Introduction
As reported by the World Health Organization in 2007
(1), glioma is a common malignant
tumor of the central nervous system, categorized in four grades
based on histological features. Grade IV glioma, also termed
glioblastoma multiforme (GBM), is the most common primary malignant
brain tumor, resulting in a poorly demarcated interface between
tumor and healthy brain tissues with no clear boundaries, which
represents a serious obstacle in conventional treatment (2). Approximately 50% of gliomas occupy
more than one lobe of a hemisphere or are bilateral (3). Through finger-like tentacles,
gliomas often spread to other parts of the brain and cause fatal
damage to healthy brain tissues (4). The survival rate of patients with
malignant glioma treated with conventional methods remains very low
due to poor prognosis. Even when combining radiation therapy and
concurrent chemotherapy with temozolomide, the median patient
survival is limited to 14.6 months (5). Therefore, the identification of
genes for effective targeting of malignant glioma is a priority in
current research related to the treatment of this type of
tumor.
As a protein with a key role in cytoskeletal
rearrangements, the mammalian diaphanous-related formin 1 (mDRF1),
also termed DIAPH1, is indispensable in microtubule formation and
in tumor cell migration (6). The
mDRF1 protein has two domains categorized as members of the formin
family, formin homology 1 (FH1) and 2 (FH2) (7). The protein is involved in
morphogenesis, cytokinesis, the formation of cell polarity, cell
adhesion and migration, and shows anomalous expression during tumor
differentiation (8,9), as well as during the invasion and
metastasis of MDA-MB-231 cells (10). However, very little is known about
its roles in human glioma.
In highly invasive tumor cells, the actin-rich
protrusions with extracellular matrix (ECM) proteolytic activity
are termed invadopodia (11).
Invadopodia have been found in a number of invasive malignant
neoplasms, including head and neck squamous cell carcinoma, breast
carcinoma and melanoma (12–14). Invadopodia that can accelerate the
metastasis and invasion of malignant tumor cells in 3D Matrigel
serve as the most important target in cancer treatment (15). Cortactin and phosphotyrosine are
abundant in the invadopodia of malignant tumor cells (16). The potential association of
invadopodia with mDRF1 remains unconfirmed.
In this study, we demonstrate that, as a vital
component of glioma cells, mDRF1 plays a pivotal role during the
invasion, metastasis, proliferation, apoptosis and invadopodia
formation of human malignant glioma cells.
Materials and methods
Cell lines, human tissues and
reagents
The U87 human malignant glioma (MG) cell line was
purchased from the Chinese Academy of Sciences. Cells were cultured
in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10%
fetal bovine serum (FBS) (both from Invitrogen Life Technologies,
Carlsbad, CA, USA) at 37°C in a 5% CO2 incubator.
Human glioma (grade IV) and healthy brain tissues
were obtained from patients registered at the Renji Hospital of
Shanghai Jiaotong University (Shanghai, China). Healthy brain
tissues (mostly from the cortex) were collected from patients with
physical brain injuries. Written informed consent was obtained from
all patients. The use of human tissue was approved by the ethics
committees of the hospital.
Nude mice were obtained from B&K Universal Group
Ltd. (Shanghai, China). All animals were treated in accordance with
the principles and procedures of The Animal Care and Experimental
Committee of the School of Medicine of Shanghai Jiao Tong
University.
Rabbit monoclonal antibody for mDRF1 was purchased
from Abcam Biosciences (Cambridge, MA, USA). Monoclonal mouse
anti-β-actin, anti-cortactin and anti-phosphotyrosine antibodies
were obtained from Cell Signaling Technology (Danvers, MA,
USA).
Reagents and kits were purchased from:
FuGENE® HD Transfection reagent from Promega Corp.
(Madison, WI, USA), Transwell kit for cell invasion assays from BD
Biosciences (Franklin Lakes, NJ, USA), Annexin V-Cy3 apoptosis
detection kit from BioVision Inc. (Milpitas, CA, USA), Cell
Counting kit-8 (CCK-8) for proliferation assays from Dojindo
Molecular Technologies Inc. (Kumamoto, Japan) and Radius™ 24-Well
Cell Migration Assay kit from Cell Biolabs, Inc. (San Diego, CA,
USA).
Constructs and transfection
The pGPU6-DIAPH1-1 plasmid (Shanghai GenePharma Co.,
Shanghai, China) was constructed and transfected into the U87 MG
cells to knock down the expression of mDRF1. A random siRNA
plasmid, pGPU6-DIAPH1-NC (Shanghai GenePharma Co.) was used as the
negative control. Cells were seeded in 6-well plates at
1×106/well and allowed to attach for 24 h prior to
transfection. Subsequently, the pGPU6-DIAPH1-1 and pGPU6-DIAPH1-NC
plasmids were transfected using the FuGENE® HD
Transfection reagent according to the manufacturer’s instructions.
Following incubation for 48 h at 37°C, cells were collected to
study the expression of mDRF1 by western blot analysis.
Atomic force microscopy (AFM)
The atomic force microscope [also referred to as the
scanning force microscope (SFM)] uses a microcantilever system to
detect the atomic or molecular forces between the probe tip and the
samples to generate images of samples. We used the 5500 AFM/SFM
(Agilent Technologies, Santa Clara, USA) in its contacting mode in
air at room temperature to obtain the topography AFM images.
Rectangular nitrate silicon probes were used with nominal spring
constant around 2.5 N/m (Agilent Technologies) and microcantilever
length of 120 μm. The microcantilever resonance frequency was
approximately 30 kHz. The rms free amplitude of the cantilever was
on the order of 15 nm and the relative set-point above 97% of the
free amplitude. Images were recorded with a slow scan rate (below 1
Hz).
Sample preparation for AFM imaging
Three groups of U87 MG cells were prepared for AFM
imaging: untreated U87 MG cells, U87 MG cells transfected with the
pGPU6-DIAPH1-1 plasmid and U87 MG cells transfected with the
pGPU6-DIAPH1-NC plasmid. After 48 h of transfection, the medium was
carefully removed without disturbing the cells. This was followed
by the addition of 100% ethyl alcohol to the petri dish to fix the
U87 MG cells for 15–20 min. The ethyl alcohol was then removed and
the petri dish was allowed to to air dry.
Immunohistochemistry
The localization of mDRF1 in human healthy/glioma
tissue was examined by immunohistochemical assays. Following the
antibody-antigen interaction, the targeted protein was visualized
via either chromogenic detection with diaminobenzidine (DAB) or
fluorescent detection. We followed standard procedures, briefly:
the primary antibody of mDRF1 was incubated with the tissue for 2 h
at room temperature or overnight at 4°C, to allow binding to the
target antigen. A biotinylated secondary antibody, specific to
mDRF1, was incubated with the tissue for 2 h at room temperature to
allow binding to the primary antibody. A biotinylated enzyme (HRP)
was pre-incubated with free avidin to form avidin-biotin-enzyme
complexes. This solution was added to the tissue sections, and any
remaining biotin-binding sites on the avidin bound to the
biotinylated secondary antibody that had already bound to the
tissue. The colored sites were observed under a confocal
microscope.
Immunocytochemistry
After cleaning the poly-L-lysine- coated coverslips
with ethanol, we plated the U87 MG cells at a density of
5×105 cells per coverslipped well and let cells grow
overnight. The culture medium was then removed and the U87 MG cells
were fixed with 4% formaldehyde for 20 min at room temperature. The
fixative was aspirated and the cells were washed twice with PBS.
The U87 MG cells were then incubated with PBST (PBS with 0.1%
Triton X-100) for 30 min. After washing with PBS three times, we
blocked unspecific binding sites with 5% goat serum. The blocking
buffer was then removed and the primary antibody was added followed
by incubation at 4°C overnight. Subsequently, the primary antibody
was removed and the cells were washed three times with PBST for 5
min. The secondary antibody was incubated for 1 h in the dark and
washed with PBST three times. Nuclear labeling was performed by
incubating with the DNA-binding agent DAPI in dry form, followed by
three 5-min washes with PBS. Fingernail polish was applied at the
edges of the coverslip and the samples were examined under a
confocal microscope.
Western blot analysis
Forty-eight hours after transfection, an equal
number of U87 MG cells from each experimental group was used to
extract total protein using RIPA lysis buffer. Electrophoresis was
performed on a 10% SDS-PAGE gel and was followed by transfer onto a
PVDF membrane. The membrane was then blocked by incubating with
skim milk overnight at 4°C. Following incubation with primary and
secondly antibodies, the expression level of immunoreactive mDRF1
was detected using enhanced chemiluminescence (ECL) reagents (Cell
Signaling Technology).
Proliferation assay
The assay was performed using the CCK-8 kit as
follows: U87 MG cells were seeded in 96-well plates
(1×104 cells/well) with six replicate wells for each
group. Forty-eight hours after transfection, 10 μl of WST-8 reagent
{[2-(2-methoxy-4-nitrophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium,
monosodium salt]} were added to each well and the cells were
incubated in a 5% CO2 incubator at 37°C for 1 h. Cell
proliferation, expressed as the cell survival rate, was analyzed in
a microplate reader at 450 nm absorbance. Untransfected cells,
cells transfected with pGPU6-DIAPH1-NC and cells transfected with
pGPU6-DIAPH1-1 were deemed as the untreated, the mDRF1
knockdown-negative and the mDRF1 knockdown or RNAi group,
respectively.
Apoptosis assay
Following transfection for 48 or 72 h,
4×105 U87 MG cells were collected by centrifugation. The
Annexin V-Cy3 reagent was then added and the cells were incubated
at room temperature for 5 min in the dark. Apoptotic cells, the
plasma membrane of which exhibited red fluorescence, were
quantified under a fluorescent microscope in five randomly selected
areas. The results are expressed as apoptotic rates. Untransfected
cells served as the untreated negative control group, while the
cells transfected with the pGPU6-DIAPH1-1 plasmid were deemed as
the RNAi group.
Wound healing assay
The assay was performed using the Radius™ 24-Well
Cell Migration Assay kit (Cell Biolabs, Inc.). Briefly, to monitor
the migratory properties of the cells during wound healing 48 h
after transfection, the U87 MG cells were seeded in 24-well plates
until they reached 80–90% confluence. Each plate well contains a
0.68 mm non-toxic, biocompatible hydrogel spot in the center, where
cells cannot attach. Once firm cell attachment was achieved, the
hydrogel was rapidly removed to expose a cell-free region for cell
migration This cell-free region in the center was the wound. Cells
were photographed under a microscope at 0, 2, 5, 8, 11, 18 and 24 h
after wound formation. The experiments were performed in
triplicate. For the evaluation of the ‘wound closure’ area, we used
Cell Profiler™ Cell Image Analysis software (available at:
http://www.cellprofiler.org/).
Transwell assay
A cell invasion assay was performed using a 24-well
Transwell chamber containing an 8-μ pore size polyethylene glycol
terephthalate (PET) membrane with a thin layer of Matrigel basement
membrane matrix. Forty-eight hours after transfection,
1×104 U87 MG cells were trypsinized and transferred to
the upper matrigel chamber in 500 μl of serum-free DMEM medium.
DMEM medium supplemented with 10% FBS was added to the lower
chamber as the chemoattractant. Following incubation for 24 h,
cells remaining in the upper chamber were carefully removed with
cotton swabs, while invading cells were fixed with dehydrated
alcohol, stained with hematoxylin and eosin (H&E), counted and
photographed under a microscope by randomly selecting five ×200
fields for each well. The experiment was repeated three independent
times.
Tumor model in nude mice
Age-matched adult male nude mice, four weeks old,
were housed in a temperature- and light-controlled environment with
a 14/10-h light/dark cycle. The U87 MG cells were harvested in
serum-free culture medium and the concentration of the cell
suspension was adjusted to 5×107 viable cells per ml.
The cell suspension (0.2 ml) was subcutaneously injected in the
right lateral of the mice oxter. The tumor was formed within
approximately 15 days after injection. The mice were randomly
divided into three groups: i) the untreated group, where the mice
were transplanted with untransfected cells; ii) the mDRF1
knockdown-negative group, where the mice were transplanted with
cells transfected with the pGPU6-DIAPH1-NC plasmid; and iii) the
mDRF1 knockdown/RNAi group, where the mice were transplanted with
cells transfected with the pGPU6-DIAPH1-1 plasmid. Mice were
observed daily and inspected every two days for the progression of
tumor growth. After 25 days of observation, the mice were
sacrificed and an autopsy was immediately performed. The tumor
tissues were then fixed in 4% paraformaldehyde.
Statistical analysis
Data were processed using a one-way ANOVA and are
presented as the means ± standard error of the mean (SEM). The
significance threshold for all tests was set to P<0.05.
Results
Expression and distribution of mDRF1 in
U87 MG cells
The human primary glioblastoma cell line, U87 MG,
that shows epithelial morphology (grade IV) was used in our
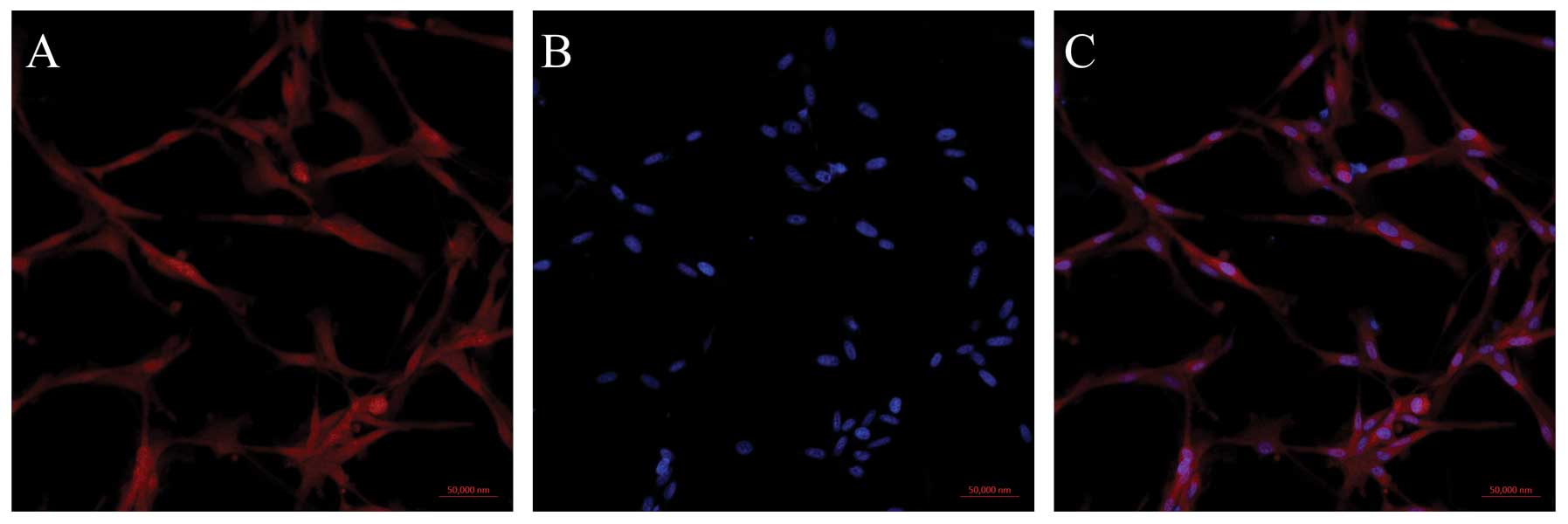
experiments. In order to analyze the distribution of mDRF1 in these
cells, we performed an immunocytochemical staining assay. By using
a biotinylated mDRF1-specific antibody, the expression and cellular
distribution of mDRF1 was indicated by red fluorescence, which we
found distributed in both the cell membrane and the cytoplasm
(Fig. 1). By contrasting the red
fluorescence associated with mDRF1 to the blue fluorescence emitted
by DNA in the nucleus, mDRF1 was detected as expressed in the
membrane as a cytoskeletal protein.
mDRF1 is overexpressed in human glioma
tissue
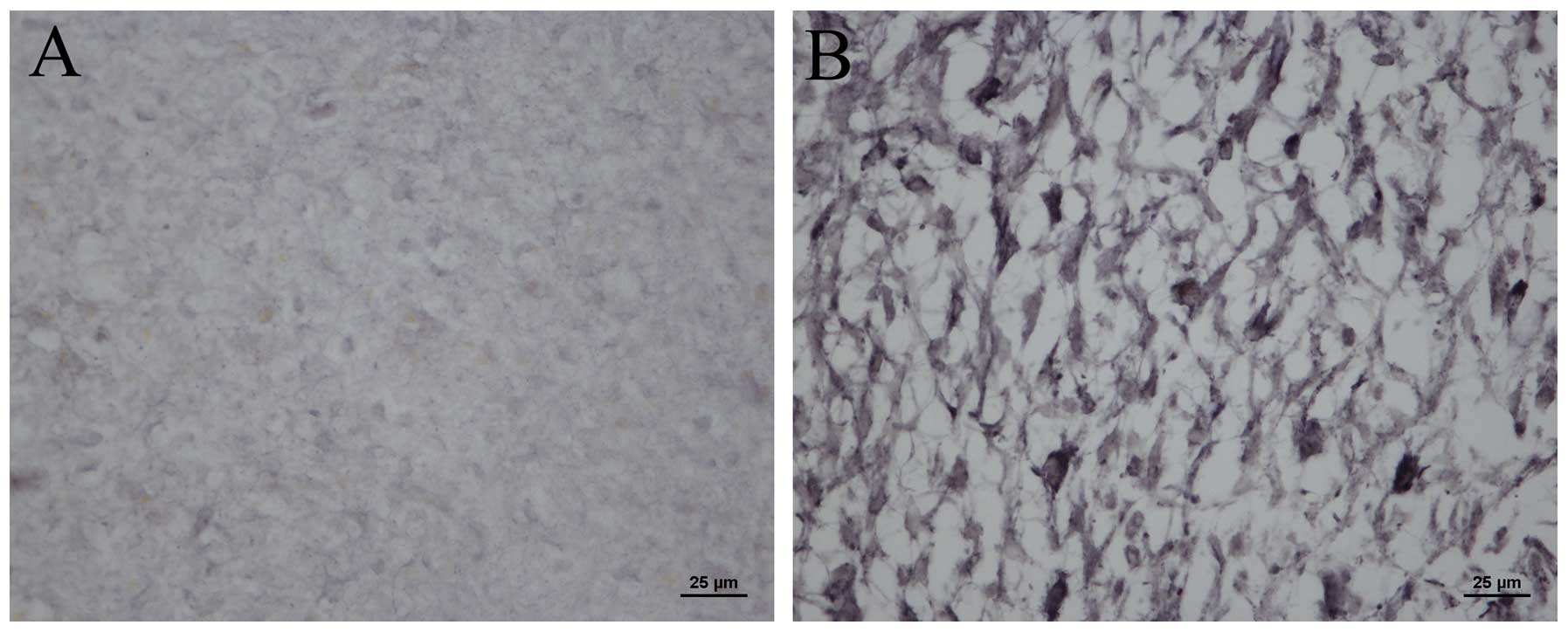
In order to determine whether mDRF1 is overexpressed
in human glioma tissue, we compared the expression levels of mDRF1
in human healthy tissue to malignant glioma brain tissue. Using the
mDRF1-specific antibody, immunohistochemical and immunofluorescence
assays were performed. Modena particles where mDRF1 was expressed
in the malignant glioma tissue (Fig.
2B) markedly outnumbered those in the healthy brain tissue
(Fig. 2A). This indicates that
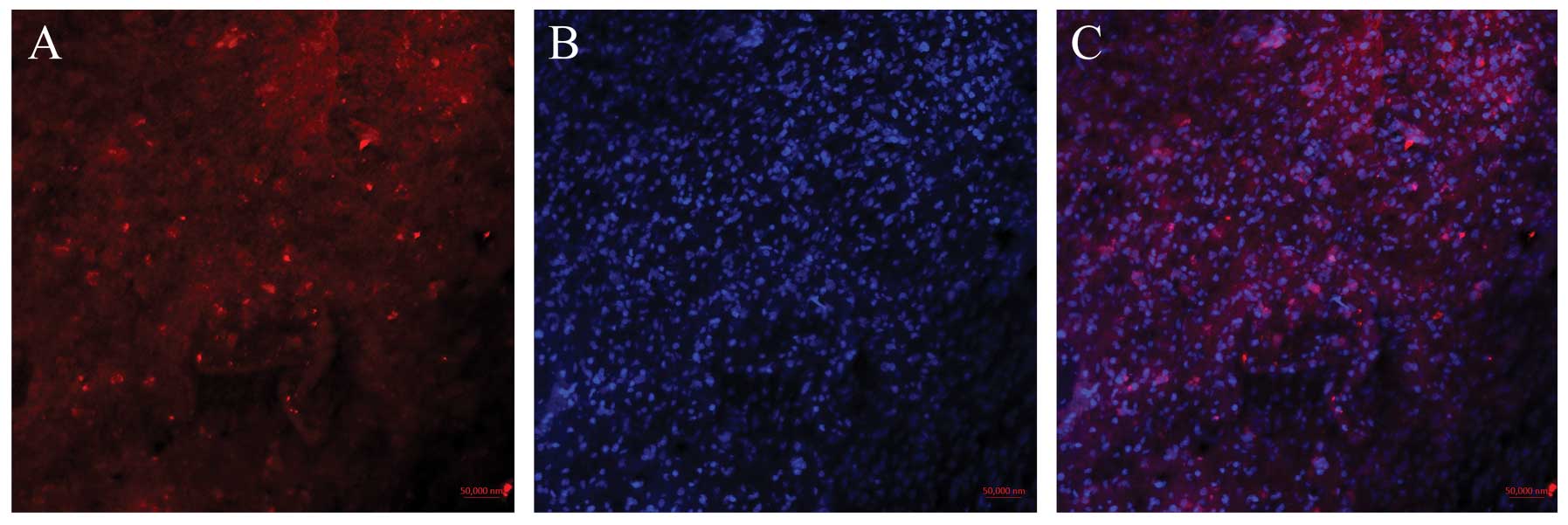
mDRF1 may be overexpressed in human glioma tissue. Fig. 3A shows the red fluorescent areas
indicating the expression of mDRF1 following incubation with the
mDRF1-specific antibody.
Successful knockdown of mDRF1 protein
expression in U87 MG cells
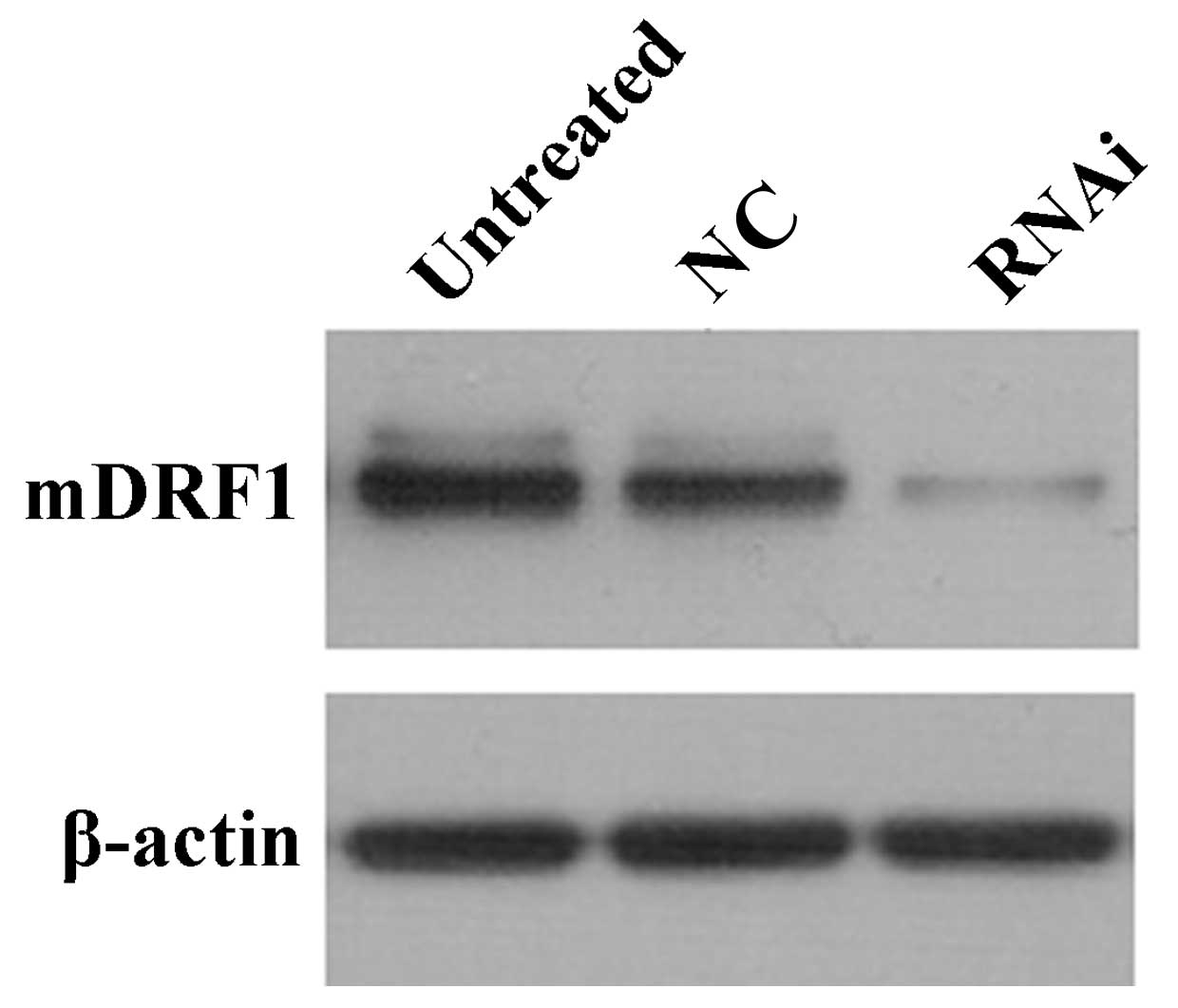
To confirm the transient transfection effects in the
three groups of cells mentioned above, we measured the protein
level of mDRF1 at 48 h after transfection. Western blot analysis
indicated that transient transfection efficiency was most
predominant at 48 h. The negative control groups (untreated and
mDRF1 knockdown-negative) showed no difference in the mDRF1 protein
level (Fig. 4). The level of the
β-actin gene was used for normalization.
Knockdown of mDRF1 inhibits cell
proliferation
After transfection for 48 h and incubation of the
cells with WTS-8 for 1 h, the proliferation of the U87 MG cells was
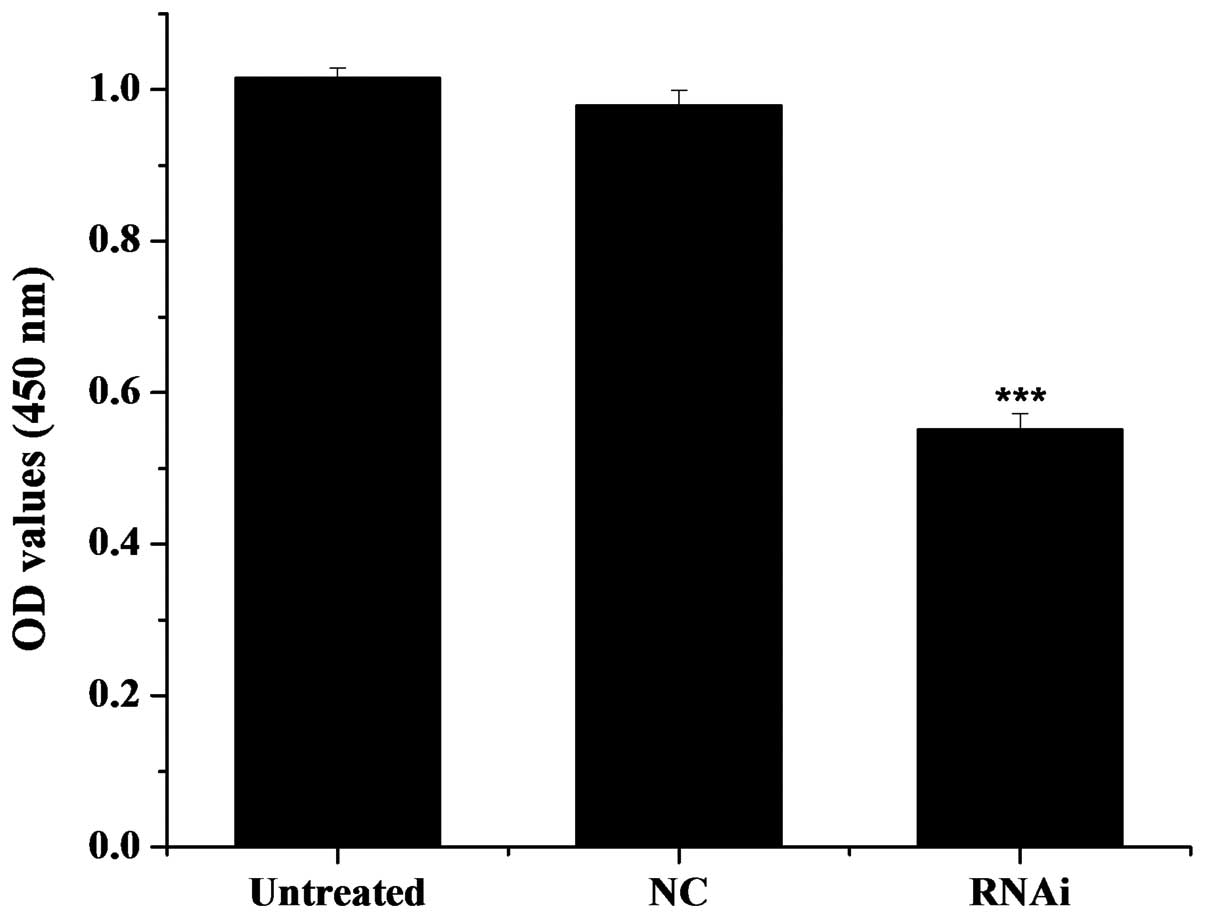
assessed. As illustrated in Fig.
5, the optical density (OD) value was 1.149 for the
untransfected cells (untreated group), 1.112 for the cells
transfected with the pGPU6-DIAPH1-NC plasmid (mDRF1
knockdown-negative group) and 0.747 for the U87 cells transfected
with the pGPU6-DIAPH1-1 plasmid (RNAi group). The OD values of the
untreated control group were significantly higher compared with the
RNAi group, i.e., compared with the cells in which mDRF1 was
knocked down. There was no obvious difference between the untreated
and the mDRF1 knockdown-negative group. In conclusion, the
knockdown of mDRF1 significantly reduced the proliferation ability
of the U87 MG cells. These results indicate that mDRF1 is involved
in U87 MG cell proliferation.
Knockdown of mDRF1 promotes cell
apoptosis
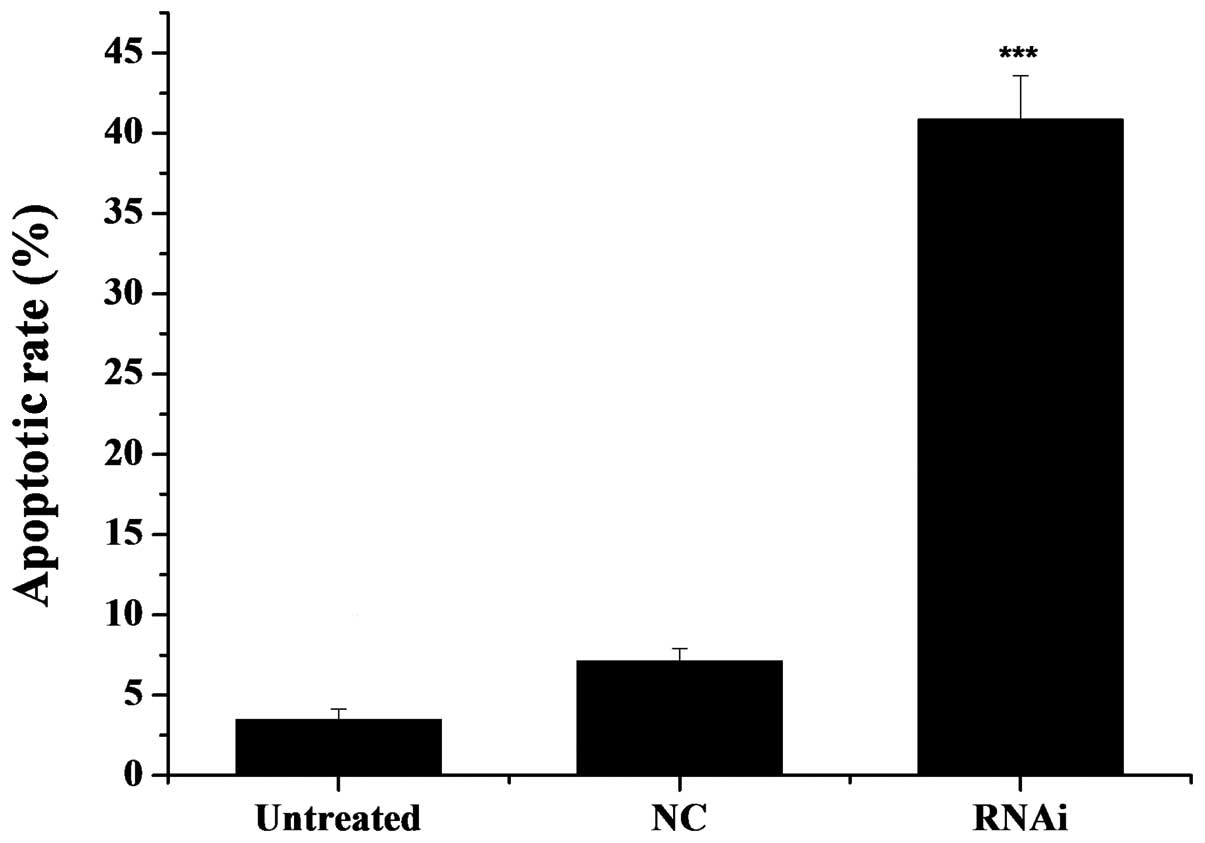
After a 48-h transfection, we detected the apoptotic
rate using the Annexin V-Cy3 apoptosis detection kit. As observed
under a fluorescence microscope, the cells with red fluorescence
were undergoing apoptosis. As illustrated in Fig. 6, the rate of apoptosis was 41.86%
in the mDRF1 knockdown group, 3.87% in the untreated group and
7.54% in the mDRF1 knockdown-negative group. In contrast to the
latter two negative control groups, the apoptotic rate was markedly
higher in the U87 MG cells transfected with the pGPU6-DIAPH1-1
plasmid (mDRF1 knockdown goup). There were limited differences
between the two negative control groups. These results reveal that
the knockdown of mDRF1 promotes apoptosis in the U87 MG cell
line.
Knockdown of mDRF1 inhibits cell
migration
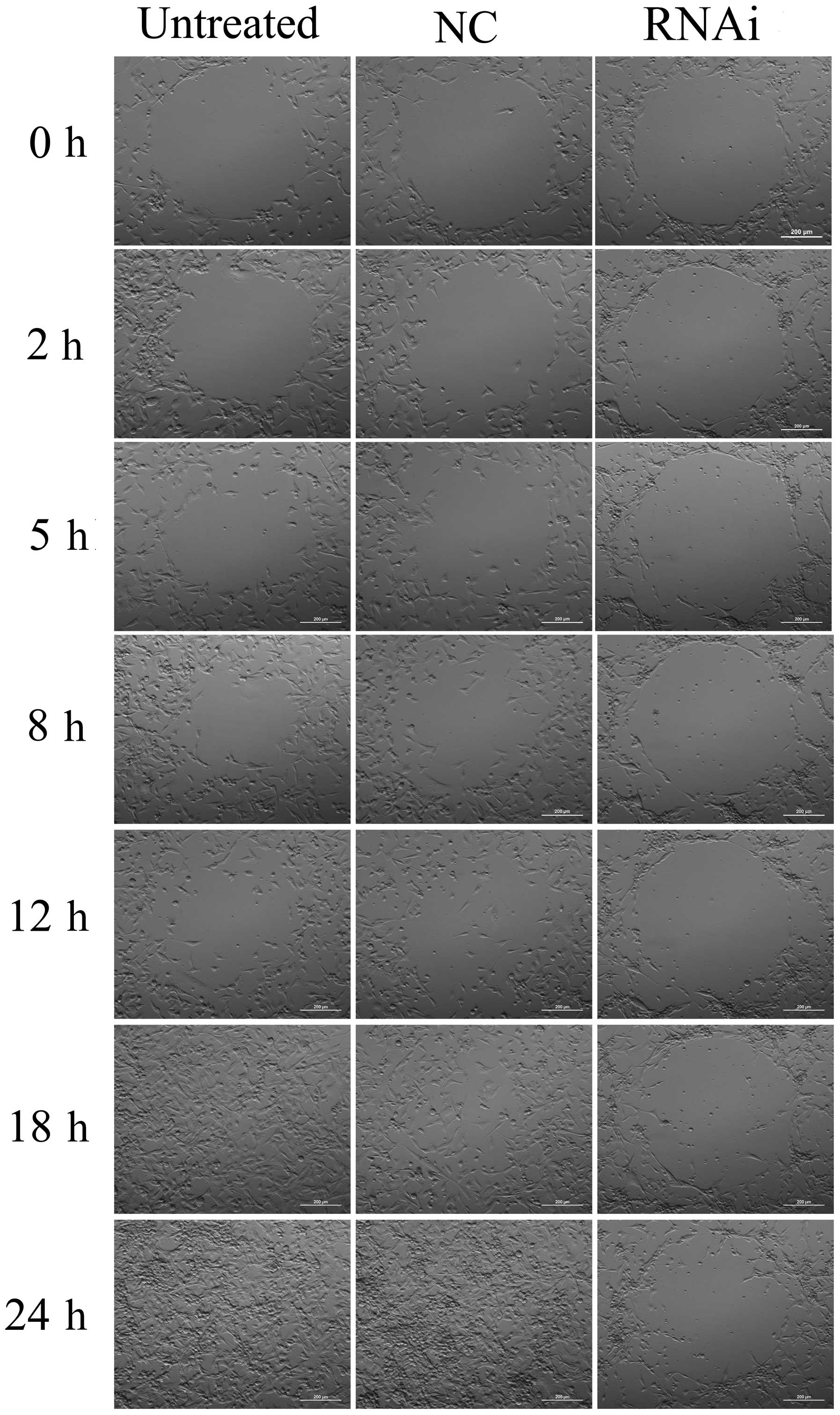
To examine the role of mDRF1 in U87 MG cell
migration, we performed a wound healing assay. As shown in Fig. 7, cells in the mDRF1 knockdown
group showed a decreased ability to migrate compared with the cells
in the negative control group. Cells in the untreated and the mDRF1
knockdown-negative control (NC) groups exhibited strong migration,
so that the open areas were completely filled and reached
saturation within 24 h. By contrast, cells in the mDRF1 knockdown
group showed a markedly slower migration rate, and even arrested
motility. Twenty-four hours later, cells with the silenced mDRF1
copy were still unable to migrate to the open area in the center of
the wells and a higher number of them died. The motility of the U87
MG cells decreased linearly with the decrease in the expression of
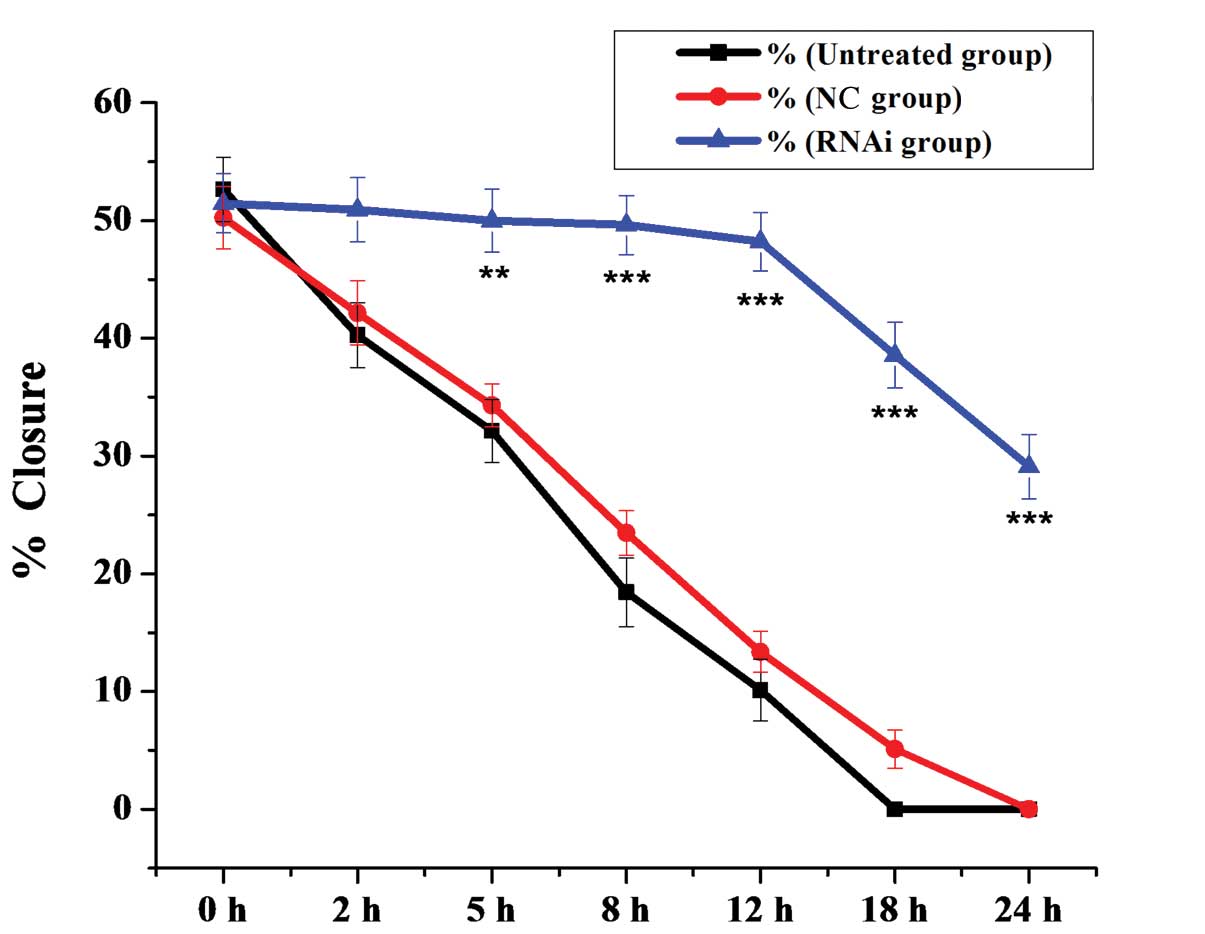
mDRF1. Statistical comparisons of cell migration rates among the
three groups, estimated from the ability of the cells to migrate
into the central area, are shown in Fig. 8. The negative control groups
showed a significantly faster migration rate than the mDRF1
knockdown group, which indicates that the silencing of mDRF1
reduces the motility of U87 MG cells.
Knockdown of mDRF1 reduces the invasive
ability of U87 MG cells
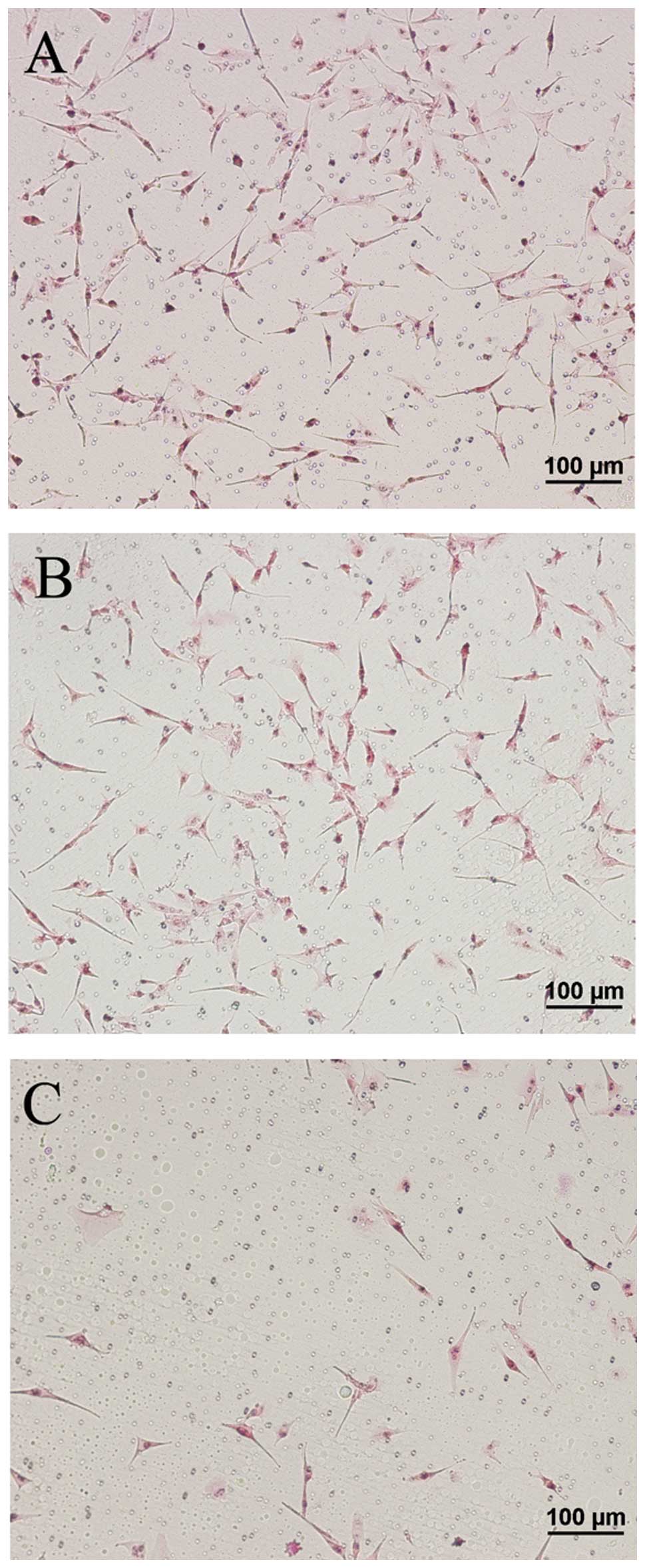
In the Transwell invasion assay, an ECM gel was used
to mimic the ECM surrounding the glioma cells, and thereby, the
in vitro tumor microenvironment. Following incubation of the
U87 MG cells in the Transwell chamber for 24 h, there was a greater
number of cells that crossed the membranes in the control groups
(untreated and mDRF1 knockdown-negative) compared with the mDRF1
knockdown/RNAi group (Fig. 9).
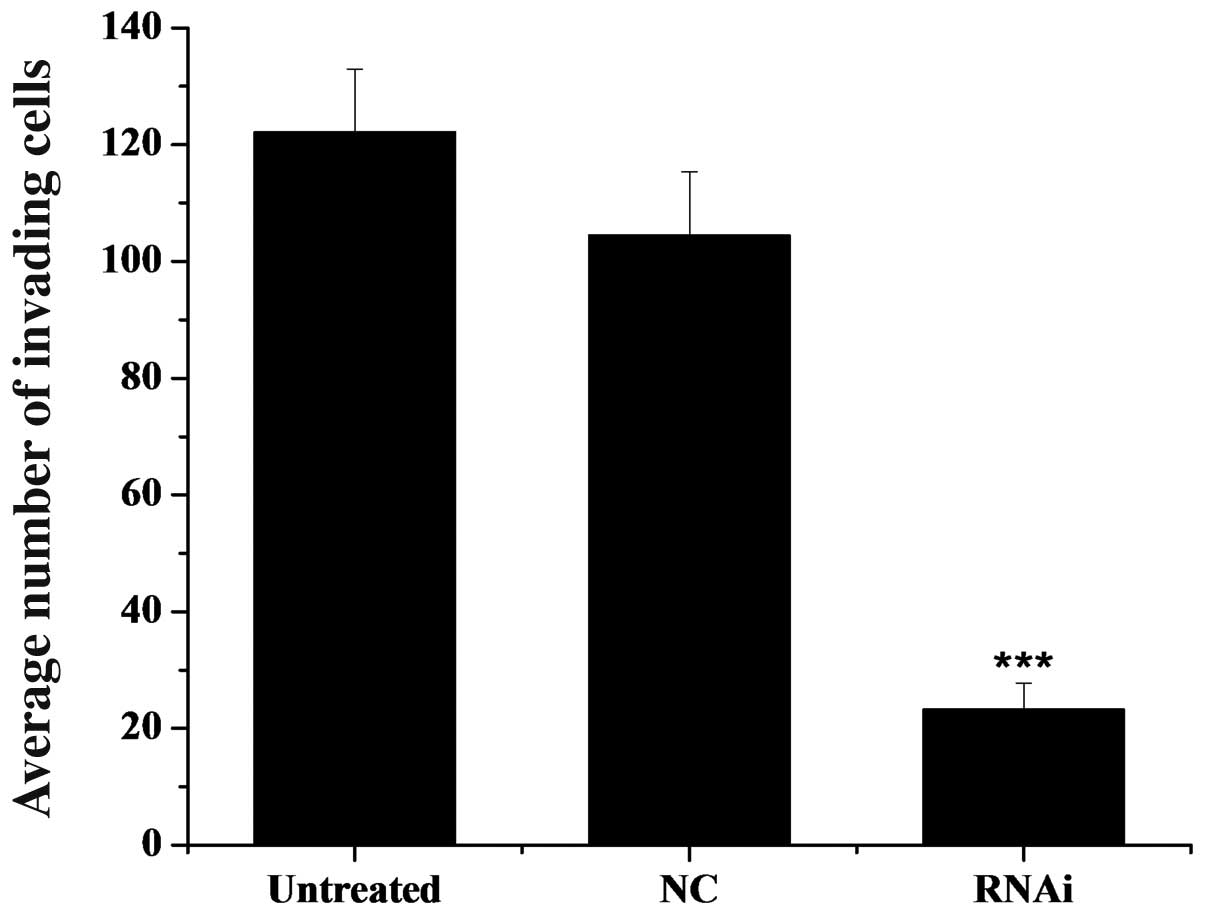
The average number of invasive cells that crossed the membrane was
approximately 125 in the untreated control group, approximately 105
in the NC (mDRF1 knockdown-negative) group and only 20 in the RNAi
group, as shown in Fig. 10.
Statistical analysis of these data revealed a significant
difference between the two negative control groups and the RNAi
group. No statistical difference was found between the two negative
control groups. We conclude that the silencing of mDRF1 can reduce
the invasive ability of the U87 MG cells and may be involved in the
process of glioma cell invasion.
Knockdown of mDRF1 reduces invadopodia
formation
The results presented above indicated that mDRF1 may
play an important role in the invasion and metastasis of human U87
MG cells. In a variety of highly invasive tumor cells, invadopodia
are the main structure degrading the ECM during invasion and
migration. We therefore explored the effects of the silencing of
mDRF1 on the formation of invadopodia in the U87 MG cell line. We
used the contact mode to scan the glioma cells by atomic force
microscopy (gas phase).
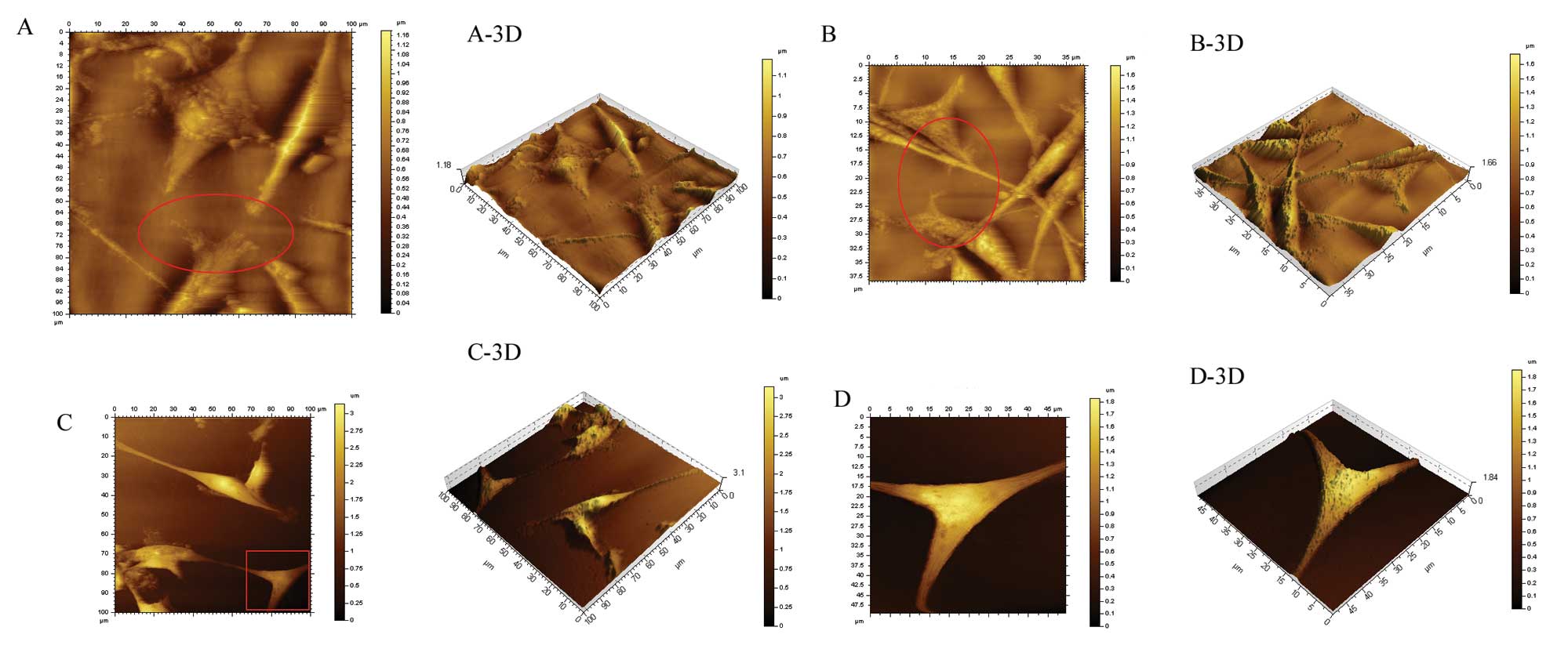
In the U87 MG cells that were not transfected, a
large number of invasive branches, showing a slender filamentous
structure, was observed at the cell edge, while cell morphology was
normal (Fig. 11A and A-3D).
Figs. 11A-, B-, C- and D-3D
display the relationship between the cell and the basement (bottom
of the cell culture dish). One can observe that the cells inlay the
basement in an ‘imbedded’ phenotype in the untreated group
(Fig. 11A-3D). Similarly, a high
number of invadopodia appeared at the cell edge in an imbedded
phenotype in the mDRF1 knockdown-negative group (Fig. 11B and B-3D). By contrast, in the
mDRF1 knockdown group, shown in Fig.
11C and D, a smooth cell edge with no invadopodia was observed.
In addition, the cell shape appeared changed and at times thinner.
Cells at the surface of the basement displayed a ‘suspended’
phenotype (Fig. 11C-3D and D-3D
). These results demonstrate that mDRF1 is involved in the
formation of invadopodia in U87 MG cells.
mDRF1 knockdown leads to reduced
expression of cortactin and phosphotyrosine
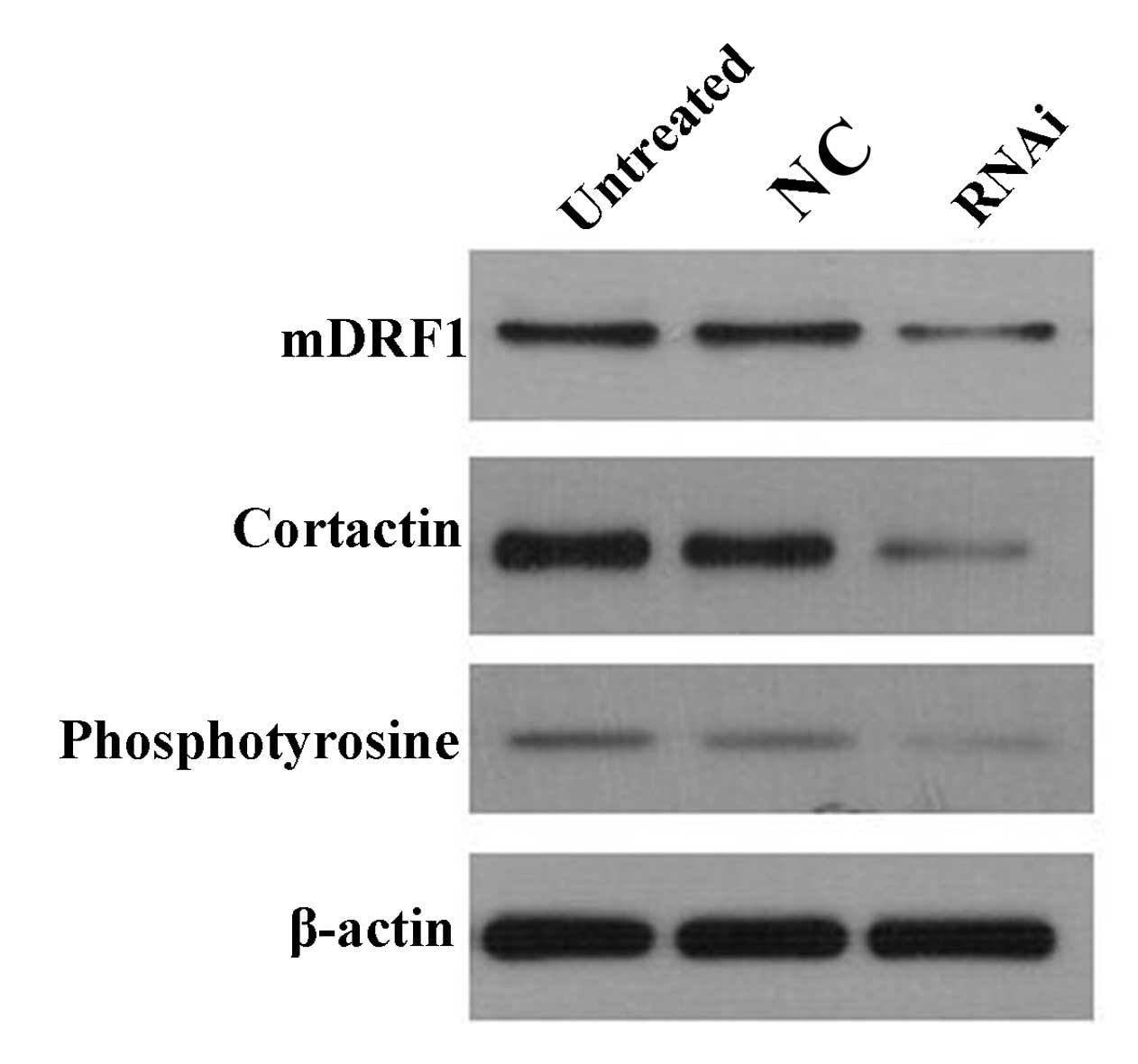
The results from atomic force microscopy showed that
the reduction in mDRF1 expression was accompanied by an absence of
invadopodia. A previous study showed that cortactin and
phosphotyrosine are abundant in the invadopodia (16). Therefore, changes in cortactin and
phosphotyrosine expression levels were examined to explore the
association between mDRF1 and these two proteins. Western blot
analysis (Fig. 12) revealed that
when the expression of mDRF1 decreased, and that the expression of
cortactin and phosphotyrosine was reduced as well. Thus, we
concluded that the silencing of the mDRF1 protein can affect the
expression of cortactin and phosphotyrosine.
Knockdown of mDRF1 expression suppresses
the growth of U87 MG-derived tumors in nude mice
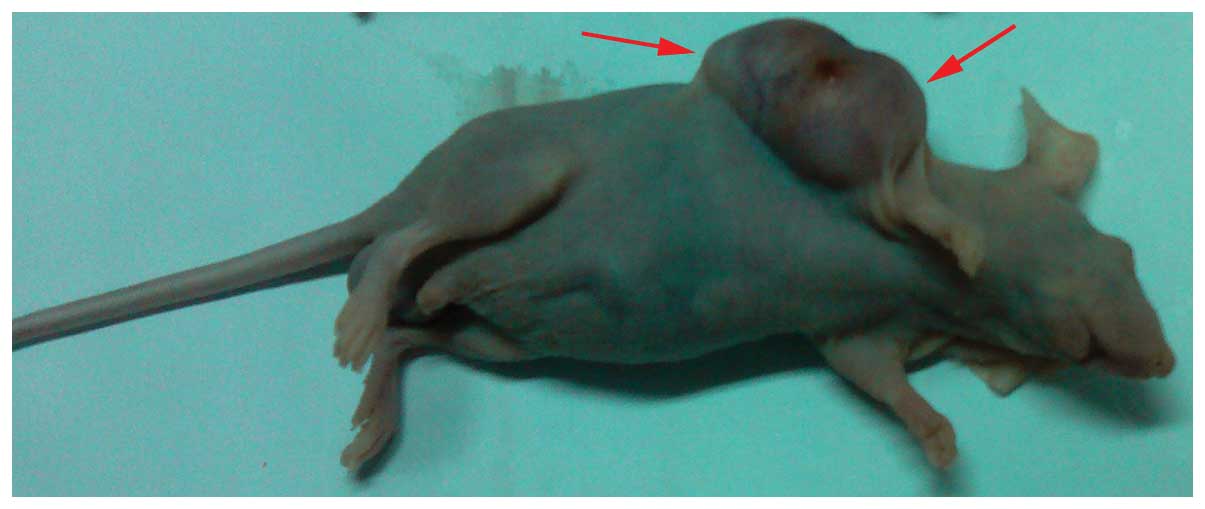
In order to further explore the function of mDRF1,
we established the transplanted tumor model using nude mice
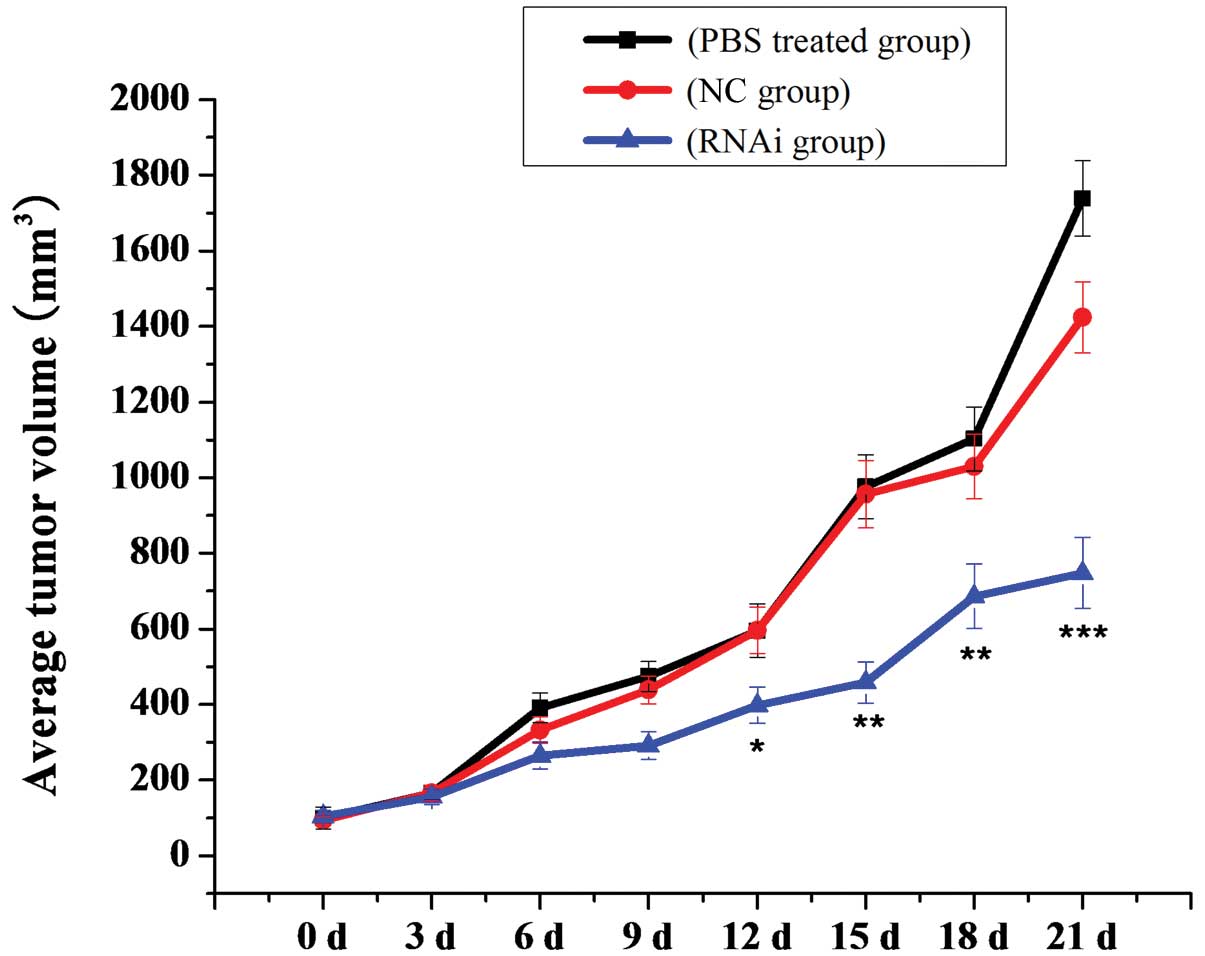
transplanted with human U87 MG cells (Fig. 13). As shown in Fig. 14, there was a significant
inhibitory effect on tumor growth in the mDRF1 knockdown group. By
contrast, rapid tumor growth was observed in the negative control
groups. Twenty-one days following these observations, the weight of
the transplanted tumor was lower in the mDRF1 knockdown group
compared with the control groups (Fig. 15). These data suggest that the
silencing of mDRF1 inhibits tumor growth in nude mice.
Discussion
mDRF1 is a multi-domain protein. A point mutation in
the gene (also termed DIAPH1) has been linked to non-syndromic
sensorineural hearing loss (17).
In addition, the abnormally high expression of mDRF1 in a variety
of cancer types plays an important role in the formation of stress
fibers, filopodia and microtubules, as well as in focal adhesion.
To our knowledge, there is no study to date that has evaluated the
roles of mDRF1 in glioma.
We previously conducted a microarray analysis of
tumor-related genes and demonstrated that the expression of
mDRF1/DIAPH1 in glioma is up to 6.583-fold higher compared
with healthy human brain tissue (18). Lizárraga et al (10) reported that upon the
downregulation of mDRF1 in malignant breast cancer cells
(MDA-MB-231), invadopodia formation was significantly reduced.
Moreover, the study by Carramusa et al (19) indicated that mDRF1 can affect
cross-linking between cells by regulating the synthesis of
E-cadherin. In malignant breast cancer cells, the silencing of
mDRF1 inhibited the E-cadherin protein located at the cell-cell
junctions and thereby significantly reduced the adhesion between
adjacent cells. Transfecting the cells with a plasmid, inducing the
high expression of mDRF1, restored cross-linking between adjacent
cells. These studies provide strong evidence that mDRF1 represents
a promising target molecule in the treatment of cancer and suggest
that it is involved in the invasion and metastasis of tumor
cells.
In the present study, we successfully engineered U87
MG cells with a silenced copy of the mDRF1 gene by transfection
with the pGPU6-DIAPH1-1 plasmid. Western blot analysis confirmed
that the protein level of mDRF1 was reduced in these cells. Kato
et al (20) reported that
the reduced expression of mDRF1 inhibited spindle assembly and the
formation of midbodies in HeLa cells. Moreover, altering the
expression of mDRF1 can disturb the cell cycle and cell
proliferation. The survival rate of the mDRF1 knockdown group in
our study was lower compared to that of the control groups,
indicating that the untreated cells had a stronger proliferative
ability compared with the mDRF1-silenced ones. Upon the silencing
of mDRF1, the number of apoptotic cells increased. The results from
both assays are consistent with those from the study by Kato et
al (20).
An earlier study demonstrated that mDRF1 expression
correlates with cell invasion, with invasive ability increasing
linearly with the increase in mDRF1 expression (10). Our results are in agreement with
this report. Following the silencing of mDRF1 in the U87 MG cells,
the cell invasive ability was reduced compared with the healthy
cells. The results from the wound healing assay, where the
migration of the mDRF1 knockdown cell group appeared reduced,
further support this conclusion.
The molecular mechanisms underlying the process of
invasion and migration in glioma cells are complex, involving
numerous intracellular signaling pathways. From these, changes in
cell morphology and invadopodia formation are believed to be
regulated by the Rho GTP pathway and particularly by the proteins,
RhoA, Rac1 and Cdc42 (21,22).
This pathway plays an important role at the onset of tumor cell
invasion and metastasis. Regulation, by mDRF1, of the invadopodia
formation process has been reported in human breast cancer cells
(10). Similarly, in our study,
the U87 MG cells in which the mDRF1 protein was silenced had a
reduced number of invadopodia. In the absence of mDRF1, the
invasive and metastatic ability of the tumor cells appeared
reduced, a result which warrants effective blocking of tumor cells
from spreading to surrounding areas. However, the signaling
pathway(s) through which mDRF1 exerts its effects remain to be
fully elucidated.
The invasion of surrounding tissue by malignant
tumor cells is the most important step initiating the process of
cancer metastasis, further facilitated by the degradation of ECM
barriers (23,24). However, since the molecular
mechanisms underlying tumor cell invasion and metastasis are
complex, mDRF1 may regulate the motility of tumor cells in
combination with other components, such as MMP-family proteins and
E-cadherins (25).
In conclusion, our study demonstrates that mDRF1 is
highly expressed in human U87 MG cells. Upon the knockdown of mDRF1
in these cells, the survival rate was reduced compared with the
controls. The apoptotic rate of the cells in which mDRF1 was
knocked down was higher than that of the control cells. The
motility and invasive ability of the control cells was higher
compared with that of the cells in which mDRF1 was knocked down.
The formation of invadopodia was reduced in the mDRF1-silenced
cells. In conclusion, mDRF1 plays a decisive role in the processes
of proliferation, apoptosis, migration, invasive ability and
invadopodia formation in U87 MG cells. These results indicate that
the mDRF1 protein may be a good target for the treatment of glioma.
Our study provides a novel prospect for the development of improved
treatment strategies for glioma, through the use of the mDRF1
protein.
Acknowledgements
This study was supported by the Innovation Program
of Shanghai Municipal Education Commission (grant no. 12ZZ100), the
National Basic Research Program of China (grant no. 2010CB529806),
the Leading Academic Discipline Project of Shanghai Municipal
Education Commission ‘Molecular Physiology’.
References
|
1
|
Louis DN, Ohgaki H, Wiestler OD, et al:
The 2007 WHO classification of tumours of the central nervous
system. Acta Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Lee Y, Scheck AC, Cloughesy TF, et al:
Gene expression analysis of glioblastomas identifies the major
molecular basis for the prognostic benefit of younger age. BMC Med
Genomics. 1:522008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Miller G: Brain cancer. a viral link to
glioblastoma? Science. 323:30–31. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mentlein R, Hattermann K and Held-Feindt
J: Lost in disruption: role of proteases in glioma invasion and
progression. Biochim Biophys Acta. 1825:178–185. 2012.PubMed/NCBI
|
|
5
|
Stupp R, Hegi ME, Mason WP, van den Bent
MJ, et al: Effects of radiotherapy with concomitant and adjuvant
temozolomide versus radiotherapy alone on survival in glioblastoma
in a randomised phase III study: 5-year analysis of the EORTC-NCIC
trial. Lancet Oncol. 10:459–466. 2009.
|
|
6
|
Goode BL and Eck MJ: Mechanism and
function of formins in the control of actin assembly. Annu Rev
Biochem. 76:593–627. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Castrillon DH and Wasserman SA: Diaphanous
is required for cytokinesis in Drosophila and shares domains
of similarity with the products of the limb deformity gene.
Development. 120:3367–3377. 1994.PubMed/NCBI
|
|
8
|
Zhu XL, Liang L and Ding YQ:
Overexpression of FMNL2 is closely related to metastasis of
colorectal cancer. Int J Colorectal Dis. 23:1041–1047. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
DeWard AD, Eisenmann KM, Matheson SF and
Alberts AS: The role of formins in human disease. Biochim Biophys
Acta. 1803:226–233. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lizárraga F, Poincloux R, Romao M, et al:
Diaphanous-related formins are required for invadopodia formation
and invasion of breast tumor cells. Cancer Res. 69:2792–2800.
2009.PubMed/NCBI
|
|
11
|
Buccione R, Caldieri G and Ayala I:
Invadopodia: specialized tumor cell structures for the focal
degradation of the extracellular matrix. Cancer Metastasis Rev.
28:137–149. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamaguchi H, Pixley F and Condeelis J:
Invadopodia and podosomes in tumor invasion. Eur J Cell Biol.
85:213–218. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stylli SS, Kaye AH and Lock P:
Invadopodia: at the cutting edge of tumour invasion. J Clin
Neurosci. 15:7252008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Carreira S, Goodall J, Denat L, et al:
Mitf regulation of Dia1 controls melanoma proliferation and
invasiveness. Genes Dev. 20:3426–3439. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schoumacher M, Louvard D and Vignjevic D:
Cytoskeleton networks in basement membrane transmigration. Eur J
Cell Biol. 90:93–99. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Bowden ET, Onikoyi E, Slack R, et al:
Co-localization of cortactin and phosphotyrosine identifies active
invadopodia in human breast cancer cells. Exp Cell Res.
312:1240–1253. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Drummond MC, Belyantseva IA, Friderici KH
and Friedman TB: Actin in hair cells and hearing loss. Hear Res.
288:89–99. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang Y, Qiu Y, Ren W, Gong J and Chen F:
An identification of stem cell-resembling gene expression profiles
in high-grade astrocytomas. Mol Carcinog. 47:893–903. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Carramusa L, Ballestrem C, Zilberman Y and
Bershadsky AD: Mammalian diaphanous-related formin Dia1 controls
the organization of E-cadherin-mediated cell-cell junctions. J Cell
Sci. 120:3870–3882. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kato T, Watanabe N, Morishima Y, Fujita A,
Ishizaki T and Narumiya S: Localization of a mammalian homolog of
diaphanous, mDia1, to the mitotic spindle in HeLa cells. J Cell
Sci. 114:775–784. 2001.PubMed/NCBI
|
|
21
|
Tominaga T, Sahai E, Chardin P, McCormick
F, Courtneidge SA and Alberts AS: Diaphanous-related formins bridge
Rho GTPase and Src tyrosine kinase signaling. Mol Cell. 5:13–25.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Narumiya S, Tanji M and Ishizaki T: Rho
signaling, ROCK and mDia1, in transformation, metastasis and
invasion. Cancer Metastasis Rev. 28:65–76. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weaver AM: Invadopodia: specialized cell
structures for cancer invasion. Clin Exp Metastasis. 23:97–105.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yamaguchi H: Pathological roles of
invadopodia in cancer invasion and metastasis. Eur J Cell Biol.
91:902–907. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kobielak A, Pasolli HA and Fuchs E:
Mammalian formin-1 participates in adherens junctions and
polymerization of linear actin cables. Nat Cell Biol. 6:21–30.
2004. View
Article : Google Scholar : PubMed/NCBI
|