Introduction
Success in reproduction is dependent on the ordered
regulation of hormone and neuropeptide production in the
hypothalamic-pituitary-gonadal (HPG) axis. The main hypothalamic
peptide gonadotropin-releasing hormone (GnRH) exerts particularly
important functions in driving this axis (1). GnRH is normally produced by the
hypophysial portal system to stimulate gonadotropin production
(1). In 2009, Tsutsui (2) first demonstrated that a previously
unknown hypothalamic neuropeptide was able to inhibit gonadotropin
production in Japanese quail, which was referred to as
gonadotropin-inhibitory hormone (GnIH). Subsequently, existence of
GnIH orthologs, such as RFamide-related peptide (RFRP)-1 and -3,
were revealed in numerous vertebrates. RFRP-3 serves a particularly
key role and has been extensively studied (3). GnIH exerts an important role in the
suppression of reproduction, specifically by acting on GnRH neurons
and pituitary glands (4,5), through G protein-coupled receptor 147
(GPR147) (4,5). GnIH/RFRP-3 has been previously
suggested to inhibit the immediate peripheral effects on vertebrate
gonads (6). The expression of GnIH
and its receptor can be detected within germ and steroidogenic
cells in bird or mammal gonads, where it potentially suppresses
gonadal steroid production to prevent the maturation and
differentiation of germ cells in either an autocrine or paracrine
manner (6). The uterus is an
important reproductive organ in female mammals. Han et al
(7) previously suggested that the
intracerebroventricular (ICV) injection of RFRP-3 can delay uterine
development. Therefore, it would be of interest to examine the
molecular mechanism by which GnIH can regulate uterus function.
According to previous studies, GnIH can directly
regulate the function of pro-opiomelanocortin (POMC), neuropeptide
Y (NPY) and kisspeptin neurons of opioid melanocytes, by inhibiting
the expression of POMC and kisspeptin whilst increasing that of NPY
(8-10).
In addition, POMC, NPY and kisspeptin neurons have been shown to
directly mediate effects on the GnRH neurons and modulate the
reproductive function by regulating GnRH synthesis (11-13).
Therefore, GnIH appear to confer both direct and indirect effects
on hypothalamic GnRH neurons, mainly by reducing pituitary
gonadotropin release through inhibiting POMC and kisspeptin neurons
whilst promoting NPY neurons. This ultimately affects the HPG axis
and downstream target organs, such as the uterus. However, it
remains unclear if GnIH can regulate the uterus through the HPG
reproductive axis.
Endometrial cancer (EC) is one of the most common
malignancies of the female reproductive system that can be lethal
(14). In recent years, alongside
the increasing incidence of obesity, the worldwide incidence of EC
has also been increasing on an annual basis, particularly among
younger women (15). Treatment of
EC is difficult due to the simultaneous need for preserving patient
fertility (16). Therefore,
hormone adjuvant therapy has been garnering attention due to its
advantages of low toxicity, reduced severity of side effects,
maneuverability and favorable safety profiles (14,15).
A previous study has found that GnRH analogues can stimulate the
HPG axis to reduce the secretion of luteinizing hormone (LH) and
follicle-stimulating hormone (FSH), which suppresses gonadal
function and indirectly inhibits tumor growth (17). In addition, GnRH agonists can
reduce the proliferation of human EC cells in a dose- and
time-dependent manner (18,19).
By contrast, GnRH antagonists can induce apoptosis in human EC
cells by activating p38 MAPK and JNK signaling, in addition to the
proapoptotic protein Bax (20).
This strategy of reversible medical has been successfully
introduced into the treatment regimen of EC, where the underlying
mechanism is consistent with the biological characteristics of GnIH
(17-20).
Therefore, GnIH may provide a novel avenue for developing a
treatment strategy for EC.
In the present study, the potential effects of
GnIH/RFRP-3 injection through the lateral ventricle on the uterus
of ovariectomized estrogen-primed (OEP) rats were investigated
using liquid chromatography (LC)-tandem mass spectrometry (MS/MS)
analysis. Using these data, the differentially expressed proteins
(DEPs) were selected, which were subjected to Gene Ontology (GO)
functional annotation and Kyoto Encyclopedia of Genes and Genomes
(KEGG) enrichment analyses. Subsequently, their molecular mechanism
and clinical values were investigated through protein-protein
interaction (PPI) network analysis. Cell Counting Kit-8 (CCK-8)
assay, Annexin V-FITC/PI double staining and western blot analysis
were used to assess the effects of RFRP-3 on the apoptosis of
HEC-1A cells. Findings from the present study may provide novel
ideas for examining the effects of GnIH on reproduction.
Materials and methods
Study animals and model
establishment
In total, 30 female Sprague-Dawley rats (age, 6-7
weeks; weight, 180-220 g) were provided by Beijing Huafukang
Biotechnology Co., Ltd. (animal certification no. 11401300067446).
These animals were fed with normal diet and maintained at 22-25˚C,
in a natural light/dark cycle (12/12 h), the relative humidity of
the animal facility was 40-70%, with free access to food and water
for 1 week before animal experimentation commenced. Animal
experimental procedures were performed in accordance with the
ethical standards of and approved by The Experimental Animal
Welfare Ethics Committee of Chengde Medical University (Chengde,
China).
The rat ovariectomized estrogen-primed (OEP) model
was established. Briefly, the rats were anesthetized with 10%
chloral hydrate (300 mg/kg) before being bilaterally ovariectomized
or in all anesthetic procedures under sterile conditions. None of
the rats exhibited any signs of peritonitis, pain or discomfort
during the operation. After 15 days, the animals were subjected to
a subcutaneous injection of estradiol benzoate [Shanghai full woo
Biotechnology (Zhumadian) Co., Ltd.; 1 mg/kg/day] for 5 days. This
drug was given to keep the estrogen level in each rat consistent
and eliminate the influence of estrogen on GnIH. Animal status was
monitored daily. The animals were then randomly divided into the
following two groups: i) RFRP-3 group (n=15), in which RFRP-3 (16
µl/kg; final concentration, 2 µg/µl; solvent, normal saline; Bachem
AG) was injected into the lateral ventricle of the rats following
anesthesia as aforementioned; and ii) normal saline injection group
(n=15), in which saline was injected into the lateral ventricle of
rats. The uterine fluid of five rats from the experiment and
control groups was mixed and divided into three parts, which were
stored in liquid nitrogen. Euthanasia was performed by cervical
dislocation, before which the animals were anesthetized, and 6 h
after administration, uterine fluid samples were collected. The
chest was opened to verify death of each animal, where absence of
any fluctuations in the chest caused by breathing and heartbeat was
verified.
The humane endpoints used to determine when the
animal must be immediately euthanized were as follows: i) Rapid
weight loss of 15-20%; ii) lack of food and drinking water intake;
iii) in the state of non-anesthesia or sedation, when animal mental
depression is accompanied by hypothermia; and iv) abnormal central
nervous responses (convulsions, tremors, paralysis, head tilt).
Cell lines
Human EC HEC-1A cells were provided by Procell Life
Science & Technology Co., Ltd. HEC-1A cells were cultured in
McCoy 5A medium [Zhongke Maichen (Beijing) Technology Co., Ltd.],
supplemented with 15% FBS (Sartorius AG) and 1%
penicillin/streptomycin, at 37˚C with 5% CO2.
Determination of optimal concentration
of RFRP-3
RFRP-3 (Tocris Bioscience) was diluted with PBS to
obtain a final concentration of 2 mg/ml as the stock solution and
stored at -20˚C. HEC-1A cells were seeded into 96-well plates in
triplicates. The cells were then treated with 0 (control), 0.1, 1,
10, 100, 1,000 and 10,000 ng/ml RFRP-3. Cells in the control group
were subjected to no treatments, whilst the cells in the treatment
groups were incubated with RFRP-3 at the indicated concentrations
for 24 h.
Identification of DEPs
Peptides were separated via high-performance LC
(Agilent 1260 Infinity II LC; Agilent Technologies, Inc.) from
uterine fluid. Samples were separated on a XTerra RP18 column
(4.6x50 mm; 5 µm; Waters Corporation) at 35˚C. Mobile phases A and
B were comprised of 1,000:1 (v/v) methanol/formic acid and 1,000:1
(v/v) water/formic acid, respectively. The flow rate was 0.4
ml/min. The injection volume was 3 µl and the total running time
was 2 min.
Peptides were identified via MS/MS. Multiple
reaction monitoring was performed with an Agilent 6460 triple
quadrupole MS/MS fitted with the Agilent Jet Stream Electrospray
Ionization probe (Agilent Technologies, Inc.) in the positive ion
mode. Data were analyzed using the full scan mass spectra (300-1800
M/z). The mass spectrometry ion source was a nano-ESI ion source,
so without parameters such as nitrogen gas temperature, nebuliser
pressure and flow rate. Experiments were performed in triplicates.
Finally, the mass spectrometry data were analyzed using the
Proteome Discoverer software (version 1.4.0.288; Thermo Fisher
Scientific, Inc.). DEPs were screened using P<0.05 and Fold
Change (FC)=2 as criteria. Protein name, gene ID, molecular weight
and other information were obtained using the UniProt knowledgebase
(http://www.uniprot.org/).
Bioinformatics analysis
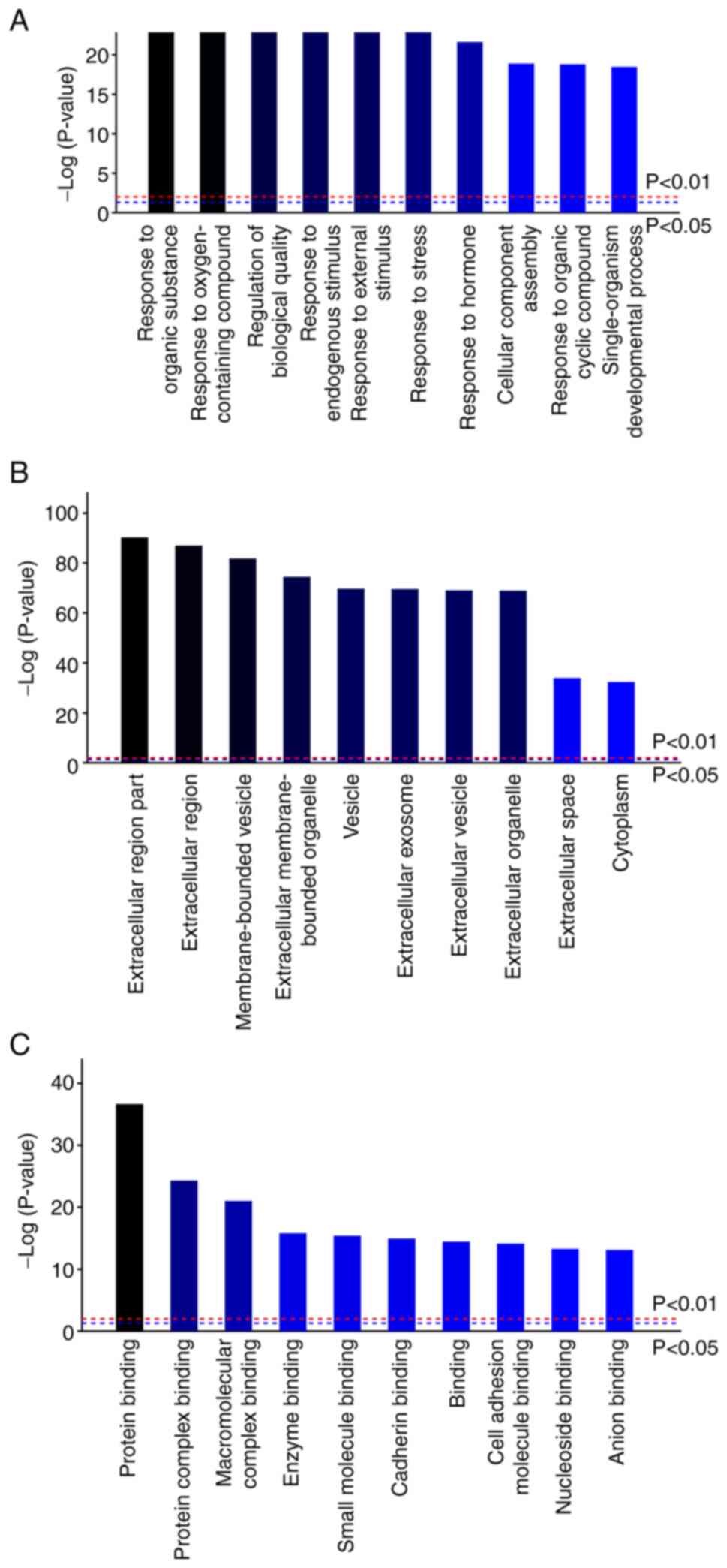
GO analysis was performed, including the biological
process (BP), cell component (CC) and molecular function (MF),
using the GO (http://www.geneontology.org/) online tool. KEGG
pathways associated with the DEPs were analyzed using the KEGG
(http://www.KEGG.jp) online tool. The dashed blue line
indicates P<0.05, the dashed red line indicates P<0.01. The
top 10 items were selected according to P<0.01 based on their
corresponding enrichment results.
PPI network of DEPs
The interaction network of DEPs was searched against
the Search Tool for the Retrieval of Interacting Genes/Proteins
(STRING; http://string-db.org) database. The
STRING database was also utilized to construct the PPI networks of
DEPs. Statistical significance was deemed as interaction with a
pooled score of >0.4. The PPI network was drawn using the
multi-omics data analysis tool, OmicsBean V1.0 (http://www.omicsbean.cn/). The interaction between
proteins is shown as a solid line, and proteins and KEGG terms are
linked by a dotted line. The protein that interacts most closely
with other proteins in the PPI network was selected as the hub
protein. It can be seen from the PPI network that the DEPs Gna 13,
Gnaq and Gnai3 participate in the platelet activation pathway. In
addition, KEGG pathway analysis (https://www.kegg.jp/kegg/) was performed (‘platelet
activation’ was searched for in the PATHWAY database).
University of Alabama cancer (UALCAN)
database analysis
The UALCAN database (http://ualcan.path.uab.edu/index.html); public
datasets name, TCGA Gene (http://ualcan.path.uab.edu/analysis.html); TCGA
dataset, uterine corpus endometrial carcinoma public datasets) was
used to explore the internal relationship between endometrial
cancer and hub proteins (Gna13, Gnaq, Gnai3, Kras and MMP9) that
were screened using the PPI network. Gna13, Gnaq, Gnai3, Kras and
MMP9 expression was compared between human endometrial carcinoma
tissues and normal tissues using The Cancer Genome Atlas (TCGA)
database. In the results, only the hub proteins with statistical
differences were displayed. The number of normal tissues was 35 and
the number of EC tissues was 546.
CCK-8 assay
HEC-1A cells were seeded into the 96-well plate at a
density of 1x104/well. After 24 h, 100 µl RFRP-3 at the
indicated concentrations (0, 0.1, 1, 10, 100, 1,000 and 10,000
ng/ml) was used to treat the cells for 24 h. After removing the
medium, a solution containing 10% CCK-8 (APeXBIO Technology LLC) in
McCoy 5A medium was added into each well. The reaction was
conducted at 37˚C for 1 h, before the optical density value at 450
nm was measured and recorded. The calculation formula of the
viability rate was: [(RFRP-3 group-blank)/(Control-blank)-1]
x100%.
Detection of apoptosis
Apoptosis of HEC-1A cells was detected by flow
cytometry using an Annexin V-FITC/PI apoptosis kit (BD
Biosciences). HEC-1A cells were seeded into 6-well plates at a
density of 3x105/well and treated with either complete
McCoy 5A medium or complete McCoy 5A medium with 10,000 ng/ml
RFRP-3 for 24 h. The cells were then suspended in binding buffer
and stained with 5 µl Annexin V-FITC reagent and 5 µl PI solution
at room temperature in dark for 15 min. The stained cells were then
analyzed by Coulter ELITEesp flow cytometry (Beckman Coulter, Inc.)
and data analysis was performed using FloMax 2.7 software (Sysmex
Partec GmbH). In the present study, the sum of late and early
apoptotic cells was used as the readout for the apoptotic rate.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from treated HEC-1A cells
(control and 10,000 ng/ml RFRP-3 groups) using the Takara MiniBEST
Universal RNA Extraction kit (Takara Biotechnology Co., Ltd.),
according to the manufacturer's protocol. Concentration of total
RNA was determined using NanoDrop™ 2000 (Thermo Fisher Scientific,
Inc.), before the RT reaction was performed using the Prime Script™
RT reagent Kit (Takara Bio, Inc.) according to the manufacturer's
instructions. qPCR was performed using the TB Green™ Premix ExTaq™
II kit (Takara Bio, Inc.) on a CFX96 Real-Time PCR Detection System
(Bio-Rad Laboratories, Inc.). The thermocycling conditions were as
follows: Initial denaturation step at 95˚C for 30 sec, followed by
40 cycles of denaturation at 95˚C for 5 sec, and annealing and
extension at 60˚C for 30 sec. The primer sequences were as follows:
PI3K forward, 5'-GGAATGCTGCAAGATCAAGAAA-3' and reverse,
5'-TTGGTGGTAATGGAAGAGGAAGA-3'; AKT forward,
5'-CTTGCTTTCAGGGCTGCTCA-3' and reverse,
5'-TACACGTGCTGCCACACGATAC-3'; mTOR forward,
5'-CTTGCTGAACTGGAGGCTGATGG-3' and reverse,
5'-CCGTTTTCTTATGGGCTGGCTCTC-3'; LC3-II forward,
5'-gtCagCgTctCcACACCAATCTC-3' and reverse,
5'-TCCTGGGAGGCATAGaCCATGTAC-3'; and GAPDH forward,
5'-GCACCGTCAAGGCTGAGAAC-3' and reverse, 5'-TGGTGAAGACGCCAGTGGA-3'.
The relative expression levels of target genes were calculated
using the 2-∆∆Cq (Livak) method (21). GAPDH was used as internal
reference.
Western blot analysis
HEC-1A cells (control and RFRP-3 groups) were lysed
using RIPA lysis buffer (Beijing Solarbio Science & Technology
Co., Ltd.). Protein concentrations were measured using a BCA kit. A
total of 30 µg protein per lane was separated by 10-12% SDS-PAGE
and then transferred onto PVDF membranes. The membranes were then
blocked with 5% skimmed milk at room temperature for 2 h, before
being incubated with primary antibodies against GAPDH (rabbit;
1:6,000; cat. no. ab9485; Abcam), Kras (rabbit; 1:2,000; cat. no.
ab191595; Abcam), Bcl-2 (rabbit; 1:800; cat. no. ab59348; Abcam),
Bax (rabbit; 1:800; cat. no. ab32503; Abcam), PI3K (rabbit; 1:500;
cat. no. AP0231; Bioworld Technology, Inc.), AKT (rabbit; 1:10,000;
cat. no. ab179463; Abcam), phosphorylated (p)-AKT (rabbit; 1:2,000;
cat. no. 4060; Cell Signaling Technology, Inc.), mTOR (rabbit;
1:2,000; cat. no. ab32028; Abcam), LC3B (rabbit; 1:2,000; cat. no.
ab192890; Abcam), p62 (rabbit; 1:1,000; cat. no. A19700; ABclonal
Biotech Co., Ltd.) and GPR147 (also referred to as NPFF-1 receptor;
rabbit; 1:5,00; cat. no. bs-12018R; Beijing Bioss Biotechnology
Co., Ltd.) at 4˚C overnight. After TBST washing, the membranes were
incubated with the corresponding HRP-conjugated secondary
antibodies (rabbit; 1:8,000; cat. no. ab205718; Abcam) for another
1 h at room temperature. Immune-reactive proteins were visualized
with the ECL luminescence reagent (cat. no. MA0186; Dalian Meilun
Biology Technology Co., Ltd.). Protein band intensities were
analyzed and normalized to GAPDH that was used as loading control.
A Tanon 5200 imaging system (Tanon Science and Technology Co.,
Ltd.) was used to detect the bands, and ImageJ 1.52a software
(National Institutes of Health) was used to semi-quantify the
integrated density.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Each experiment was carried out three times. Statistical analysis
was performed using the SPSS 22 software (IBM Corp.). Comparisons
among multiple groups were conducted using one-way ANOVA followed
by Dunnett's post hoc test. An unpaired t-test was used to compare
the differences between two groups for the results of RT-qPCR and
western blotting. P<0.05 was considered to indicate a
statistically significant difference.
Results
Protein profile of uterine fluid is
changed by RFRP-3
Using P<0.05 and FC=2 as thresholds, a total of
417 DEPs were identified in the uterine fluid of OEP rats treated
with RFRP-3 compared with control rats, including 279 upregulated
and 138 downregulated proteins (Tables
I and II).
 | Table ITop 10 upregulated differentially
expressed proteins. |
Table I
Top 10 upregulated differentially
expressed proteins.
| UniProt ID | Gene name | Protein name (short
name, full name) | Log2 fold
change | Organism |
|---|
| P0CC09 | Hist2h2aa3 | H2A2A_RAT, histone
H2A type 2-A | 13.78 | Rattus norvegicus
(Rat) |
| P84245 | H3f3b | H33_RAT, histone
H3.3 | 13.31 | Rattus norvegicus
(Rat) |
| P36953 | Afm | AFAM_RAT,
afamin | 12.73 | Rattus norvegicus
(Rat) |
| Q62714 | Np4 | DEF4_RAT,
neutrophil antibiotic peptide NP-4 | 12.35 | Rattus norvegicus
(Rat) |
| P15684 | Anpep | AMPN_RAT,
aminopeptidase N | 12.16 | Rattus norvegicus
(Rat) |
| Q9QYK2 | Ppargc1a | PRGC1_RAT,
peroxisome proliferator-activated receptor gamma coactivator
1-alpha | 12.13 | Rattus norvegicus
(Rat) |
| P30349 | Lta4h | LKHA4_RAT,
leukotriene A-4 hydrolase | 12.02 | Rattus norvegicus
(Rat) |
| Q9R063 | Prdx5 | PRDX5_RAT,
peroxiredoxin-5, mitochondrial | 11.81 | Rattus norvegicus
(Rat) |
| Q5XHY1 | Carmil3 | LR16B_RAT,
leucine-rich repeat-containing protein 16B | 11.79 | Rattus norvegicus
(Rat) |
| P20762 | P20762 | IGG2C_RAT, Ig
gamma-2C chain C region | 11.72 | Rattus norvegicus
(Rat) |
 | Table IITop 10 downregulated differentially
expressed proteins. |
Table II
Top 10 downregulated differentially
expressed proteins.
| UniProt ID | Gene name | Protein name (short
name, full name) | Log2 fold
change | Organism |
|---|
| P04218 | Cd200 | OX2G_RAT, OX-2
membrane glycoprotein | -18.03 | Rattus norvegicus
(Rat) |
| Q8K3K9 | Gimap4 | GIMA4_RAT, GTPase
IMAP family member 4 | -12.8 | Rattus norvegicus
(Rat) |
| B4F795 | Slc44a2 | CTL2_RAT, choline
transporter-like protein 2 | -10.95 | Rattus norvegicus
(Rat) |
| P14925 | Pam | AMD_RAT,
peptidyl-glycine alpha-amidating monooxygenase | -10.65 | Rattus norvegicus
(Rat) |
| A9UMV8 | H2afj | H2AJ_RAT, histone
H2A.J | -10.47 | Rattus norvegicus
(Rat) |
| P33568 | Rb1 | RB_RAT,
retinoblastoma-associated protein | -10.05 | Rattus norvegicus
(Rat) |
| Q9ESV6 | Gapdhs | G3PT_RAT,
glyceraldehyde-3-phosphate dehydrogenase, testis-specific | -9.75 | Rattus norvegicus
(Rat) |
| Q08849 | Stx3 | STX3_RAT,
syntaxin-3 | -9.41 | Rattus norvegicus
(Rat) |
| Q3B8N7 | Tsc22d4 | T22D4_RAT, TSC22
domain family protein 4 | -9.41 | Rattus norvegicus
(Rat) |
| Q3KRC4 | Gprc5c | GPC5C_RAT,
G-protein coupled receptor family C group 5 member C | -9.2 | Rattus norvegicus
(Rat) |
Analysis of the biological function of
DEPs after the ICV injection of RFRP-3
To analyze the BP, CC and MF terms of DEPs, GO
functional annotation was performed (Fig. 1). In terms of BP, DEPs were found
to be mostly enriched in ‘response to organic substance’ (GO ID:
GO0010033), ‘response to oxygen-containing compound’ (GO ID:
GO1901700), ‘regulation of biological quality’ (GO ID: GO0065008)
and ‘response to external stimulus’ (GO ID: 0009605) (Fig. 1A). For MF, the DEPs were
particularly enriched in ‘protein binding’ (GO ID: GO0005515),
‘protein complex binding’ (GO ID: GO0032403) and ‘macromolecular
complex binding’ (GO ID: GO0044877) (Fig. 1C). In terms of CC, the DEPs were
found to be associated with ‘extracellular region’ (GO ID:
GO0005576) and ‘membrane-bounded vesicle’ (GO ID: GO0031988)
(Fig. 1B). The aforementioned
results imply that these DEPs participate in protein, polypeptide
and carbohydrate metabolism. Furthermore, these proteins exhibited
an association with antioxidation and were indicated to regulate
cell proliferation, apoptosis, cell migration, cell adhesion and
angiogenesis.
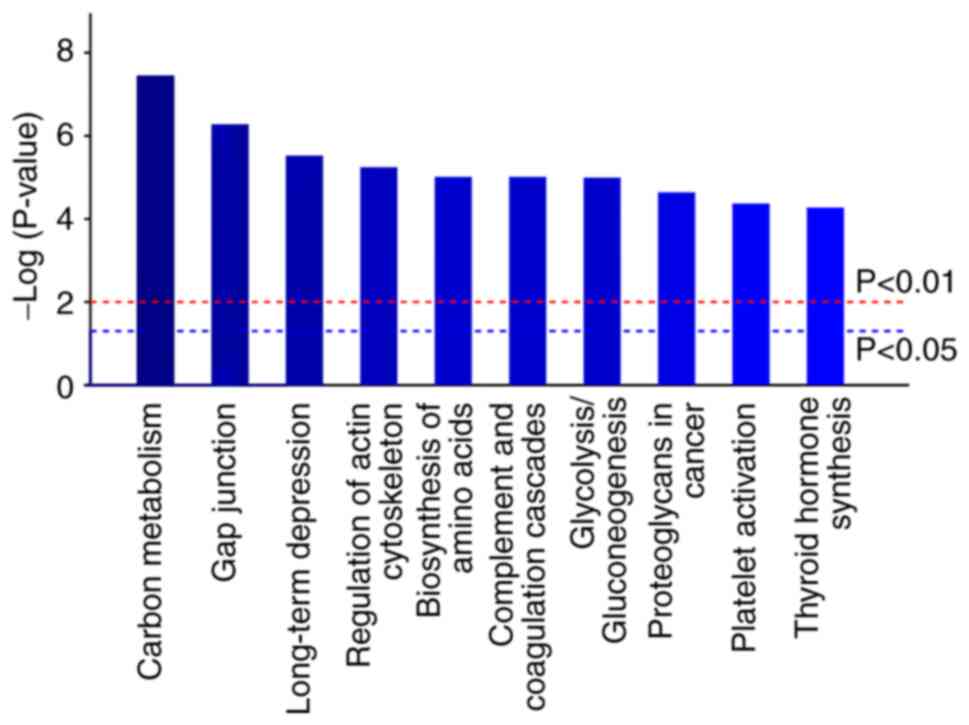
Analysis of signal pathways associated
with the DEPs after the ICV injection of RFRP-3
To further assess the possible functions of DEPs
caused by RFRP-3 treatment, signaling pathway enrichment analysis
was performed using KEGG pathway analysis (Fig. 2). These DEPs were observed to be
enriched in ‘carbon metabolism’ (pathway ID: rno01200), ‘gap
junction’ (pathway ID: rno04540), ‘long-term depression’ (pathway
ID: rno04730), ‘regulation of actin cytoskeleton’ (pathway ID:
rno04810), ‘biosynthesis of amino acids’ (pathway ID: rno01230),
‘complement and coagulation cascades’ (pathway ID: rno04610),
‘glycolysis/gluconeogenesis’ (pathway ID: rno00010), ‘proteoglycans
in cancer’ (pathway ID: rno05205), ‘platelet activation’ (pathway
ID: rno04611) and ‘thyroid hormone synthesis’ (pathway ID:
rno04918).
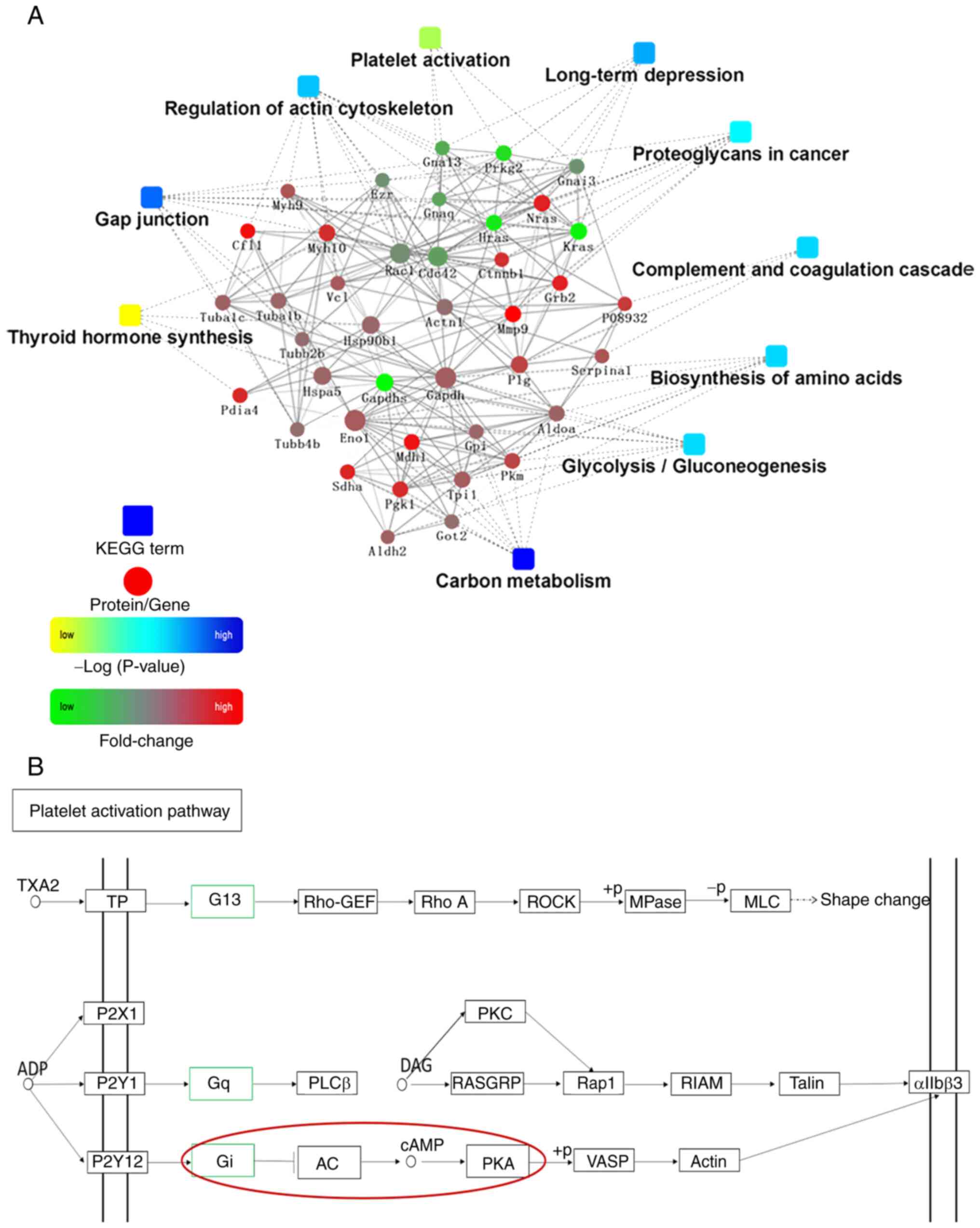
PPI network of the DEPs after the ICV
injection of RFRP-3
PPI analysis was then performed using the STRING
database, which was used to build the PPI network (Fig. 3A). OmicsBean was utilized for the
visualization of this network (Fig.
3A). In total, five proteins were identified to be hub
proteins, namely G protein subunit α (Gna)13, Gnaq, Gnai3, Kras and
MMP9. Among them, the protein expression levels of Gna 13, Gnaq,
Gnai3 and Kras were decreased, while the protein expression levels
of MMP9 were increased. In addition, based on KEGG pathway database
analysis, it can be seen that the platelet activation pathway
activated the AC/cAMP/PKA signaling pathway (Fig. 3B).
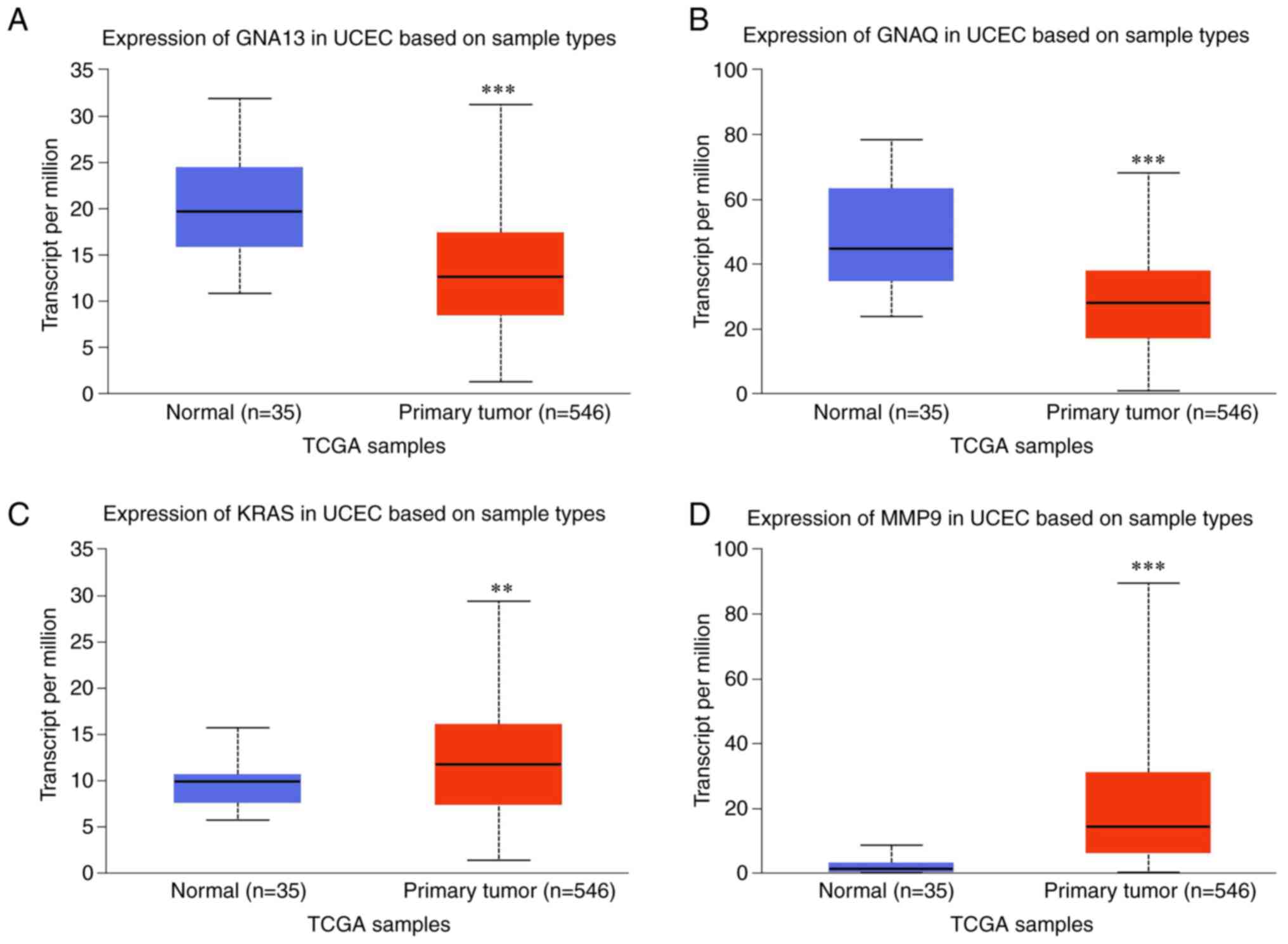
Expression of Gna13, Gnaq, Kras and
MMP9 is significantly changed in EC
Using the UALCAN database platform, The Cancer
Genome Atlas database was screened. The expression of the hub
proteins Gna13, Gnaq, Kras and MMP9 was found to be significantly
different between EC and the normal tissues (Fig. 4). Specifically, compared with that
in normal samples, Kras and MMP9 expression was increased, whereas
Gnaq and Gna13 expression was decreased in EC tissues.
RFRP-3 reduces the expression of Kras
in HEC-1A cells
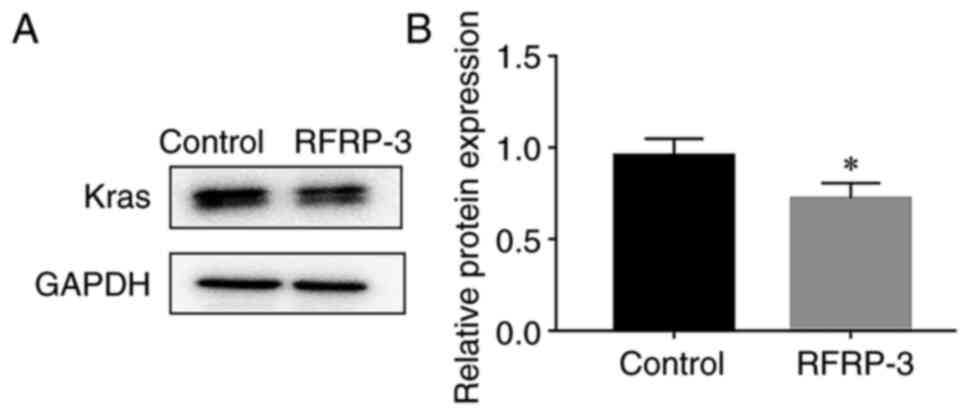
The expression levels of Kras were examined. As
shown in Fig. 5, compared with
those in the control group, the expression levels of Kras were
significantly reduced in the RFRP-3 group.
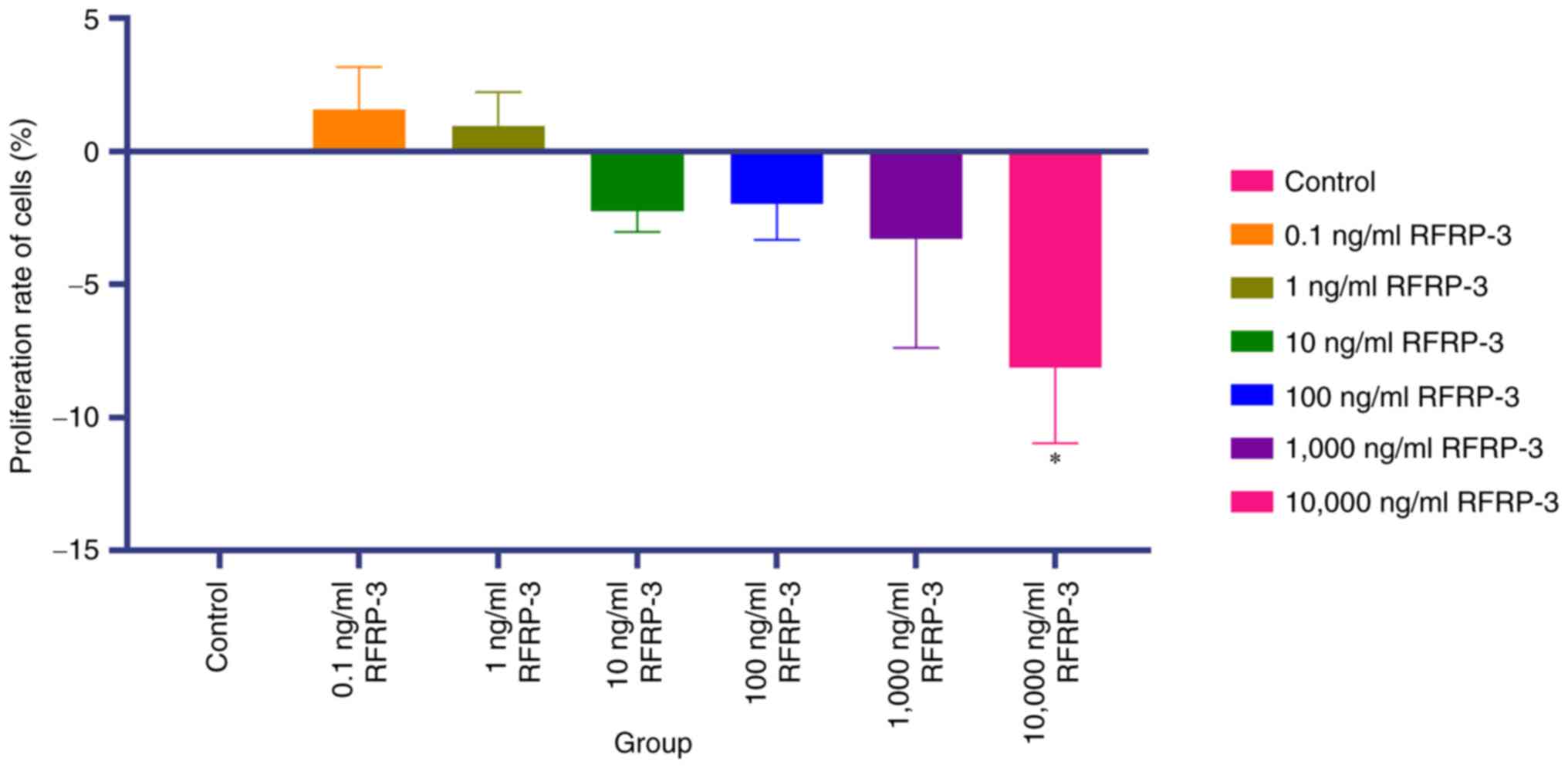
RFRP-3 inhibits the viability of
HEC-1A cells
The potential effects of RFRP-3 on the viability of
HEC-1A cells were next investigated using the CCK-8 assay. As shown
in Fig. 6 and Table III, compared with that in the
control group, cell viability was significantly decreased in the
10,000 ng/ml RFRP-3 treatment group (P<0.05). However, the
differences in cell viability were not significant between the
control group and the 0.1, 1, 10, 100 and 1,000 ng/ml RFRP-3
groups. These results suggest that RFRP-3 at 10,000 ng/ml was able
to suppress EC cell viability. Therefore, in subsequent experiments
HEC-1A cells were treated with 10,000 ng/ml RFRP-3.
 | Table IIIEffects of RFamide-related peptide 3
on cell viability of HEC-1A cells. |
Table III
Effects of RFamide-related peptide 3
on cell viability of HEC-1A cells.
| Group | Viability rate of
cells, % | P-value |
|---|
| Control | 0.000 | |
| 0.1 ng/ml
RFRP-3 | 1.560±1.598 | - |
| 1 ng/ml RFRP-3 | 0.960±1.254 | - |
| 10 ng/ml
RFRP-3 | -2.250±0.782 | - |
| 100 ng/ml
RFRP-3 | -1.983±1.356 | - |
| 1,000 ng/ml
RFRP-3 | -3.297±4.085 | - |
| 10,000 ng/ml
RFRP-3 | -8.127±2.837 |
P<0.05a |
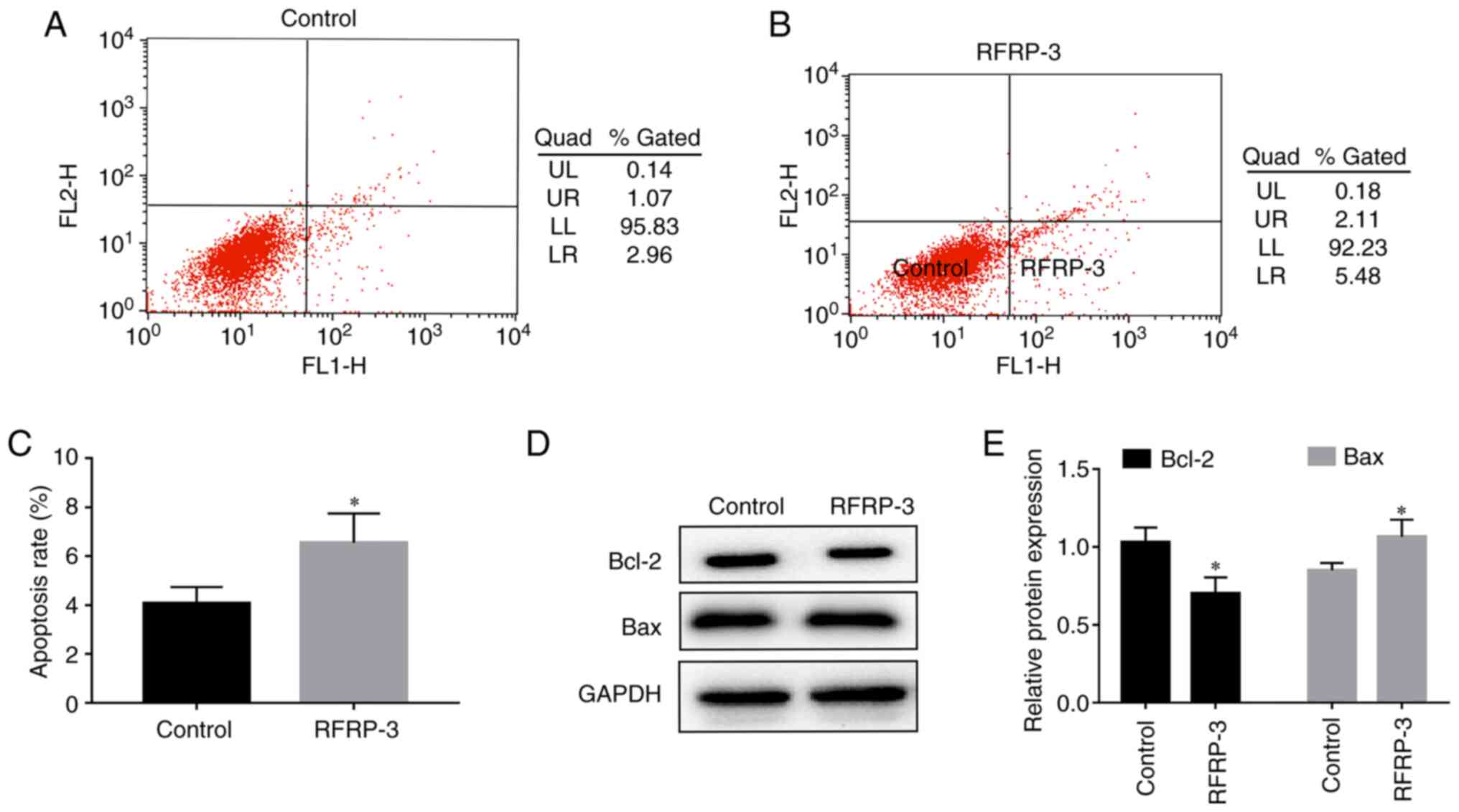
RFRP-3 induces apoptosis in HEC-1A
cells
To investigate the effects of RFRP-3 on HEC-1A cell
apoptosis, flow cytometry was performed. As shown in Fig. 7, the upper right quadrant
represents late apoptotic cells, whilst the lower right quadrant
represents early apoptotic cells. The proportion of apoptotic cells
(%) in the RFRP-3 group was found to be higher compared with that
in the control group (Fig. 7A-C).
Results from the western blot analysis revealed that compared with
those in the control group, Bcl-2 protein expression was
significantly lower whereas Bax protein expression was
significantly higher in the RFRP-3 group (Fig. 7D and E). These results suggest that RFRP-3 can
induce apoptosis in HEC-1A cells.
RFRP-3 inhibits the PI3K/AKT/mTOR
pathway in HEC-1A cells
To investigate the effects of RFRP-3 on the
PI3K/AKT/mTOR pathway in HEC-1A cells, the expression levels of the
associated components were detected using RT-qPCR and western blot
analysis. As shown in Fig. 8A,
compared with those in the control group, the expression levels of
PI3K/AKT/mTOR genes were significantly lower in the RFRP-3 group.
Treatment with 10,000 ng/ml RFRP-3 significantly reduced the
protein expression levels of PI3K and mTOR whilst also reducing the
phosphorylation of AKT, compared with those in the control group
(Fig. 8B and C). The expression levels of AKT were
similar between the control and RFRP-3 groups. These results
suggest that RFRP-3 can inhibit the PI3K/AKT/mTOR pathway to
potentially promote tumor apoptosis.
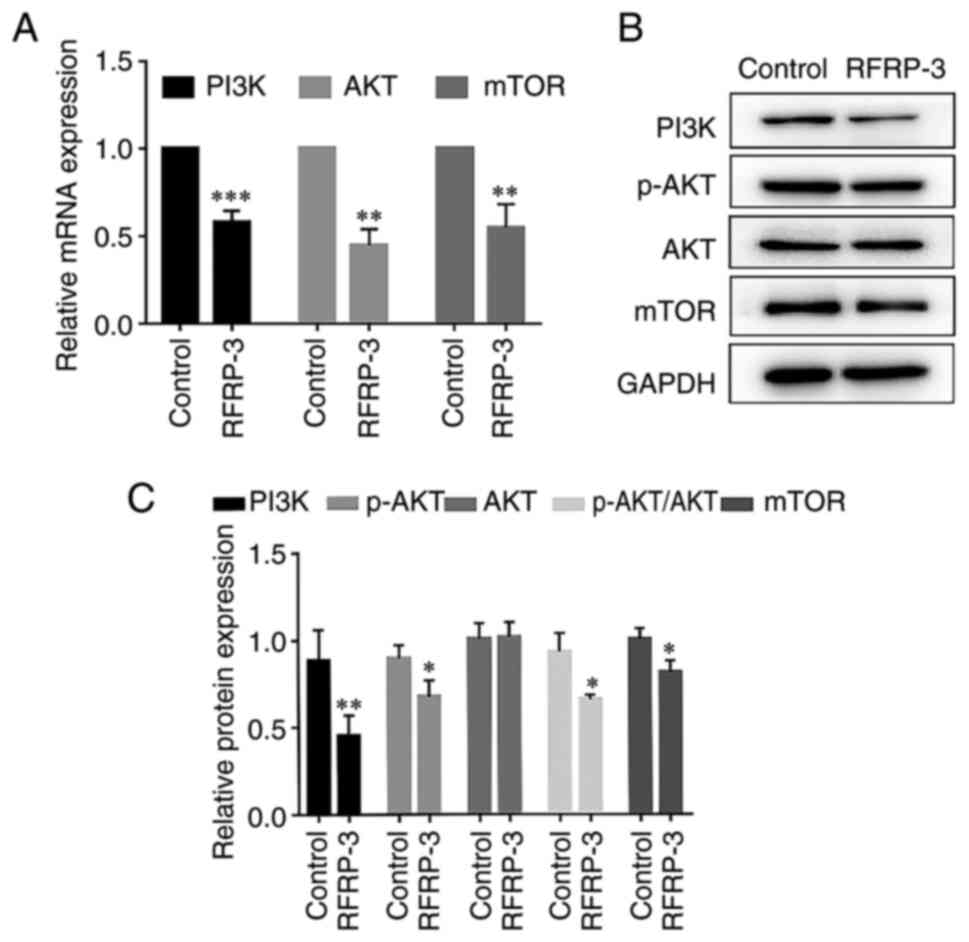 | Figure 8Effects of RFRP-3 on the
PI3K/AKT/mTOR pathway in HEC-1A cells. HEC-1A cells in the RFRP-3
group were treated with 10,000 ng/ml RFRP-3 for 24 h. (A) The mRNA
expression levels of PI3K, AKT and mTOR were measured by reverse
transcription-quantitative PCR. (B) The protein expression levels
of PI3K, AKT and mTOR, in addition to AKT phosphorylation, were
measured using western blot analysis. (C) Semi-quantification of
PI3K, AKT and mTOR expression, as well as AKT phosphorylation,
presented as relative expression. *P<0.05,
**P<0.01 and ***P<0.001 vs. control.
RFRP-3, RFamide-related peptide 3; p, phosphorylated. |
RFRP-3 induces autophagy in HEC-1A
cells
mTOR serves a key role in regulating autophagy.
Therefore, the expression levels of associated genes were detected
using RT-qPCR and western blot analysis. As shown in Fig. 9A, compared with those in the
control group, the mRNA expression levels of LC3-II were
significantly higher in the RFRP-3 group. Treatment with 10,000
ng/ml RFRP-3 significantly increased the LC3-II/I protein
expression ratio compared with those in the control group. In
addition, RFRP-3 treatment significantly reduced p62 expression
compared with that in the control group (Fig. 9B and C). The concurrent accumulation of LC3-II
and degradation of p62 induced by RFRP-3 suggests that it increased
autophagic activity.
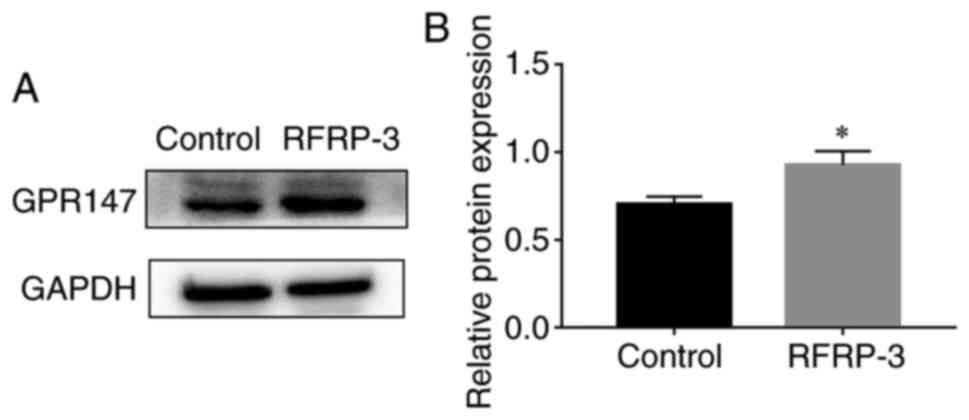
RFRP-3 increases GPR147 protein
expression in HEC-1A cells
To explore the underlying molecular mechanism of
RFRP-3, the protein expression of GPR147, one of the potential
receptors for RFRP-3, was examined using western blot analysis. As
shown in Fig. 10, compared with
that in the control group, the expression levels of GPR147 were
significantly increased in the RFRP-3 group.
Discussion
GnIH is a relatively novel hypothalamic
neuropeptide, discovered in 2000(2). GnIH has been previously shown to show
a high degree of conservation in vertebrates, from agnatha to
humans (2). In addition, orthologs
of GnIH have also been discovered in mammals, referred to as RFRP-1
and RFRP-3(2). In particular,
RFRP-3 has been found to mediate important roles in various
biological processes, especially in energy balance, feeding and
reproduction (22-27).
For GnIH neurons, the cell bodies are localized to the dorsomedial
hypothalamic (DMH) area and paraventricular nucleus in the majority
of mammals (28). RFRP-3 mRNA
expression has previously been detected in the hypothalamus, eyes,
ovaries and uterus of juvenile sheep (29). In addition, GnIH is highly
expressed in the hypothalamus, pituitary, ovaries, testis and
epididymis (30). Li et al
(31) reported that in sows, GnIH
mRNA can be detected in the central nervous system, specifically in
the hypothalamus and medulla oblongata. Furthermore, it can be
found in the peripheral tissues, such as the ovaries and uterus
(31). RFRP mRNA transcripts have
also been detected in the hypothalamus of rats and humans (32). GnIH neuronal cell bodies have been
observed in the DMH area of the human hypothalamus, where their
neural fibers can project into the median eminence external zone
(27).
RFRPs are mainly localized in the normal human
ovaries, corpus luteum and the large pre-ovulatory follicle
granulosa cell layer (27,33). The GnIH receptor mainly mediates
the function of GnIH. Bonini et al (34) previously detected two G-coupled
protein receptors of neuropeptide FF (NPFF). There are C-terminal
PQRFamide motifs in NPFF1 (the same as GPR147), NPFF2 (the same as
GPR74). GPR147 is considered to be the major GnIH receptor
(34). GnIH can exert direct
effects on hypothalamic GnRH and pituitary neurons through the GnIH
receptor to inhibit the production of gonadotropin and the
reproductive function (33,35).
It has been shown that the mRNA expression levels of GnIH receptors
in rabbits are high, whereas GPR147 mRNA is mainly expressed in the
hypothalamus, pituitary, ovaries, testis and uterus (30). Hinuma et al (32) found that the GPR147 mRNA expression
can be detected in the ovaries and uterus of rats. In the human
hypothalamus, pituitary gland and in hormone cells, GPR147 mRNA can
also be detected by in situ hybridization (27,33).
Another study reported that GPR147 is expressed in normal human
ovaries and granulosa-luteinizing cells (27). The localization pattern of
RFRP-3/GPR147 in mammals therefore provides a basis for its
participation in regulating reproductive function. RFRP-3 mRNA can
be detected in the uterus of juvenile sheep (29) and sows (31), where it may exert functional
effects on the reproductive function and the development of uterus.
GPR147 is extensively expressed throughout the mammalian uterus,
suggesting that GnIH can regulate uterus function and ultimately
reproductive function.
GnIH has been shown to mediate a number of functions
on energy balance, nutrient availability and reproduction. To
ensure species continuity, successful reproduction relies on the
tightly regulated balance of hormones in the HPG axis (1). Various factors, including the
external environment, individual energy state and social
relationships, can influence the HPG axis and reproductive function
through various neuroendocrine systems. GnRH has been recognized to
be a key stimulus for normal reproduction in animals. This is
achieved by stimulating gonadotropin secretion, enhancing gonadal
development, promoting the synthesis of sex steroid hormones and
facilitating gamete production (4-6).
By contrast, GnIH serves an important role in suppressing
reproduction in the central nervous system. It can act directly on
both hypothalamic GnRH and pituitary neurons through GPR147 to
inhibit the production of gonadotropins, such as FSH and LH, and
suppress reproduction (33,34).
Johnson et al (36)
previously revealed that administering GnIH in male rats through
the ICV route can suppress sexual behaviors. In addition, Piekarski
et al (37) reported that
the ICV administration of GnIH can decrease the extent of vaginal
scent marking and sexual motivation in female hamsters.
Administering GnIH has also been shown to change Fos expression
levels in the medial amygdala, medial preoptic area, critical
neural loci and the stria terminalis bed nucleus, which are
associated with female sexual behaviors (37). These aforementioned findings
suggest that GnIH is heavily involved in modulating reproductive
and feeding behaviors. Qi et al (38) found that GnIH neurons can project
onto NPY, POMC and melanin-concentrating hormone neurons to
regulate the feeding behavior (38). Consistent with this observation,
administering GnIH through the ICV route can elevate food
consumption in rats (36) and
sheep (39). In addition, ICV
injection of RFRPs has been found to promote the release of
adrenocorticotropic hormone and oxytocin, which induces anxious
behavior in rats (40). These
findings suggest that GnIH can act on neurons in the brain to
regulate various physiological functions. RFRP-3 is a HPG axis
inhibitor that has been previously demonstrated to exert various
regulatory functions on mammalian reproduction (1). However, information on RFRP-3 remains
limited and the molecular mechanism of RFRP-3 in the female
reproductive organs remain to be fully elucidated.
The uterus is an important reproductive organ in
females. In particular, the uterine epithelium can be modulated by
steroids to synthesize and/or secrete specific proteins into the
uterine fluid, which serve important roles in reproductive
processes, such as fertilization, embryo implantation and pregnancy
maintenance (41,42). One method of studying the
reproductive process is by analyzing the protein components
contained within the uterine fluid and their function. To explore
the role of GnIH on protein synthesis in the uterus and in
reproduction, the DEPs in the uterine fluid samples were screened
using LC-MS/MS. Compared with the those in OEP rats without
GnIH/RFRP-3 injection, GnIH/RFRP-3 injection through the lateral
ventricle exerted an important effect on the protein composition
and expression in the uterine fluid of OEP rats, suggesting that
GnIH/RFRP-3 can act on the uterus by inhibiting the HPG axis. To
explore the mechanism by which GnIH/RFRP-3 regulates uterus
function on a molecular level, bioinformatics analysis was
performed to analyze the role of DEPs. According to GO analysis,
these DEPs were mostly enriched in ‘extracellular region’ and
‘membrane-bounded vesicle’ (CC), ‘protein binding’, ‘protein
complex binding’ and ‘macromolecular complex binding’ (MF). In
terms of BP, these DEPs mainly participated in ‘response to organic
substance’, ‘regulation of biological quality’ and ‘response to
oxygen-containing compound’. These results suggest that GnIH/RFRP-3
treatment can affect protein binding. In addition, these DEPs were
found to participate in protein, polypeptide and carbohydrate
metabolism. Furthermore, these proteins showed association with
antioxidation and were indicated to regulate cell proliferation,
apoptosis, cell migration, cell adhesion and angiogenesis.
According to PPI network analysis, the DEPs may participate in ‘gap
junction’ and ‘regulation of actin cytoskeleton’. In addition,
these DEPs are likely to be involved in
‘glycolysis/gluconeogenesis’, ‘carbon metabolism’, ‘biosynthesis of
amino acids’, ‘long-term depression’, ‘thyroid hormone synthesis’
and ‘platelet activation’. Glycolysis, carbon metabolism and amino
acid biosynthesis serve to provide energy for cells. After
GnIH/RFRP-3 treatment, glycolysis becomes the main method of energy
metabolism in cells, suggesting a close relationship with
tumorigenesis (43,44). In addition, glycolysis has been
shown to sperm maturation, motility and ultimately male
reproduction by providing lactic acid or ATP (45,46).
In addition, glycolysis can significantly affect the nuclear
maturation in oocytes (47),
suggesting that GnIH/RFRP-3 can regulate the development of uterus
through the glycolytic pathway. Platelet activation is beneficial
to tumor growth, angiogenesis and metastasis (48), suggesting that increased
GnIH/RFRP-3 can affect the occurrence and development of tumors,
which may have various clinical implications. GnIH/RFRP-3 treatment
was found to affect the thyroid hormone synthesis pathway. Thyroid
hormone can regulate metabolism and development in the ovary,
uterus and placenta (49). It has
been previously found that the GnIH mRNA expression is increased in
the hypothalamus region of female hypothyroid mice (50), suggesting that GnIH/RFRP-3
treatment can regulate the development of uterus and female
reproduction by affecting the synthesis of thyroid hormone.
In total, five hub proteins, namely Gna13, Gnaq,
Gnai3, Kras and MMP9, were obtained based on PPI network analysis
in the present study. The Ras family of genes, Hras, Kras and Nras
represent the most commonly mutated oncogenes in human malignancies
(51). Kras mutations have been
associated with poor prognosis and treatment resistance in tumors
(51). Accumulating evidence has
shown that the Kras mutation is associated with cell proliferation
and apoptosis, which upregulates estrogen receptor levels in
endometrial cells (51-53).
Furthermore, Kras appears to be directly associated with type I EC
(51-53).
In the present study, GnIH/RFRP-3 treatment reduced the expression
levels of Kras. Therefore, it can be hypothesized that GnIH/RFRP-3
can downregulate the expression levels of Kras, thereby affecting
the uterus and the occurrence/development of EC through the HPG
axis. These five hub proteins were also associated with the
platelet activation signaling pathway. It can be seen from the PPI
network that the DEPs Gna 13, Gnaq and Gnai3 participate in the
platelet activation pathway. When searching ‘platelet activation’
in the KEGG pathway database, the results showed that the DEPs were
mostly associated with the ‘platelet activation pathway’, the
activation of which affects the activity of adenylyl cyclase
(AC)/cAMP/protein kinase A (PKA) signaling. Previous findings have
shown that kisspeptin is co-expressed with GnIH/RFRP-3 within the
neurons in the DMH area of Sprague-Dawley rats, where the direct
interaction between recombinant RFRP-3 and kisspeptin was found
using surface plasmon resonance (54). Therefore, GnIH/RFRP-3 may inhibit
the expression of kisspeptin by inhibiting the AC/cAMP/PKA
signaling pathway, further inhibiting the HPG reproductive axis to
the uterus. GnIH has been shown to suppress AC/cAMP/PKA signal
transduction (28,55,56).
In addition, it has been found that there is an association between
kisspeptin and ERK and cAMP/PKA pathway activation (57,58).
The formation and development of EC is a complex process that
involves interactions among multiple signal transduction pathways
and genetic abnormalities (51-53).
Therefore, it is necessary to perform further experiments to
determine the effects of GnIH/RFRP-3 on EC. In the present study,
to verify if the effects of RFRP-3 on the viability and apoptosis
of HEC-1A cells were mediated by the hub proteins, further
experiments were performed.
Using the UALCAN database to assess the expression
profiles of the hub proteins, the expression of Gna13, Gnaq, Kras
and MMP9 was found to be significantly changed in EC compared with
that in normal samples. Gna13 is a member of the G protein G12
subfamily and is highly expressed in breast and prostate cancer
(59). In addition, it has been
previously associated with tumor metastasis and progression
(60). Muralidharan et al
(61) found the expression of
Gna13 in EC tissues to be increased compared with that in normal
tissues. However, the opposite result was obtained in UCLUAN
database analysis. Thus, the role of Gna13 in EC remains
controversial (60), which
warrants further study. By contrast, Padol et al (62) found a significant decrease in Gnaq
protein expression in the uterus of hypercholesterolemic mice
(62), a condition that has been
found to indirectly increase the risk of EC (63). Gnai3 is expressed to varying
degrees in the mammalian uterus, the decreased expression of which
has been indicated to result in the abnormal asymmetric division of
stem cells in vivo (64).
The disturbance of this balance not only affects normal development
and growth but can also lead to cancer-associated cell
hyperproliferation (64-67).
MMP9 is associated with tumor growth, apoptosis, angiogenesis and
metastasis; it has been reported to be overexpressed in EC, where
it is also associated with disease progression (68-71).
The Kras gene is one of the most frequently mutated oncogenes in
cancer, which is regularly associated with poor prognosis and
treatment resistance (51).
Previous studies (52,53) have suggested that Kras mutations
are associated with cell proliferation and apoptosis, thereby
upregulating the expression of estrogen receptors in endometrial
cells. In addition, Kras appears to be directly associated with
type I EC (52,53).
By contrast, Gnaq and MMP9 may be associated with
proliferation in EC (62,68-71),
which is inconsistent with the results of CCK-8 assay in the
present study, where GnIH reduced HEC-1A cell viability. Indeed,
RFRP-3 at 10,000 ng/ml inhibited the viability of HEC-1A cells.
Compared with that in the control group, the apoptotic rate in
RFRP-3 group was increased, whereas the expression of Bcl-2 was
decreased and that of Bax was increased. Kras was selected for
further study, since its protein expression levels were increased
in the tumor compared with normal tissues in TCGA database
analysis, whereas it was decreased in OEP rat uterine fluid after
the ICV injection of RFRP-3.
Western blotting revealed that RFRP-3 at 10,000
ng/ml inhibited Kras expression. RFRP-3 reduced the expression of
components in the PI3K/AKT/mTOR pathway in both mRNA and protein
levels in HEC-1A cells. It has been shown that hyperactivation of
the PI3K/AKT/mTOR pathway is associated with the pathogenesis of
EC, whereas inhibition of the PI3K/AKT/mTOR pathway has important
therapeutic significance (72).
Therefore, RFRP-3 may inhibit the viability of HEC-1A EC cells
whilst promoting apoptosis by inhibiting the PI3K/AKT/mTOR pathway.
mTOR serves an important role in regulating autophagy in tumor
cells (73). As in the present
study RFRP-3 was found to decrease mTOR, the expression of
autophagy markers LC3-II and p62 were detected (74). The results showed that 10,000 ng/ml
RFRP-3 increased the ratio of LC3-II/ I whilst decreasing the
expression of p62, suggesting enhanced autophagy activity (74). We hypothesize that the inhibitory
effects of RFRP-3 on HEC-1A cells may be mediated by activating the
GPR147 receptor, which promotes the hub protein Kras to activate
the downstream PI3K/AKT/mTOR pathway, thereby reducing the
expression of Bcl-2, whilst promoting the expression of Bax and
inducing autophagy.
There are a number of limitations in the present
study. For the validation of the proteomics analysis, several of
the proteins identified via LC-MS/MS analysis should have been
confirmed by western blot analysis. In addition, although
GnIH/RFRP-3 may inhibit the immediate peripheral effects on
vertebrate gonads, the specificity and sensitivity of GnIH/RFRP-3
in tissues and cells needs to be further verified. To enhance the
consistency between in vivo and in vitro experiments,
primary human endometrial epithelial cell lines should be used
instead of EC cells. The cell cycle progression,
immunohistochemistry, LC3 puncta and autophagic flux should also be
measured.
In summary, the present study showed that RFRP-3
affects the uterus by upregulating MMP9 whilst downregulating
Gna13, Gnaq, Gnai3 and Kras expression. In addition, RFRP-3
inhibited the viability but promoted the apoptosis of HEC-1A cells.
These results can encourage the further exploration into the
underlying molecular mechanism of the effects of GnIH/RFRP-3 on the
uterus and provide novel ideas for the treatment of patients with
EC.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the National Natural
Science Foundation of China (grant nos. 81441133 and 81703001),
Natural Science Foundation of Hebei Province (grant no.
H2013406115), the Plan Project of Hebei Provincial Science and
Technology Department (grant no. 08276101D-20), Hebei Higher
Education Research Project (grant no. QN2015121) and Key Discipline
of College and Universities in Hebei Province [grant. no. (2013)04;
title, Pathogenic Biology].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request. The proteomics datasets generated and/or analyzed during
the current study are available in the iProX database (https://www.iprox.cn/; Project ID, IPX0005261000;
https://www.iprox.cn//page/project.html?id=IPX0005261000).
Authors' contributions
LC and SY directed and designed the study. XZ wrote
the manuscript. XZ, MW, LN, FW, LS and XL performed the
experiments. XZ and LS confirm the authenticity of all the raw
data. ZC and YQ analyzed the bioactive compounds using
high-performance liquid chromatography. All authors have read and
approved the final manuscript.
Ethics approval and consent to
participate
All experiments involving animals were approved by
The Experimental Animal Welfare Ethics Committee of Chengde Medical
University (Chengde, China; approval no. CDMULAC-201808-003).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang X, Guo GL, Zhang X, Li M, Xiao K, Hu
C and Li X: Effect of RFRP-3, the mammalian ortholog of GnIH, on
the epididymis of male rats. Theriogenology. 118:196–202.
2018.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Tsutsui K: A new key neurohormone
controlling reproduction, gonadotropin inhibitory hormone (GnIH):
Biosynthesis, mode of action and functional significance. Prog
Neurobiol. 88:76–88. 2009.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Kriegsfeld LJ, Mei DF, Bentley GE, Ubuka
T, Mason AO, Inoue K, Ukena K, Tsutsui K and Silver R:
Identification and characterization of a gonadotropin-inhibitory
system in the brains of mammals. Proc Natl Acad Sci USA.
103:2410–2415. 2006.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Tsutsui K, Bentley GE, Bedecarrats G,
Osugi T, Ubuka T and Kriegsfeld LJ: Gonadotropin-inhibitory hormone
(GnIH) and its control of central and peripheral reproductive
function. Front Neuroendocrinol. 31:284–295. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Kriegsfeld LJ, Jennings KJ, Bentley GE and
Tsutsui K: Gonadotrophin-inhibitory hormone and its mammalian
orthologue RFamide-related peptide-3: Discovery and functional
implications for reproduction and stress. Neuroendocrinol.
30(e12597)2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Tsutsui K, Ubuka T, Son YL, Bentley GE and
Kriegsfeld LJ: Contribution of GnIH research to the progress of
reproductive neuroendocrinology. Front Endocrinol (Lausanne).
6(179)2015.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Han X, He Y, Zeng G, Wang Y, Sun W, Liu J,
Sun Y and Yu J: Intracerebroventricular injection of RFRP-3 delays
puberty onset and stimulates growth hormone secretion in female
rats. Reprod Biol Endocrinol. 15(35)2017.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Chen L, Si LN, Wei M, Guo S, Yang SH, Chen
ZH, Cheng LY and Qiao YB: Effects of GnIH on hypothalamic POMC
positive neurons in ovariectomized rats supplemented with estrogen.
Chin J Neuroanatomy. 35:57–63. 2019.(In Chinese).
|
|
9
|
Su W, Wei M, Feng C, Yang ZJ, Chen XY,
Cheng LY, Yang SH and Qiao YB: Changes of neuropeptide Y mRNA and
protein expression in hypothalamus of rats after
intracerebroventricular injection of GnIH. Shandong Med J.
56:27–28. 2016.(In Chinese).
|
|
10
|
Wei M, Si LN, Chen L, Yang SH, Cheng LY
and Qiao YB: Regulating effects of GnIH on POMC in hypothalamus of
OEP rats. J Chengde Med College. 34:458–460. 2017.(In Chinese).
|
|
11
|
Chen WP, Witkin JW and Silverman AJ:
Beta-Endorphin and gonadotropin-releasing hormone synaptic input to
gonadotropin-releasing hormone neurosecretory cells in the male
rat. J Comp Neurol. 286:85–95. 1989.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Fetissov SO, Kopp J and Hokfelt T:
Distribution of NPY receptors in the hypothalamus. Neuropeptides.
38:175–188. 2004.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Morelli A, Marini M, Mancina R, Luconi M,
Vignozzi L, Fibbi B, Filippi S, Pezzatini A, Forti G, Vannelli GB
and Maggi M: Sex steroids and leptin regulate the ‘first Kiss’
(KiSS 1/G-protein-coupled receptor 54 system) in human
gonadotropin-releasing-hormone-secreting neuroblasts. J Sex Med.
5:1097–1113. 2008.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Braun MM, Overbeek-Wager EA and Grumbo RJ:
Diagnosis and management of endometrial cancer. Am Fam Physician.
93:468–474. 2016.PubMed/NCBI
|
|
15
|
Gompel A: Progesterone and endometrial
cancer. Best Pract Res Clin Obstet Gynaecol. 69:95–107.
2020.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Corzo C, Barrientos Santillan N, Westin SN
and Ramirez PT: Updates on conservative management of endometrial
cancer. J Minim Invasive Gynecol. 25:308–313. 2018.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Emons G and Gründker C: The role of
gonadotropin-releasing hormone (GnRH) in endometrial cancer. Cells.
10(292)2021.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Grundker C, Gunthert AR, Westphalen S and
Emons G: Biology of the gonadotropin-releasing hormone system in
gynecological cancers. Eur J Endocrinol. 146:1–14. 2002.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Emons G, Grundker C, Gunthert AR,
Westphalen S, Kavanagh J and Verschraegen C: GnRH antagonists in
the treatment of gynecological and breast cancers. Endocr Relat
Cancer. 10:291–299. 2003.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Fister S, Günthert AR, Aicher B, Paulini
KW, Emons G and Gründker C: GnRH-II antagonists induce apoptosis in
human endometrial, ovarian, and breast cancer cells via activation
of stress-induced MAPKs p38 and JNK and proapoptotic protein Bax.
Cancer Res. 69:6473–6481. 2009.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Ukena K and Tsutsui K: A new member of the
hypothalamic RF-amide peptide family, LPXRF-amide peptides:
Structure, localization and function. Mass Spectrom Rev.
24:469–486. 2005.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Tsutsui K and Ukena K: Hypothalamic
LPXRF-amide peptides in vertebrates: Identification, localization
and hypophysiotropic activity. Peptides. 27:1121–1129.
2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Tsutsui K: How to contribute to the
progress of neuroendocrinology: New insights from discovering novel
neuropeptides and neurosteroids regulating pituitary and brain
functions. Gen Comp Endocrinol. 227:3–15. 2016.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Tsutsui K and Ubuka T: GnIH control of
feeding and reproductive behaviors. Front Endocrinol (Lausanne).
7(170)2016.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Ukena K, Iwakoshi E, Minakata H and
Tsutsui K: A novel rat hypothalamic RFamide-related peptide
identified by immunoaffinity chromatography and mass spectrometry.
FFBS Lett. 512:255–258. 2002.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Ubuka T, Morgan K, Pawson AJ, Osugi T,
Chowdhury VS, Minakata H, Tsutsui K, Millar RP and Bentley GE:
Identification of human GnIH homologs, RFRP-1 and RFRP-3, and the
cognate receptor, GPR147 in the human hypothalamic pituitary axis.
PLoS One. 4(e8400)2009.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Ubuka T, Inoue K, Fukuda Y, Mizuno T,
Ukena K, Kriegsfeld LJ and Tsutsui K: Identification, expression,
and physiological functions of Siberian hamster
gonadotropin-inhibitory hormone. Endocrinology. 153:373–385.
2012.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Li H: The expression of RFRP-3 in lamb
tissue uses and its role between nutrition and reproduction
(unpublished PhD thesis). Nanjing Agricultural University,
2014.
|
|
30
|
Liu J: The cloning mRNA expression of GnIH
and its receptors in rabbit (unpublished PhD thesis). Nanjing
Agricultural University, 2011.
|
|
31
|
Li X, Su J, Lei Z, Zhao Y, Jin M, Fang R,
Zheng L and Jiao Y: Gonadotropin-inhibitory hormone (GnIH) and its
receptor in the female pig: cDNA cloning, expression in tissues and
expression pattern in the reproductive axis during the estrous
cycle. Peptides. 36:176–185. 2012.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Hinuma S, Shintani Y, Fukusumi S, Iijima
N, Matsumoto Y, Hosoya M, Fujii R, Watanabe T, Kikuchi K, Terao Y,
et al: New neuropeptides containing carboxy-terminal RFamide and
their receptor in mammals. Nat Cell Biol. 2:703–708.
2000.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Ubuka T, Son YL, Bentley GE, Millar RP and
Tsutsui K: Gonadotropin-inhibitory hormone (GnIH), GnIH receptor
and cell signaling. Gen Comp Endocrinol. 190:10–17. 2013.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Bonini JA, Jones KA, Adham N, Forray C,
Artymyshyn R, Durkin MM, Smith KE, Tamm JA, Boteju LW, Lakhlani PP,
et al: Identification and characterization of two G protein-coupled
receptors for neuropeptide FF. J Biol Chem. 275:39324–39331.
2000.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Son YL, Ubuka T, Millar RP, Kanasaki H and
Tsutsui K: Gonadotropin-inhibitory hormone inhibits GnRH-induced
gonadotropin subunit gene transcriptions by inhibiting
AC/cAMP/PKA-dependent ERK pathway in LβT2 cells. Endocrinology.
153:2332–2343. 2012.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Johnson MA, Tsutsui K and Fraley GS: Rat
RFamide-related peptide-3 stimulates GH secretion, inhibits LH
secretion, and has variable effects on sex behavior in the adult
male rat. Horm Behav. 51:171–180. 2007.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Piekarski DJ, Zhao S, Jennings KJ, Iwasa
T, Legan SJ, Mikkelsen JD, Tsutsui K and Kriegsfeld LJ:
Gonadotropin-inhibitory hormone reduces sexual motivation but not
lordosis behavior in female Syrian hamsters (Mesocricetus auratus).
Horm Behav. 64:501–510. 2013.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Qi Y, Oldfield BJ and Clarke IJ:
Projections of RFamide-related peptide-3 neurons in the ovine
hypothalamus, with special reference to rigions regulating energy
balance and reproduction. J Neuroendocrinol. 21:690–697.
2009.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Clarke IJ, Smith JT, Henry BA, Oldfield
BJ, Stefanidis A, Millar RP, Sari IP, Chng K, Fabre-Nys C, Caraty
A, et al: Gonadotropin-inhibitory hormone is a hypothalamic peptide
that provides a molecular switch between reproduction and feeding.
Neuroendocrinology. 95:305–316. 2012.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Kaewwongse M, Takayanagi Y and Onaka T:
Effects of RFamide-related Peptide (RFRP)-1 and RFRP-3 on oxytocin
release and anxiety-related behaviour in rats. J Neuroendocrinol.
23:20–27. 2011.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Lee RS, Wheeler TT and Peterson AJ:
Large-format, two-dimensional polyacrylamide gel electrophoresis of
ovine periimplantation uterine luminal fluid proteins:
Identification of aldose reductase, cytoplasmic actin, and
transferrin as conceptus-synthesized proteins. Biol Reprod.
59:743–752. 1998.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Cheng LY, Yang SH, Sun TC, Wang XQ, Hu W,
Qi XW and Yu HM: Identification of an unknown uterine estrogen
reactive protein ULF-250. Chin J Biochem Mol Biology. 25:60–64.
2009.(In Chinese).
|
|
43
|
Fuller GG and Kim JK: Compartmentalization
and metabolic regulation of glycolysis. J Cell Sci.
134(jcs258469)2021.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Vettore L, Westbrook RL and Tennant DA:
New aspects of amino acid metabolism in cancer. Br J Cancer.
122:150–156. 2020.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Narisawa S, Hecht NB, Goldberg E,
Boatright KM, Reed JC and Millán JL: Testis-specific cytochrome
c-null mice produce functional sperm but undergo early testicular
atrophy. Mol Cell Biol. 22:5554–5562. 2002.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Boussouar F and Benahmed M: Lactate and
energy metabolism in male germ cells. Trends Endocrinol Metab.
15:345–350. 2004.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Alvarez GM, Casiró S, Gutnisky C, Alvarez
G, Dalvit GC, Sutton-McDowall ML, Thompson JG and Cetica PD:
Implications of glycolytic and pentose phosphate pathways on the
oxidative status and active mitochondria of the porcine oocyte
during IVM. Theriogenology. 86:2096–2106. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Ren J, He J, Zhang H, Xia Y, Hu Z,
Loughran P, Billiar T, Huang H and Tsung A: Platelet TLR4-ERK5 axis
facilitates NET-Mediated capturing of circulating tumor cells and
distant metastasis after surgical stress. Cancer Res. 81:2373–2385.
2021.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Silva JF, Ocarino NM and Serakides R:
Thyroid hormones and female reproduction. Biol Reprod. 99:907–921.
2018.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Haraguchi S, Hara S, Ubuka T, Mita M and
Tsutsui K: Possible role of pineal allopregnanolone in Purkinje
cell survival. Proc Natl Acad Sci USA. 109:21110–21115.
2012.PubMed/NCBI View Article : Google Scholar
|
|
51
|
Sideris M, Emin EI, Abdullah Z, Hanrahan
J, Stefatou KM, Sevas V, Emin E, Hollingworth T, Odejinmi F,
Papagrigoriadis S, et al: The Role of KRAS in endometrial cancer: A
mini-review. Anticancer Res. 39:533–539. 2019.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Bulsa M and Urasińska E: Triple negative
endometrial cancer. Ginekol Pol. 88:212–214. 2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Vitale SG, Valenti G, Gulino FA, Cignini P
and Biondi A: Surgical treatment of high stage endometrial cancer:
Current perspectives. Updates Surg. 68:149–154. 2016.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Cheng L, Yang S, Si L, Wei M, Guo S, Chen
Z, Wang S and Qiao Y: Direct effect of RFRP-3 microinjection into
the lateral ventricle on the hypothalamic kisspeptin neurons in
ovariectomized estrogen-primed rats. Exp Ther Med.
23(24)2022.PubMed/NCBI View Article : Google Scholar
|
|
55
|
Son YL, Ubuka T and Tsutsu K: Molecular
mechanisms of gonadotropin-inhibitory Hormone(GnIH) actions in
target cells and regulation of GnIH expression. Front Endocrinol
(Lausanne). 10(110)2019.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Son YL, Ubuka T, Soga T, Yamamoto K,
Bentley GE and Tsutsui K: Inhibitory action of
gonadotropin-inhibitory hormone on the signaling pathways induced
by kisspeptin and vasoactive intestinal polypeptide in GnRH
neuronal cell line,GT1-7. FASEB J. 30:2198–2210. 2016.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Sukhbaatar U, Kanasaki H, Mijiddorj T,
Oride A and Miyazaki K: Kisspeptin induces expression of
gonadotropin-releasing hormone receptor in GnRH-producing GT1-7
cells overexpressing G protein-coupled receptor 54. Gen Comp
Endocrinol. 194:94–101. 2013.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Hara T, Kanasaki H, Tumurbaatar T, Oride
A, Okada H and Kyo S: Role of Kisspeptin and Kiss1R in the
regulation of prolactin gene expression in aat somatolactotroph GH3
cells. Endocrine. 63:101–111. 2019.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Rasheed SAK, Teo CR, Beillard EJ,
Voorhoeve PM and Casey PJ: MicroRNA-182 and microRNA-200a control
G-protein subunit α-13(GNA13) expression and cell invasion
synergistically in prostate cancer cells. J Biol Chem.
288:7986–7995. 2013.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Muhammad S, Tang Q, Wei L, Zhang Q, Wang
G, Muhammad BU, Kaur K, Kamchedalova T, Gang Z, Jiang Z, et al:
miRNA-30d serves a critical function in colorectal cancer
initiation, progression and invasion via directly targeting the
GNA13 gene. Exp Ther Med. 17:260–272. 2019.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Jayaraman M, Radhakrishnan R, Mathews CA,
Yan M, Husain S, Moxley KM, Song YS and Dhanasekaran DN:
Identification of novel diagnostic and prognostic miRNA signatures
in endometrial cancer. Genes Cancer. 8:566–576. 2017.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Padol AR, Sukumaran VS, Sadam A, Kesavan
M, Arunvikram K, Verma AD, Srivastava V, Panigrahi M, Singh TU,
Telang AG, et al: Hypercholesterolemia impairs oxytocin-induced
uterine contractility in late pregnant mouse. Reproduction.
53:565–576. 2017.PubMed/NCBI View Article : Google Scholar
|
|
63
|
Staff S, Aaltonen M, Huhtala H,
Pylvänäinen K, Mecklin JP and Mäenpää J: Endometrial Cancer risk
factors among lynch syndrome women: A retrospective cohort study.
Br J Cancer. 115:375–381. 2016.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Martin-Belmonte F and Perez-Moreno M:
Epithelial cell polarity, stem cells and cancer. Nat Rev Cancer.
12:23–38. 2012.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Knoblich JA: Asymmetric cell division:
Recent developments and their implications for tumour biology. Nat
Rev Mol Cell Biol. 11:849–860. 2010.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Gonzalez C: Spindle orientation,
asymmetric division and tumour suppression in Drosophila stem
cells. Nat Rev Genet. 8:462–472. 2007.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Williams ES, Ratliff AL, Postiglione MP,
Knoblich JA and Fuchs E: Par3-mInsc and Gαi3 cooperate to promote
oriented epidermal cell divisions through LGN. Nat Cell Biol.
16:758–769. 2014.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Karahan N, Güney M, Baspinar S, Oral B,
Kapucuoglu N and Mungan T: Expression of gelatinase (MMP-2 and
MMP-9) and cyclooxygenase-2(COX-2) in endometrial cancer. Eur J
Gynaecol Oncol. 28:184–188. 2007.PubMed/NCBI
|
|
69
|
Iurlaro M, Loverro G, Vacca A, Cormio G,
Ribatti D, Minischetti M, Ria R, Bruno M and Selvaggi L:
Angiogenesis extent and expression of matrix metalloproteinase-2
and -9 correlate with upgrading and myometrial invasion in
endometrial cancer. Eur J Clin Invest. 29:793–801. 1999.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Graesslin O, Cortez A, Fauvet R, Lorenzato
M, Birembaut P and Daraï E: Metalloproteinase-2, -7 and -9 and
tissue inhibitor of metalloproteinase-1 and -2 expression in
normal, hyperplastic and neoplastic endometrium: A
clinical-pathological correlation study. Ann Oncol. 17:637–645.
2006.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Srdelić Mihalj S, Kuzmić-Prusac I,
Zekić-Tomaš S, Šamija-Projić I and Čapkun V: Lipocalin-2 and matrix
metalloproteinase-9 expression in high-grade endometrial cancer and
their prognostic value. Histopathology. 67:206–215. 2015.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Barra F, Evangelisti G, Ferro Desideri L,
Di Domenico S, Ferraioli D, Vellone VG, De Cian F and Ferrero S:
Investigational PI3K/AKT/mTOR inhibitors in development for
endometrial cancer. Expert Opin Investig Drugs. 28:131–142.
2019.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Wullschleger S, Loewith R and Hall MN: TOR
signaling in growth and metabolism. Cell. 124:471–484.
2006.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Vishnupriya S, Priya Dharshini LC,
Sakthivel KM and Rasmi RR: Autophagy markers as mediators of lung
injury-implication for therapeutic intervention. Life Sci.
260(118308)2020.PubMed/NCBI View Article : Google Scholar
|