Introduction
The nuclear receptor (NR) superfamily is comprised of 59 member classes, all of which are ligand-dependent transcription factors that are involved in several important biological processes, such as the control of embryonic development, organ physiology, cell differentiation and homeostasis (1,2). In humans, only 48 members of the superfamily have been found and genetic mutations in these NRs have been proven to cause rare diseases, such as cancer, diabetes, rheumatoid arthritis, asthma and hormone resistance syndromes (3). Bearing that in mind and the fact that nuclear hormone receptors possess internal pockets, that bind to hydrophobic, drug-like molecules, they are considered ideal drug targets for the treatment of several diseases. The main limitation, though, in using NRs for drug discovery, is the fact that they control numerous genes and are involved in a number of complex pathways. The major goal of analyzing this protein family is to discover ligands that control the desired, limited subset of genes for therapeutic purposes (4).
As proteins, NRs consist of four functional domains (A/B, C, D and E), each with a separate crucial function (Fig. 1). The N-terminal, NTD domain, also referred to as the activation-function-1 (AF-1) domain, varies in length between NRs and is implicated in transactivation in a cell- and promoter-specific manner. The DNA-binding domain (DBD) is the most highly conserved region among the receptors. Although the precise mechanism of DNA interaction varies from receptor to receptor, a common theme is that direct contact of the NR to DNA is made through two consecutive zinc fingers. Zinc fingers are DNA-binding motifs that contain the key amino acids necessary for the association of the NR to the major groove of an appropriate DNA response element that is located within the promoter segment of a target gene. The domain at the C-terminus is by far the most important to the drug discovery efforts of the pharmaceutical industry. This region, known as the ligand-binding domain (LBD), contains all the binding determinants of the small molecule hormone or synthetic ligand (5). Apart from binding to ligands, this domain also interacts directly with coregulator proteins through a region in the C-terminal part, termed AF2-AD which has a ligand-dependent autonomous activating function. Upon ligand binding, the LBD domain undergoes major structural rearrangements which relieve the binding of co-repressors, unmask AF2-AD, allow the binding of co-activators and render the receptor able to regulate transcription. There is also another domain known as the hinge region or D domain, which is a short, flexible linker between the DBD and the LBD. This region has the least sequence and size conservation between NRs and in numerous cases, it harbors nuclear localization signals (6,7). Last but not least, some but not all NRs have a C-terminal domain or CTD domain, which seems to play an important role in the transcriptional activation and dimerization of the receptor (6,7).
 |
Figure 1
Structural domains of the NR superfamily. Each domain is represented in a different color. NTD in green, DBD in blue, HR in yellow, LBD in purple and CTD in red. NTD, N-terminal domain; DBD, DNA-binding domain; HR, hinge region; LBD, ligand-binding domain; CTD, C-terminal domain.
|
NR members are involved in a number of biological pathways of human physiology and pathology, serving as sensors of stimuli, dominant regulators of controlling molecular events and hubs governing complex gene regulatory networks. The importance of the NR superfamily in control development, differentiation and homeostasis has led to substantial efforts being made to target this family therapeutically (6,7). In the present study, the sequence and structural information of both LBD and DBD were analyzed with an aim of drawing conclusions regarding NR conserved motifs and novel pharmacological targets.
Data and methods
Dataset collection and filtering
The amino acid sequences that are related to the NR proteins were selected from the NCBI database (ncbi.nlm.nih.gov), using the representative keywords as described in Table I. Protein sequences that responded to the query, but did not include NR members were eliminated from the primary dataset, by using related keywords and regular expression techniques in the header information, and local alignments with reference protein sequences. Furthermore, a final dataset for each member class was produced by using internal protein alignments and protein identity score. Duplicated protein sequences in each phylum that were found to share <90% protein identity within the dataset were removed. In total, 109,613 NR protein sequences were identified from several members and species, and a dataset containing 333 representative, unique, non-duplicate protein sequences of phylum and kingdom was created.
 |
Table I
Nuclear receptor members analyzed in the present study.
|
Table I
Nuclear receptor members analyzed in the present study.
| Nuclear receptor |
Abbreviation |
Sample |
| Thyroid hormone receptor |
TRα, TRβ |
7,800 |
| Retinoic acid receptor |
RARα, RARβ, RARγ |
13,200 |
| Peroxisome proliferator activated receptor |
PPARα, PPARβ, PPARγ |
7,500 |
| V-ErbA-related protein |
Rev-ErbΑ, Rev-ErbΒ |
1,160 |
| Ecdysone-induced protein 78C |
Eip78C |
400 |
| RAR related orphan receptor |
RORα, RORβ, RORγ |
4,500 |
| Steroid hormone receptor cnr14 |
Cnr14 |
8 |
| Ecdysone receptor |
EcR |
3,370 |
| Liver X receptor |
LXRα, LXRβ |
2,080 |
| Farnesoid X receptor |
FXRα, FXRβ |
3,450 |
| Vitamin D receptor |
VDR |
2,160 |
| Pregnane X receptor |
PXR |
590 |
| Constitutive androstane receptor |
CAR |
880 |
| Nuclear receptor HR96, HR8 and HR48 |
HR96, HR8, HR48 |
390 |
| Nuclear receptor HR1 |
HR1 |
26 |
| Hepatocyte nuclear factor 4 |
HNF4α, HNF4γ |
3,800 |
| Retinoid X receptor |
RXRα, RXRβ, RXRγ |
5,040 |
| Ultraspiracle |
USP |
3,790 |
| Testicular receptor |
TR2, TR4 |
3,080 |
| Tailes-related receptor |
TLX |
1,400 |
| Photoreceptor specific nuclear receptor |
PNR |
1,660 |
| COUP transcription factor |
COUP-TF1, COUP-TF2 |
80 |
| V-erbA-related protein 2 |
EAR-2 |
35 |
| Estrogen receptor |
ERα, ERβ |
13,740 |
| Estrogen related receptor |
ERRα, ERRβ, ERRγ |
5,244 |
| Glucocorticoid receptor |
GR |
6,240 |
| Mineralocorticoid receptor |
MR |
1,640 |
| Progesterone receptor |
PR |
4,150 |
| Androgen receptor |
AR |
4,670 |
| Nerve growth factor IB |
NGFIB |
45 |
| Nuclear receptor related 1 |
NURR1 |
1,590 |
| Neuron-derived orphan receptor 1 |
NOR-1 |
22 |
| Steroidogenic factor 1 |
SF-1 |
1,120 |
| Liver receptor homolog 1 |
LRH-1 |
2,360 |
| Nuclear hormone receptor FTZ-F1β |
FTZ-F1β |
420 |
| Germ cell nuclear factor |
GCNF |
1,100 |
| Zygotic gap protein knirps |
kni |
105 |
| Dosage-sensitive sex reversal |
DSS |
18 |
| Small heterodimer partner |
SHP |
750 |
| Total |
|
109,613 |
Structural identification of NR protein domains
A more specialized analysis was performed in order to identify the protein domains in each NR member. The analysis was performed using the InterPro Database and a representative protein sequence from each NR member class (8,9). In the majority of cases, the Homo sapiens species protein sequence was used as a representative, except in cases where no human sequences existed and thus, another species was selected.
Multiple sequence alignment (MSA)
MSA was performed using the MATLAB Bioinformatics Toolbox (https://www.mathworks.com/products/bioinfo.html), utilizing a guide tree and the progressive MSA method as previously described (9,10). Pairwise distances among sequences were estimated based on the pairwise alignment and followed by calculating the differences between each pair of sequences. The Neighbor-Joining method was used towards to estimating the guide tree by assuming equal variance and independence of evolutionary distance estimates (11). A more specific analysis was performed using the MATLAB Bioinformatics toolbox towards comparing the MSA results and the protein domains structural features from the previous step.
Exploration of conserved motifs
Consensus sequence was calculated and visualized through the Jalview platform (12) using the MSA result and parameters including amino acid conservation (9,10,12). The commentary section of Jalview, which presents the amino acid conservation using logos and histograms, was further observed to uncover innovative motifs within the protein domains.
Phylogenetic analysis of steroid hormones (SRs)
The representative dataset of the steroid hormone members was used for the construction of the phylogenetic analysis. The primary dataset was filtered, and duplicated protein sequences in each species that were found to share <95% protein identity within the dataset were removed. MSA was performed using the MATLAB Bioinformatics Toolbox (13) alignment methods. Only unambiguous homologous regions were retained for phylogenetic analysis; manual masking, trimming and consensus multiple alignments were performed in MATLAB (13). The phylogenetic analysis was performed using the MATLAB bioinformatics toolbox. The distances between the sequences of the SR dataset were measured with the Jukes-Cantor pairwise distance method (14,15) and phylogenetic analysis was performed utilizing the average distance statistical method (16,17) with 100 bootstrap replicates and visualized using MEGA software radiation option (18).
Structural and functional characterization of conserved motifs
The structural and functional characterization of the LBD domain conserved motifs was analyzed using representative 3D structures of NRs within the steroid hormone subfamily. In the present study, the 3D structures of the glucocorticoid receptor (GR) LBD [4P6X (19) and 5NFT (20)] and the androgen receptor (AR) LBD [6NWL (21)] from the Protein Data Bank (https://www.rcsb.org) were used. The structural and functional analysis at SR structures was gained using MOE (20,22-24), where all the interactions sites with the corresponding proteins and ligands were identified and studied. Specifically, each PDB entry was examined for ligand interaction using the ligand interaction function. MOE showcased the LBD amino acids that interacted with said ligands or co-activators.
Results
NR protein domains
The NR proteins are composed of a number of domains and sub-domains which are differentially conserved between the NR members. The NRs protein domains play diverse roles, including the variable N-terminal region, the conserved DBD, the variable hinge region, the conserved LBD and the variable C-terminal region. The representative protein sequences were selected for the analysis of NR domains belonging to the species Homo sapiens, except in a few members from which this species was absent, including FXRβ, Eip78C, Cnr14, EcR, HR96, USP, FTZ-F1β and kni. The FXRβ NR representative was selected from the species Mus musculus, and Eip78C, Cnr14, EcR, HR96, USP, FTZ-F1β and kni from the Drosophila melanogaster species. Sequence analysis of the NR protein domains revealed a clear separation between them (Fig. 2). The characteristic five-protein domains of the NR family were identified in the majority of the members. NR protein domains vary in length among members, and in a few cases, some of them are missing (Fig. 2). Based on these results, some members appear without a C-terminal region, while others do not even have a DBD (SHP, DSS) or LBD (kni) (Fig. 2).
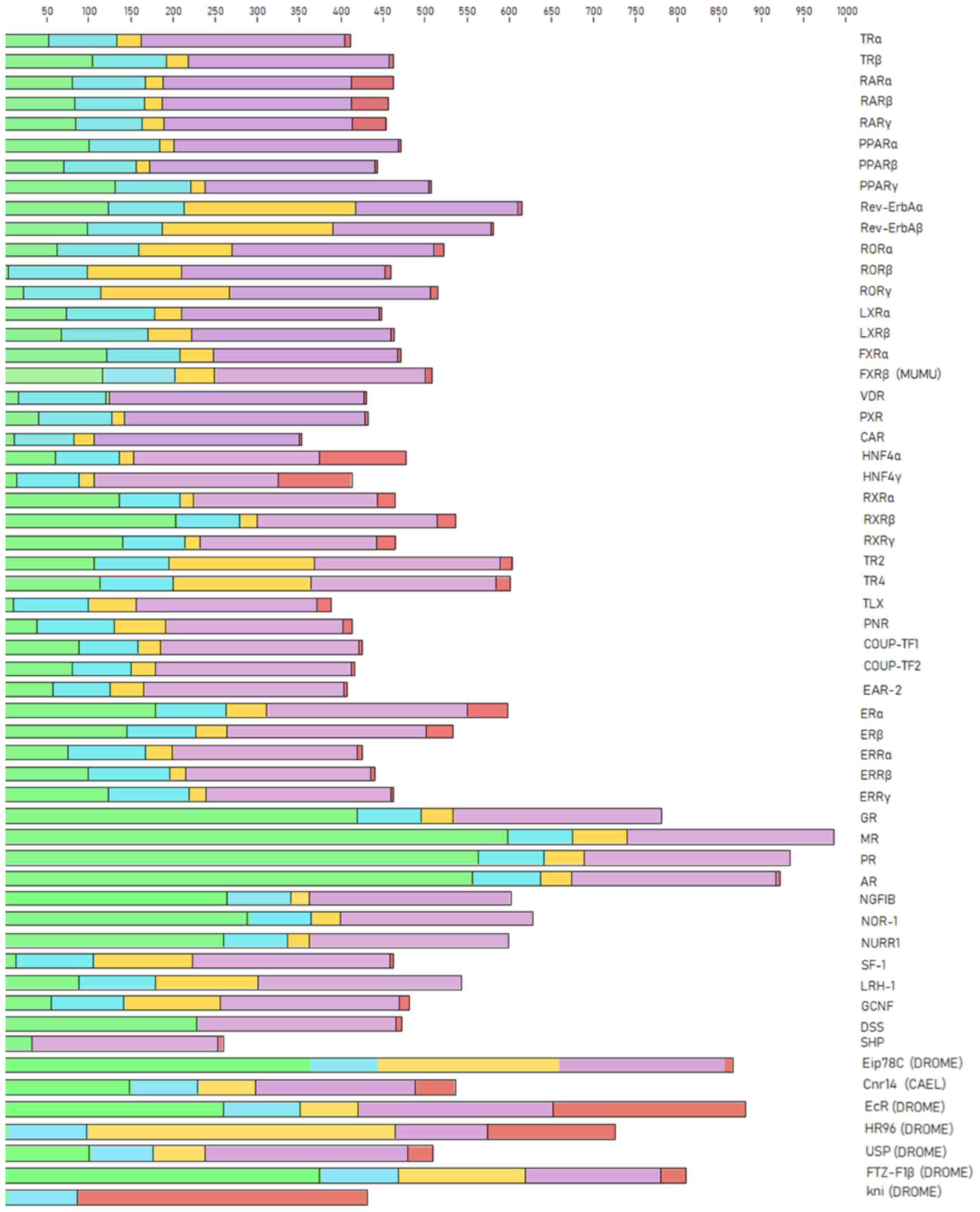 |
Figure 2
Protein domains in nuclear receptors. The N-terminal region is shown in green, the DBD is shown in blue, the hinge region is shown in yellow, the LBD is shown in purple, and the C-terminal region is shown in red. DBD, DNA-binding domain; LBD, ligand-binding domain.
|
MSA and conserved motifs
MSA of proteins sequences from the NR superfamily, as well as the subfamily of SRs, was performed to identify highly conservative regions within all organisms of the animal kingdom. The visualization and analysis of the MSA results in both cases were performed using Jalview. As is known, the majority of NRs consist of five structural domains, including the N-terminal, DBD, LBD, C-terminal domains and the hinge region. Sequence analysis of the SR protein sequences dataset has revealed clear conservation in all species. The DBD is the most highly conserved region in the NR superfamily, as observed in the MSA of both NRs and SRs. Two zinc finger motifs can be identified, each of which consisted of a zinc ion linked to four cysteine residues. In the first zinc finger, a conserved pattern, termed P-box, can be identified, containing amino acid residues necessary for the specialized identification and binding of the receptor to the DNA of the target-gene (22). In the second zinc finger, another pattern of amino acids, can be identified, which is involved in the dimerization of the receptors and is termed D-box (23). The amino acids of the D-box appear to be quite diverse in the NR superfamily (Fig. 3), whilst in the SRs, they are highly conserved (Fig. 4) (23).
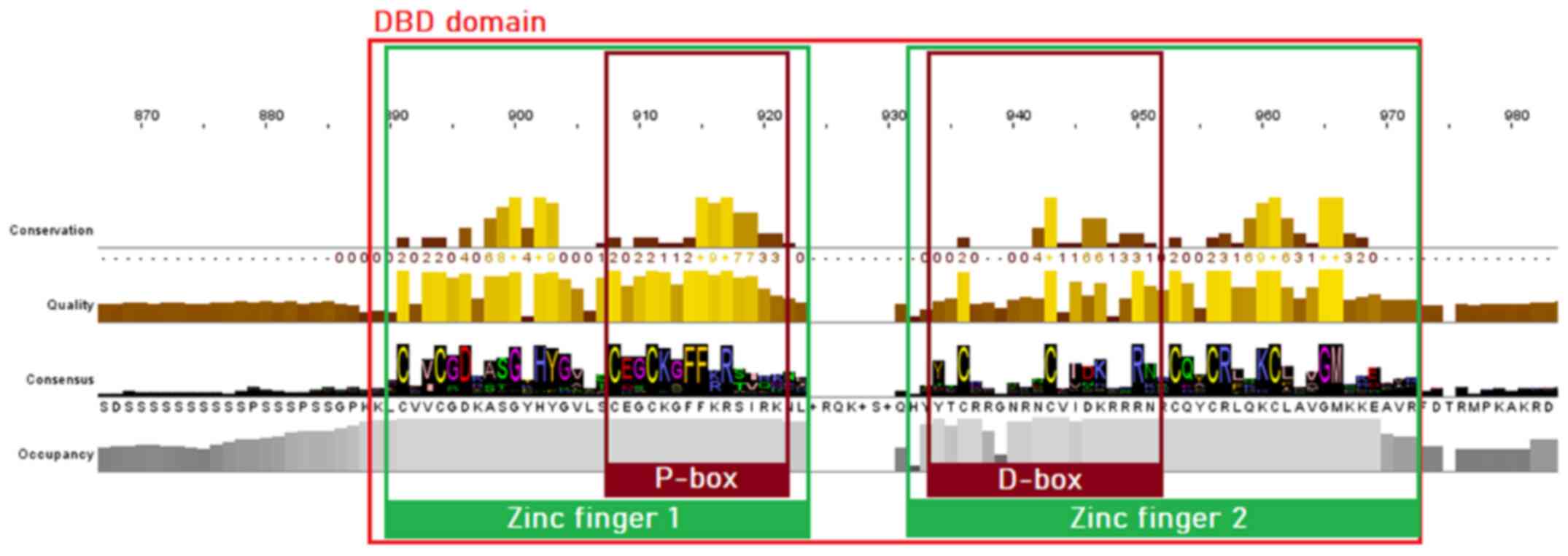 |
Figure 3
Highly conserved motifs of the nuclear receptor superfamily in the DBD, based on the MSA of 333 protein sequences. The two zinc fingers with the preserved cysteine residues are colored green, whilst the P-box and D-box are highlighted with a dark red color. DBD, DNA-binding domain; MSA, multiple sequence alignment.
|
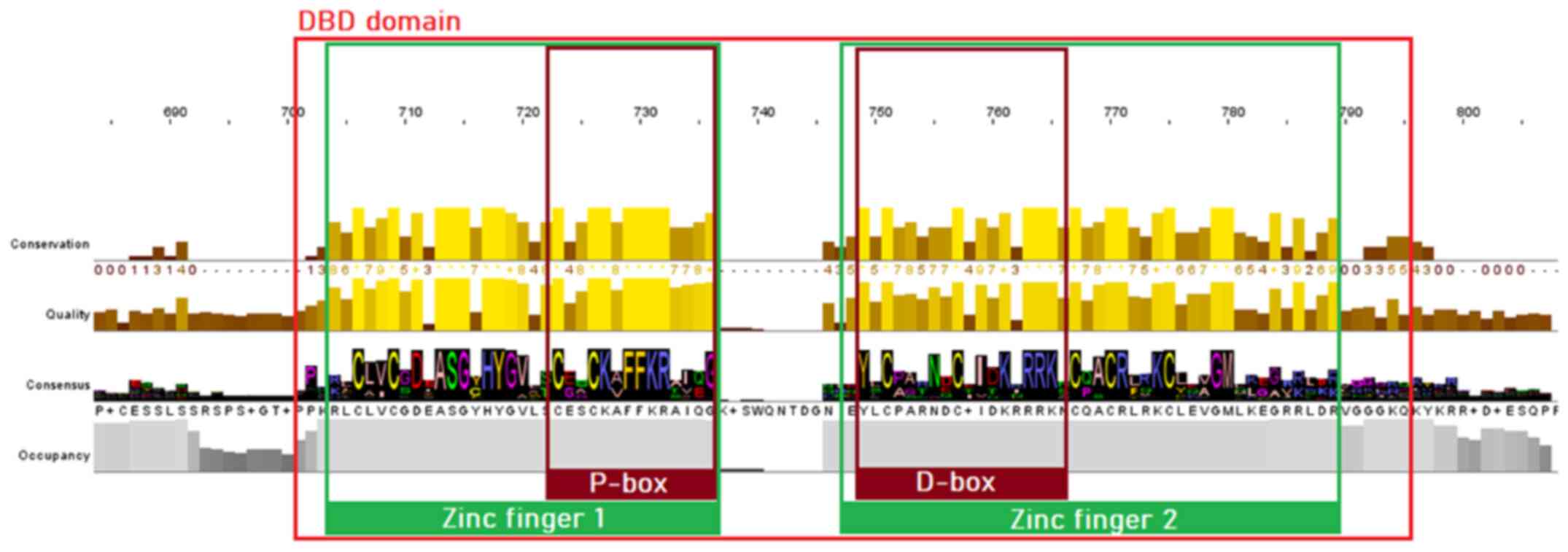 |
Figure 4
Highly conserved motifs of the steroid receptor family in the DBD. The two zinc fingers with the preserved cysteine residues are colored green, whilst the P-box and D-box are highlighted with a dark red color. DBD, DNA-binding domain.
|
The LBD appears to be less conserved; nevertheless, some important conserved motifs can be observed, particularly within the SRs. Motif A can be found in positions 944-949 of the steroid receptor MSA (Fig. 5). This is an LLxxL motif (where L represents a leucine residue and x denotes any amino acid), which is an inverse NR box (LxxLL) (24). The NR box (NR interacting box) plays an important role in the interaction of NRs with co-activators and therefore, in the regulation of transcription. Motif B, occupying positions 960-970 of the SR alignment (Fig. 5) and motif C, occupying positions 972-982 (Fig. 5), as reported in the literature, also appear to be regions necessary for co-activator function (25). Motif D, which occupies positions 1051-1063 of the steroid receptor multiple alignment (Fig. 5), appears to be highly conserved and is an area of utmost significance for ligand binding, as mutations in this area have been proven to lead to the complete inability of the receptor to bind (22,24,25). Finally, in alignment positions 1115-1122, a LxxLL motif is observed (motif E), whereas in alignment positions 1144-1153, an inverse NR box (LLxxL) can be found, named motif F (Fig. 5) (24).
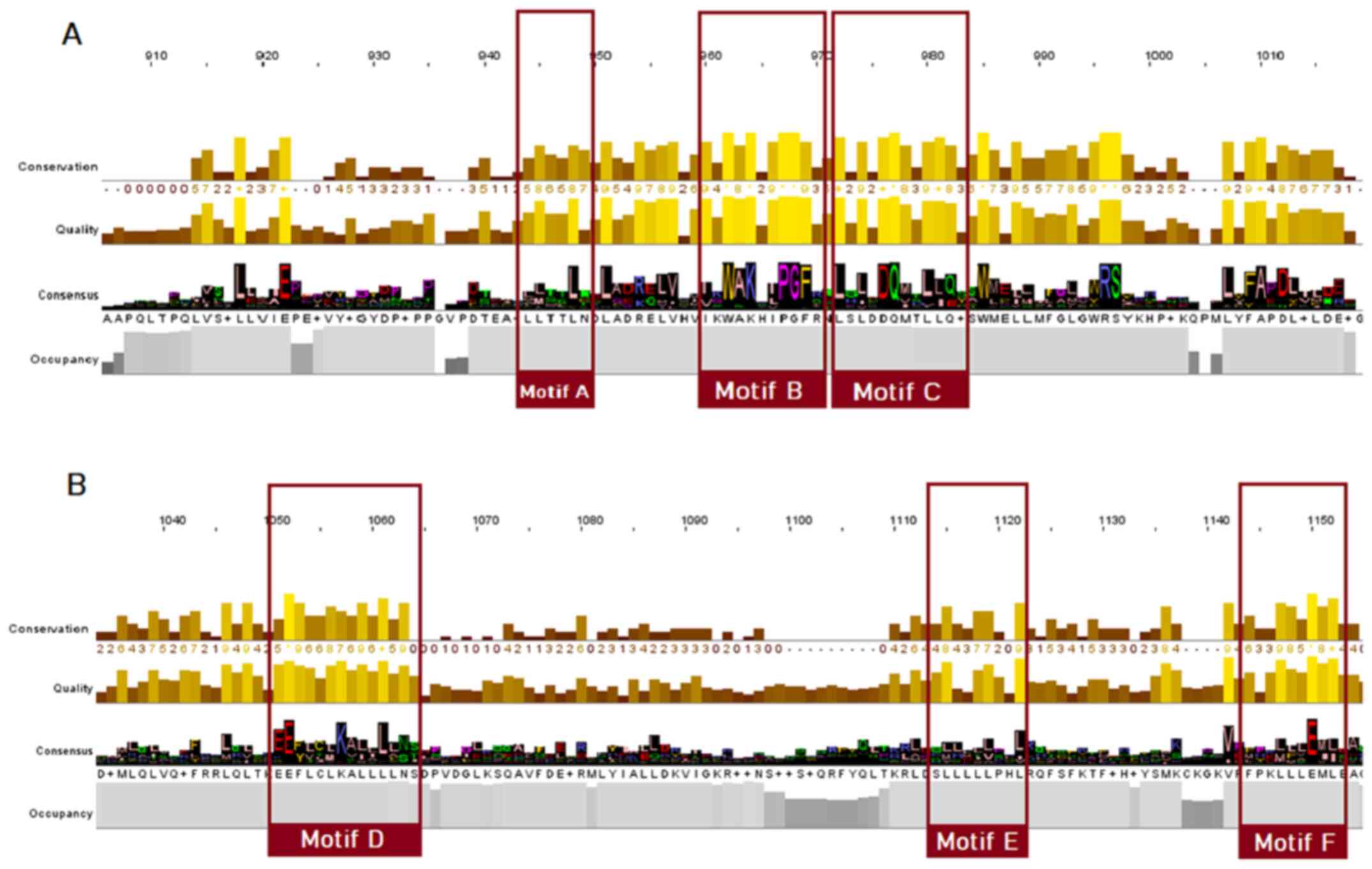 |
Figure 5
Highly conserved LBD motifs, based on the steroid receptors MSA. Motifs A, B, C, D, E and F are highlighted (colored red). (A) Motifs A-C; (B) motifs D-F. LBD, ligand-binding domain; MSA, multiple sequence alignment.
|
As for the NR superfamily (Fig. 6), only motifs B, C, D and E seem to be conserved, whilst in the alignment positions that motifs A and F would be located, a great amino acid variability is observed.
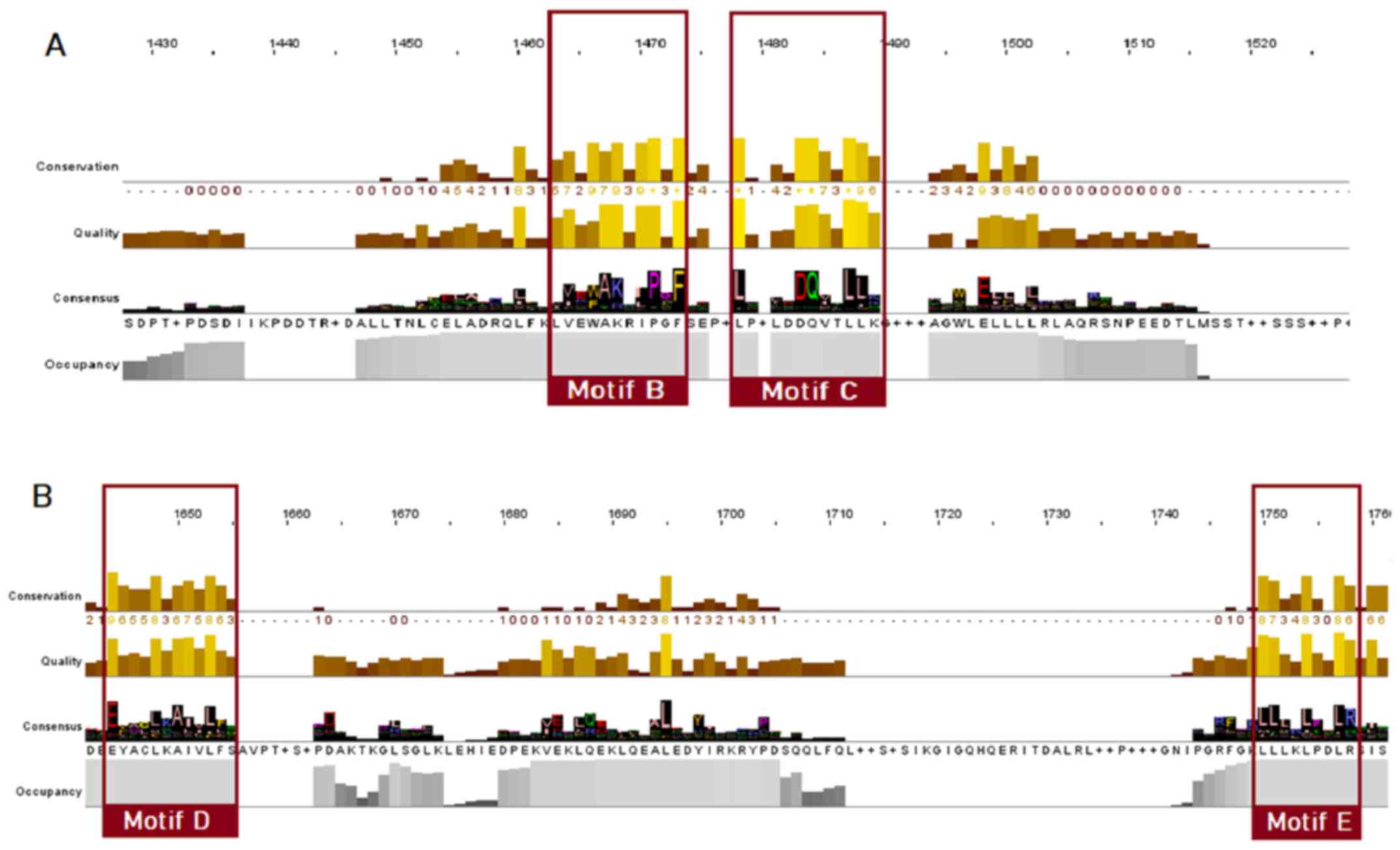 |
Figure 6
Highly conserved LBD motifs of the nuclear receptor superfamily, based on the MSA of 333 protein sequences. Motifs B, C, D and E are highlighted with red. (A) Motifs B and C; (B) motifs D and E. LBD, ligand-binding domain; MSA, multiple sequence alignment.
|
Phylogenetic analysis
The phylogenetic tree of the steroid hormones comprises of five major monophyletic sub-clusters including the GR, mineralocorticoid receptor (MR), AR, progesterone receptor (PR) and estrogen receptor (ER) (Fig. 7). Based on the tree topology, steroid receptors have been clustered in distinct evolutionary clades. In the phylogenetic analyses, representative members from several kingdoms and phyla were identified, including mammals, Aves (birds), Pisces (fish), insectum (arthropods) and Reptilia.
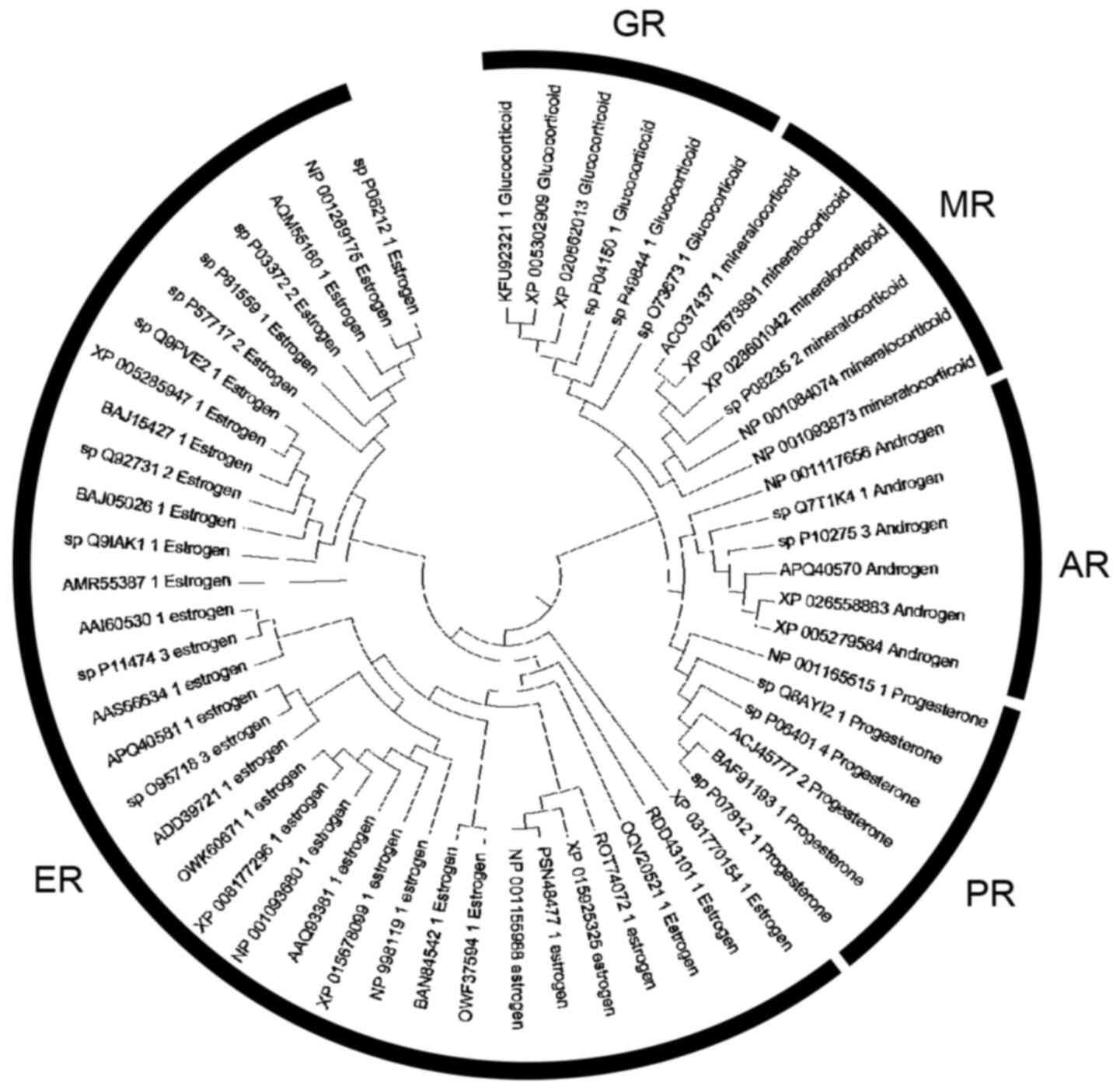 |
Figure 7
Phylogenetic analysis of the steroid hormone receptors.
|
Conserved motif exploration
In the present study, eight candidate conserved motifs that were identified in both DBD and LBD are summarized. Although no structural information is provided for the DBD of the NR, some ‘key’ amino acid residues have been identified and provided within the P-box and B-box motifs, which are mainly termed ‘inc-coordinating motif’ (Figs. 3 and 4) (24,26). The zinc-coordinating motif is a potential pharmacological target of the DBD of the NRs. It is characterized by two anti-parallel helices connected by loops at their amino-terminal ends, from which each helix-loop combine coordinates a single zinc ion using four ‘key’ cysteines (26). Through this specific mechanism, NR members are able to change conformation and activate the selection mechanism towards binding DNA molecules (Fig. 8).
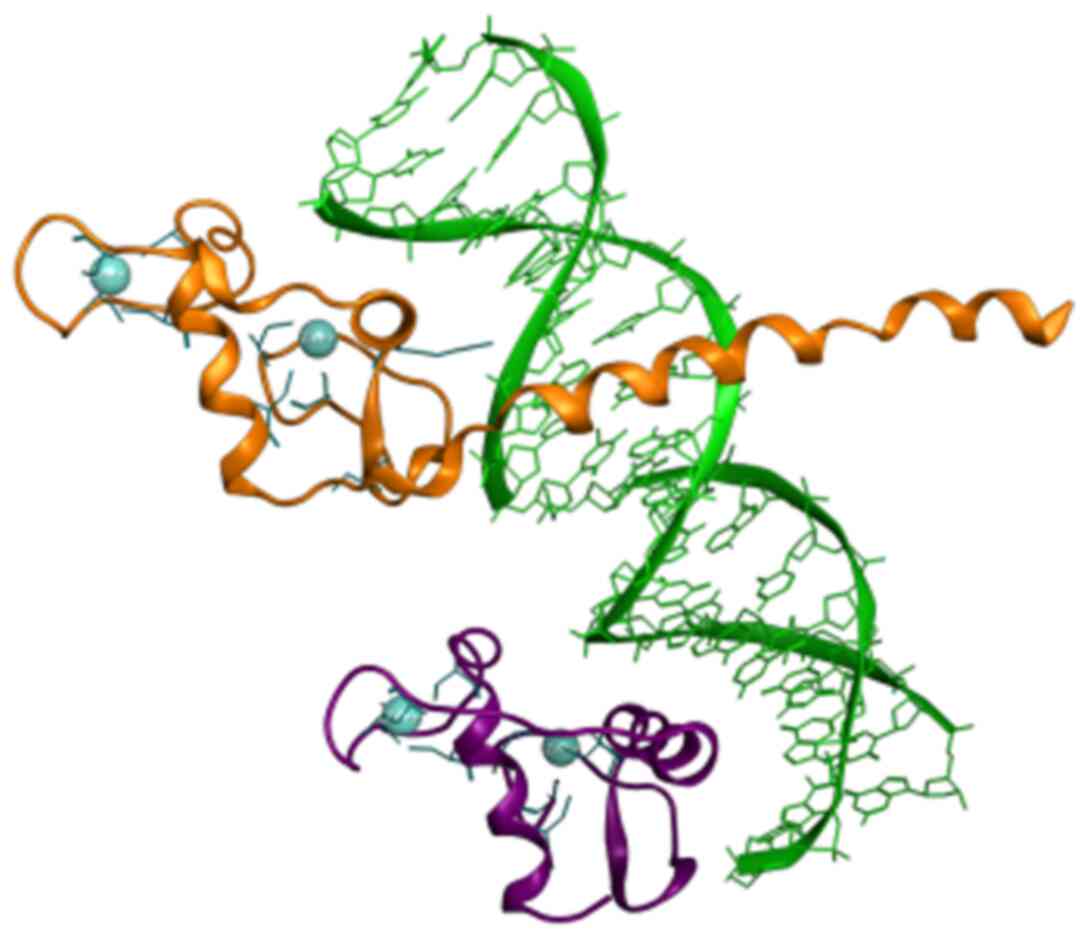 |
Figure 8
Rippon representation of the RXR (colored purple) and the THR (colored orange) DBD heterodimer, which is bound to thyroid response element DNA (colored green). Zinc-coordinating motif cysteines residues are colored blue. Crystal structure from the PDB (ID: 2NLL). RXR, retinoid X receptor; DBD, DNA-binding domain; PDB, Protein Data Bank.
|
On the other hand, conserved motifs that are formed the interaction site of the LBD of NR are of a major interest as potent pharmacological targets. The majority of these directly interact with ligands, and some others are contained critical patterns, including the ‘LLxxL’ pattern in back and forward direction (Fig. 9). The ‘LLxxL’ pattern is repeated >3 times in the LBD of the majority of the NR members and constitutes a critical conserved region that is directly related to their biological function (24,27). Mutations in the ‘LxxLL’ pattern, have been shown to be strongly associated with incorrect signaling in NR members, particularly in motif D (28). Mitsis et al (24) provided a structural and chemical analysis of the LBD of the NRs, which explains the possible role of this pattern and its significant as a potent pharmacological target.
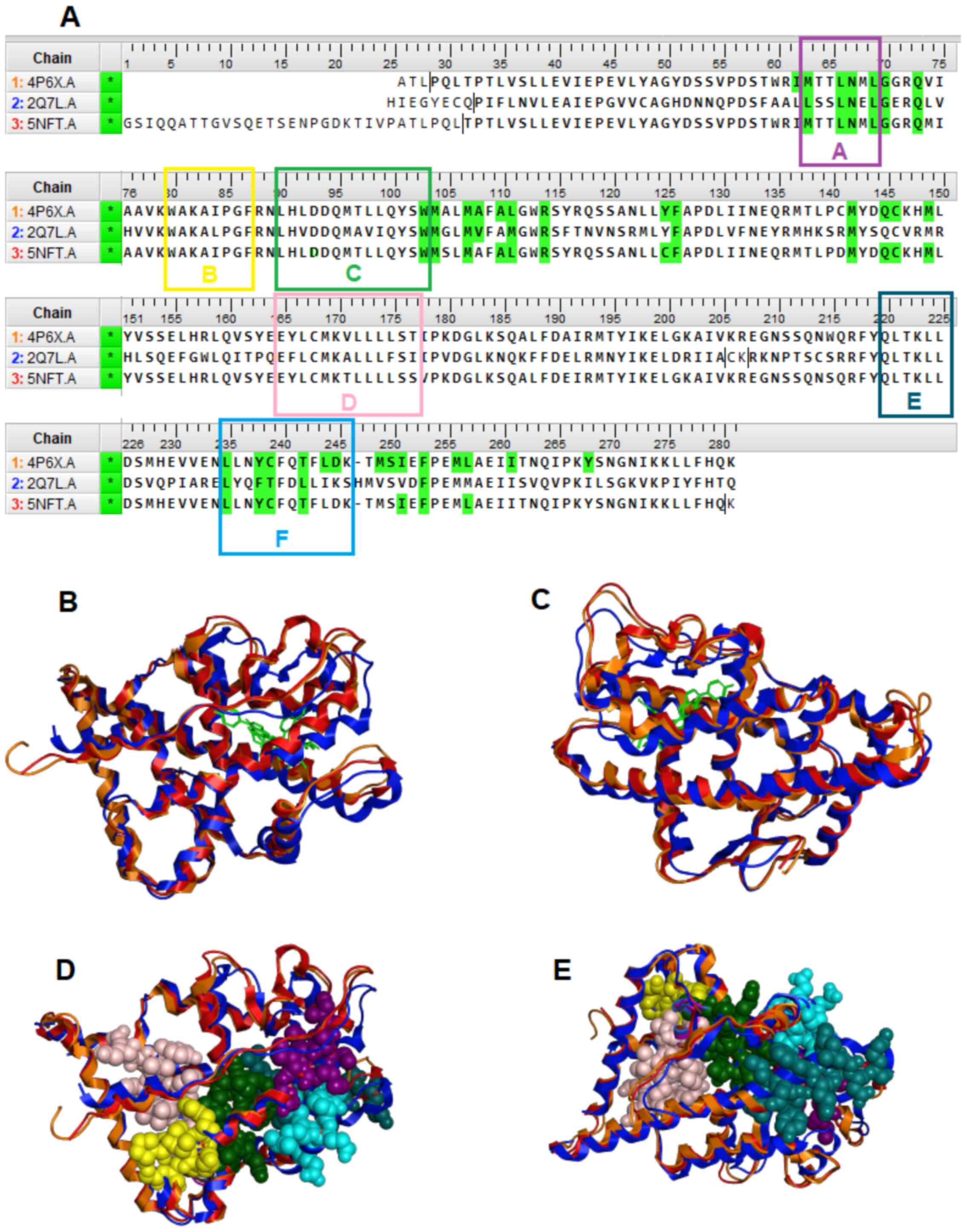 |
Figure 9
(A) Multiple sequence alignment of the GR (4PGX, 2Q7L) and AR (5NFT) protein sequences. In color green are highlighted the ‘key’ amino acids between each NR and their ligand. The five conserve motifs A-F are presented in different colors. (B and C) Structural superposition of the of the LBD of the GR (colored red and orange) and the AR (colored blue) in ribbon representation. (D and E) Conserved motifs A-F and interaction site of the LBD of the GR and the AR. GR, glucocorticoid receptor; AR, androgen receptor; NR, nuclear receptor; LBD, ligand-binding domain.
|
Discussion
The NR superfamily consists of transcription factors that influence gene expression in a positive or negative manner (6). Although >300 transcription factors have been recognized as members of this superfamily, the human genome encodes only 48 NRs, whereas mice and rats have 49 and 47 NR superfamily members, respectively (29-31). These DNA-binding proteins play a crucial role in several physiological functions, such as proliferation, differentiation, apoptosis, immune and stress response (32,33). The first founding members of the NR superfamily, which were sequenced, were the human GR and ERα (34,35). The analysis of their full-length sequence demonstrated similar structural domains, indicating the presence of a receptor family (36). Several other receptors, including the MR, AR, PR, and receptors for vitamin A and D, have been shown to have high amino acid sequence similarity (37). Indeed, NRs consists of a less conserved N-terminal or immunogenic domain, a highly conserved DBD, and a LBD (36).
The NR superfamily consists of six families of transcription factors (38). A significant number of NRs have not yet been shown to bind to a known ligand (39,40). On the other hand, SRs form the third subfamily of steroid-activated NRs consisting of the ER and AR, which both control reproduction and sexual development; the PR, which plays a key role in female reproduction; the MR that maintains electrolyte balance; and the GR, which plays fundamental role in maintaining basal and stress-related homeostasis (Fig. 7) (41). Since this class of transcription factors substantially influences numerous aspects of human physiology, they have been early identified as potential pharmacological targets (42). Currently, a number of steroid receptor agonists and antagonists are widely used in the therapeutic management of numerous benign and malignant diseases (43). Importantly, synthetic glucocorticoids are frequently prescribed for several inflammatory diseases due to their potent anti-inflammatory and immunosuppressive actions (44).
Evolution is a complex biological process, which is characterized by the elaboration and optimization of the inherited traits from one generation to the next, leading to the emergence of new divergent functions (45,46). During the evolutionary process, genes are duplicated, rapidly mutated and naturally selected for each function (47,48). NRs are ancient in origin and diversified prior to the arthropod/vertebrate split (49). Escriva et al (50) demonstrated that NRs are specific to metazoans (cnidarians and acoelomates). Of note, they demonstrated that the NR superfamily was evolved through two serial waves of gene duplications; the first wave occurred before the cnidarians diverged from bilaterians, whereas the second wave took place in vertebrates (50). Thornton (45) further elucidated the mechanisms underlying the evolution of steroid hormone receptors. Using an extensively parallel PCR screen, Thornton (45) found specific sequences of steroid receptors in the sea lamprey Petromyzon marinus, a jawless fish that diverged from gnathostomes ~450 million years ago. According to his study, Thornton (45) identified the lamprey PR, ER and CR, and reconstructed the two first ancestral proteins (AncSR1 and AncSR2) according to sequencing analyses of lamprey receptors and subsequent alignment with 70 other available steroid and related receptors (9). The AncSR1 was 71% identical to the human ERα, while the AncSR2 was 77% similar to the human PR (35). Further phylogenetic analysis revealed that steroid receptors evolved through two serial genomic expansions, which are likely to have occurred one before the lamprey-gnathostome divergence and one after (30). The first created an ER and a 3-ketosteroid receptor, whereas the second produced a receptor for 3-ketogonadal steroids (progestins, androgens or both) and a corticoid receptor that finally gave rise to MR and GR (Fig. 7) (6).
Considering that a number of NRs participate in diverse physiological functions, it would be of great interest to further examine the evolutionary story of these proteins. In the present study, eight different conserved motifs were identified and are subject to further investigation. These significant key regions, in combination with the structural features of the protein domains they are contained will provide beneficial knowledge towards understanding the evolution of NRs. In addition, the delineation of the evolutionary pathway of NRs may further explain the interactions between environment and the expressed genome, since each NR has a different group of target genes.
In conclusion, the present study provides a comprehensive sequence analysis study of the NR superfamily. Using several filtering techniques, the present study was able to select representative protein sequences from different phyla and kingdoms for each NR member. Therefore, the present study dealt with the ‘big data’ of the NR family. NRs and in particular, SRs, are involved in numerous biological processes, constituting the basic regulators of the human body. The main aim of the present study was to identify conserved motifs or patterns that play major roles in biological processes in which NRs are involved. Based on these results, eight ‘key’ motifs in the two main protein domains of the NRs were recognized, namely the LBD and the DBD. Moreover, all the identified conserved motifs were associated with the NR function. In addition, evolutionary differences between steroid hormones receptors and NRs were identified in these conserved regions, leading to an updated phylogenetic tree of the steroid hormones receptors that indicates the ideal candidate groups for the development of new pharmacological targets.
Acknowledgements
Not applicable.
Funding
The present study received funding from the project ‘INspIRED-The National Research Infrastructures on Integrated structural Biology, Drug screening Efforts and Drug Target Functional characterization’ (grant no. MIs 5002550) and by the project: ‘OpENscREENgR An Open-Access Research Infrastructure of chemical Biology and Target-Based screening Technologies for Human and Animal Health, Agriculture and the Environment’ (grant no. MIs 5002691), which are implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational program ‘competitiveness, Entrepreneurship and Innovation’ (NsRF 2014-2020) and co‑financed by Greece and the European Union (European Regional Development Fund). The present study also received funding from the ‘competitiveness, Entrepreneurship and Innovation, EpAnEK 2nd cycle’ Operational program (grant no. Τ2ΕΔΚ‑02222, MIS 5074548), which was co‑funded by greece and the European Union.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
GPC, EE and DV conceived the present study. LP, LS, AE, FB, GPC, EE and DV wrote, drafted, revised, edited and reviewed the manuscript, and were involved in data collection. GPC, EE and DV confirm the authenticity of all the raw data. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
GPC is the Editor in Chief of the journal, and DV and EE are Editors of the journal. However, they had no personal involvement in the reviewing process, or any influence in terms of adjudicating on the final decision, for this article.
References
|
1
|
Novac N and Heinzel T: Nuclear receptors: Overview and classification. Curr Drug Targets Inflamm Allergy. 3:335–346. 2004.PubMed/NCBI View Article : Google Scholar
|
|
2
|
Shi Y: Orphan nuclear receptors in drug discovery. Drug Discov Today. 12:440–445. 2007.PubMed/NCBI View Article : Google Scholar
|
|
3
|
Ottow E and Weinmann H (eds): Nuclear Receptors as Drug Targets. In: Methods and Principles in Medicinal Chemistry. John Wiley & Sons, Inc., Hoboken NJ, 2008.
|
|
4
|
Huang P, Chandra V and Rastinejad F: Structural overview of the nuclear receptor superfamily: Insights into physiology and therapeutics. Annu Rev Physiol. 72:247–272. 2010.PubMed/NCBI View Article : Google Scholar
|
|
5
|
Pearce KH, Iannone MA, Simmons CA and Gray JG: Discovery of novel nuclear receptor modulating ligands: An integral role for peptide interaction profiling. Drug Discov Today. 9:741–751. 2004.PubMed/NCBI View Article : Google Scholar
|
|
6
|
Laudet V: Evolution of the nuclear receptor superfamily: Early diversification from an ancestral orphan receptor. J Mol Endocrinol. 19:207–226. 1997.PubMed/NCBI View Article : Google Scholar
|
|
7
|
Weikum ER, Liu X and Ortlund EA: The nuclear receptor superfamily: A structural perspective. Protein Sci. 27:1876–1892. 2018.PubMed/NCBI View Article : Google Scholar
|
|
8
|
Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al: InterPro: The integrative protein signature database. Nucleic Acids Res. 37 (Database):D211–D215. 2009.PubMed/NCBI View Article : Google Scholar
|
|
9
|
Vlachakis D, Papageorgiou L, Papadaki A, Georga M, Kossida S and Eliopoulos E: An updated evolutionary study of the Notch family reveals a new ancient origin and novel invariable motifs as potential pharmacological targets. PeerJ. 8(e10334)2020.PubMed/NCBI View Article : Google Scholar
|
|
10
|
Papageorgiou L, Loukatou S, Sofia K, Maroulis D and Vlachakis D: An updated evolutionary study of Flaviviridae NS3 helicase and NS5 RNA-dependent RNA polymerase reveals novel invariable motifs as potential pharmacological targets. Mol Biosyst. 12:2080–2093. 2016.PubMed/NCBI View Article : Google Scholar
|
|
11
|
Papageorgiou L, Megalooikonomou V and Vlachakis D: Genetic and structural study of DNA-directed RNA polymerase II of Trypanosoma brucei, towards the designing of novel antiparasitic agents. PeerJ. 5(e3061)2017.PubMed/NCBI View Article : Google Scholar
|
|
12
|
Waterhouse AM, Procter JB, Martin DM, Clamp M and Barton GJ: Jalview Version 2 - a multiple sequence alignment editor and analysis workbench. Bioinformatics. 25:1189–1191. 2009.PubMed/NCBI View Article : Google Scholar
|
|
13
|
Sobie EA: An introduction to MATLAB. Sci Signal. 4(tr7)2011.PubMed/NCBI View Article : Google Scholar
|
|
14
|
Thuillard M: Minimizing contradictions on circular order of phylogenic trees. Evol Bioinform Online. 3:267–277. 2007.PubMed/NCBI
|
|
15
|
Crow JF: Thomas H. Jukes (1906-1999). Genetics. 154:955–956. 2000.PubMed/NCBI
|
|
16
|
Gibbons FD and Roth FP: Judging the quality of gene expression-based clustering methods using gene annotation. Genome Res. 12:1574–1581. 2002.PubMed/NCBI View Article : Google Scholar
|
|
17
|
Leach A: Molecular modelling: Principles and applications. 2nd edition. Pearson, London, 2021.
|
|
18
|
Kumar S, Stecher G and Tamura K: MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 33:1870–1874. 2016.PubMed/NCBI View Article : Google Scholar
|
|
19
|
He Y, Yi W, Suino-Powell K, Zhou XE, Tolbert WD, Tang X, Yang J, Yang H, Shi J, Hou L, et al: Structures and mechanism for the design of highly potent glucocorticoids. Cell Res. 24:713–726. 2014.PubMed/NCBI View Article : Google Scholar
|
|
20
|
Hemmerling M, Nilsson S, Edman K, Eirefelt S, Russell W, Hendrickx R, Johnsson E, Kärrman Mårdh C, Berger M, Rehwinkel H, et al: Selective nonsteroidal glucocorticoid receptor modulators for the inhaled treatment of pulmonary diseases. J Med Chem. 60:8591–8605. 2017.PubMed/NCBI View Article : Google Scholar
|
|
21
|
Liu X, Wang Y and Ortlund EA: First high-resolution crystal structures of the glucocorticoid receptor ligand-binding domain-peroxisome proliferator-activated γ coactivator 1-α complex with endogenous and synthetic glucocorticoids. Mol Pharmacol. 96:408–417. 2019.PubMed/NCBI View Article : Google Scholar
|
|
22
|
Weikum ER, Liu X and Ortlund EA: The nuclear receptor superfamily: A structural perspective. Protein Sci. 27:1876–1892. 2018.PubMed/NCBI View Article : Google Scholar
|
|
23
|
Pawlak M, Lefebvre P and Staels B: General molecular biology and architecture of nuclear receptors. Curr Top Med Chem. 12:486–504. 2012.PubMed/NCBI View Article : Google Scholar
|
|
24
|
Mitsis T, Papageorgiou L, Efthimiadou A, Bacopoulou F, Vlachakis D, Chrousos GP and Eliopoulos E: A comprehensive structural and functional analysis of the ligand binding domain of the nuclear receptor superfamily reveals highly conserved signaling motifs and two distinct canonical forms through evolution. World Acad Sci J. 1:264–274. 2019.
|
|
25
|
Porter BA, Ortiz MA, Bratslavsky G and Kotula L: Structure and function of the nuclear receptor superfamily and current targeted therapies of prostate cancer. Cancers (Basel). 11(E1852)2019.PubMed/NCBI View Article : Google Scholar
|
|
26
|
Pace NJ and Weerapana E: Zinc-binding cysteines: Diverse functions and structural motifs. Biomolecules. 4:419–434. 2014.PubMed/NCBI View Article : Google Scholar
|
|
27
|
Plevin MJ, Mills MM and Ikura M: The LxxLL motif: A multifunctional binding sequence in transcriptional regulation. Trends Biochem Sci. 30:66–69. 2005.PubMed/NCBI View Article : Google Scholar
|
|
28
|
Loinder K and Söderström M: Functional analyses of an LXXLL motif in nuclear receptor corepressor (N-CoR). J Steroid Biochem Mol Biol. 91:191–196. 2004.PubMed/NCBI View Article : Google Scholar
|
|
29
|
Evans RM: The steroid and thyroid hormone receptor superfamily. Science. 240:889–895. 1988.PubMed/NCBI View Article : Google Scholar
|
|
30
|
Barish GD, Downes M, Alaynick WA, Yu RT, Ocampo CB, Bookout AL, Mangelsdorf DJ and Evans RM: A Nuclear Receptor Atlas: Macrophage activation. Mol Endocrinol. 19:2466–2477. 2005.PubMed/NCBI View Article : Google Scholar
|
|
31
|
Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM and Mangelsdorf DJ: Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 126:789–799. 2006.PubMed/NCBI View Article : Google Scholar
|
|
32
|
Sonoda J, Pei L and Evans RM: Nuclear receptors: Decoding metabolic disease. FEBS Lett. 582:2–9. 2008.PubMed/NCBI View Article : Google Scholar
|
|
33
|
Raftopoulou S, Nicolaides NC, Papageorgiou L, Amfilochiou A, Zakinthinos SG, George P, Eliopoulos E, Chrousos GP and Vlachakis D: Structural Study of the DNA: Clock/Bmal1 Complex Provides Insights for the Role of Cortisol, hGR, and HPA Axis in Stress Management and Sleep Disorders. Adv Exp Med Biol. 1195:59–71. 2020.PubMed/NCBI View Article : Google Scholar
|
|
34
|
Weinberger C, Hollenberg SM, Ong ES, Harmon JM, Brower ST, Cidlowski J, Thompson EB, Rosenfeld MG and Evans RM: Identification of human glucocorticoid receptor complementary DNA clones by epitope selection. Science. 228:740–742. 1985.PubMed/NCBI View Article : Google Scholar
|
|
35
|
Greene GL, Gilna P, Waterfield M, Baker A, Hort Y and Shine J: Sequence and expression of human estrogen receptor complementary DNA. Science. 231:1150–1154. 1986.PubMed/NCBI View Article : Google Scholar
|
|
36
|
Krust A, Green S, Argos P, Kumar V, Walter P, Bornert JM and Chambon P: The chicken oestrogen receptor sequence: Homology with v-erbA and the human oestrogen and glucocorticoid receptors. EMBO J. 5:891–897. 1986.PubMed/NCBI
|
|
37
|
Weinberger C, Giguère V, Hollenberg SM, Thompson C, Arriza J and Evans RM: Human steroid receptors and erb-A gene products form a superfamily of enhancer-binding proteins. Clin Physiol Biochem. 5:179–189. 1987.PubMed/NCBI
|
|
38
|
Owen GI and Zelent A: Origins and evolutionary diversification of the nuclear receptor superfamily. Cell Mol Life Sci. 57:809–827. 2000.PubMed/NCBI View Article : Google Scholar
|
|
39
|
Shiau AK, Coward P, Schwarz M and Lehmann JM: Orphan nuclear receptors: From new ligand discovery technologies to novel signaling pathways. Curr Opin Drug Discov Devel. 4:575–590. 2001.PubMed/NCBI
|
|
40
|
Kliewer SA, Lehmann JM and Willson TM: Orphan nuclear receptors: Shifting endocrinology into reverse. Science. 284:757–760. 1999.PubMed/NCBI View Article : Google Scholar
|
|
41
|
Aranda A and Pascual A: Nuclear hormone receptors and gene expression. Physiol Rev. 81:1269–1304. 2001.PubMed/NCBI View Article : Google Scholar
|
|
42
|
Chen T: Nuclear receptor drug discovery. Curr Opin Chem Biol. 12:418–426. 2008.PubMed/NCBI View Article : Google Scholar
|
|
43
|
Giannini A, Russo E, Mannella P and Simoncini T: Selective steroid receptor modulators in reproductive medicine. Minerva Ginecol. 67:431–455. 2015.PubMed/NCBI
|
|
44
|
Rhen T and Cidlowski JA: Antiinflammatory action of glucocorticoids - new mechanisms for old drugs. N Engl J Med. 353:1711–1723. 2005.PubMed/NCBI View Article : Google Scholar
|
|
45
|
Thornton JW: Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci USA. 98:5671–5676. 2001.PubMed/NCBI View Article : Google Scholar
|
|
46
|
Bridgham JT, Carroll SM and Thornton JW: Evolution of hormone-receptor complexity by molecular exploitation. Science. 312:97–101. 2006.PubMed/NCBI View Article : Google Scholar
|
|
47
|
Magadum S, Banerjee U, Murugan P, Gangapur D and Ravikesavan R: Gene duplication as a major force in evolution. J Genet. 92:155–161. 2013.PubMed/NCBI View Article : Google Scholar
|
|
48
|
Baker ME: Steroid receptor phylogeny and vertebrate origins. Mol Cell Endocrinol. 135:101–107. 1997.PubMed/NCBI View Article : Google Scholar
|
|
49
|
Laudet V, Hänni C, Coll J, Catzeflis F and Stéhelin D: Evolution of the nuclear receptor gene superfamily. EMBO J. 11:1003–1013. 1992.PubMed/NCBI
|
|
50
|
Escriva H, Safi R, Hänni C, Langlois MC, Saumitou-Laprade P, Stehelin D, Capron A, Pierce R and Laudet V: Ligand binding was acquired during evolution of nuclear receptors. Proc Natl Acad Sci USA. 94:6803–6808. 1997.PubMed/NCBI View Article : Google Scholar
|























