Introduction
SPC24 is a component of the nuclear division cycle
80 (NDC80) kinetochore complex and is involved in the formation of
the kinetochore complex along with three other proteins, NUF2,
NDC80 and SPC25(1). The NDC80
kinetochore complex mediates microtubule binding and is anchored
onto the inner kinetochore via the SPC24-SPC25 protein complex. In
addition, the SPC24-SPC25 protein complex mediates dynamic
interactions between nuclear spindle microtubules and kinetochores,
which ensures faithful and accurate chromosomal segregation during
mitosis (2,3). Abnormal mitosis is a common hallmark
of cancer (4). It has also been
reported that mutation of SPC24 may lead to a chromosome
segregation defect (5). Moreover,
simultaneous disruption of both SPC24 and SPC25 may cause a spindle
checkpoint defect in the cell, allowing the cell to bypass mitosis
(6). These findings suggest that
dysregulation of SPC24 may lead to genomic instability and disrupt
control of the cell cycle, which can ultimately promote
malignancy.
Prostate cancer (PCa) has been identified as the
second most common cause of malignant tumor driven death in men
(7). Dietary factors,
lifestyle-related factors, region and genetic polymorphism have
been recognized as contributors to the risk of PCa (8,9).
Specific risk factors, such as prostatitis (inflammation/swelling
of the prostate gland) and benign prostatic hypertrophy (BPH;
proliferation of the cellular elements of the prostate, leading to
an enlarged prostate gland) have been reported to have a strong
association with the development of prostate cancer and a higher
relative risk of prostate cancer was estimated in men with both of
these conditions (10). BPH has
shown a high correlation with prostate cancer in Asian populations
(11). There is a 30% probability
of biochemical relapse [prostate specific antigen (PSA) <0.2
ng/ml twice in a row with no recurrent or metastatic lesions found
on radiography] in prostate cancer patients within the first 5
years after radical prostatectomy (12). In the clinic, detection of PSA is
often used for early diagnosis of prostate cancer. However, PSA is
not specific to malignant PCa and has a high false-positive and
false-negative rate of ~15% (13).
Therefore, a better PCa biomarkerwould be beneficial for accurate
diagnosis.
High levels of SPC24 have been found to be expressed
in various cancers, including breast, lung, thyroid and liver
cancer and osteosarcoma (14-17).
A recent study also indicated that increased SPC24 expression is
associated with advanced stage lung tumors (15). An additional study indicated that
when endogenous SPC24 was knocked down, the growth and invasion
ability of anaplastic thyroid cancer cells was inhibited and cell
apoptosis was promoted (16).
However, the role played by SPC24 in PCa is unclear.
In the present study, RNA-Seq data from The Cancer
Genome Atlas (TCGA) database were used to analyze SPC24 messenger
RNA (mRNA) levels in PCa tissues and normal tissues. Subsequently,
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) and western blot analysis were conducted to measure SPC24
expression in PCa and adjacent tissues, along with BPH tissues.
SPC24 expression was also detected in 40 clinical BPH tissues, 35
PCa tissues, 35 paired adjacent tissues, 12 prostatitis tissues and
9 normal prostate tissues by immunohistochemistry (IHC). In these
tissues, the association of SPC24 with clinicopathological features
was also assessed. Gene ontology (GO) analyses, Reactome pathway
analyses and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
analysis of the identified genes were conducted to find
correlations between SPC24 and its interactors in biological
processes, which were confirmed by qPCR.
Materials and methods
Data collection and analysis of TCGA
database
RNA sequencing (RNA-seq) data from selected genes
(SPC24 and its interacting partners obtained from literature review
and online database prediction) in PCa and clinicopathological
information of patients were downloaded from the TCGA Prostate
Cancer (PRAD) database (https://xena.ucsc.edu/public/; version 2017-10-13).
This dataset indicates the gene-level transcription estimates, as
log2(x+1) transformed RSEM (RNA-Seq by expectation-maximization)
normalized counts. The 3rd level data (normalized and
quantified raw data) was obtained from the TCGA data coordination
center (cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga).
Patients and specimens
From 2016 to 2018, a total of 96 patients (age,
46-87 years) including 35 patients with PCa (median age, 65 years),
40 patients with BPH (median age, 64 years), 12 patients with
prostatitis (median ages, 68 years) and 9 patients without prostate
disease (median age, 62 years) participated in the present study.
Written informed consent from patients and permission from the
Ethics Committee of The First Affiliated Hospital of Guangxi
Medical University were acquired. No preoperative treatment was
performed before surgery. All samples were provided by the First
Affiliated Hospital of the Guangxi Medical University. BPH tissues
were collected from patients with transurethral plasmakinetic
resection of prostate. PCa tissues and adjacent tissues were
obtained from patients that underwent radical prostatectomy, while
prostatitis tissues and normal tissues were obtained from patients
that underwentradical cystectomy. All prostate samples with
hematoxylin and eosin staining (H&E) were diagnosed by two
expert pathologists. Tumor grades were calculated in accordance
with Gleason classification. Clinical features of patients
including ethnicity, age, smoking, drinking, PSA, urea, creatinine,
lithic acid, HCO3-, and creatinine clearance
(CrCl) were obtained by Medical History Taking, PSA Test, Blood Gas
Test, and Renal Panel.
Quantitative real-time polymerase
chain reaction
TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to extract total RNA from the
clinical BPH, PCa and adjacent tissues. A NanoDrop
spectrophotometer (Thermo Fisher Scientific, Inc.) was used to
assess the purity and quantity of the isolated RNA. Complementary
DNA (cDNA) was synthesized using the Applied Biosystems StepOne
(Thermo Fisher Scientific, Inc.) with qPCR kit and SYBR-Green
master mix (Roche Diagnostics.). According to the instructions of
the RT kit (PrimeScript™ RT Master Mix; Takara), RT was
performed as follows: Initiation at 37˚C for 15 min, then 85˚C for
5 sec. The qPCR conditions included an initiation at 95˚C for 10
min, followed by 40 cycles of 95˚C for 15 sec and 60˚C for 1 min.
The 2-ΔΔCq method (18)
was performed to analyze relative quantities for the level of mRNA
expression. The specific primer sequences were as follows:
5'-TGGCCTCAGCTAGGTAACCA-3' (forward) and 5'-GTACCTGGGAGCTGTCATCG-3'
(reverse) for SPC24. 5'-GAAGCGCAGTTCAGTTTCC-3' (forward) and
5'-GGTTTCTCTTTGGTTTGAGGG-3' (reverse) for NDC80.
5'-TGGGAAAGATACATACAGTGG-3' (forward) and
5'-GAATCTTGGGTCATTGTGGT-3' (reverse) for BUB1.
Western blot analysis
Clinical PCa tissues and paired adjacent tissues
were obtained, washed twice with 0.01 M PBS and lysed on ice with
radioimmunoprecipitation assay (RIPA, Beijing Solarbio Science
& Technology Co., Ltd.) a cell lysis reagent for 30 min.
Tissues were centrifuged at 16,000 x g for 15 min at 4˚C to remove
insoluble proteins. The concentration of extracted protein was
determined by bicinchoninic acid (BCA) protein assay kit (Leagene,
Inc.). A total of 50 µg of each total protein extract was separated
using 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis
and transferred onto PVDF membranes (Bio-Rad Laboratories, Inc.).
After being blocked at 23˚C for 1 h with 5% milk solution,
membranes were washed three times with Tris-buffered saline-tween
(0.1% Tween-20) solution. The membranes were then incubated with
primary antibodies, rabbit anti-human SPC24 (Abcam; 1:1,000; cat.
no. ab169786) and β-actin (Proteintech Group, Inc.; 1:1,000; cat.
no. HRP-60008) at 4˚C overnight. After being washed with TBST, the
membranes were incubated with horseradish peroxidase-labeled
goat-anti-rabbit IgG secondary antibody (Abcam; 1:4,000; cat. no.
ab6721) at 37˚C for 1 h. The membranes were processed for protein
detection using the BeyoECL Plus Kit (Beyotime Institute of
Biotechnology).
Immunohistochemistry
All tissues were embedded in paraffin and then cut
into 4-µm-thick sections. Immunostaining was conducted using the
Elivision™ Plus method (EliVision™ Super KIT9922; Fuzhou Maixin
Biotech. Co., Ltd.), with the procedure performed according to the
kit instructions. All samples were deparaffinized with xylene and
rehydrated with a graded ethanol series (100, 95, 90 and 85%).
Citrate buffer (pH 6.0) was used for antigen retrieval. Methanol
containing 3% H2O2 solution was used to block
endogenous peroxidase activity (at 23˚C for 15 min without light),
before 10% goat serum (OriGene Technologies, Inc.) was used to
block nonspecific proteins (at 23˚C for 15 min). Anti-SPC24
antibody (cat. no. ab169786) primary antibody was added at a
dilution of 1:250 and the sections were subsequently incubated
overnight at 4˚C. After washing in PBS, all sections were incubated
with biotinylated secondary antibodies (1:100, cat. no. SP-9000;
OriGene Technologies, Inc.) at 23˚C for 25 min. All sections were
stained with horseradish-labeled chains of ovalbumin working fluid
(OriGene Technologies, Inc.) at 23˚C for 15 min, then were washed
in PBS for 5 min (repeated 3 times). The images were allowed to
develop in diaminobenzidine DAB substrate Kit (ZLI9018; Beijing
Zhongshan Golden Bridge Biotechnology Co., Ltd.). Finally, all
sections were re-dyed with hematoxylin for 1 min at 23˚C and
mounted with neutral resin. The degree of immunohistochemistry
(IHC) staining was evaluated by two independent blinded
pathologists (light microscope; cat. no. BX53+DP80; Olympus
Corporation) observing ≥20 fields of view in each slide. The
results were analyzed by an immunoreactivity score (IRS) system
based on staining intensity and cell staining proportion data. The
staining intensity was scored from 0-3 as follows: 0=unstained;
1=weakly stained; 2=moderately stained and 3=strongly stained. The
proportion of cell staining was scored on a scale from 0-4 as
follows: 0=negative; 1=1-10%; 2=11-50%; 3=51-80% and 4>80%. The
intensity score was multiplied by the proportion of staining to
obtain an immunoreactivity score. A total score >3 was
considered to be a high level of expression. A total score of 1-3
was considered as low expression and 0 was considered no
expression.
Functional and pathway enrichment
analysis
To analyze the identified genes at the functional
level, Gene ontology (GO) analyses, Reactome pathway analyses
(reactome.org/) and Kyoto Encyclopedia of Genes
and Genomes (KEGG) pathway analyses of the identified genes were
conducted. The biological online database tool adopted in this
study for GO and KEGG analysis was the Search Tool for the
Retrieval of Interacting Genes (STRING, Version: 11.0, https://string-db.org/). Identified genes were
submitted and Homo sapiens was selected in the species
column. Finally, the GO terms or KEGG pathways with the cut-off
criteria (P<0.05) were identified to be associated with the
input genes.
Statistical analysis
Software including SPSS 21.0 (IBM Corp.), GraphPad
Prism 5 (Graph Pad Software, Inc.) and MedCalc. v9.2.0.1 (MedCalc,
Inc.) were used to perform statistical analyses. Unpaired t-tests
were used to compare quantitative sample data from the TCGA. Data
are presented in the form of the mean ± SD. Two paired-group
(PCa/BPH/prostatitis tissues and adjacent/normal tissues)
comparisons were conducted using χ2 test with Fisher's
exact test. Association between SPC24 expression and
clinicopathological factors was also determined using χ2
test. A P-value of <0.05 was considered statistically
significant. Overall survival (OS) and Disease-free survival (DFS)
times were stratified according to median expression level (high
and low) of SPC24 (conducted in GEPIA version 2017, http://gepia.cancer-pku.cn/). Survival analyses were
conducted and visualized using Kaplan-Meier analysis and log-rank
test. The diagnostic ability was evaluated using receiver operating
characteristic curve (ROC). The sensibility and specificity were
obtained at an optimal cut-off with the max Youden index. Based on
mRNA data (quantified as RSEM to estimate gene and isoform
expression levels from RNA-Seq data; deweylab.github.io/RSEM/README.html) extracted from
TCGA database, diagnostic ability of two combined models was also
analyzed by ROC curves using binary logistic regression. Combined
models acquired using binary logistic regression were: SPC24 +
BUB1: Y=SPC24+BUB1*0.849/1.230; SPC24 + NDC80:
Y=SPC24+NDC80*(-1.022)/1.486
Results
SPC24 analyses in TCGA dataset
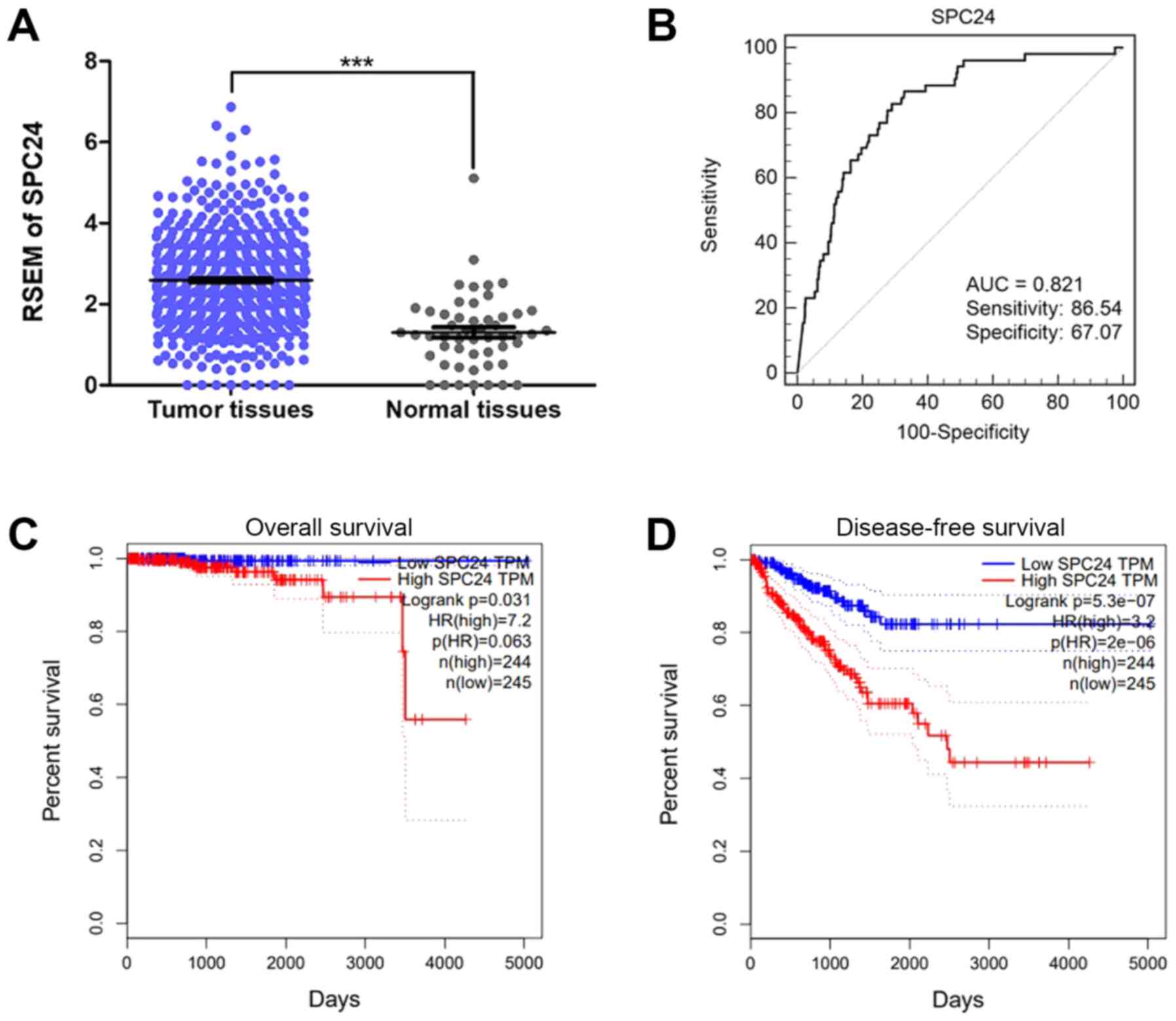
The expression of SPC24 in PCa was significantly
higher than that in normal tissues (P<0.0001; Fig. 1A). In addition, higher SPC24
expression was found in older PCa patients (>60), higher Gleason
scores (8-10),
and with higher PSA values (>0.1; Table I). Increased SPC24 was also
associated with lymph node metastasis (Table I). An ROC curve was plotted to
further evaluate the diagnostic value of SPC24 and the AUC value of
SPC24 was 0.821 (P<0.05). Based on the maximum Youden index
discriminating patients with PCa from controls, an optimal cut-off
value of 2.06 was obtained, and its sensitivity and specificity for
predicting PCa were calculated to be 86.54 and 67.07%, respectively
(Fig. 1B). Survival analyses also
suggested that high SPC24 expression was associated with negative
outcomes in PCa patients (P<0.05: Fig. 1C and D).
 | Table IAssociation between SPC24 and
clinicopathologic characteristics in The Cancer Genome Atlas PRAD
database (n=498). |
Table I
Association between SPC24 and
clinicopathologic characteristics in The Cancer Genome Atlas PRAD
database (n=498).
| Clinical
feature | Number of
cases | SPC24 (RSEM; mean ±
SD) | P-value |
|---|
| Sample | | |
<0.05a |
|
PCa | 498 | 2.590±1.179 | |
|
Normal | 52 | 1.303±0.928 | |
| Age at diagnosis
(years) | | |
<0.05a |
|
≤60 | 223 | 2.405±1.128 | |
|
>60 | 275 | 2.740±1.200 | |
| Gleason score | | |
<0.05a |
|
2-4 | 0 | | |
|
5-7 | 292 | 2.247±0.969 | |
|
8-10 | 206 | 3.077±1.277 | |
| Laterality | | | >0.05 |
|
Bilateral | 433 | 2.599±1.193 | |
|
Unilateral | 65 | 2.534±1.083 | |
| PSA value | | |
<0.05a |
|
≤0.1 | 329 | 2.456±1.125 | |
|
>0.1 | 112 | 2.824±1.24 | |
|
Unknown | 27 | | |
| Tumor level | | | >0.05 |
|
Apex | 292 | 2.66±1.181 | |
|
Middle|Base | 63 | 2.588±1.153 | |
|
Unknown | 143 | | |
| Lymph node | | |
<0.05a |
|
N0 | 345 | 2.533±1.122 | |
|
N1 | 80 | 3.103±1.313 | |
|
Unknown | 73 | | |
| Metastasis | | | |
|
M0 | 456 | | |
|
M1 | 3 | | |
|
Unknown | 39 | | |
Expression of SPC24 in BPH, PCa,
prostatitis and adjacent/normal tissues
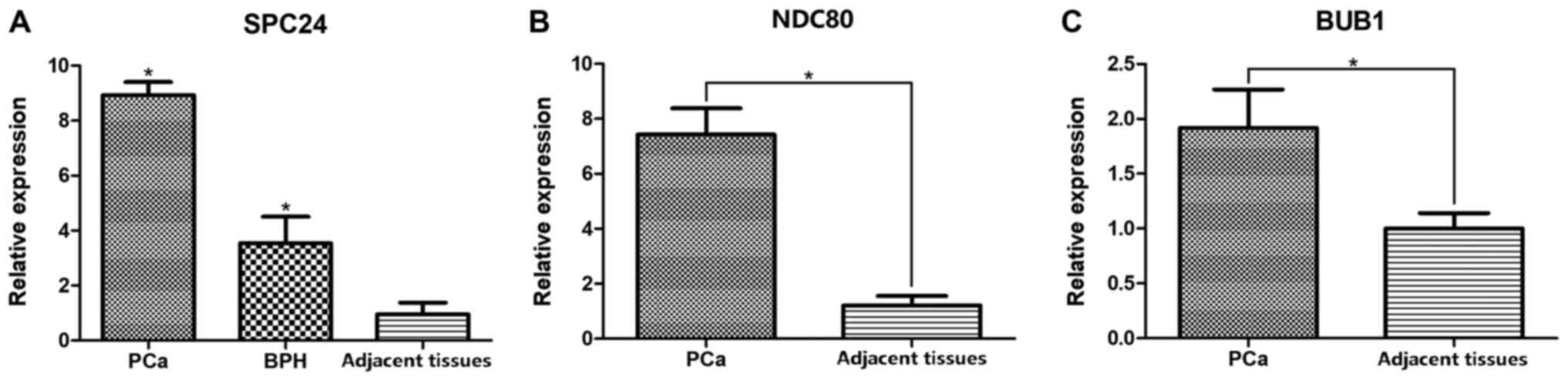
To further confirm the expression of SPC24 in PCa,
qPCR, western blot and IHC were performed in 35 PCa tissues and
paired adjacent tissues from Chinese patients with PCa. Increased
SPC24 expression was found in PCa tissues compared with adjacent
tissues (P<0.05: Figs. 2 and
3). qPCR analysis also suggested
that SPC24 expression was higher in PCa than in BPH and adjacent
tissues (P<0.05; Fig. 2A). IHC
(Fig. 3B) indicated that 13 of the
35 PCa patients had high SPC24 expression, while the remaining 22
had low expression. Of the 40 BPH patients, 11 had high SPC24
expression and the remaining 29 had low or no expression.
Additionally, 5 of the 12 patients with prostatitis had high levels
of SPC24 expression, while the remaining had no expression. SPC24
was expressed at low levels or not expressed in both the adjacent
tissues (35/35) and the normal tissues (9/9; P<0.05; Fig. 3B; Table
II). These results indicated that SPC24 expression was higher
in PCa tissues than in adjacent tissues, which was also confirmed
by western blot (Fig. 3A). Higher
SPC24 levels in BPH tissues than in the adjacent tissues were also
found, while there was no difference between PCa and BPH in SPC24
expression (Fig. 3; Table II).
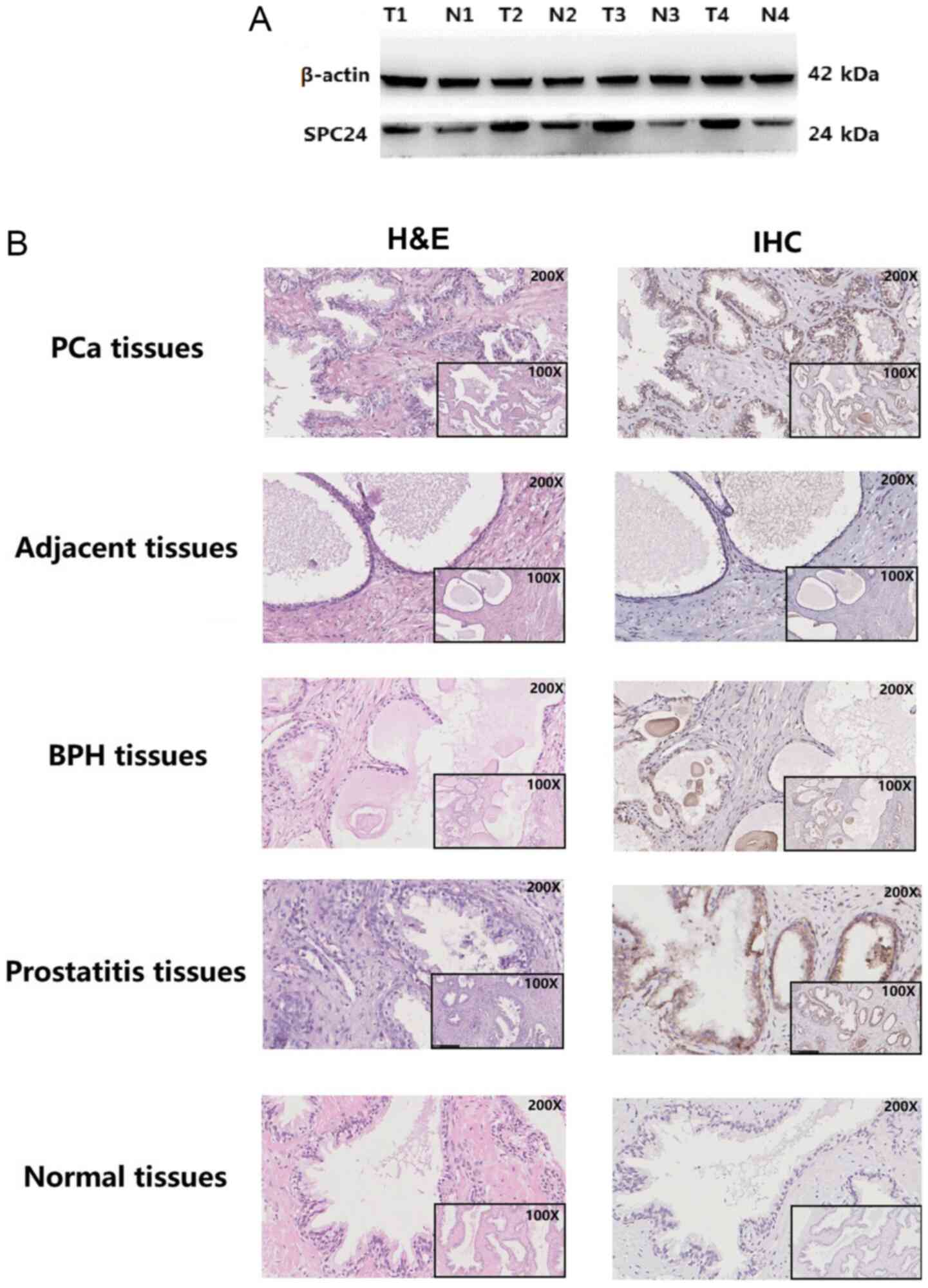 | Figure 3Protein level of SPC24 in different
prostate samples. (A) Highly expressed SPC24 was found in PCa
tissues compared to the adjacent tissues detected by western
blotting. (B) Expression of SPC24 in different prostate samples
including PCa tissues, paired-adjacent tissues, BPH tissues,
prostatitis tissues, and normal tissues. Positive staining was
mostly concentrated in the epithelial cells of prostate gland. BPH,
benign prostatic hypertrophy; H&E, hematoxylin-eosin staining;
IHC, immunohistochemistry; T, PCa tissue; N, paired adjacent
tissues; PCa, prostate cancer. |
 | Table IIExpression of SPC24 in PCa tissues,
adjacent tissues, BPH tissues, prostatitis tissues,and normal
prostate tissues. |
Table II
Expression of SPC24 in PCa tissues,
adjacent tissues, BPH tissues, prostatitis tissues,and normal
prostate tissues.
| A, Benign prostatic
hypertrophy and adjacent tissues |
|---|
| | SPC24 level | |
|---|
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
|---|
| Disease tissue | 40 | 29 | 11 | 0.001a |
| Adjacent
tissue | 35 | 35 | 0 | |
| B, Prostate cancer
tissues and adjacent tissues |
| | SPC24 level | |
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
| Disease tissue | 35 | 22 | 13 |
<0.001a |
| Adjacent
tissue | 35 | 35 | 0 | |
| C, Prostate cancer
tissue and benign prostatic hypertrophy tissues |
| | SPC24 level | |
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
| PCa tissue | 35 | 22 | 13 | 0.459 |
| BPH tissue | 40 | 29 | 11 | |
| D, Prostatitis
tissue and normal tissue |
| | SPC24 level | |
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
| Prostatitis
tissue | 12 | 7 | 5 | 0.045a |
| Normal tissue | 9 | 9 | 0 | |
Association between SPC24 expression
and clinical features of patients with PCa/BPH/prostatitis
In Chinese patients with PCa, high expression of
SPC24 was associated with high Gleason stage (IV and V; P<0.05).
Other clinical features in PCa, including ethnicity, age, smoking,
drinking, PSA, urea, creatinine, lithic acid,
HCO3-, and creatinine clearance, were not
correlated with SPC24 expression (Table III). In BPH tissues, SPC24 levels
were associated with ethnicity and CrCl (P<0.05; Table IV). However, there was no
association between SPC24 and other clinical factors in
prostatitis.
 | Table IIIAssociation between SPC24 and
clinicopathological characteristics in prostate cancer tissues
(n=35). |
Table III
Association between SPC24 and
clinicopathological characteristics in prostate cancer tissues
(n=35).
| | SPC24 level | |
|---|
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
|---|
| Urea | | | | |
|
Positive | 0 | 0 | 0 | |
|
Negative | 35 | 22 | 13 | |
| Ethnicity | | | | 0.431 |
|
Han | 26 | 15 | 11 | |
|
Zhuang | 9 | 7 | 2 | |
| Age (years) | | | | 0.721 |
|
≤70 | 22 | 13 | 9 | |
|
>70 | 13 | 9 | 4 | |
| Smoking | | | | 1.000 |
|
Yes | 7 | 4 | 3 | |
|
No | 28 | 18 | 10 | |
| Drinking | | | | 0.274 |
|
Yes | 4 | 4 | 0 | |
|
No | 31 | 18 | 13 | |
| tPSA | | | | 0.014a |
|
Positive
(>4.0 ng/ml) | 31 | 22 | 9 | |
|
Negative
(≤4.0 ng/ml) | 4 | 0 | 4 | |
| Creatinine | | | | 0.519 |
|
Positive | 2 | 2 | 0 | |
|
Negative | 33 | 20 | 13 | |
| Lithic acid | | | | 1.000 |
|
Positive | 9 | 6 | 3 | |
|
Negative | 26 | 16 | 10 | |
|
HCO3- | | | | 1.000 |
|
Positive | 9 | 6 | 3 | |
|
Negative | 26 | 16 | 10 | |
| CrCl | | | | 0.175 |
|
Positive
(<80 ml/min) | 20 | 15 | 5 | |
|
Negative
(>80 ml/min) | 15 | 7 | 8 | |
| Gleason stage | | | | 0.035a |
|
I+III | 17 | 14 | 3 | |
|
IV+V | 18 | 8 | 10 | |
 | Table IVAssociation between SPC24 and
clinicopathological characteristics in benign prostatic
hypertrophic tissues (n=40). |
Table IV
Association between SPC24 and
clinicopathological characteristics in benign prostatic
hypertrophic tissues (n=40).
| | SPC24 level | |
|---|
| Clinical
feature | Number of
cases | Low/negative | High | P-value |
|---|
| Urea | | | | 1.000 |
|
Positive | 7 | 5 | 2 | |
|
Negative | 33 | 24 | 9 | |
| Ethnicity | | | | 0.031a |
|
Han | 20 | 11 | 9 | |
|
Zhuang | 20 | 18 | 2 | |
| Age (years) | | | | 0.293 |
|
≤70 | 19 | 12 | 7 | |
|
>70 | 21 | 17 | 4 | |
| Smoking | | | | 0.286 |
|
Yes | 22 | 14 | 8 | |
|
No | 18 | 15 | 3 | |
| Drinking | | | | 0.369 |
|
Yes | 7 | 4 | 3 | |
|
No | 33 | 25 | 8 | |
| tPSA | | | | 0.455 |
|
Positive
(>4.0 ng/ml) | 29 | 22 | 7 | |
|
Negative
(≤4.0 ng/ml) | 11 | 7 | 4 | |
| Creatinine | | | | 1.000 |
|
Positive | 9 | 7 | 2 | |
|
Negative | 31 | 22 | 9 | |
| Lithic acid | | | | 1.000 |
|
Positive | 16 | 12 | 4 | |
|
Negative | 24 | 17 | 7 | |
|
HCO3- | | | | 1.000 |
|
Positive | 20 | 15 | 5 | |
|
Negative | 20 | 14 | 6 | |
| CrCl | | | | 0.020a |
|
Positive
(<80 ml/min) | 27 | 23 | 4 | |
|
Negative
(>80 ml/min) | 13 | 6 | 7 | |
| Gleason stage | | | | 1.000 |
|
I+III | 7 | 5 | 2 | |
|
IV+V | 33 | 24 | 9 | |
Gene ontology and pathway functional
enrichment analysis
As shown in Table V,
GO annotation and pathway enrichment analysis suggested that SPC24
and its interactors were related to several biological processes,
including cell division and cell cycle. In addition, SPC24 and its
interactors were related to terms suggesting they were located in
the nucleus, cytosol and intracellular organelle components of the
cell. Reactome pathway analysis indicated that SPC24 participated
in many pathways, as it was associated with the terms resolution of
sister chromatid cohesion, amplification of signal from unattached
kinetochores via a MAD2 inhibitory signal, cell cycle checkpoints,
RHO GTPases activate formins, separation of sister chromatids and
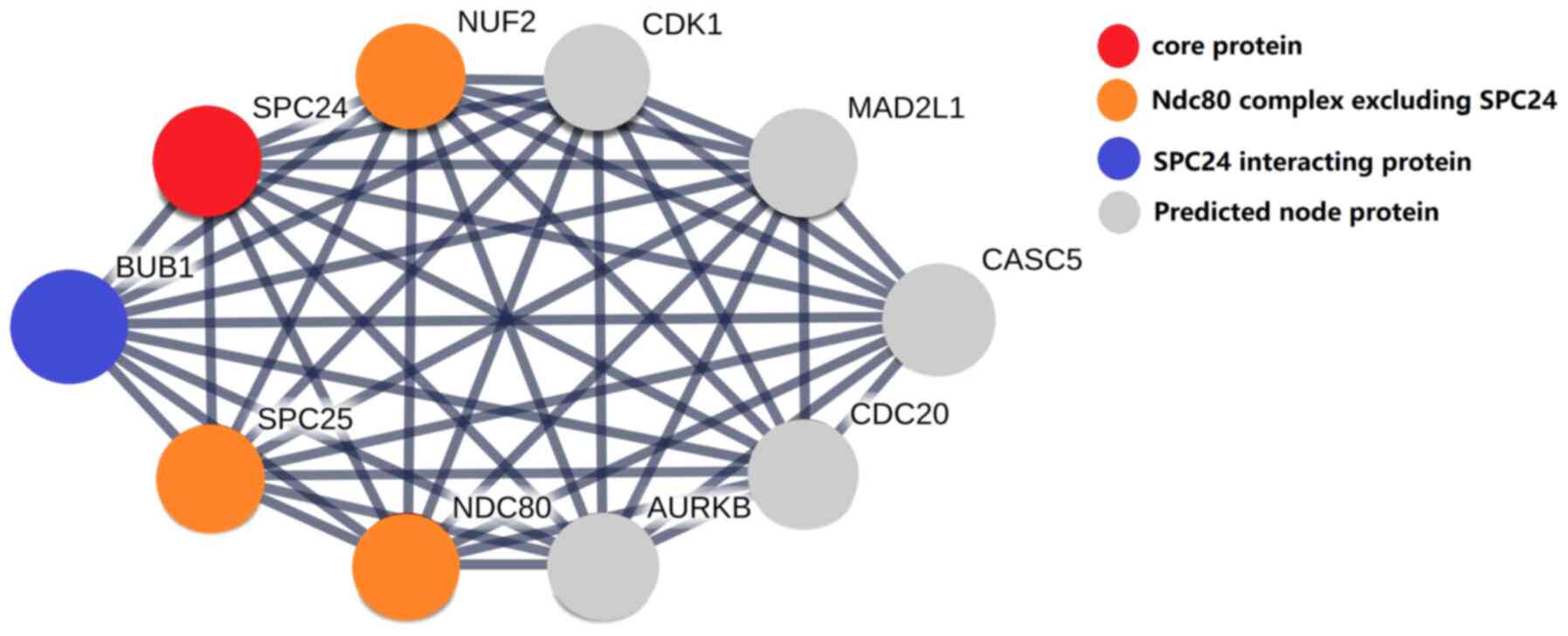
signal transduction. The protein-protein interaction (PPI) network
is shown in Fig. 4.
 | Table VGene ontology analyses, reactome
pathway analyses and Kyoto Encyclopedia of Genes and Genomes
pathway enrichment analyses of SPC24. |
Table V
Gene ontology analyses, reactome
pathway analyses and Kyoto Encyclopedia of Genes and Genomes
pathway enrichment analyses of SPC24.
| A, Biological
processes |
|---|
| Pathway ID | Term
description | Observed gene
count | Background gene
count | False discovery
rate | Matching
proteins |
|---|
| GO:0051301 | Cell division | 10 | 483 |
6.22x10-14 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0007049 | Cell cycle | 10 | 1263 |
2.89x10-10 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| B, Cell
components |
| Pathway ID | Term
description | Observed gene
count | Background gene
count | False discovery
rate | Matching
proteins |
| GO:0000779 | Condensed
chromosome, centromeric region | 8 | 117 |
8.92x10-15 | AURKB, BUB1, CASC5,
MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0000776 | kinetochore | 8 | 130 |
1.01x10-14 | AURKB, BUB1, CASC5,
MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0000777 | Condensed
chromosome kinetochore | 7 | 104 |
3.35x10-13 | BUB1, CASC5,
MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0031262 | Ndc80 complex | 4 | 4 |
3.12x10-11 | NDC80, NUF2, SPC24,
SPC25 |
| GO:0043232 | Intracellular
non-membrane-bounded organelle | 10 | 4005 |
1.19x10-06 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0032991 | Protein-containing
complex | 10 | 4792 |
5.48x10-06 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0005829 | Cytosol | 10 | 4958 |
7.15x10-06 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0005634 | Nucleus | 10 | 6892 | 0.00013 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| GO:0031981 | Nuclear lumen | 8 | 4030 | 0.00037 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, SPC24 |
| GO:0044446 | Intracellular
organelle part | 10 | 8882 | 0.0012 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| C, Reactomes |
| Pathway ID | Term
description | Observed gene
count | Background gene
count | False discovery
rate | Matching
proteins |
| HSA-2500257 | Resolution of
Sister Chromatid Cohesion | 10 | 118 |
1.02x10-20 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| HSA-141444 | Amplification of
signal from unattached kinetochores via a MAD2 inhibitory
signal | 9 | 90 |
5.05x10-19 | AURKB, BUB1, CASC5,
CDC20, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| HSA-69620 | Cell Cycle
Checkpoints | 10 | 265 |
4.32x10-18 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| HSA-5663220 | RHO GTPases
activate Formins | 9 | 131 |
5.46x10-18 | AURKB, BUB1, CASC5,
CDC20, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| HSA-2467813 | Separation of
Sister Chromatids | 9 | 178 |
6.13x10-17 | AURKB, BUB1, CASC5,
CDC20, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
| HSA-162582 | Signal
Transduction | 10 | 2605 |
1.14x10-08 | AURKB, BUB1, CASC5,
CDC20, CDK1, MAD2L1, NDC80, NUF2, SPC24, SPC25 |
Increased expression of SPC24
interactors
In PCa samples, qPCR results showed that expression
of SPC24 interacting proteins (BUB1 and NDC80) was higher than that
in the adjacent tissues (Fig. 2B
and C). Based on the mRNA data from
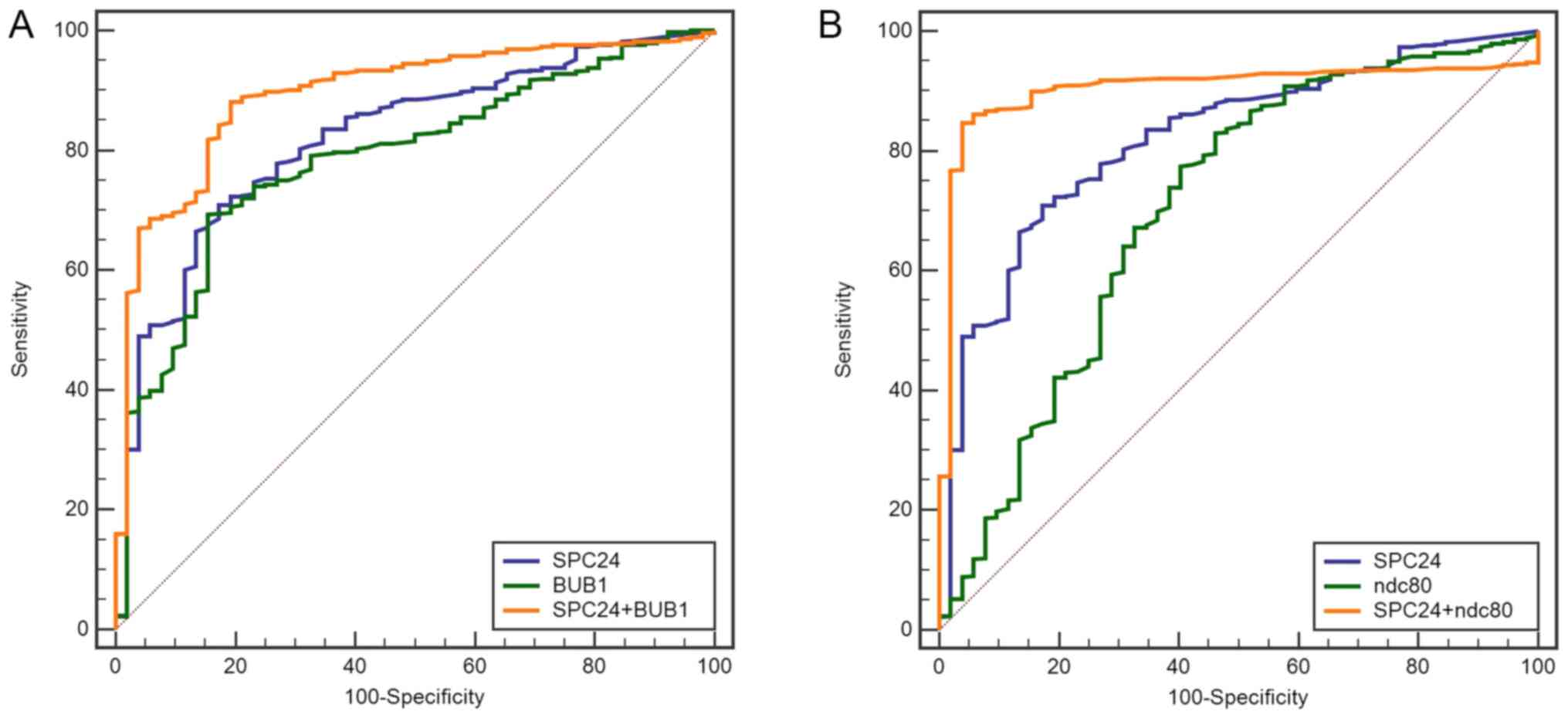
TCGA database, two combined models via SPC24 were built to obtain
better diagnosis efficiency for PCa using binary logistic
regression (Fig. 5). The ROC curve
of combined model of SPC24 and BUB1 was generated, and the
sensitivity and specificity were 88.2 and 80.8%, respectively. The
AUC was 0.894. The ROC curve of combined model of SPC24 and NDC80
had a AUC of 0.905 and the sensitivity and specificity were 84.7
and 96.2%, respectively (Table
VI).
 | Table VISensitivity and specificity to
diagnose prostate cancer with SPC24 and its interacting
proteins. |
Table VI
Sensitivity and specificity to
diagnose prostate cancer with SPC24 and its interacting
proteins.
| Model | AUC | Cut off | Sensitivity
(%) | Specificity
(%) |
|---|
| SPC24 | 0.821 | 1.91 | 70.9 | 82.7 |
| SPC24 + BUB1 | 0.894 | 4.71 | 88.2 | 80.8 |
| SPC24 + NDC80 | 0.905 | -3.10 | 84.7 | 96.2 |
Discussion
As a subunit of the NDC80 complex, NUF2 is highly
expressed in a variety of tumors and has also been shown to
correlate with poor prognosis (19,20).
It has been reported that SPC24 and SPC25 are required to establish
and maintain kinetochore-microtubule attachments and metaphase
alignment and are essential for chromosomal movement to the spindle
poles during anaphase (21). High
expression of SPC25 has been found in PCa (22); however, the impact of SPC24 in PCa
remains to be elucidated.
The present study analyzed the TCGA database to
evaluate the possible diagnostic value of SPC24 in PCa and the
associations between SPC24 and clinical characteristics. The
results of the TCGA dataset analysis indicated that SPC24 was
highly expressed in PCa tissues compared to normal tissues.
Further, results demonstrated that the high expression of SPC24 was
associated with increased age, PSA value, lymph node metastasis and
Gleason score, without an association with tumor level and
laterality. Furthermore, analysis of a subset of clinical specimens
by RT-qPCR, western blotting and immunohistochemistry demonstrated
that SPC24 was significantly up-regulated in PCa tissues and BPH
tissues compared to the adjacent tissues. In prostatitis tissues,
SPC24 was also expressed at an increased level when compared to
that in normal tissues. Future studies should include more prostate
samples of patients from different nationality. The results of the
present study suggested that increased SPC24 may participate in
many prostatic diseases, including prostatitis, BPH and PCa.
In order to explore the potential mechanism of SPC24
and its interacting proteins in the development of prostatic
diseases, bioinformatics analysis was necessary. In the present
study, GO and pathway functional enrichment analysis suggested that
SPC24 and its interacting proteins may participate in the cell
cycle and cell division, which indicated that abnormal expression
of SPC24 and its interacting proteins may interfere with the cell
cycle and cell division. As loss of cell cycle control is one of
the most important hallmarks of cancer (23), increased expression of SPC24 and its
interacting proteins may also support the concept that there is an
irregular cell cycle in PCa cells. Furthermore, multiple predicted
pathways containing SPC24 determined from analysis in the study
also hinted that SPC24, cooperating with its interacting proteins,
could take part in many other biological processes. Validation of
these predicted pathways needs to be performed in the future. Both
SPC24 and SPC25 are linked with these predicted pathways. In terms
of structure of protein complex, SPC24 and SPC25 fold tightly
together into a single globular entity with pseudo-2-fold symmetry
in vivo (24,25). In terms of protein function, SPC24
is required for meiotic kinetochore-microtubule attachment and
production of euploid eggs and SPC25 is required for chromosome
alignment, spindle formation and proper spindle checkpoint
signaling during meiosis (26,27).
These studies imply that SPC24 and SPC25 are closely related, not
only in structure, but also in function. In the present study,
multiple predicted pathways which all contained SPC24 and
SPC25suggest a close interaction between SPC24 and SPC25. However,
the effect of SPC24/SPC25 complex in prostatic diseases remains
unclear. The potential role of SPC24/SPC25 complex in prostatic
diseases also requires further study. Dysregulation of the
kinetochore may result in chromosome aneuploidy and instability,
resulting from a guidance defect for proper and accurate chromosome
segregation during mitosis (28,29).
NDC80 is known to be the core component of the NDC80 centromere
complex and the protein interaction partner of SPC24(1). In addition, high-level expression of
NDC80 was found in Human PCa tissues and increased NDC80 expression
can also enhance the proliferation, migration, and invasion ability
of PCa cell lines (30). It has
been reported that SPC24 expression is upregulated in
NDC80-overexpressing cancer cells (31). Therefore, in the present study
experiments were conducted to verify NDC80 expression levels in the
clinical specimens. The qPCR results showed that the level of NDC80
mRNA was higher in PCa tissues than in adjacent tissues, which was
also related to expression of SPC24. The formation of proper
kinetochore-microtubule attachments facilitates the procedural
separation of chromosomes. The NDC80 protein is responsible for
lateral attachment of microtubules, helping establish end-on
attachment by indirectly interacting with the microtubule plus end
through recruitment of microtubule-associated proteins (32). However, overproduction of NDC80
leads to the absorption of its interacting proteins by binding to
the internal loop, blocking its interacting proteins from
connecting to microtubules. This sequestration impedes mitotic
progression and leads to chromosome mis-segregation, resulting in
the formation of aneuploid progenies. Aneuploidy promotes
tumorigenesis (33). In the present
study, increased BUB1 mRNA was detected in PCa specimens. It is
known that there is a surveillance system in the eukaryotic cell,
the spindle assembly checkpoint (SAC), which can identify
incorrectly attached or unattached kinetochores to ensure faithful
chromosome segregation. The SAC consists of six components (MPH1,
MAD1, MAD2, MAD3, BUB1 and BUB3). Upon SAC activation, BUB1 and
other SAC components arrive at the kinetochore, generating a
‘wait-anaphase’ signal to prevent mitotic progression until all
kinetochores are properly attached to microtubules emanating from
the opposite spindle poles (34,35).
However, when BUB1 is over-expressed, it causes mitotic errors of
chromosome misalignment and lagging and resulted in near-diploid
aneuploidies and tumor formation (36). These results suggest coordination
among NDC80, BUB1, and SPC24 in PCa-genesis. Patients with elevated
serum PSA levels are not diagnosed accurately with PCa (13). To diagnose PCa more accurately, two
combined models via SPC24 mRNA were established. The AUC of two
combined ROC curves was bigger than that of single SPC24, which
demonstrated that combined models between SPC24 and its interacting
partners could provide a discriminatory ability to diagnose PCa. At
an optimal cut-off point with the maximum Youden index, the
sensitivity and specificity of two combined ROC curves were
different. A combined model of SPC24 and BUB1 had the highest
sensitivity, while a combined model of SPC24 and NDC80 had the
highest specificity. This result might be caused by different
diagnostic ability of single BUB1 and NDC80, which will require
further study. Compared to other previously reported PCa diagnosis
markers, such as serum PSA (AUC: 0.52; sensitivity: 58%;
specificity: 72%), prostate cancer antigen 3 (PCA3) mRNA (AUC:
0.865; sensitivity: 94.9%; specificity: 60.1%), and a combined
model of PSA, PCA3, human glandular kallikrein 2 (HK2) (AUC: 0.667;
sensitivity: 55.8%; specificity: 82.5%) (37-39),
a diagnostic model of SPC24 combining BUB1/NDC80 could provide
better diagnostic ability for PCa with a bigger AUC (>0.89),
higher sensitivity and higher specificity (both over 80%). To the
best of our knowledge, this study was the first to demonstrate that
SPC24 was significantly up-regulated in BPH, prostatitis and PCa.
SPC24 and its interactors may participate in some biological
processes and many reactome pathways, including cohesion and
separation of sister chromatids. In conclusion, SPC24 may serve a
carcinogenic role in the development of PCa and may act as a new
diagnostic and potential therapeutic target for PCa. The diagnostic
and prognostic values of SPC24 and its possible therapeutic
applications are worthy of further investigation.
Acknowledgements
Prof. Jian Qin from school of public health, Guangxi
Medical University was invited to check the statistical methods in
our study.
Funding
Funding: This work was supported by the National Natural Science
Foundation of China (grant nos. 81560130 and 21505025) and the
Guangxi Nature Science Foundation (grant nos. 2018GXNSFAA281033,
2016GXNSFDA380010 and 2013GXNSFGA019005).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL and XY made substantial contributions to
conception and design and agreed to be accountable for all aspects
of the work in ensuring that questions related to the accuracy or
integrity of any part of the work were appropriately investigated
and resolved. SC, XW and SZ wrote and revised the manuscript,
performed the experiments and analyzed and interpreted scientific
data. SC and SQ confirmed the authenticity of all the raw data. HL,
SQ, JL and WJ performed IHC, qPCR and western blot analysis. MS,
YT, HL, WS and SL participated in data collection and analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The First Affiliated Hospital of Guangxi Medical
University (Nanning, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
DeLuca JG and Musacchio A: Structural
organization of the kinetochore-microtubule interface. Curr Opin
Cell Biol. 24:48–56. 2012.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Cheeseman IM and Desai A: Molecular
architecture of the kinetochore-microtubule interface. Nat Rev Mol
Cell Biol. 9:33–46. 2008.PubMed/NCBI View
Article : Google Scholar
|
|
3
|
Wilson-Kubalek EM, Cheeseman IM, Yoshioka
C, Desai A and Milligan RA: Orientation and structure of the Ndc80
complex on the microtubule lattice. J Cell Biol. 182:1055–1061.
2008.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Dominguez-Brauer C, Thu KL, Mason JM,
Blaser H, Bray MR and Mak TW: Targeting mitosis in cancer: Emerging
strategies. Mol Cell. 60:524–536. 2015.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Ma L, McQueen J, Cuschieri L, Vogel J and
Measday V: Spc24 and Stu2 promote spindle integrity when DNA
replication is stalled. Mol Biol Cell. 18:2805–2816.
2007.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Bharadwaj R and Yu H: The spindle
checkpoint, aneuploidy, and cancer. Oncogene. 23:2016–2027.
2004.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nelson WG, De Marzo AM and Isaacs WB:
Prostate cancer. N Engl J Med. 349:366–381. 2003.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Gutiérrez-González E, Castelló A,
Fernández-Navarro P, Castaño-Vinyals G, Llorca J, Salas D,
Salcedo-Bellido I, Aragonés N, Fernández-Tardón G, Alguacil J, et
al: Dietary zinc and risk of prostate cancer in Spain: MCC-Spain
Study. Nutrients. 11(18)2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Hung SC, Lai SW, Tsai PY, Chen PC, Wu HC,
Lin WH and Sung FC: Synergistic interaction of benign prostatic
hyperplasia and prostatitis on prostate cancer risk. Br J Cancer.
108:1778–1783. 2013.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Dai X, Fang X, Ma Y and Xianyu J: Benign
prostatic hyperplasia and the risk of prostate cancer and bladder
cancer: A meta-analysis of observational studies. Medicine
(Baltimore). 95(e3493)2016.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Boorjian SA, Eastham JA, Graefen M,
Guillonneau B, Karnes RJ, Moul JW, Schaeffer EM, Stief C and Zorn
KC: A critical analysis of the long-term impact of radical
prostatectomy on cancer control and function outcomes. Eur Urol.
61:664–675. 2012.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sharma N and Baruah MM: The microRNA
signatures: Aberrantly expressed miRNAs in prostate cancer. Clin
Transl Oncol. 21:126–144. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Zhou J, Pei Y, Chen G, Cao C, Liu J, Ding
C, Wang D, Sun L, Xu P and Niu G: SPC24 Regulates breast cancer
progression by PI3K/AKT signaling. Gene. 675:272–277.
2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Zhou J, Yu Y, Pei Y, Cao C, Ding C, Wang
D, Sun L and Niu G: A potential prognostic biomarker SPC24 promotes
tumorigenesis and metastasis in lung cancer. Oncotarget.
8:65469–65480. 2017.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Yin H, Meng T, Zhou L, Chen H and Song D:
SPC24 is critical for anaplastic thyroid cancer progression.
Oncotarget. 8:21884–21891. 2017.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sheng J, Yin M, Sun Z, Kang X, Liu D,
Jiang K, Xu J, Zhao F, Guo Q and Zheng W: SPC24 promotes
osteosarcoma progression by increasing EGFR/MAPK signaling.
Oncotarget. 8:105276–105283. 2017.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Tokuzumi A, Fukushima S, Miyashita A,
Nakahara S, Kubo Y, Yamashita J, Harada M, Nakamura K, Kajihara I,
Jinnin M and Ihn H: Cell division cycle-associated protein 1 as a
new melanoma-associated antigen. J Dermatol. 43:1399–1405.
2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Obara W, Sato F, Takeda K, Kato R, Kato Y,
Kanehira M, Takata R, Mimata H, Sugai T, Nakamura Y and Fujioka T:
Phase I clinical trial of cell division associated 1 (CDCA1)
peptide vaccination for castration resistant prostate cancer.
Cancer Sci. 108:1452–1457. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
McCleland ML, Kallio MJ, Barrett-Wilt GA,
Kestner CA, Shabanowitz J, Hunt DF, Gorbsky GJ and Stukenberg PT:
The vertebrate Ndc80 complex contains Spc24 and Spc25 homologs,
which are required to establish and maintain
kinetochore-microtubule attachment. Curr Biol. 14:131–137.
2004.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Cui F, Hu J, Fan Y, Tan J and Tang H:
Knockdown of spindle pole body component 25 homolog inhibits cell
proliferation and cycle progression in prostate cancer. Oncol Lett.
15:5712–5720. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Hanahan D and Weinberg RA: The hallmarks
of cancer: The next generation. Cell. 144:646–674. 2011.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Wei RR, Sorger PK and Harrison SC:
Molecular organization of the Ndc80 complex, an essential
kinetochore component. Proc Natl Acad Sci USA. 102:5363–5367.
2005.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Wei RR, Schnell JR, Larsen NA, Sorger PK,
Chou JJ and Harrison SC: Structure of a central component of the
yeast kinetochore: The Spc24p/Spc25p globular domain. Structure.
14:1003–1009. 2006.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Sun SC, Lee SE and Xu YN: Perturbation of
SPC25 expression affects meiotic spindle organization, chromosome
alignment and spindle assembly checkpoint in mouse oocytes. Cell
cycle. 9:4552–4559. 2010.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhang T, Zhou Y, Wang HH, Meng TG, Guo L,
Ma XS, Shen W, Schatten H and Sun QY: SPC24 is required for meiotic
kinetochore-microtubule attachment and production of euploid eggs.
Oncotarget. 7:71987–71997. 2016.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Charters GA, Stones CJ, Shelling AN,
Baguley BC and Finlay GJ: Centrosomal dysregulation in human
metastatic melanoma cell lines. Cancer Genet. 204:477–485.
2011.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Tomonaga T, Matsushita K, Ishibashi M,
Nezu M, Shimada H, Ochiai T, Yoda K and Nomura F: Centromere
protein H is up-regulated in primary human colorectal cancer and
its overexpression induces aneuploidy. Cancer Res. 65:4683–4689.
2005.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang H, Gao X, Lu X, Wang Y, Ma C, Shi Z,
Zhu F, He B, Xu C and Sun Y: The mitotic regulator Hec1 is a
critical modulator of prostate cancer through the long non-coding
RNA BX647187 in vitro. Biosci Rep. 35(e00273)2015.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Huang LY, Chang CC, Lee YS, Chang JM,
Huang JJ, Chuang SH, Kao KJ, Lau GM, Tsai PY, Liu CW, et al:
Activity of a novel Hec1-targeted anticancer compound against
breast cancer cell lines in vitro and in vivo. Mol Cancer Ther.
13:1419–1430. 2014.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Chmielewska AE, Tang NH and Toda T: The
hairpin region of Ndc80 is important for the kinetochore
recruitment of Mph1/MPS1 in fission yeast. Cell Cycle. 15:740–747.
2016.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Tang NH and Toda T: MAPping the Ndc80 loop
in cancer: A possible link between Ndc80/Hec1 overproduction and
cancer formation. Bioessays. 37:248–256. 2015.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Musacchio A: The molecular biology of
spindle assembly checkpoint signaling dynamics. Curr Biol.
25:R1002–R1018. 2015.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Lara-Gonzalez P, Westhorpe FG and Taylor
SS: The spindle assembly checkpoint. Curr Biol. 22:R966–R980.
2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Ricke RM, Jeganathan KB and van Deursen
JM: Bub1 overexpression induces aneuploidy and tumor formation
through Aurora B kinase hyperactivation. J Cell Biol.
193:1049–1064. 2011.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Marks LS, Fradet Y, Deras IL, Blasé A,
Mathis J, Aubin SM, Cancio AT, Desaulniers M, Ellis WJ, Rittenhouse
H and Groskopf J: Pca3 molecular urine assay for prostate cancer in
men undergoing repeat biopsy. Urology. 69:532–535. 2007.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Merola R, Tomao L, Antenucci A, Sperduti
I, Sentinelli S, Masi S, Mandoj C, Orlandi G, Papalia R,
Guaglianone S, et al: Pca3 in prostate cancer and tumor
aggressiveness detection on 407 high-risk patients: A national
cancer institute experience. J Exp Clin Cancer Res.
34(15)2015.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Mao Z, Ji A, Yang K, He W, Hu Y, Zhang Q,
Zhang D and Xie L: Diagnostic performance of PCA3 and hK2 in
combination with serum PSA for prostate cancer. Medicine
(Baltimore). 97(e12806)2018.PubMed/NCBI View Article : Google Scholar
|