Introduction
Stroke is one of the leading causes of mortality
worldwide, and ischemic stroke accounts for 80-85% of all strokes
(1). Ischemic stroke is associated
with multiple physiological and pathological processes, including
neuronal apoptosis, inflammation, oxidative stress and
excitotoxicity, ultimately leading to neuronal death (2,3).
However, only 20% of patients with stroke receive effective
treatment due to its increased risk of symptomatic intracerebral
hemorrhage (4). Increasing evidence
has revealed that ischemia typically involves a range of neural
activities, including hypoxia, oxidative stress and inflammation
(5), and these processes lead to
necrosis, autophagy and apoptosis in the ischemic brain (6). Ischemic stroke severely compromises
the quality of life of the patient, but there is currently no
effective medication to accelerate rehabilitation (7). Therefore, the development of effective
therapies is urgently required.
Long non-coding RNA (lncRNAs) are considered as
pseudogenes that compete for microRNA (miRNA/miR) binding, as well
as serving a key role in gene regulation and cell development
(8,9). LncRNAs and miRNAs also present
competing endogenous RNA (ceRNA) activity (10). It was reported that lncRNAs are
involved in numerous complicated biological processes, including
ischemic stroke. For example, lncRNA-1810034E14Rik exerts
anti-inflammatory effects in ischemic stroke (11). Moreover, metastasis-associate lung
adenocarcinoma transcript 1 (MALAT1) promotes cerebral
ischemia-reperfusion injury by competitively binding miR-145 by
affecting aquaporin 4 expression (12).
It has been noted that lncRNA 319 (LINC00319) has a
role in a variety of tumors. For instance, LINC00319 promotes the
proliferation and invasion of lung cancer cells (13,14).
Furthermore, LINC00319 exerts oncogenic roles by repressing
miR-3127 in bladder cancer progression (15,16),
but its detailed role in ischemic brain injury is yet to be
elucidated.
The aim of the present study was to investigate the
role of LINC00319 in ischemic stroke. The current data suggested
that LINC00319 expression was significantly upregulated in ischemic
stroke. Furthermore, LINC00319 promoted the progression of ischemic
stroke by modulating miR-200a-3p. Thus, the present study provides
a novel insight into the underlying molecular mechanism of ischemic
stroke.
Materials and methods
Human samples
Patients with acute ischemic stroke (n=50) admitted
to The First Affiliated Hospital of Jiamusi University (Jiamusi,
China) between January 2018 and January 2020 were selected for the
study. Blood samples were collected from patients with acute
ischemic stroke (males, n=27; females, n=23; age range, 31-72
years; median age, 51.34 years). The etiology of stroke was
classified according to the TOAST classification criteria (17). Patients with neurological diseases,
cardiac embolism, transient ischemic attack, hemorrhagic
infarction, occult cerebrovascular malformation or traumatic
cerebrovascular disease were excluded. The present study was
approved by the Ethics Committee of Jiamusi University. All
procedures were in agreement with the Declaration of Helsinki
(18). Written informed consent was
obtained from all patients. The study was approved by the
Institutional Review Board of The First Affiliated Hospital of
Jiamusi University (approval no. JUIRBR-2019-301).
Analysis of differentially expressed
lncRNAs
The microarray data for the current study was
obtained from the Gene Expression Omnibus (GEO) database
(https://www.ncbi.nlm.nih.gov/geo/)
using the accession nos. GSE46266, GSE58294 and GSE22255(19). Normalized analysis of differentially
expressed genes was performed using RStudio 3.5.1 software and
related R software packages (Bioconductor, RStudio, Inc.;
https://www.rstudio.com/). LncRNAs with a
fold-change ≥2 and P<0.01 were identified as differentially
expressed. Gene Ontology (GO) analysis was used to evaluate
LINC00319 positive-associated genes in GEO datasets using the
Database for Annotation, Visualization and Integrated Discovery
v6.8 (https://david.ncifcrf.gov/).
Primary neuron culture
Neonatal clean grade Sprague-Dawley rats (n=50; age,
<24 h) were obtained from the Experimental Animal Center of
Jiamusi University. Primary culture of cortical neurons in rats was
performed using a previously described method (20). The rats were placed in an anesthesia
induction box and anesthetized with isoflurane (induced
concentration, 3-4%) for 2-3 min and anesthesia was maintained
using 1-1.5% isoflurane. The rats were decapitated, and the heads
were soaked with 75% alcohol for 1 min under aseptic conditions,
and the whole brain was dissected. Ophthalmic scissors were used to
repeatedly cut the cortex into pieces. Subsequently, the tissues
were incubated with 2 ml 0.25% trypsin (cat. no. C0201; Beyotime
Institute of Biotechnology) at 37˚C for 30 min. Next, the tissue
mass was prepared into a suspension via digestion. Finally,
1x107 cells were inoculated in a six-well culture plate.
After 24 h, the complete culture medium [DMEM/F12 (cat. no. 670087;
Gibco; Thermo Fisher Scientific, Inc.) supplemented with 2% B27
(cat. no. 27010; Engreen Biosystem Co., Ltd.)] was changed and
cells were cultured for 3 days. The Directive 2010/63/EU on the
protection of animals used for scientific purposes stipulates that
a competent person shall perform killing using a method that is
appropriate for respective species. All applicable international,
national and/or institutional guidelines or the care and use of
animals were followed. The animal study was approved by
Institutional Review Board of The First Affiliated Hospital of
Jiamusi University (approval no. JUIRBR-2019-017).
To inhibit the proliferation of glial cells and
hybrid cells, primary neurons were cultured in cytosine arabidopsis
culture medium (final concentration, 2.5 µg/ml, half solution; cat.
no. C0525; Tokyo Chemical Industry Co., Ltd.) for 3 days. The
complete culture medium was changed once every 3 days, and half
volume of the medium was changed each time.
Establishment of an oxygen-glucose
deprivation (OGD) model
An OGD model can simulate in vivo cerebral
ischemia and is also used as an in vitro ischemic model
(21). Neurons were cultured in
DMEM with low glucose (cat. no. 11885076; Gibco; Thermo Fisher
Scientific, Inc.) in a wet incubator at 37˚C, 95% N2 and
5% O2 for 6 h. During reperfusion, the neurons were
cultured in DMEM with high glucose (cat. no. 10313021; Gibco;
Thermo Fisher Scientific, Inc.) replenished with 2% B-27 Plus
Supplement (cat. no. A3582801; Gibco; Thermo Fisher Scientific,
Inc.) and incubated in a constant oxygen incubator for 12, 24 or 48
h. Cells kept in normal medium and normal conditions were used as
controls.
Glucose uptake assay
To measure changes in glucose metabolism, neurons
were inoculated in a 96-well plate. Neurons were then treated with
or without 100 nM/l insulin (cat. no. P3376; Beyotime Institute of
Biotechnology) for 20 min. Subsequently, 0.01 mM 2-deoxyglucose
(2DG; cat. no. ST1024; Beyotime Institute of Biotechnology) was
added to cells and incubated for 20 min at 37˚C. Cells were washed
with PBS three times, harvested and 2DG uptake was assessed using
the Glucose Uptake Assay kit (Colorimetric) (cat. no. ab136955;
Abcam) following the manufacturer's instructions.
Caspase-3 activity detection
Neurons were seeded in six-well plates. The cell
supernatant was obtained and centrifuged at 500 x g at room
temperature for 5 min. Caspase-3 activity was detected with a
Caspase-3 Assay kit (cat. no. ab39383; Abcam), according to the
manufacturer's instructions. The activity of caspase-3 was measured
at a wavelength of 405 nm with a Tecan microplate reader (Infinite
F50; Tecan Group, Ltd.).
Cell transfection
Neurons were transfected with 20 µM of LINC00319
overexpression plasmid (OE-LINC00319), small interfering (si) RNA
LINC00319 (si-LINC00319), miR-200a-3p mimic, miR-inhibitor and
corresponding negative controls (NC) with scramble sequence (si-NC
or miR-NC/miR-inhibitor NC) or empty vector (OE-NC) at room
temperature, which were synthesized by Shanghai GenePharma Co.,
Ltd. Lipofectamine® 2000 (cat. no. 11668030; Invitrogen;
Thermo Fisher Scientific, Inc.) was used to transfect neurons
according to manufacturer's protocol. Cells were incubated at 37˚C
for 6 h. The culture medium was then changed with fresh DMEM
containing 10% FBS. The plasmid sequences are listed in Table I. After 48 h of transfection, the
cells were harvested for subsequent experiments.
 | Table IPlasmid sequences. |
Table I
Plasmid sequences.
| Plasmid | Sequence
(5'-3') |
|---|
|
siRNA-LINC00319 | F:
GCTGTAATGTGCTGTGACT |
| | R:
AGTCACAGCACATTACAGC |
| OE-LINC00319 | F:
GACUAAACAAGGUCUUAAUTT |
| | R:
AUUAAGACCUUGUUUAGUCTT |
| si-NC | F:
UUCUCCGAACGUGUCACGU |
| | R:
ACGUGACACGUUCGGAGAA |
| miR-200a-3p
mimic | F:
UAACACUGUCUGGUAACGAU |
| | R:
GCGGGUCACCUUUGAACAUC |
| miR-200a-3p
inhibitor | F:
AUUGUGACAGACCAUUGCU |
| | R:
UGGGGUCGUCGGUCUAGGG |
| miR-200a-3p mimic
NC | F:
UUCUCCGAACGUGUCACGU |
| | R:
ACGUGACACGUUCGGAGAA |
| miR-200a-3p
inhibitor NC | F:
GAACAGGUAGUCUGAACACUG |
| | R:
CAGUACUUUUGUGUAGUACAA |
RNA extraction and reverse
transcription-quantitative (RT-q)PCR
After transfection, neuron cells were collected.
Total RNA was extracted and lysed using TRIzol® reagent
(cat. no. 15596026; Invitrogen; Thermo Fisher Scientific, Inc.),
and then reverse transcribed into cDNA utilizing a PrimeScript
RT-PCR kit (cat. no. RR014A; Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. qPCR was performed
using a SYBR-Green PCR Master Mix (cat. no. 4309155; Invitrogen;
Thermo Fisher Scientific, Inc.) on a 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 95˚C for 10 min, followed
by 40 cycles at 95˚C for 15 sec, and 60˚C for 60 sec. GAPDH and U6
were used as controls. The primer sequences used were as follows:
LINC00319 forward, 5'-GAACCTCAGTTCCTGGCCTC-3' and reverse,
5'-GAGGCCAGGAACTGAGGTTC-3'; GAPDH forward,
5'-AGCCACATCGCTCAGACAC-3' and reverse, 5'-GCCCAATACGACCAAATCC-3';
miR-200a-3p forward, 5'-TAACACTGTCTGGTAACGATGT-3' and reverse,
5'-CAGGTCCAGTTTTTTTTTTTTTT-3'; and U6 forward,
5'-CTCGCTTCGGCAGCACA-3' and reverse, 5'-AACGCTTCACGAATTTGCGT-3'.
Relative gene expression levels were calculated by comparing to the
expression level of the internal standard using the
2-ΔΔCq method (22).
Luciferase reporter assay
The interaction between LINC00319 and miR-200a-3p
was predicted using TargetScan (http://www.targetscan.org/vert_72/) and starBase v2.0
(http://starbase.sysu.edu.cn/). Among all
the statistically relevant miRNAs, miR-335-5p, miR-141-3p and
miR-200a-3p were obtained from two databases and selected for
further experiments.
A luciferase reporter assay was used to evaluate
potential binding between LINC00319 and miR-200a-3p. The psiCHECK2
vector (GeneChem, Inc.) was conducted to construct a LINC00319
3'-untranslated region (UTR)-containing reporter. The 3'-UTRs of
LINC00319, as well as its wild-type (WT) and mutant (MUT) binding
sites with miR-200a-3p, were amplified and cloned into luciferase
reporter vectors. miR-200a-3p mimics and plasmids (20 µM) were
co-transfected for 48 h using Lipofectamine® 2000. The
luciferase activity was analyze using a Dual-Luciferase Reporter
Assay System (cat. no. E1910; Promega Corporation) according to the
manufacturer's instructions. Renilla luciferase intensity
was utilized as an internal control.
RNA immunoprecipitation (RIP)
assay
RIP was performed with EZ-Magna RIP™ RNA-Binding
Protein Immunoprecipitation kit (cat. no. 17-701; EMD Millipore)
according to the manufacturer's protocols. RNA fractions extracted
from RIP were analyzed via qPCR.
Cell viability and cell apoptosis
Cell proliferation was measured using a Cell
Counting Kit-8 (CCK-8; cat. no. C0037; Beyotime Institute of
Biotechnology) assay. Neuron cells were collected 24 h
post-transfection and were seeded into 96-well plates at the
density of 2x103 cells/well. Subsequently, CCK-8
solution was added and incubated for 2 h at 37˚C at days 1, 2 and 3
post seeding. The absorbance was measured at a wavelength of 450 nm
with a Tecan microplate reader (Infinite F50; Tecan Group,
Ltd.).
Apoptosis was analyzed using a Hoechst 33258
detection assay kit (cat. no. C1017; Beyotime Institute of
Biotechnology) in accordance with the manufacturer's instructions.
Neurons (1x105) were collected and seeded in cell slides
for 24 h, which were precoated with 0.1% poly-L-lysine at 4˚C for
12 h. Subsequently, the glass slides were fixed with 4%
paraformaldehyde at room temperature for 15 min. The nuclei were
stained with Hoechst 33258 (10 µg/ml) for 20 min and observed under
a fluorescence microscope (Olympus Corporation) at x200
magnification.
Western blotting
Total protein was extracted from neuronal cells with
RIPA lysis buffer (cat. no. C05-01001; BIOSS). Protein
concentration was measured using a BCA protein assay kit (Beyotime
Institute of Biotechnology). Proteins (30 µg/lane) were separated
via 10% SDS-PAGE gel and transferred onto PVDF membranes (cat. no.
3010040001; EMD Millipore). After blocking in 5% BSA (cat. no.
ST025; Beyotime Institute of Biotechnology) for 1 h at room
temperature, membranes were incubated with the following primary
antibodies at 4˚C overnight: Rabbit anti-caspase-3 (rabbit
polyclonal antibody; cat. no. 66470-2-lg; 1:500; Wuhan Sanying
Biotechnology) and mouse anti-GAPDH (mouse monoclonal antibody;
1:1,000; cat. no. SC-47724; Santa Cruz Biotechnology, Inc.).
Following primary antibody incubation, membranes were incubated
with horseradish peroxidase-conjugated anti-mouse (cat. no. ab6728)
or anti-rabbit (cat. no. ab6721; both at 1:2,000; both from Abcam)
secondary antibodies for 1 h at room temperature. Protein bands
were visualized via ECL detection (cat. no. P0018S; Beyotime
Institute of Biotechnology) with a ChemiDoc™ MP Imaging detection
System (Bio-Rad Laboratories, Inc.) and analyzed by Image Lab
software (version 3.0; Bio-Rad Laboratories, Inc.).
Statistical analysis
Statistical analyses were conducted using SPSS
version 21.0 (IBM Corp.) and graphs were plotted using GraphPad
Prism 6.0 (GraphPad Software, Inc.). Data are presented as the mean
± SD. Comparisons between groups were determined using a two-tailed
Student's t-test or one-way ANOVA with Tukey's post hoc test.
Aberrantly expressed lncRNAs were examined based on the
Benjamini-Hochberg method (23).
The association between the expression of LINC00319 and the
expression of miR-200a-3p was analyzed using Pearson's correlation.
Experiments were independently performed three times. P<0.05 was
considered to indicate a statistically significant difference.
Results
LINC00319 is significantly upregulated
in ischemic stroke
To identify vital lncRNAs involved in ischemic
stroke, differentially expressed lncRNAs were analyzed via
bioinformatics analysis in a middle cerebral artery occlusion
(MCAO) rat model in the GSE46266 dataset (Fig. 1A). The top 20 aberrantly expressed
lncRNAs were selected with cut-off values of log2
fold-change level (FC) >2 and false discovery rate (FDR)
<0.01. It was observed that LINC00319 was significantly
overexpressed in MCAO rat models compared with normal models
(Fig. 1B). For further analysis, a
Venn diagram was conducted using FunRich to visualize the
overlapping lncRNAs among GSE46266, GSE58294 and GSE22255 datasets.
In total, five intersecting lncRNAs were identified and
investigated according to their logFC level (Fig. 1C). LncRNA-LINC00319 was present on
the list of significantly upregulated lncRNAs (Fig. 1C). Gene Ontology (GO) analysis
demonstrated that LINC00319 was closely associated with ‘Apoptotic
process’ (Fig. 1D).
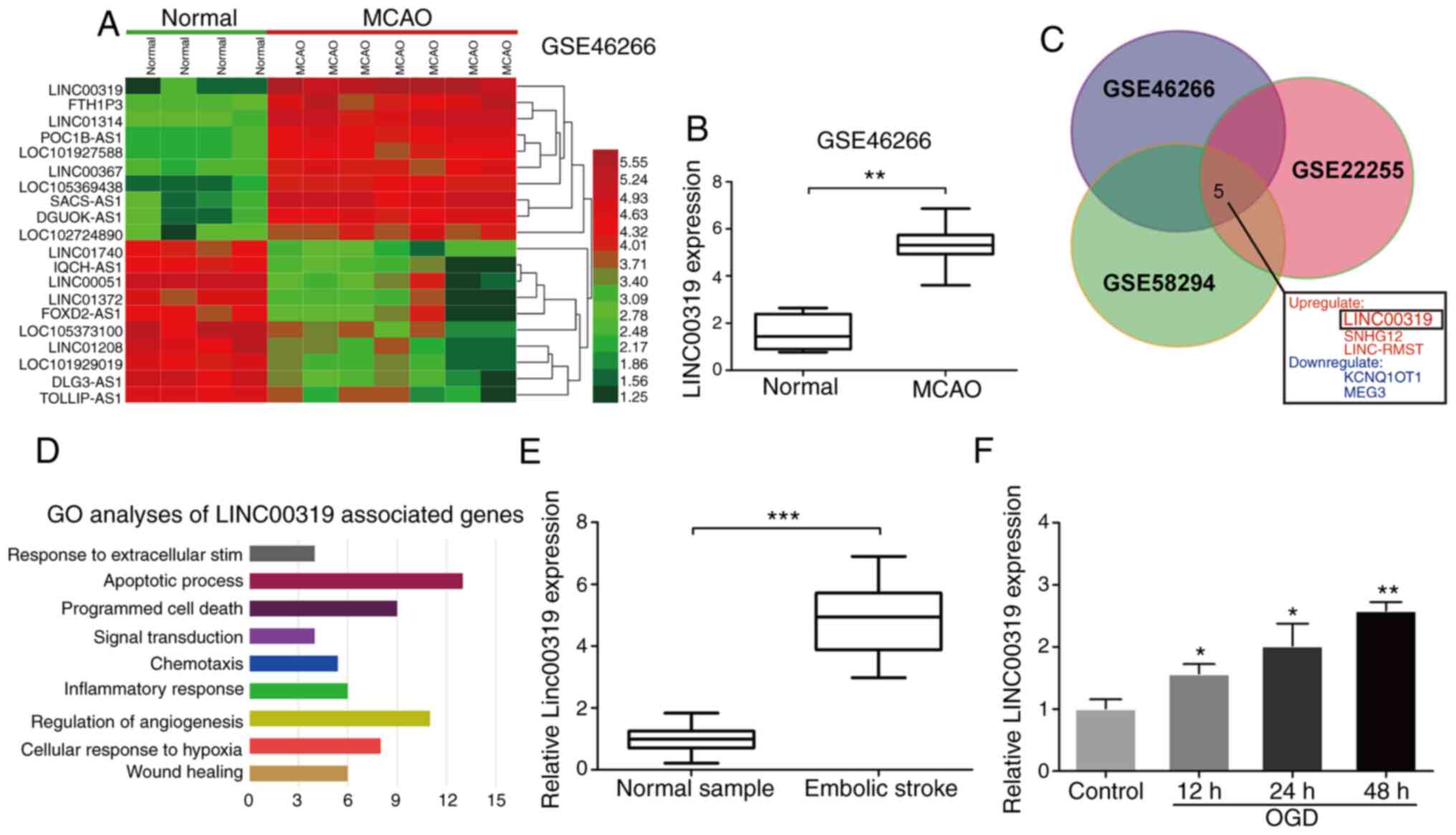 | Figure 1Upregulated LINC00319 in ischemic
stroke. (A) Heatmaps were constructed using the differential lncRNA
between normal and MCAO rat model detected, as by significance
analysis in GSE46266 database. (B) GSE46266 dataset demonstrated
that LINC00319 was markedly upregulated in MCAO rats compared with
normal group. (C) A total of five significantly
differential-expressed lncRNAs were identified among GSE46266,
GSE22255 and GSE58294 datasets. (D) GO analysis was performed using
the LINC00319 positive-associated genes in GEO datasets. (E)
Reverse transcription-quantitative PCR analysis indicated that
LINC00319 was upregulated in blood samples of patients with
ischemic stroke (n=50), as compared with healthy group. (F)
LINC00319 expression was upregulated by OGD induction in primary
neurons, and was gradually increased with the prolongation of
reperfusion (compared with control). *P<0.05,
**P<0.01, ***P<0.001. Data are
presented as the mean ± SD of three independent experiments. GO,
Gene Ontology; GEO, Gene Expression Omnibus; MCAO, middle cerebral
artery occlusion; lncRNA, long non-coding RNA; OGD, oxygen-glucose
deprivation. |
Peripheral blood samples were collected from
patients with ischemic stroke and physical examination volunteers
from the First Affiliated Hospital of Jiamusi University.
Consistently. LINC00319 expression was significantly upregulated in
patients with ischemic stroke compared with control samples
(Fig. 1E). Next, ischemic stroke
models were established via OGD in primary neurons in vitro.
The results demonstrated that LINC00319 was upregulated under
ischemia-reperfusion induction and increased gradually with the
extension of reperfusion time (Fig.
1F).
LINC00319 participates in OGD-induced
ischemic stroke
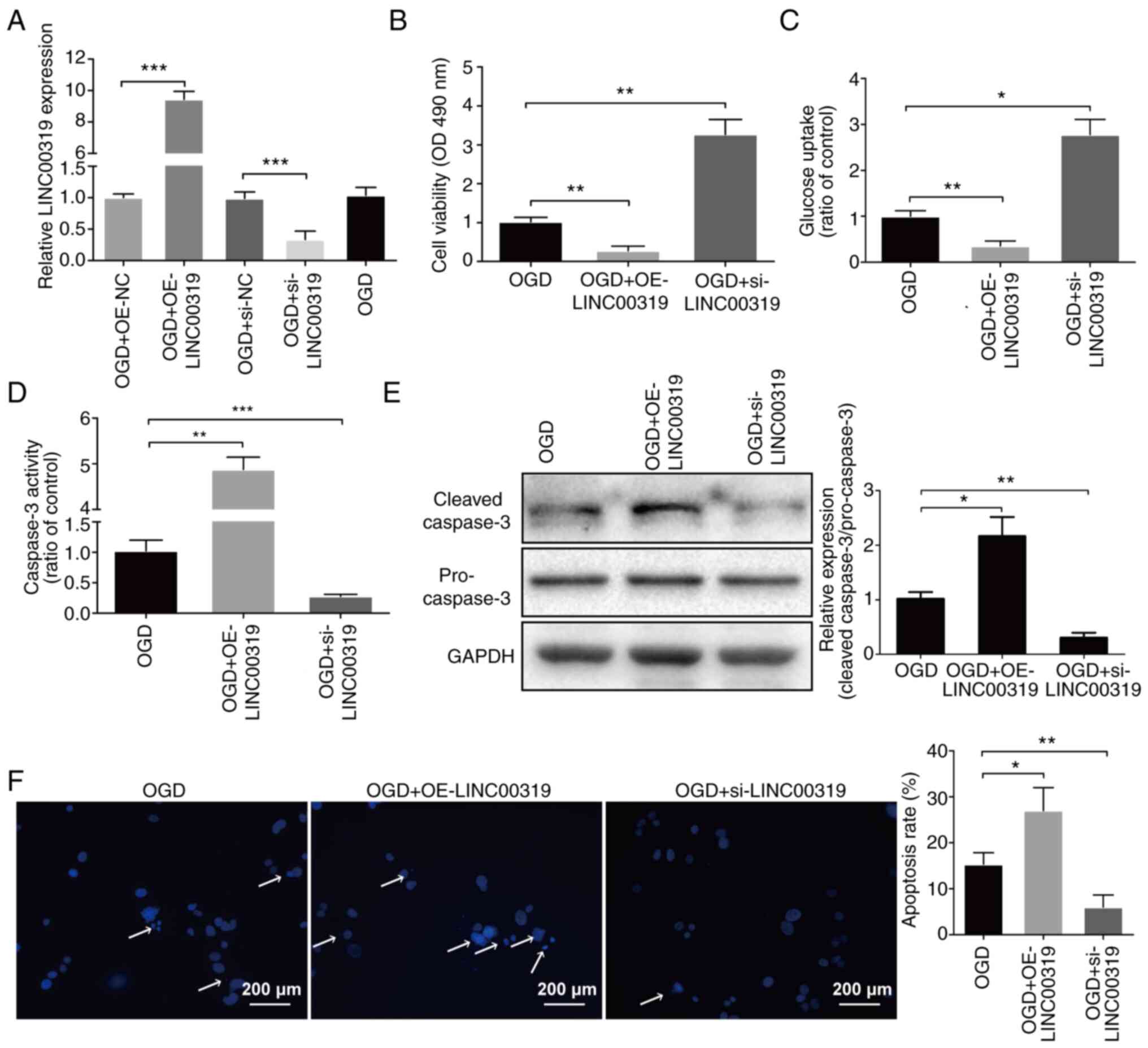
To investigate the biological function of LINC00319
in ischemic stroke, LINC00319 was overexpressed with pcDNA3.1
(OE-LINC00319) and silenced with siRNA (si-LINC00319) in primary
neurons (Fig. 2A). LINC00319
overexpression in ischemic stroke significantly decreased cell
viability, while LINC00319 knockdown significantly increased cell
viability (Fig. 2B). Furthermore,
LINC00319 knockdown significantly increased glucose uptake
(Fig. 2C). Caspase-3 activity and
protein level measurement results suggested that LINC00319
knockdown significantly inhibited neuronal apoptosis (Fig. 2D and E). The apoptosis assay also demonstrated
similar results (Fig. 2F). Thus,
the results indicated that LINC00319 participated in OGD-induced
ischemic injury.
Correlation between LINC00319 and
miR-200a-3p
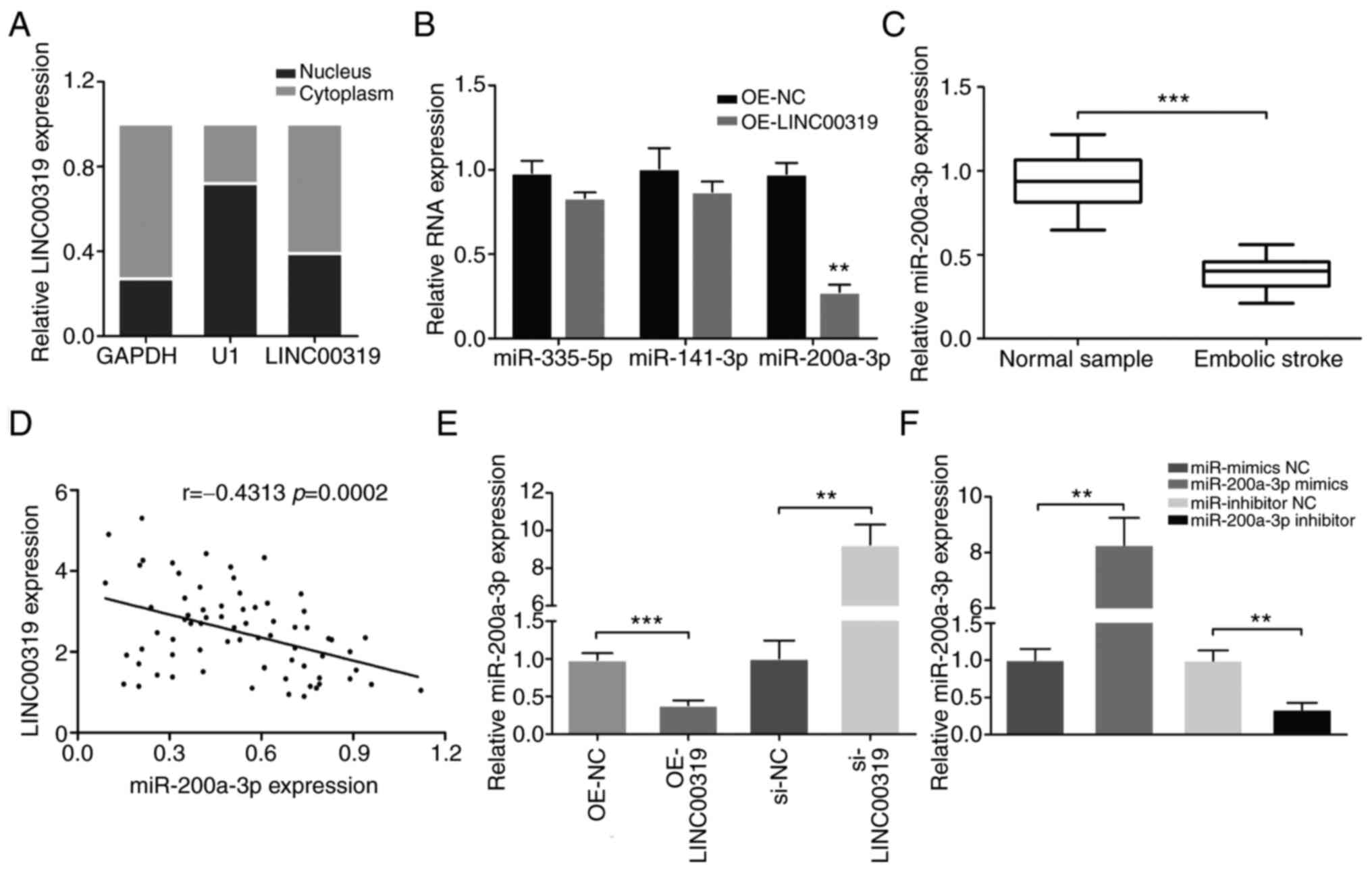
To detect the subcellular localization of LINC00319,
the nuclear and cytoplasmic fractions of neurons were collected.
LINC00319 expression was measured in different subcellular
fractions, and it was found that LINC00319 expression was higher in
cytoplasmic fractions compared with that in the nucleus (Fig. 3A). These results indicated that
LINC00319 may exert both transcriptional and post-transcriptional
regulatory roles in neuron cells.
Using TargetScan and starBase v2.0 database blast
prediction, it was demonstrated that LINC00319 contained a putative
targeting site for miR-335-5p, miR-141-3p and miR-200a-3p.
Furthermore, the expression levels of miR-335-5p, miR-141-3p and
miR-200a-3p were measured in primary neurons overexpressing
LINC00319. The results demonstrated that LINC00319 overexpression
significantly decreased the expression of miR-200a-3p (Fig. 3B). Moreover, miR-200a-3p expression
was significantly decreased in the blood samples of patients with
ischemic stroke compared with the healthy control group (Fig. 3C). The mRNA expression of
miR-200A-3p was negatively correlated with LINC00319 in patients
with ischemic stroke (r=-0.4313, P<0.001; Fig. 3D).
Subsequently, the effects of LINC00319 on
miR-200a-3p in vitro were examined. The results demonstrated
that LINC00319 knockdown promoted miR-200a-3p expression (Fig. 3E). In the follow-up experiments,
miR-200a-3p mimics and inhibitor were successfully transfected into
OGD-induced primary neurons (Fig.
3F). Collectively, a negative interaction was identified
between LINC00319 and miR-200a-3p.
LINC00319 directly binds to
miR-200a-3p
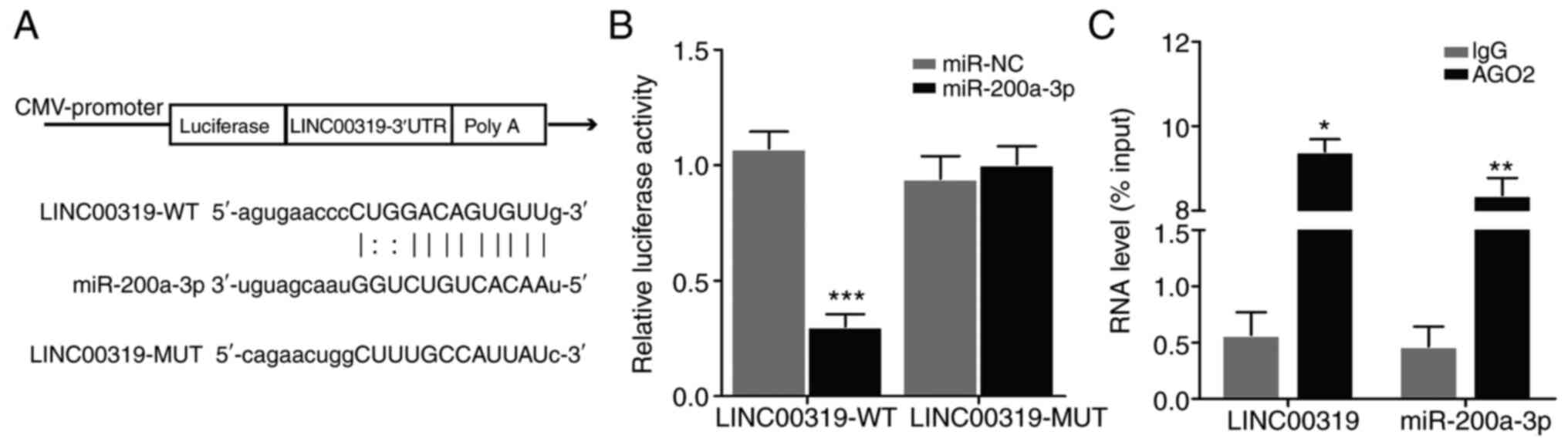
To elucidate the functional relationship between
LINC00319 and miR-200a-3p, bioinformatics analyses were performed.
Using starBase v2.0 (http://starbase.sysu.edu.cn/mirMrna.php) and
TargetScan (http://www.targetscan.org), the
binding pattern between LINC00319 and miR-200a-3p was investigated.
In addition, a structural mutation was induced in LINC00319
(Fig. 4A). The luciferase reporter
gene was then detected. The results demonstrated that miR-200a-3p
inhibited the activity of LINC00319-WT reporter (Fig. 4B), indicating that LINC00319 and
miR-200a-3p interacted directly at this binding site. Furthermore,
the RIP assay results suggested that LINC00319 could directly bind
to miR-200a-3p (Fig. 4C).
miR-200a-3p overexpression facilitates
OGD-induced cerebral ischemic injury
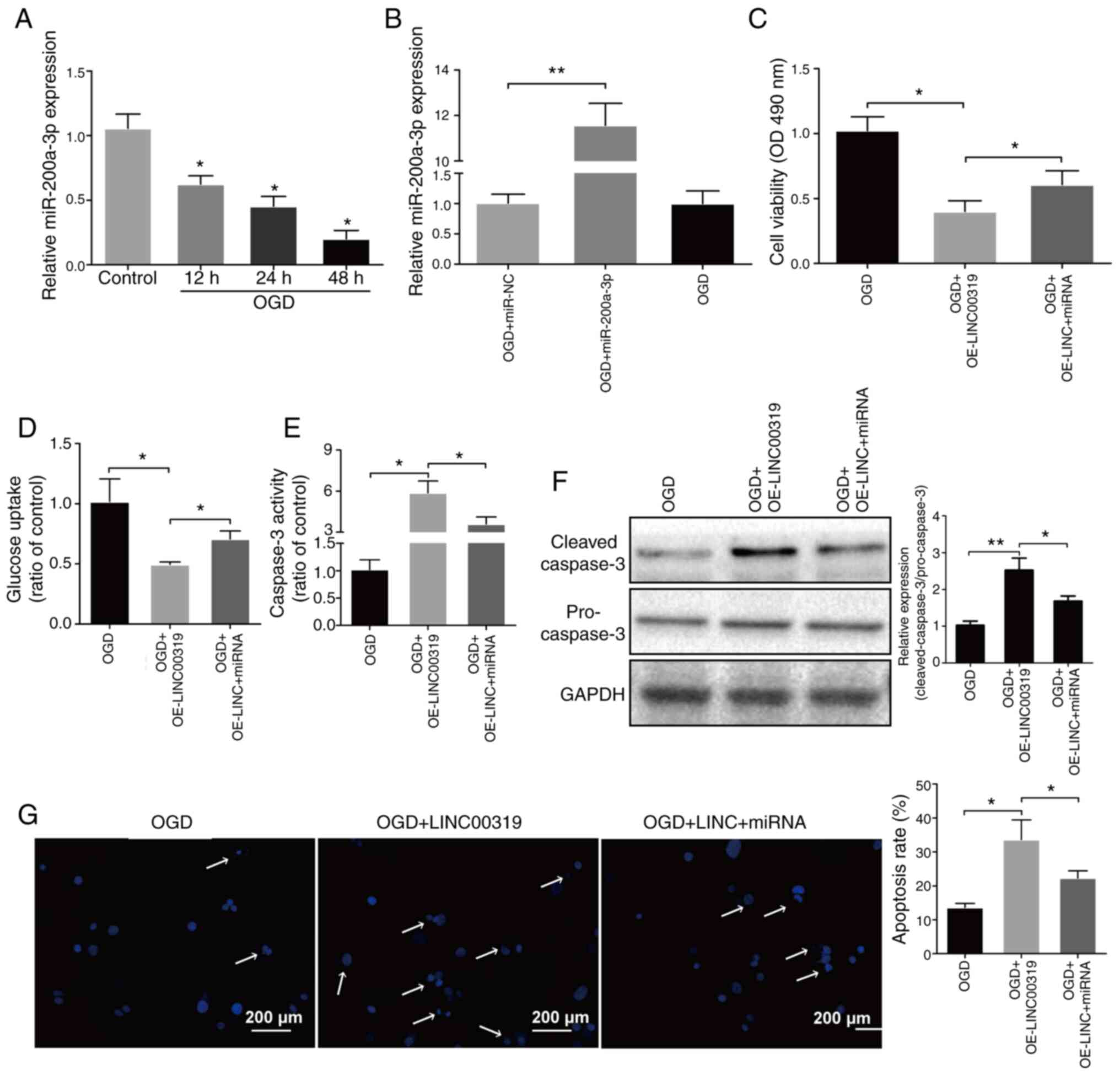
To investigate the effects of miR-200a-3p in
ischemic stroke, its expression levels were detected in neurons.
miR-200a-3p expression significantly declined with the prolongation
of reperfusion (Fig. 5A).
Subsequently, OGD-induced neurons were transfected with miR-200a-3p
(Fig. 5B). It was found that the
declining cell viability and glucose uptake due to LINC00319
overexpression were partially reversed by miR-200a-3p (Fig. 5C and D). In addition, caspase-3 activity,
protein levels and apoptosis rates were increased after LINC00319
overexpression, but were decreased when co-transfected with
miR-200a-3p (Fig. 5E-G). Therefore,
the results indicated that miR-200a-3p could reverse the effects of
LINC00319 in OGD-induced cerebral ischemic injury.
Discussion
Ischemic stroke is a major public health issue
resulting in mortality and disability and is more difficult to
treat compared with other cerebral diseases (24). Ischemic stroke is considered one of
the most serious diseases worldwide, threatening the health and
life of aging populations (25). It
is well known that lncRNAs are present in large numbers in the
central nervous system (26).
Moreover, lncRNAs serve a crucial role in cerebral function and in
nervous system diseases, particularly in central lesions (27-29).
The dysregulation of lncRNAs is associated with dysplasia and
pathological process of the brain (29). Previous studies have reported that
lncRNAs can interact with miRNAs, DNA and proteins to adjust and
control gene expression (30).
LncRNAs can also promote apoptosis and angiogenesis, as well as
cause inflammation and cell death in ischemic stroke (31). It has been revealed that lncRNAs
MALAT1, H19, taurine-upregulated gene 1, maternally expressed gene
3, small nucleolar RNA host gene 14, C2dat1, ANRIL and FosDT are
upregulated in cerebral ischemia (32).
miRNAs, which are short non-coding RNAs (18-21
nucleotides), can inhibit the function of protein-coding
transcripts, leading to changes in cell structure and function
(33,34). Previous studies have observed that
miRNAs regulate responses involved in curative effects in the
conditions of stroke and neuroinflammation. Furthermore, miRNAs are
key regulators of inflammation in ischemic stroke. A previous study
proposed that miR-669c-3p exerts a protective role in ischemic
stroke via the inhibition of MyD88 signaling (35). Moreover, miR-665-3p protects
microglia from OGD-induced apoptosis and inflammation (36). miR-98 decreases the infiltration of
proinflammatory factors and ameliorates the neurological outcome of
ischemic/reperfusion stroke in mice (37), while intracerebral injection of
miR-494 agomir decreases neuronal apoptosis and cerebral infarct
volume in the acute phase of MCAO (38).
Recent studies have shown that lncRNA host gene 15
(SNHG15) was abnormally expressed in ischemic stroke and inhibited
miR-18a expression and activated the ERK/MEK pathway to promote
ischemic stroke progression (39).
The researchers found that lncRNA HULC was associated with high
risk, severe illness, and poor prognosis in patients with acute
ischemic stroke (40). LncRNA
MACC1-AS1 alleviated microvascular endothelial cell injury,
promoted angiogenesis by regulating mir-6867-5p/TWIST1, and played
a protective role in ischemic stroke (41). In addition, the researchers found
that lncRNA RMRP had a protective effect on ischemic stroke by
activating the PI3K/Akt signaling pathway through valproate
(42). It was found that lncRNA
Nespas played an anti-inflammatory and anti-apoptotic role in
ischemic stroke by inhibiting TAK1 (transforming growth
factor-β-activated kinase 1) (43).
Another study showed that the knockdown of lncRNA KCNQ1OT1
increased cell activity and inhibited the autophagy induced by
ischemic stroke through the miR-200a/FOXO3/ATG7 pathway (25).
In the present study, LINC00319 was screened via
bioinformatics analysis. The expression of LINC00319 was
significantly increased in ischemic stroke. To the best of our
knowledge, there has been no previous research regarding the role
of LINC00319 in ischemic stroke. The present bioinformatics
analysis suggested that LINC00319 may be involved in apoptosis and
the regulation of angiogenesis. Furthermore, it was demonstrated
that LINC00319 could aggravate OGD-induced cerebral ischemic injury
by competitively binding miR-200a-3p.
lncRNAs and miRNAs serve crucial roles in oxidative
stress-induced brain injury (44).
For instance, it has been revealed that lncRNA-KCNQ1OT1 is
significantly elevated and promotes autophagy via the
miR-200a/FOXO3/autophagy related 7 axis in ischemic stroke
(25). In addition, lncRNA-Meg3
acts as a ceRNA by targeting the miR-21/programmed cell death 4
signaling pathway to accommodate ischemic neuron death (45). Another study reported that
downregulated expression of MALAT1 weakened neuronal death by
regulating miR-30a expression to suppress autophagy in cerebral
ischemic stroke (46).
Collectively, these aforementioned studies revealed that lncRNAs
could serve vital roles as a ceRNA for miRNAs in cerebral ischemic
stroke.
In conclusion, the present study demonstrated that
LINC00319 contributed to brain damage and promoted apoptosis in
cerebral ischemic stroke. Furthermore, LINC00319 exacerbated the
progression of ischemic stroke by binding miR-200a-3p to inhibit
neuronal proliferation and decrease glucose uptake. The present
data suggested that the LINC00319/miR-200a-3p axis may facilitate
the development of novel therapeutic strategies to intervene in
ischemic stroke.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Heilongjiang
Health and Health Committee Scientific Research (grant no. 2018377)
and the Basic Research Project of Jiamusi University (grant no.
JMSUJCMS2016-032).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HY and HW designed the study and performed the
experiments. HY, HL and XZ collected the data. XZ, HY, HL and YY
performed the data processing, statistical analysis and
bioinformatics investigations. HY and HL prepared the manuscript.
HY and HW confirm the authenticity of the raw data. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Institutional Review Board of Jiamusi University
(Jiamusi, China; approval no. JUIRBR-2019-301) and all procedures
were performed in accordance with national (D.L.n.26, March 4th,
2014) and international laws and policies (directive 2010/63/EU).
Written informed consent was provided by all patients prior to the
study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Powers WJ, Rabinstein AA, Ackerson T,
Adeoye OM, Bambakidis NC, Becker K, Biller J, Brown M, Demaerschalk
BM, Hoh B, et al: Guidelines for the early management of patients
with acute ischemic stroke: 2019 update to the 2018 guidelines for
the early management of acute ischemic stroke: A guideline for
healthcare professionals from the American Heart
Association/American Stroke Association. Stroke. 50:e344–e418.
2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Muth CC: Long-term outcomes after
thrombolytic therapy for acute ischemic stroke. JAMA.
323:2184–2185. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Prabhakaran S, Ruff I and Bernstein RA:
Acute stroke intervention: A systematic review. JAMA.
313:1451–1462. 2015.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Li T, Qin JJ, Yang X, Ji YX, Guo F, Cheng
WL, Wu X, Gong FH, Hong Y, Zhu XY, et al: The ubiquitin E3 ligase
TRAF6 exacerbates ischemic stroke by ubiquitinating and activating
Rac1. J Neurosci. 37:12123–12140. 2017.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Mehta SL, Manhas N and Raghubir R:
Molecular targets in cerebral ischemia for developing novel
therapeutics. Brain Res Rev. 54:34–66. 2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gu J, Gui S, Hu L, Kong L, Di M and Wang
Y: Downregulated miRNA-324-5p aggravates neuronal injury induced by
oxygen-glucose deprivation via modulating RAN. Exp Ther Med.
19:658–664. 2020.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Yang L, Han B, Zhang Z, Wang S, Bai Y,
Zhang Y, Tang Y, Du L, Xu L, Wu F, et al: Extracellular
vesicle-mediated delivery of CircSCMH1 promotes functional recovery
in rodent and nonhuman primate ischemic stroke models. Circulation.
142:556–574. 2020.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Ulitsky I and Bartel DP: lincRNAs:
Genomics, evolution, and mechanisms. Cell. 154:26–46.
2013.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Quinn JJ and Chang HY: Unique features of
long non-coding RNA biogenesis and function. Nat Rev Genet.
17:47–62. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang X, Zhu XL, Ji BY, Cao X, Yu LJ,
Zhang Y, Bao XY, Xu Y and Jin JL: LncRNA-1810034E14Rik reduces
microglia activation in experimental ischemic stroke. J
Neuroinflammation. 16(75)2019.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Wang H, Zheng X, Jin J, Zheng L, Guan T,
Huo Y, Xie S, Wu Y and Chen W: LncRNA MALAT1 silencing protects
against cerebral ischemia-reperfusion injury through miR-145 to
regulate AQP4. J Biomed Sci. 27(40)2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhou B, Yuan W and Li X: Long intergenic
noncoding RNA 319 (linc00319) promotes cell proliferation and
invasion in lung cancer cells by directly downregulating the tumor
suppressor MiR-32. Oncol Res: Aug 11, 2017 doi:
10.3727/096504017X15016337254650 (Epub ahead of print).
|
|
14
|
Qi G, Kong W, Mou X and Wang S: A new
method for excavating feature lncRNA in lung adenocarcinoma based
on pathway crosstalk analysis. J Cell Biochem. 120:9034–9046.
2019.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Wang X, Meng R and Hu QM:
LINC00319-mediated miR-3127 repression enhances bladder cancer
progression through upregulation of RAP2A. Front Genet.
11(180)2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yang Y, Zhang F, Huang H, Xie Z, Huang W,
Xie H and Wang F: Long noncoding RNA LINC00319 regulates ROMO1
expression and promotes bladder cancer progression via
miR-4492/ROMO1 axis. J Cell Physiol. 235:3768–3775. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Lang Q, Zhou M, Feng H, Guo J, Chen N and
He L: Research on the relationship between fibrinogen level and
subtypes of the TOAST criteria in the acute ischemic stroke. BMC
Neurol. 13(207)2013.PubMed/NCBI View Article : Google Scholar
|
|
18
|
General Assembly of the World Medical
Association: World medical association declaration of Helsinki.
Ethical principles for medical research involving human subjects. J
Am Coll Dent. 81:14–18. 2014.PubMed/NCBI
|
|
19
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 41 (Database
Issue):D991–D995. 2013.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Zhang ZH, Li JJ, Wang QJ, Zhao WQ, Hong J,
Lou SJ and Xu XH: WNK1 is involved in Nogo66 inhibition of OPC
differentiation. Mol Cell Neurosci. 65:135–142. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Ryou MG and Mallet RT: An in vitro
oxygen-glucose deprivation model for studying ischemia-reperfusion
injury of neuronal cells. Methods Mol Biol. 1717:229–235.
2018.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Chen X, Robinson DG and Storey JD: The
functional false discovery rate with applications to genomics.
Biostatistics. 28(kxz010)2019.
|
|
24
|
Schönenberger S, Hendén PL, Simonsen CZ,
Uhlmann L, Klose C, Pfaff JAR, Yoo AJ, Sørensen LH, Ringleb PA,
Wick W, et al: Association of general anesthesia vs procedural
sedation with functional outcome among patients with acute ischemic
stroke undergoing thrombectomy: A systematic review and
meta-analysis. JAMA. 322:1283–1293. 2019.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Yu S, Yu M, He X, Wen L, Bu Z and Feng J:
KCNQ1OT1 promotes autophagy by regulating miR-200a/FOXO3/ATG7
pathway in cerebral ischemic stroke. Aging Cell.
18(e12940)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Riva P, Ratti A and Venturin M: The long
non-coding RNAs in neurodegenerative diseases: Novel mechanisms of
pathogenesis. Curr Alzheimer Res. 13:1219–1231. 2016.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang L and Wang H: Long non-coding RNA in
CNS injuries: A new target for therapeutic intervention. Mol Ther
Nucleic Acids. 17:754–766. 2019.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Briggs JA, Wolvetang EJ, Mattick JS, Rinn
JL and Barry G: Mechanisms of long non-coding RNAs in mammalian
nervous system development, plasticity, disease, and evolution.
Neuron. 88:861–877. 2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ng SY, Lin L, Soh BS and Stanton LW: Long
noncoding RNAs in development and disease of the central nervous
system. Trends Genet. 29:461–468. 2013.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Paraskevopoulou MD and Hatzigeorgiou AG:
Analyzing MiRNA-LncRNA interactions. Methods Mol Biol.
1402:271–286. 2016.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Barangi S, Hayes AW, Reiter R and Karimi
G: The therapeutic role of long non-coding RNAs in human diseases:
A focus on the recent insights into autophagy. Pharmacol Res.
142:22–29. 2019.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Bao MH, Szeto V, Yang BB, Zhu SZ, Sun HS
and Feng ZP: Long non-coding RNAs in ischemic stroke. Cell Death
Dis. 9(281)2018.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Treiber T, Treiber N and Meister G:
Regulation of microRNA biogenesis and its crosstalk with other
cellular pathways. Nat Rev Mol Cell Biol. 20:5–20. 2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Brennan GP and Henshall DC: MicroRNAs as
regulators of brain function and targets for treatment of epilepsy.
Nat Rev Neurol. 16:506–519. 2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Kolosowska N, Gotkiewicz M, Dhungana H,
Giudice L, Giugno R, Box D, Huuskonen MT, Korhonen P, Scoyni F,
Kanninen KM, et al: Intracerebral overexpression of miR-669c is
protective in mouse ischemic stroke model by targeting MyD88 and
inducing alternative microglial/macrophage activation. J
Neuroinflammation. 17(194)2020.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Zhang X, Feng Y, Li J, Zheng L, Shao Y,
Zhu F and Sun X: MicroRNA-665-3p attenuates oxygen-glucose
deprivation-evoked microglial cell apoptosis and inflammatory
response by inhibiting NF-κB signaling via targeting TRIM8. Int
Immunopharmacol. 85(106650)2020.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Bernstein DL, Zuluaga-Ramirez V, Gajghate
S, Reichenbach NL, Polyak B, Persidsky Y and Rom S: miR-98 reduces
endothelial dysfunction by protecting blood-brain barrier (BBB) and
improves neurological outcomes in mouse ischemia/reperfusion stroke
model. J Cereb Blood Flow Metab. 40:1953–1965. 2020.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Zhao H, Li G, Zhang S, Li F, Wang R, Tao
Z, Ma Q, Han Z, Yan F, Fan J, et al: Inhibition of histone
deacetylase 3 by MiR-494 alleviates neuronal loss and improves
neurological recovery in experimental stroke. J Cereb Blood Flow
Metab. 39:2392–2405. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Guo T, Liu Y, Ren X, Wang W and Liu H:
Promoting role of long non-coding RNA small nucleolar RNA host gene
15 (SNHG15) in neuronal injury following ischemic stroke via the
MicroRNA-18a/CXC chemokine ligand 13 (CXCL13)/ERK/MEK Axis. Med Sci
Monit. 26(e923610)2020.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Chen X, Zhang X, Su C and Huang S: Long
noncoding RNA HULC in acute ischemic stroke: Association with
disease risk, severity, and recurrence-free survival and relation
with IL-6, ICAM1, miR-9, and miR-195. J Clin Lab Anal.
34(e23500)2020.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Yan G, Zhao H and Hong X: LncRNA MACC1-AS1
attenuates microvascular endothelial cell injury and promotes
angiogenesis under hypoxic conditions via modulating
miR-6867-5p/TWIST1 in human brain microvascular endothelial cells.
Ann Transl Med. 8(876)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Li X and Sui Y: Valproate improves middle
cerebral artery occlusion-induced ischemic cerebral disorders in
mice and oxygen-glucose deprivation-induced injuries in microglia
by modulating RMRP/PI3K/Akt axis. Brain Res.
1747(147039)2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Deng Y, Chen D, Wang L, Gao F, Jin B, Lv
H, Zhang G, Sun X, Liu L, Mo D, et al: Silencing of long noncoding
RNA nespas aggravates microglial cell death and neuroinflammation
in ischemic stroke. Stroke. 50:1850–1858. 2019.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Wang C, Wan H, Wang Q, Sun H, Sun Y, Wang
K and Zhang C: Safflor yellow B attenuates ischemic brain injury
via downregulation of long noncoding AK046177 and inhibition of
MicroRNA-134 expression in rats. Oxid Med Cell Longev.
2020(4586839)2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Yan H, Rao J, Yuan J, Gao L, Huang W, Zhao
L and Ren J: Long non-coding RNA MEG3 functions as a competing
endogenous RNA to regulate ischemic neuronal death by targeting
miR-21/PDCD4 signaling pathway. Cell Death Dis.
8(3211)2017.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Guo D, Ma J, Yan L, Li T, Li Z, Han X and
Shui S: Down-regulation of Lncrna MALAT1 attenuates neuronal cell
death through suppressing beclin1-dependent autophagy by regulating
Mir-30a in cerebral ischemic stroke. Cell Physiol Biochem.
43:182–194. 2017.PubMed/NCBI View Article : Google Scholar
|