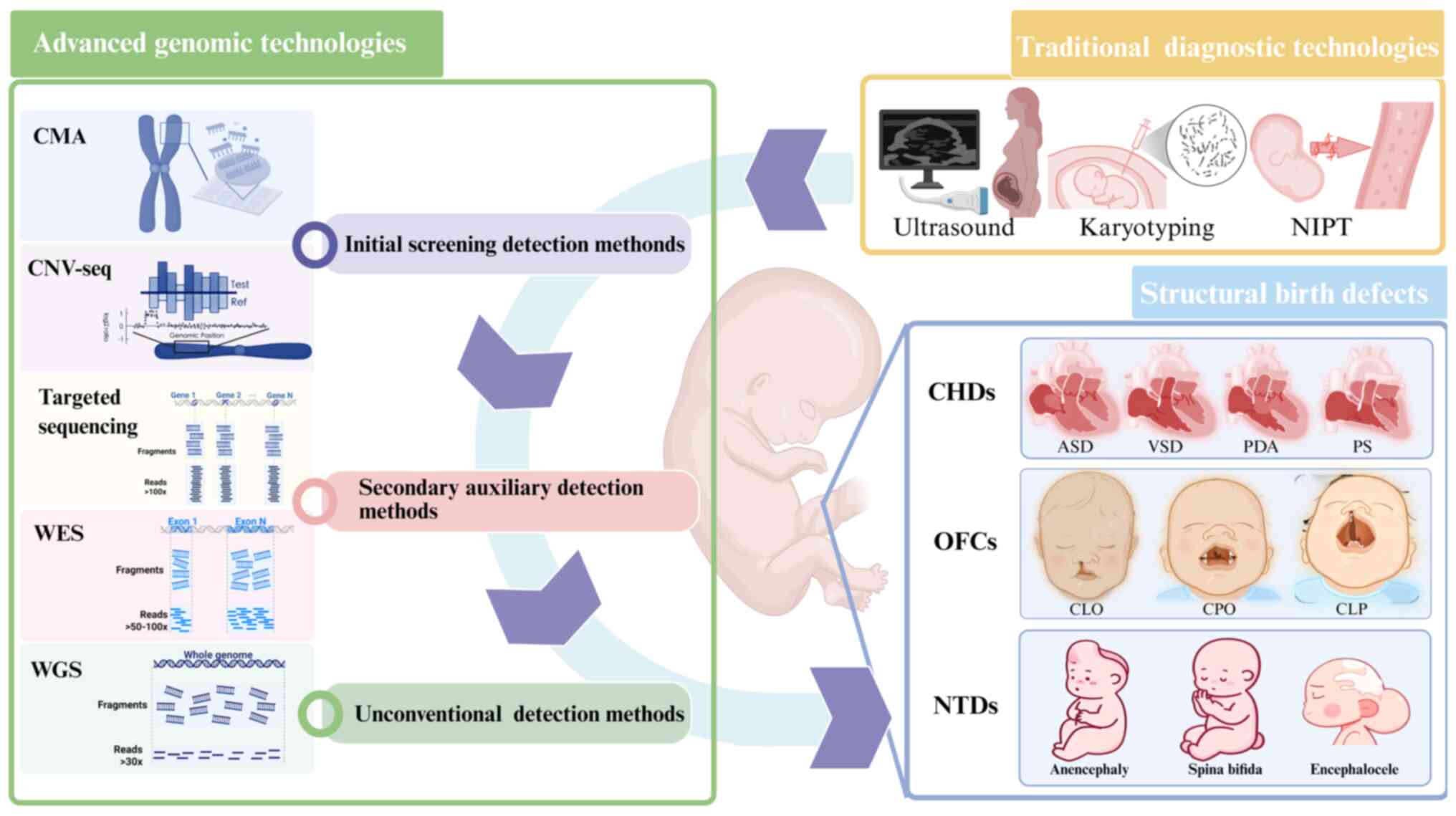
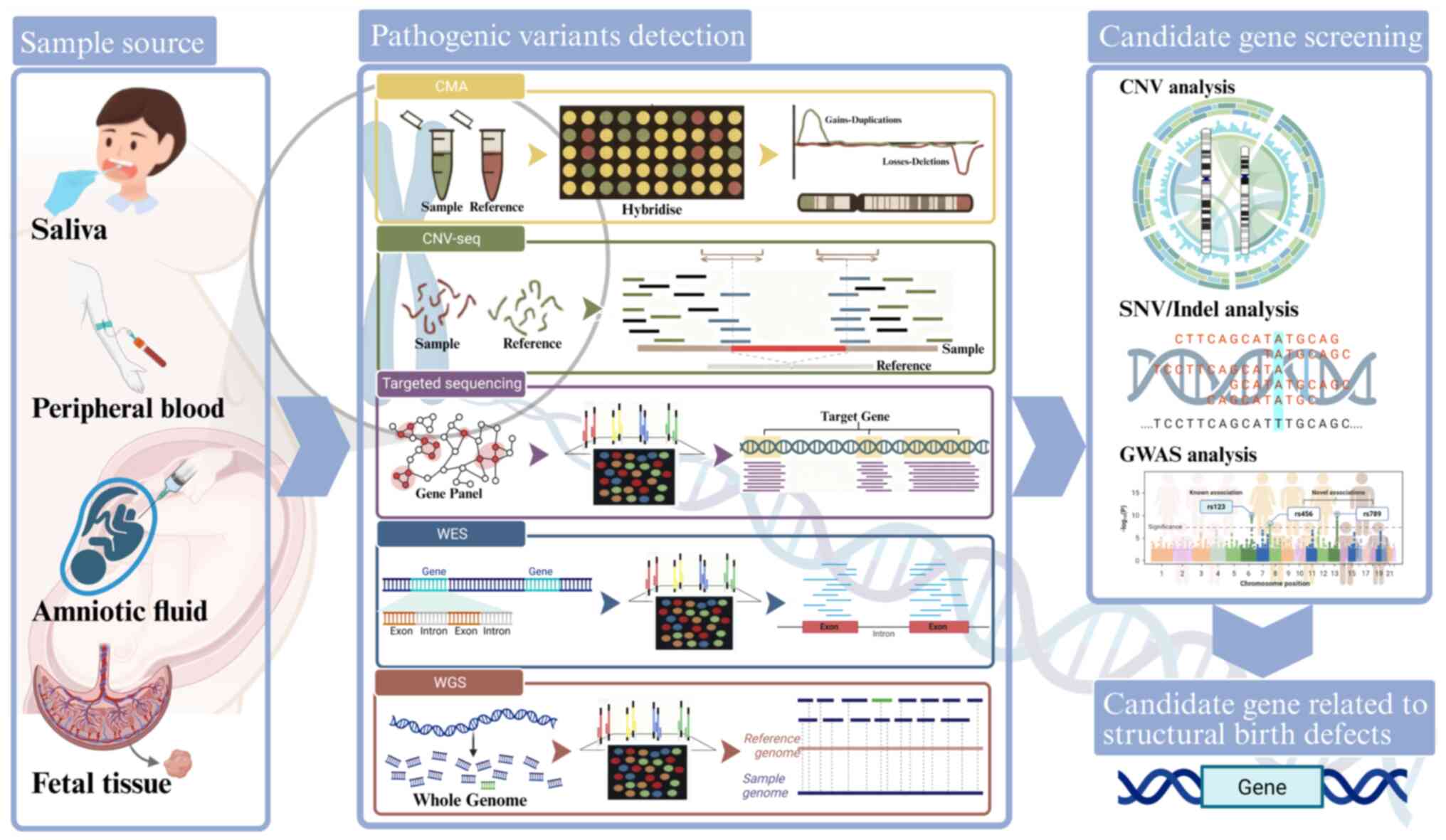
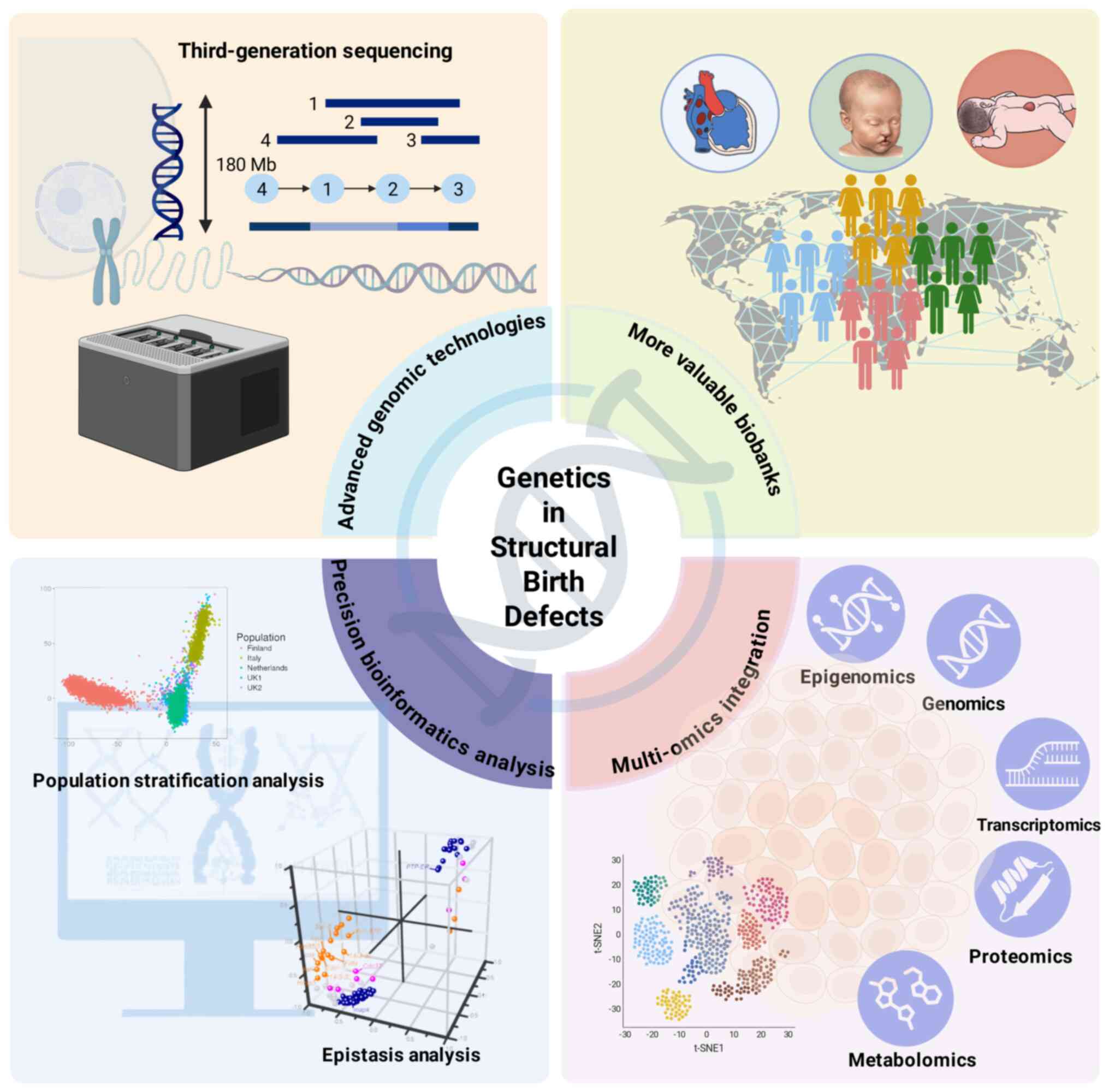
Structural birth defects (SBDs) are defined as
congenital abnormalities in physical structure that arise during
fetal development. Globally, these defects affect 3-6% of live-born
infants (1). The most prevalent
forms include congenital heart defects (CHDs) (0.33%), orofacial
clefts (OFCs) (0.14%) and neural tube defects (NTDs) (0.13%)
(2-4). These conditions are associated with
substantial perinatal mortality and long-term morbidity, imposing
notable burdens on affected families and society (5).
Current clinical screening for SBDs primarily relies
on prenatal ultrasound, maternal peripheral blood testing and
amniocentesis. While prenatal ultrasound can detect >80% of
fetal structural abnormalities (6), its diagnostic accuracy is limited
by operator expertise and equipment resolution, frequently failing
to identify subtle or complex malformations (7). Peripheral blood biochemical
analysis provides risk assessment for high-risk congenital
anomalies; however, current methodologies exhibit relatively high
rates of both false-positive and false-negative results (8-10). By contrast, genomics-based
approaches, including amniotic fluid karyotyping and conventional
non-invasive prenatal testing (NIPT) of peripheral blood, directly
identify chromosomal aneuploidies and
microdeletion/microduplication syndromes and address the diagnostic
limitations of ultrasound and enhance structural anomaly detection
rates (11,12). Nevertheless, these methods
currently elucidate the genetic factors in merely 19.1% of SBDs
cases, leaving 79.8% undiagnosed (13). Consequently, incorporating
advanced, comprehensive genomic technologies into both research and
clinical practice is essential for deciphering the complex genetic
architecture underlying SBDs.
Genomics, an interdisciplinary field investigating
genomic structure, function, evolution and interactions within
biological systems, seeks to elucidate fundamental biological
principles through comprehensive analysis of genetic information
(14,15). Recent advances in genomic
technologies have notably enhanced both research and clinical
applications for SBDs. These innovations facilitate not only more
precise identification of pathogenic variants and the discovery of
novel disease-associated genes, but also the refinement of prenatal
diagnostic approaches, thereby strengthening the scientific basis
for clinical decision-making and genetic counseling.
The present review discusses the genomic
technologies currently prevalent in genetic research, as well as
their application in prenatal diagnosis and candidate gene
screening for SBDs (Fig. 1),
while assessing the advantages and limitations of each method.
Furthermore, the present review explores potential future
directions within the context of reproductive health applications.
The aim of the present review was to establish a comprehensive
framework to equip clinicians with actionable technical and
analytical guidelines for evaluating the clinical translation of
these technologies as precision medicine modalities, with the
ultimate goal of improving diagnostic accuracy and therapeutic
strategies for SBDs.
In SBDs, genomic variations primarily present as
copy number variations (CNVs) and single nucleotide variations
(SNVs)/insertions-deletions (indels) (16,17). The precise detection of these
variants within the vast human genome constitutes the foundation
for elucidating the underlying genetic factors of SBDs. Advanced
genomic screening approaches primarily rely on high-throughput
technologies, including chromosomal microarray analysis (CMA) and
next-generation sequencing (NGS). The NGS platform encompasses
various methodologies such as CNV-sequencing (CNV-seq), whole-exome
sequencing (WES), whole-genome sequencing (WGS) and targeted
sequencing. These technologies have markedly improved the detection
rate of pathogenic variations in SBDs, providing important evidence
support for clinical diagnosis (Table I).
CMA is a high-throughput microarray-based
technology. Through specifically designed probes that hybridize
with sample DNA, CMA detects various genomic variations including
CNVs, single nucleotide polymorphisms (SNPs), loss of
heterozygosity and uniparental disomy through fluorescence signal
intensity analysis (18). At
present, CMA is recommended as the effective genomic test for
fetuses with SBDs in clinical practice (19,20).
In fetuses with structurally abnormal findings
detected by ultrasound, CMA can effectively enable high-resolution
genomic screening to identify chromosomal abnormalities
undetectable by conventional karyotyping, including clinically
relevant microdeletions and microduplications (21-23). Besides, for fetuses with
ultrasound-detected multi-system structural abnormalities, CMA
demonstrates a notably higher detection rate of 41.2% for
chromosomal abnormalities, markedly surpassing the rates observed
in isolated system anomalies (such as 25.4% for cardiovascular
system abnormalities and 18.6% for central nervous system
abnormalities) (24).
Furthermore, CMA offers particular diagnostic value by identifying
~1.2% of clinically relevant abnormalities in low-risk pregnancies
with normal NIPT results (25).
Furthermore, CMA technology is relatively mature and easy to
operate, making it conducive to promoting prenatal genetic testing
in regions with limited medical resources (26).
However, CMA has inherent technical limitations in
clinical practice. The methodology carries detection risks for
variants <50 kb and cannot identify balanced translocations or
low-level mosaicism (26).
Furthermore, CMA lacks diagnostic capability for balanced
chromosomal rearrangements such as inversions and translocations
(27).
CNV-seq is an NGS technique that utilizes
low-coverage WGS (0.1-1x depth) to analyze fetal DNA for
chromosomal abnormalities and CNVs (28). With the development of NGS
technology, CNV-seq is emerging as a viable alternative to CMA for
the prenatal diagnosis (29).
CNV-seq demonstrates superior detection efficiency
for both CNVs and chromosomal abnormalities compared with NIPT in
SBDs (30,31). For fetuses with
ultrasound-detected abnormalities, CNV-seq exhibits a 2-4% higher
sensitivity compared with karyotyping in identifying chromosomal
aberrations (32,33). Moreover, CNV-seq provides
superior resolution compared with CMA, allowing for precise
detection of microdeletions, microduplications and low-level
mosaicism (<20%) (34,35).
Notably, CNV-seq not only detects all chromosomal aneuploidies and
large-scale genomic rearrangements identifiable by CMA but also
uncovers an additional 34.88% of clinically relevant variants
(34). In clinical practice, due
to its faster detection speed and smaller sample requirement,
CNV-seq can serve as a preliminary rapid test for early clinical
intervention. Meanwhile, its high compatibility with other NGS
platforms greatly simplifies testing workflows and reduces costs,
making the combination of CNV-seq with other NGS technologies a
preferred strategy in genetic testing for SBDs (36).
However, in clinical testing, CNV-seq cannot detect
balanced chromosomal translocations or inversions and has
restricted diagnostic utility in identifying polyploidy and
uniparental disomy associated with syndromic SBDs (37). Furthermore, CNV-seq exhibits
reduced sensitivity and undetermined specificity in CNV detection
when benchmarked against CMA (30,38).
Targeted sequencing represents a precision detection
methodology that employs NGS technology. This hypothesis-driven
approach utilizes customized probe or primer panels to selectively
capture and enrich predefined genomic regions for high-throughput
analysis, focusing primarily on functionally characterized regions
with established or putative disease associations (39). Through target enrichment,
targeted sequencing achieves notably enhance sequencing depth,
enabling sensitive detection of rare pathogenic SNVs/indels and
CNVs in known disease-related genes among fetuses with SBDs
(40,41). Therefore, in clinical practice,
specific genes or gene combinations can be selected based on fetal
phenotypes to perform targeted deep sequencing analysis and
diagnosis, while avoiding the blind use of WES or WGS (42).
In prenatal diagnosis, targeted sequencing serves as
a supplementary diagnostic tool for CMA-negative cases and is
particularly advantageous for fetuses with specific phenotypes,
detecting ~13.6% of pathogenic variants in those with SBDs
(42,43). Besides, the technology
demonstrates particular utility in diagnosing genetically
heterogeneous SBDs, allowing simultaneous analysis of multiple
candidate genes (44). Targeted
sequencing offers a cost-effective diagnostic solution for SBDs,
with core advantages of lower cost and short turnaround time. When
prenatal examinations suggest a fetus may have specific types of
structural abnormalities, this method can quickly confirm relevant
genetic subtypes and provide key evidence for genetic counseling
and clinical decision-making (41,45,46).
However, targeted sequencing has notable limitations
inherent to its design, which limits its application in certain
clinical scenarios; its diagnostic scope remains constrained by
current genomic knowledge, potentially missing novel pathogenic
genes or functionally relevant non-coding variants beyond
established disease associations (47). Furthermore, its capacity for
systematic investigation of complex structural variations is
intrinsically limited by restricted genomic coverage, which impedes
comprehensive genome-wide analyses (48).
WES employs an NGS technology coupled with
probe-based hybridization capture systems to specifically target
and enrich exonic regions and adjacent splice sites (±20 bp).
Although these regions constitute merely 1-2% of the human genome,
they harbor ~85% of known pathogenic variants. WES of these regions
enables the identification of clinical relevance for the majority
of SNVs/indels and CNVs in the human genome (49). Clinically, trio-WES is the most
effective exome testing method, as it enhances diagnostic rates,
elucidates variant origins and permits accurate recurrence risk
assessment and early prenatal diagnosis (50).
In SBDs, WES serves as a supplementary diagnostic
tool following inconclusive initial tests (such as CMA/CNV-seq) or
in special cases such as recurrent or lethal fetal abnormalities,
offering comprehensive exonic coverage that enhances the detection
of pathogenic variants compared with these methods (50,51). For fetuses with isolated SBDs
(such as CHDs or NTDs), WES offers an incremental diagnostic yield
of 10-50%, increasing to ~33% in cases with multi-system SBDs
(52-55). This approach is particularly
advantageous for disorders exhibiting high genetic and phenotypic
heterogeneity. Moreover, in fetuses with ultrasound-detected
cardiac or central nervous system abnormalities but negative CMA
and karyotyping results, WES effectively identifies recessive or
de novo dominant pathogenic mutations undetectable by
conventional methods, demonstrating superior diagnostic sensitivity
(56,57).
However, as WES relies on short-read sequencing
platforms, it may lead to inaccurate alignment in genomic regions
with high sequence homology, such as repetitive elements or
pseudogenes, potentially generating false-positive variant calls
(58,59). Additionally, emerging evidence
implicates that non-coding variations (including deep intronic
variants, long non-coding RNA regulatory elements and mitochondrial
DNA structural alterations) are also involved in the occurrence and
development of SBDs, while WES is mainly restricted to exonic
regions and adjacent splice sites, exhibiting markedly reduced
sensitivity for detecting these non-coding variants (60).
WGS shares technical principles with WES but extends
beyond WES's limitation to exonic regions. By enabling systematic
identification of SNVs/indels and CNVs across the entire genome,
WGS provides comprehensive genome-wide detection (49).
In fetuses with SBDs, WGS enables detection of
subtle structural variations (such as <10 kb) and yields more
precise genetic characterization (61,62). Beyond structural variants, WGS
identifies regulatory mechanisms underlying gene expression
dysregulation, including promoter/enhancer variants and deep
intronic splice-altering mutations. Notably, WGS precisely maps
complex genomic rearrangements such as intronic balanced
translocation breakpoints that disrupt normal mRNA splicing
(63,64). For fetuses exhibiting SBDs, WGS
demonstrates superior diagnostic capability by identifying both
variants detectable through CMA and WES, along with additional
pathogenic variants (65). This
comprehensive approach enhances diagnostic yield by 10-20% compared
with conventional methods. Currently, WGS offers the most
comprehensive diagnostic performance for SBDs, making it a
potential 'one-stop' test that reduces the need for repeated
testing (66).
Despite its superior sensitivity for structural
variation detection, WGS presents technical challenges. First,
genome-wide coverage generates extensive datasets with typically
low per-base sequencing depth, increasing the risk of
false-positive variant interpretation due to reduced confidence in
mutation calling (67,68). Second, the substantial costs
associated with per-sample sequencing and bioinformatic analysis
create economic barriers to implementation WGS in routine prenatal
screening programs (69). Thus,
the application of WGS in prenatal diagnosis is restricted to
supplementary roles in select cases such as fetuses with organ
system abnormalities, a history of recurrent adverse pregnancies
and negative CMA and WES results. However, with advancing
sequencing technology and falling costs, WGS is expected to become
a key tool in prenatal genetic diagnosis (70).
In genomic research, candidate genes are key genetic
elements identified through integrated bioinformatic analyses of
their potential associations with specific genetic disorders or
phenotypes. The CNVs and SNVs/indels identified through advanced
genomic technologies have precisely narrowed the search scope for
pathogenic variants via CNV analysis and SNVs/indels analysis,
while also advancing the development of genetic diagnostic methods.
Furthermore, genome-wide association studies (GWAS) have expanded
the candidate gene spectrum by further analyzing the polygenic
inheritance effects of SBDs. These findings provide critical
insights into deciphering the genetic basis of SBDs (Fig. 2).
CNVs represent a class of genomic structural
variations involving DNA segment duplications or deletions
typically >50 kb. These variations can influence gene expression
through dosage effects, gene structure disruption or regulatory
element interference (71,72). To uncover such candidate genes
related to SBDs, comprehensive bioinformatic analysis of CNV
regions and their functionally associated gene networks provides a
crucial approach (73,74) (Table II).
CNVs contribute to 10-15% of CHD cases. Recurrent
CNVs in specific genomic regions, such as 1q21.1, 22q11.2 and
16p11.2, are frequently observed in patients with a CHD (75). These regions harbor
dosage-sensitive genes critical for cardiac development,
underscoring their role as a major genetic etiology of CHDs.
TBX1 haploinsufficiency caused by 22q11.2 microdeletions
represents one of the well-characterized core genetic mechanisms
(76). A WES study by Zhao et
al (77) further revealed
that chromatin regulatory genes within the 22q11.2 region,
including EP400, KAT6A, KMT2C, KMT2D, NST1,
CHD7 and PHF21A, exhibit markedly higher mutation
frequencies in patients with a CHD compared with controls, which
may contribute to abnormal cardiac development through epigenetic
regulatory mechanism. Additionally, a case-control study using CMA
identified SLC2A3 duplications in 5.0% (18/346) of 22q11.2
deletion syndrome patients with CHDs and aortic arch abnormalities,
a frequency markedly higher than in deletion-positive individuals
with normal cardiac anatomy. This finding implicates SLC2A3
duplication as a potential genetic modifier of cardiac phenotypes
in 22q11.2 deletion syndrome (78). CNV studies of other pathogenic
regions have revealed additional candidate genes. For instance, Lin
et al (79) performed CMA
on a cohort of 1,118 Chinese fetuses with CHDs, identifying a
notable association between proximal 16p11.2 deletions (BP4-BP5
region) and CHDs; moreover, this association may be mediated
through TBX6 gene dysfunction. Another investigation of 78
non-22q11.2 deletion patients identified 15q21.1 and 2p22.3
duplications as novel pathogenic CNVs, with multiple implicated
genes (IRX4, BMPR1A, SORBS2, ID2, ROCK2, E2F6, GATA4, SOX7,
SEMAD6D, FBN1 and LTBP1) known to participate in cardiac
development (80). Mak et
al (81) performed a CMA
analysis using a large control cohort comprising 3,987 Caucasian
and 1,945 Singaporean Chinese subjects. The study identified 10
large rare CNVs, with further analysis revealing that nucleoredoxin
in the 17p13.3 region, COL4A1 and COL4A2 in 13q33.3,
and ZEB2 in the 2q22.3 region were strongly associated with
CHDs. Dasouki et al (82)
conducted CNV-seq analysis in 134 Saudi Arabian patients with CHDs,
detecting 21 copy number gains and 11 losses. The genomic variants
were primarily clustered in chromosomal regions 17q21.31, 8p11.21,
22q11.23 and 16p11.2. Functional and network analyses identified
NPHP1, PLCB1, KANSL1 and NR3C1 as potential candidate
genes associated with CHD pathogenesis.
Genetic investigations of OFCs have demonstrated the
crucial role of CNVs and their associated genes in disease
pathogenesis. Lansdon et al (83) performed CMA on cohorts from the
Philippines (n=869) and Europe (n=233) with non-syndromic cleft
lip/palate (nsCL/P), identifying recurrent 2q31.1 duplications (11
cases), 22q11.2 CNVs (4 cases) and 3q29 deletions (2 cases).
Further analyses implicated COBLL1, RIC1 and
ARHGEF38 as candidate genes, all involved in Rho/Rab GTPase
signaling pathways associated with OFCs and craniofacial anomalies.
Additionally, a separate multi-center study involving 270 orofacial
clefts cases identified a rare duplication variant at 19p13.12,
suggesting its potential as a pathogenic locus. The variant affects
SYDE1, BRD4 and AKAP8, which exhibit
spatiotemporally specific expression patterns in the branchial
arches and frontonasal processes of E10.5 mouse embryos. These
findings indicate that this gene cluster may represent a novel
susceptibility factor for nsCL/P (84). Furthermore, in a CMA analysis of
467 OFC trios (comprising 1,375 subjects) and 391 control trios
(902 subjects), Younkin et al (85) identified a genome-wide
significant 62 kb non-coding region at 7p14.1 and proposed
TARP as a candidate gene for the early prenatal diagnosis of
OFCs, suggesting a potential role of T-cell receptors in OFC
pathogenesis. A further study on this cohort identified two
deletions: A 67 kb deletion in MGAM on chromosome 7q34 and a
206 kb deletion spanning ADAM3A and ADAM5 in the 8p11
region. The frequency of these deletions was markedly higher in
affected families compared with in unaffected controls, further
supporting their potential role as candidate genes (86).
CNV analysis has been used to identify associations
among common and rare genetic variants and NTD risks and has
identified several promising candidate genes. A WES study analyzed
patients with meningomyelocele and identified 6 cases harboring a
22q11.2 deletion, which included an LCR22C-D deletion within the
low-copy repeat region. Furthermore, the study pinpointed
AIFM3, CRKL and PI4KA as key candidate genes within
this locus (87). Additionally,
a WGS-based analysis involving cohorts from the United States and
Qatar (comprising 140 patients with spina bifida and 183 controls)
expanded the gene network associated with NTD-related CNVs. Rare
coding CNVs were identified in pathways critical to NTD
pathogenesis, including cell cycle regulation (SH3GL3 and
PARD3), mitochondrial metabolism (DMGDH and
FOXRED1), transmembrane transport (SLC44A2 and
SLC44A3) and signal transduction (VAV2 and
DOCK10) (88). Tian et
al (89) performed CNV
analysis of all exonic regions in planar cell polarity (PCP)
pathway-related genes (VANGL1, VANGL2, CELSR1, SCRIB, DVL2,
DVL3 and PTK7) in 11 NTD probands, identifying 16 CNVs.
The CNVs were predominantly located in DVL2, VANGL1 and
VANGL2, suggesting these genes as strong candidates for NTD
pathogenesis. These findings not only delineate the multi-pathway
pathogenic framework of NTDs but also provide novel insights into
the genetic association between NTDs and CNVs.
In summary, CNVs are notably associated with the
incidence of SBDs. CNV analysis can elucidate the genetic effects
of large-fragment deletion mutations, holding substantial relevance
for identifying key pathogenic genes and exploring potential gene
dosage-phenotype associations.
Small variants typically encompass SNVs, short
insertions and short deletions (<50 bp) (16). These variants can impact the
function of critical genes, thereby contributing to the development
of SBDs (90,91). The detection and analysis of
SNVs/indels have notably enhanced the understanding of these subtle
genetic alterations in SBDs and have effectively expanded the
candidate gene spectrum for these conditions (Table III).
Through SNV/indel analyses have served a key role in
unraveling the complex genetic heterogeneity characteristic of
CHDs. In a landmark study, Sevim et al (92) performed a systematic WES analysis
on 559 families with CHDs, identifying 23 novel candidate
pathogenic genes. Notably, HSP90AA1, IQGAP1 and
TJP2 showed strong associations with isolated CHDs, whereas
ROCK2, APBB1, KDM5A and CHD4 were
primarily associated with non-isolated CHDs. Among these,
HSP90AA1, ROCK2, IQGAP1 and CHD4
exhibited the highest relevance in pathogenicity prediction models,
underscoring their central roles in disease pathogenesis. An
additional WES study involving 52 Qatari families (178 individuals)
identified four pathogenic or likely pathogenic variants: ROBO1,
SLC2A10, SMAD6 and CHD7. Notably, ROBO1 was found
to functionally interact with classical CHD-related genes such as
TBX1, TBX5, NOTCH1 and NKX2-5 in interaction
networks. Mechanistic studies using a chick heart development model
demonstrated that SMAD6 contributes to CHD pathogenesis
through synergistic regulation with NKX2-5 (93). Substantial progress has also been
made in subtype-specific CHD research. Shi et al (94) analyzed WES data from 100 patients
with pulmonary artery atresia and 100 healthy controls, integrating
network analysis and gene expression validation to establish
associations between DNAH10, DST, FAT1, HMCN1, HNRNPC, TEP1
and TYK2 with the pathological mechanisms of pulmonary
artery atresia for the first time. In the research of total
anomalous pulmonary venous connection, WES analysis of 78 sporadic
cases and 100 controls identified seven candidate genes: CLTCL1,
CST3, GXYLT1, HMGA2, SNAI1, VAV2 and ZDHHC8. Functional
verification and zebrafish model interaction network analysis
further validated SNAI1, HMGA2 and VAV2 as key
candidate genes driving total anomalous pulmonary venous connection
pathogenesis (95).
By examining protein-altering variations such as
missense and frameshift mutations, SNV/indel analysis has revealed
how coding sequence modifications induce functional changes
(loss/gain-of-function), thereby offering novel perspectives on OFC
candidate genes. WES of 84 affected individuals from 46 families
with nsCL/P identified rare deleterious variants in TP63,
TBX1, LRP6 and GRHL3, genes previously associated
with syndromic forms (96).
Mangold et al (24)
performed targeted sequencing in a cohort of 576 European patients
with nsCL/P and 96 non-syndromic cleft palate only cases,
identifying four novel truncating GRHL3 mutations. Notably,
all nine mutation carriers exclusively presented with non-syndromic
cleft palate only, providing compelling evidence for the notable
pathogenic contribution of GRHL3 to non-syndromic cleft
palate only. A subsequent WES study of 132 non-syndromic cleft
palate only cases and 623 multiethnic controls identified three
novel candidate genes (ACACB, PTPRS and MIB1), along
with de novo variants in GRHL3 and CREBBP,
genes associated with Van der Woude syndrome and Rubinstein-Taybi
syndrome, respectively (97). A
trio-WES analysis of 130 African nsCL/P families identified
pathogenic AFDN missense mutations (p.E485K and p.G1703R)
that may disrupt connexin binding, potentially impairing cell
polarity regulation and mesenchymal migration during facial
morphogenesis (98). A WES study
from Eastern Chinese cohorts demonstrated LAMA5 mutations in
nsCL/P cases, with further investigations revealing the crucial
role of LAMA5 in palatal development, establishing it as a
strong candidate gene (99).
Kumari et al (100)
identified variants in TGFβ3, MSX1 and MMP3 through
WES analysis of 245 Indian nsCL/P cases and validated these genes
as strong candidates for OFCs.
The application of SNV/indel analysis has
facilitated the identification of candidate genes within the
underlying pathways of NTDs. WES performed on 23 myelomeningocele
cases in France revealed that de novo variants in crucial
genes of the vitamin B12 metabolism pathway (LRP2, MMAA and
TCN2), genes related to folate metabolism (FPGS), the
core regulator of choline metabolism (BHMT) and GLI3,
a key regulatory element of the Sonic Hedgehog signaling pathway,
were notably linked to genetic susceptibility to NTDs (101). A WGS study involving 140
isolated spina bifida samples further identified eight rare
missense variants in the CIC gene, which were validated
through functional analysis. These CIC missense variants
found in NTD cases notably suppressed folate receptor 1 protein
expression levels and disrupted the transduction of the PCP
signaling pathway (102).
Additionally, analysis of trio-WES in 43 cases of sporadic
myelomeningocele/anencephaly and their parents identified seven
core candidate genes: SHROOM3, PAX3, GRHL3, PTPRS, WBSCR28,
MFAP1 and DDX3X. Among them, SHROOM3 is noted for
its high degree of evolutionary conservation, the knockout of its
double isoform resulted in 100% incidence of exencephaly and a 23%
penetrance rate of spina bifida phenotypes in mouse embryos. As a
key regulator of neural crest differentiation, rare variants
c.218C>A in PAX3 can potentially lead to NTD phenotypes
by disrupting neural crest cell migration during neural tube
closure (103). Building on
this understanding, Chen et al (104) employed targeted sequencing
technology to perform a comprehensive analysis of the SHROOM
gene family in a cohort of 343 individuals with NTDs and 206
control subjects in China. The findings revealed a markedly higher
frequency of damaging missense variants in the SHROOM2 gene
among the case group compared with the control group. These
mutations lead to the failure of neural tube closure by disrupting
the interaction between SHROOM2 and ROCK1 proteins, thereby
interfering with the cytoskeletal remodeling mediated by the planar
cell polarity pathway.
SNV/indel analysis systematically investigates
subtle genetic alterations overlooked in CNV analysis, precisely
mapping associated loci of SBDs and revealing the impact of
nucleotide-level modifications on gene function. This approach
notably establishes a crucial molecular foundation for optimizing
precision prevention strategies of SBDs and exploring molecular
pathways for targeted therapies.
The genetic basis of most SBDs stems from the
cumulative effects of multiple minor-effect genes rather than
single-gene mutations (105).
Consequently, identifying candidate genes involved in polygenic
synergistic pathogenesis is crucial for elucidating the genetic
factors underlying these complex disorders. GWAS analysis represent
a pivotal approach in contemporary genomics research, employing SNP
arrays or NGS technologies to systematically examine genome-wide
genetic variations. Through multivariate data analysis, GWAS
analysis elucidates associations between these variations and
phenotypic traits, thereby revealing the genetic architecture of
complex traits (106,107). Advances in GWAS applications
for SBDs have led to the discovery of numerous potential candidate
genes, representing a notable breakthrough in understanding the
molecular pathogenesis of these conditions (Table IV).
GWAS analysis has provided crucial insights into the
complex genetic architecture of CHDs. A large-scale GWAS involving
40,000 UK Biobank samples identified 130 loci notably associated
with right heart phenotypes, a number of which are located near
known regulatory genes such as NKX2-4, TBX5/TBX3, WNT9B and
GATA4. The pivotal role of these genes in cardiac
morphogenesis has been further substantiated (108). Additionally, a single-trait
GWAS analysis of 29,506 individuals with right ventricular
structural abnormalities revealed 12 candidate genes shared across
different right ventricular phenotypes, including TTN, ATXN2,
PTPN11 and ACTN4. Notably, FHOD3, MYH6,
MYL4 and TMEM43 exhibit functional overlap with
Mendelian cardiomyopathy genes, with their encoded proteins
primarily involved in myocardial contraction and cell adhesion
processes (109). Lahm et
al (110) identified
MACROD2, GOSR2, WNT3 and MSX1 as critical genes in
embryonic cardiac development, which were significantly associated
with SNPs based on CMA and GWAS analyses of 4,034 Caucasian
patients with CHDs and 8,486 healthy controls. Jin et al
(111) expanded this research
by performing WES combined with GWAS on 2,871 CHD probands. The
findings demonstrated that MYH6 mutations were notably
associated with an elevated risk of ventricular dysfunction,
whereas dominant mutations in FLT4 were specifically linked
to tetralogy of Fallot. Furthermore, rare inherited and de
novo heterozygous variants were markedly enriched in genes such
as CHD7, KMT2D, PTPN11, RBFOX2, FLT4, SMAD6 and
NOTCH1 among patients with complex CHDs, highlighting novel
candidate targets for further investigation.
GWAS analysis has identified multiple candidate
genes potentially involved in OFCs. A GWAS of a Maya population
(149 patients with nsCL/P and 303 controls) revealed notable
associations between nsCL/P and genetic variants in IRF6, as
well as loci at 8q24, 10q25 and 17q22. Single-marker analysis
further confirmed these associations, particularly for IRF6,
8q24 and 10q25 (112). A
pleiotropy-informed conditional false discovery rate analysis,
incorporating 814 nsCL/P cases, 205 nsCPO cases and 2,159 controls
from an African population GWAS, along with mouse facial
development expression data, identified 19 potential candidate
genes. Among these, MDN1, MAP3K7, KMT2A and ARCN1
emerged as top candidates, supported by evidence from mouse models
or individuals harboring pathogenic variants in these genes
(113). In a separate GWAS of
oral clefts in a Polish cohort, eight single-nucleotide
polymorphisms in PAX7 were strongly associated with nsCL/P,
with the etiological role of the gene independently validated in a
cohort of 247 patients and 445 controls (114). These findings not only expand
the repertoire of candidate genes implicated in OFCs but also
provide a molecular framework for understanding how genetic
background influences craniofacial development across diverse
populations. The application of GWAS has notably advanced the
identification of candidate genes for SBDs, with large sample sizes
enhancing result accuracy.
GWAS analyses enhance the understanding of SBDs
with polygenic inheritance patterns by analyzing genome-wide
associations between genetic variants and these defects while
quantifying the cumulative effects of multiple variants.
Furthermore, GWAS analysis provides a comprehensive assessment of
gene-gene interactions throughout the genome, filling the research
gap in the study of polygenic effects in CNV analysis or SNV/indel
analysis, and greatly expanding the potential candidate gene pool
associated with SBDs.
The clinical evaluation of SBDs has been
revolutionized by continuous innovations in genomic technologies,
establishing a tiered diagnostic pathway. For fetuses with
ultrasound anomalies, CMA or CNV-seq serves as first-tier testing.
In cases with negative results, particularly those with a complex
phenotype or adverse pregnancy history, NGS methodologies such as
WES or targeted sequencing provide a subsequent high-yield option.
This evolving diagnostic paradigm enhances variant detection,
facilitates precise genetic diagnosis and ultimately informs
prenatal counseling, prognosis and recurrence risk assessment. WGS,
through comprehensive detection of the entire genome, further
improves the detection rate of pathogenic genes; although its
clinical application is still limited due to technical constraints
and cost factors, WGS has the potential to become the 'ultimate'
prenatal diagnostic method.
Notwithstanding notable progress in genomics that
has advanced research on SBDs, numerous challenges remain in
pathogenic variant detection and candidate genes screening
strategies. Current genomic approaches exhibit technical
limitations in sensitivity and specificity when detecting specific
genetic variants (61,116,117). CMA fails to identify certain
complex structural variations, while NGS inherently misaligns
sequences in GC-rich regions, resulting in undetected mutations
within these GC-rich areas and consequent false-negative findings
(118,119). Additionally, the short-read
(<300 bp) nature of NGS presents inherent limitations,
demonstrating particularly poor mapping quality for homologous
region sequences, with frequent read misalignments from homologous
segments leading to insufficient coverage. For repetitive genomic
regions, reliable read mapping requires the presence of unique
flanking sequences adjacent to the repetitive elements (120). Single-omics-based candidate
gene screening strategies impose additional limitations on genetic
dissection of SBDs. Genomics-dependent analyses cannot
authentically validate the functional disruption and regulatory
network imbalance caused by pathogenic variants. Moreover, numerous
detected genetic variants remain classified as variants of
uncertain relevance, substantially constraining variant
interpretation and accurate classification (121,122). Furthermore, insufficient cohort
sizes and a lack of ethnic diversity in current studies not only
constrain the discovery of pathogenic variants and analysis of
candidate genes, but also compromise the generalizability of
diagnostic findings, particularly hindering the clinical
translation of candidate gene research outcomes across diverse
populations (105). Therefore,
it is necessary to utilize large-scale, multi-ethnic biobanks
established through international collaboration, which provide
statistically robust cohorts for screening and validating candidate
genes while reducing the impact of population representation
bias.
Therefore, improving the identification of
pathogenic variants and candidate gene screening strategies
represents a critical challenge for advancing the understanding of
the genetics of SBDs (Fig. 3).
In recent years, high-precision genomic technologies, particularly
third-generation sequencing (TGS) and optical genome mapping (OGM),
have emerged as powerful tools in genetic disorder exploration
(120). TGS platforms, with
their long-read (>10 kb) sequencing capabilities, effectively
address short-read sequencing limitations by enabling precise
characterization of structurally complex genomic regions, including
telomeres and centromeres (123-125). TGS has demonstrated promising
application prospects in neonatal genetic disorders but has not yet
been applied to research on SBDs (126). OGM demonstrates exceptional
capabilities in detecting cryptic structural variations through
fluorescent labeling of DNA fragments >150 kb (123,127). In the field of NTDs research,
Sahajpal et al (128)
pioneered the application of OGM technology to perform
comprehensive analysis of genome-wide structural variations in 104
patient samples with NTD. The findings not only identified notable
associations between NTDs and genes including RMND5A, HNRNPC,
FOXD4 and RBBP4, but also extended for the first time
the phenotypic spectrum of AMER1 and TGIF1 genes to
encompass NTDs. This groundbreaking study demonstrated the crucial
role of genomic structural variations in the pathogenesis of such
SBDs, providing essential molecular genetic evidence for subsequent
research in this field. Furthermore, the rapid advancement of
bioinformatics has notably optimized the genomic-based analysis
pipeline for pathogenic variants. For instance, epistasis analysis
enables the examination of joint genetic effects across multiple
loci, helping to elucidate a portion of the 'missing heritability'
that encompasses marginal genetic effects undetectable by
conventional GWAS (129,130).
Population stratification methods, such as principal component
analysis or linear mixed models with random effects, can
effectively mitigate confounding effects from population
stratification and structure in GWAS (131,132). Multi-omics approaches enable
mechanistic investigations into how candidate genes disrupt
cellular pathways, proving particularly valuable for elucidating
the functions of regulatory elements in non-coding regions that
were previously overlooked in genomic studies (133). Therefore, it has further
refined candidate gene screening strategies, providing novel
perspectives for deciphering the genetic basis of SBDs (134).
In summary, the present review delineates current
advanced genomic technologies applied in SBD research and
diagnostics, while providing a comparative analysis of their
respective advantages and limitations. The present review further
consolidates candidate genes identified through these technological
approaches, with these clinically-relevant candidates being derived
from human clinical specimens that more faithfully recapitulate
disease pathogenesis compared with animal or cellular models. The
investigation of genetic contributors to SBDs has now entered a
transformative era marked by multi-omics convergence and
technological synergy. Through the implementation of
higher-resolution platforms, expanded cohort sizes, refined
bioinformatics analyses and sophisticated multi-omics integration,
researchers are now empowered to more systematically and precisely
identify pathogenic variants associated with SBDs, thereby
uncovering clinically actionable candidate genes.
Not applicable.
RX and WH conceived the idea; RX contributed to
writing the initial draft and preparing the figures; HR designed
the tables; WH and HG contributed to the improvement of the initial
draft and revision; RX, ZY and HR helped proofread and improved the
manuscript; RX, WH, ZY and HG critically reviewed and edited the
manuscript. All authors have read and approved the final version of
the manuscript. Data authentication is not applicable.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
This work was supported by the National Natural Science
Foundation of China (grant no. 82502070), National Natural Science
Foundation of China (grant no. 82271730) and Shenyang Science and
Technology Planning Project (grant no. 22-321-33-22).
|
1
|
Hobbs CA, Chowdhury S, Cleves MA, Erickson
S, MacLeod SL, Shaw GM, Shete S, Witte JS and Tycko B: Genetic
epidemiology and nonsyndromic structural birth defects: From
candidate genes to epigenetics. Jama Pediatr. 168:371–377. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mone F, Quinlan-Jones E, Ewer AK and Kilby
MD: Exome sequencing in the assessment of congenital malformations
in the fetus and neonate. Arch Dis Child Fetal Neonatal Ed.
104:F452–F456. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kang L, Guo Z, Shang W, Cao GY, Zhang YP,
Wang QM, Shen HP, Liang WN and Liu M: Perinatal prevalence of birth
defects in the Mainland of China, 2000-2021: A systematic review
and meta-analysis. World J Pediatr. 20:669–681. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Basu M, Zhu J, LaHaye S, Majumdar U, Jiao
K, Han Z and Garg V: Epigenetic mechanisms underlying maternal
diabetes-associated risk of congenital heart disease. JCI Insight.
2:e950852017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Nasreddine G, El HJ and Ghassibe-Sabbagh
M: Orofacial clefts embryology, classification, epidemiology, and
genetics. Mutat Res Rev Mutat. 787:1083732021. View Article : Google Scholar
|
|
6
|
Buijtendijk MF, Bet BB, Leeflang MM, Shah
H, Reuvekamp T, Goring T, Docter D, Timmerman MG, Dawood Y,
Lugthart MA, et al: Diagnostic accuracy of ultrasound screening for
fetal structural abnormalities during the first and second
trimester of pregnancy in low-risk and unselected populations.
Cochrane Database Syst Rev. 5:CD147152024.
|
|
7
|
van Nisselrooij AEL, Teunissen AKK, Clur
SA, Rozendaal L, Pajkrt E, Linskens IH, Rammeloo L, van Lith JMM,
Blom NA and Haak MC: Why are congenital heart defects being missed?
Ultrasound Obstet Gynecol. 55:747–757. 2020. View Article : Google Scholar :
|
|
8
|
Hematian MN, Hessami K, Torabi S and Saleh
M, Nouri B and Saleh M: A prospective cohort study on association
of first-trimester serum biomarkers and risk of isolated foetal
congenital heart defects. Biomarkers. 26:747–751. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang X, Yang X, Huang P, Meng X, Bian Z
and Meng L: Identification of maternal serum biomarkers for
prenatal diagnosis of nonsyndromic orofacial clefts. Ann NY Acad
Sci. 1510:167–179. 2022. View Article : Google Scholar
|
|
10
|
Lupo PJ, Archer NP, Harris RD, Marengo LK,
Schraw JM, Hoyt AT, Tanksley S, Lee R, Drummond-Borg M, Freedenberg
D, et al: Newborn screening analytes and structural birth defects
among 27,000 newborns. PLoS One. 19:e3042382024. View Article : Google Scholar
|
|
11
|
Li L, He Z, Huang X, Lin S, Wu J, Huang L,
Wan Y and Fang Q: Chromosomal abnormalities detected by karyotyping
and microarray analysis in twins with structural anomalies.
Ultrasound Obstet Gynecol. 55:502–509. 2020. View Article : Google Scholar
|
|
12
|
Jayashankar SS, Nasaruddin ML, Hassan MF,
Dasrilsyah RA, Shafiee MN, Ismail NAS and Alias E: Non-invasive
prenatal testing (NIPT): Reliability, challenges, and future
directions. Diagnostics (Basel). 13:25702023. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Feldkamp ML, Carey JC, Byrne JLB, Krikov S
and Botto LD: Etiology and clinical presentation of birth defects:
Population based study. BMJ. 357:j22492017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang J: What has genomics taught an
evolutionary biologist? Genom Proteom Bioinf. 21:1–12. 2023.
View Article : Google Scholar
|
|
15
|
Green ED, Gunter C, Biesecker LG, Di
Francesco V, Easter CL, Feingold EA, Felsenfeld AL, Kaufman DJ,
Ostrander EA, Pavan WJ, et al: Strategic vision for improving human
health at The Forefront of Genomics. Nature. 586:683–692. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kosugi S, Momozawa Y, Liu X, Terao C, Kubo
M and Kamatani Y: Comprehensive evaluation of structural variation
detection algorithms for whole genome sequencing. Genome Biol.
20:1172019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Audano PA, Sulovari A, Graves-Lindsay TA,
Cantsilieris S, Sorensen M, Welch AE, Dougherty ML, Nelson BJ, Shah
A, Dutcher SK, et al: Characterizing the major structural variant
alleles of the human genome. Cell. 176:663–675. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Levy B and Wapner R: Prenatal diagnosis by
chromosomal microarray analysis. Fertil Steril. 109:201–212. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Committee Opinion No. 682 Summary:
Microarrays and Next-Generation Sequencing Technology: The use of
advanced genetic diagnostic tools in obstetrics and gynecology.
Obstet Gynecol. 128:1462–1463. 2016. View Article : Google Scholar
|
|
20
|
Hu P, Zhang Q, Cheng Q, Luo C, Zhang C,
Zhou R, Meng L, Huang M, Wang Y, Wang Y, et al: Whole genome
sequencing vs chromosomal microarray analysis in prenatal
diagnosis. Am J Obstet Gynecol. 229:301–302. 2023. View Article : Google Scholar
|
|
21
|
Lan L, Luo D, Lian J, She L, Zhang B,
Zhong H, Wang H and Wu H: Chromosomal abnormalities detected by
chromosomal microarray analysis and karyotype in fetuses with
ultrasound abnormalities. Int J Gen Med. 17:4645–4658. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rodriguez-Revenga L, Madrigal I, Borrell
A, Martinez JM, Sabria J, Martin L, Jimenez W, Mira A, Badenas C
and Milà M: Chromosome microarray analysis should be offered to all
invasive prenatal diagnostic testing following a normal rapid
aneuploidy test result. Clin Genet. 98:379–383. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xia M, Yang X, Fu J, Teng Z, Lv Y and Yu
L: Application of chromosome microarray analysis in prenatal
diagnosis. BMC Pregnancy Childbirth. 20:6962020. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Mangold E, Böhmer AC, Ishorst N, Hoebel
AK, Gültepe P, Schuenke H, Klamt J, Hofmann A, Gölz L, Raff R, et
al: Sequencing the GRHL3 coding region reveals rare truncating
mutations and a common susceptibility variant for nonsyndromic
cleft palate. AM J Hum Genet. 98:755–762. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maya I, Salzer SL, Brabbing-Goldstein D,
Matar R, Kahana S, Agmon-Fishman I, Klein C, Gurevitch M,
Basel-Salmon L and Sagi-Dain L: Residual risk for clinically
significant copy number variants in low-risk pregnancies, following
exclusion of noninvasive prenatal screening-detectable findings. Am
J Obstet Gynecol. 226:562.e1–562.e8. 2022. View Article : Google Scholar
|
|
26
|
Liu X, Liu S, Wang H and Hu T: Potentials
and challenges of chromosomal microarray analysis in prenatal
diagnosis. Front Genet. 13:9381832022. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee CL, Lee CH, Chuang CK, Chiu HC, Chen
YJ, Chou CL, Wu PS, Chen CP, Lin HY and Lin SP: Array-CGH increased
the diagnostic rate of developmental delay or intellectual
disability in Taiwan. Pediatr Neonatol. 60:453–460. 2019.
View Article : Google Scholar
|
|
28
|
Xie C and Tammi MT: CNV-seq, a new method
to detect copy number variation using high-throughput sequencing.
BMC Bioinformatics. 10:802009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang J, Chen L, Zhou C, Wang L, Xie H,
Xiao Y, Zhu H, Hu T, Zhang Z, Zhu Q, et al: Prospective chromosome
analysis of 3429 amniocentesis samples in China using copy number
variation sequencing. AM J Obstet Gynecol. 219:287.e1–287.e18.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang L, Yang J, Bu G, Han R, Rezhake J and
La X: Efficiency of Non-invasive prenatal testing in detecting
fetal copy number variation: A retrospective cohort study. Int J
Womens Health. 16:1661–1669. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wang X, Sha J, Han Y, Pang M, Liu M, Liu
M, Zhang B and Zhai J: Efficiency of copy number variation
sequencing combined with karyotyping in fetuses with congenital
heart disease and the following outcomes. Mol Cytogenet. 17:122024.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Huang Y, Fu S, Shao D, Yao Y, Wu F and Yao
M: Comprehensive chromosomal abnormality detection: Integrating
CNV-Seq with traditional karyotyping in prenatal diagnostics. BMC
Med Genomics. 18:812025. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Luo H, Wang Q, Fu D, Gao J and Lu D:
Additional diagnostic value of CNV-seq over conventional
karyotyping in prenatal diagnosis: A systematic review and
meta-analysis. J Obstet Gynaecol Res. 49:1641–1650. 2023.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ma N, Xi H, Chen J, Peng Y, Jia Z, Yang S,
Hu J, Pang J, Zhang Y, Hu R, et al: Integrated CNV-seq, karyotyping
and SNP-array analyses for effective prenatal diagnosis of
chromosomal mosaicism. BMC Med Genomics. 14:562021. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhao X and Fu L: Efficacy of copy-number
variation sequencing technology in prenatal diagnosis. J Perinat
Med. 47:651–655. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Chen X, Jiang Y, Chen R, Qi Q, Zhang X,
Zhao S, Liu C, Wang W, Li Y, Sun G, et al: Clinical efficiency of
simultaneous CNV-seq and whole-exome sequencing for testing fetal
structural anomalies. J Transl Med. 20:102022. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen Y, Han X, Hua R, Li N, Zhang L, Hu W,
Wang Y, Qian Z and Li S: Copy number variation sequencing for the
products of conception: What is the optimal testing strategy. Clin
Chim Acta. 557:1178842024. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yao R, Zhang C, Yu T, Li N, Hu X, Wang X,
Wang J and Shen Y: Evaluation of three read-depth based CNV
detection tools using whole-exome sequencing data. Mol Cytogenet.
10:302017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pei XM, Yeung MHY, Wong ANN, Tsang HF, Yu
ACS, Yim AKY and Wong SCC: Targeted sequencing approach and its
clinical applications for the molecular diagnosis of human
diseases. Cells. 12:4932023. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hu P, Qiao F, Wang Y, Meng L, Ji X, Luo C,
Xu T, Zhou R, Zhang J, Yu B, et al: Clinical application of
targeted next-generation sequencing in fetuses with congenital
heart defect. Ultrasound Obstet Gynecol. 52:205–211. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gong B, Li D, Łabaj PP, Pan B,
Novoradovskaya N, Thierry-Mieg D, Thierry-Mieg J, Chen G, Bergstrom
Lucas A, LoCoco JS, et al: Targeted DNA-seq and RNA-seq of
reference samples with Short-read and Long-read sequencing. Sci
Data. 11:8922024. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Vora NL and Norton ME: Prenatal exome and
genome sequencing for fetal structural abnormalities. Am J Obstet
Gynecol. 228:140–149. 2023. View Article : Google Scholar :
|
|
43
|
Allen VM, Schollenberg E, Aberg E and
Brock JK: Use of Clinically informed strategies and diagnostic
yields of genetic testing for fetal structural anomalies following
a non-diagnostic microarray result: A Population-based cohort
study. Prenatal Diag. 45:318–325. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Pangalos C, Hagnefelt B, Lilakos K and
Konialis C: First applications of a targeted exome sequencing
approach in fetuses with ultrasound abnormalities reveals an
important fraction of cases with associated gene defects. PeerJ.
4:e19552016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Li Y, Anderson LA, Ginns EI and Devlin JJ:
Cost effectiveness of karyotyping, chromosomal microarray analysis,
and targeted Next-generation sequencing of patients with
unexplained global developmental delay or intellectual disability.
Mol Diagn Ther. 22:129–138. 2018. View Article : Google Scholar
|
|
46
|
Li AH, Hanchard NA, Furthner D, Fernbach
S, Azamian M, Nicosia A, Rosenfeld J, Muzny D, D'Alessandro LCA,
Morris S, et al: Whole exome sequencing in 342 congenital cardiac
left sided lesion cases reveals extensive genetic heterogeneity and
complex inheritance patterns. Genome Med. 9:952017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Tarozzi M, Bartoletti-Stella A, Dall'Olio
D, Matteuzzi T, Baiardi S, Parchi P, Castellani G and Capellari S:
Identification of recurrent genetic patterns from targeted
sequencing panels with advanced data science: A case-study on
sporadic and genetic neurodegenerative diseases. BMC Med Genomics.
15:262022. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Iyer SV, Goodwin S and McCombie WR:
Leveraging the power of long reads for targeted sequencing. Genome
Res. 34:1701–1718. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Mandlik JS, Patil AS and Singh S:
Next-generation sequencing (NGS): Platforms and applications. J
Pharm Bioallied Sci. 16(Supp 1): S41–S45. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Flores WG and Pereira WC: A contrast
enhancement method for improving the segmentation of breast lesions
on ultrasonography. Comput Biol Med. 80:14–23. 2017. View Article : Google Scholar
|
|
51
|
Jelin AC and Vora N: Whole Exome
sequencing: Applications in prenatal genetics. Obstet Gyn Clin N
Am. 45:69–81. 2018. View Article : Google Scholar
|
|
52
|
Chen M, Chen J, Wang C, Chen F, Xie Y, Li
Y, Li N, Wang J, Zhang VW and Chen D: Clinical application of
medical exome sequencing for prenatal diagnosis of fetal structural
anomalies. Eur J Obstet Gynecol Reprod Biol. 251:119–124. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Emms A, Castleman J, Allen S, Williams D,
Kinning E and Kilby M: Next generation sequencing after invasive
prenatal testing in fetuses with congenital malformations: Prenatal
or neonatal investigation. Genes (Basel). 13:15172022. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Qin Y, Yao Y, Liu N, Wang B, Liu L, Li H,
Gao T, Xu R, Wang X, Zhang F and Song J: Prenatal whole-exome
sequencing for fetal structural anomalies: A retrospective analysis
of 145 Chinese cases. BMC Med Genomics. 16:2622023. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Pauta M, Martinez-Portilla RJ and Borrell
A: Diagnostic yield of exome sequencing in fetuses with multisystem
malformations: Systematic review and meta-analysis. Ultrasound
Obstet Gynecol. 59:715–722. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Reches A, Hiersch L, Simchoni S, Barel D,
Greenberg R, Ben Sira L, Malinger G and Yaron Y: Whole-exome
sequencing in fetuses with central nervous system abnormalities. J
Perinatol. 38:1301–1308. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Li M, Ye B, Chen Y, Gao L, Wu Y and Cheng
W: Analysis of genetic testing in fetuses with congenital heart
disease of single atria and/or single ventricle in a Chinese
prenatal cohort. BMC Pediatr. 23:5772023. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Jaillard S, McElreavy K, Robevska G,
Akloul L, Ghieh F, Sreenivasan R, Beaumont M, Bashamboo A,
Bignon-Topalovic J, Neyroud AS, et al: STAG3 homozygous missense
variant causes primary ovarian insufficiency and male
non-obstructive azoospermia. Mol Hum Reprod. 26:665–677. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Chong AS, Chong G, Foulkes WD and Saskin
A: Reclassification of a frequent African-origin variant from PMS2
to the pseudogene PMS2CL. Hum Mutat. 41:749–752. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Beaman MM, Yin W, Smith AJ, Sears PR,
Leigh MW, Ferkol TW, Kearney B, Olivier KN, Kimple AJ, Clarke S, et
al: Promoter deletion leading to allele specific expression in a
genetically unsolved case of primary ciliary dyskinesia. Am J Med
Genet A. 197:e638802025. View Article : Google Scholar
|
|
61
|
Sarwal V, Niehus S, Ayyala R, Kim M,
Sarkar A, Chang S, Lu A, Rajkumar N, Darfci-Maher N, Littman R, et
al: A comprehensive benchmarking of WGS-based deletion structural
variant callers. Brief Bioinform. 23:bbac2212022. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Lindstrand A, Eisfeldt J, Pettersson M,
Carvalho CMB, Kvarnung M, Grigelioniene G, Anderlid BM, Bjerin O,
Gustavsson P, Hammarsjö A, et al: From cytogenetics to
cytogenomics: Whole-genome sequencing as a first-line test
comprehensively captures the diverse spectrum of disease-causing
genetic variation underlying intellectual disability. Genome Med.
11:682019. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Reurink J, Weisschuh N, Garanto A, Dockery
A, van den Born LI, Fajardy I, Haer-Wigman L, Kohl S, Wissinger B,
Farrar GJ, et al: Whole genome sequencing for USH2A-associated
disease reveals several pathogenic deep-intronic variants that are
amenable to splice correction. HGG Adv. 4:1001812023.PubMed/NCBI
|
|
64
|
Fadaie Z, Whelan L, Ben-Yosef T, Dockery
A, Corradi Z, Gilissen C, Haer-Wigman L, Corominas J, Astuti GDN,
de Rooij L, et al: Whole genome sequencing and in vitro splice
assays reveal genetic causes for inherited retinal diseases. NPJ
Genom Med. 6:972021. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Goncalves A, Fortuna A, Ariyurek Y,
Oliveira ME, Nadais G, Pinheiro J, den Dunnen JT, Sousa M, Oliveira
J and Santos R: Integrating Whole-genome sequencing in clinical
genetics: A novel disruptive structural rearrangement identified in
the dystrophin gene (DMD). Int J Mol Sci. 23:592021. View Article : Google Scholar
|
|
66
|
Iurillo AM, Sharifi S and Jafferany M:
Artificial intelligence in dermatology: Ethical dilemmas and
diagnostic decisions. J Am Acad Dermatol. Aug 25–2025.Epub ahead of
print. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kishikawa T, Momozawa Y, Ozeki T,
Mushiroda T, Inohara H, Kamatani Y, Kubo M and Okada Y: Empirical
evaluation of variant calling accuracy using ultra-deep
whole-genome sequencing data. Sci Rep. 9:17842019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Dippenaar A, Ismail N, Heupink TH,
Grobbelaar M, Loubser J, Van Rie A and Warren RM: Droplet based
whole genome amplification for sequencing minute amounts of
purified Mycobacterium tuberculosis DNA. Sci Rep. 14:99312024.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Testard Q, Vanhoye X, Yauy K, Naud ME,
Vieville G, Rousseau F, Dauriat B, Marquet V, Bourthoumieu S,
Geneviève D, et al: Exome sequencing as a first-tier test for copy
number variant detection: Retrospective evaluation and prospective
screening in 2418 cases. J Med Genet. 59:1234–1240. 2022.
View Article : Google Scholar : PubMed/NCBI
|
|
70
|
International Society for Prenatal
Diagnosis; Society for Maternal Fetal Medicine; Perinatal Quality
Foundation: Joint Position Statement from the International Society
for Prenatal Diagnosis (ISPD), the Society for Maternal Fetal
Medicine (SMFM), and the Perinatal Quality Foundation (PQF) on the
use of genome-wide sequencing for fetal diagnosis. Prenatal Diag.
38:6–9. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Haller M, Au J, O'Neill M and Lamb DJ:
16p11.2 transcription factor MAZ is a dosage-sensitive regulator of
genitourinary development. Proc Natl Acad Sci U S A.
115:E1849–E1858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Flottmann R, Kragesteen BK, Geuer S, Socha
M, Allou L, Sowińska-Seidler A, Bosquillon de Jarcy L, Wagner J,
Jamsheer A, Oehl-Jaschkowitz B, et al: Noncoding copy-number
variations are associated with congenital limb malformation. Genet
Med. 20:599–607. 2018. View Article : Google Scholar
|
|
73
|
Costain G, Silversides CK and Bassett AS:
The importance of copy number variation in congenital heart
disease. NPJ Genom Med. 1:160312016. View Article : Google Scholar
|
|
74
|
Chen X, Shen Y, Gao Y, Zhao H, Sheng X,
Zou J, Lip V, Xie H, Guo J, Shao H, et al: Detection of copy number
variants reveals association of cilia genes with neural tube
defects. PLoS One. 8:e544922013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Ehrlich L and Prakash SK: Copy-number
variation in congenital heart disease. Curr Opin Genet Dev.
77:1019862022. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Fulcoli FG, Franzese M, Liu X, Zhang Z,
Angelini C and Baldini A: Rebalancing gene haploinsufficiency in
vivo by targeting chromatin. Nat Commun. 7:116882016. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Zhao Y, Wang Y, Shi L, McDonald-McGinn DM,
Crowley TB, McGinn DE, Tran OT, Miller D, Lin JR, Zackai E, et al:
Chromatin regulators in the TBX1 network confer risk for
conotruncal heart defects in 22q11.2DS. NPJ Genom Med. 8:172023.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Mlynarski EE, Sheridan MB, Xie M, Guo T,
Racedo SE, McDonald-McGinn DM, Gai X, Chow EW, Vorstman J, Swillen
A, et al: Copy-number variation of the glucose transporter gene
SLC2A3 and congenital heart defects in the 22q11.2 deletion
syndrome. Am J Hum Genet. 96:753–764. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lin S, Shi S, Lu J, He Z, Li D, Huang L,
Huang X, Zhou Y and Luo Y: Contribution of genetic variants to
congenital heart defects in both singleton and twin fetuses: A
Chinese cohort study. Mol Cytogenet. 17:22024. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Molck MC, Simioni M, Paiva VT, Sgardioli
IC, Paoli Monteiro F, Souza J, Fett-Conte AC, Félix TM, Lopes
Monlléo I and Gil-da-Silva-Lopes VL: Genomic imbalances in
syndromic congenital heart disease. J Pediatr (Rio J). 93:497–507.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Mak C, Chow PC, Liu APY, Chan KYK, Chu
YWY, Mok GTK, Leung GKC, Yeung KS, Chau AKT, Lowther C, et al: De
novo large rare copy-number variations contribute to conotruncal
heart disease in Chinese patients. NPJ Genom Med. 1:160332016.
View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Dasouki MJ, Wakil SM, Al-Harazi O,
Alkorashy M, Muiya NP, Andres E, Hagos S, Aldusery H, Dzimiri N,
Colak D, et al: New insights into the impact of Genome-wide copy
number variations on complex congenital heart disease in Saudi
Arabia. OMICS. 24:16–28. 2020. View Article : Google Scholar
|
|
83
|
Lansdon LA, Dickinson A, Arlis S, Liu H,
Hlas A, Hahn A, Bonde G, Long A, Standley J, Tyryshkina A, et al:
Genome-wide analysis of copy-number variation in humans with cleft
lip and/or cleft palate identifies COBLL1, RIC1, and ARHGEF38 as
clefting genes. Am J Hum Genet. 110:71–91. 2023. View Article : Google Scholar :
|
|
84
|
Cao Y, Li Z, Rosenfeld JA, Pursley AN,
Patel A, Huang J, Wang H, Chen M, Sun X, Leung TY, et al:
Contribution of genomic copy-number variations in prenatal oral
clefts: A multi-center cohort study. Genet Med. 18:1052–1055. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Younkin SG, Scharpf RB, Schwender H,
Parker MM, Scott AF, Marazita ML, Beaty TH and Ruczinski I: A
genome-wide study of de novo deletions identifies a candidate locus
for non-syndromic isolated cleft lip/palate risk. BMC Genet.
15:242014. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Younkin SG, Scharpf RB, Schwender H,
Parker MM, Scott AF, Marazita ML, Beaty TH and Ruczinski I: A
genome-wide study of inherited deletions identified two regions
associated with nonsyndromic isolated oral clefts. Birth Defects
Res A Clin Mol Teratol. 103:276–283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Vong KI, Lee S, Au KS, Crowley TB, Capra
V, Martino J, Haller M, Araújo C, Machado HR, George R, et al: Risk
of meningomyelocele mediated by the common 22q11.2 deletion.
Science. 384:584–590. 2024. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Wolujewicz P, Aguiar-Pulido V, AbdelAleem
A, Nair V, Thareja G, Suhre K, Shaw GM, Finnell RH, Elemento O and
Ross ME: Genome-wide investigation identifies a rare copy-number
variant burden associated with human spina bifida. Genet Med.
23:1211–1218. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Tian T, Lei Y, Chen Y, Guo Y, Jin L,
Finnell RH, Wang L and Ren A: Rare copy number variations of planar
cell polarity genes are associated with human neural tube defects.
Neurogenetics. 21:217–225. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Zhai Y, Zhang Z, Shi P, Martin DM and Kong
X: Incorporation of exome-based CNV analysis makes trio-WES a more
powerful tool for clinical diagnosis in neurodevelopmental
disorders: A retrospective study. Hum Mutat. 42:990–1004. 2021.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Yang L, Wei Z, Chen X, Hu L, Peng X, Wang
J, Lu C, Kong Y, Dong X, Ni Q, et al: Use of medical exome
sequencing for identification of underlying genetic defects in
NICU: Experience in a cohort of 2303 neonates in China. Clin Genet.
101:101–109. 2022. View Article : Google Scholar
|
|
92
|
Sevim BC, Zhang P, Tristani-Firouzi M,
Gelb BD and Itan Y: De novo variants in exomes of congenital heart
disease patients identify risk genes and pathways. Genome Med.
12:92020. View Article : Google Scholar
|
|
93
|
Okashah S, Vasudeva D, El Jerbi A,
Khodjet-El-Khil H, Al-Shafai M, Syed N, Kambouris M, Udassi S,
Saraiva LR, Al-Saloos H, et al: Investigation of genetic causes in
patients with congenital heart disease in Qatar: Findings from the
Sidra cardiac Registry. Genes (Basel). 13:13692022. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Shi X, Zhang L, Bai K, Xie H, Shi T, Zhang
R, Fu Q, Chen S, Lu Y, Yu Y and Sun K: Identification of rare
variants in novel candidate genes in pulmonary atresia patients by
next generation sequencing. Comput Struct Biotechnol J. 18:381–392.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Shi X, Huang T, Wang J, Liang Y, Gu C, Xu
Y, Sun J, Lu Y, Sun K, Chen S and Yu Y: Next-generation sequencing
identifies novel genes with rare variants in total anomalous
pulmonary venous connection. EBioMedicine. 38:217–227. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Basha M, Demeer B, Revencu N, Helaers R,
Theys S, Bou Saba S, Boute O, Devauchelle B, Francois G, Bayet B
and Vikkula M: Whole exome sequencing identifies mutations in 10%
of patients with familial non-syndromic cleft lip and/or palate in
genes mutated in well-known syndromes. J Med Genet. 55:449–458.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Hoebel AK, Drichel D, van de Vorst M,
Böhmer AC, Sivalingam S, Ishorst N, Klamt J, Gölz L, Alblas M,
Maaser A, et al: Candidate genes for nonsyndromic cleft palate
detected by exome sequencing. J Dent Res. 96:1314–1321. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Awotoye W, Mossey PA, Hetmanski JB, Gowans
LJJ, Eshete MA, Adeyemo WL, Alade A, Zeng E, Adamson O, James O, et
al: Damaging mutations in AFDN contribute to risk of nonsyndromic
cleft lip with or without cleft palate. Cleft Palate Craniofac J.
61:697–705. 2024. View Article : Google Scholar
|
|
99
|
Fu Z, Yue J, Xue L, Xu Y, Ding Q and Xiao
W: Using whole exome sequencing to identify susceptibility genes
associated with nonsyndromic cleft lip with or without cleft
palate. Mol Genet Genomics. 298:107–118. 2023. View Article : Google Scholar
|
|
100
|
Kumari P, Singh SK and Raman R: TGFβ3,
MSX1, and MMP3 as Candidates for NSCL+/-P in an Indian population.
Cleft Palate-Cran J. 56:363–372. 2019. View Article : Google Scholar
|
|
101
|
Renard E, Chery C, Oussalah A, Josse T,
Perrin P, Tramoy D, Voirin J, Klein O, Leheup B, Feillet F, et al:
Exome sequencing of cases with neural tube defects identifies
candidate genes involved in one-carbon/vitamin B12 metabolisms and
Sonic Hedgehog pathway. Hum Genet. 138:703–713. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Han X, Cao X, Aguiar-Pulido V, Yang W,
Karki M, Ramirez PAP, Cabrera RM, Lin YL, Wlodarczyk BJ, Shaw GM,
et al: CIC missense variants contribute to susceptibility for spina
bifida. Hum Mutat. 43:2021–2032. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
103
|
Lemay P, Guyot MC, Tremblay E,
Dionne-Laporte A, Spiegelman D, Henrion É, Diallo O, De Marco P,
Merello E, Massicotte C, et al: Loss-of-function de novo mutations
play an important role in severe human neural tube defects. J Med
Genet. 52:493–497. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Chen Z, Kuang L, Finnell RH and Wang H:
Genetic and functional analysis of SHROOM1-4 in a Chinese neural
tube defect cohort. Hum Genet. 137:195–202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Lupo PJ, Mitchell LE and Jenkins MM:
Genome-wide association studies of structural birth defects: A
review and commentary. Birth Defects Res. 111:1329–1342. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Abdellaoui A, Yengo L, Verweij KJH and
Visscher PM: 15 years of GWAS discovery: Realizing the promise. Am
J Hum Genet. 110:179–194. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Tam V, Patel N, Turcotte M, Bosse Y, Pare
G and Meyre D: Benefits and limitations of genome-wide association
studies. Nat Rev Genet. 20:467–484. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Pirruccello JP, Di Achille P, Nauffal V,
Nekoui M, Friedman SF, Klarqvist MDR, Chaffin MD, Weng LC,
Cunningham JW, Khurshid S, et al: Genetic analysis of right heart
structure and function in 40,000 people. Nat Genet. 54:792–803.
2022. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Aung N, Vargas JD, Yang C, Fung K, Sanghvi
MM, Piechnik SK, Neubauer S, Manichaikul A, Rotter JI, Taylor KD,
et al: Genome-wide association analysis reveals insights into the
genetic architecture of right ventricular structure and function.
Nat Genet. 54:783–791. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Lahm H, Jia M, Dreßen M, Wirth F, Puluca
N, Gilsbach R, Keavney BD, Cleuziou J, Beck N, Bondareva O, et al:
Congenital heart disease risk loci identified by genome-wide
association study in European patients. J Clin Invest.
131:e1418372021. View Article : Google Scholar :
|
|
111
|
Jin SC, Homsy J, Zaidi S, Lu Q, Morton S,
DePalma SR, Zeng X, Qi H, Chang W, Sierant MC, et al: Contribution
of rare inherited and de novo variants in 2,871 congenital heart
disease probands. Nat Genet. 49:1593–1601. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Rojas-Martinez A, Reutter H,
Chacon-Camacho O, Leon-Cachon RB, Munoz-Jimenez SG, Nowak S, Becker
J, Herberz R, Ludwig KU, Paredes-Zenteno M, et al: Genetic risk
factors for nonsyndromic cleft lip with or without cleft palate in
a Mesoamerican population: Evidence for IRF6 and variants at 8q24
and 10q25. Birth Defects Res A Clin Mol Teratol. 88:535–537. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Alade A, Peter T, Busch T, Awotoye W,
Anand D, Abimbola O, Aladenika E, Olujitan M, Rysavy O, Nguyen PF,
et al: Shared genetic risk between major orofacial cleft phenotypes
in an African population. Genet Epidemiol. 48:258–269. 2024.
View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Gaczkowska A, Biedziak B, Budner M,
Zadurska M, Lasota A, Hozyasz KK, Dąbrowska J, Wójcicki P,
Szponar-Żurowska A, Żukowski K, et al: PAX7 nucleotide variants and
the risk of non-syndromic orofacial clefts in the Polish
population. Oral Dis. 25:1608–1618. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Tindula G, Issac B, Mukherjee SK,
Ekramullah SM, Arman DM, Islam J, Suchanda HS, Sun L, Rockowitz S,
Christiani DC, et al: Genome-wide analysis of spina bifida risk
variants in a case-control study from Bangladesh. Birth Defects
Res. 116:e23312024. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
King DA, Sifrim A, Fitzgerald TW, Rahbari
R, Hobson E, Homfray T, Mansour S, Mehta SG, Shehla M, Tomkins SE,
et al: Detection of structural mosaicism from targeted and
whole-genome sequencing data. Genome Res. 27:1704–1714. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
VanOudenhove J, Yankee TN, Wilderman A and
Cotney J: Epigenomic and transcriptomic dynamics during human heart
organogenesis. Circ Res. 127:e184–e209. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Suhaimi SA, Zulkipli IN, Ghani H and
Abdul-Hamid M: Applications of next generation sequencing in the
screening and diagnosis of thalassemia: A mini-review. Front
Pediatr. 10:10157692022. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Bajaj LM, Agarwal S, Paliwal P, Saviour P,
Joshi A, Joshi A, Mahajan S, Bijarnia-Mahay S, Dua Puri R and Verma
IC: Prenatal diagnosis by chromosome microarray analysis, an Indian
experience. J Obstet Gynaecol India. 71:156–167. 2021. View Article : Google Scholar
|
|
120
|
Yin Y, Butler C and Zhang Q: Challenges in
the application of NGS in the clinical laboratory. Hum Immunol.
82:812–819. 2021. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Povysil G, Petrovski S, Hostyk J, Aggarwal
V, Allen AS and Goldstein DB: Rare-variant collapsing analyses for
complex traits: guidelines and applications. Nat Rev Genet.
20:747–759. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Giacopuzzi E, Popitsch N and Taylor JC:
GREEN-DB: A framework for the annotation and prioritization of
non-coding regulatory variants from whole-genome sequencing data.
Nucleic Acids Res. 50:2522–2535. 2022. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Pei Y, Tanguy M, Giess A, Dixit A, Wilson
LC, Gibbons RJ, Twigg SRF, Elgar G and Wilkie AOM: A comparison of
structural variant calling from Short-read and Nanopore-based
Whole-genome sequencing using optical genome mapping as a
benchmark. Genes (Basel). 15:9252024. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Lu H, Giordano F and Ning Z: Oxford
nanopore MinION sequencing and genome assembly. Genomics Proteomics
Bioinformatics. 14:265–279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
van Dijk EL, Jaszczyszyn Y, Naquin D and
Thermes C: The third revolution in sequencing technology. Trends
Genet. 34:666–681. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Hassan S, Bahar R, Johan MF, Mohamed
Hashim EK, Abdullah WZ, Esa E, Abdul Hamid FS and Zulkafli Z:
Next-generation sequencing (NGS) and Third-Generation Sequencing
(TGS) for the diagnosis of thalassemia. Diagnostics (Basel).
13:3732023. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Jeffet J, Kobo A, Su T, Grunwald A, Green
O, Nilsson AN, Eisenberg E, Ambjörnsson T, Westerlund F, Weinhold
E, et al: Super-resolution genome mapping in silicon nanochannels.
ACS Nano. 10:9823–9830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Sahajpal NS, Dean J, Hilton B, Fee T,
Skinner C, Hastie A, DuPont BR, Chaubey A, Friez MJ and Stevenson
RE: Optical genome mapping identifies rare structural variants in
neural tube defects. Genome Res. 35:798–809. 2025. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Tang D, Freudenberg J and Dahl A:
Factorizing polygenic epistasis improves prediction and uncovers
biological pathways in complex traits. Am J Hum Genet.
110:1875–1887. 2023. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Zhang W, Dai X, Wang Q, Xu S and Zhao PX:
PEPIS: A pipeline for estimating epistatic effects in quantitative
trait locus mapping and genome-wide association studies. PLoS
Comput Biol. 12:e10049252016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Wang S, Ge S, Sobkowiak B, Wang L,
Grandjean L, Colijn C and Elliott LT: Genome-Wide association with
uncertainty in the genetic similarity matrix. J Comput Biol.
30:189–203. 2023. View Article : Google Scholar
|
|
132
|
Bose A, Burch M, Chowdhury A, Paschou P
and Drineas P: Structure-informed clustering for population
stratification in association studies. BMC Bioinformatics.
24:4112023. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Zhang D, Zhou S, Zhou Z, Jiang X, Chen D,
Sun HX, Huang J, Qu S, Yang S, Gu Y, et al: BDdb: A comprehensive
platform for exploration and utilization of birth defect
multi-omics data. BMC Med Genomics. 14:2602021. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Kernohan KD and Boycott KM: The expanding
diagnostic toolbox for rare genetic diseases. Nat Rev Genet.
25:401–415. 2024. View Article : Google Scholar : PubMed/NCBI
|