Introduction
Preterm birth affects >15 million infants
worldwide (1). These infants are
at a significantly increased risk of developing acute adverse
health outcomes, such as life-long metabolic (2) and immune health complications
(3). Breast milk confers
significant benefits to infant health, including a decreased risk
of sudden infant death syndrome, sepsis and necrotizing
enterocolitis (4). It has also
been associated with improved neurodevelopmental outcomes and
protection against metabolic diseases, including obesity and
diabetes mellitus type 1 and 2 during the later stages of life
(5,6). The latter is further illustrated by
recent studies demonstrating the modulatory effects of
breastfeeding on the expression of genes, such as the fat mass and
obesity-associated (FTO) gene, which interacts with
mRNAs/long non-coding RNAs (lncRNAs) to influence body mass index
and fat tissue growth (7-9).
Human milk composition is unique; however, it is influenced by
maternal factors, including stress, nutrition, presence of
inflammation and preterm birth (10,11). Apart from nutrients, human breast
milk contains a heterogeneous array of non-nutritive components,
including enzymes, hormones, growth factors, immune cells, stem
cells, bacteria, antibodies, cytokines, antimicrobial peptides and
extracellular vesicles (EVs), that may play significant roles in
the development of infants (12,13).
EVs comprise a vast spectrum of microparticles and
based on the corresponding size range and biogenesis, can be
broadly classified into three distinct classes: Microvesicles,
exosomes and apoptotic bodies (14). Microvesicles have a diameter
ranging from 50 nm to 1 µm and are released by cell membrane
budding. Exosomes are particles with a diameter ranging from 30-150
nm and are derived via a targeted mechanism from the cell endocytic
compartment through the formation of multivesicular bodies (MVBs)
(15). Exosomes are small
lipid-bound vesicles released from all cells into the extracellular
space or biological fluids (16). They are highly heterogeneous as
regards their size, content and function to recipient cells and
cells of origin, and they can induce a repertoire of biological
responses. Exosomes are considered as an integral part of the
senescence-associated secretory phenotype (17) and depending on their cargo or
surface composition, they may function as specific
signals/mediators of systemic stress between cells or between
tissues (14). Breast milk is a
biofluid enriched in exosomes. Previous studies have indicated the
protective role of human milk-derived exosomes, focusing on their
role against necrotizing enterocolitis in preterm infants (9,18). Furthermore, exosomes in breast
milk may be central epigenetic regulators as regards the expression
of developmental genes, such as FTO, insulin (INS)
and insulin like growth factor 1 (IGF1) (9).
A number of studies have suggested that exosomes
harbor a variety of active or non-autonomous biomolecules,
including proteins, lipids, DNA, mRNAs, microRNAs (miRNAs/miRs) and
lncRNAs. Thus, owing to their unique potential to group multiple
signals together, exosomes constitute an alternative and largely
unexplored mode of communication between neighboring and distant
cells, differing from the conventional hormone-mediated mechanisms
(14,15,19,20). Likewise, regulatory non-coding
RNAs (ncRNAs) have already been established as an appealing new
source of novel 'genetic hormones' and biomarkers, with an
increased sensitivity and specificity for an unprecedented range of
diseases, conditions or cell states (20). There is a growing body of
literature that recognizes the crucial roles of lncRNAs in gene
regulation as they can interact with proteins, DNA and RNA, and
modulate mRNA expression, chromatin function and signaling pathways
(21). Of note, the RNA cargo in
exosomes does not simply reflect the tissue and cell state, nor
does it represent the RNA composition of the cell of origin, but
rather a selective sorting and loading of specific RNAs into EVs.
lncRNAs are at the forefront of both basic biological and clinical
research due to their immense predictive value as novel biomarkers
in precision medicine, as well as their significant therapeutic
potential, given that lncRNAs are considered easier to be targeted
for disease prevention and therapy, compared to protein-coding
genes. Previous studies have revealed the presence of miRNAs
(22) and lncRNAs in EVs in
breast milk (12,23). Karlsson et al (12) detected exosomal lncRNAs in breast
milk, including colorectal neoplasia differentially expressed
(CRNDE) gene, differentiation antagonizing non-protein
coding RNA (DANCR), growth arrest-specific 5 (GAS5),
steroid receptor RNA activator 1 (SRA1) and ZNFX1 antisense
RNA 1 (ZFAS1), that may represent epigenetic regulators
involved in child development.
The present study demonstrates that exosomes
circulating in human breast milk consistently carry two sets of
lncRNAs with well-known functions in the inflammatory response and
auto-immunity that exhibit opposite loading patterns corresponding
to two potential sets of epigenetic exosomal cargo in human breast
milk. The specificity and reproducibility of the present study is
ensured by the simultaneous co-detection of 'reference lncRNAs'
previously identified in the study by Karlsson et al
(12) in breast milk-derived
exosomes (e.g., GAS5). A novel lncRNA, LINC00657 [non-coding RNA
activated at DNA damage (NORAD)] was consistently
co-detected with GAS5, which exhibited the highest detection signal
among exosomes in all breast milk samples analyzed. Furthermore,
the present study represents the first differential expression
analysis of breast milk exosomal lncRNAs in breast milk of mothers
who gave birth preterm vs. term. The results presented herein
indicate that the loading levels of NORAD in breast milk-derived
exosomes are suppressed at least 2-fold in mothers who gave birth
preterm compared with those who delivered at term, in a
statistically significant manner.
Materials and methods
Study participants
Participants (mothers) between 27-40 years of age
without major health issues (e.g., diabetes, toxemia, etc.) and any
medication treatment (apart from intake of vitamins, calcium and
other supplements) were recruited at the Neonatal Unit of the First
Department of Pediatrics of National and Kapodistrian University of
Athens at 'Aghia Sophia' Children's Hospital (Athens, Greece)
during a 6-month period (January, 2020 to June, 2020). Overall, 20
breast milk samples were collected, 10 from mothers who delivered
preterm (gestational age, <37 weeks) and 10 from mothers who
delivered at term (gestational age, ≥37 weeks). All participants
provided written informed consent prior to enrolment, and the study
was approved by the Ethics Committee of 'Aghia Sophia' Children's
Hospital (protocol code, 24814; date of approval, October 30,
2017).
Breast milk sample collection
Breast milk samples (20-40 ml) were collected using
a manual pump at the end of the first month postpartum. Samples
were kept at 4°C until their transport to the laboratory on the
same day. Upon arrival, the samples were centrifuged at 1,500 × g
for 15 min at 4°C and the skim milk fraction was collected through
a disposable needle syringe of 5 ml. The skim milk sample was
centrifuged again at 3,000 × g for 30 min, at 4°C to remove the
remaining fat globules and cell debris, and stored in a 1 ml
aliquot at −80°C until further analysis.
Isolation of EVs
Breast milk aliquots were thawed on ice and
centrifuged at 3,000 × g for 15 min at 4°C. The fat layer was
discarded using a vacuum and the supernatant was carefully
aspirated, mixed with 2 volumes of 1X PBS and filtered using a
0.8-µm membrane unit (EMD Millipore) to remove large
aggregates and any residual cell debris. To isolate intact exosomes
for western blot analysis, the exoRNeasy Serum/Plasma MaxiKit
(Qiagen, Inc.) was used according to the provided protocol. Intact
exosomes were extracted from 2 ml of pre-filtered skim milk and
were eluted in 140 µl of 2X elution buffer (Qiagen,
Inc.).
Western blot analysis
Exosome samples were first lysed using RIPA buffer
[cat. no. 89900, Thermo Fisher Scientific, Inc.; 1.0% (v/v) NP-40
or Triton X-100, 0.5% (w/v) sodium deoxycholate, 0.1% (w/v) SDS, 50
mM Tris, pH 8.0] and the protein content was quantified using BCA
Protein assay (Thermo Fisher Scientific, Inc.). A total of 30
µg of each sample were electrophoretically separated on 10%
(w/v) SDS-gel and transferred onto a nitrocellulose membrane (0.2
nm). The membrane was blocked using 5% non-fat milk diluted in 1X
TBST (20 mM Tris-base, 150 mM NaCl, 0,1% w/v Tween-20), for 1 h at
room temperature. To visualize protein markers, the membrane was
incubated with mouse anti-CD63 monoclonal IgG1 antibody (cat. no.
sc-5275), mouse anti-CD9 monoclonal IgG1 antibody (cat. no.
sc-13118) and mouse anti-cytochrome b5 type b monoclonal IgG1
antibody (cat. no. sc-390876) (all from Santa Cruz Biotechnology,
Inc.). All antibodies were diluted 1/500 in 5% non-fat milk. The
samples were incubated for 2 h at room temperature with anti-mouse
secondary IgG Fc antibody HRP-conjugated (cat. no. sc-525409; Santa
Cruz Biotechnology, Inc.) diluted 1/2,000 in 5% non-fat milk.
Visualization was performed using Immobilon Forte Western HRP
Substrate (cat. no. WBLUF0020; Merck Millipore).
Electron microscopy
An aliquot of 3 µl from each sample was added
to a grid with a carbon supporting film for 5 min. The excess
solution was soaked off using a filter paper, the grid was rinsed
by the addition of 5 µl distilled water for 10 sec, soaked
off and stained with 1% uranyl acetate in water for 10 sec and then
air-dried. The samples were examined in a Morgagni 268 transmission
electron microscope (FEI Company) at 60 kV. Digital images were
obtained using a Veleta camera (Olympus Soft Imaging Solutions
GmbH).
Reverse transcription-quantitative PCR
(RT-qPCR)
For the extraction of the total RNA content
encapsulated in exosomes, the exoRNeasy Serum/Plasma MaxiKit (cat
no. 77164; Qiagen, Inc.) was used according to the manufacturer's
protocol. RNA was extracted from 4 ml of pre-filtered skim milk and
was eluted in 14 µl of RNAse-free water. The quality of the
isolated total RNA content was assessed using a NanoDrop (ND-1000)
spectrophotometer and its concentration was determined using the
Qubit RNA assay kit and the Qubit 3.0 fluorometer (Thermo Fisher
Scientific, Inc.). The expression of exosomal lncRNAs was
determined using a custom 96-well RT2 PCR Array (Qiagen,
cat. no. CLAH00049). The array was based on the Human Inflammatory
Response and Autoimmunity RT2 lncRNA PCR array (cat. no.
LAHS-004Z; Qiagen, Inc.) consisting of 84 lncRNAs
(RT2lncRNA PreAMP primer mix; Qiagen GmbH; contains
pre-dispensed, laboratory verified, gene-specific primer pairs for
84 genes) involved in autoimmune and inflammatory immune responses,
as well as three lncRNAs that were reported previously by Karlsson
et al (12) to be
consistently detected in human breast milk-derived EVs (SRA1, CRNDE
and DANCR). This panel also included a set of controls to evaluate
genomic DNA contamination, as well as PCR and reverse transcription
performance. cDNA synthesis was performed using the RT2 First
Strand kit (ID:330401, custom cat. no. CLAH00049-LAHS-004Z; Qiagen,
Inc.) according to the manufacturer's protocol that includes a DNA
elimination step prior the reverse transcription. For the cDNA
synthesis, 1 µg of RNA was diluted in RNase-free
H2O to a final volume of 8 µl (without a
pre-amplification step) was used as an input. The synthesized cDNA
was then mixed with RT2 SYBR-Green MasterMix and RNase-free water
to yield the PCR component, which was distributed in 25 µl
aliquots to each well of the lncRNA PCR plate. qPCR was performed
on a Light cycler 480 II PCR machine (96-well; Roche Diagnostics)
and the PCR cycling conditions consisted of an initial step of 10
min at 95°C for HotStart Activation, followed by 40 cycles of 15
sec at 95°C and 1 min at 60°C. A Cq value <40 was considered as
the limit for the detection of expression. Normalized expression
levels were calculated using inverse Min-max feature scaling,
1-[NorCqtarget
lncRNA-MIN(NorCq)]/[MAX(NorCq)-MIN(NorCq)], where
NorCq=Cqtarget lncRNA/CqReference, while
CqReference=(CqRPLP0 + CqACTB)/2.
To investigate the differences between the expression of lncRNAs in
the breast milk of mothers who delivered infants at term and those
who delivered preterm, raw data were analyzed and the relative
expression level of target lncRNAs was determined using the
2−ΔCq method, where ΔCq=Cqtarget
lncRNA-CqReference (24). Ribosomal protein lateral stalk
subunit P0 (RPLPO) and actin beta (ACTB) were
selected as reference genes to minimize the standard deviation (SD)
of expression. lncRNAs were considered differentially expressed if
the fold change was >2 or <0.5.
Statistical analysis
P-values were calculated between two groups (10
preterm and 10 full-term breastmilk samples) using the t-test in
programming language R (package 'stats', version 4.2.0) and the
statistical significance threshold was set to 0.05. The correlation
of expression was estimated using Pearson's coefficient which
reflects the linear correlation between two variables accounting
for differences in their mean and SD.
Results
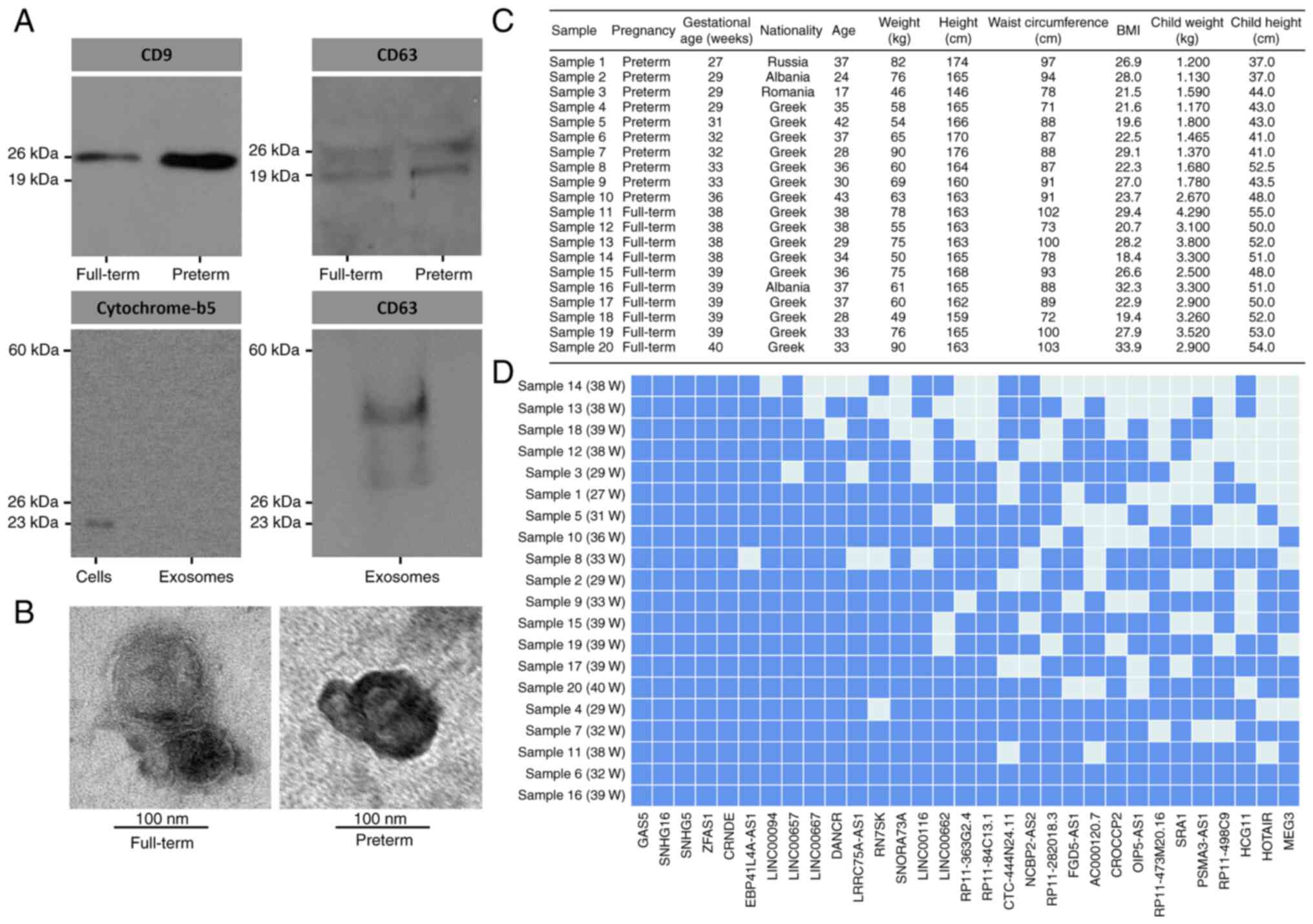
Isolated exosomes from human breastmilk samples from
mothers who gave birth at term or preterm were labeled as positive
for two common exosomal tetraspanins, CD9 and CD63, using western
blot analysis (Fig. 1A). To
examine exosome purity, western blot analysis was performed for
cytochrome-b5 type b, a common cell marker of the outer
mitochondrial membrane. The results revealed that the cells were
positive for cytochrome-b5 type b, while the exosomes exhibited no
signal, thus suggesting no evidence of cell contamination. To
further characterize the composition of the exosomes in the present
study, western blot analysis was performed using an antibody
against CD63 as an additional positive tetraspanin marker for
exosomes. Again, the exosome lysates were positive for CD63. Of
note, the CD9 protein levels in the exosomes derived from the
breastmilk of mothers who delivered preterm were markedly higher
compared with those in the breastmilk from mothers who delivered at
term, even though equal protein quantities of whole exosome lysates
were loaded to the gel. Subsequently, exosomal RNA extraction from
all samples was performed. To address the issue regarding the size
and morphology of exosomes derived from breastmilk from mothers who
delivered at term and preterm, electron micros-copy was performed.
The results indicated that the exosomes isolated from breastmilk
exhibited the classical 'doughnut' morphology due to the
surrounding lipid bilayer and their size was between 50-100 nm
(Fig. 1B). The demographic and
anthropometric characteristics of the mothers, as well their
children, are presented in Fig.
1C.
In order to study the lncRNA cargo of human
breastmilk-derived exosomes, 20 RNA samples were isolated from
exosomes of 10 breastmilk samples obtained from mothers who
delivered at term (≥37 weeks of gestation) and an equal number of
samples from mothers with preterm birth (<37 weeks of
gestation). Subsequently, RNA samples were analyzed using RT-qPCR.
The analysis was performed using a customized panel of targets
against lncRNAs with well documented roles in the inflammatory
response and auto-immunity, as well as selected lncRNAs that were
previously reported to be significantly detected in human
breastmilk-derived exosomes (12). Following RT-qPCR, 76 out of 88
lncRNAs were detected in at least one sample. Furthermore, 31
lncRNAs were detected as an exosomal load in ≥50% of the samples
(Fig. 1D). Importantly, 13
lncRNAs were consistently detected in the breastmilk-derived
exosomes in ≥85% of the samples (Table I). The biological functions of
these lncRNAs are presented in detail at Table SI.
 | Table IlncRNA expression levels detected in
>85% of the breastmilk samples from mothers who delivered
preterm and or at full-term. |
Table I
lncRNA expression levels detected in
>85% of the breastmilk samples from mothers who delivered
preterm and or at full-term.
| lncRNA ID | Mean Δcq preterm (±
SD) | Mean Δcq full-term
(± SD) | Sample no. | Log2
fold change | P-value |
|---|
| LINC00657 | 3.78 (±0.61) | 2.79 (±1.44) | 19/20 | −0.996 | 0.034 |
| LRRC75A-AS1 | 2.90 (±0.65) | 3.62 (±0.95) | 17/20 | 0.713 | 0.045 |
| CRNDE | 3.35 (±1.00) | 2.20 (±1.83) | 20/20 | −1.144 | 0.052 |
| SNHG16 | 4.55 (±0.60) | 4.13 (±0.78) | 20/20 | −0.413 | 0.102 |
| ZFAS1 | 1.78 (±0.76) | 1.24 (±1.09) | 20/20 | −0.541 | 0.108 |
| SNHG5 | 1.49 (±0.57) | 0.86 (±1.79) | 20/20 | −0.629 | 0.156 |
| SNORA73A | 6.73 (±1.88) | 6.12 (±1.77) | 17/20 | −0.617 | 0.251 |
| DANCR | 5.51 (±1.17) | 5.77 (±0.85) | 18/20 | 0.256 | 0.299 |
| LINC00667 | 5.84 (±1.55) | 6.06 (±1.30) | 18/20 | 0.221 | 0.373 |
| GAS5 | 1.64 (±1.92) | 1.39 (±1.55) | 20/20 | −0.251 | 0.376 |
| EBP41L4A-AS1 | 5.61 (±0.99) | 5.44 (±1.63) | 19/20 | −0.169 | 0.393 |
| LINC00094 | 6.17 (±1.42) | 6.25 (±0.94) | 19/20 | 0.080 | 0.443 |
| RN7SK | 4.89 (±1.24) | 4.84 (±1.12) | 17/20 | −0.047 | 0.468 |
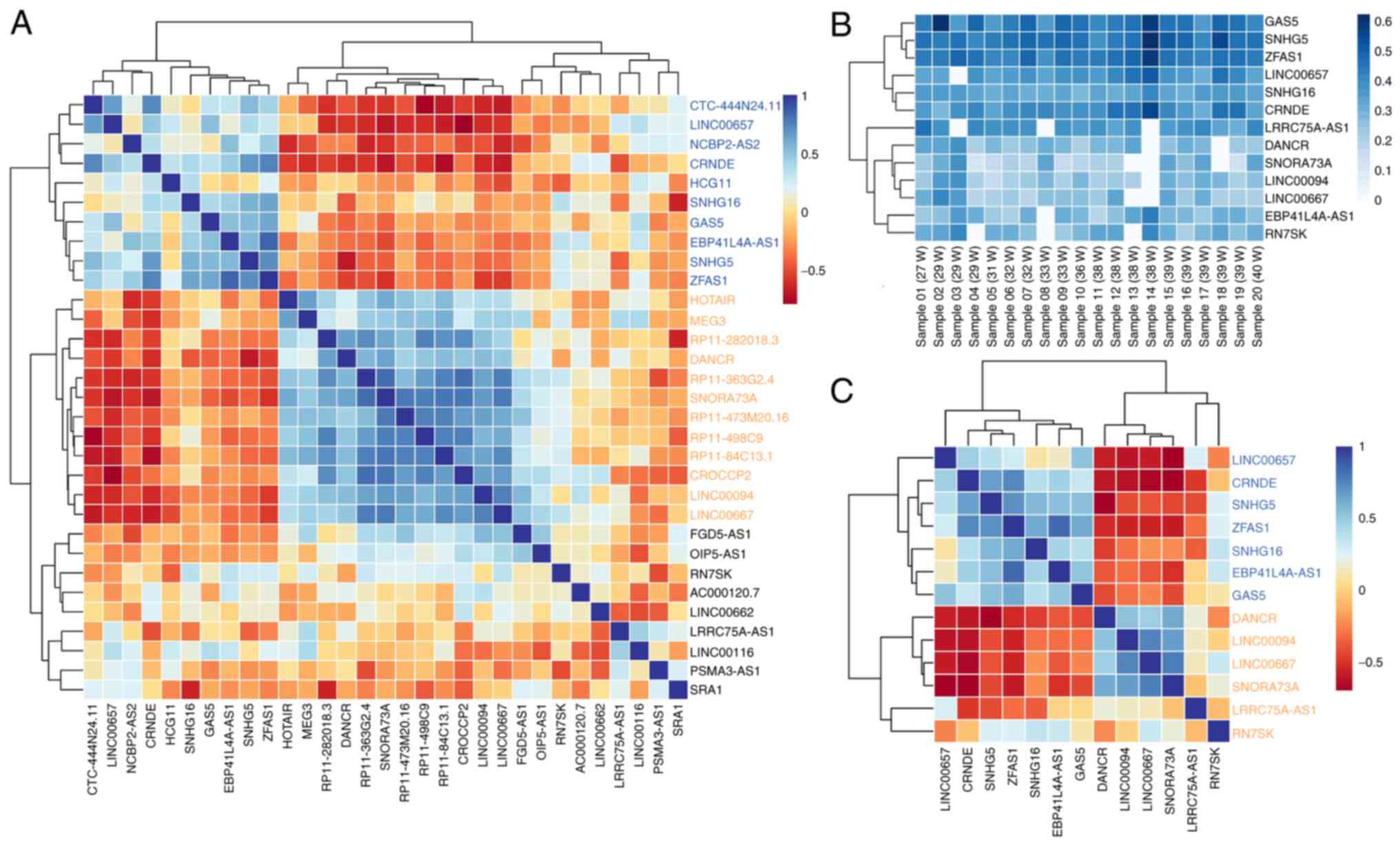
In the heatmap analysis of the normalized expression
levels of selected lncRNAs detected in ≥50% of the samples
(Fig. S1), GAS5 lncRNA
exhibited the highest degree of loading levels in the breastmilk
exosomes among all the lncRNAs analyzed. Following the correlation
heatmap analysis of the normalized expression data, two clusters of
lncRNAs were identified as differentially co-detected across
samples (Fig. 2A). The first
cluster (blue color) included 10 lncRNAs [CTC-444N24.11,
LINC00657, NCBP2-AS2, CRNDE, HCG11,
small nucleolar RNA host gene (SNHG16), GAS5,
EBP41L4A-AS1, SNHG5 and ZFAS1] exhibiting a
positive intergroup expression correlation and a negative
correlation with the majority of the remaining lncRNAs. In the
aforementioned group, 7 out of the 10 lncRNAs co-loaded in the
breastmilk exosomes were detected consistently across the samples
(≥85%). The second cluster (orange color) included a much larger
subset of lncRNAs (HOTAIR, MEG3,
RP11-282018.3, DANCR, RP11-363G2.4,
SNORA73A, RP11-473M20.16, RP11-498C9,
RP11-84C13.1, CROCCP2, LINC00094,
LINC00667, LRRC75A-AS1 and RN7SK); six of
these (DANCR, LINC00094, LINC00667,
SNORA73A, LRRC75A-AS1 and RN7SK) were
consistently detected in ≥85% of the samples.
This study then aimed to cross-validate these two
novel opposing correlation patterns by analyzing only the 13
lncRNAs with the highest frequency across samples (≥85% of samples)
(Fig. 2B). Correlation analysis
again revealed the existence of two distinct loading lncRNA
patterns in the breastmilk-derived exosomes with a strong inverted
correlation. More precisely, six lncRNAs (GAS5,
SNHG5, ZFAS1, LINC00657, SNHG16 and
CRNDE) formed a prominent group of abundantly co-detected
lncRNAs that exhibited strong negative correlation with a subset of
four different lncRNAs (DANCR, LINC00094,
LINC00667 and SNORA73A) that formed the core of the
second cluster of abundantly co-detected lncRNAs (Fig. 2C). The identification of two
differentially co-detected sub-sets of lncRNA cargo in
breastmilk-derived exosomes may indicate their co-regulation and
common loading in the same breastmilk-derived exosomes, in a
distinct and mutual exclusive manner across samples.
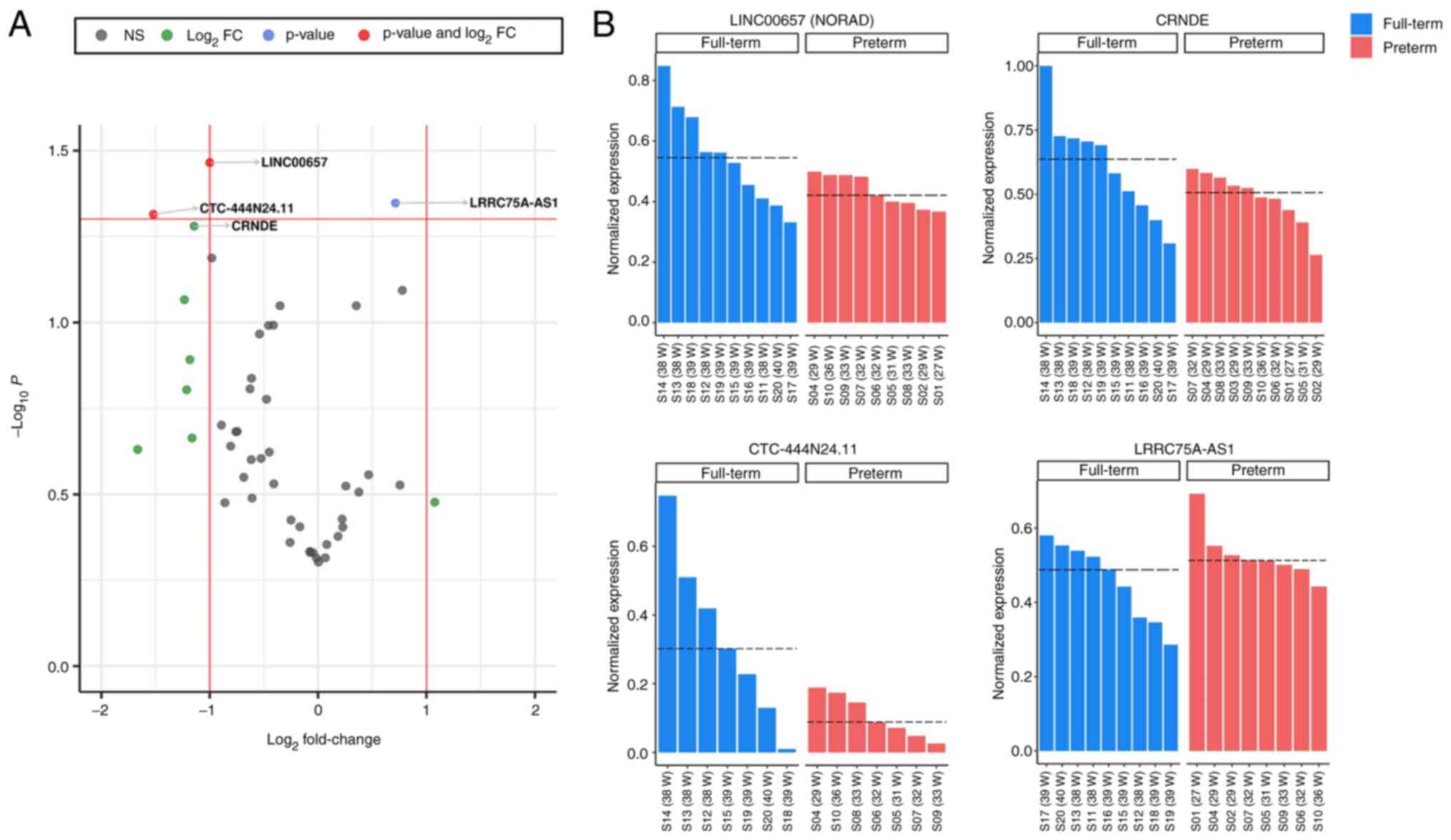
A differential expression analysis of the lncRNAs
loaded in the exosomes of breastmilk samples from mothers who
delivered preterm compared to those who delivered at term was
conducted. The results were obtained as a volcano plot of the log2
fold change of lncRNAs (Fig. 3A)
and corresponding values for statistical significance (P-value;
−log10P) (up- and downregulated lncRNAs are presented in
Tables SII and SIII,
respectively). In total, nine lncRNAs exhibited at least ≥1
log2 fold change (one upregulated and eight
down-regulated lncRNAs), which indicated that their levels were at
least 2-fold up- or downregulated in the breastmilk of mothers who
delivered prematurely compared to those who gave birth at term. In
addition, one lncRNA (LRRC37BP1) was upregulated at least 2-fold,
although with no statistically significant difference (t-test
P-value=0.333, log2 fold change=1.076). By contrast, out
of the eight downregulated lncRNAs with ≥1 log2 fold
change, two exhibited statistically significant differences (t-test
P-value ≤0.05). Thus, the levels of exosomal lncRNAs appeared to be
mostly downregulated in the breastmilk of mothers who delivered
preterm. LINC00657 (NORAD) (P-value=0.034,
log2 fold change=−1.00) was the only lncRNA in the
current panel that was consistently detected in ≥85% of the
samples, clustering well together with previously reported lncRNAs
in breastmilk-derived exosomes and exhibiting a statistically
significant difference (≥2-fold downregulation).
Likewise, lncRNA CRNDE was downregulated in
the exosomes of breastmilk-from mothers who delivered preterm,
although with a fractionally acceptable P-value of 0.052
(log2 fold change=−1,14), while CTC-444N24.11 was
downregulated with a log2 fold change of -1.521 and a
P-value of 0.048. However, CTC-444N24.11 was detected only
in 50% of the samples. By contrast, LRRC75A-AS1 was
upregulated with a statistically significant difference
(P-value=0.045), albeit it exhibited a <2-fold chance
(log2 fold change=0.713). More specifically, the
expression levels of the four lncRNAs which exhibited statistically
significant differences (LINC00657, CTC-444N24.11,
CRNDE and LRRC75A-AS1) across all samples, in both
the term and preterm group, are presented in bar plots in Fig. 3B. The lncRNAs LINC00657
(NORAD) and CRNDE were both found to be: i) Detected
in ≥85% of the samples; ii) downregulated ≥2-fold in exosomes in
breastmilk from mothers who delivered preterm compared to those who
delivered at term; and iii) positively correlated with a signature
of abundantly detected lncRNAs composed of previously reported, as
well as newly identified lncRNAs. Based on the aforementioned
results, it was reported that lncRNA NORAD was downregulated
in a statistically significant manner in exosomes in breastmilk
from mothers who gave birth prematurely while it was, consistently
and specifically, co-detected in breastmilk-derived exosomes. The
findings described above suggest that lncRNA NORAD
represents a specific loading signature of lncRNA cargo in
breastmilk-derived exosomes (lncRNAs positively and negatively
correlated with NORAD levels in breastmilk exosomes are
presented in Table SIV).
Discussion
Breastmilk is abundant in exosomes/EVs that have a
higher propensity for therapeutic effects, while also having the
potential to be easily administered clinically (25). In addition, milk-derived exosomes
exhibit an increased survivability following simulated
gastric/pancreatic digestion (26). There is emerging evidence to
indicate that exosome-encapsulated RNAs can regulate target cell
pathways related to cellular growth, division, differentiation,
stress response, survival, apoptosis, metabolism and immunity
(14). Recent discoveries have
revealed the presence of lncRNAs in breastmilk-derived exosomes,
with well-documented roles in development (12). In addition, it has been
demonstrated that the protein levels of CD9, CD63 and CD81
tetraspanins are consistently higher in exosomes in breastmilk from
mothers who deliver preterm compared to those in breastmilk from
mothers who delivered at term. The increased CD9 expression
observed in breastmilk from mothers who deliver preterm may
constitute an additional signature of preterm birth that reflects
the difference in EV constitution and it remains to be determined
if it plays a functional role in mother-child communications
(27). In the present study, 13
key lncRNAs were consistently and robustly detected in exosomes
across 85% of all breastmilk samples, reaffirming the findings of
the study by Karlsson et al (12), but also significantly expanding
the list to include lncRNAs with well-documented roles in
inflammation, auto-immunity, metabolism, cell cycle control and
cell differentiation, and thus in the development of neonates.
Among these, ten lncRNAs (CTC-444N24.11, LINC00657,
NCBP2-AS2, CRNDE, HCG11, SNHG16,
GAS5, EBP41L4A-AS1, SNHG5 and ZFAS1)
were highly expressed concurrently in all samples, and importantly,
they belong to the same gene cluster, suggesting their highly
positive correlation. The correlation of expression regarding
lncRNAs detected in ≥50% of the samples revealed two distinct
clusters that negatively correlated with each other. Namely, the
increased expression levels of the first cluster signify decreased
levels of the second one. The lncRNAs of each cluster exhibited a
positive correlation among them, reflecting a potential regulatory
mechanism of lncRNA loading to breastmilk exosomes. These results
are likely to be related to the function of each lncRNA, as it can
be hypothesized that lncRNAs of the same cluster employ similar
function at recipient cells, in this case, contributing to the
development of infants.
GAS5 was the most highly expressed lncRNA in
breastmilk-derived exosomes. GAS5 is known to be activated
in response to growth arrest and cellular starvation due to lack of
nutrients or growth factors. It functions as a riborepressor,
regulating glucocorticoid receptor, by binding to its DNA binding
domain through glucocorticoid response element (28). Importantly, GAS5 is involved in
immunity and plays a vital role in moderating the normal growth
arrest of T-cells and non-transformed lymphocytes, as well as
macrophage polarization (29,30). Consequently, GAS5 appears to play
an essential role in programming the neonatal immune system.
Furthermore, the present study provides new insight on the
differential content of lncRNAs in EVs from breastmilk obtained
from mothers who delivered at term compared to those who delivered
preterm. In total, eight lncRNAs were downregulated, having at
least ≥1 log2 fold change, while only one lncRNA was
found to be upregulated in the breastmilk of mothers who delivered
prematurely. LRRC75A-AS1 was only marginally upregulated,
but exhibited a statistically significant difference
(P-value=0.045). LRRC75A-AS1, also known as SNHG29, has previously
been shown to be associated with spontaneous preterm birth.
According to the study by Jiang et al (31), oxidative stress regulates SNHG29,
which in turn accelerates cellular senescence and triggers
pro-inflammatory cytokine release from senescent cells.
SNHG29 was found to be upregulated in the placentas of women
who underwent preterm labor (31). Thus, the results of the present
study further strengthen the association between SNHG29 and preterm
birth.
Among the lncRNAs found to be downregulated in the
exosomes in breastmilk from mothers who delivered prematurely,
three (LINC00657, CTC-444N24.11 and CRNDE)
were downregulated in a statistically significant manner; notably,
they belong to the same cluster, thus indicating that a common
suppression mechanism(s) may be responsible for their differential
regulation in exosomes in breastmilk from mothers who delivered
preterm. lncRNA CTC-444N24.11 was found to be significantly
downregulated with a ≥2-fold change (P-value=0.048) in the
breastmilk of mothers who delivered preterm birth compared to those
who delivered at term. To date, the role of CTC-444N24.11 remains
unknown, while by reviewing the literature no data were found
exclusively for CTC-444N24.11. In a recent bioinformatics
analysis of miRNA expression in retinopathy of prematurity (ROP),
CTC-444N24.11 was indicated as a target of miRNA-128-3p,
which was downregulated in premature infants with ROP (32). CRNDE was also found to
display a ≥2-fold differential expression and can be referred to as
a marginally significant (P-value=0.052) differentially expressed
lncRNA. CRNDE is a central regulator of glucose and lipid
metabolism, while it is regulated by insulin and insulin growth
factors (33). It regulates
cyclin D1 and consequently, normal cell division (34). Its role in normal cellular
developmental and pluripotency (35) possibly indicates the importance
of its presence in breastmilk in normal infant's development.
Only LINC00657 was found to be ubiquitously
detected in breastmilk-derived exosomes and was also significantly
downregulated (≥2 fold change) in breastmilk form mothers who
delivered preterm. LINC00657 is a long intergenic ncRNA named
NORAD (HGNC ID), for non-coding RNA activated by DNA damage.
NORAD is a cytoplasmic lncRNA, and is one of the most abundant and
highly conservative lncRNAs among mammalian cells and species,
respectively (36). Of note, due
to tandem sequence duplication that occurred during evolutional
processes, NORAD consists of 12 repeated units, which facilitate
its function (37). NORAD is
referred to as the guardian of genome, while mounting evidence
suggests its crucial role in maintaining genome stability. NORAD
binds Pumilio RNA-binding proteins (PUM 1 and PUM 2), functioning
as a decoy, and inhibits them from repressing their mRNA targets,
which are key regulators of DNA repair and replication, mitosis and
mitochondrial homeostasis (38,39). In addition, NORAD functions as
scaffold, interacting with RNA binding motif protein X-linked
(RBMX), a component of DNA damage response, and mediates the
assembly of a ribonucleoprotein complex [NORAD-activated
ribonucleoprotein complex 1 (NARC1)], which contains the known
suppressors of genomic instability topoisomerase I (TOP1), Aly/REF
export factor (ALYREF) and the pre-mRNA processing factor 19
(PRPF19)-cell division cycle 5 like (CDC5L) complex (40). Another study revealed the role of
NORAD in regulating nuclear translocation and signal transduction.
More precisely, NORAD interacts with importin-β1 and therefore,
regulates Smad translocation into the nucleus and TGF-β signaling
(41).
The existence of lncRNA NORAD in exosomes may serve
as an important signaling molecule in the adaptive responses of the
infant after birth to hypoxic conditions. NORAD lncRNA has been
found to be upregulated during hypoxic conditions (42) and as such, NORAD can participate
in the adaptive mechanism of the newborns during the perinatal
period; i.e., to combat the high levels of oxygen to which they are
exposed after birth. Failure to adapt to higher-than-normal levels
of oxygen leads to the induction of oxidative stress and the
generation of reactive oxygen species (43). Critically, previous research
point towards a higher level of oxidative stress in preterm
newborns than those born full-term (43) due to immature and not fully
developed respiratory, digestive, immune and antioxidant defense
systems. Thus, the downregulation of lncRNA NORAD in the breastmilk
of mothers who delivered prematurely in the present study, is in
line with the findings of previous research reporting elevated
levels of oxidative stress biomarkers (8-OHdG, hydroperoxide,
malondialdehyde, etc.) and a lower activity of antioxidant enzymes
(superoxide dismutase, glutathione peroxidase, etc.) in preterm
babies. Importantly, the formula feeding of preterm infants has
been associated with higher levels of oxidative stress than
breastmilk feeding and, as such, the existence of NORAD in exosomes
together with the known nutritional antioxidant content of breast
milk may help the newborn to adapt to the higher levels of
oxidative stress (43). Thus,
NORAD may be an important therapeutic target with notable
consequences for premature infants who exhibit higher levels of OS
(43).
It is important to take into consideration the
limitations of the present study. The main limitation is the small
number of samples used. However, these are preliminary results.
Larger numbers of breastmilk samples are required to increase the
statistical validity of the results presented herein. Another
limitation in determining lncRNA expression levels involves the
lack of an accurate exosomal reference gene. Nevertheless, two
genes Actin and RPLP0 were used as reference genes in order to
diminish the possibility of false results (i.e., based on
randomness). The currents findings provide some insight into the
upcoming field of exosomal research, breaking new ground and
providing promising evidence in understanding the role of
epigenetic mechanisms, providing a good starting point for
discussion and future research. Integrated high-throughput
technologies can complement the results of the present study by
elucidating the type of physiological complexity with higher
precision. Translating these technologies into clinical practice in
the form of a non-invasive tests will help realize in the near
future, the personalization of medicine. Even though the miRNA
transcriptome of exosomes in human breastmilk has been uncovered
(13,22,23) and the miRNA expression dynamics
between full-term and preterm breastmilk's lipid fractions have
been characterized (26,44), the exosome-enriched lncRNA
populations in the two groups mentioned above remain unknown.
Not with standing these limitations, the present
study validates and expands previous findings that specific lncRNAs
are loaded in EVs derived from breastmilk, possibly by a common
regulatory mechanism, and are thus, detected together in a highly
correlated manner. NORAD appears to belong to the same
loading/regulatory mechanism; however, it represents the first
lncRNA to date, whose levels are differentially regulated and
significantly suppressed in human breastmilk from mothers who
delivered prematurely. The absence of NORAD among others, such as
impaired mitotic division, mitochondrial dysfunction, premature
cell aging and neuronal dysfunction, contributes to
replication-related stress and DNA damage. If further validated as
true, a significant downregulation of NORAD exosomal cargo in the
breastmilk of mothers who deliver preterm may serve as an exosomal
biomarker for replication-related stress and may be associated with
inflammatory and immune responses in preterm infants. Finally,
NORAD may be directly used as a therapeutic agent. It could be
reverse-engineered to be enriched in exosomes from human mammary
stem cells isolated from mother's own breastmilk and added even on
a given milk formula.
In conclusion, breastmilk-derived exosomes are
important mediators of communication between mother and child,
while lncRNAs may play a crucial role in neonatal growth and
development, in a short- and long-term manner. Collectively, the
data presented herein reveal the consistent co-detection of lncRNAs
that are implicated in several key processes, such as immune system
development, metabolism and cell cycle control, as well as
providing a potential mechanism regarding their load in
breastmilk-derived EVs in a highly correlated manner. To the best
of our knowledge, this is the first study to analyze the
differential expression of lncRNAs in breastmilk between term and
preterm mothers. NORAD is a lncRNA involved in the DNA damage
response and repair pathway, referred to as the guardian of the
human genome and among others, found to be significantly
downregulated at least 2-fold in the breastmilk of preterm mothers.
Broadly translated, this result indicates that the absence of NORAD
may serve as an exosomal biomarker for replication-related stress
in preterm infants. Notably, NORAD and other lncRNAs could be
produced by reverse-engineering and added to breastmilk or other
milk formulas and may function therapeutically in preterm infants.
Future research is required to analyze lncRNA expression and
function in breastmilk from mothers who deliver at term compared to
those who deliver preterm on a larger scale. This will possibly
shed more light on lncRNA involvement and impact on children's
development and health.
Supplementary Data
Availability of data and materials
The RT-qPCR data [Cq values (raw data)]
corresponding to exosomes from breastmilk samples that support the
findings of this study are available in figshare public databases
(https://doi.org/10.6084/m9.figshare.16635592).
Authors' contributions
TS, AG, AS and GPC were involved in the
conceptualization of the study. AG, NM, DV, MP, TS and GPC were
involved in the study methodology. AG, NM, EK, AK, GL, GB, MT and
AT were involved in formal analysis (data acquisition, data
analysis and in the experiments). AG, TS, AS, NM, EK, DV and GPC
were involved in the writing and preparation of the original draft.
AG, MT, AT, GB, DV and GPC were involved in the writing, reviewing
and editing of the manuscript. GPC and AG were responsible for the
acquisition of funding and confirm the authenticity of all the raw
data. All authors have read and agreed to the published version of
the manuscript.
Ethics approval and consent to
participate
The study was conducted according to the guidelines
of the Declaration of Helsinki and approved by the Institutional
Review Board of General Hospital 'Agia Sophia' (protocol code,
24814; date of approval, October 30, 2017). Informed consent was
obtained from all subjects involved in the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Walani SR: Global burden of preterm birth.
Int J Gynaecol Obstet. 150:31–33. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Markopoulou P, Papanikolaou E, Analytis A,
Zoumakis E and Siahanidou T: Preterm birth as a risk factor for
metabolic syndrome and cardiovascular disease in adult life: A
systematic review and meta-analysis. J Pediatr. 210:69–80.e5. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Parets SE, Bedient CE, Menon R and Smith
AK: Preterm birth and its long-term effects: Methylation to
mechanisms. Biology (Basel). 3:498–513. 2014.
|
|
4
|
Lyons KE, Ryan CA, Dempsey EM, Ross RP and
Stanton C: Breast milk, a source of beneficial microbes and
associated benefits for infant health. Nutrients. 12:10392020.
View Article : Google Scholar :
|
|
5
|
ESPGHAN Committee on Nutrition; Agostoni
C, Braegger C, Decsi T, Kolacek S, Koletzko B, Michaelsen KF,
Mihatsch W, Moreno LA, Puntis J, et al: Breast-feeding: A
commentary by the ESPGHAN committee on nutrition. J Pediatr
Gastroenterol Nutr. 49:112–125. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim KU, Kim WH, Jeong CH, Yi DY and Min H:
More than nutrition: Therapeutic potential of breast milk-derived
exosomes in cancer. Int J Mol Sci. 21:73272020. View Article : Google Scholar :
|
|
7
|
Cheshmeh S, Nachvak SM, Rezvani N and
Saber A: Effects of breastfeeding and formula feeding on the
expression level of FTO, CPT1A and PPAR-α genes in healthy infants.
Diabetes Metab Syndr Obes. 13:2227–2237. 2020. View Article : Google Scholar :
|
|
8
|
Horta BL, Victora CG, França GV, Hartwig
FP, Ong KK, Rolfe EL, Magalhães EI, Lima NP and Barros FC:
Breastfeeding moderates FTO related adiposity: A birth cohort study
with 30 years of follow-up. Sci Rep. 8:25302018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Melnik B and Schmitz G: Milk's role as an
epigenetic regulator in health and disease. Diseases. 5:122017.
View Article : Google Scholar
|
|
10
|
Boix-Amorós A, Collado MC, Van't Land B,
Calvert A, le Doare K, Garssen J, Hanna H, Khaleva E, Peroni DG,
Geddes DT, et al: Reviewing the evidence on breast milk composition
and immunological outcomes. Nutr Rev. 77:541–556. 2019. View Article : Google Scholar
|
|
11
|
Seferovic MD, Mohammad M, Pace RM, Engevik
M, Versalovic J, Bode L, Haymond M and Aagaard KM: Maternal diet
alters human milk oligosaccharide composition with implications for
the milk metagenome. Sci Rep. 10:220922020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Karlsson O, Rodosthenous RS, Jara C,
Brennan KJ, Wright RO, Baccarelli AA and Wright RJ: Detection of
long non-coding RNAs in human breastmilk extracellular vesicles:
Implications for early child development. Epigenetics. 11:721–729.
2019. View Article : Google Scholar
|
|
13
|
Kim SY and Yi DY: Components of human
breast milk: From macronutrient to microbiome and microRNA. Clin
Exp Pediatr. 63:301–309. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar :
|
|
15
|
van Niel G, D'Angelo G and Raposo G:
Shedding light on the cell biology of extracellular vesicles. Nat
Rev Mol Cell Biol. 19:213–228. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Soares Martins T, Catita J and Martins
Rosa I: Exosome isolation from distinct biofluids using
precipitation and column-based approaches. PLoS One.
13:e01988202018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Vlachakis D, Mitsis T, Nicolaides N,
Efthimiadou A, Giannakakis A, Bacopoulou F and Chrousos GP:
Functions, pathophysiology and current insights of exosomal
endocrinology (Review). Mol Med Rep. 23:262021.
|
|
18
|
Madden JW: Human breast milk exosomes may
protect against necrotizing enterocolitis in preterm infants.
Pediatr Res. 90:244–245. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Huang X, Yuan T, Tschannen M, Sun Z, Jacob
H, Du M, Liang M, Dittmar RL, Liu Y, Liang M, et al:
Characterization of human plasma-derived exosomal RNAs by deep
sequencing. BMC Genomics. 14:3192013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Valadi H, Ekström K, Bossios A, Sjöstrand
M, Lee JJ and Lötvall JO: Exosome-mediated transfer of mRNAs and
microRNAs is a novel mechanism of genetic exchange between cells.
Nat Cell Biol. 9:654–659. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Statello L, Guo CJ, Chen LL and Huarte M:
Gene regulation by long non-coding RNAs and its biological
functions. Nat Rev Mol Cell Biol. 22:96–118. 2021. View Article : Google Scholar
|
|
22
|
Zhou Q, Li M, Wang X, Li Q, Wang T, Zhu Q,
Zhou X, Wang X, Gao X and Li X: Immune-related microRNAs are
abundant in breast milk exosomes. Int J Biol Sci. 8:118–123. 2021.
View Article : Google Scholar
|
|
23
|
Bozack AK, Colicino E, Rodosthenous R,
Bloomquist TR, Baccarelli AA, Wright RO, Wright RJ and Lee AG:
Associations between maternal lifetime stressors and negative
events in pregnancy and breast milk-derived extracellular vesicle
microRNAs in the programming of intergenerational stress mechanisms
(PRISM) pregnancy cohort. Epigenetics. 16:389–404. 2021. View Article : Google Scholar :
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Galley JD and Besner GE: The therapeutic
potential of breast milk-derived extracellular vesicles. Nutrients.
12:7452020. View Article : Google Scholar :
|
|
26
|
Kahn S, Liao Y, Du X, Xu W, Li J and
Lönnerdal B: Exosomal MicroRNAs in milk from mothers delivering
preterm infants survive in vitro digestion and are taken up by
human intestinal cells. Mol Nutr Food Res. 62:e17010502018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhou Y, Yu Z, Wang X, Chen W, Liu Y, Zhang
Y, Yin J and Han S: Exosomal circRNAs contribute to intestinal
development via the VEGF signalling pathway in human term and
preterm colostrum. Aging (Albany NY). 13:11218–11233. 2021.
View Article : Google Scholar
|
|
28
|
Gharesouran J, Taheri M, Sayad A,
Ghafouri-Fard S, Mazdeh M and Omrani MD: The growth arrest-specific
transcript 5 (GAS5) and nuclear receptor subfamily 3 group C member
1 (NR3C1): Novel markers involved in multiple sclerosis. Int J Mol
Cell Med. 7:102–110. 2018.PubMed/NCBI
|
|
29
|
Ahmad I, Valverde A, Naqvi RA and Naqvi
AR: Long non-coding RNAs RN7SK and GAS5 regulate macrophage
polarization and innate immune responses. Front Immunol.
11:6049812020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mourtada-Maarabouni M, Hedge VL, Kirkham
L, Farzaneh F and Williams GT: Growth arrest in human T-cells is
controlled by the non-coding RNA growth-arrest-specific transcript
5 (GAS5). J Cell Sci. 121:939–946. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang J, Hu H, Chen Q, Zhang Y, Chen W,
Huang Q, Chen X, Li J and Zhong M: Long non-coding RNA SNHG29
regulates cell senescence via p53/p21 signaling in spontaneous
preterm birth. Placenta. 103:64–71. 2021. View Article : Google Scholar
|
|
32
|
Yang Y, Pan JJ, Zhou XG, Zhou XY and Cheng
R: Differentially expressed miRNAs in premature infants with
retinopathy-a bioinformatics analysis. Int J Ophthalmol.
11:773–779. 2018.PubMed/NCBI
|
|
33
|
Ellis BC, Graham LD and Molloy PL: CRNDE,
a long non-coding RNA responsive to insulin/IGF signaling,
regulates genes involved in central metabolism. Biochim Biophys
Acta. 1843:372–386. 2014. View Article : Google Scholar
|
|
34
|
Huan J, Xing L, Lin Q, Xui H and Qin X:
Long noncoding RNA CRNDE activates Wnt/β-catenin signaling pathway
through acting as a molecular sponge of microRNA-136 in human
breast cancer. Am J Transl Res. 9:1977–1989. 2017.
|
|
35
|
Ellis BC, Molloy PL and Graham LD: CRNDE:
A long non-coding RNA involved in CanceR, neurobiology, and
development. Front Genet. 3:2702012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tichon A, Gil N, Lubelsky Y, Havkin
Solomon T, Lemze D, Itzkovitz S, Stern-Ginossar N and Ulitsky I: A
conserved abundant cytoplasmic long noncoding RNA modulates
repression by pumilio proteins in human cells. Nat Commun.
7:122092016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tichon A, Perry RBT, Stojic L and Ulitsky
I: SAM68 is required for regulation of pumilio by the NORAD long
noncoding RNA. Genes Dev. 32:70–78. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kopp F, Elguindy MM, Yalvac ME, Zhang H,
Chen B, Gillett FA, Lee S, Sivakumar S, Yu H, Xie Y, et al: Pumilio
hyperactivity drives premature aging of Norad-deficient mice.
Elife. 8:e426502019. View Article : Google Scholar :
|
|
39
|
Lee S, Kopp F, Chang TC, Sataluri A, Chen
B, Sivakumar S, Yu H, Xie Y and Mendell JT: Noncoding RNA NORAD
regulates genomic stability by sequestering pumilio proteins. Cell.
164:69–80. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Munschauer M, Nguyen CT, Sirokman K,
Hartigan CR, Hogstrom L, Engreitz JM, Ulirsch JC, Fulco CP,
Subramanian V, Chen J, et al: The NORAD lncRNA assembles a
topoisomerase complex critical for genome stability. Nature.
561:132–136. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Kawasaki N, Miwa T, Hokari S, Sakurai T,
Ohmori K, Miyauchi K, Miyazono K and Koinuma D: Long noncoding RNA
NORAD regulates transforming growth factor-β signaling and
epithelial-to-mesenchymal transition-like phenotype. Cancer Sci.
109:2211–2220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li H, Wang X, Wen C, Huo Z, Wang W, Zhan
Q, Cheng D, Chen H, Deng X, Peng C and Shen B: Long noncoding RNA
NORAD, a novel competing endogenous RNA, enhances the
hypoxia-induced epithelial-mesenchymal transition to promote
metastasis in pancreatic cancer. Mol Cancer. 16:1692017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Martin A, Faes C, Debevec T, Rytz C,
Millet G and Pialoux V: Preterm birth and oxidative stress: Effects
of acute physical exercise and hypoxia physiological responses.
Redox Biol. 17:315–322. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Carney MC, Tarasiuk A, DiAngelo SL,
Silveyra P, Podany A, Birch LL, Paul IM, Kelleher S and Hicks SD:
Metabolism-related microRNAs in maternal breast milk are influenced
by premature delivery. Pediatr Res. 82:226–236. 2017. View Article : Google Scholar : PubMed/NCBI
|