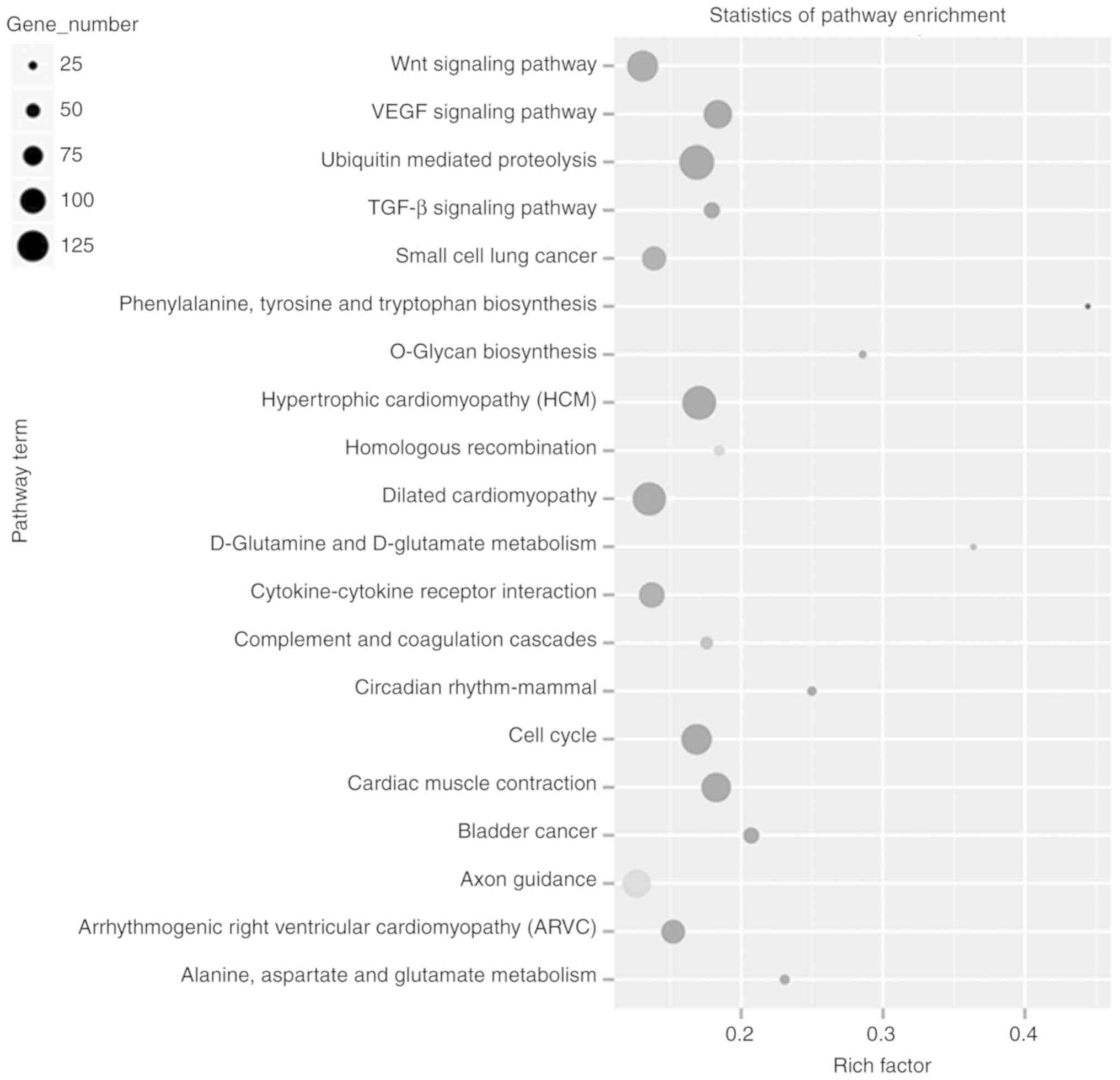
Subsequently, pathway analysis was performed using the 5,094 genes
that were differentially expressed by LPS damage. The results
showed that these genes were classified into 20 different pathways,
such as the ‘Wnt signaling pathway’, ‘VEGF signaling pathway’,
‘small cell lung cancer’, and ‘cardiac muscle contraction’ pathways
(Fig. 4).
Wnt signaling is involved in
LPS-induced heart damage
As Wnt signaling genes were regulated by LPS
injection in mice, the role of Wnt signaling in sepsis development
was further investigated. Wnt signaling serves a key role in cell
survival, proliferation and differentiation (25). To test the role of Wnt signaling in
sepsis regulation, Wnt3a, an inducer of Wnt signaling, and IWR, an
inhibitor of Wnt signaling were used. First, an inducer of Wnt
signaling was injected into mice to determine the function of Wnt
signaling in LPS-induced heart damage. After 4 days of injection,
LPS induced PAI-1 expression, but Wnt3a treatment partially
inhibited the LPS-mediated induction of PAI-1. In addition,
LPS treatment suppressed FZD8 expression, whereas Wnt3a
injection inhibited the LPS suppression of FZD8 (Fig. 5A). Heart tissues were evaluated
further by HE staining. LPS injection damaged heart tissues by
rearranging muscle, which exhibited deconstruction, whereas Wnt3a
protected against LPS-induced cardiac damage (Fig. 5B). In contrast, IWR, a Wnt signaling
inhibitor, was injected into the mice. After 4 days of injection,
PAI-1 expression was significantly induced in the heart (Fig. 5C). Furthermore, IWR injection damaged
heart tissues, similar to LPS injection, which exhibited aberrant
cardiac muscle (Fig. 5D).
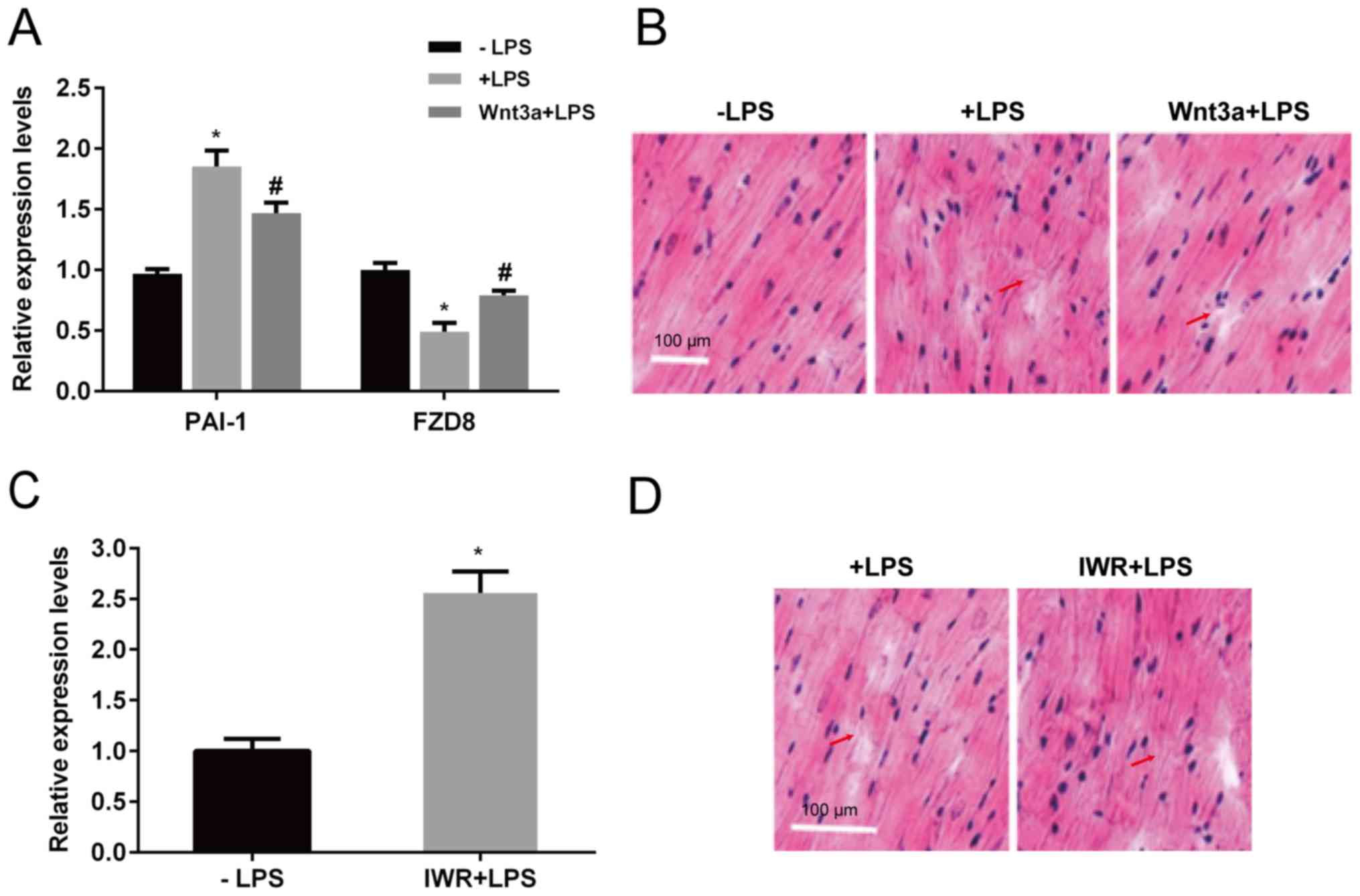 | Figure 5Effects of Wnt3a and IWR on
expression of gene markers of heart tissue structure. LPS or Wnt3a
together with LPS was injected, and the mouse hearts were
extracted. (A) PAI-1 and FZD8 expression levels were
determined by reverse transcription-quantitative PCR, and (B) heart
tissue structure was analyzed by staining tissues with hematoxylin
and eosin and observing under a light microscope. Scale bar, 100
µm. IWR, a Wnt signaling inhibitor, was injected, and (C) the PAI-1
expression level and (D) heart tissue structure were analyzed.
Arrows indicated sites of myocardial fiber fracture. Scale bars,
100 µm. *P<0.05 vs. the control group;
#P<0.05 vs. the LPS group. LPS, lipopolysaccharide;
PAI-1, plasminogen activator inhibitor 1; FZD8,
frizzled class receptor 8. |
Discussion
Sepsis is caused by the imbalanced response of
humans to pathogen infection, and this may result in severe damage
to various organs, including the heart. A previous clinical study
demonstrated that sepsis during the early stages is associated with
a high incidence of organic damage to the myocardium, which causes
hypotension, heart failure and arrhythmia (42). Currently, ~40% of patients with
sepsis suffer from myocardial injury, ~60% of patients in intensive
care units have clinical manifestations of myocardial injury, and
the mortality of patients with sepsis can reach 70-90% (43). However, no therapeutic strategies are
available for patients with sepsis, and therefore, this issue has
attracted attention worldwide. Exploring the molecular basis of
sepsis will be useful for isolating target genes or proteins for
therapy. However, studies regarding sepsis are still limited to
date.
In the present study, bacterial endotoxin, including
LPS, which is a potential inducer of inflammation, was injected
into a mouse model of sepsis, and transcriptome analysis was
performed to understand the molecular mechanism of sepsis in the
mouse heart model. Initially, LPS was injected; which markedly
induced an inflammatory response, based on evaluation of
inflammatory response markers by western blot analysis.
Additionally, histological analysis demonstrated that LPS injection
severely damaged cardiac tissues by rearranging muscle order. This
may have been caused by either the injection of low-concentration
LPS or the LPS tolerance response (44); however, further analyses are required
to verify this observation. The inflammatory response was also
induced by the detection of IL-6, IκBα, p-IκBα, PAI-1 and TNF-α
levels using western blot analysis. LPS treatment significantly
increased the expression levels of these protein, The critical step
in NF-kB activation is the phosphorylation of IkBα (45), suggesting that TNF-α and NF-κB
signaling was activated in the mouse heart. In a subsequent
RNA-seq-based transcriptome study, the expression levels of several
genes were altered by injection of LPS in mouse hearts. Among
these, 3,326 and 1,769 genes were upregulated and downregulated,
respectively. These differentially expressed genes were classified
into 41 GO terms and 20 KEGG pathways. Among the 20 enriched
pathways, ‘Wnt signaling pathway’, ‘VEGF signaling pathway’, ‘TFG-β
signaling pathway’ and ‘cytokine-cytokine receptor interaction’
pathways, and activity of certain metabolic pathways was altered.
The western blot analysis suggested that TNF-α and NF-κB signaling
was significantly increased by LPS injection. However, these
signaling pathway genes were not enriched in the KEGG analysis.
This difference in results may be due to the induction time point
for transcripts or proteins, as 4 days is a relatively long period,
and the occurrence of transcriptional changes is normally fast, or
the increase in the number of these pathway genes may be lower.
Nevertheless, IL-8 was detected and induced by LPS
treatment. TNF-α and IL proteins are cytokines that serve important
roles during the inflammatory response (46), and the KEGG analysis also showed that
the cytokine-cytokine receptor interaction term was enriched. VEGF,
TGF-β and small lung cancer pathways are known to serve roles
during cancer development (47,48).
Notably, another key signaling pathway in cancer biology, the Wnt
signaling pathway, was enriched, and the Wnt genes were suppressed
by LPS treatment, suggesting that these signaling pathways may be
associated with inflammation and sepsis. Furthermore, the role of
Wnt signaling was analyzed by activation or inhibition experiments
in the present study. The function of Wnt signaling in sepsis has
not been previously reported to the best of our knowledge.
Furthermore, Wnt3a was added to induce Wnt signaling, to determine
the role of Wnt signaling during sepsis. Wnt3a treatment inhibited
LPS-mediated PAI-1 and FZD8 gene expression, and also
prevented damage to cardiac tissue structure caused by LPS. Wnt3a
treatment together with LPS reduced the LPS-induced damage to the
myocardium. A Wnt signaling inhibitor, IWR, was subsequently
injected; IWR injection increased the PAI-1 expression
levels and resulted in damage to the heart tissues, which exhibited
abnormal cardiac organization. These data suggest that Wnt
signaling protects against LPS-induced heart damage by reducing the
inflammatory response. In addition, LPS treatment reduced
expression of the Wnt signaling gene FZD8, suggesting that
the LPS-mediated inflammatory response signaling may function
upstream of Wnt signaling. Wnt3a and IWR treatment also inhibited
and induced PAI-1 expression, respectively, suggesting that
Wnt signaling functioned upstream of the inflammatory responsive
gene PAI-1. These data suggest that Wnt and inflammatory
response signals are associated with each other, and the crosstalk
should be further analyzed in future studies. FGF21, a fibroblast
growth factor, was also induced by LPS damage. FGF21 is an
important hormone that protects organs from different damages
(49,50). Testing FGF21 function in further
studies may be useful. However, the present study has its own
limitations. Key regulators of Wnt signaling GSK-3β and β-catenin,
were not measured following treatment with LPS, Wnt3a and IWR.
In conclusion, RNA-Seq analysis was performed to
determine the molecular mechanism of a sepsis-damaged heart using a
mouse model generated by LPS injection. The data suggested that
sepsis damage in the heart may be caused by complicated processes,
but LPS inhibited Wnt signaling to induce heart injury. This
finding may be important for further exploration of sepsis-mediated
damage to different tissues.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated during the current study are
not publicly available because the investigator team is still
actively analyzing and publishing study results, but are available
from the corresponding author on reasonable request.
Authors' contributions
CC, JW and DF performed the experiments, wrote the
manuscript, as well as the analysis and interpretation of data. JC
collected and analyzed the data. MC designed the study. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All animal studies were performed according to
international ethical guidelines and the National Institutes of
Health Guide Concerning the Care and Use of Laboratory Animals with
The Approval of The Animal Experimentation Ethics Committee of The
Affiliated Hospital of Putian University.
Patient consent for participation
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Michalek SM, Moore RN, McGhee JR,
Rosenstreich DL and Mergenhagen SE: The primary role of
lymphoreticular cells in the mediation of host responses to
bacterial endotoxim. J Infect Dis. 141:55–63. 1980.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Beeson PB: Technical Assistance of
Elizabeth Roberts: Tolerance to bacterial pyrogens: I. Factors
influencing its development. J Exp Med. 86:29–38. 1947.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Singer M, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (sepsis-3). JAMA.
315:801–810. 2016.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Seymour CW, Liu VX, Iwashyna TJ,
Brunkhorst FM, Rea TD, Scherag A, Rubenfeld G, Kahn JM,
Shankar-Hari M, Singer M, et al: Assessment of clinical criteria
for sepsis: For the third international consensus definitions for
sepsis and septic shock (sepsis-3). JAMA. 315:762–774.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Engel C, Brunkhorst FM, Bone HG,
Brunkhorst R, Gerlach H, Grond S, Gruendling M, Huhle G, Jaschinski
U, John S, et al: Epidemiology of sepsis in Germany: Results from a
national prospective multicenter study. Intensive Care Med.
33:606–618. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Angus DC and Wax RS: Epidemiology of
sepsis: An update. Crit Care Med. 29 (7 Suppl):S109–S116.
2001.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Cohen J: The immunopathogenesis of sepsis.
Nature. 420:885–891. 2002.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Drosatos K, Lymperopoulos A, Kennel PJ,
Pollak N, Schulze PC and Goldberg IJ: Pathophysiology of
sepsis-related cardiac dysfunction: Driven by inflammation, energy
mismanagement, or both? Curr Heart Fail Rep. 12:130–140.
2015.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Harbarth S, Holeckova K, Froidevaux C,
Pittet D, Ricou B, Grau GE, Vadas L and Pugin J: Geneva Sepsis
Network: Diagnostic value of procalcitonin, interleukin-6, and
interleukin-8 in critically ill patients admitted with suspected
sepsis. Am J Respir Crit Care Med. 164:396–402. 2001.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Simon L, Gauvin F, Amre DK, Saint-Louis P
and Lacroix J: Serum procalcitonin and C-reactive protein levels as
markers of bacterial infection: A systematic review and
meta-analysis. Clin Infect Dis. 39:206–217. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
11
|
Bozza FA, Salluh JI, Japiassu AM, Soares
M, Assis EF, Gomes RN, Bozza MT, Castro-Faria-Neto HC and Bozza PT:
Cytokine profiles as markers of disease severity in sepsis: A
multiplex analysis. Crit Care. 11(R49)2007.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Vaschetto R, Nicola S, Olivieri C, Boggio
E, Piccolella F, Mesturini R, Damnotti F, Colombo D, Navalesi P,
Della Corte F, et al: Serum levels of osteopontin are increased in
SIRS and sepsis. Intensive Care Med. 34:2176–2184. 2008.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Brenner T, Rosenhagen C, Steppan J,
Lichtenstern C, Weitz J, Bruckner T, Martin EO, Hoffmann U, Weigand
MA and Hofer S: Redox responses in patients with sepsis: High
correlation of thioredoxin-1 and macrophage migration inhibitory
factor plasma levels. Mediators Inflamm.
2010(985614)2010.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Bae JS: Role of high mobility group box 1
in inflammatory disease: Focus on sepsis. Arch Pharm Res.
35:1511–1523. 2012.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wu Y, Wang F, Fan X, Bao R, Bo L, Li J and
Deng X: Accuracy of plasma sTREM-1 for sepsis diagnosis in systemic
inflammatory patients: A systematic review and meta-analysis. Crit
Care. 16(R229)2012.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Backes Y, van der Sluijs KF, Mackie DP,
Tacke F, Koch A, Tenhunen JJ and Schultz MJ: Usefulness of suPAR as
a biological marker in patients with systemic inflammation or
infection: A systematic review. Intensive Care Med. 38:1418–1428.
2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Rautanen A, Mills TC, Gordon AC, Hutton P,
Steffens M, Nuamah R, Chiche JD, Parks T, Chapman SJ, Davenport EE,
et al: Genome-wide association study of survival from sepsis due to
pneumonia: An observational cohort study. Lancet Respir Med.
3:53–60. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Scherag A, Schöneweck F, Kesselmeier M,
Taudien S, Platzer M, Felder M, Sponholz C, Rautanen A, Hill AVS,
Hinds CJ, et al: Genetic factors of the disease course after
sepsis: A genome-wide study for 28 day mortality. EbioMedicine.
12:239–246. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Wang L, Ko ER2, Gilchrist JJ, Pittman KJ,
Rautanen A, Pirinen M, Thompson JW, Dubois LG, Langley RJ, Jaslow
SL, et al: Human genetic and metabolite variation reveals that
methylthioadenosine is a prognostic biomarker and an inflammatory
regulator in sepsis. Sci Adv. 3(e1602096)2017.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Brown MA and Jones WK: NF-kappaB action in
sepsis: The innate immune system and the heart. Front Biosci.
9:1201–1217. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Li Y, Ge S, Peng Y and Chen X:
Inflammation and cardiac dysfunction during sepsis, muscular
dystrophy, and myocarditis. Burns Trauma. 1:109–121.
2013.PubMed/NCBI View Article : Google Scholar
|
|
22
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gordon MD and Nusse R: Wnt signaling:
Multiple pathways, multiple receptors, and multiple transcription
factors. J Biol Chem. 281:22429–22433. 2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Shtutman M, Zhurinsky J, Simcha I,
Albanese C, D'Amico M, Pestell R and Ben-Ze'ev A: The cyclin D1
gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad
Sci USA. 96:5522–5527. 1999.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Moon RT, Kohn AD, De Ferrari GV and Kaykas
A: WNT and β-catenin signalling: Diseases and therapies. Nat Rev
Genet. 5:691–701. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
26
|
Clevers H: Wnt/β-catenin signaling in
development and disease. Cell. 127:469–480. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Brack AS, Conboy MJ, Roy S, Lee M, Kuo CJ,
Keller C and Rando TA: Increased Wnt signaling during aging alters
muscle stem cell fate and increases fibrosis. Science. 317:807–810.
2007.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Aberle H, Bauer A, Stappert J, Kispert A
and Kemler R: Beta-catenin is a target for the ubiquitin-proteasome
pathway. EMBO J. 16:3797–3804. 1997.PubMed/NCBI View Article : Google Scholar
|
|
29
|
National Research Council: Guide for the
care and use of laboratory animals. National Academies Press,
Washington, DC, 1996.
|
|
30
|
Leary S: AVMA Guidelines for the
Euthanasia of Animals: 2013 Edition. American Veterinary Medical
Association. J Am Veterinary Med Association, 2013.
|
|
31
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Rani B: RNA Sequencing (RNA-Seq): Method
and Applications. Int J Pure App Biosci. 6:167–173. 2018.
|
|
33
|
Kim D, Pertea G, Trapnell C, Pimentel H,
Kelley R and Salzberg SL: TopHat2: Accurate alignment of
transcriptomes in the presence of insertions, deletions and gene
fusions. Genome Biol. 14(R36)2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Langmead B and Salzberg SL: Fast
gapped-read alignment with Bowtie 2. Nat Methods. 9:357–359.
2012.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Gene Ontology Consortium. The gene
ontology (GO) project in 2006. Nucleic Acids Res 34: D322-D326,
2006.
|
|
36
|
Dupuy D, Bertin N, Hidalgo CA, Venkatesan
K, Tu D, Lee D, Rosenberg J, Svrzikapa N, Blanc A, Carnec A, et al:
Genome-scale analysis of in vivo spatiotemporal promoter activity
in Caenorhabditis elegans. Nat Biotechnol. 25:663–668.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
37
|
Dunnick JK, Brix A, Cunny H, Vallant M and
Shockley KR: Characterization of polybrominated diphenyl ether
toxicity in Wistar Han rats and use of liver microarray data for
predicting disease susceptibilities. Toxicol Pathol. 40:93–106.
2012.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Kanehisa M, Goto S, Kawashima S and Nakaya
A: The KEGG databases at Genome Net Nucleic Acids. Res. 30:42–46.
2002.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Nishimura D: BioCarta. Biotech Software
Internet Rep. 2:117–120. 200.
|
|
40
|
Matthews L, Gopinath G, Gillespie M, Caudy
M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B,
et al: Reactome knowledgebase of human biological pathways and
processes. Nucleic Acids Res. 37 (Database Issue):D619–D622.
2009.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Draghici S, Khatri P, Tarca AL, Amin K,
Done A, Voichita C, Georgescu C and Romero R: A systems biology
approach for pathway level analysis. Genome Res. 17:1537–1545.
2007.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Turner A, Tsamitros M and Bellomo R:
Myocardial cell injury in septic shock. Crit Care Med.
27:2035–2036. 1999.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hochstadt A, Meroz Y and Landesberg G:
Myocardial dysfunction in severe sepsis and septic shock: More
questions than answers? J Cardiothorac Vasc Anesth. 25:526–535.
2011.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Dios S, Balseiro P, Costa MM, Romero A,
Boltaña S, Roher N, Mackenzie S, Figueras A and Novoa B: The
involvement of cholesterol in sepsis and tolerance to
lipopolysaccharide highlighted by the transcriptome analysis of
zebrafish (Danio rerio). Zebrafis. 11:421–433. 2014.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Traenckner EB, Pahl HL, Henkel T, Schmidt
KN, Wilk S and Baeuerle PA: Phosphorylation of human I kappa
B-alpha on serines 32 and 36 controls I kappa B-alpha proteolysis
and NF-kappa B activation in response to diverse stimuli. EMBO J.
14:2876–2883. 1995.PubMed/NCBI
|
|
46
|
Jaffer U, Wade RG and Gourlay T: Cytokines
in the systemic inflammatory response syndrome: A review. HSR Proc
Intensive Care Cardiovasc Anesth. 2:161–175. 2010.PubMed/NCBI
|
|
47
|
Carmeliet P: VEGF as a key mediator of
angiogenesis in cancer. Oncology. 69:4–10. 2005.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Derynck R, Akhurst RJ and Balmain A: TGF-β
signaling in tumor suppression and cancer progression. Nature
Genet. 29:117–129. 2001.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Planavila A, Redondo-Angulo I, Ribas F,
Garrabou G, Casademont J, Giralt M and Villarroya F: Fibroblast
growth factor 21 protects the heart from oxidative stress.
Cardiovasc Res. 106:19–31. 2015.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Singhal G, Fisher FM, Chee MJ, Tan TG El
Ouaamari A, Adams AC, Najarian R, Kulkarni RN, Benoist C, Flier JS
and Maratos-Flier E: Fibroblast Growth Factor 21 (FGF21) protects
against high fat diet induced inflammation and islet hyperplasia in
pancreas. PLoS One. 11(e0148252)2016.PubMed/NCBI View Article : Google Scholar
|