Introduction
Breast cancer (BC) is a common malignancy in
females, with an estimated 2.1 million new cases (11.6%) and
626,679 deaths (6.6%) in 2018 (1,2).
Triple-negative BC (TNBC) often develops in young women and is
phenotypically defined as the lack of receptors for estrogen (ER),
progesterone (PR) and human epidermal growth factor receptor 2
(HER-2) (3). Cells from TNBC are
more invasive, prone to lymph node metastasis and are associated
with poor clinical outcomes, when compared with cells from non-TNBC
(4-6).
TNBC cells respond poorly to endocrine and anti-HER-2 therapeutic
strategies, as they lack the receptors targeted by these therapies
and, thus, surgical treatment is preferred, followed by
postoperative chemotherapy or radiotherapy (2). Ongoing clinical studies are evaluating
the efficacy of targeting PI3K and inhibiting EGFR (7), but their efficacies remain
unknown.
BC cells invade and migrate to surrounding or
distant tissues via the process of epithelial-mesenchymal
transition (EMT) (8), which
converts cancerous epithelial cells to mesenchymal cells under
defined physiologic or pathological conditions (9). EMT is characterized by the decreased
expression of epithelial markers, such as E-cadherin (E-Ca) and
increased expression of mesenchymal markers, such as N-cadherin
(N-Ca) and vimentin (10,11). While being homologous and belonging
to the same cadherin superfamily, E-Ca is primarily expressed on
epithelial cells and is reported to inhibit cancer cell invasion
(12,13), while N-Ca is one of the major
cadherins expressed on mesenchymal cells. Therefore, switching
expression from E-Ca to N-Ca is a key cellular signature of EMT.
Thus, suppressing the expression of N-Ca could potentially decrease
the invasion and migration of cancer cells (14). Vimentin is one of the most widely
expressed and highly conserved proteins in the type III
intermediate filament protein family (15), and is known to promote the invasion
and migration of cancer cells. Furthermore, EMT markers are
associated with the biological features of tumors, such as the
characteristics of the tumor, and its invasive and migratory
abilities. Detection of changes in the expression levels of EMT
markers could assist in investigating the differences in the
biological features and the changes in protein expression levels.
This method has been used to evaluate the prognosis of patients
with hepatocellular carcinoma, prostate cancer and BC (8,16,17).
The coatomer protein complex subunit β2
(COPB2) gene located in chromosome 3q2.3 encodes the 102-kDa
nucleoprotein COPB2, which is a member of the 7 protein Golgi
coatomer complex. COPB2 is essential for budding and vesicular
trafficking between the endoplasmic reticulum and Golgi membrane.
It is therefore essential in maintaining cellular homeostasis,
including the transcriptional regulation and signal transduction of
cells (18-22).
Previous studies have revealed that the expression
of COPB2 is significantly upregulated in prostate cancer,
cholangiocarcinoma, lung cancer and colon cancer cells, and that it
enhances proliferation (23-26).
Furthermore, COPB2 regulates the proliferation of colon cancer
cells via the JNK/c-Jun signaling pathway (26). Moreover, silencing the COPB2
gene decreases the expression levels of proteins associated with
the receptor-tyrosine kinase (RTK) signaling pathway in gastric
cancer cells, such as EGFR, HER-2, fms related RTK 3 and
phosphorylated (p)-AKT (27).
The role of COPB2 in the pathogenesis of TNBC
remains poorly understood. However, circumstantial evidence
reported in the abovementioned literature suggests that COPB2 may
be a key contributor to the phenotype of TNBC cells and that its
action may be mediated via the AKT signaling pathway. The present
study hypothesized that the increased expression level of COPB2 in
TNBC cells may contribute to the occurrence of EMT, which could
promote the migration and invasion capacities of TNBC cells in
vitro. This effect may be mediated via the AKT signaling
pathway.
Materials and methods
Cell lines and culture conditions
The clonal TNBC HS-578T and non-TNBC MCF-7 cell
lines (The Type Culture Collection of The Chinese Academy of
Sciences) were cultured at 37˚C with 5% CO2 in DMEM
(Hyclone; Cytiva) supplemented with 10 and 15% FBS (Biological
Industries).
Lentiviral transduction of HS-578T
cells
HS-578T cells were seeded in 6-well culture plates
(4x104 cells/well) and incubated at 37˚C until 30%
confluent. Cells were then divided into three groups: Infection
with lentiviral vector (Lv)-short hairpin (sh)RNA COPB2 with a
green fluorescent protein (GFP) tag (sh-COPB2 from Shanghai
GenePharma Co., Ltd.); infection with empty lentiviruses with a GFP
tag (sh-Control; Shanghai GenePharma Co., Ltd.); or no infection.
sh-COPB2 (AGATTAGAGTGTTCAATTA) was inserted into Gv248 lentiviral
vectors (Shanghai GenePharma Co., Ltd.) and the cells were infected
as previously described (26).
Then, 96 h after the final infection, the cells were analyzed for
infection efficiency by counting the number of GFP-positive cells
under a fluorescence microscope (ECLIPSE 80i; Nikon
Corporation).
Immunoblots
HS-578T (three experimental groups) and MCF-7 cells
were lysed in a RIPA buffer (Beijing Solarbio Science &
Technology Co., Ltd.) containing 1 mM PMSF (Applygen Technologies,
Inc.). The Cell lysates (60 µg total proteins/lane) were separated
via 10% SDS-PAGE and transferred to a PVDF membrane (EMD
Millipore). The membrane was blocked with 5% BSA (Beijing Solarbio
Science & Technology Co., Ltd.) at room temperature for 1 h and
then probed with antibodies (1:1,000 dilution) against COPB2 (cat.
no. HPA036867; MilliporeSigma), E-Ca (cat. no. 3195S; Cell
Signaling Technology, Inc.), N-Ca (cat. no. 13116S; Cell Signaling
Technology, Inc.), vimentin (cat. no. 5741S; Cell Signaling
Technology, Inc.), p-AKT (cat. no. 4060T; Cell Signaling
Technology, Inc.), AKT (cat. no. 4685S; Cell Signaling Technology,
Inc.) and AKT agonist SC79 (cat. no. HY-18749; MedChemExpress).
GAPDH (cat. no. YM3040; ImmunoWay Biotechnology Company) served as
the protein loading control and was probed using a monoclonal
antibody. ImageJ 1.51j8 software (National Institutes of Health)
was used to analyze the density of the bands.
MTT cell proliferation assay
HS-578T and MCF-7 cells were seeded at a cell
density of 2x103 cells/well in 96-well plates and
cultured at 37˚C with 5% CO2. Cell densities were
analyzed on day 1, 2, 3, 4 and 5 after seeding. At each time point,
the cells were incubated with 20 µl MTT solution for 4 h at 37˚C,
and then cells were treated with 150 µl DMSO with constant
agitation. After a 10-min incubation at room temperature, the
supernatant was collected and MTT signals at an optical density
(OD) of 490 nm were quantitatively detected using a microplate
reader (Bio-Rad Laboratories, Inc.).
Colony formation assay
HS-578T and MCF-7 cells were seeded in 6-well plates
at a density of 1x103 cells/well and cultured at 37˚C
with 5% CO2 for 10 days. Cells were then stained for 10
min with crystal violet and fixed with 4% paraformaldehyde for 30
min at room temperature. The number of cell colonies was counted,
and images were captured under a light microscope.
Cellular migration and invasion
assay
Cell migration and invasion assays were performed
using a 24-well Transwell chamber (pore size, 8 µm; Corning, Inc.)
as previously described (26). For
the invasion assay, the upper chambers were coated with 40 µl
Matrigel (BD Biosciences) diluted in DMEM for 30 min at 37˚C.
HS-578T and MCF-7 cells were cultured in the upper chamber at a
density of 2x105 cells/ml (200 or 160 µl/well for
migration and invasion assays, respectively). The upper chambers
were submerged into the lower chamber containing 15%
FBS-supplemented medium (500 µl). After 24 and 48 h in culture at
37˚C (5% CO2), non-migrated cells that remained in the
upper chambers were removed using a cotton swab. The migrated or
invaded cells on the opposite side of the membrane were stained
with 0.1% crystal violet for 10 min at room temperature, and
counted under an ECLIPSE 80i fluorescence microscope.
Statistical analysis
The dataset regarding COPB2 gene expression
and histological grade of BC was obtained from The Cancer Genome
Atlas (TCGA) database. The differential expression analysis was
performed using the R package, DESeq2 (https://www.bioconductor.org/packages/devel/bioc/html/DESeq2.html).
Quantitative data from multiple independent experiments were
expressed as the means ± SD. The quantitative data were analyzed
using Student's unpaired t-test to compare between two groups. To
compare the data of three or more groups, one-way ANOVA and Tukey's
post hoc test was used. All statistical analysis was performed
using GraphPad Prism 8.0 statistical software (GraphPad Software,
Inc.) and SPSS 19.0 software (IBM Corp.) and P<0.05 was
considered to indicate a statistically significant difference.
Results
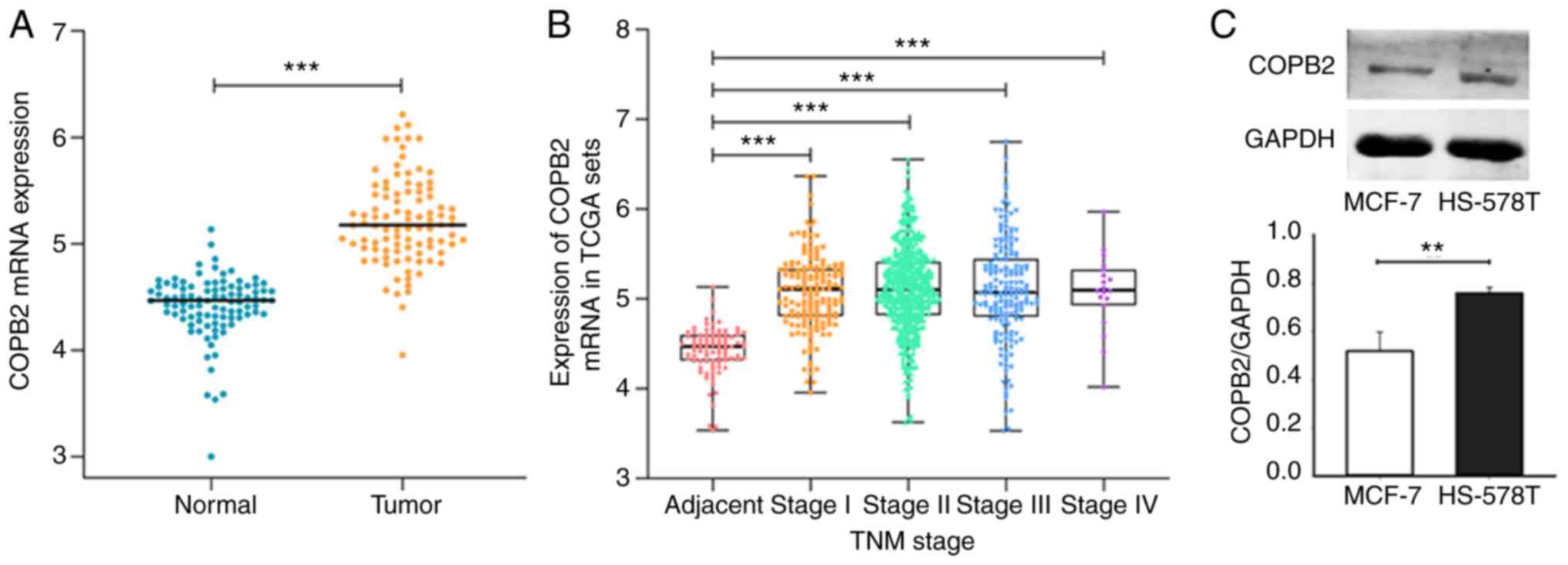
COPB2 mRNA is upregulated in TNBC
COPB2 mRNA expression was evaluated using the
data of 110 patients with BC obtained from TCGA database. It was
found that mRNA expression levels of COPB2 in BC tissue were
significantly higher when compared with those in paracancerous
tissue (Fig. 1A). Furthermore, the
COPB2 mRNA expression level progressively increased in line
with the pathological stage of the cancer (Fig. 1B). As the TNBC status was not
provided for patients in TCGA database, COPB2 expression was
analyzed in the cultured TNBC-derived HS-578T cells and the
non-TNBC-derived MCF-cells. The HS-578T cells expressed more COPB2
protein when compared with the non-TNBC MCF-7 cells (Fig. 1C). These results suggest that COPB2
expression was increased in all BC cells, but the increase was
significantly greater in TNBC cells. Thus, differential COPB2
expression may contribute to the distinct phenotypes between TNBC
and non-TNBC cells.
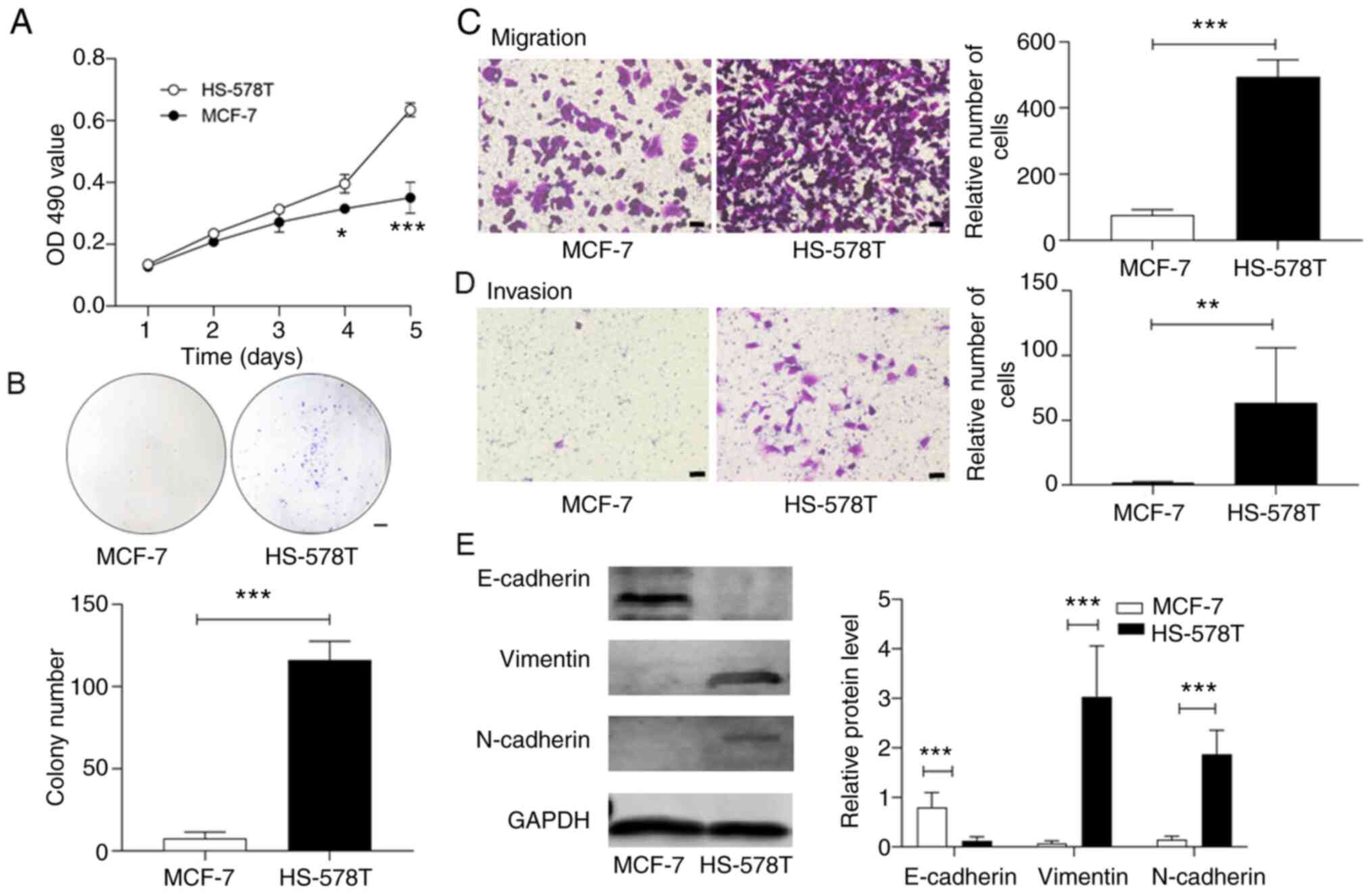
HS-578T and MCF-7 cells exhibit
differential proliferative rates
HS-578T cells demonstrated a greater rate of
proliferation in culture when compared with MCF-7 cells, as
detected by the MTT assay (Fig.
2A). Moreover, HS-578T cells formed significantly more cellular
colonies (Fig. 2B). Using the
Transwell chamber system as previously described (26), it was identified that HS-578T cells
invaded and transmigrated through the extracellular matrix of the
Matrigel significantly faster than the MCF-7 cells (Fig. 2C and D).
To investigate the regulatory pathways responsible
for the distinct phenotypes of HS-578T and MCF-7 cells, the
expression levels of factors involved in EMT were detected. The
results demonstrated that epithelial E-Ca was predominantly
expressed in MCF-7 cells, whereas the homologous mesenchymal N-Ca
was mostly detected in HS-578T cells (Fig. 2E). The cytoskeletal protein
vimentin, another EMT marker, was also increased in HS-578T cells.
These data indicated that the TNBC HS-578T and non-TNBC MCF-7 cells
had different proliferative potentials that were likely determined
by their EMT states.
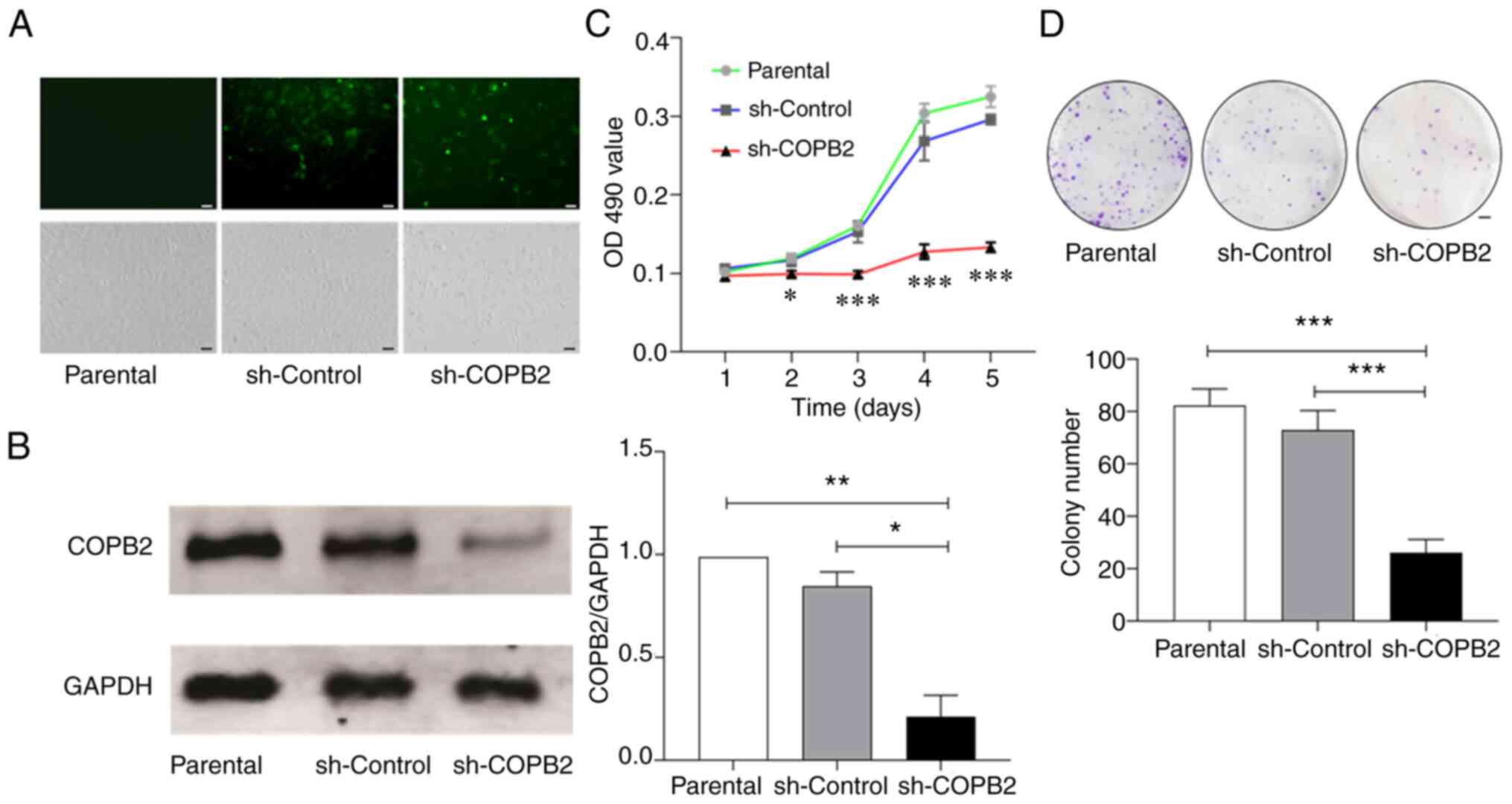
Silencing COPB2 decreases the
proliferation of HS-578T cells
As high COPB2 expression was detected in tissue
samples collected from patients with BC, and differential
expressions of COPB2 was observed in the clonal TNBC HS-578T and
non-TNBC MCF-7 cells (Fig. 1); the
role of COPB2 in the formation of TNBC was subsequently
investigated. The present study examined the phenotypic changes of
the TNBC HS-578T cells after the COPB2 gene was silenced
using a lentivirus as the carrier, which infected >80% of cells
(Fig. 3A; upper panel) without
inducing significant cell death or detachment (Fig. 3A; lower panel). When infected with
the Lv-sh-COPB2 lentivirus, COPB2 expression was decreased by 90%,
whereas the control lentivirus achieved a similar infection
efficiency, but did not downregulate COPB2 expression (Fig. 3B).
Using these techniques, it was found that HS-578T
cells infected with the Lv-sh-COPB2 lentivirus had a significantly
decreased rate of proliferation when compared with parental cells
and the cells infected with the sh-Control lentivirus (Fig. 3C). The inhibition of proliferation
was detected primarily on days 3, 4 and 5 after infection.
Consistent with the results from the MTT assay, colony formation
was also reduced in the HS-578T cells infected with Lv-sh-COPB2
lentivirus (Fig. 3D).
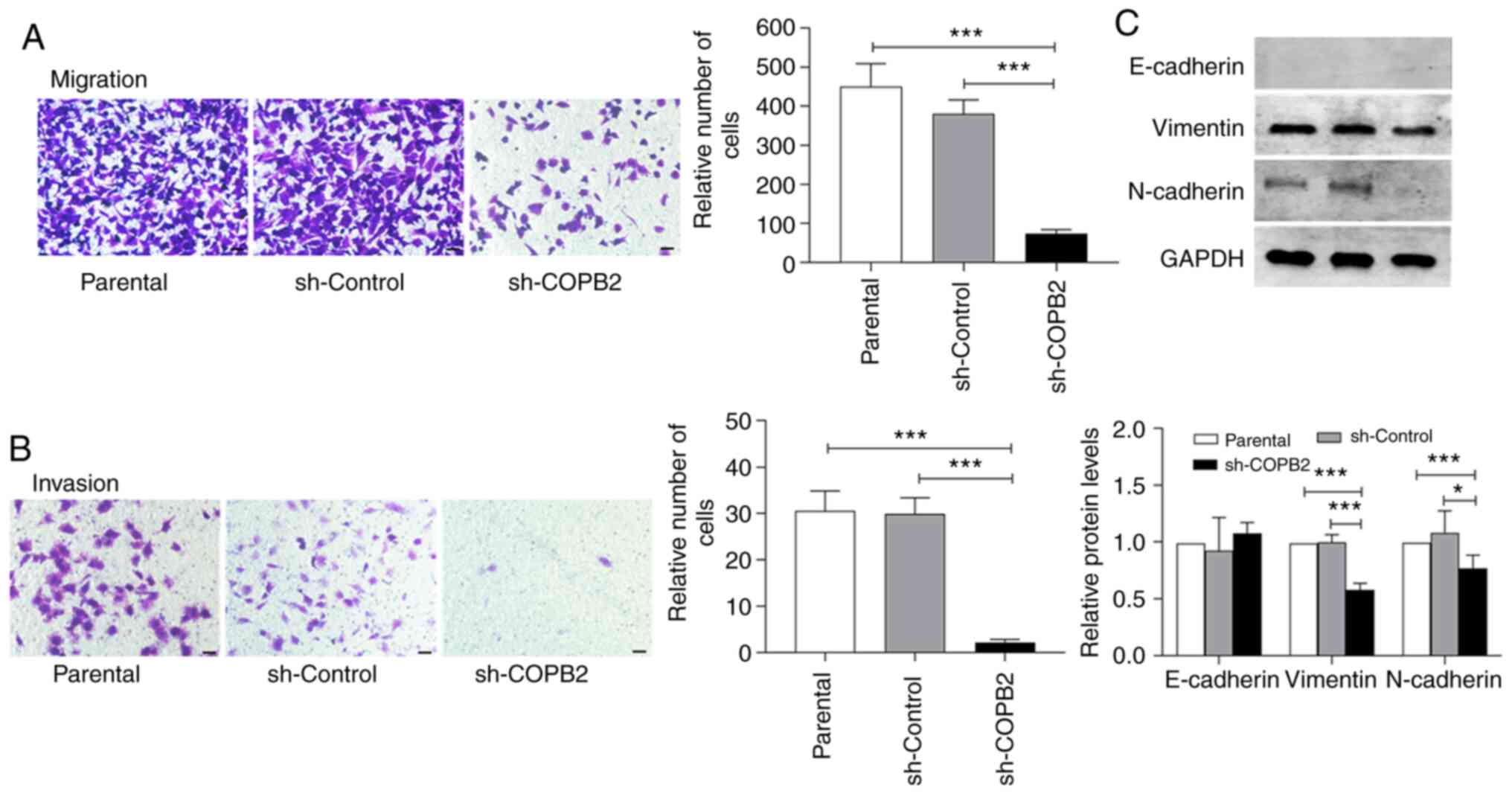
The ability of HS-578T cells to invade and migrate
via the subendothelial matrix was reduced after infection with
Lv-sh-COPB2, but this was not observed in the control lentivirus
group (Fig. 4A and B). These data suggest that silencing COPB2
expression decreased the proliferation of the TNBC HS-578T cells.
Moreover, the expression levels of vimentin and N-Ca were decreased
in HS-578T cells when the COPB2 gene was silenced, while
E-Ca expression (which was low in HS-578T cells, as shown in
Fig. 2E) and the quantitative
analysis remained unchanged (with the parental cell serving as a
baseline; Fig. 4C). These results
suggest that silencing COPB2 altered the EMT status of HS-578T
cells, leading to changes in the proliferative characteristics of
these cells.
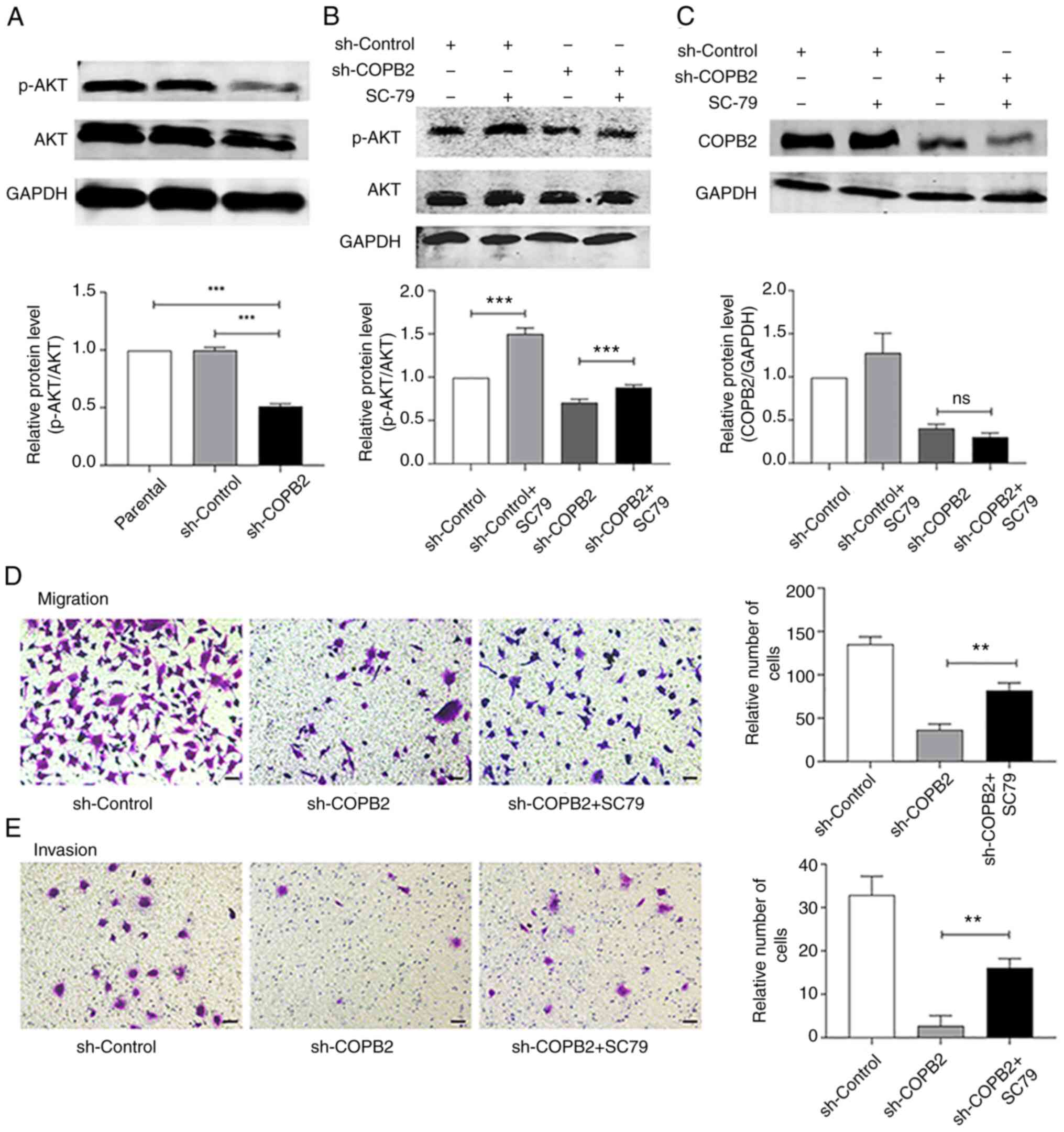
COPB2 silencing alters AKT signaling
in HS-578T cells
The AKT signaling pathway is a major pathway that
regulates the proliferation, migration and invasion of cancer cells
(28). It was identified that
silencing the COPB2 gene significantly decreased the
phosphorylation of AKT (Fig. 5A).
The AKT agonist, SC79 (5 µg/ml) served as a control, and increased
the rate of AKT phosphorylation (Fig.
5B) without significantly changing the expression of COPB2
(Fig. 5C). SC79 also promoted the
invasion and migration of HS-578T cells, as detected in the
Transwell assay (Fig. 5D and
E). These results indicated that
COPB2 may regulate AKT signaling to alter the proliferative rate of
HS-578T cells.
Discussion
Tumor targeting therapy is increasingly recognized
as an effective method of improving the efficacy and reducing the
cytotoxicity of anti-cancer drugs (2). As TNBC cells lack receptors targeted
by anti-HER-2 receptor, and anti-ER and anti-PR drugs, patients
with TNBC have fewer therapeutic options than those with non-TNBC.
Therefore, developing novel targeted therapeutic strategies for
TNBC is highly desirable. The present study identified a key role
of COPB2 in the EMT transition of TNBC cells.
In the present study, it was found that BC tissue
samples from patients and clonal cells in culture expressed high
levels of COPB2. These findings differ from a previous study, which
observed that the expression level of COPB2 was decreased in
cervical cancer cells (29).
However, the current findings were similar to a study by Bhandari
et al (30) who identified
that COPB2 was upregulated in BC. Thus, it was suggested that the
expression of COPB2 in BC is an important prognostic (30). The present study also found that the
TNBC cells expressed COPB2 at a level significantly higher than the
non-TNBC MCF-7 cells, indicating that COPB2 expression was
associated with TNBC. Notably, high COPB2 expression levels
resulted in greater rates of proliferation, migration and invasion
of the HS-578T cells in culture, when compared with the MCF-7
cells, and silencing COPB2 decreased the rate of EMT
transformation. The current findings are consistent with the EMT
state of HS-578T cells, showing a transition from expressing
epithelial E-Ca to expressing mesenchymal N-Ca and vimentin.
Moreover, these findings are consistent with a previous study, that
HS-578T cells are deficient in E-Ca expression and that MCF-7 cells
lack vimentin and N-Ca expression (31).
The present study used lentiviruses carrying a COPB2
inhibitory RNA sequence to decrease COPB2 transcription. Using this
approach, the present study was able to silence COPB2 expression by
>90% without affecting cell survival. Furthermore, silencing the
COPB2 gene decreased the proliferative, invasive and
migratory rates of HS-578T cells in vitro. These functional
effects of COPB2 silencing appeared to be mediated via
downregulation of the EMT proteins, N-Ca and vimentin, with a
minimal impact on E-Ca expression. These in vitro findings
justify the requirement for further investigation into the
potential of gene therapies targeting COPB2.
The present study demonstrated that the levels of
AKT phosphorylation were elevated in HS-578T cells and were reduced
by COPB2 silencing. In reciprocal experiments, the AKT agonist,
SC79 increased the migration and invasion of HS-578T cells. These
observations are consistent with the proposal that the AKT
signaling pathway promotes EMT transition and the proliferation of
cancer cells (32,33). The present results are also
consistent with those of various previous studies (34-36).
For example, AKT phosphorylation is downregulated after silencing
COPB2 in gastric cancer cells (27). The AKT agonist, SC79 only partially
increased the migration and invasion of HS-578T cells to a level
similar to that before silencing, suggesting that COPB2 may also
regulate cell invasion and migration via other signaling pathways,
such as the RTK signaling pathway and the inflammatory
immune-related pathway (27,37).
Furthermore, SC79 increased AKT phosphorylation, but did not change
the expression of COPB2, indicating that AKT phosphorylation is a
downstream event that COPB2 regulates. However, this experimental
study was only validated in triple-negative breast cancer HS-578T
cells, therefore subsequent experiments are required to further
verify this in the pathogenesis of BC. Additionally, the
correlation between the effect of COPB2 and the AKT signaling
pathway requires further investigation in the future.
Thus, the present study demonstrated that COPB2
expression was upregulated in BC cells and that the increase was
greater in TNBC cells when compared with non-TNBC cells.
Furthermore, higher levels of COPB2 expression enhance the
proliferation of TNBC cells. However, when COPB2 was silenced, EMT
transition was blocked and the proliferation of TNBC cells was
decreased. These findings suggested that COPB2 may be involved in
the clinical progression of TNBC and its underlying mechanism may
be associated with upregulation of the AKT signaling pathway.
Acknowledgements
The authors would like to thank Dr Jingfei Dong
(Bloodworks Research Institute, Seattle, WA, USA) for the critical
reading of the manuscript.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 81871919 and 81672399) and
the Fundamental Research Funds for the Central Universities (grant
nos. lzujbky-2017-136 and lzujbky-2019-it12) and the Lanzhou
Science and Technology planning project (grant nos. 2018-3-44).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML and FZ participated in conceiving the study, the
design of the experiments, interpretation of the results and
drafting of the manuscript. ML and FZ confirmed the authenticity of
all the raw data. WW performed the experiment, data analysis and
wrote part of the manuscript. CW contributed to data analysis,
wrote part of the manuscript and prepared the figures. FW, YW, YJ,
JL, MW, CZ and SW collected the majority of the data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
National Health Commission of the People's
Republic of China. Chinese guidelines for diagnosis and treatment
of breast cancer 2018 (English version). Chin J Cancer Res.
31:259–277. 2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948.
2010.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Prat A, Adamo B, Cheang MC, Anders CK,
Carey LA and Perou CM: Molecular characterization of basal-like and
non-basal-like triple-negative breast cancer. Oncologist.
18:123–133. 2013.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Blows FM, Driver KE, Schmidt MK, Broeks A,
van Leeuwen FE, Wesseling J, Cheang MC, Gelmon K, Nielsen TO,
Blomqvist C, et al: Subtyping of breast cancer by
immunohistochemistry to investigate a relationship between subtype
and short and long term survival: A collaborative analysis of data
for 10,159 cases from 12 studies. PLoS Med.
7(e1000279)2010.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Perou CM and Børresen-Dale AL: Systems
biology and genomics of breast cancer. Cold Spring Harb Perspect
Biol. 3(a003293)2011.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Dent R, Trudeau M, Pritchard KI, Hanna WM,
Kahn HK, Sawka CA, Lickley LA, Rawlinson E, Sun P and Narod SA:
Triple-negative breast cancer: Clinical features and patterns of
recurrence. Clinical Cancer Res. 13:4429–4434. 2007.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Blick T, Widodo E, Hugo H, Waltham M,
Lenburg ME, Neve RM and Thompson EW: Epithelial mesenchymal
transition traits in human breast cancer cell lines. Clin Exp
Metastasis. 25:629–642. 2008.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Hay ED: An overview of
epithelio-mesenchymal transformation. Acta Anat (Basel). 154:8–20.
1995.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Scanlon CS, Van Tubergen EA, Inglehart RC
and D'Silva NJ: Biomarkers of epithelial-mesenchymal transition in
squamous cell carcinoma. J Dent Res. 92:114–121. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Turley EA, Veiseh M, Radisky DC and
Bissell MJ: Mechanisms of disease: Epithelial-mesenchymal
transition-does cellular plasticity fuel neoplastic progression?
Nat Clin Pract Oncol. 5:280–290. 2008.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Nose A, Nagafuchi A and Takeichi M:
Isolation of placental cadherin cDNA: Identification of a novel
gene family of cell-cell adhesion molecules. EMBO J. 6:3655–3661.
1987.PubMed/NCBI
|
|
13
|
Wijnhoven BPL, Dinjens WN and Pignatelli
M: E-cadherin-catenin cell-cell adhesion complex and human cancer.
Br J Surg. 87:992–1005. 2000.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Li K, He W, Lin N, Wang X and Fan QX:
N-cadherin knock-down decreases invasiveness of esophageal squamous
cell carcinoma in vitro. World J Gastroenterol. 15:697–704.
2009.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Chang L and Goldman RD: Intermediate
filaments mediate cytoskeletal crosstalk. Nat Rev Mol Cell Biol.
5:601–613. 2004.PubMed/NCBI View
Article : Google Scholar
|
|
16
|
Hu L, Lau SH, Tzang CH, Wen JM, Wang W,
Xie D, Huang M, Wang Y, Wu MC, Huang JF, et al: Association of
Vimentin overexpression and hepatocellular carcinoma metastasis.
Oncogene. 23:298–302. 2004.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Wei J, Xu G, Wu M, Zhang Y, Li Q, Liu P,
Zhu T, Song A, Zhao L, Han Z, et al: Overexpression of vimentin
contributes to prostate cancer invasion and metastasis via src
regulation. Anticancer Res. 28:327–334. 2008.PubMed/NCBI
|
|
18
|
Li D and Roberts R: Human genome and
diseases: WD-repeat proteins: Structure characteristics, biological
function, and their involvement in human diseases. Cell Mol Life
Sci CMLS. 58:2085–2097. 2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Beck R, Ravet M, Wieland FT and Cassel D:
The COPI system: Molecular mechanisms and function. FEBS Lett.
583:2701–2709. 2009.PubMed/NCBI View Article : Google Scholar
|
|
20
|
De Baere E, Speleman F, Van Roy N, Mortier
K, De Paepe A and Messiaen L: Assignment of the cellular
retinol-binding protein 2 gene (RBP2) to human chromosome band 3q23
by in situ hybridization. Cytogenet Cell Genet. 83:240–241.
1998.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Presley JF, Ward TH, Pfeifer AC, Siggia
ED, Phair RD and Lippincott-Schwartz J: Dissection of COPI and Arf1
dynamics in vivo and role in Golgi membrane transport. Nature.
417:187–193. 2002.PubMed/NCBI View
Article : Google Scholar
|
|
22
|
Lee MC, Miller EA, Goldberg J, Orci L and
Schekman R: Bi-directional protein transport between the ER and
Golgi. Annu Rev Cell Dev Biol. 20:87–123. 2004.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Mi Y, Yu M, Zhang L, Sun C, Wei B, Ding W,
Zhu Y, Tang J, Xia G and Zhu L: COPB2 is upregulated in prostate
cancer and regulates PC-3 cell proliferation, cell cycle, and
apoptosis. Arch Med Res. 47:411–418. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Li ZS, Liu CH, Liu Z, Zhu CL and Huang Q:
Downregulation of COPB2 by RNAi inhibits growth of human
cholangiocellular carcinoma cells. Eur Rev Med Pharmacol Sci.
22:985–992. 2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wang XL, Shi J, Niu Z, Wang J and Zhang W:
MiR-216a-3p regulates the proliferation, apoptosis, migration, and
invasion of lung cancer cells via targeting COPB2. Biosci
Biotechnol Biochem. 84:2014–2027. 2020.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Wang Y, Chai Z, Wang M, Jin Y, Yang A and
Li M: COPB2 suppresses cell proliferation and induces cell cycle
arrest in human colon cancer by regulating cell cycle-related
proteins. Exp Ther Med. 15:777–784. 2018.PubMed/NCBI View Article : Google Scholar
|
|
27
|
An C, Li H, Zhang X, Wang J, Qiang Y, Ye
X, Li Q, Guan Q and Zhou Y: Silencing of COPB2 inhibits the
proliferation of gastric cancer cells and induces apoptosis via
suppression of the RTK signaling pathway. Int J Oncol.
54:1195–1208. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Yu H, Yao J, Du M, Ye J, He X and Yin L:
CDKN3 promotes cell proliferation, invasion and migration by
activating the AKT signaling pathway in esophageal squamous cell
carcinoma. Oncol Lett. 19:542–548. 2020.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tan MS, Chang SW, Cheah PL and Yap HJ:
Integrative machine learning analysis of multiple gene expression
profiles in cervical cancer. PeerJ. 6(e5285)2018.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Bhandari A, Zheng C, Sindan N, Sindan N,
Quan R, Xia E, Thapa Y, Tamang D, Wang O, Ye X and Huang D: COPB2
is up-regulated in breast cancer and plays a vital role in the
metastasis via N-cadherin and Vimentin. J Cell Mol Med.
23:5235–5245. 2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Thompson EW, Paik S, Brünner N, Sommers
CL, Zugmaier G, Clarke R, Shima TB, Torri J, Donahue S, Lippman ME,
et al: Association of increased basement membrane invasiveness with
absence of estrogen receptor and expression of vimentin in human
breast cancer cell lines. J Cell Physiol. 150:534–544.
1992.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Xu Q, Chang H, Tian X, Lou C, Ma H and
Yang X: Hypoxia-induced MFAP5 promotes tumor migration and invasion
via AKT pathway in head and neck squamous cell carcinoma. J Cancer.
11:1596–1605. 2020.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Zhang YZ, Zheng YP and Zhu GM: MiR-203a-3p
targets PTEN to promote hepatocyte proliferation by regulating
PI3K/Akt pathway in BRL-3A cells. Biosci Biotechnol Biochem.
84:725–733. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Umemura S, Yoshida S, Ohta Y, Naito K,
Osamura RY and Tokuda Y: Increased phosphorylation of Akt in
triple-negative breast cancers. Cancer Sci. 98:1889–1892.
2007.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Shao Z, Ma X, Zhang Y, Sun Y, Lv W, He K,
Xia R, Wang P and Gao X: CPNE1 predicts poor prognosis and promotes
tumorigenesis and radioresistance via the AKT singling pathway in
triple-negative breast cancer. Mol Carcinog. 59:533–544.
2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang Y, Zhao Z, Li S, Dong L, Li Y, Mao
Y, Liang Y, Tao Y and Ma J: Inhibition of miR-214 attenuates the
migration and invasion of triple-negative breast cancer cells. Mol
Med Rep. 19:4035–4042. 2019.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Zhou Y, Wang X, Huang X, Li XD, Cheng K,
Yu H, Zhou YJ, Lv P and Jiang XB: High expression of COPB2 predicts
adverse outcomes: A potential therapeutic target for glioma. CNS
Neurosci Ther. 26:309–318. 2020.PubMed/NCBI View Article : Google Scholar
|