Introduction
Acute lung injury (ALI) is an acute progressive
hypoxic respiratory failure caused by external and external
pathogenic factors other than cardiogenesis. ALI is a syndrome of
increased lung inflammation and permeability (1). On one hand, LPS stimulation increases
the permeability of human pulmonary microvascular endothelial cells
(HPMVECs) and causes protein exudation and edema. On the other
hand, lipopolysaccharide (LPS) stimulates endothelial cells to
secrete inflammatory cytokines and other adhesion molecules, which
induces aggregation of a large number of neutrophils and leads to
oxidative damage and the inflammatory response in lung tissue,
promoting the development of lung injury (2-4).
TNF-α, IL-1β and IL-6 are the primary pro-inflammatory factors in
the early development of ALI, which not only directly cause lung
injury, but also activate other signaling pathways and promote
expression of inflammatory factors, leading to lung injury
(5).
Inactive rhomboid-like protein 2 (iRHOM2) is a
member of the rhomboid protein family, of which >10 members have
been identified (6). iRHOM2
promotes atherosclerosis by inducing macrophage inflammation and
oxidative stress; interfering with iRHOM2 decreases obesity by
increasing thermogenesis (7,8). In
addition, iRHOM2 is involved in promoting particulate matter
2.5-induced chronic kidney injury and promotes lupus nephritis via
TNFα and EGFR signaling (9).
iRHOM2 silencing decreases LPS-induced inflammatory release of
cardiomyocytes (10). A previous
study demonstrated that iRHOM2 alleviates lung injury caused by
intestinal ischemia-reperfusion (11). Therefore, the present study aimed
to explore if iRHOM2 is involved in lung microvascular endothelial
cell injury.
Materials and methods
Bioinformatics analysis. Data
source
The National Center for Biotechnology
Information-Gene Expression Omnibus database (ncbi.nlm.nih.gov/geo/) is a free database of
microarray/gene profile and next-generation sequencing data from
which gene expression profile data (accession no. GSE5883) of human
lung microvascular endothelial cells exposed to LPS were obtained
in the present study.
Identification of differentially expressed genes
(DEGs). DEGs were identified between the LPS 4 hrs and CTL 4
hrs groups in the aforementioned dataset using the ‘limma’ R
package as previously described (12). Statistically significant DEGs were
determined with adjusted P-value (adj.P-value)<0.01 as the
cut-off criterion. The analysis results were presented using a
volcano plot and heatmap.
Biological function analysis. Using Gene
Ontology (GO) enrichment analysis (http://geneontology.org/docs/go-enrichment-analysis/),
the biological function, pathway or cell location of enriched genes
was identified. The GO annotation contains three aspects of
biological content: i) Biological process (BP); ii) cellular
component (CC) and iii) molecular function (MF). By analyzing the
genes in the Kyoto Encyclopedia of Genes and Genomes (KEGG,
https://www.kegg.jp/) signaling pathways, the
pathways that were altered under disease conditions were
identified. The present study used R package ‘Org.Hs.eg.db’
(13) to perform GO functional
annotation and KEGG pathway analysis of differential expressed
immune-related genes (DEIRGs).
Construction of the protein-protein interaction
(PPI) network. Search Tool for the Retrieval of Interacting
Genes/Proteins (STRING) (14)
online database was used to develop the PPI network. iRHOM2 and
C-X3-C motif chemokine ligand 1 (CX3CL1) were uploaded to the
STRING database and isolated nodes were removed from the
network.
Cell culture
Immortalized HPMVECs were purchased from
Sigma-Aldrich (cat. no. 540-05A; Merck KGaA) and cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with
1% penicillin-streptomycin and 10% fetal bovine serum (HyClone;
Cytiva) at 37˚C with 5% CO2. The cells were treated with
LPS (10 µg/ml) for 12, 24 or 48 h at 37˚C.
Cell transfection
For knockdown of iRHOM2, the specific small
interfering (si)RNA targeting iRHOM2 (siRNA-iRHOM2#1,
5'-GCGUGAGAUGGUUGGUUAAGG-3'; siRNA-iRHOM2#2,
5'-GACGAUGUCUCAUGGAUUAAA-3') and corresponding negative control
(NC) siRNA (5'-UUCUCCGAACGUGUCACGU-3') were synthesized by Shanghai
GenePharma Co., Ltd. To overexpress CX3CL1, pc-DNA3.1 vector
containing the whole length of CX3CL1 (Ov-CX3CL1) and empty vector
(Ov-NC) were synthesized by Shanghai GeneChem Co., Ltd. A total of
100 nM recombinants were transfected into HPMVECs for 48 h at 37˚C
using Lipofectamine® 2000 reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. After 48 h, cells were collected for further
assay.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was used to detect mRNA expression levels.
Total RNAs were extracted from 1x104 HPMVECs using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. All primers were
designed and synthesized by Shanghai Sangong Pharmaceutical Co.,
Ltd. RT of first-strand cDNAs was performed by using PrimeScript™
RT Master Mix (Perfect Real Time; Takara Bio, Inc.), according to
the manufacturer's instructions. Amplification of cDNA was
performed using qPCR using the SYBR Premix Ex Taq™ II kit (Takara
Bio, Inc.). The following thermocycling conditions were used for
qPCR: Initial denaturation at 95˚C for 2 min, followed by 40 cycles
of denaturation at 95˚C for 15 sec, amplification at 53˚C for 20
sec and extension at 60˚C for 3 sec. The following primer pairs
were used: iRHOM2: Forward, 5'-TGGCCTGGAGCTGTCTATCT-3' and reverse,
5'-GTGATGGAGAGGTTGGGTGG-3'; TNFα: Forward,
5'-CTGGGCAGGTCTACTTTGGG-3' and reverse, 5'-CTGGAGGCCCCAGTTTGAAT-3';
IL-1β: Forward, 5'-AACCTCTTCGAGGCACAAGG-3' and reverse,
5'-AGATTCGTAGCTGGATGCCG-3'; IL-6: Forward,
5'-TCCACAAGCGCCTTCGGTC-3' and reverse, 5'-GGTCAGGGGTGGTTATTGCAT-3';
CX3CL1: Forward, 5'-TCCGATATCTCTGTCGTGGC-3' and reverse,
5'-TGTCTCGTCTCCAAGCAGC-3' and GAPDH: Forward,
5'-GGGAAACTGTGGCGTGAT-3' and reverse, 5'-GAGTGGGTGTCGCTGTTGA-3'.
GAPDH was used as an internal reference and relative expression
levels of target mRNAs were quantified using the 2-ΔΔCq
method (15).
Western blotting
Total protein of HPMVECs was extracted using RIPA
buffer (Hunan Auragene Biotech Co., Ltd) and quantified using the
BCA Protein Assay Kit (Beijing Dingguo Changsheng Biotechnology
Co., Ltd.). The protein (20 µg per lane) in cell lysates was
separated by 8% SDS-PAGE (Bio-Rad Laboratories, Inc.) and
transferred onto PVDF membranes (MilliporeSigma; Merck KGaA).
Subsequently, membranes were blocked in 5% non-fat milk for 1.5 h
at room temperature and incubated with primary antibodies against
iRHOM2 (1:1,000; cat. no. MAB10048; R&D Systems China Co.,
Ltd.), TNFα (1:1,000; cat. no. ab183218; Abcam), IL-1β (1:1,000;
cat. no. ab216995; Abcam), IL-6 (1:1,000; cat. no. ab233706;
Abcam), phosphorylated p65 (p-p65; 1:1,000; cat. no. ab76302;
Abcam), p65 (1:1,000; cat. no. ab32536; Abcam), Bax (1:1,000; cat.
no. ab32503; Abcam), Bcl-2 (1:1,000; cat. no. ab32124; Abcam),
Cleaved caspase 3 (1:500; cat. no. ab32042; Abcam), zonula
occludens-1 (ZO-1; 1:1,000; cat. no. ab221547; Abcam),
vascular-endothelial (VE)-cadherin (1:1,000; cat. no. ab205336;
Abcam), occludin (1:1,000, cat. no. ab242202; Abcam), CX3CL1
(1:1,000, cat. no. ab25088; Abcam) or GAPDH (1:1,000, cat. no.
ab8245; Abcam) overnight at 4˚C. Following primary antibody
incubation, membranes were incubated with HRP-conjugated anti-mouse
(1:2,000; cat. no. ab6789; Abcam) or anti-rabbit (1:2,000; cat. no.
ab6721; Abcam) secondary antibodies at 37˚C for 2 h. Then, protein
bands were visualized using ECL Detection Reagent (Shanghai Yeasen
Biotechnology Co., Ltd.) and densitometry analysis was performed
using ImageJ software (version 1.49; National Institutes of
Health).
Cell Counting Kit-8 (CCK-8) assay
HPMVECs were added to 96-well plates at a density of
5x103 cells/well. The cells were placed in an incubator
at 37˚C with 5% CO2 for 24 h. Subsequently, 20 µl CCK-8
reagent (Beyotime Institute of Biotechnology) was added to each
well. After 4 h, the absorbance at 450 nm was measured using a
microplate reader.
TUNEL assay
TUNEL assay was used to detect apoptotic cells using
a TUNEL kit (cat. no. C1082; Beyotime Institute of Biotechnology),
according to the manufacturer's protocol. HPMVECs were fixed with
4% paraformaldehyde at room temperature for 15 min and permeated
with 0.1% Triton X-100 for 5 min at room temperature. Subsequently,
cells were incubated with TUNEL at 37˚C for 1 h and cell nuclei
were counterstained with DAPI (10 mg/ml; Koritai Biotechnology) for
5 min at room temperature. A total of five fields of view were
randomly selected from each slice. The number of positive cells was
mounted with fluorescent mounting media (Beijing Solarbio Science
& Technology Co., Ltd.) and the observation was conducted under
a fluorescence microscope (Nikon Corporation). The cell apoptosis
rate (%) was calculated as follows: Number of apoptotic positive
cells/total number of cells.
Measurement of endothelial barrier
permeability
Transendothelial electrical resistance (TEER) is a
key indicator for the detection of the integrative function of the
monolayer cell barrier. It reflects the change in monolayer cell
permeability by detecting the voltage difference between the inside
and outside of the cell (16).
TEER of HPMVECs was detected using a Millicdl ESR-2 cell resistance
instrument (Millipore). TEER was determined using the following
formula: TEER=(TEERA-TEERblank) x
Smembrane, where A is the TEER value for each Transwell
filter and Smembrane is the area of the filter membrane
(S=π x r; r=6 mm).
Immunofluorescence (IF) assay
HPMVECs were fixed with 4% paraformaldehyde at 4˚C
for 15 min and permeabilized with Triton X-100 at 37˚C for 30 min.
Subsequently, cells were incubated with primary antibodies against
ZO-1 (1:100; cat. no. ab221547; Abcam) at 4˚C overnight, and then
incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG
H&L secondary antibodies (1:500; cat. no. ab150077; Abcam) for
1 h at room temperature. Nuclei were stained with 1 µg/ml DAPI
solution at room temperature for 15 min. The images were observed
under a fluorescence microscope (magnification, x100).
Immunoprecipitation (IP) assay
The association between iRHOM2 and CX3CL1 was
detected using IP assay. HPMVECs were collected and lysed using
RIPA lysis buffer (Thermo Fisher Scientific, Inc.). Antibodies
against iRHOM2 (1:500; cat. no. orb386934, Biorbyt, Ltd.), CX3CL1
(1:500; cat. no. 14-7986-81; Invitrogen; Thermo Fisher Scientific,
Inc.) and IgG (1:200; cat. no. ab205718; Abcam) were added to the
lysate for incubation at 4˚C overnight. Then, 40 µl Protein A/G
PLUS-Agarose beads (Invitrogen; Thermo Fisher Scientific, Inc.)
were added and incubated at room temperature for another 2 h. Then,
the beads were rinsed with lysis buffer for three times and the
collected by centrifugation at 12,000 x g for 2 min at 4˚C.
Following the final wash, the supernatant was aspirated and
discarded, then the precipitated proteins were re-suspended in 2 x
SDS-PAGE loading buffer, boiled for 5min, and rinsed from the
beads. The products from immunoprecipitation were collected for the
analysis of iRHOM2 and CX3CL1 expression.
Statistical analysis
GraphPad Prism 8.0 statistical software (GraphPad
Software, Inc.; Dotmatics) was used for statistical analysis. Data
are presented as the mean ± standard deviation of three independent
experiments. Differences between multiple groups were analyzed
using one-way ANOVA followed by Tukey's post hoc test. R software
version 3.6.3 was used for bioinformatics analysis (17). P<0.05 was considered to indicate
a statistically significant difference.
Results
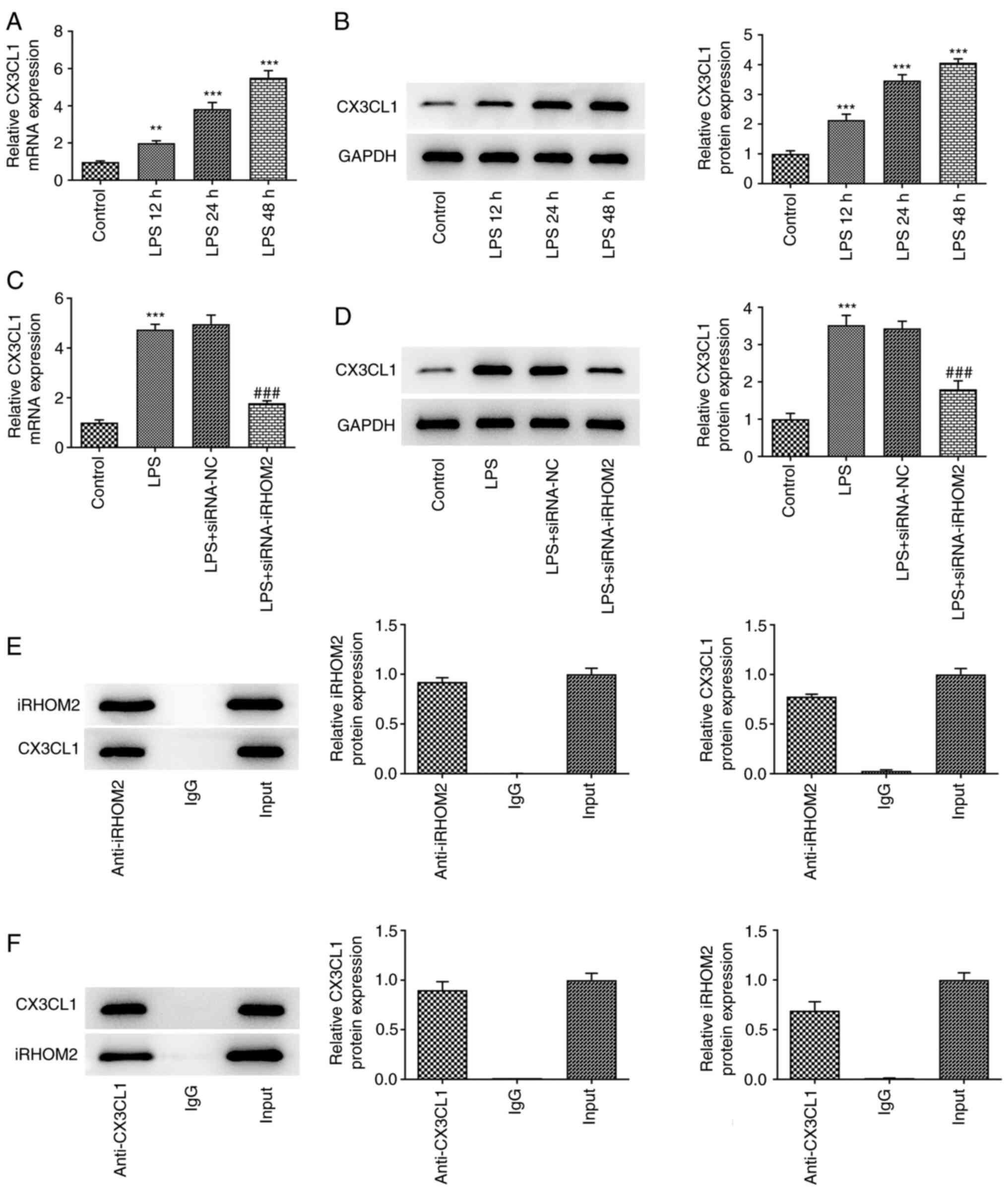
Identification of DEGs
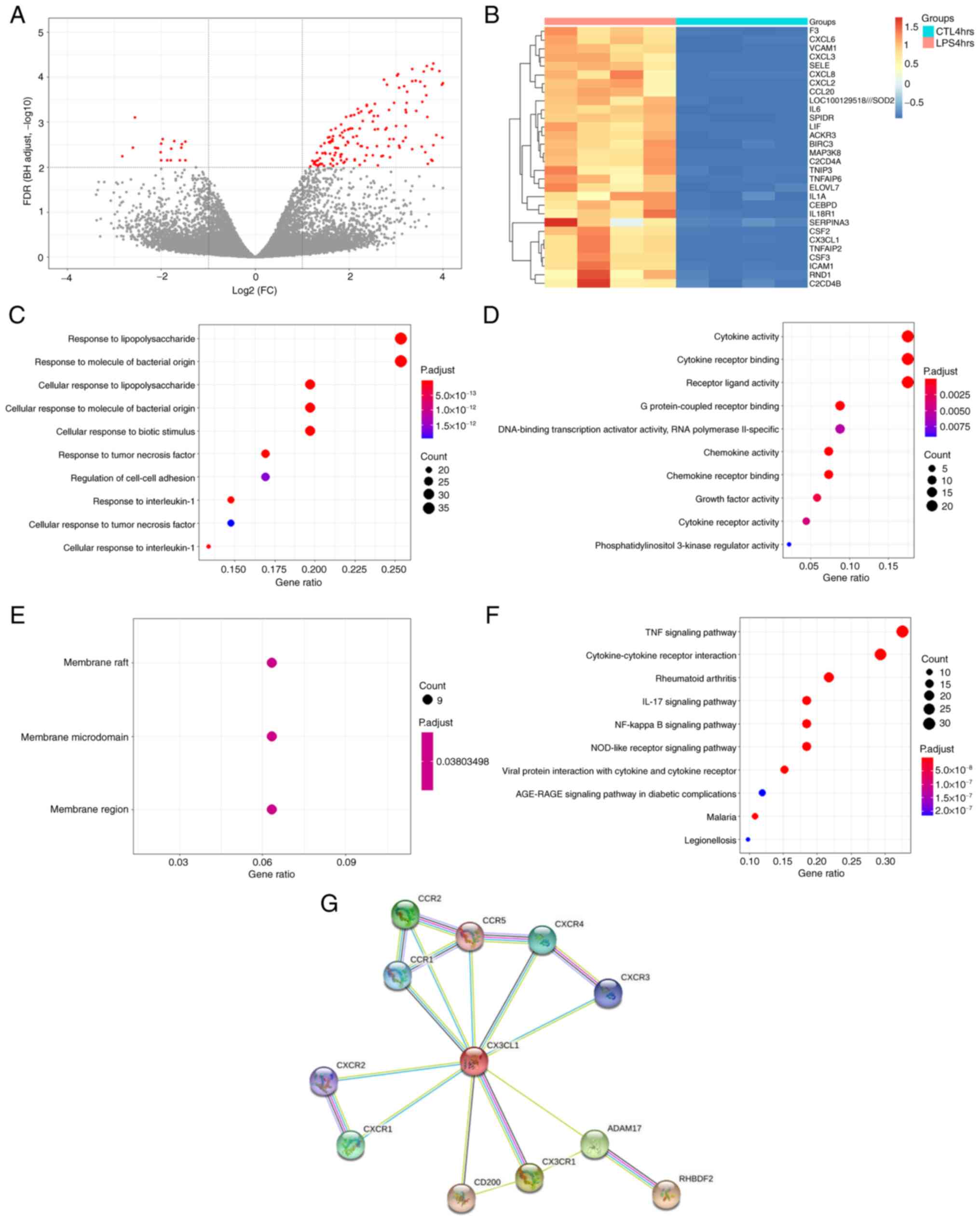
Using adj. P-value <0.01 as the cut-off
criterion, 203 DEGs were extracted from the expression profile
dataset GSE5883, including 187 up- and 16 downregulated genes in
the LPS 4 hrs group compared with the CTL 4 hrs group (Fig. 1A). A heatmap of the top 30
upregulated DEGs, which included CX3CL1, is shown in Fig. 1B. In the BP group, DEGs were mainly
enriched in ‘response to lipopolysaccharide’, ‘response to molecule
of bacterial origin’ and ‘response to tumor necrosis factor’
(Fig. 1C-F). In the CC group, DEGs
were primarily enriched in ‘membrane raft’, ‘membrane microdomain’
and ‘membrane region’. In the MF group, DEGs were primarily
enriched in ‘cytokine activity’, ‘cytokine receptor binding’ and
‘receptor ligand activity’. KEGG pathway analysis revealed that the
DEGs were primarily enriched in ‘TNF signaling pathway’,
‘cytokine-cytokine receptor interaction’, ‘Rheumatoid arthritis’,
‘IL-17 signaling pathway’ and ‘NF-kappa B signaling pathway’.
Identification of the interaction
between iRHOM2 and CX3CL1 using a PPI network
To investigate the interaction between the gene
iRHOM2 and CX3CL1, the STRING online tool was used to construct a
PPI network (Fig. 1G). There was
no direct association between iRHOM2 and CX3CL1.
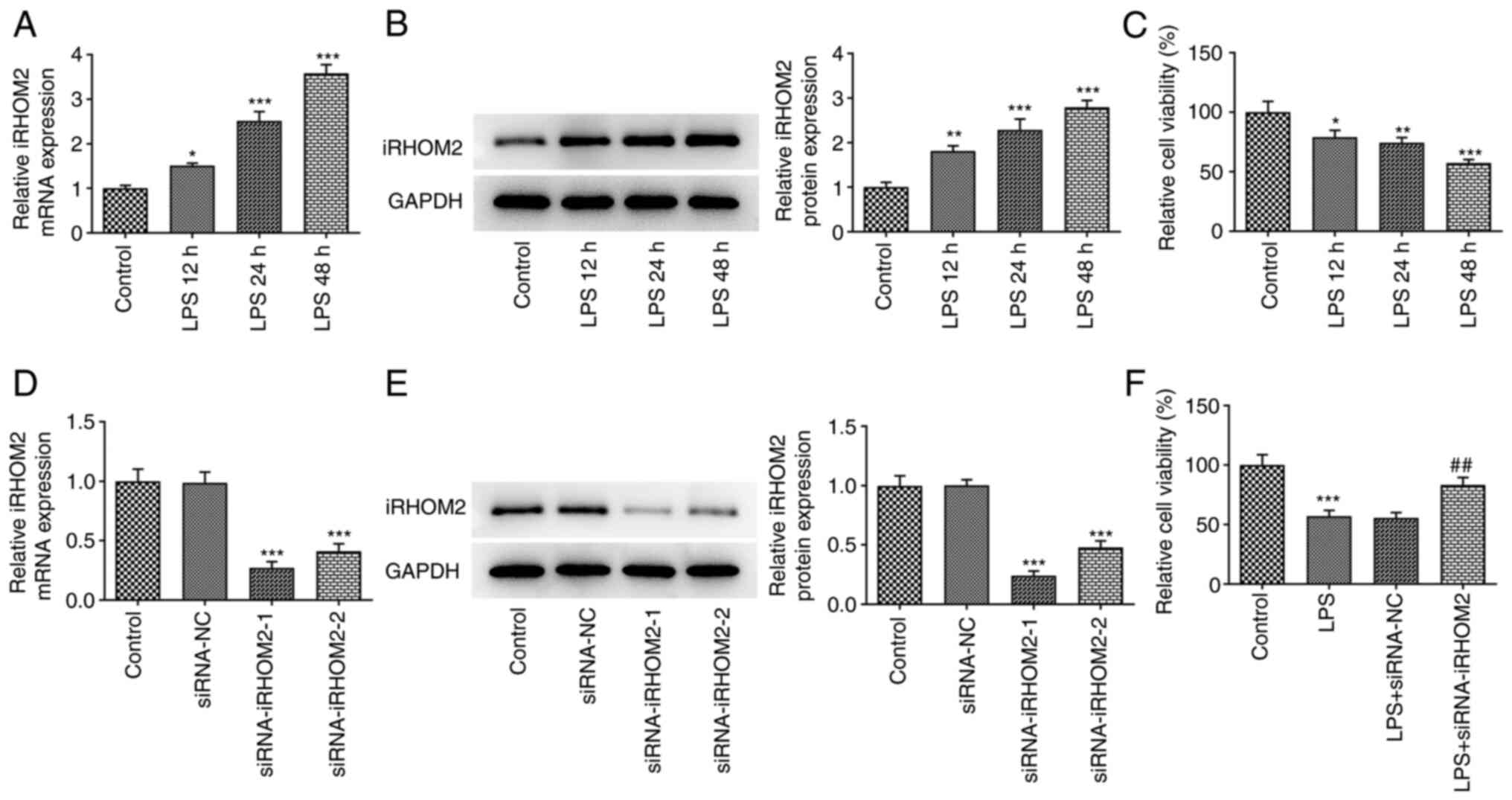
iRHOM2 silencing increases HPMVEC
viability induced by LPS
To analyze the role of iRHOM2 in ALI, HPMVECs were
treated with LPS for 12, 24 or 48 h. LPS treatment induced a
time-dependent increase in iRHOM2 expression at both mRNA and
protein levels compared with that in the control group (Fig. 2A and B). HPMVEC viability was decreased by LPS
treatment in a time-dependent manner (Fig. 2C). iRHOM2 silencing was induced in
HPMVECs using siRNA targeting iRHOM2. The mRNA and protein
expressions of iRHOM2 were decreased following iRHOM2 silencing
compared with that in the siRNA-NC group (Fig. 2D and E). HPMVEC viability following LPS
treatment was increased in the iRHOM2 silencing group compared with
that in the siRNA-NC group (Fig.
2F).
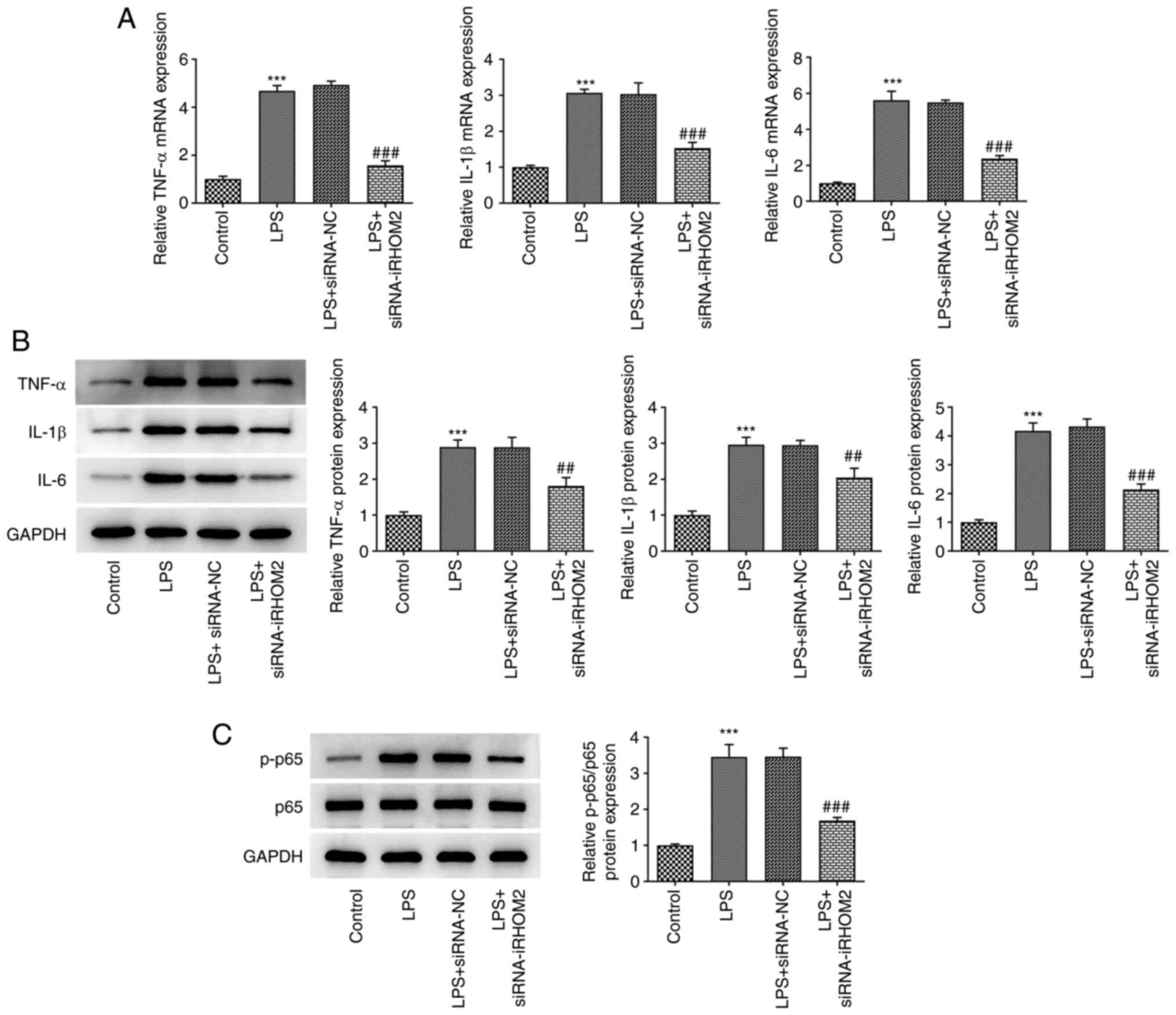
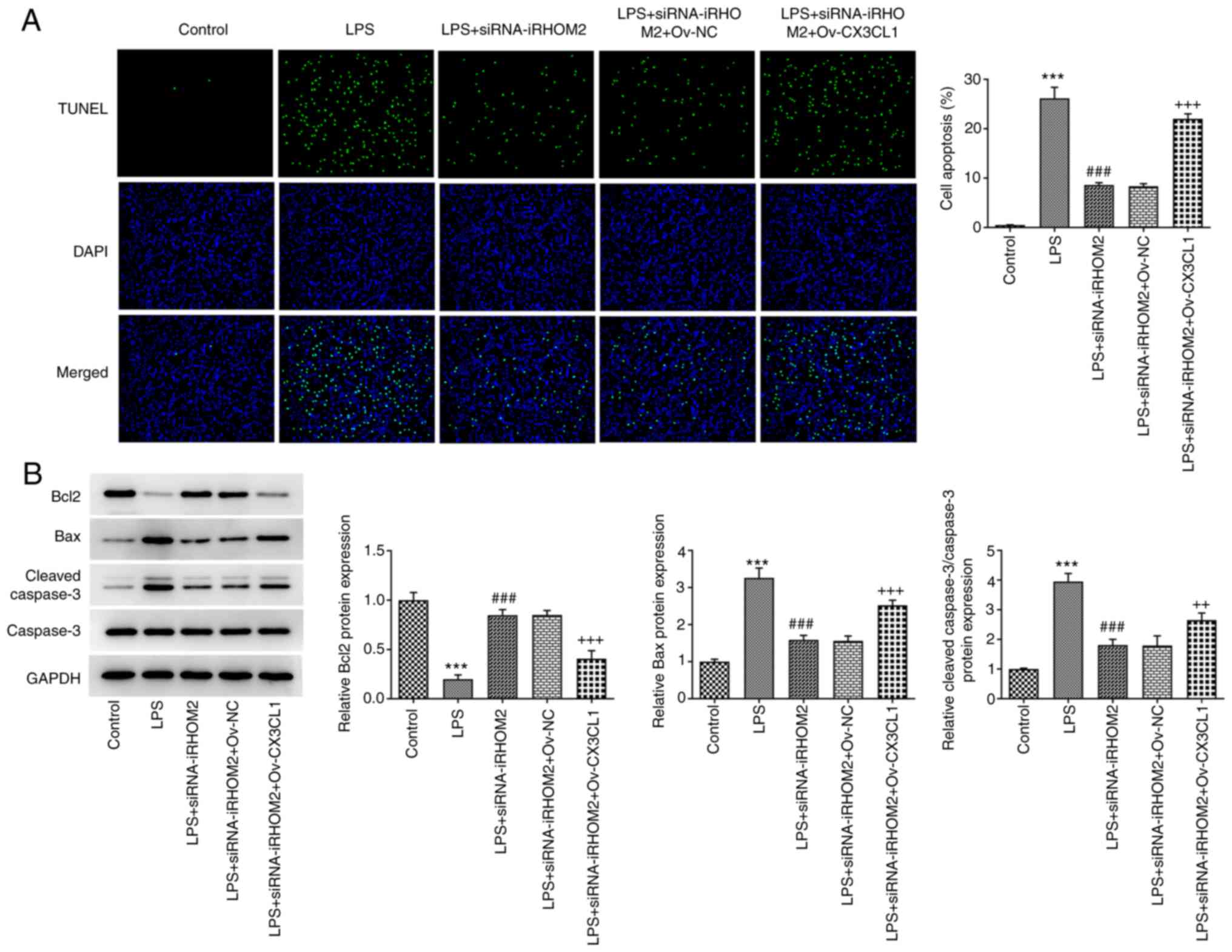
iRHOM2 silencing decreases
inflammation, apoptosis and endothelial barrier permeability in
LPS-treated HPMVECs
The present study examined if iRHOM2 serves a role
in inflammation and apoptosis associated with ALI. TNF-α, IL-1β and
IL-6 levels were measured by RT-qPCR and western blotting. iRHOM2
silencing decreased the levels of TNF-α, IL-1β and IL-6 compared
with those in the siRNA-NC group (Fig.
3A-B). As Fig. 3C depicted,
the increased expression of p-p65 in LPS group was decreased by
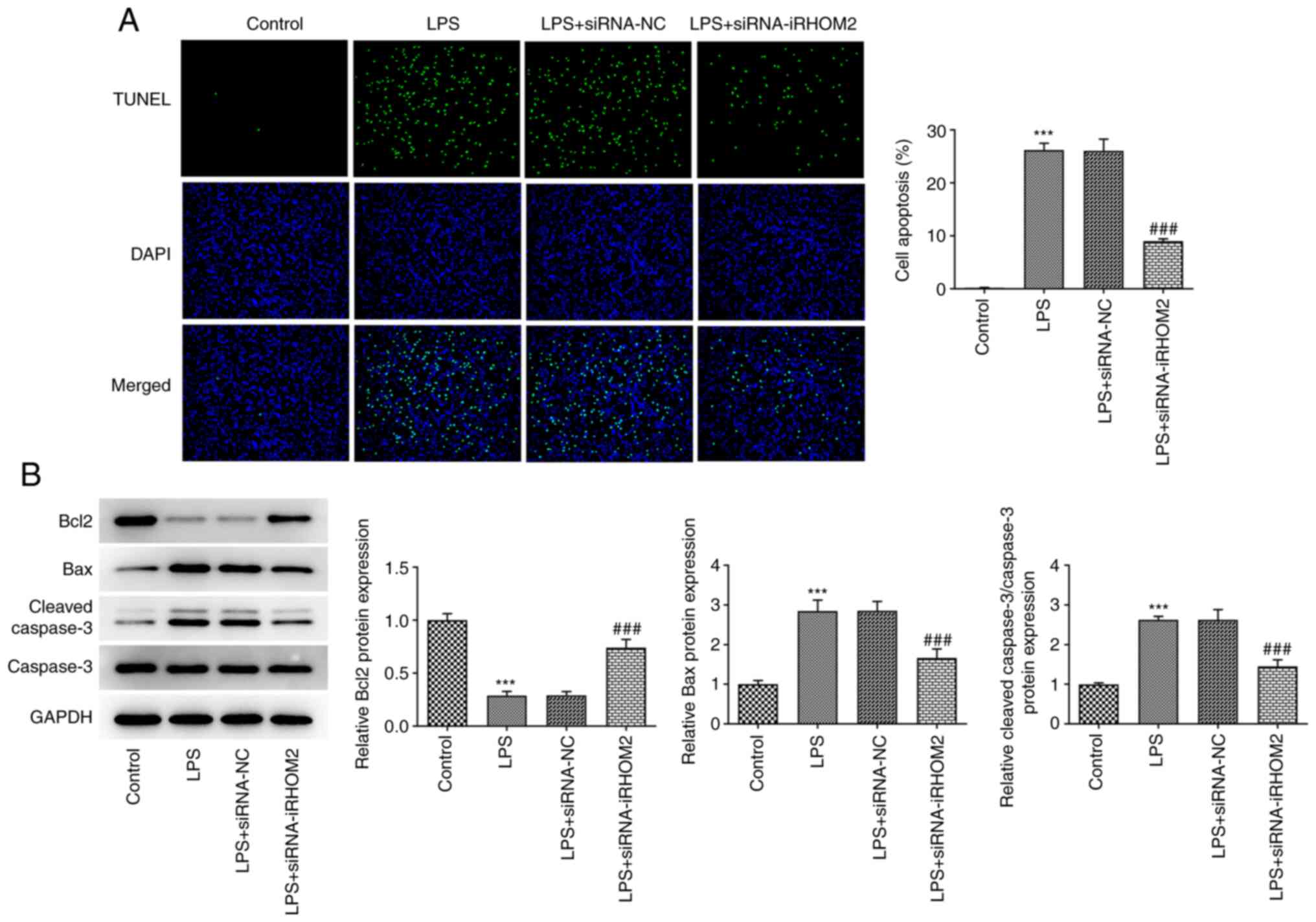
iRHOM2 silence. In addition, apoptosis levels were decreased in the
iRHOM2 silencing group compared with those in the siRNA-NC group
(Fig. 4A). Furthermore, the iRHOM2
silencing group showed increased Bcl-2 expression but decreased Bax
and cleaved caspase3 expression compared with the siRNA-NC group
(Fig. 4B). To determine the role
of iRHOM2 in endothelial barrier permeability and expression levels
of ZO-1, VE-cadherin and Occludin, endothelial permeability
measurement and an IF assay for ZO-1 expression, as well as western
blotting for the detection of tight junction proteins, were
performed. A decrease in TEER was observed following LPS induction
compared with that in the control group and this effect was
recovered by iRHOM2 silencing (Fig.
5A). Increased ZO-1 staining was observed, accompanied by the
upregulation of expression levels of ZO-1, VE-cadherin and occludin
in the LPS and siRNA-iRHOM2 co-treatment group compared with the
LPS and siRNA-NC co-treatment group (Fig. 5B and C).
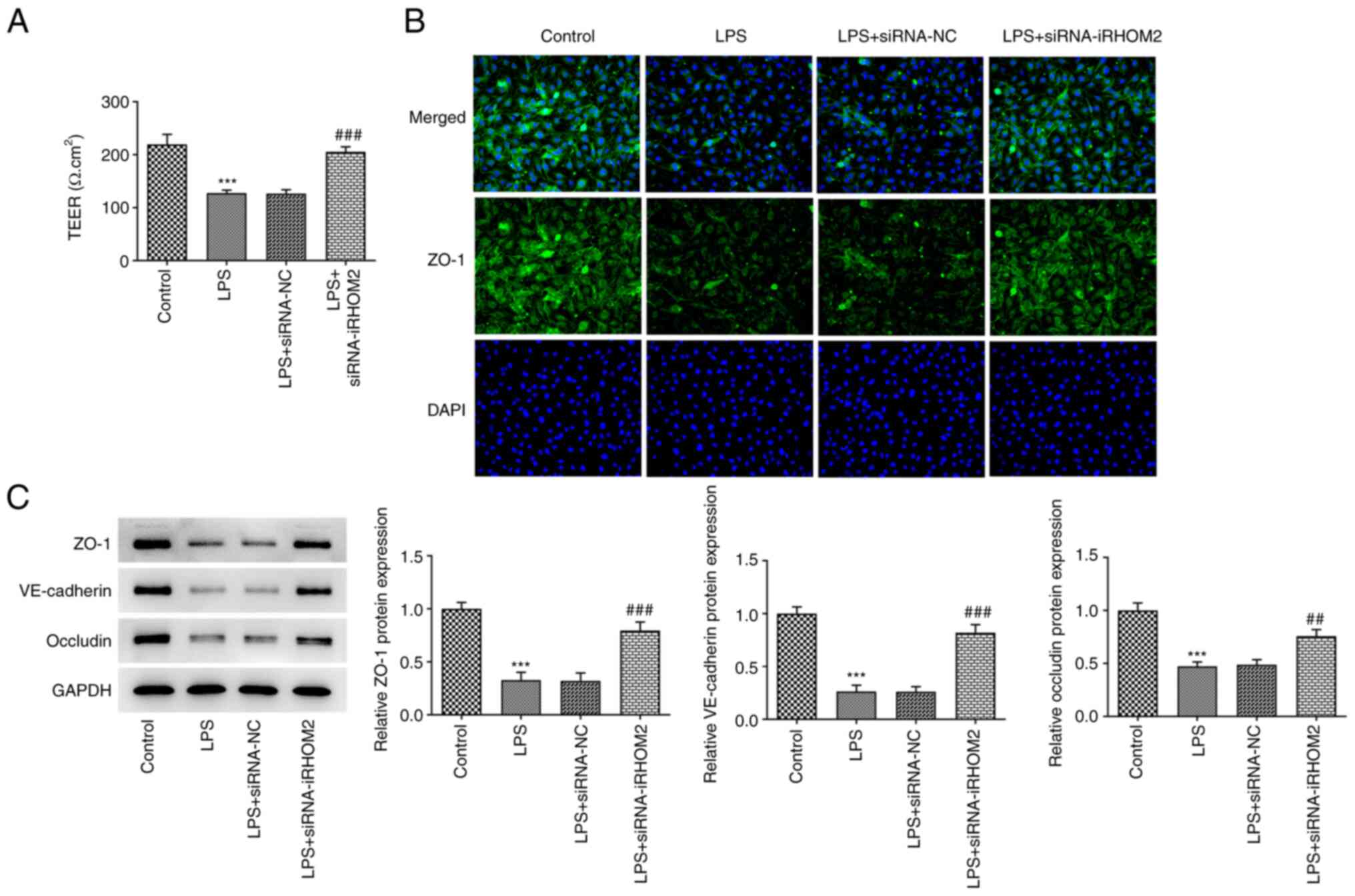 | Figure 5iRHOM2 silencing maintains endothelial
integrity. (A) Analysis of endothelial barrier permeability using
TEER measurement. (B) Immunofluorescence staining assay for ZO-1
protein expression (Magnification, x100). (C) Protein expression of
ZO-1, VE-cadherin and occludin was increased by iRHOM2 silencing
following LPS challenge. Data are presented as the mean ± standard
deviation of three independent experiments performed in triplicate.
***P<0.001 vs. control. ##P<0.01,
###P<0.001 vs. LPS + siRNA-NC. ZO-1, zonula
occludens-1; iRHOM2, inactive rhomboid-like protein 2; siRNA, small
interfering RNA; NC, negative control; LPS, lipopolysaccharide; VE,
vascular-endothelial; TEER, transendothelial electrical
resistance. |
iRHOM2 interference inhibits CX3CL1
expression in LPS-induced HPMVECs
The present study investigated if the aforementioned
effects of iRHOM2 were due to changes in CX3CL1 expression. CX3CL1
expression was measured using RT-qPCR and western blotting.
Upregulation of CX3CL1 expression at both mRNA and protein levels
was observed in HPMVECs in response to LPS treatment (Fig. 6A and B). Furthermore, iRHOM2 silencing promoted
CX3CL1 expression compared with the siRNA-NC group of LPS-treated
HPMVECs (Fig. 6C and D). Furthermore, there was an interaction
between iRHOM2 and CX3CL1 (Fig. 6E
and F).
iRHOM2 interference alters cell
viability, inflammation, apoptosis and endothelial integrity via
CX3CL1
To determine the regulatory role of iRHOM2 in cell
viability, inflammation and apoptosis, CX3CL1 overexpression was
induced by transfection of Ov-CX3CL1 into HPMVECs. Increased
expression levels of CX3CL1 were observed following Ov-CX3CL1
transfection compared with those in the control (Fig. 7A and B). The present study examined whether
CX3CL1 mediated the role of iRHOM2 in LPS-induced HPMVECs. It was
found that the viability in LPS + siRNA-iRHOM2 group was reduced
after overexpressing CX3CL1 (Fig.
7C). Compared with LPS + siRNA-iRHOM2 + Ov-NC group, CX3CL1
overexpression increased the expressions of TNF-α, IL-1β and IL-6
in LPS-induced HPMVECs transfected with siRNA-iRHOM2 (Figs. 7D-E). The decreased mRNA and
protein expressions of p-p65 in LPS-induced due to iRHOM2 silence
were increased by CX3CL1 overexpression (Fig. 7F). Compared with LPS + siRNA-iRHOM2
+ Ov-NC group, CX3CL1 overexpression promoted the cell apoptosis,
accompanied by decreased Bcl-2 expression and increased expressions
of Bax and cleaved caspase3 (Figs.
8A-B). However, this effect was reversed in the LPS,
siRNA-iRHOM2 and Ov-CX3CL1 co-treatment groups. Furthermore, CX3CL1
overexpression reversed the effect of iRHOM2 silencing on TEER
levels and expression levels of ZO-1, VE-cadherin and occludin
(Fig. 9A-C), suggesting that
iRHOM2/CX3CL1 affected endothelial integrity.
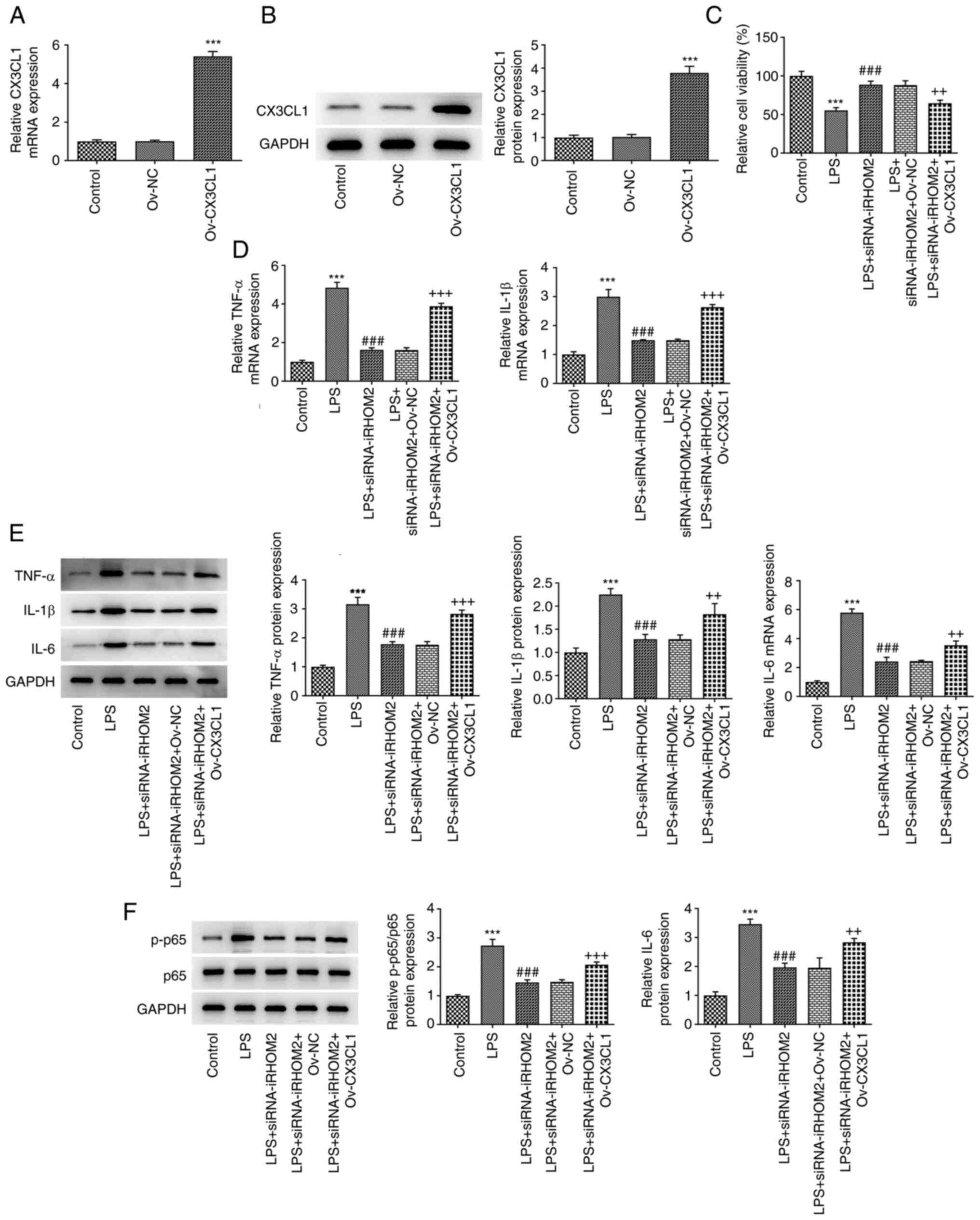 | Figure 7iRHOM2 interacts with CX3CL1 to
affect cell viability and inflammation and apoptosis in response to
LPS. (A) Reverse transcription-quantitative PCR analysis was
performed to measure mRNA expression of CX3CL1. (B) Western blot
assay was performed to measure protein levels of CX3CL1.
***P<0.001 vs. Ov-NC. (C) Cell Counting Kit-8 assay
showed cell viability. (D) RT-qPCR analysis of TNF-α, IL-1β and
IL-6 expressions. (E) Western blot analysis of TNF-α, IL-1β and
IL-6 expressions. (F) Western blot analysis of the p-p65 and p65
protein levels. Data are presented as the mean ± standard deviation
of three independent experiments performed in triplicate.
***P<0.001 vs. control. ###P<0.001 vs.
LPS. +P<0.05, ++P<0.01 and
+++P<0.01 vs. LPS + siRNA-iRHOM2 + Ov-NC. RT-qPCR,
reverse transcription-quantitative polymerase chain reaction;
CX3CL1, C-X3-C motif chemokine ligand 1; iRHOM2, inactive
rhomboid-like protein 2; siRNA, small interfering RNA; NC, negative
control; LPS, lipopolysaccharide; Ov-, overexpression; p,
phosphorylated. |
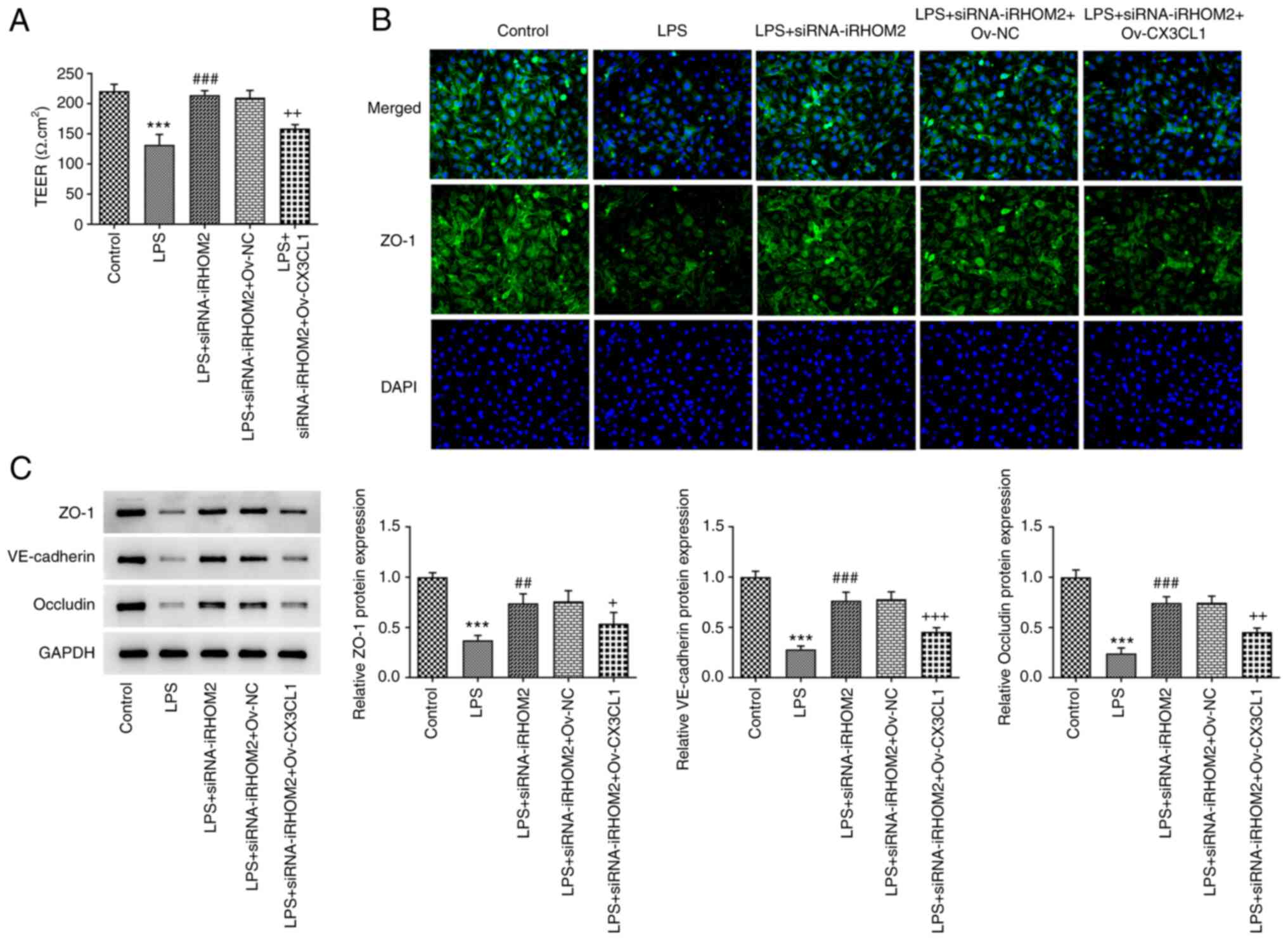 | Figure 9iRHOM2 interacts with CX3CL1 to
affect endothelial integrity in LPS-induced human pulmonary
microvascular endothelial cells. (A) Analysis of endothelial
barrier permeability using TEER measurement. (B) Immunofluorescence
staining for ZO-1 protein expression. (magnification, x100). (C)
Western blot assay was performed to analyze protein expression of
ZO-1, VE-cadherin and occludin. Data are presented as the mean ±
standard deviation of three independent experiments performed in
triplicate. ***P<0.001 vs. control.
##P<0.01 and ###P<0.001 vs. LPS.
+P<0.05, ++P<0.01 and
+++P<0.01 vs. LPS + siRNA-iRHOM2 + Ov-NC. ZO-1,
zonula occludens-1; iRHOM2, inactive rhomboid-like protein 2;
siRNA, small interfering RNA; NC, negative control; LPS,
lipopolysaccharide; VE, vascular-endothelial; TEER,
transendothelial electrical resistance. |
Discussion
CX3CL1 is a unique chemokine that can exist in a
soluble form, as a chemotactic cytokine, or in a membrane-attached
form that serves as a binding molecule (18). A previous study demonstrated that
Baicalin, a type of flavonoid, exerts protective effects against
LPS-induced ALI by regulating the CX3CL1/CX3CR1 axis and NF-κB
pathway in CX3CL1-knockout mice (19). The present bioinformatics analysis
showed that CX3CL1 expression was upregulated in the LPS group
compared with that in the control group. KEGG pathway analysis
demonstrated that the TNF signaling pathway affected by iRHOM2 and
cytokine-cytokine receptor interaction, including CX3CL1, was
involved in ALI. To the best of our knowledge, however, the
specific roles of CX3CL1 in ALI and the association between CX3CL1
and iRHOM2 have not reported yet. iRHOM2 is a key cofactor for
TNF-α-converting enzyme, the metalloprotease that removes both the
proinflammatory cytokine TNF-α and TNF receptors (TNFRs) from the
cell surface (20). PPI network
analysis revealed no direct association between iRHOM2 and CX3CL1.
However, the present study showed that iRHOM2 could interact with
CX3CL1 by IP assay. The present study demonstrated that, following
LPS induction and iRHOM2 silencing, HPMVECs exhibited enhanced cell
viability, decreased pro-inflammatory factor expression and
apoptosis levels and improved endothelial barrier permeability
compared with HPMVECs only treated with LPS. However, CX3CL1
overexpression counteracted the effects of iRHOM2 silencing on
LPS-induced HPMVECs. Given the interaction of iRHOM2 and CX3CL1,
LPS treatment led to inflammation, apoptosis and the alteration of
endothelial barrier permeability that were potentially associated
with iRHOM2 upregulation and CX3CL1 downregulation. The present
study used the total form of VE-cadherin to evaluate VE cell
permeability (21-23).
p-VE-cadherin may also be a marker of VE cell permeability and
should be detected in future studies (24,25).
The present study demonstrated that iRHOM2 silencing
led to decreased levels of TNFα, IL-1β and IL-6 in HPMVECs
stimulated with LPS, accompanied by decreased phosphorylation of
p65. Previous studies have shown that iRHOM2 participates in the
regulation of inflammation (7,26,27).
A recent report demonstrated that inhibition of iRHOM2/NF-κB
signaling is involved in suppressing inflammation (28). The present study identified an
interaction between iRHOM2 and CX3CL1 and found that CX3CL1
overexpression inhibited the protective effects of iRHOM2 silencing
against LPS-induced cell injury. In response to LPS stimulation,
endothelial cells overproduce pro-inflammatory chemokines,
including CX3CL1(29). Adult
patients with sepsis exhibit increased CX3CL1 expression, which is
positively associated with inflammatory cytokines, such as IL-6,
IL-1β and TNF-α (30). CX3CL1
blockade leads to alleviation of lung injury in cecal ligation and
puncture-induced sepsis (30).
However, the present study did not use CX3CL receptor inhibitors;
use of CX3CL receptor inhibitors to block the effects of CX3CL
should be explored in future studies. Moreover, the effects of
CX3CL on iRHOM2-regulated ALI should also be investigated in
further study. In addition, to the best of our knowledge, the
majority of LPS-associated experiments use 12-48 h for LPS
treatment to treat cells (31-33).
Thus, the present study used 12, 24 and 48 h as LPS treatment
duration. Moreover, the present study found that iRHOM2 regulated
CX3CL1 expression and iRHOM2 silencing affected inflammation and
endothelial barrier permeability via CX3CL1 but did not test the
expression of iRHOM2 in CX3CL1-overexpressing cells. Therefore,
further studies on the association between iRHOM2 and CX3CL1 and
the role of iRHOM2 in CX3CL1-regulated cells should be performed in
future.
In conclusion, the present data indicated that
targeting iRHOM2/CXCL1 therapeutically may ameliorate the
inflammation and improve endothelial barrier permeability. Further
study of the mechanism of iRHOM2/CXCL1 will determine the
regulatory role of these molecules in inflammation and endothelial
barrier permeability.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HLY and HYY designed the study and drafted and
revised the manuscript. HLY, JSW and HYY analyzed the data,
searched the literature and performed experiments. HLY and HYY
confirm the authenticity of all the raw data All authors have read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Schmidt GA: Managing Acute Lung Injury.
Clin Chest Med. 37:647–658. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Lu Z, Li Y, Ru JH, Lopes-Virella MF, Lyons
TJ and Huang Y: Interaction of palmitate and LPS regulates cytokine
expression and apoptosis through sphingolipids in human retinal
microvascular endothelial cells. Exp Eye Res. 178:61–71.
2019.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Yang J, Ruan F and Zheng Z: Ripasudil
attenuates lipopolysaccharide (LPS)-mediated apoptosis and
inflammation in pulmonary microvascular endothelial cells via
ROCK2/eNOS Signaling. Med Sci Monit. 24:3212–3219. 2018.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Zyrianova T, Lopez B, Liao A, Gu C, Wong
L, Ottolia M, Olcese R and Schwingshackl A: BK channels regulate
LPS-induced CCL-2 release from human pulmonary endothelial cells.
Am J Respir Cell Mol Biol. 64:224–234. 2021.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Wang X, Yan J, Xu X, Duan C, Xie Z, Su Z,
Ma H, Ma H, Wei X and Du X: Puerarin prevents LPS-induced acute
lung injury via inhibiting inflammatory response. Microb Pathog.
118:170–176. 2018.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Freeman M: Rhomboid proteases and their
biological functions. Annu Rev Genet. 42:191–210. 2008.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Chaohui C, Wei H, Hongfeng W, Yueliang Z,
Xiaoqin P, Pingli Z and Zhibing A: iRhom2 promotes atherosclerosis
through macrophage inflammation and induction of oxidative stress.
Biochem Biophys Res Commun. 503:1897–1904. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Badenes M, Amin A, González-García I,
Félix I, Burbridge E, Cavadas M, Ortega FJ, de Carvalho É, Faísca
P, Carobbio S, et al: Deletion of iRhom2 protects against
diet-induced obesity by increasing thermogenesis. Mol Metab.
31:67–84. 2020.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Xu MX, Qin YT, Ge CX, Gu TT, Lou DS, Li Q,
Hu LF, Li YY, Yang WW and Tan J: Activated iRhom2 drives prolonged
PM2.5 exposure-triggered renal injury in Nrf2-defective
mice. Nanotoxicology. 12:1045–1067. 2018.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Lu XL, Zhao CH, Zhang H and Yao XL: iRhom2
is involved in lipopolysaccharide-induced cardiac injury in vivo
and in vitro through regulating inflammation response. Biomed
Pharmacother. 86:645–653. 2017.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Kim JH, Kim J, Chun J, Lee C, Im JP and
Kim JS: Role of iRhom2 in intestinal ischemia-reperfusion-mediated
acute lung injury. Sci Rep. 8(3797)2018.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43(e47)2015.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang C, Zheng Y, Li X, Hu X, Qi F and Luo
J: Genome-wide mutation profiling and related risk signature for
prognosis of papillary renal cell carcinoma. Ann Transl Med.
7(427)2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
von Mering C, Huynen M, Jaeggi D, Schmidt
S, Bork P and Snel B: STRING: A database of predicted functional
associations between proteins. Nucleic Acids Res. 31:258–261.
2003.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Srinivasan B, Kolli AR, Esch MB, Abaci HE,
Shuler ML and Hickman JJ: TEER measurement techniques for in vitro
barrier model systems. J Lab Autom. 20:107–126. 2015.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Li S, Gao P, Dai X, Ye L, Wang Z and Cheng
H: New prognostic biomarker CMTM3 in low grade glioma and its
immune infiltration. Ann Transl Med. 10(206)2022.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Liu W, Jiang L, Bian C, Liang Y, Xing R,
Yishakea M and Dong J: Role of CX3CL1 in Diseases. Arch Immunol
Ther Exp (Warsz). 64:371–383. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Ding XM, Pan L, Wang Y and Xu QZ: Baicalin
exerts protective effects against lipopolysaccharide-induced acute
lung injury by regulating the crosstalk between the CX3CL1-CX3CR1
axis and NF-κB pathway in CX3CL1-knockout mice. Int J Mol Med.
37:703–715. 2016.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Badenes M and Adrain C: iRhom2 and TNF:
Partners or enemies? Sci Signal. 12(eaaz0444)2019.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Yokota Y, Noda T, Okumura Y, Kobayashi S,
Iwagami Y, Yamada D, Tomimaru Y, Akita H, Gotoh K, Takeda Y, et al:
Serum exosomal miR-638 is a prognostic marker of HCC via
downregulation of VE-cadherin and ZO-1 of endothelial cells. Cancer
Sci. 112:1275–1288. 2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Jiang W, Sun Y, Wang H, Hu Z, Song J, Meng
C, Duan S, Jiang Z, Yu Y and Hu D: HIF-1α Enhances Vascular
Endothelial Cell Permeability Through Degradation and Translocation
of Vascular Endothelial Cadherin and Claudin-5 in Rats With Burn
Injury. J Burn Care Res. 42:258–268. 2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Gomez Perdiguero E, Liabotis-Fontugne A,
Durand M, Faye C, Ricard-Blum S, Simonutti M, Augustin S, Robb BM,
Paques M, Valenzuela DM, et al: ANGPTL4-αvβ3 interaction
counteracts hypoxia-induced vascular permeability by modulating Src
signalling downstream of vascular endothelial growth factor
receptor 2. J Pathol. 240:461–471. 2016.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Smith RO, Ninchoji T, Gordon E, André H,
Dejana E, Vestweber D, Kvanta A and Claesson-Welsh L: Vascular
permeability in retinopathy is regulated by VEGFR2 Y949 signaling
to VE-cadherin. Elife. 9(e54056)2020.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Liu J, Miao G, Wang B, Zheng N, Ma L, Chen
X, Wang G, Zhao X and Zhang L and Zhang L: Chlamydia pneumoniae
infection promotes monocyte transendothelial migration by
increasing vascular endothelial cell permeability via the tyrosine
phosphorylation of VE-cadherin. Biochem Biophys Res Commun.
497:742–748. 2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Zhou C, Chen R, Gao F, Zhang J and Lu F:
4-Hydroxyisoleucine relieves inflammation through iRhom2-dependent
pathway in co-cultured macrophages and adipocytes with LPS
stimulation. BMC Complement Med Ther. 20(373)2020.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Zhou C, Qin Y, Chen R, Gao F, Zhang J and
Lu F: Fenugreek attenuates obesity-induced inflammation and
improves insulin resistance through downregulation of iRhom2/TACE.
Life Sci. 258(118222)2020.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Chenxu G, Xianling D, Qin K, Linfeng H,
Yan S, Mingxin X, Jun T and Minxuan X: Fisetin protects against
high fat diet-induced nephropathy by inhibiting inflammation and
oxidative stress via the blockage of iRhom2/NF-κB signaling. Int
Immunopharmacol. 92(107353)2021.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Jiang R, Wei L, Zhu M, Wu J and Wang L:
Aspirin Inhibits LPS-Induced Expression of PI3K/Akt, ERK, NF-κB,
CX3CL1, and MMPs in human bronchial epithelial cells. Inflammation.
39:643–650. 2016.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Chen X, Wei Q, Hu Y and Wang C: Role of
Fractalkine in promoting inflammation in sepsis-induced multiple
organ dysfunction. Infect Genet Evol. 85(104569)2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Zhang Y, Yan M, Yu QF, Yang PF, Zhang HD,
Sun YH, Zhang ZF and Gao YF: Puerarin Prevents LPS-Induced
osteoclast formation and bone loss via inhibition of Akt
activation. Biol Pharm Bull. 39:2028–2035. 2016.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Abarca-Vargas R and Petricevich VL:
Extract from Bougainvillea xbuttiana (Variety Orange) Inhibits
Production of LPS-Induced inflammatory mediators in macrophages and
exerts a protective effect in vivo. Biomed Res Int.
2019(2034247)2019.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Suzuki K, Okada H, Takemura G, Takada C,
Kuroda A, Yano H, Zaikokuji R, Morishita K, Tomita H, Oda K, et al:
Neutrophil elastase damages the pulmonary endothelial glycocalyx in
lipopolysaccharide-induced experimental endotoxemia. Am J Pathol.
189:1526–1535. 2019.PubMed/NCBI View Article : Google Scholar
|