Introduction
Medulloblastoma (MB) is the most common pediatric
malignant brain tumor. Historically, MB has been classified into
many variants based on its histopathology and cytogenetic
biomarkers (1). Different
pathways have been implicated in the development of MB and the
regulation of the malignant phenotype of MB cells (2–5).
Patients with high-risk or recurrent MB respond poorly to current
therapies and they exhibit a higher related mortality (6). Thus, promising clinical targets are
required for MB to facilitate the development of novel therapeutics
(7).
At present, several targeted molecular therapies are
under clinical investigation in patients with MB (6,8).
To further understand this malignant disease, high-throughput and
genome-wide microarrays have been applied for molecular
classification (9), thereby
providing novel insight into the underlying biological mechanisms
of MB (10). Various cell lines
are available for use as experimental models of this neoplastic
brain disease (11). Genetic data
related to MB are available in an international functional genomics
data repository, i.e., the Gene Expression Omnibus (GEO), which
were derived from the MB cell line, Daoy (12,13). Thus, the study of a combination of
genomic scale analyses of the data available in GEO and other
whole-genome microarrays may facilitate the identification of
underlying biomarkers, which may provide insight into the
pathogenesis of brain tumors (14–16).
In the present study, various MB microarray data
were extracted and compared. Significant genes that had similar
expression patterns in MB according to different microarray systems
were selected following gene annotation. Enrichment analysis of the
gene set was also performed to elucidate the biological pathways
and processes that are affected in patients with MB. Archived and
formalin-fixed/paraffin-embedded human MB specimens were also used
to validate the significant candidates. A crucial molecular pathway
related to MB was identified by this whole-genome survey.
Materials and methods
Patients and MB cell lines
The archived specimens were derived from previously
formalin-fixed/paraffin-embedded biopsies, which were confirmed as
MB by the Department of Pathology at Cathay General Hospital,
Taipei, Taiwan, and their characteristics are described in Table I. The Institutional Review Board
of Cathay General Hospital approved this study. Written form of
consent was obtained from the participants or their guardians. MB
cell lines, i.e., Daoy (HTB-186™), D341 Med (HTB-187™) and VGH-Med
(60375), were purchased from the American Type Culture Collection
(Manassas, VA, USA) and the Bioresource Collection and Research
Center (Hsinchu, Taiwan). All the cultured cells were maintained in
the indicated medium with 10% (for Daoy) or 20% (for D341 Med and
VGH-Med) fetal bovine serum (Biological Industries, Kibbutz Beit
Haemek, Israel) according to routine protocols. Briefly, the
adherent Daoy and the suspended D341 Med cells were cultivated in
Eagle's minimum essential medium (Cat. no. 41500-034), whereas the
adherent VGH-Med cells were cultured in Dulbecco's modified Eagle's
medium (12100-046) (both from Life Technologies, Grand Island, NY,
USA).
 | Table ICharacteristics of patients with
medulloblastoma. |
Table I
Characteristics of patients with
medulloblastoma.
| Patient no. | Gender/onset age
(years) | Symptom | Image analysis | Pathology | Outcome |
|---|
| p't 19 | F/8 | Dizziness only |
2*2*2 tumor
(cm3) | MB,
desmoplastic | Deceased, due to
meta in 2003 |
| p't 31 | M/6 | HA, vomiting, neck
tilting, dysmetria | R't CT, H(−) | MB,
hemispheric | Still alive in Aug,
2009 |
| p't 69 | M/9 | Vomiting, HA for 1
week, nystagmus(+), papilledema(−) | CT, H | MB | Still alive in Feb,
2009 |
| p't 83 | M/46 | Dizziness, HA,
ataxia for over 20 days, slurred speech | L't HT, CHH | MB | Still alive in Feb,
2009 |
Significantly and differentially
expressed genes in MB according to microarray hybridizations
Different microarray systems were used in the
analyses, as shown in Fig. 1.
Briefly, 15 (GSE7578) and 3 (GSE22139) expression datasets for Daoy
cells (from accession series GSE7578 and GSE22139) and human normal
brains (from accession series accession series GSE30563), which
were obtained using Affymetrix microarrays (Human Genome U133 Plus
2.0 array, HG-U133 Plus 2; Affymetrix, Santa Clara, CA, USA), were
extracted from the GEO website (http://www.ncbi.nlm.nih.gov/geo). The differentially
expressed genes in MB were also determined based on comparisons
with MB cell lines (Daoy and VGH-Med) and various normal brain
tissues (two males aged 47 and 55 years, Cat. no. 636530; Clontech,
Mountain View, CA, USA; 13 males and 10 females aged 23–86 years,
Cat. no. AM7000; Ambion, Austin, TX, USA) using an Agilent Human
Whole Genome Oligo 4×44K microarray system (Agilent Technologies,
Santa Clara, CA, USA) according to the manufacturer's standard
procedures. We deposited the Agilent microarray data onto a public
domain (GSE74947). Common genes with potentially significant
changes (>1.50- or <0.67-fold) and clear expression trends in
MB were compared and extracted from the above-mentioned Affymetrix
and Agilent microarray systems, as previously described (17,18). The minimized difference in
expression (GSE7578 vs. GSE30563; GSE22139 vs. GSE30563;
Daoy/VGH-Med vs. normal brains from Clontech/Ambion) between
separate microarrays (standard deviation <0.75) was used to
filter genes with significant differences (p<0.05, Student's
t-test) in MB (19), and the top
genes obtained were annotated based on their gene ontology (GO)
(http://david.abcc.ncifcrf.gov) (20). Finally, enrichment analysis
(http://compbio.charite.de) was performed
to select the significant GO terms (21,22).
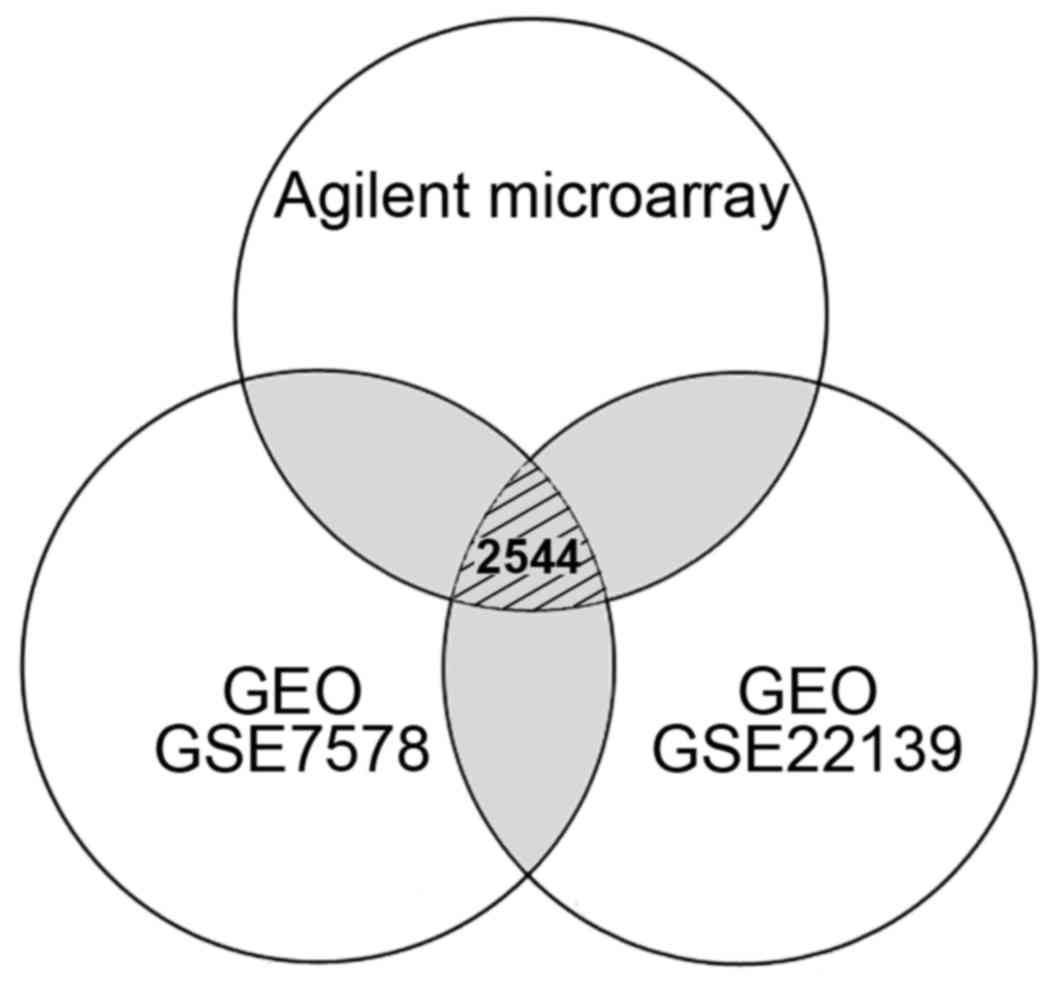 | Figure 1Venn diagram indicating the number of
differentially expressed genes in medulloblastoma from the
comparisons of different microarray systems. A total of 4,596
(1,656 up- and 2,940 downregulated) genes from Agilent microarray
system, 12,262 (8,306 up- and 3,956 downregulated) genes from GEO
GSE7578, and 14,257 (11,274 up- and 2,983 downregulated) genes from
GEO GSE22139 were respectively selected according to their
differential expression. Gray regions indicated the genes repeated
in different experiments. Only 2,544 genes with consistent
expression level differences were determined by comparing the data
obtained from Agilent and Affymetrix (GEO GSE7578 and GSE22139)
microarrays. GEO, Gene Expression Omnibus; GSE, GEO series. |
Relative gene expression levels of
synaptosomal-associated protein 25 (SNAP25) in MB cells
Total RNA from the normal brain tissue purchased
from Clontech (Cat. no. 636530) was also used for gene
quantification. Cellular RNA was extracted from each MB cell line
using TRIzol reagent according to the manufacturer's instructions
(Invitrogen, Carlsbad, CA, USA). The expression levels of total
SNAP25 or its two variants (variant 1: NM_003081, and variant 2:
NM_130811) in each MB cell line relative to those in the normal
brain were determined by reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) in the presence of specific
TaqMan probes and TaqMan Master Mix (Roche Diagnostics, Mannheim,
Germany), according to the manufacturer's instructions. Briefly,
amplification primer pairs for the total and variant SNAP25s (the
locations are mapped in Fig. 2B)
and glyceraldehyde-3-phosphate dehydrogenase (GAPDH: NM_002046),
which was employed as an internal control, were designed using the
Roche website (http://www.roche-applied-science.com) (Table II). LightCycler (version 4.05;
Roche Diagnostics) was used to analyze the PCR kinetics. Each run
also included an appropriate and predetermined diluted human
reference cDNA (Clontech) as a positive control.
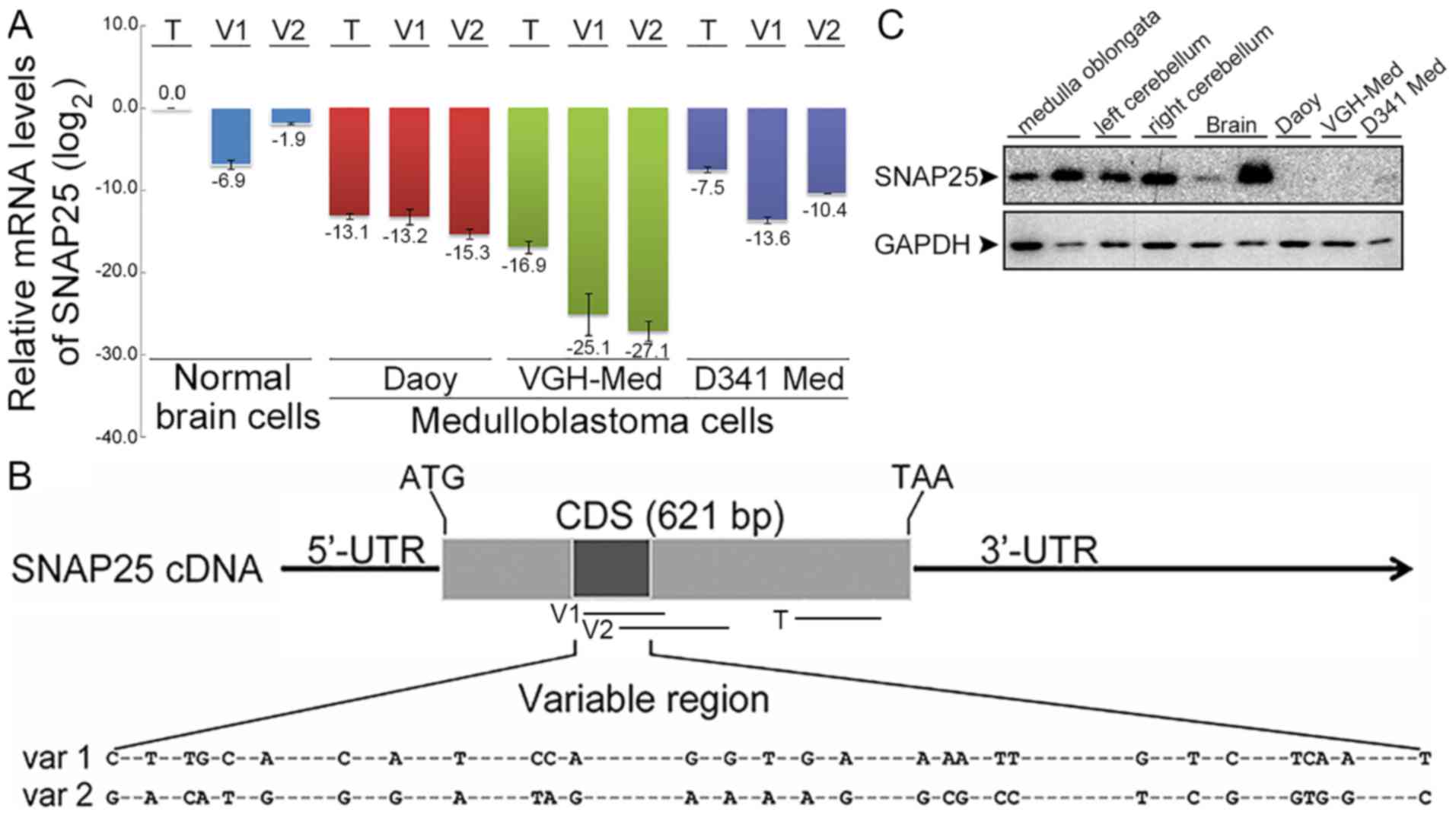 | Figure 2Expression differences of
synaptosomal-associated protein 25 (SNAP25) between medulloblastoma
and normal brain cells. (A) Comparative mRNA levels of total SNAP25
and variants by RT-qPCR. SNAP25 mRNA levels were quantified in
three medulloblastoma (MB) cell lines (Daoy cells: sonic hedgehog
subgroup; VGH-Med; D341 Med: group 3 subgroup) and normal brain
cells. Each SNAP25 mRNA level in MB cell lines was normalized with
their endogenous glyceraldehyde-3-phosphate dehydrogenenase (GAPDH)
mRNA level and relative to normal brain cells. All data were then
log2 transformed [log2(total SNAP25 of normal
brains), 0]. (B) Map of primers for quantifications of total SNAP25
and variants. -, indicates the conserved sequences. (C) Endogenous
SNAP25 protein in MB cell lines. Protein levels of SNAP25 (25 kDa)
and GAPDH (36 kDa) were detected by western blot analysis. Lanes 1
and 2, normal medulla oblongata from BioChain and Abcam,
respectively; lanes 3 and 4, left and right cerebellum from
BioChain; lanes 5 and 6, normal brains from Abcam and ProSci; lanes
7 to 9, Daoy, VGH-Med and D341 Med MB cell lines. T, V1, and V2
indicated the amplified regions. T, total SNAP25; V1 or var 1,
variant 1; V2 or var 2, variant 2. CDS, coding sequence; 5′-UTR,
5′-untranslated region; 3′-UTR, 3′-untranslated region; ATG, start
codon; TAA, stop codon. |
 | Table IIPrimers and probes for used for
RT-qPCR. |
Table II
Primers and probes for used for
RT-qPCR.
| Gene name
(variant) | Primer
sequence | Accession no. | Universal probe
no. |
|---|
SNAP25
(Total) | F:
ATCGGGAACCTCCGTCAC | NM_003081 | #81 |
| R:
AATTCTGGTTTTGTTGGAATCAG | NM_130811 | |
SNAP25
(variant 1) | F:
GGCATGAACCATATCAACCA | NM_003081 | #75 |
| R:
TTTTGTAAGCATCACTTGATTTAAGC | | |
SNAP25
(variant 2) | F:
CTTTGTGTGTGTCCCTGTAACAA | NM_130811 | #20 |
| R:
GTTCGTCCACTACACGAGCA | | |
| GAPDH | F:
CTCTGCTCCTCCTGTTCGAC | NM_002046 | #60 |
| R:
ACGACCAAATCCGTTGACTC | | |
Protein levels of SNAP25 in MB cells
The protein levels of SNAP25 in MB cells were
immunodetected by standard western blot analysis. In addition, 6
protein extracts of normal medulla oblongata (female aged 82 years,
Cat. no. P1234057 from BioChain, San Leandro, CA, USA; Cat. no.
ab29859 from Abcam, Cambridge, UK), normal cerebellum (male aged 66
years, cat. no. P1234040; and female aged 70 years, Cat. no.
P1234041; both from BioChain), and normal brain (Cat. no. ab29466
from Abcam; female aged 38 years, Cat. no. 1303 from ProSci, Poway,
CA, USA) were used in western blot analysis. Briefly, 10 μg
of each protein extract was diluted with reducing NuPAGE sodium
dodecyl sulfate (SDS) sample buffer (Invitrogen) and heated for 10
min at 95°C. The heat-denatured extracts were then electrophoresed
by 12% SDS-polyacrylamide gel electrophoresis and transferred
electrophoretically onto a polyvinylidene fluoride membrane
(Millipore, Billerica, MA, USA) using an electroblot system (Hofer
TE70 Semi-Dry Transfer Unit; GE Healthcare, Fairfield, CT, USA)
with a constant current of 0.8 mA/cm2. The membrane was
then blocked with 5% (w/v) skim milk in TBS-T (50 mmol/l Tris-HCl,
pH 7.5; 150 mmol/l NaCl; 0.1% Tween-20) and probed with anti-SNAP25
IgG (1:1,000 in blocking solution; Cat. no. HPA001830; Sigma, St.
Louis, MO, USA), according to a standard procedure. This antibody
cannot distinguish between different variants as its antigenic
peptide is located within a conserved region. The blot was then
incubated with horseradish peroxidase (HRP)-conjugated anti-rabbit
IgG (1:5,000 in TBS-T; Cat. no. ab6721; Abcam) and developed using
the enhanced chemiluminescence method (ECL system; GE Healthcare).
The GAPDH protein level in each sample was also immunodetected as
an internal control using anti-GAPDH IgG (1:5,000 in TBS-T; Cat.
no. AM4300; Ambion) and HRP-conjugated anti-mouse IgG (1:5,000 in
TBS-T; Cat. no. ab6789; Abcam). Images of the blots were acquired
using the FluorChem FC2 system (Alpha Innotech, San Leandro, CA,
USA).
Immunohistochemical analysis of tissue
arrays and clinical tissues
A routine immunohistochemical procedure was applied
after hybridizing individual tissue sections. Immunohistochemical
staining was performed using the biotin-streptavidin-peroxidase
method with a VECTASTAIN® Elite ABC kit (Vector
Laboratories, Burlingame, CA, USA). During epitope retrieval, the
slides with tissue sections were immersed in Tris-EDTA buffer (10
mM Tris base, 1 mM EDTA solution, 0.05% Tween-20, pH 9.0) and
incubated for 20 min on a hot plate (95–99°C), or boiled, before
cooling to room temperature for 20 min. The tissues on the slides
were blocked with blocking solution (VECTASTAIN® Elite
ABC kit) for 2 h at room temperature and probed with anti-SNAP25
(1:1,000 in blocking solution; Cat. no. HPA001830; Sigma) antibody
overnight at 4°C. The hybridized tissue sections were then washed 3
times with phosphate-buffered saline and endogenous peroxidase
production was blocked with 0.3% hydrogen peroxide in distilled
water for 15 min. The specific signals in the tissue arrays were
acquired by incubation with the biotin-labeled secondary antibody
(VECTASTAIN® Elite ABC kit) for 1 h and developed using
a peroxidase substrate solution until the development of the
desired stain intensity. Before dehydration and mounting,
hematoxylin was used as an appropriate counterstain. In addition,
the tumor regions of individual tissue sections were identified by
routine hematoxylin and eosin (H&E) staining, which was
performed at the Department of Pathology, Sijhih Cathay General
Hospital, New Taipei, Taiwan. Finally, independent pathologists
diagnosed the imaging results.
Generation of stable
SNAP25-overexpressing MB cells and morphological changes of cells
with SNAP25 expression by immunofluorescence
A human cDNA that coded for a protein related to
SNAP25 was amplified by qualitative PCR from a Human Reference cDNA
(Clontech) using an appropriate primer pair (forward,
CTCGAGATGGCCGAAGACGCAGACAT; reverse, GGATCCACCACTTCCCAGCATCTTTG).
PCR bands with the predicted size were isolated routinely and
sequenced (ABI 3100) using an ABI Prism Big Dye Terminator Cycle
Sequencing Ready reaction kit (both from Applied Biosystems,
Framingham, MA, USA) to confirm the gene identities. The green
fluorescent protein (GFP)-containing vector, pEGFP C3, was used as
the expression vector (Invitrogen). Both pEGFP C3 and pEGFP
C3-SNAP25 (the SNAP25-containing pEGFP C3) were isolated with a
Genopure Plasmid Midi Maxi kit (Roche Diagnostics). In total,
7×104 MB cells (Daoy) were seeded into each well of a
6-well plate. On the second day, cells at 80–90% confluence were
transfected with the purified plasmid using FuGENE® HD
(Roche Diagnostics) according to the following procedures: 2
μg of plasmid DNA (in 10 μl ddH2O) was
mixed with 100 μl of serum-free medium; the transfection
reagent, 7 μl FuGENE® HD, was then added to the
100 μl serum-free medium and incubated for 20 min at room
temperature. The resulting complex mixture was subsequently added
to the cells. The transfected cells were placed into selection
medium containing 500 μg/ml G418 (also known as Geneticin™;
BioShop, Oakville, ON, Canada) the following day. Next, increasing
concentrations of G418 (up to 1,000 μg/ml) were used to
select stable SNAP25-overexpressing clones over the next 30 days.
The increasing SNAP25 mRNA levels in the stable clones were
quantified by RT-qPCR and protein was immunoblotted by western blot
analysis, as described above. The GFP was also immunodetected with
anti-EGFP IgG (1:1,000 in blocking solution; Cat. no. ab184601;
Abcam). Normal neural cells were acquired by a differentiation of
placenta-derived multipotent cells (PDMCs) under a 3-day incubation
with 0.4 mM 3-isobutyl-1-methylxanthine (IBMX) (23). The expression pattern of SNAP25
(green) was immunostained with rabbit anti-SNAP25 antibody (1:100;
ab109105; Abcam) and DyLight 488-conjugated goat anti-rabbit IgG
antibody (1:200; A120-101D2; Bethyl Laboratories, Montgomery, TX,
USA). The glial fibrillary acidic protein (GFAP), the Tau protein,
and pan neuronal markers (PNM) were respectively probed using
anti-GFAP (1:200; MAB3402; Millipore), anti-Tau (1:100, ab80579;
Abcam) and anti-PNM antibodies (1:25; MAB2300, Millipore). Finally,
the Cy3-conjugated goat anti-mouse antibody (1:200; AP124C;
Millipore) was used as the secondary antibody. For the
immunofluorescence analysis, 4′,6-diamidino-2-phenylindole (DAPI)
was used to counterstain the nucleus and exhibited blue color. The
fluorescent samples were then dehydrated, mounted, and analyzed
using a Nikon Eclipse 80i fluorescence microscope (Nikon
Instruments, Melville, NY, USA).
Determination of the cytotoxicity of
arabinofuranosyl cytidine (Ara-C) in SNAP25-expressing MB
cells
MB cells (Daoy) without or with SNAP25
overexpression were cultured at 3×103 cells/well in
96-well plates for 24 h. Cell viability was examined using a Cell
Counting kit-8 (CCK-8; Sigma) in the presence of various
concentrations (from 0.1 nM to 10 mM) of Ara-C (C1768; Sigma)
following incubation for 48 h, according to the manufacturer's
instructions. Briefly, the absorbance of each well was read at 450
nm using a microplate reader (iMark; Bio-Rad, Richmond, CA, USA)
after 2–4 h of incubation until a brown precipitate formed.
Statistical analysis
The differences in SNAP25 expressions, numbers of
dendrites, and Ara-C concentrations were compared using the
Student's t-test. Statistical analysis was performed using SPSS
13.0 (SPSS, Inc., Somers, NY, USA). The data were representative of
at least 3 experiments with similar results, and a value of
p<0.05 was considered to indicate a statistically significant
difference.
Results
Significant candidates with differential
expression levels in MB
Differentially expressed genes in MB were selected
using Agilent and Affymetrix microarrays (Fig. 1). We identified 4,596 candidates
based on their significantly up- (>1.50-fold) or downregulated
(<0.67-fold) expression levels in MB cells using Agilent
oligonucleotide microarrays. In addition, over 10,000 genes with
differential expression levels (>1.50- or <0.67-fold) were
obtained from the public database. As shown in Fig. 1, 2,544 genes with consistent expression
level differences were determined by comparing the data obtained
from different microarray systems (Affymetrix and Agilent), but
only 278 genes (130 upregulated and 148 downregulated: link to
download these significant genes in MB cells, https://drive.google.com/file/d/0Bw-QVez2CXs5YWhaMER1Zl9Kd1k/view?usp=sharing)
exhibited similar expression patterns. From the 278 genes, 86
candidates (41 upregulated and 45 downregulated) were also
identified based on consistent differences in their expression
levels according to different microarrays (standard deviation
<0.75; p<0.05). We found that many genes had statistically
different expression levels according to the whole-genome
microarrays, where 35 GO terms were enriched, including vesicle,
neuron part and synapse. Among the candidates, SNAP25 was
associated with 6 GO terms (GO0042995, cell projection; GO0044763,
single-organism cellular process; GO0097458, neuron part;
GO0044699, single-organism process; GO0045202, synapse; GO0016020,
membrane) and its expression level was reduced in MB cells. Soluble
N-ethylmaleimide-sensitive factor attachment protein
receptors (SNAREs) and their associated proteins play critical
roles in vesicle docking, priming, fusion, and the synchronization
of neurotransmitter release via exocytosis pathways (24,25). Briefly, the significant GO terms
with functional similarity, including molecular function,
biological process and cellular component (Table III), were explored based on
their GO annotations and using enrichment analysis. In particular,
genes for SNAREs annotated with the function 'vesicle' were
identified and one of these proteins, SNAP25, had significantly
lower expression levels in the MB cells in all of the microarrays.
It should be noted that the probe used for SNAP25 in the Agilent
microarrays was the probe A_23_P210756 near the stop codon (TAA),
which indicated the total SNAP25 expression level.
 | Table IIIThe significant gene ontology terms
with similarity. |
Table III
The significant gene ontology terms
with similarity.
| GO no. | GO name | p-value |
|---|
| Molecular
function |
| 0016667 | Oxidoreductase
activity, acting on a sulfur group of donors | <0.005 |
| 0016853 | Isomerase
activity | <0.005 |
| 0005543a | Phospholipid
binding | <0.005 |
| Biological
process |
| 0044763b | Single-organism
cellular process | <0.005 |
| 0044699b | Single-organism
process | <0.005 |
| 0032502 | Developmental
process | <0.005 |
| 0050687 | Negative regulation
of defense response to virus | <0.005 |
| 0018904 | Ether metabolic
process | <0.005 |
| 2000374 | Regulation of
oxygen metabolic process | <0.01 |
| 0002832 | Negative regulation
of response to biotic stimulus | <0.01 |
| 2000376 | Positive regulation
of oxygen metabolic process | <0.01 |
| 0032956 | Regulation of actin
cytoskeleton organization | <0.01 |
| 0060138 | Fetal process
involved in parturition | <0.01 |
| 0097212 | Lysosomal membrane
organization | <0.01 |
| 0097350 | Neutrophil
clearance | <0.01 |
| 0030155 | Regulation of cell
adhesion | <0.01 |
| 0048519 | Negative regulation
of biological process | <0.01 |
| 0048133 | Male germ-line stem
cell division | <0.01 |
| 0097213 | Regulation of
lysosomal membrane permeability | <0.01 |
| 0051611 | Regulation of
serotonin uptake | <0.01 |
| 0002698 | Negative regulation
of immune effector process | <0.01 |
| Cellular
component |
| 0042995b | Cell
projection | <0.005 |
| 0098563 | Intrinsic component
of synaptic vesicle membrane | <0.005 |
| 0030285 | Integral component
of synaptic vesicle membrane | <0.005 |
| 0097458b | Neuron part | <0.005 |
| 0043230 | Extracellular
organelle | <0.005 |
| 0065010 | Extracellular
membrane-bounded organelle | <0.005 |
| 0031982 | Vesicle | <0.005 |
| 0043256 | Laminin
complex | <0.005 |
| 0005856 | Cytoskeleton | <0.005 |
| 0005788 | ER lumen | <0.005 |
| 0005793 | ER-Golgi
intermediate compartment | <0.01 |
| 0045202b | Synapse | <0.01 |
| 0005605 | Basal lamina | <0.01 |
| 0016020a,b | Membrane | <0.01 |
Decreased protein expression levels of
SNARE and SNAP25 in MB cells
The differences in the mRNA and protein levels also
validated the differential expression of the SNARE protein. Two
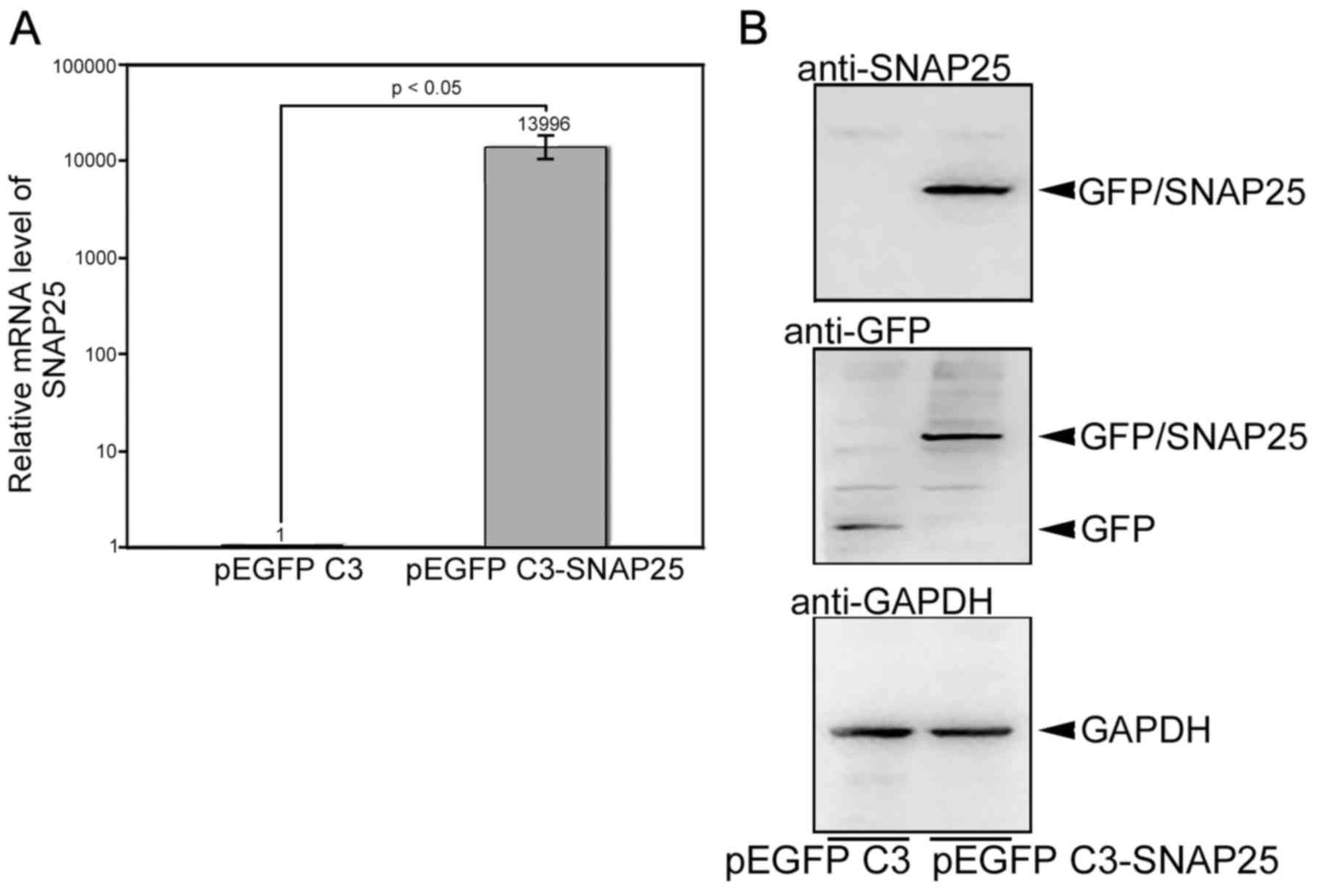
SNAP25 variants have been reported previously (26). For the total SNAP25 or individual
variants, lower mRNA levels were detected in each MB cell line
compared with those in normal brain cells (Fig. 2A). Moreover, we detected different
expression patterns for variants 1 and 2. Normal brain cells and
D341 Med (group 3 subgroup) expressed relatively more of variant 2,
whereas lower levels of variant 2 were detected in Daoy cells
[sonic hedgehog (SHH) subgroup] and VGH-Med cells. The very high
difference in the gene expression level of total SNAP25 was also
replicated in the protein levels compared with the normal brain
tissues, as well as in other normal tissues in the brain stem, such
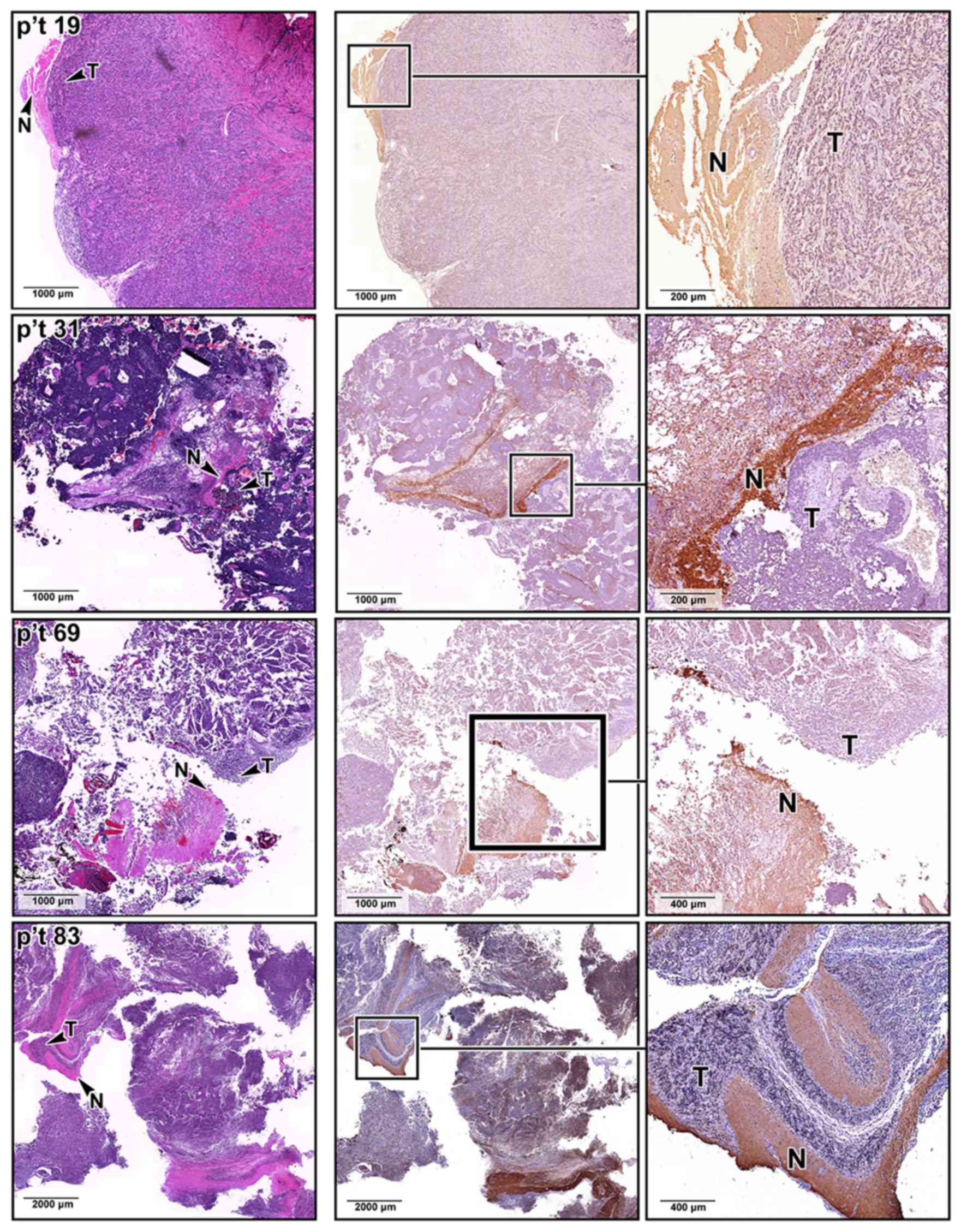
as the medulla oblongata and cerebellum (Fig. 2C). The difference in SNAP25
expression was also confirmed by immunohistochemical staining
analyses of our archived MB sections, where the MB lesions
identified by H&E staining expressed less SNAP25 protein
(Fig. 3). By contrast, SNAP25 was
expressed at normal levels in the non-tumor regions of the same MB
tissue sections.
Increased dendrite density and
chemotherapeutic effects on SNAP25-expressing MB cells
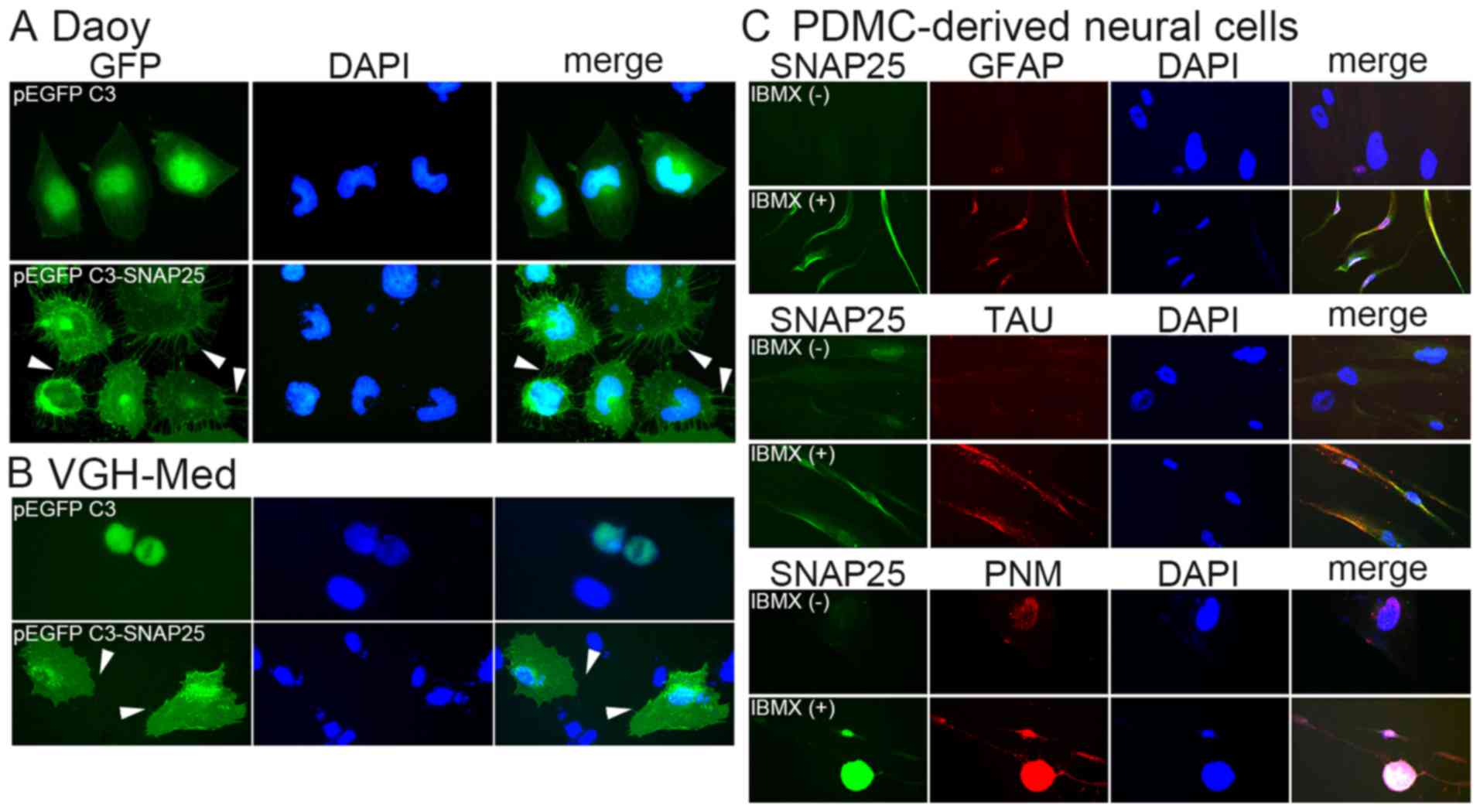
The stable clones of Daoy cells with pEGFP C3 or
pEGFP C3-SNAP25 were selected as described above. The mRNA level of
SNAP25 in pEGFP C3-SNAP25-transfected Daoy cells was extremely
higher (>1,0000 fold) than in cells transfected with only pEGFP
C3 (Fig. 4A). This overexpressed
SNAP25 was also specifically immunodetected, wheras only GFP could
be hybridized in the cells with pEGFP C3 (Fig. 4B). A morphological change in the
MB cells was associated with SNAP25 expression. Daoy cells and
VGH-Med cells with only GFP, but low endogenous SNAP25 expression,
had a smooth cell membrane (Fig.
5). None of the cells which only expressed GFP exhibited
significant dendrites. By contrast, many dendrites were observed on
the membrane of each cell that overexpressed GFP/SNAP25 fusion
protein (the lower panels of Fig. 5A
and B). This was also supported by the neural differentiation
analysis, which showed that markedly positive levels of SNAP25 were
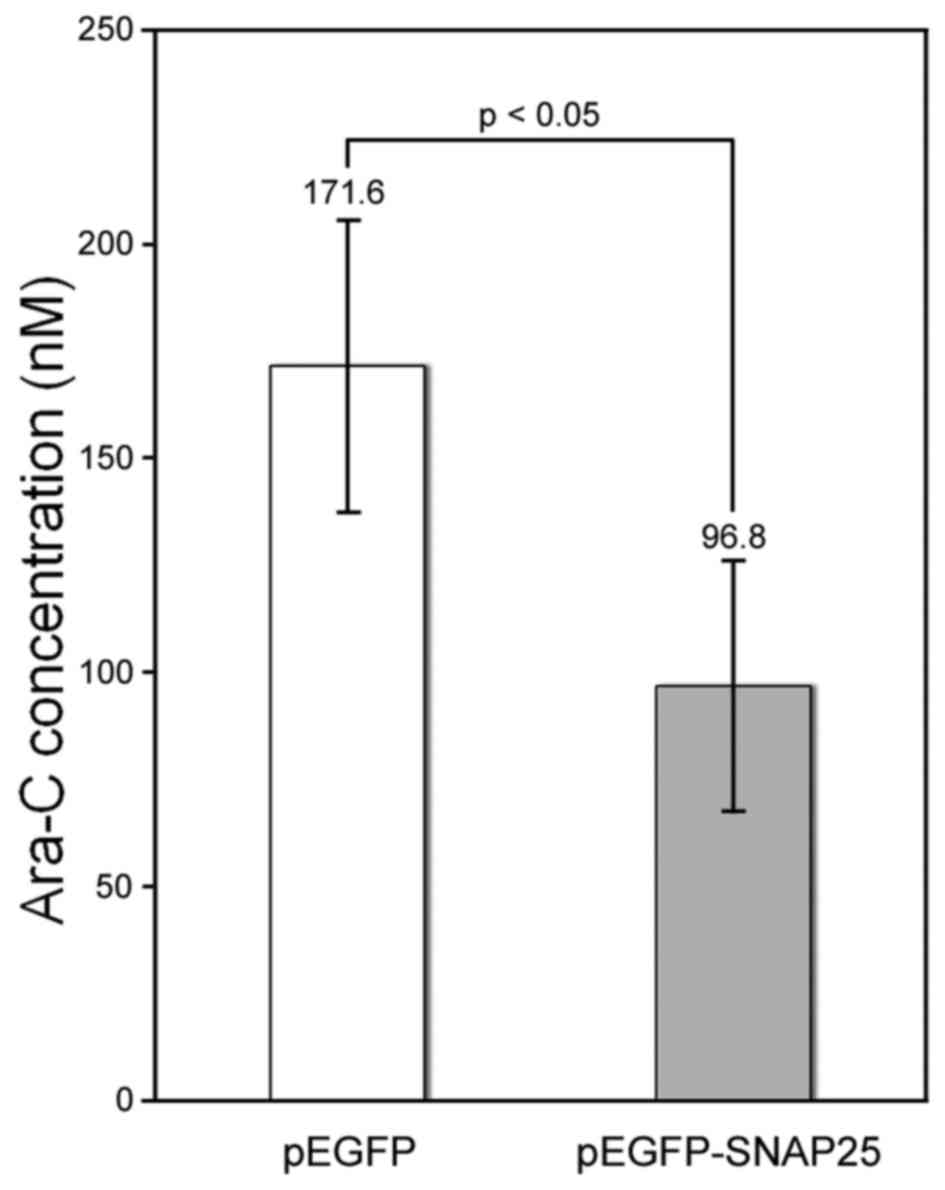
detected in differentiated neural cells (Fig. 5C). Currently, Ara-C is being
developed as a chemical agent for the clinical treatment of MB
(27,28). We found that the protein
expression of SNAP25 in Daoy cells increased susceptibility to
Ara-C. The half-maximal inhibitory concentration of Ara-C was
significantly lower for the SNAP25-expressing Daoy cells (96.8±29.5
nM) compared with that in cells that had low endogenous SNAP25
levels (171.6±34.0 nM) (p<0.05) (Fig. 6).
Discussion
Cancer, including MB, is generally attributed to
genetic aberrations in tumor tissues (29,30). Previous studies have explored the
genetic alterations in MB to determine their associations with the
clinical prognosis, thereby facilitating the development of
therapeutic approaches based on analyses of high-throughput
databases (29,31). Moreover, internet-based tools have
been used to determine the effectiveness of treatments for
childhood MB (32). In the
present study, we performed comparative genome-wide surveys to
determine the molecular characteristics of MB and to identify
potential targets for MB treatment.
The synaptic vesicle membrane glycoprotein,
synaptophysin, has been studied in the context of MB (30). Over the past decade, the
subcellular localizations of SNAREs and their ability to form the
so-called SNARE complex have been shown to play important roles in
the minimal fusion machinery and in cell-to-cell signaling
(33,34). In addition, SNAREs may also be
involved in the functional regulation of some ion channels
(35). Ramakrishnan et al
suggested that deficits in the expression of SNAREs may impair the
functions of the brain and sense organs (25). A deficiency of SNAP25 may block
normal vesicle fusion in brain cells (36,37). SNAP25 is known to be involved in
endocytosis or exocytosis during vesicle mobilization, where its
role is facilitated by other SNAREs and syntaxins (34,38,39). Furthermore, SNAP25 may interact
with various proteins that drive the spontaneous
calcium-independent fusion of synaptic vesicles (40). In addition to SNAP25, another
SNARE protein, synaptic vesicle protein synaptophysin, has been
shown to have differential expression levels in MB (41). This suggests that a complex
mechanism affects the production of synaptic vesicles during MB
tumorigenesis and that SNAREs are critical for normal vesicle
fusion (42).
Similarly, reduced levels of SNAP25 have also been
detected in the hippocampus of patients with schizophrenia
(43,44). Moreover, Dubuc et al
reported that the WNT subgroup expressed higher levels of variant 2
(26). Similarly, high levels of
variant 2 were detected in normal brain cells and in D341 Med cells
(group 3 subgroup). By contrast, lower levels were expressed in
Daoy cells (SHH subgroup) and VGH-Med cells. This implies that the
expression patterns of SNAP25 variants may be subgroup specific.
Recent efforts at stratifying MBs based on their molecular features
have revolutionized our understanding of this morbidity (9). We believe that the knowledge of
classification may contribute to the development of novel molecular
therapies targeting a specific subgroup of MBs (45).
A number of clinical studies have shown that SNAP25
participates in synaptic plasticity and it may be associated with a
separate synaptic pathology (43,46). These hypotheses were also
supported by our cytological results, where an increased dendrite
density was observed in Daoy cells and VGH-Med cells when the
expression of SNAP25 was restored in the present study. Thus, nerve
impulses may be conducted via these dendrites (47). The molecular significance of
SNAP25 in brain cells may be related to protein-protein
interactions with many other proteins, such as syntaxin 1A and
syntaxin-binding protein 1 (38,48,49). Moreover, dendrite instability has
been detected in many neuronal diseases (50). The decreased dendrite density
associated with reduced levels of SNAP25 and the presence of MB
cells without dendrites may be related to MB tumorigenesis.
Therefore, the SNARE complex, which includes SNAP25, may play
various complex roles when synaptic vesicle fusion occurs after a
nerve impulse reaches the synapse. However, the SNAP25-restored
dendrites may be more important in the SHH subgroup of MB. Other
than the SHH subgroup, three principle subgroups (WNT, Group 3 and
4) were also defined by their distinct molecules (51,52). In the present study, only the Daoy
cells (SHH subgroup) (53) and an
undefined MB subgroup, VGH-Med cells, could restore dendrites in
the presence of SNAP25. These implied that the restoration of
dendrites caused by SNAP25 expression may lead cells to the
terminal differentiation and then loss of tumor-igenicity (54). By contrast, the D341 Med cells
classified in the Group 3 subgroup (55) did not produce similar results
(data not shown). Therefore, we hypothesized that SNAP25 may play a
critically significant role in MB, particularly for the SHH
subgroup, where Daoy cells and VGH-Med cells exhibited similar
molecular patterns.
Alterations in synaptic vesicle fusion or dendrite
density may be associated with MB chemotherapy. In 1998, Hodel
reported that a reduction of SNAP25 in the brain impaired neuronal
dopamine signaling, thereby providing a target for the development
of therapeutic treatments (56).
An important morphogen for neural differentiation, retinoic acid,
can enhance MB chemosensitivity (57). These data indicate that optimal
synaptic vesicle fusion and a normal dendrite density may improve
the chemotherapeutic outcomes in this specific neuronal disorder.
The results of our analysis of Ara-C treatment in MB cells also
support this hypothesis. Thus, restoring the expression of SNAP25
in MB cells can increase the sensitivity to Ara-C, which is an
intrathecal chemotherapeutic that is used as an antineoplastic
agent in children (58).
Genomic variation affects the structure and
expression of genes; thus, genetic analyses may enhance our
understanding of the predisposition, biology and clinical response
to therapy in various diseases (59). Methods are needed that highlight
biomarkers with potential clinical utility in the diagnosis and
treatment of the common types of primary pediatric brain tumors
(15). In the present study,
SNAP25 was less abundant in the tissue arrays and in our enrolled
patients. Thus, we suggest that the reduced levels of SNAP25 may
have implications for clinical treatment. In addition, the dendrite
density was restored when MB cells expressed SNAP25 protein, which
may rebuild the synaptic junctions between neuronal cells. The
pyrimidine analogue, Ara-C, has been reported to be one of the most
effective agents in inducing the apoptosis of many tumor cells
(60). However, the
chemosensitivity to Ara-C was greatly increased when MB cells
expressed SNAP25.
In conclusion, our results suggest that a SNARE
complex that includes SNAP25 is crucial for the dendrite formation
and is associated with the effects of targeted chemotherapy. A
downregulation of SNAP25 may impede targeted chemotherapy as the
SNAREs in brain cells may not form correctly. The detection of
SNAP25 expression in MB cells may be essential for the
chemotherapeutic application of Ara-C.
Abbreviations:
|
MB
|
medulloblastoma
|
|
SNAREs
|
soluble
N-ethylmaleimide-sensitive factor attachment protein
receptors
|
|
SNAP25
|
synaptosomal-associated protein 25
|
|
Ara-C
|
arabinofuranosyl cytidine
|
|
GEO
|
Gene Expression Omnibus
|
|
GO
|
Gene Ontology
|
|
RT-qPCR
|
reverse transcription-quantitative
polymerase chain reaction
|
|
SDS
|
sodium dodecyl sulfate
|
|
HRP
|
horseradish peroxidase
|
|
GFP
|
green fluorescent protein
|
Acknowledgments
This study was supported by grants from the Cathay
General Hospital and Taipei Medical University (no. 103CGH-TMU-02)
and the Cathay General Hospital (no. CGH-MR-10122).
References
|
1
|
Shih DJ, Northcott PA, Remke M, Korshunov
A, Ramaswamy V, Kool M, Luu B, Yao Y, Wang X, Dubuc AM, et al:
Cytogenetic prognostication within medulloblastoma subgroups. J
Clin Oncol. 32:886–896. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Anne SL, Govek EE, Ayrault O, Kim JH, Zhu
X, Murphy DA, Van Aelst L, Roussel MF and Hatten ME: WNT3 inhibits
cerebellar granule neuron progenitor proliferation and
medulloblastoma formation via MAPK activation. PLoS One.
8:e817692013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Lastowska M, Al-Afghani H, Al-Balool HH,
Sheth H, Mercer E, Coxhead JM, Redfern CP, Peters H, Burt AD,
Santibanez-Koref M, et al: Identification of a neuronal
transcription factor network involved in medulloblastoma
development. Acta Neuropathol Commun. 1:352013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rajurkar M, Huang H, Cotton JL, Brooks JK,
Sicklick J, McMahon AP and Mao J: Distinct cellular origin and
genetic requirement of Hedgehog-Gli in postnatal rhabdomyosarcoma
genesis. Oncogene. 33:5370–5378. 2014. View Article : Google Scholar
|
|
5
|
Unland R, Kerl K, Schlosser S, Farwick N,
Plagemann T, Lechtape B, Clifford SC, Kreth JH, Gerss J, Mühlisch
J, et al: Epigenetic repression of the dopamine receptor D4 in
pediatric tumors of the central nervous system. J Neurooncol.
116:237–249. 2014. View Article : Google Scholar
|
|
6
|
MacDonald TJ, Aguilera D and Castellino
RC: The rationale for targeted therapies in medulloblastoma. Neuro
Oncol. 16:9–20. 2014. View Article : Google Scholar
|
|
7
|
Robinson G, Parker M, Kranenburg TA, Lu C,
Chen X, Ding L, Phoenix TN, Hedlund E, Wei L, Zhu X, et al: Novel
mutations target distinct subgroups of medulloblastoma. Nature.
488:43–48. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gerber NU, Mynarek M, von Hoff K,
Friedrich C, Resch A and Rutkowski S: Recent developments and
current concepts in medulloblastoma. Cancer Treat Rev. 40:356–365.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jahn R and Fasshauer D: Molecular machines
governing exocytosis of synaptic vesicles. Nature. 490:201–207.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ma HI, Kao CL, Lee YY, Chiou GY, Tai LK,
Lu KH, Huang CS, Chen YW, Chiou SH, Cheng IC, et al: Differential
expression profiling between atypical teratoid/rhab and
medulloblastoma tumor in vitro and in vivo using microarray
analysis. Childs Nerv Syst. 26:293–303. 2010. View Article : Google Scholar
|
|
11
|
Wybranska I, Polus A, Mikolajczyk M, Knapp
A, Sliwa A, Zapala B, Staszel T and Dembinska-Kiec A:
Apoptosis-related gene expression in glioblastoma (LN-18) and
medulloblastoma (Daoy) cell lines. Hum Cell. 26:137–148. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fiaschetti G, Castelletti D, Zoller S,
Schramm A, Schroeder C, Nagaishi M, Stearns D, Mittelbronn M,
Eggert A, Westermann F, et al: Bone morphogenetic protein-7 is a
MYC target with prosurvival functions in childhood medulloblastoma.
Oncogene. 30:2823–2835. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiederschain D, Chen L, Johnson B, Bettano
K, Jackson D, Taraszka J, Wang YK, Jones MD, Morrissey M, Deeds J,
et al: Contribution of polycomb homologues Bmi-1 and Mel-18 to
medulloblastoma pathogenesis. Mol Cell Biol. 27:4968–4979. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kong B, Yang T, Chen L, Kuang YQ, Gu JW,
Xia X, Cheng L and Zhang JH: Protein-protein interaction network
analysis and gene set enrichment analysis in epilepsy patients with
brain cancer. J Clin Neurosci. 21:316–319. 2014. View Article : Google Scholar
|
|
15
|
Russell MD, Young AM and Karri SK:
Biomarkers of pediatric brain tumors. Front Pediatr. 1:72013.
View Article : Google Scholar
|
|
16
|
Rustici G, Kolesnikov N, Brandizi M,
Burdett T, Dylag M, Emam I, Farne A, Hastings E, Ison J, Keays M,
et al: ArrayExpress update - trends in database growth and links to
data analysis tools. Nucleic Acids Res. 41:D987–D990. 2013.
View Article : Google Scholar
|
|
17
|
Glatt SJ, Everall IP, Kremen WS, Corbeil
J, Sásik R, Khanlou N, Han M, Liew CC and Tsuang MT: Comparative
gene expression analysis of blood and brain provides concurrent
validation of SELENBP1 up-regulation in schizophrenia. Proc Natl
Acad Sci USA. 102:15533–15538. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tarnopolsky M, Phillips S, Parise G,
Varbanov A, Demuth J, Stevens P, Qu A, Wang F and Isfort R: Gene
expression, fiber type, and strength are similar between left and
right legs in older adults. J Gerontol A Biol Sci Med Sci.
62:1088–1095. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Adler P, Kolde R, Kull M, Tkachenko A,
Peterson H, Reimand J and Vilo J: Mining for coexpression across
hundreds of datasets using novel rank aggregation and visualization
methods. Genome Biol. 10:R1392009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alvo M, Liu Z, Williams A and Yauk C:
Testing for mean and correlation changes in microarray experiments:
An application for pathway analysis. BMC Bioinformatics. 11:602010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bauer S, Grossmann S, Vingron M and
Robinson PN: Ontologizer 20. - a multifunctional tool for GO term
enrichment analysis and data exploration. Bioinformatics.
24:1650–1651. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yen BL, Chien CC, Chen YC, Chen JT, Huang
JS, Lee FK and Huang HI: Placenta-derived multipotent cells
differentiate into neuronal and glial cells in vitro. Tissue Eng
Part A. 14:9–17. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Northcott PA, Dubuc AM, Pfister S and
Taylor MD: Molecular subgroups of medulloblastoma. Expert Rev
Neurother. 12:871–884. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ramakrishnan NA, Drescher MJ and Drescher
DG: The SNARE complex in neuronal and sensory cells. Mol Cell
Neurosci. 50:58–69. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dubuc AM, Morrissy AS, Kloosterhof NK,
Northcott PA, Yu EP, Shih D, Peacock J, Grajkowska W, van Meter T,
Eberhart CG, et al: Subgroup-specific alternative splicing in
medulloblastoma. Acta Neuropathol. 123:485–499. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cefalo MG, De Ioris MA, Cacchione A, Longo
D, Staccioli S, Arcioni F, Bernardi B and Mastronuzzi A: Wernicke
encephalopathy in pediatric neuro-oncology: Presentation of 2 cases
and review of literature. J Child Neurol. 29:NP181–NP185. 2014.
View Article : Google Scholar
|
|
28
|
Nygaard R and Kivivuori SM: Treatment for
recurrent medulloblastoma with intrathecal liposomal cytarabine and
systemic metronomic combination therapy. Anticancer Drugs.
23:342–346. 2012. View Article : Google Scholar
|
|
29
|
Chen P, Fan Y, Man TK, Hung YS, Lau CC and
Wong ST: A gene signature based method for identifying subtypes and
subtype-specific drivers in cancer with an application to
medulloblastoma. BMC Bioinformatics. 14(Suppl 18): S12013.
View Article : Google Scholar :
|
|
30
|
He XM, Wikstrand CJ, Friedman HS, Bigner
SH, Pleasure S, Trojanowski JQ and Bigner DD: Differentiation
characteristics of newly established medulloblastoma cell lines
(D384 Med, D425 Med, and D458 Med) and their transplantable
xenografts. Lab Invest. 64:833–843. 1991.PubMed/NCBI
|
|
31
|
Schroeder K and Gururangan S: Molecular
variants and mutations in medulloblastoma. Pharmgenomics Pers Med.
7:43–51. 2014.PubMed/NCBI
|
|
32
|
Gudrunardottir T, Lannering B, Remke M,
Taylor MD, Wells EM, Keating RF and Packer RJ: Treatment
developments and the unfolding of the quality of life discussion in
childhood medulloblastoma: A review. Childs Nerv Syst. 30:979–990.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Duman JG and Forte JG: What is the role of
SNARE proteins in membrane fusion? Am J Physiol Cell Physiol.
285:C237–C249. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Hiersemenzel K, Brown ER and Duncan RR:
Imaging large cohorts of single ion channels and their activity.
Front Endocrinol (Lausanne). 4:1142013.
|
|
35
|
Jiménez-Garduño AM, Mitkovski M,
Alexopoulos IK, Sánchez A, Stühmer W, Pardo LA and Ortega A: KV10.1
K(+)-channel plasma membrane discrete domain partitioning and its
functional correlation in neurons. Biochim Biophys Acta.
1838:921–931. 2014. View Article : Google Scholar
|
|
36
|
Mohrmann R, de Wit H, Connell E, Pinheiro
PS, Leese C, Bruns D, Davletov B, Verhage M and Sørensen JB:
Synaptotagmin interaction with SNAP-25 governs vesicle docking,
priming, and fusion triggering. J Neurosci. 33:14417–14430. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Manca P, Mameli O, Caria MA,
Torrejón-Escribano B and Blasi J: Distribution of SNAP25, VAMP1 and
VAMP2 in mature and developing deep cerebellar nuclei after
estrogen administration. Neuroscience. 266:102–115. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ramakrishnan NA, Drescher MJ and Drescher
DG: Direct interaction of otoferlin with syntaxin 1A, SNAP-25, and
the L-type voltage-gated calcium channel Cav1.3. J Biol Chem.
284:1364–1372. 2009. View Article : Google Scholar :
|
|
39
|
Xu J, Luo F, Zhang Z, Xue L, Wu XS, Chiang
HC, Shin W and Wu LG: SNARE proteins synaptobrevin, SNAP-25, and
syntaxin are involved in rapid and slow endocytosis at synapses.
Cell Reports. 3:1414–1421. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Woodbury DJ and Rognlien K: The t-SNARE
syntaxin is sufficient for spontaneous fusion of synaptic vesicles
to planar membranes. Cell Biol Int. 24:809–818. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bühren J, Christoph AH, Buslei R, Albrecht
S, Wiestler OD and Pietsch T: Expression of the neurotrophin
receptor p75NTR in medulloblastomas is correlated with distinct
histological and clinical features: Evidence for a medulloblastoma
subtype derived from the external granule cell layer. J Neuropathol
Exp Neurol. 59:229–240. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Südhof TC and Rizo J: Synaptic vesicle
exocytosis. Cold Spring Harb Perspect Biol. 3:a0056372011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fatemi SH, Earle JA, Stary JM, Lee S and
Sedgewick J: Altered levels of the synaptosomal associated protein
SNAP-25 in hippocampus of subjects with mood disorders and
schizophrenia. Neuroreport. 12:3257–3262. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Thompson PM, Egbufoama S and Vawter MP:
SNAP-25 reduction in the hippocampus of patients with
schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry.
27:411–417. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
DeSouza RM, Jones BR, Lowis SP and Kurian
KM: Pediatric medulloblastoma - update on molecular classification
driving targeted therapies. Front Oncol. 4:1762014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Selak S, Paternain AV, Aller MI, Picó E,
Rivera R and Lerma J: A role for SNAP25 in internalization of
kainate receptors and synaptic plasticity. Neuron. 63:357–371.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lesch KP and Schmitt A: Antidepressants
and gene expression profiling: How to SNARE novel drug targets.
Pharmacogenomics J. 2:346–348. 2002. View Article : Google Scholar
|
|
48
|
Hamdan FF, Piton A, Gauthier J, Lortie A,
Dubeau F, Dobrzeniecka S, Spiegelman D, Noreau A, Pellerin S, Côté
M, et al: De novo STXBP1 mutations in mental retardation and
nonsyndromic epilepsy. Ann Neurol. 65:748–753. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hussain S: Developing a PPI
inhibitor-based therapy for STXBP1 haploinsufficiency-associated
epileptic disorders. Front Mol Neurosci. 7:62014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Murmu RP, Li W, Holtmaat A and Li JY:
Dendritic spine instability leads to progressive neocortical spine
loss in a mouse model of Huntington's disease. J Neurosci.
33:12997–13009. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Northcott PA, Jones DT, Kool M, Robinson
GW, Gilbertson RJ, Cho YJ, Pomeroy SL, Korshunov A, Lichter P,
Taylor MD, et al: Medulloblastomics: The end of the beginning. Nat
Rev Cancer. 12:818–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Perreault S, Ramaswamy V, Achrol AS, Chao
K, Liu TT, Shih D, Remke M, Schubert S, Bouffet E, Fisher PG, et
al: MRI surrogates for molecular subgroups of medulloblastoma. AJNR
Am J Neuroradiol. 35:1263–1269. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sengupta R, Dubuc A, Ward S, Yang L,
Northcott P, Woerner BM, Kroll K, Luo J, Taylor MD, Wechsler-Reya
RJ, et al: CXCR4 activation defines a new subgroup of Sonic
hedgehog-driven medulloblastoma. Cancer Res. 72:122–132. 2012.
View Article : Google Scholar
|
|
54
|
Zhang X, Cruz FD, Terry M, Remotti F and
Matushansky I: Terminal differentiation and loss of tumorigenicity
of human cancers via pluripotency-based reprogramming. Oncogene.
32:2249–2260. e1–21. 2013. View Article : Google Scholar
|
|
55
|
Snuderl M, Batista A, Kirkpatrick ND, Ruiz
de Almodovar C, Riedemann L, Walsh EC, Anolik R, Huang Y, Martin
JD, Kamoun W, et al: Targeting placental growth factor/neuropilin 1
pathway inhibits growth and spread of medulloblastoma. Cell.
152:1065–1076. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hodel A: Snap-25. Int J Biochem Cell Biol.
30:1069–1073. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Liu J, Guo L, Jun-Wei L, Liu N and Li H:
All-trans retinoic acid modulates fas expression and enhances
chemosensitivity of human medulloblastoma cells. Int J Mol Med.
5:145–149. 2000.PubMed/NCBI
|
|
58
|
Ruggiero A, Conter V, Milani M, Biagi E,
Lazzareschi I, Sparano P and Riccardi R: Intrathecal chemotherapy
with antineoplastic agents in children. Paediatr Drugs. 3:237–246.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Hudson TJ: Genome variation and
personalized cancer medicine. J Intern Med. 274:440–450. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Sreenivasan Y, Sarkar A and Manna SK:
Mechanism of cytosine arabinoside-mediated apoptosis: Role of Rel A
(65) dephosphorylation. Oncogene. 22:4356–4369. 2003. View Article : Google Scholar : PubMed/NCBI
|