Introduction
Over the last decade, numerous naturally-occurring
or plant products, especially bioactive constituents of different
phytochemical classes, have been reported to be viable antiviral
agents against several pathogenic viruses, such as human
immunodeficiency virus (HIV), herpes simplex virus (HSV), influenza
virus (INV), Dengue virus (DENV), Chikungunya virus (CHIKV) and
hepatitis C virus (HCV) (1-3).
Globally, >300 million individuals have hepatitis B virus (HBV)
infection, who may progress into developing chronic diseases, like
liver cirrhosis or carcinoma. In addition, some cases may even
result in mortality (4). Although
efficacious nucleoside analog-based drugs, including the HBV
polymerase (POL) inhibitors [such as lamivudine (LAM), adefovir and
famciclovir], are available, the emergence of drug-resistance due
to mutations in HBV POL due to long-term therapy restricts their
further use (5). Alternatively, a
range of herbal formulations consisting of isolated
phytoconstituents like alkaloids, flavonoids, terpenes and
anthraquinones have been widely identified as promising anti-HBV
therapeutics with no such resistance in vitro or in
vivo (2,3).
Rhazya stricta Decne (family: Apocynaceae) is
a medicinal plant that is distributed in arid South Asian regions,
including the Arabian Peninsula (6,7).
Traditionally, the leaves, fruits and roots of R. stricta
have been used to treat various diseases, such as diabetes, fever,
sore-throat, helminthiasis, syphilis, rheumatism, inflammation,
tooth ache, chest pain, conjunctivitis and constipation (7-9).
In addition, pharmacological properties of this plant, such as
antioxidant, anticancer, antidiabetic and antihypertensive
activities, have also been experimentally validated using in
vitro and animal model systems (9). Furthermore, its total ethanolic
extracts have been previously shown to exert in vitro
anti-fungal and anti-bacterial activities (10). Its isolated alkaloids have also
been reported to have antitumor, anti-hypertensive and
anti-microbial properties in cancer cell lines and microbial
culture systems (10). Recently,
antiviral activities of the Saudi-grown R. stricta leaf
extract against INV (11) and
severe acute respiratory syndrome coronavirus 2(12) have been reported. However, to the
best of our knowledge, the potential effects of R. stricta
extracts or phytoconstituents on HBV characteristics remain to be
elucidated.
Several phytochemical studies on R. stricta
have led to the isolation of >100 alkaloids from its roots,
flavonoids from the stem, tannins and phenolic compounds from the
leaves (7-10,13,14).
Among the bioactive flavonoids, rutin, quercetin (QRC), kaempferol,
catechin and acacetin (ACT), all of which have already known
antiviral activities, have also been identified in R.
stricta leaves (14,15). However, whilst in vitro
anti-HBV efficacies of rutin, QRC, kaempferol and catechin have
already been reported by previous studies (16-21),
the anti-HBV potential of ACT remains elusive. In addition, despite
having a unique alkaloid profile (22), β-carboline alkaloids such as
1-acetyl-β-carboline have not been previously reported in R.
stricta.
Therefore, to the best of our knowledge, the present
study reports for the first time the isolation of ABC along with
ACT from R. stricta, before assessing their anti-HBV
activities.
Materials and methods
Plant material and extraction
The fresh aerial parts of Rhazya stricta
Decne (locally known as Harmal) was collected in June 2021 from the
Raudhat AI-Khafs region near Riyadh, Saudi Arabia. The plant
material was identified (voucher specimen no. RS-0721) by a
phytotaxonomist at the College of Pharmacy, King Saud University
(Riyadh, Saudi Arabia). The air-dried powder of the plant material
(~3 kg) was extracted with double-distilled water followed by
methanol (MeOH) under reflux for 24 h. Both the extracts were
filtered through Whatman's paper (no. 1). The water and methanol
(MeOH) extracts were obtained by evaporating the solvent in a
rotatory vacuum evaporator (Rotavapor R-220; BUCHI Labortechnik AG)
at ±40˚C under reduced pressure. The methanol extract
(leftover residue) was re-suspended in water and then sequentially
fractionated at room temperature (RT) with chloroform
(CHCl3) and ethyl acetate (EtOAc). Each fraction was
individually filtered and distilled using the rotary vacuum
evaporator to furnish the CHCl3-fraction (~102 g) and
EtOAc-fraction (~142 g), before being stored at 4˚C until further
analysis. All analytical grade solvents were procured from
Sigma-Aldrich; Merck KGaA.
Isolation and chemical
characterization
The EtOAc-fraction was subjected to column
chromatography on a silica gel and eluted with a mixture of solvent
(10% MeOH in CHCl3). All eluates (25 drops/min) were
pooled together based on their thin-layer chromatography (TLC)
patterns under the mobile-phase CHCl3:MeOH (95:5; v/v),
producing two spots on the plate. The combined sub-fraction was
further subjected to column chromatography and eluted with
CHCl3 by gradually varying the percentage (1-5%) of
MeOH. The elutes (25 drops/min) showing similar TLC patterns under
mobile-phase CHCl3:MeOH (97:3, v/v) were pooled and
re-crystallized with MeOH at RT for slow evaporation for 24 h to
furnish compound 1 in its purest form (97%). The elutes obtained
under mobile-phase CHCl3:MeOH (95:5, v/v) and repeated
re-crystallization with MeOH at RT ultimately furnished compound 2
in its purest form. The NMR measurements of the compounds were
performed on Bruker Avance 700 spectrometer (Bruker Corporation)
with standard pulse sequences operating at 700 and 175 MHz for
1H and 13C NMR, respectively. Therein,
tetramethylsilane and DMSO-d6 were used as internal standard and
solvent, respectively. EIMS was performed on a Micromass Autospec
model (70 eV) spectrometer. HRESIMS spectra were obtained in the
positive-ionization mode using an LCT Premier XE Micromass
spectrometer (Waters Corporation). Silica gel 60 (230-400 mesh;
Merck KGaA) was used for column chromatography, while precoated
silica gel plates 60 GF254 (0.25 mm thickness; Merck KGaA) were
used for analytical TLC. The chemical structures were determined by
comparing their published physical and spectral data (23-25).
Cell cultures and drug
HepG2.2.15, the HBV-reporter cells (derivative of
the human hepatoma HepG2 cell line), were a kind gift from Dr
Shahid Jameel (Virology Lab. International Centre for Genetic
Engineering and Biotechnology, New Delhi, India). The cells were
maintained in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc.), supplemented with 10% heat-inactivated bovine serum (Gibco;
Thermo Fisher Scientific, Inc.), 1X penicillin-streptomycin mix
(HyClone; Cytiva) and 1X sodium pyruvate (HyClone; Cytiva) at 37˚C
under 5% CO2. The HBV POL-inhibitor drug LAM (2 µM;
Sigma-Aldrich; Merck KGaA) and the anti-HBV flavonoid QRC (12.5
µg/ml; Sigma-Aldrich; Merck KGaA) were used as standards or
positive controls, as described previously (26,27).
Cytotoxicity assay of isolated
compounds
Potential cytotoxicity of compounds 1 and 2 isolated
from R. stricta was tested on HepG.2.2.15 cells, using an
MTT assay, to determine their maximally safe concentrations (doses)
for subsequent antiviral assays. The test compounds were dissolved
in 50 µl dimethyl sulfoxide (DMSO) and re-dissolved in RPMI 1640 to
furnish the stock concentration (1 mg/ml), followed by further
reconstitution in RPMI to produce the various desired doses. DMSO
concentration did not exceed 0.1% even in the maximal dose
prepared.
Briefly, HepG2.2.15 cells were seeded
(0.5x105 cells/100 µl/well) in a 96-well culture plate
and grown overnight at 37˚C. On the next day, cells were treated
(in triplicate) with four different doses (6.25, 12.5, 25 and 50
µg/ml) of compounds 1 and 2, including vehicle or negative control
(0.1% DMSO). Following 72 h of incubation at 37˚C, cells were then
treated with an MTT reagent (10 µl/well) and incubated for 3.5 h at
RT in the dark. Upon the appearance of a purple color, the
kit-supplied detergent solution (100 µl/well) was added for cell
lysis, followed by 1 h incubation at 37˚C. The optical density (OD;
λ=570 nm) was recorded in a microplate reader and analyzed in
relation to the negative control, using non-linear regression
(Excel 10.0, Microsoft Corporation) to determine the 50% cytotoxic
concentration (CC)50 values.
Dose-dependent analysis of hepatitis B
surface antigen (HBsAg) inhibition
Compounds 1 and 2 isolated from R. stricta at
the selected non-cytotoxic concentrations (12.5, 25 and 50 µg/ml)
were first assessed to determine their optimal inhibitory dose at
day 2 (a single time point). HepG2.2.15 cells were grown in a
96-well plate (0.5x105/well) overnight at 37˚C. On the
next day, the culture media was replenished with fresh media
containing three doses (12.5, 25 and 50 µg/ml) of compounds 1 and
2, positive controls (QRC and LAM) or negative (DMSO) controls in
triplicates and incubated at 37˚C for 2 days. The culture
supernatants of each sample were then collected and quantitatively
analyzed for secreted HBsAg using the ELISA kit (Monolisa HBsAg
Ultra; cat. no. 72348; Bio-Rad Laboratories, Inc.) according to the
protocols of the manufacturer. The OD (λ=460 nm) was recorded using
microplate reader and analyzed to determine the inhibition of HBsAg
expression (%) in relation to that of the negative control (100%
expression). The inhibitory activity (%) of compounds 1 and 2 in
comparison to LAM (positive control) was presented.
Time-course analysis of HBsAg
inhibition
Following dose-dependent inhibition assay, a
time-dependent (days 1, 3 and 5) inhibition (%) analysis of HBsAg
at the selected optimal dose (25 µg/ml) of compounds 1 and 2 in
relation to the negative control was performed as aforementioned.
The inhibitory activity (%) of compounds 1 and 2 in comparison to
LAM (positive control) was presented.
Time-course analysis of hepatitis B
pre-core-antigen (HBeAg) inhibition
The selected optimal dose (25 µg/ml) of compounds 1
and 2 was subjected to a time-course (days 1, 3 and 5) quantitative
analysis of HBeAg secretion in the culture supernatants using ELISA
(HBeAg/Anti-HBe Elisa kit; cat. no. KAPG4BNE3; DiaSource
ImmunoAssays S.A.) according to the manufacturer's instructions.
The OD (λ=460 nm) was recorded using a microplate reader and
analyzed to determine the inhibition of HBeAg expression (%) in
relation to the negative control (100% expression). The inhibitory
activity (%) of compounds 1 and 2 in comparison to LAM (positive
control) was presented.
Structure-based in silico molecular
docking analysis
The structure-based interactions of HBV proteins
(wt-POL ‘YMDD’ and mut-POL ‘YIDD’) with the anti-HBV active
compounds 1 and 2 and the standard LAM were analyzed by molecular
docking in AutoDock 4.2(20).
Previously, the 3D structures of HBV wt-POL and mut-POL were
modelled using HIV-polymerase/reverse transcriptase (protein data
bank ID: 1RTD) as template (16,20).
The modeled proteins (targets) were prepared by deleting water
molecules or any attached hetero atoms, and by adding hydrogens.
Kollman charges were assigned and the protein structures were
energy-minimized using the Merck molecular force field, as
described previously (20). The 2D
structures of the test ligands [ACT and alkaloid acetyl-β-carboline
(ABC)] and the control ligand (LAM) were drawn in ChemDraw Pro 8.0
(PerkinElmer, Inc.), which were then prepared for docking by
assigning bond orders and angles, and by defining Gasteiger partial
charges. Furthermore, the ligands were energy-minimized by
Universal force field, as described previously (20).
Docking was performed inside a defined grid box in
the protein structure, which includes the key amino acid residues
in the active-site/binding-motif. For wt-POL catalytic residues
(Tyr, Met, Asp, and Asp of ‘YMDD’ motif) and mut-POL residues (Tyr,
Ile, Asp, and Asp of ‘YIDD’ motif), dimensions of the grid boxes
were adjusted to 28x28x28 Å and centered at 47x31x35 Å, with 0.375
Å spacing. The Lamarck genetic algorithm was employed for the
global search, whereas the Solis-Wets method was used for the local
search, as previously described (16,20).
A total of 2.5x106 energy calculations was computed for
each run, where a total of 10 docking runs were performed. The
following conditions were set: Population size, 150; translational
step, 0.2; quaternions, 5; and torsions, 5. The van der Waals’ and
electrostatic parameters were also calculated with using the
distance-dependent dielectric function. The docking affinity
(Kd) of each ligand for the protein targets was
estimated from its docking energy (∆G) using the following formula:
∆G=-2.303RT log10Kd, where R (1.987 cal/mol)
was the universal gas constant and T (298 K) was the temperature
(28).
Statistical analysis
All data of the analyzed samples were presented as
the mean ± SEM of three determinants (i.e., values of triplicated
samples). In a set of data, total variation was determined by
performing one-way analysis of variance (ANOVA), followed by
Dunnett's test (Excel 2010; Microsoft Corporation). P<0.05 was
considered to indicate a statistically significant difference.
Results
Isolation of bioactive compounds from
R. stricta
In total, two known compounds 1 and 2 isolated from
the EtOAc-fraction of the aerial parts of R. stricta were
identified after comparing their physical and spectral data
available in the literature (23-25).
Compound 1 was a yellow, needle-like crystallized
substance in MeOH that was soluble in DMSO, with the general
chemical formula of C16H12O5.
ESI-HRMS: m/z 285.12 (M+H)+. Its 1H-NMR
(DMSO-d6, 700 MHz) δ are as follows: 12.82 (1H, s,
5-OH), 9.76 (1H,br s, 7-OH), 7.94 (2H, d, J=9.0 Hz, H-2',6'), 7.09
(2H, d, J=8.8 Hz, H-3',5'), 5.98 (1H, s, H-3), 5.41 (1H, d, J=1.8
Hz, H-8) and 5.19 (1H, d, H-3, J=2.4 Hz, H-6). Its
13C-NMR (DMSO-d6, 125 MHz) δ were as follows: 182.6
(C-4), 165.1 (C-7), 164.2 (C-2), 163.2 (C-4'), 162.3 (C-5), 156.2
(C-9), 127.2 (C2',6'), 121.6 (C-1'), 115.3 (C-3',5'), 102.9 (C-10),
102.6 (C-3), 98.6 (C-6) and 92.0 (C-8). Based on the available
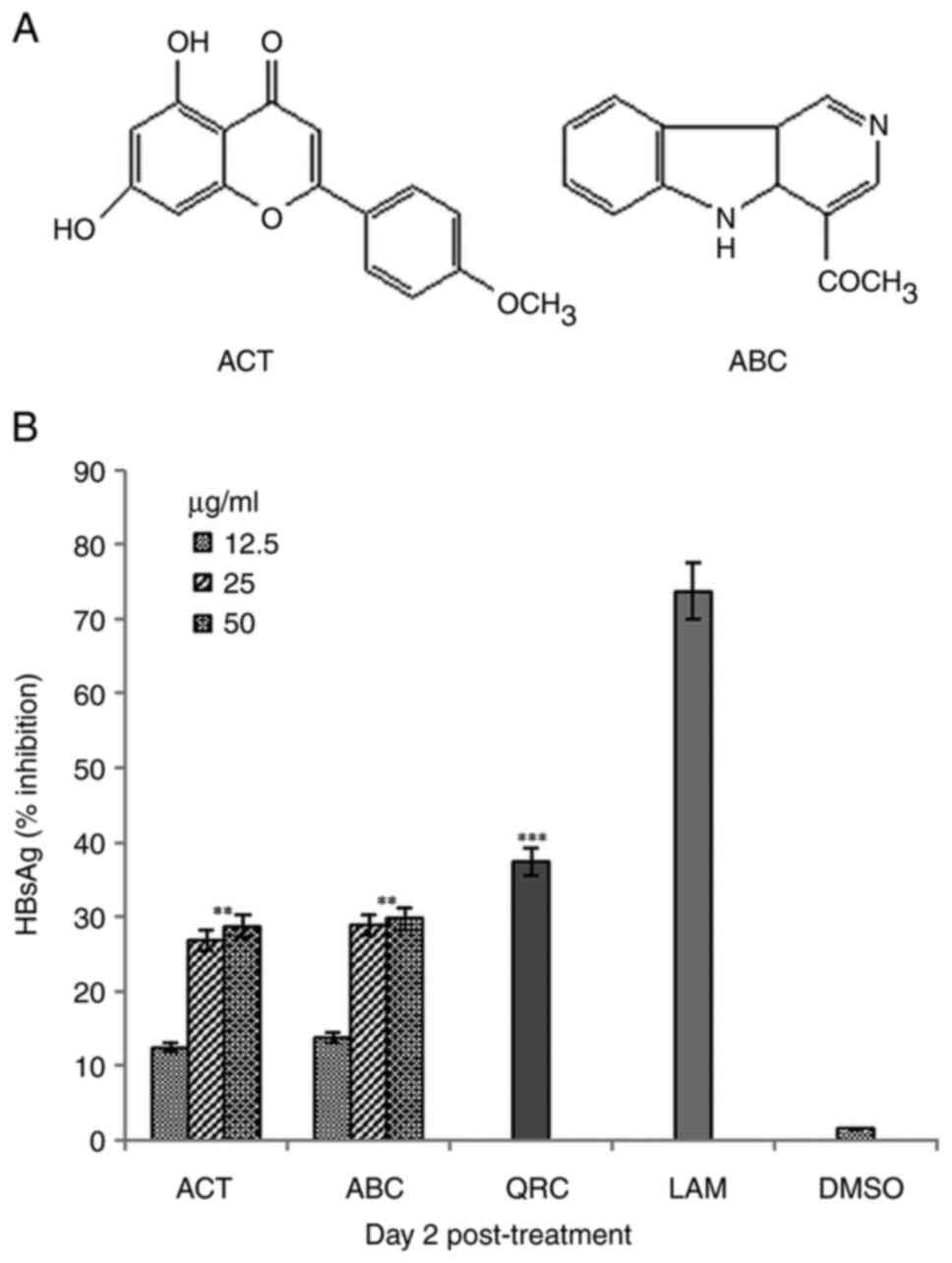
structural data (23,24), it was identified as ACT, a
flavonoid (Fig. 1A).
Compound 2 was a yellow amorphous solid substance
that was also soluble in DMSO, with the general chemical formula of
C12H10N2. ESI-HRMS: m/z 183.3
(M+H)+. Its 1H-NMR (DMSO-d6, 700
MHz) δ are as follows: 8.13 (1H, d, 5.3 Hz, H-3), 6.98 (1H, d, 5.6
Hz, H-4), 7.98 (1H, d, 8.0 Hz, H-5), 6.92 (1H, t, 6.8 Hz, H-6),
7.45 (1H, t, 7.2 Hz, H-7), 7.65 (1H, d, 8.0 Hz, H-8), 10.65 (1H, s,
br, H-10) and 2.79 (3H, s, CH3). It 13CNMR
(DMSO-d6, 125 MHz) δ were as follows: 143.0 (C-1), 138.9 (C-3),
113.2 (C-4), 122.3 (C-5), 120.3 (C-6), 128.6 (C-7), 112.7 (C-8),
135.6 (C-9) and 21.4 (C-11). Based on the structural data (23), it was identified as ABC, an
alkaloid (Fig. 1A).
Lack of cytotoxicity exerted by ACT
and ABC
Νeither of ACT nor ABC tested up to 50 µg/ml showed
any sign of toxicity on HepG2.2.15 cells at 72 h, as determined by
MTT assay (data not shown). This was in agreement with the observed
morphological integrity of the treated cells, when compared with
untreated or control cells under the microscope. The
CC50 value was therefore not determined.
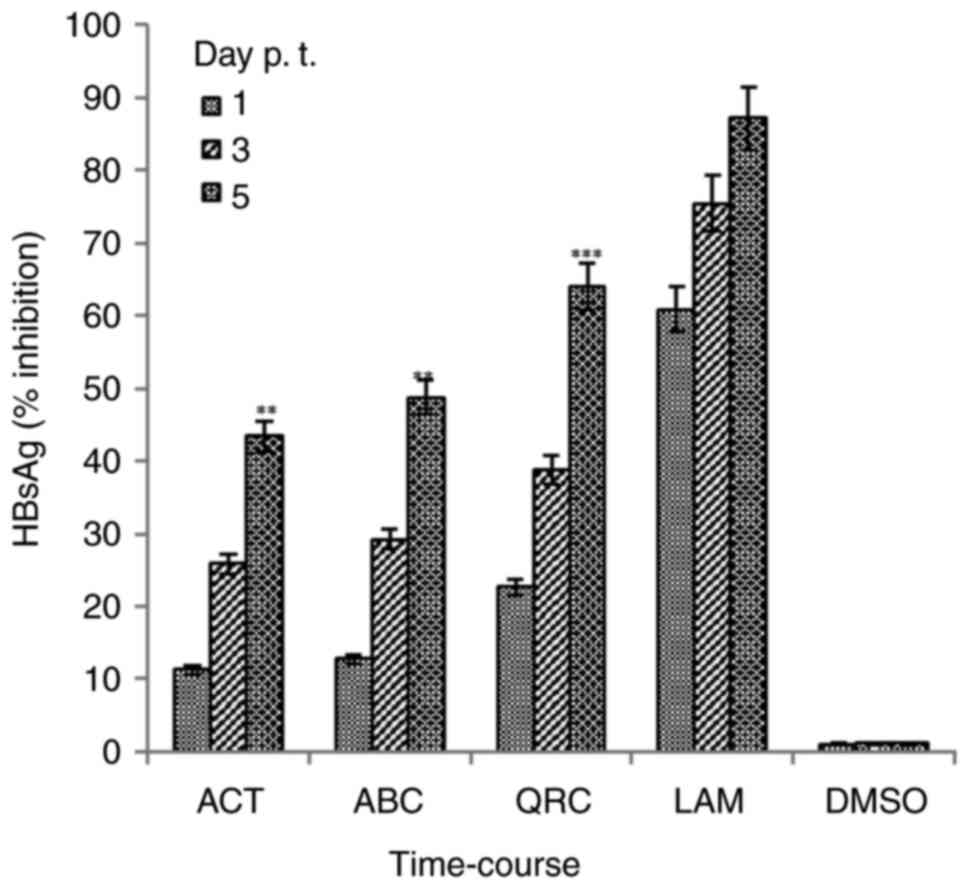
Dose- and time-dependent inhibition of
HBsAg synthesis by ACT and ABC
When tested for dose-dependent effects at a single
time-point (day 2), ACT and ABC exerted maximal inhibition of HBsAg
synthesis at 25 µg/ml, since 50 µg/ml could not significantly
potentiate the inhibition of HBsAg any further (Fig. 1B). Therefore, using a single-time
point, a single maximal dose of 25 µg/ml was selected for the
time-course analysis. Among the three time-points (days 1, 3 and
5), the observed maximal inhibition values of HBsAg production by
ACT and ABC were 43.4% (P<0.01 vs. LAM) and 48.7% (P<0.01 vs.
LAM) on day 5, respectively. The corresponding value for QRC was
63.9% (P<0.001), compared with that of LAM (87.1%) on day 5
(Fig. 2). Since further culturing
with the selected optimal dose resulted in cell over-growth and
apoptotic cell death, the assay was not extended beyond day 5.
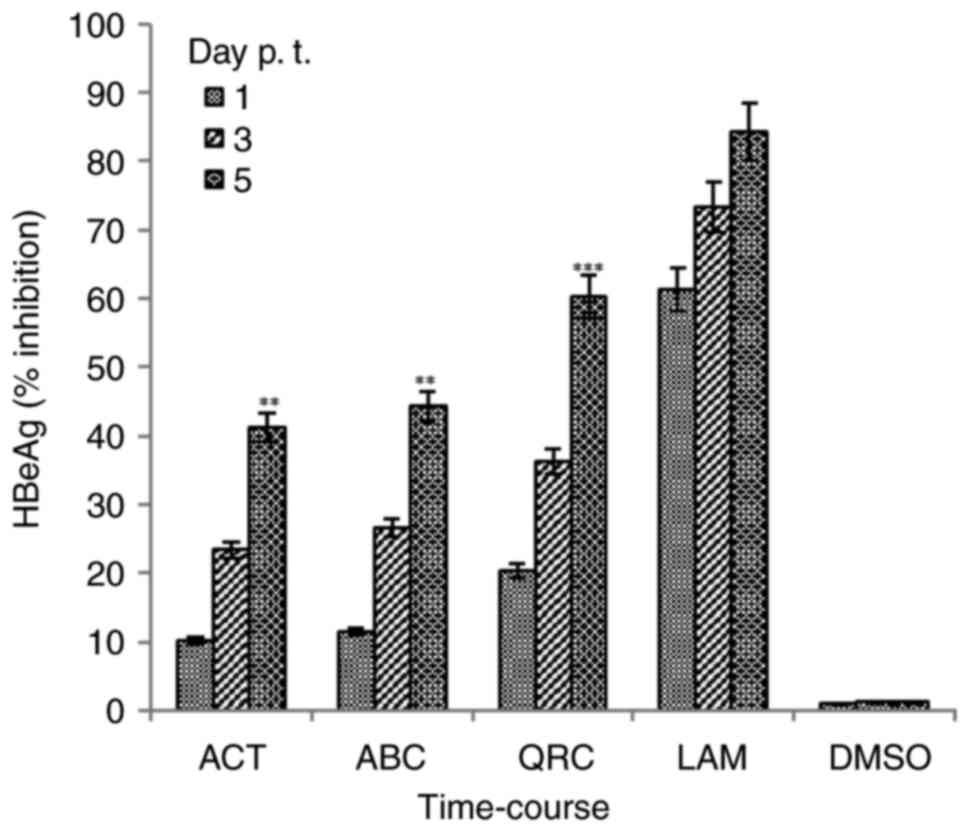
Downregulation of virus replication by
ACT and ABC
Synthesis of HBeAg is a clinical serological marker
of active HBV DNA replication (5,26).
Therefore, the optimal 25 µg/ml dose was also tested for its
effects on HBeAg expression. Among the three time-points tested
(days 1, 3 and 5), the maximal inhibition values of HBeAg synthesis
by ACT and ABC were 41.2% (P<0.01 vs. LAM) and 44.2% (P<0.01
vs. LAM) on day 5, respectively. The corresponding value for QRC
was 60.2% (P<0.001), compared with that of LAM (84.3%) at day 5
(Fig. 3). Since further culturing
with the selected optimal doses led to cell over-growth and
apoptotic cell death, the assay was not extended beyond day 5.
Structure-based interactions of the
isolated compounds with HBV wt-POL
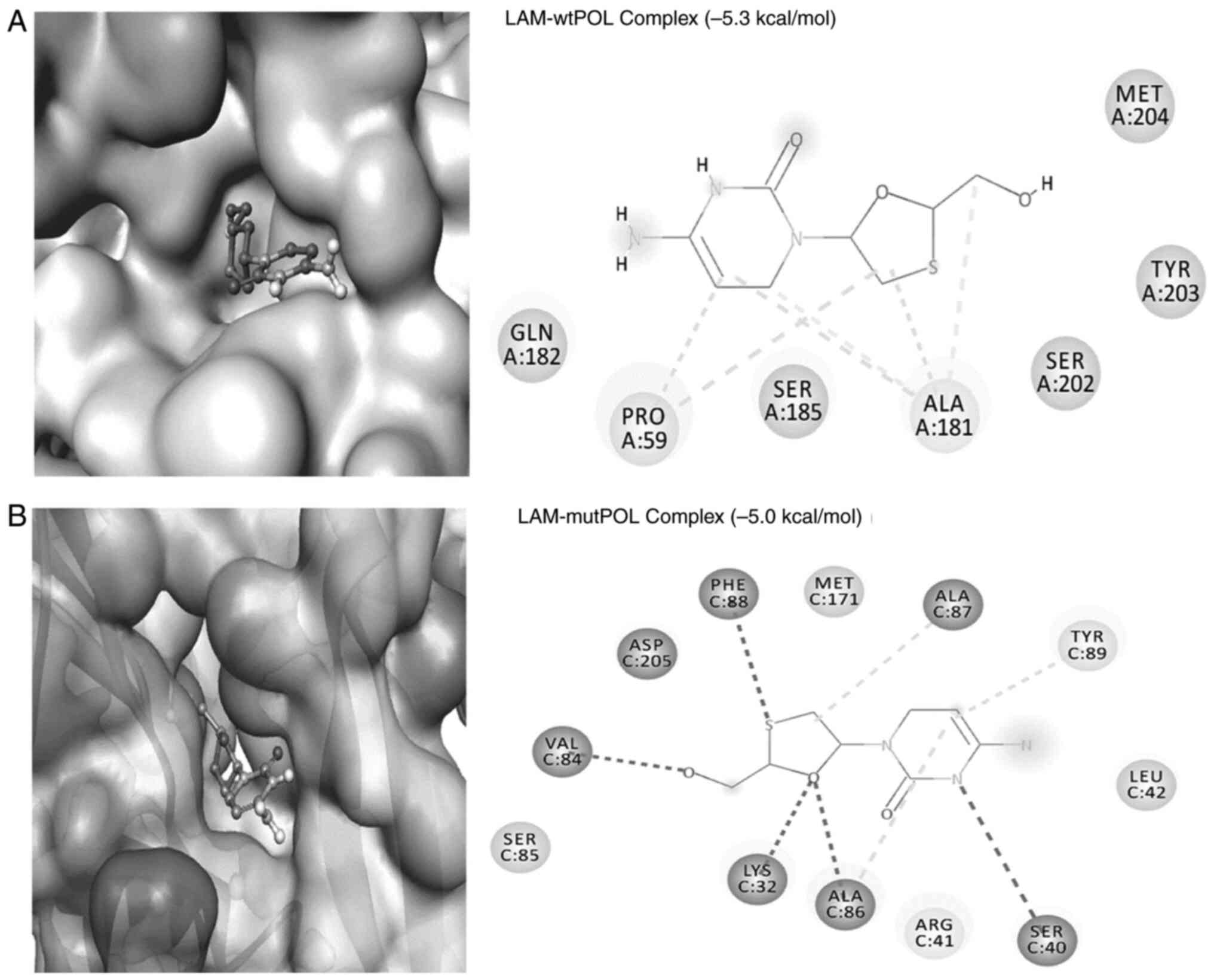
Molecular docking analysis showed formation of a
stable complex between LAM (reference drug) and the HBV wt-POL
active-site residues (Fig. 4A).
LAM formed two conventional H-bonds with Ala181:O (3.51 Å and 3.66
Å) and four hydrophobic interactions with Pro59 (5.20 Å and 4.42 Å)
and Ala181 (5.12 Å and 3.84 Å; Fig.
4A). Notably, in addition to the classical bonding of LAM with
the POL ‘YMDD (Tyr, Met, Asp, Asp)’ motif's residues Tyr203 and
Met204, its van der Waals' interactions with Gln182 and Ser185
further strengthened the complex. The binding free energy and the
corresponding binding affinity of the LAM-wt-POL complex were
estimated to be-5.3 kcal/mol and 7.71x103/M,
respectively (Table I).
 | Table IMolecular docking parameters for the
interaction of hepatitis B virus wild-type polymerase (‘YMDD’
motif) with ACT and ABC. |
Table I
Molecular docking parameters for the
interaction of hepatitis B virus wild-type polymerase (‘YMDD’
motif) with ACT and ABC.
| Interaction between
donor and acceptor atoms | Type of
interaction | Distance (Å) | Docking energy
(ΔG), kcal/mol | Dissociation
constant (Kd/mol) |
|---|
| Lamivudine | | | -5.3 |
7.71x103 |
|
LIG:C-ALA181:O | Carbon H-bond | 3.51 | | |
|
LIG:C-ALA181:O | Carbon H-bond | 3.66 | | |
|
PRO59-LIG | Hydrophobic
(alkyl) | 4.42 | | |
|
PRO59-LIG | Hydrophobic
(alkyl) | 5.20 | | |
|
ALA181-LIG | Hydrophobic
(alkyl) | 5.12 | | |
|
ALA181-LIG | Hydrophobic
(alkyl) | 3.84 | | |
| ACT | | | -6.8 |
9.72x104 |
|
LYS32:HZ1-LIG:O | Conventional
H-bond | 2.08 | | |
|
LYS32:HZ1-LIG:O | Conventional
H-bond | 2.75 | | |
|
ARG41:HH22-LIG:O | Conventional
H-bond | 2.28 | | |
|
LIG:C-ASP206:OD2 | Carbon H-bond | 3.55 | | |
|
ASP205:OD1-LIG | Electrostatic
(π-anion) | 3.44 | | |
|
ALA86:HN-LIG | H-bond
(π-donor) | 2.86 | | |
|
LIG-ALA86 | Hydrophobic
(π-alkyl) | 4.78 | | |
| ABC | | | -6.8 |
9.72x104 |
|
LYS32:HZ1-LIG:O | Conventional
H-bond | 2.20 | | |
|
LIG:HN-ASP205:OD2 | Conventional
H-bond | 2.04 | | |
|
LIG:C-VAL84:O | Carbon H-bond | 3.59 | | |
|
ARG41:NH2-LIG | Electrostatic
(π-cation) | 4.07 | | |
|
ARG41:CG-LIG | Hydrophobic
(π-σ) | 3.76 | | |
|
LIG-MET171 | Hydrophobic
(π-alkyl) | 5.05 | | |
|
LIG-ARG41 | Hydrophobic
(π-alkyl) | 4.97 | | |
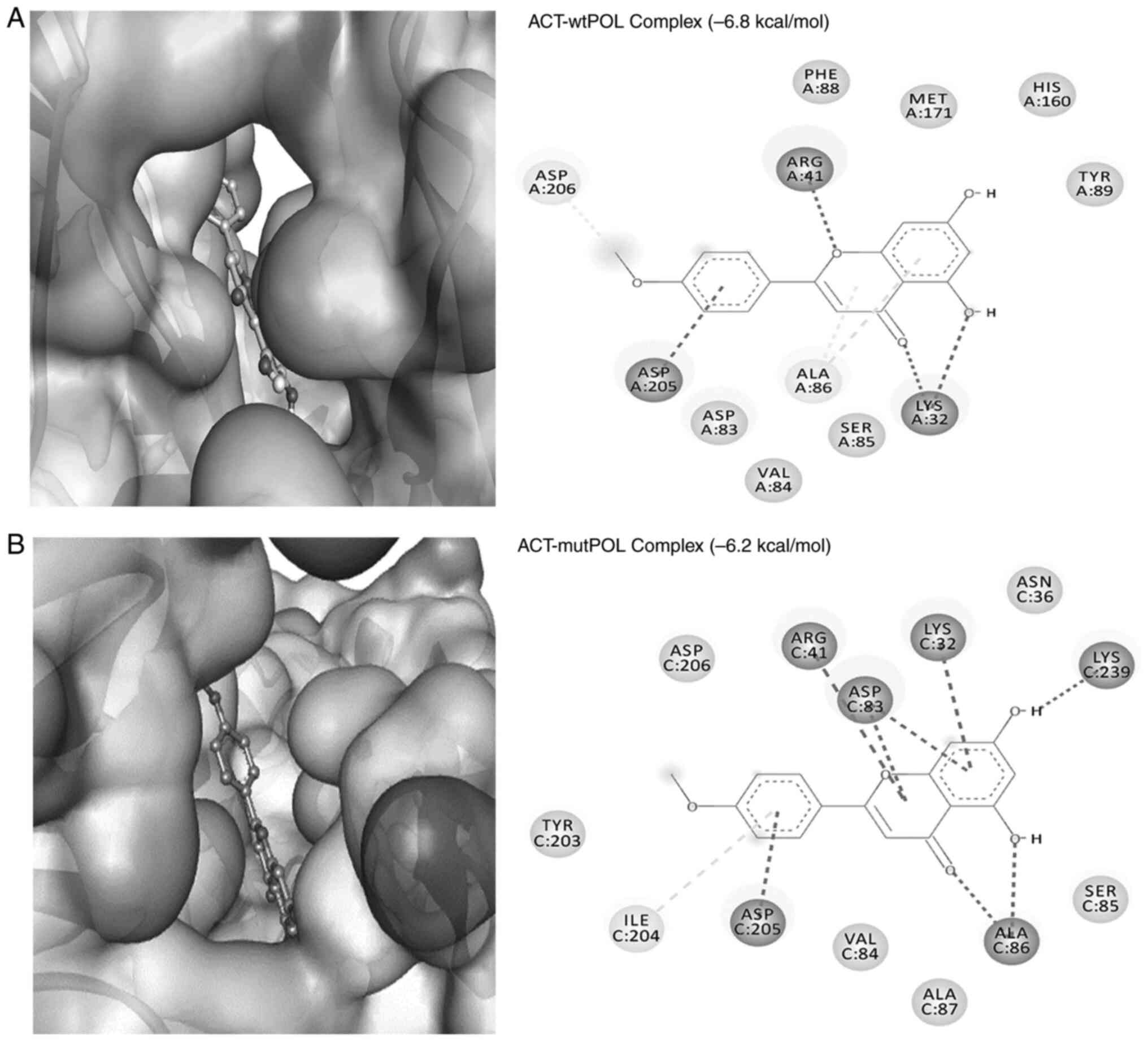
The flavonoid ACT also formed a stable complex with
the target protein wt-POL through hydrogen bonds and hydrophobic
interactions (Fig. 5A). The wt-POL
residue Lys:HZ1 formed two conventional H-bonds with the O-atom of
ACT (2.08 Å and 2.75 Å), whilst Arg41:HH22 interacted with the
O-atom of ACT through another conventional H-bond (2.28 Å; Table I). A C-H bond was also formed
between ACT and Asp206:OD2 (3.55 Å). In addition, electrostatic
(π-anion) interaction with Asp205:OD1 (3.44 Å) and a π-donor
hydrogen bond with Ala86:HN (2.86 Å) were also involved in
stabilizing the ACT-wt-POL complex. ACT also formed hydrophobic
interactions (π-alkyl; 4.78 Å) with Ala86 (Fig. 5A). The ACT-wt-POL complex was
further strengthened by van der Waals' interactions with Asp83,
Val84, Ser85, Phe88, Tyr89, His160 and Met171. The ACT-wtPOL
complex binding energy and docking affinity was estimated to be
-6.8 kcal/mol and 1.15x105/M, respectively (Table I).
The alkaloid ABC also formed a stable complex with
the target protein wt-POL through two conventional hydrogen bonds
involving Lys32:HZ1 and LIG:O (2.20 Å), alongside LIG:HN and
Asp205:OD2 (2.04 Å), with a C-H bond between LIG:C and Val84:O
(3.59 Å; Table I). In the complex,
an electrostatic (π-cation) interaction between Arg41:NH2 and LIG
(4.07 Å) was also observed (Table
I; Fig. 6A). Furthermore, ABC
formed a π-σ hydrophobic interaction with Arg41:CG (3.76 Å) and two
π-alkyl hydrophobic interactions with Met71 (5.05 Å) and Arg41
(4.97 Å; Fig. 6A). The residue
Ser85 was found to be involved in an unfavorable acceptor-acceptor
interaction with the O-atom of ABC. Additionally, the van der
Waals' interactions were also established by the wt-POL residues
Ser40, Leu42, Asp83, Val84, Aal87, Phe88, Tyr89 and His160. The
docking free energy and binding affinity of the ABC-wt-POL complex
were estimated to be -6.8 kcal/mol and 1.15x105/M,
respectively (Table I).
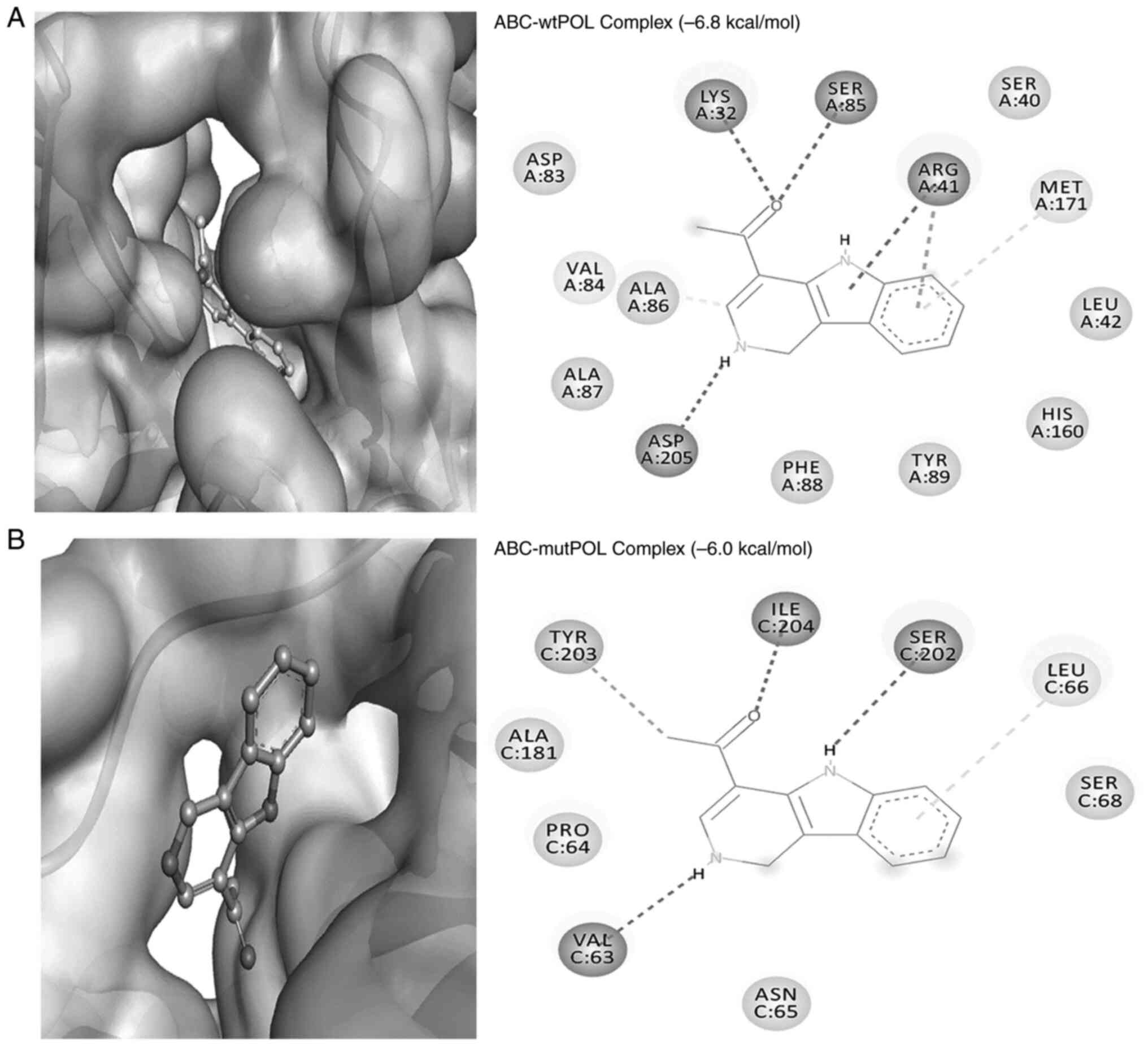 | Figure 6Molecular docking analysis of ABC
onto the hepatitis B virus POL protein. (A) Structure-based
interaction of ABC with (A) wt-POL (‘YMDD’ motif's Tyr, Met, Asp,
and Asp residues) and, (B) mut-POL (‘YIDD’ motif's Tyr, Ile, Asp,
and Asp residues). ABC, acetyl-β-carboline; wt-POL, wild-type
polymerase; mut-POL, mutant polymerase. |
Structure-based interactions of the
isolated compounds with HBV mut-POL
In the case of mut-POL, LAM was found to form weak
interactions with the active-site (Fig. 4B). LAM interacted through four
H-bonds with Lys32:HZ1 (2.60 Å), Ser40:O (2.27 Å), Val84:O (1.91 Å)
and Ala86:HN (2.94 Å). It also formed a π-Sigma bond with Phe88
(5.45 Å) and three hydrophobic interactions with Ala86 (4.54 Å),
Ala87 (5.06 Å) and Tyr89 (4.85 Å; Fig.
4B). Specifically, LAM formed van der Waals' interactions with
the ‘Tyr-Ile-Asp-Asp’ motif's Asp205, as well as residues Arg41,
Leu42, Ser85 and Met171. In addition, the binding free energy and
binding affinity of LAM-mut-POL complex were calculated as -5.0
kcal/mol and 4.65x103/M, respectively (Table II).
 | Table IIMolecular docking parameters for the
interaction of hepatitis B virus mutant polymerase (‘YIDD’ motif)
with ACT and ABC. |
Table II
Molecular docking parameters for the
interaction of hepatitis B virus mutant polymerase (‘YIDD’ motif)
with ACT and ABC.
| Interaction between
donor and acceptor atoms | Type of
interaction | Distance (Å) | Docking energy
(ΔG); kcal/mol | Dissociation
constant (Kd/mol) |
|---|
| Lamivudine | | | -5.0 |
4.65x103 |
|
LYS32:HZ1-LIG:O | Conventional
H-bond | 2.60 | | |
|
ALA86:HN-LIG:O | Conventional
H-bond | 2.94 | | |
|
LIG:N-SER40:O | Conventional
H-bond | 2.27 | | |
|
LIG:O-VAL84:O | Conventional
H-bond | 1.91 | | |
|
LIG:S-PHE88 | π-sulfur | 5.45 | | |
|
ALA86-LIG | Hydrophobic
(alkyl) | 4.54 | | |
|
ALA87-LIG | Hydrophobic
(alkyl) | 5.06 | | |
|
TYR89-LIG | Hydrophobic
(π-alkyl) | 4.85 | | |
| ACT | | | -6.2 |
3.53x104 |
|
ALA86:HN-LIG:O | Conventional
H-bond | 2.05 | | |
|
ALA86:HN-LIG:O | Conventional
H-bond | 2.30 | | |
|
LYS32:NZ-LIG | Electrostatic
(π-cation) | 3.89 | | |
|
ARG41:NH2-LIG | Electrostatic
(π-cation) | 4.34 | | |
|
ASP83:OD1-LIG | Electrostatic
(π-anion) | 3.41 | | |
|
ASP83:OD1-LIG | Electrostatic
(π-anion) | 4.13 | | |
|
ASP205:OD1-LIG | Electrostatic
(π-anion) | 3.29 | | |
|
LIG-ILE204 | Hydrophobic
(π-alkyl) | 5.42 | | |
| ABC | | | -6.0 |
2.52x104 |
|
ILE204:HN-LIG:O | Conventional
H-bond | 1.85 | | |
|
LIG:N-SER202:O | Conventional
H-bond | 3.01 | | |
|
LIG:N-VAL63:O | Conventional
H-bond | 2.87 | | |
|
LIG:C-TYR203 | Hydrophobic
(π-σ) | 3.99 | | |
|
LIG-LEU66 | Hydrophobic
(π-alkyl) | 5.25 | | |
The interaction between ACT and the target protein
mut-POL was favored by two H-bonds with Ala86:HN (2.05 and 2.30 Å),
two π-cation electrostatic interactions with Lys32:NZ (3.89 Å) and
Arg41:NH2 (4.34 Å), in addition to two π-anion electrostatic
interactions with Asp83:OD1 (3.41 and 4.13 Å) and Asp205:OD1 (3.29
Å; Fig. 5B). Notably, ACT formed a
hydrophobic interaction with Ile204 (5.42 Å) and van der Waals'
interactions with Tyr203 and Asp206 of the ‘Tyr-Ile-Asp-Asp’ motif.
The complex was further strengthened by van der Waals' interactions
involving with Asn36, Val84, Ser85 and Ala87. The estimated binding
free energy and binding affinity of the ACT-mut-POL complex were
-6.2 kcal/mol and 3.53x104/M, respectively (Table II).
Docking analysis of ABC and the target protein
mut-POL revealed that the complex was mainly stabilized by H-bonds
and hydrophobic interactions (Table
II). Notably, the NH-group of Ile204 in the ‘Tyr-Ile-Asp-Asp’
motif formed a H-O bond with ABC (1.85 Å). Similarly, N-atoms of
ABC formed two H-bonds with the O-atom of Ser202 (3.01 Å) preceding
Tyr203 and the O-atom of Val63 (2.87 Å; Fig. 6B). Furthermore, ABC formed a π-σ
and a π-alkyl hydrophobic interaction with Tyr203 and Leu66
residues, respectively. In addition, van der Waals' interactions
were also formed with Pro64, Asn65, Ser68 and Ala181 residues in
the complex. The estimated docking free energy and binding affinity
of ABC-mutPOL complex were -6.0 kcal/mol and 2.52x104/M,
respectively (Table II).
Discussion
Among the known classes of phytochemicals with
antiviral potential (2,3), flavonoids are particular noted for
their significantly higher activities against several viruses
(29,30), including HBV (19-21,27,31,32).
Organ or liver toxicity is known to be caused by some antiviral
herbal products, notably the Chinese traditional medicines
(33). To minimize this risk, the
R. stricta-derived ACT and ABC were first tested for
potential hepatotoxicity in cultured HepG2.2.15 cells. Both
compounds were not found to be cytotoxic up to the tested maximal
concentration of 50 µg/ml. Therefore, they were then assessed for
their anti-HBV efficacies. Notably, due to the close similarity of
the genome replication mechanism used by HBV with that of HSV and
HIV, the majority of the anti-HSV/HIV drugs are also viable against
HBV (26). ACT (100 µg/ml)
isolated from Scoparia dulcis has been previously reported
to inhibit HSV in cultured Vero cells (34). In addition, ACT has been
demonstrated to suppress HIV gene expression in vitro
(35). Consistent with this
finding, to the best of our knowledge, the present study reported
for the first time the anti-HBV activity of R.
stricta-derived ACT in cultured HepG2.2.15 cells. ACT (25
µg/ml) maximally inhibited the production of HBsAg by 43.4% and
HBeAg by 41.2% compared with those in the untreated control.
Comparatively, the standards LAM and QRC suppressed synthesis of
HBsAg by 87.1 and 63.9%, respectively, and HBeAg by 84.3 and 64.2%,
respectively. Notably, the concentration used to maximally inhibit
HBV was 25% less of that used against HSV elsewhere (34).
β-carboline alkaloids are natural compounds with
documented antiviral activities (22). Previously, anti-HSV active
β-carboline compounds eudistomin C and E have been isolated from
the Caribbean tunicate (36-38).
Caribbean deep-sea sponge-derived imidazole alkloids topsentin and
bromotopsentin have also demonstrated bioactivities against HSV and
the human coronavirus A59 (HCoV-A59), whereas
dihydrodeoxybromotopsentin was shown to confer activity against
HCoV-A59 only (39). In addition,
β-carboline alkaloids, such as dercitin, from other marine sources,
were found to have both anti-HSV and anti-HIV properties (40). Acarnidines, polyandrocarpidines and
didemnins were reported to be active against HSV, coxackie virus,
equine rhinovirus, paraINV, Rift Valley fever virus, Venezuelan
equine encephalomyelitis virus and yellow fever virus (41). Consistent with these observation, a
recent study has shown the anti-HBV efficacy of solanopubamine
(β-amino-5,22,25-solanidan-23-ol), a steroidal alkaloid isolated
from Solanum schimperianum (20). In the present study, to the best of
our knowledge, the anti-HBV activity of ABC in HepG2.2.15 cells was
found for the first time. ABC (25 µg/ml) maximally inhibited the
production of HBsAg by 48.7% and HBeAg by 44.2% compared with those
in the untreated control. Notably, in a previous study, LAM and QRC
have been tested for non-cytotoxicity and their doses were
optimized, following which a gene reporter assay was also conducted
to confirm their lack of direct or indirect interference with host
proteins involved in the production of viral HBsAg and HBeAg
(16,17,26).
LAM, the cytidine analog
(2',3'-dideoxy-3'-thiacytidine) and other such nucleotide analogs
are approved anti-viral drugs for effectively treating chronic
hepatitis B (4). The HBV POL
catalyzes its DNA replication, where the conserved POL residues
Tyr, Met, Asp, and Asp of the ’YMDD’ motif incorporates new
nucleot(s)ides into its growing single-strand DNA (5). Incorporation of LAM blocks the
addition of new nucleosides, resulting in the termination of DNA
strand elongation (5). In patients
with chronic HBV on long-term LAM therapy, HBV resistance to LAM
emerges due to the substitutions of Met to Ile or Val in the ‘YMDD’
motif. This ‘YMDD’ to ’YI/VDD’ change significantly affects the
tertiary structure and hence, the activity of POL (5), leading to inefficient or no
incorporation of LAM into the DNA strand, allowing the elongation
of the DNA strand and viral replication (5). Molecular docking analyses of the
tested anti-HBV active compounds have demonstrated formations of
stable complexes with HBV drug-sensitive (wt-POL: ‘YMDD’) and
drug-resistant (mut-POL: ‘YIDD’) POLs. Both ACT and ABC showed
bonding with the catalytic ‘Tyr-Met/Ile-Asp-Asp’ motif residues,
including other interactions in their respective complexes.
Notably, ACT interacted with wt-POL Asp205 and Asp206, as well as
mut-POL Tyr203 and Asp206, whilst ABC interacted with wt-POL
Asp205, as well as mutPOL Tyr203 and Ile204. Inhibition of HBV POL
activity leads to the downregulation of viral RNA transcription and
therefore, translation of its proteins, including HBsAg and HBeAg
(5,26). In conclusion, to the best of our
knowledge, the present study shows promising in vitro
anti-HBV efficacies of ACT and ABC, endorsed by in silico
data, for the first time. However, further molecular and
pharmacological studies are required to validate their pre-clinical
therapeutic potential.
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by the Researchers
Supporting Project (grant no. RSP2023R379), King Saud University,
Riyadh, Saudi Arabia.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
MKP and MSA conceptualized the study, performed
in vitro assays, collected and analyzed data, and wrote the
manuscript. TAA, MA and HMA isolated and characterized the
compounds, and contributed to writing. ARA performed the
statistical analysis. MTR and MFA performed the in silico
analysis and contributed to writing. All authors read and approved
the final manuscript. MKP and TAA confirm the authenticity of all
the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kapoor R, Sharma B and Kanwar SS:
Antiviral phytochemicals: An overview. Biochem Physiol.
6(220)2017.
|
|
2
|
Ben-Shabat S, Yarmolinsky L, Porat D and
Dahan A: Antiviral effect of phytochemicals from medicinal plants:
Applications and drug delivery strategies. Drug Deliv Transl Res.
10:354–367. 2020.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Parvez MK, Arbab AH and Al-Dosari MS: An
update on natural or herbal drugs against hepatitis B virus. In:
Advances in Medicine and Biology. Berhardt LV (ed). Vol 179. NOVA
Science Publishers, Inc., New York, NY pp159-184, 2021.
|
|
4
|
World Health Organization (WHO). Hepatitis
B. WHO, Geneva, 2022. https://www.who.int/news-room/fact-sheets/detail/hepatitis-b.
|
|
5
|
Devi U and Locarnini S: Hepatitis B
antivirals and resistance. Curr Opin Virol. 3:495–500.
2013.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Marwat SK, Usman K, Shah SS, Anwar N and
Ullah I: A review of phytochemistry, bioactivities and ethno
medicinal uses of Rhazya stricta decsne (apocynaceae). Afr J
Microbiol Res. 6:1629–1641. 2012.
|
|
7
|
Gilani SA, Kikuchi A, Khan ZS, Khattak ZI
and Watanabe KN: Phytochemical, pharmacological and ethnobotanical
studies of Rhazya stricta decne. Phytother Res. 21:301–307.
2007.PubMed/NCBI View
Article : Google Scholar
|
|
8
|
Ali BH, Al-Qarawi AA, Bashir AK and Tanira
MO: Phytochemistry, pharmacology and toxicity of Rhazya
stricta decne: A review. Phytother Res. 14:229–234.
2000.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Albeshri A, Baeshen NA, Bouback TA and
Aljaddawi AA: A review of Rhazya stricta decne
phytochemistry, bioactivities, pharmacological activities,
toxicity, and folkloric medicinal uses. Plants (Basel).
10(2508)2021.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Sultana N and Khalid A: Phytochemical and
enzyme inhibitory studies on indigenous medicinal plant Rhazya
stricta. Nat Prod Res. 24:305–314. 2010.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Albeshri A, Baeshen NA, Bouback TA and
Aljaddawi AA: Evaluation of cytotoxicity and antiviral activity of
Rhazya stricta decne leaves extract against influenza
A/PR/8/34 (H1N1). Saudi J Biol Sci. 29(03375)2022.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Baeshen MN, Attar R, Bouback TA, Albeshri
AO, Baeshen NN, Karkashan A, Abbas B, Aljaddawi AA, Almulaikyd YQ,
Mahmoude SH, et al: Assaying for antiviral activity of the
folkloric medicinal desert plant Rhazya stricta on
coronavirus SARS-CoV-2. Biotech Biotechnol Equip. 36:68–74.
2022.
|
|
13
|
Bukhari NA, Al-Otaibi RA and Ibhrahim MM:
Phytochemical and taxonomic evaluation of Rhazya stricta in
Saudi Arabia. Saudi J Biol Sci. 24:1513–1521. 2017.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Lanjwani AH, Ganghro AB and Khuhawar TMJ:
Phytochemical analysis and biological activity of different parts
of Rhazya stricta. Rawal Med J. 43:532–535. 2018.
|
|
15
|
Badshah SL, Faisal S, Muhammad A, Poulson
BG, Emwas AH and Jaremko M: Antiviral activities of flavonoids.
Biomed Pharmacother. 140(11596)2021.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Parvez MK, Tabish Rehman M, Alam P,
Al-Dosari MS, Alqasoumi SI and Alajmi MF: Plant-derived antiviral
drugs as novel hepatitis B virus inhibitors: Cell culture and
molecular docking study. Saudi Pharm J. 27:89–400. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Parvez MK, Al-Dosari MS, Alam P, Rehman
MT, Alajmi MF and Alqahtani AS: The anti-hepatitis B virus
therapeutic potential of anthraquinones derived from Aloe vera.
Phytother Res. 33:1960–1970. 2019.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Parvez MK, Al-Dosari MS, Arbab AH,
Al-Rehaily AJ and Abdelwahid MAS: Bioassay-guided isolation of
anti-hepatitis B virus flavonoid myricetin-3-O-rhamnoside along
with quercetin from Guiera senegalensis leaves. Saudi Pharm J.
28:550–559. 2020.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Parvez MK, Ahmed S, Al-Dosari MS,
Abdelwahid MAS, Arbab AH, Al-Rehaily AJ and Al-Oqail MM: Novel
anti-hepatitis B virus activity of Euphorbia schimperi and its
quercetin and kaempferol derivatives. ACS Omega. 6:29100–29110.
2021.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Parvez MK, Al-Dosari MS, Rehman MT,
Al-Rehaily AJ, Alqahtani AS and Alajmi MF: The anti-hepatitis B
virus and anti-hepatotoxic efficacies of solanopubamine, a rare
alkaloid from Solanum schimperianum. Saudi Pharm J. 30:359–368.
2022.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Wang G, Zhang L and Bonkovsky HL: Chinese
medicine for treatment of chronic hepatitis B. Chin J Integr Med.
18:253–255. 2012.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Szabó T, Volk B and Milen M: Recent
advances in the synthesis of β-carboline alkaloids. Molecules.
26(663)2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Qu GR, Liu J, Li XX, Wang SX, Wu LJ and Li
X: Flavonoids constituents of Sonchus arvensis L. Zhong Cao Yao.
26:233–235. 1995.
|
|
24
|
Gong FJ, Wang GL and Wang YW: Chemical
constituents of the flowers of Dendranthema indicum var.
aromaticum. Wuhan Zhiwuxue Yanjiu. 23:610–612. 2005.
|
|
25
|
Cao R, Peng W, Wang Z and Xu A:
beta-Carboline alkaloids: Biochemical and pharmacological
functions. Curr Med Chem. 14:479–500. 2007.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Parvez MK, Sehgal D, Sarin SK, Basir SF
and Jameel S: Inhibition of hepatitis B virus DNA replicative
intermediate forms by recombinant interferon-gamma. World J
Gastroenterol. 12:3006–3014. 2006.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Parvez MK, Al-Dosari MS, Basudan OA and
Herqash RN: The anti-hepatitis B virus activity of sea buckthorn is
attributed to quercetin, kaempferol and isorhamnetin. Biomed Rep.
17(89)2022.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Rehman MT, Ahmed S and Khan AU:
Interaction of meropenem with ‘N’ and ‘B’ isoforms of human serum
albumin: A spectroscopic and molecular docking study. J Biomol
Struct Dyn. 34:1849–1864. 2016.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Zakaryan H, Arabyan E, Oo A and Zandi K:
Flavonoids: Promising natural compounds against viral infections.
Arch Virol. 162:2539–2551. 2017.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Wang L, Song J, Liu A, Xiao B, Li S, Wen
Z, Lu Y and Du G: Research progress of the antiviral bioactivities
of natural flavonoids. Nat Prod Bioprospect. 10:271–283.
2020.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Ahmed S, Parvez MK, Zia K, Nur-e-Alam M,
Ul-Haq Z, Al-Dosari MS and Al-Rehaily AJ: Natural anti-hepatitis B
virus flavones isolated from Stachys schimperi Vatke growing in
Saudi Arabia. Pharmacog Mag. 18:386–392. 2022.
|
|
32
|
Parvez MK, Al-Dosari MS, Abdelwahid MAS,
Alqahtani AS and Alanzi AR: Novel anti-hepatitis B virus-active
catechin and epicatechin from Rhus tripartita. Exp Ther Med.
23(398)2022.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Parvez MK and Rishi V: Herb-Drug
interactions and hepatotoxicity. Curr Drug Metab. 20:275–282.
2019.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Singh S, Gupta P, Meena A and Luqman S:
Acacetin, a flavone with diverse therapeutic potential in cancer,
inflammation, infections and other metabolic disorders. Food Chem
Toxicol. 145(111708)2020.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Hayashi K, Hayashl T, Arisawa M and Morita
N: Antiviral agents of plant origin. Antiherpetic activity of
acacetin. Antiviral Chem Chemother. 4:49–53. 1993.
|
|
36
|
Critchfield JW, Butera ST and Folks TM:
Inhibition of HIV activation in latently infected cells by
flavonoid compounds. AIDS Res Hum Retroviruses. 12:39–46.
1996.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Rinehart KL, Kobayashi J, Harbour GC,
Hughes RG Jr, Mizsak SA and Scahill TA: Eudistomins C, E, K, and L,
potent antiviral compounds containing a novel oxathiazepine ring
from the Caribbean tunicate Eudistoma olivaceum. J Am Chem Soc.
106:1524–1526. 1984.
|
|
38
|
Rinehart KL Jr, Kobayashi J, Harbour GC,
Gilmore J, Mascal M, Holt TG, Shield LS and Lafargue F: Eudistomins
A-Q, beta-carbolines from the antiviral Caribbean tunicate
Eudistoma olivaceum. J Am Chem Soc. 109:3378–3387. 1987.
|
|
39
|
Tsujii S, Rinehart KL, Gunasekera SP,
Kashman Y, Cross SS, Lui MS, Pomponi SA and Diaz MC: Topsentin,
bromotopsentin, and dihydrodeoxybromotopsentin, antiviral and
antitumor bis(indo1yl)imidazoles from Caribbean deepsea sponges of
the family Halichondriidae. Structural and synthetic studies. J Org
Chem. 33:5446–5453. 1998.
|
|
40
|
Gunawardana GP, Kohmoto S, Gunasekera SP,
McConnell OJ and Koehn FE: Dercitin, a new biologically active
acridine alkaloid from a deep water marine sponge, Dercitus sp. J
Am Chem Soc. 110:4856–4858. 1988.
|
|
41
|
Rinehart KL Jr, Gloer JB, Cook JC Jr,
Mizsak SA and Scahill TA: Structures of the didemnins, antiviral
and cytotoxic depsipeptides from a Caribbean tunicate. J Am Chem
Soc. 103:1857–1859. 1981.
|