Introduction
Prostate cancer (Pca) is a common malignant tumor in
men and is also the second most serious malignant tumor that
threatens mens' health (1,2).
Patients with Pca do not exhibit clear symptoms in the early stages
of the disease, resulting in ~20% of patients presenting with
metastasis at the time of diagnosis, thereby missing the optimal
window for surgical treatment (3,4).
Therefore, the identification of novel methods for the early
diagnosis of Pca and the corresponding treatments have attracted
much attention.
von Willebrand factor C and EGF domain-containing
protein (URG11), located on the long arm of human chromosome 1,
encodes a 70 kDa protein containing 5 von Willebrand factor type C
domains; of these, the primary domain is 1 C-type lectin (5). The function of URG11 is to regulate
cell adhesion, migration and interaction, and it is closely
associated with signal transduction (6,7).
Previous studies have confirmed that URG11 is not only expressed in
various tumor cells and tissues including hepatocellular carcinoma
(8), and gastric (9) and pancreatic cancer (10), and may promote the occurrence and
development of tumors, it is considered to serve as a regulator of
cell growth (10,11). In a previous study, URG11 was
overexpressed in Pca tissues and detected by immunohistochemistry,
and was demonstrated to be positively correlated with Gleason score
and clinical stage of Pca, and closely associated with the
development of Pca (12).
Epithelial-mesenchymal transition (EMT) refers to the process
through which epithelial cells gradually lose their epithelial
differentiation characteristics and obtain an interstitial
phenotype (13). In this process,
adhesion between epithelial cells and basement membrane gradually
disappears; instead, changes in the cytoskeleton and shape are
observed. In addition, an increase in podoplanin and motor
abilities, and enhanced migratory and invasive abilities have been
identified (14,15). In the process of embryonic
development and the formation of organs, cells may diffuse from the
primary tissue through EMT, and migrate to secondary tissue sites
to continue to grow and differentiate (16,17). EMT is activated again during wound
healing, tissue fibrosis and tumor metastasis (18,19).
Previous studies have indicated that one of the most
important pathways in the development of EMT in epithelial cells is
the Wnt/β-catenin signaling pathway (20). The activation of Wnt/β-catenin
signaling induces motility, invasiveness, cell fate determination
and maintenance of self-renewal potential (21). Additionally, it was suggested that
Wnt/β-catenin signaling is required for the biological processes of
metastasis in Pca (22). In the
present study, the corresponding agonists and inhibitors of
Wnt/β-catenin signaling were used to explore the role of
Wnt/β-catenin signaling in Pca. The results provided a basis for a
treatment method for Pca by overexpressing or silencing the
regulation of URG11 in Pca cells.
Materials and methods
Reagents
The 293 cell line, human normal prostate epithelial
RWPE-1 cell line, and human Pca DU-145, PC-3 and LNCaP cell lines
were obtained from the American Type Culture Collection (Manassas,
VA, USA). FH55 [F5682; high performance liquid chromatography
(HPLC) purity ≥98%] and LiCl (746460; HPLC ≥99%) were purchased
from Sigma-Aldrich; Merck KGaA (Darmstadt, Germany).
Cell culture
All cells were placed in RPMI-1640 medium (HyClone;
GE Healthcare Life Sciences, Logan, UT, USA), and cultured with 5%
CO2 at 37°C, under saturated humidity. The cells in
logarithmic growth phase were detected.
Cell transfection
Lipofectamine® 3000 (Invitrogen; Thermo
Fisher Scientific, Inc., Waltham, MA, USA) was performed following
the manufacturer's protocol. In brief, 2 µl
Lipofectamine® 3000, 40 pmol small interfering (si) RNA
targeting URG11 and the negative control (siNC; Shanghai GenePharma
Co., Ltd., Shanghai, China) were mixed separately in 50 µl
serum-free medium and incubated at room temperature for 15 min. The
lipid compounds were diluted in 300 µl serum-free medium and
600 µl medium containing fetal bovine serum (FBS) to produce
a 1 ml volume mixture, and incubated with the LNCaP cells at 37°C
with 5% CO2. The URG11-targeting siRNA sequence and NC
siRNA were 5′-CAGACGGAUUGCUGUACUU-3′, and
5′-UUCUCCGAACGUGUCACGUTT-3′, respectively. For the expression
vectors used to upregulate URG11 (pcDNA3.1-URG11) and the
corresponding NC (pcDNA3.1-NC), 5 µl
Lipofectamine® 3000 and 2 µg vector (Shanghai
GenePharma Co., Ltd.) were mixed in 125 µl Dulbecco's
modified Eagle's medium (DMEM; Corning Inc., Corning, NY, USA) and
incubated at room temperature for 5 min. A total of 4 µl
Lipofectamine® 3000 and 125 µl DMEM were mixed,
and incubated at room temperature for 5 min. Then, the
Lipofectamine® 3000 + DNA mixture was combined with the
Lipofectamine® 3000 + DMEM solution. After 5 min, the
LNCaP cells were treated with the combined mixture at 37°C with 5%
CO2. The cells were transfected for 48 h for subsequent
experiments.
Transwell assay detects cell
invasion
The LNCaP cells of each treatment group were
collected, and 1×105 cells/well were counted. Following
the addition of Matrigel (BD Biosciences, Franklin Lakes, NJ, USA)
into the Transwell chamber for 6 h at 37°C, the cells was
resuspended in serum-free medium and added to the chamber of the
Transwell cell culture plate. Following incubation with 5%
CO2 for 24 h at 37°C, the chamber was removed, and cells
were fixed in 4% paraformaldehyde for 20 min at 4°C and washed once
with PBS. The cells were then stained with 0.1% crystal violet for
10 min at room temperature, washed once with PBS, and observed
under a light microscope. Images of the cells were captured and the
number of cells that had passed through the membrane was counted in
5 fields using a light microscope at magnification, ×200. The
number of cells per field was calculated, and experiments were
repeated 3 times.
Cell migration assay
LNCaP cells in the logarithmic growth phase
(~1×109 cells/well) were collected, and the suspension
was uniformly inoculated into a 6-well culture plate and cultured
in an incubator with 5% CO2 at 37°C. The control group
was cultured in serum-free RMPI-1640 medium, and the experimental
group was treated with drug-containing (20 µM FH535 or 20 mM
LiCl) serum-free RMPI-1640 medium for 48 h at 37°C. Following
scratching, images of the samples were captured at 0 and 48 h. The
scratch healing was observed under an inverted light microscope at
magnification, ×200. The scratch widths at 8 different sites in the
groups were measured by Image-Pro Plus 6.0 software (Media
Cybernetics Inc., Rockville, MD, USA) and the cell migration rate
in each group was calculated.
Cell Counting kit-8 (CCK-8) assay
A CCK-8 assay (Beyotime Institute of Biotechnology,
Haimen, China) was utilized to assess cell viability according to
the manufacturer's protocols. Briefly, transfected and
un-transfected LNCaP cells were transferred to 96-well plates
(3×103 cells/well). Following incubation for 2 h at 37°C
in 10 µl CCK-8, the absorbance in every well was evaluated
using a microplate reader at 450 nm (Tecan Infinite M200 Micro
Plate Reader; Tecan Group, Ltd., Männedorf, Switzerland).
Apoptosis assay
Following transfection of the LNCaP cells for 48 h,
1×106 cells were collected, and 1 ml trypsin was used to
digest the cells. The mixture was then gently shaken. Following
removal of the trypsin and incubation for 1 min at room
temperature, the digestion was terminated by adding DMEM containing
10% FBS (Corning Inc.). The cells were centrifuged at 1,000 × g for
3 min at 4°C and the supernatant was removed. The cells were then
washed twice with pre-cooled PBS and resuspended in 1X Annexin V
binding buffer. According to the protocol of the manufacturer of
the Annexin V-fluorescein isothiocyanate (FITC) cell apoptosis
detection kit (K201-100; BioVision, Inc., Milpitas, CA, USA), the
cells was stained with 1.25 µl Annexin V-FITC and 10
µl propidium iodide (PI) for 10 min at room temperature, and
measured by flow cytometry (FACSCalibur™; BD Biosciences) using
FlowJo v10.0 software (FlowJo LLC, Ashland, OR, USA)
Cell cycle analysis
Following transfection for 48 h, 1×106
LNCaP cells/well were collected, washed twice with PBS, then fixed
with 70% ethanol at 4°C overnight. Then, 500 µl PBS
containing 50 mg/l PI, 100 mg/l RNase A and 0.2% Triton X-100 was
added to the cells, which were incubated for 30 min at 4°C in the
dark. Flow cytometry was performed, and the results were analyzed
using the cell cycle fitting software ModFit LT™ software v2.0 (BD
Biosciences).
RNA extraction and reverse transcription
quantitative polymerase chain reaction (RT-qPCR)
Following culture of the RWPE-1, DU-145, PC-3 and
LNCaP cells for 24 h in the medium, the culture solution was
discarded and the cells were washed twice with PBS. Then, 500
µl TRIzol (Thermo Fisher Scientific, Inc.) was added to the
wells of each 6-well plate, pipetted several times, and then the
lysate was transferred to 1.5 ml RNase-free EP tubes. Then, 200
µl chloroform per 1 ml TRIzol was added into the EP tube,
and incubated on ice for 10 min. Following centrifugation at 12,000
× g for 15 min at 4°C, the upper aqueous phase was carefully
pipetted into a second RNase-free EP tube; the same volume of
isopropanol was the added to the EP tube, and the solution was
mixed by inversion several times, and incubated on ice for 10 min.
Following centrifugation at 10,000 × g for 5 min at 4°C, the
supernatant was discarded, an equal volume of 75% ethanol was
added, and the sediment at the bottom was gently agitated,
centrifuged at 7,500 × g for 5 min at 4°C, and the RNA was
air-dried at the vent. Subsequently, 10 µl of DEPC water was
added and the RNA concentration and purity were measured using a
NanoDrop 2000 spectrophotometer (NanoDrop Technologies; Thermo
Fisher Scientific, Inc., Wilmington, DE, USA). A total of 1
µg RNA was used for reverse transcription into cDNA using a
reverse transcription cDNA kit (Thermo Fisher Scientific, Waltham,
MA, USA). The reaction conditions were as follows: 42°C for 60 min
and 70°C for 5 min, followed by preservation at 4°C. SYBR-Green PCR
Master Mix (Roche Diagnostics, Basel, Switzerland) was used to
conduct the qPCR experiment using an Opticon RT-PCR Detection
System (ABI 7500; Thermo Fisher Scientific, Inc.), The PCR
thermocycler conditions were as follows: Pretreatment at 95°C for
10 min; followed by 40 cycles at 94°C for 15 sec, 60°C for 1 min
and 60°C for 1 min, and then preservation at 4°C. The comparative
cycle threshold (2−ΔΔCq) method was employed to analyze
the expression of mRNA (23).
GAPDH expression was used for normalization. The primer sequences
used are summarized in Table
I.
 | Table IPrimers for quantitative polymerase
chain reaction. |
Table I
Primers for quantitative polymerase
chain reaction.
| Genes | Forward
(5′-3′) | Reverse
(5′-3′) |
|---|
| URG11 |
TGAATCAAGGAGTCGCTGGAC |
GCATCTCACTGGAACACAAG |
| E-cadherin |
TCGACACCCGATTCAAAGTG |
GTCCCAGGCGTAGACCAAGA |
| Vimentin |
TGCCCTTAAAGGAACCAATGAG |
AGGCGGCCAATAGTGTCTTG |
| α-SMA |
CTGTTCCAGCCATCCTTCAT |
CCGTGATCTCCTTCTGCATT |
| Cyclin D1 |
CACACGGACTACAGGGGAGT |
CACAGGAGCTGGTGTTCCAT |
| c-Myc |
TCAAGAGGCGAACACACAAC |
GGCCTTTTCATTGTTTTCCA |
| GAPDH |
CGCTACAGTCGTTGCCATCA |
ACGACGAGGAAGCCATCTTG |
Western blot analysis
Total proteins were collected by
radioimmunoprecipitation assay lysis buffer (Cell Signaling
Technology, Inc., Danvers, MA, USA). BCA Protein Assay kit (Pierce;
Thermo Fisher Scientific, Inc.) was applied to measure the
concentration of proteins, which were adjusted to a concentration
of 6 µg/µl using 1X loading buffer and DEPC water.
Then, 5 µl samples was separated by 10% SDS-PAGE gels and
then transferred onto polyvinylidene fluoride membranes (EMD
Millipore, Billerica, MA, USA). Following blocking of the membranes
in 5% nonfat milk in PBST (0.1% Tween-20 in PBS) for 1 h at room
temperature, the membranes were probed with the primary antibodies
overnight at 4°C. The membranes were then washed 3 times with PBST
and then incubated with horseradish peroxidase (HRP)-conjugated
goat anti-mouse (cat. no. sc-516102; 1:2,000) and HRP-rabbit IgG
(cat. no. sc-2357; 1:2,000; both Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA) secondary antibodies at room temperature for 2 h.
Then, the membranes were washed 3 times with PBST. The EZ-ECL kit
(Biological Industries, Kibbutz Beit Haemek, Israel) was used to
visualize the gels, and the gray values were analyzed and counted
using ImageJ software (v5.0; Bio-Rad Laboratories, Inc., Hercules,
CA, USA). The antibodies used were mouse anti-GAPDH (1:1,000; cat.
no. LS-B1625; LifeSpan BioSciences, Inc.), rabbit anti-URG11
(1:1,000; cat. no. ab109232), mouse anti-epithelial cadherin
(E-cadherin; 1:1,000; cat. no. ab1416), rabbit anti-Vimentin
(1:1,000; cat. no. ab92547) and rabbit anti-α-smooth muscle actin
(α-SMA; 1:1,000; cat. no. ab5694), mouse anti-cyclin D1 (1:1,000;
cat. no. ab134175) and rabbit anti-MYC proto-oncogene protein
(c-Myc; 1:1,000; cat. no. ab32072; all Abcam, Cambridge, MA,
USA).
Statistical analysis
The results are presented as the mean ± standard
deviation. Statistical software Prism 7 (GraphPad Software, Inc.,
La Jolla, CA, USA) was used for statistical analysis. Comparisons
between two groups were performed using un-paired Student's
t-tests, and the comparisons between multiple groups were performed
by one-way analysis of variance followed by Dunnett's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
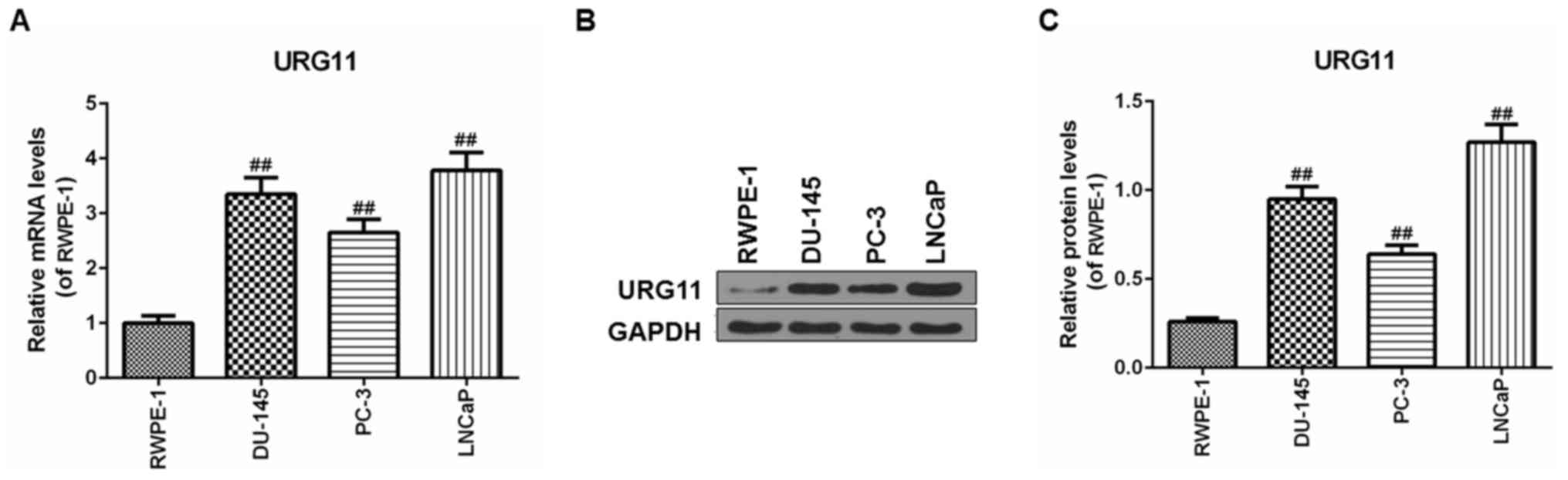
URG11 is expressed in cancer cells
The expression levels of URG11 were first
investigated in Pca cells lines, including DU145, PC3 and LNCaP
cell lines, and in the nontumor prostate epithelial RWPE-1 cell
line. The URG11 mRNA levels in human prostate cells were
significantly increased compared with that in the epithelial cells
(Fig. 1A). Furthermore, western
blot analysis was used to determine the URG11 protein level in the
Pca cell lines. Similarly, the URG11 protein level exhibited the
same pattern of expression as the mRNA level (Fig. 1B and C). Specifically, the LNCaP
cells exhibited increased URG11 mRNA and protein expression levels
in comparison with DU145 cells and PC3 cells. Therefore, LNCaP
cells were used for the subsequent experiments.
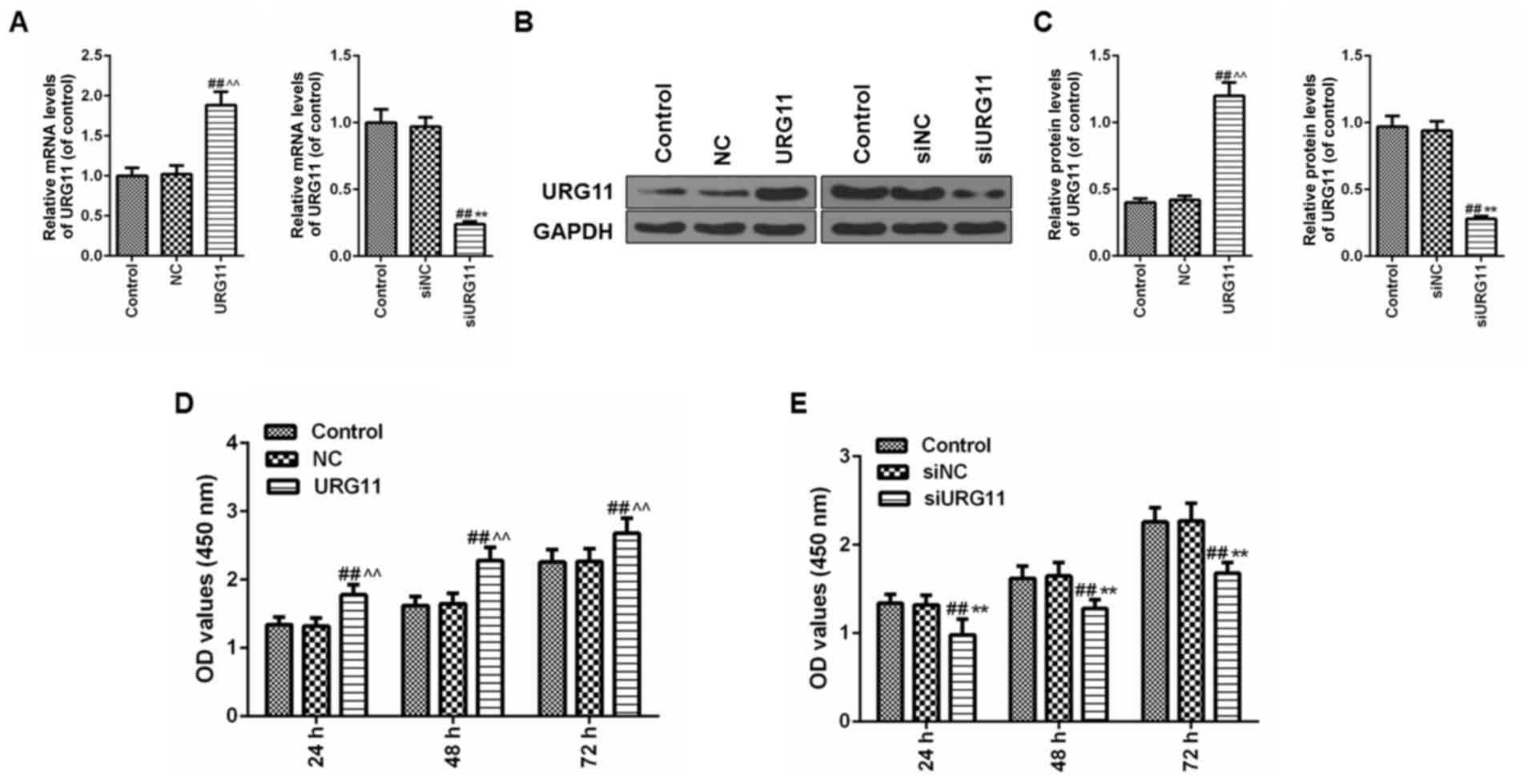
Overexpression of URG11 promotes cells
viability of cultured LNCaP cells and siURG11 elicits the opposite
effect
To additionally explore the function of URG11 in Pca
cells, overexpression plasmids and siRNA were applied to
overexpress and silence URG11, respectively. Following the
transfection of the URG11 plasmids and siURG11 fragments into LNCaP
prostate cells, the expression level of URG11 was significantly
increased in the URG11 overexpression group but suppressed in the
siURG11 group compared with their corresponding control and NC
groups at the mRNA (Fig. 2A) and
protein levels (Fig. 2B and C).
Following confirmation of the efficacy of the overexpression URG11
vector and siURG11 fragments, a CCK-8 assay was then used to detect
the effects of URG11 overexpression and silencing on cell
viability. As demonstrated in Fig.
2D, the cells in the URG11 overexpression group exhibited
increased growth compared with the normal culture and NC groups at
24, 48 and 72 h, while siURG11 decreased cell viability compared
with the normal culture and NC groups at 24, 48 and 72 h (Fig. 2E).
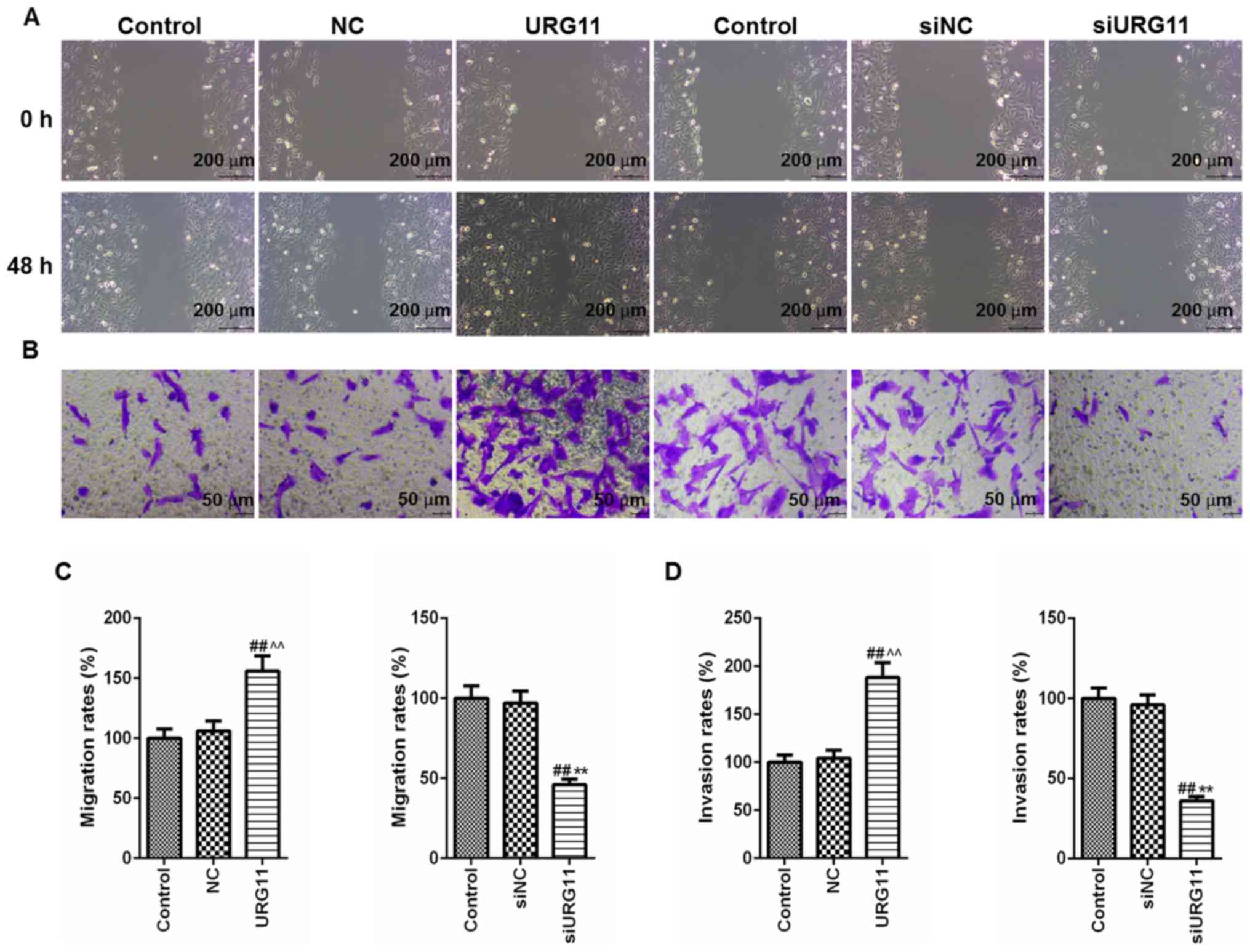
Overexpression of URG11 promotes
migration and invasion of LNCaP cells and siURG11 elicits the
opposite effects
The specific role of URG11 in the invasion and
migration of Pca cells was then determined. Cell migratory and
invasive abilities were determined by wound healing and Transwell
assays, respectively. The wounds generated in the cells in the
URG11 overexpression group were almost healed at 48 h, at which
time the wounds in normal culture and control groups were
significantly wider. Treatment with siURG11 exhibited the opposite
effects (Fig. 3A). Furthermore,
the Transwell invasion assays revealed that the overexpression of
URG11 significantly promoted the invasion of LNCaP Pca cells
compared with those of in the normal culture and NC control groups,
and genetic knockout of URG11 elicited the opposite effects
(Fig. 3B). The quantified data
from the migration and invasion assays are presented in Fig. 3C and D.
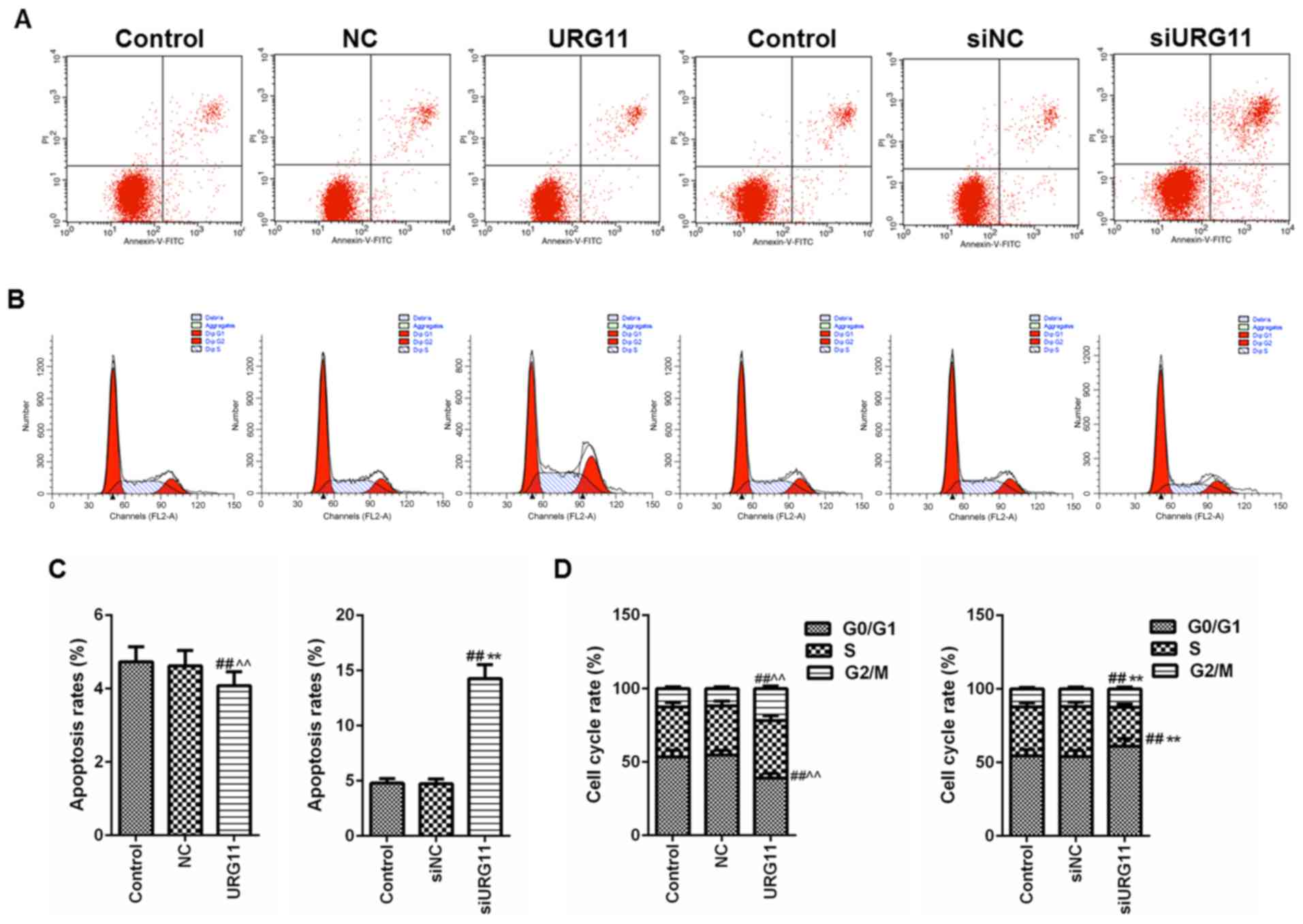
Overexpression of URG11 inhibits
apoptosis and induces cell cycle progression, while siURG11
exhibits the opposite effects
To improve understanding of the role of URG11 in Pca
metastasis and the underlying mechanism of action, flow cytometry
was performed with PI/Annexin V staining for apoptosis and cell
cycle analyses. Using flow cytometric analysis with PI/Annexin V
staining, overexpression of URG11 was observed to significantly
suppress cell apoptosis, compared with that in the normal culture
and NC groups (Fig. 4A).
Furthermore, overexpression of URG11 significantly decreased the
number of cells in G0/G1 phase and increased the number of cells in
S phase, compared with that in the normal culture and NC control
groups (Fig. 4B). By contrast,
siURG11 elicited the opposite effects (Fig. 4A-D).
Overexpression of URG11 inhibits the
level of E-cadherin and increases the levels of Vimentin and α-SMA,
while siURG11 elicits the opposite effects
Due to the importance of EMT in the development of
LNCaP cells, the effect of URG11/siURG11 on EMT markers in LNCaP
cells was assessed. The mRNA and protein levels of EMT markers were
examined by RT-qPCR and western blot analysis, respectively. As
indicated in Fig. 5A,
overexpression URG11 treatment significantly decreased E-cadherin
mRNA levels and increased vimentin and α-SMA mRNA levels compared
with the controls (Fig. 5A).
Furthermore, western blot analysis was used to determine the
protein levels of those EMT markers, and the results demonstrated
that the effects of URG11 on protein levels of E-cadherin, vimentin
and α-SMA were in accordance with mRNA levels (Fig. 5B and C). However, siURG11 elicited
the opposite effects (Fig.
5D-F).
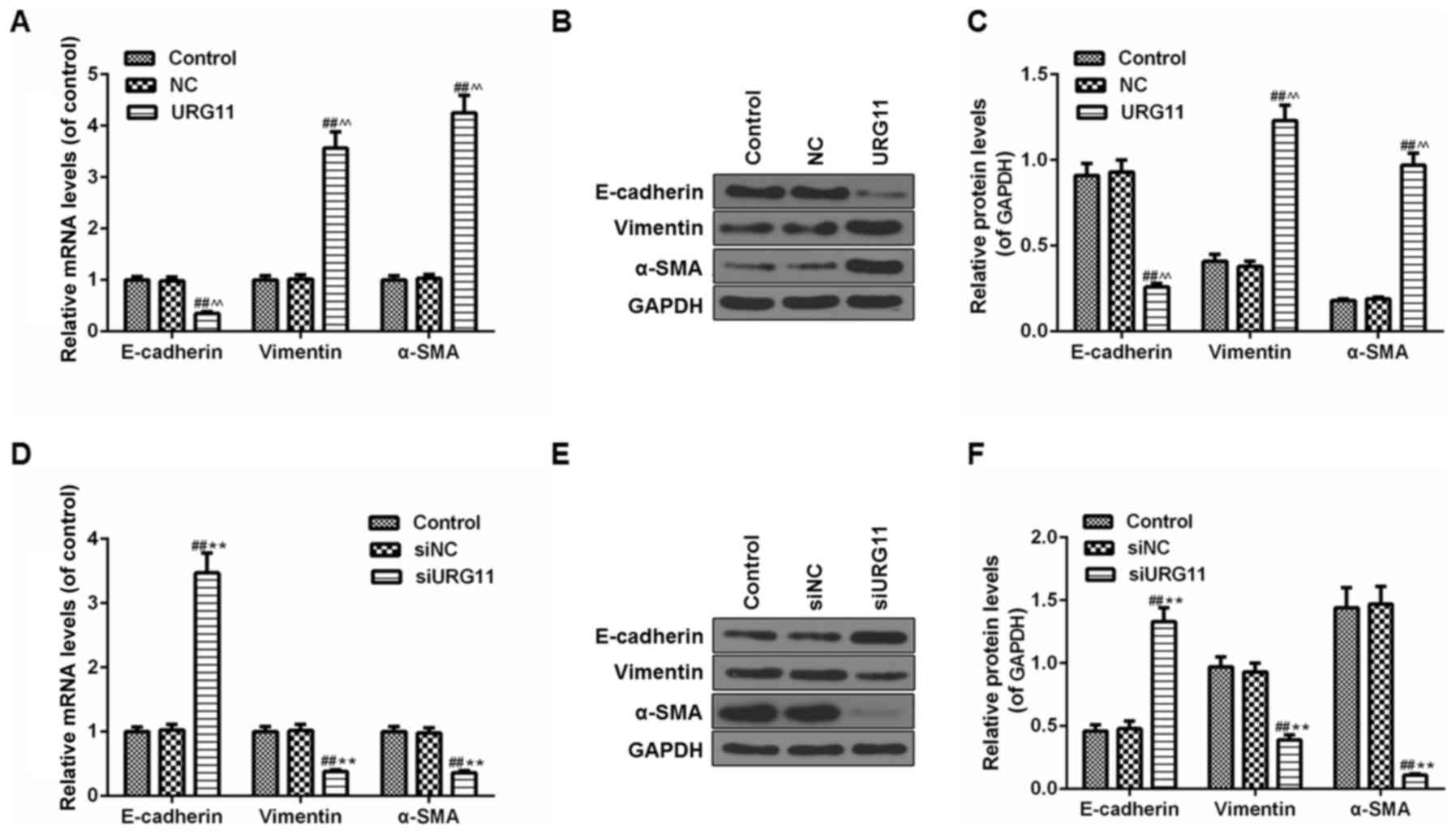 | Figure 5Effects of URG11 and siURG11 on EMT
in LNCaP cells. (A) The mRNA levels of EMT markers (E-cadherin,
vimentin and α-SMA) in URG11-overexpressing cells were determined
by RT-qPCR. (B) The protein levels of EMT markers (E-cadherin,
vimentin and α-SMA) in URG11-overexpressing cells were quantified
by western blot analysis. (C) Densitometric analysis of the EMT
protein levels in URG11-overexpressing cells, normalized to GAPDH.
(D) The mRNA levels of EMT markers (E-cadherin, vimentin and α-SMA)
in URG11-knockout cells were determined by RT-qPCR. (E) The protein
levels of EMT markers (E-cadherin, vimentin and α-SMA) in
URG11-knockout cells were quantified by western blot analysis. (F)
Densitometric analysis of the EMT protein levels in URG11-knockout
cells, normalized to GAPDH. Error bars represent standard
deviation. ##P<0.01 vs. control;
^^P<0.01 vs. NC; **P<0.01 vs. siNC. si,
small interfering; URG11, von Willebrand factor C and EGF
domain-containing protein; EMT, epithelial-mesenchymal transition;
RT-qPCR, reverse transcription quantitative polymerase chain
reaction; E-cadherin, epithelial cadherin; α-SMA, α smooth muscle
actin; NC, negative control. |
URG11 significantly increases the
expression of cyclin D1 and c-Myc in LNCaP cells, while siURG11
inhibits the levels of cyclin D1 and c-Myc
As the overexpression of URG11 and siURG11
significantly affected proliferation and cell cycle arrest in human
LNCaP cells, western blot analysis was performed to examine the
protein expression levels of cyclin D1 and c-Myc, which are
involved in the regulation of cell proliferation and the cell
cycle. It was identified that the transfection of LNCaP cells with
URG11 overexpression plasmid vectors significantly promoted the
expression of cyclin D1 and c-Myc at the mRNA (Fig. 6A) and protein levels (Fig. 6B and C). However, gene knockdown
of URG11 markedly inhibited the levels of cyclin D1 and c-Myc at
the mRNA (Fig. 6D) and protein
levels (Fig. 6E and F).
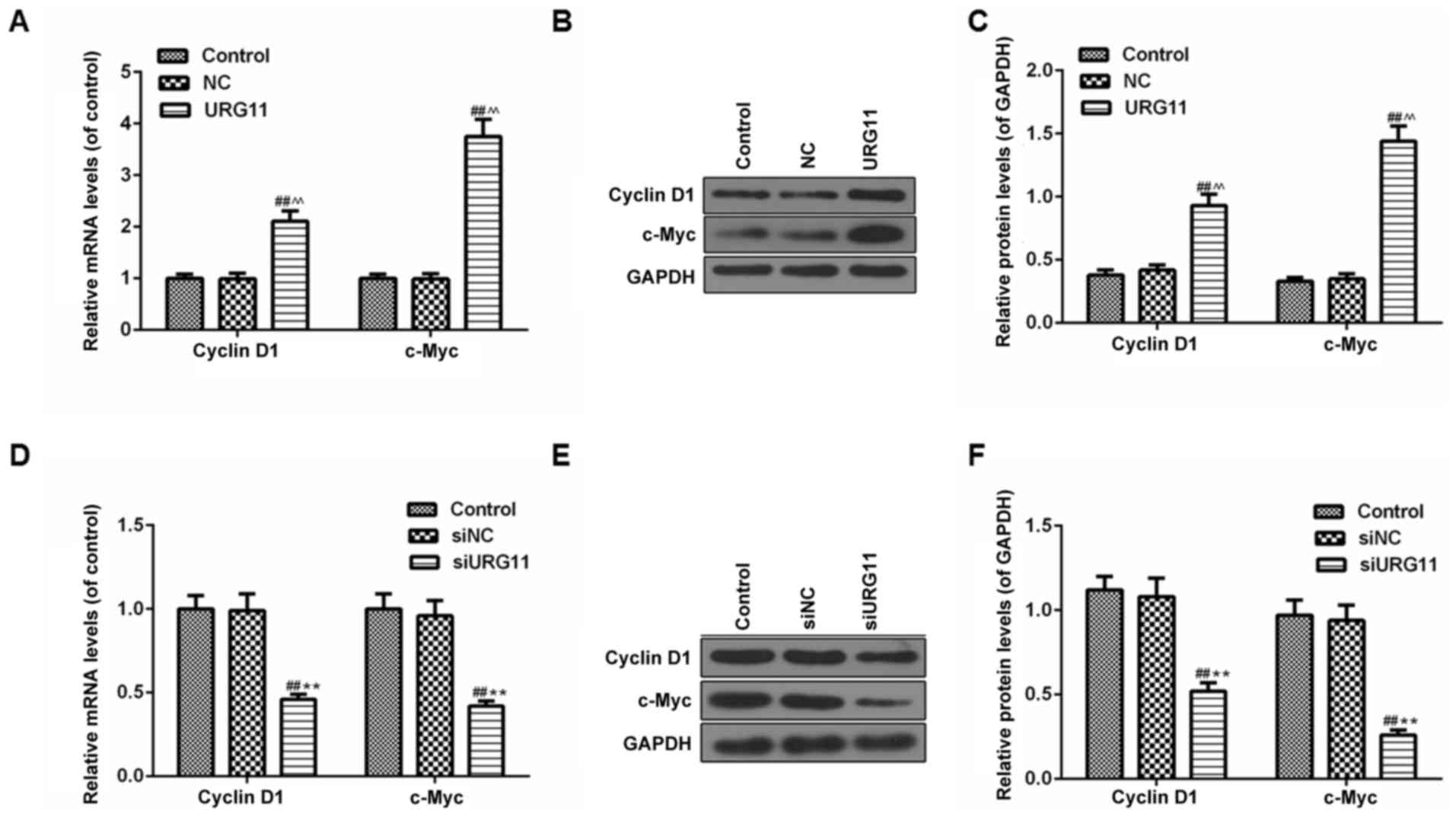 | Figure 6URG11 significantly increases the
expression of cyclin D1 and c-Myc in LNCaP cells, while siURG11
elicits the opposite effect. (A) The mRNA levels of cyclin D1 and
c-Myc in URG11-overexpressing cells were determined by RT-qPCR. (B)
The protein levels of cyclin D1 and c-Myc in URG11-overexpressing
cells were quantified by western blot analysis. (C) Densitometric
analysis of the cyclin D1 and c-Myc protein levels in
URG11-overexpressing cells, normalized to GAPDH. (D) The mRNA
levels of cyclin D1 and c-Myc in URG11-knockout cells were
determined by RT-qPCR. (E) The protein levels of cyclin D1 and
c-Myc in URG11-knockout cells were quantified by western blot
analysis. (F) Densitometric analysis of the cyclin D1 and c-Myc
protein levels in URG11-knockout cells, normalized to GAPDH. Error
bars represent standard deviation. ##P<0.01 vs.
control, ^^P<0.01 vs. NC, **P<0.01 vs.
siNC. URG11, von Willebrand factor C and EGF domain-containing
protein; c-Myc, MYC proto-oncogene protein; si, small interfering;
RT-qPCR, reverse transcription quantitative polymerase chain
reaction; NC, negative control. |
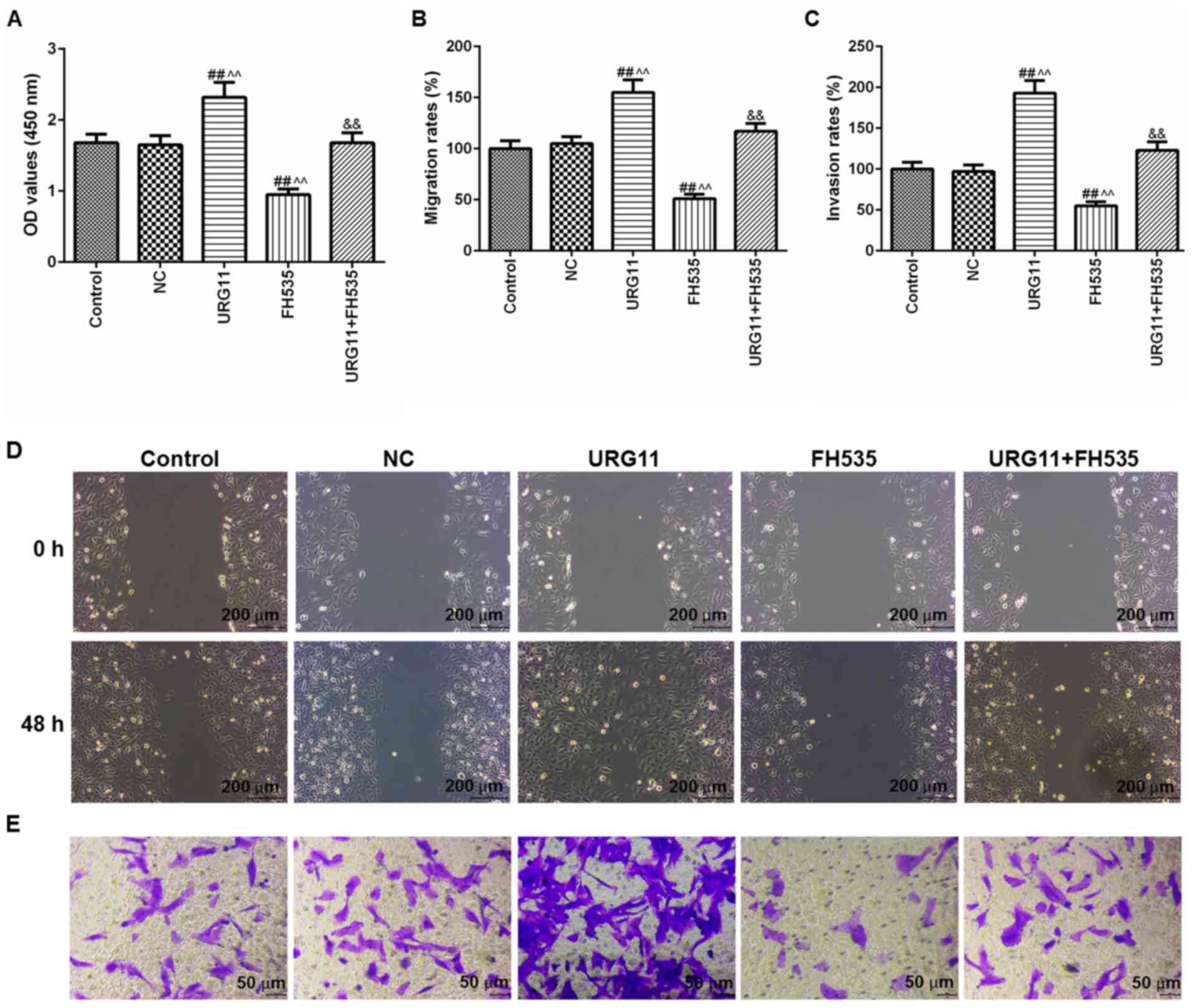
URG11 promotes cell viability, migration
and invasion, which is reversed by FH535
Due to the importance of Wnt/β-catenin signal
pathway in URG11-induced cell viability, migration and invasion in
LNCaP cells, the cells were treated with URG11 overexpression
plasmids and the Wnt/β-catenin inhibitor FH535 (20 µM),
together and individually. Then, the cell viability, migration and
invasion were respectively determined by CCK-8, wound healing and
Transwell assays. With these functional experiments, it was
identified that FH535 treatment significantly inhibited the plasmid
vector URG11 transfection-induced effects on cell viability
(Fig. 7A), migration (Fig. 7B and D) and invasion (Fig. 7C and E). The images of migration
and invasion were presented in Fig.
7D and E, respectively.
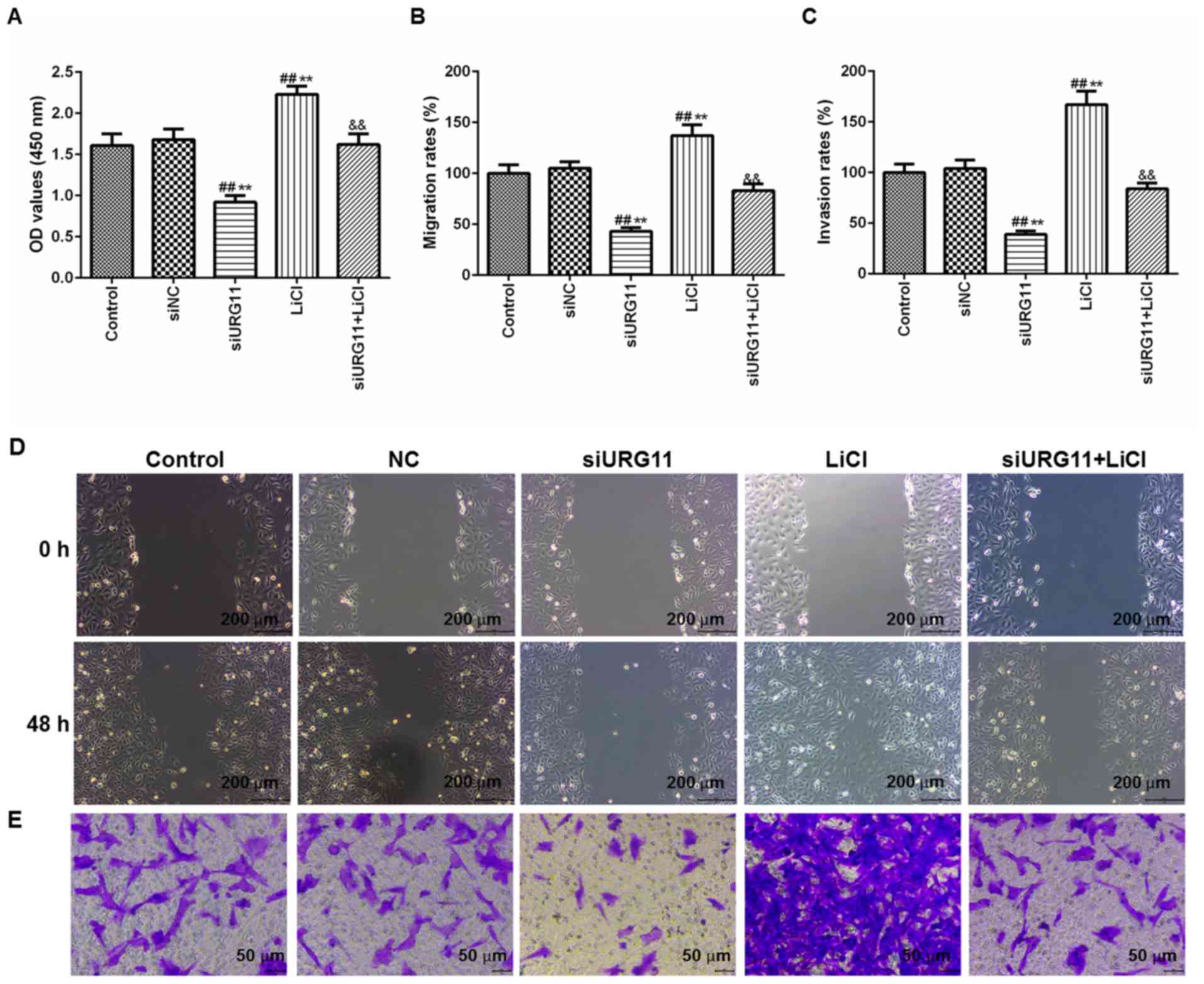
URG11 knockdown suppresses cell
viability, migration and invasion, which is reversed by LiCl
In order to additionally validate the role of the
Wnt/β-catenin signaling pathway in LNCaP cells proliferation, the
cells were treated with siURG11 and the Wnt/β-catenin agonist LiCl
(20 µM), together and individually. CCK-8, wound healing and
Transwell assays were also conducted to determine the effects on
cell viability, migration and invasion, respectively. Notably, it
was identified that that the cell viability (Fig. 8A), migration (Fig. 8B) and invasion (Fig. 8C) were increased in the siURG11+
LiCl group compared with those in the siURG11 group. The images of
migration and invasion were presented in Fig. 8D and E.
Discussion
Pca is the most common malignant tumor of the male
genitourinary system, and exhibited the fifth highest incidence
rate in 2008 worldwide (24).
Numerous studies have identified various agents that are able to
treat cancer cells; however, the majority of clinical trials have
failed to provide promising treatment options due to their
inefficiency or unexpected side effects (25,26). Therefore, studies investigating
novel molecules that may serve key roles in the development of Pca
will assist in developing novel therapeutic methods and targets,
which may be crucial for the improvement of the treatment and
prognoses of patients with Pca.
As an effector of hepatitis B virus X protein, URG11
is upregulated in various types of human cancer, including
hepatocellular carcinoma (8), and
gastric (9) and colon cancer
(12). In 2018, Pan et al
(12) identified that URG11 was
significantly upregulated in Pca. These studies indicated that
URG11 served an important role in the development of these types of
cancer. However, the underlying mechanisms of the URG11 gene in Pca
cells remain unknown.
According to a previous study, Peng et al
(10) identified that URG11
promoted pancreatic cancer invasion through EMT, leading to poor
prognosis. Fan et al (6)
demonstrated that the inhibition of URG11 on hepatocellular
carcinoma cells inhibited cell proliferation by downregulating G1-S
phase-associated proteins, and induced apoptosis by downregulating
B cell lymphoma 2. Gene knockdown by URG11 inhibited proliferation
of pancreatic cancer cells and suppressed invasion (10). Consistent with previous studies,
the data from the present study indicated that URG11 was
significantly upregulated in Pca cell lines, and that the
overexpression of URG11 promoted cell viability, migration and
invasion, and inhibited apoptosis and cell cycle arrest, whereas
inhibition of URG11 expression by interference RNA suppressed cell
viability, metastasis and invasion, and induced apoptosis and cell
cycle arrest. These data suggested that URG11 may be involved in
the development of Pca, as demonstrated by its effects in LNCaP
cells.
EMT is widely regarded as one of the important
factors that contribute to tumor invasion and metastasis (27). Downregulation of epithelial tissue
markers and upregulation of mesenchymal tissue markers are
important molecular events in the development of EMT (28). Silencing URG11 expression
inhibited EMT by altering E-cadherin, neural cadherin and vimentin
levels in prostatic hyperplasia cells (29). Overexpression of URG11 promoted
EMT accompanied by a downregulation of the epithelial marker
E-cadherin and upregulation of the mesenchymal markers vimentin and
α-SMA in a human proximal tubule cell line (30). The present study identified that
overexpression of URG11 attenuated the expression of E-cadherin and
increased the expression levels of vimentin and α-SMA in LNCaP
cells, while URG11 knockdown by siRNA effectively reversed this
effect on the EMT-associated proteins in the LNCaP cells. These
data demonstrated that URG11 accelerated the progression of Pca by
activating EMT. Therefore, targeting EMT may be a promising
treatment strategy for the management of Pca.
Wnt/β-catenin signaling pathway is an important
mechanism of action in various tumorigenesis and development
processes (31). The
Wnt/β-catenin pathway controls the expression of a number of
downstream target genes including cyclin D1 and c-Myc, thereby
promoting tumorigenesis (32,33). At present, β-catenin mutations or
dysregulation have been identified in various types of tumors
including colorectal (34), renal
(35), gastric (36) and liver cancer (37), and they participate in
tumorigenesis and malignant progression. A previous study suggested
that knockdown of URG11 inhibited β-catenin expression in non-small
cell lung cancer cells (11).
Accumulating studies have indicated that aberrant activation of
Wnt/β-catenin pathway is implicated in Pca tumorigenesis (38-40). In the present study, it was
identified that the mRNA and protein levels of cyclin D1 and c-Myc
were increased following URG11 overexpression. However, knockdown
of UGR11 effectively inhibited the expression of cyclin D1 and
c-Myc. LNCaP cells were treated with URG11 overexpression plasmids
and Wnt/β-catenin pathway inhibitor FH535, and with siURG11 and
Wnt/β-catenin pathway agonist LiCl; the results indicated that cell
viability, migration and invasion may be reversed in comparison
with the URG11 and siURG11 group, respectively. These results
suggested that the regulation of URG11 in Pca may be associated
with the Wnt/β-catenin signaling pathway. However, certain aspects
of the present study require additional investigation, including
the role of URG11 in Pca in vivo.
In conclusion, the present study provided evidence
that URG11 was positively associated with Pca tumorigenesis and
metastasis, and that the overexpression of URG11 promoted
proliferation, migration and invasion, and was involved in the
Wnt/β-catenin signaling pathway in Pca cells, while treatment with
siURG11 elicited the opposite effects. Taken together, these data
suggested that URG11 may serve an oncogene role in Pca, and that
URG11 may be involved in the early development and progression of
Pca. URG11 may be a potential novel clinical target for Pca.
Funding
No funding was received.
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
CS made substantial contributions to the conception
and design of the study. GZ and ML were responsible for data
acquisition, data analysis and interpretation. SC and HQ drafted
the article and critically revised it for important intellectual
content. All authors provided final approval of the version to
publish. DL and CS agree to be accountable for all aspects of the
work in ensuring that questions related to the accuracy or
integrity of the work are appropriately investigated and
resolved.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
VanderWalde A and Hurria A: Aging and
osteoporosis in breast and prostate cancer. CA Cancer J Clin.
61:139–156. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nelson BA, Shappell SB, Chang SS, Wells N,
Farnham SB, Smith JA Jr and Cookson MS: Tumour volume is an
independent predictor of prostate-specific antigen recurrence in
patients undergoing radical prostatectomy for clinically localized
prostate cancer. BJU Int. 97:1169–1172. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang HF, Wang HL, Xu N, Li SW, Ji GY, Li
XM, Pan YZ, Zhang L, Zhao XJ and Gao HW: Mass screening of 12,027
elderly men for prostate carcinoma by measuring serum prostate
specific antigen. Chin Med J (Engl). 117:67–70. 2004.
|
|
5
|
Jin Y, Chen Y, Jiang Y and Xu M: Proteome
analysis of the silkworm (Bombyx mori. L) colleterial gland during
different development stages. Arch Insect Biochem Physiol.
61:42–50. 2006. View Article : Google Scholar
|
|
6
|
Fan R, Li X, Du W, Zou X, Du R, Zhao L,
Luo G, Mo P, Xia L, Pan Y, et al: Adenoviral-mediated RNA
interference targeting URG11 inhibits growth of human
hepatocellular carcinoma. Int J Cancer. 128:2980–2993. 2011.
View Article : Google Scholar
|
|
7
|
Lian Z, Liu J, Li L, Li X, Tufan NL,
Clayton M, Wu MC, Wang HY, Arbuthnot P, Kew M, et al: Upregulated
expression of a unique gene by hepatitis B x antigen promotes
hepatocellular growth and tumorigenesis. Neoplasia. 5:229–244.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xie H and Liu J: Increased expression
URG11 in hepatocellular carcinoma tissues promotes the growth of
hepatocellular carcinoma cells. Xi bao yu fen zi mian yi xue za
zhi. 31:1523–1527. 2015.In Chinese. PubMed/NCBI
|
|
9
|
Du R, Xia L, Sun S, Lian Z, Zou X, Gao J,
Xie H, Fan R, Song J, Li X, et al: URG11 promotes gastric cancer
growth and invasion by activation of beta-catenin signalling
pathway. J Cell Mol Med. 14:621–635. 2010.
|
|
10
|
Peng W, Zhang J and Liu J: URG11 predicts
poor prognosis of pancreatic cancer by enhancing
epithelial-mesenchymal transition-driven invasion. Med Oncol.
31:642014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu ZL, Wu J, Wang LX, Yang JF, Xiao GM,
Sun HP and Chen YJ: Knockdown of Upregulated Gene 11 (URG11)
Inhibits Proliferation, Invasion, and β-Catenin Expression in
Non-Small Cell Lung Cancer Cells. Oncol Res. 24:197–204. 2016.
View Article : Google Scholar
|
|
12
|
Pan B, Ye Y, Liu H, Zhen J, Zhou H, Li Y,
Qu L, Wu Y, Zeng C and Zhong W: URG11 regulates prostate cancer
cell proliferation, migration, and invasion. BioMed Res Int.
2018:40607282018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen Y, Tan W and Wang C: Tumor-associated
macrophage-derived cytokines enhance cancer stem-like
characteristics through epithelial-mesenchymal transition.
OncoTargets Ther. 11:3817–3826. 2018. View Article : Google Scholar
|
|
14
|
Yan L, Xu F and Dai CL: Relationship
between epithelial-to-mesenchymal transition and the inflammatory
microenvironment of hepatocellular carcinoma. J Exp Clin Cancer
Res. 37:2032018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Brennen WN and Isaacs JT: Mesenchymal stem
cells and the embryonic reawakening theory of BPH. Nat Rev Urol.
15:703–715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Usova EV, Kopantseva MR, Egorov VI,
Kopantzev EP and Sverdlov ED: SNAI1 and SNAI2 - transcriptional
master-regulators of epithelial-mesenchimal transition. Patol
Fiziol Eksp Ter. 59:76–87. 2015.In Russian. PubMed/NCBI
|
|
17
|
Sung CO, Park CK and Kim SH:
Classification of epithelial- mesenchymal transition phenotypes in
esophageal squamous cell carcinoma is strongly associated with
patient prognosis. Modern pathology: An official journal of the
United States and Canadian Academy of Pathology Inc. 24:1060–1068.
2011. View Article : Google Scholar
|
|
18
|
Inoue T, Umezawa A, Takenaka T, Suzuki H
and Okada H: The contribution of epithelial-mesenchymal transition
to renal fibrosis differs among kidney disease models. Kidney Int.
87:233–238. 2015. View Article : Google Scholar
|
|
19
|
Lim SH, Becker TM, Chua W, Ng WL, de Souza
P and Spring KJ: Circulating tumour cells and the epithelial
mesenchymal transition in colorectal cancer. J Clin Pathol.
67:848–853. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Giles RH, van Es JH and Clevers H: Caught
up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta.
1653:1–24. 2003.PubMed/NCBI
|
|
21
|
Rabbani SA, Arakelian A and Farookhi R:
LRP5 knockdown: Effect on prostate cancer invasion growth and
skeletal metastasis in vitro and in vivo. Cancer Med. 2:625–635.
2013.
|
|
22
|
Dai J, Hall CL, Escara-Wilke J, Mizokami
A, Keller JM and Keller ET: Prostate cancer induces bone metastasis
through Wnt-induced bone morphogenetic protein-dependent and
independent mechanisms. Cancer Res. 68:5785–5794. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Ferlay J, Shin HR, Bray F, Forman D,
Mathers C and Parkin DM: Estimates of worldwide burden of cancer in
2008: Globocan 2008. Int J Cancer. 127:2893–2917. 2010. View Article : Google Scholar
|
|
25
|
Chang L, Graham PH, Hao J, Bucci J, Cozzi
PJ, Kearsley JH and Li Y: Emerging roles of radioresistance in
prostate cancer metastasis and radiation therapy. Cancer Metastasis
Rev. 33:469–496. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Alberti C: Prostate cancer:
Radioresistance molecular target-related markers and foreseeable
modalities of radiosensitization. Eur Rev Med Pharmacol Sci.
18:2275–2282. 2014.PubMed/NCBI
|
|
27
|
Singh A and Settleman J: EMT, cancer stem
cells and drug resistance: An emerging axis of evil in the war on
cancer. Oncogene. 29:4741–4751. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bronsert P, Enderle-Ammour K, Bader M,
Timme S, Kuehs M, Csanadi A, Kayser G, Kohler I, Bausch D, Hoeppner
J, et al: Cancer cell invasion and EMT marker expression: A
three-dimensional study of the human cancer-host interface. J
Pathol. 234:410–422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang G, Zhu F, Han G, Li Z, Yu Q, Li Z
and Li J: Silencing of URG11 expression inhibits the proliferation
and epithelial mesenchymal transition in benign prostatic
hyperplasia cells via the RhoA/ROCK1 pathway. Mol Med Rep.
18:391–398. 2018.PubMed/NCBI
|
|
30
|
Du R, Huang C, Bi Q, Zhai Y, Xia L, Liu J,
Sun S and Fan D: URG11 mediates hypoxia-induced
epithelial-to-mesenchymal transition by modulation of E-cadherin
and β-catenin. Biochem Biophys Res Commun. 391:135–141. 2010.
View Article : Google Scholar
|
|
31
|
Gupta A, Verma A, Mishra AK, Wadhwa G,
Sharma SK and Jain CK: The Wnt pathway: Emerging anticancer
strategies. Recent Pat Endocr Metab Immune Drug Discov. 7:138–147.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Vallée A, Lecarpentier Y, Guillevin R and
Vallée JN: Thermodynamics in gliomas: Interactions between the
canonical WNT/beta-catenin pathway and PPAR gamma. Front Physiol.
8:3522017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tang X, Wang Y, Fan Z, Ji G, Wang M, Lin
J, Huang S and Meltzer SJ: Klotho: A tumor suppressor and modulator
of the Wnt/β-catenin pathway in human hepatocellular carcinoma. Lab
Invest. 96:197–205. 2016. View Article : Google Scholar
|
|
34
|
Pandurangan AK, Divya T, Kumar K,
Dineshbabu V, Velavan B and Sudhandiran G: Colorectal
carcinogenesis: Insights into the cell death and signal
transduction pathways: A review. World J Gastrointest Oncol.
10:244–259. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li YL, Jin YF, Liu XX and Li HJ: A
comprehensive analysis of Wnt/β-catenin signaling pathway-related
genes and crosstalk pathways in the treatment of As2O3 in renal
cancer. Ren Fail. 40:331–339. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang XZ, Cheng TT, He QJ, Lei ZY, Chi J,
Tang Z, Liao QX, Zhang H, Zeng LS and Cui SZ: LINC01133 as ceRNA
inhibits gastric cancer progression by sponging miR-106a-3p to
regulate APC expression and the Wnt/β-catenin pathway. Mol Cancer.
17:1262018. View Article : Google Scholar
|
|
37
|
Adebayo Michael AO, Ko S, Tao J, Moghe A,
Yang H, Xu M, Russell JO, Pradhan-Sundd T, Liu S, Singh S, et al:
Inhibiting glutamine-dependent mTORC1 activation ameliorates Liver
cancers driven by β-catenin mutations. Cell Metab: Jan. 28:2019Epub
ahead of print.
|
|
38
|
Ren W, Wang D, Li C, Shu T, Zhang W and Fu
X: Capn4 expression is modulated by microRNA-520b and exerts an
oncogenic role in prostate cancer cells by promoting
Wnt/beta-catenin signaling. Biomed Pharmacother. 108:467–475. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhang Z, Cheng L, Li J, Farah E, Atallah
NM, Pascuzzi PE, Gupta S and Liu X: Inhibition of the Wnt/β-catenin
pathway overcomes resistance to enzalutamide in
castration-resistant prostate cancer. Cancer Res. 78:3147–3162.
2018.PubMed/NCBI
|
|
40
|
Sha J, Han Q, Chi C, Zhu Y, Pan J, Dong B,
Huang Y, Xia W and Xue W: PRKAR2B promotes prostate cancer
metastasis by activating Wnt/β-catenin and inducing
epithelial-mesenchymal transition. J Cell Biochem. 119:7319–7327.
2018. View Article : Google Scholar : PubMed/NCBI
|