Introduction
Liver cancer is one of the most frequently occurring
malignant tumors and is the second major cause of tumor-related
mortalities worldwide (1). The
incidence of liver cancer in China accounts for 55% of all
worldwide cases (2,3). Although strategies of treating liver
cancer have improved, including surgical resection,
transplantation, ablation, transarterial chemoembolisation, and the
tyrosine-kinase inhibitors sorafenib, lenvatinib, and regorafenib
(4), the efficacies remain
unsatisfactory (5). The
recurrence of HCC remains a major problem after curative treatment,
reaching an incidence of >70% at 5 years (6,7).
The development of molecular biology can give rise to improving
liver cancer treatment at the molecular level (8).
Hematopoietic cell-specific protein 1-associated
protein X-1 (HAX1) is a multifunctional protein ~35 kDa in size and
is involved in anti-apoptosis, migration, adhesion and endocytosis
regulation (9-11). A previous study found that
highly-expressed HAX1 is associated with poor survival in human
colorectal cancer (12),
cutaneous squamous cell carcinoma (13), laryngeal carcinoma (14) and multiple myeloma (15). Additionally, HAX1 is involved in
the metastasis and genesis of various types of tumors (16). Previous studies reported that HAX1
was overexpressed in liver cancer samples (17,18), suggesting that dysregulation of
HAX-1 expression plays a key role in liver cancer development.
However, the underlying mechanism of HAX1 in the progression of
liver cancer and its effects remain unclear.
MicroRNAs (miRs/miRNAs), which are a class of
endogenous non-coding genes ~22 nucleotides in size (19), modulate gene expression by
directly binding to the 3′-untranslated region (3′-UTR) of the
target mRNA at the post-transcriptional level (20). MiRNAs can act as oncogenes or
tumor suppressors in tumor occurrence and progression, including
liver cancer (21-23). It was found that miR-125a-5p is a
tumor suppressor in the development of various cancers, including
in human gastric cancer (24),
hepatitis B virus-related liver cancer (25) and gastric cancer (26). Notably, a previous report
demonstrated that miR-125a-5p expression is downregulated in liver
cancer tissues and cell lines, and lowly expressed miR-125a-5p is
correlated with aggressive pathological features (27). However, whether miR-125a-5p
participates in the regulation of HAX1 in liver cancer remains to
be elucidated.
In the present study, HAX1 expression in liver
cancer tissues and cells, and its association with the prognosis of
patients with liver cancer were examined. The effects of HAX1 on
the development of liver cancer were also explored. Furthermore,
the present study determined whether miR-125a-5p was involved in
the development of liver cancer via regulating HAX1 expression.
Materials and methods
Ethics statement
The present study was approved by the Ethics
Committee of the Luoyang Central Hospital Affiliated to Zhengzhou
University (approval no. LC20170416022). All patients signed
informed consent.
Clinical specimens
A total of 40 primary liver tumor tissues and
adjacent non-cancerous samples were obtained from patients with
liver cancer (age range, 28-75; 22 males; 18 females) who received
tumor resection in Luoyang Central Hospital Affiliated to Zhengzhou
University between May 2012 and May 2014. All patients with liver
cancer did not receive radiotherapy or chemotherapy prior to the
surgery.
Cell lines
Human normal liver cell line (THLE-2) and liver
cancer cell lines (Hep3B, PLC/PRF/5, SK-Hep1, SNU-182 and SNU-387)
were obtained from American Type Culture Collection. The cells were
cultured in DMEM (cat. no. 12100; Beijing Solarbio Science &
Technology Co., Ltd.) supplemented with 10% FBS (cat. no.
11011-8611; Beijing Solarbio Science & Technology Co., Ltd.)
and penicillin-streptomycin liquid (100 U/ml penicillin, 100 mg/ml
streptomycin; cat. no. P1400; Beijing Solarbio Science &
Technology Co., Ltd.) at 37°C with 5% CO2.
Cell transfection
To construct plasmids expressing HAX1, the
full-length human HAX1 sequence was synthesized by Guangzhou
RiboBio Co., Ltd. and ligated into the pcDNA3.1 plasmid (cat. no.
V79020; Thermo Fisher Scientific, Inc.). For cell transfection, 2
mg/ml HAX1 vector, 50 pmol/ml miR-125a mimics (cat. no.
miR10000443-1-5; Guangzhou RiboBio Co., Ltd.), 50 pmol/ml mimics
negative control (mimics NC; cat. no. miR1N0000001-1-5; Guangzhou
RiboBio Co., Ltd.), 50 pmol/ml miR-223 mimics (cat. no.
miR10000280-1-5; Guangzhou RiboBio Co., Ltd.), 50 pmol/ml miR-125a
inhibitor (cat. no. miR20000443-1-5; Guangzhou RiboBio Co., Ltd.),
50 pmol/ml inhibitor NC (cat. no. miR2N0000001-1-5; Guangzhou
RiboBio Co., Ltd.), small interfering RNA (siRNA) against HAX1
(siHAX1; cat. no. siG000010456A-1-5; Guangzhou RiboBio Co., Ltd.)
and siRNA negative control (siNC; cat. no. siN0000002-1-5,
Guangzhou RiboBio Co., Ltd.) were transfected into SK-Hep1 and
SNU-387 cells. Cell transfection was conducted using Lipofectamine™
2000 (cat. no. 11668; Invitrogen; Thermo Fisher Scientific, Inc.).
In addition, untreated cells were used as control. SNU-387 cells
transfected with pcDNA3.1 empty plasmid were used as the NC group,
while SK-Hep1 cells transfected with siRNA negative control used as
the siNC group.
Prognosis analysis
The association between patient survival and HAX1
expression in patients with liver cancer was plotted using
Kaplan-Meier curves and log-rank tests based on data obtained from
Kaplan-Meir Plotter (http://kmplot.com/anal-ysis/index.php?p=service&cancer=liver_rnaseq)
(28) and data from follow-up
time (60 months).
Immunohistochemistry
Immunohistochemistry was performed as previously
described (29). The sections
were incubated with primary antibody against HAX1 (rabbit; cat. no.
ab137613; 1:1,000; Abcam) overnight at 4°C followed by secondary
antibody incubation (goat anti-rabbit; cat. no. ab205718; 1:2,000;
Abcam). The sections were dyed with 3,3-diaminobenzidine
horseradish peroxidase color development kit (cat. no. P0203;
Beyotime Institute of Biotechnology), counter-stained with
hematoxylin, added to coverslips and observed under a light
microscope (Olympus Corporation).
Reverse transcription-quantitative
real-time PCR (RT-qPCR)
Total RNAs were extracted from the collected tissues
and cells (SK-Hep1 and SNU-387) using TRIzol® reagent
(Invitrogen; Thermo Fisher Scientific, Inc.). The reverse
transcription of non-miRNAs was performed using the PrimeScript RT
reagent kit (cat. no. RR036B; Takara Bio, Inc.) according to the
manufacturer's instructions. The reverse transcription of miRNAs
was performed using a stem-loop RT primer and the PrimeScript RT
reagent kit (Takara Bio, Inc.) according to the manufacturer's
instructions. cDNAs were amplified on a 7500 Fast Real-Time PCR
system (Applied Biosystems; Thermo Fisher Scientific, Inc.) using
TB Premix Ex Taq (cat. no. RR820L; Takara Bio, Inc.). The following
thermocycling conditions were used for the qPCR: Initial
denaturation at 94°C for 30 sec, followed by 40 cycles of
denaturation at 94°C for 5 sec, annealing at 55°C for 30 sec and
final extension at 72°C for 30 sec. Gene expression was normalized
to GAPDH or U6 as appropriate (Sangon Biotech Co., Ltd.). The
expression was calculated and quantified using the
2−ΔΔCq method (30).
The primer sequences are listed in Table I.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Primer sequences
(5′-3′) |
|---|
| HAX1 | F:
CAGGAGGAGGGATACGTTTCC |
| R:
CCCATATCGCTGAAGATGCTATT |
| miR-125a-5p | RT:
5′-GTCGTATCCAGTGCGTGTCG |
|
TGGAGTCGGCAATTGCACTGGATA |
| C | GACTCACAGGT-3′ |
| F:
TGTGAGTCGTATCCAGTGCAA |
| R:
GTATCCAGTGCGTGTCGTGG |
| miR-223-3p | F:
CCCAGTCGTATCCAGTGCAA |
| R:
GTCGTATCCAGTGCGTGTCG |
| p53 | F:
CAGCACATGACGGAGGTTGT |
| R:
TCATCCAAATACTCCACACGC |
| VEGF | F:
AGGGCAGAATCATCACGAAGT |
| R:
AGGGTCTCGATTGGATGGCA |
| E-cadherin | F:
CGAGAGCTACACGTTCACGG |
| R:
GGGTGTCGAGGGAAAAATAGG |
| N-cadherin | F:
TCAGGCGTCTGTAGAGGCTT |
| R:
ATGCACATCCTTCGATAAGACTG |
| Vimentin | F:
GACGCCATCAACACCGAGTT |
| R:
CTTTGTCGTTGGTTAGCTGGT |
| GADPH | F:
GGAGCGAGATCCCTCCAAAAT |
| R:
GGCTGTTGTCATACTTCTCATGG |
| U6 | F:
CTCGCTTCGGCAGCACA |
| R:
ACGCTTCACGAATTTGCGT |
Western blotting
Total proteins were extracted from the tissues and
cells (SK-Hep1 and SNU-387) using RIPA buffer (cat. no. R0010;
Beijing Solarbio Science & Technology Co., Ltd.). Protein
concentration was detected using a BCA protein assay kit (cat. no.
PC0020; Beijing Solarbio Science & Technology Co., Ltd.). Whole
cell lysates (30 µg/lane) were separated by 12.5% SDS-PAGE
and then transferred to PVDF membranes (cat. no. FFP32; Beyotime
Institute of Biotechnology) with an electroblotting apparatus.
Subsequently, the membranes were incubated in 5% (w/v) skimmed milk
for 1 h at 37°C to block non-specific binding, and then treated
with the appropriate primary antibodies overnight at 4°C (Table II). The membranes were then
treated with goat anti-mouse secondary antibody (cat. no. ab205719;
Abcam) or goat anti-rabbit secondary antibody (cat. no. ab205718;
Abcam) at 1:2,000 dilution for 1 h at 37°C. Protein signals were
visualized using an ECL detection kit (cat. no. P0018FS; Beyotime
Institute of Biotechnology) and normalized to GAPDH. The target
bands were visualized by a gel documentation system (C-DiGit Blot
Scanner; LI-COR Biosciences).
 | Table IIList of primary antibodies used for
western blotting. |
Table II
List of primary antibodies used for
western blotting.
| Protein | Antibody | Cat. no. | Company | Antibody
dilution |
|---|
| HAX1 | Rabbit anti-HAX1
antibody | ab78939 | Abcam | 1:1,000 |
| p53 | Mouse anti-p53
antibody | ab26 | Abcam | 1:1,000 |
| E-cadherin | Rabbit
anti-E-cadherin antibody | ab40772 | Abcam | 1:500 |
| VEGF | Rabbit anti-VEGF
antibody | ab1316 | Abcam | 1:1,000 |
| N-cadherin | Rabbit
anti-N-cadherin antibody | ab18203 | Abcam | 1:1,000 |
| Vimentin | Rabbit
anti-vimentin antibody | ab92547 | Abcam | 1:1,000 |
| GAPDH | Mouse anti-GAPDH
antibody | ab8245 | Abcam | 1:1,000 |
Cell Counting Kit-8 (CCK-8) assay
Cell viability was detected using a CCK-8 kit (cat.
no. CK04; Dojindo Molecular Technologies, Inc.) according to the
manufacturer's protocol. Transfected SK-Hep1 and SNU-387 cells
(1×104 cell/well) were cultured in 96-well plates
containing DMEM at 37°C with 5% CO2 for 0, 24 and 48 h.
CCK-8 solution (10 ml) was added into the cells and incubated for
another 4 h. A micro-plate reader (Sunrise Microplate Reader; Tecan
Group, Ltd.) was used to detect the absorbance at a wavelength of
450 nm.
Scratch wound healing assay
Cell migration was determined using a scratch
wound-healing assay. In brief, the SK-Hep1 and SNU-387 cells were
cultured in six-well plates to 80% confluence. Subsequently, DMEM
was discarded and the cells in the six-well plates were scratched
with a 10-µl tip and incubated with serum-free DMEM. The
cells were further cultured for 24 h. Cell migration was analyzed
by counting migrated cells under an inverted microscope (x100
magnification; Ts2R-FL; Nikon Corporation) using ImageJ 1.8.0
(National Institutes of Health).
Transwell invasion assay
Cell invasion was detected using a Transwell assay.
In brief, transfected SK-Hep1 and SNU-387 cells (5×105
cell/well) were added into the upper chamber (precoated with
Matrigel matrix), which contained 200 ml DMEM without serum, while
the lower Transwell chamber contained with 600 µl DMEM
supplemented with 20% FBS. Following culture for 24 h, non-invasive
cells remaining in the upper chamber were gently scraped off with
cotton swabs, while the medium in the lower chamber were aspirated.
Cells were fixed with 4% paraformaldehyde for 15 min at room
temperature and stained with 0.1% crystal violet for 20 min at room
temperature. Cells were counted from five random fields under an
inverted microscope (x200 magnification; Ts2R-FL; Nikon
Corporation).
Colony formation unit assay
SK-Hep1 and SNU-387 cells were seeded into six-well
plates at a density of 1×103 cells/well. DMEM was
replaced every 4 days. After culture for 14 days, the cells were
fixed with 4% paraformaldehyde for 15 min at room temperature and
stained with crystal violet for 15 min at room temperature. The
cell clones were photographed using a camera (Nikon D90; Nikon
Corporation) and the relative colony formation rates were
calculated.
Cell apoptosis detection
The apoptosis of SK-Hep1 and SNU-387 cells was
determined by flow cytometry using an Annexin V/PI kit (cat. no.
KGA108; Nanjing KeyGen Biotech Co., Ltd.) according to the
manufacturer's instructions. In brief, treated SK-Hep1 and SNU-387
cells were collected and incubated with Annexin V/PI for 15 min at
room temperature in the dark. Finally, the fluorescence of cells
was detected and analyzed by fluorescence-activated cell sorting
(FACSCalibur; BD Biosciences).
Dual-luciferase activity assay
TargetScan (http://www.targetscan.org/vert_72/) predicted that
miR-125a-5p and miR-223-3p were the target genes for HAX1. The
sequence of the 3′-UTR of HAX1 with the binding sites for
has-miR-125a-5p or has-miR-223-3p was synthesized by Guangzhou
RiboBio Co., Ltd. and cloned into a luciferase reporter gene vector
(cat. no. E1330; Promega Corporation). In this experiment, the
mutation (mut) refers to the change of sequence of the putative
binding site of HXA1 in miR-125a-5p or miR-223-3p. HXA1-mut
contained a mutation in the predicted binding sites of miR-125a-5p
or miR-223-3p. The sequence of HXA1-mut was as follows:
5′-AGCUUCUCUUGCCACCUAGCCAG-3′ or 5′-UUUGUCACUCACCCAAAGGCGC-3′. The
HAX1-3′-UTR mut was purchased from Guangzhou RiboBio Co., Ltd. and
cloned into a luciferase reporter gene vector (cat. no. E1330;
Promega Corporation). For the dual-luciferase reporter assay, the
reporter vector plasmid and miR-125a mimics or miR-223 mimics were
co-transfected into 293T cells (cat. no. CRL-11268; American Type
Culture Collection) using Lipofectamine™ 2000 (cat. no. 11668;
Invitrogen; Thermo Fisher Scientific, Inc.). Following transfection
for 48 h, the luciferase activities of different groups were
measured using the Dual-Luciferase Reporter Assay system (cat. no.
E1910; Promega Corporation). The luciferase activities were
normalized to Renilla luciferase activity.
Statistical analysis
The data are represented as the mean ± SD and
analyzed using SPSS 19.0 software (IBM Corp.). Kaplan-Meier plots
were analyzed with log-rank test. Paired t-test was used for
analysis of paired samples. Comparison between two groups was
performed by Student's t-test, whereas comparison among multiple
groups was performed by one-way ANOVA analysis followed by Tukey's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
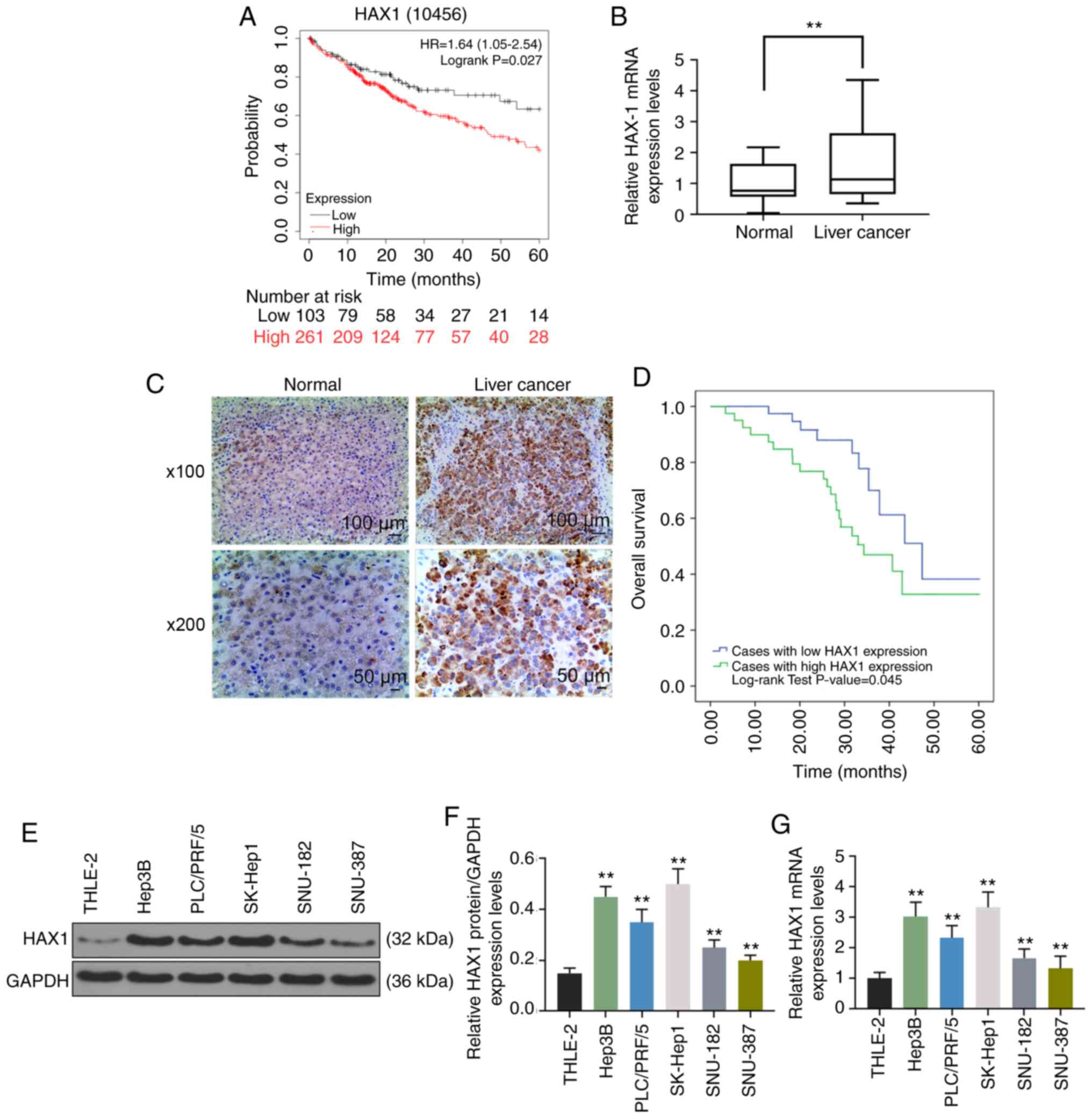
High HAX1 expression is observed in liver
cancer tissues and cells and associated with prognosis of
patients
Based analysis of data obtained from Kaplan-Meier
Plotter (http://kmplot.com/analysis/index.php?p=service&cancer=liver_rnaseq),
Kaplan-Meier plots revealed that high HAX1 expression in liver
cancer cases was associated with lower overall survival compared
with low HAX1 expression (Fig.
1A; P<0.05), and HAX1 showed significantly higher expression
in liver cancer tissues compared with normal tissues (Fig. 1B; P<0.01). HXA1 protein
staining from two representative cases are shown in Fig. 1C.
The overall survival of liver cancer patients with
high HAX1 expression was significantly lower compared with patients
with low HAX1 expression (Fig.
1D; P<0.05) obtained from Luoyang Central Hospital
Affiliated to Zhengzhou University. Additionally, significantly
higher protein (Fig. 1E and F;
P<0.01) and mRNA (Fig. 1G;
P<0.01) expression of HAX1 were observed in human liver cancer
cell lines (Hep3B, PLC/PRF/5, SK-Hep1, SNU-182 and SNU-387)
compared with human normal liver cell line THLE-2. HAX1 expression
was lower in SNU-387 cells compared with other liver cancer cell
lines (Fig. 1E-G), while HAX1
expression was higher in SK-Hep1 cells than other liver cancer cell
lines (Fig. 1E–G). Therefore,
SNU-387 and SK-Hep1 cells were used in the following
experiments.
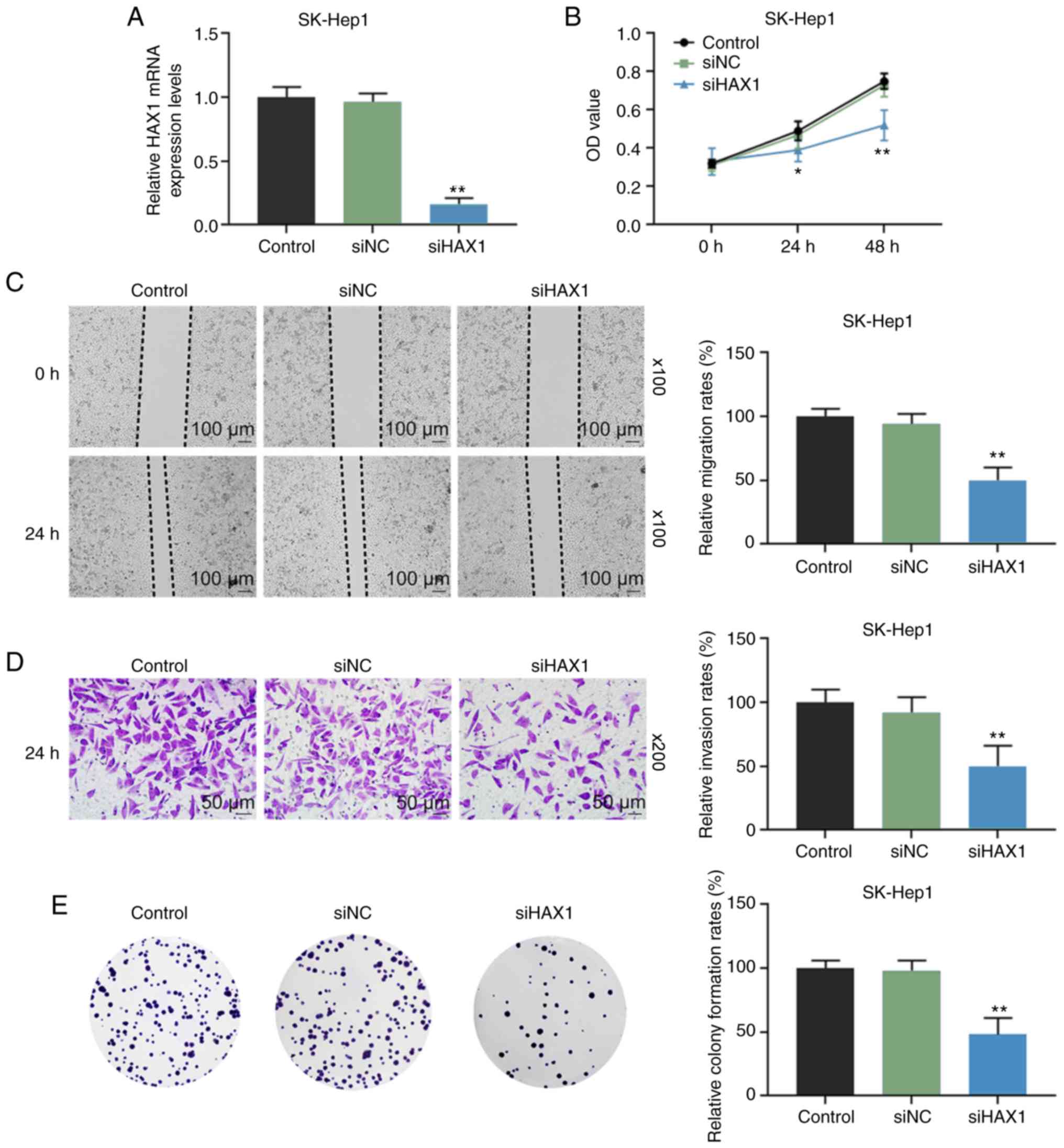
HAX1 silencing suppresses the viability,
migration, invasion and colony formation of SK-Hep1 cells
To determine the biological role of HAX1 in the
development of liver cancer, SK-Hep1 cells were transfected with
siHAX1 to downregulate HAX1 expression. RT-qPCR results showed that
HAX1 expression was significantly lower in the siHAX1 group
compared with the siNC group (Fig.
2A; P<0.01). As shown in Fig.
2B–E, compared with the siNC group, HAX1 silencing
significantly suppressed the cell viability at 24 and 48 h
(Fig. 2B; P<0.05 or
P<0.01), migration at 24 h (Fig.
2C; P<0.01), invasion at 24 h (Fig. 2D; P<0.01) and colony formation
at 24 h (Fig. 2E; P<0.01).
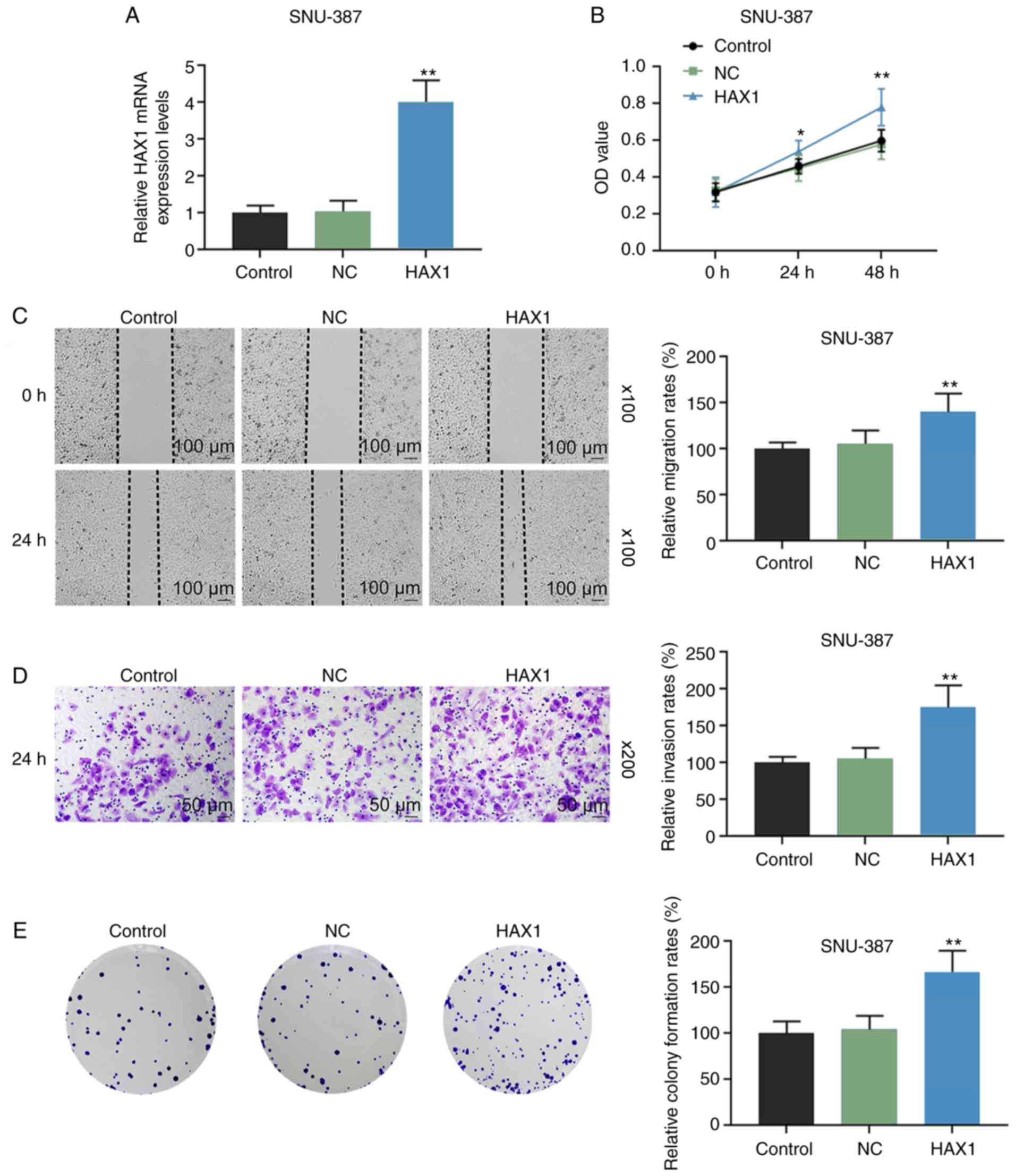
HAX1 overexpression promotes the
viability, migration, invasion and colony formation of SNU-387
cells
SNU-387 cells were transfected with pcDNA3.1-HAX1
plasmid to upregulate HAX1 expression, and cells transfected with
pcDNA3.1 empty plasmid as were used as the negative control.
RT-qPCR results revealed that HAX1 expression was significantly
higher in the HAX1 group compared with the NC group (Fig. 3A; P<0.01). As shown in Fig. 3B-E, compared with the NC group,
HAX1 overexpression significantly promoted cell viability at 24 and
48 h (Fig. 3B; P<0.05 or
P<0.01), migration at 24 h (Fig.
3C; P<0.01), invasion at 24 h (Fig. 3D; P<0.01) and colony formation
at 24 h (Fig. 3E; P<0.01).
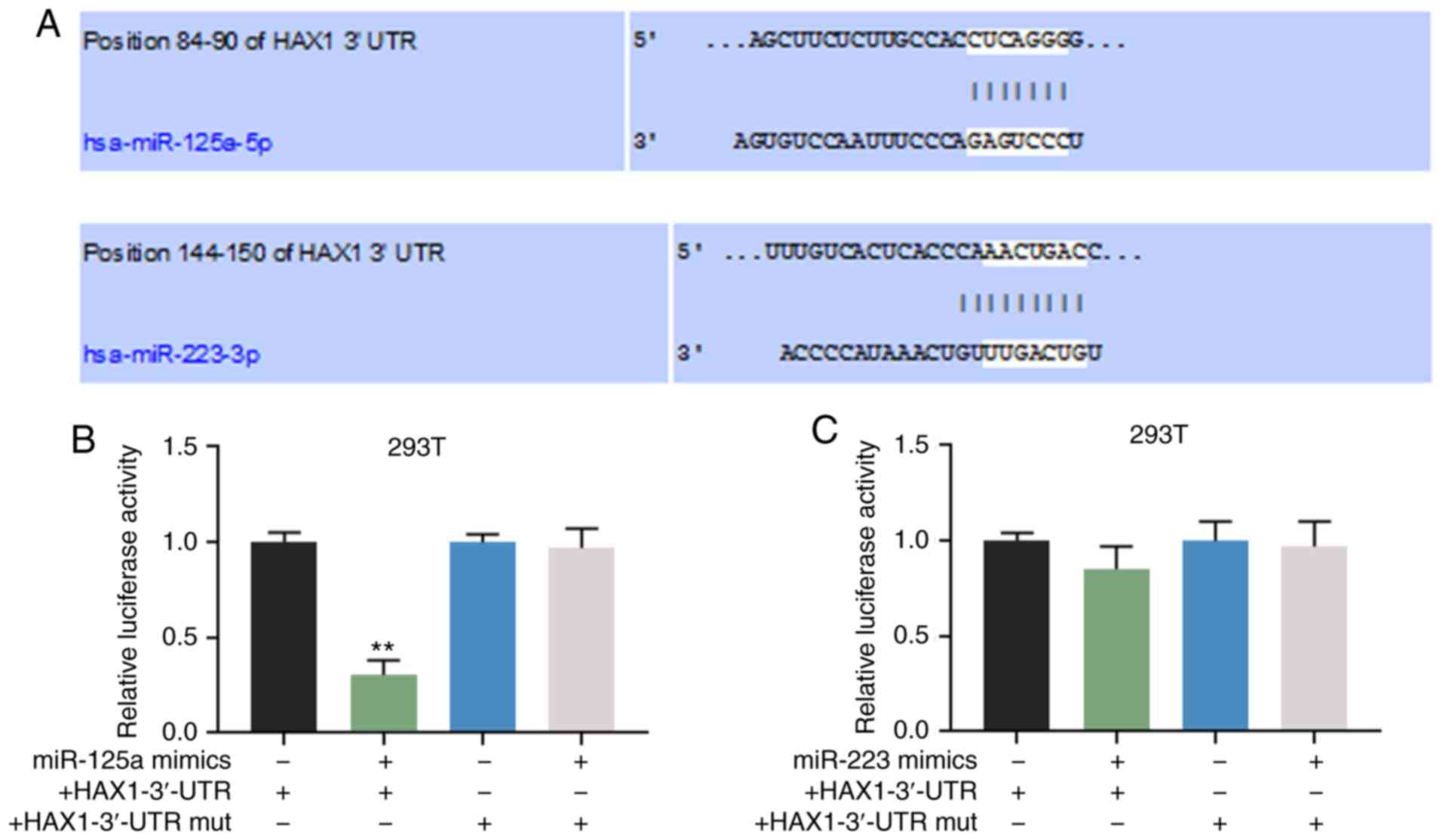
HAX1 is a target gene of miR-125a-5p
TargetScan predicted that miR-125a-5p and miR-223-3p
could bind to the HAX1 3′-UTR (Fig.
4A). Dual-luciferase reporter assay demonstrated that
miR-125amimics significantly inhibited the luciferase activity of
the HAX1-3′-UTR (Fig. 4B;
P<0.01), but did not affect HAX1-3′-UTR mut (Fig. 4B), whereas miR-223 mimics only
slightly altered the luciferase activity of HAX1-3′-UTR (Fig. 4C). This indicated that the binding
capacity of miR-125a to HAX1-3′-UTR was stronger compared with
miR-223, thus miR-125a was seen as the target gene and investigated
in subsequent experiments.
HAX1 reverses the effects of miR-125a-5p
on the viability, migration, invasion and colony formation of
SK-Hep1 cells
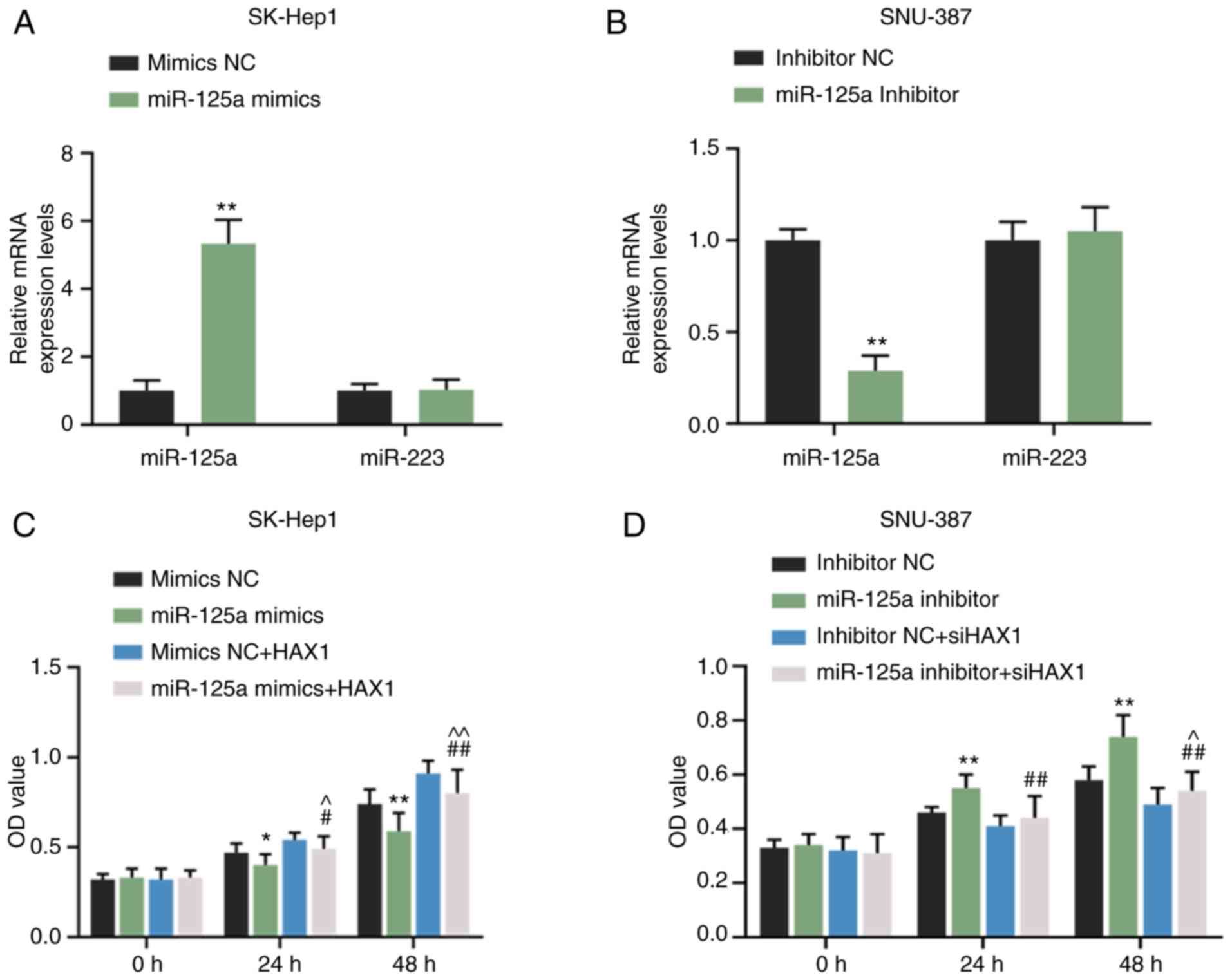
RT-qPCR results revealed that miR-125a mimics
significantly promoted miR-125a expression in SK-Hep1 cells
(Fig. 5A; P<0.01) but did not
affect miR-223 expression (Fig.
5A). miR-125a inhibitor significantly inhibited miR-125a
expression in SNU-387 cells (Fig.
5B; P<0.01) but exerted no effects on miR-223 expression
(Fig. 5B). Compared with mimic
NC, miR-125a mimics inhibited the cell viability of SK-Hep1 cells
at 24 (Fig. 5C; P<0.05) and 48
h (Fig. 5C, P<0.01). However,
the effects were reversed by HAX1 overexpression (Fig. 5C; P<0.05 or P<0.01).
miR-125a inhibitor promoted the cell viability of SNU-387 cells at
24 and 48 h (Fig. 5C; P<0.01),
but the effect was reversed by HAX1 silencing (Fig. 5C; P<0.01). Compared with the
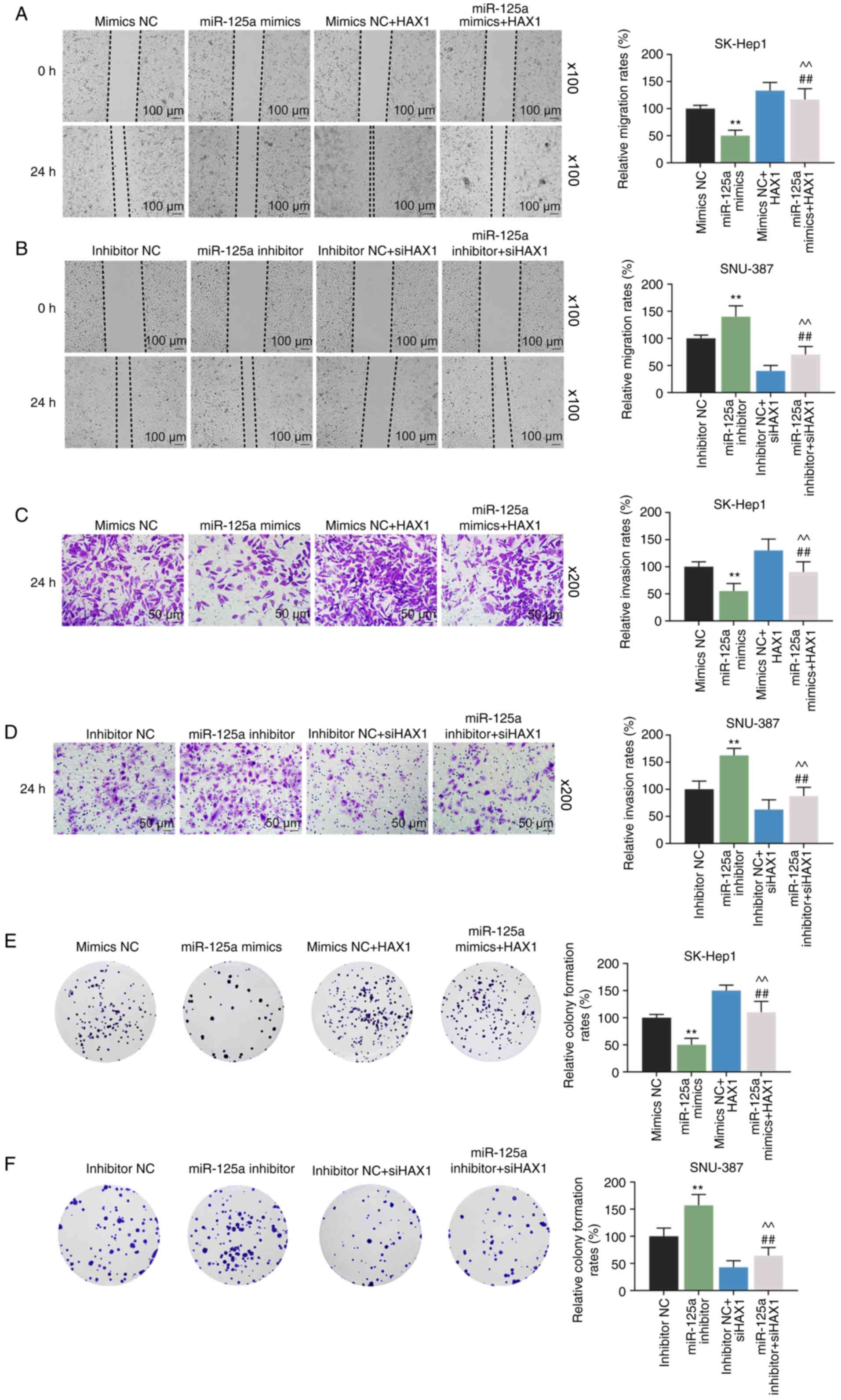
mimics NC group, miR-125a mimics significantly inhibited cell
migration (Fig. 6A; P<0.01),
inva-sion (Fig. 6C; P<0.01)
and colony formation (Fig. 6E;
P<0.01), which were significantly reversed by HAX1
overexpression (Fig. 6A, C and E;
P<0.01). miR-125a inhibitor significantly increased cell
migration (Fig. 6B; P<0.01),
invasion (Fig. 6D; P<0.01) and
colony formation (Fig. 6F;
P<0.01) compared with the inhibitor NC groups. However, these
effects were significantly reversed by HAX1 silencing (Fig. 6B, D and F; P<0.01).
HAX1 reverses the effects of miR-125a-5p
on cell apoptosis
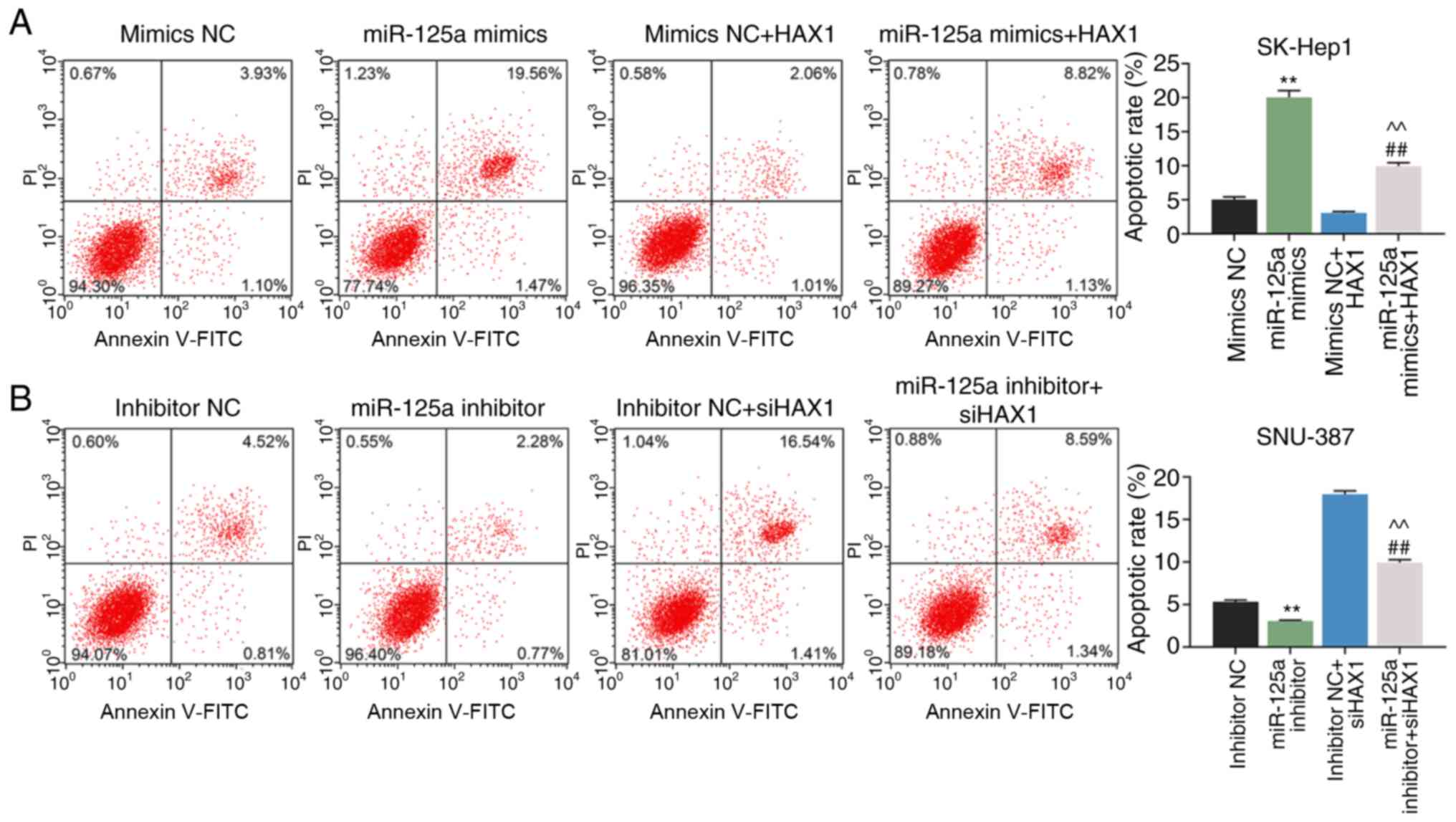
The effects of miR-125a-5p on cell apoptosis was
detected using flow cytometry. Compared with the mimics NC group,
the results showed that miR-125a-5p overexpression significantly
increased the apoptosis of the SK-Hep1 cells, which was partially
reserved by HAX1 overexpression (Fig.
7A; P<0.01). The apoptosis of SNU-387 cells was decreased in
the miR-125a-5p inhibitor group compared with the inhibitor NC
group (Fig. 7B; P<0.01), and
silencing HAX1 significantly increased the apoptosis in the
miR-125a-5p inhibitor + siHAX1 group compared with the miR-125a-5p
inhibitor group (Fig. 7B;
P<0.01).
HAX1 reverses the effects of miR-125a-5p
on the expression of p53, VEGF, E-cadherin, N-cadherin and vimentin
in SK-Hep1 and SNU-387 cells
As shown in Fig.
8A-E, compared with the mimic NC group, miR-125a-5p
overexpression significantly suppressed the protein and mRNA
expression of HAX1 (P<0.01), VEGF (P<0.01), N-cadherin
(P<0.01) and vimentin (P<0.01), and increased the protein and
mRNA expression of p53 (P<0.01) and E-cadherin (P<0.01) in
SK-Hep1 cells. However, these effects were significantly reversed
by HAX1 overexpression (P<0.01). As shown in Fig. 8F-J, miR-125a-5p silencing
significantly increased the protein and mRNA levels of HAX1
(P<0.01), VEGF (P<0.01), N-cadherin (P<0.01) and vimentin
(P<0.01), and decreased the protein and mRNA levels of p53
(P<0.01) and E-cadherin (P<0.01) in SNU-387 cells. However,
the effects were significantly reversed by downregulation of HAX1
(P<0.01).
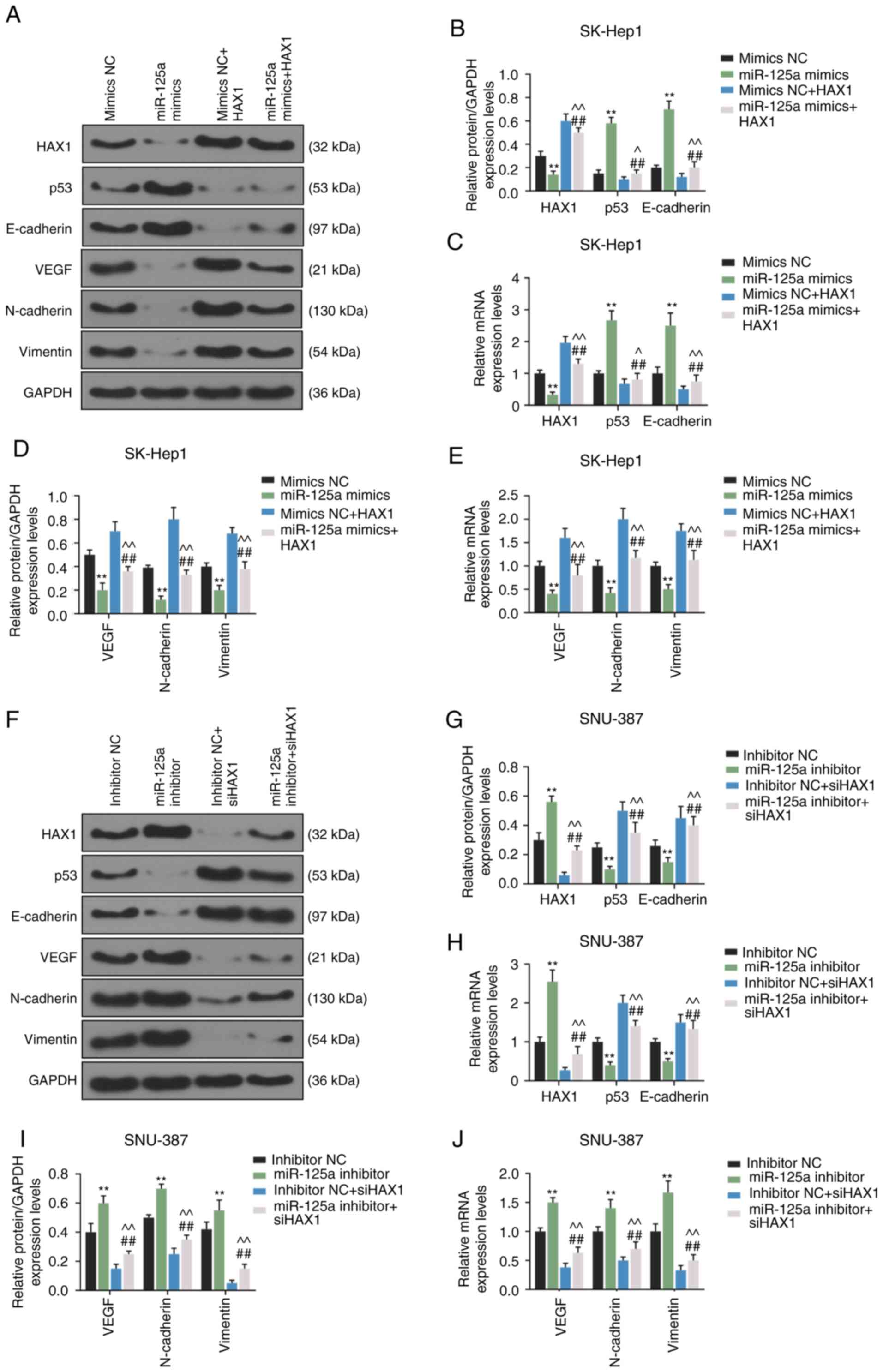 | Figure 8HAX1 reverses the effects of
miR-125a-5p on the expression of p53, VEGF, and E-cadherin,
N-cadherin, and vimentin in SK-Hep1 and SNU-387 cells. (A) The
protein expression of HAX1, p53, E-cadherin, VEGF, N-cadherin and
vimentin in SK-Hep1 cells transfected with miR-125a mimics or HAX1
or co-transfected with miR-125a mimics and HAX1 were determined by
western blotting. (B) Quantification of HAX1, p53 and E-cadherin
protein expression. (C) The mRNA expression of HAX1, p53 and
E-cadherin in SK-Hep1 cells transfected with miR-125a mimics or
HAX1 or co-transfected with miR-125a mimics and HAX1 were detected
by RT-qPCR. (D) Quantification of VEGF, N-cadherin and vimentin
protein expression. (E) The mRNA expression of VEGF, N-cadherin and
vimentin in SK-Hep1 cells transfected with miR-125a mimics or HAX1
or co-transfected with miR-125a mimics and HAX1 were detected by
RT-qPCR. **P<0.01 vs. mimics NC.
##P<0.01 vs. miR-125a mimics. ^P<0.05
and ^^P<0.01 vs. mimics NC + HAX1. (F) The protein
expression of HAX1, p53, E-cadherin, VEGF, N-cadherin and vimentin
in SNU-387 cells transfected with miR-125a inhibitor or siHAX1 or
co-transfected with miR-125a inhibitor and siHAX1 were determined
by western blotting. (G) Quantification of HAX1, p53 and E-cadherin
protein expression. (H) The mRNA expression of HAX1, p53 and
E-cadherin in SNU-387 cells transfected with miR-125a inhibitor or
siHAX1 or co-transfected with miR-125a inhibitor and siHAX1 were
determined by RT-qPCR. (I) Quantification of VEGF, N-cadherin and
vimentin protein expression (J) The mRNA expression of VEGF,
N-cadherin and vimentin in SNU-387 cells transfected with miR-125a
inhibitor or siHAX1 or co-transfected with miR-125a inhibitor and
siHAX1 were determined by RT-qPCR. GAPDH was used as an internal
control. **P<0.01 vs. inhibitor NC.
##P<0.01 vs. miR-125a inhibitor.
^^P<0.01 vs. inhibitor NC + siHAX1. Data are shown as
the mean ± standard deviation. HAX1,
hematopoietic-substrate-1-associated protein X-1; miR, microRNA;
si, small interfering RNA NC, negative control; RT-qPCR, reverse
transcription-quantitative PCR. |
Discussion
HAX1 is a prognostic factor and is abnormally
expressed in several types of cancer (31-33). Similar to previous results
(34), the present data suggested
that high HAX1 expression was associated with poorer prognosis in
the liver cancer tissues and cell lines examined. The data
demonstrated that HAX1 might be an oncogene for liver cancer
progression and have diagnostic and prognostic values for liver
cancer.
Tumor cell migration refers to directed cell
movement within the body, while cancer invasion is the penetration
of tumor cells through tissue barriers (35). The migration and invasion of
cancer cells into surrounding tissues and vasculatures are
important factors for initiating cancer metastasis (36). Tumor metastasis will result in
less desired treatment outcomes of patients with liver cancer
(37). The present data suggested
that upregulated HAX1 expression promoted the growth, migration and
invasion of liver cancer cells, whereas HAX1 knockdown produced the
opposite effects, which were in accordance with the results of
previous studies (17,34). The present data further confirmed
that HAX1 is a tumor oncogene of liver cancer development and might
be an underlying target for the treatment of liver cancer.
The upstream target of HAX1 in the progression of
liver cancer was examined. Different miRNAs may participate in the
control of a same mRNA molecule (38). Bioinformatics analysis predicted
that both miR-125a-5p and miR-223-3p could bind to HAX1. However,
the results of the dual-luciferase reporter assay revealed that
only miR-125a-5p can bind to HAX1, which verified that HAX1 was the
target of miR-125a-5p. Several reports demonstrated that
miR-125a-5p functions as a tumor suppressor in different cancers by
downregulating the expression of its downstream target (39-42). In prostate carcinoma, Fu and Cao
(43) indicated that miR-125a-5p
modulates cancer cell proliferation and migration via targeting
nuclear apoptosis-inducing factor 1. Tang et al (44) reported that miR-125a-5p suppresses
EMT, invasion and migration of colorectal cancer cells via
targeting transcriptional activator with PDZ-binding domain. In
human cervical carcinoma, Qin et al (45) suggested that miR-125a-5p regulates
the proliferation and migration of human cervical carcinoma cells
by targeting tyrosine-protein kinase ABL2. To the best of our
knowledge, the present study was the first to report that
upregulated miR-125a-5p suppressed cell growth, migration and
invasion of SK-Hep1 cells through inhibiting HAX1 expression,
whereas downregulated miR-125a-5p produced the opposite results in
SNU-387 cells through promoting HAX1 expression. These novel
findings extended our previous understanding on liver cancer. In
addition, Potenza et al (46) found that upregulation of miR-125a
suppressed the proliferation of liver cancer cells by inhibition of
sirtuin-7, which is a NAD(+)-dependent deacetylase, and induced
cell cycle arrest in the G1 phase; Kim et al (47) also found that ectopic expression
of miR-125a-5p and miR-125b caused growth retardation by cell cycle
arrest. However, the effect of miR-125a/HAX1 on THE cell cycle of
liver cancer was not detected in the current study.
p53 is a tumor suppressor that suppresses VEGF
expression, tumor growth and metastasis (48). Zhou et al (49) demonstrated that miR-141-3p
promoted glioma cell growth via targeting p53. VEGF is considered
as a potent angiogenic mitogen and able to induce tumor
angiogenesis (50). Angiogenesis
is a process of developing and forming new blood vessels, and plays
an important role in tumor growth and metastasis (51). In laryngeal cancer, Zhang et
al (52) demonstrated that
downregulated miR-206 promoted the proliferation and invasion of
cancer cells by modulating VEGF expression. Wu et al
(53) suggested that miR-125
suppressed cell growth of RKO colorectal cancer cells via targeting
VEGF. It was reported that the EMT process is involved in the
migration and invasion of cancer cells and plays a critical role in
cancer metastasis (54,55). Decreased E-cadherin expression and
increased N-cadherin expression are indicative of EMT (56). Vimentin, an EMT marker, is widely
found in normal mesenchymal cells and maintains cellular integrity
(57). The expression of p53,
VEGF, E-cadherin, N-cadherin and vimentin were detected to assess
the mechanism underlying the effects of miR-125a-5p on cell growth,
migration and invasion via targeting HAX1. The present findings
showed that miR-125a-5p overexpression increased the expression of
p53 and E-cadherin, and suppressed the expression of VEGF,
N-Cadherin, and Vimentin in SK-Hep1 cells via inhibiting HAX1
expression, while down-regulated miR-125a-5p produced the opposite
effects in SNU-387 cells via promoting HAX1 expression. The results
suggested that miR-125a-5p regulated cell growth, migration and
invasion via directly targeting HAX1, which might be dependent on
the regulation of p53 and VEGF expression and the EMT process.
The present results revealed the effects and the
underlying molecular mechanism of HAX1 on liver cancer progression.
However, the present study also has some limitations. Although the
present findings developed the current under-standing the
pathogenesis of liver cancer and discovered a novel target for the
treatment of liver cancer, the function of the miR-125a/HAX1 axis
should be further verified in vivo. Moreover, whether
mir-125a/HAX1 can affect the role of hepatocellular carcinoma drugs
by regulating certain signaling pathways remain to be
determined.
Taken together, HAX1 acts as a tumor oncogene in
liver cancer. miR-125a-5p regulated the viability, colony
formation, migration and invasion of liver cancer cells by
negatively regulating HAX1 expression. The process also involves
the expression of p53, VEGF and EMT.
Funding
This work was supported by the Joint Project of
Provincial Health Department (grant no. LHGJ20191222).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZZ designed the study. JL performed the experiments
and contributed to data analysis. ZZ drafted and revised the
article. All authors gave final approval of the version to be
published and agree to be accountable for all aspects of the
work.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Luoyang Central Hospital Affiliated to Zhengzhou
University (approval no. LC20170416022). All patients signed
informed consent in any experimental work involving human
samples.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
References
|
1
|
Costentin C: Hepatocellular carcinoma
surveillance. Presse Med. 46:381–385. 2017.In French. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Alsaied OA, Sangwan V, Banerjee S, Krosch
TC, Chugh R, Saluja A, Vickers SM and Jensen EH: Sorafenib and
triptolide as combination therapy for hepatocellular carcinoma.
Surgery. 156:270–279. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhu ZX, Huang JW, Liao MH and Zeng Y:
Treatment strategy for hepatocellular carcinoma in China:
Radiofrequency ablation versus liver resection. Jpn J Clin Oncol.
46:1075–1080. 2016.PubMed/NCBI
|
|
4
|
Forner A, Reig M and Bruix J:
Hepatocellular carcinoma. Lancet. 391:1301–1314. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhang Y, Huang F, Wang J, Peng L and Luo
H: MiR-15b mediates liver cancer cells proliferation through
targeting BCL-2. Int J Clin Exp Pathol. 8:15677–15683. 2015.
|
|
6
|
Liu CY, Chen KF and Chen PJ: Treatment of
liver cancer. Cold Spring Harb Perspect Med. 5:a0215352015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Llovet JM, Schwartz M and Mazzaferro V:
Resection and liver transplantation for hepatocellular carcinoma.
Semin Liver Dis. 25:181–200. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo M, Zhang H, Zheng J and Liu Y:
Glypican-3: A new target for diagnosis and treatment of
hepatocellular carcinoma. J Cancer. 11:2008–2021. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Balcerak A, Trebinska-Stryjewska A, Wakula
M, Chmielarczyk M, Smietanka U, Rubel T, Konopinski R,
Macech-Klicka E, Zub R and Grzybowska EA: HAX1 impact on collective
cell migration, cell adhesion, and cell shape is linked to the
regulation of actomyosin contractility. Mol Biol Cell.
30:3024–3036. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bidwell PA, Liu GS, Nagarajan N, Lam CK,
Haghighi K, Gardner G, Cai WF, Zhao W, Mugge L, Vafiadaki E, et al:
HAX-1 regulates SERCA2a oxidation and degradation. J Mol Cell
Cardiol. 114:220–233. 2018. View Article : Google Scholar :
|
|
11
|
Fadeel B and Grzybowska E: HAX-1: A
multifunctional protein with emerging roles in human disease.
Biochim Biophys Acta. 1790:1139–1148. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wei XJ, Li SY, Yu B, Chen G, Du JF and Cai
HY: Expression of HAX-1 in human colorectal cancer and its clinical
significance. Tumour Biol. 35:1411–1415. 2014. View Article : Google Scholar
|
|
13
|
Li X, Li T, You B, Shan Y, Shi S, Cao X
and Qian L: Expression and function of HAX-1 in human cutaneous
squamous cell carcinoma. J Cancer. 6:351–359. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
You Y, Yao H, You B, Li X, Ni H, Shi S,
Shan Y and Cao X: Clinical significance of HAX-1 expression in
laryngeal carcinoma. Auris Nasus Larynx. 42:299–304. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Feng X, Kwiecinska A, Rossmann E, Bottai
M, Ishikawa T, Patarroyo M, Österborg A, Porwit A, Zheng C and
Fadeel B: HAX-1 overexpression in multiple myeloma is associated
with poor survival. Br J Haematol. 185:179–183. 2019. View Article : Google Scholar
|
|
16
|
Ramsay AG, Keppler MD, Jazayeri M, Thomas
GJ, Parsons M, Violette S, Weinreb P, Hart IR and Marshall JF:
HS1-associated protein X-1 regulates carcinoma cell migration and
invasion via clathrin-mediated endocytosis of integrin alphavbeta6.
Cancer Res. 67:5275–5284. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Y, Huo X, Cao Z, Xu H, Zhu J, Qian L,
Fu H and Xu B: HAX-1 is overexpressed in hepatocellular carcinoma
and promotes cell proliferation. Int J Clin Exp Pathol.
8:8099–8106. 2015.PubMed/NCBI
|
|
18
|
Banerjee A, Saito K, Meyer K, Banerjee S,
Ait-Goughoulte M, Ray RB and Ray R: Hepatitis C virus core protein
and cellular protein HAX-1 promote 5-fluorouracil-mediated
hepatocyte growth inhibition. J Virol. 83:9663–9671. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu W, Cui Z and Zan X: Identifying
cancer-related microRNAs based on subpathways. IET Syst Biol.
12:273–278. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gandhi NS, Tekade RK and Chougule MB:
Nanocarrier mediated delivery of siRNA/miRNA in combination with
chemotherapeutic agents for cancer therapy: Current progress and
advances. J Control Release. 194:238–256. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Gallach S, Calabuig-Farinas S,
Jantus-Lewintre E and Camps C: MicroRNAs: Promising new
antiangiogenic targets in cancer. Biomed Res Int. 2014:8784502014.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li E, Ji P, Ouyang N, Zhang Y, Wang XY,
Rubin DC, Davidson NO, Bergamaschi R, Shroyer KR, Burke S, et al:
Differential expression of miRNAs in colon cancer between African
and Caucasian Americans: Implications for cancer racial health
disparities. Int J Oncol. 45:587–594. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nishida N, Mimori K, Fabbri M, Yokobori T,
Sudo T, Tanaka F, Shibata K, Ishii H, Doki Y and Mori M:
MicroRNA-125a-5p is an independent prognostic factor in gastric
cancer and inhibits the proliferation of human gastric cancer cells
in combination with trastuzumab. Clin Cancer Res. 17:2725–2733.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li G, Zhang W, Gong L and Huang X:
MicroRNA 125a-5p inhibits cell proliferation and induces apoptosis
in hepatitis B virus-related hepatocellular carcinoma by
downregulation of ErbB3. Oncol Res. 27:449–458. 2019. View Article : Google Scholar
|
|
26
|
Cai M, Chen Q, Shen J, Lv C and Cai L:
Epigenetic silenced miR-125a-5p could be self-activated through
targeting Suv39H1 in gastric cancer. J Cell Mol Med. 22:4721–4731.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Bi Q, Tang S, Xia L, Du R, Fan R, Gao L,
Jin J, Liang S, Chen Z, Xu G, et al: Ectopic expression of MiR-125a
inhibits the proliferation and metastasis of hepatocellular
carcinoma by targeting MMP11 and VEGF. PLoS One. 7:e401692012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Menyhárt O, Nagy Á and Győrffy B:
Determining consistent prognostic biomarkers of overall survival
and vascular invasion in hepatocellular carcinoma. R Soc Open Sci.
5:1810062018. View Article : Google Scholar
|
|
29
|
Ke Q, Ji J, Cheng C, Zhang Y, Lu M, Wang
Y, Zhang L, Li P, Cui X, Chen L, et al: Expression and prognostic
role of Spy1 as a novel cell cycle protein in hepatocellular
carcinoma. . Exp Mol Pathol. 87:167–172. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
31
|
Sheng C and Ni Q: Expression of HAX1 and
Ki-67 in breast cancer and its correlations with patient's
clinicopathological characteristics and prognosis. Int J Clin Exp
Med. 8:20904–20910. 2015.
|
|
32
|
Li M, Tang Y, Zang W, Xuan X, Wang N, Ma
Y, Wang Y, Dong Z and Zhao G: Analysis of HAX-1 gene expression in
esophageal squamous cell carcinoma. Diagn Pathol.
8:472013.PubMed/NCBI
|
|
33
|
Deng X, Song L, Wei Y and Guo XB: Analysis
of the expression of HAX-1 gene in human glioma. Neurosci Lett.
657:189–193. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wu Z, Ai X, Hu H, Wang S, Wang Y, Kang F,
Ouyang C and Zhu J: Hematopoietic-substrate-1 associated protein
X-1 (HAX-1) regulates liver cancer cells growth, metastasis, and
angiogenesis through Akt. Cancer Biol Ther. 20:1223–1233. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kramer N, Walzl A, Unger C, Rosner M,
Krupitza G, Hengstschläger M and Dolznig H: In vitro cell migration
and invasion assays. Mutat Res. 752:10–24. 2013. View Article : Google Scholar
|
|
36
|
Duff D and Long A: Roles for RACK1 in
cancer cell migration and invasion. Cell Signal. 35:250–255. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hua L, Wang CY, Yao KH, Chen JT, Zhang JJ
and Ma WL: High expression of long non-coding RNA ANRIL is
associated with poor prognosis in hepatocellular carcinoma. Int J
Clin Exp Pathol. 8:3076–3082. 2015.PubMed/NCBI
|
|
38
|
Shan H, Zhou X and Chen C: MicroRNA214
suppresses the viability, migration and invasion of human
colorectal carcinoma cells via targeting transglutaminase 2. Mol
Med Rep. 20:1459–1467. 2019.PubMed/NCBI
|
|
39
|
Yan L, Yu MC, Gao GL, Liang HW, Zhou XY,
Zhu ZT, Zhang CY, Wang YB and Chen X: MiR-125a-5p functions as a
tumour suppressor in breast cancer by downregulating BAP1. J Cell
Biochem. 119:8773–8783. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Yang X, Qiu J, Kang H, Wang Y and Qian J:
miR-125a-5p suppresses colorectal cancer progression by targeting
VEGFA. Cancer Manag Res. 10:5839–5853. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang Y, Zhang D, Lv J, Wang S and Zhang
Q: MiR-125a-5p suppresses bladder cancer progression through
targeting FUT4. Biomed Pharmacother. 108:1039–1047. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhong L, Sun S, Shi J, Cao F, Han X and
Chen Z: MicroRNA-125a-5p plays a role as a tumor suppressor in lung
carcinoma cells by directly targeting STAT3. Tumour Biol.
39:10104283176975792017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fu Y and Cao F: MicroRNA-125a-5p regulates
cancer cell proliferation and migration through NAIF1 in prostate
carcinoma. Onco Targets Ther. 8:3827–3835. 2015. View Article : Google Scholar
|
|
44
|
Tang L, Zhou L, Wu S, Shi X, Jiang G, Niu
S and Ding D: miR-125a-5p inhibits colorectal cancer cell
epithelial-mesenchymal transition, invasion and migration by
targeting TAZ. Onco Targets Ther. 12:3481–3489. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Qin X, Wan Y, Wang S and Xue M:
MicroRNA-125a-5p modulates human cervical carcinoma proliferation
and migration by targeting ABL2. Drug Des Devel Ther. 10:71–79.
2015.
|
|
46
|
Potenza N, Mosca N, Zappavigna S,
Castiello F, Panella M, Ferri C, Vanacore D, Giordano A, Stiuso P,
Caraglia M and Russo A: MicroRNA-125a-5p is a downstream effector
of sorafenib in its antiproliferative activity toward human
hepato-cellular carcinoma cells. J Cell Physiol. 232:1907–1913.
2017. View Article : Google Scholar
|
|
47
|
Kim JK, Noh JH, Jung KH, Eun JW, Bae HJ,
Kim MG, Chang YG, Shen Q, Park WS, Lee JY, et al: Sirtuin7
oncogenic potential in human hepatocellular carcinoma and its
regulation by the tumor suppressors MiR-125a-5p and MiR-125b.
Hepatology. 57:1055–1067. 2013. View Article : Google Scholar
|
|
48
|
Yu YF, Zhang Y, Shen N, Zhang RY and Lu
XQ: Effect of VEGF, P53 and telomerase on angiogenesis of gastric
carcinoma tissue. Asian Pac J Trop Med. 7:293–296. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou X, Wu W, Zeng A, Nie E, Jin X, Yu T,
Zhi T, Jiang K, Wang Y, Zhang J and You Y: MicroRNA-141-3p promotes
glioma cell growth and temozolomide resistance by directly
targeting p53. Oncotarget. 8:71080–71094. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Leung DW, Cachianes G, Kuang WJ, Goeddel
DV and Ferrara N: Vascular endothelial growth factor is a secreted
angiogenic mitogen. Science. 246:1306–1309. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Folkman J: What is the evidence that
tumors are angiogenesis dependent? J Natl Cancer Inst. 82:4–6.
1990. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Zhang T, Liu M, Wang C, Lin C, Sun Y and
Jin D: Down-regulation of MiR-206-promotes proliferation and
invasion of laryngeal cancer by regulating VEGF expression.
Anticancer Res. 31:3859–3863. 2011.PubMed/NCBI
|
|
53
|
Wu QB, Chen J, Zhu JW, Yin X, You HY, Lin
YR and Zhu HQ: MicroRNA-125 inhibits RKO colorectal cancer cell
growth by targeting VEGF. Int J Mol Med. 42:665–673.
2018.PubMed/NCBI
|
|
54
|
Jakobsen KR, Demuth C, Sorensen BS and
Nielsen AL: The role of epithelial to mesenchymal transition in
resistance to epidermal growth factor receptor tyrosine kinase
inhibitors in non-small cell lung cancer. Transl Lung Cancer Res.
5:172–182. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiao Z, Chen M, Yang J, Yang C, Lü X, Tian
H and Liu C: MTBP regulates migration and invasion of prostate
cancer cells in vitro. Nan Fang Yi Ke Da Xue Xue Bao. 39:6–12.
2019.In Chinese. PubMed/NCBI
|
|
56
|
Ramamurthy VP, Ramalingam S, Gediya LK and
Njar VCO: The retinamide VNLG-152 inhibits f-AR/AR-V7 and MNK-eIF4E
signaling pathways to suppress EMT and castration-resistant
prostate cancer xenograft growth. FEBS J. 285:1051–1063. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Satelli A and Li S: Vimentin in cancer and
its potential as a molecular target for cancer therapy. Cell Mol
Life Sci. 68:3033–3046. 2011. View Article : Google Scholar : PubMed/NCBI
|