Introduction
Hepatitis B virus (HBV) is a noncytopathic virus
that can establish a lifelong chronic infection in humans.
Currently, 3.5% of the global population suffers from chronic HBV
infection, and therefore have a high risk of developing liver
fibrosis, cirrhosis and hepatocellular carcinoma (HCC), which cause
nearly 1 million annual deaths (1,2).
Patients with chronic hepatitis B (CHB) exhibit a suppression of
virus-specific immune responses that is closely associated with
viral persistence and reduced responsiveness to interferon-based
therapies (3-5).
However, to the best of our knowledge, the underlying mechanisms
for this antiviral immune hypo-responsiveness are not yet well
defined.
HBV typically encodes four antigens, hepatitis B
core antigen (HBcAg), hepatitis B envelope antigen (HBeAg),
hepatitis B X antigen (HBxAg) and hepatitis B surface antigen
(HBsAg). Patients with CHB are known to have persistently high
quantities of the viral antigens HBsAg and HBeAg in their
peripheral blood (6,7). HBeAg, a secretory form of the
nucleocapsid antigen, is not required for viral infection or
replication (8-10).
However, HBeAg clearance or seroconversion in CHB patients is
closely related to the restoration of antiviral T cell function
following effective treatment (11,12).
Secreted HBeAg has been shown to suppress the HBV-specific immune
response and promote HBV persistence (12,13).
Diverse suppressive pathways have been implicated in
the dysfunctional antiviral responses in CHB (6). Among the proposed mechanisms,
regulatory T cell (Treg) activation in CHB is a major concern.
Tregs, an immunoregulatory T cell subpopulation, are characterized
by high expression of CD25 and transcription factor forkhead box P3
(Foxp3) compared to conventional CD4+ T cells and
capable of suppressing the immune functions of numerous cell types,
including CD4+ and CD8+ T cells, B cells,
natural killer cells, natural killer T cells and dendritic cells
(14). Patients with CHB harbor an
increased percentage of Tregs in their peripheral blood that is
positively correlated with serum HBV DNA load, the reduction of
antiviral and treatment responses and increased risk of HCC
(15-19).
Furthermore, patients with HBeAg-positive CHB exhibited a higher
percentage of Tregs in peripheral blood than those with
HBeAg-negative CHB (15,19,20).
However, to the best of our knowledge, whether and how HBeAg
manipulates the host immune system to induce the conversion of T
cells to Tregs remains to be elucidated.
The present manuscript describes a preliminary study
to investigate the involvement of HBeAg in the activation of Tregs
using mouse model. In vitro and in vivo studies
indicated that HBeAg may convert naive T cells into Tregs,
potentially due to increased transforming growth factor (TGF)-β
production induced by HBeAg. The results of the present study may
provide further evidence of the effect of HBeAg on Tregs and of the
benefit of the development of novel HBeAg-based immunotherapy for
CHB.
Materials and methods
Animal studies
Animal experiments were conducted in strict
accordance with the Regulations for the Administration of Affairs
Concerning Experimental Animals (date issued, November
14th, 1988; http://en.pkulaw.cn). All efforts were made to
minimize suffering, and all procedures for the use of laboratory
animals were approved by the Institutional Animal Care and Use
Committee (IACUC) of Nanjing Medical University (Permit Number:
IACUC-1601123).
Specific pathogen-free 7-8-week-old female BALB/c
mice (22~25 g body weight) were purchased from the Laboratory
Animal Center of Nanjing Medical University (Nanjing, China) and
bred in a specific pathogen-free animal facility. At the end of the
experiments, mice were sacrificed by cervical dislocation under
isoflurane anesthesia (5% in oxygen).
MHC class II binding prediction
The Immune Epitope Database (IEDB 2.22; http://tools.iedb.org/mhcii/) was used to predict
human and mouse MHC class II binding peptides in HBeAg (GenBank
accession no. AUR80753.1) from the HLA-DRB1*03:01 and
H2-IAb/H2-IAd MHC class II alleles. In the prediction, percentile
ranks ranged from 0-100, and low percentile ranks were good MHC II
binders. As predicted by IEDB, MHC class II epitopes with a
percentile rank below 10 were considered good binders.
Cytokine assays
The amounts of total and active TGF-β in the
supernatants of spleen cells were determined with commercial ELISA
kits (cat. nos. 437707 and 436707; BioLegend, Inc.) according to
the manufacturer's instructions. The cytokine concentration in each
supernatant was extrapolated from a standard curve.
Cell isolation
Spleens from 7-8-week-old female BALB/c mice were
pressed through nylon nets to prepare single-cell suspensions.
Cells were washed with PBS containing 1% fetal bovine serum (Gibco;
Thermo Fisher Scientific, Inc.) and 0.2 M EDTA, counted after
lysing the red blood cells, and then used to isolate
CD4+CD25– T cells by using a mouse regulatory
T cell isolation kit (Miltenyi Biotec GmbH) and a magnetic
activated cell sorter (MACS; Miltenyi Biotec GmbH) according to the
manufacturer's instructions, achieving ~90% purity as determined by
flow cytometry (FCM).
Antigen-presenting cells (APCs) were prepared from
spleen cell suspensions by negative selection using CD90.2 magnetic
microbeads (Miltenyi Biotec GmbH) to deplete T cells. Isolated APCs
were treated with mitomycin-C (Sigma-Aldrich; Merck KGaA) at 50
µg/ml for 30 min and then washed thoroughly before incorporation
into cocultures.
FCM analysis
To analyze
CD4+CD25+Foxp3+ Tregs, the Mouse
Regulatory T Cell Staining kit (eBioscience, Thermo Fisher
Scientific, Inc.) was used according to the manufacturer's
instructions. In brief, cells from mice spleens or cell cultures
were surface stained with anti-CD3-PerCP-Cy5.5 (clone 145-2C11),
anti-CD4-FITC (clone RM4-5) and anti-CD25-APC (clone PC61.5) and
subsequently permeabilized with cold Fix/Perm Buffer. The Fc
receptors were then blocked with anti-CD16/32 for. A PE-labeled
anti-Foxp3 (clone FJK16s) was then added. Following staining, the
cells were examined using a FACS Verse flow cytometer (BD
Biosciences) and data were analyzed using FlowJo (v10.0.7; FlowJo
LLC).
In vitro cell cultures
To determine the in vitro activation of Tregs
by HBeAg, spleen cells from normal BALB/c mice were cultured in
96-well round-bottom plates at a density of 5x105
cells/well in complete RPMI 1640 medium (Gibco; Thermo Fisher
Scientific, Inc.). Corresponding concentrations of HBeAg (0.2, 1, 5
or 10 µg/ml; Shanghai Yuan Mu Biotechnology Co., Ltd) or Grade VII
chicken egg ovalbumin (OVA; 1 µg/ml; Sigma-Aldrich; Merck KGaA)
were added to each culture in four wells per condition. An
inhibitor of transforming growth factor-β type 1 receptor (TGF-βRI
signaling; 20 µM; SB-431542; Sigma-Aldrich; Merck KGaA) was added
to a proportion of the wells. After 3 days, cells were harvested
from cultures and washed twice by centrifugation (300xg, 10 min,
4˚C) in PBS. After counting, cells were stained for FCM analysis,
and supernatants were collected for cytokine detection by
ELISA.
To investigate the Treg conversion of naive T cells
induced by HBeAg in vitro, MACS-isolated
CD4+CD25- T cells (2x105
cells/well) were cocultured with mitomycin-C-treated APCs
(1x105 cells/well) in triplicate in 96-well round-bottom
culture plates and stimulated with HBeAg (1 µg/ml) or OVA (1
µg/ml). After 3 days, cells were collected for FCM analysis.
Immunization protocol
In each experiment, 10 normal BALB/c mice were
randomly divided into two groups consisting of 5 mice per group.
Mice were subcutaneously (inguinal region) injected with 100 µl of
a 1:1 (v/v) mixture of antigen (25 µg of HBeAg or PBS alone) and
incomplete Freund's adjuvant (Sigma-Aldrich; Merck KGaA). Each
mouse was injected twice with a 14-d internal injection. Immunized
mice were sacrificed under isoflurane anesthesia 10 days after the
last injection and their spleens were collected for analysis.
Statistical methods
All statistical calculations were performed by using
the SPSS program (v26 for Windows; IBM Corp.). The comparisons
between more than two groups were analyzed with one-way analysis of
variance (ANOVA) followed by an LSD post hoc test (n<4 groups)
or a Tukey's post hoc test (n≥4 groups). P<0.05 was considered
to indicate a statistically significant difference.
Results
Prediction of potential
CD4+ T cell epitopes in the HBeAg
The IEDB consensus binding prediction platform was
employed to predict the potential CD4+ T cell epitopes
of HBeAg. Putative epitopes with a percentile rank <10% were
considered MHC class II binders. As recommended by IEDB, 38 human
and 20 mouse potential CD4+ T cell epitopes were
identified in HBeAg from the HLA-DRB1*03:01 and
H2-IAb/H2-IAd MHC class II alleles, respectively (Table I and II). These data suggest that HBeAg has
multiple high-affinity agonist peptides and may potentially induce
CD4+ T cell activation.
 | Table IPrediction of human MHC class II
epitopes for HBeAg. |
Table I
Prediction of human MHC class II
epitopes for HBeAg.
| MHC class II
Epitopes | Peptides | Percentile
ranka |
|---|
| 1 |
SYVNVNMGLKIRQLL | 1 |
| 2 |
VNVNMGLKIRQLLWF | 1.04 |
| 3 |
YVNVNMGLKIRQLLW | 1.07 |
| 4 |
NVNMGLKIRQLLWFHI | 1.19 |
| 5 |
PASRELVVSYVNVNM | 1.94 |
| 6 |
SRELVVSYVNVNMGL | 1.94 |
| 7 |
ASRELVVSYVNVNMG | 2.07 |
| 8 |
DPASRELVVSYVNVN | 2.6 |
| 9 |
IRDLLDTASALYREA | 3.26 |
| 10 |
RDLLDTASALYREAL | 3.55 |
| 11 |
PSDFFPSIRDLLDTA | 3.64 |
| 12 |
QLFHLCLIISCSCPTVQAS | 3.93 |
| 13 |
SIRDLLDTASALYRE | 3.97 |
| 14 |
TTVVRRRGRSPRRRT | 4.1 |
| 15 |
TVVRRRGRSPRRRTP | 4.14 |
| 16 |
ETTVVRRRGRSPRRR | 4.2 |
| 17 |
PETTVVRRRGRSPRR | 4.26 |
| 18 |
LPSDFFPSIRDLLDT | 4.45 |
| 19 |
LPETTVVRRRGRSPR | 4.54 |
| 20 |
SCLTFGRETVLEYLV | 5.73 |
| 21 |
PSIRDLLDTASALYR | 5.76 |
| 22 |
SCPTVQASKLCLGWL | 5.85 |
| 23 |
CLTFGRETVLEYLVS | 5.86 |
| 24 |
CLIISCSCPTVQASK | 5.87 |
| 25 |
CPTVQASKLCLGWLW | 5.92 |
| 26 |
SDFFPSIRDLLDTAS | 5.94 |
| 27 |
SCSCPTVQASKLCLG | 6.04 |
| 28 |
CSCPTVQASKLCLGW | 6.24 |
| 29 |
LIISCSCPTVQASKL | 6.33 |
| 30 |
FLPSDFFPSIRDLLD | 6.37 |
| 31 |
ISCLTFGRETVLEYL | 6.56 |
| 32 |
LLSFLPSDFFPSIRDLL | 7.64 |
| 33 |
LSFLPSDFFPSIRDL | 7.71 |
| 34 |
HISCLTFGRETVLEY | 8.48 |
| 35 |
MNLATWVGSNLEDPA | 8.5 |
| 36 |
NLATWVGSNLEDPASRELV | 8.84 |
| 37 |
DFFPSIRDLLDTASA | 9.53 |
| 38 |
LTFGRETVLEYLVSF | 9.83 |
 | Table IIPrediction of mouse MHC class II
epitopes for HBeAg. |
Table II
Prediction of mouse MHC class II
epitopes for HBeAg.
| MHC class II
Epitopes | Peptides | Percentile
ranka |
|---|
| 1 |
RTPPAYRPPNAPILST | 0.53 |
| 2 |
PPAYRPPNAPILSTLP | 0.65 |
| 3 |
IRTPPAYRPPNAPIL | 0.81 |
| 4 |
AYRPPNAPILSTLPET | 2.1 |
| 5 |
CSPHHTALRQAILCW | 4.33 |
| 6 |
WIRTPPAYRPPNAPI | 4.76 |
| 7 |
SPHHTALRQAILCWGE | 4.89 |
| 8 |
HCSPHHTALRQAILC | 5.09 |
| 9 |
VWIRTPPAYRPPNAP | 5.65 |
| 10 |
SFGVWIRTPPAYRPP | 7.02 |
| 11 |
IDPYKEFGASVELLS | 7.32 |
| 12 |
HHTALRQAILCWGEL | 7.36 |
| 13 |
GVWIRTPPAYRPPNA | 7.46 |
| 14 |
VSFGVWIRTPPAYRP | 7.57 |
| 15 |
FGVWIRTPPAYRPPN | 7.98 |
| 16 |
FGRETVLEYLVSFGV | 8.16 |
| 17 |
DPYKEFGASVELLSF | 8.39 |
| 18 |
MDIDPYKEFGASVELL | 9.02 |
| 19 |
LTFGRETVLEYLVSFG | 9.32 |
| 20 |
CLTFGRETVLEYLVS | 10 |
HBeAg converts naive T cells to Tregs
in spleen cells in vitro
HBeAg was further examined for its ability to induce
the conversion of naive T cells into Tregs in spleen cells cultured
for 3 days in vitro. FCM analysis suggested that there were
no significant differences in the proportions of CD4+ T
cells among all the groups (Fig. 1A
and B). But notably, FCM analysis
revealed that the percentage of CD25+Foxp3+
Tregs within the CD4+ T cell population of HBeAg-treated
spleen cells increased over the 3-day period in a dose-dependent
manner (Fig. 1A and 1C). By contrast, no significant increase
was observed within OVA-treated spleen cells (Fig. 1A and 1C). Different doses (0.2, 1, 5, and 10
µg/ml) were used to test the ability of HBeAg to induce spleen cell
differentiation into Tregs in vitro. In these experiments,
the medium doses (1 and 5 µg/ml) of HBeAg increased the population
of Tregs, but the low dose (0.2 µg/ml) and the high dose (10 µg/ml)
appeared unable to trigger this increase of Tregs (Fig. 1A and 1C).
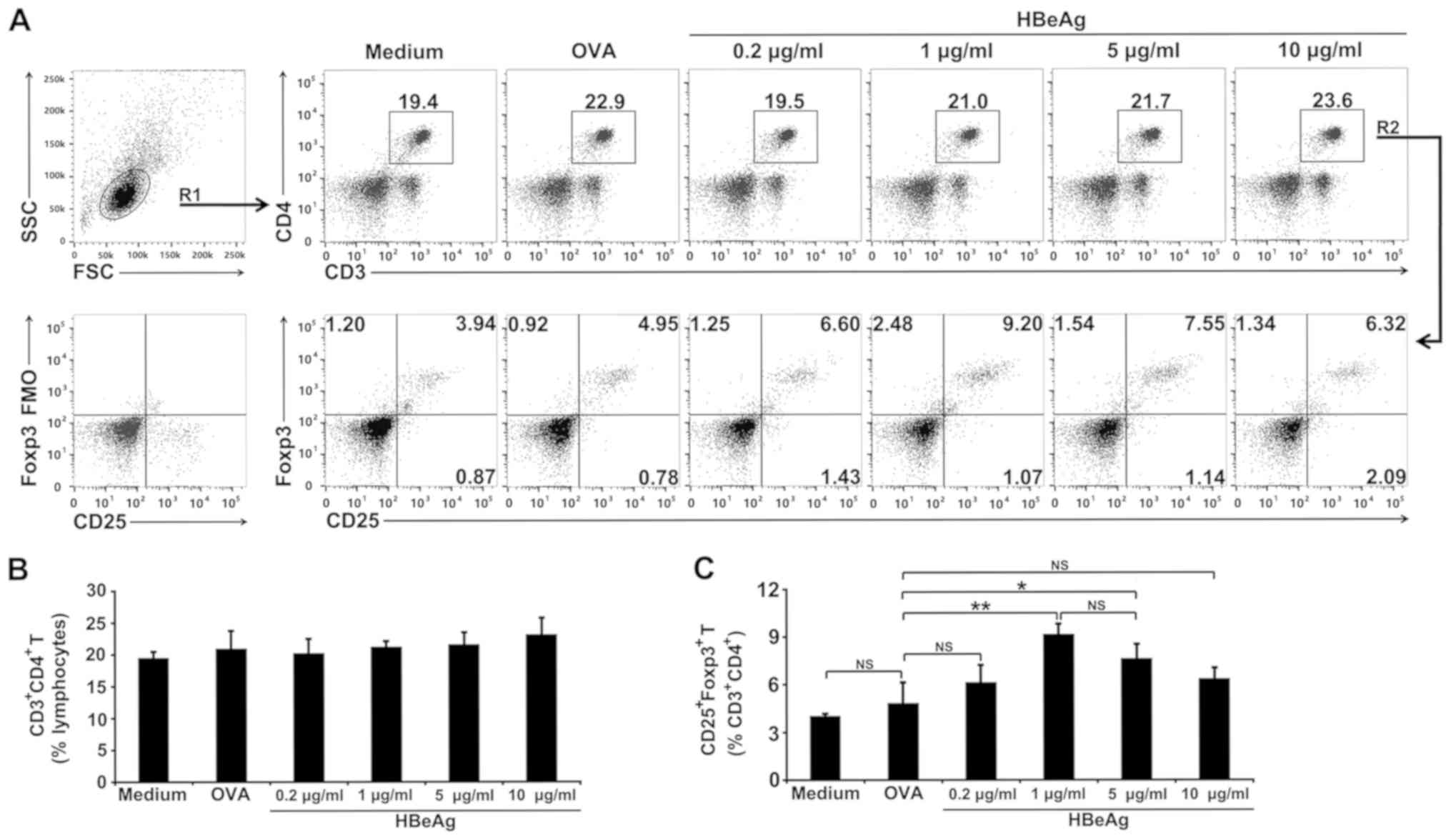 | Figure 1HBeAg treatment increases the
proportion of Tregs in spleen cells in vitro. (A) Total
spleen cells from normal mice were cultured in medium alone or in
the presence of 1 µg/ml of OVA or different concentrations of HBeAg
(0.2, 1, 5 or 10 µg/ml). Cells were collected, and flow cytometry
was performed to identify
CD3+CD4+CD25+Foxp3+
Tregs 3 days later. The gating strategy shows total lymphocytes
(R1) and total CD3+CD4+ T cells (R2). Dot
plots of CD3 vs. CD4 gated lymphocytes, or CD25 vs. Foxp3
expression, gated on CD3+CD4+ T cells, are
representative of one of three independent experiments. Numbers on
plots represent the percentage of cells for each quadrant in gated
cell populations. Percentages of CD3+CD4+ T
cells in (B) lymphocytes or (C) CD25+Foxp3+
Tregs in CD3+CD4+ T cells are shown as the
mean ± SD of triplicate cultures and representative of three
independent experiments. *P<0.05,
**P<0.01. HBeAg, hepatitis B envelope antigen; Foxp3,
forkhead box protein 3; NS, not significant; Tregs, T regulatory
cells. |
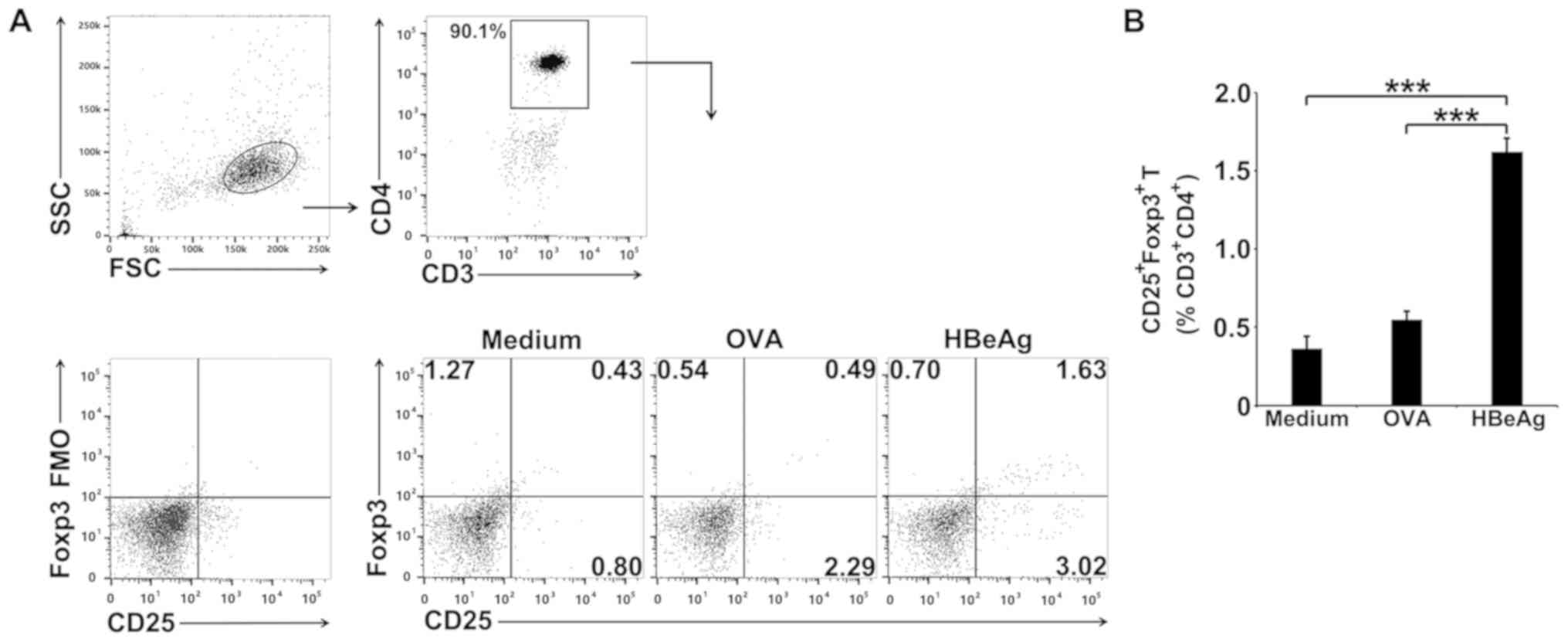
HBeAg converts
CD4+CD25- T cells to
CD4+CD25+Foxp3+ Tregs in
vitro
To determine whether the increase in Treg numbers
induced by HBeAg could be due to de novo induction of Foxp3
expression in activated T cells, MACS-isolated
CD4+CD25- T cells from spleens of normal
BALB/c mice were cultured with HBeAg in the presence of APCs. The
number of Tregs was determined 3 days later by FCM, and the results
suggested that HBeAg significantly converted
CD4+CD25- T cells to
CD4+CD25+Foxp3+ Tregs when
compared to the medium or OVA control groups (Fig. 2A and 2B).
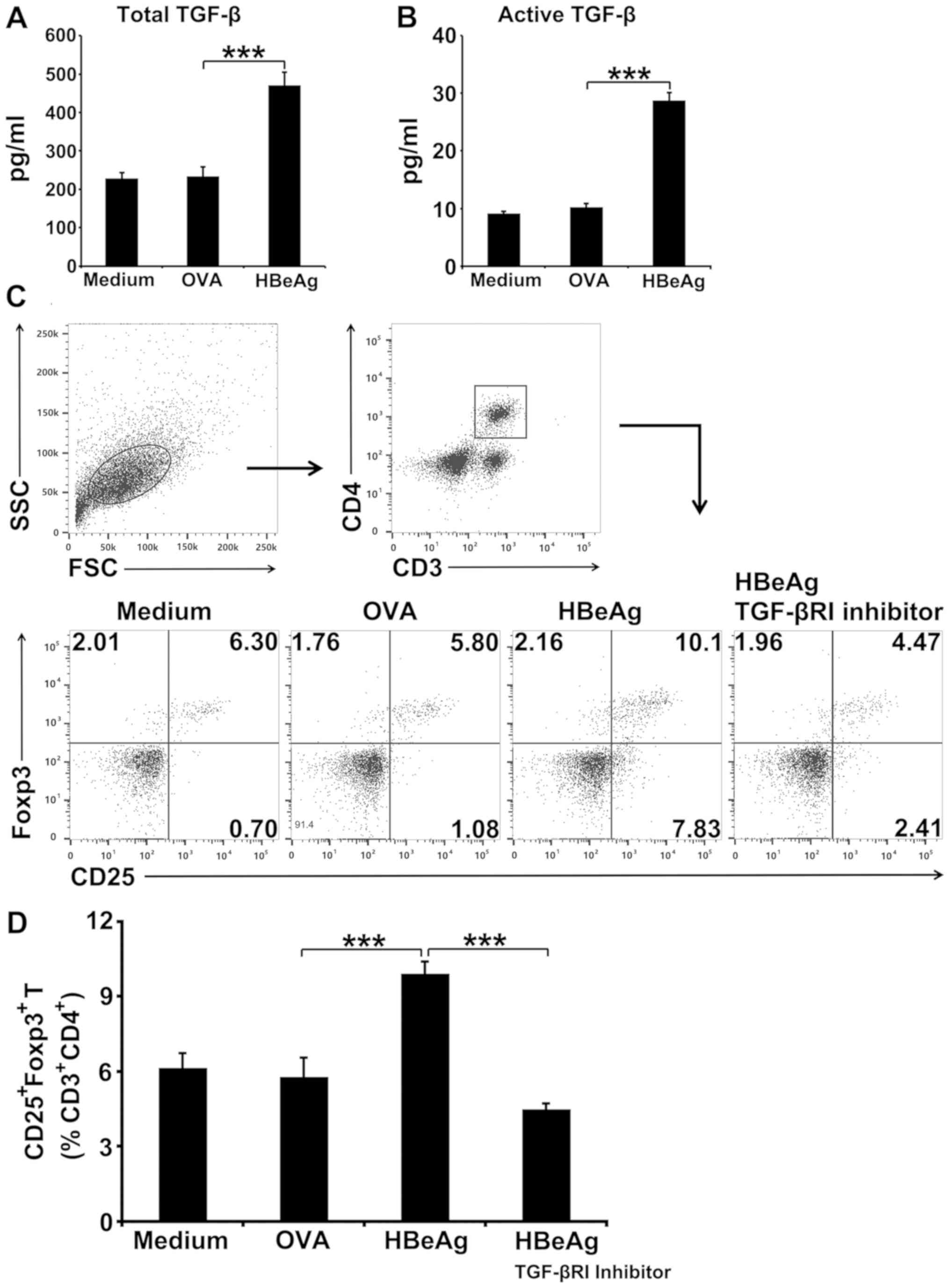
HBeAg induces conversion of T cells to
Tregs in spleen cells in vitro by triggering TGF-β production
As TGF-β plays a critical role in the conversion of
peripheral T cells to Tregs by promoting Foxp3 expression (21,22),
whether HBeAg could induce TGF-β production in murine spleen cells
in vitro was analyzed. Indeed, HBeAg induced a two-fold
increase in total TGF-β production by spleen cells compared with
the OVA-treated control (Fig. 3A).
The presence of biologically active TGF-β was also assessed and a
3-fold increase in active TGF-β release from HBeAg-treated spleen
cells was observed when compared to OVA-treated cultures (Fig. 3B). To further investigate whether
TGF-β was required for HBeAg to induce conversion to Tregs, studies
with TGF-βRI inhibitor in spleen cell culture were performed. It
was observed that blocking of TGF-β signaling almost completely
abolished the ability of HBeAg to induce conversion of T cells to
Tregs (Fig. 3C and 3D). Thus, TGF-β is required to enable HBeAg
to convert T cells to Tregs.
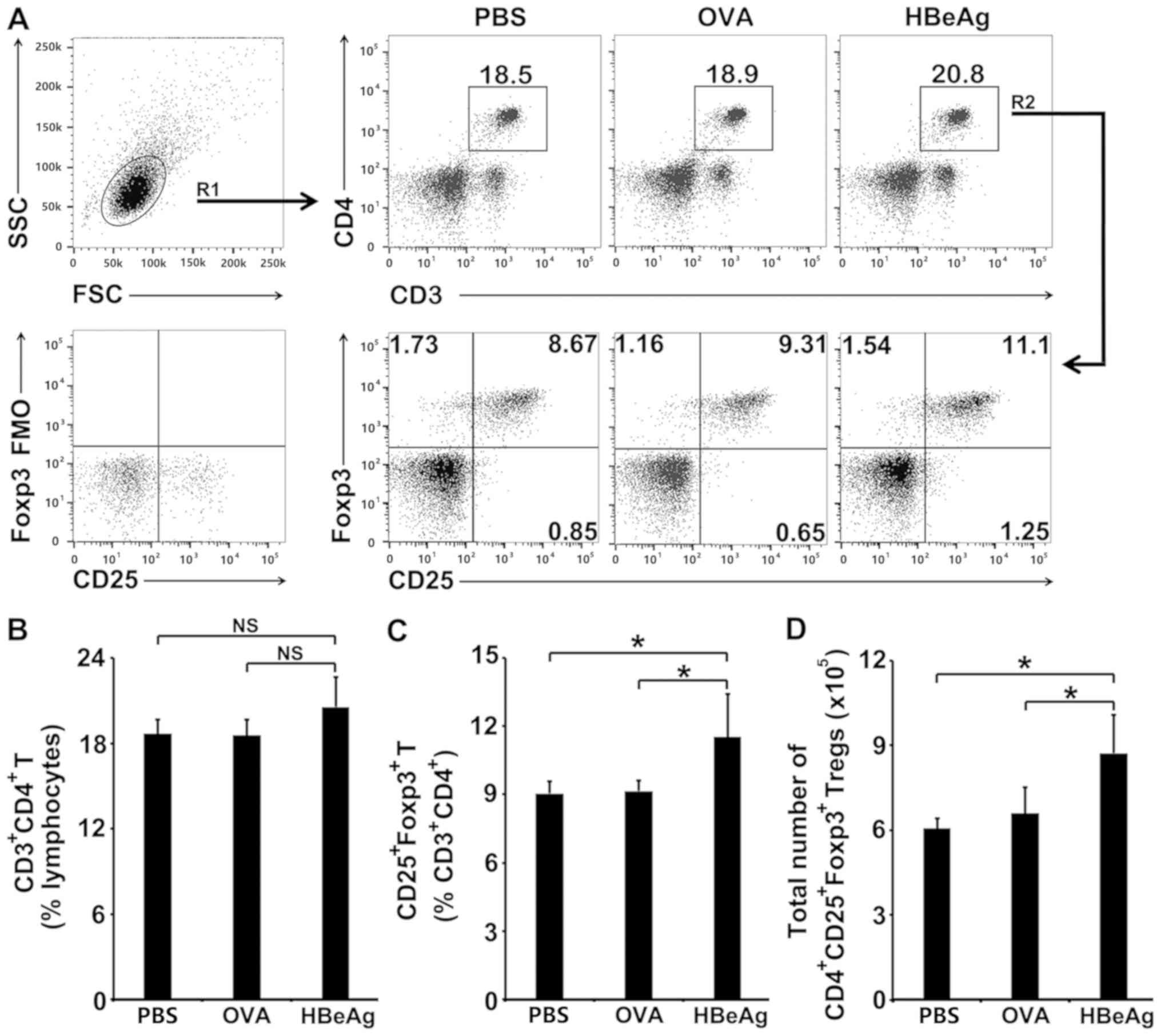
HBeAg converts T cells to Tregs in
vivo
Whether HBeAg had the capacity to convert peripheral
T cells into Tregs in vivo was then examined. HBeAg was
subcutaneously injected into normal BALB/c mice. As shown in
Fig. 4A and 4B, OVA or HBeAg treatment in vivo
did not significantly change the percentage of CD4+ T
cells in total lymphocytes. HBeAg treatment in vivo induced
a pronounced increase in the percentage of Tregs within the
CD4+ T cell population when compared to either OVA- or
PBS-treated control (Fig. 4A and
4C). In addition, absolute numbers
of Tregs in the spleens of HBeAg-injected mice increased
consistently (Fig. 4D).
Discussion
CHB is characterized by HBeAg positivity. HBeAg has
been shown to regulate the host immune response to maintain a
tolerant state and promote HBV persistence in natural infection
(13,23), but the mechanism by which HBeAg
induces immune tolerance remains unclear. The mouse is an
appropriate animal model for immunological studies (24), and the present study does not involve
the investigation of HBV pathogenesis; thus, we used a murine
experimental system to explore immunoregulatory function of
HBeAg.
Among immunoregulatory cell populations, Tregs
remain paramount in CHB patients (14). CHB patients, especially those with
HBeAg positivity, exhibit a higher percentage of Tregs in their
peripheral blood than those with HBeAg-negative CHB (15,19,20).
Therefore, it was initially hypothesized that HBeAg was involved in
the conversion of T cells to Tregs in CHB patients. A combination
of in vitro and in vivo assays appeared to validate
the hypothesis that HBeAg induced a significant increase in the
proportion of Tregs isolated from mouse spleens. To the best of our
knowledge, the current study is the first to provide preliminary
evidence for the contribution of HBeAg to Treg generation in
patients with CHB. In addition, previous studies have suggested
that efficient Treg induction requires low doses of antigens, which
is possibly related to weak T cell receptor signaling (25-27).
Consistently, in in vitro study, a dose of 1 µg/ml HBeAg was
found to be more effective in the conversion of T cells to Tregs
than that of 5 µg/ml, although the difference between the two was
not statistically significant. The high dose (10 µg/ml) of HBeAg
was found unable to induce the increase the number of Tregs,
probably due to the induction of a strong T cell receptor
stimulation.
Peripheral Treg differentiation is induced upon T
cell activation with high-affinity agonist antigens (28,29).
Predictions indicated multiple high-affinity potential MHC class
II-binding epitopes on HBeAg. However, currently it was not
possible to verify the most immunogenic epitopes that are predicted
to be potentially responsible for naive CD4+ T cell
activation and subsequent peripheral Treg differentiation in the
present study. The naive CD4+ T cell population
expresses a huge repertoire of receptors that are highly diverse in
their epitope-binding sites (30).
Only few naive CD4+ T cells can be reactive to a single
HBeAg epitope, so it is likely that very few CD4+ T
cells could be induced to differentiate into Tregs by a single
epitope. HBeAg-mediated differentiation of T cells into Tregs may
require a combination of multiple epitopes, in a complex process
which merits further study.
Previous studies have revealed that high amounts of
TGF-β are required for foreign antigen-mediated induction of Foxp3
expression in peripheral naive CD4+ T cells (29,31).
Considering these findings, the present data suggested that HBeAg
was able to trigger TGF-β production and indeed required to enable
HBeAg to induce T cell conversion into Tregs in mouse spleen.
Previous research has demonstrated that patients with CHB exhibit
significantly higher serum levels of TGF-β than healthy people
(32) and HBeAg may contribute to
the elevated serum levels of TGF-β in patients with CHB. Diverse
varieties of immune cells, such as macrophages, monocytes, NK cells
and CD4+ T cells, are involved in the production of
TGF-β during chronic HBV infection (33). However, which cells in the spleen
that are primarily responsible for HBeAg-increased TGF-β production
requires further investigation. The present data suggest that HBV
exploits immune cells to create an TGF-β-rich microenvironment for
peripheral Treg differentiation and HBV persistence, and are
consistent with the notion that HBeAg can condition innate immune
cells into anti-inflammatory types (34,35).
Although HBeAg is not required for HBV assembly,
replication or viral infection, various studies have shown that
HBeAg is capable of impairing innate immune responses or
inactivating HBeAg-specific T cells by clonal deletion or anergy
(34,36). The present study demonstrated the
additional ability of HBeAg to convert
CD4+CD25- T cells to
CD4+CD25+Foxp3+ Tregs in
vitro and suggest a mechanistic explanation for HBeAg, as an
immune tolerogen, to modulate the host immune response and promote
HBV chronicity. However, current data in mice are preliminary, and
further investigation is needed to validate the T cell
differentiating capabilities of HBeAg in HBV transgenic mice and to
explore the underlying molecular mechanisms in future works.
In summary, the present murine experimental data
indicate that HBeAg is able to convert T cells into Tregs in mouse
spleen and suggest this may be due to the increased TGF-β
production induced by HBeAg.
Acknowledgements
Not applicable.
Funding
This work was supported by grants from the Natural
Science Foundation of Jiangsu Province (grant no. BK20170105) and
the National Natural Science Foundation of China (grant no.
81971963) to Professor Sha Zhou and from the National Natural
Science Foundation of China (grant no. 81871675) to Professor Chuan
Su.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
SZ and CS conceived and designed the study. RT, ZL,
XW, QQ, JH, DL, XW and YL performed the experiments. SZ, XC, JZ and
CS analyzed the data. SZ and CS wrote the paper. All the authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures for the use of laboratory animals
were approved by the Institutional Animal Care and Use Committee
(IACUC) of Nanjing Medical University (permit no.
IACUC-1601123).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Seto WK, Lo YR, Pawlotsky JM and Yuen MF:
Chronic hepatitis B virus infection. Lancet. 392:2313–2324.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Yuen MF, Chen DS, Dusheiko GM, Janssen
HLA, Lau DTY, Locarnini SA, Peters MG and Lai CL: Hepatitis B virus
infection. Nat Rev Dis Primers. 4(18035)2018.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Maini MK and Pallett LJ: Defective T-cell
immunity in hepatitis B virus infection: Why therapeutic
vaccination needs a helping hand. Lancet Gastroenterol Hepatol.
3:192–202. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Park JJ, Wong DK, Wahed AS, Lee WM, Feld
JJ, Terrault N, Khalili M, Sterling RK, Kowdley KV, et al:
Hepatitis B virus--specific and global T-cell dysfunction in
chronic hepatitis B. Gastroenterology. 150:684–695.e5.
2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Bertoletti A and Naoumov NV: Translation
of immunological knowledge into better treatments of chronic
hepatitis B. J Hepatol. 39:115–124. 2003.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bertoletti A and Ferrari C: Innate and
adaptive immune responses in chronic hepatitis B virus infections:
Towards restoration of immune control of viral infection. Gut.
61:1754–1764. 2012.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Bertoletti A and Ferrari C: Kinetics of
the immune response during HBV and HCV infection. Hepatology.
38:4–13. 2003.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chang C, Enders G, Sprengel R, Peters N,
Varmus HE and Ganem D: Expression of the precore region of an avian
hepatitis B virus is not required for viral replication. J Virol.
61:3322–3325. 1987.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chen HS, Kew MC, Hornbuckle WE, Tennant
BC, Cote PJ, Gerin JL, Purcell RH and Miller RH: The precore gene
of the woodchuck hepatitis virus genome is not essential for viral
replication in the natural host. J Virol. 66:5682–5684.
1992.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Schlicht HJ, Salfeld J and Schaller H: The
duck hepatitis B virus pre-C region encodes a signal sequence which
is essential for synthesis and secretion of processed core proteins
but not for virus formation. J Virol. 61:3701–3709. 1987.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Boni C, Laccabue D, Lampertico P, Giuberti
T, Viganò M, Schivazappa S, Alfieri A, Pesci M, Gaeta GB, et al:
Restored function of HBV-specific T cells after long-term effective
therapy with nucleos(t)ide analogues. Gastroenterology.
143:963–973.e9. 2012.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Chen M, Sällberg M, Hughes J, Jones J,
Guidotti LG, Chisari FV, Billaud JN and Milich DR: Immune tolerance
split between hepatitis B virus precore and core proteins. J Virol.
79:3016–3027. 2005.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Milich D and Liang TJ: Exploring the
biological basis of hepatitis B e antigen in hepatitis B virus
infection. Hepatology. 38:1075–1086. 2003.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Manigold T and Racanelli V: T-cell
regulation by CD4 regulatory T cells during hepatitis B and C virus
infections: Facts and controversies. Lancet Infect Dis. 7:804–813.
2007.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Stoop JN, van der Molen RG, Baan CC, van
der Laan LJ, Kuipers EJ, Kusters JG and Janssen HL: Regulatory T
cells contribute to the impaired immune response in patients with
chronic hepatitis B virus infection. Hepatology. 41:771–778.
2005.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Aalaei-Andabili SH and Alavian SM:
Regulatory T cells are the most important determinant factor of
hepatitis B infection prognosis: A systematic review and
meta-analysis. Vaccine. 30:5595–5602. 2012.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lin CY, Tsai MC, Huang CT, Hsu CW, Tseng
SC, Tsai IF, Chen YC, Yeh CT, Sheen IS and Chien RN: Liver injury
is associated with enhanced regulatory T-cell activity in patients
with chronic hepatitis B. J Viral Hepat. 14:503–511.
2007.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Stoop JN, van der Molen RG, Kuipers EJ,
Kusters JG and Janssen HL: Inhibition of viral replication reduces
regulatory T cells and enhances the antiviral immune response in
chronic hepatitis B. Virology. 361:141–148. 2007.PubMed/NCBI View Article : Google Scholar
|
|
19
|
El-Badawy O, Sayed D, Badary MS,
Abd-Alrahman ME, El-Feky MA and Thabit AG: Relations of regulatory
T cells with hepatitis markers in chronic hepatitis B virus
infection. Hum Immunol. 73:335–341. 2012.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Peng G, Li S, Wu W, Sun Z, Chen Y and Chen
Z: Circulating CD4+ CD25+ regulatory T cells correlate with chronic
hepatitis B infection. Immunology. 123:57–65. 2008.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Chen W, Jin W, Hardegen N, Lei KJ, Li L,
Marinos N, McGrady G and Wahl SM: Conversion of peripheral
CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta
induction of transcription factor Foxp3. J Exp Med. 198:1875–1886.
2003.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Peng Y, Laouar Y, Li MO, Green EA and
Flavell RA: TGF-beta regulates in vivo expansion of
Foxp3-expressing CD4+CD25+ regulatory T cells responsible for
protection against diabetes. Proc Natl Acad Sci USA. 101:4572–4577.
2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Maruyama T, Iino S, Koike K, Yasuda K and
Milich DR: Serology of acute exacerbation in chronic hepatitis B
virus infection. Gastroenterology. 105:1141–1151. 1993.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Masopust D, Sivula CP and Jameson SC: Of
mice, dirty mice, and men: Using mice To understand human
immunology. J Immunol. 199:383–388. 2017.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Turner MS, Kane LP and Morel PA: Dominant
role of antigen dose in CD4+Foxp3+ regulatory T cell induction and
expansion. J Immunol. 183:4895–4903. 2009.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Shevach EM: From vanilla to 28 flavors:
Multiple varieties of T regulatory cells. Immunity. 25:195–201.
2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Kang J, Huddleston SJ, Fraser JM and
Khoruts A: De novo induction of antigen-specific CD4+CD25+Foxp3+
regulatory T cells in vivo following systemic antigen
administration accompanied by blockade of mTOR. J Leukoc Biol.
83:1230–1239. 2008.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Gottschalk RA, Corse E and Allison JP: TCR
ligand density and affinity determine peripheral induction of Foxp3
in vivo. J Exp Med. 207:1701–1711. 2010.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li MO and Rudensky AY: T cell receptor
signalling in the control of regulatory T cell differentiation and
function. Nat Rev Immunol. 16:220–233. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Moon JJ, Chu HH, Pepper M, McSorley SJ,
Jameson SC, Kedl RM and Jenkins MK: Naive CD4(+) T cell frequency
varies for different epitopes and predicts repertoire diversity and
response magnitude. Immunity. 27:203–213. 2007.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Josefowicz SZ, Lu LF and Rudensky AY:
Regulatory T cells: Mechanisms of differentiation and function.
Annu Rev Immunol. 30:531–564. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Khorramdelazad H, Hassanshahi G, Nasiri
Ahmadabadi B and Kazemi Arababadi M: High serum levels of TGF-β in
Iranians with chronic HBV infection. Hepat Mon.
12(e7581)2012.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Li H, Zhai N, Wang Z, Song H, Yang Y, Cui
A, Li T, Wang G, Niu J, Crispe IN, et al: Regulatory NK cells
mediated between immunosuppressive monocytes and dysfunctional T
cells in chronic HBV infection. Gut. 67:2035–2044. 2018.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Tsai KN, Kuo CF and Ou JJ: Mechanisms of
hepatitis B virus persistence. Trends Microbiol. 26:33–42.
2018.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Yang F, Yu X, Zhou C, Mao R, Zhu M, Zhu H,
Ma Z, Mitra B, Zhao G, et al: Hepatitis B e antigen induces the
expansion of monocytic myeloid-derived suppressor cells to dampen
T-cell function in chronic hepatitis B virus infection. PLoS
Pathog. 15(e1007690)2019.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ciupe SM: Modeling the dynamics of
hepatitis B infection, immunity, and drug therapy. Immunol Rev.
285:38–54. 2018.PubMed/NCBI View Article : Google Scholar
|