Introduction
Retinal ischemia is a major cause of vision loss,
which has been implicated in the pathogenesis of several ocular
diseases, such as acute angle-closure glaucoma, retinal vascular
occlusion and diabetic retinopathy (1,2).
Ischemia-induced retinal neurodegeneration is characterized by
neural apoptosis, glial activation, abnormal electroretinograms and
decreased retinal layer thickness (3,4).
Previous studies have revealed that oxidative stress, abnormal
neurotrophic factor release, inflammation, metabolic changes and
genetic/epigenetic changes are tightly associated with the
pathogenesis of retinal neurodegeneration (5,6).
Currently, intraocular pressure (IOP)-lowering treatment remains
the primary focus of glaucoma management, and anti-angiogenic
treatment is the main therapeutic method for diabetic retinopathy
management. The established treatment for retinal neurodegeneration
is adjunctive neuroprotective therapy for the management of
glaucoma or diabetic retinopathy; however, these neuroprotective
treatments are only partially or transiently effective as they
neither repair the degenerated retinal neurons nor arrest retinal
neurodegeneration (7,8) Therefore, further studies are
required to identify the mechanism underlying retinal
neurodegeneration.
Retinal neurodegeneration is a complicated
pathological process, which encompasses a complex interplay of
various molecular regulators. Non-coding RNAs account for a large
part of the transcriptome and have no protein-coding potential;
however, they do generate non-coding transcripts that are involved
in numerous biological processes and human diseases (9,10).
Circular RNAs (circRNAs) are a group of endogenous RNAs in
eukaryotes that are characterized by covalently closed loops with
tissue-specific and cell-specific expression patterns (11). Previous studies have shown that
circRNAs are involved in the pathogenesis of several
neurodegenerative diseases, such as Alzheimer's disease, traumatic
brain injury and stroke (12,13). The retina and optic nerve are
known as the direct extension of the diencephalon during embryonic
development. The brain and the eye have several common
characteristics, including a similar vasculature and underlying
gene regulatory network (14,15). The present study hypothesized that
circRNAs may serve an important role in the pathogenesis of retinal
neurodegeneration.
An important facet of retinal ischemic injury is
ischemia/reperfusion (I/R) injury. The present study investigated
the role of a circRNA in I/R-induced retinal neurodegeneration.
circRNA-ZYG11B (circZYG11B; hsa_circ_0003739) is a circRNA located
at chr1:53236691-53245665, which has a conserved homologous gene in
the mouse genome (mmu_circ_0011029). Moreover, circZYG11B has been
reported to be dysregulated in I/R-injured retinas (16). The present study aimed to
investigate the effects of circZYG11B silencing on retinal ganglion
cell (RGC) injury and I/R-induced retinal neurodegeneration. In
addition, it aimed to unveil the molecular mechanisms of
circZYG11B-mediated RGC injury and retinal neurodegeneration.
Materials and methods
Primary mouse RGC isolation and
culture
A total of 32 C57BL/6J pregnant mice (age, 14 weeks,
weight 32-36 g) were purchased from Shanghai SLAC Laboratory Animal
Co., Ltd. and maintained in our laboratory at a controlled
temperature of 23±2°C and a relative humidity of 50±10% under a
12-h light/dark cycle with free access to food and water. For RGC
isolation, approximately eight postnatal day 5 mice were used. Mice
were anesthetized with a mixture of ketamine (80 mg/kg) and
xylazine (4 mg/kg), and then euthanized in a chamber via 100%
CO2 exposure at a displacement rate of 50% volume/min
for 50 min (17). After
euthanasia, the eyes were immediately enucleated in cold dissection
buffer containing HBSS (cat. no. 14025134; Gibco; Thermo Fisher
Scientific, Inc.) and 2 mM HEPES (cat. no. H3784; MilliporeSigma).
Subsequently, the retinas were separated from the sclera and
pigment epithelium under a binocular dissecting microscope (MZ7.5;
Leica Microsystems GmbH). The retinas were placed in dissection
buffer and digested with 5 mg/ml papain for 0.5 h at 37°C. The
suspensions of retinal cells were incubated with anti-macrophage
antiserum (1:100; cat. no. AIA31240; Accurate Chemical &
Scientific Corporation) for 1 h at 37°C to eliminate macrophages
and microglial cells. The non-adherent cells were transferred to
100-mm petri dishes pre-conjugated with anti-Thy1.2 antibody
(pre-conjugation for 3 h at room temperature; 1:20 dilution; cat.
no. MCA02R; Bio-Rad Laboratories, Inc) to purify RGCs for 1 h at
37°C. The dishes were then rinsed with PBS to remove the
non-adherent cells. The adherent cells were released using trypsin
(cat. no. T4799; MilliporeSigma) and triturated with a pipette.
RGCs were cultured in DMEM (cat. no. 21013024; Gibco; Thermo Fisher
Scientific, Inc.) supplemented with insulin (2.0 mM; cat. no.
I6634; MilliporeSigma), progesterone (40 nM; cat. no. P0130;
MilliporeSigma), selenite (60 nM; cat. no. S5261; MilliporeSigma),
transferrin (100 nM; cat. no. T8158; MilliporeSigma), CNTF (40
ng/ml; cat. no. 450-13; PeproTech, Inc.) and forskolin (6
µM; cat. no. F6886; MilliporeSigma) at 37°C and in an
atmosphere containing 5% CO2. For hypoxic induction,
RGCs were cultured in a NAPCO 7001 incubator (The Precision
Scientific Company) containing 1% O2, 5% CO2
and 94% N2 at 37°C for 24 h. For oxidative stress
induction, RGCs were exposed to H2O2 (50
µM) to mimic oxidative stress at 37°C for 24 h. The group
without hypoxic induction or oxidative stress induction was taken
as the control group.
Cell Counting Kit (CCK)-8 assay
The viability of RGCs was determined by CCK-8 assays
(cat. no. CK04; Dojindo Laboratories, Inc.) according to the
manufacturer's protocol. Briefly, 5×103 RGCs/well were
seeded into a 96-well plate. After transfection and exposure to
hypoxic stress or oxidative stress, RGCs were incubated with 20
µl CCK-8 solution in each well for 3 h at 37°C. The
absorbance was then detected at a wavelength of 450 nm using a
microplate reader (Molecular Devices, LLC).
Caspase-3 activity assay
Caspase-3 activity was determined using the
caspase-3 assay kit (cat. no. C1168S; Beyotime Institute of
Biotechnology) according to the manufacturer's protocol. Briefly,
RGCs were cultured at a density of 5×103 cells/well in a
96-well plate. After the required treatment, cells were incubated
with 100 µl caspase-3 substrate/well for 1 h at 37°C.
Caspase-3 activity was determined under 490 nm excitation
wavelength and 525 nm emission wavelength. Values were obtained
from the blank group (caspase 3 substrate and cell culture medium,
without RGCs) and subtracted from the experimental group values.
The change in caspase-3 activity was calculated relative to the
value obtained from the untreated cell group. Each sample was
measured in duplicate.
Retinal I/R injury model
A total of 35 male C57BL/6 mice (age, 8 weeks;
weight, 25-30 g) were used for detecting circZYG11B expression
change during retinal neurodegeneration and investigating the role
of circZYG11B in retinal neurodegeneration. The mice were purchased
from Shanghai SLAC Laboratory Animal Co., Ltd. The mice were housed
in a pathogen-free environment (temperature, 23±2°C; humidity,
50±5%) under a 12-h light/dark cycle with free access to food and
water. All operations were conducted under anesthesia via an
intra-peritoneal injection of ketamine (80 mg/kg) and xylazine (4
mg/kg) (18,19). During I/R injury, the body
temperature was maintained at 37°C using a temperature-controlled
heating pad. The pupils were dilated using 0.5% phenylephrine and
0.5% tropicamide (Santen). Oxybuprocaine hydrochloride (Santen) was
then applied to the corneas, followed by injection with a 30-gauge
needle connected to a sterile bag containing 0.9% sodium chloride.
To detect circZYG11B expression changes during I/R-induced retinal
neurodegeneration, the anterior chamber of one eye was cannulated
with a 30-gauge needle connected to the saline bag, which was
located 120 cm above the eye, leading to a high IOP of ~90 mmHg.
After removing the needle from the anterior chamber for 1 h, the
IOP returned to normal. The contralateral eye was connected to the
saline bag, which was not raised above the eye, thus a normal IOP
was maintained as the control. The needle was withdrawn from the
eye after 1 h. The retinas were collected at 0, 3 and 7 days after
I/R injury (n=5 mice/time point).
To investigate whether silencing of circZYG11B
affected retinal neurodegeneration, the mice were randomly
separated into the following four groups: i) Control group: One eye
was injected with the needle without elevating IOP and the other
eye was left untreated (n=5 mice); ii) I/R group: One eye underwent
I/R injury and the other eye was left untreated (n=5 mice); iii)
I/R + control shRNA group: One eye was injected with scrambled
(Scr) short hairpin (sh)RNA and underwent I/R injury and the other
eye was left untreated (n=5 mice); and iv) I/R + circZYG11B shRNA:
One eye was injected with circZYG11B shRNA and underwent I/R injury
and the other eye was left untreated (n=5 mice). A total of 7 days
after I/R injury, the mice were euthanized in a chamber via 100%
CO2 exposure at a displacement rate of 50% volume/min.
The mice received an intravitreous injection of 1.0 µl
(7.5×10−5 µg/ml) circZYG11B shRNA or equal
amounts of Scr shRNA 14 days before I/R injury. The mice were
injected once and sacrificed 21 days after injection. The sequence
for circZYG11B shRNA was 5′-GGA CAC TTG AAG GAG GAA GCC TTC AAG AGA
GGC TTC CTC CTT CAA GTG TCC TTT TTG-3′; the sequence for negative
control shRNA was 5′-TCC TAA GGT TAA GTC GCC CTC GCT CGA GCG AGG
GCG ACT TAA CCT TAG GTT TTT G-3′. The shRNAs (OBio Technology) were
inserted into an AAV-2 vector (Stratagene; Agilent Technologies,
Inc.) under the control of RNA polymerase III promoter U6, and then
packaged with pHelper and pAAV-RC to produce recombinant
AAV2-circZYG11B shRNA or AAV2-Scr shRNA. The target site for
circZYG11B shRNA was located at the junctional site of circZYG11B
spanning 9 nt sequence on one side and 12 nt on the other side.
Cell transfection
RGCs were cultured onto 6-well plates at 37°C
overnight at a density of 1×105 cells/well. When cell
confluence had reached 80%, RGCs were transfected with circZYG11B
small interfering (si)RNA, negative control siRNA, Scr mimic,
miR-620 mimic, pcDNA 3.1 (Vector) or PTEN-pcDNA 3.1 (all purchased
from Sangon Biotech Co., Ltd.) using Lipo6000™ transfection reagent
(cat. no. C0526; Beyotime Institute of Biotechnology) according to
the manufacturer's protocol. circZYG11B siRNA, negative control
siRNA, Scr mimic, miR-620 mimic, pcDNA 3.1 and PTEN-pcDNA 3.1 were
diluted in Opti-MEM (Gibco; Thermo Fisher Scientific, Inc.), added
to a culture plate and mixed gently 3-5 times. Lipo6000™
transfection reagent was also diluted in Opti-MEM and incubated for
5 min at room temperature. Subsequently, the diluted siRNA, mimics
or plasmids, and transfection reagent were combined and incubated
at room temperature for 20 min. The mixture was then added to each
well for transfection. After 12 h at 37°C, the medium was replaced
and cells were cultured at 37°C for an additional 12 h for the
subsequent experiments. The sequences were as follows: circZYG11B
siRNA, sense 5′-GGA CAC UUG AAG GAG GAA GCC TdT d-3′, anti-sense
5′-GG CUU CCU CCU UCA AGU GCU CCT dTd-3′; Scr siRNA, sense 5′-UUC
UCC GAA CGU GUC ACG UTd Td-3′, anti-sense 5′-ACG UGA CAC GUU CGG
AGA ATd TdT-3′; miR-620 mimic, 5′-AUG GAG AUA GAU AUA GAA AUU U-3′;
negative control miRNA mimic, 5′-UCGCUUGGUGCAGGUCGGG-3′. During
transfection, the concentration of miRNA mimics and siRNAs was 50
nM, and the concentration of plasmids was 4.0 µg.
Hematoxylin and eosin (H&E)
staining
Eyes obtained from the four groups of mice were
enucleated and fixed in 4% paraformaldehyde at 4°C overnight. The
eyes were then dehydrated in graded concentrations of ethanol and
embedded in paraffin. Sections (5 µm) were cut vertically
through the optic nerve head. Subsequently, the paraffin-embedded
tissue sections were dewaxed with xylene and rehydrated in graded
concentrations of ethanol. After washing with ddH2O, the
sections were stained with hematoxylin for 5 min at room
temperature, stained with eosin for 45 sec at room temperature, and
mounted with neutral resin at room temperature for long-term
storage (cat. no. C0105S; Beyotime Institute of Biotechnology). The
images were captured using an Olympus IX-73 bright-field microscope
(Olympus Corporation).
Immunofluorescence staining
The retinas were dissected in PBS and fixed in 4%
paraformaldehyde for 12 h at room temperature. Subsequently, they
were immersed in 30% sucrose solution for another 12 h and embedded
in optimum cutting temperature compound mounting media. Sections
(10 µm) were cut and placed on gelatin-coated slides. After
rehydration with PBS, retinal sections were blocked with 5% bovine
serum albumin (cat. no. ST025; Beyotime Institute of Biotechnology)
for 30 min at 37°C to minimize nonspecific labeling. Retinal
sections were incubated with primary antibodies against GFAP
(1:200; cat. no. ab7260; Abcam), NeuN (1:300; cat. no. ab128886;
Abcam) or anti-tubulin β3 (TUJ1; 1:300; cat. no. 801213; Biolegend,
Inc.) overnight at 4°C. The slides were then washed with PBS
containing 0.1% Tween 20 and incubated with Alexa Fluor 594 8
secondary antibodies (1:500; cat. nos. ab150080 and ab150116;
Abcam) overnight at 4°C. After washing with PBS, the nuclei were
labeled with DAPI (cat. no. D9542; MilliporeSigma) for 15 min at
room temperature. The staining signaling was observed in six
selected visual fields using an Olympus IX-73 fluorescence
microscope (Olympus Corporation). Adobe Photoshop CS2 (Adobe
Systems, Inc.) software was used for the preprocessing the
fluorescent images.
TUNEL staining assay
The apoptosis of RGCs was determined using the TUNEL
Apoptosis Detection kit (cat. no. C11026; Guangzhou RiboBio Co.,
Ltd.) according to the manufacturer's protocol. RGCs were fixed
with 4% paraformaldehyde at 4°C for 30 min and were blocked with
the blocking buffer (3% H2O2 in
CH3OH) at room temperature for 15 min. Subsequently,
RGCs were permeabilized with 0.1% Triton X-100 for 10 min on ice,
incubated with the TUNEL reaction mixture for 1 h at 37°C and
stained with DAPI for 5 min at room temperature to label cell
nuclei. The staining signaling was observed in six selected visual
fields using an Olympus IX-73 fluorescence microscope (Olympus
Corporation).
Luciferase reporter assay
circZYG11B sequence was inserted into the
KpnI and HindIII sites of pGL3 vector (Promega
Corporation) to generate the Luc-circZYG11B vector.
circZYG11B-interacting miRNAs were predicted using the circular RNA
Interactome database (https://circinteractome.nia.nih.gov/) (20). RGCs were seeded in 96-well plates
at a density of 5×103 cells/well. After 24 h culture,
RGCs were transfected with different miRNA mimics (final
concentration: 50 nM) using Lipo6000™ transfection reagent
according to the manufacturer's protocol. After 12 h, the medium
was replaced and RGCs were cultured at 37°C for additional 12 h for
luciferase activity assay. All miRNA mimics were synthesized by
Sangon Biotech Co., Ltd. The mRNA mimics used were as follows:
miR-149 mimic, 5′-UCU GGC UCC GUG UCU UCA CUC CCU U-3′; miR-186
mimic, 5′-CAA AGA AUU CUC CUU UUG GGC UUU-3′; miR-543 mimic, 5′-AAA
CAU UCG CGG UGC ACU UCU UUU-3′; miR-545 mimic, 5′-UCAGCA AAC AUU
UAU UGU GUG CUU-3′; miR-568 mimic, 5′-AUG UAU AAA UGU AUA CAC ACU
U-3′; miR-607 mimic, 5′-GUU CAA AUC CAG AUC UAU AAC UU-3′; miR-616
mimic, 5′-AGU CAU UGG AGG GUU UGA GCA GUU-3′; miR-587 mimic, 5′-UUU
CCA UAG GUG AUG AGU CAC UU-3′; miR-620 mimic, 5′-AUG GAG AUA GAU
AUA GAA AUU U-3′; miR-624 mimic, 5′-CAC AAG GUA UUG GUA UUA CCU
UU-3′; miR-634 mimic, 5′-AAC CAG CAC CCC AAC UUU GGA CUU-3′;
miR-654 mimic, 5′-UGG UGG GCC GCA GAA CAU GUG CUU-3′; miR-936
mimic, 5′-ACA GUA GAG GGA GGA AUC GCA GUU-3′; miR-95 mimic, 5′-UCA
AUA AAU GUC UGU UGA AUU UU-3′; The luciferase activity was detected
using a Dual-Luciferase Reporter Assay kit (Promega Corporation).
The vector pRL-TK expressing Renilla luciferase was used as
the internal control for transfection. The empty vector pGL3-basic
was used as the negative control. For comparisons, the luciferase
activity of the pGL3 vector was normalized with Renilla
luciferase activity.
Biotin-coupled miRNA capture
RGCs (~1×106) were transfected with
biotinylated miR-620 mimics (5′-AUG GAG AUA GAU AUA GAA AUU U-3′)
or control mimics (miR-95 mimic: 5′-UCA AUA AAU GUC UGU UGA AUU
UU-3′), which were synthesized by Sangon Biotech Co., Ltd., using
Lipo6000™ transfection reagent according to the manufacturer's
protocol. The final concentration of miRNA mimics was 50 nM. After
transfection for 24 h, the cells were lysed in 500 µl lysis
buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 1% Triton X-100; 1%
sodium deoxycholate; 0.1% SDS). Subsequently, 50 µl washed
streptavidin magnetic beads (Thermo Fisher Scientific, Inc.) were
blocked in 1% bovine serum albumin (cat. no. ST025; Beyotime
Institute of Biotechnology) for 2 h (21). These beads were then added to the
reaction tubes and incubated with 300 µl lysates for 3 h at
4°C to pull down the biotin-coupled RNA complex. After washing the
beads with ice-cold lysis buffer, RNA was isolated with
TRIzol® reagent (cat. no. 15596018; Invitrogen; Thermo
Fisher Scientific, Inc.) and the expression levels of circZYG11B
and circZNF236 (negative control) were detected by reverse
transcription-quantitative PCR (RT-qPCR).
RNA immunoprecipitation (RIP) assay
RIP assays were conducted using the EZ-Magna RIP kit
(MilliporeSigma) according to the manufacturer's protocol. Briefly,
RGCs were lysed in the RIP lysis buffer and the cell lysate was
incubated with magnetic beads conjugated with anti-Ago2 or anti-IgG
antibody for 8 h at 4°C. The beads were then incubated with
proteinase K (Invitrogen; Thermo Fisher Scientific, Inc.) for 30
min at 58°C to remove proteins. Finally, the immunoprecipitated
RNAs were extracted using TRIzol reagent according to the
manufacturer's instructions and subjected for RT-qPCR to determine
the expression levels of circZYG11B.
Target gene prediction
The target gene information of miR-620 was predicted
using TargetScan software (http://www.targetscan.org/) (22).
RT-qPCR
Total RNAs were extracted from retinal tissues or
RGCs using TRIzol reagent according to the manufacturer's
instructions. For mRNA expression detection, total RNAs were
reverse transcribed using the SuperScript III first-strand
synthesis kit (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The temperature protocols
for RT were as follows: 37°C for 60 min, 95°C for 5 min and
maintenance at 4°C. Subsequently, qPCR assays were conducted using
the PowerUp™ SYBR™ Green Master Mix (cat. no. A25742; Applied
Biosystems; Thermo Fisher Scientific, Inc.) in a PikoReal 96 cycler
(Thermo Fisher Scientific, Inc.) using gene-specific primers
(Table I) according to the
manufacturer's protocol. The thermocycling protocols were as
follows: Initial denaturation at 95°C for 3 min, followed by 42
cycles at 95°C for 10 sec, 56°C for 30 sec and 72°C for 30 sec,
followed by final extension at 72°C for 60 sec. GAPDH was used as a
housekeeping gene.
 | Table IList of quantitative PCR primers. |
Table I
List of quantitative PCR primers.
| Gene | Sequences
(5′-3′) |
|---|
| Mouse
circZYG11B | F:
CTTGCCCAATCTTGTGTCCC |
| R:
CAGGGTTCCATCTTGGCGT |
| Mouse ZYG11B | F:
AACTTTCTCCCGCCTTTCCA |
| R:
TGTGTCCTAAAGAGGCACCC |
| Human
circZYG11B | F:
GGGCATTCTTTGTTCGGGAA |
| R:
CACCTGTGGCATCAAGTTCC |
| Human ZYG11B | F:
CCCACTGTCAAATTCTCACTGAC |
| R:
GCCATAGTGAGAACAGTGCCT |
| Mouse PTEN | F:
AGGATACGCGCTTGGGC |
| R:
ACAGCGGCTCAACTCTCAAA |
| Human GAPDH | F:
AAGACGGGCGGAGAGAAACC |
| R:
CGTTGACTCCGACCTTCACC |
| Mouse GAPDH | F:
AGGTCGGTGTGAACGGATTTG |
| R:
TGTAGACCATGTAGTTGAGGTCA |
| miR-620 | F:
GCCGAGATGGAGATAGATAT |
| R: CTCAACTGG
TGTCGTGGA |
| U6 | F:
CTCGCTTCGGCAGCACATATACT |
| R:
ACGCTTCACGAATTTGCGTGTC |
For miR-620 detection, RT was conducted using the
miScript Reverse Transcription kit (Qiagen GmbH) according to the
manufacturer's protocol in a final volume of 20 µl
containing 5 µl total RNA, 5 µl 5X miScript RT buffer
and 1 µl miScript Reverse Transcriptase Mix. The temperature
protocols for RT were as follows: 37°C for 60 min, at 95°C for 5
min and maintenance at 4°C. qPCR was conducted using the miScript
SYBR-Green PCR kit (Qiagen GmbH) in a PikoReal 96 cycler (Thermo
Fisher Scientific, Inc.) using gene-specific primers (Table I) according to the manufacturer's
protocol. The thermocycling protocols were as follows: Initial
denaturation at 95°C for 3 min, followed by 42 cycles at 95°C for
10 sec, 56°C for 30 sec and 72°C for 30 sec, followed by final
extension at 72°C for 60 sec. Small nuclear RNA U6 was used as a
housekeeping gene. Relative mRNA or miR-620 expression was
calculated using the 2−ΔΔCq method (23).
Clinical sample collection
Aqueous humor (AH) samples were obtained from Eye
& ENT Hospital (Shanghai, China) between November 2018 and
March 2019. Written informed consent was obtained from the patients
involved. Glaucoma AH samples were obtained from patients diagnosed
with acute angle-closure glaucoma (12 male and eight female
patients; age range, 53-66 years). Control AH samples were obtained
from patients diagnosed with cataracts without acute angle-closure
glaucoma (11 male and nine female patients; age range, 50-63
years). The patients were diagnosed at the hospital through ocular
examination, including anterior segment photography, Goldmann
applanation tonometry, fundus examination, visual field, optic disc
photography and ultrasound biomicroscopy examination. The patients
with glaucoma were diagnosed as having acute angle-closure glaucoma
without cataracts and required filtration surgery. Patients were
excluded if they had other ophthalmic or systemic diseases, had
accepted systemic and local steroids therapy for glaucoma
treatment, or had accepted ophthalmic surgery or intravitreous
injections of anti-VEGF drugs.
ELISA
The levels of PTEN in AH samples were determined
using a human PTEN ELISA Kit (cat. no. ab206979; Abcam) according
to the manufacturer's protocol. Briefly, the standard samples and
AH samples were added to the pre-coated 96-well plates and
incubated for 1 h at room temperature, followed by the addition of
the antibody cocktail to each well. Subsequently, the plates were
sealed and incubated for 1 h at room temperature. After washing the
plates three times, the TMB Development Solution was added to each
well and incubated for 10 min in the dark on a plate shaker set to
400 rpm. Finally, the Stop Solution was added to each well to
terminate the enzyme-substrate reaction. After the reaction was
terminated for 3 min, the absorbance value of each well was
measured at a wavelength of 450 nm. A standard curve was plotted
for determining the concentration of PTEN in each sample. All
measurements were performed in duplicate.
Statistical analysis
All data were analyzed using GraphPad Prism 5
(GraphPad Software, Inc.). The experiments were independently
repeated three times and the data are presented as the mean ±
standard deviation. Comparisons between two independent samples
were analyzed using the paired Student's t-test (Comparison of
circZYG11B expression between I/R retinas and normal retinas) or
unpaired Student's t-test (comparison of circZYG11B, miR-620 and
PTEN levels in AH samples between patients with glaucoma and
patients with cataracts), and multiple comparisons were made using
one-way ANOVA followed by Bonferroni's test. P<0.05 was
considered to indicate a statistically significant difference.
Results
circZYG11B expression is increased during
I/R-induced retinal neurodegeneration in vivo and in vitro
Models of retinal I/R injury are important for
studying retinal neurodegeneration (16). Retinal I/R injury was induced by
an acute increase in IOP. The present study used a mouse model of
retinal I/R injury and detected the expression levels of
circ-ZYG11B between I/R retinas and control retinas. IOP in one eye
was raised to ~90 mm Hg for 1 h, and the contralateral eye was
cannulated and maintained at normal IOP as the control. The needle
was withdrawn from the eye after 1 h. The mice were sacrificed at
0, 3 and 7 days after I/R injury. The results demonstrated that
circZYG11B was significantly increased in the retinas at day 3 or
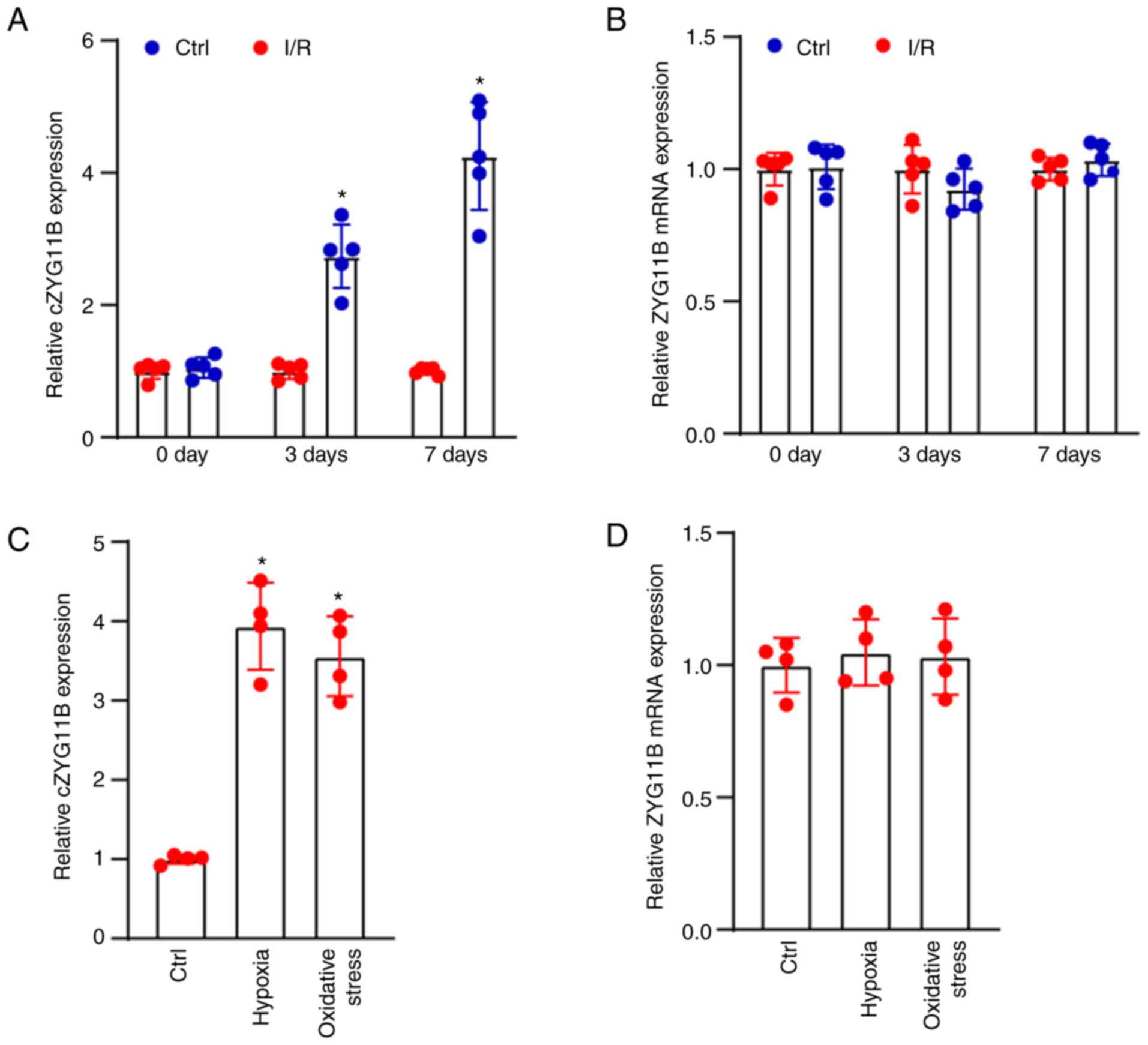
day 7 after I/R injury (Fig. 1A).
By contrast, the mRNA expression levels of ZYG11B were not altered
in the retinas after I/R injury (Fig.
1B). Retinal neurons, particularly RGCs, have been shown to be
highly sensitive to I/R injury (3). In the present study, the isolated
RGCs were exposed to H2O2 (50 µM) or
1% O2 at 37°C for 24 h to mimic oxidative stress or
hypoxic stress. The group without exposure to
H2O2 or 1% O2 was taken as the
control group. The results revealed that compared with in the
control group, exposure to H2O2 or 1%
O2 increased the expression levels of circZYG11B. By
contrast, exposure to H2O2 or 1%
O2 did not alter the mRNA expression levels of ZYG11B
(Fig. 1C and D).
Silencing of circZYG11B alleviates
hypoxic stress-induced RGC injury
The present study then investigated the role of
circZYG11B in RGCs in vitro. RT-qPCR confirmed that
transfection with circZYG11B siRNA decreased the expression levels
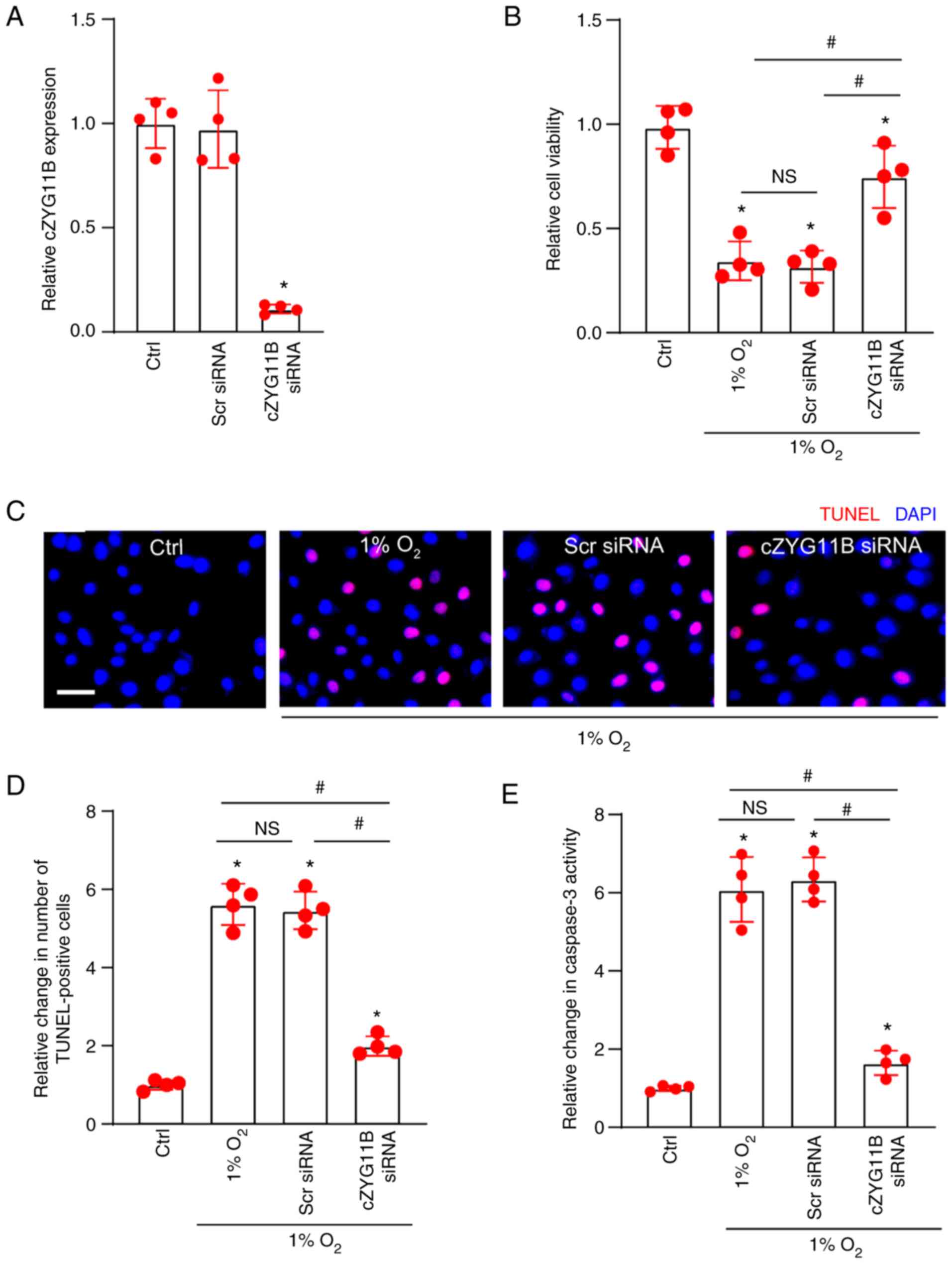
of circZYG11B compared with in the control group (Fig. 2A). Hypoxic stress is an important
pathological factor in retinal I/R injury (24). The non-transfected RGCs, or RGCs
transfected with circZYG11B siRNA or Scr siRNA were exposed to 1%
O2 to mimic hypoxic stress in vitro. The
non-transfected RGCs without 1% O2 treatment was taken
as the control group. CCK-8 assays revealed that compared with in
the control group without 1% O2 exposure, hypoxic stress
induced by 1% O2 led to decreased RGC viability.
Compared with the 1% O2 group, transfection of
circZYG11B siRNA, but not Scr siRNA, significantly alleviated
hypoxic stress-induced reduction of RGC viability (Fig. 2B). TUNEL assays revealed that
compared with the control group without 1% O2 exposure,
1% O2 treatment led to an increased number of apoptotic
RGCs. The number of apoptotic RGCs in the circZYG11B group was
lower than that in the 1% O2 group or in the 1%
O2 + Scr siRNA group (Fig.
2C and D). Caspase-3 activity assays revealed that hypoxic
stress enhanced the activity of caspase-3. Compared with in the 1%
O2 group, transfection with circZYG11B siRNA, but not
Scr siRNA, reduced the activity of caspase-3 induced by hypoxic
stress (Fig. 2E). Collectively,
silencing of circZYG11B may have a protective role in RGCs against
hypoxic stress.
Silencing of circZYG11B alleviates
oxidative stress-induced RGC injury
Oxidative stress is another pathological factor in
retinal I/R injury (24). The
non-transfected RGCs or RGCs transfected with circZYG11B siRNA or
Scr siRNA were exposed to H2O2 (50
µmol/l) to mimic oxidative stress in vitro. CCK-8
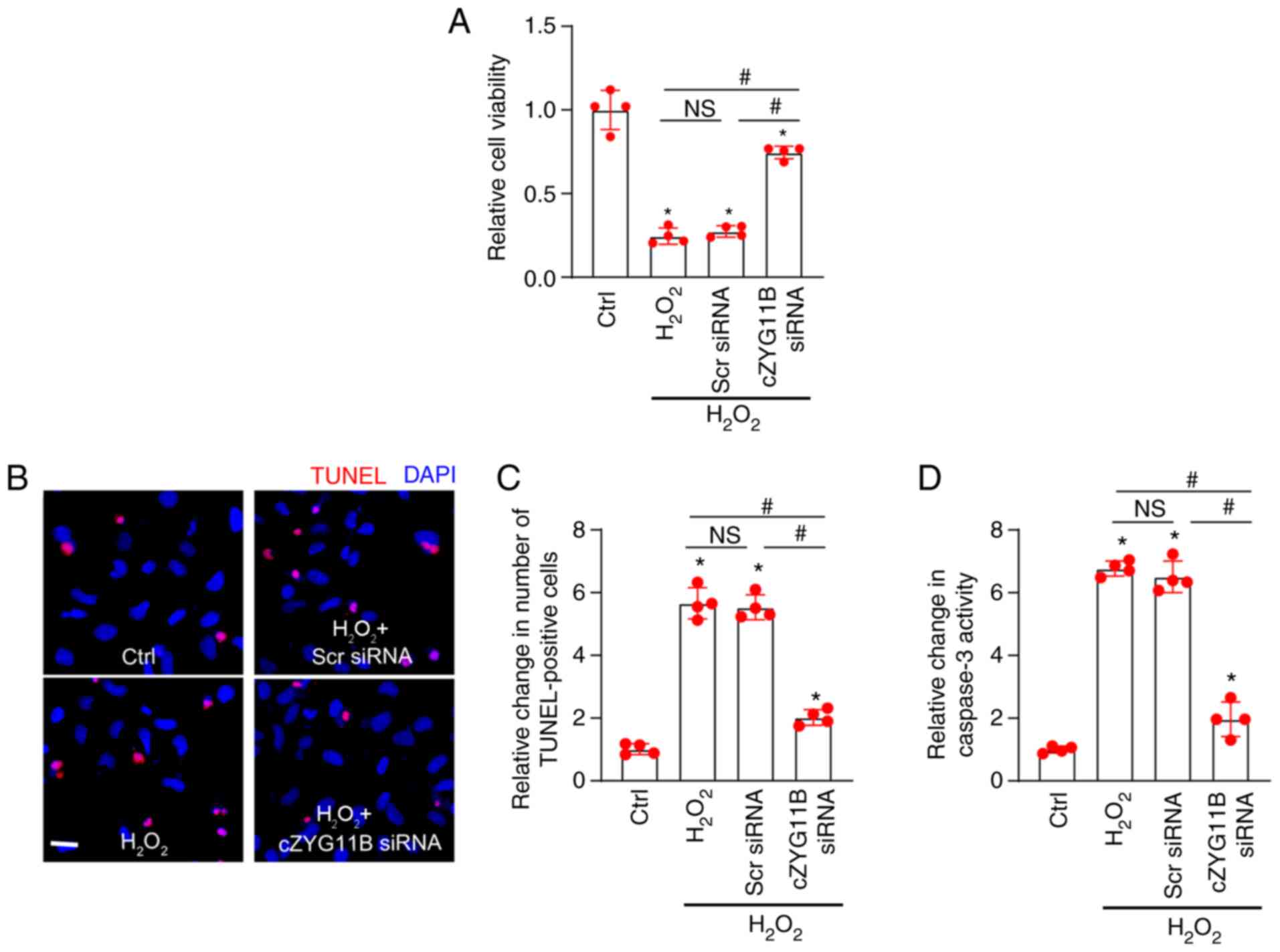
assays revealed that compared with in the control group without
H2O2 exposure, H2O2
treatment-mediated oxidative stress led to reduced viability of
RGCs. Compared with in the H2O2 group,
transfection with circZYG11B siRNA, but not Scr siRNA, alleviated
the oxidative stress-induced reduction of RGC viability (Fig. 3A). TUNEL assays revealed that
compared with in the control group without
H2O2 exposure, treatment with
H2O2 (50 µmol/l) led to an increased
number of apoptotic RGCs. However, the number of apoptotic RGCs was
significantly decreased post-transfection with circZYG11B siRNA,
but not Scr siRNA (Fig. 3B).
Oxidative stress led to enhanced caspase-3 activity in RGCs.
Compared with in the H2O2 group, transfection
with circZYG11B siRNA, but not Scr siRNA, significantly decreased
the activity of caspase-3 induced by H2O2
(Fig. 3C). These findings
suggested that silencing of circZYG11B could exert protective
effects on RGCs against oxidative stress.
Silencing of circZYG11B alleviates
I/R-induced retinal injury in vivo
The present study next determined the role of
circ-ZYG11B in I/R-induced retinal injury in vivo. C57BL/6
mice received an intravitreous injection of circZYG11B shRNA to
knockdown the expression of circZYG11B. Injection with circZYG11B
shRNA significantly reduced the expression levels of circZYG11B
compared with in the control group, but did not alter the mRNA
expression levels of ZYG11B in the retinas (Fig. 4A and B). H&E staining revealed
that silencing of circZYG11B could partially reverse the reduction
in I/R injury-induced retinal thickness and reduce I/R-induced
neurodegeneration in the ganglion cell layer (GCL) (Fig. 4C and 4D). Retinas were also extracted and the
surviving RGCs were quantified at day 7 after I/R injury. The
density of surviving RGCs in the I/R group was decreased by ~40%
compared with in the control group. Silencing of circZYG11B exerted
protective effects on RGC survival and protected against
I/R-induced RGC injury (Fig. 4E and
F).
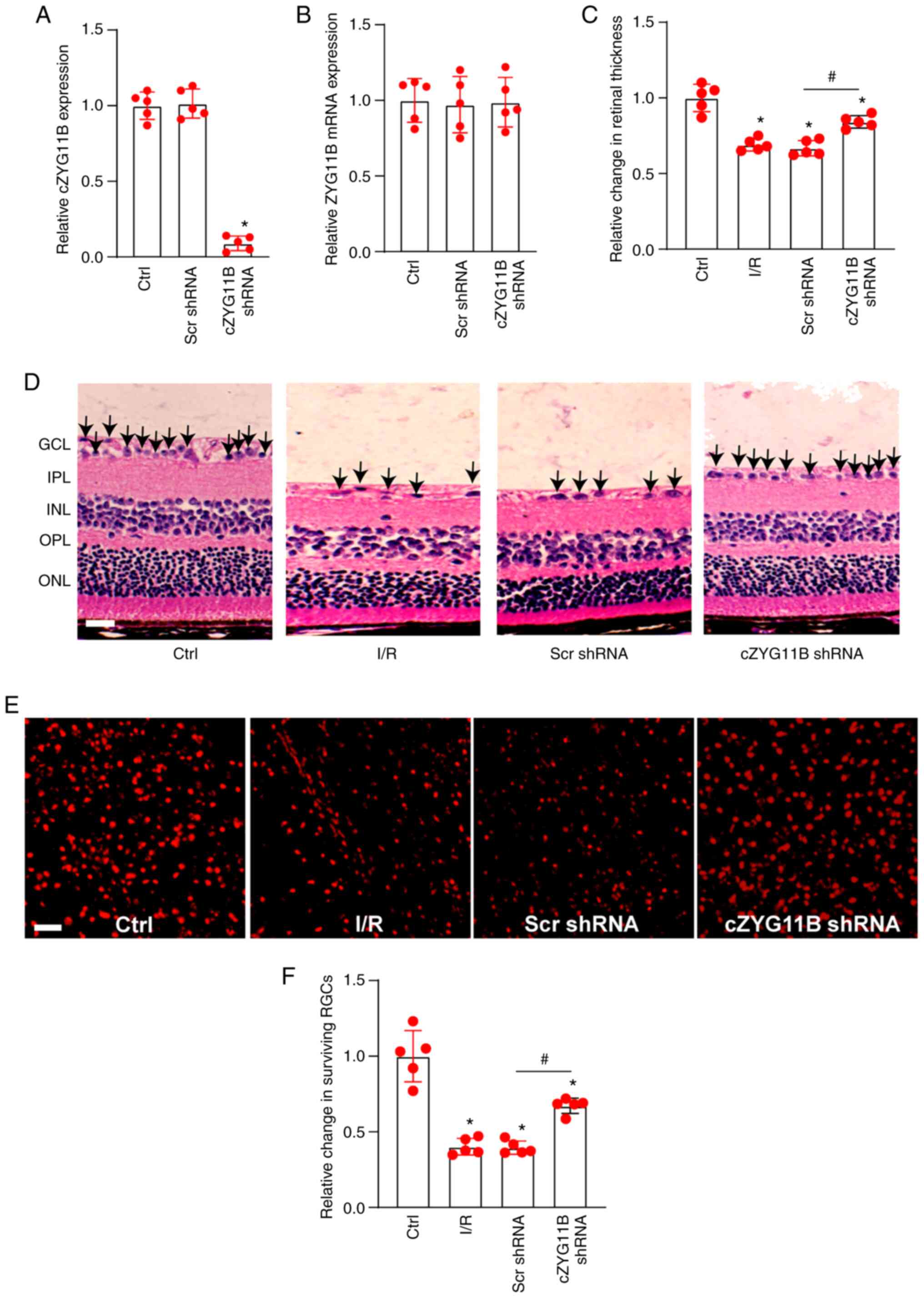 | Figure 4Silencing of cZYG11B alleviates
I/R-induced retinal injury in vivo. (A and B) C57BL/6 mice
received an intravitreous injection of cZYG11B shRNA, Scr shRNA or
were untreated for 7 days. Reverse transcription-quantitative PCR
was conducted to detect the expression levels of (A) cZYG11B and
(B) ZYG11B mRNA. (C and D) Hematoxylin and eosin staining of
retinal sections and semi-quantification analysis was conducted to
detect changes in retinal thickness in normal retinas (Ctrl), I/R
retinas, I/R retinas injected with Scr shRNA or I/R retinas
injected with cZYG11B shRNA. Scale bar, 50 µm. Arrows
indicate the nuclei of RGCs. (E and F) Normal retinas (Ctrl), I/R
retinas, I/R retinas injected with Scr shRNA or I/R retinas
injected with cZYG11B shRNA were stained with TUJ1 to label the
surviving RGCs at day 7 after I/R injury. Scale bar, 20 µm.
*P<0.05 vs. Ctrl group; #P<0.05 as
indicated; Kruskal-Wallis test followed by the post hoc Bonferroni
test. n=5. Ctrl, control; cZYG11B, circZYG11B; I/R,
ischemia/reperfusion; RGC, retinal ganglion cell; Scr, scrambled;
shRNA, short hairpin RNA; GCL, ganglion cell layer; IPL, inner
plexiform layer; INL, inner nuclear layer; OPL, outer plexiform
layer; ONL, outer nuclear layer. |
Silencing of circZYG11B suppresses
I/R-induced RGC injury and retinal reactive gliosis
The present study further determined the role of
circZYG11B in I/R-induced retinal degeneration in vivo.
Immunofluorescence staining with NeuN was conducted to identify
RGCs in the GCL. Compared with in the control retinas, I/R injury
led to reduced expression of NeuN. Compared with in the Scr
shRNA-injected retinas, injection of circZYG11B shRNA alleviated
I/R-induced RGC injury as shown by increased NeuN staining
(Fig. 5A and B). The retinas were
then stained with GFAP to detect retinal reactive gliosis. Compared
with in the control retinas, I/R injury led to activation of
retinal reactive gliosis. Silencing of circZYG11B could alleviate
I/R-induced retinal reactive gliosis as shown by decreased GFAP
staining (Fig. 5C and D). These
findings indicated that circZYG11B silencing could alleviate
I/R-induced retinal neurodegeneration.
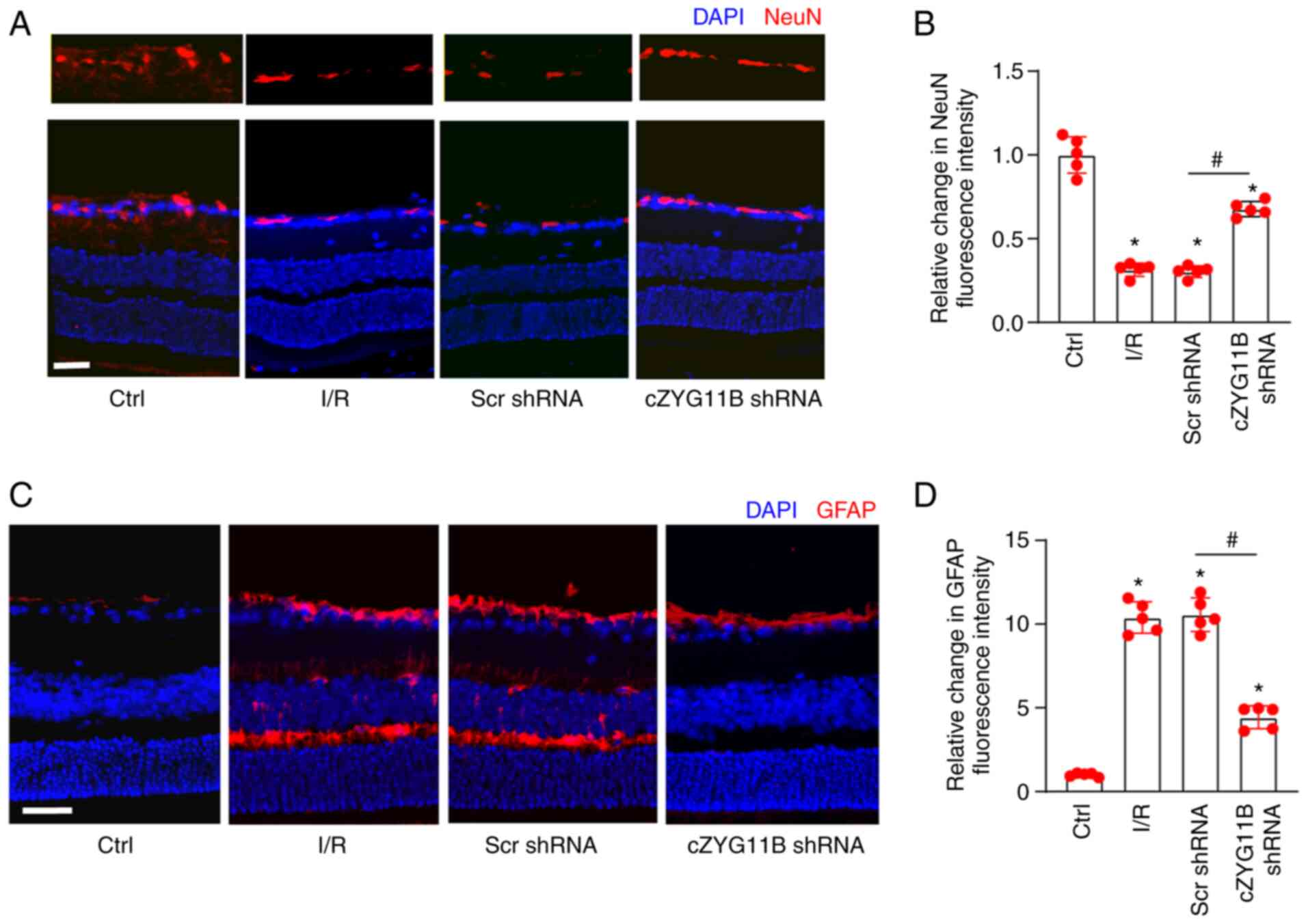 | Figure 5Silencing of cZYG11B suppresses
I/R-induced RGC injury and retinal reactive gliosis in vivo.
(A-D) Mice received an intravitreal injection of cZYG11B shRNA or
Scr shRNA. A total of 3 weeks after intravitreal injection, I/R
model was constructed to study retinal neurodegeneration.
Immunofluorescence staining with (A and B) NeuN or (C and D) GFAP
was conducted to detect retinal neurodegeneration at day 7 after
I/R injury. The representative images and semi-quantitative results
were shown. Nuclei, blue; NeuN-positive cells, red; GFAP-positive
cells, red. Scale bar, 50 µm. *P<0.05 vs. Ctrl
group; #P<0.05 as indicated; Kruskal-Wallis test
followed by the post hoc Bonferroni test. n=5. Ctrl, control;
cZYG11B, circZYG11B; I/R, ischemia/reperfusion; RGC, retinal
ganglion cell; Scr, scrambled; shRNA, short hairpin RNA. |
circZYG11B regulates RGC function by
acting as a miR-620 sponge
The present study next investigated the underlying
mechanism of circZYG11B in retinal neurodegeneration. The Ago2
protein is a core component of the RNA-induced silencing complex
that binds miRNA complexes to target genes. miRNAs associate with
the Ago2 protein to post-transcriptionally suppress target gene
expression (20). The RIP assays
indicated that circZYG11B could be pulled down by Ago2 antibody but
not IgG, suggesting that circZYG11B may serve a role by acting as a
miRNA sponge (Fig. 6A).
Luciferase activity assay demonstrated that transfection with
miR-620 mimics or miR-654 mimics, but not the other miRNA mimics,
reduced the activity of Luc-circZYG11B (Fig. 6B). Due to the greater silencing
efficiency of miR-620 compared with miR-654 on the activity of
Luc-circZYG11B, the present study focused on the role of miR-620 in
RGCs. Because miR-95 mimics did not reduce the luciferase activity
of Luc-circZYG11B, it was suggested that miR-95 could not directly
bind to circZYG11B; therefore, miR-95 was selected as the negative
control for RNA pull-down assays. RNA pull-down assays revealed
that circZYG11B but not circZNF236 was greatly enriched in the
miR-620-captured fraction. By contrast, there was no difference
between the amount of circZYG11B and circZNF236 in the
miR-95-captured fraction (negative control) (Fig. 6C). Subsequently, the potential
target genes of miR-620 were predicted using TargetScan software
(http://www.targetscan.org/) (22). The candidate gene, PTEN, was
assessed further due to its role in cell apoptosis.
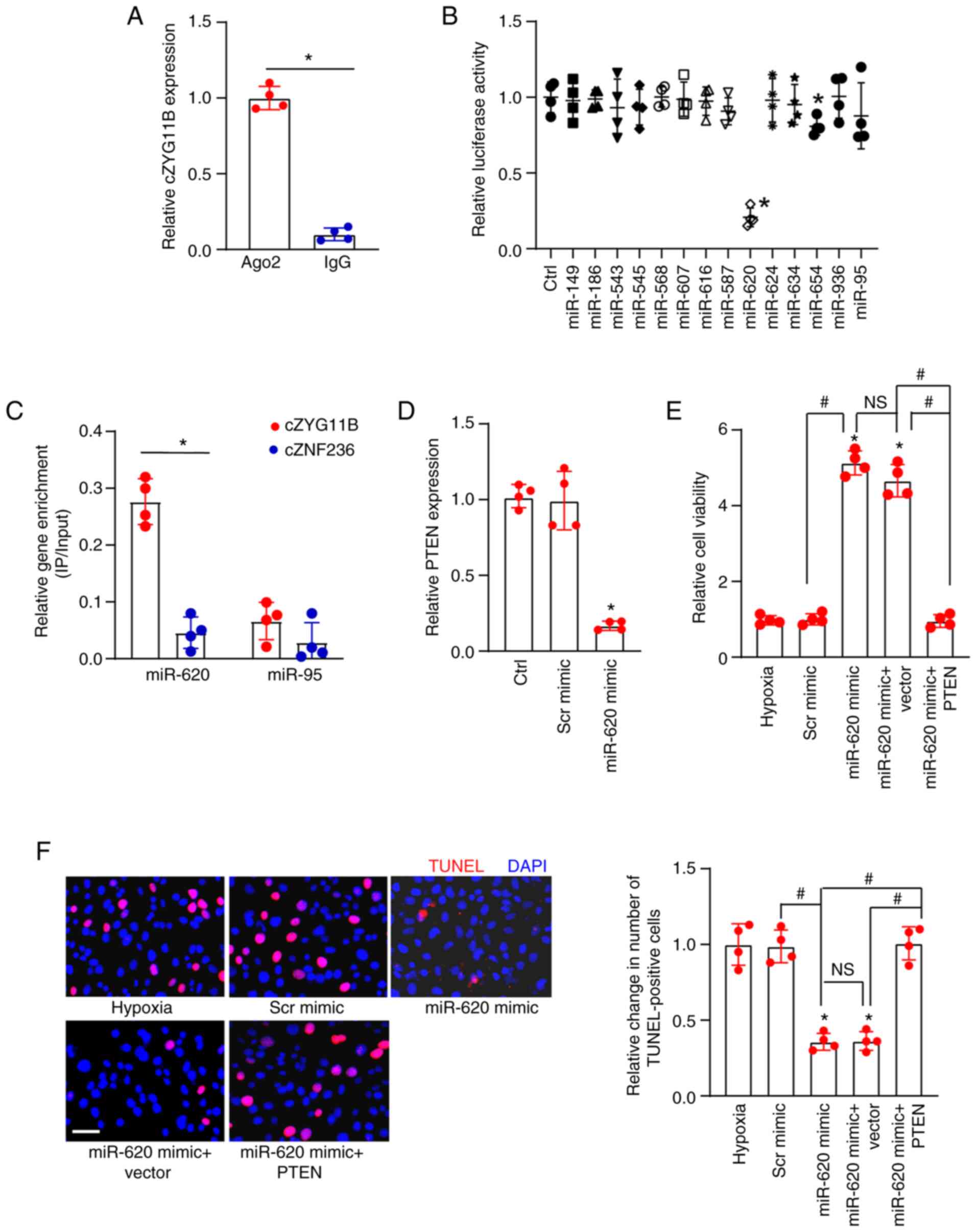 | Figure 6cZYG11B regulates RGC function by
acting as a miR-620 sponge. (A) Total cellular fractions were
isolated from RGCs and immunoprecipitated using Ago2 or IgG
antibody. The amount of cZYG11B in the IP was determined by
RT-qPCR. *P<0.05 as indicated. (B) RGCs were
transfected with pGL3-Basic (Ctrl) or Luc-cZYG11B with various
miRNA mimics and pRL-TK vector (internal transfection control).
Luciferase activity was detected at 24 h post-transfection using
the Dual-Luciferase Reporter Assay kit. *P<0.05 vs.
Ctrl. (C) Biotinylated miR-620 or miR-95 were transfected into
RGCs. After streptavidin capture, the amount of cZYG11B and cZNF236
(negative control) in the input and bound fractions were detected
by RT-qPCR. The relative IP/input ratios were plotted.
*P<0.05 as indicated. (D) RGCs were transfected with
Scr mimic, miR-620 mimic or were untreated. RT-qPCR was conducted
to detect PTEN expression. *P<0.05 vs. Ctrl. (E and
F) RGCs were left untreated or transfected with Scr mimic, miR-620
mimic, miR-620 mimic + pcDNA 3.1 (Vector) or PTEN-pcDNA 3.1 (PTEN)
for 12 h. The medium was replaced and cells were cultured for an
additional 12 h at 37°C. These groups were then exposed to 1%
O2 to mimic hypoxic stress for 24 h at 37°C. (E) CCK-8
assays were conducted to detect the viability of RGCs. (F) TUNEL
staining assays and semi-quantification analysis were conducted to
detect the apoptosis of RGCs. Scale bar, 50 µm.
*P<0.05 vs. hypoxia group; #P<0.05 as
indicated; NS, no significant difference. All significant
differences were evaluated by (A and C) Student's t-test or (B,
D-F) one-way ANOVA followed by the post hoc Bonferroni test. n=4.
Ctrl, control; cZYG11B, circZYG11B; IP, immunoprecipitated; miR,
microRNA; RGC, retinal ganglion cell; RT-qPCR, reverse
transcription-quantitative PCR; Scr, scrambled. |
RT-qPCR revealed that transfection with a miR-620
mimic led to increased expression of miR-620 but decreased
expression of PTEN (Figs. S1 and
6D). The present study then
investigated the role of miR-620 in RGC biology in vitro.
All experimental groups were exposed to 1% O2 to mimic
hypoxic stress. The non-transfected group was taken as the control
group (hypoxia group). CCK-8 and TUNEL assays demonstrated that
compared with in the control group, transfection with the miR-620
mimic, but not Scr mimic, led to increased cell viability and
reduced number of TUNEL-positive cells, thus suggesting that the
miR-620 mimic exerted protective effects on RGC function (Fig. 6E and F). Western blotting
demonstrated that transfection with pcDNA3.1-PTEN increased the
protein expression levels of PTEN in RGCs (Fig. S2). Notably, overexpression of
PTEN could interrupt the protective effects of miR-620 on RGC
function (Fig. 6E and F).
Collectively, these results suggested that circZYG11B may regulate
RGC function via the circZYG11B/miR-620/PTEN signaling axis.
Clinical relevance of circZYG11B-mediated
signaling in I/R-related ocular disease
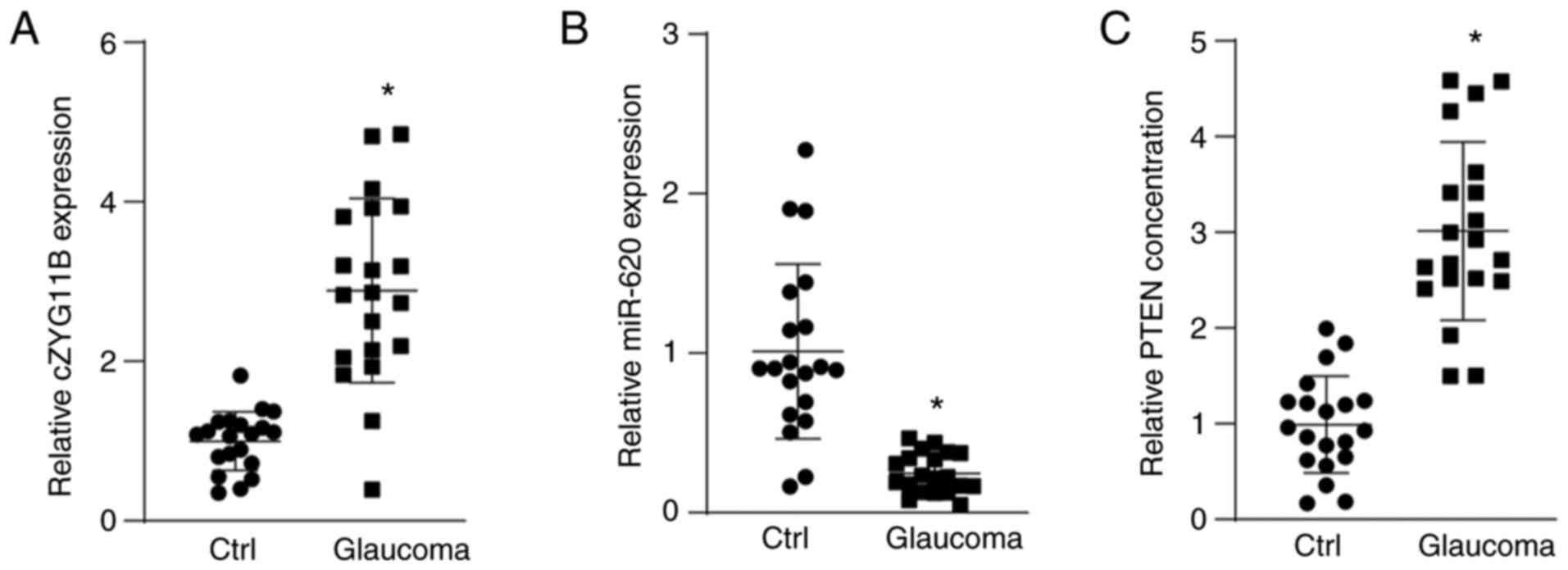
The present study also investigated the clinical
significance of circZYG11B-mediated signaling. Acute angle-closure
glaucoma is an I/R-related ocular disease (23). AH samples were collected from
patients with glaucoma (n=20) and patients with cataracts (control;
n=20). circZYG11B expression was significantly higher in the AH
samples from patients with glaucoma compared with that in control
patients (Fig. 7A). By contrast,
miR-620 expression was significantly downregulated in the AH
samples from patients with glaucoma compared with control patients
(Fig. 7B). The concentrations of
PTEN in AH samples were determined using ELISA. The results
revealed that PTEN concentration was markedly increased in the AH
samples of patients with glaucoma compared with the control
patients, showing a similar pattern as circZYG11B (Fig. 7C). Collectively, these results
suggested that circZYG11B-mediated signaling is potentially
involved in I/R-related ocular disease.
Discussion
Retinal ischemia is a major cause of vision
impairment and a common pathological basis of several ocular
diseases, including glaucoma, diabetic retinopathy and central
retinal artery occlusion (25).
Prevention of I/R injury could slow the development of retinal
neurodegeneration (26). The
present study demonstrated that silencing of circZYG11B could
reduce retinal reactive gliosis and decrease RGC injury in
vivo. Knockdown of circZYG11B could also alleviate hypoxic
stress- and oxidative stress-induced RGC injury. The present study
provides a novel circRNA-mediated mechanism underlying retinal
ischemic diseases.
Retinal I/R injury is involved in a wide spectrum of
pathological conditions, such as retinal neurodegeneration and
retinal vascular dysfunction (25). Previous studies have shown that
this process is regulated by a complex regulatory network,
including coding RNAs and non-coding RNAs (27-29). circRNAs are a novel class of
endogenous non-coding RNA generated by back-splicing, which have
emerged as novel regulators of gene expression and are known to be
involved in the pathogenesis of diabetic retinopathy, glaucoma and
central retinal artery occlusion (30-33). Therefore, circRNAs have been
highlighted as promising biomarkers for the diagnosis and
assessment of disease progression and prognosis. The present study
demonstrated that circZYG11B was significantly upregulated during
retinal neurodegeneration in vivo and in vitro. Acute
angle-closure glaucoma is an I/R-related ocular disease, and in the
present study, circZYG11B expression levels were significantly
upregulated in the AH samples of patients with glaucoma compared
with those in the control patients with cataracts.
circZYG11B-mediated signaling was also dysregulated in I/R-related
ocular disease. Thus, circZYG11B may be considered a promising
biomarker for the diagnosis and prognosis of retinal I/R-related
diseases.
I/R-related retinal diseases are major causes of
vision impairment worldwide and there are currently no available
treatments (34). Oxidative
stress and hypoxic stress are the key drivers of disease
progression and RGC injury during retinal I/R (35). I/R injury can lead to the
production of reactive oxygen species, inflammation and retinal
injury, including vascular injury and neurodegeneration. Retinal
neurons, particularly RGCs, are highly sensitive to retinal
ischemia (25,36). In response to hypoxic or oxidative
stress, silencing of circ-ZYG11B could protect RGCs against the
relevant injuries in vitro. Moreover, knockdown of
circZYG11B could alleviate I/R injury-induced retinal
neurodegeneration in vivo. Thus, circZYG11B silencing may be
a promising neuroprotective strategy for the treatment of
I/R-related ocular diseases.
Previous studies have shown that circRNAs generated
from exons often reside in the cytoplasm and may act as miRNA
sponges (37,38). The present study demonstrated that
circ-ZYG11B could bind to miR-620 and regulate RGC function by
acting as a miRNA sponge. circZYG11B acted as a binding platform
for Ago2 and miR-620. Notably, PTEN was verified as a target gene
of miR-620; PTEN has been reported to act as a mediator of
apoptosis and to have an important role in neurons (39,40). In hippocampal cells, knockdown of
PTEN has been shown to significantly protect against
staurosporine-induced apoptosis as shown, by decreased reactive
oxygen species production, reduced release of cytochrome c
and decreased activation of caspase 9 (39). PTEN is normally localized in the
cytosol, and can be recruited to specific microdomains of the
plasma membrane and contribute to the development of
lactacystin-induced neuronal apoptosis (40). In prostate cancer cells, PTEN was
able to suppress constitutive Akt activation and enhance cell
apoptosis. PTEN could also sensitize cells to death
receptor-mediated apoptosis and non-receptor mediated apoptosis
(41). Overexpression of PTEN
could also sensitize prostate cells to cell death induced by
staurosporine, doxorubicin and vincristine. By contrast, loss of
PTEN could enhance the mRNA and protein expression levels of Bcl-2,
an anti-apoptotic protein, leading to decreased death in prostate
cancer cells (42). Therefore,
circZYG11B could act as a sponge for miR-620, thus releasing the
suppressive effect of miR-620 on PTEN. This regulatory mechanism
could provide a novel insight into RGC injury and I/R-induced
retinal neurodegeneration.
In conclusion, the present study investigated the
expression pattern of circZYG11B and the relevant mechanism in
I/R-induced retinal neurodegeneration. The results revealed that
circZYG11B was increased in retinal neurodegeneration in
vivo and in vitro. By contrast, silencing of circZYG11B
could protect against I/R-induced RGC injury and alleviate
I/R-induced retinal neurodegeneration. The present study provides a
novel perspective on the mechanism of retinal I/R injury and a
potential target for treating I/R-related ocular diseases.
Supplementary Data
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
BY and JLD designed the study. CM, MDY, ZHS and XYH
conducted the experiments, and participated in the design,
statistics, drafting and revising of the manuscript. CM, MDY and
ZHS analyzed the data. All authors read and approved the final
manuscript. BY and JLD confirm the authenticity of all the raw
data.
Ethics approval and consent to
participate
The animals were maintained according to the
guidelines of the Care and Use of Laboratory Animals (published by
the National Institutes of Health) (43) and handled according to the ARVO
Statement for the Use of Animals in Ophthalmic and Vision Research
(44). Animal experiments were
approved by the Animal Care and Use Committee of Eye & ENT
Hospital. Clinical experiments were approved by the Ethics
Committee of Eye & ENT hospital. Written informed consent was
received from the patients involved.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was generously supported by grants from the
National Natural Science Foundation of China (grant no.
81470594).
References
|
1
|
Piano I, Di Paolo M, Corsi F, Piragine E,
Bisti S, Gargini C and Di Marco S: Retinal neurodegeneration:
Correlation between nutraceutical treatment and animal model.
Nutrients. 13:7702021. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Usategui-Martin R and Fernandez-Bueno I:
Neuroprotective therapy for retinal neurodegenerative diseases by
stem cell secretome. Neural Regen Res. 16:117–118. 2021. View Article : Google Scholar :
|
|
3
|
Carrasco E, Hernández C, Miralles A,
Huguet P, Farrés J and Simó R: Lower somatostatin expression is an
early event in diabetic retinopathy and is associated with retinal
neurodegeneration. Diabetes Care. 30:2902–2908. 2007. View Article : Google Scholar
|
|
4
|
Carrasco E, Hernández C, de Torres I,
Farrés J and Simó R: Lowered cortistatin expression is an early
event in the human diabetic retina and is associated with apoptosis
and glial activation. Mol Vis. 14:1496–1502. 2008.PubMed/NCBI
|
|
5
|
Fukumoto M, Nakaizumi A, Zhang T, Lentz
SI, Shibata M and Puro DG: Vulnerability of the retinal
microvasculature to oxidative stress: Ion channel-dependent
mechanisms. Am J Physiol Cell Physiol. 302:C1413–C1420. 2012.
View Article : Google Scholar :
|
|
6
|
Kowluru RA, Engerman RL, Case GL and Kern
TS: Retinal glutamate in diabetes and effect of antioxidants.
Neurochem Int. 38:385–390. 2001. View Article : Google Scholar
|
|
7
|
Baltmr A, Duggan J, Nizari S, Salt TE and
Cordeiro MF: Neuroprotection in glaucoma-Is there a future role?
Exp Eye Res. 91:554–566. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Simó R, Stitt AW and Gardner TW:
Neurodegeneration in diabetic retinopathy: Does it really matter?
Diabetologia. 61:1902–1912. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Zelinger L and Swaroop A: RNA biology in
retinal development and disease. Trends Genet. 34:341–351. 2018.
View Article : Google Scholar :
|
|
10
|
Song J and Kim YK: Targeting non-coding
RNAs for the treatment of retinal diseases. Mol Ther Nucleic Acids.
24:284–293. 2021. View Article : Google Scholar :
|
|
11
|
Kristensen LS, Andersen MS, Stagsted LV,
Ebbesen KK, Hansen TB and Kjems J: The biogenesis, biology and
characterization of circular RNAs. Nat Rev Genet. 20:675–691. 2019.
View Article : Google Scholar
|
|
12
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Esteller M: Non-coding RNAs in human
disease. Nat Rev Genet. 12:861–874. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheung CY, Ikram MK, Chen C and Wong TY:
Imaging retina to study dementia and stroke. Prog Retin Eye Res.
57:89–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jiang Q, Su DY, Wang ZZ, Liu C, Sun YN,
Cheng H, Li XM and Yan B: Retina as a window to cerebral
dysfunction following studies with circRNA signature during
neurodegeneration. Theranostics. 11:1814–1827. 2021. View Article : Google Scholar :
|
|
16
|
Yao MD, Zhu Y, Zhang QY, Zhang HY, Li XM,
Jiang Q and Yan B: CircRNA expression profile and functional
analysis in retinal ischemia-reperfusion injury. Genomics.
113:1482–1490. 2021. View Article : Google Scholar
|
|
17
|
Pritchett K, Corrow D, Stockwell J and
Smith A: Euthanasia of neonatal mice with carbon dioxide. Comp Med.
55:275–281. 2005.PubMed/NCBI
|
|
18
|
Zhang J, Sio SW, Moochhala S and Bhatia M:
Role of hydrogen sulfide in severe burn injury-induced inflammation
in mice. Mol Med. 16:417–424. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bae GS, Park KC, Choi SB, Jo IJ, Choi MO,
Hong SH, Song K, Song HJ and Park SJ: Protective effects of
alpha-pinene in mice with cerulein-induced acute pancreatitis. Life
Sci. 91:866–871. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Dudekula DB, Panda AC, Grammatikakis I, De
S, Abdelmohsen K and Gorospe M: CircInteractome: A web tool for
exploring circular RNAs and their interacting proteins and
microRNAs. RNA Biol. 13:34–42. 2016. View Article : Google Scholar :
|
|
21
|
Jiang L, Luirink J, Kooijmans SA, van
Kessel KP, Jong W, van Essen M, Seinen CW, de Maat S, de Jong OG,
Gitz-François JF, et al: A post-insertion strategy for surface
functionalization of bacterial and mammalian cell-derived
extracellular vesicles. Biochim Biophys Acta Gen Subj.
1865:1297632021. View Article : Google Scholar
|
|
22
|
Witkos TM, Koscianska E and Krzyzosiak WJ:
Practical aspects of microRNA target prediction. Curr Mol Med.
11:93–109. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Li SY, Fu ZJ and Lo AC: Hypoxia-induced
oxidative stress in ischemic retinopathy. Oxid Med Cell Longev.
2012:4267692012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Osborne NN, Casson RJ, Wood JP, Chidlow G,
Graham M and Melena J: Retinal ischemia: Mechanisms of damage and
potential therapeutic strategies. Prog Retin Eye Res. 23:91–147.
2004. View Article : Google Scholar
|
|
26
|
Tezel G: Oxidative stress in glaucomatous
neurodegeneration: Mechanisms and consequences. Prog Retin Eye Res.
25:490–513. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wan P, Su W, Zhang Y, Li Z, Deng C, Li J,
Jiang N, Huang S, Long E and Zhuo Y: LncRNA H19 initiates
microglial pyroptosis and neuronal death in retinal
ischemia/reperfusion injury. Cell Death Differ. 27:176–191. 2020.
View Article : Google Scholar :
|
|
28
|
Ge Y, Zhang R, Feng Y and Li H: Mbd2
mediates retinal cell apoptosis by targeting the lncRNA
Mbd2-AL1/miR-188-3p/Traf3 axis in ischemia/reperfusion injury. Mol
Ther Nucleic Acids. 19:1250–1265. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ghafouri-Fard S, Shoorei H and Taheri M:
Non-coding RNAs participate in the ischemia-reperfusion injury.
Biomed Pharmacother. 129:1104192020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Cui G, Wang L and Huang W: Circular RNA
HIPK3 regulates human lens epithelial cell dysfunction by targeting
the miR-221-3p/PI3K/AKT pathway in age-related cataract. Exp Eye
Res. 198:1081282020. View Article : Google Scholar
|
|
31
|
Wu PC, Zhang DY, Geng YY, Li R and Zhang
YN: Circular RNA-ZNF609 regulates corneal neovascularization by
acting as a sponge of miR-184. Exp Eye Res. 192:1079372020.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Guo N, Liu XF, Pant OP, Zhou DD, Hao JL
and Lu CW: Circular RNAs: Novel promising biomarkers in ocular
diseases. Int J Med Sci. 16:513–518. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Liu C, Ge HM, Liu BH, Dong R, Shan K, Chen
X, Yao MD, Li XM, Yao J, Zhou RM, et al: Targeting
pericyte-endothelial cell crosstalk by circular RNA-cPWWP2A
inhibition aggravates diabetes-induced microvascular dysfunction.
Proc Natl Acad Sci USA. 116:7455–7464. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Biousse V, Nahab F and Newman NJ:
Management of acute retinal ischemia: Follow the guidelines!
Ophthalmology. 125:1597–1607. 2018. View Article : Google Scholar
|
|
35
|
Fortmann SD and Grant MB: Molecular
mechanisms of retinal ischemia. Curr Opin Physiol. 7:41–48. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Neufeld AH, Kawai Si, Das S, Vora S,
Gachie E, Connor JR and Manning PT: Loss of retinal ganglion cells
following retinal ischemia: The role of inducible nitric oxide
synthase. Exp Eye Res. 75:521–528. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kulcheski FR, Christoff AP and Margis R:
Circular RNAs are miRNA sponges and can be used as a new class of
biomarker. J Biotechnol. 238:42–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lavenniah A, Luu TD, Li YP, Lim TB, Jiang
J, Ackers-Johnson M and Foo RS: Engineered circular RNA sponges act
as miRNA inhibitors to attenuate pressure overload-induced cardiac
hypertrophy. Mol Ther. 28:1506–1517. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu Y, Hoell P, Ahlemeyer B and
Krieglstein J: PTEN: A crucial mediator of mitochondria-dependent
apoptosis. Apoptosis. 11:197–207. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Choy MS, Bay BH, Cheng HC and Cheung NS:
PTEN is recruited to specific microdomains of the plasma membrane
during lactacystin-induced neuronal apoptosis. Neurosci Lett.
405:120–125. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yuan XJ and Whang YE: PTEN sensitizes
prostate cancer cells to death receptor-mediated and drug-induced
apoptosis through a FADD-dependent pathway. Oncogene. 21:319–327.
2002. View Article : Google Scholar
|
|
42
|
Huang H, Cheville JC, Pan Y, Roche PC,
Schmidt LJ and Tindall DJ: PTEN induces chemosensitivity in
PTEN-mutated prostate cancer cells by suppression of Bcl-2
expression. J Biol Chem. 276:38830–38836. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
National Research Council (US): Committee
for the update of the guide for the care and use of laboratory
animals: Guide for the care and use of laboratory animals. 8th
edition. National Academies Press; Washington, DC: 2011
|
|
44
|
Seitz R, Hackl S, Seibuchner T, Tamm ER
and Ohlmann A: Norrin mediates neuroprotective effects on retinal
ganglion cells via activation of the Wnt/beta-catenin signaling
pathway and the induction of neuroprotective growth factors in
Muller cells. J Neurosci. 30:5998–6010. 2010. View Article : Google Scholar : PubMed/NCBI
|