Introduction
Gliomas, which originate from neuroepithelial
tissue, account for 31% of all central nervous system (CNS) tumors
and 81% of all malignant CNS tumors; they are the most common type
of primary brain tumors worldwide (1,2).
Great progress has been made in the treatment of gliomas, with the
most commonly used treatments including tumor resection, adjuvant
radiotherapy, chemotherapy and immunotherapy. However, gliomas,
particularly glioblastoma (GBM), which is a grade IV tumor
according to the glioma grading standards of the World Health
Organization (WHO) (1), are highly
malignant, strongly invasive and prone to recurrence. As a result,
patients with GBM have a median survival time of only 1.5 years,
and a 5-year survival rate of only 5.5% (3,4).
Gliomas severely affect the quality of life and health of patients;
therefore, it is important to identify novel therapeutic mechanisms
and possible therapeutic targets for gliomas.
Replication factor A (RPA) is a heterotrimer that
comprises RPA1 (70 kDa), RPA2 (32 kDa) and RPA3 (14 kDa). RPA1 is
the main subunit of the RPA complex and the main contributor to its
biological function; it also contributes to the stabilization of
the complex formed by subunits RPA2 and RPA3 (5-8). RPA1 has the
following important roles: i) Maintains the correct replication of
DNA (7); ii) participates in DNA
repair, homologous recombination and DNA damage monitoring
(8,9); and iii) participates in the
occurrence and development of numerous kinds of tumors (10-13). To date, a
number of studies on the association of RPA1 with tumors have shown
that RPA1 serves as an oncogene, promoting the occurrence and
development of liver cancer, bladder urothelial carcinoma,
nasopharyngeal carcinoma, esophageal cancer, gastrointestinal
cancer and other tumors (10-13). However, to
the best of our knowledge, there have been no studies on the
associations of RPA1 expression with glioma cell proliferation or
the prognosis of patients with glioma.
The main aim of the present study was to explore the
expression of RPA1 in glioma and its clinical relevance in the
clinicopathology and prognosis of glioma. In addition, the
biological functions and signal transduction pathways mediated by
RPA1 were predicted using bioinformatics analysis to provide basic
theoretical support for further mechanistic research and in
vivo experiments.
Materials and methods
Bioinformatics analysis
All data used in the bioinformatics analysis were
obtained from and analyzed using the following public databases:
the UALCAN (http://ualcan.path.uab.edu/index.html), The Cancer
Genome Atlas (TCGA; https://cancergenome.nih.gov/), STRING (https://string-db.org/) and the DAVID online analysis
tool (https://davidbioinformatics.nih.gov). Gene Ontology
(GO) and Kyoto Encyclopedia of Genome (KEGG) via the DAVID online
analysis tool. Gene expression levels were compared and
Kaplan-Meier survival analyses performed using the UALCAN online
tool, which provides access to cancer transcriptome data from
TCGA.
RPA1-related data were retrieved from TCGA, and
differentially expressed genes were screened using the limma
package in R (3.56.0; https://bioconductor.org/packages/limma/). The
differentially expressed genes were subjected to protein-protein
interaction (PPI) network analysis using the STRING database, which
provides known and predicted protein interactions. The results were
visualized using Cytoscape, and the molecular complex detection
(MCODE) plug-in was used to identify clustered sub-networks within
the PPI network In the GO and KEGG gene enrichment analysis
conducted via DAVID, the significance threshold was set at
P<0.05.
Cell culture
The HA1800 normal astrocyte cell line, two low-grade
glioma (LGG) cell lines, namely U251 and SF295, and two GBM cell
lines, namely A172 and TG905, were acquired. The normal astrocyte
cell lines HA1800 and human glioma cell lines U251, SF295, A172 and
TG905 were purchased from the Cell Bank of the Chinese Academy of
Science. All cell lines were cultured in Dulbecco's modified
Eagle's medium (DMEM) (HyClone; Cytiva) supplemented with 10% fetal
bovine serum (HyClone; Cytiva), 100 g/ml streptomycin and 100 U/ml
penicillin (Gibco; Thermo Fisher Scientific, Inc.) in a humidified
atmosphere of 5% CO2 at 37˚C. The cells were collected
and homogenized, and the cellular proteins and RNA were extracted
as described below.
Sample acquisition and general data
collection
Glioma tissue samples (WHO grades I-IV) were
collected by neurosurgeons from 70 patients who underwent brain
glioma resection and were diagnosed with glioma by postoperative
histopathological examination at Linyi People's Hospital (Linyi,
China) between June 2014 and December 2016 (Table I). In addition, 10 normal brain
tissue samples were obtained from patients with craniocerebral
trauma during frontal lobe decompression at Linyi People's Hospital
(Linyi, China) between June 2014 and December 2016 (male:female,
6:4; 3 were <44 years old and 7 were ≥44 years old). Basic
patient characteristics and clinical diagnosis information were
obtained from all participants. The inclusion criteria comprised
the availability of imaging data and complete details of the
specific treatment process. Patients with a clear imaging diagnosis
and postoperative pathological diagnosis were included in the
experimental group. The study was approved by the Human Ethics
Committee of Linyi People's Hospital (approval no. 30036), and all
patients provided written informed consent.
 | Table IAnalysis of the associations between
RPA1 expression and clinicopathological parameters in 70 patients
with glioma. |
Table I
Analysis of the associations between
RPA1 expression and clinicopathological parameters in 70 patients
with glioma.
| | RPA1 expression, n
(%) | |
|---|
| Clinicopathological
parameters | Cases, n (%) | Low | High | χ2 | P-value |
|---|
| Sex | | | | 0.043 | 0.836 |
|
Male | 36 (51.4) | 15 (41.7) | 21 (58.3) | | |
|
Female | 34 (48.6) | 15 (44.1) | 19 (55.9) | | |
| Age, years | | | | 0.019 | 0.890 |
|
<44 | 38 (54.3) | 16 (42.1) | 22 (57.9) | | |
|
≥44 | 32 (45.7) | 14 (43.8) | 18 (56.2) | | |
| Tumor diameter,
cm | | | | 2.337 | 0.126 |
|
<4 | 17 (24.3) | 10 (58.9) | 7 (41.1) | | |
|
≥4 | 53 (75.7) | 20 (37.7) | 33 (62.3) | | |
| Tumor location | | | | 2.705 | 0.259 |
|
Frontal
lobe | 33 (47.1) | 16 (48.5) | 17 (51.5) | | |
|
Temporal
lobe | 21 (30.0) | 10 (47.6) | 11 (52.4) | | |
|
Other | 16 (22.9) | 4 (25.0) | 12 (75.0) | | |
| KPS | | | | 1.544 | 0.214 |
|
<80 | 36 (51.4) | 18 (50.0) | 18 (50.0) | | |
|
≥80 | 34 (48.6) | 12 (35.3) | 22 (64.7) | | |
| WHO grade | | | | 8.4 | 0.037 |
|
Ⅰ-Ⅱ | 35 (50.0) | 21 (60.0) | 14 (40.0) | | |
|
Ⅲ-Ⅳ | 35 (50.0) | 9 (25.7) | 26 (74.3) | | |
| Ki-67
expression | | | | 11.748 | 0.001 |
|
Low | 26 (37.1) | 18 (69.2) | 8 (30.8) | | |
|
High | 44 (62.9) | 12 (27.3) | 32 (62.7) | | |
| p53 mutation
status | | | | 7.099 | 0.008 |
|
Wild
type | 25 (35.7) | 16 (64.0) | 9 (36.0) | | |
|
Mutant | 45 (64.3) | 14 (31.1) | 31 (68.9) | | |
Western blot analysis
Western blot analysis was performed to determine the
RPA1 expression levels of the cell lines. Total protein was
extracted from the cell homogenate in RIPA lysis buffer (Wanleibio)
containing protease inhibitors, and the protein concentration was
determined using the bicinchoninic acid method. Following
electrophoresis (25 µg per lane; 10% SDS-PAGE), the proteins were
transferred to a PVDF membrane. After blocking with 5% skimmed milk
for 2 h at a room temperature (20-25˚C), the membrane was incubated
with an RPA1 antibody (1:400; cat. no. DF6172; Wuhan Sanying
Biotechnology) overnight at 4˚C. GAPDH mAb (1:2,000; cat. no.
AF7021; Wuhan Sanying Biotechnology) was used as a control. The
membrane was then washed by PBST (0.05% Tween-20) and incubated
with horseradish peroxidase-labeled secondary antibody (1:2,500;
cat. no. S0001; Cell Signaling Technology, Inc.) for 2 h at a room
temperature. After washing the membrane three times, an ECL
Supersensitive Detection kit (Shanghai Biyuntian Biotechnology Co.,
Ltd.) was used to visualize the protein bands. QuantityOne
densitometric analysis software (version 4.6; Bio-Rad Laboratories,
Inc.) was used to calculate the expression level of RPA1 using
GAPDH as the loading control. Each western blot experiment was
repeated at least three times to ensure the reproducibility of the
results.
Immunohistochemical (IHC)
staining
The glioma tissue was embedded in paraffin with 4%
paraformaldehyde for 24 h at room temperature (20-25˚C), sectioned
(4 µm) and dried at 70˚C for 45 min. The paraffin sections were
routinely dewaxed and hydrated, followed by antigen retrieval in
citrate buffer (PH 6.0) for 15 min at 95-100˚C. The sections were
then subjected to quenching with a peroxidase blocker (3%
H2O2), washed with PBS, and blocked with 5%
normal goat serum (Vector Laboratories, Inc.) for 1 h at 4˚C. The
sections were then incubated with primary RPA1 antibody (1:400;
cat. no. DF6172; Wuhan Sanying Biotechnology), anti-Ki67 (1:400;
cat. no.27309-1-AP; Proteintech Group, Inc.) and anti-p53mut
(1:300; cat. no. AF0879; Affinity Biosciences) overnight at 4˚C.
After washing with PBS, the sections were incubated with a goat
anti-mouse IgG (H+L) HRP secondary antibody (1:2,000; cat. no.
S0002; Affinity Biosciences) for 1 h at room temperature (20-25˚C).
Finally, the sections were counterstained with hematoxylin (5 min
at normal temperature), digested with hydrochloric acid, washed,
stained with ammonia bluing solution, dehydrated in an alcohol
gradient, cleared and mounted. The IHC staining was interpreted
independently by two pathologists using a light microscope.
Positive expression of RPA1 was identified based on the appearance
of brown granules in the cytoplasm and/or nucleus. The counting
method was to count all tumor cells in the field of view one by
one, and calculate the percentage of positive cells in the total
cells, such as ’the RPA1-positive cell rate was ~30%’. An
expression score was determined based on the percentage of positive
cells and the staining intensity, which were scored as follows: 0
points, no positive cells in any visual fields; 2 points, ++
staining intensity and <10% positive cells; 3 points, + staining
intensity and >50% positive cells; 4 points, ++ staining
intensity and <50% positive cells); 5 points, ++ staining
intensity and >50% positive cells; and 6 points, +++ staining
intensity and 100% positive cells. A score of >4 was defined as
high RPA1 expression and a score of ≤4 was defined as low RPA1
expression (14). In this
immunohistochemistry of glioma cells, the high/low Ki-67 groups
were classified based on the proportion of Ki-67 positive cells to
total cells. Glioma samples with a positive cell proportion ≤5%
were defined as the low Ki-67 groups, while those with a positive
cell proportion >5% were classified as the high Ki-67
groups.
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from cells using an RNeasy
Mini Kit (Takara Bio, Inc.), and the quality of the RNA was
assessed using a NanoDrop ND-1000 instrument. cDNA was synthesized
from 1 µg RNA using a High-Capacity cDNA Reverse Transcription Kit
(Thermo Fisher Scientific, Inc.) was used according to the
manufacturer's protocol. qPCR was then performed using the
StepOnePlus system (Applied Biosystems; Thermo Fisher Scientific,
Inc.) with SYBR Green Master Mix (Thermo Fisher Scientific, Inc.)
and gene-specific primers (Fuzhou Phygene Biotechnology Co., Ltd.).
The following PCR conditions were used: 95˚C for 10 min followed by
40 cycles of 95˚C for 15 sec and 60˚C for 1 min, and melting curve
analysis. Relative gene expression was determined via the
2-ΔΔCq method (15)
using GAPDH as the reference gene. Triplicate reactions were
performed, and a standard curve was used to verify 100% primer
efficiency. The forward and reverse primer sequences were as
follows: RPA1 forward, 5'-AAGTGGAGACCTACAACGAC-3' and reverse
5'-ACAACCACCTGAGCGTAT-3'; and GAPDH forward,
5'-GACCTCAACTACATGGTTTA-3' and reverse, 5'-AATGAGCCCCAGCCTTCTCC-3'.
Primer synthesis was performed by Sangon Biotech Co., Ltd. The
experiments were repeated three times under the same
conditions.
Statistical methods
SPSS 22.0 (IBM Corporation) statistical analysis
software was used. Unpaired Student's t-test was used to compare
RPA1 expression between two groups. One-way ANOVA followed by
Tukey's post hoc tests was used for the comparison of multiple
groups. χ2 test was used to analyze the associations of
RPA1 expression with clinicopathological parameters. The
relationship between RPA1 expression and overall survival (OS) in
patients with glioma was evaluated using Kaplan-Meier survival
analysis, and survival differences were compared using the log-rank
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
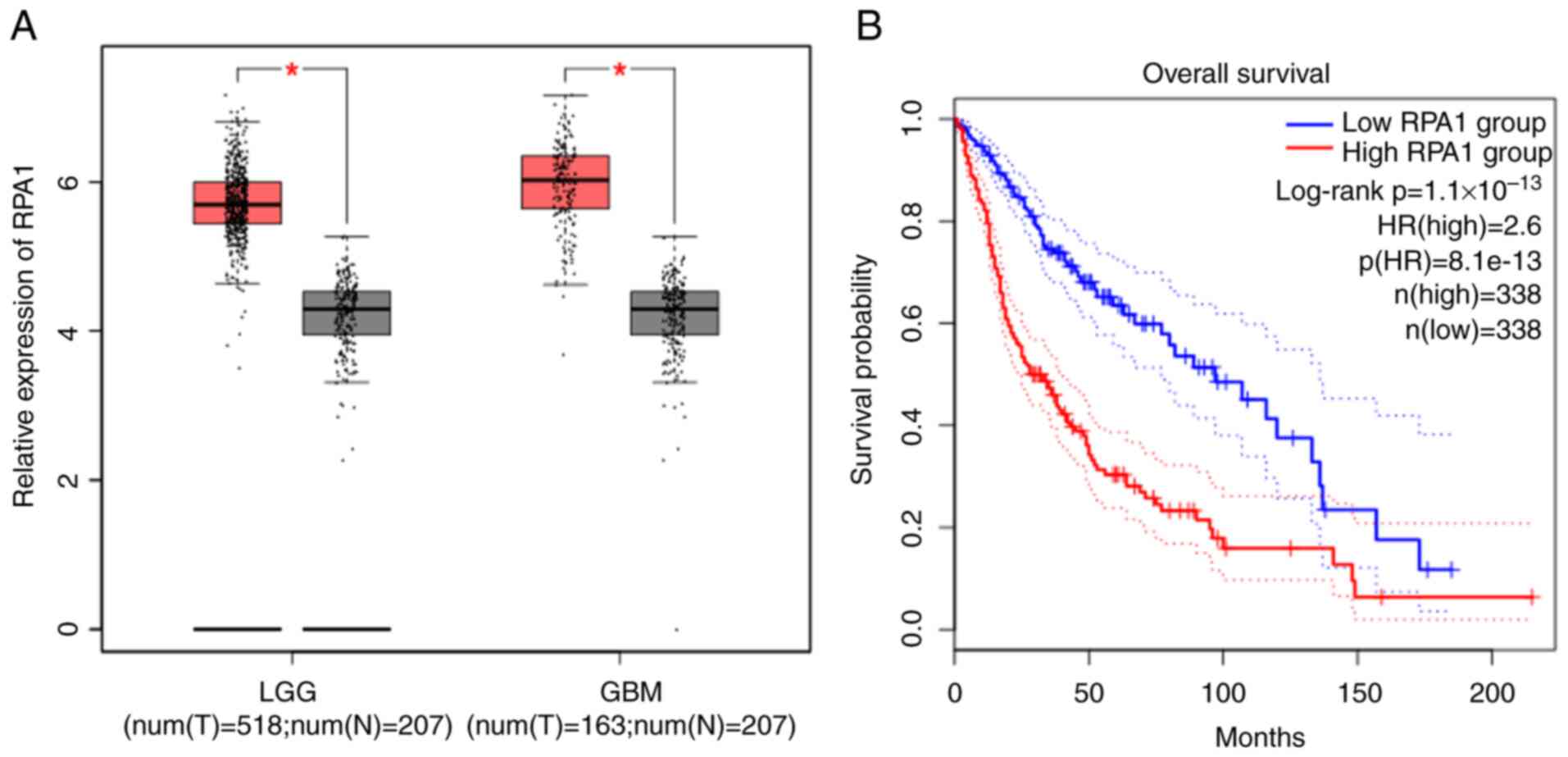
Differential expression analysis and
survival analysis based on RPA1 gene expression data from TCGA
To clarify the relationships between RPA1 and glioma
features, TCGA database and the UALCAN online tool were used to
analyze the differences in RPA1 expression among 518 LGGs, 163
high-grade gliomas (HGGs) and 207 normal brain tissues. This
revealed that the expression levels of RPA1 in both types of glioma
were significantly higher than those in normal brain tissues. In
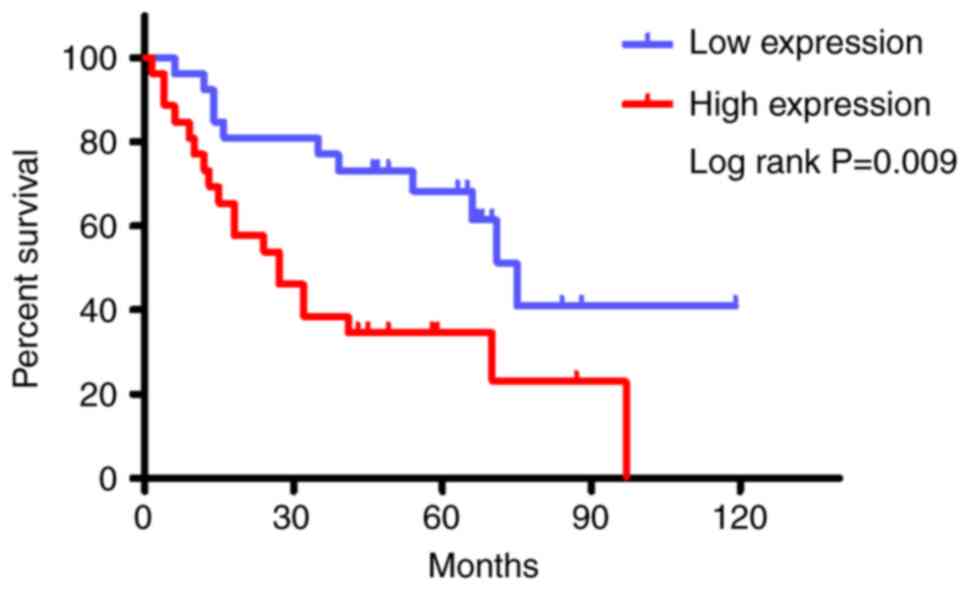
addition, Kaplan-Meier OS analysis was performed for patients with
glioma and high or low RPA1 expression (n=338/group). This analysis
revealed that high RPA1 expression was associated with a
significant shortening of the duration of OS in patients with
glioma (Fig. 1).
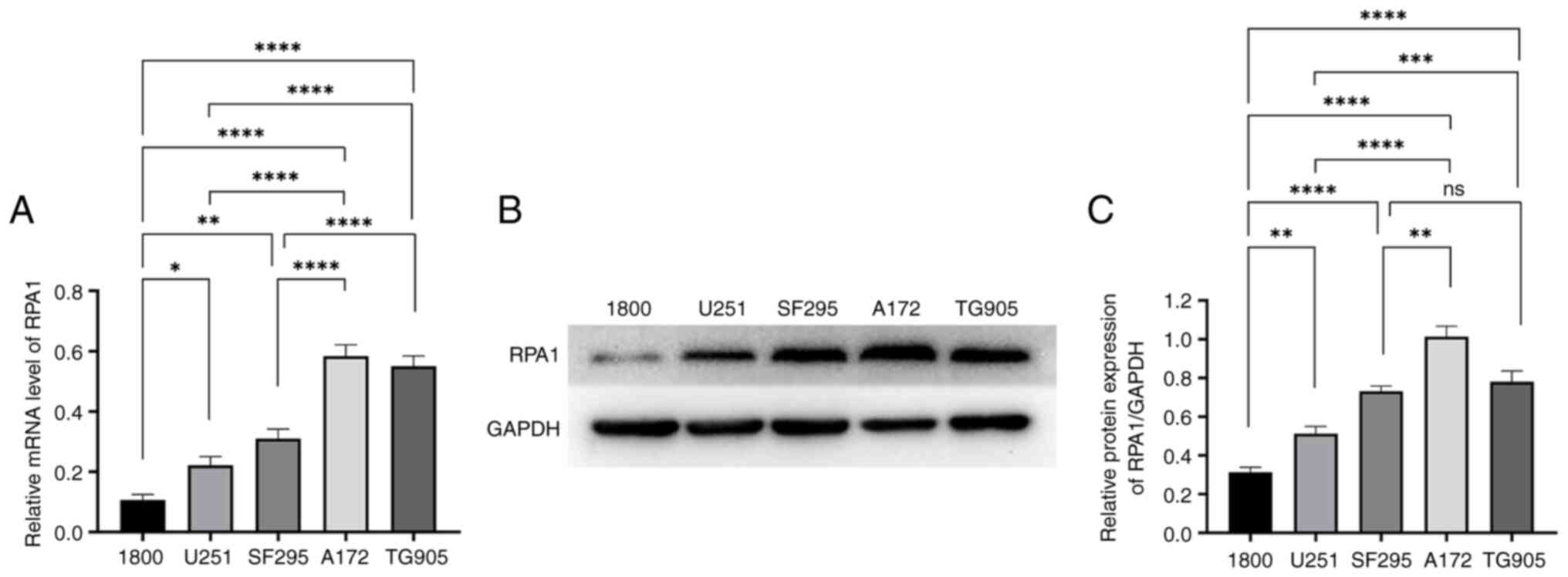
Expression of RPA1 in glioma
cells
The expression of RPA1 in HA1800 normal astrocyte
cell line, U251 and SF295 glioma cell lines and A172 and TG905 GBM
cell lines was analyzed. According to the WHO tumor grading
criteria, gliomas are classified into grades I-IV, based on their
cytological characteristics and molecular biological features,
where a higher grade indicates greater malignancy. The U251 and
SF295 glioma cell lines are considered LGGs while the A172 and
TG905 glioma cell lines are considered HGGs. RT-qPCR analysis
revealed that the mRNA levels of RPA1 in the glioma cell lines were
significantly greater than those in the normal astrocyte cell line,
and significantly higher in the A172 and TG905 GBM cell lines
compared with those in the U251 and SF295 LGG cell lines (Fig. 2A). These findings indicate that the
mRNA expression levels of RPA1 in glioma cells were greater than
those in normal astrocyte cells and increased with the degree of
glioma malignancy. The upregulation of RPA1 in HGGs suggests that
RPA1 may play a role in the progression of glioma.
Similarly, western blot analysis revealed that the
protein expression levels of RPA1 in the glioma and GBM cells were
significantly greater than that in normal astrocyte cells.
Furthermore, the expression levels of RPA1 in the A172 and TG905
GBM cell lines were greater than that in the U251 glioma cell line,
and the expression level of RPA1 in the A172 glioblastoma cell line
was greater than that in the SF295 glioma cell lines. These
observations support the mRNA results in suggesting that the RPA1
expression level is positively associated with the WHO grade in
glioma cells (Fig. 2B and C).
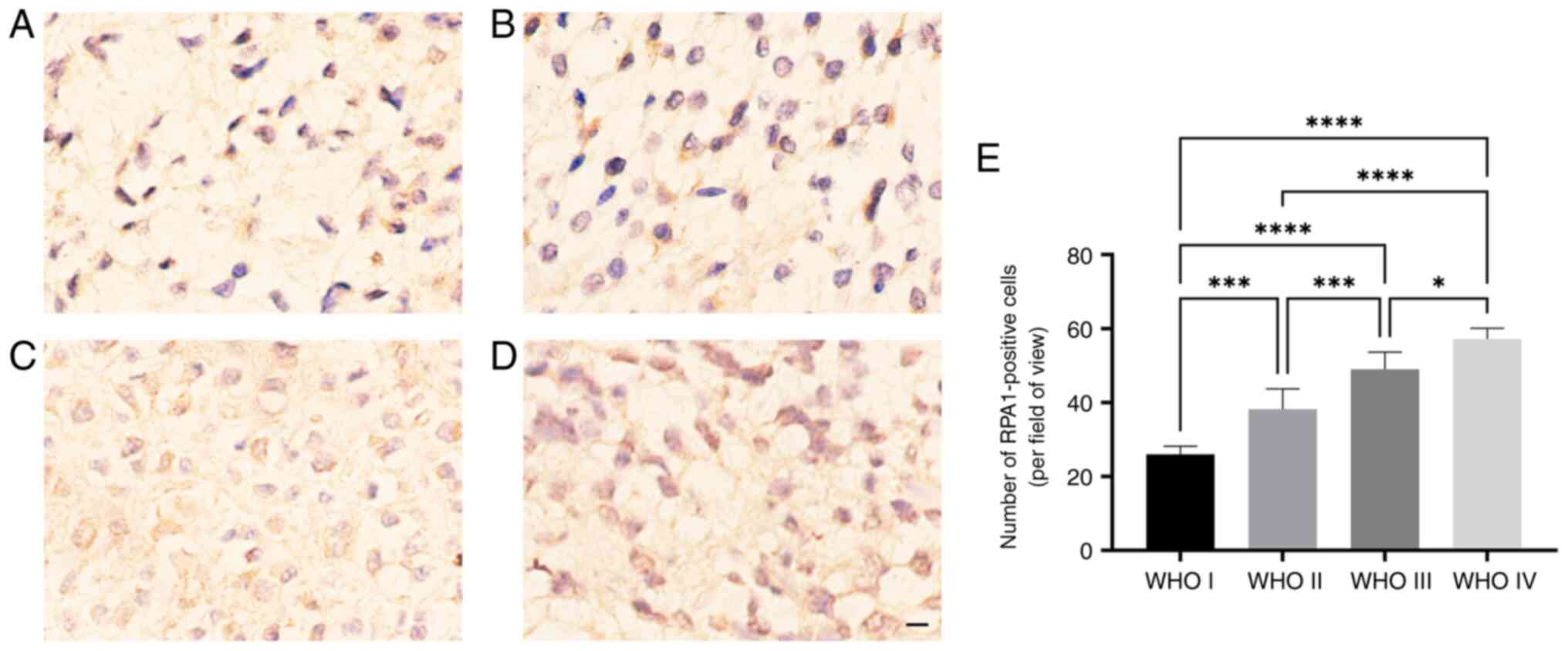
Analysis of the association between
RPA1 expression and clinicopathological parameters in patients with
gliomas
RPA1 IHC staining was performed on 70 glioma tissue
samples from patients with glioma of WHO grades I-IV. The results
revealed that the expression of RPA1 increased as the degree of
glioma malignancy increased. In addition, the expression of RPA1 in
HGG was significantly greater than that in LGG (Fig. 3). The 70 glioma samples were
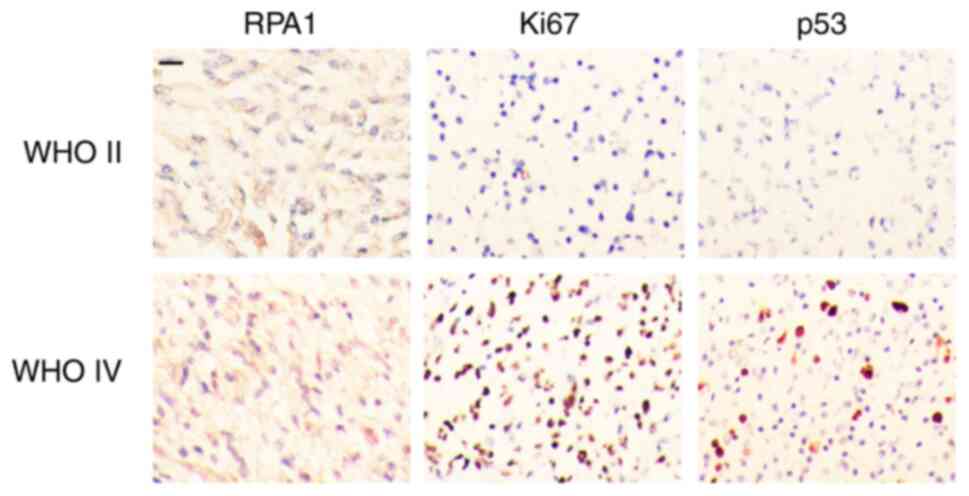
divided into high and low RPA1 groups, and the associations between
the expression of RPA1 and various clinicopathological parameters
were analyzed (Table I, Fig. 4). The results revealed that RPA1
expression was significantly associated with tumor WHO grade, Ki-67
and p53mut (P<0.05). However, no associations were found between
RPA1 expression and patient sex distribution, age, tumor size,
Karnofsky Performance Status (KPS; a commonly used tool for
evaluating the overall health status and functional level of
patients) score, or tumor location in 70 patients with glioma
(Table I; Fig. 4).
Association between RPA1 expression
and the prognosis of patients with gliomas
The survival of the 70 patients with gliomas was
analyzed. KaplanMeier analysis revealed that low RPA1 expression
was associated with a significantly prolonged OS (Fig. 5). These findings are consistent
with the results of the initial database analysis, further
confirming that high RPA1 expression is associated with the degree
of malignancy of glioma.
Bioinformatics analysis of
RPA1-related gene enrichment
To further explore the role of RPA1 in glioma
proliferation, bioinformatics analysis was performed to predict the
signaling pathways mediated by RPA1. A PPI network involving RPA1
was constructed using the STRING database (Fig. 6). Following identification of the
top 10 genes associated with RPA1 (Table II), gene set enrichment analysis
was performed using the DAVID online analysis tool. The results
indicate that RPA1 is mainly involved in GO pathways associated
with DNA replication monitoring, including ‘DNA replication
initiation’, ‘regulation of mitotic cell cycle’ and ‘nucleosome
assembly’ through its target genes (Table III). KEGG pathway enrichment
analysis was also performed to explore pathways associated with the
differentially expressed genes. The main enriched pathways were
‘DNA replication’, ‘mismatch repair’, ‘Fanconi anemia pathway’,
‘homologous recombination’, ‘nucleotide excision repair’ and ‘cell
cycle’ (Table IV). The results of
the bioinformatics analysis provide a theoretical basis for further
study of the functional role of RPA1 in gliomas.
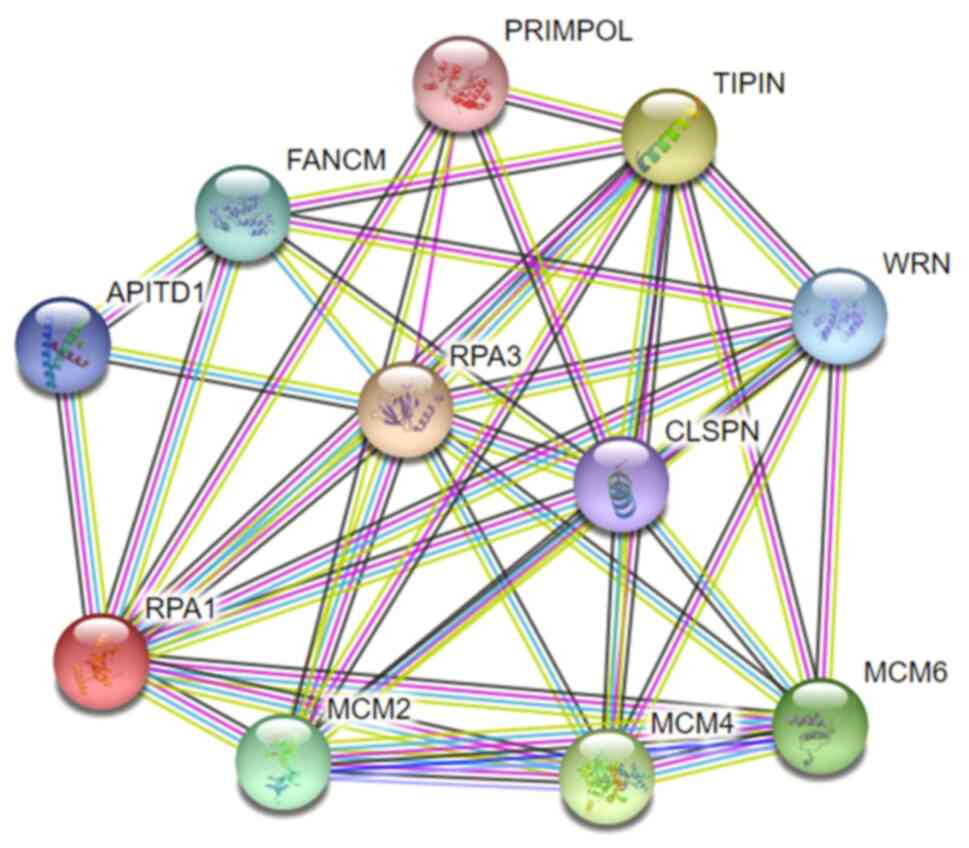 | Figure 6Protein-protein interaction network
showing the RPA1 interaction network, generated using the STRING
database. RPA1/3, replication protein A1/3; PRIMPOL, primase and
DNA-directed polymerase; TIPIN, TIMELESS-interacting protein;
FANCM, Fanconi anemia complementation group M protein; APITD1,
apoptosis-inducing, TAF9-like domain-containing protein 1; WRN,
Werner syndrome ATP-dependent helicase; RPA3, replication protein
A1; CLSPN, claspin; MCM2/4/6, minichromosome maintenance complex
component 2/4/6. |
 | Table IIGene set enrichment analysis of the
top 10 genes in the RPA1 interaction network. |
Table II
Gene set enrichment analysis of the
top 10 genes in the RPA1 interaction network.
| Node 1 | Node 2 | Combined score |
|---|
| RPA1 | POLD3 | 0.76 |
| RPA1 | ELAVL1 | 0.75 |
| HAT1 | RPA1 | 0.75 |
| NEDD1 | RPA1 | 0.74 |
| RPA1 | USP1 | 0.73 |
| RPA1 | SF3A3 | 0.73 |
| RBBP4 | RPA1 | 0.73 |
| TMEM237 | RPA1 | 0.72 |
| RPA1 | MCM3 | 0.72 |
| RPA1 | BAZ1B | 0.72 |
 | Table IIIGO biological function cluster
analysis of the top 10 replication protein A1-related genes. |
Table III
GO biological function cluster
analysis of the top 10 replication protein A1-related genes.
| ID | Description | Count | P-value |
|---|
| GO.0006268 | DNA unwinding
involved in DNA replication | 3 |
3.9x10-7 |
| GO.0045005 | DNA-dependent DNA
replication maintenance of fidelity | 5 |
1.1x10-9 |
| GO.0000712 | Resolution of
meiotic recombination intermediates | 2 |
2.4x10-4 |
| GO.0031297 | Replication fork
processing | 4 |
8.7x10-8 |
| GO.0000076 | DNA replication
checkpoint | 2 |
2.9x10-4 |
| GO.0006270 | DNA replication
initiation | 3 |
7.4x10-6 |
| GO.0032201 | Telomere
maintenance via semi-conservative replication | 2 |
4.0x10-4 |
| GO.0007346 | Regulation of
mitotic cell cycle | 3 |
1.5x10-2 |
| GO.0000731 | DNA synthesis
involved in DNA repair | 4 |
3.3x10-7 |
| GO.0006334 | Nucleosome
assembly | 2 |
1.0x10-2 |
 | Table IVKyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of genes associated with
replication protein A1. |
Table IV
Kyoto Encyclopedia of Genes and
Genomes pathway enrichment analysis of genes associated with
replication protein A1.
| Pathway | Description | Count in
network | P-value |
|---|
| Hsa03030 | DNA
replication | 5 |
8.6x10-11 |
| Hsa03430 | Mismatch
repair | 2 |
1.3x10-4 |
| Hsa03460 | Fanconi anemia
pathway | 4 |
5.4x10-8 |
| Hsa03440 | Homologous
recombination | 2 |
2.9x10-4 |
| Hsa03420 | Nucleotide excision
repair | 2 |
3.2x10-4 |
| Hsa04110 | Cell cycle | 3 |
8.3x10-5 |
Discussion
Glioma is the most common type of primary
intracranial malignant tumor. Current treatment strategies include
maximum tumor resection combined with radiotherapy, chemotherapy
and immunotherapy; however, treatment outcomes remain
unsatisfactory due to the invasive nature of gliomas, the
protective effect of the blood-brain barrier, tumor drug resistance
and recurrence (2,16). Patients with GBM have a
particularly poor prognosis, with an average median survival time
of 1.5 years and a 5-year survival rate of only 5.5%. Thus, gliomas
are a major cause of cancer-related deaths in adults worldwide.
Future directions for tumor therapy include molecular targeted
therapy aimed at key tumor-related genes that affect tumor cell
proliferation, invasion, angiogenesis and apoptosis through a
variety of regulatory pathways.
RPA1 is the main subunit of the RPA complex in terms
of biological function. It plays a central role in the maintenance
of genome stability and is involved in multiple types of DNA
metabolism (5-7). RPA interacts
with numerous proteins, primarily through the N-terminal
interaction of RPA1, which facilitates protein exchange and is
important for DNA damage signaling (17,18).
In addition, RPA1 binds to a number of DNA repair proteins and
checkpoint regulators during DNA replication. Following DNA
unwinding, RPA assists in the recruitment and function of DNA
polymerase during the initiation of DNA replication. RPA1 also
interacts with DNA2 helicases and endonucleases, which help to
maintain cell viability (19,20).
These findings underscore the important role of RPA1 in the
maintenance of accurate DNA replication.
Previous studies have suggested that RPA1 functions
as an oncogene and is significantly negatively associated with OS
in patients with various types of cancer. For example, Zhang et
al (10) demonstrated that
RPA1 overexpression is associated with progression, metastasis and
poor prognosis in nasopharyngeal carcinoma. Knockdown of RPA1 was
found to impair DNA damage repair, inhibit cell proliferation and
induce cell cycle arrest in the G2/M phase. In addition,
in a xenograft model using nude mice, RPA1 knockdown increased the
radiosensitivity of nasopharyngeal carcinoma cells and prolonged
the DNA damage repair time (10).
Similarly, Ni et al (21)
have demonstrated that the suppression of RPA1 expression
significantly disrupts DNA replication, thereby suppressing the
proliferation of non-small cell lung cancer cells.
Few studies have investigated the effect of RPA1 on
glioma. In the present study, differential expression analysis
using TCGA data revealed that the expression level of RPA1 in
glioma tissues was significantly higher than that in normal brain
tissue. In addition, high RPA1 expression was significantly
associated with a shorter OS time in patients with glioma. These
data suggest that RPA1 is a tumor-promoting factor that contributes
to the occurrence and progression of glioma. RT-qPCR and western
blot analyses confirmed that RPA1 expression in glioma cell lines
was significantly increased at both the mRNA and protein levels
compared with that in a normal astrocyte cell line, and that the
expression level increased as the degree of glioma malignancy
increased. IHC staining of glioma tissue samples from 70 patients
revealed that the number of RPA1-positive cells in the tissues
increased as the pathological grade of the glioma increased.
Statistical analysis also revealed a significant association of
RPA1 expression level with the WHO tumor grade, Ki-67 proliferation
index and p53 mutation status. Furthermore, Kaplan-Meier survival
analysis with a long-term follow-up was performed to monitor the
survival of 70 patients with glioma. The findings were consistent
with those of the bioinformatics analysis, and support the
suggestion that RPA1 may serve as an important prognostic marker
for predicting the prognosis and survival of patients with
gliomas.
RPA1 is known to directly or indirectly regulate
multiple downstream target genes in various types of tumors,
contributing to tumor proliferation, apoptosis and drug resistance
through signaling pathways associated with DNA metabolism (22,23).
Previous studies have shown that RPA1 acts as an oncogene in
gastrointestinal tumors, promoting tumorigenesis and proliferation.
For example, a study on colorectal cancer demonstrated that RPA1
knockout significantly inhibited the formation of tumor cell
colonies and arrested the cell cycle in the G1 phase. In
addition, silencing RPA1 expression increased the sensitivity of
malignant colorectal tumor cells to 5-fluorouracil compared with
that of control cells (13).
Mechanistically, RPA1 silencing decreases the phosphorylation level
of extracellular signal-regulated kinase, upregulates pro-apoptotic
caspase 3 expression, and induces apoptosis in colorectal tumor
cells (24,25). In cancer cells, hypoxia and
DNA-dependent protein kinase induce the phosphorylation of p53,
thereby disrupting the p53-RPA1 complex, which enhances
RPA1-mediated NER/nonhomologous end joining and suppresses
apoptosis (26). In addition, the
study also showed that DNA damage in tumor cells is associated with
a significant upregulation of RPA1 expression. RPA1 blocks p53
activity, allowing damaged DNA to evade cell cycle arrest (26). In colorectal cancer, the
overexpression of RPA1 has been shown to decrease the activity of
p53 and promote tumor progression (13,27),
suggesting that targeting RPA1 may be a novel strategy for cancer
treatment.
To further explore the function and molecular
mechanisms of RPA1 in glioma, GO and KEGG gene enrichment analyses
of RPA1-associated genes were performed in the present study using
bioinformatics methods. The GO analysis indicated that RPA1
primarily affects processes associated with DNA replication
surveillance, including ‘DNA replication initiation’, ‘regulation
of mitotic cell cycle’ and ‘nucleosome assembly’ through its target
genes. In addition, KEGG pathway analysis revealed enrichment of
the ‘DNA replication’, ‘Fanconi anemia pathway’, ‘mismatch repair’,
‘homologous recombination’, ‘nucleotide excision repair’ and ‘cell
cycle’ pathways, suggesting that RPA1 may mediate the
proliferation, invasion and apoptosis of glioma cells through these
pathways.
In conclusion, the present study demonstrated that
the expression of RPA1 was significantly increased in glioma,
especially GBM, and was strongly associated with malignancy.
Therefore, RPA1 may serve as a promising target for reducing
malignancy to increase the benefit of conventional multimodal
therapies for human gliomas. These data can be further utilized
through in vitro techniques and in vivo models to
elucidate the role of RPA1 in differentiation, proliferation and
other malignancy-related biological behaviors.
Acknowledgements
Not applicable.
Funding
Funding: No funding was received.
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
ZZ, YS and JZ participated in all aspects of the
experimental design and research procedure and wrote the
manuscript. PY was involved in conducting and analyzing all
experimental measurements. QZ participated in sample preparation
and the molecular biology experiments. ZZ and JZ confirm the
authenticity of all the raw data. All authors read and approved the
final version of the manuscript.
Ethics approval and consent to
participate
All written informed consent was obtained from all
patients. The experimental protocol was established in accordance
with the ethical guidelines of the Declaration of Helsinki and
approved by the Human Ethics Committee of Linyi People's Hospital
(approval no. 30036).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Louis DN, Perry A, Reifenberger G, von
Deimling A, Figarella-Branger D, Cavenee WK, Ohgaki H, Wiestler OD,
Kleihues P and Ellison DW: The 2016 World Health Organization
classification of tumors of the central nervous system: A summary.
Acta Neuropathol. 131:803–820. 2016.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Ostrom QT, Gittleman H, Xu J, Kromer C,
Wolinsky Y, Kruchko C and Barnholtz-Sloan JS: CBTRUS statistical
report: Primary brain and other central nervous system tumors
diagnosed in the United States in 2009-2013. Neuro Oncol. 18 (Suppl
5):v1–v75. 2016.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Xie Y, Ding W, Xiang Y, Wang X and Yang J:
Calponin 3 acts as a potential diagnostic and prognostic marker and
promotes glioma cell proliferation, migration, and invasion. World
Neurosurg. 165:e721–e731. 2022.PubMed/NCBI View Article : Google Scholar
|
|
4
|
Scaringi C, Agolli L and Minniti G:
Technical advances in radiation therapy for brain tumors.
Anticancer Res. 38:6041–6045. 2018.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Liu T and Huang J: Replication protein A
and more: Single-stranded DNA-binding proteins in eukaryotic cells.
Acta Biochim Biophys Sin (Shanghai). 48:665–670. 2016.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Awate S and Brosh RM Jr: Interactive roles
of DNA helicases and translocases with the single-stranded DNA
binding protein RPA in nucleic acid metabolism. Int J Mol Sci.
18(1233)2017.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Byrne BM and Oakley GG: Replication
protein A, the laxative that keeps DNA regular: The importance of
RPA phosphorylation in maintaining genome stability. Semin Cell Dev
Biol. 86:112–120. 2019.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Shi B, Xue J, Yin H, Guo R, Luo M, Ye L,
Shi Q, Huang X, Liu M, Sha J and Wang PJ: Dual functions for the
ssDNA-binding protein RPA in meiotic recombination. PLoS Genet.
15(e1007952)2019.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Taniguchi JB, Kondo K, Fujita K, Chen X,
Homma H, Sudo T, Mao Y, Watase K, Tanaka T, Tagawa K, et al: RpA1
ameliorates symptoms of mutant ataxin-1 knock-in mice and enhances
DNA damage repair. Hum Mol Genet. 25:4432–4447. 2016.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Zhang Z, Huo H, Liao K, Wang Z, Gong Z, Li
Y, Liu C and Hu G: RPA1 downregulation enhances nasopharyngeal
cancer radiosensitivity via blocking RAD51 to the DNA damage site.
Exp Cell Res. 371:330–341. 2018.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Zhang DJ, Xiang J, Wang X, Wang J, Xiao
JC, Xu W, Xu H, Xin Y, Zhang LZ, Pei DS, et al: RPA1 expression in
esophageal carcinoma and its influence on radiosensitivity of
esophageal carcinoma TE-1 cells. Panminerva Med. 57:183–189.
2015.PubMed/NCBI
|
|
12
|
Fourtziala E, Givalos N, Alexakis N,
Griniatsos J, Alevizopoulos N, Kavantzas N, C Lazaris A,
Korkolopoulou P and Gakiopoulou H: Replication protein A (RPA1,
RPA2 and RPA3) expression in gastric cancer: correlation with
clinicopathologic parameters and patients' survival. J BUON.
25:1482–1489. 2020.PubMed/NCBI
|
|
13
|
Li S, Xu K, Gu D, He L, Xie L, Chen Z, Fan
Z, Zhu L, Du M, Chu H, et al: Genetic variants in RPA1 associated
with the response to oxaliplatin-based chemotherapy in colorectal
cancer. J Gastroenterol. 54:939–949. 2019.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Kaur S, Ramdzan ZM, Guiot MC, Li L, Leduy
L, Ramotar D, Sabri S, Abdulkarim B and Nepveu A: CUX1 stimulates
APE1 enzymatic activity and increases the resistance of
glioblastoma cells to the mono-alkylating agent temozolomide. Neuro
Oncol. 20:484–493. 2018.PubMed/NCBI View Article : Google Scholar
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408.
2001.PubMed/NCBI View Article : Google Scholar
|
|
16
|
Xu S, Tang L, Li X, Fan F and Liu Z:
Immunotherapy for glioma: Current management and future
application. Cancer Lett. 476:1–12. 2020.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Jacobs DM, Lipton AS, Isern NG, Daughdrill
GW, Lowry DF, Gomes X and Wold MS: Human replication protein A:
Global fold of the N-terminal RPA-70 domain reveals a basic cleft
and flexible C-terminal linker. J Biomol NMR. 14:321–331.
1999.PubMed/NCBI View Article : Google Scholar
|
|
18
|
Maréchal A and Zou L: RPA-coated
single-stranded DNA as a platform for post-translational
modifications in the DNA damage response. Cell Res. 25:9–23.
2015.PubMed/NCBI View Article : Google Scholar
|
|
19
|
Zou L and Stillman B: Assembly of a
complex containing Cdc45p, replication protein A, and Mcm2p at
replication origins controlled by S-phase cyclin-dependent kinases
and Cdc7p-Dbf4p kinase. Mol Cell Biol. 20:3086–3096.
2000.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Bae SH, Bae KH, Kim JA and Seo YS: RPA
governs endonuclease switching during processing of Okazaki
fragments in eukaryotes. Nature. 412:456–461. 2001.PubMed/NCBI View
Article : Google Scholar
|
|
21
|
Ni Z, Yao C, Zhu X, Gong C, Xu Z, Wang L,
Li S, Zou C and Zhu S: Ailanthone inhibits non-small cell lung
cancer cell growth through repressing DNA replication via
downregulating RPA1. Br J Cancer. 117:1621–1630. 2017.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Haring SJ, Humphreys TD and Wold MS: A
naturally occurring human RPA subunit homolog does not support DNA
replication or cell-cycle progression. Nucleic Acids Res.
38:846–858. 2010.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Fanning E, Klimovich V and Nager AR: A
dynamic model for replication protein A (RPA) function in DNA
processing pathways. Nucleic Acids Res. 34:4126–4137.
2006.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Zhu Y, Yi Y, Bai B, Li L, You T, Sun W and
Yu Y: The silencing of replication protein A1 induced cell
apoptosis via regulating caspase 3. Life Sci. 201:141–149.
2018.PubMed/NCBI View Article : Google Scholar
|
|
25
|
A RE, El-Mesery M, El-Karef A and Eissa
LA: Vitamin D potentiates anti-tumor activity of 5-fluorouracil via
modulating caspase-3 and TGF-β1 expression in hepatocellular
carcinoma-induced in rats. Can J Physiol Pharmacol. 96:1218–1225.
2018.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Madan E, Gogna R and Pati U: p53 Ser15
phosphorylation disrupts the p53-RPA70 complex and induces
RPA70-mediated DNA repair in hypoxia. Biochem J. 443:811–820.
2012.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Givalos N, Gakiopoulou H, Skliri M,
Bousboukea K, Konstantinidou AE, Korkolopoulou P, Lelouda M,
Kouraklis G, Patsouris E and Karatzas G: Replication protein A is
an independent prognostic indicator with potential therapeutic
implications in colon cancer. Mod Pathol. 20:159–166.
2007.PubMed/NCBI View Article : Google Scholar
|