Introduction
Acute myeloid leukemia (AML) is a hematologic
malignancy featured by the abnormal differentiation of myeloid
cells and growth inhibition of normal hematopoietic cells (1,2).
Similar to other types of cancer, AML is also associated with a
high morbidity and mortality rate (3). Currently, allogeneic stem cell
transplantation and chemotherapy are the main treatment methods for
AML (4,5). Although some advances in AML therapy
have been made, the survival rate of patients with AML remains
discouraging and the underlying molecular mechanisms of AML have
not yet been fully elucidated. Therefore, the identification of
novel drug targets and the development of accurate and rapid
therapeutic strategies is of utmost importance, as this would help
to improve the clinical curative effects and quality of life of
patients with AML.
Long non-coding RNAs (lncRNAs) are a class of
conserved non-coding RNAs that contain >200 nucleotides and
participate in the regulation of multifarious human diseases,
including cancers (6,7). Accumulating evidence has indicated
that lncRNAs play a critical role in the pathogenesis and
progression of AML (8). Previous
studies have found that lncRNA zinc finger E-box binding homeobox 2
(ZEB2)-antisense RNA 1 (AS1) may serve as a valuable biomarker for
the diagnosis and treatment of cancers, and that it promotes cell
proliferation and invasion in numerous types of cancer, including
gastric cancer (9), lung cancer
(10) and pancreatic cancer
(11). Moreover, a previous study
demonstrated that ZEB2-AS1 expression was notably upregulated in
AML, and that the elevated level of ZEB2-AS1 was closely associated
with a shorter overall survival; thus, ZEB2-AS1 may be regarded as
a prognostic indicator for AML (12). However, the mechanisms through
which ZEB2-AS1 regulates various biological processes in AML have
not yet been determined.
MicroRNAs (miRNAs or miRs) are a type of short
non-coding RNAs containing 21-25 nucleotides that play critical
roles in carcinogenesis (13,14). Increasing evidence supports the
hypothesis that deregulated miRNAs are intimately linked to the
occurrence and progression of various types of cancer, including
AML (8,15). Studies have also disclosed that
miR-122 expression is decreased and that this miRNA may be regarded
as a tumor suppressor in AML (16,17). Polo-like kinase 1 (PLK1) is a
vital mitotic regulator belonging to the polo-like kinase family
(18), and the overexpression of
PLK1 has been demonstrated to be a marker of a poor prognosis in a
number of types of cancer (19).
Therefore, it has become an attractive target for anticancer
therapy over the years. Furthermore, PLK1 has been shown to be
highly expressed in AML and the inhibition of PLK1 has been shown
to result in cell apoptosis in pre-clinical models (19,20). In addition, lncRNAs can act as
miRNA 'sponges' by sharing common miRNA response elements (MREs),
inhibiting normal miRNA targeting activity on messenger RNAs
(mRNAs) (21). However, the
underlying mechanisms of miR-122-5p in tumorigenesis, and the
correlation between lncRNA ZEB2-AS1, miR-122-5p and PLK1 in AML
need to be further clarified.
In the present study, the levels of ZEB2-AS1,
miR-122-5p and PLK1 in AML tissues and cells were investigated, and
an lncRNA-miRNA-mRNA competing endogenous RNA (ceRNA) network was
constructed. Furthermore, the effects of ZEB2-AS1 on the
proliferation and apoptosis of AML cells, as well as the underlying
mechanisms, were verified by gain- and loss-of-function
experiments.
Materials and methods
Patients and clinical tissues
Bone marrow specimens of 36 patients (17 females and
19 males; range, 3-12 years; mean age, 6.67 years) with AML at
diagnosis and 14 healthy controls (8 females and 6 males; range,
6-10 years; mean age, 8.29 years) enrolled in the present study
were collected from Heze Medical College (from January, 2017 to
February, 2019). The present study was approved by the Ethics
Committee of Heze Medical College. Moreover, the parents/legal
guardians of all the participants gave informed consents and
patients with AML did not receive any medical treatment prior to
the study.
Cells and cell culture
The human bone marrow stromal cell line, HS-5, and
the AML cell lines, HL-60, THP-1, U-937 and Kasumi-1, were obtained
from the American Type Culture Collection (ATCC). The AML cells
were grown in RPMI-1640 medium (Invitrogen; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum (FBS;
Gibco; Thermo Fisher Scientific, Inc.), and the HS-5 cells were
maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco;
Thermo Fisher Scientific, Inc.) supplemented with 10% FBS. All cell
lines were cultured at 37°C in an incubator with 5%
CO2.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was isolated from the AML tissues and
cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and quantified using a spectrophotometer (Bio-Rad
Laboratories, Inc.). The first strand of cDNA was reverse
transcribed from RNA using the PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd.). qPCR was adopted to examine the levels of
ZEB2-AS1, miR-122-5p and PLK1 using the Platinum SYBR-Green qPCR
SuperMix-UDG kit (Invitrogen; Thermo Fisher Scientific, Inc.) on a
7300 PCR System (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Using the following thermocycling conditions for PCR:
Starting at a high temperature of 95°C for 10 min, followed by
denaturation at 95°C for 10 sec, annealing at 60°C for 60 sec, and
extension at 72°C for 30 sec, with a total of 45 cycles. The
expression of ZEB2-AS1, miR-122-5p and PLK1 was quantified in
triplicate and calculated using the 2−ΔΔCq method
(22). U6 and
glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were served as
internal references. All the primers were obtained from Sangon
Biotech Co., Ltd. and are listed in Table I.
 | Table ISequences of primers used in the
present study. |
Table I
Sequences of primers used in the
present study.
| Gene | Forward primer | Reverse primer |
|---|
| ZEB2-AS1 |
5′-GGCTGGATAGCAAAGGAC-3′ |
5′-ACACTCTTGGCGAGGT-3′ |
| miR-122-5p |
5′-GTCACAATGGTGGAATGTGG-3′ |
5′-TAAGAATGTCATCTCCTTGAGGA-3′ |
| PLK1 |
5′-AAGAGATCCCGGAGGTCCTA-3′ |
5′-TCATTCAGGAAAAGGTTGCC-3′ |
| U6 |
5′-AACGCTTCACGAATTTGCGT-3′ |
5′-CCAAGCTTATGACAGCCATCATC-3′ |
| GAPDH |
5′-CAAGGTCATCCATGACAACTTTG-3′ |
5′-GTCCACCACCCTGTTGCTGTAG-3′ |
Cell transfection
ZEB2-AS1 or PLK1 overexpression vector
(pcDNA3.1-ZEB2-AS1 or pcDNA3.1-PLK1) and corresponding negative
control (pcDNA3.1-NC), small interfering RNA (siRNA) against
ZEB2-AS1 (si-ZEB2-AS1#1, #2, #3) and corresponding negative control
(si-NC), miR-122-5p mimic (miR-122-5p) and corresponding negative
control (miR-NC), miR-122-5p inhibitor (anti-miR-122-5p) and
corresponding negative control (anti-miR-NC) were acquired from
RiboBio Co., Ltd. Short hairpin small interfering RNA (shRNA) for
ZEB2-AS1 (sh-ZEB2-AS1) and negative control (sh-NC) were obtained
from Shanghai Genechem Co., Ltd. These plasmids (100 nM) and
si-ZEB2-AS1 (50 nM), miR-122-5p mimic (50 nM), miR-122-5p inhibitor
(50 nM), sh-ZEB2-AS1 (50 nM) and their controls were transfected
into AML cell lines using Lipofectamine 2000 reagent (Invitrogen;
Thermo Fisher Scientific, Inc.) referring to the user guide. Cells
(1×105/well) were reaped after 48 h for use in
subsequent experiments.
Methyl thiazolyl tetrazolium (MTT)
assay
The proliferation of the HL-60 and THP-1 cells was
examined by MTT assay. Transfected cells were cultivated in 96-well
plates for 1-3, or 4 days and 15 µl of MTT solution (Sangon
Biotech) at a 5 mg/ml concentration was then added to each well
followed by culture at 37°C for 4 h. Subsequently, the medium
containing MTT solution was discarded and 150 µl dimethyl
sulfoxide (DMSO; Sangon Biotech) was used to dissolve the product
of formazan. The absorbance was detected at 490 nm using microplate
reader (Bio-Rad Laboraroties, Inc.).
Apoptosis assay
The Annexin V-fluorescein isothiocyanate/propidium
iodide (Annexin V-FITC/PI) apoptosis assay kit (Invitrogen; Thermo
Fisher Scientific, Inc.) was employed to evaluate the apoptosis of
the HL-60 and THP-1 cells refer-ring to the directions of the
manufacturer. The apoptotic cells were assessed using a flow
cytometer (model: DxP 10; Cytek Biosciences).
Dual-luciferase reporter assay
The targeted sequence between miR-122-5p and
ZEB2-AS1 or PLK1 was predicted using the LncBase Predicted v.2
database (http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php?r=lncbasev2%2Findex)
or StarBase database (https://bio.tools/starbase; http://starbase.sysu.edu.cn/), indicating that
miR-122-5p was a target of ZEB2-AS1 and miR-122-5p targeted PLK1.
The luciferase reporter plasmids of wild-type ZEB2-AS1 (ZEB2-AS1
WT) or wild-type PLK (PLK WT) containing the potential binding
sequence of miR-122-5p and mutant type ZEB2-AS1 (ZEB2-AS1 MUT) or
mutant type PLK (PLK MUT) were constructed. For dual-luciferase
reporter assay, the HL-60 and THP-1 cells were maintained in
96-well plates and co-transfected with ZEB2-AS1 WT, ZEB2-AS1 MUT,
PLK WT, or PLK MUT, together with miR-122-5p or miR-NC using
Lipofectamine 2000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Following 36 h of transfection, HL-60 and THP-1 cells were
harvested and lysed to examine the luciferase activity using the
dual-luciferase reporter assay system (Promega Corp.). The
normalization of Firefly luciferase activity utilized the
Renilla luciferase activity as the control.
Western blot analysis
Total protein was segregated from the HL-60 and
THP-1 cells using RIPA Lysis buffer (Sangon Biotech) and quantified
using a Bradford Protein Assay kit (Sangon Biotech). The equal
amounts of protein (20 µg/lane) was isolated by 10% SDS-PAGE
and later transferred to a PVDF membrane (Sangon Biotech).
Subsequently, 5% non-fat milk was employed to block the membrane
for 2 h followed by the addition of the primary antibodies and
incubation for 24 h at 4°C, followed by the addition of horseradish
peroxidase (HRP)-conjugated secondary antibody for 1 h at 37°C. The
protein bands were visualized using an EasyBlot ECL kit (Sangon
Biotech). Finally, the protein expression levels were quantified
using by ImageJ software and normalized to GAPDH. The antibodies
used are listed as follows: PLK1 antibody (orb315655; 1:1,000;
Biorbyt), Bcl-2 associated X protein (Bax) antibody (orb4655;
1:500; Biorbyt), B-cell lymphoma/leukemia-2 (Bcl-2) antibody
(orb135113; 1:1,000; Biorbyt), cleaved caspase-3 (c-caspase-3)
antibody (orb106556; 1:1,000; Biorbyt) and GAPDH antibody
(orb38655; 1:1,000; Biorbyt).
Mouse tumor xenograft model
The animal experiments were approved by the
Committee on the Use and Care of Animals of Heze Medical College. A
total of 14 BALB/c male nude mice (5-6 weeks old, weighing 16±2 g)
were acquired from the Guangdong Medical Laboratory Animal Center.
Mice were maintained under standard housing conditions, with a
temperature of 18-23°C, 12 h light/dark cycle, 45-65% humidity, and
free access to water and food. Firstly, the THP-1 cells
(1×106 cell/ml) were stably injected into the flanks of
the nude mice (7 mice/group). After 7 days of the first cell
injection, the THP-1 cells stably transfected with sh-ZEB2-AS1 or
negative control (sh-NC) in 20 µl of phosphate-buffered
saline (PBS) were injected into the implanted tumors every 3 days
for 7 times. After the injection, the tumor size was estimated
every week using a Vernier caliper, and tumor volume was estimated
following the formula: Volume=length x width2/2. Mice
were examined daily to ensure that they were able to eat, drink,
defecate and ambulate normally. If any mice exhibited evidence of
physical deficits, such as tacky mucous membranes, dry or dull
eyes, a decrease in ambulation, in dyspnea or cachexia (loss of 15%
of animal original body weight), they were considered to have met
the criteria for euthanasia. No mouse died prior to the completion
of the experiment. At 42 days after the first injection, the mice
were sacrificed by cervical dislocation following sedation with
isoflurane (2%). The tumors were excised from all the mice and
collected for subsequent RT-qPCR assay. Each individual animal had
a subcutaneous tumor, and the maximum diameter/volume of a single
subcutaneous tumor was 11.2 mm/568.60 mm3.
Statistical analysis
All data are presented as the means ± standard
deviation (SD) based on >3 replicates. Statistical analysis was
performed with the Student's t test and one-way analysis of
variance (ANOVA) followed by Tukey's post-hoc test using SPSS 21.0
software. The correlation between the expression of ZEB2-AS1,
miR-122-5p and PLK1 in AML tissues was analyzed using Pearson's
correlation coefficient. A Kaplan-Meier survival curve was obtained
by using the log-rank test to assess the cumulative (Cum) survival.
P<0.05 was considered to indicate a statistically significant
difference.
Results
ZEB2-AS1 expression is upregulated in
AML
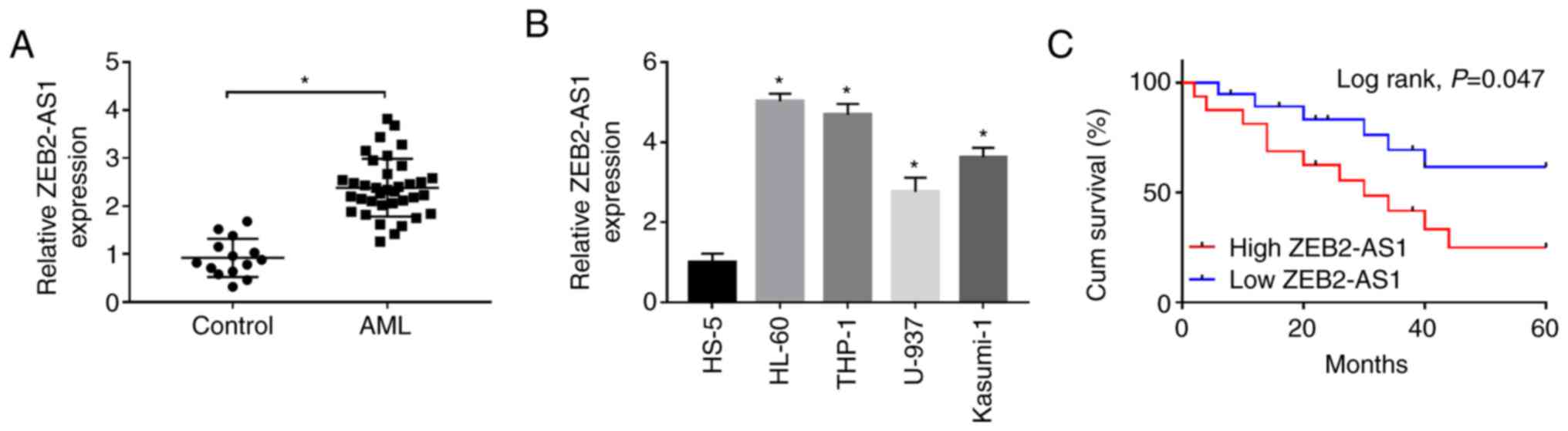
Firstly, the expression of ZEB2-AS1 in AML tissues
(n=36) and healthy control tissues (n=14) was examined by RT-qPCR.
The results revealed that the expression level of ZEB2-AS1 was
significantly increased in AML tissues compared with healthy
tissues (Fig. 1A). Moreover, the
expression level of ZEB2-AS1 in the human bone marrow stromal cell
line, HS-5, was visibly lower than that in the AML cell lines,
HL-60, THP-1, U-937 and Kasumi-1 (Fig. 1B). To examine the effects of
ZEB2-AS1 on survival rate, the patients were divided into the high
ZEB2-AS1 expression group and low ZEB2-AS1 expression group based
on ZEB2-AS1 expression, with the median of ZEB2-AS1 as the
threshold. As depicted in Fig.
1C, the Kaplan-Meier survival curve demonstrated that patients
with AML with a high ZEB2-AS1 expression exhibited a clearly
shorter cum survival (P= 0.047) in comparison with that of patients
in the low ZEB2-AS1 expression group. From these data, it was thus
suggested that a high expression of ZEB2-AS1 is associated with a
poor survival rate of patients with AML. The HL-60 and THP-1 cells
were selected for use in subsequent experiments as they exhibited
the highest expression of ZEB2-AS1.
Knockdown of ZEB2-AS1 inhibits the
proliferation and promotes the apoptosis of AML cells
To explore the role of ZEB2-AS1 in the proliferation
and apoptosis of AML cells, the HL-60 and THP-1 cells were
transfected with si-ZEB2-AS1 (#1, #2, #3) or negative control
si-NC. The results revealed that the expression level of ZEB2-AS1
in HL-60 and THP-1 cells following ZEB2-AS1 knockdown was markedly
decreased compared with the negative control si-NC group, which
indicated that the transfection was successful (Fig. 2A). Moreover, si-ZEB2-AS1#1 which
produced the most prominent decrease in the expression of ZEB2-AS1
was selected for use in follow-up experiments. The proliferation of
the HL-60 and THP-1 cells following transfection with si-ZEB2-AS1#1
was markedly lower than that of the si-NC group, as revealed by MTT
assay (Fig. 2B and C).
Simultaneously, flow cytometric analysis revealed that the
apoptotic rate of the HL-60 and THP-1 cells transfected with
si-ZEB2-AS1#1 was markedly increased, as demonstrated using the
Annexin V-FITC/PI apoptosis assay kit (Fig. 2D). To further examine the effects
of ZEB2-AS1 on the levels of the apoptosis-associated proteins,
Bcl-2, Bax and c-caspase-3, western blot analysis was performed.
The results revealed that the expression levels of the
pro-apoptotic proteins, Bax and c-caspase-3, were increased, while
those of the anti-apoptotic protein, Bcl-2, were decreased in the
HL-60 and THP-1 cells transfected with si-ZEB2-AS1#1 compared with
the si-NC group (Fig. 2E). These
data suggested that ZEB2-AS1 silencing suppressed the proliferation
and promoted the apoptosis of AML cells.
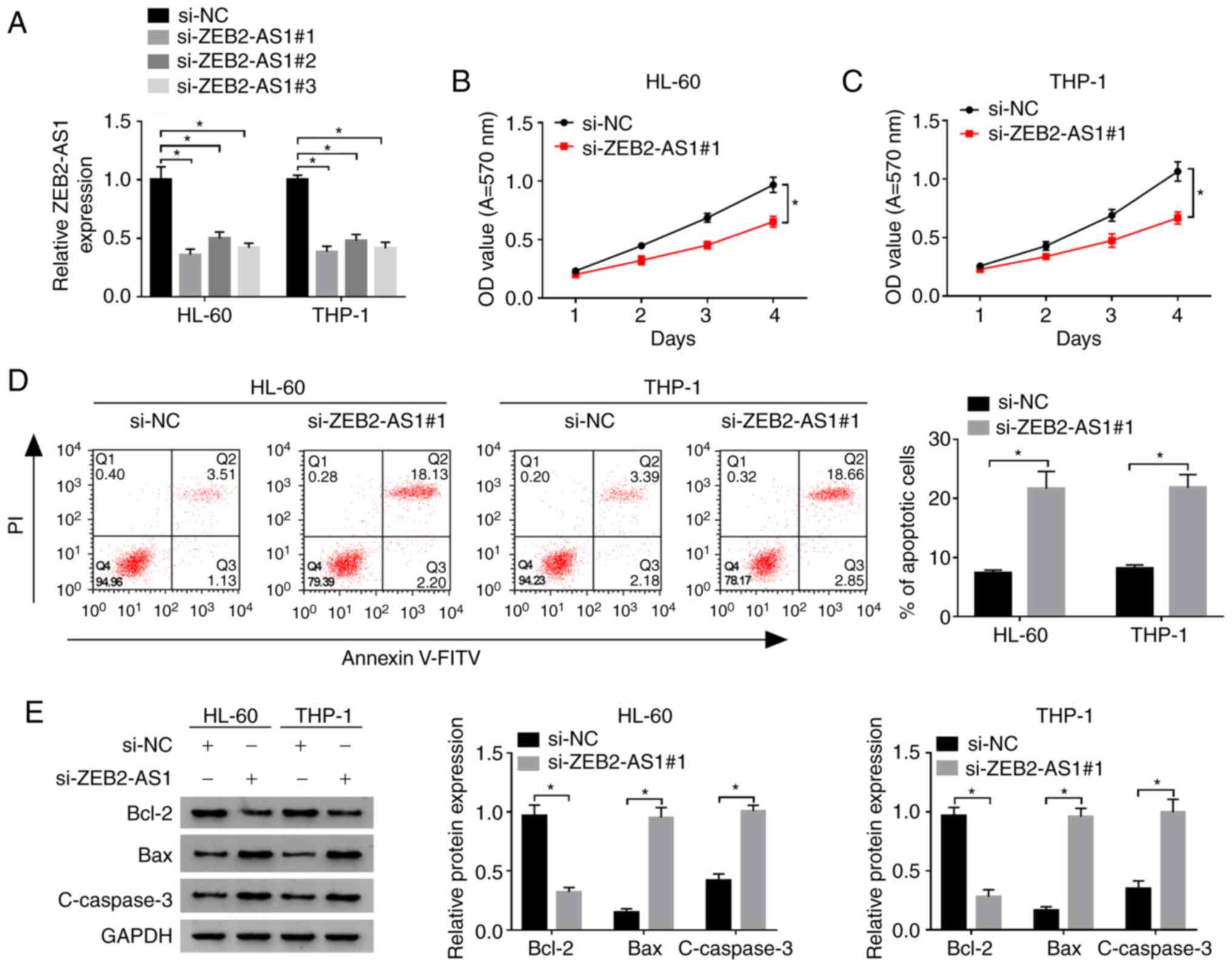 | Figure 2Effects of ZEB2-AS1 on the
proliferation and apoptosis of AML cells. (A) The expression of
ZEB2-AS1 in HL-60 and THP-1 cells transfected with si-ZEB2-AS1 (#1,
#2, #3) or control si-NC was detected by RT-qPCR. (B and C) The
proliferation of HL-60 and THP-1 cells transfected with
si-ZEB2-AS1#1 or si-NC was examined by MTT assay. (D) The apoptotic
rate of the HL-60 and THP-1 cells transfected with si-ZEB2-AS1#1 or
si-NC was detected by flow cytometry. (E) The expression of
apoptosis-associated proteins in HL-60 and THP-1 cells transfected
with si-ZEB2-AS1#1 or si-NC was examined by western blot analysis.
*P<0.05. ZEB2-AS1, zinc finger E-box binding homeobox
2-antisense RNA 1; si, small interfering; NC, negative control; OD,
optical density; V-FITC, V-fluorescein isothiocyanate; PI,
propidium iodide; Bcl-2, B-cell lymphoma/leukemia-2; Bax, Bcl-2
Associated X Protein; c-caspase-3, cleaved caspase-3; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase. |
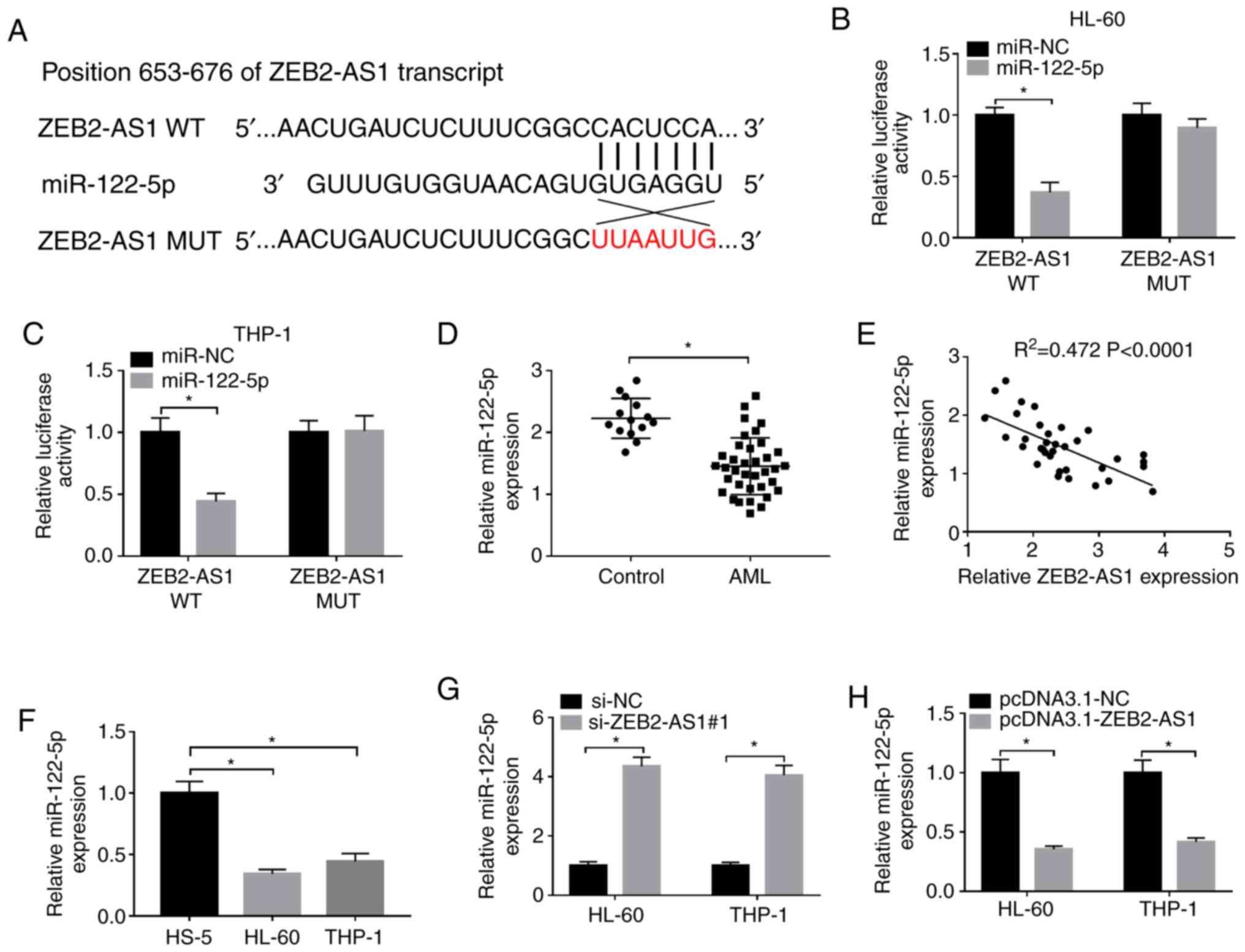
ZEB2-AS1 targets miR-122-5p and
negatively regulates the expression of miR-122-5p in AML
To explore the association between ZEB2-AS1 and
miR-122-5p, the binding sites were predicted using the LncBase
Predicted v.2 database (Fig. 3A).
The related luciferase reporter plasmids containing the binding
sites of ZEB2-AS1 WT or ZEB2-AS1 MUT were then constructed. The
results indicated that miR-122-5p markedly decreased the
lucif-erase activity of ZEB2-AS1 WT in both the HL-60 and THP-1
cells, while the luciferase activity of ZEB2-AS1 MUT was unaffected
following transfection with miR-122-5p (Fig. 3B and C). The expression level of
miR-122-5p in the AML tissues and cells was notably decreased,
detected by RT-qPCR (Fig. 3D and
F). Moreover, correlation analysis revealed that a negative
correlation between the expression levels of miR-122-5p and
ZEB2-AS1 in AML tissues (Fig.
3E). Moreover, the expression level of miR-122-5p in the HL-60
and THP-1 cells transfected with si-ZEB2-AS1#1 was evidently
increased, while it was evidently downregulated following the
overexpression of ZEB2-AS1 (Fig. 3G
and H). The above data confirmed that miR-122-5p was a direct
target of ZEB2-AS1 and ZEB2-AS1 downregulated the expression of
miR-122-5p in AML.
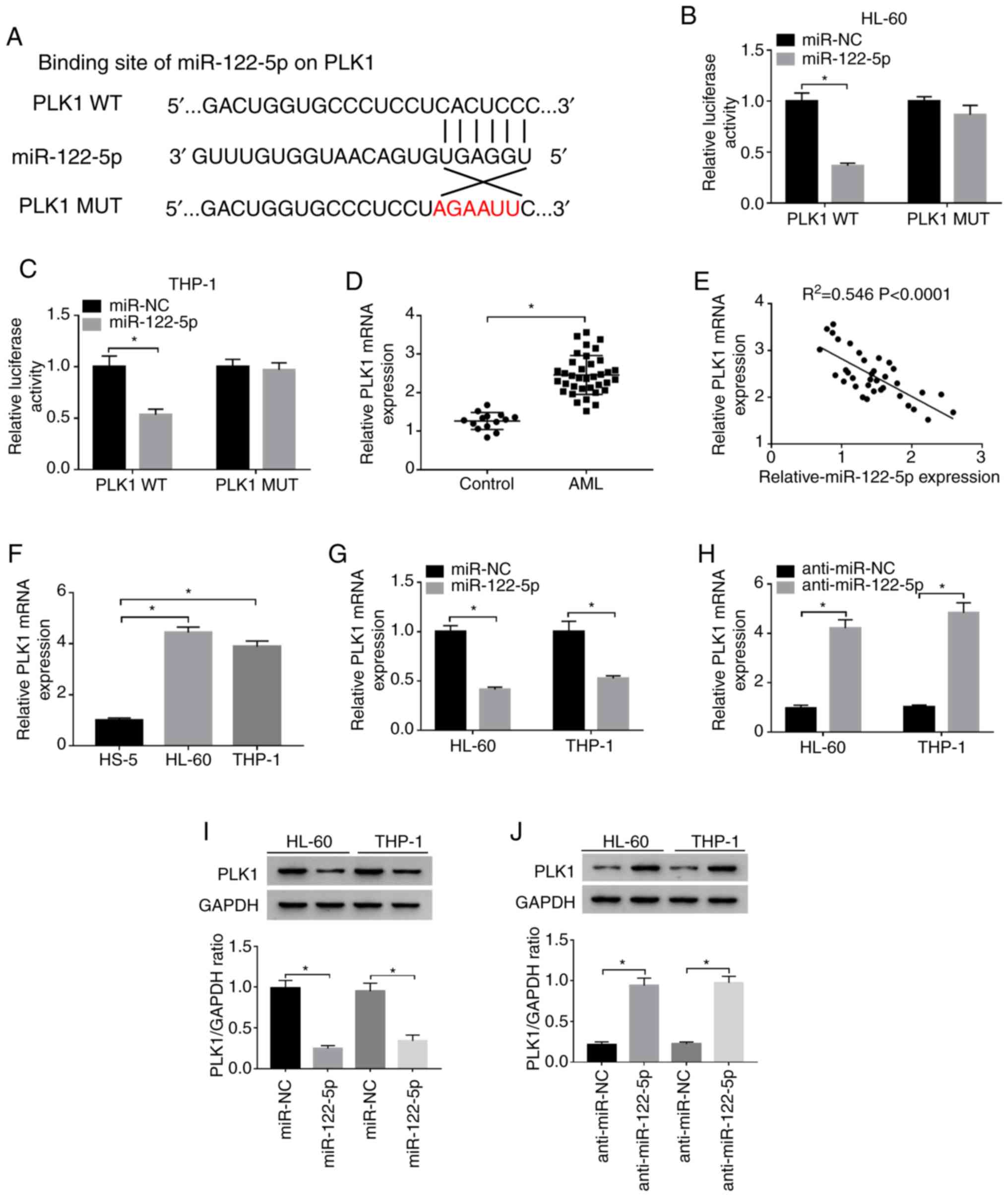
PLK1 is a target of miR-122-5p in
AML
Furthermore, miR-122-5p was predicted to include the
binding sequence of PLK1 using the StarBase database (Fig. 4A). Moreover, the luciferase
activity of the HL-60 and THP-1 cells co-transfected with
miR-122-5p and PLK1 WT was significantly suppressed, whereas the
luciferase activity was only minimally altered following
transfection with PLK1 MUT, as determined by dual-luciferase
reporter assay (Fig. 4B and C).
The mRNA expression of PLK1 was markedly upregulated and negatively
correlated with the expression of miR-122-5p in AML tissues,
detected by RT-qPCR and correlation analysis, respectively
(Fig. 4D and E). Similarly, the
mRNA expression of PLK1 in the HL-60 and THP-1 cells was markedly
increased compared with that of the HS-5 cells (Fig. 4F). To confirm the regulatory
effects between miR-122-5p and PLK1, the mRNA and protein
expression levels of PLK1 in the HL-60 and THP-1 cells transfected
with miR-122-5p, miR-NC, anti-miR-122-5p or anti-miR-NC were
examined by RT-qPCR or western blot analysis. As depicted in
Fig. 4G-J, the expression of PLK1
was markedly downregulated in the HL-60 and THP-1 cells following
the overexpression of miR-122-5p, while the expression of PLK1 was
evidently upregulated following the knockdown of miR-122-5p. Taken
together, these data indicated that PLK1 was a target of miR-122-5p
in AML.
Overexpression of PLK1 reverses the
effects of miR-122-5p overexpression on the proliferation and
apoptosis of AML cells
To explore the association between miR-122-5p and
PLK1 as regards the proliferation and apoptosis of AML cells, the
HL-60 and THP-1 cells were transfected with miR-NC, miR-122-5p,
miR-122-5p + pcDNA3.1-NC or miR-122-5p + pcDNA3.1-PLK1. The mRNA
expression of PLK1 in HL-60 and THP-1 cells transfected with
miR-122-5p or miR-122-5p + pcDNA3.1-NC was significantly lower than
that in the miR-NC group, as shown by RT-qPCR, while the expression
of PLK1 was evidently enhanced in the miR-122-5p + pcDNA3.1-PLK1
group (Fig. 5A). In addition, the
proliferation of the HL-60 and THP-1 cells following the
overexpression of miR-122-5p was inhibited compared with the miR-NC
group, as shown by MTT assay, whereas this effect was partially
reversed by co-transfection with miR-122-5p and pcDNA3.1-PLK1
(Fig. 5B and C). In accordance
with this, the increase in the cell apoptosis of the HL-60 and
THP-1 cells induced by transfection with miR-122-5p or miR-122-5p +
pcDNA3.1-NC was partly reversed in the miR-122-5p + pcDNA3.1-PLK1
group, as shown by flow cytometry (Fig. 5D). Synchronously, the expression
of Bcl-2 was markedly decreased, and the expression levels of Bax
and c-caspase-3 were notably increased in the HL-60 and THP-1 cells
following the overexpression of miR-122-5p; these effects were
partly eliminated by transfection with pcDNA3.1-PLK1 (Fig. 5E). Collectively, these findings
confirmed that miR-122-5p inhibited the proliferation and induced
the apoptosis of AML cells by targeting PLK1.
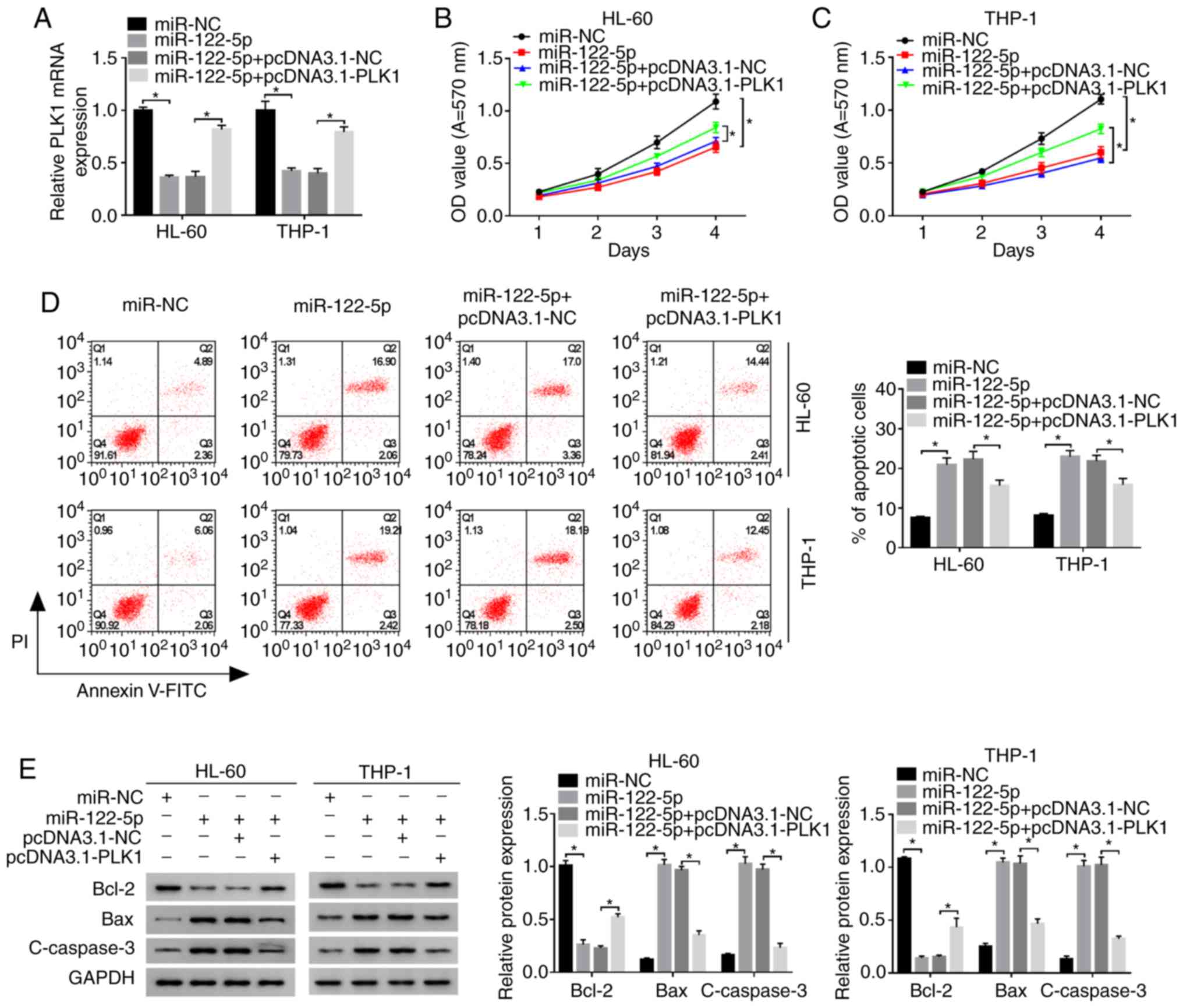 | Figure 5miR-122-5p regulates the
proliferation and apoptosis of AML cells by targeting PLK1. HL-60
and THP-1 cells were transfected with miR-122-5p, miR-122-5p +
pcDNA3.1-PLK1 or corresponding negative controls. (A) The mRNA
expression of PLK1 was detected by RT-qPCR. (B and C) The
proliferation of HL-60 and THP-1 cells following transfection was
detected by MTT assay. (D) The apoptotic rate was detected by flow
cytometry using an Annexin V-FITC/PI apoptosis assay kit. (E) The
expression levels of apoptosis-associated proteins in HL-60 and
THP-1 cells following transfection were examined by western blot
analysis. *P<0.05. PLK1, polo-like kinase 1; mRNA,
messenger RNA; miR, microRNA; NC, negative control; OD, optical
density; Bcl-2, B-cell lymphoma/leukemia-2; Bax, Bcl-2-associated X
protein; c-caspase-3, cleaved caspase-3; GAPDH,
glyceraldehyde-3-phosphate dehydrogenase. |
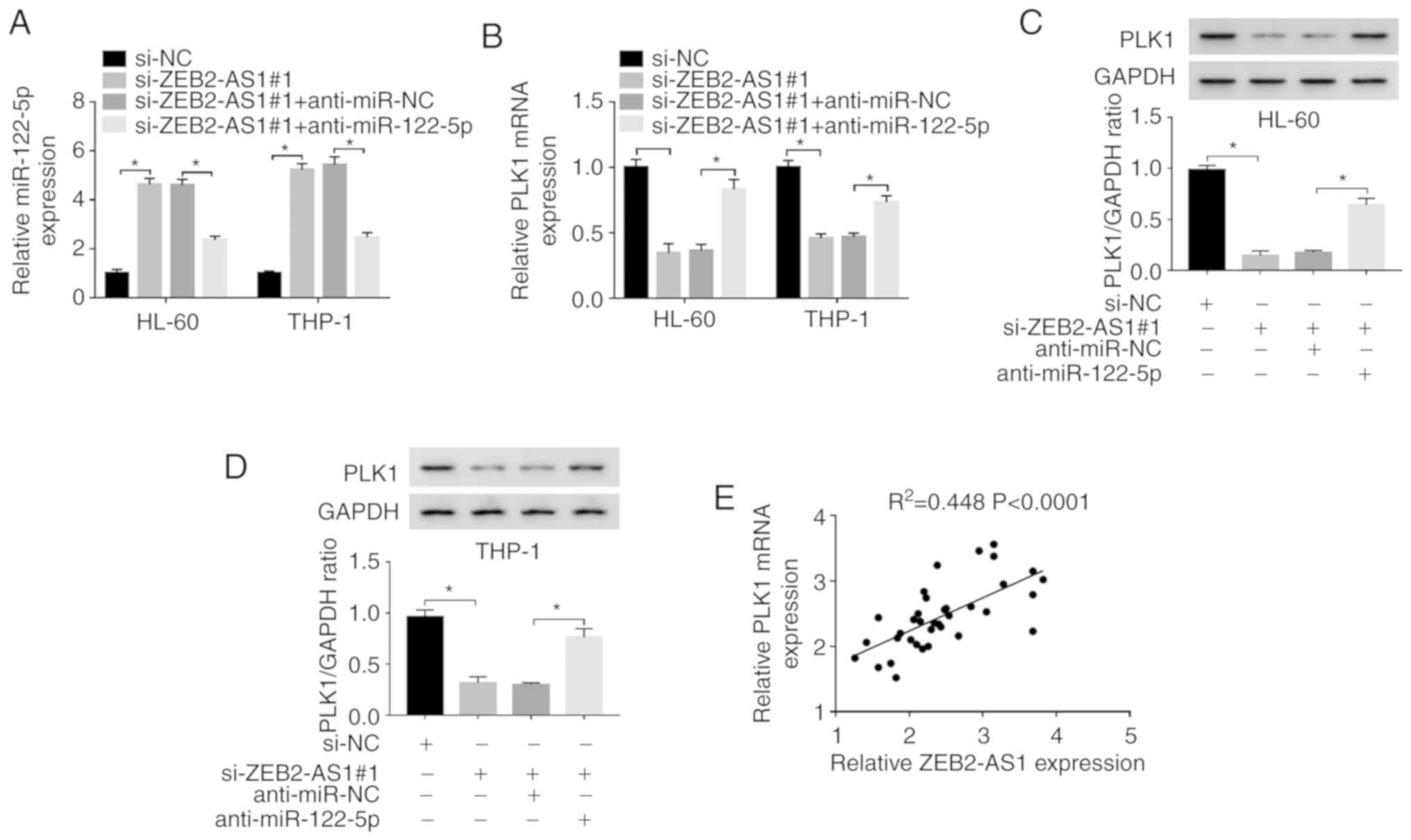
ZEB2-AS1 regulates the expression of PLK1
via miR-122-5p in AML cells
Based on the above-mentioned results, the
association between ZEB2-AS1 and PLK1 was further explored.
Firstly, the HL-60 and THP-1 cells were transfected with si-NC,
si-ZEB2-AS1#1, si-ZEB2-AS1#1 + anti-miR-NC, or si-ZEB2-AS1#1 +
anti-miR-122-5p. The expression of miR-122-5p was overtly increased
in the HL-60 and THP-1 cells following transfection with
si-ZEB2-AS1#1 or si-ZEB2-AS1#1 + anti-miR-NC, whereas it was
effectively decreased in the si-ZEB2-AS1#1 + anti-miR-122-5p group
(Fig. 6A). Moreover, the mRNA and
protein expression levels of PLK1 were markedly downregulated in
the HL-60 and THP-1 cells following the knockdown of ZEB2-AS1,
while this downregulation was efficiently attenuated by
anti-miR-122-5p, as determined by RT-qPCR and western blot
analysis, respectively (Fig.
6B-D). Finally, it was confirmed that there was a positive
correlation between the expression of ZEB2-AS1 and PLK1 in AML
tissues by a correlation analysis (Fig. 6E). Taken together, it was
concluded that ZEB2-AS1 positively regulated the expression of PLK1
by sponging miR-122-5p in AML cells.
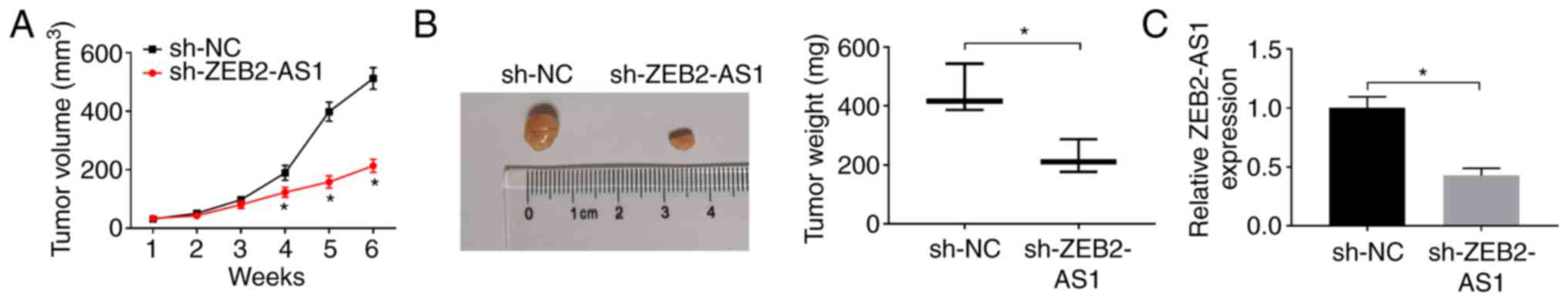
Knockdown of ZEB2-AS1 inhibits tumor
growth in vivo
In the present study, it was demonstrated that the
knockdown of ZEB2-AS1 impeded the proliferation and promoted the
apoptosis of AML cells. To further confirm this result, a THP-1
cell xenograft mouse model was established. THP-1 cells transfected
with sh-ZEB2-AS1 or sh-NC were injected into nude mice in order to
observe the effects of ZEB2-AS1 on tumor formation. As was
expected, the mice injected with si-ZEB2-AS1 exhibited a smaller
tumor volume and a lower tumor weight compared with those injected
with sh-NC control cells in vivo (Fig. 7A and B). Moreover, the expres-sion
of ZEB2-AS1 was significantly downregulated in the tumor tissues
(Fig. 7C), as determined by
RT-qPCR. Thus, it was concluded that the knockdown of ZEB2-AS1
suppressed tumor growth in vivo.
Discussion
AML is a malignant type of blood cancer featured by
the abnormal proliferation of myeloid cells and the growth
inhibition of normal hematopoietic cells (23-25). In recent years, in spite of some
progress being made in the treatment of AML and the availability of
several treatment methods, the prognosis of patients with AML
remains unsatisfactory (25).
Moreover, the pathogenesis of AML is very complicated and the
precise mechanisms in the occurrence and development of AML have
not yet been completely illuminated. Therefore, it is imperative to
explore the potential mechanisms responsible for the development of
AML and to identify novel drug targets and therapeutic strategies.
In the present study, the expression trend and effects of ZEB2-AS1
on the proliferation and apoptosis of AML cells were first
confirmed. The ZEB2-AS1/miR-122-5p/PLK1 regulatory axis was then
constructed to illuminate the mechanisms of action of ZEB2-AS1 in
AML.
lncRNAs have been indicated to play a pivotal role
in the cancer progression by modulating the expression of miRNAs or
target proteins (26).
Furthermore, a number of studies have revealed that lncRNAs play an
important role in the proliferation, apoptosis and differentiation
of leukemia cells (8). Recently,
emerging evidence has indicated that ZEB2-AS1 is a novel
tumor-associated lncRNA which accelerates cancer progression
(11). Certain studies have
demonstrated that ZEB2-AS1 expression is notably elevated, and
promotes cell proliferation and metastasis in numerous types of
cancer, such as pancreatic cancer (11) breast cancer (27) and bladder cancer (28). Recent research has also suggested
that the level of ZEB2-AS1 is notably enhanced in AML and that
ZEB2-AS1 silencing effectively decreases the invasion and migration
of AML cells; furthermore, the increased ZEB2-AS1 level is strongly
associated with a shorter overall survival (12). In accordance with this, the
present study demonstrated that ZEB2-AS1 expression was upregulated
in AML and was associated with a poor survival rate of patients
with AML. In addition, the silencing of ZEB2-AS1 suppressed the
proliferation and accelerated the apoptosis of AML cells.
Simultaneously, the silencing of ZEB2-AS1 also hampered tumor
growth in vivo in a THP-1 tumor xenograft mouse model. These
data certified that ZEB2-AS1 functioned as a tumor-promoting gene
in AML and that it may promote the progression of AML by promoting
the proliferation and inhibiting the apoptosis of AML cells.
miR-122 has been reported as a tumor-inhibiting
factor in variety of human cancers (29). An increasing amount of evidence
suggests that miR-122-5p is significantly downregulated, and
inhibits cell proliferation and metastasis in numerous types of
cancer, including nasopharyngeal carcinoma (30), hepatocellular carcinoma (31) and bile duct carcinoma (32). Moreover, some studies have
indicated that the level of miR-122 is significantly decreased in
bone marrow samples from patients with AML and is associated with a
lower shorter overall survival rate (16,17). The present study indicated that
the level of miR-122-5p was decreased in AML tissues and cells,
which was consistent with the above-mentioned findings. However, in
contrast to the findings of these other studies, the present study
found that ZEB2-AS1 targeted miR-122-5p, and that the levels of
ZEB2-AS1 and miR-122-5p were inversely correlated in AML.
Simultaneously, ZEB2-AS1 adversely regulated the expression of
miR-122-5p in AML cells. Furthermore, it was verified that
miR-122-5p targeted PLK1 and that there was an inverse correlation
between the levels of miR-122-5p and PLK1. Accordingly, it was
hypothesized that the interaction of miR-122-5p and PLK1 may play a
pivotal role in the progression of AML.
Recent studies have indicated that lncRNAs and mRNAs
function as ceRNAs that competitively bind to MREs to perform
specific biological functions during tumorigenesis, forming the
post-transcriptional ceRNA network to regulate mRNA expression
(21,33). PLK1 is a pivotal kinase for the
progression of mitotic cell cycle and is associated with a poor
prognosis in cancers (34). PLK1
overexpression is regarded as a marker for poor prognosis in a
number of types of cancer, and PLK1 has become an attractive target
for anticancer therapy over the past several years (19). Studies have demonstrated that PLK1
is overexpressed in a variety of cancers, and that it regulates
proliferation, migration and apoptosis on various cancer types,
such as gastric cancer (35),
non-small cell lung cancer (36)
and pancreatic cancer (37).
Moreover, it has been suggested that PLK1 silencing may lead to
cell cycle arrest and apoptosis in AML (20). Similarly, the mRNA expression of
PLK1 was upregulated and negatively correlated with the expression
of miR-122-5p in AML tissues in the present study. Moreover, the
overexpression of PLK1 reversed the effects of miR-122-5p
overexpression on the proliferation and apoptosis of AML cells,
which verified that miR-122-5p impedes the progression of AML by
sponging PLK1. Finally, loss-of-function experiments demonstrated
that the downregulation of PLK1 expression induced by ZEB2-AS1
silencing was effectively reversed by anti-miR-122-5p. Thus, all
these data indicate that ZEB2-AS1 regulates the proliferation and
apoptosis of AML cells by functioning as a ceRNA of miR-122-5p to
increase the expression of PLK1.
Certain limitations to the present study should be
stated, however. For instance, apart from dual-luciferase reporter
assay, RNA immunoprecipitation (RIP) assay and RNA pull-down assay
need to carry out to further verify the binding association between
miR-122-5p and ZEB2-AS1 or PLK1. Another limitation of the present
study was the small sample size of the subjects. Therefore, the
authors aim to further explore the above-mentioned results in
future studies.
In conclusion, the present study verified the
involvement of ZEB2-AS1, PLK1 and miR-122-5p in AML tissues and
cells, and constructed the ZEB2-AS1/miR-122-5p/PLK1 ceRNA network.
ZEB2-AS1 regulates the proliferation and apoptosis of AML cells by
sponging miR-122-5p, thus reinforcing the protein level of PLK1.
Therefore, the findings of the present study may enhance our
understanding of the underlying mechanisms of ZEB2-AS1 in the
progression of AML, and provide potential novel molecular targets
for the treatment of AML.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JG conceived and designed the study. PL, AW and BW
performed the data analyses and interpretations. JG and BW wrote
the manuscript. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Heze Medical College. All parents/legal guardians of
the patients involved in the study provided informed consents. The
animal experiments were approved by the Committee on the Use and
Care of Animals of Heze Medical College.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Short NJ, Rytting ME and Cortes JE: Acute
myeloid leukaemia. Lancet. 392:593–606. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Burnett A, Wetzler M and Löwenberg B:
Therapeutic advances in acute myeloid leukemia. J Clin Oncol.
29:487–494. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
De Kouchkovsky I and Abdul-Hay M: Acute
myeloid leukemia: A comprehensive review and 2016 update. Blood
Cancer J. 6:e4412016. View Article : Google Scholar
|
|
4
|
Roboz GJ: Current treatment of acute
myeloid leukemia. Curr Opin Oncol. 24:711–719. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bullinger L, Döhner K and Döhner H:
Genomics of acute myeloid leukemia diagnosis and pathways. J Clin
Oncol. 35:934–946. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ernst C and Morton CC: Identification and
function of long non-coding RNA. Front Cell Neurosci. 7:1682013.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Liu Y, Cheng Z, Pang Y, Cui L, Qian T,
Quan L, Zhao H, Shi J, Ke X and Fu L: Role of microRNAs, circRNAs
and long noncoding RNAs in acute myeloid leukemia. J Hematol Oncol.
12:512019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu C, Cui H, Li H, Wu Y, An H and Guo C:
Long non-coding RNA ZEB2-AS1 expression is associated with disease
progression and predicts outcome in gastric cancer patients. J
BUON. 24:663–671. 2019.PubMed/NCBI
|
|
10
|
Guo Y, Hu Y, Hu M, He J and Li B: Long
non-coding RNA ZEB2-AS1 promotes proliferation and inhibits
apoptosis in human lung cancer cells. Oncol Lett. 15:5220–5226.
2018.PubMed/NCBI
|
|
11
|
Gao H, Gong N, Ma Z, Miao X, Chen J, Cao Y
and Zhang G: LncRNA ZEB2-AS1 promotes pancreatic cancer cell growth
and invasion through regulating the miR-204/HMGB1 axis. Int J Biol
Macromol. 116:545–551. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi X, Li J, Ma L, Wen L, Wang Q, Yao H,
Ruan C, Wu D, Zhang X and Chen S: Overexpression of ZEB2-AS1 lncRNA
is associated with poor clinical outcomes in acute myeloid
leukemia. Oncol Lett. 17:4935–4947. 2019.PubMed/NCBI
|
|
13
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kavitha N, Vijayarathna S, Jothy SL, Oon
CE, Chen Y, Kanwar JR and Sasidharan S: MicroRNAs: Biogenesis,
roles for carcinogenesis and as potential biomarkers for cancer
diagnosis and prognosis. Asian Pac J Cancer Prev. 15:7489–7497.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang J, Yuan Y, Yang X, Hong Z and Yang L:
Decreased expression of microRNA-122 is associated with an
unfavorable prognosis in childhood acute myeloid leukemia and
function analysis indicates a therapeutic potential. Pathol Res
Pract. 213:1166–1172. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang TJ, Qian Z, Wen XM, Zhou JD, Li XX,
Xu ZJ, Ma JC, Zhang ZH, Lin J and Qian J: Lower expression of bone
marrow miR-122 is an independent risk factor for overall survival
in cytogenetically normal acute myeloid leukemia. Pathol Res Pract.
214:896–901. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Colicino EG and Hehnly H: Regulating a key
mitotic regulator, polo-like kinase 1 (PLK1). Cytoskeleton
(Hoboken). 75:481–494. 2018. View
Article : Google Scholar
|
|
19
|
Weng Ng WT, Shin JS, Roberts TL, Wang B
and Lee CS: Molecular interactions of polo-like kinase 1 in human
cancers. J Clin Pathol. 69:557–562. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Brandwein JM: Targeting polo-like kinase 1
in acute myeloid leukemia. Ther Adv Hematol. 6:80–87. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language. Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Khwaja A, Bjorkholm M, Gale RE, Levine RL,
Jordan CT, Ehninger G, Bloomfield CD, Estey E, Burnett A,
Cornelissen JJ, et al: Acute myeloid leukaemia. Nat Rev Dis
Primers. 2:160102016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath
SD, et al: Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Young AL, Tong RS, Birmann BM and Druley
TE: Clonal haematopoiesis and risk of acute myeloid leukemia.
Haematologica. 104:2410–2417. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rathinasamy B and Velmurugan BK: Role of
lncRNAs in the cancer development and progression and their
regulation by various phytochemicals. Biomed Pharmacother.
102:242–248. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang G, Li H, Sun R, Li P, Yang Z, Liu Y,
Wang Z, Yang Y and Yin C: Long non-coding RNA ZEB2-AS1 promotes the
proliferation, metastasis and epithelial mesenchymal transition in
triple-negative breast cancer by epigenetically activating ZEB2. J
Cell Mol Med. 23:3271–3279. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wu X, Yan T, Wang Z, Wu X, Cao G and Zhang
C: LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and
inhibits apoptosis by regulating miR-27b. Biomed Pharmacother.
96:299–304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan Y, Dong Y, Dang R, Hu Z, Yang Y, Hu Y
and Cheng J: MiR-122 inhibits epithelial mesenchymal transition by
regulating P4HA1 in ovarian cancer cells. Cell Biol Int.
42:1564–1574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Liu YH, Liu JL, Wang Z, Zhu XH, Chen XB
and Wang MQ: MiR-122-5p suppresses cell proliferation, migration
and invasion by targeting SATB1 in nasopharyngeal carcinoma. Eur
Rev Med Pharmacol Sci. 23:622–629. 2019.PubMed/NCBI
|
|
31
|
Zhang L, Wang Y, Sun J, Ma H and Guo C:
LINC00205 promotes proliferation, migration and invasion of HCC
cells by targeting miR-122-5p. Pathol Res Pract. 215:1525152019.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Xu Z, Liu G, Zhang M, Zhang Z, Jia Y, Peng
L, Zhu Y, Hu J, Huang R and Sun X: MiR-122-5p inhibits the
proliferation, invasion and growth of bile duct carcinoma cells by
targeting ALDOA. Cell Physiol Biochem. 48:2596–2606. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Ann Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Z, Sun Q and Wang X: PLK1, a potential
target for cancer therapy. Transl Oncol. 10:22–32. 2017. View Article : Google Scholar
|
|
35
|
Dang SC, Fan YY, Cui L, Chen JX, Qu JG and
Gu M: PLK1 as a potential prognostic marker of gastric cancer
through MEK-ERK pathway on PDTX models. Onco Targets Ther.
11:6239–6247. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xu C, Li S, Chen T, Hu H, Ding C, Xu Z,
Chen J, Liu Z, Lei Z, Zhang HT, et al: MiR-296-5p suppresses cell
viability by directly targeting PLK1 in non-small cell lung cancer.
Oncol Rep. 35:497–503. 2016. View Article : Google Scholar
|
|
37
|
Li J, Wang R, Schweickert PG, Karki A,
Yang Y, Kong Y, Ahmad N, Konieczny SF and Liu X: Plk1 inhibition
enhances the efficacy of gemcitabine in human pancreatic cancer.
Cell Cycle. 15:711–719. 2016. View Article : Google Scholar : PubMed/NCBI
|