Introduction
Sepsis can cause poor tissue hypoperfusion and a
series of life-threatening organ dysfunctions, which are common
causes of death in hospitalized patients. Sepsis remains a
significant concern globally, with an estimated 48.9 million cases
and 11 million deaths occurring worldwide in 2017(1). Septic shock refers to sepsis with
persisting low blood pressure requiring vasopressors to maintain
the mean arterial pressure (2). It
has been clinically confirmed that sepsis is caused by bacteria or
foci of infection (3-5).
Sepsis is the pathological process through which the body responds
to infectious factors (6).
Septic shock belongs to a subset of sepsis in which
underlying circulatory, cellular, and metabolic abnormalities are
associated with a greater risk of death than sepsis alone (7,8). The
underlying pathogenesis of sepsis is still not clear, and it
involves complex systemic inflammatory network effects, gene
polymorphisms, immune dysfunction, abnormal blood coagulation,
tissue damage and abnormal host responses to different infectious
pathogenic microorganisms and their toxins (9-11).
Sepsis is closely related to the pathophysiological changes of
multiple systems and organs in the body, and the pathogenesis of
sepsis still needs to be further clarified (12-14).
Sepsis is a systemic inflammatory response syndrome caused by
infection and is a dangerous condition. Elderly individuals are
prone to shock and multiple organ failure, especially acute kidney
injury (AKI), due to their weakened immunity (15). Early evaluation and timely
treatment of sepsis are particularly important; however, since the
clinical manifestations of sepsis are more diverse and nonspecific
compared with fever and tachycardia, especially in elderly patients
with sepsis, the common signs of sepsis in the elderly are changes
in mental status (delirium, lethargy or coma), gastrointestinal
dysfunction and shortness of breath (16,17).
Fever and tachycardia are relatively easy to be detected by
doctors. Sepsis in the elderly is likely to be absent from fever
and tachycardia, so sepsis symptoms in the elderly are atypical and
more difficult to detect early. There are several biological
indicators related to sepsis, such as C-reactive protein,
procalcitonin, soluble triggering receptor expressed on myeloid
cells 1 (TREM-1) and various inflammatory factors, including IL-6,
IL-8, TNF-α and caspase-11; however, their specificity and
sensitivity are not ideal (18,19).
Therefore, novel biomarkers that can be used to diagnose sepsis and
assess prognosis, as well as potential therapeutic targets, are
required (20-22).
AKI is a clinical syndrome characterized by a rapid
decline in renal function. Sepsis is one of the common causes of
AKI in hospitalized and intensive care unit (ICU) patients
(23). Sepsis-associated AKI
increases the risk of developing chronic comorbidities and is
associated with high mortality (24,25).
A prospective observational study of 1,753 patients at 54 hospitals
in 23 countries found that septic AKI had a higher in-hospital
case-fatality rate compared with non-septic AKI (70.2 vs. 51.8%;
P<0.001). After adjustment for covariates, septic AKI remained
associated with higher risk of mortality (1.48; 95% Confidence
Interval (CI) 1.17 to1.89; P=0.001) (26). Therefore, it is of great
significance to find novel biomarkers for the early, reliable and
noninvasive diagnosis of sepsis-associated AKI.
MicroRNAs (miRNAs/miRs) are a class of noncoding
single-stranded RNA molecules with a length of ~22 nucleotides
encoded by endogenous genes (27-30).
They are involved in post-transcriptional gene expression
regulation in animals and plants. miRNAs are involved in a series
of important life processes, including early development (31), cell proliferation, apoptosis, cell
death (32), fat metabolism
(33) and cell differentiation
(34). In 2008, it was reported
for the first time that circulating miRNAs have the potential to
become a novel marker of solid tumors (35). Subsequently, the noninvasive
acquisition of circulating miRNAs through plasma or serum attracted
the attention of researchers. Studies have reported that miRNAs can
exist in a variety of body fluids, such as blood, urine, saliva,
sweat, tears, cerebrospinal fluid, semen and milk (36-40).
During organ damage, miRNAs are usually released into biological
fluids and are stably expressed (41). Therefore, miRNAs in blood or urine
can be used as noninvasive biomarkers to detect renal disease and
toxicity, and the detection method is simple, economical and
efficient (42). Thus, miRNA
provides a novel platform and ideas for the diagnosis and treatment
of diseases and may become a useful tool in the field of precision
medicine. Studies have demonstrated that a variety of miRNAs are
involved in the inflammatory process of sepsis, and they serve an
important role by targeting the toll-like receptor/NF-κB signaling
pathway (43,44). Circulating miR-150 was the first
miRNA reported as a biomarker for sepsis (45). miR-146a, miR-143 (46,47),
miR-25(48), miR-15a/16 (49,50),
miR-1333a, miR-297 and miR-574-5p have been indicated to be useful
as markers for the diagnosis of sepsis (51-54).
Currently, sepsis is a significant cause of death in the ICU
(55-57),
and effective prevention and diagnostic techniques are still
lacking.
However, there are few studies on the relationship
between miRNAs and sepsis-associated AKI in elderly individuals. In
the present study, the differential expression of miRNAs in the
urine of elderly patients with sepsis was screened to explore the
value of miRNAs in the noninvasive diagnosis of elderly patients
with sepsis-associated AKI.
Materials and methods
Study population
Elderly patients (>65 years old) diagnosed with
sepsis were enrolled as the research subjects. For enrollment,
patients had to meet the international diagnostic criteria for
sepsis. The diagnosis of AKI was based on the 2012 Kidney Disease
Improving Global Outcomes diagnostic criteria (58). These criteria involve a sharp
decline in renal function within 48 h, manifested by an increase in
serum creatinine >0.3 mg/dl (26.5 µmol/l) or an increase >50%
(According to KDIGO, AKI is defined as an increase in serum
creatinine levels by at least 0.3 mg/dl within 48 h with 1.5-fold
being the baseline), patient age >65 years, expected survival
time >3 days, and pathogen culture or laboratory test results
showing gram-negative bacteria. The diagnostic criteria for sepsis
were based on the Third International Consensus on the Management
of Sepsis and Septic Shock (Sepsis-3) in 2016, which entails a
joint diagnosis by >2 attending physicians. The exclusion
criteria were as follows: i) Patients with tumor, acute stroke,
rheumatic immune system disease and mental illness; ii) patients
with viral myocarditis; iii) patients with severe hepatitis and
cirrhosis; iv) patients who received anti-infective treatment
before enrollment; v) patients with end-stage renal diseases; vi)
patients who died or were discharged within 48 h after admission;
and vii) patients who do not have complete clinical records or do
not cooperate with urine sample collection.
The control group included healthy elderly
individuals [elderly people with no previous history of chronic
disease, age (75.29±5.46), female (58.82%)] who underwent a
physical examination during the same period. All subjects were
recruited between August 2020 and December 2021. The current study
was approved by the Ethics Committee of Huadong Hospital Affiliated
to Fudan University (Shanghai, China). All patients or their family
members (some older people lose the ability to write) signed
informed consent forms before enrollment.
Collection of clinical samples
Urine samples were collected within 24 h after the
onset of sepsis in elderly patients admitted to Huadong Hospital
Affiliated to Fudan University (Shanghai, China). Urine samples
were collected in the morning of the physical examination day in
healthy subjects. All urine samples were centrifuged at 845 x g for
10 min at 4˚C, and the supernatant was aliquoted into 1.8-ml
Eppendorf tubes and frozen within 4 h of collection at -80˚C.
miRNA-sequencing
Total RNA was extracted using the
mirVana™ miRNA Isolation kit (cat. no. AM1561; Thermo
Fisher Scientific, Inc.) and the samples were extracted for total
RNA according to the standard procedure provided by the
manufacturer, and the extracted total RNA was electrophoresed by an
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) for quality
control. The samples were then prepared for use by Agilent 2100
Bioanalyzer (Agilent Technologies, Inc.). The purified total RNA
was subjected to 3' end-joining, 5' end-joining, reverse
transcription, amplification, cDNA library size selection and
purification according to the experimental instructions to complete
the library construction of the sequenced samples. Total RNA was
separated using 17% denaturing polyacrylamide gels and small RNAs
between 10 and 60 nucleotides (nt) were collected. Then, 5'- and
3'-RNA adaptors were ligated to the small RNAs, followed by reverse
transcription to produce cDNAs. These cDNAs were subsequently
amplified by PCR and subjected to Solexa/Illumina sequencing by
Shanghai Biotechnology Corporation. The libraries were created
using the Qubit 2.0 Fluorometer (; Thermo Fisher Scientific, Inc.)
for concentration and the Agilent 2100 for library size. Cluster
generation and first-way sequencing primer hybridization were
performed on the Illumina HiSeq sequencer's cBot (Illumina, Inc.)
according to the appropriate procedure shown in the cBot User
Guide. Sequencing reagents are prepared according to the Illumina
User Guide and flow cells with clusters are loaded onto the
machine. Single-ended sequencing was performed using the
single-read program. The sequencing process was controlled by
Illumina's data collection software (Illumina, Inc.) and real-time
data analysis was performed. FastX software (https://anaconda.org/biobuilds/fastx-toolkit,fastx-toolkit
0.0.14) was used to preprocess the original reads for sequencing,
remove linker sequences and low-quality sequences (including
ambiguous base N sequences with a base quality <10 nt and length
<18 nt), and provide (statistical analysis based on the
processed results table and length distribution diagram. The
sequences obtained through the Sanger miRBase database (https://www.mirbase.org; such as those of known
ribosomal RNA, transfer RNA and repeat regions), RefSeq database
(https://www.ncbi.nlm.nih.gov/refseq/)
and other noncoding RNA databases, including the non-coding RNA,
PIWI-interacting RNA (https://www.smallrnagroup.uni-mainz.de/piRNAclusterDB)
and Rfam databases (https://rfam.xfam.org/), were compared, and the known
miRNAs were annotated. The sequence obtained by sequencing was
compared with the genome database corresponding to the species, the
annotated reads were classified and counted, and the known miRNAs
and various other types of small RNA molecules were identified and
counted. The DEGseq R language package combined with Perl script
was used to group samples according to the current requirements
(such as the control and experimental groups) for comparative
analysis of miRNA expression. In the differential analysis, the
transcripts per million (TPM) formula (single miRNA reads x
106/total reads) was used to present the data.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA was extracted using TransZol Up reagent
(Beijing Transgen Biotech Co., Ltd.). Briefly, the process was as
follows: A total of 1 ml TransZol UP reagent was added into 500 µl
of urine sample, followed by mixing with 200 µl chloroform and
centrifugation at 4˚C for 15 min at 10,000 x g. The aqueous phase
containing the RNA was transferred to a new Eppendorf tube and the
same volume (~500 µl) isopropyl alcohol was added. A total of 1 ml
pre-cooled (4˚C) 75% ethanol was added, and centrifugation at 4˚C
for 5 min at 7,500 x g. The RNA precipitate was air-dried, followed
by dissolution in RNA solution buffer. cDNA synthesis was conducted
with TransScript miRNA First-Strand cDNA Synthesis SuperMix
(TransGen Biotech Co., Ltd.). The RT kit was used according to the
manufacturer's protocol. The reactions were performed in a PCR
instrument and the reaction program was set to 37˚C for 1 h and
85˚C for 5 sec. The Hieff qPCR SYBR Green Master Mix kit (Shanghai
Yeasen Biotechnology Co., Ltd.) was used to perform RT-qPCR assays.
The qPCR cycling conditions were 95˚C for 10 sec, 55˚C for 30 sec
and 72˚C for 30 sec for 40 cycles. Relative quantification of
hsa-miR-31-5p, hsa-miR-151a-3p, hsa-miR-142-5p and hsa-miR-16-5p
was performed by normalization to U6 small nuclear (sn)RNA
expression levels. The 2-ΔΔCq method was used to analyze
miRNA levels (59). The primer
sequences used are presented in Table
SI.
Bioinformatics analysis
Bioinformatics analysis was performed to preprocess
sequencing data and analyze the results. Bioinformatics analyses
included miRNA expression quantitative analysis, expression
correlation analysis, miRNA differential expression analysis,
differential miRNA target gene prediction, and Gene Ontology (GO;
http://www.geneontology.org) and Kyoto
Encyclopedia of Genes and Genomes (KEGG; http://www.kegg.jp/kegg/pathway.html) enrichment
analysis of different miRNA target genes. The results of the GO
enrichment analysis were displayed in plots, where Rich Factor=(the
number of miRNA target genes in a GO term/the number of all target
genes that can correspond to the GO database)/(the number of genes
contained in a GO term/the total number of genes that can
correspond to the GO database). The greater the Rich Factor is, the
greater the degree of enrichment, while the smaller the Q-value is,
the more significant the enrichment (60).
Statistical analysis
All the experimental results in this study were
verified by three biological repetitions to ensure the accuracy of
the experimental results. Data analysis was performed using SPSS 23
software (IBM Corp.). Data are presented as the mean ± SEM. Each
experiment, controlling a single variable and setting up two
experimental groups (the AKI and non-AKI group) and a control
group, had at least three biological repetitions. EdgeR (http://www.R-project.org/) was used to analyze the
difference in miRNAs between samples. After obtaining the P-value,
multiple hypothesis test correction was performed, and the P-value
threshold was determined by controlling the false discovery rate,
thereby providing the Q-value. Fold-change was calculated as the
differential expression based on the TPM value. The screening
conditions for differential genes were as follows: Q-value ≤0.05;
fold-change ≥2. One-way ANOVA followed by Dunnett's multiple
comparisons test was used to compare the groups. Receiver operating
characteristic (ROC) curves were plotted to analyze the predictive
value of miR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p for the
prognosis and 28-day mortality of elderly patients with sepsis. The
ROC curve analysis and the derived area under the curve (AUC)
statistic provide a global and standardized appreciation of the
accuracy of a marker or a composite score for predicting an event
(61). ROC curves were generated
by plotting sensitivity against 1-specificity. P<0.05 was
considered to indicate a statistically significant difference.
Results
Patient demographic and baseline
characteristics
Patients were diagnosed with sepsis, and the main
infection sites were the lung, urinary system and gastrointestinal
tract. According to the general clinical data, the 74 study
subjects included 17 healthy elderly patients, 29 septic patients
with AKI and 28 septic patients without AKI. The mean age was 81
years, with a range of 65-97 years. After 28 days of follow-up in
the observation group, 18 patients of the 57 patients with sepsis
succumbed, accounting for 31.58% (Table SII).
Differential expression of miRNAs in
the sepsis AKI vs. sepsis non-AKI group
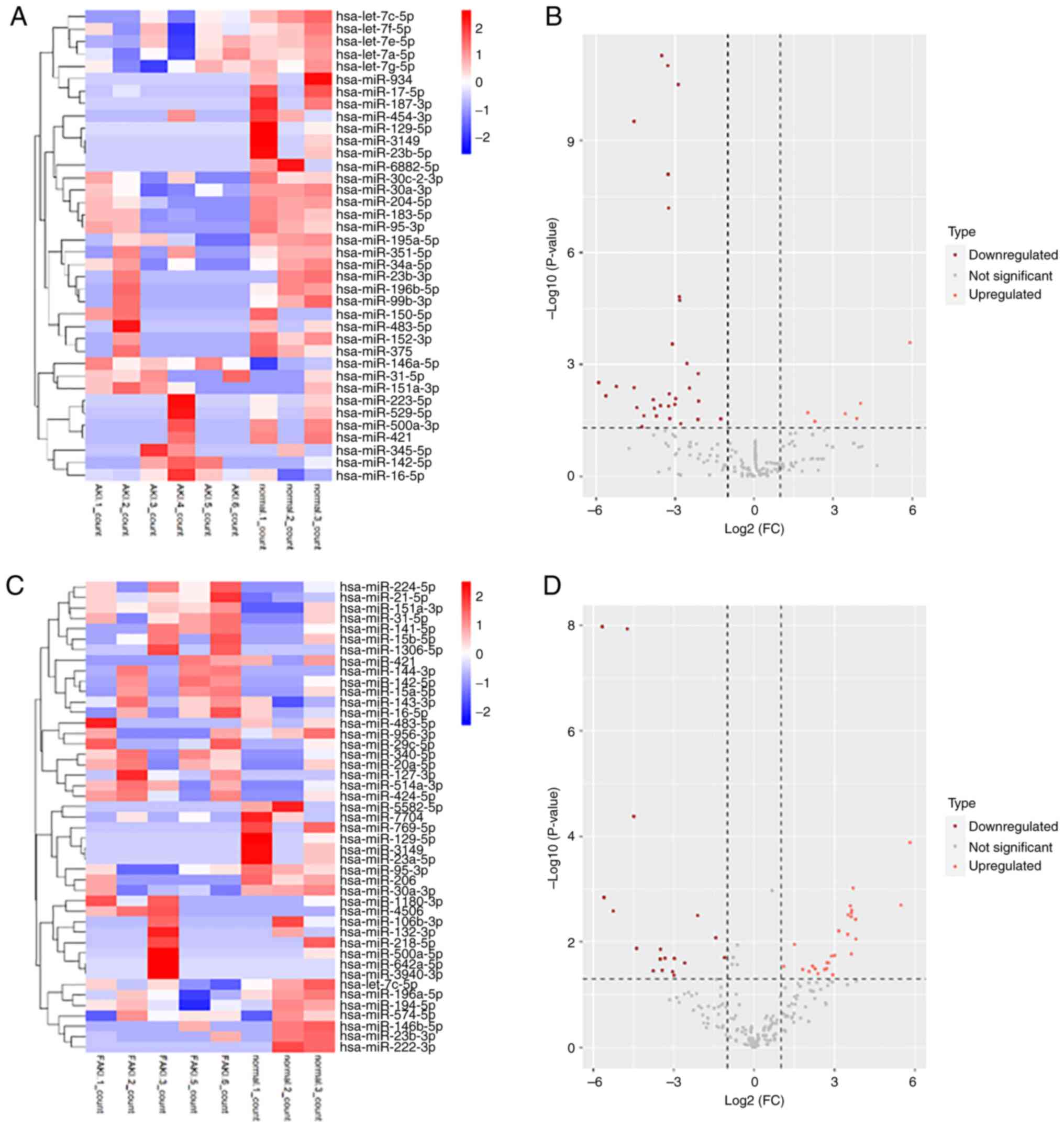
Details of the patients are provided in Table I. Heatmap and cluster analysis
demonstrated that there were differentially expressed miRNAs
between the sepsis AKI and non-AKI groups. The sepsis AKI and
non-AKI groups were compared with the normal group. Among the
differentially expressed miRNAs in the sepsis AKI group, six miRNAs
were upregulated compared with the normal group (Fig. 1A). Among the differentially
expressed miRNAs in the sepsis non-AKI group, 28 miRNAs were
upregulated compared with the normal group (Fig. 1C). The volcano plots show the
differentially expressed miRNAs under the two different conditions
(AKI vs. non-AKI groups) (Fig. 1B
and D).
 | Table IClinical data of the control, sepsis
AKI and sepsis non-AKI groups. |
Table I
Clinical data of the control, sepsis
AKI and sepsis non-AKI groups.
| | Control (n=3) | Non-AKI (n=5) | AKI (n=6) |
|---|
| Characteristic | 1 | 2 | 3 | 1 | 2 | 3 | 4 | 5 | 1 | 2 | 3 | 4 | 5 | 6 |
|---|
| Sex | Female | Male | Female | Female | Male | Male | Female | Male | Female | Male | Female | Male | Male | Female |
| Age | 82 | 81 | 76 | 80 | 77 | 85 | 86 | 89 | 97 | 87 | 77 | 67 | 88 | 83 |
| Lung | no | no | no | yes | no | no | yes | no | yes | yes | yes | no | no | yes |
| Urine | no | no | no | yes | yes | yes | no | yes | no | no | yes | yes | no | yes |
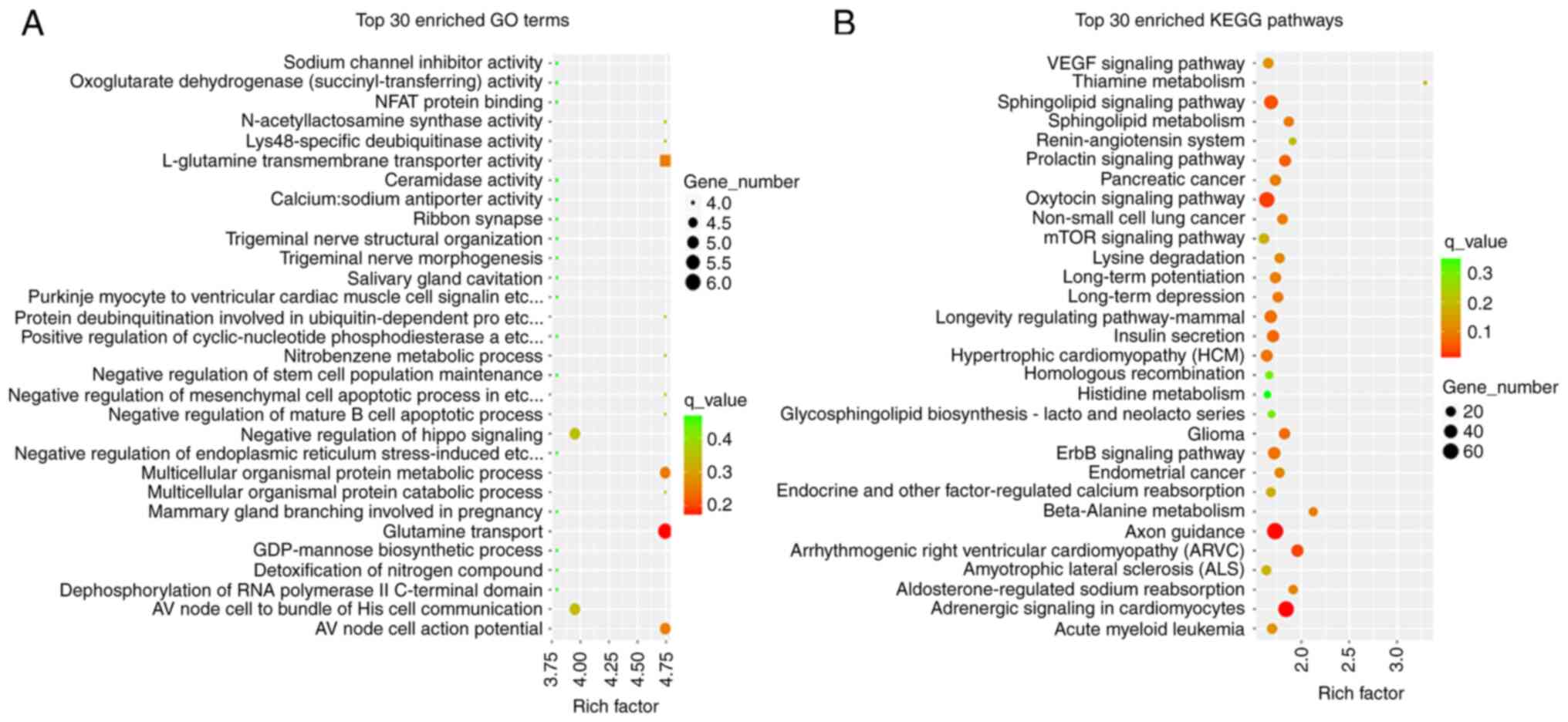
GO function and KEGG signaling pathway
analysis of differentially expressed miRNA target genes
Firstly, the number of target genes corresponding to
the three GO elements, biological process, cellular component and
molecular function, was counted. Only the top 30 GO entries are
shown in Fig. 2A. Using the same
principle as for GO enrichment analysis, KEGG pathway enrichment
analysis was also performed for target genes of differentially
expressed miRNAs, and the results are shown in Fig. 2B.
miRNAs with differentially upregulated
expression in patients with sepsis in the AKI and non-AKI
groups
There were six upregulated miRNAs in the AKI group
(Table II) and 27 in the non-AKI
group (Table SIII) compared with
the control. Analysis of the data revealed that several miRNAs in
the sepsis AKI and non-AKI groups were differentially expressed and
upregulated compared with the control. A high expression trend was
found in the sepsis AKI and non-AKI groups.
 | Table IIUpregulated miRNAs in the sepsis
acute kidney injury group compared with the control group (partial
results). |
Table II
Upregulated miRNAs in the sepsis
acute kidney injury group compared with the control group (partial
results).
| Name | LogFC | Average
expression | t-value | P-value | Adjusted
P-value | B |
|---|
| hsa-miR-345-5p | 5.91748 | 5.52460616 | 6.261657 | 0.000262745 | 0.007849 | -0.70176 |
| hsa-miR-31-5p | 4.029956 | 7.20973673 | 3.300482 | 0.011146469 | 0.139838 | -2.57026 |
|
hsa-miR-151a-3p | 3.458328 | 7.02248642 | 2.87484 | 0.021092684 | 0.200744 | -3.19779 |
| hsa-miR-142-5p | 3.898493 | 7.01479061 | 2.682209 | 0.028311824 | 0.235373 | -3.45029 |
|
hsa-miR-146a-5p | 2.308655 | 10.8501514 | 2.562092 | 0.034059476 | 0.261123 | -3.47134 |
| hsa-miR-16-5p | 2.04111 | 16.2923015 | 2.914069 | 0.019872937 | 0.19589 | -3.71548 |
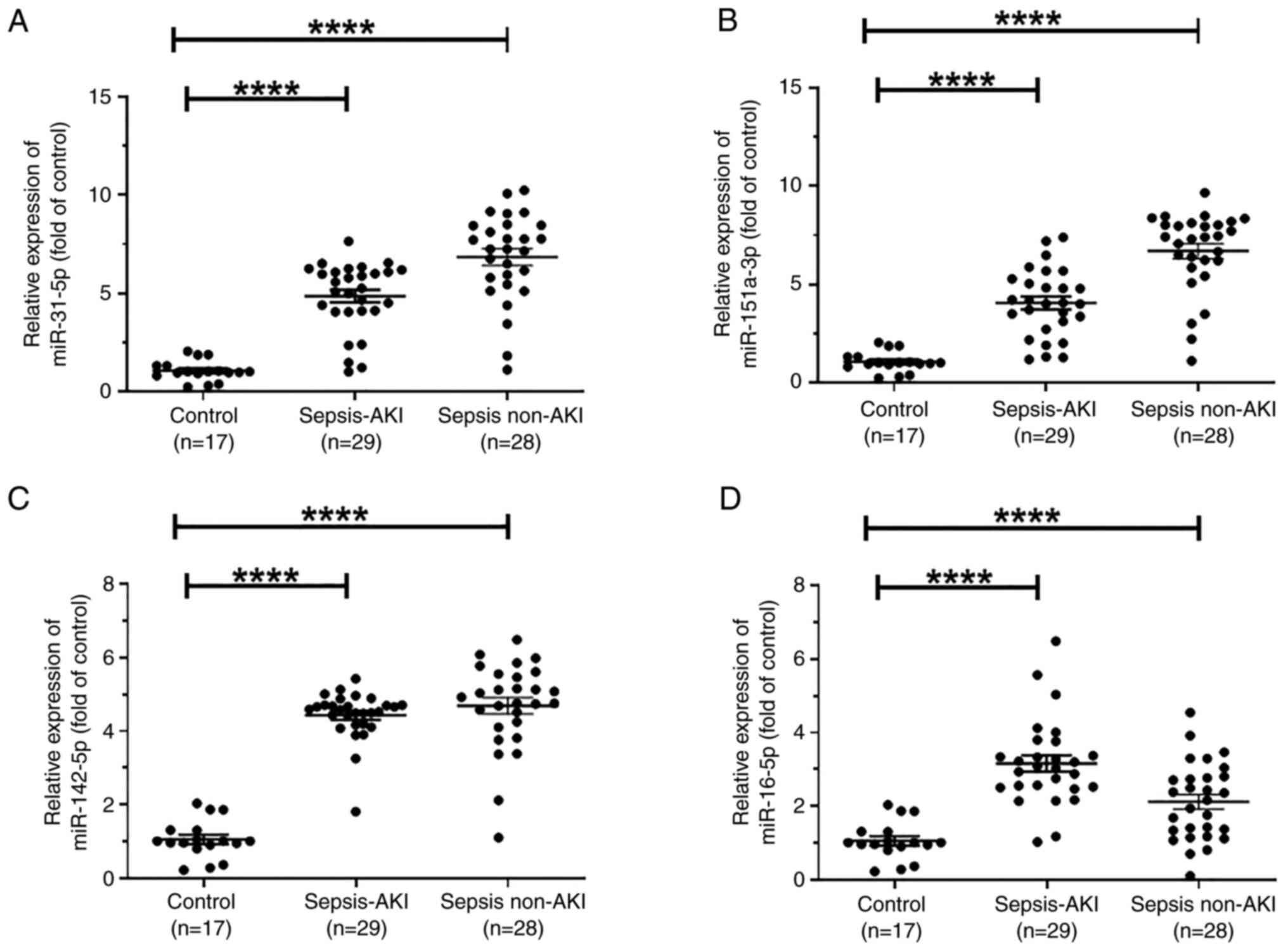
Differentially expressed miRNAs
verified by RT-qPCR
To verify the expression levels of these four miRNAs
in sepsis, 17 samples from healthy controls, 29 samples from
patients with sepsis and AKI and 28 samples from patients with
sepsis without AKI were collected. Details of the patients are
provided in Table SII. Compared
with those in the control group, the expression levels of
miR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p were significantly
increased in the sepsis AKI and sepsis non-AKI groups (Fig. 3A-D).
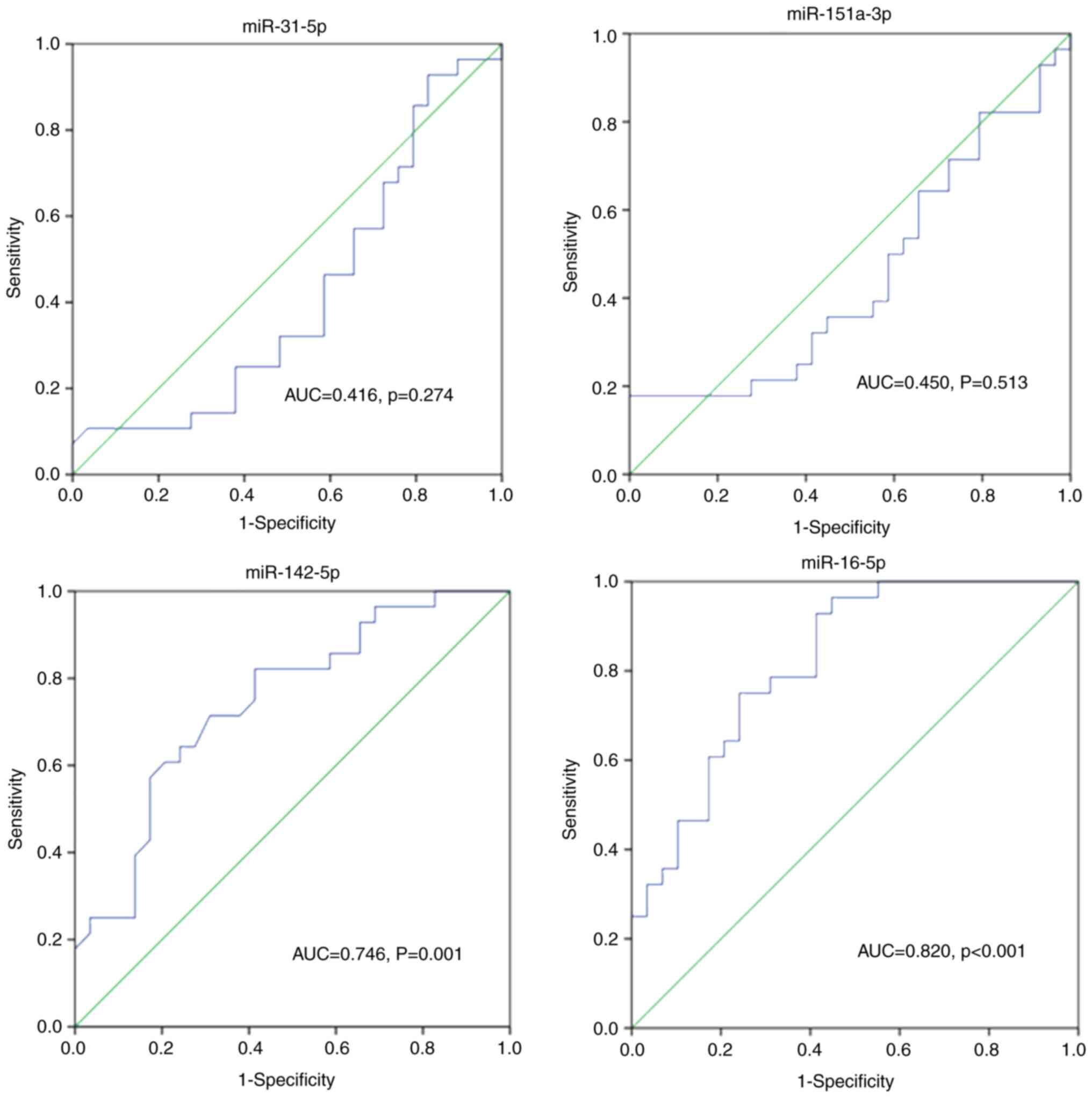
Clinical predictive value of miRNAs
for AKI occurrence in elderly patients with sepsis
ROC curve analysis of miR-31-5p, miR-151a-3p,
miR-142-5p and miR-16-5p was performed to assess their predictive
value in the diagnosis of AKI in elderly patients with sepsis.
Fig. 4 shows that the AUC for
miR-142-5p and miR-16-5p expression was 0.746 and 0.820,
respectively, indicating a good predictive value of miR-142-5p and
miR-16-5p for patients with sepsis-induced AKI. The AUC for
miR-31-5p and miR-151-3p expression was 0.416 (P=0.274) and 0.450
(P=0.513) respectively, with no statistical significance.
Prognostic value of miRNAs for the
28-day survival of 57 septic patients
ROC curves were generated to evaluate the predictive
value of miR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p for the
28-day mortality in patients with sepsis (Fig. 5). The AUC for each miRNA was 0.668,
0.747,0.714 and 0.838, respectively. These results indicated a good
predictive value of miR-31-5p, miR-151a-3p, miR-142-5p and
miR-16-5p in the prognosis of 57 patients with sepsis.
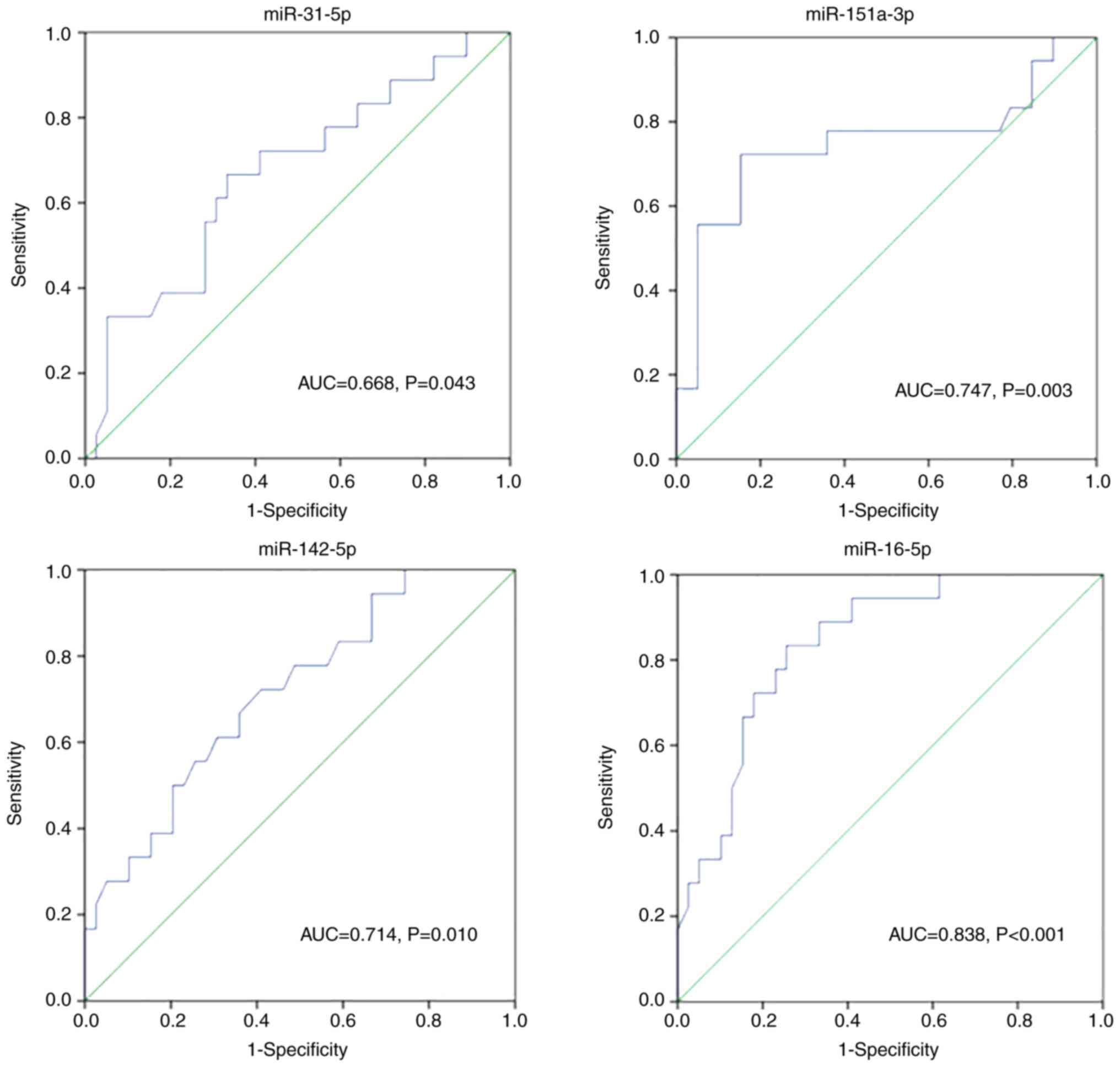 | Figure 5Receiver operating characteristic
curve analysis of miR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p
in predicting 28-day mortality in elderly patients with sepsis. The
sensitivity of miR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p was
66.7, 72.2, 72.2 and 83.3%, and the specificity was 66.7, 84.6,
59.0 and 74.4%, respectively. AUC, area under the curve; miR,
microRNA. |
Discussion
Sepsis is an important clinical area in the
emergency and critical care medicine field. Clinical management of
sepsis remains a major challenge (62). Sepsis can lead to life-threatening
multiple organ dysfunction and has a high fatality rate; therefore,
it is important to reduce the incidence and fatality rate of sepsis
(63). With the in-depth study of
its pathogenesis, biomarkers for the prediction of the prognosis of
sepsis have emerged (64).
Circulating miRNA can be used as a novel candidate biomarker for
the clinical diagnosis and treatment of sepsis (61). miRNAs have previously been
considered as biomarkers in different diseases, such as lung cancer
and sepsis. However, several issues should be investigated before
their use in clinical practice (65-67).
miRNA detection is convenient and fast and its clinical diagnostic
and prognostic value for patients with sepsis and related
complications has gained increasing attention (68,69).
The complex pathogenesis and diagnostic value of miRNAs in sepsis
need to be further explored, as the identification of specific
miRNAs will help to further clarify the pathogenesis of the disease
and provide a way to screen novel clinical diagnostic indicators or
explore molecular targeted therapies (70).
Previous studies show that there are distinct miRNA
regulation models in the different cohorts of patients with sepsis.
This is due to a lack of standardization of sample collection, data
normalization and analysis methods. There is still no optimal
normalization strategy for miRNA analysis from serum, urine or
other samples. Usually, miR-16 or U6 snRNA is used as an internal
gene for normalization. However, U6 snRNA is differentially
regulated between healthy subjects and septic patients (71). Aomatsu et al (72) demonstrated that the upregulation of
miRNA-5100 may inhibit the development of AKI at least partially by
regulating multiple apoptotic pathways, and miRNA-5100 can be used
as a diagnostic biomarker of AKI. Zhao et al (73) found that inhibition of miR-16-5p
could reduce the symptoms of AKI in mice with
ischemia-reperfusion-induced AKI. In addition, it has been reported
that serum miR-16-5p (74) and
serum miR-142-5p (75) are
downregulated in septic patients with AKI.
The present study screened four miRNAs in urine,
namelymiR-31-5p, miR-151a-3p, miR-142-5p and miR-16-5p, as
potential biological markers in patients with sepsis-induced AKI.
These four miRNAs were confirmed by RT-qPCR to be specific markers
for predicting secondary AKI in elderly patients with sepsis.
Therefore, the present study provides potential diagnostic
biomarkers for the early diagnosis, disease staging and prognosis
of elderly patients with sepsis. Future studies are required to
further examine and verify the accuracy and specificity of the four
miRNAs as diagnostic markers in sepsis, laying a foundation for
clinical application. Originally diagnosed and treated according to
conventional methods, our study offers the possibility of early
diagnosis and prognostic judgement. Based on the present study
results and ROC curve analysis, miR-16-5p showed the best
diagnostic results among the four genes examined. Further studies
will verify the regulatory mechanism of miR-16-5p, its relationship
with the major gene of pyroptosis and gasdermin D and its
regulatory mechanism in AKI.As circulating miRNAs have several
advantages, such as the easy and noninvasive sample collection from
patients, it is possible for them to have a wide use in the clinic
(76). The four miRNAs examined in
the present study can facilitate the rapid initiation of directed
treatment in sepsis and infection.
In summary, the present study indicated that
specific miRNAs, and especially miR-16-5p, represent novel
candidates for the clinical management of patients with sepsis.
Supplementary Material
The sequence of the primers used in
the present study.
The sample information of 74 cases
patients
Sepsis group non-AKI vs. control group
up-regulate miRNAs (partial results).
Acknowledgements
Not applicable.
Funding
Funding: The present study was supported by Shanghai Innovation
Project (grant no. 20S11901300).
Availability of data and materials
The datasets generated and/or analyzed during the
current study are available in the Sequence Read Archive repository
(accession no. PRJNA906749; https://www.ncbi.nlm.nih.gov/sra/PRJNA906749).
Authors' contributions
ZM and ZB conceived the experiments. RH and WL
developed the methodology. RH, WL, HT, YZ, HZ and WP performed the
experiments. WP, XW and LX organized and analyzed the data. RH and
ZM wrote the manuscript. WP and ZM confirmed the authenticity of
all the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The current study was approved by the Ethics
Committee of Huadong Hospital Affiliated to Fudan University
(approval no. 2020K039; Shanghai, China).
Patient consent for publication
All patients or their family members signed informed
consent forms before enrollment.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rudd KE, Johnson SC, Agesa KM, Shackelford
KA, Tsoi D, Kievlan DR, Colombara DV, Ikuta KS, Kissoon N, Finfer
S, et al: Global, regional, and national sepsis incidence and
mortality, 1990-2017: Analysis for the global burden of disease
study. Lancet. 395:200–211. 2020.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Singer M, Deutschman CS, Seymour CW,
Shankar-Hari M, Annane D, Bauer M, Bellomo R, Bernard GR, Chiche
JD, Coopersmith CM, et al: The third international consensus
definitions for sepsis and septic shock (sepsis-3). JAMA.
315:801–810. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Cerceo E, Rachoin JS, Gaughan J and
Weisberg L: Association of gender, age, and race on renal outcomes
and mortality in patients with severe sepsis and septic shock. J
Crit Care. 61:52–56. 2021.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Chinai B, Gaughan J and Schorr C:
Implementation of the affordable care Act: A comparison of outcomes
in patients with severe sepsis and septic shock using the national
inpatient sample. Crit Care Med. 48:783–789. 2020.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Gong Y, Li D, Cheng B, Ying B and Wang B:
Increased neutrophil percentage-to-albumin ratio is associated with
all-cause mortality in patients with severe sepsis or septic shock.
Epidemiol Infect. 148(e87)2020.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Gazmuri RJ, Añez de Gomez CI, Siddiqui M,
Schechtman J and Nadeem AUR: Severe sepsis and septic shock early
management bundle risks aiding vasopressor misuse. Crit Care Med.
47(e717)2019.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Shankar-Hari M, Phillips GS, Levy ML,
Seymour CW, Liu VX, Deutschman CS, Angus DC, Rubenfeld GD and
Singer M: Sepsis Definitions Task Force. Developing a new
definition and assessing new clinical criteria for septic shock:
For the third international consensus definitions for sepsis and
septic shock (sepsis-3). JAMA. 315:775–787. 2016.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Angus DC, Seymour CW, Coopersmith CM,
Deutschman CS, Klompas M, Levy MM, Martin GS, Osborn TM, Rhee C and
Watson RS: A framework for the development and interpretation of
different sepsis definitions and clinical criteria. Crit Care Med.
44:e113–e121. 2016.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Chang JC: Sepsis and septic shock:
Endothelial molecular pathogenesis associated with vascular
microthrombotic disease. Thromb J. 17(10)2019.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Wang T, Zhang X, Liu Z, Yao T, Zheng D,
Gan J, Yu S, Li L, Chen P and Sun J: Single-cell RNA sequencing
reveals the sustained immune cell dysfunction in the pathogenesis
of sepsis secondary to bacterial pneumonia. Genomics.
113:1219–1233. 2021.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Hashemian SM, Pourhanifeh MH, Fadaei S,
Velayati AA, Mirzaei H and Hamblin MR: Non-coding RNAs and
exosomes: Their role in the pathogenesis of sepsis. Mol Ther
Nucleic Acids. 21:51–74. 2020.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Sungurlu S, Kuppy J and Balk RA: Role of
antithrombin III and tissue factor pathway in the pathogenesis of
sepsis. Crit Care Clin. 36:255–265. 2020.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Zhang H, Feng YW and Yao YM: A profound
understanding of the pathogenesis network in sepsis. Zhonghua Yi
Xue Za Zhi. 100:881–885. 2020.PubMed/NCBI View Article : Google Scholar : (In Chinese).
|
|
14
|
Shenoy S: Coronavirus (Covid-19) sepsis:
Revisiting mitochondrial dysfunction in pathogenesis, aging,
inflammation, and mortality. Inflamm Res. 69:1077–1085.
2020.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Nie C, Qian KJ, Wang LQ, Liu F, Zeng ZG,
Zhu F, Xia L and Zhan YA: A clinical study on organ protective
effect of early high-volume hemofiltration (HVHF) in patients with
multiple organ dysfunction syndrome (MODS) complicated by acute
kidney injury (AKI). Zhongguo Wei Zhong Bing Ji Jiu Yi Xue.
23:605–607. 2011.PubMed/NCBI(In Chinese).
|
|
16
|
Erlanger D, Assous MV, Wiener-Well Y,
Yinnon AM and Ben-Chetrit E: Clinical manifestations, risk factors
and prognosis of patients with Morganella morganii sepsis. J
Microbiol Immunol Infect. 52:443–448. 2019.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Kasahara E and Inoue M: Cross-talk between
HPA-axis-increased glucocorticoids and mitochondrial stress
determines immune responses and clinical manifestations of patients
with sepsis. Redox Rep. 20:1–10. 2015.PubMed/NCBI View Article : Google Scholar
|
|
18
|
El-Hefnawy SM, Mostafa RG, El Zayat RS,
Elfeshawy EM, Abd El-Bari HM and El-Monem Ellaithy MA: Biochemical
and molecular study on serum miRNA-16a and miRNA-451 as neonatal
sepsis biomarkers. Biochem Biophys Rep. 25(100915)2021.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Fabri-Faja N, Calvo-Lozano O, Dey P,
Terborg RA, Estevez MC, Belushkin A, Yesilköy F, Duempelmann L,
Altug H, Pruneri V and Lechuga LM: Early sepsis diagnosis via
protein and miRNA biomarkers using a novel point-of-care photonic
biosensor. Anal Chim Acta. 1077:232–242. 2019.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Li Z, Huang B, Yi W, Wang F, Wei S, Yan H,
Qin P, Zou D, Wei R and Chen N: Identification of potential early
diagnostic biomarkers of sepsis. J Inflamm Res. 14:621–631.
2021.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Kyriazopoulou E, Poulakou G and
Giamarellos-Bourboulis EJ: Biomarkers in sepsis: Can they help
improve patient outcome? Curr Opin Infect Dis. 34:126–134.
2021.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Zhang M, Xie M, Wang Y, Li J and Zhou J:
Combination value of biomarkers in discriminating adult onset
Still's disease and sepsis. Wien Klin Wochenschr. 133:118–122.
2021.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Poston JT and Koyner JL: Sepsis associated
acute kidney injury. BMJ. 364(k4891)2019.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Hoste EA, Bagshaw SM, Bellomo R, Cely CM,
Colman R, Cruz DN, Edipidis K, Forni LG, Gomersall CD, Govil D, et
al: Epidemiology of acute kidney injury in critically ill patients:
The multinational AKI-EPI study. Intensive Care Med. 41:1411–1423.
2015.PubMed/NCBI View Article : Google Scholar
|
|
25
|
Fabbian F, Savriè C, De Giorgi A,
Cappadona R, Di Simone E, Boari B, Storari A, Gallerani M and
Manfredini R: Acute kidney injury and in-hospital mortality: A
retrospective analysis of a nationwide administrative database of
elderly subjects in Italy. J Clin Med. 8(1371)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Bagshaw SM, Uchino S, Bellomo R, Morimatsu
H, Morgera S, Schetz M, Tan I, Bouman C, Macedo E, Gibney N, et al:
Septic acute kidney injury in critically ill patients: Clinical
characteristics and outcomes. Clin J Am Soc Nephrol. 2:431–439.
2007.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Vasilescu C, Dragomir M, Tanase M, Giza D,
Purnichescu-Purtan R, Chen M, Yeung SJ and Calin GA: Circulating
miRNAs in sepsis-A network under attack: An in-silico prediction of
the potential existence of miRNA sponges in sepsis. PLoS One.
12(e0183334)2017.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Dumache R, Rogobete AF, Bedreag OH,
Sarandan M, Cradigati AC, Papurica M, Dumbuleu CM, Nartita R and
Sandesc D: Use of miRNAs as biomarkers in sepsis. Anal Cell Pathol
(Amst). 2015(186716)2015.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Ahmad S, Ahmed MM, Hasan PMZ, Sharma A,
Bilgrami AL, Manda K, Ishrat R and Syed MA: Identification and
validation of potential miRNAs, as biomarkers for sepsis and
associated lung injury: A network-based approach. Genes (Basel).
11(1327)2020.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Zhang Y, Li M, Bao L and Hu P: A
case-control study on the relationship between miRNAs single
nucleotide polymorphisms and sepsis risk. Medicine (Baltimore).
98(e16744)2019.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Reinhart K and Carlet J: Procalcitonin-a
new marker of severe infection and sepsis. Intensive Care Med. 26
(Suppl 2)(S145)2000.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Stark A, Brennecke J, Russell RB and Cohen
SM: Identification of Drosophila MicroRNA targets. PLoS Biol.
1(E60)2003.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Xu P, Vernooy SY, Guo M and Hay BA: The
Drosophila microRNA Mir-14 suppresses cell death and is required
for normal fat metabolism. Curr Biol. 13:790–795. 2003.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Jeker LT and Bluestone JA: MicroRNA
regulation of T-cell differentiation and function. Immunol Rev.
253:65–81. 2013.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Mitchell PS, Parkin RK, Kroh EM, Fritz BR,
Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant
KC, Allen A, et al: Circulating microRNAs as stable blood-based
markers for cancer detection. Proc Natl Acad Sci USA.
105:10513–10518. 2008.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Pigati L, Yaddanapudi SC, Iyengar R, Kim
DJ, Hearn SA, Danforth D, Hastings ML and Duelli DM: Selective
release of microRNA species from normal and malignant mammary
epithelial cells. PLoS One. 5(e13515)2010.PubMed/NCBI View Article : Google Scholar
|
|
37
|
Mir R, Elfaki I, Khullar N, Waza AA, Jha
C, Mir MM, Nisa S, Mohammad B, Mir TA, Maqbool M, et al: Role of
selected miRNAs as diagnostic and prognostic biomarkers in
cardiovascular diseases, including coronary artery disease,
myocardial infarction and atherosclerosis. J Cardiovasc Dev Dis.
8(22)2021.PubMed/NCBI View Article : Google Scholar
|
|
38
|
Gessner I, Fries JWU, Brune V and Mathur
S: Magnetic nanoparticle-based amplification of microRNA detection
in body fluids for early disease diagnosis. J Mater Chem B. 9:9–22.
2021.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Demircan T, Sibai M, Avşaroğlu ME,
Altuntaş E and Ovezmyradov G: The first report on circulating
microRNAs at Pre- and Post-metamorphic stages of axolotls. Gene.
768(145258)2021.PubMed/NCBI View Article : Google Scholar
|
|
40
|
Tan PPS, Hall D, Chilian WM, Chia YC, Mohd
Zain S, Lim HM, Kumar DN, Ching SM, Low TY, Md Noh MF and Pung YF:
Exosomal microRNAs in the development of essential hypertension and
its potential as biomarkers. Am J Physiol Heart Circ Physiol.
320:H1486–H1497. 2021.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Cui C and Cui Q: The relationship of human
tissue microRNAs with those from body fluids. Sci Rep.
10(5644)2020.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Harrill AH and Sanders AP: Urinary
MicroRNAs in environmental health: Biomarkers of emergent kidney
injury and disease. Curr Environ Health Rep. 7:101–108.
2020.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Dos Santos CC, Amatullah H, Vaswani CM,
Maron-Gutierrez T, Kim M, Mei SHJ, Szaszi K, Monteiro APT, Varkouhi
AK, Herreroz R, et al: Mesenchymal stromal (stem) cell therapy
modulates miR-193b-5p expression to attenuate sepsis-induced acute
lung injury. Eur Respir J. 59(2004216)2022.PubMed/NCBI View Article : Google Scholar
|
|
44
|
Jiao Y, Zhang T, Zhang C, Ji H, Tong X,
Xia R, Wang W, Ma Z and Shi X: Exosomal miR-30d-5p of neutrophils
induces M1 macrophage polarization and primes macrophage pyroptosis
in sepsis-related acute lung injury. Crit Care.
25(356)2021.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Roderburg C, Luedde M, Vargas Cardenas D,
Vucur M, Scholten D, Frey N, Koch A, Trautwein C, Tacke F and
Luedde T: Circulating microRNA-150 serum levels predict survival in
patients with critical illness and sepsis. PLoS One.
8(e54612)2013.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Roderburg C, Koch A, Benz F, Vucur M,
Spehlmann M, Loosen SH, Luedde M, Rehse S, Lurje G, Trautwein C, et
al: Serum levels of miR-143 predict survival in critically Ill
patients. Dis Markers. 2019(4850472)2019.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Han Y, Dai QC, Shen HL and Zhang XW:
Diagnostic value of elevated serum miRNA-143 levels in sepsis. J
Int Med Res. 44:875–881. 2016.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Zhu C, Chen T and Liu B: Inhibitory
effects of miR-25 targeting HMGB1 on macrophage secretion of
inflammatory cytokines in sepsis. Oncol Lett. 16:5027–5033.
2018.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Moon HG, Yang J, Zheng Y and Jin Y:
miR-15a/16 regulates macrophage phagocytosis after bacterial
infection. J Immunol. 193:4558–4567. 2014.PubMed/NCBI View Article : Google Scholar
|
|
50
|
Wang X, Wang X, Liu X, Wang X, Xu J, Hou
S, Zhang X and Ding Y: miR-15a/16 are upreuglated in the serum of
neonatal sepsis patients and inhibit the LPS-induced inflammatory
pathway. Int J Clin Exp Med. 8:5683–5690. 2015.PubMed/NCBI
|
|
51
|
Arroyo AB, Águila S, Fernández-Pérez MP,
Reyes-García AML, Reguilón-Gallego L, Zapata-Martínez L, Vicente V,
Martínez C and González-Conejero R: miR-146a in cardiovascular
diseases and sepsis: An additional burden in the inflammatory
balance? Thromb Haemost. 121:1138–1150. 2021.PubMed/NCBI View Article : Google Scholar
|
|
52
|
Pfeiffer D, Roßmanith E, Lang I and
Falkenhagen D: miR-146a, miR-146b, and miR-155 increase expression
of IL-6 and IL-8 and support HSP10 in an in vitro sepsis model.
PLoS One. 12(e0179850)2017.PubMed/NCBI View Article : Google Scholar
|
|
53
|
Bai X, Zhang J, Cao M, Han S, Liu Y, Wang
K, Han F, Li X, Jia Y, Wang X, et al: MicroRNA-146a protects
against LPS-induced organ damage by inhibiting Notch1 in
macrophage. Int Immunopharmacol. 63:220–226. 2018.PubMed/NCBI View Article : Google Scholar
|
|
54
|
Möhnle P, Hirschberger S, Hinske LC,
Briegel J, Hübner M, Weis S, Dimopoulos G, Bauer M,
Giamarellos-Bourboulis EJ and Kreth S: MicroRNAs 143 and 150 in
whole blood enable detection of T-cell immunoparalysis in sepsis.
Mol Med. 24(54)2018.PubMed/NCBI View Article : Google Scholar
|
|
55
|
SepNet Critical Care Trials Group.
Incidence of severe sepsis and septic shock in German intensive
care units: The prospective, multicentre INSEP study. Intensive
Care Med. 42:1980–1989. 2016.PubMed/NCBI View Article : Google Scholar
|
|
56
|
Baykara N, Akalın H, Arslantaş MK, Hancı
V, Çağlayan Ç, Kahveci F, Demirağ K, Baydemir C and Ünal N: Sepsis
Study Group. Epidemiology of sepsis in intensive care units in
Turkey: A multicenter, point-prevalence study. Crit Care.
22(93)2018.PubMed/NCBI View Article : Google Scholar
|
|
57
|
Zhou J, Qian C, Zhao M, Yu X, Kang Y, Ma
X, Ai Y, Xu Y, Liu D, An Y, et al: Epidemiology and outcome of
severe sepsis and septic shock in intensive care units in mainland
China. PLoS One. 9(e107181)2014.PubMed/NCBI View Article : Google Scholar
|
|
58
|
Mizuno T, Sato W, Ishikawa K, Shinjo H,
Miyagawa Y, Noda Y, Imai E and Yamada K: KDIGO (kidney disease:
Improving global outcomes) criteria could be a useful outcome
predictor of cisplatin-induced acute kidney injury. Oncology.
82:354–359. 2012.PubMed/NCBI View Article : Google Scholar
|
|
59
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
60
|
Cui M, Cheng C and Zhang L:
High-throughput proteomics: A methodological mini-review. Lab
Invest. 102:1170–1181. 2022.PubMed/NCBI View Article : Google Scholar
|
|
61
|
Cheng H, Xu JH, Kang XH, Wu CC, Tang XN,
Chen ML, Lian ZS, Li N and Xu XL: Nomograms for predicting overall
survival and cancer-specific survival in elderly patients with
epithelial ovarian cancer. J Ovarian Res. 16(75)2023.PubMed/NCBI View Article : Google Scholar
|
|
62
|
Hegamyer E, Smith N, Thompson AD and
Depiero AD: Treatment of suspected sepsis and septic shock in
children with chronic disease seen in the pediatric emergency
department. Am J Emerg Med. 44:56–61. 2021.PubMed/NCBI View Article : Google Scholar
|
|
63
|
D'Onofrio V, Meersman A, Vijgen S,
Cartuyvels R, Messiaen P and Gyssens IC: Risk factors for
mortality, intensive care unit admission, and bacteremia in
patients suspected of sepsis at the emergency department: A
prospective cohort study. Open Forum Infect Dis.
8(ofaa594)2020.PubMed/NCBI View Article : Google Scholar
|
|
64
|
Barichello T, Generoso JS, Singer M and
Dal-Pizzol F: Biomarkers for sepsis: More than just fever and
leukocytosis-a narrative review. Crit Care. 26(14)2022.PubMed/NCBI View Article : Google Scholar
|
|
65
|
Nassar FJ, Msheik ZS, Itani MM, Helou RE,
Hadla R, Kreidieh F, Bejjany R, Mukherji D, Shamseddine A, Nasr RR
and Temraz SN: Circulating miRNA as biomarkers for colorectal
cancer diagnosis and liver metastasis. Diagnostics (Basel).
11(341)2021.PubMed/NCBI View Article : Google Scholar
|
|
66
|
Smolarz M and Widlak P: Serum exosomes and
their miRNA load-a potential biomarker of lung cancer. Cancers
(Basel). 13(1373)2021.PubMed/NCBI View Article : Google Scholar
|
|
67
|
Wang M, Zheng S, Li X, Ding Y, Zhang M,
Lin L, Xu H, Cheng Y, Zhang X, Xu H and Li S: Integrated analysis
of lncRNA-miRNA-mRNA ceRNA network identified lncRNA EPB41L4A-AS1
as a potential biomarker in non-small cell lung cancer. Front
Genet. 11(511676)2020.PubMed/NCBI View Article : Google Scholar
|
|
68
|
Liu S, Liu C, Wang Z, Huang J and Zeng Q:
microRNA-23a-5p acts as a potential biomarker for sepsis-induced
acute respiratory distress syndrome in early stage. Cell Mol Biol
(Noisy-le-grand). 62:31–37. 2016.PubMed/NCBI
|
|
69
|
Bukauskas T, Kairytė M, Mickus R,
Puleikytė L and Macas A: Values of circulating molecular biomarkers
(microRNAs) for the evaluation of renal failure during urgent
abdominal sepsis anaesthesia. Acta Med Litu. 26:17–24.
2019.PubMed/NCBI View Article : Google Scholar
|
|
70
|
Hollen MK, Stortz JA, Darden D, Dirain ML,
Nacionales DC, Hawkins RB, Cox MC, Lopez MC, Rincon JC, Ungaro R,
et al: Myeloid-derived suppressor cell function and epigenetic
expression evolves over time after surgical sepsis. Crit Care.
23(355)2019.PubMed/NCBI View Article : Google Scholar
|
|
71
|
Benz F, Roderburg C, Vargas Cardenas D,
Vucur M, Gautheron J, Koch A, Zimmermann H, Janssen J,
Nieuwenhuijsen L, Luedde M, et al: U6 is unsuitable for
normalization of serum miRNA levels in patients with sepsis or
liver fibrosis. Exp Mol Med. 45(e42)2013.PubMed/NCBI View Article : Google Scholar
|
|
72
|
Aomatsu A, Kaneko S, Yanai K, Ishii H, Ito
K, Hirai K, Ookawara S, Kobayashi Y, Sanui M and Morishita Y:
MicroRNA expression profiling in acute kidney injury. Transl Res.
244:1–31. 2022.PubMed/NCBI View Article : Google Scholar
|
|
73
|
Zhao W, Zhang Y, Zhang M, Zhi Y, Li X and
Liu X: Effects of total glucosides of paeony on acute renal injury
following ischemia-reperfusion via the lncRNA HCG18/miR-16-5p/Bcl-2
axis. Immunobiology. 227(152179)2022.PubMed/NCBI View Article : Google Scholar
|
|
74
|
Pan W, Zhang J, Hu L and Huang Z:
Evaluation value of serum miR-4299 and miR-16-5p in risk
stratification of sepsis-induced acute kidney injury. Biomed Res
Int. 2022(5165892)2022.PubMed/NCBI View Article : Google Scholar
|
|
75
|
Tang B, Li W, Ji T, Li X, Qu X, Feng L,
Zhu Y, Qi Y, Zhu C and Bai S: Downregulation of XIST ameliorates
acute kidney injury by sponging miR-142-5p and targeting PDCD4. J
Cell Physiol. 235:8852–8863. 2020.PubMed/NCBI View Article : Google Scholar
|
|
76
|
Reithmair M, Buschmann D, Märte M,
Kirchner B, Hagl D, Kaufmann I, Pfob M, Chouker A, Steinlein OK,
Pfaffl MW and Schelling G: Cellular and extracellular miRNAs are
blood-compartment-specific diagnostic targets in sepsis. J Cell Mol
Med. 21:2403–2411. 2017.PubMed/NCBI View Article : Google Scholar
|