Introduction
Thyroid cancer (THCA) is one of the most common
malignant tumors in the head and neck, and endocrine system
(1). The incidence rate of THCA is
increasing rapidly and this disease is projected to become the
fourth major type of cancer worldwide (2). It has been reported that the
incidence rate of THCA in females is higher compared with that in
males (2-4 times), and thyroid nodules often occur in young
individuals (3,4). The vast majority of THCA cases are
considered to be indolent tumors; however, a small number of
patients have a poor prognosis (5). To date, the knowledge of the
molecular mechanism underlying the development of THCA has
primarily focused on the roles of various genes and oncogenes
(6,7). However, the detailed mechanism of the
initiation and progression of THCA remains poorly understood, and
there only a few available biomarkers used to diagnose and treat
patients with THCA (8-10).
The fox transcription factor family have a
C-terminal winged-helix/Forkhead DNA binding domain, which is
involved in cell proliferation and differentiation, and organism
development (11). Forkhead box P2
(FOXP2) is one member of the fox transcription factor family,
located on chromosome 7q31 and is involved in embryonic
development, the cell cycle and organ development, including the
heart, the lungs and the central nervous system (12-14).
In addition, FOXP2 expression levels were reported to be decreased
in various types of cancer, such as gastric and lung cancer
(15,16). Sun et al (17) revealed that FOXP2 expression levels
were decreased in THCA samples using integrated microarray and
bioinformatics analysis. However, the role and specific mechanism
of FOXP2 in THCA remains unclear. In the current study, the
biological roles and mechanisms of FOXP2 in THCA cell growth, and
apoptosis was investigated. The results revealed that the
FOXP2/ribosomal protein S6 kinase A6 (RPS6KA6) axis could be a
novel therapeutic target for the treatment THCA.
Materials and methods
Bioinformatic analysis
The GEPIA database (http://gepia.cancer-pku.cn) was used to analyze the
mRNA expression levels of FOXP2 and RPS6KA6 in THCA tissues and
normal tissues (The original image downloaded from GEPIA database
displayed in the form of log transformation). The association
between FOXP2 and RPS6KA6 was characterized using the LinkedOmics
(http://www.linkedomics.org) and GEPIA
databases. The JASPAR (http://jaspar.genereg.net) database was used to
predict the binding site of FOXP2 with the RPS6KA6 promotor.
Cell culture
The normal human Nthy-ori3-1 thyroid cell line and
the SW579, CGTH-W3 and TPC-1 THCA cell lines were purchased from
the Cell Bank of Shanghai Institute of Biological Sciences,
cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.),
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.),
100 µg/ml streptomycin, and 100 U/ml penicillin at 37˚C in a
humidified incubator with 5% CO2.
Cell transfection
A pcDNA3.1 expression vector containing full-length
FOXP2 [overexpression (Ov)-FOXP2] and a negative control (Ov-NC)
were constructed by Shanghai GenePharma Co., Ltd. The small
interfering (si)RNA oligonucleotides targeting RPS6KA6
(si-RPS6KA6-1/2) and corresponding control (si-NC) were purchased
from GeneCopoeia, Inc. Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) was used for
transfection. The following siRNA sequences were used:
si-RPS6KA6-1, 5'-GGAUGAAGAUGAAAUUAAAUG-3'; si-RPS6KA6-2,
5'-GCUACUACUGCUACUACUACU-3'; si-NC, 5'-UUCUCCGAACGUGUCACGU-3'.
After 48 h, the cells were harvested for subsequent
experiments.
Cell Counting Kit (CCK)-8 assay
After transfection, the cells were seeded into a
96-well plate at the density of 5x103 cells/well and
cultured at 37˚C. After 24, 48 and 72 h, 10 µl CCK-8 solution
(Beyotime Institute of Biotechnology) was added to each well. The
plates were incubated for 2 h and the absorbance was measured with
a microplate reader (Thermo Fisher Scientific, Inc.) at 450 nm.
Colony formation assay
TPC-1 cells were seeded in triplicate in 6-well
plates at 500 cells/well and cultured in DMEM, with 10% FBS at
37˚C. The cells were fixed in 4% paraformaldehyde at room
temperature for 15 min and stained with 0.1% crystal violet for 30
min at room temperature 2 weeks later. The number of visible
colonies (defined as >50 cells/colony) were counted using a
light microscope (Olympus Corporation).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from THCA tissues and cells
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's instructions. Then, 0.5 µg
RNA was converted into cDNA at 37˚C for 1 h using PrimeScript RT
MasterMix (Takara Bio, Inc.). qPCR was performed using ChamQTM
SYBR® qPCR MasterMix (Vazyme Biotech, Co., Ltd.). The
following primer sequences were used: FOXP2 forward,
5'-AGTGCAAGACGAGACAGCTC-3' and reverse,
5'-GCCGTATTTTTCATCACACTCA-3'; RPS6KA6 forward,
5'-CTCCTGTTTGAGTGCTCCTGA-3' and reverse,
5'-ACTGGAGTAGTACGCAGTCG-3'; GAPDH forward, 5'-GGGAAACTGTGGCGTGAT-3'
and reverse, 5'-GAGTGGGTGTCGCTGTTGA-3'. The following thermocycling
conditions were used: Initial denaturation at 95˚C for 3 min,
followed by 35 cycles at 95˚C for 30 sec, 60˚C for 30 sec and 72˚C
for 1 min, and 72˚C final extension for 7 min. GAPDH was used as an
internal reference and the 2-ΔΔCq method (18) was used to calculate the relative
quantification.
TUNEL assay
A TUNEL assay was used to analyze cell apoptosis
using an apoptosis detection kit (Roche Diagnostics, GmbH).
Fluorescein isothiocyanate (FITC; green) and 4',
6-diamidino-2-phenylindole (DAPI; blue) were used to stain the
apoptotic cells and the nuclei for 10 min at room temperature in
the dark, respectively. The labeled cells were washed with PBS and
visualized using a fluorescence microscope (Olympus BX53; Olympus
Corporation) and at least 10 fields of view for each sample were
examined.
Dual-luciferase reporter assay
The wild-type (WT) and corresponding mutant (MUT)
RPS6KA6 promotor fragments, including the putative FOXP2 sites,
were cloned into the pGL3-basic vector (Promega Corporation). The
TPC-1 cell line was co-transfected with the constructed luciferase
reporter vectors and Ov-FOXP2/Ov-NC. Luciferase activity was then
detected using a Dual-Luciferase Reporter Assay kit (Promega
Corporation) after transfection for 48 h using
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Luciferase activities were normalized against
Renilla luciferase.
Chromatin immunoprecipitation
(ChIP)
ChIP experiments were performed according to the
method previously described (19).
The cells were cross-linked with 1% formaldehyde for 10 min at 37˚C
and quenched with 2.5 M glycine for 5 min at room temperature.
After being immunoprecipitated from the cell lysates using a FOXP2
antibody (1:200; cat. no. #5337; Cell Signaling Technology, Inc.)
for incubation at 4˚C overnight, the precipitated DNA was obtained
via phenol/chloroform extraction and ethanol precipitation, and PCR
was performed as aforementioned to amplify the FOXP2 binding site.
The data obtained were normalized to the corresponding DNA
precipitated by IgG. The ssequences used for PCR were as follows:
FOXP2 forward, 5'-AGTGCAAGACGAGACAGCTC-3', and reverse,
5'-GCCGTATTTTTCATCACACTCA-3'; RPS6KA6 forward,
5'-CTCCTGTTTGAGTGCTCCTGA-3' and reverse,
5'-ACTGGAGTAGTACGCAGTCG-3'.
Western blot analysis
Total protein was extracted from THCA tissues and
cells using RIPA buffer (Changsha Auragene Biological Technology
Co., Ltd.) and quantified using a BCA Protein Assay kit (Beijing
Dingguo Changsheng Biotechnology, Co., Ltd.). The lysates were
incubated at 95˚C for 5 min, separated using 10% SDS-PAGE (Bio-Rad
Laboratories) and transferred onto PVDF membranes (MilliporeSigma).
After being blocked with 5% skimmed milk, primary antibodies
targeting FOXP2 (1:1,000; cat. no. ab16046; Abcam), Ki67 (1:1,000;
cat. no. ab92742; Abcam), PCNA (1:1,000; cat. no. ab29; Abcam),
Bcl-2 (1:1,000; cat. no. ab32124; Abcam), Bax (1:1,000; cat. no.
ab32503; Abcam), cleaved (C)-caspase 3 (1:500; cat. no. ab32042;
Abcam), RPS6KA6 (1:1,000; cat. no. ab76117; Abcam), phosphorylated
(p)-PI3K (1:1,000; cat. no. ab182651; Abcam), PI3K (1:1,000; cat.
no. ab86714; Abcam), p-Akt (1:1,000; cat. no. ab38449; Abcam), Akt
(1:500; cat. no. ab8805; Abcam) or GAPDH (1:1,000; cat. no. ab8245;
Abcam) were added and incubated overnight at 4˚C. Subsequently, the
blots were incubated with horseradish peroxidase-conjugated
secondary antibody (1:2,000; cat. no. ab6721; Abcam) at room
temperature for 1 h. The protein bands were visualized using an ECL
detection system and analyzed using ImageJ software (version 1.46;
National Institutes of Health).
Statistical analysis
All the data are presented as the mean ± SD.
Statistical analysis was performed using SPSS v13.0 statistical
software (SPSS, Inc.) or GraphPad Prism v5.0 (GraphPad Software,
Inc.). Significant differences between groups were analyzed using
an unpaired Student's t-test or one-way ANOVA followed by a
Bonferroni's post hoc test. P<0.05 was considered to indicate a
statistically significant difference.
Results
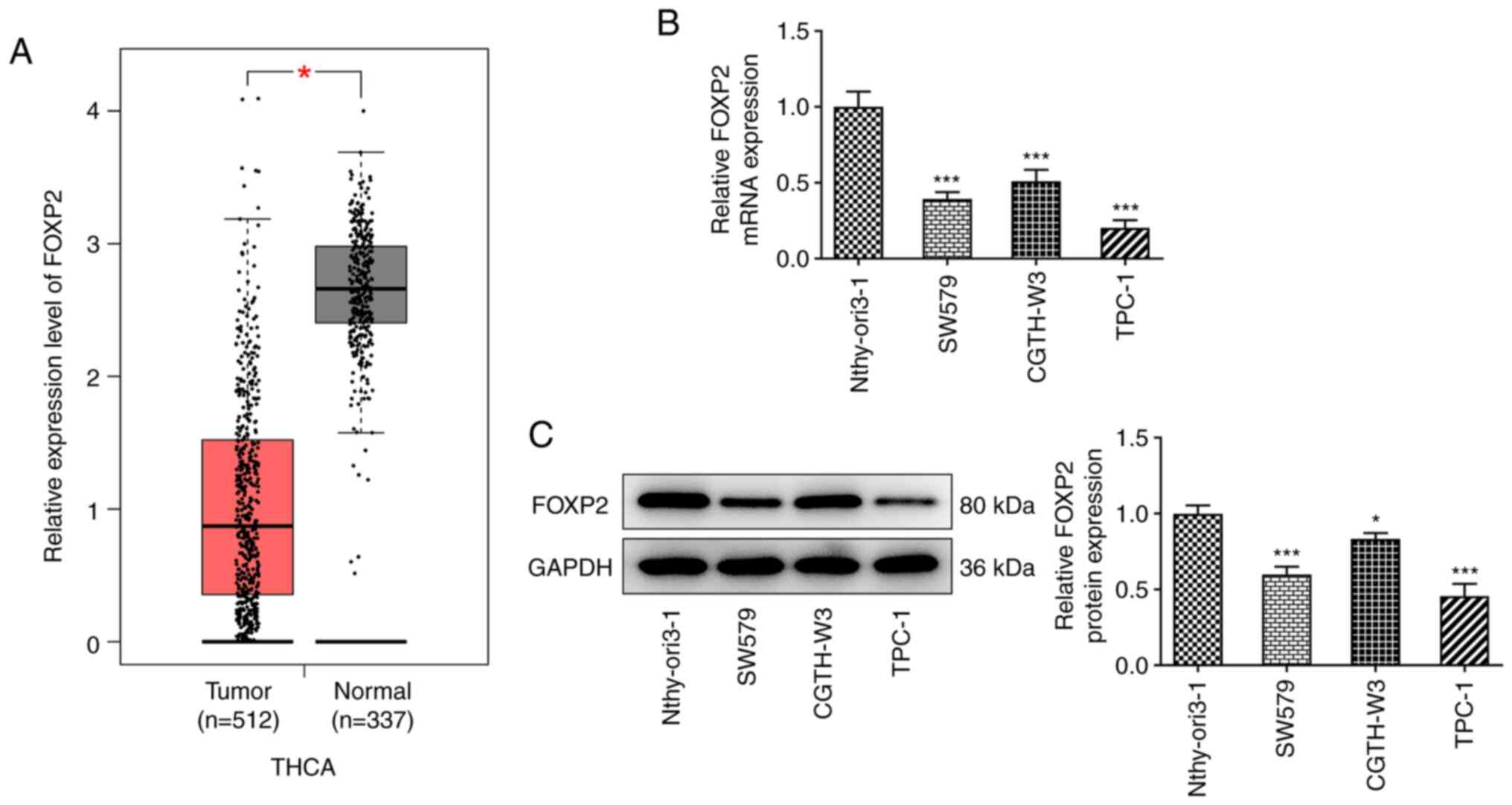
FOXP2 expression levels are decreased
in THCA tissues and cells
To investigate the role of FOXP2 in THCA
progression, the mRNA and protein expression levels of FOXP2 in
THCA was analyzed. As shown in Fig.
1A, FOXP2 mRNA expression levels were significantly decreased
in patients with THCA compared with that in the normal tissues from
healthy individuals, based on data from the GEPIA database. In
addition, the results from RT-qPCR and western blot analysis
revealed that the mRNA and protein expression levels of FOXP2 in
THCA cells were markedly decreased compared with that in the normal
thyroid cell line (Fig. 1B and
C). Among the THCA cell lines,
TPC-1 showed the lowest mRNA and protein expression levels of
FOXP2; therefore, this was selected for the subsequent
experiments.
Overexpression of FOXP2 inhibits cell
proliferation and promotes apoptosis in the TPC-1 cell line
Subsequently, FOXP2 overexpression vectors were
designed and transfected into the TPC-1 cell line to overexpress
FOXP2. RT-qPCR and western blot analysis was used to determine the
transfection efficiency (Fig. 2A
and B). Subsequently, cell
proliferation was evaluated using CCK-8 and colony formation
assays, as well as western blot analysis. The CCK-8 results showed
that the optical density values at three time points were reduced
in the Ov-FOXP2 group compared with that in the NC group (Fig. 2C). Consistently, the number of
colonies in the Ov-FOXP2 group was markedly lower compared with
that in the NC group (Fig. 2D). In
addition, the results from the TUNEL assay revealed that the
apoptosis rate in the TPC-1 cell line was significantly increased
following FOXP2 overexpression (Fig.
2E). Furthermore, the western blot results revealed that the
protein expression levels of Ki67, PCNA and Bcl-2 were decreased,
while the protein expression levels of Bax and C-caspase 3 were
increased in the cells transfected with Ov-FOXP2 (Fig. 2F).
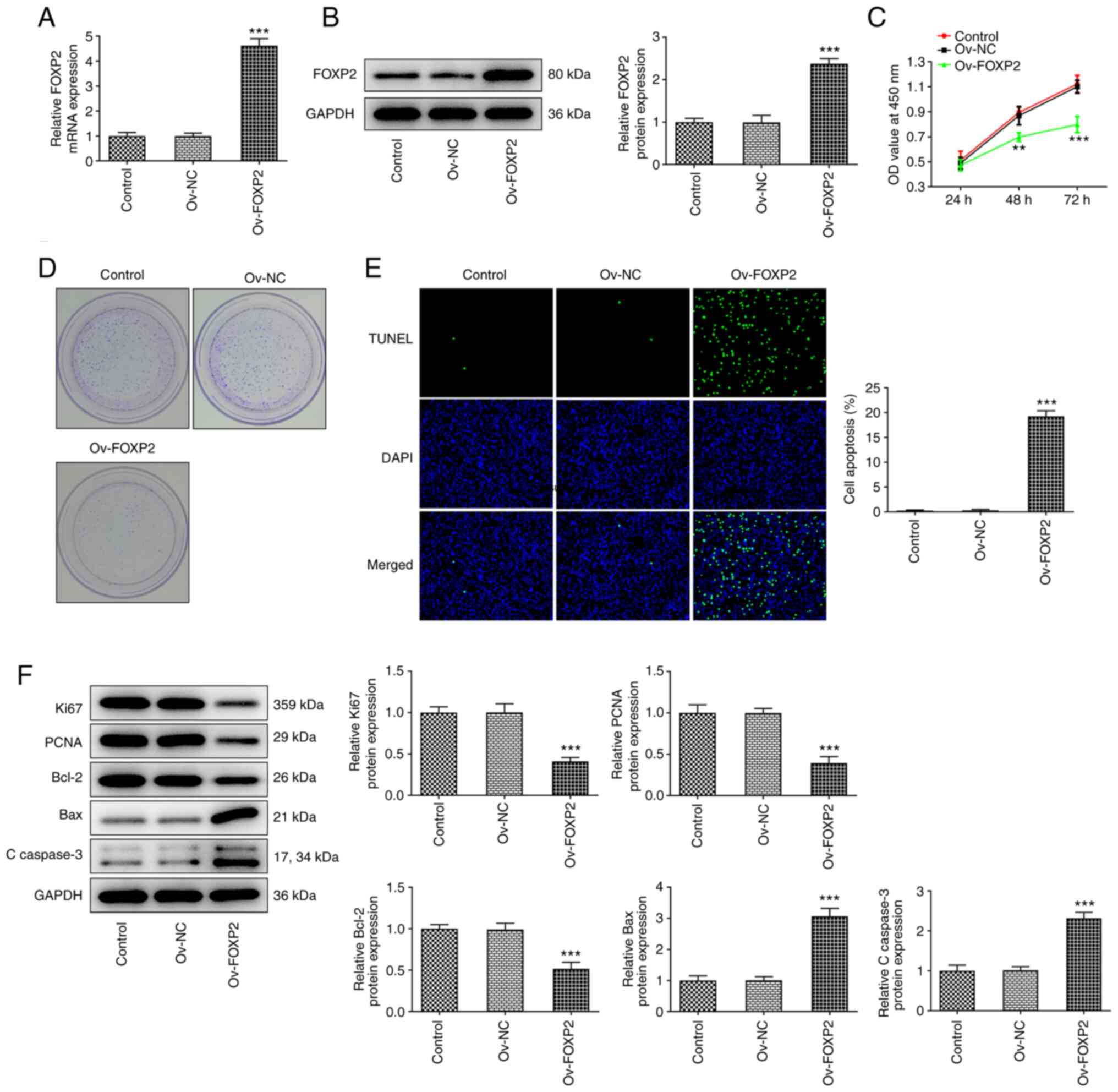 | Figure 2Effects of FOXP2 overexpression on
TPC-1 cell proliferation and apoptosis. (A) mRNA and (B) protein
expression level of FOXP2 in TPC-1 cells were measured using
reverse transcription-quantitative PCR and western blot analysis,
respectively. (C) Cell proliferation was evaluated using Cell
Counting Kit-8 assay. (D) Cell colony number was examined using
colony formation assay. (E) Cell apoptosis was detected using TUNEL
assay. (F) Western blot analysis was used to assess the protein
expression levels of Ki67, PCNA, Bcl-2, Bax and C-caspase 3. Data
are expressed as the mean ± SD. **P<0.01,
***P<0.001 vs. Ov-NC. Ov, overexpression; NC,
negative control; C, cleaved; OD, optical density; FOXP2, forkhead
box P2. |
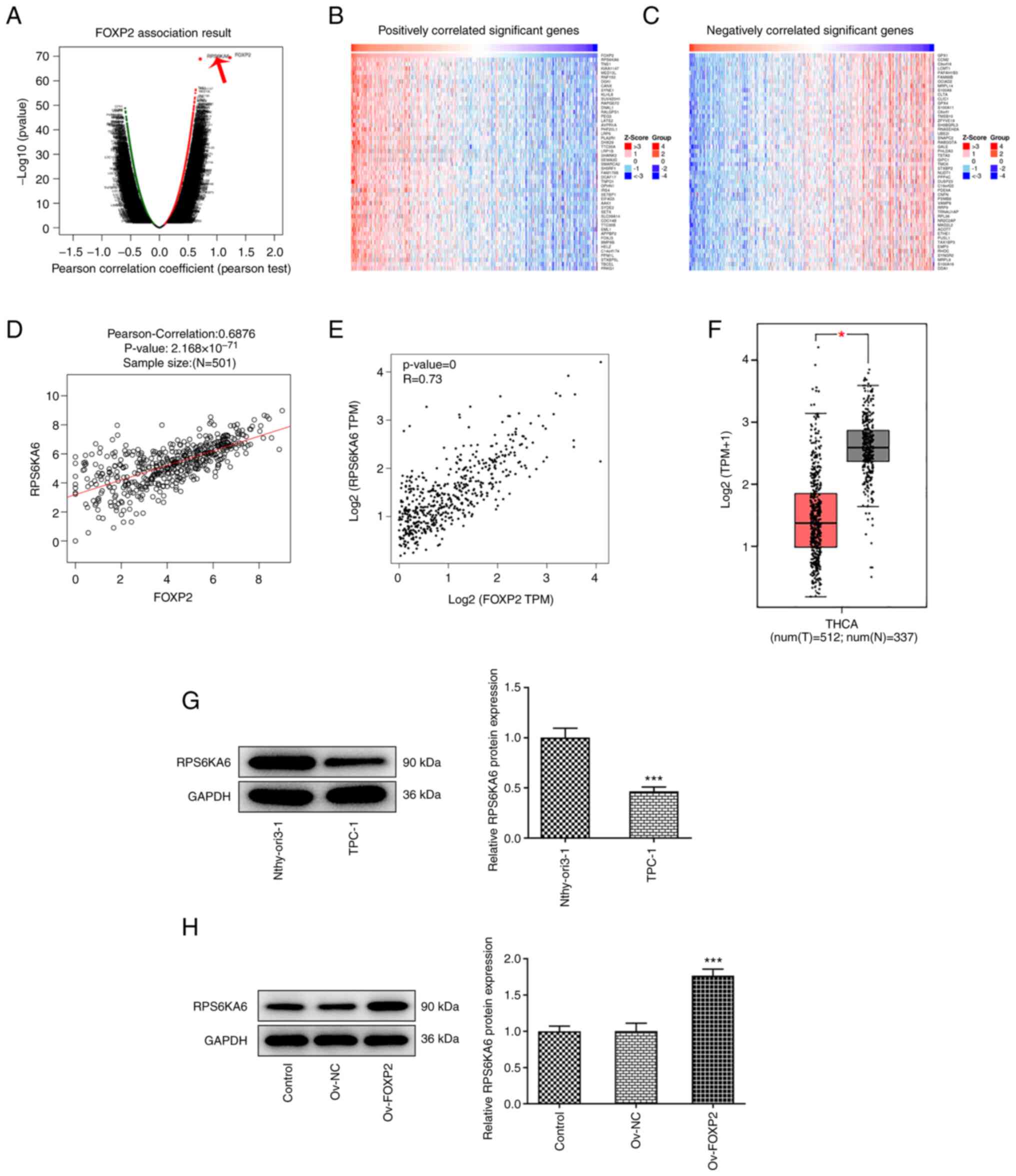
RPS6KA6 is correlated with FOXP2
To further investigate the underlying mechanism of
FOXP2 in the development of THCA, bioinformatics analysis was
performed to identify the potential targets of FOXP2. Data from the
LinkedOmics database revealed that there was an association between
RPS6KA6 and FOXP2 (Fig. 3A-C). In
addition, data from the GEPIA database also showed that RPS6KA6
expression levels were correlated with FOXP2 expression levels
(Fig. 3D and E). Next, RPS6KA6 mRNA and protein
expression levels were found to be decreased in THCA tissues based
on data from the GEPIA database and in THCA cells using western
blot analysis, respectively (Fig.
3F and G). The effects of
FOXP2 overexpression on RPS6KA6 protein expression levels in the
TPC-1 cells were subsequently investigated. The results revealed
that RPS6KA6 protein expression levels were increased following
FOXP2 overexpression, indicating an association between RPS6KA6 and
FOXP2 (Fig. 3H).
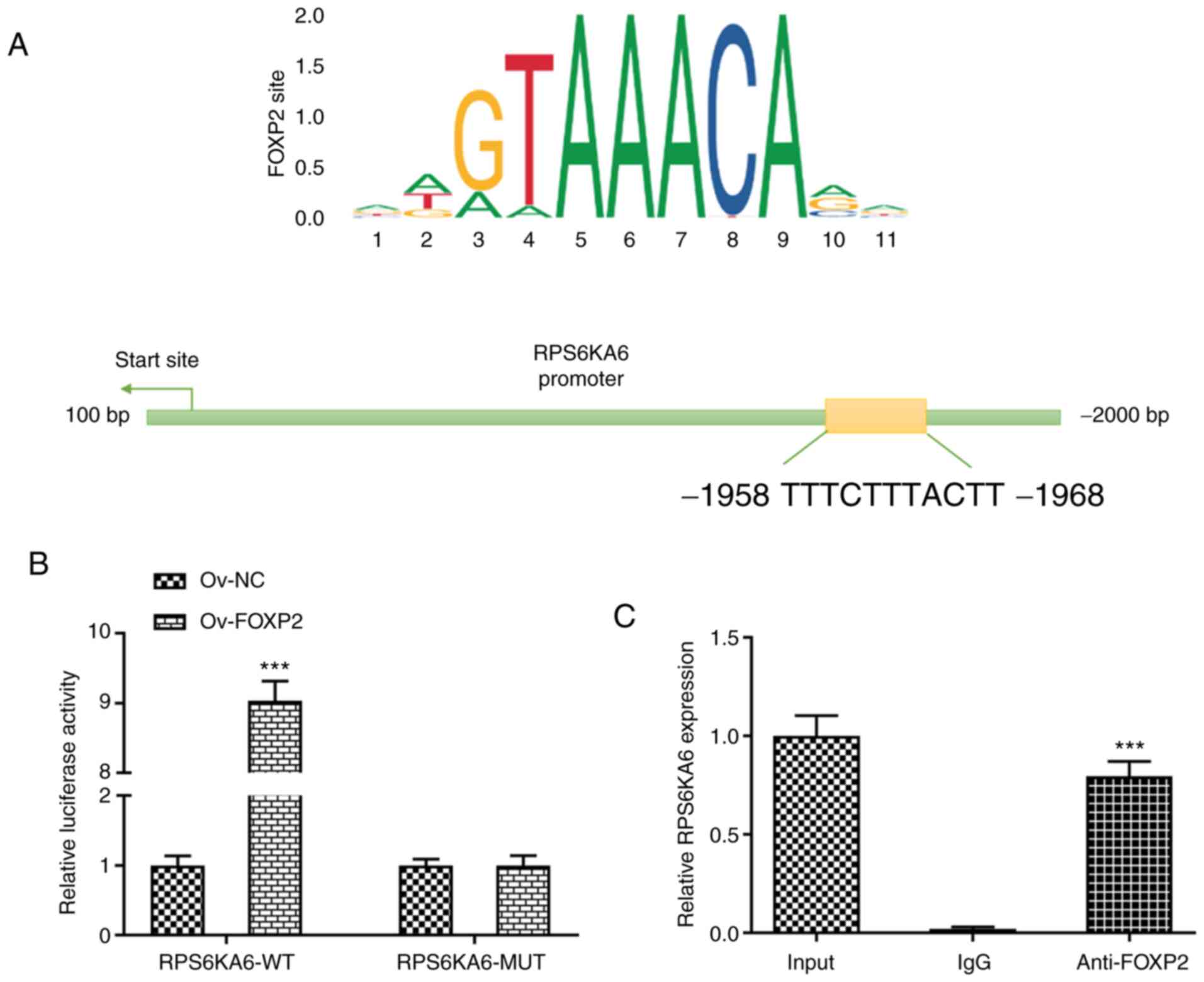
RPS6KA6 is a direct transcriptional
target of FOXP2
To investigate how FOXP2 targets RPS6KA6 in the
TPC-1 cell line, the consensus sequences between FOXP2 and the
promotor region of RPS6KA6 was predicted using the JASPAR database
(Fig. 4A). To confirm the direct
binding of FOXP2 with the RPS6KA6 promotor region, a
dual-luciferase reporter assay was performed. The results showed
that the luciferase activity of the WT RPS6KA6 promotor was
significantly increased following FOXP2 overexpression, while there
were no notable changes in luciferase activity in the other groups
(Fig. 4B). To further verify the
interaction between FOXP2 and RPS6KA6 promoter, a ChIP assay was
performed. The results showed that FOXP2 binds to the predicted
binding sites of RPS6KA6 (Fig.
4C).
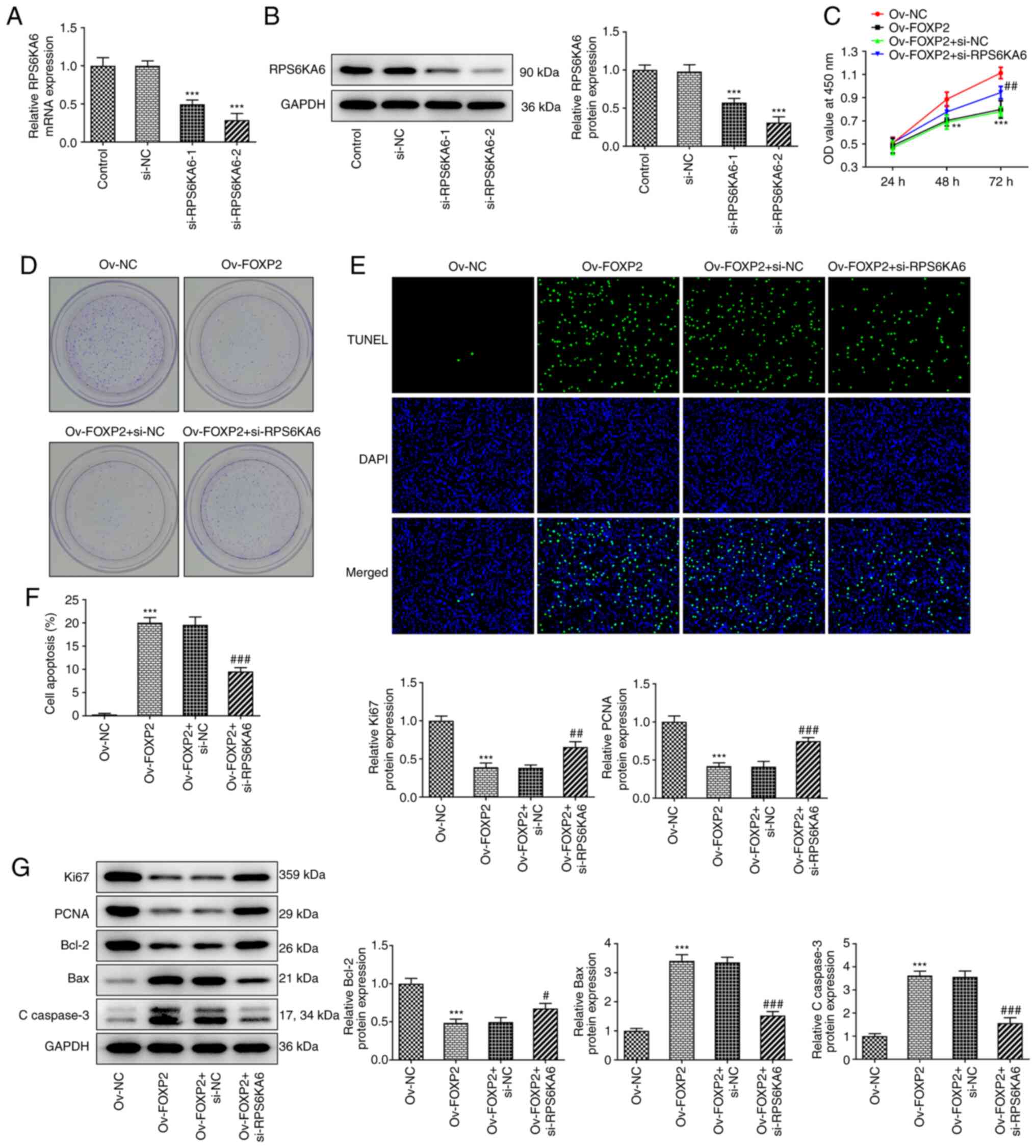
FOXP2 regulates the proliferation and
apoptosis of the TPC-1 cell line by targeting RPS6KA6
Then, si-RPS6KA6-1/2 was transfected into the TPC-1
cell line to knockdown the expression of RPS6KA6. The results from
RT-qPCR and western blot analysis showed that both si-RPS6KA6-1 and
si-RPS6KA6-2 decreased the mRNA and protein expression level of
RPS6KA6, respectively. In addition, si-RPS6KA6-2 showed more
significant interference efficiency; therefore, si-RPS6KA6-2 was
selected for subsequent experiments (Fig. 5A and B). Next, it was found that knockdown of
RPS6KA6 reversed the effects of FOXP2 overexpression on cell
proliferation and colony formation (Fig. 5C and D). Furthermore, the promoted apoptosis
due to FOXP2 overexpression was also reduced following knockdown of
RPS6KA6 expression (Fig. 5E and
F). Lastly, western blot analysis
demonstrated that the decreased levels of Ki67, PCNA and Bcl-2
following FOXP2 overexpression were increased following knockdown
of RPS6KA6 expression. Also, FOXP2 depletion-induced protein levels
of Bax and C-caspase 3 were decreased after RPS6KA6 was silenced
(Fig. 5G).
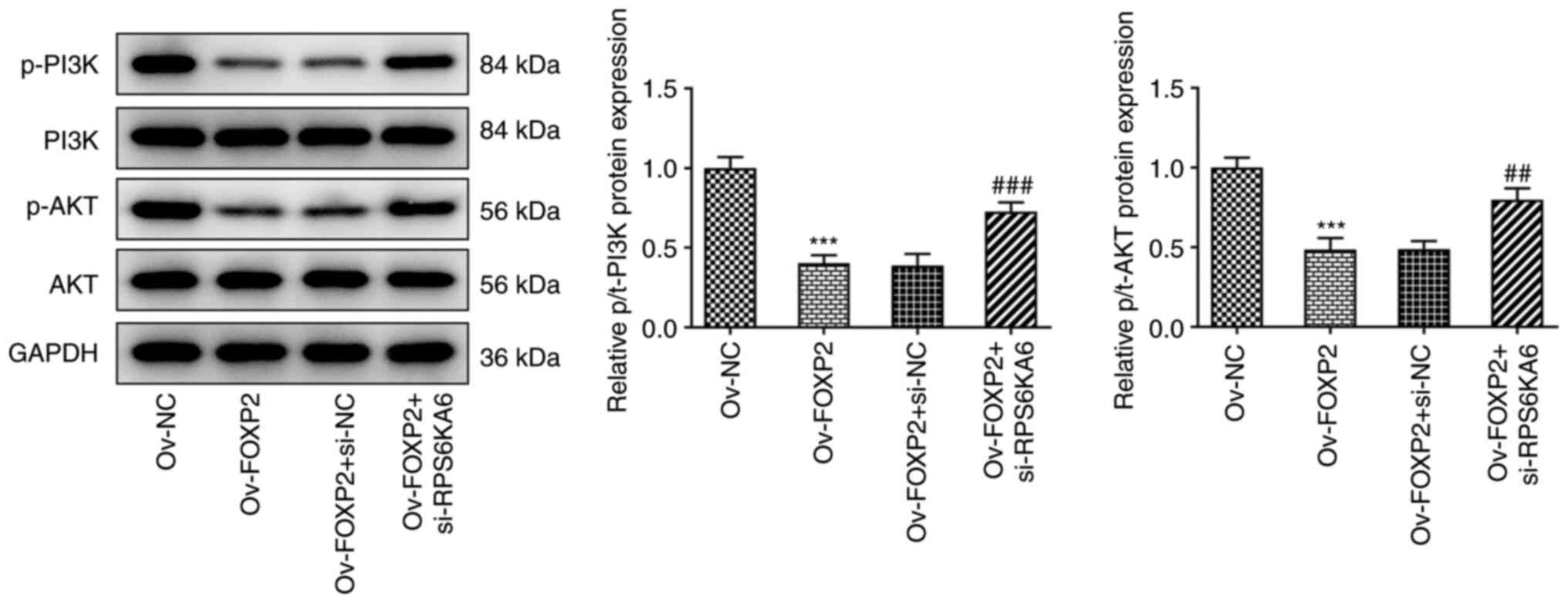
FOXP2 inactivates the PI3K/AKT pathway
in the TPC-1 cells by targeting RPS6KA6
It is well-known that the PI3K/AKT signaling pathway
participates in THCA progression (20,21).
Moreover, previous studies have reported that RPS6KA6 exerted
biological roles by regulating the phosphorylation activation of
PI3K/AKT pathway (22,23). Thus, the effects of transcriptional
activation of RPS6KA6 by FOXP2 on the PI3K/AKT signaling pathway
was investigated. As shown in Fig.
6, the results from western blot analysis revealed that
phosphorylation of PI3K and AKT was reduced following
overexpression of FOXP2, but the effects were reversed following
knockdown of RPS6KA6 expression. At the same time, the expression
levels of total PI3K and AKT protein were not significantly changed
in each group. These results indicate that RPS6KA6 participates in
the regulation of FOXP2 via the PI3K/AKT pathway in the TPC-1 cell
line.
Discussion
Emerging evidence has indicated the crucial role of
FOXP2 in the initiation and progression of numerous types of
cancer, including breast cancer, hepatocellular carcinoma, gastric
cancer and multiple myeloma (24,25).
However, the functional role and underlying molecular mechanisms of
FOXP2 in THCA cell growth and apoptosis have not been completely
clarified. In the current study, it was found that FOXP2 mRNA and
protein expression levels were decreased in THCA cells, and FOXP2
played an inhibitory role in cell proliferation and a promoting
role in cell apoptosis. Furthermore, we found that RPS6KA6 mRNA
expression levels were correlated with FOXP2 mRNA expression levels
and was activated by FOXP2 transcriptionally. Lastly, it was
revealed that the PI3K/AKT signaling pathway was associated with
FOXP2-mediated transcriptional activation of RPS6KA6 in the THCA
cell line. These findings suggested that a FOXP2/RPS6KA6 axis
exerts a tumor-suppressing role in THCA.
FOXP2, a transcription factor, is known to be
essential for language and memory function, and has been associated
with an increased susceptibility to schizophrenia (26-28).
Recent studies reported the dysregulation of FOXP2 in multiple
types of cancers (16,29,30).
Chen et al (31) revealed
that low expression levels of FOXP2 were associated with poor
relapse-free survival times in breast cancer, and FOXP2 inhibited
breast cancer cell migration, invasion and epithelial-mesenchymal
transition. In the present study, it was found that the FOXP2
expression level was decreased in patients with THCA from the GEPIA
database. The in vitro experiments also proved the decreased
expression levels of FOXP2 in THCA cell lines, which is consistent
with a previous report that FOXP2 mRNA expression levels were
decreased in THCA using microarray analysis (17). However, it is controversial that
some studies found that FOXP2 was expressed at low levels in
several tumors, such as breast cancer, hepatocellular carcinoma and
gastric cancer biopsies (15,32,33),
while FOXP2 was found to be overexpressed in some other types of
cancers, including multiple myelomas, several subtypes of
lymphomas, osteosarcoma, neuroblastomas, and ERG fusion-negative
prostate cancers (34-36).
Thus, FOXP2 cannot be defined simply to act as a tumor suppressor
or an oncogene. Based on the downregulation of FOXP2 in THCA
tissues and cells, FOXP2 was overexpressed to observe its role in
THCA. Functional experiments revealed that FOXP2 overexpression
significantly suppressed THCA cell proliferation and induced cell
apoptosis, indicating FOXP2 may exert suppressive effects on THCA
progression.
It is well-known that transcription factors bind to
specific sequences of a gene to regulate protein expression at a
specific intensity and time, by repressing or activating
transcription of target genes (37,38).
Thus, we intend to study the mechanism how FOXP2 affects THCA as a
transcription factor. RPS6KA6, also known as X-linked ribosomal S6
kinase 4 (RSK4; one member of RSK family), is a ribosomal protein
and associated with ‘P53 dependent proliferation arrest’, which
can; therefore, act as a tumor suppressor (39,40).
RPS6KA6 expression levels were found to be decreased in several
types of cancer, such as breast and ovarian cancers (41,42).
Mei et al (22) revealed
that overexpression of RPS6KA6 suppressed migration and invasion,
and promoted apoptosis in drug resistant breast cancer cells. In
addition, Hu et al (43)
reported that knockdown of RPS6KA6 expression inhibited cell
apoptosis and promoted cell proliferation, migration, and invasion
in gastric cancer. These data indicate that RPS6KA6 may play
inhibitory role in tumors. Data from the LinkedOmics database
demonstrated that the RPS6KA6 gene was associated with FOXP2. The
GEPIA database showed that its mRNA expression levels were
decreased in THCA tissues and cells, and RPS6KA6 and FOXP2
expression levels were correlated in THCA. In support of this view
that FOXP2 interacts with RPS6KA6, the binding sites between FOXP2
and the RPS6KA6 promotor regions were predicted using the JASPAR
database and verified using dual-luciferase reporter and ChIP
assays. Rescue experiments also showed that knockdown of RPS6KA6
expression facilitated cell growth and reduced cell apoptosis in
the THCA cells by reversing the effects of FOXP2
overexpression.
A previous study has shown that the activation of
the PI3K/AKT signaling pathway promotes the transcription of
downstream genes, including CDK4, cyclin D1 and Bax, participating
in the regulation of cell proliferation, apoptosis and other
cellular processes in cancer (44). The PI3K/AKT signaling pathway is
also an important regulatory pathway in THCA (45). In addition, a recent study has
confirmed that overexpression of RPS6KA6 in breast cancer cells
reversed Adriamycin-resistance by inhibiting the PI3K/AKT signaling
pathway (22). RPS6KA6 also
functions as an endogenous inhibitor of the MAPK pathway and
represses the phosphorylation of AKT (46). In the present study, knockdown of
RPS6KA6 expression reversed the FOXP2-mediated reduced protein
expression levels of p-PI3K and p-AKT, suggesting that RPS6KA6 may
be associated with the regulation of FOXP2 in THCA cells by
blocking the PI3K/AKT pathway. Notably, RSK has also been reported
to be a downstream target of PI3K/AKT in breast cancer (47). We hypothesize that RPS6KA6 and
PI3K/AKT may adjust to each other or RPS6KA6 yields different
functions on PI3K/AKT in different type of cancers. Moreover, it
was preliminarily revealed the inhibitory effects of FOXP2 and
RPS6KA6 on the phosphorylation of PI3K/AKT, but the specific
mechanism was not investigated. PI3K and AKT phosphorylation are
usually activated by receptor tyrosine kinases and
G-protein-coupled receptors (48).
Based on this, the molecular mechanism by which RPS6KA6 inhibits
the PI3K/AKT pathway or interacts with this pathway in THCAwill be
investigated in a further study.
In conclusion, the data from the present study
indicated the essential inhibitory role of the FOXP2/RPS6KA6 axis
in THCA and revealed the important role of RPS6KA6 in FOXP2-driven
THCA cell proliferation, and apoptosis. This suggests that the
FOXP2/RPS6KA6 axis could be an independent prognostic marker and a
promising therapeutic strategy for patients with THCA.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
FY and ZX designed the study, drafted and revised
the manuscript. ZX and SZ analyzed the data and searched the
literature. FY, ZX and SZ performed the experiments. All authors
read and approved the final manuscript. FY and ZX confirm the
authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Olson E, Wintheiser G, Wolfe KM, Droessler
J and Silberstein PT: Epidemiology of thyroid cancer: A review of
the national cancer database, 2000-2013. Cureus.
11(e4127)2019.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Kim J, Gosnell JE and Roman SA: Geographic
influences in the global rise of thyroid cancer. Nat Rev
Endocrinol. 16:17–29. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Nettore IC, Colao A and Macchia PE:
Nutritional and environmental factors in thyroid carcinogenesis.
Int J Environ Res Public Health. 15(1735)2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Cabanillas ME, McFadden DG and Durante C:
Thyroid cancer. Lancet. 388:2783–2795. 2016.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Hahn LD, Kunder CA, Chen MM, Orloff LA and
Desser TS: Indolent thyroid cancer: Knowns and unknowns. Cancers
Head Neck. 2(1)2017.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Kreissl MC, Janssen MJR and Nagarajah J:
Current treatment strategies in metastasized differentiated thyroid
cancer. J Nucl Med. 60:9–15. 2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Khatami F, Larijani B, Nikfar S, Hasanzad
M, Fendereski K and Tavangar SM: Personalized treatment options for
thyroid cancer: Current perspectives. Pharmgenomics Pers Med.
12:235–245. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Nixon AM, Provatopoulou X, Kalogera E,
Zografos GN and Gounaris A: Circulating thyroid cancer biomarkers:
Current limitations and future prospects. Clin Endocrinol (Oxf).
87:117–126. 2017.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Li Q, Li H, Zhang L, Zhang C, Yan W and
Wang C: Identification of novel long non-coding RNA biomarkers for
prognosis prediction of papillary thyroid cancer. Oncotarget.
8:46136–46144. 2017.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Chou CK, Liu RT and Kang HY:
MicroRNA-146b: A novel biomarker and therapeutic target for human
papillary thyroid cancer. Int J Mol Sci. 18(636)2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Lam EW, Brosens JJ, Gomes AR and Koo CY:
Forkhead box proteins: Tuning forks for transcriptional harmony.
Nat Rev Cancer. 13:482–495. 2013.PubMed/NCBI View
Article : Google Scholar
|
|
12
|
Chiu YC, Li MY, Liu YH, Ding JY, Yu JY and
Wang TW: Foxp2 regulates neuronal differentiation and neuronal
subtype specification. Dev Neurobiol. 74:723–738. 2014.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Tsui D, Vessey JP, Tomita H, Kaplan DR and
Miller FD: FoxP2 regulates neurogenesis during embryonic cortical
development. J Neurosci. 33:244–258. 2013.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Co M, Anderson AG and Konopka G: FOXP
transcription factors in vertebrate brain development, function and
disorders. Wiley Interdiscip Rev Dev Biol. 9(e375)2020.PubMed/NCBI View
Article : Google Scholar
|
|
15
|
Jia WZ, Yu T, An Q, Yang H, Zhang Z, Liu X
and Xiao G: MicroRNA-190 regulates FOXP2 genes in human gastric
cancer. Onco Targets Ther. 9:3643–3651. 2016.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Ren T, Liu C, Hou J and Shan F:
Hsa_circ_0043265 suppresses proliferation, metastasis, EMT and
promotes apoptosis in non-small cell lung cancer through
miR-25-3p/FOXP2 pathway. Onco Targets Ther. 13:3867–3880.
2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Sun T, Guan Q, Wang Y, Qian K, Sun W, Ji
Q, Wu Y, Guo K and Xiang J: Identification of differentially
expressed genes and signaling pathways in papillary thyroid cancer:
A study based on integrated microarray and bioinformatics analysis.
Gland Surg. 10:629–644. 2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ni Z, Lu W, Li Q, Han C, Yuan T, Sun N and
Shi Y: Analysis of the HNF4A isoform-regulated transcriptome
identifies CCL15 as a downstream target in gastric carcinogenesis.
Cancer Biol Med. 18:530–546. 2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Manfredi GI, Dicitore A, Gaudenzi G,
Caraglia M, Persani L and Vitale G: PI3K/Akt/mTOR signaling in
medullary thyroid cancer: A promising molecular target for cancer
therapy. Endocrine. 48:363–370. 2015.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Petrulea MS, Plantinga TS, Smit JW,
Georgescu CE and Netea-Maier RT: PI3K/Akt/mTOR: A promising
therapeutic target for non-medullary thyroid carcinoma. Cancer
Treat Rev. 41:707–713. 2015.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Mei Y, Liao X, Zhu L and Yang H:
Overexpression of RSK4 reverses doxorubicin resistance in human
breast cancer cells via PI3K/AKT signalling pathway. J Biochem.
167:603–611. 2020.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Serra V, Eichhorn PJ, García-García C,
Ibrahim YH, Prudkin L, Sánchez G, Rodríguez O, Antón P, Parra JL,
Marlow S, et al: RSK3/4 mediate resistance to PI3K pathway
inhibitors in breast cancer. J Clin Invest. 123:2551–2563.
2013.PubMed/NCBI View
Article : Google Scholar
|
|
24
|
Katoh M and Katoh M: Human FOX gene family
(review). Int J Oncol. 25:1495–1500. 2004.PubMed/NCBI
|
|
25
|
Herrero MJ and Gitton Y: The untold
stories of the speech gene, the FOXP2 cancer gene. Genes Cancer.
9:11–38. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Lang X, Zhang W, Song X, Zhang G, Du X,
Zhou Y, Li Z and Zhang XY: FOXP2 contributes to the cognitive
impairment in chronic patients with schizophrenia. Aging (Albany
NY). 11:6440–6448. 2019.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Wu J, Liu P, Tang H, Shuang Z, Qiu Q,
Zhang L, Song C, Liu L, Xie X and Xiao X: FOXP2 promotes tumor
proliferation and metastasis by targeting GRP78 in triple-negative
breast cancer. Curr Cancer Drug Targets. 18:382–389.
2018.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Plata-Bello J, Fariña-Jerónimo H, Betancor
I and Salido E: High expression of FOXP2 is associated with worse
prognosis in glioblastoma. World Neurosurg. 150:e253–e278.
2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Nishida K, Kuwano Y and Rokutan K: The
MicroRNA-23b/27b/24 cluster facilitates colon cancer cell migration
by targeting FOXP2. Cancers (Basel). 12(174)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Diao H, Ye Z and Qin R: miR-23a acts as an
oncogene in pancreatic carcinoma by targeting FOXP2. J Investig
Med. 66:676–683. 2018.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Chen MT, Sun HF, Li LD, Zhao Y, Yang LP,
Gao SP and Jin W: Downregulation of FOXP2 promotes breast cancer
migration and invasion through TGFβ/SMAD signaling pathway. Oncol
Lett. 15:8582–8588. 2018.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Yan X, Zhou H and Zhang T, Xu P, Zhang S,
Huang W, Yang L, Gu X, Ni R and Zhang T: Downregulation of FOXP2
promoter human hepatocellular carcinoma cell invasion. Tumour Biol.
36:9611–9619. 2015.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Cuiffo BG, Campagne A, Bell GW, Lembo A,
Orso F, Lien EC, Bhasin MK, Raimo M, Hanson SE, Marusyk A, et al:
MSC-regulated microRNAs converge on the transcription factor FOXP2
and promote breast cancer metastasis. Cell Stem Cell. 15:762–774.
2014.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Wong KK, Gascoyne DM, Soilleux EJ, Lyne L,
Spearman H, Roncador G, Pedersen LM, Møller MB, Green TM and Banham
AH: FOXP2-positive diffuse large B-cell lymphomas exhibit a poor
response to R-CHOP therapy and distinct biological signatures.
Oncotarget. 7:52940–52956. 2016.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Khan FH, Pandian V, Ramraj S, Natarajan M,
Aravindan S, Herman TS and Aravindan N: Acquired genetic
alterations in tumor cells dictate the development of high-risk
neuroblastoma and clinical outcomes. BMC Cancer.
15(514)2015.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Gascoyne DM, Spearman H, Lyne L, Puliyadi
R, Perez-Alcantara M, Coulton L, Fisher SE, Croucher PI and Banham
AH: The forkhead transcription factor FOXP2 is required for
regulation of p21WAF1/CIP1 in 143B osteosarcoma cell growth arrest.
PLoS One. 10(e0128513)2015.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Garcia-Alonso L, Iorio F, Matchan A,
Fonseca N, Jaaks P, Peat G, Pignatelli M, Falcone F, Benes CH,
Dunham I, et al: Transcription factor activities enhance markers of
drug sensitivity in cancer. Cancer Res. 78:769–780. 2018.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Bushweller JH: Targeting transcription
factors in cancer-from undruggable to reality. Nat Rev Cancer.
19:611–624. 2019.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Thakur A, Rahman KW, Wu J, Bollig A,
Biliran H, Lin X, Nassar H, Grignon DJ, Sarkar FH, Liao JD, et al:
Aberrant expression of X-linked genes RbAp46, Rsk4 and Cldn2 in
breast cancer. Mol Cancer Res. 5:171–181. 2007.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Houles T and Roux PP: Defining the role of
the RSK isoforms in cancer. Semin Cancer Biol. 48:53–61.
2018.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Li Q, Jiang Y, Wei W, Ji Y, Gao H and Liu
J: Frequent epigenetic inactivation of RSK4 by promoter methylation
in cancerous and non-cancerous tissues of breast cancer. Med Oncol.
31(793)2014.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Niskakoski A, Kaur S, Staff S,
Renkonen-Sinisalo L, Lassus H, Järvinen HJ, Mecklin JP, Bützow R
and Peltomäki P: Epigenetic analysis of sporadic and
Lynch-associated ovarian cancers reveals histology-specific
patterns of DNA methylation. Epigenetics. 9:1577–1587.
2014.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Hu C, Dai J, Lin X, Meng Y and Liang H:
Effect of RSK4 on biological characteristics of gastric cancer.
Cancer Manag Res. 12:611–619. 2020.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Guerrero-Zotano A, Mayer IA and Arteaga
CL: PI3K/AKT/mTOR: Role in breast cancer progression, drug
resistance and treatment. Cancer Metastasis Rev. 35:515–524.
2016.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Wang N, Li Y, Wei J, Pu J, Liu R, Yang Q,
Guan H, Shi B, Hou P and Ji M: TBX1 functions as a tumor suppressor
in thyroid cancer through inhibiting the activities of the PI3K/AKT
and MAPK/ERK pathways. Thyroid. 29:378–394. 2019.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Myers AP, Corson LB, Rossant J and Baker
JC: Characterization of mouse Rsk4 as an inhibitor of fibroblast
growth factor-RAS-extracellular signal-regulated kinase signaling.
Mol Cell Biol. 24:4255–4266. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Grunt TW and Mariani GL: Novel approaches
for molecular targeted therapy of breast cancer: Interfering with
PI3K/AKT/mTOR signaling. Curr Cancer Drug Targets. 13:188–204.
2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Arcaro A and Guerreiro AS: The
phosphoinositide 3-kinase pathway in human cancer: Genetic
alterations and therapeutic implications. Curr Genomics. 8:271–306.
2007.PubMed/NCBI View Article : Google Scholar
|