Introduction
Orthotopic liver transplantation (OLT) is one of the
best and most effective methods for the treatment of end-stage
liver disease or liver-based metabolic diseases (1–3).
However, due to the dramatic imbalance between the limited number
of available donors and the patients who require transplantation,
an increasingly high number of patients are unable to find a
suitable liver for transplantation. On the other hand, the
complications associated with OLT surgery hamper its clinical
application (4,5). Several studies have demonstrated
that cell-based transplantation strategies have great potential for
repair and functional recovery following acute liver damage,
becoming a realistic option for the treatment of liver diseases
(6–8).
A reliable cell source is required for this
effective clinical therapy. Isolated mature hepatocytes are
difficult to manipulate and cannot be expanded in vitro to
obtain a large cell population. The viability of these cells and
hepatic markers are easily lost under common culture conditions
in vitro (9). Certain
studies have found that hepatic stem cells (HSCs) have a
multilineage differentiation potential, as well as
self-proliferative capabilities and can differentiate into a mature
hepatic cell lineage; furthermore, they have been shown to exhibit
certain phenotypes and functions characteristic of hepatocytes
in vitro (10). Moreover,
HSCs showing a greater regenerative capacity than adult
hepatocytes, participate in liver tissue repair and reconstruction
following injury. It has been reported that HSC transplantation may
be utilized to substitute OLT, having a definite therapeutic effect
on patients with end-stage liver disease (11,12).
Thus far, there are no definite distinguishing
features as regards HSCs. Most scholars hypothesize that HSCs
should be composed of two main categories, extrahepatic and
intrahepatic stem cells. The former includes embryonic stem cells,
hematopoietic stem cells and bone marrow mesenchymal stem cells.
The latter includes embryonic HSCs, hepatic oval cells and small
hepatocyte-like progenitor cells (13,14). HSCs derived from the fetal liver,
also known as hepatic progenitor cells (HPCs), exhibit bipotential
capacity and are characterized by an intermediary phenotype between
biliary epithelial cells and hepatocytes. HPCs act as an important
liver cell source of hepatocyte transplantation and biological
artificial liver (15,16).
The present study aimed to investigate the
phenotype, proliferation and differentiation capacity of primary
embryonic HPCs compared with mature hepatocytes. HPCs exhibit
better growth capability and function as mature hepatocytes
following induction in vitro. The study of HPCs may aid in
the understanding of the process of artificial liver development
and promote its clinical application.
Materials and methods
Cells and chemicals
The Hepa1–6 line was purchased from the American
Type Culture Collection (Manassas, VA, USA) and maintained in
complete Dulbecco’s modified Eagle’s medium (DMEM) supplemented
with 10% fetal bovine serum (FBS, HyClone, Logan, UT, USA), 100
U/ml penicillin, and 100 μg/ml streptomycin at 37°C in 5%
CO2. Unless indicated otherwise, all chemicals were
purchased from Sigma-Aldrich (St. Louis, MO, USA). Hepatic
differentiation induction medium was composed of DMEM supplemented
with 2% horse serum (HS), 0.1 μmol/l dexamethason (Dex), 10 ng/ml
hepatic growth factor (HGF) and 20 ng/ml fibroblast growth factor 4
(FGF4).
Isolation of primary cells
Primary cells were obtained from the livers of mouse
embryos at 14.5 days post coitus [hepatic progenitor 14.5d
(HP14.5d) cells], as well as from the livers of 3-month-old mice
[liver cells 3m (LC3m)]. Briefly, adult livers were perfused with
phosphate-buffered saline (PBS) and collagenase type IV through the
inferior vena cava, and then cut open to gently scrape off the
cells. The embryronic livers were cut into small sections and
digested with collagenase type IV at 37°C for 10 min. Isolated cell
clumps were dissociated by pipetting in 10 ml DMEM, then the
solution was filtered through a 100-μm cell strainer and
centrifuged at 50 × g for 3 min. The cells were gently resuspended
in complete DMEM and plated on 100-mm dishes that were coated with
type I collagen and incubated at 37°C. After 24 h, all non-adherent
cells were removed and the medium was changed every 3 days. The
HP14.5d cells at 90% of confluence were trypsinized and
passaged.
Cell viability
Trypan blue staining and crystal violet staining
were performed to assess cell viability. As previously described
(17), the cells were incubated
in 24-well plates at 2.0×104 cells per well and cell
viability was measured at the indicated time points. For trypan
blue staining, both adherent and suspended cells were collected and
mixed with 0.4% 2X trypan blue buffer (Beyotime, Nantong, China). A
total of 10 μl of cell mixture (106 cells/ml) was added
into the hemocytometer and observed under a microscope (TE2000-S;
Nikon, Tokyo, Japan). Blue-stained cells were counted as dead
cells. Three independent experiments were performed in duplicate.
The mean and standard deviation were calculated. The viable cells
were unstained, while dead cells were stained blue. For crystal
violet staining, the culture medium was removed, the cells were
fixed with 4% paraformaldehyde at room temperature for 10 min and
then stained with 0.05% crystal violet for 30 min. The cells were
then washed with tap water, after which the water was removed and
the cells were dried out on filter paper. After the plates were
photographed, blue dye was dissolved in 500 μl of methanol and
emission spectra were measured at an excitation wavelength of 540
nm using a Multimode Microplate Reader (Thermo Fisher Scientific,
Waltham, MA, USA). Three independent experiments were performed in
duplicate. The mean and standard deviation were calculated.
Transfection of albumin (ALB)
promoter-driven reporter gene and Gaussia luciferase reporter
assay
The plasmid of the ALB promoter-driven luciferase
reporter gene (pSEB-ALB-Gluc) was constructed in our previous study
(18). The cells were engrafted
on 24-well plates at an initial confluence of 60%. The
pSEB-ALB-Gluc plasmid was transfected into the cells using
Lipofectamine® 2000 (Invitrogen, Carslbad, CA, USA).
After 48 h, the cells were treated with hepatic differentiation
induction medium for 12 days. Gaussia luciferase reporter assay was
carried out at each indicated time point. Briefly, 20 μl of cell
medium were collected and mixed with 10 μl of fresh prepared
luciferin substrate solution. Gaussia luciferase acitivity was
immediately measured using a Single Tube Multimode Reader (Promega,
Madison, WI, USA). Each assay condition was performed in
triplicate. Data are expressed as the means ± SD.
RNA isolation and semiquantitative
reverse transcription (RT)-PCR analysis
As previously described (18,19), total RNA was extracted by using an
RNA Extraction kit (Bioteke Corp., Beijing, China) and reverse
transcribed into cDNA using Superscript II reverse transcriptase
(Invitrogen). The cDNA samples were 5- to 10-fold diluted and
subjected to PCR amplification. The sequences of all primers listed
in Table I were designed using
the Primer3 program. A touch-down PCR protocol was performed under
the following programmed conditions: 95°C × 3 min, and then 92°C ×
20 sec, 65°C × 20 sec, 72°C × 20 sec, 9 cycles, with 1°C degree
decrease per cycle, followed by 94°C × 20 sec, 55°C × 20 sec, 72°C
× 20 sec for 25–30 cycles and 72°C × 3 min. The PCR products for
RT-PCR were electrophoresed on a 1.5% agarose gel. Glyceraldehyde
3-phosphate dehydrogenase (GAPDH) expression in each sample was
used to normalize the template concentration.
 | Table IPrimers used for RT-PCR. |
Table I
Primers used for RT-PCR.
| Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| GAPDH |
GGCTGCCCAGAACATCAT |
CGGACACATTGGGGGTAG |
| DLK |
GCTGGGACGGGAAATTCT |
AACCCAGGTGTGCAGGAG |
| CD34 |
AGGGAAAGGCCAATGTGAC |
CCACCCAACCAAATCACAG |
| AFP |
ACGAGGAAAGCCCCTCAG |
GCCATTCCCTCACCACAG |
| CK19 |
GCCCTAGAGCAGGCCAAT |
ATCTTGTCGCGCAAGTCC |
| ALB |
CCAGACATTCCCCAATGC |
CAAGTTCCGCCCTGTCAT |
| CK18 |
CTGGGCTCTGTGCGAACT |
ACAGAGCCACCCCAGACA |
| TAT |
ACCTTCAATCCCATCCGA |
TCCCGACTGGATAGGTAG |
| ApoB |
CATGTGATCCCCACAGCA |
TCCCAGGACCATGGAAAA |
Western blot analysis
Cells were lysed in RIPA buffer with
phenylmethylsulfonyl fluoride (PMSF) to extract the total protein.
Approximately 20 μg of total protein per lane was loaded onto a 10%
SDS-polyacrylamide gel for electrophoretic separation. Thereafter,
proteins were transferred onto a polyvinylidene fluoride (PVDF)
membrane. The membrane was blocked with 5% fat-free skimmed milk at
room temperature for 1 h and incubated with primary antibody to UDP
glucuronosyltransferase 1 family, polypeptide A complex locus
(UGT1A) and β-actin (Santa Cruz Biotechnology, Inc., Santa Cruz,
CA, USA) at 4°C overnight. The membranes were then probed with the
appropriate second antibody conjugated with horseradish peroxidase
(Santa Cruz Biotechnology, Inc.) for 1 h. The immunoreactive bands
of protein were developed using enhanced chemiluminescent substrate
(Kaiji Bio Co., Nanjing, China) and exposed using the G:BOX iChemi
XR gel documentation system (Syngene, Cambridge, UK). All the
abovementioned steps were carried out at room temperature unless
otherswise specified.
Immunofluorescence staining
The cells were fixed with methanol at -20°C for 15
min. They were then blocked with 5% goat serum for 1 h.
Subsequently, the cells were incubated with delta-like homolog
(DLK), alpha fetoprotein (AFP), ALB, UGT1A and CK18 primary
antibody (Santa Cruz Biotechnology, Inc.) at 4°C overnight,
followed by probing with DyLight® 594- or 488-conjugated
secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West
Grove, PA, USA) for 30 min. The nuclei were stained with DAPI. The
presence of proteins was ascertained under a fluorescence
microscope (TE2000-S; Nikon). The cells were washed twice with PBS
after each step; all the abovementioned steps were carried out at
room temperature unless otherwise specified.
Indocyanine green (ICG) uptake and
release
The cultured cells in 24-well plates were washed
twice with PBS and incubated with DMEM supplemented with ICG at a
final concentration of 1 mg/ml for 1 h at 37°C in 5%
CO2. After DMEM was removed and the cells were gently
washed several times with PBS, green-stained cells were
photographed under a microscope. Complete medium was then added and
the cells were incubated for >6 h; the cells were then observed
under a microscope to ensure ICG release. At least 10
non-overlapping fields of vision were recorded.
Periodic acid-Schiff (PAS) staining
The cells were cultured in 24-well plates as
described above. Paraformaldehyde (4%) was added to fix the cells
for 10 min, followed by incubation with 0.5% periodic acid solution
for 5 min. After rinsing with tap water, the cells were incubated
in Schiff’s solution for 15 min and counterstained with hematoxylin
solution for 2 min, and finally rinsed with flowing water for
clarification. All steps were carried out at room termperature. The
positive cells were stained purple. More than 10 non-overlapping
fields of vision in each group were recorded under a
microscope.
Statistical analysis
All data are presented as the means ± standard
deviation (SD) and calculated using SPSS 15.0 statistic software. A
two-tailed Student’s t-test was used to evaluate significant
differences between 2 groups. A P-value <0.05 was considered to
indicate a statistically significant difference.
Results
Morphology of primary and passaged
cells
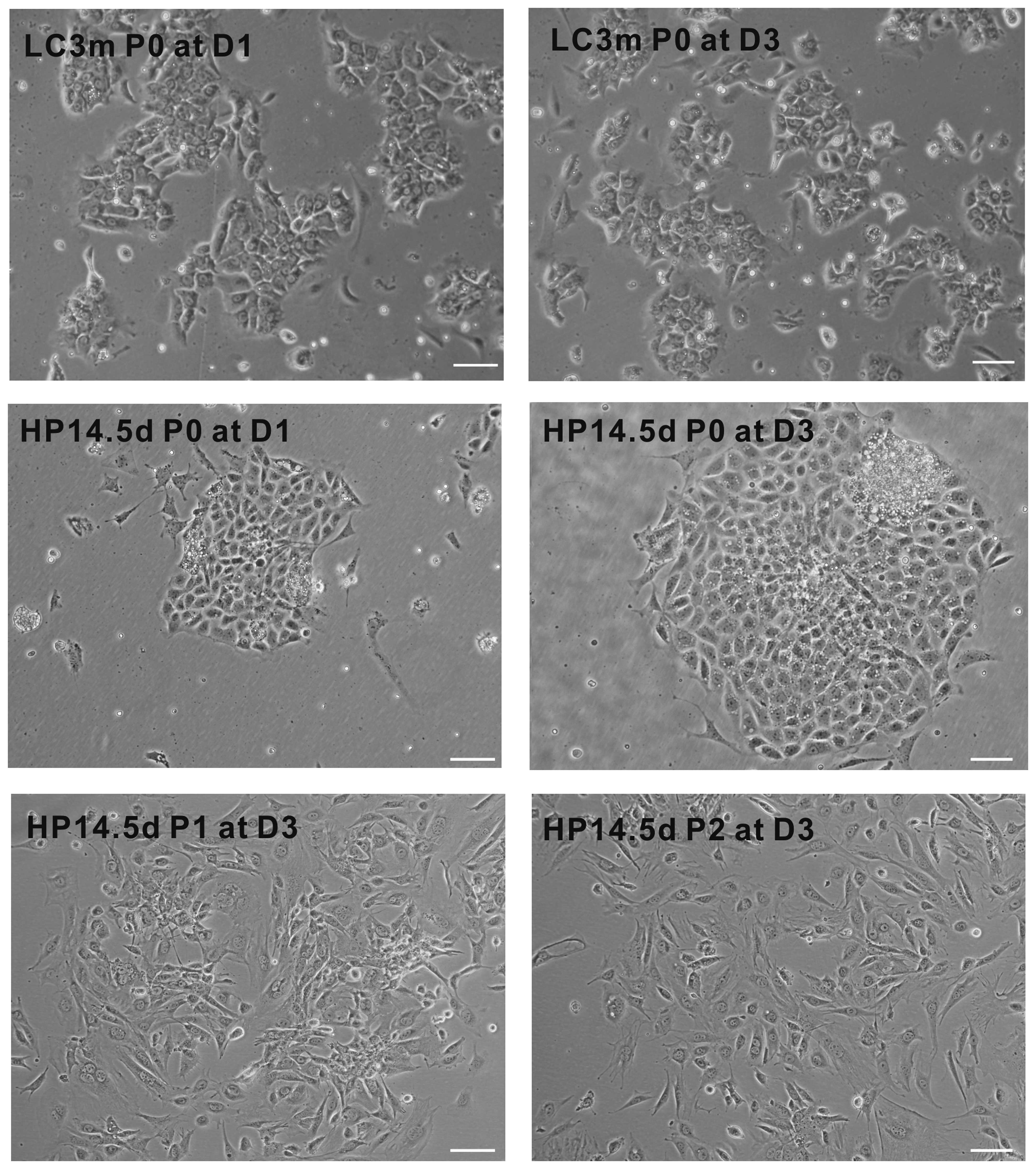
As shown in Fig.
1, we found that the primary LC3m cells were gathered together
to form sac-like glands and exhibited a typical cubic cell shape of
hepatocytes. The confluence of LC3m cells did not differ between
days 1 and 3 after induction; few of the LC3m cells were adherent
following passage. The primary HP14.5d cells displayed cluster
growth and were typically mononucleate with a single, spherical,
central nucleus and high nucleus/cytoplasm (N/C) ratio; however, a
quarter of them were binucleate. Following passage, the majority of
the HP14.5d cells exhibited an elongated morphology. These results
indicate that the morphology of HPCs is not completely similar to
that of mature hepatocytes; the passaged HP14.5d cells displayed
different cell shapes; possibly due to the cell diversity of the
primary isolated cells.
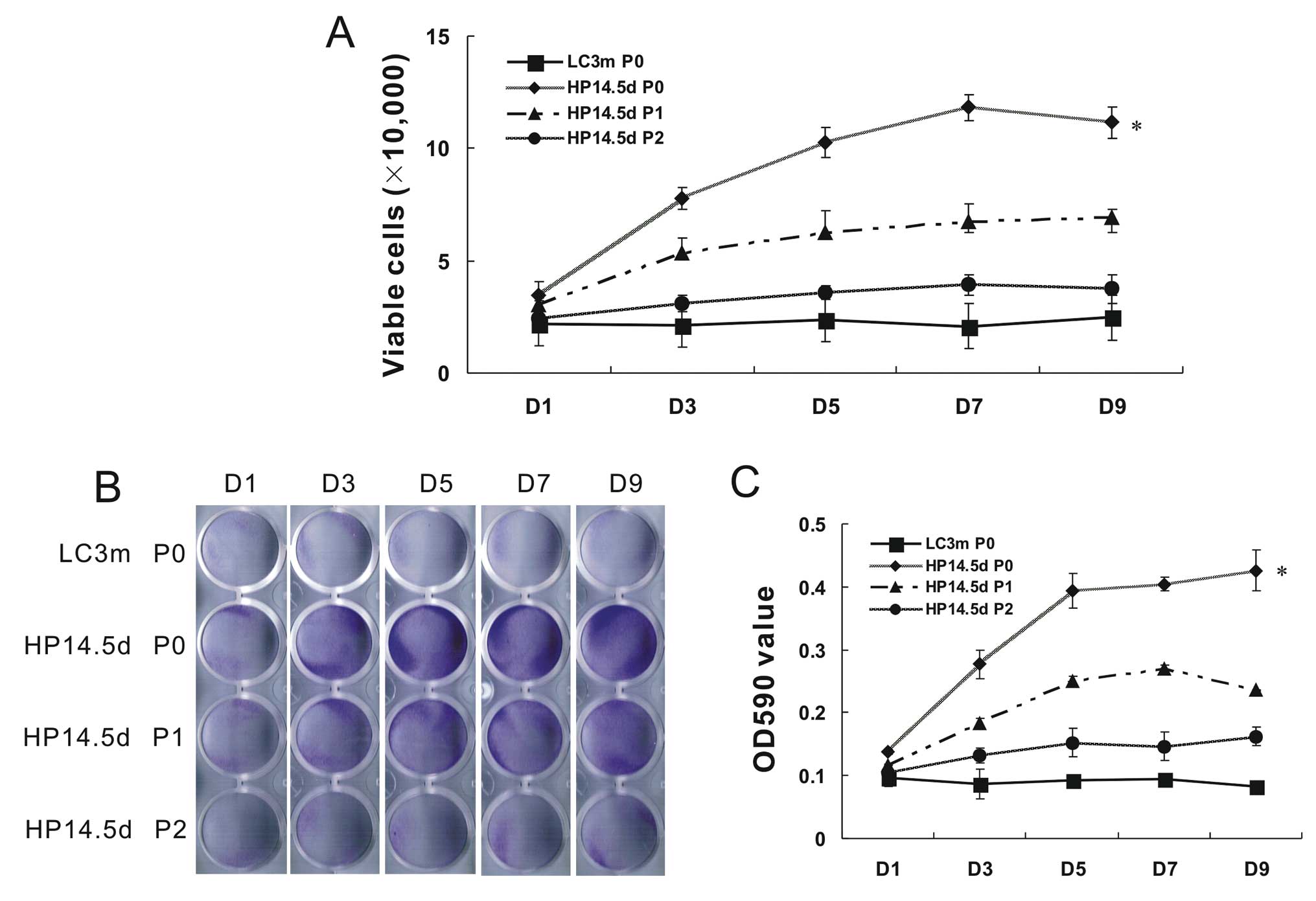
Proliferation of primary and passaged
cells
Due to the difference in cell growth status between
the HP14.5d and LC3m cells, we wished to further detect the cell
proliferation within 9 days of incubation following isolation or
passage. Compared with the non-proliferative LC3m cells, the
primary HP14.5d cells proliferated actively. The number of HP14.5d
cells on day 7 increased by >5-fold, but slighlty decreased on
day 9 due to limited growth space in the plate. The number of
viable HP14.5d cells at passage (P)1 was significantly lower than
the number of viable primary HP14.5d cells. The cell proliferation
of the HP14.5d cells at P2 was almost the same as that of the
primary LC3m cells (Fig. 2A). The
number of crystal violet-stained cells was similar to the number of
viable cells (Fig. 2B and C).
Therefore, the low success rate of primary culture of mature
hepatocytes may due to their limited growth capabilities. HPCs
primarily display active proliferation; however, their
characteristics of proliferation and the hepatic phenotype become
unstable following culture in vitro.
Identification of HP14.5d and LC3m
cells
The expression of hepatic progenitor markers and
mature hepatocyte markers was examined to identify the HP14.5d and
LC3m cells. As shown by the RT-PCR and immunofluorescence staining
results (Fig. 3), the early
hepatic marker, DLK (20), and
the pluripotent progenitor cell marker, CD34 (21), were readily detectable in the
HP14.5d cells and minimally detected in the LC3m and Hepa1–6 cells.
The tumor marker, AFP, the fetal form of serum albumin (22) and CK19, a non-specific marker for
liver stem cells (23), were
expressed at higher levels in the HP14.5d cells compared with the
LC3m cells. Albumin and CK18, as liver-specific markers, were
expressed at lower levels in the HP14.5d cells compared with the
LC3m and Hepa1–6 cells. The expression of the mature hepatic
markers, apolipoprotein B (ApoB), tyrosine aminotransferase (TAT)
and UGT1A (24–26), was undetectable in the HP14.5d
cells. Taken together, these results suggest that the LC3m and
HP14.5d cells represent mature hepatocytes and HPCs,
respectively.
 | Figure 3Identification of LC3m cells and
HP14.5d cells. Hepa1–6 cells were used as the controls. (A) RT-PCR
analysis of the hepatic-related genes, DLK, CD34, AFP, CK19, ALB,
CK18, TAT and ApoB. The RT-PCR results were confirmed in at least 3
batches of independent experiments, and representative results are
shown. (B) Immunofluorescent staining of AFP, ALB and UGT1A
markers. Scale bar, 200 μm. |
Induction of hepatic differentiation
We then wished to investigate the differentiation
potential of the 2 candidate cell lines. The combination of 2% HS +
0.1 μM Dex + 10 ng/ml HGF + 20 ng/ml FGF4 has been demonstrated to
induce the differentiation of progenitor cells into functional
hepatocytes in our previous study (27). In this study, we compared the
induction of hepatic differentiation between the 2 candidate cell
lines. Through ALB-driven luciferase reporter assay (Fig. 4A), we found that the basal level
of ALB-Gluc activity in the LC3m cells was higher than that in the
HP14.5d cells. As culture time progressed without induction, the
ALB-Gluc activity slightly increased in the HP14.5 cells but not in
the LC3m cells. ALB-Gluc activity was significantly increased after
the induction of differentiation. At 12 days of induction, the
ALB-Gluc activity of the HP14.5d cells was comparable to that of
LC3m cells. The mRNA expression levels of DLK, CD34, AFP, ALB, TAT
and ApoB in the induced HP14.5d cells were similar to those
observed in the induced LC3m cells (Fig. 4B). Western blot analysis and
immunofluorescence staining revealed that the protein expression of
UGT1A and CK18 in the induced HP14.5d cells was increased to levels
equivalent to those observed in the same induced LC3m cells
(Fig. 4C and D). Thus, our
results demonstrate that the HP14.5d cells can easily be induced to
differentiate into hepatocytes with an expression profile of
hepatic-related markers similar to that of mature hepatocytes.
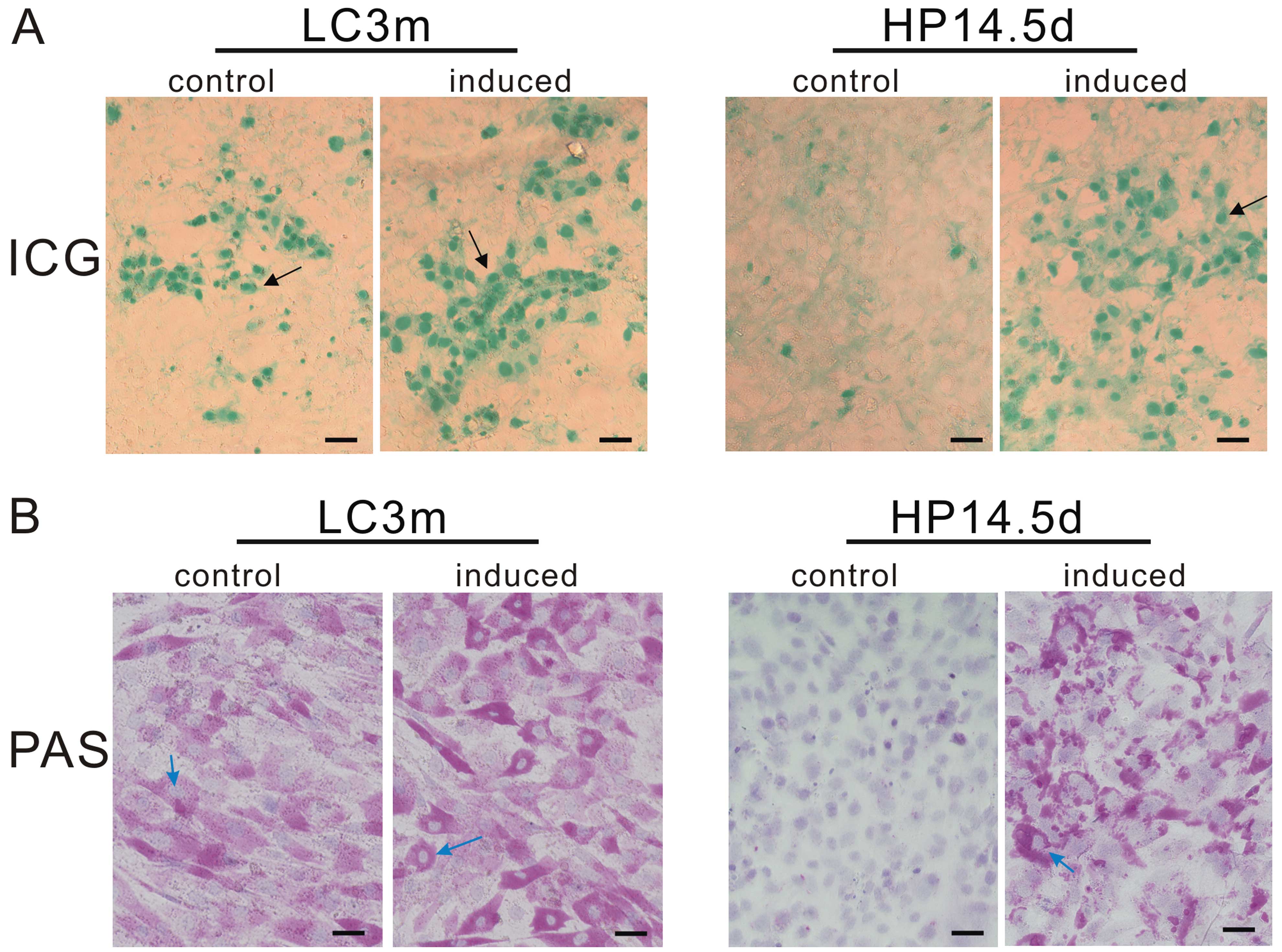
Function of induced cells
The evaluation of ICG uptake is a common way to
estimate liver function (28). As
mature hepatocytes, approximately 30% of the LC3m cells absorbed
ICG and exhibited a green-stained nucleus, whereas no ICG-positive
cells were observed in the untreated HP14.5d cells. Following 12
days of induction, the ratio of ICG-positive cells in both groups
increased to 60%, even though the green color in the HP14.5d cells
was lighter than that in the LC3m cells (Fig. 5A). Mature hepatocytes have the
ability of glycogen synthesis and storage. The PAS staining method
is used for the detection of glycogen which is displayed by a
purple color in the cytoplasm (29). The majority of LC3m cells was
positive for PAS staining, and a deeper purple color was observed
in the induced LC3m cells. Undifferentiated HP14.5d cells were not
stained; however, following treatment in the hepatic induction
medium, the number of PAS-stained cells significantly increased and
was equivalent to the number of LC3m-stained cells (Fig. 5B). The abovementioned results
indicate that the induced HP14.5d cells not only express hepatic
markers but also display functions similar to those of mature
hepatocytes.
Discussion
Compared with the strategies of liver
transplantation in the treatment of liver diseases, cell-based
transplantation therapies have the advantages of low-risk clinical
operation, low immunogenicity and good functional recovery, showing
tremendous potential for their clinical application (6–8,30,31). The cell source for liver cell
transplantation should have the characteristics of effective
proliferation capabilities to obtain an adequate population of
cells, as well as hepatic functions to replace the damaged liver
mass (13,32). To date, a variety of cell sources
have been reported in the exploration for liver cell
transplantation. Mature hepatocytes have good metabolism functions,
but their low availability and expansion efficiency in vitro
are major issues of hepatocyte transplantation (9,33).
Stem cells have the characteristics of self-renewal,
multi-potential differentiation and easy amplification in
vitro; however, their limited differentiation capabilities to a
hepatic lineage restrict their functional recovery (34,35). HPCs have the ability of
bipotential differentiation into mature hepatocytes and biliary
epithelial cells, along with self-renewal capacity. In addition,
the immunogenicity of HPCs derived from embryos is lower than that
of HPCs derived from adult livers, demonstrating their potential
for use in clinical practice (36–38). In the present study, we
investigated the proliferation, differentiation and function of
HPCs compared with mature liver cells, to identify the potential
value of HPCs.
As 3-month-old mice are at the adult period of their
life-span, the majority of liver cells are at a terminally
differentiated stage to perform normal functions. At this stage,
primary LC3m cells hardly proliferated and could not be passaged
in vitro. Hepatoblasts begin to differentiate into
hepatocytes and cholangiocytes on embryonic day 14 (E14) of mouse
liver development (39). In this
study, we isolated HPCs from the livers of mice on post coitus day
14.5. The majority of the HP14.5d cells belong to stem/progenitor
cells, and the freshly isolated HPCs exhibited active
proliferation. Following passage, however, their growth capability
significantly decreased, and the cells displayed different
morphologies. This may due to the various cell types found in liver
tissue, such as hepatic parenchymal cells, stellate cells, Kupffer
cells and liver fibroblasts (40). It is conceivable that the cell
pool may contain HPCs with different proliferative capacities,
differentiation potentials or may even have bipotential
capabilities. Thus, to obtain a reliable cell source for liver cell
transplantation, the characterization and identification of
cellular candidates should be further examined. As a following
step, we aim to establish individual progenitor clones.
Hepa1–6 is a hepatocarcinoma cell line which
expresses high levels of AFP and ALB (41). Using Hepa1–6 cells as the
controls, we found that LC3m cells had high expression levels of
the mature hepatic marker genes, ALB, CK18, ApoB, TAT and UGT1A
(24–26), while the HP14.5d cells exhibited
relative high expression levels of pluripotent progenitor cell
markers or early hepatic marker genes (DLK, CD34, AFP and CK19)
(20–23), suggesting that LC3m cells have the
genetic characteristics of mature hepatocytes, while the HP14.5d
cells retain most, if not all of the HPC phenotype. Even though it
has been reported that stem/progenitor cells can replace mature
liver cells in cell transplantation, their differentiation and
function should be detected prior to their in vivo
application. In the present study, we used the combination of 2% HS
+ 0.1 μM Dex + 10 ng/ml HGF + 20 ng/ml FGF4 to induce the hepatic
differentiation of 2 candidate cell lines (27). ALB-driven Gluc activity can
indirectly reflect the expression level of ALB, which is used to
dynamically measure the hepatic differentiation capalities of liver
cells (18,27). The slight increased in ALB-Gluc
activity in the untreated HP14.5d cells, indicated the spontaneous
differentiation of progenitor cells. Following culture in hepatic
induction medium, the expression of hepatic stem/progenitor and
late marker genes in the HP14.5d cells reached levels comparable to
those in the induced LC3m cells. Of note, though the LC3m cells
were derived from adult mouse livers, ALB-Gluc activity and the
expression of late hepatic markers increased following hepatic
induction, which suggested that not all cells in the LC3m pool were
at a mature status. Nevertheless, we demonstrated the
differentiation potential of the HP14.5d cells into mature
hepatocytes in vitro.
Synthesis and metabolism are the most important
functions of the liver. To be a reliable cell source for cell
transplantation, the candidate cell should have normal functions.
As a fluorescent dye, ICG is metabolized microsomally in the liver
and is only excreted from the circulation by the liver and bile
ducts, this characteristic is commonly used as an indicator
substance in hepatic function diagnostics. The liver is responsible
for glycogenesis (the formation of glycogen from glucose) (42,43). In this study, we performed ICG
uptake analysis and PAS staining to detect the function of ICG
metabolism and glycogen synthesis/storage, respectively. The
untreated HP14.5d cells did not have any hepatic functions, whereas
almost 100% of the LC3m cells were stained positive following PAS
staining; only approximately 30% of the LC3m cells were
ICG-positive. ICG green-stained cells gathered together, indicating
that ICG metabolism may be involved in the confluence and
co-operation among cells. On day 12 following culture in hepatic
induction medium, the HP14.5d cells showed a positive ratio of ICG-
and PAS-stained cells similar to that of the LC3m cells. However,
the green or purple color of the HP14.5d cells was lighter than
that of the LC3m cells, and the PAS staining was not distributed in
the whole area of the cell plasma. As a result, HPCs performed some
hepatic functions following induction, although not at the exact
same levels as mature hepatocytes. The treatment time and different
induction methods should be further investigated.
In conclusion, we isolated HPCs from embryonic
livers and compared their proliferation, differentiation and
function with mature liver cells. HPCs displayed active growth
capability, good differentiation potential and normal function
following treatment in hepatic induction medium, showing an
alternative to mature liver cells in the cell-based transplantation
strategy for liver disease (14,44). However, HPCs still display limited
proliferation capabilities which continue to weaken following
passage. Therefore, in future studies, immortalized HPCs should be
constructed and in vivo models should be used to further
evaluate the effectiveness of HPCs and mature liver cells in cell
transplantation for the treatment of liver diseases (45,46).
Acknowledgements
This study was supported by grant from the National
Natural Science Foundation of China (no. 81100309 to Y.B.).
References
|
1
|
Clavien PA, Lesurtel M, Bossuyt PM, Gores
GJ, Langer B and Perrier A: OLT for HCC Consensus Group:
Recommendations for liver transplantation for hepatocellular
carcinoma: an international consensus conference report. Lancet
Oncol. 13:e11–e22. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Samuel D, Colombo M, El-Serag H, Sobesky R
and Heaton N: Toward optimizing the indications for orthotopic
liver transplantation in hepatocellular carcinoma. Liver Transpl.
17(Suppl 2): S6–S13. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sze YK, Dhawan A, Taylor RM, Bansal S,
Mieli-Vergani G, Rela M and Heaton N: Pediatric liver
transplantation for metabolic liver disease: experience at King’s
College Hospital. Transplantation. 87:87–93. 2009.PubMed/NCBI
|
|
4
|
Grant D, Fisher RA, Abecassis M, McCaughan
G, Wright L and Fan ST: Should the liver transplant criteria for
hepatocellular carcinoma be different for deceased donation and
living donation? Liver Transpl. 17(Suppl 2): S133–S138. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Katyal S, Oliver JH III, Buck DG and
Federle MP: Detection of vascular complications after liver
transplantation: early experience in multislice CT angiography with
volume rendering. AJR Am J Roentgenol. 175:1735–1739.
2000.PubMed/NCBI
|
|
6
|
Lacaille F: Liver transplantation and
liver cell transplantation. Clin Res Hepatol Gastroenterol.
36:304–307. 2012. View Article : Google Scholar
|
|
7
|
Lee SW, Wang X, Chowdhury NR and
Roy-Chowdhury J: Hepatocyte transplantation: state of the art and
strategies for overcoming existing hurdles. Ann Hepatol. 3:48–53.
2004.PubMed/NCBI
|
|
8
|
Sancho-Bru P, Najimi M, Caruso M, Pauwelyn
K, Cantz T, Forbes S, Roskams T, Ott M, Gehling U, Sokal E,
Verfaillie CM and Muraca M: Stem and progenitor cells for liver
repopulation: can we standardise the process from bench to bedside?
Gut. 58:594–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ito H, Kamiya A, Ito K, Yanagida A, Okada
K and Nakauchi H: In vitro expansion and functional recovery of
mature hepatocytes from mouse adult liver. Liver Int. 32:592–601.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gaudio E, Carpino G, Cardinale V,
Franchitto A, Onori P and Alvaro D: New insights into liver stem
cells. Dig Liver Dis. 41:455–462. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hughes RD, Mitry RR and Dhawan A: Current
status of hepatocyte transplantation. Transplantation. 93:342–347.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Souza BS, Nogueira RC, de Oliveira SA, de
Freitas LA, Lyra LG, Ribeiro dos Santos R, Lyra AC and Soares MB:
Current status of stem cell therapy for liver diseases. Cell
Transplant. 18:1261–1279. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Oertel M and Shafritz DA: Stem cells, cell
transplantation and liver repopulation. Biochim Biophys Acta.
1782:61–74. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gennero L, Mortimer P, Sperber K, Carloni
G and Ponzetto A: Stem cells: an alternative to organ
transplantation in chronic, degenerative and infectious diseases?
New Microbiol. 29:151–167. 2006.PubMed/NCBI
|
|
15
|
Dollé L, Best J, Mei J, Al Battah F,
Reynaert H, van Grunsven LA and Geerts A: The quest for liver
progenitor cells: a practical point of view. J Hepatol. 52:117–129.
2010.PubMed/NCBI
|
|
16
|
Russo FP and Parola M: Stem and progenitor
cells in liver regeneration and repair. Cytotherapy. 13:135–144.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang E, Bi Y, Jiang W, Luo X, Yang K, Gao
JL, Gao Y, Luo Q, Shi Q, Kim SH, Liu X, Li M, Hu N, Liu H, Cui J,
Zhang W, Li R, Chen X, Shen J, Kong Y, Zhang J, Wang J, Luo J, He
BC, Wang H, Reid RR, Luu HH, Haydon RC, Yang L and He TC:
Conditionally immortalized mouse embryonic fibroblasts retain
proliferative activity without compromising multipotent
differentiation potential. PLoS One. 7:e324282012. View Article : Google Scholar
|
|
18
|
Bi Y, Huang J, He Y, Zhu GH, Su Y, He BC,
Luo J, Wang Y, Kang Q, Luo Q, Chen L, Zuo GW, Jiang W, Liu B, Shi
Q, Tang M, Zhang BQ, Weng Y, Huang A, Zhou L, Feng T, Luu HH,
Haydon RC, He TC and Tang N: Wnt antagonist SFRP3 inhibits the
differentiation of mouse hepatic progenitor cells. J Cell Biochem.
108:295–303. 2009. View Article : Google Scholar
|
|
19
|
Bi Y, Gong M, Zhang X, Zhang X, Jiang W,
Zhang Y, Chen J, Liu Y, He TC and Li T: Pre-activation of retinoid
signaling facilitates neuronal differentiation of mesenchymal stem
cells. Dev Growth Differ. 52:419–431. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nishina H: hDlk-1: a cell surface marker
common to normal hepatic stem/progenitor cells and carcinomas. J
Biochem. 152:121–123. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nyamath P, Alvi A, Habeeb A, Khosla S,
Khan AA and Habibullah CM: Characterization of hepatic progenitors
from human fetal liver using CD34 as a hepatic progenitor marker.
World J Gastroenterol. 13:2319–2323. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Sell S: Alpha-fetoprotein, stem cells and
cancer: how study of the production of alpha-fetoprotein during
chemical hepatocarcinogenesis led to reaffirmation of the stem cell
theory of cancer. Tumour Biol. 29:161–180. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
He Z and Feng M: Activation, isolation,
identification and culture of hepatic stem cells from porcine liver
tissues. Cell Prolif. 44:558–566. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pan RL, Chen Y, Xiang LX, Shao JZ, Dong XJ
and Zhang GR: Fetal liver-conditioned medium induces hepatic
specification from mouse bone marrow mesenchymal stromal cells: a
novel strategy for hepatic transdifferentiation. Cytotherapy.
10:668–675. 2008. View Article : Google Scholar
|
|
25
|
Tirnitz-Parker JE, Tonkin JN, Knight B,
Olynyk JK and Yeoh GC: Isolation, culture and immortalisation of
hepatic oval cells from adult mice fed a choline-deficient,
ethionine-supplemented diet. Int J Biochem Cell Biol. 39:2226–2239.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Snykers S, Vanhaecke T, Papeleu P, Luttun
A, Jiang Y, Vander Heyden Y, Verfaillie C and Rogiers V: Sequential
exposure to cytokines reflecting embryogenesis: the key for in
vitro differentiation of adult bone marrow stem cells into
functional hepatocyte-like cells. Toxicol Sci. 94:330–341. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
He Y, Zhang WY, Gong M, Huang JY, Tang N,
Feng T, Wei GH, He TC and Bi Y: Low serum concentration facilitates
the differentiation of hepatic progenitor cells. Saudi Med J.
32:128–134. 2011.PubMed/NCBI
|
|
28
|
Yamada T, Yoshikawa M, Kanda S, Kato Y,
Nakajima Y, Ishizaka S and Tsunoda Y: In vitro differentiation of
embryonic stem cells into hepatocyte-like cells identified by
cellular uptake of indocyanine green. Stem Cells. 20:146–154. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kamo N, Yasuchika K, Fujii H, Hoppo T,
Machimoto T, Ishii T, Fujita N, Tsuruo T, Yamashita JK, Kubo H and
Ikai I: Two populations of Thy1-positive mesenchymal cells regulate
in vitro maturation of hepatic progenitor cellspopulations of
Thy1-positive mesenchymal cells regulate in vitro maturation of
hepatic progenitor cells. Am J Physiol Gastrointest Liver Physiol.
292:G526–G534. 2007. View Article : Google Scholar
|
|
30
|
Enns GM and Millan MT: Cell-based
therapies for metabolic liver disease. Mol Genet Metab. 95:3–10.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fitzpatrick E, Mitry RR and Dhawan A:
Human hepatocyte transplantation: state of the art. J Intern Med.
266:339–357. 2009. View Article : Google Scholar
|
|
32
|
Zhang Z, Liu J, Liu Y, Li Z, Gao WQ and He
Z: Generation, characterization and potential therapeutic
applications of mature and functional hepatocytes from stem cells.
J Cell Physiol. 228:298–305. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Risal P, Cho BH, Sylvester KG, Kim JC, Kim
HT and Jeong YJ: The establishment and characterization of immortal
hepatocyte cell lines from a mouse liver injury model. In Vitro
Cell Dev Biol Anim. 47:526–534. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sharma AD, Cantz T, Vogel A, et al: Murine
embryonic stem cell-derived hepatic progenitor cells engraft in
recipient livers with limited capacity of liver tissue formation.
Cell Transplant. 17:313–323. 2008. View Article : Google Scholar
|
|
35
|
Tomiyama K, Miyazaki M, Nukui M, Takaishi
M, Nakao A, Shimizu N and Huh NH: Limited contribution of cells of
intact extrahepatic tissue origin to hepatocyte regeneration in
transplanted rat liver. Transplantation. 83:624–630. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Krupnick AS, Balsara KR, Kreisel D, Riha
M, Gelman AE, Estives MS, Amin KM, Rosengard BR and Flake AW: Fetal
liver as a source of autologous progenitor cells for perinatal
tissue engineering. Tissue Eng. 10:723–735. 2004.PubMed/NCBI
|
|
37
|
Machaj EK, Grabowska I, Gajkowska A,
Jastrzewska M, Oldak T, Moraczewski J and Pojda Z: Differentiation
potential of the fetal rat liver-derived cells. Folia Histochem
Cytobiol. 43:217–222. 2005.PubMed/NCBI
|
|
38
|
Khan AA, Shaik MV, Parveen N,
Rajendraprasad A, Aleem MA, Habeeb MA, Srinivas G, Raj TA, Tiwari
SK, Kumaresan K, Venkateswarlu J, Pande G and Habibullah CM: Human
fetal liver-derived stem cell transplantation as supportive
modality in the management of end-stage decompensated liver
cirrhosis. Cell Transplant. 19:409–418. 2010.PubMed/NCBI
|
|
39
|
Zhao R and Duncan SA: Embryonic
development of the liver. Hepatology. 41:956–967. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kmieć Z: Cooperation of liver cells in
health and disease. Adv Anat Embryol Cell Biol. 161:III–XIII.
1–151. 2001.
|
|
41
|
Zhang L, Jiang G, Yao F, He Y, Liang G,
Zhang Y, Hu B, Wu Y, Li Y and Liu H: Growth inhibition and
apoptosis induced by osthole, a natural coumarin, in hepatocellular
carcinoma. PLoS One. 7:e378652012. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Lee HJ, Jung J, Cho KJ, Lee CK, Hwang SG
and Kim GJ: Comparison of in vitro hepatogenic differentiation
potential between various placenta-derived stem cells and other
adult stem cells as an alternative source of functional
hepatocytes. Differentiation. 84:223–231. 2012.
|
|
43
|
Shin KS, Lee HJ, Jung J, Cha DH and Kim
GJ: Culture and in vitro hepatogenic differentiation of
placenta-derived stem cells, using placental extract as an
alternative to serum. Cell Prolif. 43:435–444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Haridass D, Narain N and Ott M: Hepatocyte
transplantation: waiting for stem cells. Curr Opin Organ
Transplant. 13:627–632. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Gong M, Bi Y, Jiang W, Zhang Y, Chen L,
Hou N, Liu Y, Wei X, Chen J and Li T: Immortalized mesenchymal stem
cells: an alternative to primary mesenchymal stem cells in neuronal
differentiation and neuroregeneration associated studies. J Biomed
Sci. 18:872011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kakinuma S, Nakauchi H and Watanabe M:
Hepatic stem/progenitor cells and stem-cell transplantation for the
treatment of liver disease. J Gastroenterol. 44:167–172. 2009.
View Article : Google Scholar : PubMed/NCBI
|