Introduction
Cartilage defects caused by congenital abnormality,
trauma or inflammation in the weight bearing joint cause extensive
health issues for the patient. Cartilage tissue engineering enables
repair of cartilage defects, which are difficult to repair due to
the lack of regenerative ability of chondrocytes (1,2).
Although various types of scaffold for cartilage repair have been
developed, dedifferentiation of chondrocytes causes a challenge to
cartilage tissue engineering (3,4).
With continuous monolayer expansion, chondrocytes dedifferentiate
and lose phenotypical characteristics, displaying fibroblastic
phenotype, thus affecting their cartilage-forming ability (5). To reconstruct cartilage tissue, it
is necessary to develop a method to maintain the chondrocyte
phenotype during chondrocyte amplification.
Proliferation and differentiation of chondrocytes
are influenced by numerous factors, such as density of cell
inoculation (6), composition of
culture medium (7) and two or
three-dimensional culture conditions (8). Oxygen partial pressure is another
key factor affecting chondrocyte differentiation and cartilage
formation (9). The oxygen
partial pressure is 1-5% inside normal cartilage because of the
lack of blood vessels (10).
Chondrocytes produce more functional extracellular matrix in
hypoxic compared with in normoxic environments (11). However, the mechanism of
chondrocyte differentiation under low oxygen partial pressure
remains unclear.
Differentiation of chondrocytes and cartilage
formation are regulated by a variety of methylases (12). In our previous study, the lysine
methyltransferase Su(var)3-9, enhancer of zeste and trithorax (SET)
domain containing 7 (SETD7) regulated apoptosis of chondrocytes at
different oxygen partial pressures (13). SETD7 specifically identifies the
K/R-S/T-K* amino acid sequence and transfers a methyl group to
lysine residue of K* (14).
SETD7 is involved in cell cycle regulation, DNA damage response,
gene transcription and cell differentiation (15). To the best of our knowledge,
however, the role and mechanism of SETD7 in regulating chondrogenic
differentiation have not been studied.
Hippo signaling controls organogenesis and cell
differentiation (16). When
Hippo signaling is activated, a series of phosphorylation events
occur via mammalian STE20-like T cells (MST) and the linker for
activation of T cells (LAT) kinases, ultimately leading to
phosphorylation of Yes-associated Protein (YAP) (17). Phosphorylated (p-)YAP is
sequestered in the cytoplasm, which decreases transcription of
downstream genes in the nucleus (18). By contrast, inactivation of the
Hippo pathway increases nuclear translocation of YAP, where YAP
interacts with other transcription factors, such as tafazzin
(19), to activate the
transcription of target genes (18). Recently, YAP was found to be
associated with chondrocyte differentiation. YAP promotes
chondrogenic phenotype maintenance of rat growth plate chondrocytes
and cartilage development in mice (20,21).
Hypoxia inducible factor-1α (HIF-1α) is a key
regulatory factor involved in the adaptation process of cells to
different oxygen partial pressures (22). The deletion of HIF-1α leads to
chondrocyte death in a hypoxic developmental growth plate, which
suggests that HIF-1α is essential for chondrocyte survival
(23). HIF-1α also regulates
collagen synthesis and modification in chondrocyte differentiation
(24). Both YAP and HIF-1α are
methylated by SETD7 (25,26).
In hepatocarcinoma cells, YAP forms a complex with HIF-1α and binds
to the promoter of the pyruvate kinase isoenzyme 2 (PKM2) gene to
promote its transcription (27).
It was hypothesized that SETD7 may serve an
important role in chondrocyte differentiation in hypoxic condition.
SETD7, YAP and HIF-1α may form a regulatory network during
chondrocyte differentiation. Therefore, the objective of the
present study was to investigate the regulatory role of SETD7 in
chondrocyte differentiation and its mechanism.
Materials and methods
Cell culture
ATDC5 cells were purchased from the American Type
Culture Collection (ATCC) cell bank and grown in Dulbecco's
modified Eagle's medium/F12 (DMEM:F12, 1:1) supplemented with 10%
(v/v) fetal bovine serum (FBS; both Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin G and 100 mg/ml streptomycin
(HyClone; Cytiva). ATDC5 cells were cultured at 37°C with either 1%
(low) or 20% O2 (high oxygen tension).
293T cells were purchased from the ATCC cell bank
and grown in Roswell Park Memorial Institute (RPMI)-1640 medium
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) at 37°C with 5%
CO2.
Viral infection
Viral packaging and infection were performed as
described previously (11).
SETD7 short hairpin (sh)RNA and non-specific control (NC) plasmids
were purchased from Jintuosi Biological Technology Co., Ltd. The
target sequences of SETD7 shRNA and NC were 5′-GGG ATA TTA CGT GGA
CGA T-3′ and 5′-TAC TGT TAG GAT CAG G AG G-3′, respectively. shRNA
or NC plasmids (both 2.4 µg), as well as two helper vectors
(pMD2G:pSPAX2:shRNA=1:2:4, were transfected into 2nd generation
293T cells using Neofect transfection reagent [Neofect (Beijing)
Biotech Co., Ltd.] at 37°C for 48 h. The viral supernatant was
collected following centrifugation at 25°C (4,000 × g; 3 min) after
48 h and applied to transfect ATDC5 in the presence of 10
µg/ml polybrene (Sigma-Aldrich; Merck KGaA) at 37°C for 24 h
at MOI=10. The viral supernatant was removed and changed for fresh
growth medium after 24 h transfection. ATDC5 cells were inoculated
and subcultured once for subsequent experiments.
Chondrogenic induction of cells
For chondrogenic differentiation, ATDC5 cells were
seeded into 24-well plates at a density of 2×104
cells/cm2 and grown in DMEM:F12 at 37°C. At 70-90%
confluence, growth medium was replaced with chondrogenic
differentiation medium supplemented with 1x
insulin-transferrin-selenium (Corning, Inc.). The medium was
replaced every other day and cells were cultured for 14 days at
37°C. Micromass culturing was performed as previously described
(28). Briefly, ATDC5 cells were
resuspended in DMEM:F12 at a density of 2×107 cells/ml.
The cell suspension (20 µl) was transferred into the middle
of 24-well plate for 3 h at 37°C and chondrogenic differentiation
medium was added for 14 days at 37°C.
Immunofluorescence (IF) staining of
cells
For IF staining, ATDC5 cells were fixed at 25°C with
4% paraformaldehyde for 15 min, permeabilized with 0.1% Triton
X-100 for 5 min and blocked at 25°C with 0.1% BSA for 1 h. The
cells were incubated with primary antibodies against HIF-1α (1:200;
Abcam; cat. no. ab16066), YAP (1:100; Cell Signaling Technology,
Inc.; cat. no. 4912S) or SETD7 (1:50; Abcam; cat. no. ab14820)
overnight at 4°C. The cells were rinsed three times with PBS and
incubated with the corresponding secondary antibodies (Alexa
Fluor® 488- or 594-conjugated goat anti-rabbit or
anti-mouse IgG; both 1:1,000; both Abcam; cat. nos. ab150113 and
ab150080, respectively). Next, the cells were rinsed with PBS three
times and stained with 10 µg/ml DAPI for 5 min at 25°C.
Images were captured with a laser scanning confocal microscope
(×400 magnification; Zeiss GmbH; cat. no. LSM780). All experiments
were performed in triplicate.
RNA extraction and reverse
transcription-quantitative (RT-q) PCR
Total RNA from ATFC5 cells was extracted using an
RNA extraction kit (Shanghai Yishan Biological Technology Co.,
Ltd.) according to the manufacturer's instructions. Concentration
and quality of the total RNA samples were measured using a
Nanodrop2000 (Thermo Fisher Scientific, Inc.). Complementary DNA
was synthesized from 1 µg total RNA using a PrimeScript
RT-PCR kit (Takara Bio, Inc.; cat. no. RR047A) following the
manufacturer's protocol. RT-qPCR was performed using the
PrimeScript RT-PCR kit (Takara Bio, Inc.; cat. no. RR820A). The
primers are listed in Table I.
The thermocycling conditions were set as follows: Initial
denaturation for 30 sec at 95°C; followed by 40 cycles of 95°C for
5 sec, 60°C for 30 sec and 95°C for 5 sec; melting at 65°C for 60
sec and 97°C for 1 sec and cooling at 50°C for 30 sec. Relative
mRNA expression of target genes was calculated using the
2−ΔΔCq method (13).
The experiments were performed in triplicate and repeated three
times.
 | Table IPrimer sequences used for reverse
transcription-quantitative PCR. |
Table I
Primer sequences used for reverse
transcription-quantitative PCR.
| Gene | Species | Primer sequence,
5′→3′ |
|---|
| SETD7 | Mouse | Forward
TCCAAGGTAGCAGTTGGACCTAA |
| Reverse
CCAAGGAGGCACAGTACTTGG |
| YAP | Mouse | Forward
ATTTCGGCAGGCAATACGGA |
| Reverse
TGCTCCAGTGTAGGCAACTG |
| HIF-1α | Mouse | Forward
AGCACAGTTACAGTATTCCAGCAGAC |
| Reverse
TCATCAGTGGTGGCAGTGGTAGT |
| COL2A1 | Mouse | Forward
GGCCAGGATGCCCGAAAATTA |
| Reverse
GGCTGCAAAGTTTCCTCCAC |
| SOX9 | Mouse | Forward
CTCCTACTACAGCCACGCAG |
| Reverse
GCTGTGTGTAGACGGGTTGT |
| Aggrecan | Mouse | Forward
AGCTTGAGGGGGAAGTGTTC |
| Reverse
GGTTGACGATGGGGTATCGG |
| GLUT1 | Mouse | Forward
ACCACCTCACTCCTGTTACTTACCT |
| Reverse
ATCCAAACCTCCTACCCTCAATCCA |
| LDHA | Mouse | Forward
TAGGCTACAACAGGATTCTAGGTGGAG |
| Reverse
GTCAGAGGTGGCAGAACTATTTC |
| PGK1 | Mouse | Forward
GAGCCAAGTCGGTAGTCCTTATGAG |
| Reverse
CACAGTCCTTCAAGAACAGAACATCCT |
| PKM2 | Mouse | Forward
GTGCCGCCTGGACATTGATTCA |
| Reverse
AGTTCAGACGAGCCACATTCATTCC |
| 18S | Mouse | Forward
GGCGCCCCCTCGATGCTCTTAG |
| Reverse
GCTCGGGCCTGCTTTGAACACTCT |
| GAPDH | Mouse | Forward
TTGCAGTGGCAAAGTGGAGA |
| Reverse
GATGGGCTTCCCGTTGATGA |
Protein extraction and western
blotting
The cytoplasmic and nuclear proteins from ATDC5
cells were isolated using a nuclear and cytoplasmic protein
extraction kit (Beyotime Institute of Biotechnology; cat. no.
P0028) according to the manufacturer's instructions. The total
protein from ATDC5 cells was collected with RIPA buffer
(ProteinTech Group, Inc.; cat no. PR20001), mixed with loading
buffer, and heated at 95°C for 10 min. The protein concentration
was determined via bicinchoninic acid assay kit (Thermo Fisher
Scientific, Inc.; cat. no. 23227).
Proteins were separated by sodium dodecyl
sulfate-poly-acrylamide 10% gel electrophoresis (GenScript; cat.
no. M00665) and transferred to nitrocellulose membranes. The
membranes were blocked at 25°C using 5% skimmed milk or 5% BSA for
60 min and then incubated with primary antibodies against SETD7
(1:1,000; Abcam; cat. no. ab14820), HIF-1α (1:2,000; Abcam; cat.
no. ab16066), large tumor suppressor 1 (LATS1; 1:1,000; Cell
Signaling Technology, Inc.; cat. no. 3477), p-LATS1 (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 8654), YAP (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 4912s), p-YAP (1:1,000; Cell
Signaling Technology, Inc.; cat. no. 4911s), β-actin (1:1,000;
ABclonal Biotech Co., Ltd.; cat. no. AC004), Lamin B (1:1,000;
Weiao Biotechnology Co., Ltd.; cat. no. WB0199) or β-tubulin
(1:1,000; Absin; cat. No. abs830032) at 4°C overnight, followed by
secondary antibodies (horseradish peroxidase-conjugated goat
anti-rabbit or anti-mouse IgG; both 1:10,000; both ABclonal Biotech
Co., Ltd.; cat nos. AS063 and AS064, respectively) for 1 h at 25°C.
The primary and secondary antibodies were diluted in 5% BSA.
Protein bands on the membranes were visualized using a Western
Bright ECL HRP substrate kit (Advansta, Inc.). ImageJ (V 1.8.0;
National Institutes of Health) was used for densitometry. All the
experiments were performed in triplicate.
Alcian blue staining
For alcian blue staining, ATDC5 cells were cultured
in chondrogenic induction medium at a density of 2×104
cells/ml in 24-well plates for 14 days at 25°C. The cells were
fixed at 25°C with 4% (w/v) paraformaldehyde for 15 min. The cells
were washed twice with PBS and stained for 30 min at 4°C with 0.5%
alcian blue dye (Sigma-Aldrich; Merck KGaA) in 1 mol/l HCl. After
staining, cells were washed twice with distilled water and observed
under a light microscope (×40 magnification; Axio Observer Z1;
Zeiss GmbH).
Co-immunoprecipitation (Co-IP)
ATDC5 cells at a density of 2×107
cells/ml were pretreated at 25°C with 5 µM MG132 for 6 h
after transfection. The cells were then collected and incubated
with 300 µl lysis buffer (Beyotime Institute of
Biotechnology; cat. no. P0013) containing protease inhibitors for
40 min on ice. The supernatant was collected following
centrifugation at 25°C (4,000 × g; 5 min) and 2 µg HIF-1α
(1:2,000; Abcam; cat. no. ab16066), YAP (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 4912s) or IgG (1:1,000; ProteinTech
Group, Inc.; cat. no. B900620) antibody was added. The samples (300
µl) were then incubated at 4°C overnight. Next, 20 µl
protein A/G-agarose beads (Santa Cruz Biotechnology, Inc.) was
added and the samples were rocked for 3 h at 4°C. The pelleted
cells were collected following centrifugation at 25°C (4,000 × g; 5
min) and washed three times with 500 µl lysis buffer.
Finally, precipitate was boiled with 40 µl loading buffer
for 5 min and analyzed by western blotting as aforementioned. The
experiments were repeated in triplicate.
Extracellular acidification rate
(ECAR)
ECAR was measured using a Seahorse XF glycolysis
stress test kit according to the manufacturer's protocol and a
Seahorse XF 96 Extracellular Flux Analyzer (Seahorse Bioscience;
Agilent Technologies, Inc.). In brief, ATDC5 cells
(2×104 cells/well) were plated in a Seahorse XF 96-well
cell culture plate. The loaded sensor cartridge with the utility
plate was placed into the instrument for calibration, then glucose,
oligomycin and 2-deoxyglucose were sequentially added to each well
at 20, 40 and 60 min. ECAR data were assessed using Seahorse XF-96
Wave (V 2.6; Seahorse Bioscience; Agilent Technologies, Inc.)
software. The tests were performed in triplicate.
Determination of lactate and glucose
levels
Lactic acid and glucose levels in ATDC5 cells
treated at 37°C for 2 h in the presence or absence of PFI-2 (10
µM, MedChemExpress; cat. no. HY-18627A), a specific
inhibitor of SETD7, were determined using a lactic acid assay kit
II (cat. no. MAK065-1KT) and highly sensitive glucose assay kit
(both Sigma-Aldrich; Merck KGaA; cat. no. MAK181-1KT),
respectively, according to the manufacturer's instructions. ATDC5
cells at a density of 2×107 cells/ml were homogenized on
ice with a glucose test or lactic acid buffer. The supernatant was
collected following centrifugation at 25°C (3,280 × g; 5 min).
Ultrafiltration tubes (10 kDa) were used to remove proteins from
the sample. Fluorescence intensity (excitation, 535; emission, 587
nm) was measured after diluting the sample with glucose
determination buffer and the glucose level was assessed. The
absorbance value at 450 nm was measured after sample was diluted
with lactic acid determination buffer solution and lactic acid
level was assessed.
Cycloheximide (CHX) chase test
CHX was dissolved in PBS to a concentration of 100
µg/ml. ATDC5 cells at a density of 2×107 cells/ml
were treated at 25°C with CHX for 0, 2 and 4 h after being treated
in the presence or absence of verteporfin (VP, 5 µM) at 37°C
for 2 h. The cells were collected and the protein levels of HIF-1α
were analyzed using western blotting as aforementioned.
Statistical analysis
Data are presented as the mean ± SD of three
independent experimental repeats. All tests were performed in
triplicate. GraphPad Prism software (GraphPad Software, Inc.; V
8.0.1.244) was used for statistical analysis. Following tests of
normality (Shapiro-Wilk test) and Levene's test for equality of
variance, paired Student's t-test was used to assess the
differences between the control and test groups. One- and two-way
ANOVA followed by Dunnett's test or Bonferroni correction were used
to analyze the differences of aggrecan, SOX9 and COL2A1 relative
expression between multiple groups during cartilage induction and
following SETD7 knockdown. Two-way ANOVA followed by Dunnett's test
or Bonferroni's correction were used to analyze the differences
between groups all other experiments. P<0.05 was considered to
indicate a statistically significant difference.
Results
Expression of SETD7 decreases during
chondrogenic differentiation
Chondrocytes in monolayer and micromass culture
exhibited greater alcian blue staining in 1% O2 group
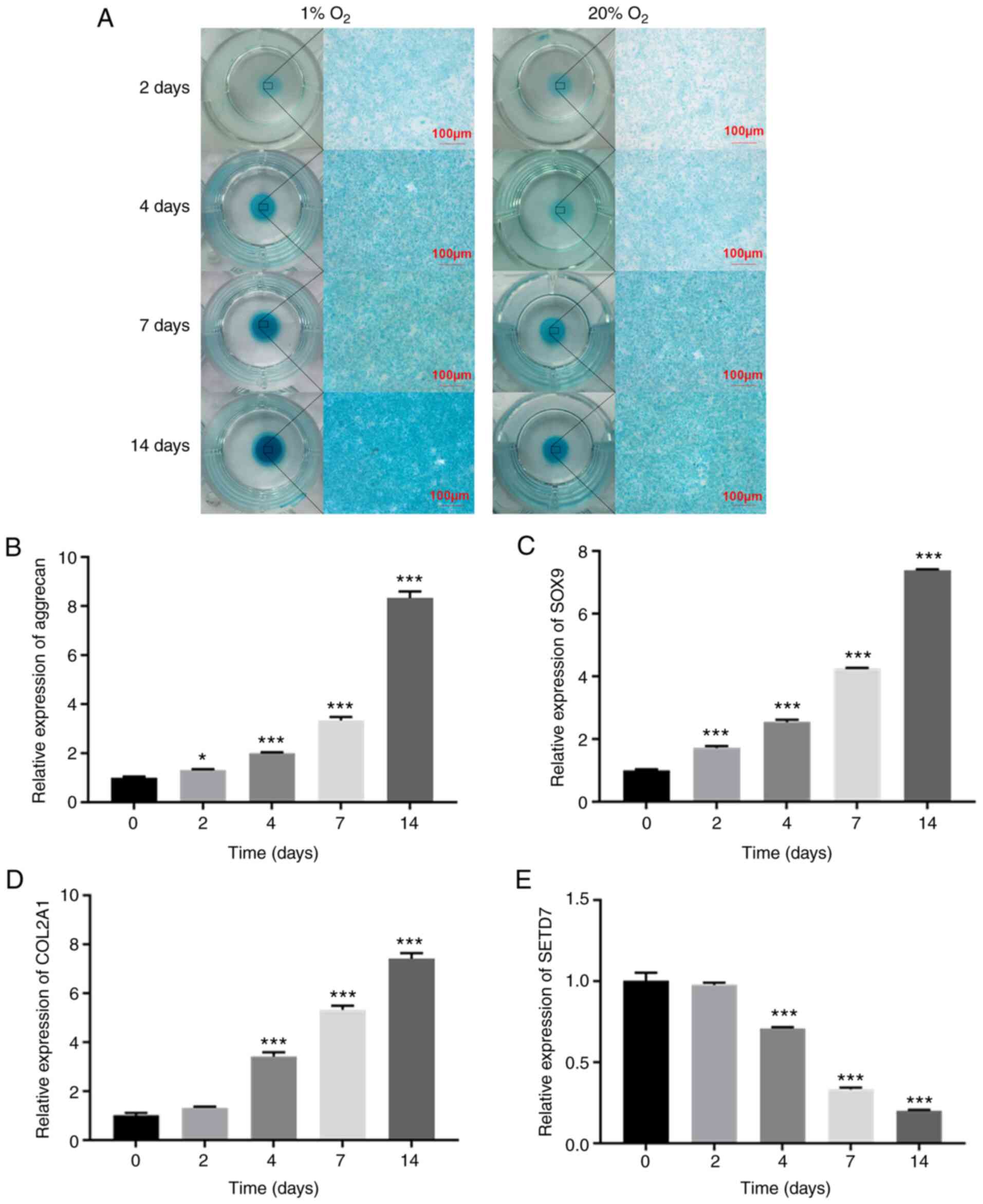
(Fig. 1A). In monolayer culture,
the expression levels of aggrecan, SRY-related box gene 9 (SOX9)
and collagen II, α1 (COL2A1; chondrocyte differentiation markers)
increased (Fig. 1B-D), while
mRNA levels of SETD7 gradually decreased (Fig. 1E) during differentiation of ATDC5
cells in 1% O2. These results indicated that expression
of SETD7 decreased during chondrogenic differentiation.
SETD7 inhibits chondrogenic
differentiation in a hypoxic environment
IF results showed that SETD7 was localized to the
cytoplasm of ATDC5 cells (Fig. 2A
and B). Following SETD7 knockdown (Fig. 2C and D), alcian staining was more
pronounced (Fig. 2E) in
monolayer and micromass culture and the expression levels of
aggrecan, SOX9 and COL2A1 increased in monolayer culture (Fig. 2F-H). These results indicated that
SETD7 inhibited chondrogenic differentiation in hypoxic
environment.
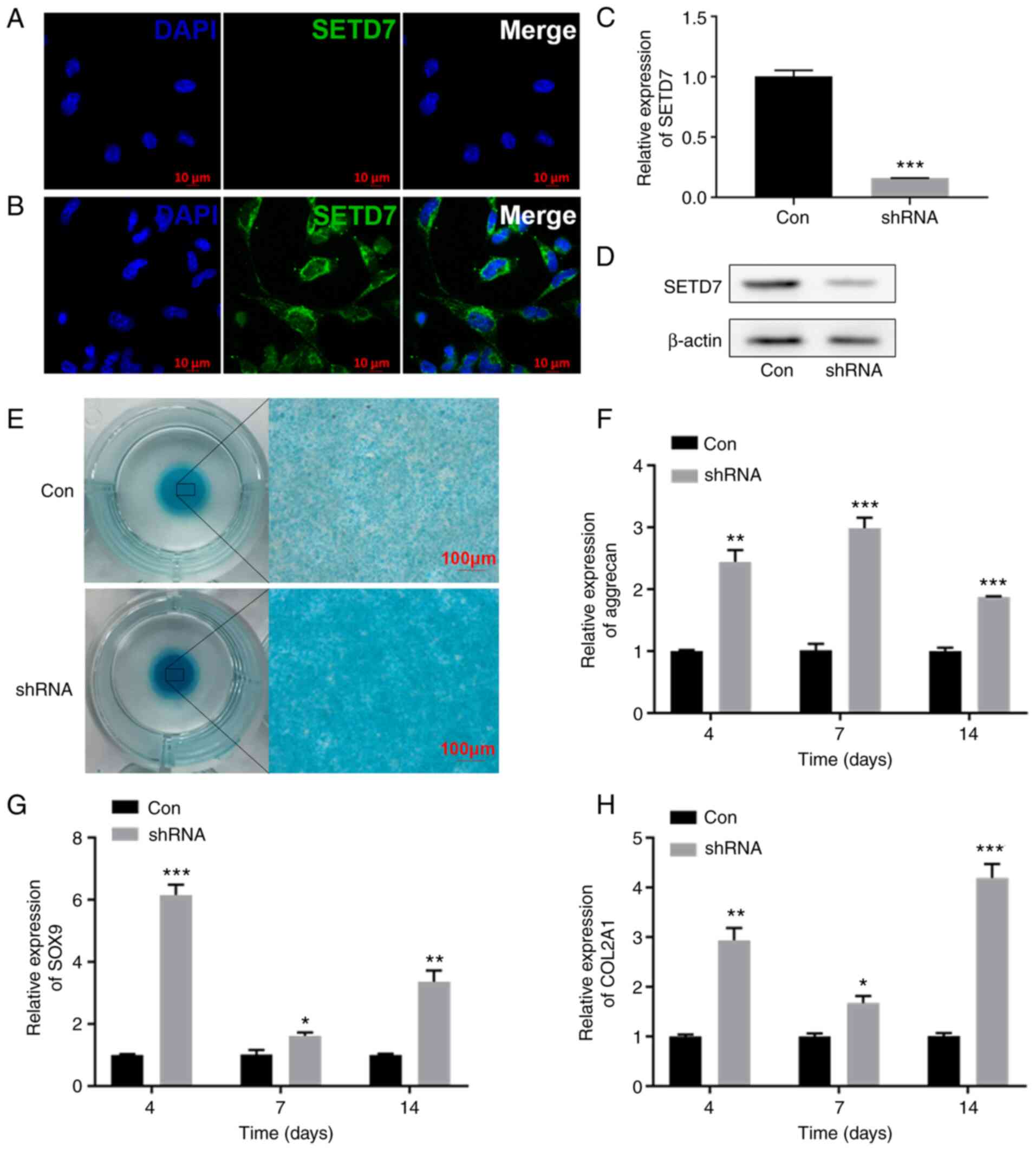 | Figure 2Knocking down SETD7 activates
chondrogenic differentiation. (A) Immunofluorescence staining of
ATDC5 cells at 1% O2 incubated with secondary antibodies
and DAPI. (B) Immunofluorescence staining with SETD7 primary
antibody, secondary antibodies and DAPI in ATDC5 cells at 1%
O2. (C) mRNA expression and (D) protein levels of SETD7
after knocking down SETD7 at 1% O2. (E) Alcian blue
staining after knocking down SETD7 in cells cultured in micromass
at 1% O2. mRNA expression of (F) aggrecan, (G) SOX9 and
(H) COL2A1 after knocking down SETD7 at 1% O2.
*P<0.05, **P<0.01,
***P<0.001 vs. con. SETD7, SET domain containing 7;
HIF-1α, hypoxia inducible factor-1α; COL2A1, collagen II, α1; SOX9,
SRY-related box gene 9; con, control; sh, short hairpin. |
SETD7 knockdown activates YAP and
HIF-1α
In monolayer culture, the mRNA expression levels of
YAP and HIF-1α showed no change following silencing of SETD7
(Fig. 3A). Protein levels of
LATS1, p-LATS1 (Ser909) and p-YAP (Ser127) decreased. Although the
protein level of total YAP remained unchanged, localization of YAP
in the nucleus increased (Fig. 3B
and C). The protein levels of total and nuclear HIF-1α in
increased in the SETD7 knockdown group (Fig. 3B and D). IF results showed that
nuclear localization of both YAP and HIF-1α in ATDC5 cells
increased following SETD7 silencing (Fig. 3E). These results indicated that
SETD7 knockdown activated YAP and HIF-1α.
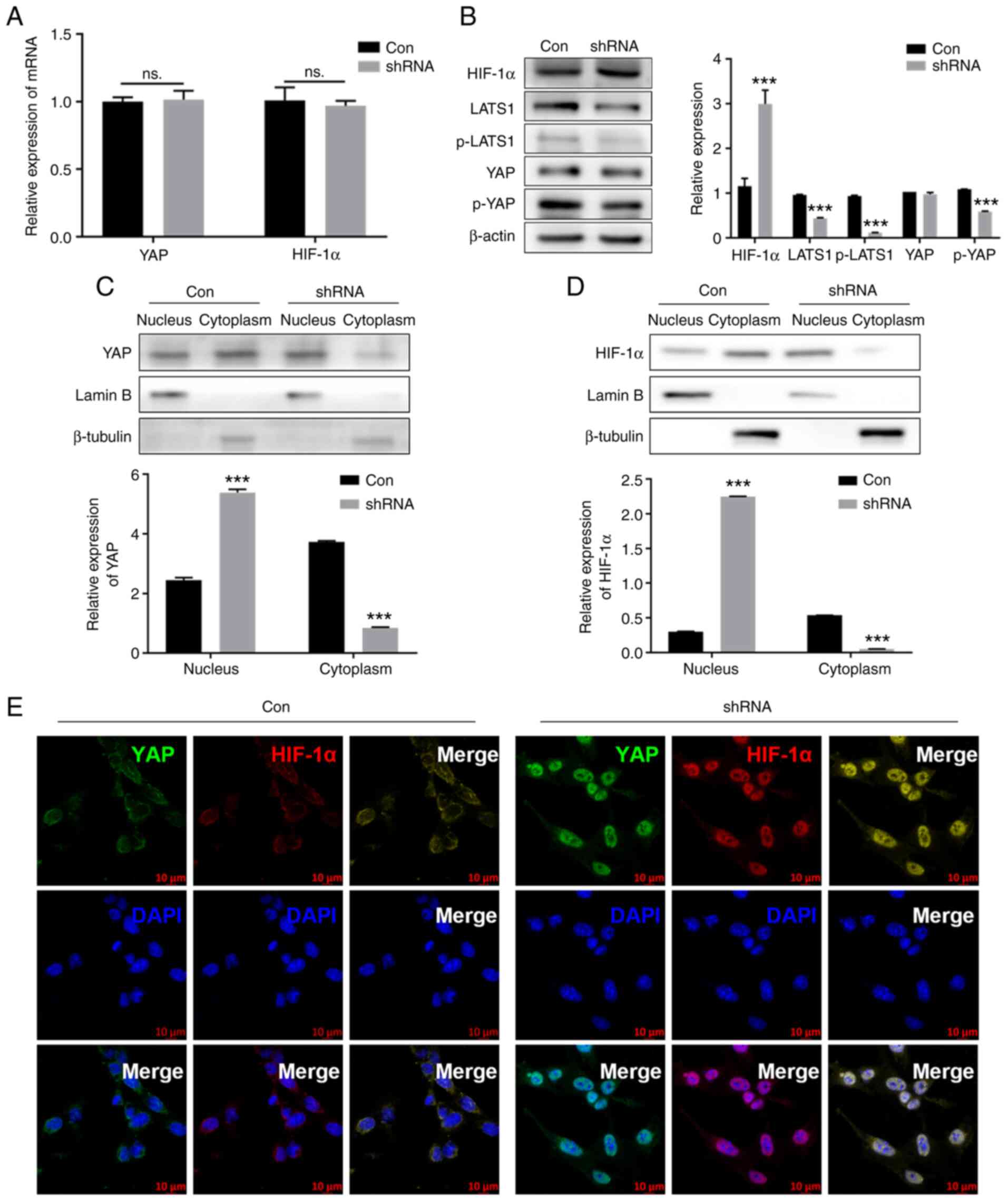 | Figure 3Knocking down SETD7 activates YAP and
HIF-1α. (A) mRNA expression of YAP and HIF-1α after knocking down
SETD7 at 1% O2. (B) Protein levels of LATS1, p-LATS1,
YAP, p-YAP and HIF-1α after knocking down SETD7 at 1%
O2. Protein levels of (C) YAP and (D) HIF-1α in the
nucleus and cytoplasm after knocking down SETD7 at 1%
O2. Lamin B and β-tubulin were used as the reference for
cytoplasmic and nuclear protein, respectively. (E)
Immunofluorescence staining of YAP and HIF-1α after knocking down
SETD7 at 1% O2. ***P<0.001 vs. con. SETD7,
SET domain containing 7; YAP, Yes-associated protein; HIF-1α,
hypoxia inducible factor-1α; LATS1, large tumor suppressor 1; p-,
phosphorylated; con, control; ns, not significant; sh, short
hairpin. |
YAP and HIF-1α form a complex
In monolayer culture, Co-IP showed that YAP bound to
HIF-1α (Fig. 4A). After
inhibiting YAP with VP, a YAP specific inhibitor (29), the HIF-1α content decreased at 2
and 4 h compared with the control group (Fig. 4B and C). These results indicated
that YAP and HIF-1α form a complex.
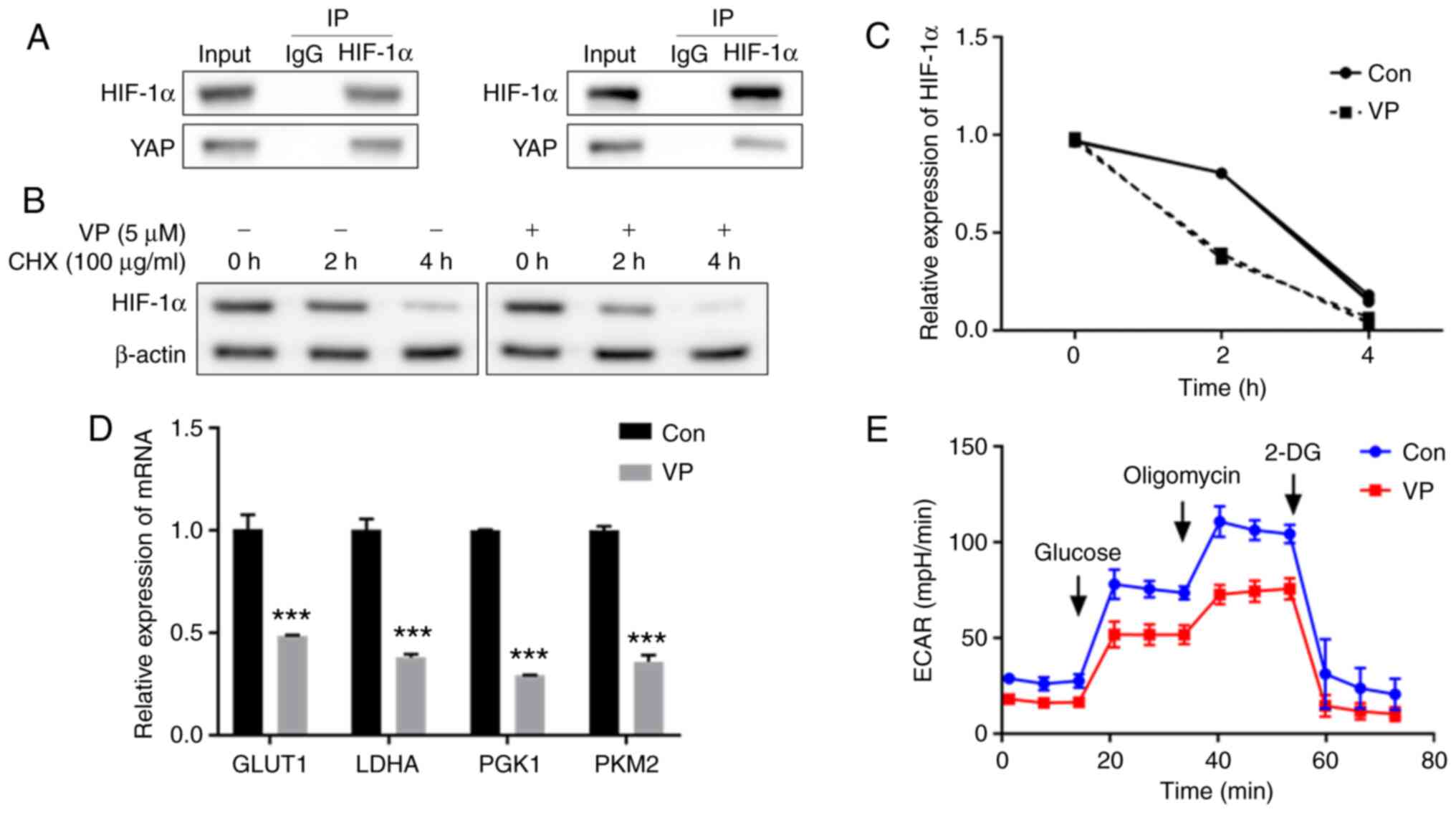 | Figure 4YAP combines with HIF-1α following
SETD7 knockdown. (A) IP results of YAP and HIF-1α after knocking
down SETD7. ATDC5 cells were pretreated with MG132 (5 µM)
for 6 h after transfection. Following ATDC5 cell pretreatment with
VP and CHX, (B) western blotting was performed and (C) protein
expression levels of HIF-1α were assessed. (D) mRNA expression of
glycolytic enzymes GLUT1, LDHA, PGK1 and PKM2 following YAP
inhibition with VP. (E) ECAR decreased following YAP inhibition.
***P<0.001 vs. con. SETD7, SET domain containing 7;
YAP, Yes-associated protein; HIF-1α, hypoxia inducible factor-1α;
VP, verteporfin; IP, immunoprecipitation; CHX, cycloheximide;
GLUT1, glucose transporter 1; LDHA, lactate dehydrogenase A; PGK1,
phosphoglycerate kinase 1; PKM2, pyruvate kinase isoenzyme 2; ECAR,
extracellular acidification rate; con, control; 2-DG,
2-deoxyglucose. |
Effect of SETD7 on chondrocyte
glycolysis
Following inhibition of YAP, mRNA levels of glucose
transporter 1 (GLUT1), lactate dehydrogenase A (LDHA),
phosphoglycerate kinase 1 (PGK1) and PKM2 in ATDC5 cells decreased
(Fig. 4D), as well as the ECAR
in monolayer culture (Fig.
4E).
After inhibiting SETD7 using PFI-2, a specific SETD7
inhibitor (30), glucose uptake,
lactic acid production rate and ECAR of ATDC5 cells all increased
in monolayer culture compared with the control (Fig. 5A-C). The mRNA expression of key
glycolytic enzymes GLUT1, LDHA, PGK1 and PKM2 also increased in the
SETD7-inhibited group in monolayer culture (Fig. 5D). These results indicated that
SETD7 inhibited chondrocyte glycolysis.
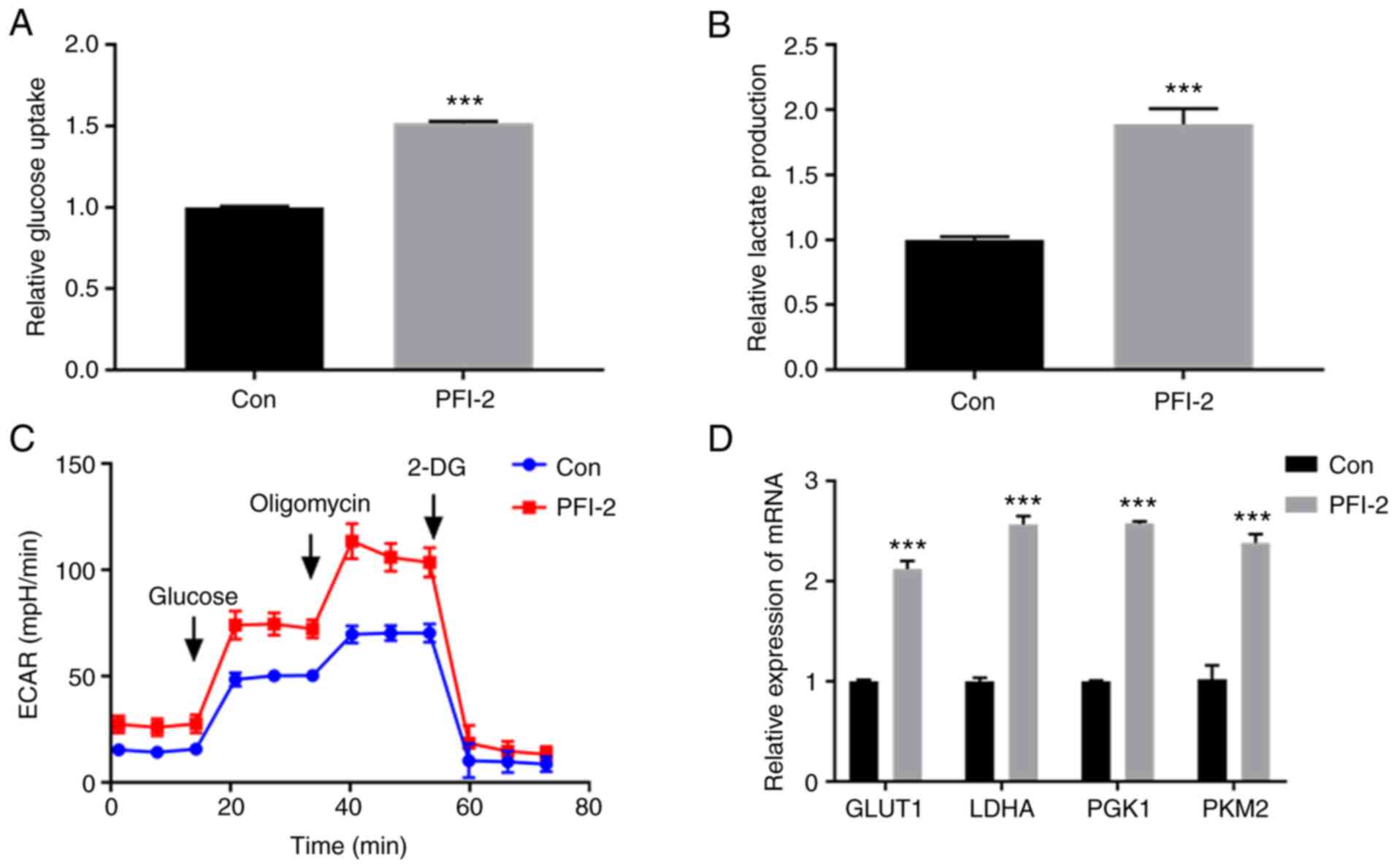 | Figure 5Effect of SETD7 on chondrocyte
glycolysis. ATDC5 cells were pretreated with PFI-2 (40 nM) for 24 h
to inhibit SETD7. (A) Glucose uptake and (B) lactic acid production
increased after SETD7 was inhibited. (C) ECAR of control and PFI-2
groups. (D) mRNA expression levels of the key glycolysis enzymes
GLUT1, LDHA, PGK1 and PKM2 of control and PFI-2 groups.
***P<0.001 vs. con. SETD7, SET domain containing 7;
GLUT1, glucose transporter 1; LDHA, lactate dehydrogenase A; PGK1,
phosphoglycerate kinase 1; PKM2, pyruvate kinase isoenzyme 2; ECAR,
extracellular acidification rate; con, control; 2-DG,
2-deoxyglucose. |
Discussion
Cartilage tissue engineering has attracted attention
for its potential to improve repair of cartilage defects (31). Chondrocytes tend to lose their
phenotype and cartilage-forming ability under normal oxygen
conditions in vitro (32). The cartilage-forming ability of
chondrocytes is essential to the success of cartilage tissue
engineering (33). Chondrocytes
produce more functional extracellular matrix in hypoxic compared
with normoxic environments (11). In the present study, chondrocytes
exhibited better differentiation under hypoxic conditions in
vitro. However, the mechanism of chondrocyte differentiation
under low oxygen partial pressure remain unclear. The present study
found that alcian staining was more pronounced in monolayer and
micromass culture and the expression levels of aggrecan, SOX9 and
COL2A1 increased in monolayer culture following SETD7 knockdown;
this indicated that chondrocyte differentiation under low oxygen
partial pressure was regulated by SETD7.
SETD7 was initially identified as a histone
methyltransferase that catalyzes methylation of H3K4, thereby
promoting transcriptional activation (34). Numerous non-histone substrates,
such as YAP (35) and E2F1
(36), of SETD7 have recently
been described as key factors in regulating cell differentiation
and proliferation (37). In the
present study, SETD7 mRNA expression gradually decreased during
chondrocyte differentiation. The regulatory mechanism of SETD7 in
chondrocyte differentiation was investigated by silencing SETD7 in
ATDC5 cells. Following SETD7 knockdown, ATDC5 cells showed improved
differentiation ability and increased expression levels of
aggrecan, SOX9 and COL2A1. These results indicated that SETD7
served a negative role in chondrogenic differentiation.
Hippo signaling pathway regulates the morphology and
cell differentiation of various organs, such as liver (38) and kidney (39). When the Hippo signal is
activated, a series of phosphorylation reactions occur via MST and
LAT kinases, culminating in YAP phosphorylation, which initiates
cytoplasmic retention and ubiquitin-mediated degradation. The
inactivation of Hippo pathway promotes YAP translocation to the
nucleus (40). After entering
the nucleus, YAP interacts with TEA domain transcription factors to
activate the transcription of target genes and promote chondrocyte
differentiation (18). Silencing
SETD7 using a lentiviral vector system demonstrated that SETD7
prevented YAP from entering the nucleus and inhibited the
chondrogenic differentiation ability of ATDC5 cells. Studies have
shown that SETD7 binds to YAP to form a complex and methylate its
lysine 494 residue, retaining it in the cytoplasm (26,35). The biological effect of SETD7
inhibition of chondrogenic differentiation via inhibiting YAP may
be caused by direct methylation of YAP or activation of Hippo
signaling by SETD7 (41). The
present study found that the expression of proteins associated with
the Hippo signaling pathway increased after silencing SETD7,
indicating that SETD7 activated the Hippo signaling pathway,
resulting in phosphorylation of YAP protein to prevent it from
entering the nucleus to perform its function, thus inhibiting
chondrogenic differentiation of ATDC5 cells.
HIF-1α is ubiquitous in human and mammalian cells,
regulating glycolytic metabolism, angiogenesis and apoptosis in the
absence of oxygen (42). When
oxygen partial pressure is <5%, HIF-1α accumulates in the
cytoplasm and transfers to the nucleus, where it binds to hypoxic
reaction elements and promotes downstream gene expression. HIF-1α
activates PDK-1 during anaerobic glycolysis, inhibits the aerobic
respiratory chain and decreases formation of reactive oxygen
species. When oxygen partial pressure >5%, prolyl hydroxylase
domain-containing enzymes specifically hydroxylate proline residues
of HIF-1α (43). The
hydroxylated HIF-1α is degraded by proteasomes following
ubiquitination (44). Recent
studies have shown that HIF-1α enhances expression of SOX9 induced
by bone morphogenetic protein 2 and inhibits the expression of
Runx2 in mesenchymal stem cells, thereby promoting chondrogenesis
and inhibiting osteogenesis (45,46). Other studies suggested that
HIF-1α binding with hypoxia response elements promotes SOX9
transcription and thus maintains chondrocyte phenotype (24,46). In the present study, HIF-1α bound
to YAP in the nucleus to form a complex, which promoted expression
of glycolysis-associated enzymes, enhanced glycolysis metabolism
and provided energy for chondrocyte proliferation and
differentiation under hypoxia. However, the effect of HIF-1α on
phenotype maintenance and chondrogenic ability of chondrocytes
remains to be further studied.
SETD7 methylates the lysine 494 residue of YAP,
retaining it in the cytoplasm (26). HIF-1α contains the R-S-K amino
acid sequence, which is also recognized by SETD7 (47). A recent study showed that YAP
forms a complex with HIF-1α in hepatocellular carcinoma cells and
binds to the promoter of the PKM2 gene to promote its transcription
(27). Therefore, a regulatory
network between SETD7, YAP and HIF-1α in chondrocytes may be formed
to regulate chondrocyte differentiation.
To investigate the association between SETD7, YAP
and HIF-1α in chondrocytes, SETD7 was silenced. The expression of
p-YAP was significantly decreased after silencing SETD7, while
total YAP was not increased; protein levels of LATS1 and p-LATS1,
upstream regulators of YAP in the Hippo pathway, were decreased.
These data implied that Hippo signaling may be inhibited by
silencing SETD7. As YAP nuclear localization is a key regulatory
mechanism for activating YAP (48), immunofluorescence staining was
performed to examine cellular localization of YAP. Silencing SETD7
triggered notable YAP nuclear translocation in ATDC5 cells.
Consistent with these data, cell fractionation showed more YAP
protein accumulation in the nuclear fraction and less YAP protein
in the cytoplasmic fraction of cells following SETD7 silencing. The
results suggest that silencing SETD7 activated YAP and induced YAP
translocation to the nucleus by inhibiting the Hippo pathway in
ATDC5 cells. However, the increase of nuclear YAP was not
significant, but the decrease in cytoplasmic YAP was significant
after silencing SETD7 in ATDC5 cells. It has been reported that
SETD7 binds to YAP in the cytoplasm to increase its stability
(26). However, cytoplasmic YAP
is degraded by hydroxylation after silencing SETD7. The reason why
the increase of YAP in the nucleus was less than the decrease in
the cytoplasm be associated with degradation of YAP after silencing
SETD7.
There was no difference in HIF-1α transcription
between ATDC5 cells in the SETD7-shRNA and control group, but
HIF-1α protein levels increased, indicating that HIF-1α protein
became more stable after silencing SETD7. This may be due to the
ability of SETD7 to degrade HIF-1α, or because YAP entry to the
nucleus makes HIF-1α more stable. The results of IF showed that
both YAP and HIF-1α were located in the nucleus after silencing
SETD7. The results of IP showed that YAP and HIF-1α bound to each
other. CHX assay showed that HIF-1α was be stabilized by YAP. Thus,
the increased HIF-1α protein level after silencing SETD7 may be
caused by the decreased SETD7, which degrades HIF-1α and
facilitates entry of YAP into the nucleus to bind to HIF-1α and
maintain its stability.
In previous studies, SETD7 was shown to be highly
expressed in chondrocytes at 20 compared with 1% O2
(13,49). The present study demonstrated
that at 1% O2, decreased SETD7 expression leads to
inhibition of Hippo signaling and an increase in YAP entering the
nucleus. YAP promotes the expression of genes associated with the
differentiation of chondrocytes and maintains chondrocyte phenotype
after entering the nucleus. It also binds and stabilizes HIF-1α to
promote glycolysis, which provides energy for chondrocyte
differentiation (Fig. 6).
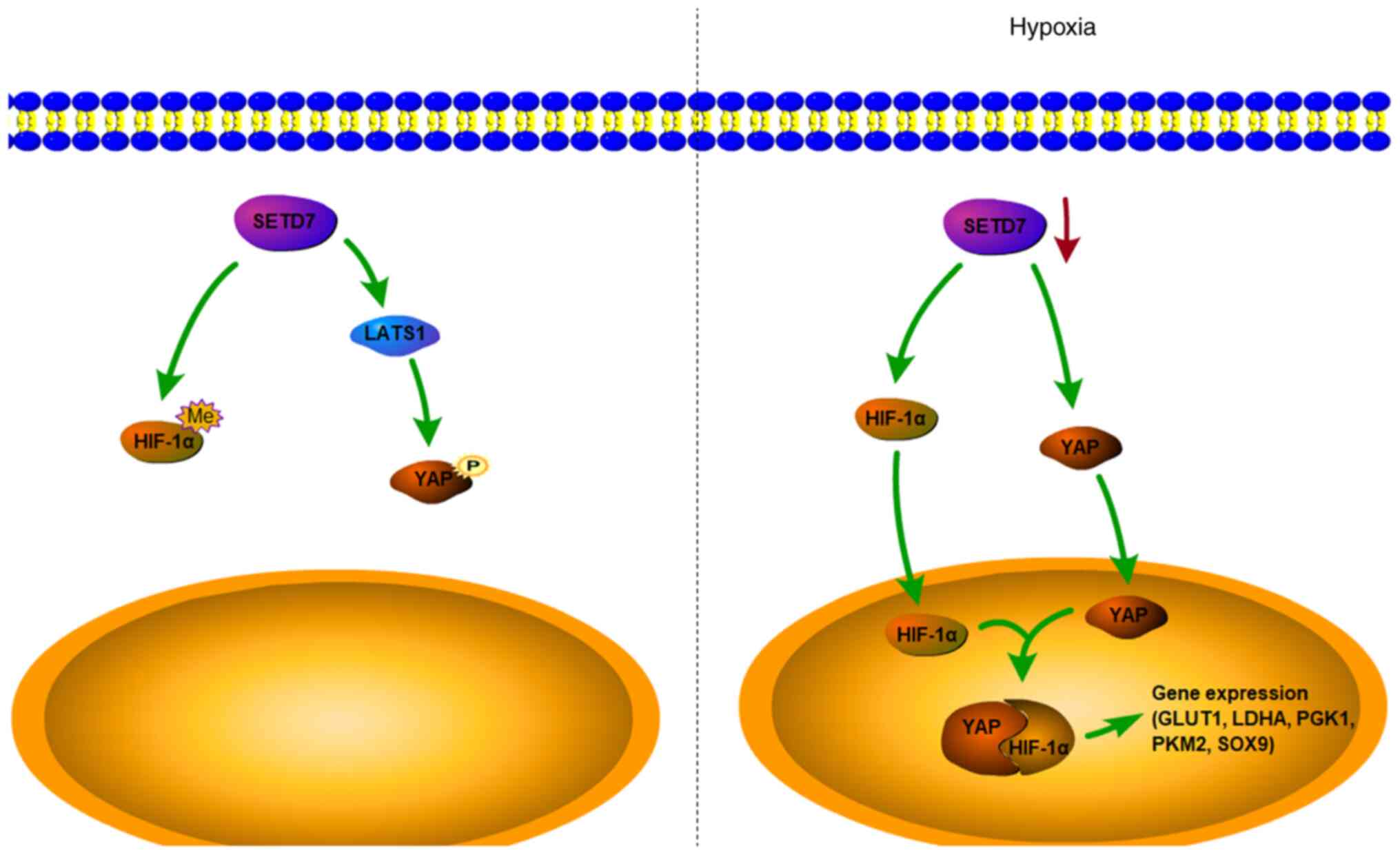 | Figure 6SETD7 regulates chondrocyte
differentiation and glycolysis via the Hippo and HIF-1α. In normal
conditions, SETD7 activates the Hippo signaling pathway, which
phosphorylates YAP and retains it in the cytoplasm. In hypoxic
conditions, expression of SETD7 is inhibited, resulting in
increased YAP and HIF-1α in the cytoplasm. The accumulated YAP and
HIF-1α translocate into the nucleus and combine to form a complex,
which further promotes expression of glycolysis-associated genes
and chondrogenic differentiation. SETD7, SET domain containing 7;
YAP, Yes-associated protein; HIF-1α, hypoxia inducible factor-1α;
GLUT1, glucose transporter 1; LDHA, lactate dehydrogenase A; PGK1,
phosphoglycerate kinase 1; PKM2, pyruvate kinase isoenzyme 2;
LATS1, large tumor suppressor 1; SOX9, SRY-related box gene 9; Me,
methylation; P, phosphorylation. |
The present study demonstrated that hypoxia was
conductive to the differentiation of chondrocytes. SETD7 inhibited
glycolysis and differentiation of chondrocytes via the Hippo
signaling pathway and HIF-1α. These findings may shed new light on
cartilage tissue engineering and provide novel therapeutic targets
of chondrogenic disease.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ML performed the experiments, collected and
visualized data and wrote the manuscript. JN and JW performed the
experiments and collected the data. QY performed the experiments,
validated the data and wrote the manuscript. KZ analyzed the data.
XJ conceptualized and supervised the study, provided resources,
performed the methodology and wrote, reviewed and edited the
manuscript. XJ and ML confirm the authenticity of all the raw data.
All authors have read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ma Q, Liao J and Cai X: Different sources
of stem cells and their application in cartilage tissue
engineering. Curr Stem Cell Res Ther. 13:568–575. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Huang K, Li Q, Li Y, Yao Z, Luo D, Rao P
and Xiao J: Cartilage tissue regeneration: The roles of cells,
stimulating factors and scaffolds. Curr Stem Cell Res Ther.
13:547–567. 2018. View Article : Google Scholar
|
|
3
|
Shi W, Sun M, Hu X, Ren B, Cheng J, Li C,
Duan X, Fu X, Zhang J, Chen H and Ao Y: Structurally and
functionally optimized silk-fibroin-gelatin scaffold using 3D
printing to repair cartilage injury in vitro and in vivo. Adv
Mater. 29:2017. View Article : Google Scholar
|
|
4
|
Park IS, Jin RL, Oh HJ, Truong MD, Choi
BH, Park SH, Park DY and Min BH: Sizable scaffold-free
tissue-engineered articular cartilage construct for cartilage
defect repair. Artif Organs. 43:278–287. 2019. View Article : Google Scholar
|
|
5
|
Ma B, Leijten JC, Wu L, Kip M, van
Blitterswijk CA, Post JN and Karperien M: Gene expression profiling
of dedifferentiated human articular chondrocytes in monolayer
culture. Osteoarthritis Cartilage. 21:599–603. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Greuel S, Hanci G, Böhme M, Miki T,
Schubert F, Sittinger M, Mandenius CF, Zeilinger K and Freyer N:
Effect of inoculum density on human-induced pluripotent stem cell
expansion in 3D bioreactors. Cell Prolif. 52:e126042019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tarahomi M, Vaz FM, Van straalen JP,
Schrauwen FAP, van Wely M, Hamer G, Repping S and Mastenbroek S:
The composition of human preimplantation embryo culture media and
their stability during storage and culture. Hum Reprod.
34:1450–1461. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Jin GZ and Kim HW: Chondrogenic potential
of dedifferentiated rat chondrocytes reevaluated in two- and
three-dimensional culture conditions. Tissue Eng Regen Med.
15:163–172. 2017. View Article : Google Scholar
|
|
9
|
Oppenheimer H, Kumar A, Meir H, Schwartz
I, Zini A, Haze A, Kandel L, Mattan Y, Liebergall M and
Dvir-Ginzberg M: Set7/9 impacts COL2A1 expression through binding
and repression of SirT1 histone deacetylation. J Bone Miner Res.
29:348–360. 2014. View Article : Google Scholar
|
|
10
|
Archer CW and Francis-West P: The
chondrocyte. Int J Biochem Cell Biol. 35:401–404. 2003. View Article : Google Scholar
|
|
11
|
Huang X, Zhong L, Hendriks J, Post JN and
Karperien M: Different response of human chondrocytes from healthy
looking areas and damaged regions to IL1β stimulation under
different oxygen tension. J Orthop Res. 37:84–93. 2019. View Article : Google Scholar
|
|
12
|
Yahiro K, HigashihoriI N and Moriyama K:
Histone methyltransferase Setdb1 is indispensable for Meckel's
cartilage development. Biochem Biophys Res Commun. 482:883–888.
2017. View Article : Google Scholar
|
|
13
|
Xiaoshi J, Maoquan L, Jjiwei W, Jinqiu N
and Ke Z: SETD7 mediates the vascular invasion in articular
cartilage and chondrocytes apoptosis in osteoarthriis. FASEB J.
35:e212832021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Herz HM, Garruss A and Shilatifard A: SET
for life: Biochemical activities and biological functions of SET
domain-containing proteins. Trends Biochem Sci. 38:621–639. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Duan B, Bai J, Qiu J, Wang J, Tong C, Wang
X, Miao J, Li Z, Li W, Yang J and Huang C: Histone-lysine
N-methyltransferase SETD7 is a potential serum biomarker for
colorectal cancer patients. EBioMedicine. 37:134–143. 2018.
View Article : Google Scholar :
|
|
16
|
Misra JR and Irvine KD: The Hippo
signaling network and its biological functions. Annu Rev Genet.
52:65–87. 2018. View Article : Google Scholar
|
|
17
|
Azad T, Nouri K, Janse van Rensburg HJ,
Maritan SM, Wu L, Hao Y, Montminy T, Yu J, Khanal P, Mulligan LM
and Yang X: A gain-of-functional screen identifies the Hippo
pathway as a central mediator of receptor tyrosine kinases during
tumorigenesis. Oncogene. 39:334–355. 2020. View Article : Google Scholar
|
|
18
|
Zhong W, Jiang H, Zou Y, Ren J, Li Z, He
K, Zhao J, Zhou X, Mou D and Cai Y: The YAP signaling pathway
promotes the progression of lymphatic malformations through the
activation of lymphatic endothelial cells. Pediatr Res. 89:110–117.
2021. View Article : Google Scholar
|
|
19
|
Kovar H, Bierbaumer L and Radic-Sarikas B:
The YAP/TAZ pathway in osteogenesis and bone sarcoma pathogenesis.
Cells. 9:9722020. View Article : Google Scholar :
|
|
20
|
Li H, Li X, Jing X, Li M, Ren Y, Chen J,
Yang C, Wu H and Guo F: Hypoxia promotes maintenance of the
chondrogenic phenotype in rat growth plate chondrocytes through the
HIF-1α/YAP signaling pathway. Int J Mol Med. 42:3181–3192.
2018.
|
|
21
|
Goto H, Nishio M, To Y, Oishi T, Miyachi
Y, Maehama T, Nishina H, Akiyama H, Mak TW, Makii Y, et al: Loss of
Mob1a/b in mice results in chondrodysplasia due to
YAP1/TAZ-TEAD-dependent repression of SOX9. Development.
145:dev1592442018. View Article : Google Scholar
|
|
22
|
Hu S, Zhang C, Ni L, Huang C, Chen D, Shi
K, Jin H, Zhang K, Li Y, Xie L, et al: Stabilization of HIF-1α
alleviates osteoarthritis via enhancing mitophagy. Cell Death Dis.
11:4812020. View Article : Google Scholar
|
|
23
|
Schipani E, Ryan HE, Didrickson S,
Kobayashi T, Knight M and Johnson RS: Hypoxia in cartilage:
HIF-1alpha is essential for chondrocyte growth arrest and survival.
Genes Dev. 15:2865–2876. 2001.PubMed/NCBI
|
|
24
|
Stegen S, Laperre K, Eelen G, Rinaldi G,
Fraisl P, Torrekens S, Van Looveren R, Loopmans S, Bultynck G,
Vinckier S, et al: HIF-1α metabolically controls collagen synthesis
and modification in chondrocytes. Nature. 565:511–515. 2019.
View Article : Google Scholar
|
|
25
|
Liu X, Chen Z, Xu C, Leng X, Cao H, Ouyang
G and Xiao W: Repression of hypoxia-inducible factor α signaling by
Set7-mediated methylation. Nucleic Acids Res. 43:5081–5098. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oudhoff MJ, Freeman SA, Couzens AL,
Antignano F, Kuznetsova E, Min PH, Northrop JP, Lehnertz B,
Barsyte-Lovejoy D, Vedadi M, et al: Control of the hippo pathway by
Set7-dependent methylation of Yap. Dev Cell. 26:188–194. 2013.
View Article : Google Scholar
|
|
27
|
Zhang X, Li Y, Ma Y, Yang L, Wang T, Meng
X, Zong Z, Sun X, Hua X and Li H: Yes-associated protein (YAP)
binds to HIF-1α and sustains HIF-1α protein stability to promote
hepatocellular carcinoma cell glycolysis under hypoxic stress. J
Exp Clin Cancer Res. 37:2162018. View Article : Google Scholar
|
|
28
|
Chen P, Vukicevic S, Sampath TK and Luyten
FP: Bovine articular chondrocytes do not undergo hypertrophy when
cultured in the presence of serum and osteogenic protein-1. Biochem
Biophys Res Commun. 197:1253–1259. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Dasari VR, Mazack V, Feng W, Nash J, Carey
DJ and Gogoi R: Verteporfin exhibits YAP-independent
anti-proliferative and cytotoxic effects in endometrial cancer
cells. Oncotarget. 8:28628–28640. 2017. View Article : Google Scholar :
|
|
30
|
Niu Y, Shi D, Li L, Guo J, Liu H and Yao
X: Revealing inhibition difference between PFI-2 enantiomers
against SETD7 by molecular dynamics simulations, binding free
energy calculations and unbinding pathway analysis. Sci Rep.
7:465472017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Armiento AR, Stoddart MJ, Alini M and
Eglin D: Biomaterials for articular cartilage tissue engineering:
Learning from biology. Acta Biomater. 65:1–20. 2018. View Article : Google Scholar
|
|
32
|
Lafont JE: Lack of oxygen in articular
cartilage: Consequences for chondrocyte biology. Int J Exp Pathol.
91:99–106. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xue K, Zhang X, Gao Z, Xia W, Qi L and Liu
K: Cartilage progenitor cells combined with PHBV in cartilage
tissue engineering. J Transl Med. 17:1042019. View Article : Google Scholar :
|
|
34
|
Wang H, Cao R, Xia L, Erdjument-Bromage H,
Borchers C, Tempst P and Zhang Y: Purification and functional
characterization of a histone H3-lysine 4-specific
methyltransferase. Mol Cell. 8:1207–1217. 2001. View Article : Google Scholar
|
|
35
|
Oudhoff MJ, Braam MJS, Freeman SA, Wong D,
Rattray DG, Wang J, Antignano F, Snyder K, Refaeli I, Hughes MR, et
al: SETD7 controls intestinal regeneration and tumorigenesis by
regulating Wnt/β-catenin and Hippo/YAP signaling. Dev Cell.
37:47–57. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gu Y, Wang X, Liu H, Li G, Yu W and Ma Q:
SET7/9 promotes hepatocellular carcinoma progression through
regulation of E2F1. Oncol Rep. 40:1863–1874. 2018.
|
|
37
|
Batista IAA and Helguero LA: Biological
processes and signal transduction pathways regulated by the protein
methyltransferase SETD7 and their significance in cancer. Signal
Transduct Target Ther. 3:192018. View Article : Google Scholar :
|
|
38
|
Pepe-Mooney BJ, Dill MT, Alemany A,
Ordovas-Montanes J, Matsushita Y, Rao A, Sen A, Miyazaki M, Anakk
S, Dawson PA, et al: Single-cell analysis of the liver epithelium
reveals dynamic heterogeneity and an essential role for YAP in
homeostasis and regeneration. Cell Stem Cell. 25:23–38.e8. 2019.
View Article : Google Scholar :
|
|
39
|
Chen J, You H, Li Y, Xu Y, He Q and Harris
RC: EGF receptor-dependent YAP activation is important for renal
recovery from AKI. J Am Soc Nephrol. 29:2372–2385. 2018. View Article : Google Scholar :
|
|
40
|
Ma S, Meng Z, Chen R and Guan KL: The
Hippo pathway: Biology and pathophysiology. Annu Rev Biochem.
88:577–604. 2019. View Article : Google Scholar
|
|
41
|
Oudhoff MJ, Antignano F, Chenery AL,
Burrows K, Redpath SA, Braam MJ, Perona-Wright G and Zaph C:
Intestinal epithelial cell-intrinsic deletion of Setd7 identifies
role for developmental pathways in immunity to helminth infection.
PLoS Pathog. 12:e10058762016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Semenza GL: Targeting HIF-1 for cancer
therapy. Nat Rev Cancer. 3:721–732. 2003. View Article : Google Scholar
|
|
43
|
Lee JW, Bae SH, Jeong JW, Kim SH and Kim
KW: Hypoxia-inducible factor (HIF-1)alpha: Its protein stability
and biological functions. Exp Mol Med. 36:1–12. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ke Q and Costa M: Hypoxia-inducible
factor-1 (HIF-1). Mol Pharmacol. 70:1469–1480. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xu G: HIF-1-mediated expression of Foxo1
serves an important role in the proliferation and apoptosis of
osteoblasts derived from children's iliac cancellous bone. Mol Med
Rep. 17:6621–6631. 2018.
|
|
46
|
Zhang C, Yang F, Cornelia R, Tang W,
Swisher S and Kim H: Hypoxia-inducible factor-1 is a positive
regulator of Sox9 activity in femoral head osteonecrosis. Bone.
48:507–513. 2011. View Article : Google Scholar
|
|
47
|
Wu Y, Zou F, Lu Y, Li X, Li F, Feng X, Sun
X and Liu Y: SETD7 promotes TNF-α-induced proliferation and
migration of airway smooth muscle cells in vitro through enhancing
NF-κB/CD38 signaling. Int Immunopharmacol. 72:459–466. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Moya IM and Halder G: Hippo-YAP/TAZ
signalling in organ regeneration and regenerative medicine. Nat Rev
Mol Cell Biol. 20:211–226. 2019. View Article : Google Scholar
|
|
49
|
Soshnikova N: Functions of SETD7 during
development, homeostasis and cancer. Stem Cell Investig. 6:262019.
View Article : Google Scholar : PubMed/NCBI
|