Introduction
The transcription factors NF-κB and Jun N-terminal
kinase (JNK) are crucial for cell survival, development, lymphocyte
activation and proliferation. Genetic deficiencies in NF-κB or its
signaling components that act upstream of NF-κB have been shown to
cause immune deficiencies, whereas over-activation of NF-κB has
been linked to autoimmunity and neoplastic disorders (1,2).
The CARMA1 signalosome, which is composed of CARMA1 [caspase
recruitment domain (CARD)-containing MAGUK protein 1], BCL10
(B-cell lymphoma 10) and MALT1 (mucosa-associated lymphoid tissue
lymphoma translocation protein 1), is the main signaling molecular
complex that performs pivotal functions in T-cell receptor (TCR)-
and B-cell receptor (BCR)-mediated NF-κB activation.
CARMA1 (also called CARD11) is a member of the MAGUK
(membrane-associated guanylate kinase) family of scaffold proteins
that assists in recruitment and assembly of signaling molecules in
the cytoplasmic membrane. CARMA1 contains an N-terminal CARD domain
followed by a coiled-coil domain and a C-terminal MAGUK domain
(3,4). The CARD domain is a subfamily of the
death domain (DD) superfamily that comprises the DD subfamily,
death effector domain (DED) subfamily, caspase recruitment domain
(CARD) subfamily and pyrin domain (PYD) subfamily. The death domain
superfamily, which is one of the largest classes of protein
interaction modules, plays a role in apoptosis, inflammation, and
innate immune signaling pathways (5–8).
MALT1 contains an N-terminal DD followed by two immunoglobulin
(IG)-like domains and a C-terminal caspase-like domain. Although
the molecular mechanism of MALT1 involvement in TCR- or
BCR-mediated NA-κB activation is unclear, several studies have
shown that MALT1 functions as an ubiquitin E3 ligase inducing
K63-linked polyubiquitination of NEMO, which is one of the
components of the IκB kinase (IKK) complex, and that it also
functions as a protease that can cleave the deubiquitinating
enzymes A20 and BCL10 (9–12). The proteolytic activity of MALT1
is required for proper NF-κB activation followed by IL-2 production
(11,12). BCL10 is composed of an N-terminal
CARD and a C-terminal Ser/Thr-rich region and has been shown to
induce apoptosis and activate NF-κB. For assembly of the CARMA1
signalosome, BCL10 functions as an adaptor protein that interacts
with CARMA1 via the CARD-CARD interaction and with MALT1 via
interaction between the C-terminal Ser/Thr-rich region of BCL10 and
the first Ig domain of MALT1 (13,14). Upon the activation of T cells, TCR
transmits an outside signal to PKCθ that can then phosphorylate
downstream target CARMA1 (15,16). This phosphorylation promotes the
assembly of the CARMA1 signalosome (16).
Due to the importance of the CARMA1 signalosome in
immune receptor signaling events, studies in these fields are of
great biological importance. Despite the biological importance of
the CARMA1 signalosome, structural and biochemical studies have
been limited as CARD-containing proteins are prone to aggregation
under physiological conditions (17–19). In the present study, we
successfully purified and characterized CARMA1 CARD and BCL10 CARD
and showed that both CARMA1 CARD and BCL10 CARD easily
self-oligomerized under physiological conditions. This
self-oligomerization of the CARD domain prevents complex formation
in the CARMA1 signalosome in vitro. Finally, we proposed an
interaction mode between CARMA1 CARD and BCL10 CARD based on a
structural-based modeling study.
Materials and methods
Materials
DNA restriction enzymes (NdeI and
XhoI), T4 DNA ligase, and Tag DNA polymerase were obtained
from NEB (New England Biolabs, Ipswich, MA, USA). IPTG, urea,
imidazole and DTT were purchased from Sigma (St. Louis, MO, USA).
E. coli BL21 (DE3) cells and plasmid pET vectors were
obtained from Novagen. A Superdex 200 gel filtration column was
purchased from GE Healthcare. Inhibitor cocktail was acquired from
Roche Diagnostics (Indianapolis, IN, USA). All concentrators were
obtained from Millipore (Billerica, MA, USA).
Expression of CARMA1 CARD and BCL10 CARD
in E. coli
The cDNA of full length mouse CARMA1 (1–1155 amino
acids) and BCL10 (1–233 amino acids) were used as templates for
PCR, while plasmid vectors pET26b and pET28a were used to add a
hexahistidine (6-His) tag to the C-terminus and the N and
C-terminus, respectively, for affinity purification. The CARD
domain of mouse CARMA1 coding for 1(Met)-109(Lys) was cloned into
pET 28a, resulting in pET (6His-CARMA1 CARD-6His). Additionally,
the CARD domain of BCL10 coding for 1(Met)-93(Asn) was cloned into
pET 26b, resulting in pET (BCL10 CARD-6His). The expression methods
for two proteins were the same as previously described (20). A brief summary of these methods is
presented below.
Each clone was streaked onto Lysogeny broth (LB)
agar plates containing appropriate antibiotics (100 μl/ml
ampicillin for the pET 26b clone and 50 μl/ml kanamycin for the pET
28a clone), after which the plates were incubated at 37°C for 18 h.
A single colony was then used to inoculate 5 ml of the LB medium,
which was subsequently incubated at 37°C in a shaking incubator.
Next, 5 ml of the overnight small-scale culture was used to
inoculate 1000 ml of LB in a 2000-ml culture flask, and the culture
growth was continued in a shaking incubator for ~4 h. When the
optical density (OD) at 600 nm reached 0.6–0.7, expression was
induced by overnight treatment with 0.5 mM isopropyl-β-D
thiogalactopyranoside (IPTG) at 20°C.
Purification of target proteins
Bacteria expressing each protein were pelleted by
centrifugation, resuspended and lysed by sonication in 50 ml of
lysis buffer [20 mM sodium citrate (pH 5.0), 500 mM NaCl and 5 mM
imidazole] supplied with a protease inhibitor-cocktail (Roche
Diagnostics). The lysate was then centrifuged at 16,000 rpm for 1 h
at 4°C, after which the supernatant fractions were applied to a
gravity column (Bio-Rad, Hercules, CA, USA) packed with Ni-NTA
affinity resin (Qiagen, Hilden, Germany). Next, the unbound
bacterial proteins were removed by rinsing with 100 ml of washing
buffer (20 mM Tris buffer at pH 8.0, 500 mM NaCl, 60 mM imidazole),
after which the target proteins were eluted from the column using
elution buffer [20 mM sodium citrate (pH 5.0), 500 mM NaCl and 250
mM imidazole], with 1 ml elution fractions being collected over a
total of 10 ml. Fractions containing >80% homogeneous target
protein as shown by SDS-PAGE were then selected and combined. In
the final purification step, the sample was loaded onto a
Superdex-200 gel-filtration column (GE Healthcare, Sweden)
equilibrated with various buffers. Fractions containing the target
protein were then pooled and stored at 4°C for further experiments.
The homogeneity of the protein was assessed by SDS-PAGE.
Circular dichroism spectroscopy
The secondary structures were measured by circular
dichroism (CD) spectroscopy using a J-715 spectropolarimeter
(Jasco, Japan). The spectra were obtained from 200 to 250 nm at
25°C in a 0.1-cm path-length quartz cuvette using a bandwidth of
1.0 nm, a speed of 50 mm/min, and a 5-sec response time. The
protein samples in the buffer containing 20 mM Tris-HCl at pH 8.0
and 150 mM NaCl were diluted to 0.1 mg/ml prior to use. Four scans
were accumulated and averaged and the α-helical content was
calculated from the molar ellipticity at 222 nm (21).
Complex assay by gel-filtration
chromatography
Purified CARMA1 was incubated with BCL10 for 1 h at
room temperature. Following incubation, the mixture was
concentrated to 10–15 mg ml−1 using a concentration kit
(Millipore). The concentrated protein mixture was then applied to a
Superdex 200 gel-filtration column 10/30 (GE Healthcare), which was
pre-equilibrated with a solution of 20 mM Tris buffer at pH 8.0 and
0 mM NaCl; 20 mM Tris buffer at pH 8.0 and 150 mM NaCl; and 20 mM
Tris buffer at pH 8.0 and 1 M NaCl. Assembly of the complex was
evaluated based on the positions of the eluted peaks monitored at
280 nm followed by SDS-PAGE.
Native-PAGE assay
Formation of the complex between CARMA1 CARD and
BCL10 CARD was assayed by native (non-denaturing) PAGE conducted on
a PhastSystem (GE Healthcare) with pre-made 8–25% acrylamide
gradient gels (GE Healthcare). Coomassie Brilliant Blue was used
for staining and detection of the band patterns. Separately
purified CARMA1 CARD and BCL10 CARD were pre-incubated for 1 h at
room temperature, after which the mixture was applied to the gel.
The assembly of the complex was evaluated based on the appearance
of newly formed bands and the disappearance of existing bands.
Homology modeling
Homology models of CARMA1 CARD and BCL10 CARD were
constructed using a homology modeling server, SWISS-MODEL (22). The previously solved Apaf-1 CARD
structure (PDB ID: 1CWW) and RAIDD CARD structure (PDB ID: 3CRD)
were used as modeling templates for CARMA1 CARD and BCL-10 CARD,
respectively. The stereochemical quality of constructed models was
validated with a Ramachandran plot generated using PROCHECK
(23).
Electrostatic surfaces and ribbon diagrams were
generated using the PyMOL program [DeDeLano WL (2002), The PyMOL
Molecular Graphics System, DeLano Scientific, San Carlos, CA,
USA].
Results
Expression, purification and
characterization of CARMA1 CARD and BCL10 CARD
CARMA1, BCL10, and MALT1 proteins are the main
components of the CARMA1 signalosome, which plays a critical role
in lymphocyte activation and proliferation through immune
cell-mediated NF-κB activation. Each protein contains one member of
the DD superfamily, CARD for CARMA1 and BCL10, and DD for MALT1 at
the N-terminus to enable homotypic protein-protein interaction
(Fig. 1A).
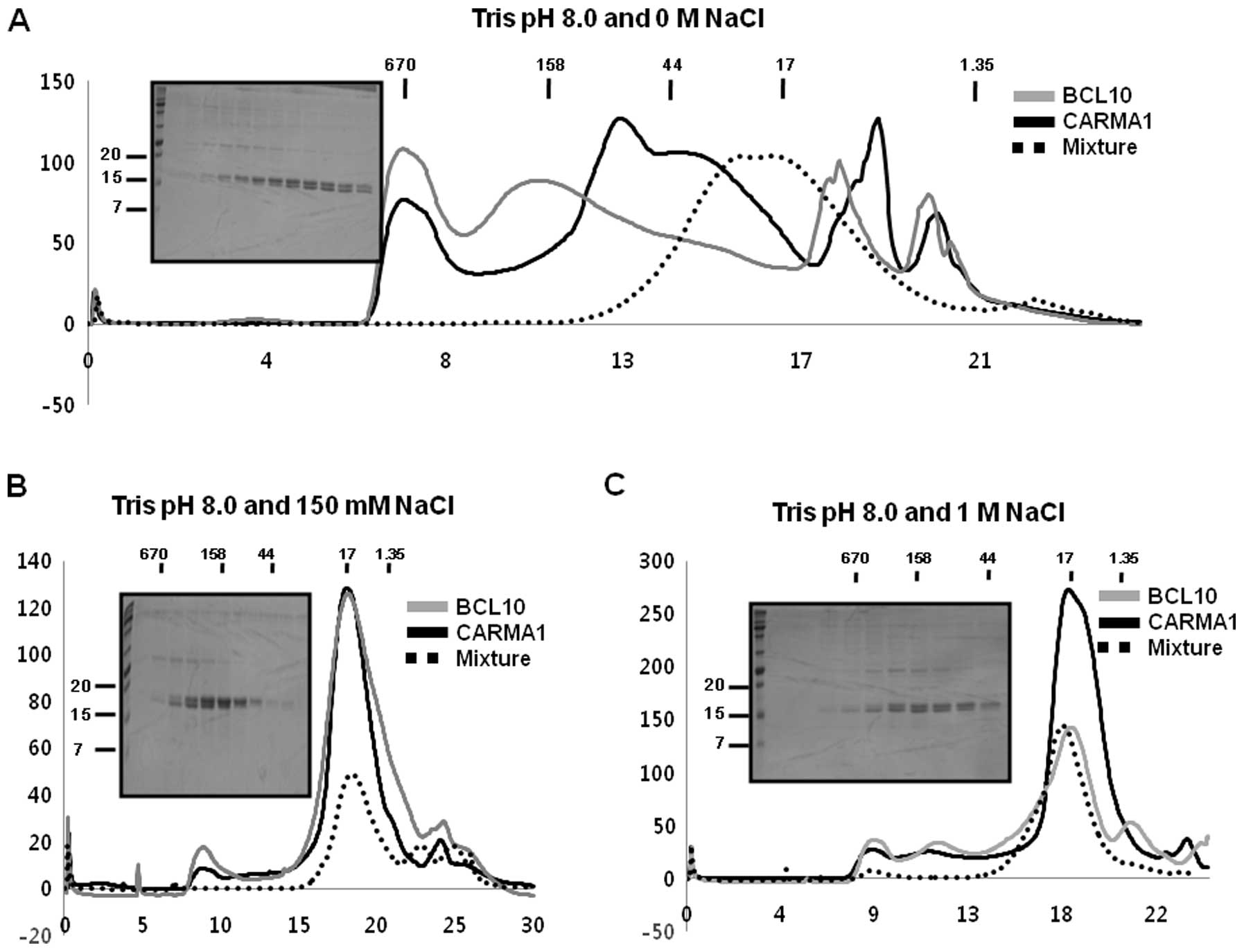
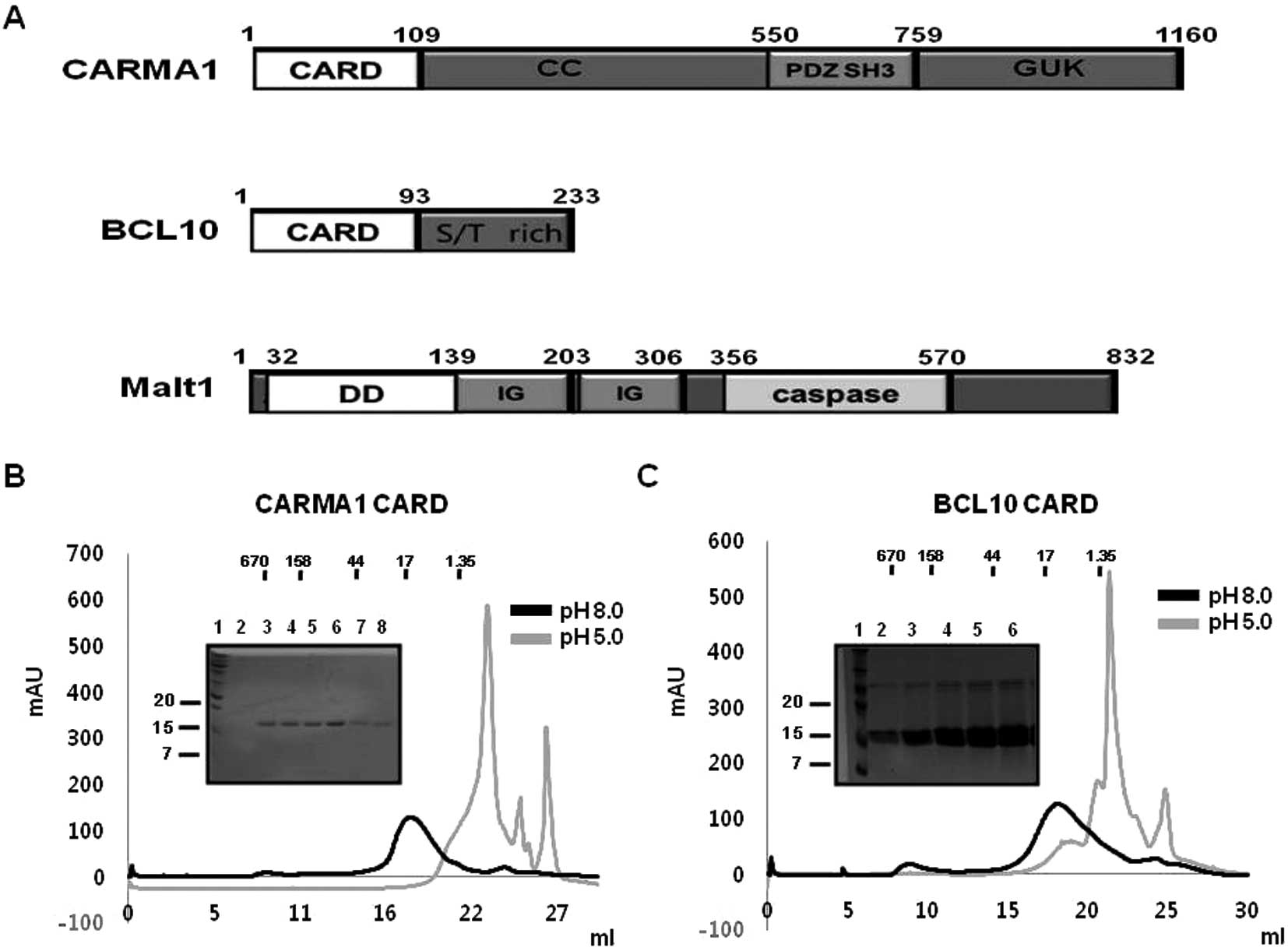 | Figure 1Purification of CARMA1 CARD and BCL10
CARD. (A) Domain boundary of protein components for the CARMA1
signalosome, CARMA1, BCL10, and MALT1. Each domain is indicated by
different colors. The number of residues of amino acids
corresponding to the various domains are indicated. CARD, caspase
recruitment domain; CC, coiled coil domain; PDZ, Psd95, Dlg1, Zo1
domain; SH3, SRC homology 3 domain; GUK, guanylate kinase domain;
DD, death domain; Ig, immunoglobulin domain. (B) Purification of
CARMA1 CARD shown by gel-filtration chromatograms and fractions.
The black line reflects the profile obtained under 150 mM NaCl
using Tris buffer at pH 8.0. The gray line shows the profile
obtained under 150 mM NaCl using citrate buffer at pH 5.0. SDS-PAGE
stained by Coomassie Blue from the fractions of gel-filtration
chromatography at 150 mM NaCl and 20 mM Tris buffer pH 8.0 is shown
at the upper left. Lane 1, marker; lanes 2–8, gel-filtration
chromatography fractions collected from 16 to 21 ml. (C)
Purification of BCL10 CARD by gel-filtration chromatograms and
fractions. The black line shows the profile obtained under 150 mM
NaCl using Tris buffer at pH 8.0. The gray line shows the profile
obtained under 150 mM NaCl using citrate buffer at pH 5.0. SDS-PAGE
stained by Coomassie Blue from the fractions of gel-filtration
chromatography at 150 mM NaCl and 20 mM Tris buffer pH 8.0 is shown
on the upper left. Lane 1, marker; lanes 2–6, gel-filtration
chromatography fractions collected from 15 to 20 ml. |
As a first step for elucidation of the molecular
basis of assembly of the CARMA1 signalosome, which is mediated by
the well known DD superfamily protein interaction module, we
attempted to express and purify CARMA1 CARD and BCL10 CARD.
Although it is well known that obtaining functional DD superfamily
members is difficult due to their insolubility under physiological
conditions (17–19), following IPTG induction bacterial
cells (BL21-DE3 strain) containing CARMA1 CARD (1–109) in pET28a
and BCL10 CARD (1–93 residue) in pET26b were found to express
recombinant proteins with molecular weights ~12 and 10 kDa,
respectively, that were detected in the soluble fraction after
sonication (Fig. 1B). Both
proteins were purified by Ni affinity chromatography followed by
gel-filtration chromatography and eluted at ~17 ml of
gel-filtration column, indicating they mainly exist as monomers in
solution (Fig. 1B and C). Based
on previous studies showing that the solubility of many DD
superfamily members is sensitive to pH and salt concentration
(20,24–26), we analyzed the characteristics of
each CARD domain of CARMA1 and BCL10. Our gel-filtration
chromatography study showed that both CARDs were well behaved at pH
8.0 and not stable at pH 5.0 (Fig. 1B
and C). Although both CARDs exhibited different characteristics
without any salt, the salt concentration did not have a great
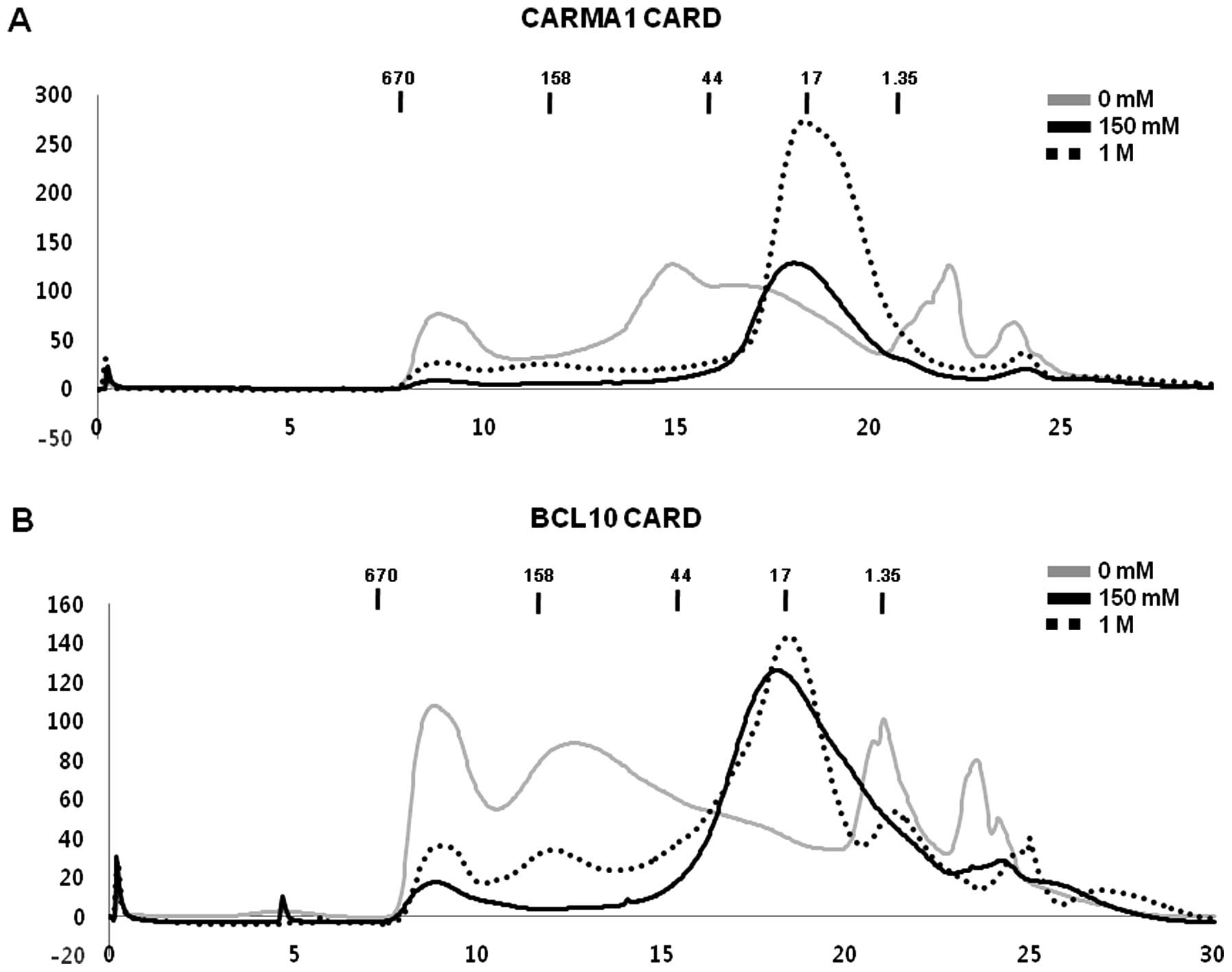
effect on the behavior of CARDs in solution (Fig. 2A and B).
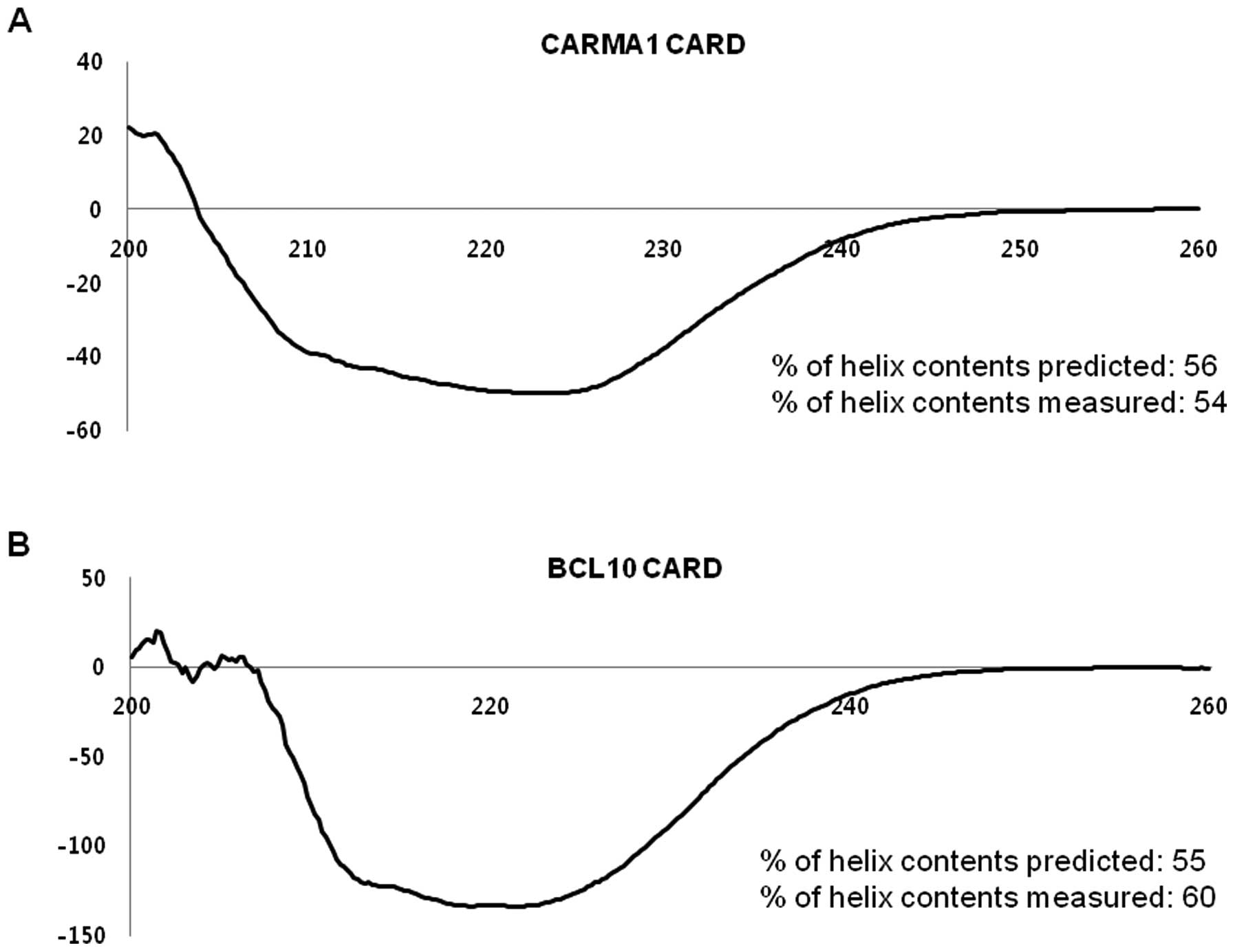
Structures of purified CARMA1 CARD and
BCL10 CARD are typical α-helix bundle folds
To confirm the correct folding and analyze the
secondary structures of CARMA1 CARD and BCL10 CARD, the α-helices
were analyzed by measuring the far UV circular dichroic spectra. As
shown in Fig. 3, both domains
showed CD spectrum patterns typical of α-helical proteins,
exhibiting two pronounced minima at 208 nm and 222 nm, and a maxima
at 195 nm, which matched well with the molecular structure of other
members of the death domain superfamily (20). The percentage of helix contents
predicted and measured agreed well, with 56% predicted vs. 54%
measured for CARMA1 and 55% predicted vs. 60% measured for
BCL10.
Purified CARMA1 CARD and BCL10 CARD did
not interact with each other in vitro
Since it is well known that CARMA1 CARD can interact
with BCL10 CARD directly for the assembly of the CARMA1 signalosome
(27,28), we tested whether purified CARMA1
CARD can interact with purified BCL10 CARD in vitro. To
accomplish this, purified CARMA1 CARD was incubated with purified
BCL10 CARD for 1 h at room temperature, after which the mixture was
applied to a Superdex 200 gel-filtration column 10/30 (GE
Healthcare) that had been pre-equilibrated with the proper
solution. Assembly of the complex was evaluated based on the
positions of the eluted peak monitored at 280 nm followed by
SDS-PAGE. Since the interaction of proteins is usually affected by
the concentration of salt, we tested the interaction between CARMA1
CARD and BCL10 CARD under no salt, low salt (150 mM NaCl) and high
salt (1 M NaCl) conditions. Because the individual components and
the protein mixture were both eluted at ~17–18 ml without any
possible new complex peak, we concluded that no complex of CARMA1
CARD and BCL10 CARD was obtained, regardless of how much salt was
added to the solution (Fig.
4A-C). Although there was a new peak generated by the protein
mixture when we used the no-salt buffer during gel filtration,
SDS-PAGE showed that CARMA1 CARD and BCL10 CARD did not co-migrate,
indicating that they had not formed a complex (Fig. 4A).
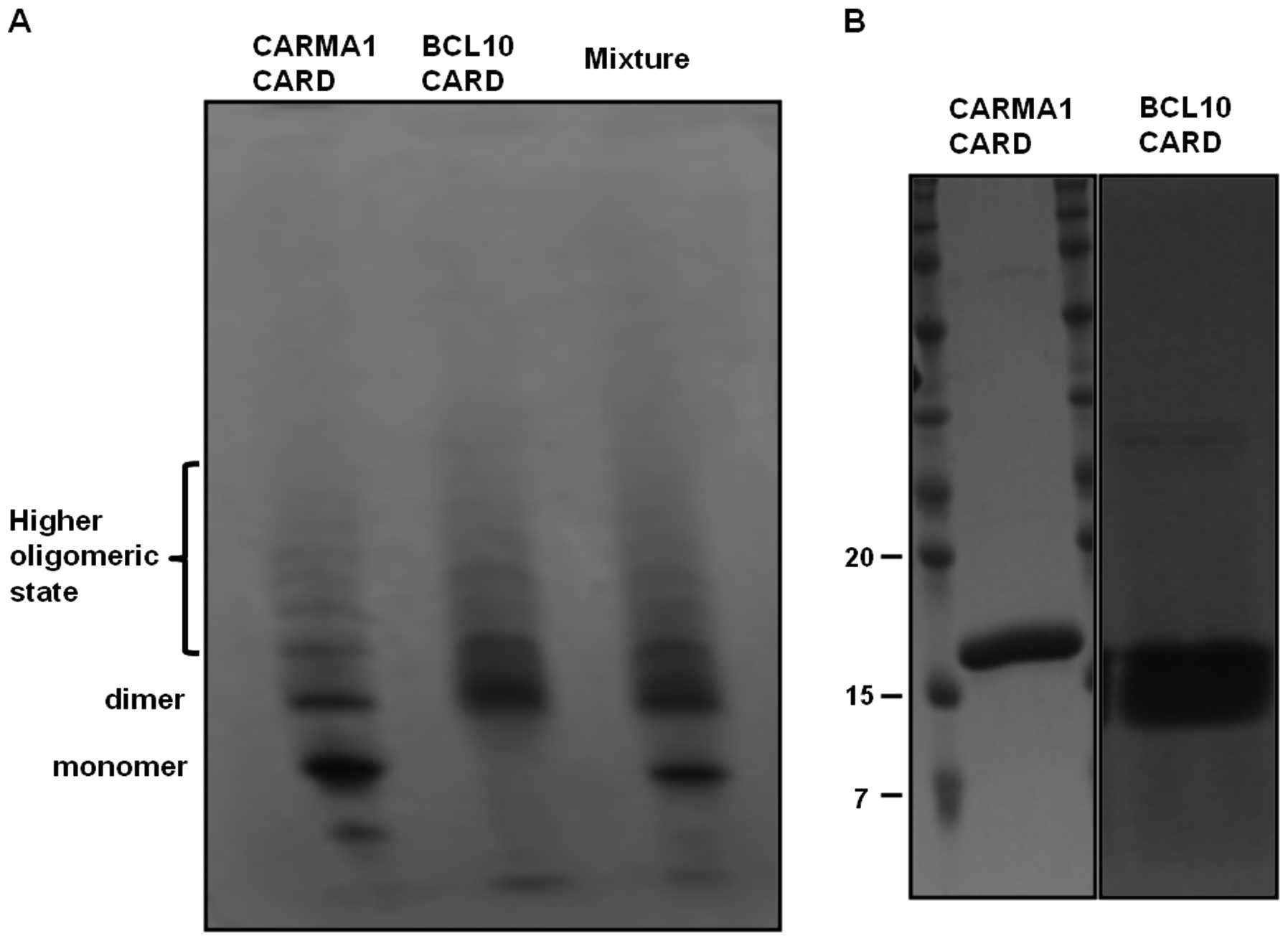
Self-oligomerization of CARD domain
prevents complex formation in the CARMA1 signalosome
Formation of a complex between CARMA1 CARD and BCL10
CARD was also assayed by native-(non-denaturing) PAGE. Separately
purified CARMA1 CARD and BCL10 CARD were incubated for 1 h at room
temperature, after which the mixture was applied to the native gel
(8–25% gradient gel from GE Healthcare). The assembly of the
complex was then evaluated based on the appearance of newly formed
bands and the disappearance of existing bands. No marked changes in
band pattern were detected upon native-PAGE, indicating that
purified CARMA1 CARD cannot form a stable complex with BCL10 CARD
(Fig. 5A). Of note, although
CARMA1 CARD and BCL10 CARD were detected as a single band upon
SDS-PAGE (Fig. 5B), they occurred
as many bands upon native-PAGE, indicating that CARMA1 CARD and
BCL10 CARD easily form a highly oligomeric form, and that this
self-oligomerization prevents complex formation between CARMA1 CARD
and BCL10 CARD.
Predicted model of the interaction
between CARMA1 CARD and BCL10 CARD
To determine whether the interaction between CARMA1
CARD and BCL10 CARD for assembly of the CARMA1 signalosome occurs
via a similar mode of interactions that were detected in the Apaf-1
CARD:caspase-9 CARD complex for assembly of the apoptosome, we
conducted homology modeling using the Apaf-1 CARD structure (PDB
ID: 1CWW) for CARMA1 CARD and the RAIDD CARD structure (PDB ID:
3CRD) for BCL10 CARD as the modeling template. Apaf-1 CARD shares
20% amino acid sequence identity with CARMA1 CARD and is the most
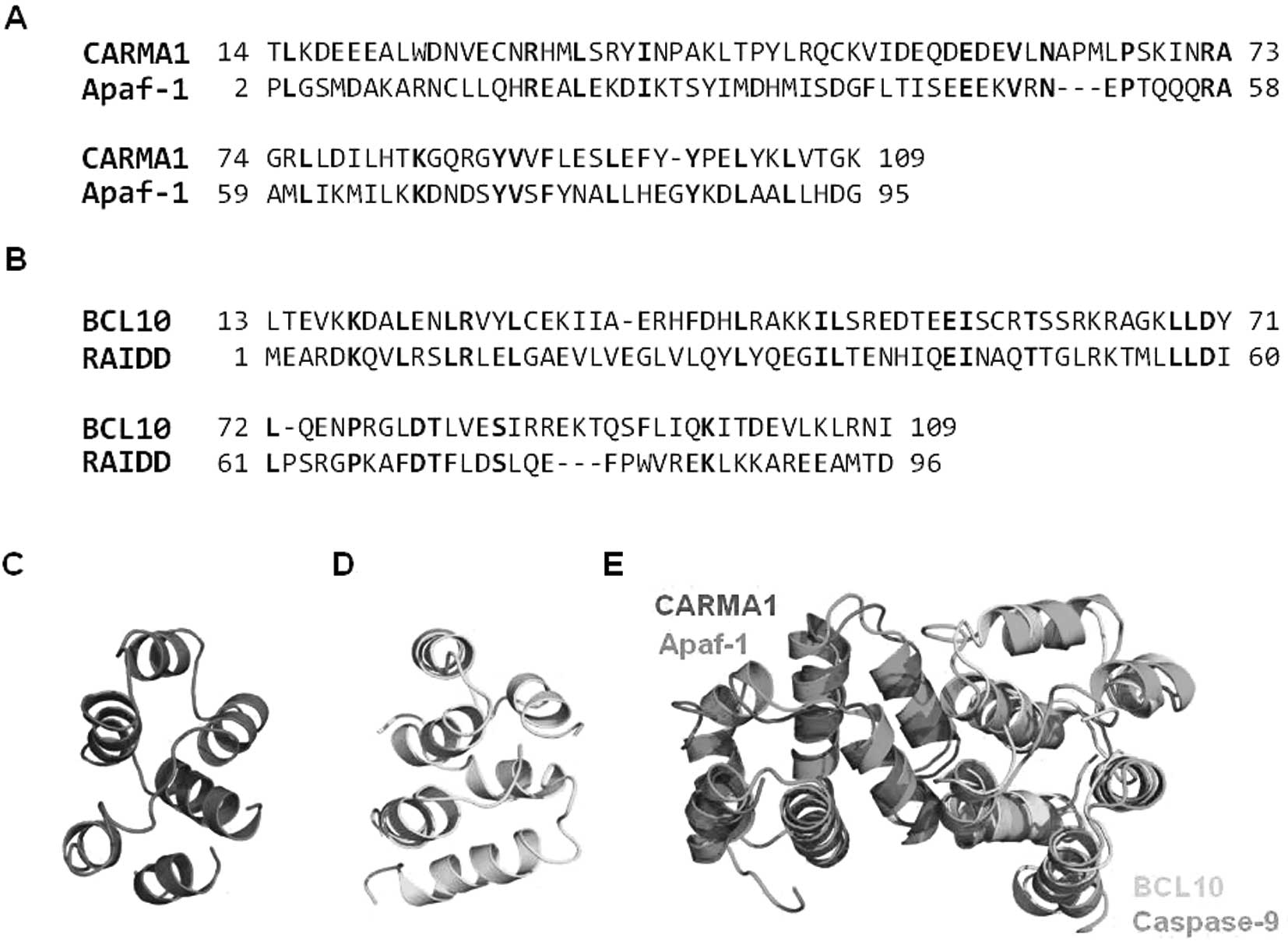
structurally similar CARD to CARMA1 CARD (Fig. 6A). RAIDD CARD shows 21% amino acid
sequence identity with BCL10 CARD (Fig. 6B). The modeled structures of
CARMA1 CARD (Fig. 6C) and BCL10
CARD (Fig. 6D) were well
constructed, possessing 6 helix bundles, which is the typical
structural composition of the DD superfamily. To analyze the
possible interaction interface, the modeling structure of CARMA1
CARD and BCL10 CARD was superimposed with the previously solved
Apaf-1 CARD:caspase-9 CARD complex structure, which is the only
available complex structure of the CARD:CARD interaction (Fig. 6E). Based on the structure of the
Apaf-1 CARD:caspase-9 CARD complex, the interaction is mainly
mediated by charged interactions. Three positively charged residues
in caspase-9 CARD, R13, R52 and R56, and two negatively charged
residues in Apaf-1 CARD, D27 and E40, are critical to this
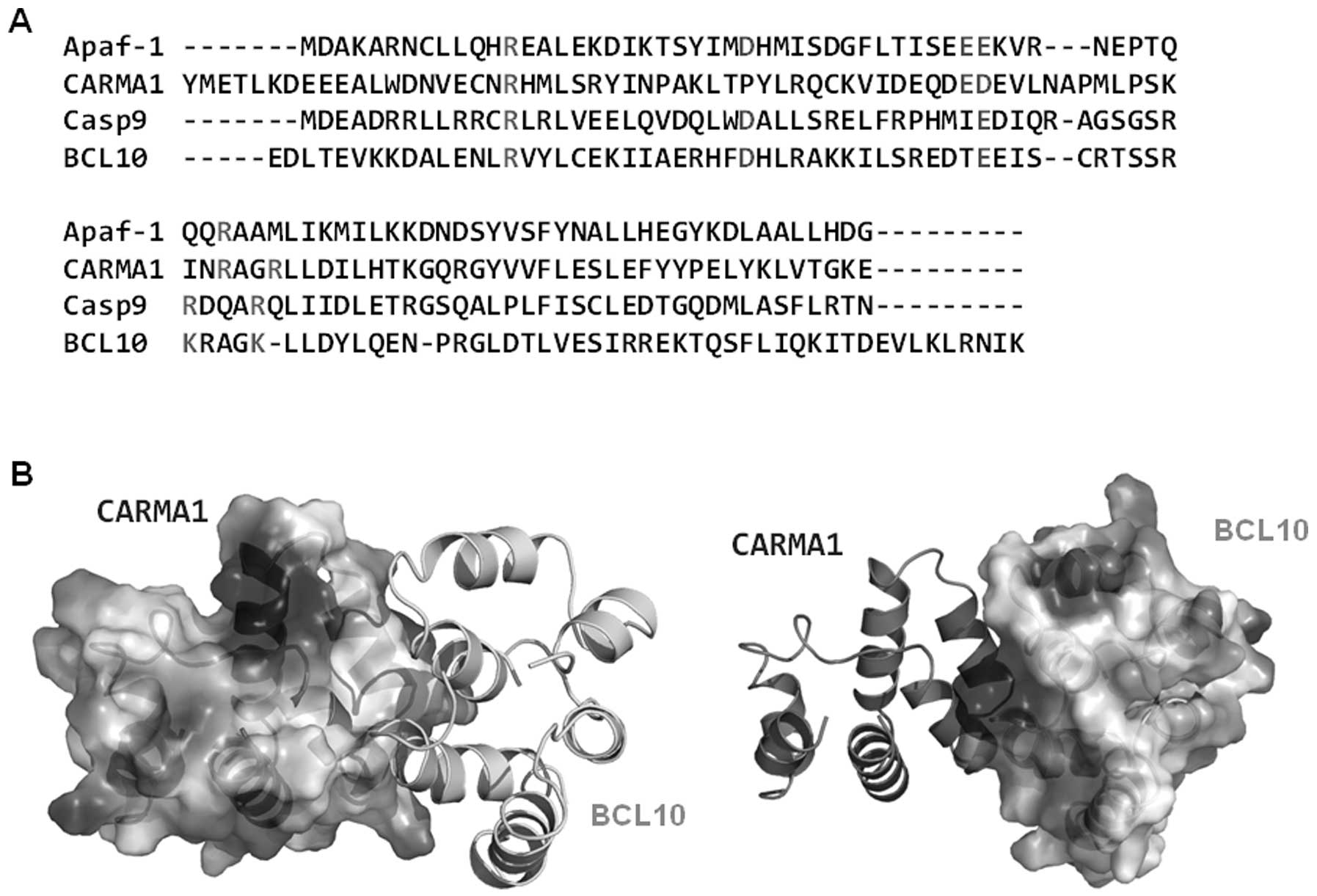
interaction. Sequence alignment revealed that all charged residues
that are critical to the interaction between Apaf-1 CARD and
caspase-9 CARD are conserved at CARMA1 CARD (E56) and BCL10 CARD
(R25, K63 and K67) (Fig. 7A). The
gross features of each electrostatic surface of CARMA1 CARD and
BCL10 CARD were very similar to those of Apaf-1 CARD and caspase-9
CARD in that one side of CARD is acidic and the other side is basic
(Fig. 7B).
Discussion
The CARMA1 signalosome, which is composed of CARMA1,
BCL10 and MALT1, is a signaling molecular complex that plays a
pivotal role in T-cell receptor (TCR)- and B-cell receptor
(BCR)-mediated NF-κB activation. Each protein contains one member
of the death domain (DD) superfamily, CARD for CARMA1 and BCL10 and
DD for MALT1, to enable homotypic protein-protein interaction. As a
first step in elucidation of the molecular basis for assembly of
the CARMA1 signalosome, we attempted to express and purify CARMA1
CARD and BCL10 CARD and to reconstitute the complex in vitro
for further structural studies. Since it is well known that
obtaining members of the functional DD superfamily including CARD
is difficult due to their insolubility under physiological
conditions and the sensitivity of many DD superfamily members to pH
and salt concentration, we analyzed the characteristics of each
CARD domain of CARMA1 and BCL10 to identify the optimal conditions
for purification and reconstitution.
The solubility of both CARDs was best at pH 8.0 and
between 150 mM and 1 M NaCl. CARMA1 CARD and BCL10 CARD contain an
α-helix, which match well with the molecular structure of other
members of the death domain superfamily. Since it is well known
that CARMA1 can interact with BCL10 directly via CARD for assembly
of the CARMA1 signalosome in the cell, we tested whether purified
CARMA1 CARD could interact with purified BCL10 CARD in vitro
and found that separately purified CARDs did not form a stable
complex. Based on the native-PAGE study, we determined that CARDs
did not interact with each other as CARMA1 CARD easily
self-oligomerized under certain conditions, preventing interaction
with BCL10 CARD, which is a proposed partner for interaction in the
cell.
Finally, the possible interaction mode between
CARMA1 and BCL10 for assembly of the CARMA1 signalosome was
proposed by sequence analysis and a modeling study. Comparison of
the amino acid sequences and the structure of the Apaf-1
CARD:caspase-9 CARD complex (the only available CARD complex
structure) revealing three positively charged residues at BCL10
CARD (R25, K63 and K67), which is well conserved in caspase-9 CARD
(R13, R52 and R56), and one negatively charged residue at CARMA1
CARD (E56), which is well conserved in Apaf-1 CARD (E40), may be
crucial for the interaction. Since the CARD domains are protein
interaction modules, their surface features dictate their mode of
interactions with partners. In the case of CARMA1 CARD and BCL10
CARD interaction for assembly of the CARMA1 signalosome, although
no structural information is available, it is possible that its
charged surface is important to the interaction. The DD, which is
in the same superfamily as CARD, forms a highly oligomeric complex
as well as a dimeric complex (29,30). Although only one dimeric complex
has been reported for CARD, DED and PYD, to date, it is still
possible that other subfamily members of the DD superfamily
including CARD, DED and PYD form more complicated oligomeric
complexes similar to DD. The advantages and disadvantages of
forming highly oligomeric complexes to transmit signals via the DD
superfamily should be investigated in detail in future studies.
Acknowledgements
This research was supported by the National Research
Foundation of Korea (NRF) grant funded by the Korean Government
(MEST) (2012-010870).
References
|
1
|
Li Q and Verma IM: NF-kappaB regulation in
the immune system. Nat Rev Immunol. 2:725–734. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Thome M: CARMA1, BCL-10 and MALT1 in
lymphocyte development and activation. Nat Rev Immunol. 4:348–359.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bertin J, Wang L, Guo Y, et al: CARD11 and
CARD14 are novel caspase recruitment domain
(CARD)/membrane-associated guanylate kinase (MAGUK) family members
that interact with BCL10 and activate NF-kappa B. J Biol Chem.
276:11877–11882. 2001. View Article : Google Scholar
|
|
4
|
Gaide O, Martinon F, Micheau O, Bonnet D,
Thome M and Tschopp J: Carma1, a CARD-containing binding partner of
Bcl10, induces Bcl10 phosphorylation and NF-kappaB activation. FEBS
Lett. 496:121–127. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Park HH, Lo YC, Lin SC, Wang L, Yang JK
and Wu H: The death domain superfamily in intracellular signaling
of apoptosis and inflammation. Annu Rev Immunol. 25:561–586. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weber CH and Vincenz C: The death domain
superfamily: a tale of two interfaces? Trends Biochem Sci.
26:475–481. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Reed JC, Doctor KS and Godzik A: The
domains of apoptosis: a genomics perspective. Sci STKE.
2004:re92004.PubMed/NCBI
|
|
8
|
Park HH, Logette E, Rauser S, et al: Death
domain assembly mechanism revealed by crystal structure of the
oligomeric PIDDosome core complex. Cell. 128:533–546. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhou H, Wertz I, O’Rourke K, et al: Bcl10
activates the NF-kappaB pathway through ubiquitination of NEMO.
Nature. 427:167–171. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Oeckinghaus A, Wegener E, Welteke V, et
al: Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell
activation. EMBO J. 26:4634–4645. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rebeaud F, Hailfinger S, Posevitz-Fejfar
A, et al: The proteolytic activity of the paracaspase MALT1 is key
in T cell activation. Nat Immunol. 9:272–281. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Coornaert B, Baens M, Heyninck K, et al: T
cell antigen receptor stimulation induces MALT1
paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat
Immunol. 9:263–271. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Uren AG, O’Rourke K, Aravind LA, et al:
Identification of paracaspases and metacaspases: two ancient
families of caspase-like proteins, one of which plays a key role in
MALT lymphoma. Mol Cell. 6:961–967. 2000.PubMed/NCBI
|
|
14
|
Lucas PC, Yonezumi M, Inohara N, et al:
Bcl10 and MALT1, independent targets of chromosomal translocation
in malt lymphoma, cooperate in a novel NF-kappa B signaling
pathway. J Biol Chem. 276:19012–19019. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sun Z, Arendt CW, Ellmeier W, et al:
PKC-theta is required for TCR-induced NF-kappaB activation in
mature but not immature T lymphocytes. Nature. 404:402–407. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Matsumoto R, Wang D, Blonska M, et al:
Phosphorylation of CARMA1 plays a critical role in T cell
receptor-mediated NF-kappaB activation. Immunity. 23:575–585. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang B, Eberstadt M, Olejniczak ET,
Meadows RP and Fesik SW: NMR structure and mutagenesis of the Fas
(APO-1/CD95) death domain. Nature. 384:638–641. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jang TH and Park HH: Purification,
crystallization and preliminary X-ray crystallographic studies of
RAIDD Death-Domain (DD). Int J Mol Sci. 10:2501–2509. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Jeong EJ, Bang S, Lee TH, Park YI, Sim WS
and Kim KS: The solution structure of FADD death domain. Structural
basis of death domain interactions of Fas and FADD. J Biol Chem.
274:16337–16342. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jang TH and Park HH: Generalized
semi-refolding methods for purification of the functional death
domain superfamily. J Biotechnol. 151:335–342. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen YH, Yang JT and Martinez HM:
Determination of the secondary structures of proteins by circular
dichroism and optical rotatory dispersion. Biochemistry.
11:4120–4131. 1972. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Schwede T, Kopp J, Guex N and Peitsch MC:
SWISS-MODEL: An automated protein homology-modeling server. Nucleic
Acids Res. 31:3381–3385. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Laskowski RA, Rullmannn JA, MacArthur MW,
Kaptein R and Thornton JM: AQUA and PROCHECK-NMR: programs for
checking the quality of protein structures solved by NMR. J Biomol
NMR. 8:477–486. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bae JY and Park HH: Crystal structure of
NALP3 protein pyrin domain (PYD) and its implications in
inflammasome assembly. J Biol Chem. 286:39528–39536. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bae JY and Park HH: Crystallization and
preliminary X-ray crystallographic studies of the PYD domain of
human NALP3. Acta Crystallogr Sect F Struct Biol Cryst Commun.
67:1421–1424. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Park HH: Structural analyses of death
domains and their interactions. Apoptosis. 16:209–220. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wegener E and Krappmann D:
CARD-Bcl10-Malt1 signalosomes: missing link to NF-kappaB. Sci STKE.
2007:pe212007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu Y, Song R, Gao Y, et al: Protein
kinase C-delta negatively regulates T cell receptor-induced
NF-kappaB activation by inhibiting the assembly of CARMA1
signalosome. J Biol Chem. 287:20081–20087. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park HH: Structural features of
caspase-activating complexes. Int J Mol Sci. 13:4807–4818. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ferrao R and Wu H: Helical assembly in the
death domain (DD) superfamily. Curr Opin Struct Biol. 22:241–247.
2012. View Article : Google Scholar : PubMed/NCBI
|