Introduction
Among malignant tumors within the female
reproductive system, ovarian cancer is recognized as the leading
cause of mortality, thereby representing a public health concern.
Among the various types of ovarian cancer, epithelial ovarian
cancer accounts for 90% of all diagnosed cases (1). The majority of patients with
epithelial ovarian cancer are diagnosed at advanced stages III or
IV, where the corresponding 5-year survival rates are 42 and 26%,
respectively (2). Despite
advancements in surgical interventions and the routine utilization
of chemotherapy and targeted therapeutics, the therapeutic efficacy
of these modalities remains limited. Consequently, there is need to
identify specific tumor markers and effective drug targets for
accurate diagnosis, treatment and monitoring of ovarian cancer.
Abnormal cell proliferation is a hallmark of
malignant tumors, and a notable association has been identified
between control of cell cycle progression and the initiation of
tumorigenesis (3). The PI3K/AKT
pathway constitutes a key intracellular signaling cascade that
regulates various physiological processes, including cell
proliferation, survival, metabolism and migration (4,5).
This pathway is ubiquitously expressed across diverse cell types
and serves a cancer-promoting role in cancer and immune cells. The
serine biosynthetic pathway is key in cancer metabolism. This
pathway is governed by three key regulatory enzymes:
Phosphoglycerate dehydrogenase (PHGDH), phosphoserine phosphatase
and phosphoserine aminotransferase 1 (PSAT1). PSAT1 is the key
enzyme in the serine synthesis pathway and catalyzes conversion of
3-phosphohydroxypyruvate to 3-phosphoserine, which is
dephosphorylated subsequently to form L-serine participating in
one-carbon metabolism and nucleic acid biosynthesis (6,7).
Hence, PSAT1 activity is associated with onset and progression of
tumorigenesis.
Our previous investigation confirmed that
overexpression of PSAT1 is associated with poor prognosis of short
overall survival time in ovarian cancer (8). Previous reports have revealed
increased expression of PSAT1 in various types of malignancies,
including colon cancer, esophageal squamous cell carcinoma and
non-small cell lung cancer (9-12). However, the functional role of
PSAT1 in ovarian carcinogenesis and the underlying molecular
mechanisms remain to be fully delineated. The present study aimed
to assess the impact of PSAT1 on development of ovarian tumors.
Materials and methods
Cell culture
CAOV3, OVCAR3 and ES2 cell lines were purchased from
Procell Life Science & Technology Co., Ltd. The A2780 cell line
was purchased from Shanghai Biowing Applied Biotechnology Co., Ltd.
The CAOV3, OVCAR3, A2780, and ES2 ovarian cancer cell lines were
cultured in RPMI-1640 medium supplemented with 10% fetal bovine
serum (both Biological Industries) at 37°C and a 5% carbon
dioxide.
Establishment of stable
PSAT1-overexpressing and transient PSAT1-knockdown cell lines
A2780 and ES2 cell lines were transfected using
PSAT1-CMV-MCS-IRES-EGFP-SV40-Neo mycin-mediated transfection
(Shanghai Genechem Co., Ltd.). The cells were inoculated into
24-well cell culture plates at a density of 2,500 cells/well.
RPMI-1640 culture medium, without serum, containing lentivirus was
added to the cells for 12-18 h at 37°C. The transfection
concentration was 1 μg/μl (MOI=20). Transfection was
performed using Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
After 12 h, complete medium containing serum was added at 37°C.
ES2-PSAT1-high expression (H), ES2-PSAT1-H-Mock, A2780-PSAT1-H and
A2780-PSAT1-H-Mock, were constructed. Subsequent experiments were
performed after 24 h.
Efficiency of three small interfering (si)RNAs were
detected by western blot. PSAT1-siRNA-3 was selected for subsequent
experiments. PSAT1-siRNA and PSAT1-MOCK-siRNA sequences are shown
in Table SI. PSAT1-siRNAs were
transfected into CAOV3 and OVCAR3 cells. The working solutions for
siRNA transfection were prepared according to the manufacturer's
instructions (Shanghai GenePharma Co., Ltd.). The cells were seeded
in 6-well plates with serum-free RPMI-1640 medium. The PSAT1
lentiviral was transfected at 37°C for 48 h. Transfection was
performed via Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions. The
cells were collected 48 h after transfection and the effect of
PSAT1-siRNA transfection was determined by western blotting.
Within 48 h of transfection, cells were used for functional
experiments and flow cytometric analysis. STAT4-siRNA was
transfected into CAOV3 and OVCAR3 cells to establish CAOV3-STAT4-L
and OVCAR3-STAT4-L, CAOV3-STAT4-Mock and OVCAR3-STAT4-Mock, CAOV3
and OVCAR3.
Western blotting
Following total protein extraction from CAOV3,
OVCAR3, A2780 and ES2 cells, the cell pellet was lysed with
precooled RIPA buffer (Beyotime Institute of Biotechnology) on ice
for 30 min and centrifuged at 4°C and 13.4 × g for 20 min. The
supernatant was evaluated via BCA method to determine the total
protein concentration in the lysate and 5X loading buffer was added
for denaturation at 100°C for 5 min. The samples were separated via
10% SDS-PAGE with 10 μg protein/lane, concentrated at 80 V
and separated at 120 V, followed by electrotransfer at 4°C and 100
V for 45-90 min to a PVDF membrane. Then, 5% skimmed milk or 5%
bovine serum albumin (Biological Industries) was used for blocking
at room temperature for 12 h. Cells were incubated with primary
antibodies against PSAT1 (1:500; Abcam, ab308512), PI3K (1:1,000;
Cell Signaling Technology, Inc.; cat. no. 4292S), phosphorylated
(p-)PI3K (1:1,000; Cell Signaling Technology, 4228S), AKT (1:1,000;
Cell Signaling Technology, 4691S), p-AKT (1:1,000; Cell Signaling
Technology, Inc.; cat. no. 4691S) and GAPDH (1:4,000; Beijing
Zhongshan Jinqiao Biotechnology, TA-08 Co., Ltd.) overnight at 4°C.
Samples were rinsed three times with 1X TBST for 10 min. Membranes
were incubated with HRP-labelled goat anti-rabbit or goat
anti-mouse (both 1:3,000; both Beijing Zhongshan Jinqiao
Biotechnology Co., Ltd.) secondary antibodies for 1 h at 37°C and
then washed again. Finally, western chemiluminescent HRP substrate
(Thermo Fisher Scientific, Inc.) was added dropwise onto the
membrane, and luminescent signals were detected via a luminometer.
Image Lab Software (Version 6.1; Bio-Rad was used for
densitometry.
Cell proliferation experiment
A total of 2,000 ES-2 cells/well were seeded in
96-well culture plates and incubated at 37°C in a 5% CO2
incubator. After cells adhered to the wall, 20 μl sterile
MTT (Beijing Solarbio Science & Technology Co., Ltd.) working
solution was added to each well, mixed and incubated at 37°C for 4
h. Subsequently, the medium was aspirated and 150 μl DMSO
was added to each well. After shaking for 5 min, the absorbance at
490 nm of each well was measured via a microplate reader.
Cell cycle analysis
The ES-2 cells in the logarithmic growth phase were
collected, washed with PBS and fixed with 70% ethanol overnight at
4°C. A total of 500 μl PI/RNase A (Nanjing KeyGen Biotech
Co., Ltd.) staining solution was added to the cell suspension
according to the manufacturer's instructions and cells were
incubated in the dark for 20 min at room temperature. The cells
were analysed via flow cytometry of BD FACSAria and FlowJo (Version
10.7.2; both BD Biosciences).
Invasion assay
Transwell insert with pore size of 8 μm was
placed in the culture plate. Matrigel (1:7.5; BD Biosciences)
pre-coating was performed at 37°C for 6 h and a serum-free
4×104 ES-2 cell suspension were added to the upper
chamber of RPMI-1640 medium and the lower chamber contained
RPMI-1640 medium supplemented with 10% fetal bovine serum medium.
After 48-72 h culture, cells were collected, fixed with 4%
paraformaldehyde for 15 min at 37°C and stained with crystal violet
for 30 min at 37°C. The number of cells in five randomly selected
fields of view was counted and the mean of three repeated
experiments was calculated. Light microscope was used to observe
invasive cells.
Migration experiment
ES-2 cells were inoculated in 6-well plates and
maintained in a 5% CO2 incubator at 37°C. After
achieving 90% confluence, the cell layer was scratched with a
100-μl pipette suction head. The cells were cultured in
serum-free medium for 24 h at 37°C and washed with PBS. Images were
obtained to monitor wound healing using a light microscope was used
with magnification of 200. Quantification analysis was performed
using ImageJ software.
In vivo xenograft model of ovarian
cancer
The temperature of nude mice was about 20-26°C and
the relative humidity should be maintained at 40-60%. The
experimental environment should maintain a 12 h light/dark cycle.
Nude mice were provided with adequate food and water to ensure
normal growth and metabolism. A total of 20 female nude mice
(Huafukang Bioscience; age, 4-6 weeks; weight, 13-17 g) were
randomly divided into two groups (n=10/group) and subcutaneously
injected with ES2-PSAT1-Mock or ES2-PSAT1-H ovarian cancer cells
(~1×106 cells in 100 μl PBS) into the armpit of
the left forelimb. Tumor progression and the overall health status
of the mice were observed every 3 days and the diameter of the
tumors and mouse weights were measured. The tumor volume was
calculated as V=(a × b2)/2, where a represents the
maximum diameter and b is the shortest diameter. At 7 days
post-injection, newly formed tumors were detected. On day 21, mice
began to show symptoms of poor health with the largest tumor
diameter of 9.6 mm and maximum volume of 186.16 mm3. The
nude mice were completely sedated using isoflurane anesthesia
(induction, 4%; maintenance, 1.5%). The animals were euthanized
through exsanguination. Tumor samples were fixed at room
temperature in 4% paraformaldehyde and embedded in paraffin for 24
h. Continuous 5-μm-thick sections were analysed via
hematoxylin and eosin (HE) or immunohistochemical staining
[including phosphorylated (p-)serine-threonine protein kinases
(C-RAF) (Cell Signaling Technology, #9427, 1:100), p-AKT (Cell
Signaling, #4060 1:200). Tissue sections were immersed in xylene,
followed by dehydration in 100 and 80% ethanol for 5 min. The
sections were rinsed in distilled water for 5 min, stained with
hematoxylin for 5 min, and washed under running water for 10 min.
Sections were counterstained with 0.5% eosin for 1-3 min and rinsed
again in distilled water for 30 sec. For dehydration, sections were
immersed in 80% ethanol for 30 sec, 95% ethanol for 1 min, and 100%
ethanol for 3 min, followed by clearing in xylene for 3 min.
Finally, the sections were mounted with neutral gum. The paraffin
sections were deparaffinised with xylene and re-hydrated with
gradient alcohol solutions, and the antigens were recovered by
heating at 100°C for 15 min. Subsequently, 3% hydrogen peroxide was
added for 30 min at 37°C, 10% goat serum blocking solution (Fuzhou
Maixin Biotech Co., Ltd., KIT-9720) for 30 min at 37°C, and
anti-p-C-RAF antibody (Cell Signaling Technology, #9427, 1:100) or
p-AKT (Cell Signaling Technology, #4060 1:200) was added overnight
at 4°C. The following day, the slices were incubated with
biotin-labeled secondary antibody in complex with
streptavidin-peroxidase (Fuzhou Maixin Biotech Co., Ltd., KIT-9720)
for 30 min at 37°C and stained using 3,3-diaminobenzidine and
hematoxylin for 10 min at 37°C.
Light microscope was used to observe. The brown
granules observed in the nucleus and cytoplasm were scored; 0, 1,
2, and 3 corresponded to no coloration, light yellow,
brownish-yellow, and tan, respectively. Additionally, scores of 0,
1, 2, 3, and 4 represented <5, 6-25%, 26-50%, 51-75%, and
>75% of the visual field, respectively. The final score was
calculated by multiplying the two aforementioned scores, with the
following interpretations: 0-2 (−), 3-4 (+), 5-8 (++), and 9-12
(+++). Quantification analysis was performed using ImageJ software
(V1.0; National Institutes of Health). The animal studies were
approved by the Shengjing Hospital Affiliated to China Medical
University Ethics Review Committee (approval no. 2021PS894K).
Signalling pathway inhibition
ES2-PSAT1-Mock or ES2-PSAT1-H ovarian cancer cells
were cultured at 37°C in the presence of PI3K inhibitor (PI3Ki;
cat. no. GDC-0941, Selleck Chemicals) for 48 h. For inhibitor
concentration selection, 5, 10, 15, 20, 25, 30, 35 and 40 μM
were used during the preliminary experiment. When the inhibitor
concentration reached 25 μM, further increases did not
enhance the inhibitory effect. Therefore, 25 μM was used for
subsequent experiments. Additionally, 0.1% DMSO served as the
negative control.
Gene expression profiling interactive
analysis (GEPIA) dataset analysis
The survival curves were generated using the GEPIA
tool (gepia.cancer-pku.cn/), which integrated RNA-seq data and
clinical survival information from The Cancer Genome Atlas) and
GTEx databases. The dataset name was 'TCGA Pan-Cancer Clinical Data
Resource', accessible via the GDC Data Portal (https://portal.gdc.cancer.gov/) under the project
accession number 'phs000178.v11.p8' (13-15). Differential mRNA expression of
PSAT1 between 426 cancerous samples and 88 normal ovarian tissue
samples was evaluated.
GeneMANIA analysis
GeneMANIA (genemania.org)
is a public database that provides a list of genes with similar
functions for construction of interactive functional association
networks and analysis of the relationships between genes and
datasets. A gene interaction network for PSAT1 was constructed on
the basis of physical interactions, predictions, colocalization,
co-expression, signalling pathway information and predicted
functions.
Promo and JASPAR database analysis
Promo
(alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)
is a tool for identifying potential transcription factor binding
sites in DNA sequences, which predicts transcription factor binding
sites by constructing a matrix of specific binding site weights
(16). JASPAR (https://jaspar.elixir.no/) is an open access database
that stores manually curated transcription factor binding
preferences as a position frequency matrix (17). Promo and JASPAR databases were
used to predict that STAT4 functions as a transcriptional regulator
of PSAT1 expression.
Statistical analysis
SPSS 22.0 (IBM Corp.) was used for the statistical
analyses. All the data are presented as the mean ± standard
deviation of three independent repeats. The data were analyzed
using χ2 and Fisher's exact probability test.
Statistical differences between two groups were determined using
the unpaired t test, and one-way analysis of variance analysis
followed by Scheffe's test was used for the comparison of >2
groups. Two-sided P<0.05 was considered to indicate a
statistically significant difference.
Results
Expression of PSAT1 is higher in cancer
tissue
Through GEPIA, in-depth analysis was conducted to
elucidate differential mRNA expression patterns of PSAT1 between
ovarian cancer and normal samples. PSAT1 mRNA was significantly
upregulated in ovarian cancer compared with normal tissue (Fig. 1A). Immunohistochemical analysis
revealed PSAT1 expression in ES2, A2780, CAOV3 and OVCAR3 cell
lines (Fig. 1B). Western
blotting demonstrated higher PSAT1 protein expression in CAOV3 and
OVCAR3 than in ES2 and A2780 cells, however this was not
significant (Fig. 1C and D).
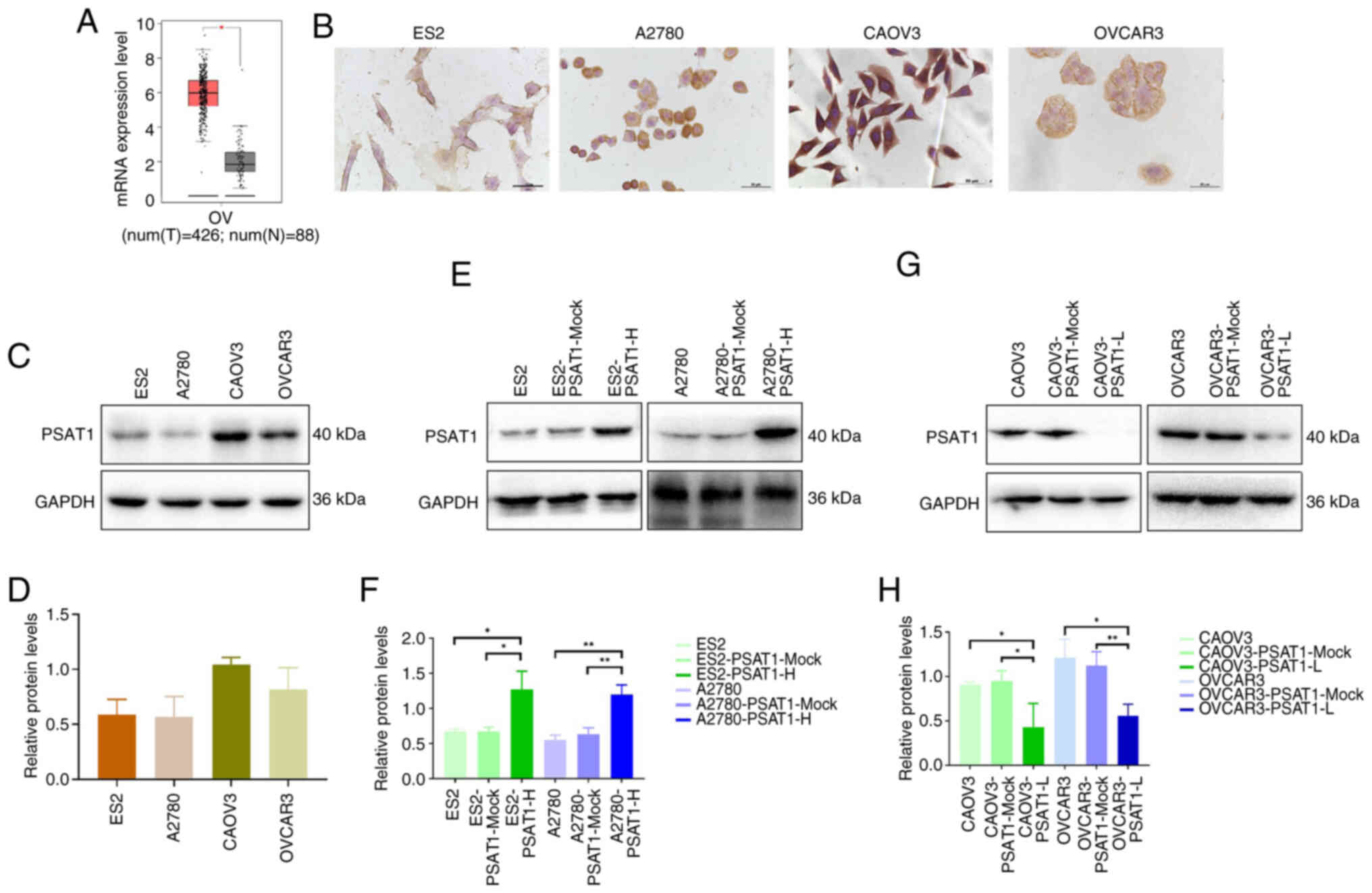 | Figure 1PSAT1 expression in ovarian cancer
tissue and cells. (A) Differential mRNA expression of PSAT1 between
ovarian cancer and normal ovarian tissues in the Gene Expression
Profiling Interactive Analysis database. (B) Expression of PSAT1 in
ES2, A2780, CAOV3 and OVCAR3 cell lines was detected by
immunohistochemistry. (C) PSAT1 expression is higher in CAOV3 and
OVCAR3 than in ES2 and A2780 cells. Magnification, ×200. (D)
Relative expression levels of PSAT1 in ES2, A2780, CAOV3, and
OVCAR3 cell lines. (E) Construction of stable and high PSAT1 of ES2
and A2780 cell lines. (F) Quantitative analysis of relative
expression levels of PSAT1 in ES2 and A2780 cell lines with PSAT1
overexpression. (G) Construction of PSAT1-L in CAOV3 and OVCAR3
cell lines. (H) Quantitative analysis of relative expression levels
of PSAT1 in CAOV3 and OVCAR3 cell lines with PSAT1 knockdown. PSAT,
phosphoserine aminotransferase; H, high; L, low; OV, OVCAR3; T,
tumor; N, normal. *P<0.05,
**P<0.01. |
To establish a cell model with decreased PSAT1
expression, CAOV3 and OVCAR3 ovarian cancer cell lines, which
exhibited relatively high PSAT1 expression were used. ES2 and A2780
ovarian cancer cell lines, characterized by comparatively low PSAT1
expression, were used to generate a cell model with overexpressed
PSAT1. Three sequences of siRNA have been screened by western blot
to obtain the one with the best knockdown effect (Fig. S1). Western blot analysis
revealed that the ES2-PSAT1-H and A2780-PSAT1-H cells presented
significantly elevated PSAT1 expression compared with control cells
(ES2-PSAT1-Mock, ES2, A2780-PSAT1-Mock and A2780). By contrast, the
PSAT1 protein levels were significantly lower in CAOV3-PSAT1-L and
OVCAR3-PSAT1-L cells than in the corresponding control cells
(CAOV3-PSAT1-Mock, CAOV3, OVCAR3-PSAT1-Mock and OVCAR3; Fig. 1E-H).
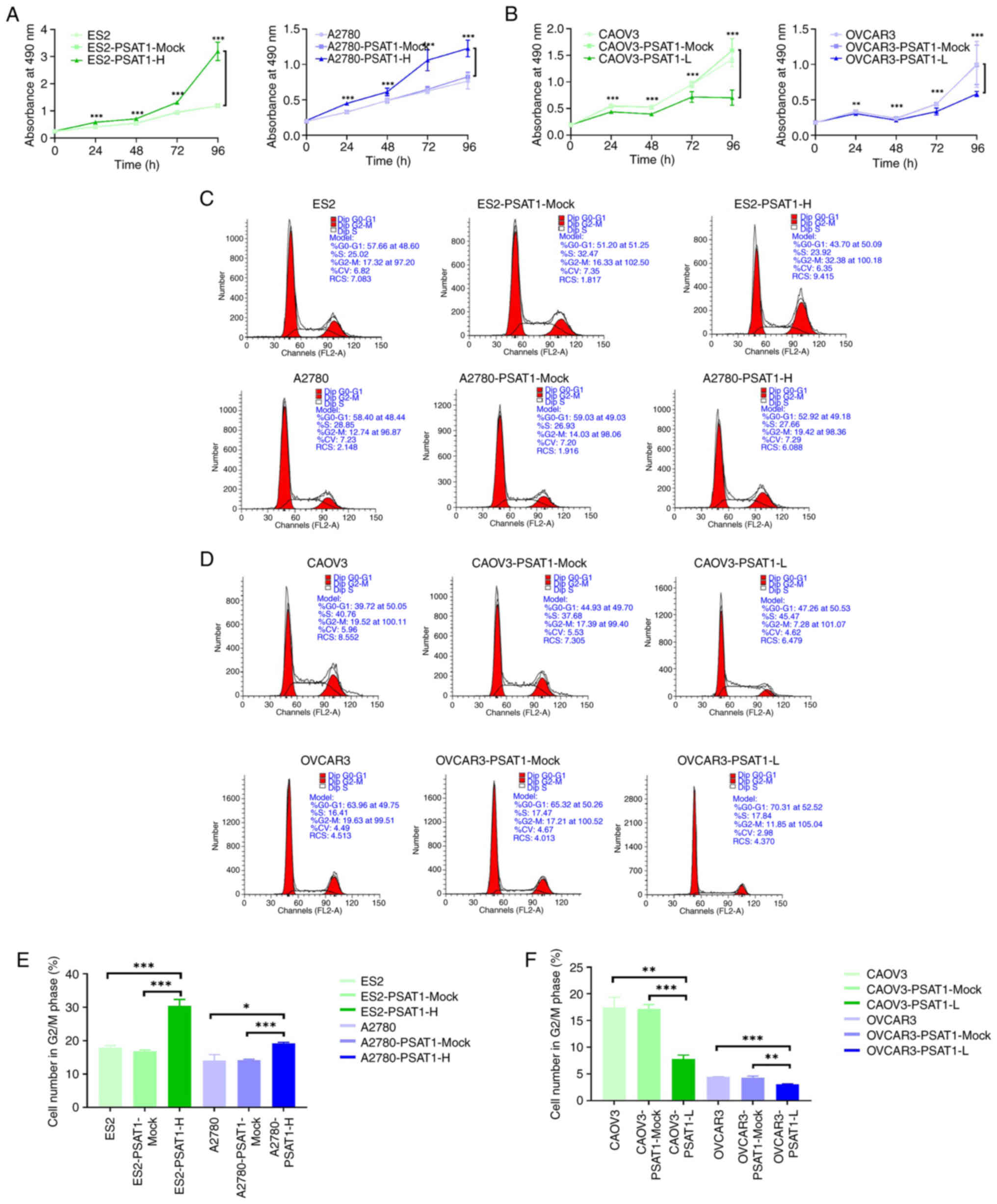
PSAT1 promotes proliferation, invasion
and migration of ovarian cancer cells
MTT assay was used to determine proliferation of
ovarian cancer cells following the overexpression or knockdown of
PSAT1. The impact of PSAT1 upregulation and suppression on the cell
cycle was assessed via flow cytometry analysis. ES2-PSAT1-H and
A2780-PSAT1-H cell lines exhibited significantly greater
proliferation and a greater proportion of cells in the G2/M phase
than their respective control (ES2-PSAT1-Mock, ES2,
A2780-PSAT1-Mock and A2780). Conversely, CAOV3-PSAT1-L and
OVCAR3-PSAT1-L lines demonstrated significantly decreased
proliferation and proportion of cells within G2/M phase relative to
the control groups (CAOV3-PSAT1-Mock, CAOV3, OVCAR3-PSAT1-Mock and
OVCAR3; Fig. 2). These findings
underscored the capacity of PSAT1 to promote proliferation of
ovarian cancer cells.
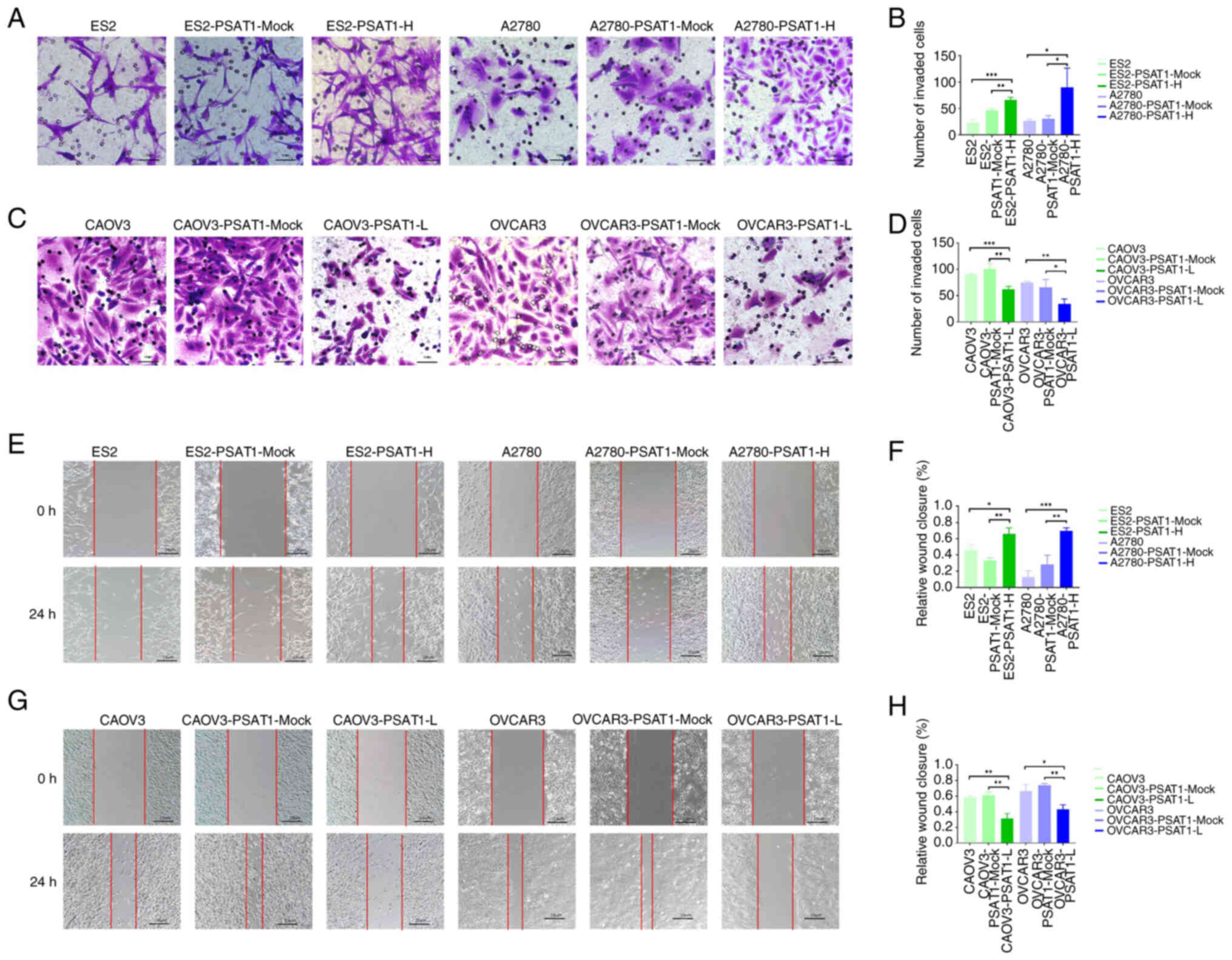
Transwell and scratch assays were conducted to
investigate the impact of PSAT1 on the invasive and migratory
capacity of ovarian cancer cells. The invasive and migratory
potential of the ES2-PSAT1-H and A2780-PSAT1-H cells was
significantly greater than that of the ES2-PSAT1-Mock, ES2,
A2780-PSAT1-Mock and A2780 control cells. By contrast,
CAOV3-PSAT1-L and OVCAR3-PSAT1-L cells exhibited significantly
decreased invasion and migration compared with CAOV3-PSAT1-Mock,
CAOV3, OVCAR3-PSAT1-Mock and OVCAR3 (Fig. 3). These findings suggested that
PSAT1 enhanced the invasive and migratory potential of ovarian
cancer cells.
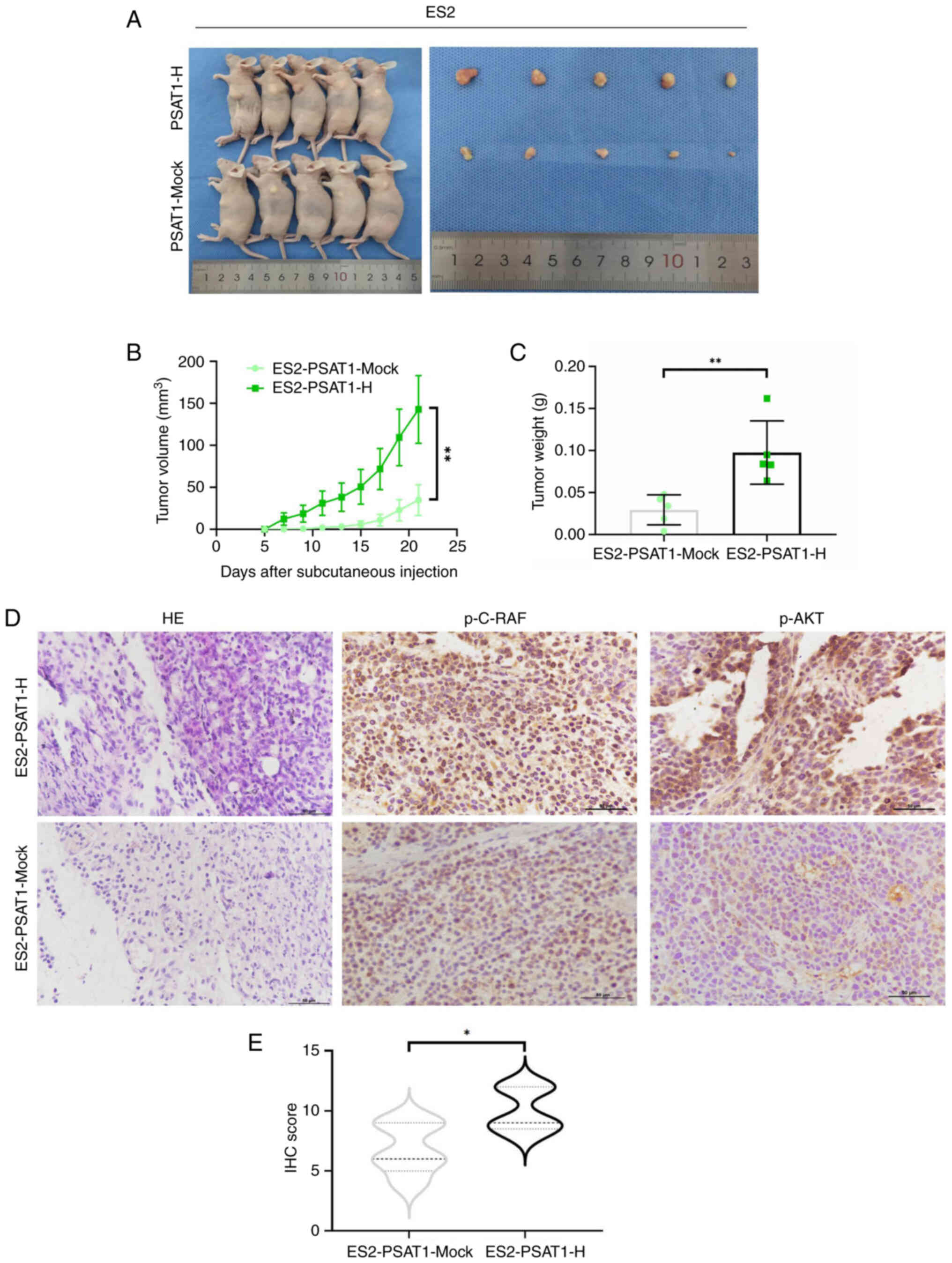
PSAT1 promotes tumor formation in
vivo
To investigate the influence of PSAT1 on the
tumorigenesis of ovarian cancer cells, ES2-PSAT1-H cells and
ES2-PSAT1-Mock cells were subcutaneously implanted into athymic
nude mice (Fig. 4A). After 21
days, the mean tumor volume in the ES2-PSAT1-H cells was
significantly greater than that in the control group. Additionally,
the mean tumor weight in the PSAT1-overexpression cohort was ~3.32
times greater than that in the control cohort (Fig. 4B and C). The tumor xenografts
from the nude mice were processed for histological analysis. The
protein expression levels of p-C-RAF and p-AKT in the ES2-PSAT1-H
group were notably elevated compared with those in the
ES2-PSAT1-Mock group, indicating that PSAT1 can facilitate tumor
cell proliferation in vivo.
PSAT1 promotes proliferation, invasion
and migration of ovarian cancer cells via the PI3K/AKT pathway
GeneMANIA database was used to construct a gene-gene
interaction network for PSAT1. The five genes demonstrating
the greatest association with PSAT1 were programmed cell
death 11, ubiquitin-like modifier activating enzyme 1,
peptidylprolyl isomerase D, heparan sulfate proteoglycan 2 and
PHGDH (Fig. 5A). Western
blotting was used to determine the expression levels of p-PI3K,
PI3K, p-AKT and AKT. Expression of p-PI3K and p-AKT was
significantly lower in CAOV3-PSAT1-L than in the control groups
CAOV3-PSAT1-Mock and CAOV3 (Fig. 5B
and C). These findings confirmed that PSAT1 modulated the
PI3K/AKT signalling pathway in ovarian cancer cells.
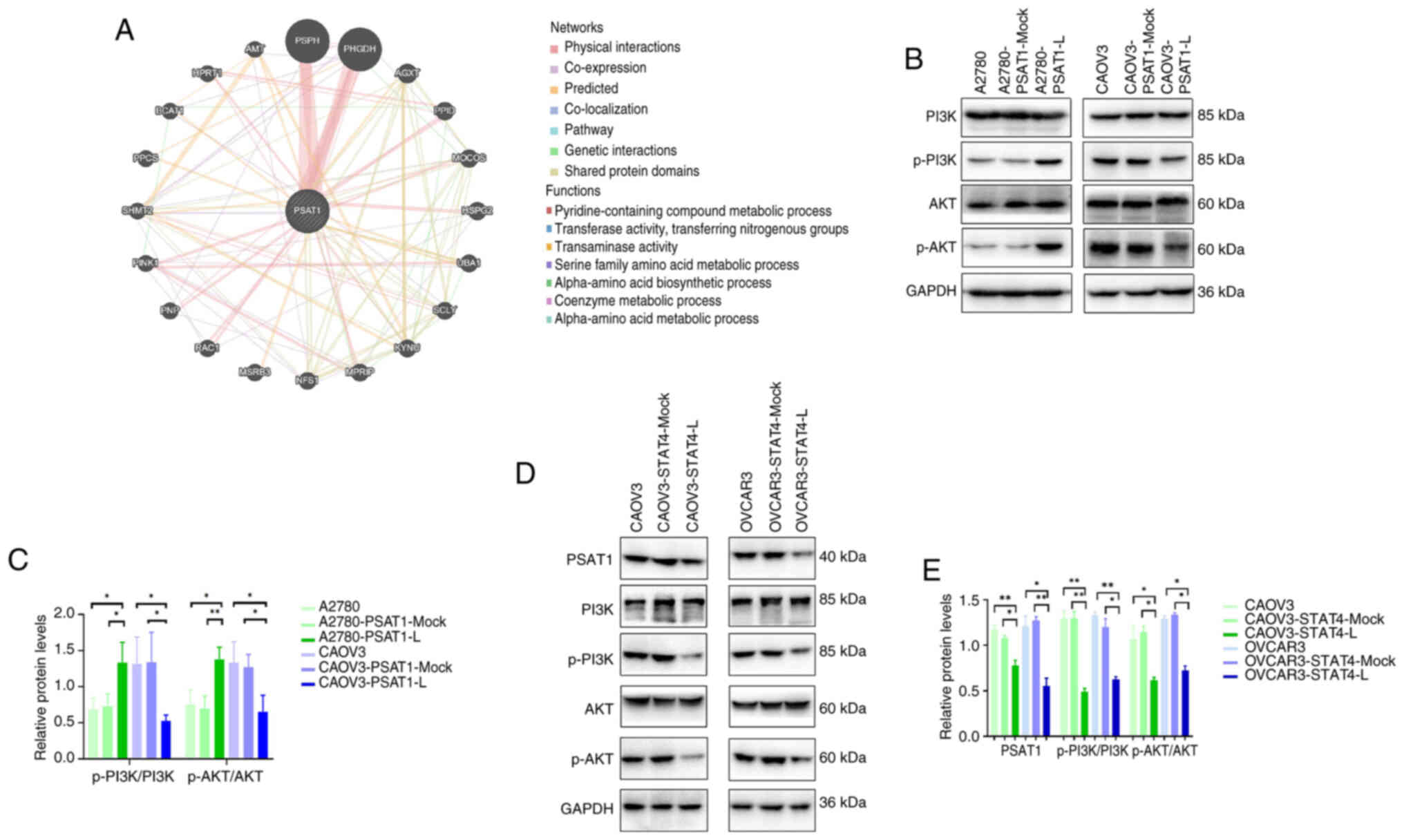 | Figure 5PSAT1 exerts cellular effects through
the PI3K/AKT signalling pathway. (A) Construction of the PSAT1 gene
interaction network using GeneMANIA. (B) Expression of p-PI3K,
PI3K, p-AKT and AKT in ovarian cancer cells. (C) Quantitative
analysis of relative expression levels of p-PI3K, PI3K, p-AKT and
AKT in ovarian cancer cells. (D) PSAT1, p-PI3K/PI3K, p-AKT/AKT
protein expression levels in CAOV3-STAT4-L and OVCAR3-STAT4-L cells
were lower than those in CAOV3-STAT4-Mock, CAOV3, OVCAR3-STAT4-Mock
and OVCAR3 cells. (E) Quantitative analysis of relative expression
of PSAT1, p-PI3K/PI3K, p-AKT/AKT in CAOV3 and OVCAR3 cell lines
with STAT4 knockdown. PSAT, phosphoserine aminotransferase; p-,
phosphorylated; L, low; H, high. *P<0.05;
**P<0.01; vs. control. |
Promo and JASPAR databases were used to predict that
STAT4 functions as a transcriptional regulator of PSAT1 expression.
Western blot analysis confirmed that protein expression levels of
PSAT1 in CAOV3-STAT4-L and OVCAR3-STAT4-L cell lines were
significantly lower than those in the CAOV3-STAT4-Mock, CAOV3,
OVCAR3-STAT4-Mock and OVCAR3 controls (Fig. 5D and E).
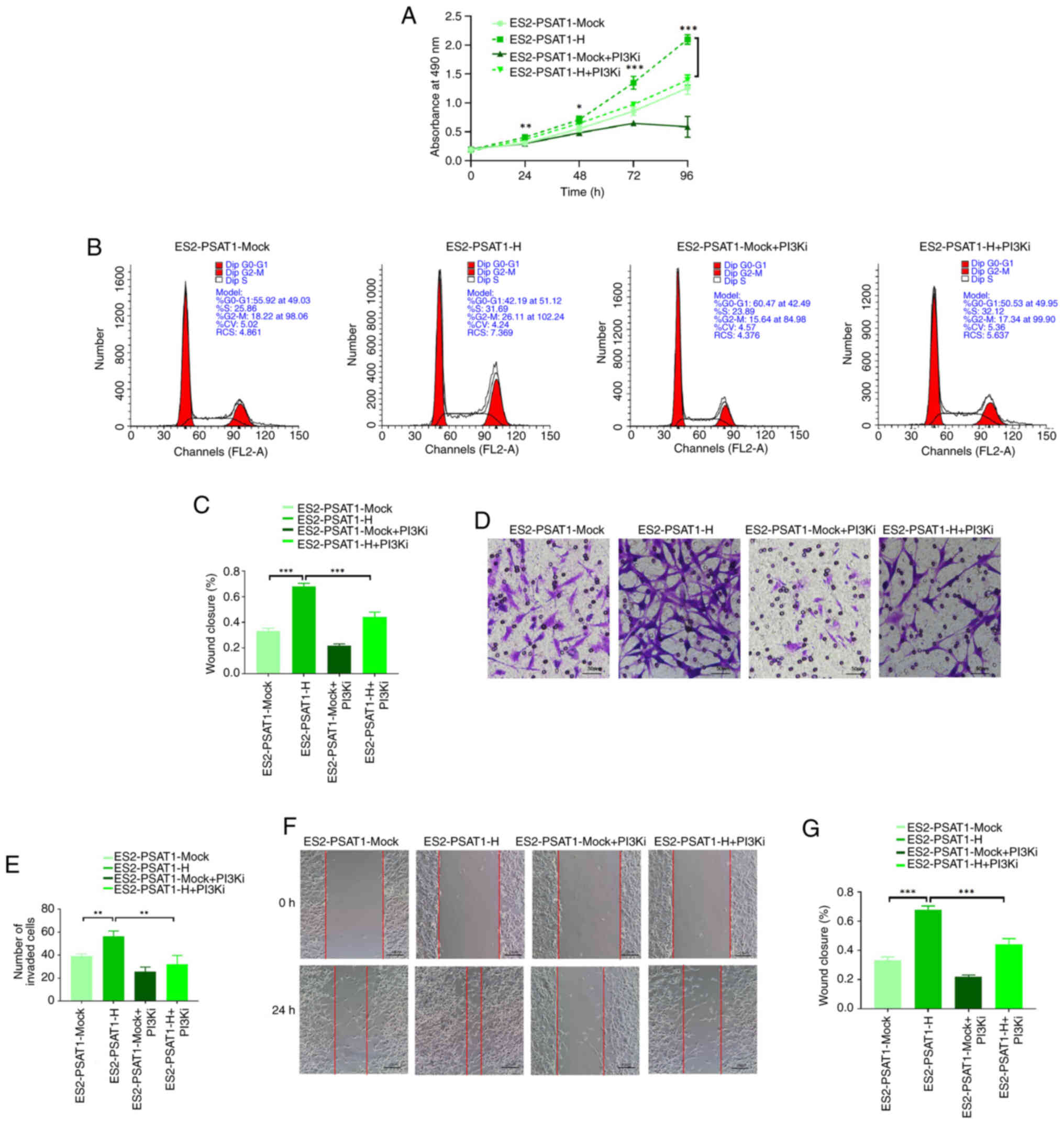
PI3K pathway inhibition decreases
proliferation, invasion and migration ability of
PSAT1-overexpressing cells
To explore the effect of PSAT1 on the PI3K pathway,
PI3Ki GDC-0941 was used. Proliferation of ES2-PSAT1-H cells was
greater than that of control cells. However, GDC-0941 significantly
diminished the increase in proliferation induced by PSAT1 (Fig. 6A). Flow cytometry revealed that
the ratio of G2/M phase cells in the ES2-PSAT1-H group was
significantly increased compared with ES2-PSAT1-Mock, and GDC-0941
significantly decreased the proportion of G2/M phase cells
(Fig. 6B and C). PSAT1 promoted
ovarian cancer cell entry into G2/M phase and promoted cell
proliferation via the PI3K signalling pathway. The PI3K inhibitor
GDC-0941 attenuated this proliferation (Fig. 6B and C). Transwell assay revealed
invasion ability of ES2-PSAT1-H cells was enhanced compared with
ES2-PSAT1-Mock, and the addition of PI3Ki significantly reduced the
invasion ability (Fig. 6D and
E). The results of the scratch assay revealed that the
migration ability of ES2-PSAT1-H cells was increased compared with
ES2-PSAT1-Mock and the addition of PI3Ki significantly decreased
the PSAT1 overexpression-induced increase in migration (Fig. 6F and G). Collectively, these
results demonstrated that PSAT1 affected the proliferation,
invasion and migration ability of ovarian cancer cells via the PI3K
signalling pathway.
Discussion
Serine is a critical component for cellular
proliferation, folate metabolism, phospholipid metabolism and the
formation of d-serine and glycine, which serve as neuromodulators
(18). Restriction of serine
availability notably affects development and functionality of the
central nervous system. Individuals with serine deficiency may
exhibit serious neurological or psychiatric disorder, including
epilepsy, severe psychomotor retardation and congenital
microcephaly (19). PSAT1, a
member of the class V pyridoxal phosphate-dependent
aminotransferase family, serves a key role in the serine
biosynthetic pathway (20).
Mutations within PSAT1, encoding the second enzyme in the serine
biosynthetic cascade, are associated with a form of serine
deficiency known as PSAT1 deficiency (21). However, clinical diagnosis of
PSAT1 deficiency solely on the basis of the measurement of serine
levels in cerebrospinal fluid is challenging because serine levels
can be influenced by multiple factors and exhibit variability
between individuals. Therefore, early detection requires additional
information such as family history or genotypic data (22,23). Understanding of the functions of
PSAT1 is very important for understanding the cancer-promoting role
of PSAT1 in tumors.
The present study demonstrated that overexpression
of PSAT1 significantly potentiated proliferation, invasion and
migration of ovarian cancer cells. Moreover, differential
expression of PSAT1 was associated with the modulation of the
malignant biological behavior. PSAT1 overexpression is a potential
biomarker of favourable prognosis in patients with low-grade glioma
(24). There is an association
between increased PSAT1 expression and advanced clinical stage in
patients with nasopharyngeal carcinoma (11). Furthermore, there is an
association between PSAT1 expression levels and aggressive
phenotypic characteristics in breast cancer, with higher expression
being predictive of a poorer prognosis in primary cases,
particularly in estrogen receptor-positive tumors (17,18). PSAT1 is upregulated in colorectal
cancer tissue and associated with resistance to chemotherapy
(25). The aforementioned
findings are consistent with the present results, which indicated
that PSAT1 promoted proliferation and invasion of cancer cells,
suggesting that PSAT1 is a biomarker of poor prognosis in various
malignancies, such as non-small cell lung cancer, oesophageal
squamous cell carcinoma, breast and colorectal cancer (26,27).
Zheng et al (8) demonstrated that genes within a
PSAT1 high expression mutant group were significantly enriched in a
variety of pathways, including the 'cytokine-cytokine receptor
interaction', 'Rap1 signalling pathway', 'chemokine signalling
pathway', 'fox signalling pathway', 'TNF signalling pathway', 'mTOR
signalling pathway' and other tumor-related signalling pathways. To
the best of our knowledge, however, the aforementioned findings
have not been experimentally corroborated in ovarian cancer tissue
or cells. The levels of PSAT1 are increased in cisplatin-resistant
cervical cancer cells (SiHa-R cells) (28). Knockdown of PSAT1 decreases the
half-maximal inhibitory concentration of cisplatin, inhibits colony
formation and promotes SiHa-R cell apoptosis (28). Suppression of PSAT1 expression
inhibits proliferation and induces apoptosis by blocking the
PI3K/AKT signalling pathway, thereby diminishing cisplatin
resistance of SiHa-R cells. Consequently, PSAT1 is a potential
therapeutic target for reversing cisplatin resistance in cervical
cancer cells. In tumor cells, alterations in the serine-glycine
amino acid biosynthetic pathway inhibit production of glutathione,
thereby diminishing the capacity of cells to withstand oxidative
stress and undergo programmed cell death (29,30). Conversely, upregulated expression
of PSAT1 has been demonstrated to increase the ratio of reduced
glutathione in ovarian cancer cells, conferring increased
resistance to oxidative damage and potentially facilitating
aggressive progression of ovarian epithelial tumors (29,31-33).
PSAT1 exerts its functional effects by targeting
proteins and transcription factors to modulate signaling pathways,
such as the GSK3β/β-catenin and PI3K/AKT/mTOR pathways and
influences biological processes by inhibiting autophagy mechanisms
(34,35). Studies on intracranial aneurysms
have demonstrated that the upregulation of microRNA (miR)-133a-3p
and downregulation of PSAT1 ameliorate endothelial cell damage and
enhance cell proliferation by suppressing the GSK3β/β-catenin
pathway in intracranial aneurysms (34,35). Moreover, overexpression of
miR-195-5p inactivates the GSK3β/β-catenin signalling pathway by
suppressing PSAT1, thereby attenuating angiogenesis, decreasing
chemoresistance to cisplatin and enhancing ovarian cancer cell
apoptosis (36,37). PSAT1, which is upregulated in
estrogen receptor-negative breast cancer, is activated by
activating transcription factor 4 and promotes cell cycle
progression by regulating the GSK3β/β-catenin/cyclin D1 pathway
(36). miR-424, which is
frequently downregulated in colorectal cancer, can inhibit the
proliferation of colorectal cancer cells and induce apoptosis.
Moreover, AKT3 and PSAT1 are direct targets of miR-424 (38). The loci of AKT3 and PSAT1 are
implicated in miR-424 regulated apoptosis, underscoring the key
role of miR-424 and its molecular targets, AKT3 and PSAT1, in the
pathogenesis of colorectal cancer. Expression of PSAT1 in
colorectal cancer is abnormally elevated due to G9A-mediated
transcriptional activation, which not only activates the serine
biosynthetic pathway but also provides α-ketoglutarate to
facilitate entry into the tricarboxylic acid cycle. Downregulation
of PSAT1 expression promotes cyclin D1 degradation through the mTOR
pathway, culminating in cell cycle arrest and programmed cell death
(39). These observations
suggeste that PSAT1 represents a promising therapeutic target for
colorectal cancer (39). In the
present study, expression levels of PSAT1, p-PI3K and AKT were
increased in ovarian cancer cells. Moreover, the invasive,
migratory and proliferative capabilities of these cells were
inhibited by PI3Ki, indicating that PSAT1 may bypass PI3K signaling
and thereby impact the aggressive biological behaviors associated
with ovarian cancer.
The STAT family comprises key cytokine-associated
signalling mediators and potential molecular targets in cancer
therapy that modulate invasion and metastasis. STAT4 expression is
associated with prognosis of hepatocellular carcinoma (40). Furthermore, the identification of
STAT4 as a regulator of KISS1 expression, which enhances the
apoptosis of ovarian granulosa cells (41), underscoress its multifaceted role
in cellular processes. Here, the suppression of STAT4 expression in
ovarian cancer cells decreased PSAT1 protein levels, implicating
STAT4 as a potential upstream transcriptional regulator of
PSAT1.
A primary objective of translational research in
precision medicine is to identify targeted therapeutics and
predictive biomarkers, thereby facilitating the development of
effective therapeutic strategies. Despite numerous inhibitors
targeting the PI3K/AKT pathway currently undergoing preclinical
trials (41,42), no PI3K or AKTi have received
clinical approval to date. PI3K exists as a heterodimer composed of
a catalytic and a regulatory subunit (43). Based on variations in catalytic
subunits, PI3Ks can be classified into four isoforms: PI3Kα, PI3Kβ,
PI3Kδ and PI3Kγ (4). Structural
similarities between AKT isoforms and off-target inhibition within
the cAMP-dependent protein kinase, cGMP-dependent protein kinase,
and protein kinase C) kinase family pose challenges to achieving
selectivity for AKTi (44). To
date, no isomer of PSAT1 has been identified. The present findings
indicated that targeting PSAT1 may offer therapeutic potential. If
the high selectivity associated with PSAT1 can be effectively
combined with potent inhibition of the PI3K/AKT pathway, this could
yield improved clinical outcomes.
In conclusion, PSAT1 is a key player in serine
metabolism that affects development and progression of tumors. The
present study demonstrated elevated expression of PSAT1 in ovarian
cancer through a combination of database and tissue-based
experiments. By modulating activity of the PI3K/AKT signalling
cascade, PSAT1 controls the proliferation, invasion and migration
of ovarian cells, influencing malignant biological behaviors. The
present results provide a theoretical foundation for PSAT1 as a
prospective target for early detection and therapeutic management
of ovarian cancer.
Supplementary Data
Availability of data and materials
The data generated in the present study may be
requested from the corresponding author.
Authors' contributions
XL, SW and BL designed the study. YH and OL
performed the experiments. XN and YW analyzed data. XL and SW
edited the manuscript. XL and BL confirm the authenticity of all
the raw data. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
The present study was approved by Shengjing Hospital
Affiliated to China Medical University Ethics Review Committee
(approval no. 2021PS894K).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural Science
Foundation of China (grant nos. 82303503, 82203702 and 82173130),
Key Research and Development Guidance Plan Project in Liaoning
Province (grant no. 2019JH8/10300022) and Shenyang Science and
Technology Program (grant nos. 22-315-6-16 and 22-321-33-19).
References
|
1
|
Arora T, Mullangi S, Vadakekut ES and
Lekkala MR: Epithelial ovarian cancer. StatPearls Treasure Island
(FL): StatPearls Publishing; 2025
|
|
2
|
Smith RA, Andrews KS, Brooks D, Fedewa SA,
Manassaram-Baptiste D, Saslow D, Brawley OW and Wender RC: Cancer
screening in the United States, 2018: A review of current American
cancer society guidelines and current issues in cancer screening.
CA Cancer J Clin. 68:297–316. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang S, Xiao X, Yi Y, Wang X, Zhu L, Shen
Y, Lin D and Wu C: Tumor initiation and early tumorigenesis:
Molecular mechanisms and interventional targets. Signal Transduct
Target Ther. 9:1492024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Peng Y, Wang Y, Zhou C, Mei W and Zeng C:
PI3K/Akt/mTOR pathway and its role in cancer therapeutics: Are we
making headway? Front Oncol. 12:8191282022. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mishra R, Patel H, Alanazi S, Kilroy MK
and Garrett JT: PI3K inhibitors in cancer: Clinical implications
and adverse effects. Int J Mol Sci. 22:34642021. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang M, Zhang H, Lu Z, Su W, Tan Y, Wang J
and Jia X: PSAT1 mediated EMT of colorectal cancer cells by
regulating Pl3K/AKT signaling pathway. J Cancer. 15:3183–3198.
2024. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang Y, Li J, Dong X, Meng D, Zhi X, Yuan
L and Yao L: PSAT1 regulated oxidation-reduction balance affects
the growth and prognosis of epithelial ovarian cancer. Onco Targets
Ther. 13:5443–5453. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng MJ, Li X, Hu YX, Dong H, Gou R, Nie
X, Liu Q, Ying-Ying H, Liu JJ and Lin B: Identification of
molecular marker associated with ovarian cancer prognosis using
bioinformatics analysis and experiments. J Cell Physiol.
234:11023–11036. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Liu B, Jia Y, Cao Y, Wu S, Jiang H, Sun X,
Ma J, Yin X, Mao A and Shang M: Overexpression of phosphoserine
aminotransferase 1 (PSAT1) predicts poor prognosis and associates
with tumor progression in human esophageal squamous cell carcinoma.
Cell Physiol Biochem. 39:395–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Vié N, Copois V, Bascoul-Mollevi C, Denis
V, Bec N, Robert B, Fraslon C, Conseiller E, Molina F, Larroque C,
et al: Overexpression of phosphoserine aminotransferase PSAT1
stimulates cell growth and increases chemoresistance of colon
cancer cells. Mol Cancer. 7:142008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pollari S, Käkönen SM, Edgren H, Wolf M,
Kohonen P, Sara H, Guise T, Nees M and Kallioniemi O: Enhanced
serine production by bone metastatic breast cancer cells stimulates
osteoclastogenesis. Breast Cancer Res Treat. 125:421–430. 2011.
View Article : Google Scholar
|
|
12
|
Li H, Wu C, Chang W, Zhong L, Gao W, Zeng
M, Wen Z, Mai S and Chen Y: Overexpression of PSAT1 is correlated
with poor prognosis and immune infiltration in non-small cell lung
cancer. Front Biosci (Landmark Ed). 28:2432023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Li C, Kang B, Gao G, Li C and
Zhang Z: GEPIA: A web server for cancer and normal gene expression
profiling and interactive analyses. Nucleic Acids Res. 45(W1):
W98–W102. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cancer Genome Atlas Research Network:
Integrated genomic analyses of ovarian carcinoma. Nature.
474:609–615. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
GTEx Consortium: Human genomics. The
genotype-tissue expression (GTEx) pilot analysis: Multitissue gene
regulation in humans. Science. 348:648–660. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Messeguer X, Escudero R, Farré D, Núñez O,
Martínez J and Albà MM: PROMO: detection of known transcription
regulatory elements using species-tailored searches.
Bioinformatics. 18:333–334. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sandelin A and Wasserman WW: Constrained
binding site diversity within families of transcription factors
enhances pattern discovery bioinformatics. J Mol Biol. 338:207–215.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Sirr A, Lo RS, Cromie GA, Scott AC,
Ashmead J, Heyesus M and Dudley AM: A yeast-based complementation
assay elucidates the functional impact of 200 missense variants in
human PSAT1. J Inherit Metab Dis. 43:758–769. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
van der Crabben SN, Verhoeven-Duif NM,
Brilstra EH, Van Maldergem L, Coskun T, Rubio-Gozalbo E, Berger R
and de Koning TJ: An update on serine deficiency disorders. J
Inherit Metab Dis. 36:613–619. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo MY, Zhou Y, Gu WM, Wang C, Shen NX,
Dong JK, Lei HM, Tang YB, Liang Q, Zou JH, et al: Metabolic and
nonmetabolic functions of PSAT1 coordinate signaling cascades to
confer EGFR inhibitor resistance and drive progression in lung
adenocarcinoma. Cancer Res. 82:3516–3531. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang SP, Chan YC, Huang SY and Lin YF:
Overexpression of PSAT1 gene is a favorable prognostic marker in
lower-grade gliomas and predicts a favorable outcome in patients
with IDH1 mutations and chromosome 1p19q Codeletion. Cancers
(Basel). 12:132019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Acuna-Hidalgo R, Schanze D, Kariminejad A,
Nordgren A, Kariminejad MH, Conner P, Grigelioniene G, Nilsson D,
Nordenskjöld M, Wedell A, et al: Neu-Laxova syndrome is a
heterogeneous metabolic disorder caused by defects in enzymes of
the L-serine biosynthesis pathway. Am J Hum Genet. 95:285–293.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Moat S, Carling R, Nix A, Henderson M,
Briddon A, Prunty H, Talbot R, Powell A, Wright K, Fuchs S and de
Koning T: Multicentre age-related reference intervals for
cerebrospinal fluid serine concentrations: Implications for the
diagnosis and follow-up of serine biosynthesis disorders. Mol Genet
Metab. 101:149–152. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liao KM, Chao TB, Tian YF, Lin CY, Lee SW,
Chuang HY, Chan TC, Chen TJ, Hsing CH, Sheu MJ and Li CF:
Overexpression of the PSAT1 gene in nasopharyngeal carcinoma is an
indicator of poor prognosis. J Cancer. 7:1088–1094. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
De Marchi T, Timmermans MA, Sieuwerts AM,
Smid M, Look MP, Grebenchtchikov N, Sweep FCGJ, Smits JG, Magdolen
V, van Deurzen CHM, et al: Phosphoserine aminotransferase 1 is
associated to poor outcome on tamoxifen therapy in recurrent breast
cancer. Sci Rep. 7:20992017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Qian C, Xia Y, Ren Y, Yin Y and Deng A:
Identification and validation of PSAT1 as a potential prognostic
factor for predicting clinical outcomes in patients with colorectal
carcinoma. Oncol Lett. 14:8014–8020. 2017.
|
|
27
|
Jia L, Li D, Wang YN, Zhang D and Xu X:
PSAT1 positively regulates the osteogenic lineage differentiation
of periodontal ligament stem cells through the
ATF4/PSAT1/Akt/GSK3β/β-catenin axis. J Transl Med. 21:702023.
View Article : Google Scholar
|
|
28
|
Fang Y, Liang X, Xu J and Cai X: miR-424
targets AKT3 and PSAT1 and has a tumor-suppressive role in human
colorectal cancer. Cancer Manag Res. 10:6537–6547. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duan W and Liu X: PSAT1 upregulation
contributes to cell growth and cisplatin resistance in cervical
cancer cells via regulating PI3K/AKT signaling pathway. Ann Clin
Lab Sci. 50:512–518. 2020.PubMed/NCBI
|
|
30
|
Hui Z, Wang X and Li X: Targeting the
serine-glycine one-carbon pathway overcomes glutathione-mediated
drug resistance in cancer. Cell Death Dis. 10:522019.
|
|
31
|
Moloney JN and Cotter TG: ROS signalling
in the biology of cancer. Semin Cell Dev Biol. 80:50–64. 2018.
View Article : Google Scholar
|
|
32
|
Smith CL, Bolton A and Nguyen G: Genomic
and epigenomic instability, fragile sites, schizophrenia and
autism. Curr Genomics. 11:447–469. 2010. View Article : Google Scholar
|
|
33
|
Butturini E, Carcereri de Prati A, Boriero
D and Mariotto S: Natural sesquiterpene lactones enhance
chemosensitivity of tumor cells through redox regulation of STAT3
signaling. Oxid Med Cell Longev. 2019:45689642019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lv H, Zhen C, Liu J, Yang P, Hu L and
Shang P: Unraveling the potential role of glutathione in multiple
forms of cell death in cancer therapy. Oxid Med Cell Longev.
2019:31501452019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gao S, Ge A, Xu S, You Z, Ning S, Zhao Y
and Pang D: PSAT1 is regulated by ATF4 and enhances cell
proliferation via the GSK3β/β-catenin/cyclin D1 signaling pathway
in ER-negative breast cancer. J Exp Clin Cancer Res. 36:1792017.
View Article : Google Scholar
|
|
36
|
Jia Q, Yan S, Huang J and Xu S: Restored
microRNA-133a-3p or depleted PSAT1 restrains endothelial cell
damage-induced intracranial aneurysm via suppressing the
GSK3β/β-catenin pathway. Nanoscale Res Lett. 15:1772020. View Article : Google Scholar
|
|
37
|
Wang H, Fang Q, You S, Wu Y and Zhang C:
miRNA-195-5p/PSAT1 feedback loop in human triple-negative breast
cancer cells. Genes Genomics. 45:39–47. 2023. View Article : Google Scholar
|
|
38
|
Wang H, Cui L, Li D, Fan M, Liu Z, Liu C,
Pan S, Zhang L, Zhang H and Zhao Y: Overexpression of PSAT1
regulated by G9A sustains cell proliferation in colorectal cancer.
Signal Transduct Target Ther. 5:472020. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang G, Chen JH, Qiang Y, Wang DZ and Chen
Z: Decreased STAT4 indicates poor prognosis and enhanced cell
proliferation in hepatocellular carcinoma. World J Gastroenterol.
21:3983–3993. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jiang Y, Xin X, Pan X, Zhang A, Zhang Z,
Li J and Yuan X: STAT4 targets KISS1 to promote the apoptosis of
ovarian granulosa cells. J Ovarian Res. 13:1352020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu L, Wei J and Liu P: Attacking the
PI3K/Akt/mTOR signaling pathway for targeted therapeutic treatment
in human cancer. Semin Cancer Biol. 85:69–94. 2022. View Article : Google Scholar
|
|
42
|
Ediriweera MK, Tennekoon KH and Samarakoon
SR: Role of the PI3K/AKT/mTOR signaling pathway in ovarian cancer:
Biological and therapeutic significance. Semin Cancer Biol.
59:147–160. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wymann MP and Pirola L: Structure and
function of phosphoinositide 3-kinases. Biochim Biophys Acta.
1436:127–150. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Quambusch L, Depta L, Landel I, Lubeck M,
Kirschner T, Nabert J, Uhlenbrock N, Weisner J, Kostka M and Levy
LM: Cellular model system to dissect the isoform-selectivity of Akt
inhibitors. Nat Commun. 12:52972021. View Article : Google Scholar : PubMed/NCBI
|