Introduction
Epidemiological studies have confirmed that
low-to-moderate alcohol consumption can reduce the incidence of
cardiovascular diseases to a certain extent. However, excessive
alcohol consumption or even alcoholism is considered a significant
cause of cardiomyopathy (1). It
has been reported that the daily consumption of 90-120 g alcohol
for ~5-15 years can increase the risk of structural and functional
changes in the heart (2,3). Non-ischemic dilated cardiomyopathy
caused by long-term heavy alcohol intake is termed alcoholic
cardiomyopathy (ACM) (4). ACM is
characterized by myocardial hypertrophy and compensatory cardiac
systolic disorder (5,6). In recent years, the incidence of ACM
has gradually increased in China, with a mortality rate of 5.1
deaths/100,000 ACM cases (7). The
onset and progression of alcohol-induced myopathic changes is
caused by multiple factors, including mitochondrial damage, the
toxicity of ethanol and its metabolites, and the accumulation of
fatty acid ethyl esters, that appear to be the direct outcome of
ethanol metabolism, which are common features of acute ethanol
exposure (5,8). Therefore, identifying effective
treatment approaches for ethanol-induced myocardial injury is of
utmost importance.
Ginseng has been used as a tonic in China for
>2,000 years. Ginsenoside Rg1 (G-Rg1), one of the most
noteworthy components of ginseng, has been shown to exert several
pharmacological effects, such as antitumor, anti-inflammatory,
antioxidant and anti-diabetic effects (9-12).
It has been reported that G-Rg1 can attenuate alcoholic liver
injury. For example, a previous study demonstrated that G-Rg1
prevented reactive oxygen species production, mitochondrial damage
and hepatocyte apoptosis in a mouse model of chronic
ethanol-induced hepatitis (13).
Additionally, G-Rg1 has been found to attenuate inflammatory
responses and to protect mice against alcohol-induced liver injury
(14,15). G-Rg1 can also ameliorate heart
injury. Therefore, previous research has suggested that G-Rg1 can
attenuate myocardial ischemia and reperfusion injury in rats
(16), while it can also
alleviate cardiac hypertrophy and fibrosis to improve cardiac
decompensation induced by abdominal aortic constriction in rats
(17). However, to the best of
our knowledge, the role of G-Rg1 in alcohol-induced myocardial
injury has not been previously investigated.
Recently, the role of endoplasmic reticulum stress
(ERS) and autophagy in the pathogenesis of ACM have received
increasing attention. The study by Li and Ren (18) first demonstrated that ERS was
activated in the heart tissues of mice exposed to ethanol,
characterized by the increased expression of cardiac
inositol-requiring transmembrane kinase/endoribonuclease 1α,
eukaryotic translation initiation factor 2α (eIF2α) and C/EBP
homologous protein (CHOP) (18),
thus suggesting that ERS may be involved in the occurrence and
development of ACM. Additionally, the ERS inhibitor, sodium
4-phenylbutyrate, has been shown to markedly attenuate
ethanol-induced cardiomyocyte injury (19), while another study demonstrated
that ERS induced by chronic alcohol intake was alleviated by
aldehyde dehydrogenase 2 upregulation (20). Furthermore, another study revealed
that ethanol-induced autophagy was suppressed by schisandrin B to
prevent acute alcohol-induced heart injury (21). Additionally, hydrogen sulfide has
been found to attenuate myocardial fibrosis in mice with ACM by
attenuating autophagy (22). A
previous study also indicated that G-Rg1 suppressed autophagy and
ERS to reduce doxorubicin-induced cardiotoxicity in mice (23). It has also been shown that G-Rg1
attenuates aldosterone-induced autophagy in NRK-52E cells (24) and high-fat diet-induced ERS in
obese rats (25). Therefore, it
was hypothesized that G-Rg1 may also regulate autophagy and ERS to
alleviate alcohol-induced myocardial cell injury.
The present study was performed to examine whether
G-Rg1 can attenuate alcohol-induced autophagy and ERS in myocardial
cells. In addition, the molecular mechanisms underlying
alcohol-induced autophagy, ERS and G-Rg1-mediated therapy were also
investigated.
Materials and methods
Cell culture and alcohol induction
H9c2 cells (cat. no. CL-0089; Procell Life Science
& Technology) were cultured in DMEM (Procell Life Science &
Technology) supplemented with 10% FBS (HyClone; Cytiva), 100 U/ml
penicillin G (HyClone; Cytiva) and 100 U/ml streptomycin (HyClone;
Cytiva) at 37°C in a humidified incubator with 5% CO2.
Briefly, following treatment with various concentrations of G-Rg1
(0, 10, 20 and 40 µM; Shanghai Kanglang Biotechnology Co.,
Ltd.) for 2 h (26-28), the H9c2 cells were co-stimulated
with 200 mmol/l ethanol (Sigma Technology China) for 24 h. When
necessary, the cells were pre-incubated with 1 mM AICAR (Selleck,
USA), an adenosine 5′-monophosphate-activated protein kinase (AMPK)
agonist, for 1 h or 10 µM CCT020312 (Selleckchem), a protein
kinase R (PKR)-like endoplasmic reticulum (ER) kinase (PERK)
agonist, for 2 h.
Cell Counting Kit-8 (CCK-8) assay
The viability of the H9c2 cells was assessed using a
CCK-8 assay kit (Dojindo Molecular Technologies, Inc.). Briefly,
the H9c2 cells were seeded in 96-well plates at a density of
5×103 cells/well and cultured for 24 h. After the
indicated treatments, each well was supplemented with 10 µl
CCK-8 solution, followed by incubation for an additional 2 h at
37°C in an incubator with 5% CO2. The absorbance at a
wavelength of 450 nm was measured using a microplate reader (Thermo
Fisher Scientific, Inc.).
Flow cytometric analysis
H9c2 cell apoptosis was measured using flow
cytometry (Beckman Coulter, Inc.) with an Annexin V-fluorescein
isothiocyanate/propidium iodide (Annexin V-FITC/PI) apoptosis
detection kit (Beijing Biosea Biotechnology Co., Ltd.). The H9c2
cells were seeded in a six-well plate at a density of
1×105 cells/well and were then treated as indicated.
Subsequently, H9c2 cells were washed with ice-cold PBS
(MilliporeSigma), re-suspended in binding buffer and mixed with 5
µl Annexin V-FITC and 10 µl PI. Finally, cell
apoptosis was analyzed using a flow cytometer (Beckman Coulter,
Inc.).
Detection of LDH and caspase-3
Following the indicated treatments, H9c2 cells were
lysed in RIPA lysis buffer (Beyotime Institute of Biotechnology),
followed by centrifugation at 600 × g for 5 min at 4°C to obtain
the supernatant. The levels of LDH and caspase-3 in the culture
supernatant were measured using the corresponding LDH (cat. no.
ab102526) and caspase-3 (cat. no. ab39401) assay kits from Abcam
based on the colorimetric method using the FLUOstar®
Omega Microplate Reader (BMG Labtech GmbH).
Western blot analysis
Total proteins were extracted from the H9c2 cells
using RIPA lysis buffer. The protein concentration was quantified
using the BCA method. Equal amounts of protein extracts (30
µg) were subjected to 10% SDS-PAGE and were then transferred
onto PVDF membranes. Subsequently, the membranes were blocked with
5% skimmed milk at room temperature for 1.5 h and were then
incubated with primary antibodies against Bcl-2 associated
X-protein (Bax; cat. no. ab32503; 1/1,000; Abcam), B-cell lymphoma
2 (Bcl-2; cat. no. ab196495; 1/1,000; Abcam), light chain 3
(LC3)-II/I (cat. no. ab192890; 1/1,000; Abcam), Beclin-1 (cat. no.
ab207612; 1/1,000; Abcam), p62 (cat. no. ab109012; 1/10,000;
Abcam), phosphorylated (p)-PERK (cat. no. #3179; 1/1,000; Cell
Signaling Technology), PERK (cat. no. ab229912; 1/1,000; Abcam),
p-eIF2α (cat. no. #3398; 1/1,000; Cell Signaling Technology), eIF2α
(cat. no. #5324; 1/1,000; Cell Signaling Technology), activating
transcription factor 4 (ATF4; cat. no. #11815; 1/1,000; Cell
Signaling Technology), CHOP (cat. no. 15204-1-AP; 1/1,000;
Proteintech Group, Inc.), caspase-12 (cat. no. ab62484; 1/1,000;
Abcam), p-AMPK (cat. no. ab133448; 1/1,000; Abcam), AMPK (cat. no.
ab207442; 1/1,000; Abcam), p-mammalian target of rapamycin (mTOR;
cat. no. ab109268; 1/1,000; Abcam), mTOR (cat. no. ab134903;
1/10,000; Abcam) and GAPDH (cat. no. ab181602; 1/10,000; Abcam) at
4°C overnight. Following washing three times with PBS/Tween-20, the
membranes were incubated with the corresponding anti-rabbit
horseradish peroxidase-conjugated secondary antibody (cat. no.
ab6721; 1/2,000; Abcam) at room temperature for 1 h. The bands were
visualized using an ECL detection kit and their intensity was
analyzed using ImageJ 1.8.0 software (National Institutes of
Health).
Green fluorescent protein (GFP)-LC3
assay
Upon reaching 90% confluency, the cells were
transfected with GFP-LC3 plasmid (Hanbio Biotechnology Co., Ltd.)
or GFP control vector (Hanbio Biotechnology Co., Ltd.) using
Lipofectamine® 2000, followed by incubation for 48 h at 37°C. The
volume of virus was 20 µl. Subsequent experiments were
implemented 48 h post-transfection. Following treatment with 0, 10,
20 and 40 µM G-Rg1 and 200 mmol/l ethanol with the presence
or absence of 1 mM AICAR or 10 µM CCT020312 and washing with
ice-cold PBS, the H9c2 cells were fixed with 4% paraformaldehyde
(Shanghai Macklin Biochemical Co., Ltd.) for 30 min at room
temperature and counterstained with 4′,6-diamidino-2-phenylindole
(DAPI; MilliporeSigma) for 10 min at room temperature. Finally, the
cells were observed under a fluorescence microscope (LSM 510 META,
Zeiss GmbH).
Immunofluorescence staining
Following the indicated treatment with 0, 10, 20 and
40 µM G-Rg1 and 200 mmol/l ethanol with the presence or
absence of 1 mM AICAR or 10 µM CCT020312, the H9c2 cells
were seeded into a 24-well plate and fixed with 4% paraformaldehyde
for 20 min at room temperature. Subsequently, the H9c2 cells were
incubated with a primary antibody against CHOP (cat. no.
15204-1-AP; 1/500; Proteintech Group, Inc.) at 4°C overnight. The
following day, the H9c2 cells were incubated with the Cy3-labelled
secondary antibody IgG (1:100; Sigma-Aldrich; Merck KGaA) in the
dark for 1 h. Following staining with DAPI for 10 min at room
temperature, CHOP-positive H9c2 cells were observed under a
fluorescence microscope (Olympus Corporation).
Molecular docking analysis
The 3D structures of AMPK (PDB ID, 4ZHX) and PERK
(PDB ID, 4G34) were downloaded from the Protein Data Bank (PDB)
website (http://www.rcsb.org/) and saved in PDB
format. Subsequently, the 3D structures in PDB format were
converted into 'pdbqt' format using AutoDockTools 1.5.6. Finally,
molecular docking was conducted using Autodock (version 4.2) and
the results were visualized with Pymol 2.5.2 (https://pymol.org/2/) software.
Statistical analysis
All statistical analyses were performed using
GraphPad Prism 8.0.1. (GraphPad Software, Inc.). The data were
compared using one-way ANOVA followed by Tukey's post hoc test. All
experiments were performed in triplicate. Data are expressed as the
mean ± standard deviation (SD). P<0.05 was considered to
indicate a statistically significant difference.
Results
G-Rg1 enhances the viability of
ethanol-stimulated H9c2 cells
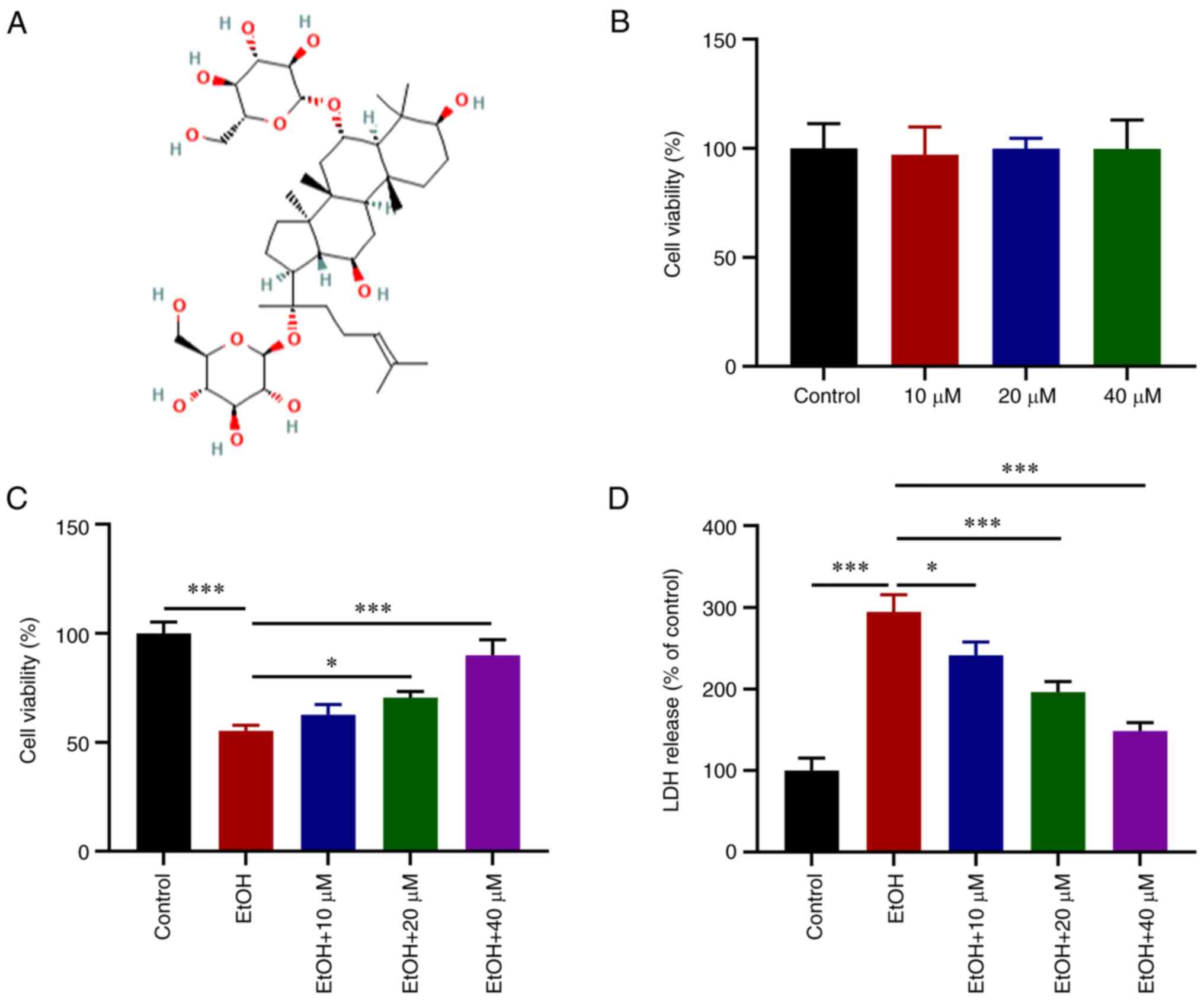
The chemical structural formula of G-Rg1 is
presented in Fig. 1A. Following
treatment of the H9c2 cells with various concentrations of G-Rg1, a
CCK-8 assay was performed. The results revealed that G-Rg1 alone
had no obvious effect on H9c2 cell viability (Fig. 1B). However, the viability of the
H9c2 cells exposed to ethanol was increased following treatment
with various concentrations of G-Rg1, and in particular at 40
µM (Fig. 1C). LDH
production was also increased in the ethanol-stimulated H9c2 cells,
which was then reduced following treatment with G-Rg1 (Fig. 1D).
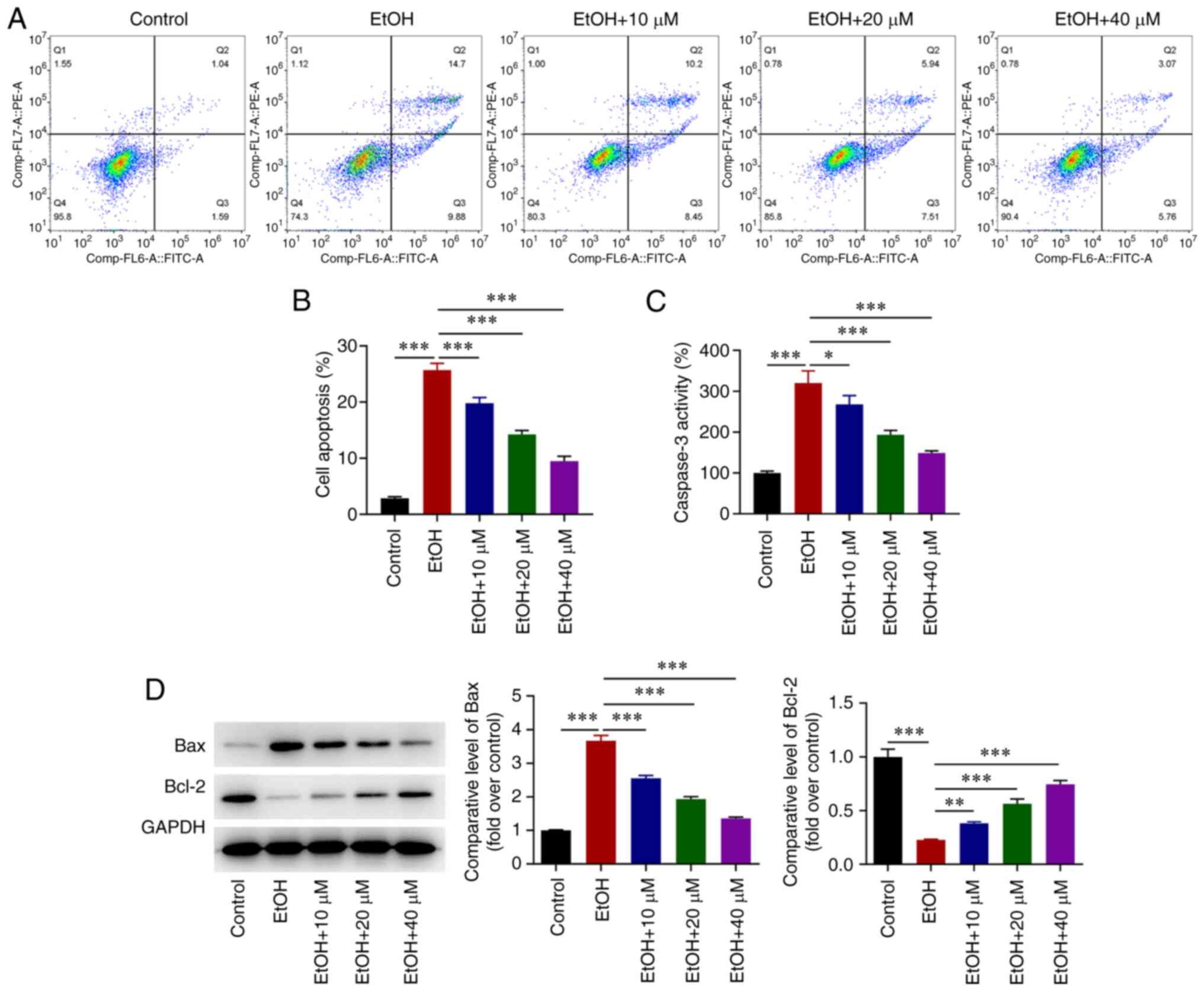
G-Rg1 inhibits the apoptosis of
ethanol-stimulated H9c2 cells
Flow cytometric analysis revealed that the apoptosis
of H9c2 cells was enhanced by stimulation with ethanol. However,
G-Rg1 gradually suppressed the apoptotic rate of the H9c2 cells in
a concentration-dependent manner, and in particular, at the G-Rg1
concentrations between 10 and 40 µM (Fig. 2A and B). Consistent with these
results, caspase-3 activity and Bax expression were increased,
while Bcl-2 was downregulated in the ethanol-stimulated H9c2 cells;
these effects were reversed by G-Rg1 (Fig. 2C and D).
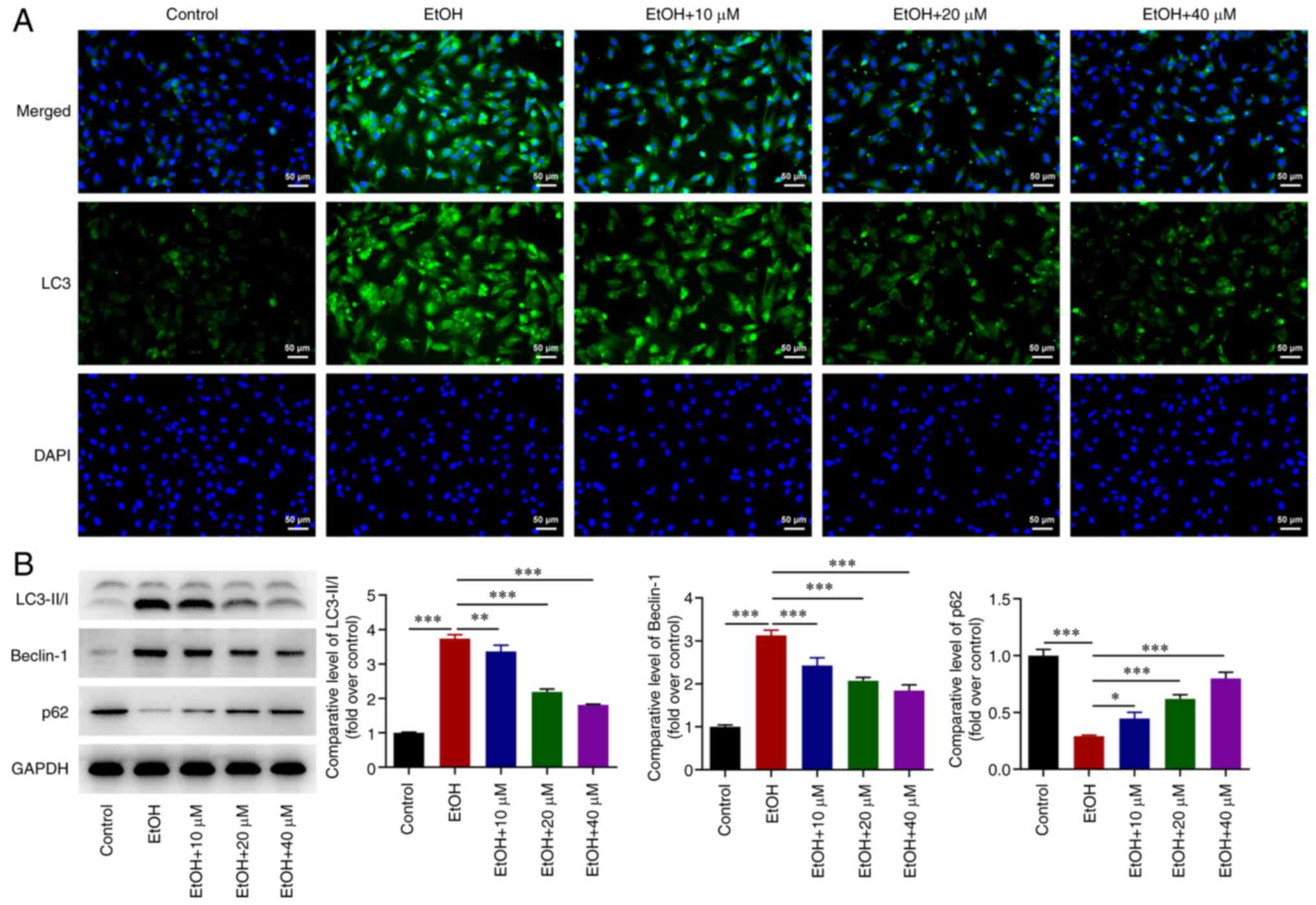
G-Rg1 inhibits the autophagy of
ethanol-stimulated H9c2 cells
Further experiments demonstrated that LC3 expression
was upregulated in ethanol-stimulated H9c2 cells. However, the
effect of ethanol on LC3 upregulation was abrogated by G-Rg1
(Fig. 3A). Additionally, LC3-II/I
and Beclin-1 expression levels were upregulated, while the
expression of p62 was downregulated in ethanol-stimulated H9c2
cells. These effects were reversed following treatment of the
ethanol-stimulated H9c2 cells with G-Rg1 (Fig. 3B).
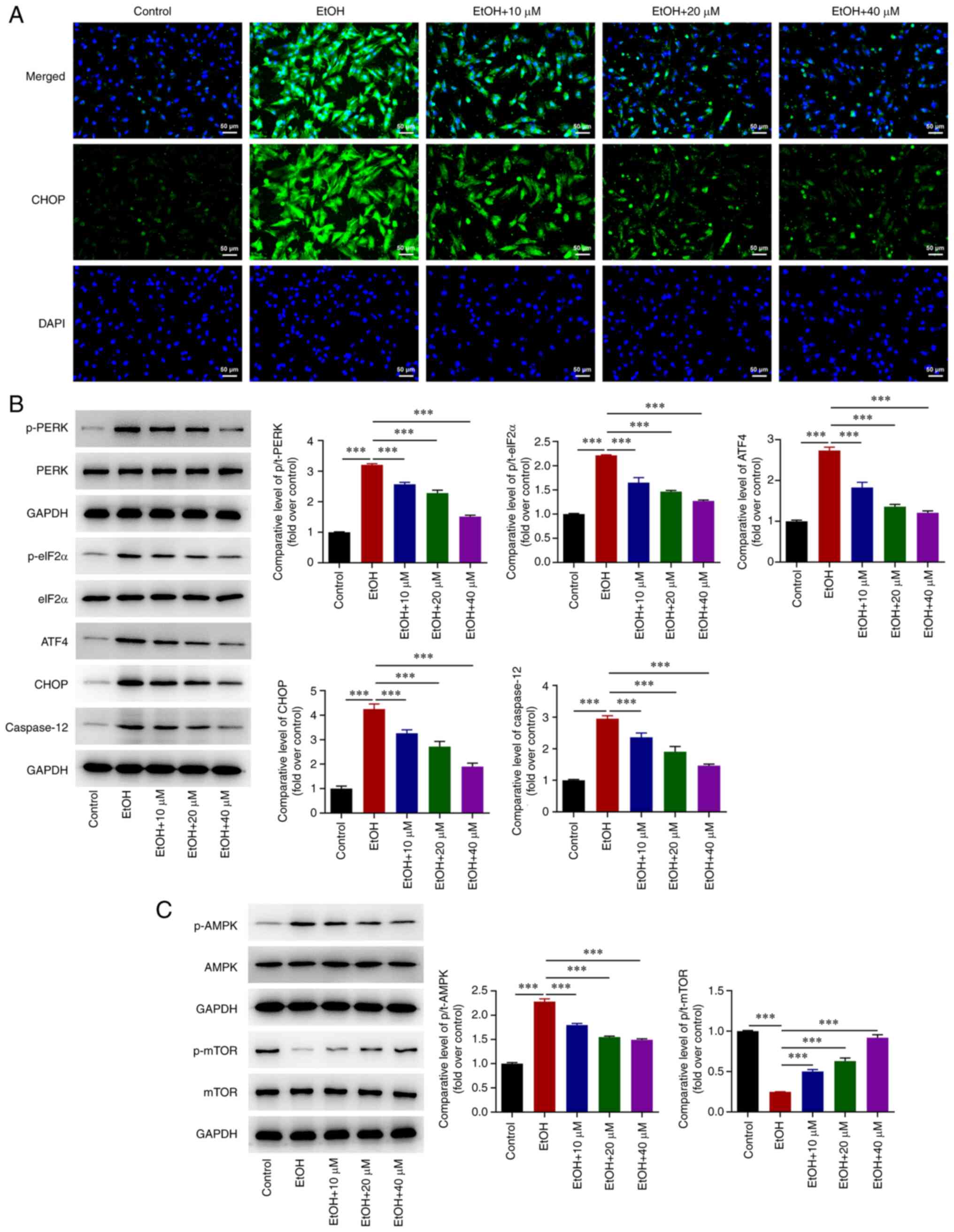
G-Rg1 inhibits ERS and modulates
AMPK/mTOR and PERK/ATF4/CHOP signaling in ethanol-stimulated H9c2
cells
Immunofluorescence staining demonstrated that the
expression of CHOP was increased in H9c2 cells stimulated with
ethanol, which was then inhibited by G-Rg1 treatment (Fig. 4A). The expression levels of
p-PERK, p-eIF2α, ATF4, CHOP and caspase-12 were enhanced in the
ethanol-stimulated H9c2 cells. However, G-Rg1 diminished the
effects of ethanol on ERS in H9c2 cells (Fig. 4B). In addition, ethanol promoted
the expression of p-AMPK and suppressed that of p-mTOR in H9c2
cells; these effects were also reversed by G-Rg1 (Fig. 4C).
Activation of AMPK/mTOR or PERK/ATF4/CHOP
signaling inhibits the viability and promotes the apoptosis of
ethanol-stimulated H9c2 cells treated with G-Rg1
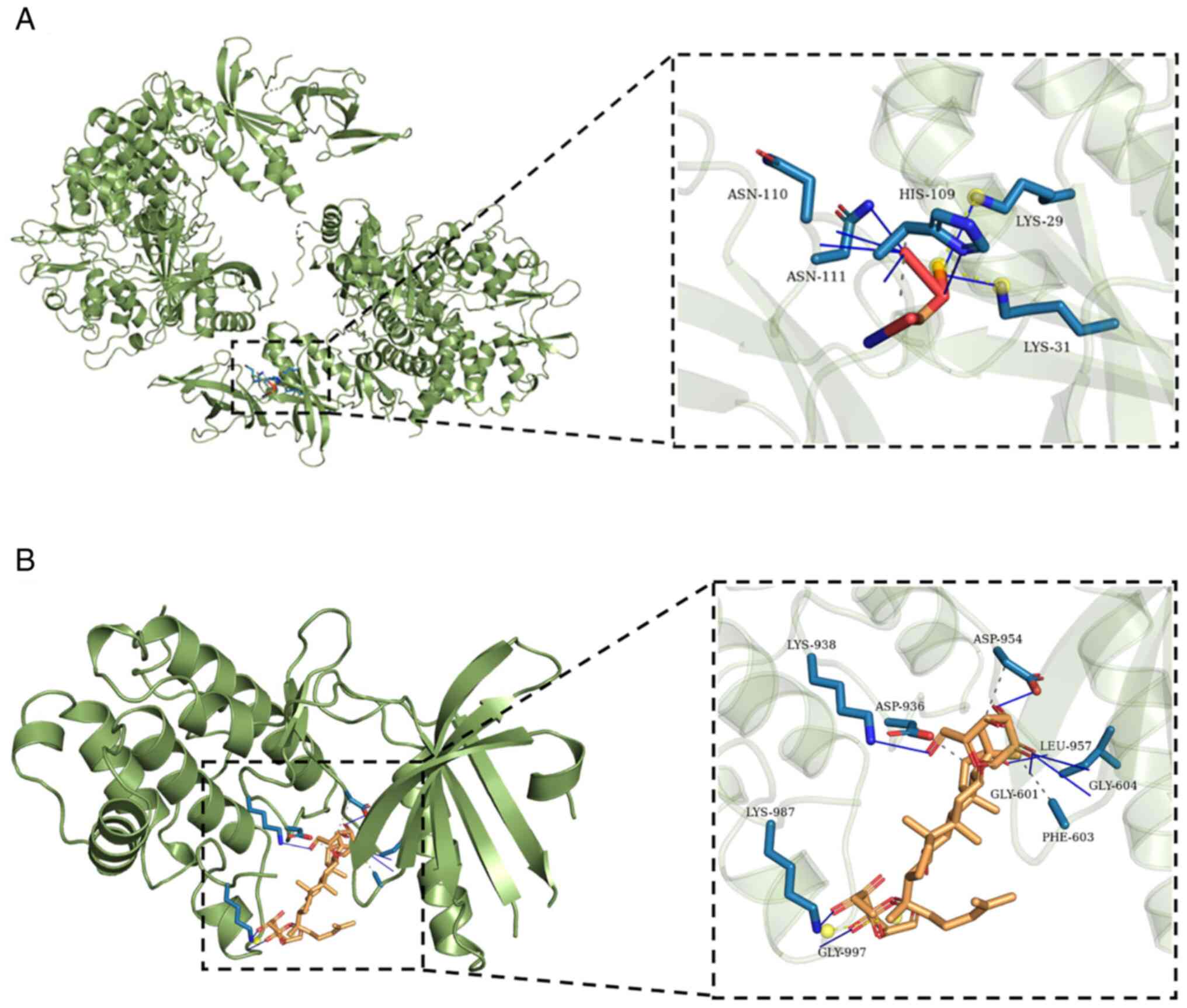
The results of molecular docking analysis revealed
that G-Rg1 exhibited strong affinity for AMPK, with a docking score
of -7.2 kcal/mol. The amino acid binding sites were as follows:
ASN-110, ASN-111, HIS-109, LYS-29 and LYS-31 (Fig. 5A). G-Rg1 also exhibited a strong
affinity for PERK, with a docking score of -6.4 kcal/mol. In this
case, the amino acid binding sites were as follows: LYS-938,
LYS-987, GLY-997, GLY-601, GLY-604, ASP-936, ASP-954, LEU-957 and
PHE-603 (Fig. 5B). In addition,
the results demonstrated that G-Rg1 increased the viability of
ethanol-stimulated H9c2 cells. This effect was abrogated following
co-treatment of the cells with AICAR or CCT020312 (Fig. 6A). The production of LDH was
decreased in ethanol-stimulated H9c2 cells treated with G-Rg1, an
effect which was also reversed following co-treatment with AICAR or
CCT020312 (Fig. 6B). In addition,
the apoptosis of ethanol-stimulated H9c2 cells was inhibited by
G-Rg1, and this effect was also reversed by AICAR or CCT020312
(Fig. 6C and D). Furthermore,
caspase-3 activity and Bax expression were decreased, and Bcl-2
expression was increased in the H9c2 cells in the ethanol + G-Rg1
group compared with the ethanol group. This effect was diminished
in the ethanol + G-Rg1 + AICAR or CCT020312 group (Fig. 6E and F).
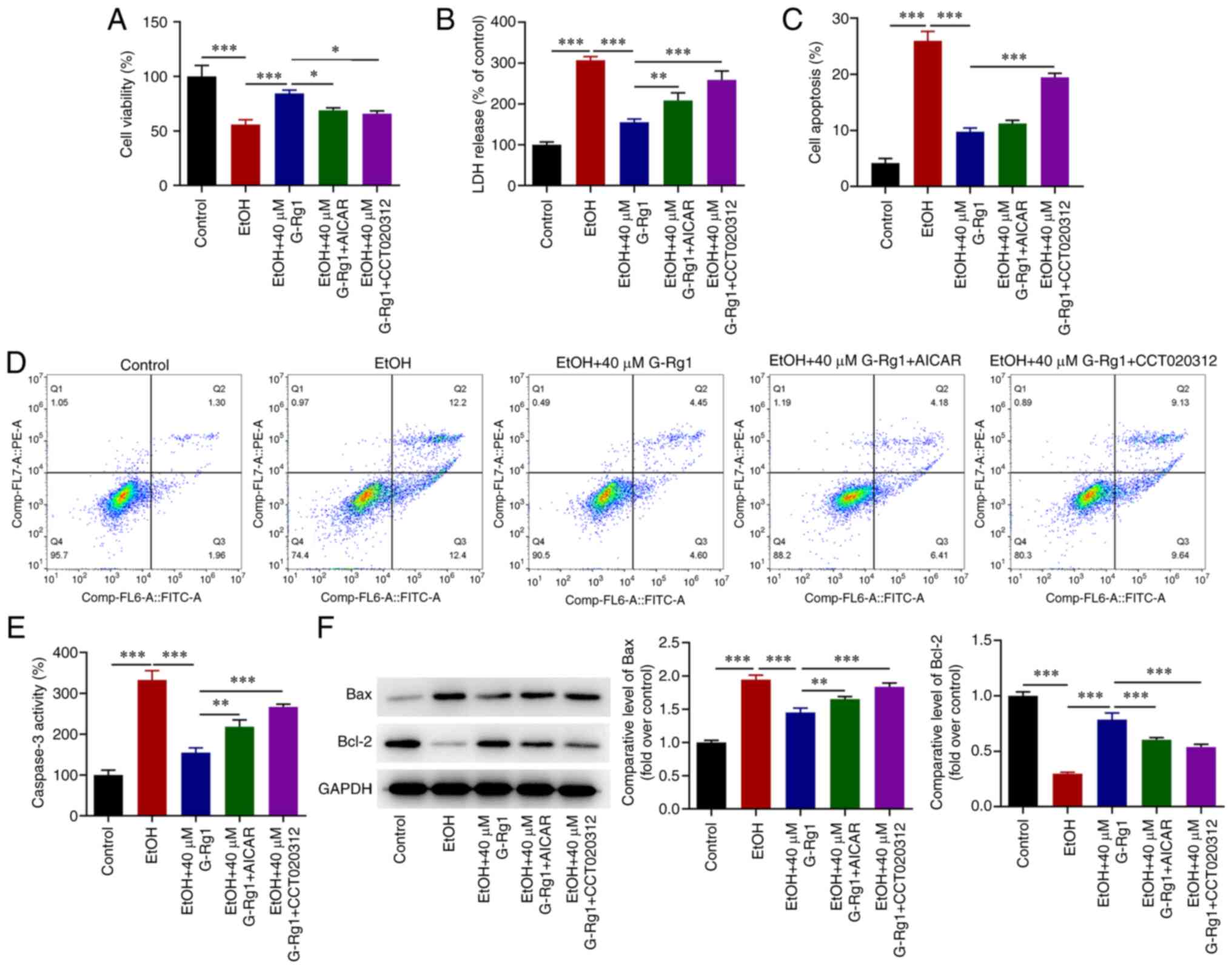 | Figure 6The activation of the AMPK/mTOR or
PERK/ATF4/CHOP signaling pathways inhibits the viability and
promotes the apoptosis of ethanol-stimulated H9c2 cells treated
with G-Rg1. (A) The viability of ethanol-stimulated H9c2 cells
co-treated with G-Rg1 and AICAR or CCT020312 was assessed using
Cell Counting Kit-8 assay. (B) LDH levels were measured in
ethanol-stimulated H9c2 cells co-treated with G-Rg1 and AICAR or
CCT020312 using the corresponding LDH kit. (C and D) The apoptotic
rate of ethanol-stimulated H9c2 cells co-treated with G-Rg1 and
AICAR or CCT020312 was determined using flow cytometric analysis.
(E) Caspase-3 activity in ethanol-stimulated H9c2 cells co-treated
with G-Rg1 and AICAR or CCT020312 was detected using the
corresponding kit. (F) The protein expression levels of Bax and
Bcl-2 were measured in ethanol-stimulated H9c2 cells co-treated
with G-Rg1 and AICAR or CCT020312 using western blot analysis. The
results are expressed as the mean ± SD. The experiments were
repeated for three times. *P<0.05,
**P<0.01 and ***P<0.001. G-Rg1,
ginsenoside Rg1; LDH, lactate dehydrogenase; AMPK, adenosine
5′-monophosphate-activated protein kinase; PERK, protein kinase R
(PKR)-like ER kinase; mTOR, mammalian target of rapamycin; CHOP,
C/EBP homologous protein; ATF4, activating transcription factor 4;
LDH, lactate dehydrogenase; EtOH, ethanol; Bax, Bcl-2 associated
X-protein; Bcl-2, B-cell lymphoma 2; EtOH, ethanol. |
Activation of the AMPK/mTOR or
PERK/ATF4/CHOP signaling pathways promotes autophagy and ERS in
ethanol-stimulated H9c2 cells treated with G-Rg1
Subsequently, the results revealed that G-Rg1
suppressed LC3 expression in the ethanol-stimulated H9c2 cells,
while LC3 expression was upregulated in the ethanol + G-Rg1 + AICAR
group (Fig. 7A). Additionally,
the LC3-II/I and Beclin-1 expression levels were downregulated, and
p62 expression was upregulated in the ethanol-stimulated H9c2 cells
treated with G-Rg1; these effects were reversed by AICAR treatment
(Fig. 7B). Furthermore, the
expression of CHOP was decreased in the ethanol + G-Rg1 group
compared with the ethanol group. However, treatment of the H9c2
cells with CCT020312 promoted CHOP expression in the ethanol +
G-Rg1 group compared with the ethanol group (Fig. 7C). Finally, the expression levels
of p-PERK, p-eIF2α, ATF4, CHOP and caspase-12 were decreased in the
ethanol + G-Rg1 group compared with the ethanol group. These
effects were all reversed by CCT020312 (Fig. 7D).
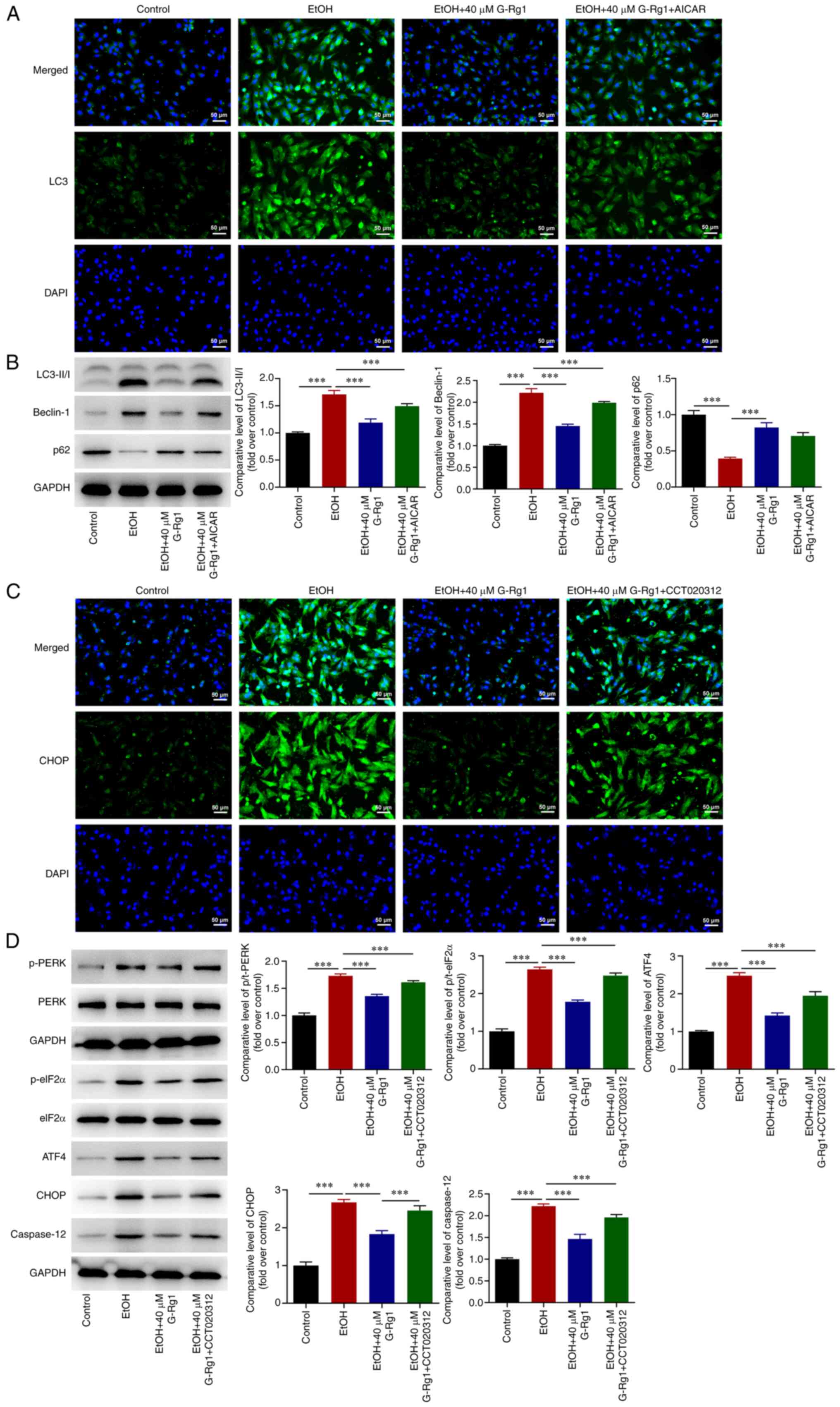 | Figure 7The activation of the AMPK/mTOR or
PERK/ATF4/CHOP signaling pathways promotes autophagy and ERS in
ethanol-stimulated H9c2 cells treated with G-Rg1. (A) The
expression of LC3 in ethanol-stimulated H9c2 cells co-treated with
G-Rg1 and AICAR was detected using GFP-LC3 assay. (B) The protein
expression levels of LC3-I, LC3-II, Beclin-1 and p62 were measured
in ethanol-stimulated H9c2 cells co-treated with G-Rg1 and AICAR
using western blot analysis. (C) The expression of CHOP was
evaluated in ethanol-stimulated H9c2 cells co-treated with G-Rg1
and CCT020312 using immunofluorescence staining. (D) The expression
levels of ERS-related proteins in ethanol-stimulated H9c2 cells
co-treated with G-Rg1 and CCT020312 were determined using western
blot analysis. The results are expressed as the mean ± SD. The
experiments were repeated three times. ***P<0.001.
G-Rg1, ginsenoside Rg1; ERS, endoplasmic reticulum stress; AMPK,
adenosine 5′-monophosphate-activated protein kinase; PERK, protein
kinase R (PKR)-like ER kinase; mTOR, mammalian target of rapamycin;
CHOP, C/EBP homologous protein; ATF4, activating transcription
factor 4; LC3, light chain 3; EtOH, ethanol. |
Discussion
ACM, a cardiac disease caused by chronic alcohol
consumption, is characterized by ventricular dilation and impaired
cardiac function (29). Patients
with ACM commonly have a history of excessive alcohol addiction for
>10 years and are characterized by diverse clinical
manifestations, such as cardiac insufficiency and arrhythmia, which
can be self-alleviated or cured following alcohol withdrawal
(30). Since uniform standards
for the outcome of ACM are lacking, there is a lack of reliable
treatment guidance (31).
Therefore, the identification of agents capable of preventing and
treating alcohol-induced myocardial injury is urgently
required.
The present study using ethanol-stimulated H9c2
cells to investigate the roles of G-Rg1 in alcohol-induced
myocardial injury; the same model has been used in previous studies
(32-34). There is accumulating evidence to
suggest that G-Rg1 protects against alcohol-induced liver injury
(13-15). In addition, it has been reported
that G-Rg1 exerts cardioprotective effects in several myocardial
injury models, including ischemia/reperfusion injury (16), adriamycin (doxorubicin)-induced
cardiotoxicity (23), diabetic
cardiomyopathy (35) and
sepsis-induced cardiac insufficiency (36). Consistent with the findings of
these previous studies, the present study demonstrated that G-Rg1
protected against alcohol-induced myocardial injury, as evidenced
by the promotion of the viability and the inhibition of the
apoptosis, autophagy and ERS of ethanol-stimulated H9c2 cells.
Long-term alcohol consumption can suppress cardiac
protein synthesis and accelerate protein degradation, thus
indicating that it can regulate catabolism and autophagy in
alcoholic heart diseases (5,37).
Of note, the expression levels of the autophagy-related markers,
LC3II and autophagy related 7, have been found to be increased in
the hearts of rats exposed to chronic alcohol, accompanied by a
reduced mTOR activity, a negative regulator of autophagy (38). Another study demonstrated that
3-methyladenine suppressed autophagy to improve alcohol-induced
myocardial injuries (39). It has
been reported that G-Rg1 is involved in the regulation of cell
autophagy. Therefore, G-Rg1 can suppress
hypoxia/reoxygenation-induced autophagy in H9c2 cells by
attenuating the activation of AMPKα, downregulating LC3II and
Beclin-1, and promoting mTOR activation (40). In addition, G-Rg1 has been shown
to markedly alleviate angiotensin II-induced podocyte autophagy via
the inactivation of the AMPK/mTOR/PI3K pathway (41). Herein, the expression levels of
the autophagy-related proteins, LC3-II and Beclin-1, were
increased, while those of p62 were decreased in the
ethanol-stimulated H9c2 cells compared with the control group.
However, the expression levels of the aforementioned proteins were
reversed following G-Rg1 treatment, thus suggesting that G-Rg1
attenuated autophagy, that was excessively activated in the
ethanol-stimulated H9c2 cells. In addition, ethanol also
upregulated p-AMPK and downregulated p-mTOR expression in H9c2
cells. This effect was also reversed by G-Rg1 treatment.
Furthermore, molecular docking analysis indicated that G-Rg1
displayed a strong affinity for AMPK, thus suggesting that G-Rg1
suppressed the AMPK/mTOR pathway to protect H9c2 cells against
ethanol-induced injury. To verify whether the cardioprotective
effect of G-Rg1 against ethanol was triggered by the inhibition of
autophagy, the cells were co-treated with AICAR, an AMPK agonist.
Therefore, the anti-autophagy effect of G-Rg1 on ethanol-stimulated
H9c2 cells was attenuated following co-treatment with AICAR.
ERS-induced apoptosis plays a critical role in the
pathogenesis of several cardiovascular diseases, such as cardiac
hypertrophy, heart failure and ischemic heart disease (42,43). To the best of your knowledge, the
findings of ERS or ERS-related events have not yet been found in
the tissues of humans with alcohol-induced myocardial injury.
Previous studies have demonstrated that ERS is activated in the
heart tissues of mice administered ethanol (18,20). However, whether ERS is involved in
ethanol-induced cardiomyocyte apoptosis remains elusive. Herein,
the results demonstrated that p-PERK, p-eIF2α, ATF4, CHOP and
caspase-12 expression levels were upregulated in ethanol-stimulated
H9c2 cells, thus suggesting that the PERK/ATF4/CHOP pathway was
involved in ethanol-induced H9c2 cell injury. More importantly,
treatment of the cells with G-Rg1 inhibited the expression of the
aforementioned ERS- and apoptosis-related proteins, while molecular
docking analysis revealed that G-Rg1 displayed a strong affinity
for PERK. To further clarify whether the cardioprotective effect of
G-Rg1 against ethanol-induced injury was mediated by ERS
inhibition, the cells were co-treated with the PERK agonist,
CCT020312. The results revealed that treatment with ethanol
promoted the ERS and apoptosis of the H9c2 cells. However,
co-treatment with CCT020312, reversed the anti-ERS effect of G-Rg1
on ethanol-stimulated H9c2 cells.
However, there are several limitations to the
present study which should be mentioned. The present study only
examined the effects of G-Rg1 on ethanol-induced myocardial injury
in an in vitro model, which may not be as clinically
relevant; thus, animal models need to be established in future
studies. Additionally, the lack of PERK and AMPK inhibitors and
other methods to detect the intracellular autophagy flow are also
limitations of the present study. Further studies are required in
the future to fully elucidate the effects and mechanisms of G-Rg1
on ethanol-induced myocardial injury.
In conclusion, the results of the present study
suggested that G-Rg1 improved the viability and inhibited
apoptosis, autophagy and ERS of ethanol-stimulated H9c2 cells via
the inactivation of the AMPK/mTOR and PERK/ATF4/CHOP signaling
pathways. Of note, AICAR or CCT020312 attenuated the
cardioprotective effects of G-Rg1 on ethanol-stimulated H9c2
cells.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ designed and conceived the study. GT and JL
conducted the experiments, and JL analyzed the experimental data.
GT drafted the manuscript, which was reviewed and edited by LZ. All
authors have read and approved the final version of the manuscript.
GT and LZ confirm the authenticity of all the raw data.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
Funding
The present study was supported by the PhD Start-up Fund of the
2022 Natural Science Foundation Project of Liaoning Province (grant
no. 2022-BS-321).
References
|
1
|
Guzzo-Merello G, Cobo-Marcos M,
Gallego-Delgado M and Garcia-Pavia P: Alcoholic cardiomyopathy.
World J Cardiol. 6:771–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
George A and Figuered VM: Alcoholic
cardiomyopathy: A review. J Card Fail. 17:844–849. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McNair P, Jones E, Truong Q and Singh H:
Incidental finding of single coronary artery in a patient with
alcoholic cardiomyopathy presenting as acute heart failure. Clin
Imaging. 42:224–227. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Naimi TS, Nelson DE and Brewer RD: The
intensity of binge alcohol consumption among U.S. adults. Am J Prev
Med. 38:201–207. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Piano MR and Phillips SA: Alcoholic
cardiomyopathy: Pathophysiologic insights. Cardiovasc Toxicol.
14:291–308. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Steiner JL and Lang CH: Etiology of
alcoholic cardiomyopathy: Mitochondria, oxidative stress and
apoptosis. Int J Biochem Cell Biol. 89:125–135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Manthey J, Imtiaz S, Neufeld M, Rylett M
and Rehm J: Quantifying the global contribution of alcohol
consumption to cardiomyopathy. Popul Health Metr. 15:202017.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Seiva FR, Amauchi JF, Rocha KK, Ebaid GX,
Souza G, Fernandes AA, Cataneo AC and Novelli EL: Alcoholism and
alcohol abstinence: N-acetylcysteine to improve energy expenditure,
myocardial oxidative stress, and energy metabolism in alcoholic
heart disease. Alcohol. 43:649–656. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim JH: Pharmacological and medical
applications of Panax ginseng and ginsenosides: A review for use in
cardiovascular diseases. J Ginseng Res. 42:264–269. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yu M, Yu X, Guo D, Yu B, Li L, Liao Q and
Xing R: Ginsenoside Rg1 attenuates invasion and migration by
inhibiting transforming growth factor-β1-induced epithelial to
mesenchymal transition in HepG2 cells. Mol Med Rep. 11:3167–3173.
2015. View Article : Google Scholar
|
|
11
|
Fan X, Zhang C, Niu S, Fan B, Gu D, Jiang
K, Li R and Li S: Ginsenoside Rg1 attenuates hepatic insulin
resistance induced by high-fat and high-sugar by inhibiting
inflammation. Eur J Pharmacol. 854:247–255. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fan X, Tao J, Zhou Y, Hou Y, Wang Y, Gu D,
Su Y, Jang Y and Li S: Investigations on the effects of
ginsenoside-Rg1 on glucose uptake and metabolism in insulin
resistant HepG2 cells. Eur J Pharmacol. 843:277–284. 2019.
View Article : Google Scholar
|
|
13
|
Yang C, He X, Zhao J and Huang W:
Hepatoprotection by Ginsenoside Rg1 in alcoholic liver disease. Int
Immunopharmacol. 92:1073272021. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Li J, Yang C, Zhang S, Liu S, Zhao L, Luo
H, Chen Y and Huang W: Ginsenoside Rg1 inhibits inflammatory
responses via modulation of the nuclear factor-κB pathway and
inhibition of inflammasome activation in alcoholic hepatitis. Int J
Mol Med. 41:899–907. 2018.
|
|
15
|
Gao Y, Chu SF, Xia CY, Zhang Z, Zhang S
and Chen NH: Rg1 Attenuates alcoholic hepatic damage through
regulating AMP-activated protein kinase and nuclear factor
erythroid 2-related factor 2 signal pathways. J Asian Nat Prod Res.
18:765–778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Pan C S, Yan L, Cui YC, Liu YY, Mu
HN, He K, Hu BH, Chang X, Sun K, et al: Ginsenoside Rg1 ameliorates
rat myocardial ischemia-reperfusion injury by modulating energy
metabolism pathways. Front Physiol. 9:782018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lu ML, Wang J, Sun Y, Li C, Sun TR, Hou XW
and Wang HX: Ginsenoside Rg1 attenuates mechanical stress-induced
cardiac injury via calcium sensing receptor-related pathway. J
Ginseng Res. 45:683–694. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li SY and Ren J: Cardiac overexpression of
alcohol dehydrogenase exacerbates chronic ethanol ingestion-induced
myocardial dysfunction and hypertrophy: Role of insulin signaling
and ER stress. J Mol Cell Cardiol. 44:992–1001. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang W, Liu T, Liu Y, Yu L, Yan X, Weng W,
Lu X and Zhang C: Astaxanthin attenuates alcoholic cardiomyopathy
via inhibition of endoplasmic reticulum stress-mediated cardiac
apoptosis. Toxicol Appl Pharmacol. 412:1153782021. View Article : Google Scholar
|
|
20
|
Li SY, Gilbert SA, Li Q and Ren J:
Aldehyde dehydrogenase-2 (ALDH2) ameliorates chronic alcohol
ingestion-induced myocardial insulin resistance and endoplasmic
reticulum stress. J Mol Cell Cardiol. 47:247–255. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tao Y, Zhou H, Huang L, Xu X, Huang Y, Ma
L, Li L, Yao X, Zhang R, Zhang Y, et al: Schisandrin B protects
against acute ethanol-induced cardiac injury by downregulating
autophagy via the NOX4/ROS pathway. Pharmacology. 106:177–188.
2021. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang B, Xiao T, Long J, Liu M, Li Z, Liu
S and Yang J: Hydrogen sulfide alleviates myocardial fibrosis in
mice with alcoholic cardiomyopathy by downregulating autophagy. Int
J Mol Med. 40:1781–1791. 2017.PubMed/NCBI
|
|
23
|
Xu ZM, Li CB, Liu QL, Li P and Yang H:
Ginsenoside Rg1 prevents doxorubicin-induced cardiotoxicity through
the inhibition of autophagy and endoplasmic reticulum stress in
mice. Int J Mol Sci. 19:36582018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang L, Mao N, Tan RZ, Wang HL, Wen J, Liu
YH, Furhad M and Fan JM: Ginsenoside Rg1 reduces
aldosterone-induced autophagy via the AMPK/mTOR pathway in NRK-52E
cells. Int J Mol Med. 36:518–526. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Yuan Z, Xiao-Wei L, Juan W, Xiu-Juan L,
Nian-Yun Z and Lei S: HIIT and MICT attenuate high-fat diet-induced
hepatic lipid accumulation and ER stress via the PERK-ATF4-CHOP
signaling pathway. J Physiol Biochem. 78:641–652. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yu H, Zhen J, Yang Y, Du J, Leng J and
Tong Q: Rg1 protects H9C2 cells from high
glucose-/palmitate-induced injury via activation of AKT/GSK-3β/Nrf2
pathway. J Cell Mol Med. 24:8194–8205. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li D, Wang J, Hou J, Fu J, Chang D,
Bensoussan A and Liu J: Ginsenoside Rg1 protects starving H9c2
cells by dissociation of Bcl-2-Beclin1 complex. BMC Complement
Altern Med. 16:1462016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lu D, Zhu LH, Shu XM, Zhang CJ, Zhao JY,
Qi RB, Wang HD and Lu DX: Ginsenoside Rg1 relieves tert-Butyl
hydroperoxide-induced cell impairment in mouse microglial BV2
cells. J Asian Nat Prod Res. 17:930–945. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shaaban A, Gangwani MK, Pendela VS and
Vindhyal MR: Alcoholic Cardiomyopathy. StatPearls. StatPearls
Publishing; Treasure Island, FL: 2022
|
|
30
|
Maisch B: Alcoholic cardiomyopathy: The
result of dosage and individual predisposition. Herz. 41:484–493.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhi H, Wang H and Ping F: Effect of
nursing intervention on patients with alcoholic cardiomyopathy
heart failure. Int J Cardiov Dis. 44:752017.
|
|
32
|
Tian G, Yu Y, Deng H, Yang L, Shi X and Yu
B: Empagliflozin alleviates ethanol-induced cardiomyocyte injury
through inhibition of mitochondrial apoptosis via a SIRT1/PTEN/Akt
pathway. Clin Exp Pharmacol Physiol. 48:837–845. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Xue Q, Zhang T, Zhu R, Qian Y, Dong X, Mo
L and Jiang Y: Inhibition of ceramide synthesis attenuates chronic
ethanol induced cardiotoxicity by restoring lysosomal function and
reducing necroptosis. Alcohol Alcohol. 58:164–174. 2023. View Article : Google Scholar
|
|
34
|
Chen Y, Zhu S, Lin Z, Zhang Y, Jin C, He
S, Chen X and Zhou X: Metformin alleviates ethanol-induced
cardiomyocyte injury by activating AKT/Nrf2 signaling in an
ErbB2-dependent manner. Mol Biol Rep. 50:3469–3478. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Yu H, Zhen J, Yang Y, Gu J, Wu S and Liu
Q: Ginsenoside Rg1 ameliorates diabetic cardiomyopathy by
inhibiting endoplasmic reticulum stress-induced apoptosis in a
streptozotocin-induced diabetes rat model. J Cell Mol Med.
20:623–631. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Z, Pan H, Zhang Y, Zheng Z, Xiao W,
Hong X, Chen F, Peng X, Pei Y, Rong J, et al: Ginsenoside-Rg1
attenuates sepsis-induced cardiac dysfunction by modulating
mitochondrial damage via the P2X7 receptor-mediated Akt/GSK-3β
signaling pathway. J Biochem Mol Toxicol. 36:e228852022. View Article : Google Scholar
|
|
37
|
Steiner JL and Lang CH: Alcoholic
cardiomyopathy: Disrupted protein balance and impaired
cardiomyocyte contractility. Alcohol Clin Exp Res. 41:1392–1401.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lang CH and Korzick DH: Chronic alcohol
consumption disrupts myocardial protein balance and function in
aged, but not adult, female F344 rats. Am J Physiol Regul Integr
Comp Physiol. 306:R23–R33. 2014. View Article : Google Scholar :
|
|
39
|
Guo R, Hu N, Kandadi MR and Ren J:
Facilitated ethanol metabolism promotes cardiomyocyte contractile
dysfunction through autophagy in murine hearts. Autophagy.
8:593–608. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang ZL, Fan Y and Liu ML: Ginsenoside
Rg1 inhibits autophagy in H9c2 cardiomyocytes exposed to
hypoxia/reoxygenation. Mol Cell Biochem. 365:243–250. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mao N, Tan RZ, Wang SQ, Wei C, Shi XL, Fan
JM and Wang L: Ginsenoside Rg1 inhibits angiotensin II-induced
podocyte autophagy via AMPK/mTOR/PI3K pathway. Cell Biol Int.
40:917–925. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kassan M, Galán M, Partyka M, Saifudeen Z,
Henrion D, Trebak M and Matrougui K: Endoplasmic reticulum stress
is involved in cardiac damage and vascular endothelial dysfunction
in hypertensive mice. Arterioscler Thromb Vasc Biol. 32:1652–1661.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Padilla J and Jenkins NT: Induction of
endoplasmic reticulum stress impairs insulin-stimulated vasomotor
relaxation in rat aortic rings: Role of endothelin-1. J Physiol
Pharmacol. 64:557–564. 2013.PubMed/NCBI
|