Introduction
Inflammatory bowel disease (IBD) is a chronic
inflammatory disease of the intestines and includes two distinct
disease categories: Crohn’s disease (CD) and ulcerative colitis
(UC). Both are associated with an increased risk of colorectal
cancer (CRC) (1,2). UC is a disease that is characterized
by chronic inflammation, rapid cell turnover and a substantial risk
of colon cancer (1,3). However, UC-associated colon cancer
differs from sporadic colon cancer in many ways (4,5).
In the case of UC patients, the risk of colon cancer
has been reported to be 10- to 20-fold higher in patients with a
disease duration of 20 years or more, even though current treatment
may have modulated this risk (6–8).
As a result of the recognition of this increased risk, annual
colonoscopic surveillance with multiple biopsies is recommended for
an early diagnosis of displasia in UC (9).
DNA methylation is one of the important epigenetic
mechanisms that controls gene expression, chromatin structure,
genome stability and X chromosome inactivation (10). Aberrant DNA methylation can lead
to a serious imbalance in the normal function of cells and can
promote pathological conditions (10). Promoter CpG island
hypermethylation of tumor-suppressor genes has been known as a
common hallmark of all human cancer (11). In addition, transcriptional
silencing associated with promoter DNA hypermethylation of genes is
an important and early event in CRC.
DNA methylation has been reported to be correlated
with the development of colitis-associated cancer. DNA methylation
level of the estrogen receptor 1 (ESR1) gene in
non-neoplastic colorectal epithelium was higher in UC patients with
neoplasia than in UC patients without neoplasia (12). In addition, E-cadherin
(CDH1)/hyperplastic polyposis protein 1 (HPP1) in
colon mucosa of UC was an early event in UC-associated
carcinogenesis (13). However,
chronic active inflammation is largely correlated with the
occurrence of dysplasia or cancer in UC as well as H.
Pyrori-associated gastritis (14,15). Therefore, active inflammation in
UC may be correlated with accumulation of methylation, resulting in
susceptibility to carcinogenesis.
In the present study, we selected 4 genes [secreted
frizzled-related protein 1 (SFRP1), transcription elongation
regulator 1-like (TCERG1L), fibrillin 2 (FBN2) and
tissue factor pathway inhibitor 2 (TFPI2)] which have
recently been identified by cDNA microarray approach using DNA
methyltransferase inhibitor (5-aza-2′-deoxycytidine) (16,17). They are cancer-specifically and
frequently methylated in CRC tumors. In addition, all 4 genes have
previously been identified as early DNA methylation biomarker
candidates during colon cancer progression (18,19). Therefore, we hypothesized that
these 4 genes are useful markers to detect, not only early-stage
colon cancer but also chronic inflammation disease such as UC. The
aim of the present study was to analyze the methylation status of
selective genes (SFRP1, TCERG1L, FBN2 and
TFPI2) as a risk marker for colon cancer in UC patients.
Materials and methods
Patient samples
Enrolled in the study were 36 patients with UC,
including 21 males and 15 females. The median age was 43.5 years
and the median clinical disease duration was 24.6 months. The
diagnosis of UC was based on standard clinical, endoscopic,
radiological, and histological criteria (20). Rectal inflammatory mucosal
specimens were obtained from all the patients during colonoscopic
biopsy and were preserved at −80°C until use. The histopathological
examinations showed mild or moderate inflammation, with no evidence
of dysplasia or neoplasia in all the cases. Clinicopathological
characteristics such as gender, age of disease onset, clinical
disease duration, lesion location and clinical type were
investigated. UC patients were also classified as proctitis, left
sided colitis or pancolitis according to the location and extension
of the inflammatory lesions as judged by endoscopic findings.
According to the clinical course, chronic UC cases were classified
into chronic relapsing, chronic continuous or only one episode of
the disease (21). Written
informed consent was obtained from all participating subjects. The
study protocol was approved by the Institutional Review Board
(IRB). Clinical characteristics of the patients are documented in
Table I.
 | Table IBasic characteristics of the UC
patient samples in this study. |
Table I
Basic characteristics of the UC
patient samples in this study.
|
Characteristics | |
|---|
| Total no. of
patients | 36 |
| Age (years) |
| Median
(range) | 43.5 (15–79) |
| Gender, n (%) |
| Male | 21 (58.3) |
| Female | 15 (41.6) |
| Age at disease
onset, n (%) |
| ≤20 years | 6 (16.7) |
| 21–40 | 12 (33.3) |
| >41 | 18 (50) |
| Duration of disease
(median, months) 24.6 |
| Lesion location, n
(%) |
| Proctitis | 12 (33.3) |
| Left sided
colitis | 14 (38.9) |
| Pancolitis | 10 (27.8) |
| Mayo endoscopic
score, n (%) |
| Normal or
inactive | 1 (2.8) |
| Mild disease | 16 (44.4) |
| Moderate
disease | 13 (36.1) |
| Severe
disease | 6 (16.7) |
| Clinical type, n
(%) |
| Only one
episode | 21 (58.3) |
| Chronic
relapsing | 15 (41.7) |
| Chronic
continuous | 0 (0) |
CRC patient samples (n=8) were also evaluated to
compare the DNA methylation level with UC patients. The mean age of
the patients was 65.8 years. Of all the patients, 3 patients had
well-differentiated and 5 had moderately differentiated carcinomas.
Based on the tumor-node-metastasis classification, stage II (n=4),
III (n=3) and IV (n=1) cases were noted.
DNA extraction and methylation
analysis
Methylation analysis was performed using the
methylation-specific polymerase chain reaction (MSP) strategy, as
previously described (22). DNA
was extracted following a standard phenol-chloroform extraction
protocol. Bisulfite modification of DNA was performed using the EZ
DNA Methylation™ Kit (Zymo Research) according to the
manufacturer’s instructions. Methylation-specific PCR was carried
out in a 25-μl reaction containing 10X MSP buffer, 10 mM dNTPs, 10
pmol of each of the methylated or unmethylated primers, 1 unit of
JumpStart™ REDTaq® DNA polymerase (Sigma) and 4 μl of
bisulfite-treated DNA. Amplification cycles were as follows: one
cycle at 95°C for 5 min followed by 35 cycles of 95°C for 30 sec,
annealing temp for 30 sec, 72°C for 30 sec and a final extension
step at 72°C for 5 min. In vitro methylated DNA (IVD) was
used as a positive control for MSP. IVD was created by treating
cell line DNA with Sassy methylate (NEB) as directed. DKO, which is
a double knockout derivative of the CRC cell line HCT 116 with
knockout of the major DNA methyltransferases
(DNMT1−/− and DNMT3b−/−) was
used as an additional negative control. DKO lacks methylation at
95% of the known CpG sites (23).
An amount of 7.5 μl of each amplification reaction was loaded and
run on 2% agarose gel containing GelStar™ Nucleic Acid Gel Stain
(Lonza) and visualized by ultraviolet illumination. All primers are
listed in Table II.
 | Table IISelected gene primers for MSP and
bisulfite sequencing analysis. |
Table II
Selected gene primers for MSP and
bisulfite sequencing analysis.
| Gene | | Sense (5′-3′) | Antisense
(5′-3′) | Ref. |
|---|
| SFRP1 | Meth |
TGTAGTTTTCGGAGTTAGTGTCGCGC |
CCTACGATCGAAAACGACGCGAACG | (16) |
| Unmeth |
GTTTTGTAGTTTTTGGAGTTAGTGTTGTGT |
CTCAACCTACAATCAAAAACAACACAAACA | |
| BS |
GTTTTGTTTTTTAAGGGGTGTTGAG |
GCCTTTTGTCCCCGGAGGTCCCTGG | |
| TFPI2 | Meth |
GTTCGTTGGGTAAGGCGTTC |
CATAAAACGAACACCCGAACCG | (18) |
| Unmeth |
CCCACATAAAACAAACACCCAAACCA |
TGGTTTGTTGGGTAAGGTGTTTG | |
| BS |
GGTTTATGGTGTAGGGG |
CAATCACTAACAAATCATTTCC | |
| FBN2 | Meth |
GGGTTTTTAAAATTTTCGCGTCGC |
CTACGAAACCGAACGAAAATACG | (31) |
| Unmeth |
GTTTTGTTGGGTTTTTAAAATTTTTGTGTTGTG |
AAATAACAACTACAAAACCAAACAAAAATACA | |
| BS |
CTTCCAACCCYACCTTC |
GTTTTTAGAAGAAGAGGAGGG | |
| TCERG1L | Meth |
GGTCGTTTGCGTCGGATTC |
CTACCCAACGCGAAACTAAAAACG | (31) |
| Unmeth |
TTTGGGGTTGTTTGTGTTGGATTTG |
CATATCCCACTACCCAACACAAAACTAAAAAC | |
| BS |
AATTTGTTTGGTTTATTTGTGTAATAGAAAT |
CTAATAACCTCTAACCCTCTAA | |
Bisulfite sequencing analysis
Genomic DNA (1 μg) from each sample was bisulfite
converted using the EZ DNA Methylation Kit following the
manufacturer’s protocol. PCR conditions and primer sequences are
provided upon request. The PCR amplicons were gel-purified and
subcloned into pCRII-TOPO vector (Invitrogen). At least 7 clones
were randomly selected and sequenced on an ABI 3730xl DNA analyzer
to ascertain the methylation patterns of each locus.
Quantitative methylation-specific PCR
(MSP) using real-time PCR
Bisulfite modification of genomic DNA was carried
out using the EZ DNA Methylation Kit. For quantitative real-time
analyses, the Maxima SYBR-Green qPCR kit (Fermentas) was used, and
the amplification conditions consisted of an initial 10-min
denaturation step at 95°C, followed by 40 cycles of denaturation at
95°C for 15 sec and annealing and extension for 30 and 60 sec,
respectively. An CFX96 real-time PCR detection system (Bio-Rad) was
used. For quantification, the comparative cycle threshold (Ct)
method was used, normalizing the Ct values for the indicated gene
to the Ct values of the unmethylated reaction relative to a
methylated reaction sample. All primer sequences are listed in
Table II.
Statistical analysis
All statistical analyses were conducted using the
STATA 9.2 software package (Stata, College Station, TX, USA). Most
analyses were conducted using a t-test, while continuous variables
were analyzed using the Mann-Whitney U test. p-values of <0.05
were considered to indicate statistically significant results.
Results and Discussion
Detection of DNA promoter
hypermethylation in UC patient samples
Hypermethylation of an increasing number of genes
has been associated with human colorectal tumorigenesis (24–27). In UC, promoter methylation seems
to precede dysplasia and occurs throughout the mucosa of colitis
(27,28), reinforcing the link between
chronic inflammation and DNA methylation (29,30).
We tested DNA methylation of selected genes,
TCERG1L, SFRP1, FBN2 and TFPI2. These
genes have been previously reported to be cancer-specifically
methylated and frequently methylated at the early stage of colon
cancer progression such as in adenomas (16,31). Previously, DNA hypermethylation of
these 4 genes have been identified from a gene expression
microarray approach (17) using
5-aza-2′-deoxycytidine in colon cancer cells. These genes were
highly methylated in CRC tumors as well as adenomas, which suggests
that these 4 genes may be candidates for use as an early detection
DNA methylation biomarker for CRC patients. To date, epigenetic
silencing of SFRP1 has been identified in a variety of
malignancies, including cancers of the colon (32), bladder (33), prostate (34), lung (35), and breast (36). Hypermethylation of the
TFPI2 gene in CRC was identified by Glöckner et al
(18), and TFPI2 was found
to have a methylation frequency of 80% in adenomas as well as in
stool from CRC patient samples, suggesting DNA methylation of the
TFPI2 gene may be an early detection biomarker of CRC.
Recently, promoter hypermethylation of the TCERG1L and
FBN2 genes was frequently noted in colon adenomas (31).
Here, we tested the methylation level of 4 genes in
36 patients with UC by MSP analysis since these 4 genes have shown
high potential to detect early-stage disease such as cancer.
Genomic DNA was extracted, and successful methylation analysis was
performed in most of the samples. We assessed the methylation
pattern of gene sets (TCERG1L, SFRP1, FBN2 and
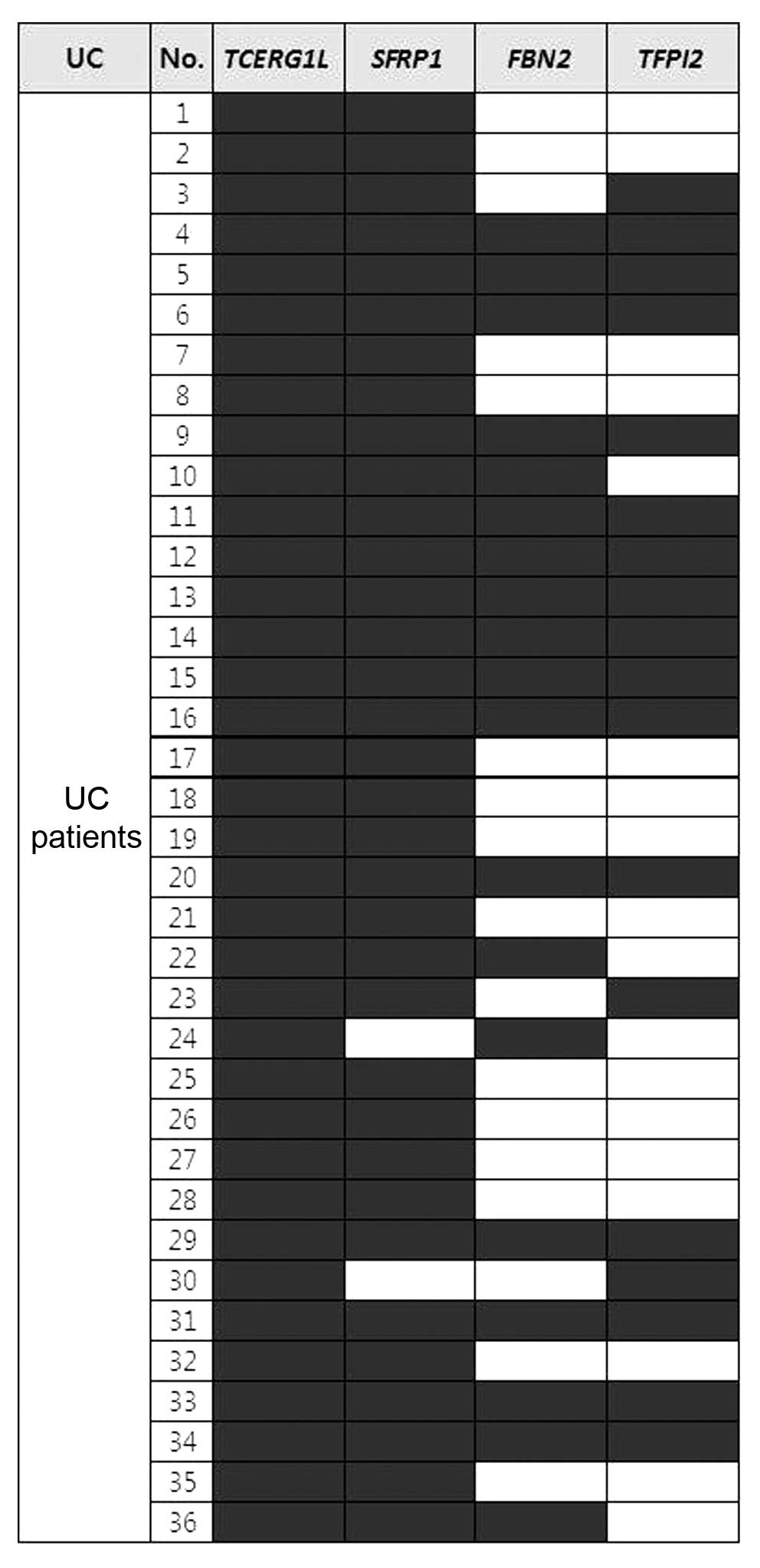
TFPI2) in the UC samples by MSP analysis. Fig. 1 shows the methylation pattern in
each UC sample tested in this study. SFRP1 and
TCERG1L genes were methylated in the majority of the samples
tested (>94%). FBN2 and TFPI2 genes showed 53 and
50% frequencies in the UC samples, respectively (Fig. 1). Fifteen out of 36 samples (42%)
showed methylation of all 4 genes. We assessed the clinical data to
determine whether a correlated was present or not, but no
correlation was noted between clinical data particularly duration
of disease with methylation of all 4 genes. In terms of the
clinical correlation with DNA methylation level, we aimed to
ascertain whether the number of DNA methylated genes was correlated
with age-related methylation since all of the UC patient samples
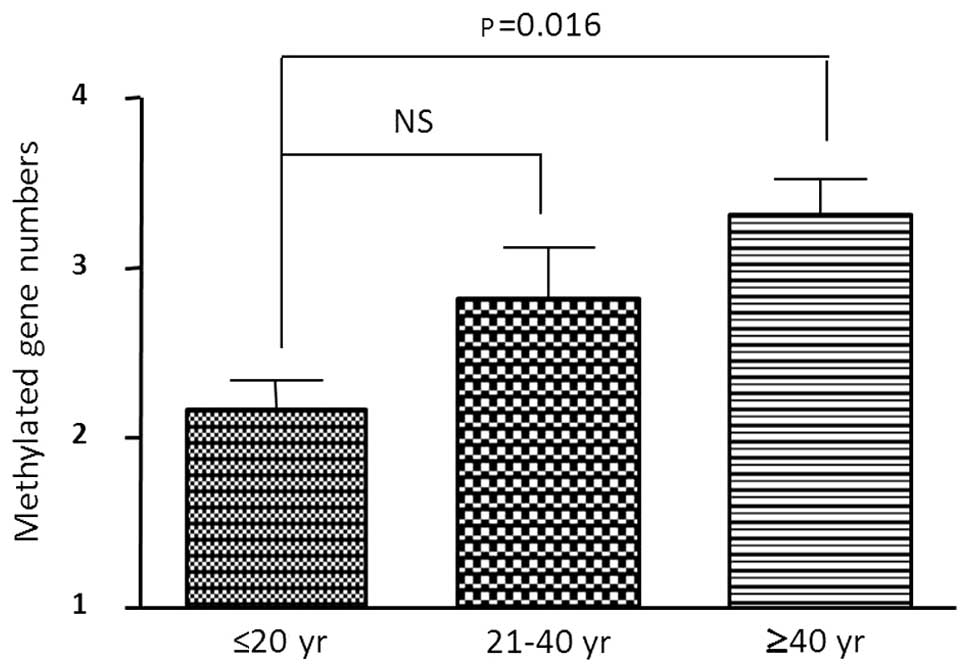
that we tested had no neoplasia. Table I indicates all of the UC patients
categorized according to 3 age groups (≤21, 21–40 and ≥41 years).
The number of methylated genes was statistically significant
between the young (≤21) group and older (≥41) group (p=0.02,
t-test) (Fig. 2). There was no
significant difference between the young (≤21) and mid-age (21–40)
group. In the older age group all 4 genes were methylated in 11 out
of 19 (58%) samples (Fig. 2).
These data suggest that age-related methylation occurred in a small
number of UC patient samples, but we need to confirm this
phenomenon using a larger sample size.
Epigenetic regulation of DNA
hypermethylation in UC patient samples
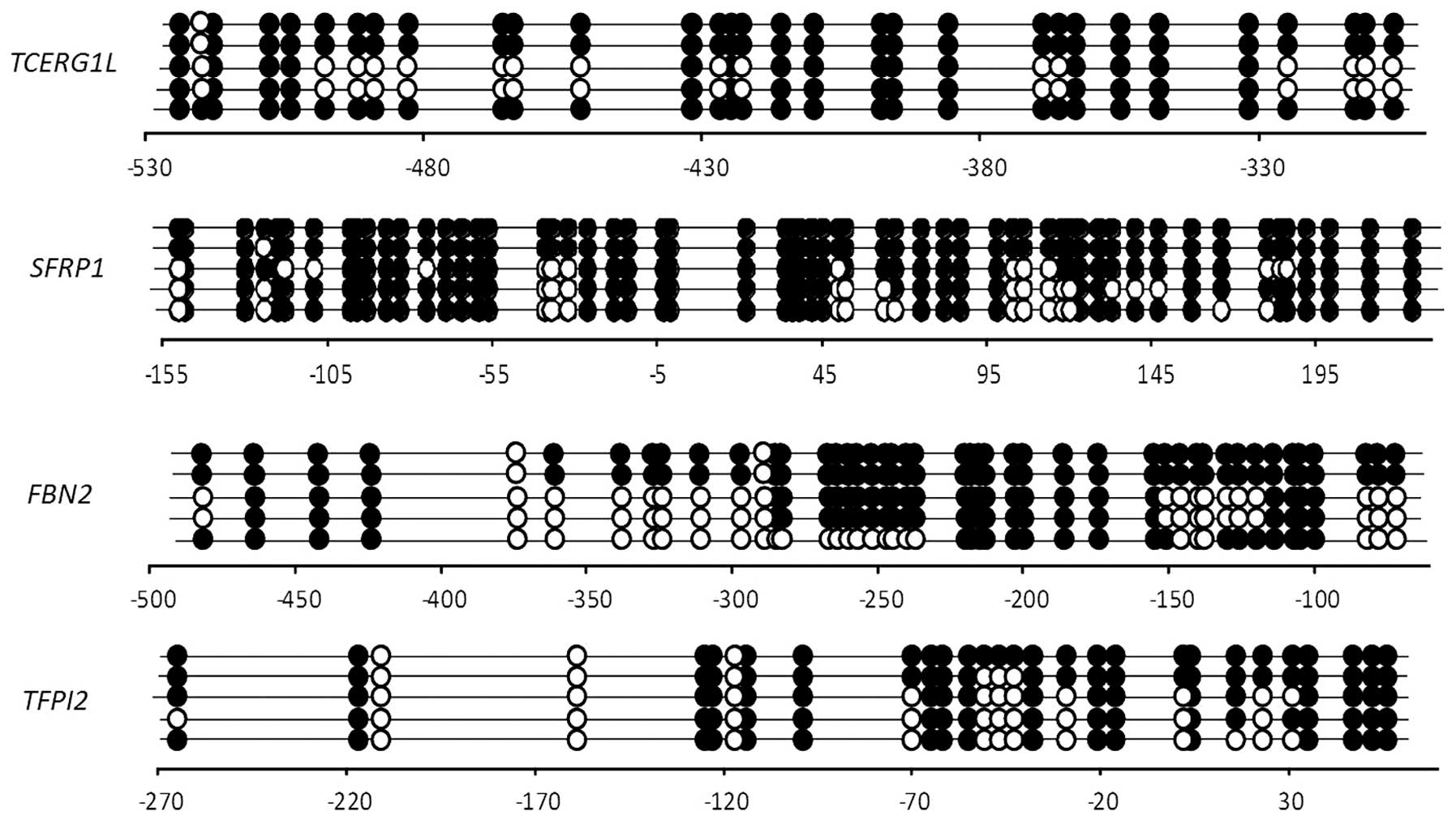
Next, we confirmed the methylation pattern in CpG
islands of the promoter regions of the 4 genes by bisulfite
sequencing analysis (Fig. 3).
TCERG1L, FBN2, TFPI2 and SFRP1 had 31,
46, 30 and 58 CpG sites, respectively, in the bisulfate sequencing
region that we amplified. TCERG1L and FBN2 genes were
previously reported to have a dense CpG methylation pattern in CRC
tumors (31). In this study, both
genes showed a dense CpG methylation pattern in s UC patient (UC4)
with 78% (methylation site per CpG site) and 71%, respectively.
TFPI2 and SFRP1 genes also were previously reported
to have a dense CpG methylation pattern in CRC tumors (18,32). Here, TFPI2 and SFRP1
showed a 71 and 84% CpG methylation frequency in UC patients, which
confirmed that both genes were definitely present in UC patients.
Our results suggest that the 4 genes are densely methylated in UC
patients, and DNA methylation of the 4 genes is sensitive enough to
detect inflammatory disease such as UC.
As previously mentioned, the 4 genes were previously
reported to be highly methylated in CRC tumors. Here, we noted a
high frequency of methylation of the 4 genes in UC patients.
Therefore, we compared the level of methylation between UC patients
and CRC tumors by quantitative real-time MSP analysis. Before
comparing the methylation level in UC patients and CRC, we used MSP
analysis on 8 CRC samples with the 4 genes to confirm methylation
in CRC. The genes were methylated in all 8 CRC samples (data not
shown).
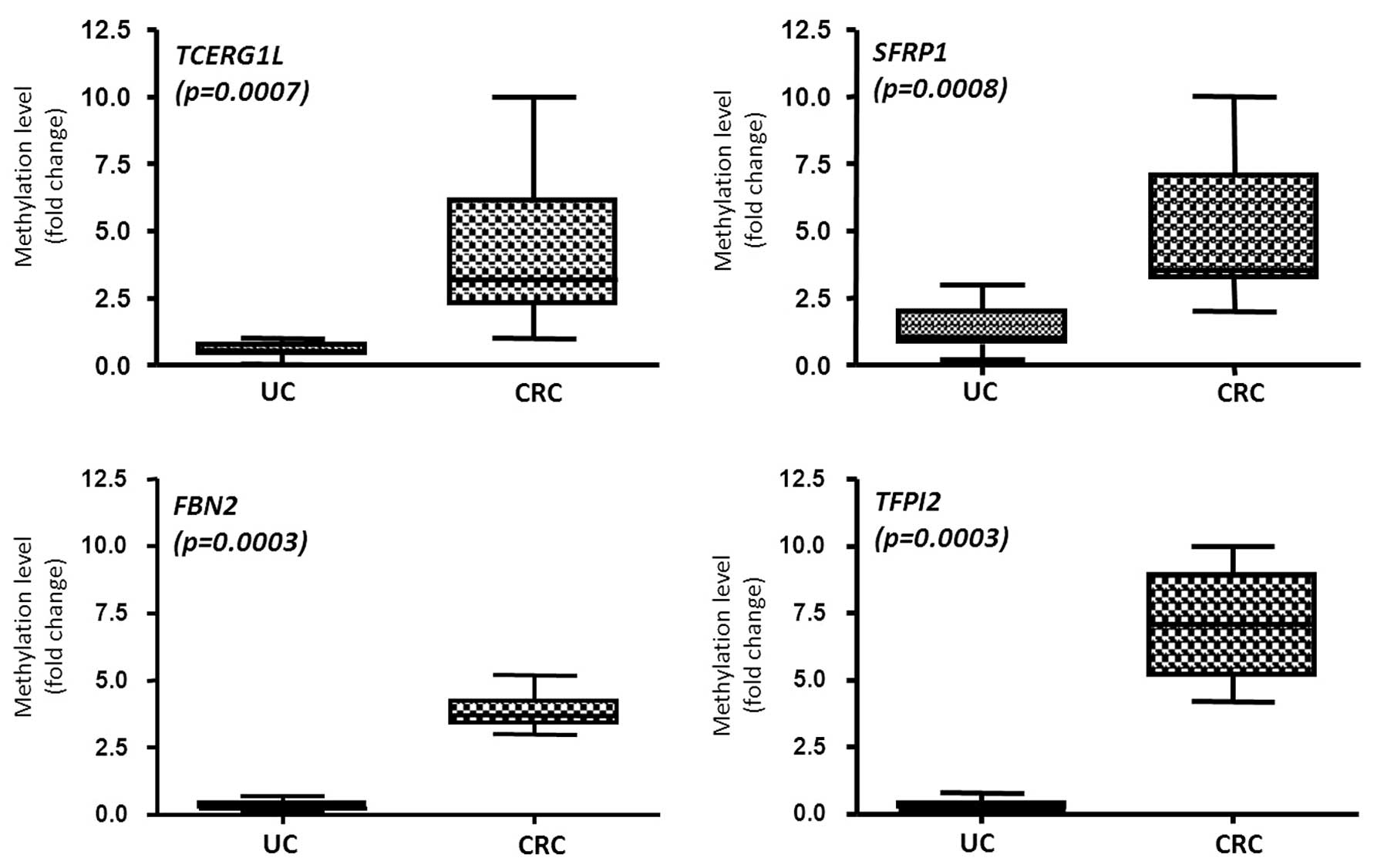
We tested 12 UC and 8 CRC samples which were
confirmed to be methylated by MSP analysis. Even though we noted
frequent methylation levels in the UC patient samples by MSP
analysis (Fig. 1), the
methylation in UC and CRC samples was significantly different. Box
plot indicates the methylation levels of all tested samples (n=13
for UC, n=8 for CRC) by real-time MSP. TCERG1L (mean, 0.81
for UC; mean, 4.28 for CRC) and SFRP1 (mean, 1.39 for UC;
mean, 4.96 for CRC) showed a significant increase in the
methylation level in UC when compared to the level in the CRC
patient samples (p<0.05) (Fig.
4). FBN2 (mean, 0.32 for UC; mean, 3.85 for CRC) and
TFPI2 (mean, 0.31 for UC; mean, 7.08 for CRC) also showed a
significant increase in the methylation level in UC when compared
to the level in the CRC patient samples (p<0.05) (Fig. 4). This suggests that DNA
methylation of TCERG1L and SFRP1 may have more
sensitivity in detecting not only CRC but also inflammatory
disease. In contrast, DNA methylation of FBN2 and
TFPI2 had a higher cancer-specific detection ability. Our
data also suggest that DNA methylation can be triggered in UC
patients, and accumulation of DNA methylation occurs during colon
cancer progression. Our data also suggest that screening UC
patients with the 4 genes may be useful to predict the risk for
CRC.
Methylation of several gene-associated CpG islands
was present in the normal-appearing epithelium from UC patients
with high-grade dysplasia or cancer (25). ESR1 methylation in
different parts of the large intestine in UC patients with and
without neoplasia confirmed that ESR1 methylation is
correlated with an increased risk of developing neoplasia (12). These reports suggest that the
methylation levels of colonic mucosa of UC vary according to the
presence or absence of neoplasia and that accumulation of
methylation finally induces cancer development.
The present study had limitations. We could not
conclude that detection of methylation in UC patients implies the
process of neoplasia since we did not compare the methylation
pattern of UC patients with and without cancer. In addition, the UC
patients that were tested in this study did not have a long
duration of disease (Table I).
However, Wang et al (28)
revealed that promoter DNA methylation of ER, p53, p14, p16,
p21 and hMLH1 genes was detected in UC patients without
neoplasia, suggesting that these genes are useful for predicting
cases at high risk of neoplasia. Our data support that detection of
DNA methylation of the 4 genes in UC samples indicates a poor
prognosis of UC patients who need routine colonoscopic surveillance
which may prevent the progression to severe disease such as
cancer.
In summary, we assessed the DNA methylation pattern
in UC patients using very sensitive DNA methylation markers which
are able to detect early-stage colon cancer such as adenomas. MSP
analysis revealed that 2 genes (TCERG1L and SFRP1)
showed a high (>95%) frequency of methylation while the other 2
genes (FBN2 and TFPI2) showed a decreased (>45%)
frequency of methylation in UC patients. We also confirmed a dense
DNA methylation status of 4 genes in UC patients by bisulfite
sequencing analysis. Notably, we compared the quantitative
methylation level between UC patient and CRC tumors even though we
noted a high frequency of methylation in UC patient by MSP
analysis. The DNA methylation level was significant higher in CRC
patients than in UC samples, which implies that DNA methylation may
be triggered by inflammation and promote abnormal DNA
hypermethylation in cancer. Our data suggest that sensitive
methylation markers may be useful to detect inflammation diseases
which have the potential risk of neoplasia. Therefore, examination
of the methylation status of our markers could predict the
progression of severe disease in UC patients.
Acknowledgements
This study was supported by the National R&D
program (50596-2013) through the Dongnam Institute of Radiological
and Medical Sciences (DIRAMS) funded by the Korean Ministry of
Education, Science and Technology. We would like to thank Khadijah
Mitchell of the Johns Hopkins School of Medicine for the critical
reading of the manuscript and providing language editing.
References
|
1
|
Ekbom A, Helmick C, Zack M and Adami HO:
Increased risk of large-bowel cancer in Crohn’s disease with
colonic involvement. Lancet. 336:357–359. 1990.
|
|
2
|
Ekbom A, Helmick C, Zack M and Adami HO:
Ulcerative colitis and colorectal cancer. A population-based study.
N Engl J Med. 323:1228–1233. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brentnall TA: Risk factors for development
of colorectal cancer in inflammatory bowel disease. Advances in
Inflammatory Bowel Disease. Norwell M: Kluwer Academic Publishers;
Norwell, MA: pp. 159–167. 1998
|
|
4
|
Brentnall TA, Crispin DA, Rabinovitch PS,
Haggitt RC, Rubin CE, Stevens AC and Burmer GC: Mutations in the
p53 gene: an early marker of neoplastic progression in ulcerative
colitis. Gastroenterology. 107:369–378. 1994.PubMed/NCBI
|
|
5
|
Rabinovitch PS, Dziadon S, Brentnall TA,
Emond MJ, Crispin DA, Haggitt RC and Bronner MP: Pancolonic
chromosomal instability precedes dysplasia and cancer in ulcerative
colitis. Cancer Res. 59:5148–5153. 1999.PubMed/NCBI
|
|
6
|
Harpaz N and Talbot IC: Colorectal cancer
in idiopathic inflammatory bowel disease. Semin Diagn Pathol.
13:339–357. 1996.PubMed/NCBI
|
|
7
|
Eaden JA, Abrams KR and Mayberry JF: The
risk of colorectal cancer in ulcerative colitis: a meta-analysis.
Gut. 48:526–535. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Rutter MD, Saunders BP, Wilkinson KH,
Rumbles S, Schofield G, Kamm MA, Williams CB, Price AB, Talbot IC
and Forbes A: Thirty-year analysis of a colonoscopic surveillance
program for neoplasia in ulcerative colitis. Gastroenterology.
130:1030–1038. 2006.PubMed/NCBI
|
|
9
|
Riddell RH, Goldman H, Ransohoff DF,
Appelman HD, Fenoglio CM, Haggitt RC, Ahren C, Correa P, Hamilton
SR, Morson BC, Sommers SC and Yardley JH: Dysplasia in inflammtory
bowel disease: standardized classification with provisional
clinical applications. Hum Pathol. 14:931–968. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Jones PA and Baylin SB: The fundamental
role of epigenetic events in cancer. Nat Rev Genet. 3:415–428.
2002.PubMed/NCBI
|
|
11
|
Esteller M: Epigenetic gene silencing in
cancer: the DNA hypermethylome. Hum Mol Genet. 16:R50–R59. 2007.
View Article : Google Scholar
|
|
12
|
Tominaga K, Fujii S, Mukawa K, Fujita M,
Ichikawa K, Tomita S, Imai Y, Kanke K, Ono Y, Terano A, Hiraishi H
and Fujimori T: Prediction of colorectal neoplasia by quantitative
methylation analysis of estrogen receptor gene in nonneoplastic
epithelium from patients with ulcerative colitis. Clin Cancer Res.
11:8880–8885. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sato F, Shibata D, Harpaz N, Xu Y, Yin J,
Mori Y, Wang S, Olaru A, Deacu E, Selaru FM, Kimos MC, Hytiroglou
P, et al: Aberrant methylation of the HPP1 gene in ulcerative
colitis-associated colorectal carcinoma. Cancer Res. 62:6820–6822.
2002.PubMed/NCBI
|
|
14
|
Maekita T, Nakazawa K, Mihara M, Nakajima
T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M,
Tamura G, Saito D, et al: High levels of aberrant DNA methylation
in Helicobacter pylori-infected gastric mucosae and its
possible association with gastric cancer risk. Clin Cancer Res.
12:989–995. 2006.PubMed/NCBI
|
|
15
|
Nakajima T, Enomoto S, Yamashita S, Ando
T, Nakanishi Y, Nakazawa K, Oda I, Gotoda T and Ushijima T:
Persistence of a component of DNA methylation in gastric mucosae
after Helicobacter pylori eradication. J Gastroenterol.
45:37–44. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suzuki H, Watkins DN, Jair KW, Schuebel
KE, Markowitz SD, Chen WD, Pretlow TP, Yang B, Akiyama Y, Van
Engeland M, Toyota M, Tokino T, et al: Epigenetic inactivation of
SFRP genes allows constitutive WNT signaling in colorectal cancer.
Nat Genet. 36:417–422. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Schuebel KE, Chen W, Cope L, Glöckner SC,
Suzuki H, Yi JM, Chan TA, Van Neste L, Van Criekinge W, van den
Bosch S, van Engeland M, Ting AH, et al: Comparing the DNA
hypermethylome with gene mutations in human colorectal cancer. PLoS
Genet. 3:1709–1723. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Glöckner SC, Dhir M, Yi JM, McGarvey KE,
Van Neste L, Louwagie J, Chan TA, Kleeberger W, de Bruïne AP, Smits
KM, Khalid-de Bakker CA, Jonkers DM, et al: Methylation of TFPI2 in
stool DNA: a potential novel biomarker for the detection of
colorectal cancer. Cancer Res. 69:4691–4699. 2009.PubMed/NCBI
|
|
19
|
Yi JM, Dhir M, Van Neste L, Downing SR,
Jeschke J, Glöckner SC, de Freitas Calmon M, Hooker CM, Funes JM,
Boshoff C, Smits KM, van Engeland M, et al: Genomic and epigenomic
integration identifies a prognostic signature in colon cancer. Clin
Cancer Res. 17:1535–1545. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Podolsky DK: Inflammatory bowel disease
(1). N Engl J Med. 325:928–937. 1991. View Article : Google Scholar
|
|
21
|
Langholz E, Munkholm P, Davidsen M and
Binder V: Course of ulcerative colitis: analysis of changes in
disease activity over years. Gastroenterology. 107:3–11.
1994.PubMed/NCBI
|
|
22
|
Herman JG, Graff JR, Myohanen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: a novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rhee I, Bachman KE, Park BH, Jair KW, Yen
RW, Schuebel KE, Cui H, Feinberg AP, Lengauer C, Kinzler KW, Baylin
SB and Vogelstein B: DNMT1 and DNMT3b cooperate to silence genes in
human cancer cells. Nature. 416:552–556. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fleisher AS, Esteller M, Harpaz N, Leytin
A, Rashid A, Xu Y, Liang J, Stine OC, Yin J, Zou TT, Abraham JM,
Kong D, et al: Microsatellite instability in inflammatory bowel
disease-associated neoplastic lesions is associated with
hypermethylation and diminished expression of the DNA mismatch
repair gene, hMLH1. Cancer Res. 60:4864–4868. 2000.
|
|
25
|
Issa JP, Ahuja N, Toyota M, Bronner MP and
Brentnall TA: Accelerated age-related CpG island methylation in
ulcerative colitis. Cancer Res. 61:3573–3577. 2001.PubMed/NCBI
|
|
26
|
Sato F, Harpaz N, Shibata D, Xu Y, Yin J,
Mori Y, Zou TT, Wang S, Desai K, Leytin A, Selaru FM, Abraham JM
and Meltzer SJ: Hypermethylation of the p14(ARF) gene in ulcerative
colitis-associated colorectal carcinogenesis. Cancer Res.
62:1148–1151. 2002.PubMed/NCBI
|
|
27
|
Fujii S, Tominaga K, Kitajima K, Takeda J,
Kusaka T, Fujita M, Ichikawa K, Tomita S, Ohkura Y, Ono Y, Imura J,
Chiba T and Fujimori T: Methylation of the oestrogen receptor gene
in non-neoplastic epithelium as a marker of colorectal neoplasia
risk in longstanding and extensive ulcerative colitis. Gut.
54:1287–1292. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang FY, Arisawa T, Tahara T, Takahama K,
Watanabe M, Hirata I and Nakano H: Aberrant DNA methylation in
ulcerative colitis without neoplasia. Hepatogastroenterology.
55:62–65. 2008.PubMed/NCBI
|
|
29
|
Lin Z, Hegarty JP, Cappel JA, Yu W, Chen
X, Faber P, Wang Y, Kelly AA, Poritz LS, Peterson BZ, Schreiber S,
Fan JB and Koltun WA: Identification of disease-associated DNA
methylation in intestinal tissues from patients with inflammatory
bowel disease. Clin Genet. 80:59–67. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Saito S, Kato J, Hiraoka S, Horii J,
Suzuki H, Higashi R, Kaji E, Kondo Y and Yamamoto K: DNA
methylation of colon mucosa in ulcerative colitis patients:
correlation with inflammatory status. Inflamm Bowel Dis.
17:1955–1965. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yi JM, Dhir M, Guzzetta AA,
Iacobuzio-Donahue CA, Heo K, Yang KM, Suzuki H, Toyota M, Kim HM
and Ahuja N: DNA methylation biomarker candidates for early
detection of colon cancer. Tumor Biol. 33:363–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Suzuki H, Gabrielson E, Chen W, Anbazhagan
R, van Engeland M, Weijenberg MP, Herman JG and Baylin SB: A
genomic screen for genes upregulated by demethylation and histone
deacetylase inhibition in human colorectal cancer. Nat Genet.
31:141–149. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Stoehr R, Wissmann C, Suzuki H, Knuechel
R, Krieg RC, Klopocki E, Dahl E, Wild P, Blaszyk H, Sauter G, Simon
R, Schmitt R, et al: Deletions of chromosome 8p and loss of sFRP1
expression are progression markers of papillary bladder cancer. Lab
Invest. 84:465–478. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lodygin D, Epanchintsev A, Menssen A,
Diebold J and Hermeking H: Functional epigenomics identifies genes
frequently silenced in prostate cancer. Cancer Res. 65:4218–4227.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fukui T, Kondo M, Ito G, Maeda O, Sato N,
Yoshioka H, Yokoi K, Ueda Y, Shimokata K and Sekido Y:
Transcriptional silencing of secreted frizzled related protein 1
(SFRP1) by promoter hypermethylation in non-small-cell lung cancer.
Oncogene. 24:6323–6327. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Suzuki H, Toyota M, Carraway H, Gabrielson
E, Ohmura T, Fujikane T, Nishikawa N, Sogabe Y, Nojima M, Sonoda T,
Mori M, Hirata K, et al: Frequent epigenetic inactivation of Wnt
antagonist genes in breast cancer. Br J Cancer. 98:1147–1156. 2008.
View Article : Google Scholar : PubMed/NCBI
|