Introduction
Venous thromboembolism (VTE) is a common and chronic
disease with a considerable risk of recurrence and can be fatal
(1,2). The crucial steps to preventing
further complications and recurrence are an accurate diagnosis and
early treatment with appropriate drugs (3). Biomarkers have been specifically
investigated for their capacity to predict VTE during the course of
disease (4). The analysis of
VTE-related molecular expression may identify potential disease
specific biomarkers associated with disease recurrence and as
potential drug targets (5). The
expression of the potent pro-coagulant protein tissue factor can
trigger thrombosis (6). Although
a number of putative mechanisms have been proposed, the mechanisms
that initiate VTE have not been completely elucidated, and
treatment options for individuals with recurrent disease in
particular remain limited (7).
The understanding of the mechanisms of VTE may lead to the
development of effective treatments for patients. Accurate
diagnosis, early prevention and the treatment of VTE with
appropriate drugs are crucial to prevent further complications and
recurrence (3). Warfarin and low
molecular weight heparin (LMWH) are the conventional drugs used for
the treatment of recurrent VTE. Kearon suggested that warfarin was
effective in the treatment of recurrent VTE (8). However, the use of warfarin for
anti-coagulation in patients with vein thrombosis does not minimize
the risk of recurrence (9). Thus,
more systematic research and drug combination strategies have been
recommended (10).
Network-based methods have been implemented to
predict drug side-effects, drug targets and new therapeutic
indications (11). The biological
functional network modules can reveal disease mechanisms and
identify drug targets (12). The
dynamic changes in gene expression would improve the understanding
of the pathological processing of complex diseases. The analysis of
gene expression using the network may be more effective in
predicting drug targets (13).
According to the different factors and differential gene expression
in easily recurrent thrombosis, a systemic method for screening
drugs and research on recurrent VTE may prove to be very good
strategy (14). The integrated
analysis of the network and high-throughput data is benefiical to
human disease research. In this study, we developed a novel module
screening method to identify the dynamic module changes by
combining human signaling networks and gene expression. We aimed to
elucidate the molecular mechanisms responsible for recurrent VTE,
and identify drug response pathways and potential drug targets with
which to improve the efficacy of VTE therapy and reduce the
recurrence of VTE.
Materials and methods
Data source
The human signaling network data were obtained from
previous studies (15,16). This included 6,287 genes and
62,239 signaling relations. The human signaling network was
integrated into the human subcellular signaling networks (HSSNs)
from the Gene Ontology (GO) database (17) and Universal Protein Resource
(UniProt) (version UniProt release 2014_03) (18).
We obtained the whole blood gene expression profile
dataset GSE19151 from the Gene Expression Omnibus database. The
GSE19151 expression profile dataset contains 3 sets of samples,
including 70 adults with one or more prior VTE treated with
warfarin (32 single and 38 recurrent) and 63 controls. The samples
were collected and screened by the Duke Anticoagulation
Satisfaction Scale (DASS) (19).
In the study, patients over 18 years had at least one prior venous
thromboembolic event. All patients administered warfarin were had
suffered from their most recent thromboembolic event ≥4 weeks
earlier. Patients who were just administered an anticoagulant
(warfarin) and patients with cancer or antiphospholipid syndrome
were excluded. The investigators who enrolled all VTE events
reviewed and confirmed the number of events, thrombus location,
thrombus type, and other clinical data. This study included
patients who suffered from spontaneous or provoked VTE for
exploratory research from a previous study (20). Blood was collected in PAXgene
tubes followed by RNA extraction and gene expression profiling by
Affymetrix arrays. The independent VTE expression profile datasets
GSE17078 from GPL96 platform contained 3 samples of patients with
VTE (VT sample) and 27 normal samples.
Recurrent risk modules (RRMs)
Considering the different conditions of recurrent
VTE under the treatment of warfarin, we developed a novel algorithm
for identifying RRMs. RRMs were selected by two steps: first, we
determined three different stages (control/single,
control/recurrent and single/recurrent) modules, respectively.
Network modules were screened through an efficient Markov
clustering algorithm of the GraphWeb (http://biit.cs.ut.ee/graphweb/) tool in each HSSN
(21). The default value of 1.8
for the Markov clustering parameter was considered. Modules with
less than four genes were excluded. The genes in each module were
both highly correlated and topologically close. The differential
stage modules were then screened by evaluating the expression
correlation differential score, which represented the significantl
changes in expression in different sets of samples at different
stages. Supposing a module M contains m edges E1…Em from the HSSNs,
thus the differential score S was expressed as follows:
wherein, X, Y and X′, Y′ denote the gene expression data of two
distinct stages, respectively. n1 and n2 denote the sample size of
two distinct stages, respectively. Ek and Ek′
denote the Pearson correlation coefficients of the edge k from the
edge E1…Em under two distinct stages, respectively. The real
differential score S was calculated between two stage samples for
each module. Subsequently, one thousand random modules were
constructed from the same HSSN with the degree conserved. The
random differential scores S1…S1000 were also calculated. If the
random differential scores were significantly less than the real
differential score, the module was considered as a differential
stage module (permutation test, p-values <0.05). The p-values
were then adjusted using the Benjamini-Hochberg method. Second, we
defined the differential stage modules existing in control/single,
control/recurrent and single/recurrent profiles as disease risk
modules to improve the reproducibility. Finally, RRMs were
identified from these disease risk modules by the
Jonckheere-Terpstra test with gene expression in different stages
of samples. The Jonckheere-Terpstra test, which is a non-parametric
test, was used to test the distribution of multiple independent
samples from the more general ones to find whether there was a
significant difference (). We
used the Jonckheere-Terpstra test ()
to evaluate the classification significance of RRMs among different
stages with a null hypothesis of no differences. The alternative
hypothesis was that the sample means changed among the disease
stages. A p-value <0.05 indicated significant differences among
disease stages. The RRMs could reveal the expression differential
among control, single and recurrent conditions.
Functional annotation analysis
In order to analyze the biological mechanisms of the
RRMs, we applied the gene-annotation enrichment analysis to measure
the functional characteristics which could increase the possibility
to identify the most relevant biological processes (23). Given an RRM with 'n' genes, the
enrichment analysis of the RRM could be efficiently measured by the
common and well-known statistical method Hypergeometric test
(24). The hypergeometric test is
executed for each RRM separately, which is defined by four
parameters: i) N is the size of human genes; ii) K is the gene
number in the annotation terms such as GO terms or pathways; iii)
'n' is the gene number of the RRM; and iv) 'c' is the common gene
number between the biological annotation terms and the RRM. The
hypergeometric test calculates the p-value by using the following
formula:
With the p-value <0.05 adjusted with
Benjamini-Hochberg method, the term is defined as a significant
enrichment. The enriched annotation terms in relation to the genes
may provide insight into understanding the biology themes behind
these genes (25).
Classifying performance with RRMs
In order to evaluate the classifying performances
and recognizing patterns for the RRMs, the Support Vector Machine
(SVM) method was employed to discriminate patients at different
disease stage s using RRMs as an input feature. Support vector
machines are supervised learning models based on relevant learning
algorithms (26). The performance
of our approach was evaluated using 5-fold cross-validation. The
stage sample dataset were equally divided into five parts. We then
performed five rounds of cross-validation. One part was used for
testing and the others were used for training for each round. We
averaged the results of the rounds to obtain the final results.
Subsequently, we applied the receiver operating characteristic
(ROC) curve to illustrate and evaluate classification performance.
The area under the ROC curve (AUC) quantifies the overall
discriminative power of the test (27).
Results
RRMs of VTE
The differential stage modules were obtained by
assessing the significantly expression changes in the gene
expression profile datasets. The disease risk modules were screened
out. Finally, seven RRMs were identified through the
Jonckheere-Terpstra test (Table
I).
 | Table IThe recurrent risk modules and
disease-related genes for VTE. |
Table I
The recurrent risk modules and
disease-related genes for VTE.
| RRMs | Genes of the
module | Localization |
|---|
| M1 | GUCY1A2, GUCY1A3,
GUCY1B3, PRKG1, TNNI1, TTN, TCAP, PDE5A, TRIM28, | Cytoplasm |
| M2 | CEBPD, MXD4, CLU,
DDIT3, E2F4, PEG10, GAPDH, MYCBP, LIN28B, LGALS1, MAX, MYC, NFYB,
NFYC, NME2, FBXW7, USP28, KIAA1524, S100A7, SMARCA2, ZFP36L1,
CDC73, TRRAP, CDCA7, MINA, PARP10, PIAS1, DDX18, HN1L, MTDH | Nucleus |
| M3 | TRIM28, PRDX3,
CKS1B, DNM1, MTOR, MYCBP, GLS, HDAC1, HMGCS2, HSPD1, HSPE1, MAOB,
MYC, NME1, NME2, GPAM, BCAT1, SHMT1, TFRC, UBE3A, STAM, TRRAP,
CAPZB, TMEM126A, NDUFAF2 | Mitochondrion |
| M4 | CKS2, DLAT, EIF5A,
ELAVL1, FDXR, GPX1, GSTP1, HMMR, HNRNPK, TRIAP1, PHB, DDIT4,
BCKDHB, TP53a, BCL2L14,
AIFM2 | Mitochondrion |
| M5 | CPB2, CRYBB2, CTSG,
A2M, F2a, F5a, F7a, F8a, F9a, F10, F11, F13A1a, F13B, FGAa, FGBa, FGG, ANGPT1, SERPIND1,
SERPINC1a, SERPINA5,
PROCa, PROS1a, TFPIa, THBDa | Extracell |
| M6 | CXCL13, FGF2, GDF2,
SERPINF1, SERPINA4, BGLAP, TNFSF11 | Extracell |
| M7 | FAS, FASLG, BID,
MAP2K4, TNFSF10, FADD, TNFRSF10D, TNFRSF10C, TNFRSF10B,
TNFRSF10A | Plasma
membrane |
The functional characteristics of the
RRMs
The correlation between RRMs and VTE was
investigated by a literature review and functional annotation
analysis (28). The functional
categories were mostly enriched in the RRMs were coagulation
cascades, blood circulation, signal transduction and cell death
(Table II), and the coagulation
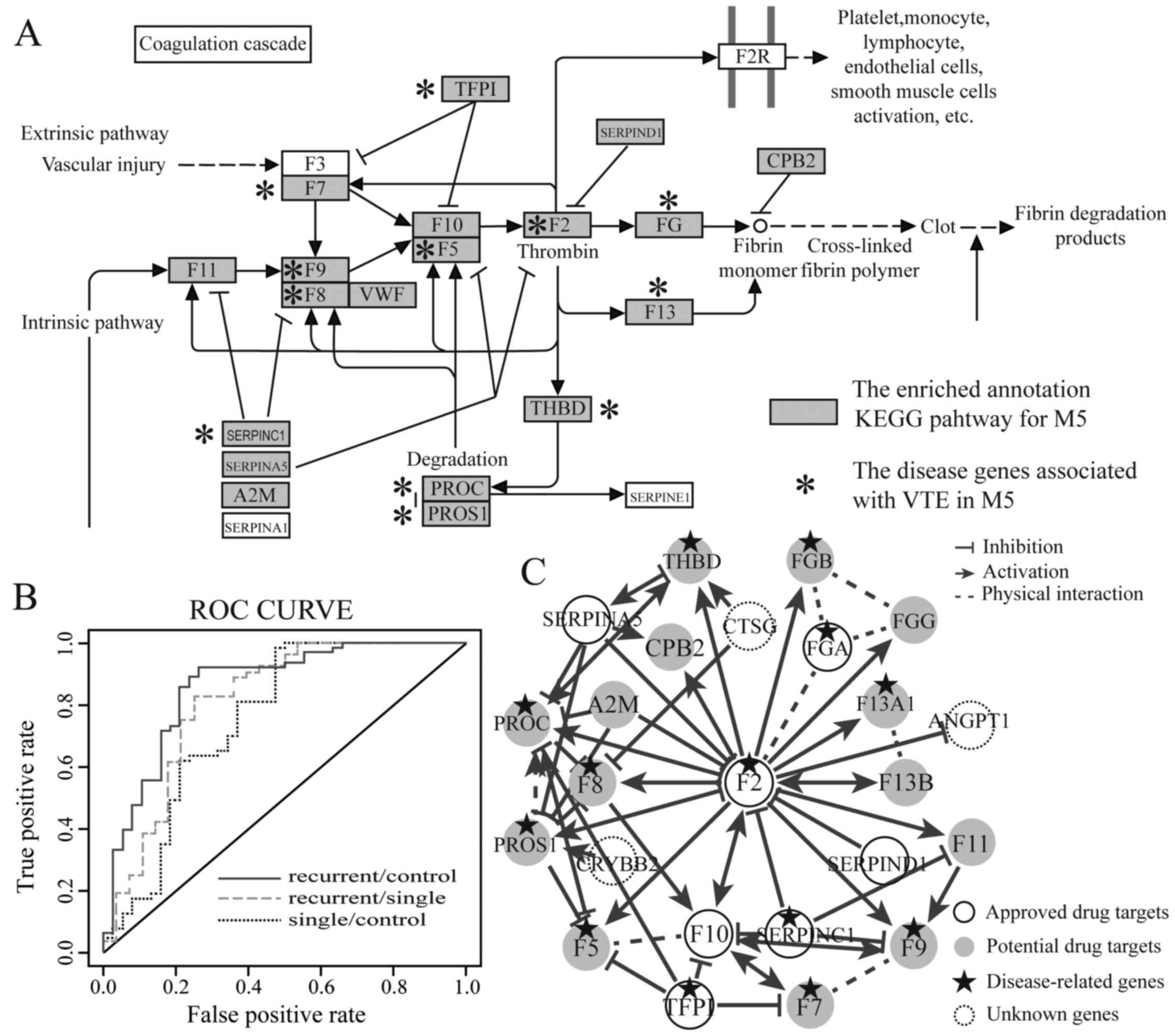
cascades pathway, apoptosis and p53 signaling pathway (Fig. 1A).
 | Table IIThe functional category of recurrent
risk modules. |
Table II
The functional category of recurrent
risk modules.
| Module | Functional category
|
|---|
| Cell death | Coagulation
cascades circulation | Blood | Signal
transduction |
|---|
| M1 | – | – | √ | – |
| M2 | – | – | – | √ |
| M3 | √ | – | – | – |
| M4 | √ | – | – | – |
| M5 | – | √ | √ | √ |
| M6 | √ | – | – | – |
| M7 | √ | – | – | √ |
Previous studies have demonstrated that these
annotated functional categories and pathways are associated with
VTE. For example, it has been shown that the alterations in both
the complement and coagulation systems are associated with VTE
(29,30). The coagulation cascade is a key
component of the hemostatic process and limits blood loss in
response to tissue injury. Derangements in this process can lead to
VTE (31). Gawaz and Vogel
indicated that apoptosis plays a critical role in platelet life and
is implicated in the pathogenicity of thrombosis (32).
Classifying the capacity of RRMs
With the rapid development of systems biology, the
modular analysis of biological networks may allow for the better
understanding of the molecular mechanisms responsible for disease
and may aid researchers in the identification of multiple module
biomarkers (33). SVM can be used
to identify recurrent-related patients and evaluate the classifying
performance with RRMs as a classification feature. The average
expression values of RRMs were the classification features of SVM
classifier to distinguish patients with different stages of VTE. We
performed a 5-fold cross-validation on VTE stage samples to
evaluate the discriminative power of RRMs. The AUC values of the
5-fold cross-validation for each two-stage samples
(recurrent/control, recurrent/single and single/control) were
evaluated, respectively (Fig.
1B). As a result, the high discriminative power was obtained
using RRMs as classification features (Table III).
 | Table IIIFive-fold cross-validation for the
RRMs of VTE in the HSSNs. |
Table III
Five-fold cross-validation for the
RRMs of VTE in the HSSNs.
| Classification
features | Datasets | AUC | TPR | FPR |
|---|
| RRMs |
Recurrent/control | 0.8584 | 0.9206 | 0.2632 |
|
Recurrent/single | 0.8118 | 0.8269 | 0.25 |
| Single/control | 0.7561 | 0.9841 | 0.4737 |
| M4 |
Recurrent/control | 0.8084 | 0.75 | 0.1842 |
| M5 |
Recurrent/control | 0.868 | 0.8254 | 0.1842 |
| M7 |
Recurrent/control | 0.8129 | 0.8571 | 0.1579 |
In order to further evaluate the effectiveness of
our RRMs, the recurrent/control samples were used for training, and
the recurrent/single and single/control samples were used for
testing in the GSE19151 dataset. The AUC values derived from this
model were 0.9062 and 0.7577, respectively (Table IV). The high discriminative power
suggested that the patients could be effectively distinguished
using RRMs as classification features.
 | Table IVClassification performances for the
RRMs of VTE in HSSNs. |
Table IV
Classification performances for the
RRMs of VTE in HSSNs.
| Classification
features | Train set | Test set | AUC | TPR | FPR |
|---|
| RRMs |
Recurrent/control |
Recurrent/single | 0.9062 | 0.9048 | 0.1875 |
|
Recurrent/control | Single/control | 0.7577 | 0.9683 | 0.4737 |
| M5 |
Recurrent/control |
Recurrent/single | 0.872 | 0.9524 | 0.2188 |
|
Recurrent/control | Single/control | 0.7657 | 0.9524 | 0.4737 |
Furthermore, each RRM from the individual datasets
as classification features was used to construct a classifier to
recognize patients with recurrent disease. The discriminative power
of each RRM was evaluated between different stages of VTE patients
with the 5-fold across-validation. Moreover, the recurrent/control
samples were used for training and the single/control and
recurrent/single samples were used for testing in the GSE19151
dataset for each RRM as classification features. We found that the
RRMs M4, M5 and M7 had good classification performance to
discriminate patients wtih recurrent disease (Tables III and IV). In particular, RRM M5 had the best
discriminative power among the RRMs. This indicated that M5 may be
used to construct an individual classifier to efficiently
distinguish the patients with recurrent disease.
The robustness and stability of our
algorithm
Naive Bayes and random forests were used to evaluate
the robustness and stability of our algorithm, which also
considered the RRM expression values as the classification features
to confirm the high classification performance for VTE stages. The
Naive Bayes classifier is based on Bayes' theorem with independence
assumptions between predictors (34). Random forests are an ensemble
learning method for classification and regression that fits a
multitude of decision tree classifiers on various sub-samples of
the dataset and uses averaging to improve the predictive accuracy
(35). The RRMs had a high
classification performance and stability, by not only Naive Bayes
but also random forests (Table
V).
 | Table VFive-fold cross-validation for the
RRMs of VTE using Naive Bayes and Random Forest. |
Table V
Five-fold cross-validation for the
RRMs of VTE using Naive Bayes and Random Forest.
| Classifier | Features | Datasets | AUC | TPR | FPR |
|---|
| Naive Bayes | RRMs |
Recurrent/control | 0.8496 | 0.8571 | 0.2368 |
| Naive Bayes | M5 |
Recurrent/control | 0.8634 | 0.9048 | 0.2105 |
| Random Forest | RRMs |
Recurrent/control | 0.8375 | 0.7937 | 0.1842 |
| Random Forest | M5 |
Recurrent/control | 0.8488 | 0.8889 | 0.2368 |
To better evaluate the discriminative power of the
RRMs for other independent datasets, the recurrent/control samples
of GSE19151 were used for training, and the VT sample/normal
samples of GSE17078 were used for testing (Table VI). High AUC values were
presented in the VTE stage datasets (the AUC values were 0.88 to
0.83). The results indicated that the RRMs had a good
classification performance for the independent datasets. In a word,
the RRMs identified by our algorithm with high AUC values have a
strong discriminative power, robustness and stability.
 | Table VIClassification performances for the
RRMs for the independent datasets. |
Table VI
Classification performances for the
RRMs for the independent datasets.
| Classification
features | Train set | Test set | AUC | TPR | FPR |
|---|
| RRMs |
Recurrent/control | VT
sample/normal | 0.83 | 0.9683 | 0.3421 |
| M5 |
Recurrent/control | VT
sample/normal | 0.8828 | 1 | 0.3438 |
Comparison with average expression value
method
The expression correlation method was compared with
the average expression value method. The RRMs for VTE were
identified using the average expression value method. Supposing a
module M' contains G genes thus the average expression value
differential score S' was expressed as follows:
where Z and Z′ denote the expression values of genes in two
distinct stages, respectively, U and V denote the numbers of
samples in two distinct stages, respectively. For a network module,
we calculated the real average expressed differential score S'.
Subsequently, one thousand random modules were constructed from the
same HSSN with the degree conserved. The random average expression
value differential scores S′1…S′1000 were also calculated. If the
random average expression value differential scores were
significantly less than the real average expressed differential
score, the module was considered as an average expression value
differential stage module (permutation test, p-values <0.05).
The p-values were adjusted using the Benjamini-Hochberg method. We
then defined the average expression value differential stage
modules existing in control/single, control/recurrent and
single/recurrent profiles as the average expression value disease
risk modules to improve the reproducibility.
However, we could not find the average expression
value disease risk modules among the VTE stage samples. The result
suggested that expression correlation method outperformed the
average expression value method, and may better reflect the
difference between VTE stage samples.
The drug target characteristics of the
RRMs
The DrugBank database is a comprehensive,
high-quality, freely accessible and online bioinformatics and
cheminformatics resource, which contains detailed information about
drugs and drug targets (36). The
drug targets associated with the pathogenesis of VTE were searched
from the DrugBank database and mapped into the RRMs. Nine approved
drug targets were identified in the RRMs (Table VII). In particular, seven drug
targets for VTE treatment are in M5 and are targeted by many of the
drugs. M5 has the significant enrichment characteristics for the
drug targets (p-value <0.01).
 | Table VIIThe approved drug targets and
therapeutic drugs of VTE in the recurrent risk modules. |
Table VII
The approved drug targets and
therapeutic drugs of VTE in the recurrent risk modules.
| Modules | Drug targets | Drug |
|---|
| M1 | PDE5A | Dipyridamole |
| M5 | F2 | Argatroban,
bivalirudin, dabigatran etexilate, lepirudin, warfarin |
| M5 | F10 | Apixaban,
certoparin sodium, edoxaban, enoxaparin sodium, fondaparinux
sodium, heparin, otamixaban, rivaroxaban |
| M5 | FGA | Alteplase,
anistreplase, reteplase |
| M5 | SERPIND1 | Ardeparin sodium,
sulodexide |
| M5 | SERPINC1 | Ardeparin sodium,
certoparin sodium, dalteparin, dalteparin sodium, enoxaparin
sodium, fondaparinux sodium, heparin, nadroparin, nadroparin
calcium, sulodexide, tinzaparin sodium |
| M5 | SERPINA5 | Urokinase |
| M5 | TFPI | Dalteparin
sodium |
| M6 | FGF2 | Phenprocoumon |
The drug target [coagulation factor II, thrombin
(F2)] of warfarin plays crucial role in M5, interacting with 19 VTE
disease-related genes (Fig. 1C).
In addition, F2 is the drug target of other therapeutic drugs for
VTE such as argatroban, dabigatran and bivalirudin. Seven of the
genes that interacted with F2 were the approved therapeutic drug
targets for VTE, revealing that these genes played an important
role in the treatment of recurrent VTE. Moreover, our results
revealed that M5 not only had a high discriminative power, but also
had significant function associated with the pathogenesis of VTE.
Therefore, we defined the genes coagulation factor XI (F11),
alpha-2-macroglobulin (A2M), coagulation factor XIII A chain
(F13A1), coagulation factor VIII (F8), coagulation factor IX (F9),
coagulation factor VII (F7), protein C, inactivator of coagulation
factors Va and VIIIa (PROC), coagulation factor XIII B chain
(F13B), fibrinogen gamma chain (FGG), thrombomodulin (THBD),
coagulation factor V (F5), fibrinogen beta chain (FGB), protein S
(alpha) (PROS1), carboxypeptidase B2 (CPB2) in M5 and the RRM M5
itself as potential drug targets (Fig. 1C). These potential drug targets
would be promising to further research, exploiting new therapeutic
drugs to treat VTE patients.
Discussion
In this study, we developed a novel strategy which
combined both dynamic gene expression and the human subcellular
signaling networks to identify RRMs. Seven RRMs in the HSSNs were
identified and mainly annotated in blood coagulation and apoptosis,
which were strongly associated with the pathogenesis of VTE
(29,30). Norris pointed out that the
activation of coagulation may predispose affected individuals to
thrombosis (37). White and Kile
indicated that the platelet activation responses contained
components of apoptotic machinery which are of particular
importance for thrombus formation, which highlighted a potential
role in apoptotic processes of thrombosis (38). Moreover, our algorithm had a good
classification performance, robustness and stability. The RRMs had
strong discriminative power to distinguish patients in different
stages of disease development. In particulaar, the M5 had the best
performance. It was suggested that the M5 was effective in
elucidating the pathogenesis of VTE, which holds promise for the
development of therapeutic strategies for the treatment of VTE.
Warfarin is an effective antithrombotic agent, but
noncompliance and cessation with warfarin therapy for over a year
are associated with a higher risk of recurrent VTE and the
therapeutic range is narrow (39). Our results revealed that nine
approved drug targets in the RRMs were targeted not only by
warfarin, but also by the other therapeutic drugs (such as apixaban
and dabigatran) for VTE. In particular, seven approved drug target
genes in the M5 were targeted by three types of drugs
(anticoagulant drugs, thrombolytic drugs and antiplatelet drugs)
for the treatment of VTE. For example, F2 is the antithrombotic and
anticoagulant drug target of not only warfarin, but also
dabigatran, argatroban, lepirudin and bivalirudin. Coagulation
factor X (F10) is the common anticoagulant drug target of apixaban,
edoxaban, heparin and enoxaparin sodium.
Among the identified RRM modules, M5 had the best
discriminative power. Thirteen disease-related genes in M5 are
associated with the pathogenesis of VTE from OMIM (http://www.omim.org/) and Genetic Association Database
(http://geneticassociationdb.nih.gov/)
(Table I). The genes, serpin
family C member 1 (SERPINC1), F2, F7 and fibrinogen alpha chain
(FGA), were not only the disease-related genes, but also the
approved drug targets. The gene SERPINC1 encodes antithrombin III,
which is a major risk factor of VTE, acting as an inhibitor of
thrombin and other coagulation proteinases (40). Many new drugs, such as dabigatran
and argatroban, targeting F2 may be used as effective anticoagulats
for the treatment of recurrent VTE (41). The genes, F10, serpin family A
member 5 (SERPINA5) and serpin family D member 1 (SERPIND1), were
the approved drug targets. The new oral anticoagulant drugs,
rivaroxaban, apixaban and edoxaban, which are direct inhibitors of
F10, have been the approved drugs or have entered clinical practice
for different clinical indications (1). The gene, F5, F7, F8, F9 and PROC,
are both disease-related genes and the potential drug targets. The
genes, F5, F9 and PROC, are involved in the blood coagulation
pathway. The dysfunction of these is associated with VTE (42). The combination of F5 with F10
activates F2 to form the effector enzyme of the coagulation cascade
(43), and the dysfunction of
this contributes to VTE (4,44).
Multiple therapeutic modes and options exist for VTE
treatment. The prevention and initial therapy of VTE usually begin
with the administration of rapidly acting parenteral
anticoagulants, such as heparin (45). Furthermore, vitamin K antagonists
(e.g., warfarin) are the commonly available oral anticoagulant
therapy used for decades (46).
However, the risk of recurrent VTE also exists following treatment
with heparin and warfarin (47,48). In the study by Deitelzweig et
al, it was demonstrated that the cessation of warfarin therapy
in 1,027 patients within three months led to recurrence in 915
(89.1%) patients (48). In order
to treat the recurrence, alternative drugs and drug combination
therapy should be considered, as well as the drugs for extended VTE
treatment (10,49). Long-term anticoagulation with low
molecular weight heparin (LMWH) compared with warfarin appears to
be more effective for the prevention of recurrent VTE (50). Compared with warfarin, dabigatran
does not require close monitoring and the adjusting of dosage for
extended therapy and has a lower rate of major or clinically
relevant bleeding (51). Pillai
et al described that therapy with dalteparin combined with
warfarin effectively prevented recurrent VTE (49). Furthermore, these effective
therapeutic drugs consistently target the crucial drug targets in
M5, which is significantly associated with VTE.
In total, 87.5% of the genes in the M5 were
significantly enriched in blood coagulation pathway. The analysis
of drug targets in M5 may contribute to elucidating the
pathogenesis of VTE. Furthermore, multiple drug targets and multi
therapeutic drugs should be considered for VTE therapy. New
therapeutic drugs may be explored by the analysis of VTE potential
drug targets, such as F5, F7, F8, F9, which may hold promise for
improving the safety and effectiveness of VTE treatment (2). In addition, the M1 and M7 were
significantly enriched in the VTE-related pathway, which may bring
new insight into VTE research, as well as the therapeutic drug
targets identification (Table
VIII). For instance, a recent study indicated that
atherosclerosis, thrombosis and myocardial infarction were
significantly associated with vascular smooth muscle contraction
and the apoptotic pathway (52).
Freedman suggested that GUCY1A3 mutations were related to
thrombosis (53). Zeng et
al indicated that miR-20a regulates the PRKG1 gene, thereby
promoting the proliferation, and contributes to thrombosis
(54). TNF receptor superfamily
members 10B, 10C, 10D and TNFSF10 linked to inflammation and
coagulation and contribute to VTE (55). Although the potential drug targets
identified by our computational biology algorithm are just
validated by the literature review theoretically, the treatment
effectiveness of these should be validated by experimental
studies.
 | Table VIIIThe enriched KEGG pathway for
RRMs. |
Table VIII
The enriched KEGG pathway for
RRMs.
| RRM | KEGG pathway | The enriched
genes | Benjamini |
|---|
| M1 | Long-term
depression | GUCY1A2, GUCY1A3,
GUCY1B3, PRKG1 | 1.64E-04 |
| M1 | Gap junction | GUCY1A2, GUCY1A3,
GUCY1B3, PRKG1 | 1.77E-04 |
| M1 | Vascular smooth
muscle contraction | GUCY1A2, GUCY1A3,
GUCY1B3, PRKG1 | 2.35E-04 |
| M1 | Purine
metabolism | PDE5A, GUCY1A2,
GUCY1A3, GUCY1B3 | 4.47E-04 |
| M5 | Complement and
coagulation cascades | F11, A2M, F10,
F13A1, F8, F9, F7, PROC, F13B, FGG, THBD, F5, FGA, FGB, SERPINA5,
F2, SERPINC1, TFPI, SERPIND1, PROS1, CPB2 | 2.73E-37 |
| M7 | Apoptosis | TNFRSF10A, BID,
TNFSF10, TNFRSF10C, TNFRSF10B, TNFRSF10D, FASLG, FADD, FAS | 9.41E-13 |
| M7 | Natural killer cell
mediated cytotoxicity | TNFRSF10A, BID,
TNFSF10, TNFRSF10C, TNFRSF10B, TNFRSF10D, FASLG, FAS | 2.47E-09 |
| M7 | Cytokine-cytokine
receptor interaction | TNFRSF10A, TNFSF10,
TNFRSF10C, TNFRSF10B TNFRSF10D, FASLG, FAS | 8.69E-06 |
| M7 | p53 signaling
pathway | BID, TNFRSF10B,
FAS | 0.02949 |
It is noteworthy that the differentially expressed
genes were commonly considered as the classification features to
discriminate patients or disease states. However, the
differentially expressed genes were not identified between
different stages of the VTE samples, which could not be used to
identify patients with recurrent disease. The RRMs identified with
the expression dynamic changes of the gene had a better advantage
for recurrent VTE treatment.
In conclusion, signaling proteins or gene products
may play a unique role in biological activity and may be intriguing
to investigators in both basic studies and drug development. The
RRMs based on the signaling network were identified with dynamic
changes related to VTE recurrence from the system level. The RRMs
(particularly M5) not only implied a good discriminative power to
distinguish recurrent patients, but also had the crucial drug
targets for VTE treatment. Most genes in M5 were proofed associated
with VTE by the literature review. This suggested that the M5 may
provide an effective guidance for the investigation of the
mechanisms responsible for recurrent VTE, and for therapeutic drug
screening and pharmaceutical studies.
Acknowledgments
This study was supported in part by the Science and
Technology Research Project of the National Natural Science
Foundation of China (grant nos. 61272388 and 31301040); the Natural
Science Foundation of Heilongjiang Province (grant no. F201237);
the Science and Technology Research Project of the Heilongjiang
Ministry of Education (grant no. 12541476); the Science Foundation
of Heilongjiang Health Department (grant no. 2012-810); and the
Master Innovation Funds of Heilongjiang Province (grant no.
YJSCX2014-18HYD).
References
|
1
|
Kyrle PA and Eichinger S: Clinical scores
to predict recurrence risk of venous thromboembolism. Thromb
Haemost. 108:1061–1064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Becattini C and Manina G: Long-term
treatment of venous thromboembolism. Curr Vasc Pharmacol.
12:384–392. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hogg K, Wells PS and Gandara E: The
diagnosis of venous thromboembolism. Semin Thromb Hemost.
38:691–701. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pabinger I, Thaler J and Ay C: Biomarkers
for prediction of venous thromboembolism in cancer. Blood.
122:2011–2018. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Perry JR: Thromboembolic disease in
patients with high-grade glioma. Neuro Oncol. 14(Suppl 4):
iv73–iv80. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mackman N: New insights into the
mechanisms of venous thrombosis. J Clin Invest. 122:2331–2336.
2012. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wells PS, Forgie MA and Rodger MA:
Treatment of venous thromboembolism. JAMA. 311:717–728. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kearon C: Long-term anticoagulation for
venous thromboembolism: Duration of treatment and management of
warfarin therapy. Clin Chest Med. 31:719–730. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zhu T, Martinez I and Emmerich J: Venous
thromboembolism: Risk factors for recurrence. Arterioscler Thromb
Vasc Biol. 29:298–310. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sharifi M, Bay C, Schwartz F and Skrocki
L: Safe-dose thrombolysis plus rivaroxaban for moderate and severe
pulmonary embolism: Drip, drug, and discharge. Clin Cardiol.
37:78–82. 2014. View Article : Google Scholar
|
|
11
|
Barabási AL and Oltvai ZN: Network
biology: Understanding the cell's functional organization. Nat Rev
Genet. 5:101–113. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Suthram S, Dudley JT, Chiang AP, Chen R,
Hastie TJ and Butte AJ: Network-based elucidation of human disease
similarities reveals common functional modules enriched for
pluripotent drug targets. PLoS Comput Biol. 6:e10006622010.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Laenen G, Thorrez L, Börnigen D and Moreau
Y: Finding the targets of a drug by integration of gene expression
data with a protein interaction network. Mol Biosyst. 9:1676–1685.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Martinelli I, De Stefano V and Mannucci
PM: Inherited risk factors for venous thromboembolism. Nat Rev
Cardiol. 11:140–156. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Newman RH, Hu J, Rho HS, Xie Z, Woodard C,
Neiswinger J, Cooper C, Shirley M, Clark HM, Hu S, et al:
Construction of human activity-based phosphorylation networks. Mol
Syst Biol. 9:6552013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Tibiche C, Fu C, Kaneko T, Moran MF,
Schiller MR, Li SS and Wang E: The human phosphotyrosine signaling
network: Evolution and hotspots of hijacking in cancer. Genome Res.
22:1222–1230. 2012. View Article : Google Scholar :
|
|
17
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al The Gene Ontology Consortium: Gene ontology: Tool for the
unification of biology. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Magrane M and Consortium U: UniProt
Knowledgebase: A hub of integrated protein data. Database (Oxford).
2011:bar0092011. View Article : Google Scholar
|
|
19
|
Samsa G, Matchar DB, Dolor RJ, Wiklund I,
Hedner E, Wygant G, Hauch O, Marple CB and Edwards R: A new
instrument for measuring anticoagulation-related quality of life:
development and preliminary validation. Health Qual Life Outcomes.
2:222004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lewis DA, Stashenko GJ, Akay OM, Price LI,
Owzar K, Ginsburg GS, Chi JT and Ortel TL: Whole blood gene
expression analyses in patients with single versus recurrent venous
thromboembolism. Thromb Res. 128:536–540. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Reimand J, Tooming L, Peterson H, Adler P
and Vilo J: GraphWeb: Mining heterogeneous biological networks for
gene modules with functional significance. Nucleic Acids Res.
36:W452–W459. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bewick V, Cheek L and Ball J: Statistics
review 10: Further nonparametric methods. Crit Care. 8:196–199.
2004. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
24
|
Huang W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar :
|
|
25
|
Curtis RK, Oresic M and Vidal-Puig A:
Pathways to the analysis of microarray data. Trends Biotechnol.
23:429–435. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Mukherjee K, Abhipriya, Vidyarthi AS and
Pandey DM: SVM based model generation for binding site prediction
on helix turn helix motif type of transcription factors in
eukaryotes. Bioinformation. 9:500–505. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bradley AP: The use of the area under the
ROC curve in the evaluation of machine learning algorithms. Pattern
Recognition. 30:1145–1159. 1997. View Article : Google Scholar
|
|
28
|
Dennis G Jr, Sherman BT, Hosack DA, Yang
J, Gao W, Lane HC and Lempicki RA: DAVID: Database for annotation,
visualization, and integrated discovery. Genome Biol. 4:P32003.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Nojima J, Suehisa E, Kuratsune H, Machii
T, Koike T, Kitani T, Kanakura Y and Amino N: Platelet activation
induced by combined effects of anticardiolipin and lupus
anticoagulant IgG antibodies in patients with systemic lupus
erythematosus - possible association with thrombotic and
thrombocytopenic complications. Thromb Haemost. 81:436–441.
1999.PubMed/NCBI
|
|
30
|
Inanç M, Donohoe S, Ravirajan CT,
Radway-Bright EL, Mackie I, Machin S and Isenberg DA:
Anti-beta2-glycoprotein I, anti-prothrombin and anticardiolipin
antibodies in a longitudinal study of patients with systemic lupus
erythematosus and the antiphospholipid syndrome. Br J Rheumatol.
37:1089–1094. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Markiewski MM, Nilsson B, Ekdahl KN,
Mollnes TE and Lambris JD: Complement and coagulation: Strangers or
partners in crime? Trends Immunol. 28:184–192. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Gawaz M and Vogel S: Platelets in tissue
repair: Control of apoptosis and interactions with regenerative
cells. Blood. 122:2550–2554. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Meunier D, Fonlupt P, Saive AL, Plailly J,
Ravel N and Royet JP: Modular structure of functional networks in
olfactory memory. Neuroimage. 95:264–275. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Maruyama O: Heterodimeric protein complex
identification by naïve Bayes classifiers. BMC Bioinformatics.
14:3472013. View Article : Google Scholar
|
|
35
|
Azar AT, Elshazly HI, Hassanien AE and
Elkorany AM: A random forest classifier for lymph diseases. Comput
Methods Programs Biomed. 113:465–473. 2014. View Article : Google Scholar
|
|
36
|
Wishart DS, Knox C, Guo AC, Cheng D,
Shrivastava S, Tzur D, Gautam B and Hassanali M: DrugBank: A
knowledgebase for drugs, drug actions and drug targets. Nucleic
Acids Res. 36:D901–D906. 2008. View Article : Google Scholar :
|
|
37
|
Norris LA: Blood coagulation. Best Pract
Res Clin Obstet Gynaecol. 17:369–383. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
White MJ and Kile BT: Apoptotic processes
in megakaryocytes and platelets. Semin Hematol. 47:227–234. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Thomé S: ACP Journal Club. Edoxaban was
noninferior to warfarin for preventing recurrent venous
thromboembolism, with less bleeding. Ann Intern Med. 160:JC42014.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Lane DA, Olds RJ and Thein SL:
Antithrombin III: Summary of first database update. Nucleic Acids
Res. 22:3556–3559. 1994.PubMed/NCBI
|
|
41
|
Hull RD and Gersh MH: The current
landscape of treatment options for venous thromboembolism: A focus
on novel oral anticoagulants. Curr Med Res Opin. 31:197–210. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Eikelboom JW, Zelenkofske SL and Rusconi
CP: Coagulation factor IXa as a target for treatment and
prophylaxis of venous thromboembolism. Arterioscler Thromb Vasc
Biol. 30:382–387. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cripe LD, Moore KD and Kane WH: Structure
of the gene for human coagulation factor V. Biochemistry.
31:3777–3785. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hoyer LW and Hemophilia A: Hemophilia A. N
Engl J Med. 330:38–47. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Geerts WH, Bergqvist D, Pineo GF, Heit JA,
Samama CM, Lassen MR and Colwell CW; American College of Chest
Physicians: Prevention of venous thromboembolism: American College
of Chest Physicians Evidence-Based Clinical Practice Guidelines
(8th Edition). Chest. 133(Suppl 6): 381S–453S. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Abo-Salem E and Becker R: Transitioning to
and from the novel oral anticoagulants: A management strategy for
clinicians. J Thromb Thrombolysis. 37:372–379. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chee CE, Ashrani AA, Marks RS, Petterson
TM, Bailey KR, Melton LJ III and Heit JA: Predictors of venous
thromboem-bolism recurrence and bleeding among active cancer
patients: A population-based cohort study. Blood. 123:3972–3978.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Deitelzweig SB, Lin J, Kreilick C, Hussein
M and Battleman D: Warfarin therapy in patients with venous
thromboembolism: Patterns of use and predictors of clinical
outcomes. Adv Ther. 27:623–633. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Pillai AR, Olujohungbe A, Evans MR, Main
NJ and Hunt BJ: The management of recurrent VTE in cancer patients
receiving therapeutic anticoagulation: The use of dual
anticoagulant therapy combined with an IVC filter. Blood Coagul
Fibrinolysis. 21:766–769. 2010.PubMed/NCBI
|
|
50
|
Akl EA, Kahale L, Barba M, Neumann I,
Labedi N, Terrenato I, Sperati F, Muti P and Schünemann H:
Anticoagulation for the long-term treatment of venous
thromboembolism in patients with cancer. Cochrane Database Syst
Rev. 7:CD0066502014.
|
|
51
|
Schulman S, Kearon C, Kakkar AK, Schellong
S, Eriksson H, Baanstra D, Kvamme AM, Friedman J, Mismetti P and
Goldhaber SZ; RE-MEDY Trial Investigators; RE-SONATE Trial
Investigators: Extended use of dabigatran, warfarin, or placebo in
venous thromboembolism. N Engl J Med. 368:709–718. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yu Y, Qin J, Chen D, Wang H, Wang J and Yu
Y: Chronic cardiovascular disease-associated gene network analysis
in human umbilical vein endothelial cells exposed to
2,3,7,8-tetrachlorodibenzo-p-dioxin. Cardiovasc Toxicol.
15:157–171. 2015. View Article : Google Scholar
|
|
53
|
Freedman JE: Inherited dysfunctional
nitric oxide signaling and the pathobiology of atherothrombotic
disease. Circ Res. 114:1372–1373. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zeng Y, Pan Y, Liu H, Kang K, Wu Y, Hui G,
Peng W, Ramchandran R, Raj JU and Gou D: MiR-20a regulates the
PRKG1 gene by targeting its coding region in pulmonary arterial
smooth muscle cells. FEBS Lett. 588:4677–4685. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Lv W, Duan Q, Wang L, Gong Z, Yang F and
Song Y: Gene expression levels of cytokines in peripheral blood
mononuclear cells from patients with pulmonary embolism. Mol Med
Rep. 7:1245–1250. 2013.PubMed/NCBI
|