Introduction
Cervical cancer is one of the most common cancers in
women. It has been estimated that more than 529,800 new cases will
be diagnosed each year, and approximately 275,100 women worldwide
will die of cervical cancer each year (1). Cervical squamous cell carcinoma
(cervical SCC) is one of the most frequent types of cervical
cancers, accounting for 80–90% of cervical cancers, and the most
important risk factor for cervical SCC is persistent human
papilloma virus (HPV) infection (2). Epidemiological studies have indicated
that more than 99% of patients with cervical SCC are positive for
high-risk HPV (HPV16, HPV18 and HPV31) (3,4). The
high-risk HPVs contain oncoproteins, i.e., E6 and E7, which
contribute to oncogenesis of cervical SCC by silencing the
tumor-suppressive p53 and Rb proteins (5–8). The
molecular mechanisms of cervical SCC initiation, development and
metastasis have not yet been fully elucidated. Therefore, an
increased understanding of the molecular targets and pathways of
cervical SCC progression and metastasis is necessary, preferably
using latest approaches in genomic analysis, including non-coding
RNA studies.
RNA can be divided into 2 categories: protein-coding
RNA and non-coding RNA (ncRNA). It is important to examine the
functions of ncRNAs and their association with human disease,
including cancer. MicroRNAs (miRNAs) are endogenous small ncRNA
molecules (19–22 bases in length) that regulate protein-coding gene
expression by repressing translation or cleaving RNA transcripts in
a sequence-specific manner (9). A
growing body of evidence suggests that miRNAs are aberrantly
expressed in many human cancers and that they play significant
roles in the initiation, development and metastasis of these
cancers (10). Some highly
expressed miRNAs can function as oncogenes by repressing tumor
suppressors, whereas low-level miRNAs can function as tumor
suppressors by negatively regulating oncogenes (11).
We previously performed miRNA expression signature
analysis of hypopharyngeal, maxillary sinus, esophageal and lung
SCCs, in addition to bladder cancer and renal cell carcinoma; these
studies indicated that miR-218 was significantly reduced in
cancer tissues compared with adjacent non-cancerous tissues,
suggesting that miR-218 is a candidate tumor-suppressive
miRNA in human cancers (12–18).
The results of past functional studies of miR-218 in various
cancers indicated that miR-218 inhibits cancer cell
proliferation and invasion through targeting oncogenic genes
(19–23). Interestingly, miR-218 was
underexpressed in HPV-positive cell lines, cervical lesions, and
cancer tissues containing HPV16 DNA, as compared to both C-33A
cells and normal cervical tissues (24).
The aim of the study was to investigate the
functional significance of miR-218 in both HPV-positive and
-negative cell lines and to identify the molecular pathways
mediating miR-218 in cervical SCC cells. Genome-wide gene
expression data for miR-218 and in silico database
analyses showed that the focal adhesion pathway was a promising
miR-218 target pathway. The laminins LAMB3 and
LAMC1 are an important and biologically active part of the
basal lamina, influencing cell differentiation, migration,
adhesion, proliferation and survival. In this study, we focused on
LAMB3 and investigation of the functional significance of
this gene in cervical SCC. The novel tumor-suppressive
miR-218-mediated cancer pathways identified herein provide
new insights into the potential mechanisms of cervical SCC
oncogenesis and metastasis.
Materials and methods
Clinical specimens
A total of 18 primary cervical SCC specimens and 11
non-cancerous specimens were collected from patients who had
undergone surgical treatment at Chiba University Hospital. The
samples were processed and stored in RNAlater (Qiagen, Valencia,
CA, USA) at −20°C until RNA extraction. Patient information is
summarized in Table I. Our study
was approved by the Bioethics Committee of Chiba University; prior
written informed consent and approval was given by each
patient.
 | Table ICharacteristics of cervical-SCC
specimens and non-cancerous specimens. |
Table I
Characteristics of cervical-SCC
specimens and non-cancerous specimens.
| No. | Age | FIGO stage | Tumor size
(cm) | Lymph node
metastasis | HPV status |
|
| 1 | 58 | IIB | 1.7×1.9 | − | 16 |
| 2 | 64 | IIB | ND | − | 16 |
| 3 | 37 | IIB | 3.5×3.0 | + | 16 |
| 4 | 41 | IB2 | 8.3×3.3 | − | 16 |
| 5 | 39 | IB1 | 3.5×3.4 | − | 16 |
| 6 | 34 | IB1 | 3.2×2.2 | − | 16 |
| 7 | 43 | IB2 | 4.0×8.0 | − | 18 |
| 8 | 56 | IIIB | 3.0×3.1 | + | 16,18 |
| 9 | 77 | IIB | 3.0×2.7 | − | 16 |
| 10 | 62 | IB1 | 3.0×2.0 | − | 16 |
| 11 | 56 | IIIA | 4.5×2.2 | + | 16 |
| 12 | 56 | IIA | 4.0×4.0 | − | 16 |
| 13 | 60 | IB1 | 4.0×4.0 | − | 16 |
| 14 | 32 | IIB | 6.0×3.0 | + | 16 |
| 15 | 38 | IB2 | 6.8×4.6 | + | 16 |
| 16 | 44 | IB1 | 3.5×2.2 | − | 16 |
| 17 | 40 | IB1 | 3.0×2.0 | − | 16 |
| 18 | 63 | IB1 | 2.7×2.4 | − | 16 |
|
| No. | Age | HPV status | | | |
|
| 1 | 44 | - | | | |
| 2 | 77 | - | | | |
| 3 | 75 | - | | | |
| 4 | 45 | - | | | |
| 5 | 47 | - | | | |
| 6 | 69 | - | | | |
| 7 | 40 | - | | | |
| 8 | 48 | - | | | |
| 9 | 41 | - | | | |
| 10 | 41 | - | | | |
| 11 | 34 | - | | | |
Cervical SCC cell lines
CaSki (HPV16-positive) and ME180 (HPV39-positive)
cells were grown in RPMI-1640 medium supplemented with 10% fetal
bovine serum. HeLa (HPV18-positive) cells were grown in E-MEM
medium supplemented with 10% fetal calf serum, and Yumoto
(HPV-negative) cells were grown in E-MEM medium supplemented with
10% fetal bovine serum. All cells were cultured in a humidified
atmosphere containing 5% CO2 at 37°C.
RNA isolation
Total RNA was isolated using TRIzol Reagent
(Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s
protocol. RNA concentration was determined spectrophotometrically.
RNA quality was confirmed using a NanoDrop 1000 Spectrophotometer
(Thermo Fisher Scientific, Wilmington, DE, USA).
DNA isolation and HPV status
Genomic DNA was extracted by QIAamp DNA mini kit
(Qiagen, Venlo, The Netherlands). Samples without HPV infection
were determined by PCR amplification using the L1 consensus primers
MY09 and MY11 as described previously (25). HPV-positive samples were analyzed
to determine the presence of DNA for HPV16 and HPV18. We designed
type-specific real-time PCR primers for the E6 and E7 regions of
HPV16 and HPV18 (Table II), and
real-time PCR was performed using a LightCyclerNano PCR System
according to the manufacturer’s protocol.
 | Table IIPrimer sequences of HPV
detection. |
Table II
Primer sequences of HPV
detection.
| Primer | Sequence (5′ to
3′) | Location | Product size
(bp) |
|---|
| HPV 16 E6-F |
GCACCAAAAGAGAACTGCAATGTT | 85–108 | 142 |
| HPV 16 E6-R |
AGTCATATACCTCACGTCGCAGTA | 203–226 | |
| HPV 16 E7-F |
CAAGTGTGACTCTACGCTTCGG | 738–759 | 81 |
| HPV 16 E7-R |
GTGCCCATTAACAGGTCTTCCAA | 796–818 | |
| HPV 18 E6-F |
CTATAGAGGCCAGTGCCATTCG | 503–524 | 79 |
| HPV 18 E6-R |
TTATACTTGTGTTTCTCTGCGTCG | 558–581 | |
| HPV 18 E7-F |
TAATCATCAACATTTACCAGCCCG | 721–744 | 113 |
| HPV 18 E7-R |
CGTCTGCTGAGCTTTCTACTACTA | 810–833 | |
Quantitative real-time RT-PCR
Stem-loop RT-PCR (TaqMan MicroRNA assays; P/N:
000521 for miR-218; Applied Biosystems, Foster City, CA,
USA) was used to quantify miRNAs according to earlier published
conditions (13). To normalize the
data for quantification of miR-218, we used RNU48
(assay ID: 001006; Applied Biosystems). TaqMan probes and primers
for LAMB3 (P/N: Hs00165078_m1) and GAPDH (P/N:
Hs02758991_g1) were obtained from Applied Biosystems. Primers for
ACTB (P/N: ACTB 533F 37546-020, ACTB 653R 37546-021) were
obtained from Sigma genetics. We used the ΔΔCt method to calculate
the fold-change.
Mature miRNA and siRNA transfections
Cervical SCC cell lines were transfected with
Lipofectamine RNAiMAX transfection reagent (Invitrogen) and
Opti-MEM (Invitrogen) with 10 nM mature miRNA or siRNA molecules.
The following RNA species were used in this study: mature miRNA,
Pre-miR miRNA Precursor (hsa-miR-218; Applied Biosystems, P/N:
AM17100), negative control miRNA (Applied Biosystems, P/N:
AM17111), small-interfering RNA (Silencer Select, Applied
Biosystems, si-LAMB3, P/N: s8075 and s8076), and negative control
siRNA (Stealth RNAi Negative Control Medium GC Duplex, Invitrogen,
P/N: 12935-300). Cells were seeded in 10-cm dishes for protein
extraction (8×105 cells per dish), 24-well plates for
mRNA extraction and Matrigel invasion assays (5×104
cells per well), and 96-well plates for XTT assays (ME180: 3,000
cells per well; CaSki and HeLa: 4,000 cells per well; and Yumoto:
2×104 cells per well).
Cell proliferation, migration, and
invasion assays
Cell proliferation was determined using an XTT assay
(Roche Applied Science, Tokyo, Japan) according to the
manufacturer’s instructions. Cell migration assays were performed
using modified Boyden Chambers (Transwells, Costar #3422, Corning
Incorporated, Corning, NY, USA) containing uncoated Transwell
polycarbonate membrane filters with 8-μm pores in 24-well
tissue culture plates. Cells were transfected with 10 nM miRNA by
reverse transfection and plated in 10-cm dishes at 8×105
cells. After 48 h, 1×105 cells were added to the upper
chamber of each migration well and were allowed to migrate for 48
h. After gentle removal of the non-migratory cells from the filter
surface of the upper chamber, the cells that migrated to the lower
side were fixed and stained with Diff-Quick (Sysmex Corporation,
Kobe, Japan). The number of cells migrating to the lower surface
was determined microscopically by counting 4 areas of constant size
per well. A cell invasion assay was carried out using modified
Boyden chambers containing transwell-precoated Matrigel membrane
filter inserts with 8-μm pores in 24-well tissue culture
plates at 1×105 cells per well (BD Biosciences, USA).
All experiments were performed in duplicate.
Pathway analysis and expression data of
putative miR-218 target genes
To obtain putative miR-218 regulated genes,
we adopted a TargetScan database searching method (http://www.targetscan.org). To identify molecular
targets and signaling pathways regulated by miR-218, in
silico and gene expression data were analyzed in the Kyoto
Encyclopedia of Genes and Genomics (KEGG) pathway (http://www.genome.jp/kegg/pathway.html)
categories using the GeneCodis program (http://genecodis.cnb.csic.es/). In this study, we
focused on the focal adhesion pathway, which included 48 genes.
Gene expression data were applied using the GEO database (accession
no. GSE6791).
Western blot analysis
Cells were harvested and lysed 72 h after
transfection. Each cell lysate (50 μg of protein) was
separated using Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA),
followed by subsequent transfer to PVDF membranes. Immunoblot
analysis was performed with polyclonal anti-LAMB3 antibodies
(HPA008069; Sigma-Aldrich, St. Louis, MO, USA). Anti-GAPDH
antibodies (ab8245; Abcam, Cambridge, UK) were used as an internal
control. The membrane was washed and incubated with anti-rabbit
IgG, HRP-linked antibodies (#7074; Cell Signaling Technology, USA).
Complexes were visualized with an Immun-Star™ WesternC
Chemiluminescence Kit (Bio-Rad), and the expression levels of these
proteins were evaluated by ImageJ software (ver.1.44; http://rsbweb.nih.gov/ij/index.html).
Plasmid construction and dual-luciferase
reporter assays
Partial sequences of the LAMB3 3′
untranslated region (3′UTR) that contain the miR-218 target
site
(ggcatgccattgaaactaagagctctcaagtcaaggaagctgggctgggcagtatcccccgcctttagttctccactggggaggaatcctggaccaagcacaaaaacttaacaaaagtgatgtaaaaatgaaaagccaaataaaatctttggaaaagagcctggaggttc)
were inserted between the XhoI and PmeI restriction
sites in the 3′UTR of the hRluc gene in the psiCHECK-2
vector (Promega, Madison, WI, USA). CaSki was then transfected with
5 ng vector, 10 nM mature miRNA. Firefly and Renilla
luciferase activities in cell lysates were determined using a
dual-luciferase assay system (E1910; Promega). Normalized data were
calculated as the quotient of Renilla/firefly luciferase
activities.
Statistical analysis
The relationships between 2 variables and numerical
values were analyzed using the Mann-Whitney U test, and the
relationships between 3 variables and the numerical values were
analyzed using the Bonferroni-adjusted Mann-Whitney U test. Expert
Stat View analysis software (ver. 4; SAS Institute Inc., Cary, NC,
USA) was used in both analyses. In the comparison of 3 variables, a
non-adjusted statistical level of significance of P<0.05
corresponded to the Bonferroni-adjusted level of P<0.0167.
Results
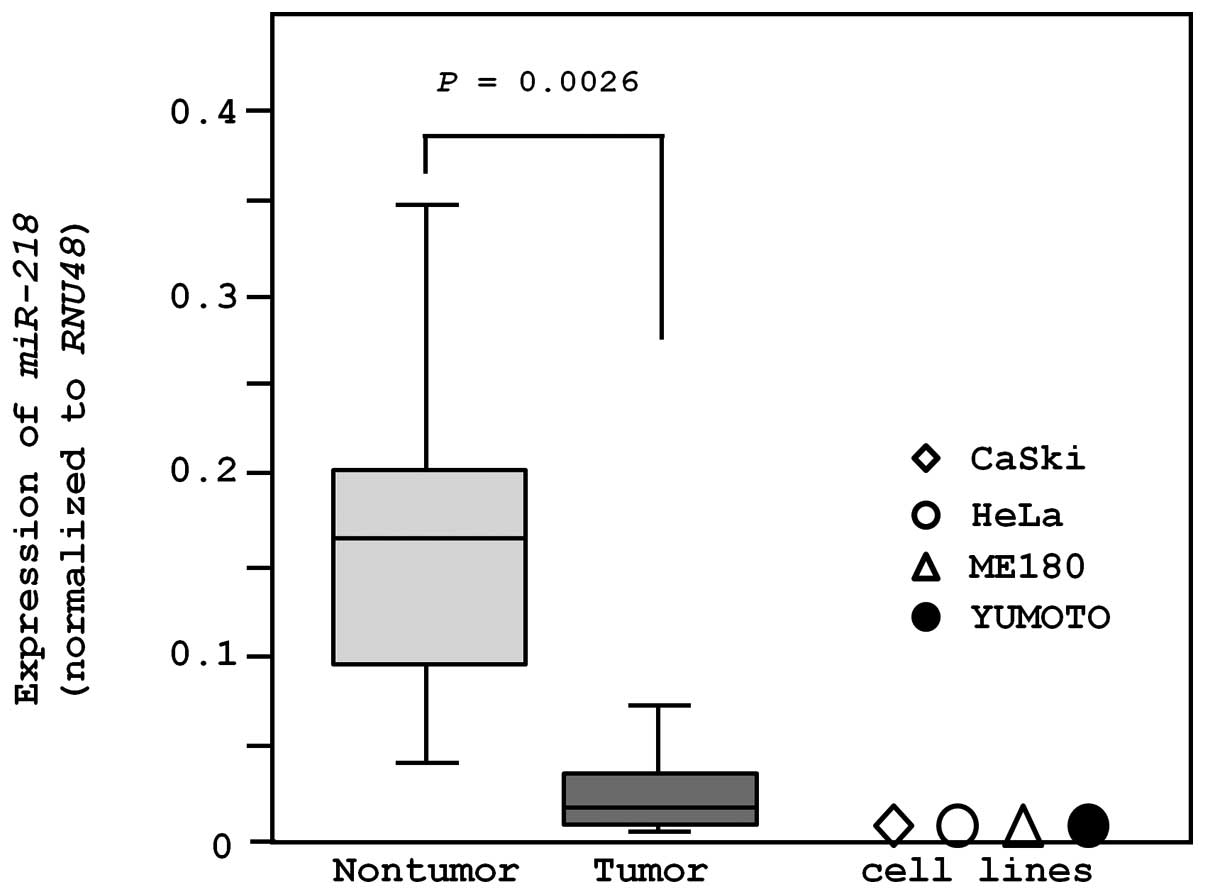
Expression levels of miR-218 in cervical
SCC clinical specimens and cell lines
The expression of miR-218 was significantly
lower in clinical cervical SCC specimens (n=18; 0.043±0.077) than
in non-cancerous specimens (n=11; 0.153±0.110, P=0.0026; Fig. 1). We also evaluated the expression
of miR-218 in cervical cancer cell lines. miR-218
expression levels in CaSki (HPV16-positive), ME180
(HPV39-positive), HeLa (HPV18-positive), and Yumoto (HPV-negative)
were significantly lower than that in non-cancerous cervical
epithelium (P<0.0001; Fig.
1).
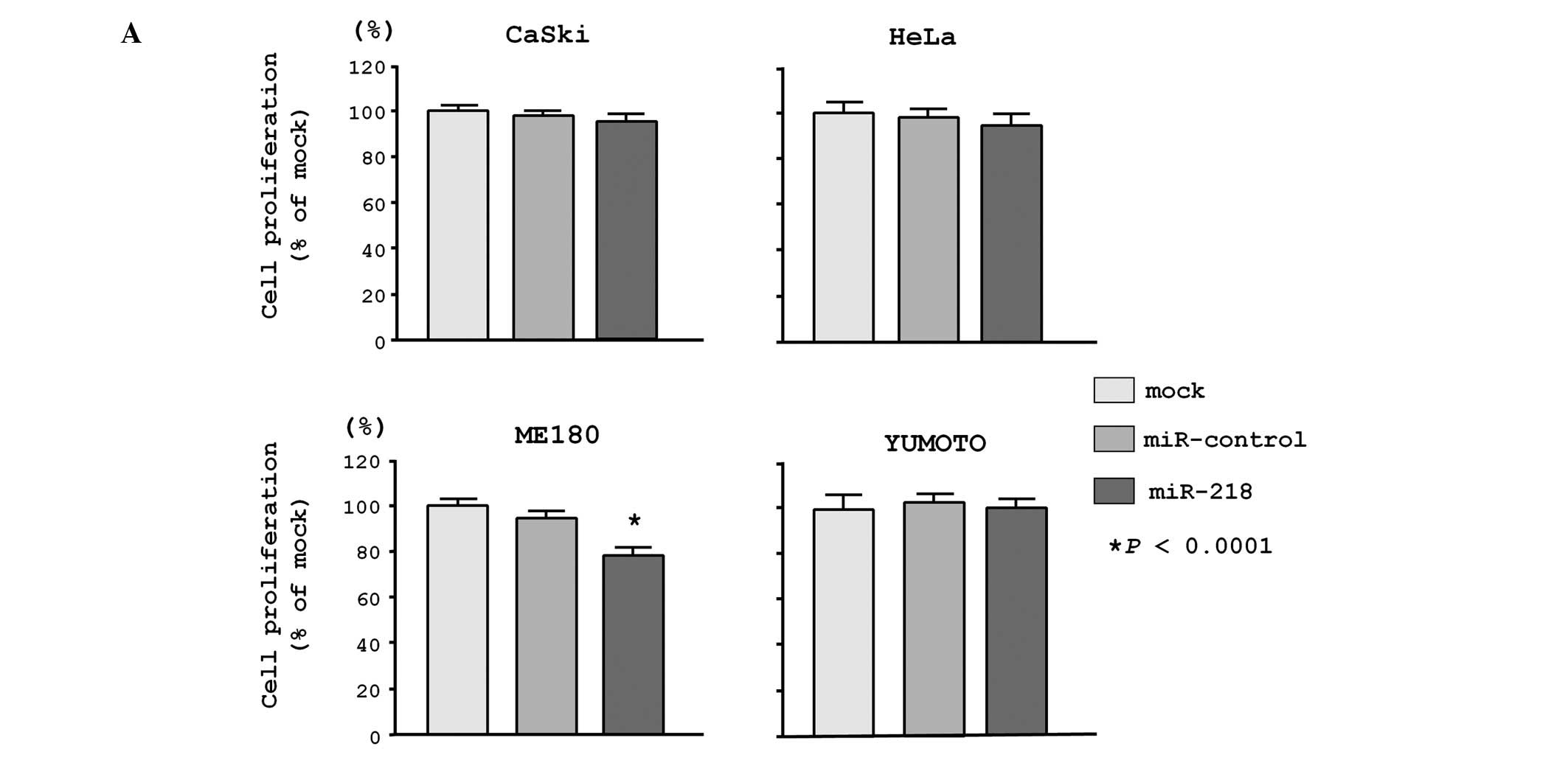
Effects of miR-218 restoration on cell
proliferation, migration and invasion in cervical SCC cell
lines
To investigate the functional role of
miR-218, we performed gain-of-function studies using cells
transfected with a precursor of miR-218. The XTT assay
revealed that cell proliferation was significantly inhibited in
miR-218 transfectants in comparison with non-transfectants
(mock) and miRNA-control transfectants (control) in ME180 cells
(78.1±3.7%, 100.0±2.5% and 96.0±3.5%, respectively; P<0.0001),
while no significant inhibition was seen in CaSki cells (94.8±3.5%,
100.0±2.3% and 97.9±2.1%, respectively; P>0.0167), HeLa cells
(93.0±5.0%, 100.0±7.5% and 97.6±4.1%, respectively; P>0.0167),
and Yumoto cells (106.3±11.1%, 100.0±3.6% and 110.8±11.8%;
P>0.0167; Fig. 2A).
Migration and Matrigel invasion assays demonstrated
that the number of invading cells significantly decreased in
miR-218 transfectants in comparison with mock and
miR-control transfectants in all cell lines tested. In fact,
migration in miR-218 transfectants was reduced to only
11.5±3.4% in CaSki cells (mock, 100.0±20.8; control, 115.4±12.2;
P<0.0001), 20.3±3.8% in ME180 cells (mock, 100.0±28.1%; control,
77.2±13.8%; P<0.0001) 49.0±5.9% in HeLa cells (mock, 100.0±6.8%;
control, 114.1±16.8%; P<0.0001), and 24.9±8.8% in Yumoto cells
(mock, 100.0±18.2%; control, 102.2±6.8%; P<0.0001; Fig. 2B).
Similarly, in the Matrigel invasion assay, the
number of invading cells was significantly decreased in
miR-218-transfectants in comparison with mock and
miR-control transfectants in all cell lines. Cell invasion was
reduced to 3.9±1.6% in CaSki cells (mock, 100.0±8.5%; control,
107.0±16.3%; P<0.0001), 37.1±9.2% in ME180 cells (mock,
100.0±14.8%; control, 71.9±18.1%; P<0.0001), 2.7±1.5% in HeLa
cells (mock, 100.0±21.6%; control, 102.7±19.0%; P<0.0001), and
14.9±6.6% for Yumoto cells (mock, 100.0±14.8%; control,
103.4±19.4%; P<0.0001; Fig.
2C).
Identification of miR-218-mediated
molecular pathways and putative miR-218 target genes
We first obtained putative miR-218 target
genes by searching the TargetScan database. According to the
database, 4,946 conserved targets, with a total of 1,865 conserved
sites and 4,372 poorly conserved sites, were deposited in this
database. These genes were analyzed and characterized in KEGG
pathway categories using the GeneCodis program. This analysis
revealed 105 signaling pathways (Table
III). In these pathways, we focused on the focal adhesion
pathway and the 48 genes contained within this pathway (Table IV). To search for genes regulated
by tumor-suppressive miR-218 in cervical SCC, we applied
gene expression profiles in the GEO database (accession no.
GSE6791). Among 48 genes, 24 genes were upregulated in cervical SCC
compared to adjacent non-cancerous tissues. The expression levels
of up- or downregulated genes in clinical specimens are shown in
Table IV. As a result of our
expression data, we identified LAMB3 as one of the most
highly upregulated genes in clinical specimens; this gene has 1
putative miR-218 binding site. LAMB3 is a laminin
that is an important and biologically active part of the basal
lamina, functioning in a variety of pathways, such as cell
differentiation, migration, adhesion, proliferation and survival.
Thus, we focused on LAMB3 as a promising target gene of
miR-218 in cervical SCC.
 | Table IIISignificantly enriched annotations
regulated by miR-218 (top 20 pathways). |
Table III
Significantly enriched annotations
regulated by miR-218 (top 20 pathways).
| No. of genes | P-value | Annotations |
|---|
| 59 | 1.55E-16 | Endocytosis |
| 40 | 4.29E-12 | Glutamatergic
synapse |
| 69 | 4.30E-11 | Pathways in
cancer |
| 58 | 4.21E-10 | MAPK signaling
pathway |
| 38 | 4.87E-10 | Insulin signaling
pathway |
| 48 | 4.98E-10 | Focal adhesion |
| 29 | 1.53E-09 | ErbB signaling
pathway |
| 25 | 3.70E-09 | Long-term
depression |
| 35 | 6.58E-09 | Axon guidance |
| 43 | 1.99E-08 | Chemokine signaling
pathway |
| 24 | 6.10E-08 | Chronic myeloid
leukemia |
| 23 | 6.35E-08 | Long-term
potentiation |
| 36 | 1.00E-07 | Wnt signaling
pathway |
| 33 | 1.06E-07 | Tight junction |
| 26 | 1.21E-07 | Prostate
cancer |
| 26 | 1.21E-07 | Gap junction |
| 21 | 2.87E-07 | Glioma |
| 26 | 2.98E-07 | FcγR-mediated
phagocytosis |
| 22 | 5.37E-07 | Adherens
junction |
| 38 | 5.54E-07 | Calcium signaling
pathway |
 | Table IVExpression of target genes by
miR-218 involved in focal adhesion pathways. |
Table IV
Expression of target genes by
miR-218 involved in focal adhesion pathways.
| Entrez gene | Gene symbol | FC | Regulation | P-value |
|---|
| 3265 | HRAS | 4.54 | Up | 4.19E-02 |
| 3914 | LAMB3 | 3.54 | Up | 4.40E-03 |
| 3676 | ITGA4 | 3.21 | Up | 6.03E-03 |
| 5062 | PAK2 | 2.40 | Up | 7.29E-05 |
| 1282 | COL4A1 | 2.33 | Up | 2.21E-02 |
| 6464 | SHC1 | 2.15 | Up | 3.05E-04 |
| 87 | ACTN1 | 2.14 | Up | 3.18E-03 |
| 1399 | CRKL | 2.10 | Up | 4.50E-04 |
| 2932 | GSK3B | 1.83 | Up | 1.36E-03 |
| 394 | ARHGAP5 | 1.75 | Up | 9.48E-04 |
| 3915 | LAMC1 | 1.75 | Up | 8.18E-03 |
| 3371 | TNC | 1.51 | Up | 3.09E-01 |
| 5501 | PPP1CC | 1.50 | Up | 6.03E-03 |
| 387 | RHOA | 1.45 | Up | 2.04E-01 |
| 1301 | COL11A1 | 1.44 | Up | 3.87E-01 |
| 5829 | PXN | 1.33 | Up | 1.27E-01 |
| 858 | CAV2 | 1.27 | Up | 3.09E-01 |
| 3480 | IGF1R | 1.26 | Up | 2.42E-01 |
| 100291393 | LOC100291393 | 1.09 | Up | 8.39E-01 |
| 25759 | LOC100291393 | 1.09 | Up | 8.39E-01 |
| 2321 | FLT1 | 1.05 | Up | 7.22E-01 |
| 5601 | MAPK9 | 1.03 | Up | 8.39E-01 |
| 896 | CCND3 | 1.02 | Up | 8.79E-01 |
| 5293 | PIK3CD | 1.02 | Up | 8.39E-01 |
| 3691 | ITGB4 | −1.02 | Down | 9.59E-01 |
| 5500 | PPP1CB | −1.07 | Down | 4.76E-01 |
| 63923 | TNN | −1.17 | Down | 4.46E-01 |
| 1289 | COL5A1 | −1.17 | Down | 6.84E-01 |
| 5170 | PDPK1 | −1.19 | Down | 7.51E-02 |
| 5058 | PAK1 | −1.23 | Down | 1.40E-01 |
| 7057 | THBS1 | −1.23 | Down | 4.46E-01 |
| 10451 | VAV3 | −1.23 | Down | 7.60E-01 |
| 5156 | PDGFRA | −1.41 | Down | 2.63E-01 |
| 10000 | AKT3 | −1.46 | Down | 6.71E-02 |
| 1956 | EGFR | −1.51 | Down | 5.99E-02 |
| 399694 | SHC4 | −1.57 | Down | 9.33E-02 |
| 55742 | PARVA | −1.58 | Down | 1.68E-02 |
| 5579 | PRKCB | −1.71 | Down | 9.50E-03 |
| 53358 | SHC3 | −1.80 | Down | 5.33E-02 |
| 5649 | RELN | −1.81 | Down | 2.52E-02 |
| 2318 | FLNC | −1.81 | Down | 2.21E-02 |
| 5295 | PIK3R1 | −1.88 | Down | 4.19E-02 |
| 1277 | COL1A1 | −2.38 | Down | 8.38E-02 |
| 8515 | ITGA10 | −2.61 | Down | 9.01E-05 |
| 6714 | SRC | −2.85 | Down | 1.36E-03 |
| 81 | ACTN4 | −2.86 | Down | 3.05E-04 |
| 3479 | IGF1 | −2.97 | Down | 8.18E-03 |
| 595 | CCND1 | −5.52 | Down | 1.14E-03 |
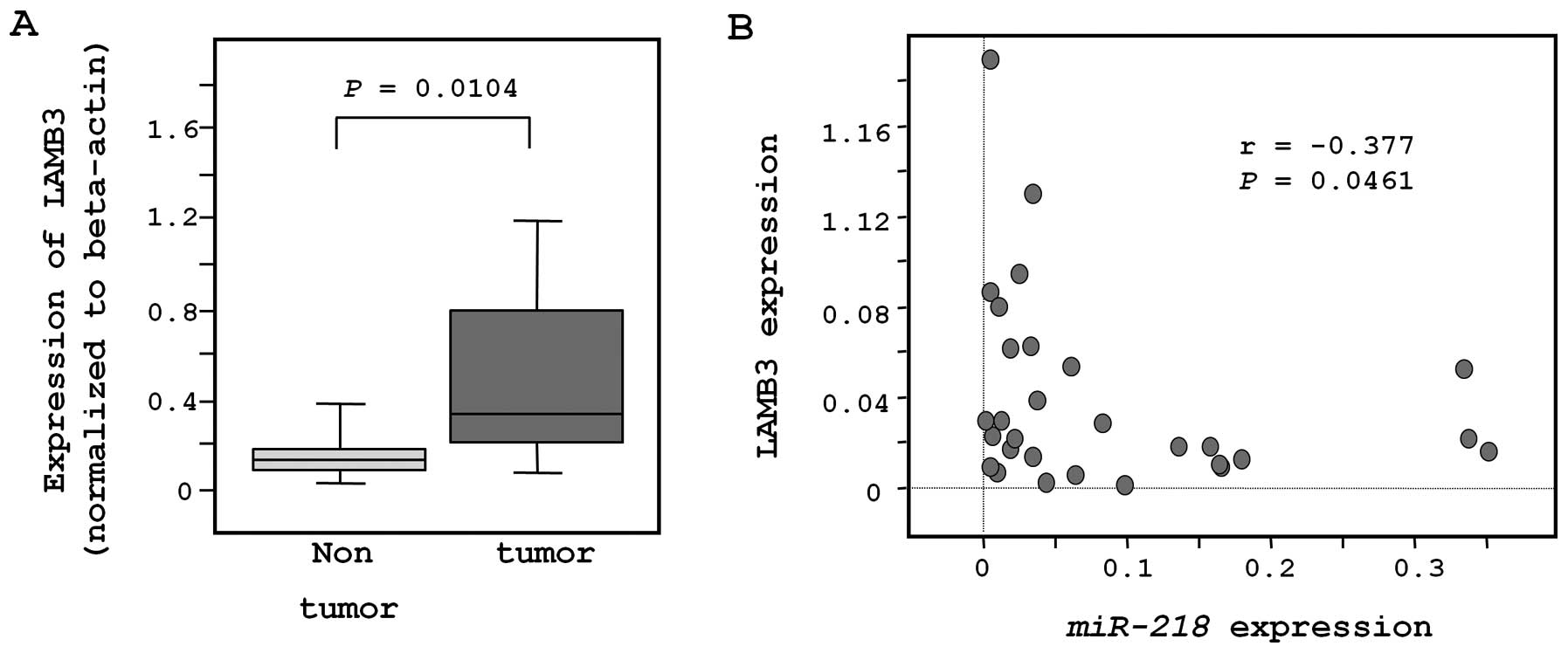
Expression of LAMB3 in cervical SCC
clinical specimens
The expression of LAMB3 was significantly
lower in clinical cervical SCC specimens (n=18; 0.053±0.049) than
in non-cancerous specimens (n=11; 0.017±0.014, P=0.0104; Fig. 3A). Moreover, LAMB3
expression was significantly inversely correlated with
miR-218 expression (r=−0.377; P=0.0461; Fig. 3B).
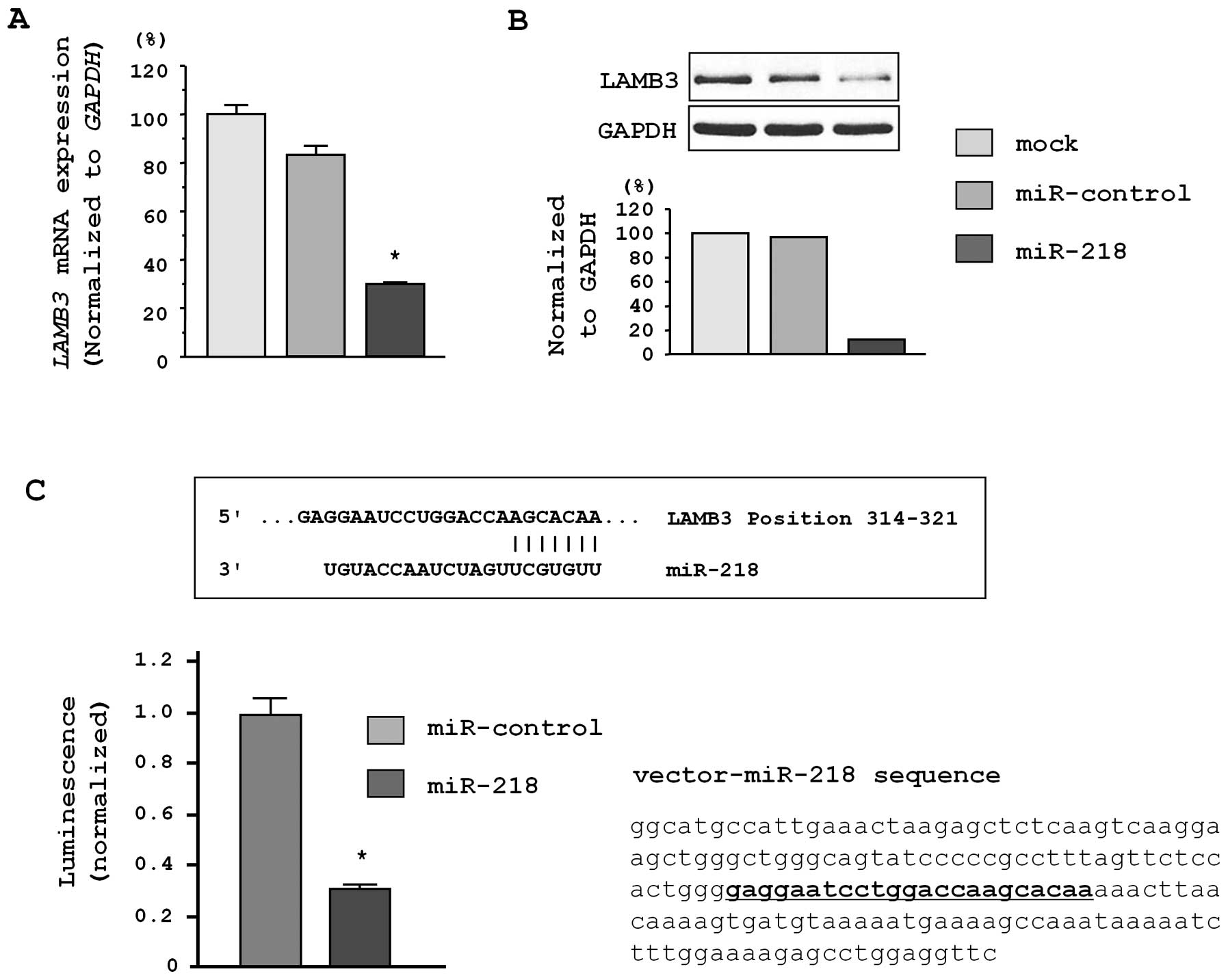
LAMB3 is a direct target of miR-218
We performed quantitative real-time RT-PCR and
western blot analysis to investigate whether LAMB3 mRNA and
protein were downregulated by restoration of miR-218.
Importantly, both LAMB3 mRNA and protein levels were
significantly repressed in miR-218-transfectants in
comparison with mock transfectants (Fig. 4A and B).
To determine whether the 3′UTR of LAMB3 had
an actual target site for miR-218, we performed a luciferase
reporter assay by using a vector encoding the 3′UTR of LAMB3
mRNA. We found that the luminescence intensity was significantly
reduced in miR-218 transfectants as compared to mock and
miRNA-control transfectants (P<0.0001; Fig. 4C).
Silencing of LAMB3 mRNA and protein in a
cervical SCC cell line
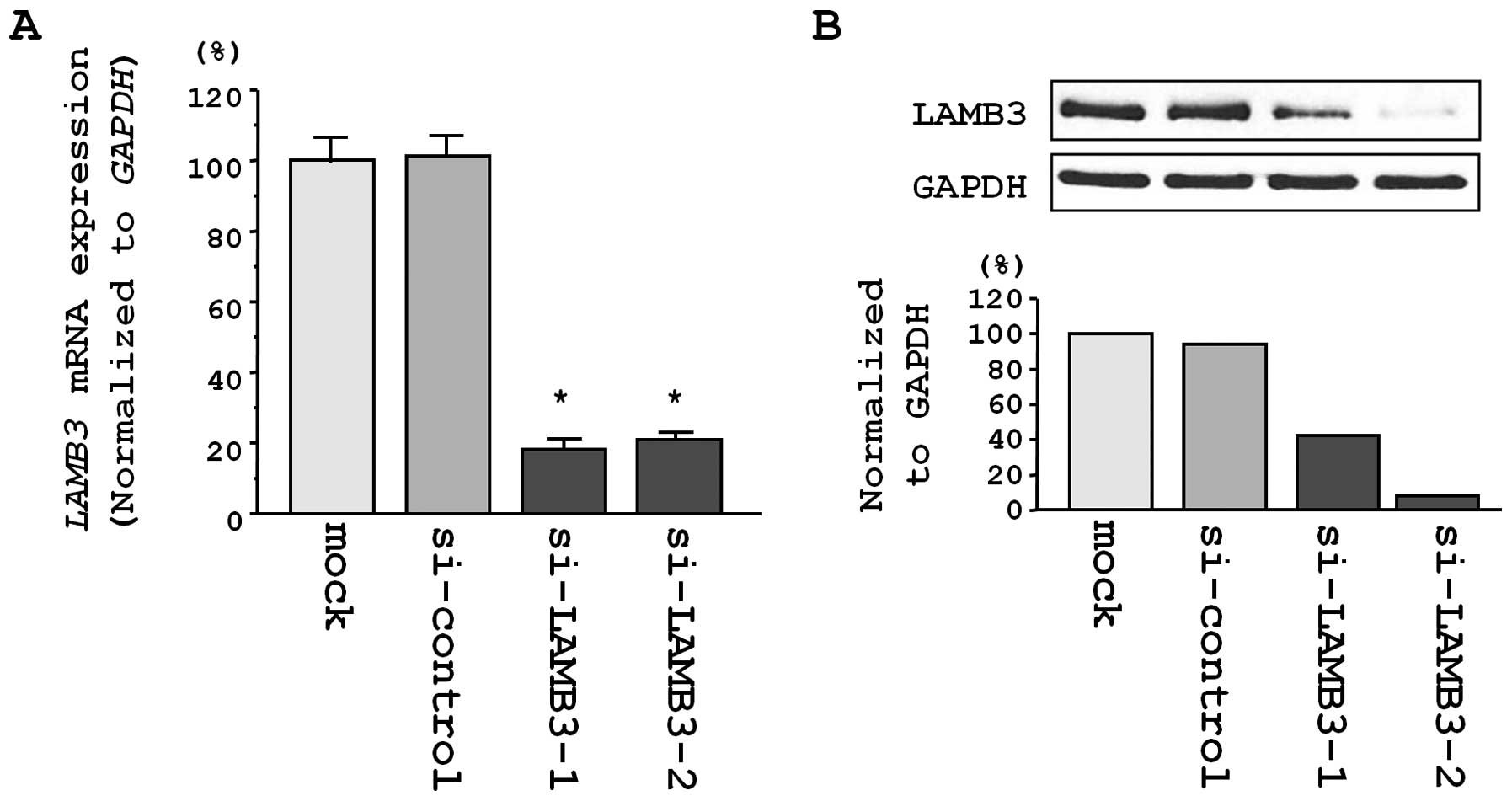
Next, we examined the impact of si-LAMB3
transfection in CaSki cells. The expression of LAMB3 mRNA
was reduced in 2 si-LAMB3 transfectants in comparison with
mock and si-control transfectants (P<0.0001; Fig. 5A). Additionally, the expression of
LAMB3 protein was reduced in si-LAMB3-1 and
si-LAMB3-2 transfectants in comparison with mock and
si-control transfectants (P<0.0001 and P=0.0002, respectively;
Fig. 5B). These results showed
that the 2 siRNAs were useful for loss-of-function assays in this
study.
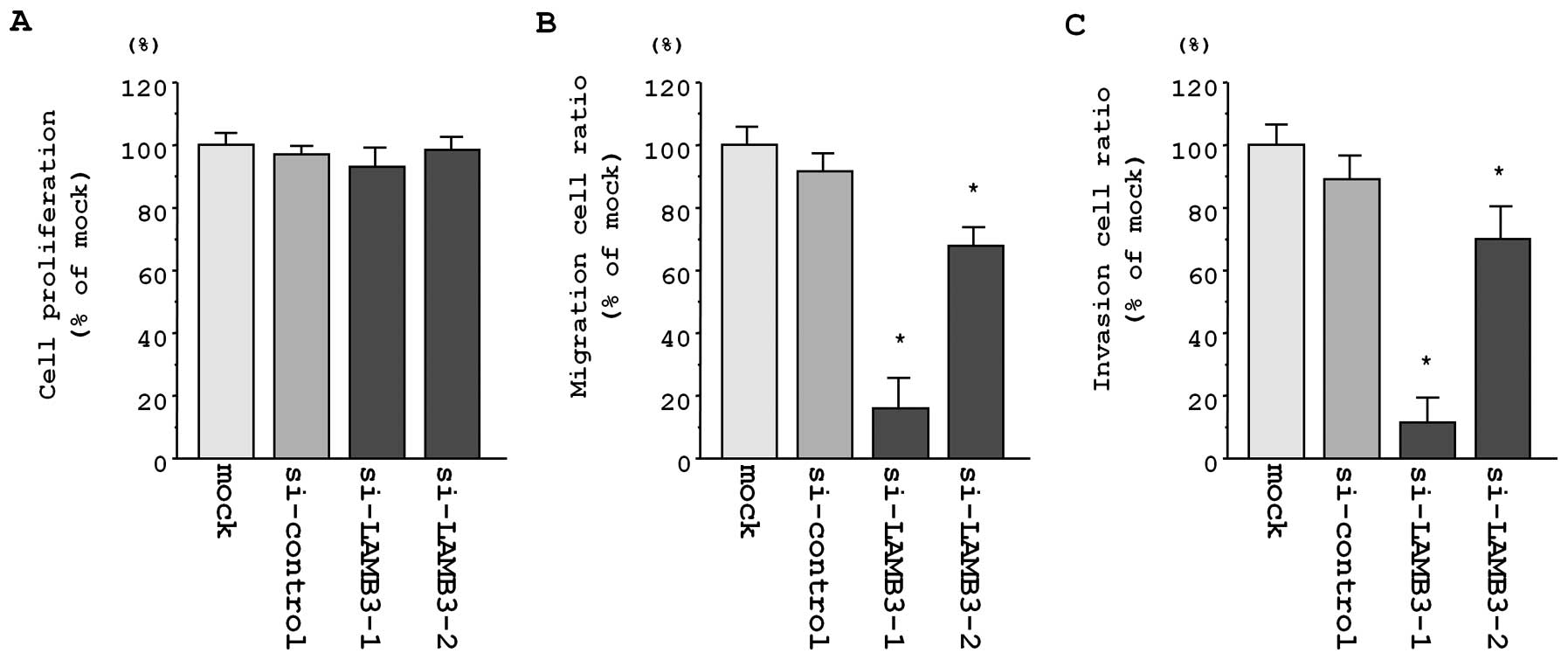
Effects of LAMB3 silencing on cell
proliferation, migration and invasion in cervical SCC cell
lines
To investigate the functional role of LAMB3,
we performed loss-of-function studies using si-LAMB3
transfectants. The XTT assay revealed that cell proliferation was
not inhibited in the 2 si-LAMB3 transfectants as compared
with mock and si-control transfectants in CaSki cells (82.4±10.7%,
100.2±6.1%, 100.0±14.6% and 95.6±11.2%; P>0.0083; Fig. 6A).
Additionally, the number of migrating cells was
significantly decreased in both si-LAMB3 transfectants as
compared with mock and si-control transfectants in CaSki cells
(10.6±2.7%, 72.2±10.7%, 100.0±6.5% and 91.8±11.0%, respectively;
P<0.0001; Fig. 6B).
The number of invading cells was also significantly
decreased in si-LAMB3 transfectants as compared with mock
and si-control transfectants in CaSki cells (13.5±4.9%, 73.1±16.2%,
100.0±13.7% and 88.4±18.0%, respectively; P<0.0001; Fig. 6C).
Discussion
The discovery of non-coding RNA during the human
genome sequencing project had a significant impact in cancer
research (26). The reconstructing
of genome-wide studies to include non-coding RNA is therefore
necessary for cancer research. miRNAs are a class of small
non-coding RNAs, and a growing body of evidence has suggested that
miRNAs also contribute to cancer initiation, development and
metastasis in many types of cancers, including cervical cancer
(10).
It is believed that normal regulatory mechanisms can
be disrupted by aberrant expression of tumor-suppressive or
oncogenic miRNAs in cancer cells. Therefore, identification of
aberrantly expressed miRNAs is the first step toward elucidating
miRNA-mediated oncogenic pathways. Based on this, we identified the
miRNA expression signatures in several human squamous cell
carcinomas, including esophageal SCC, hypopharyngeal SCC, maxillary
sinus SCC and lung SCC, allowing the elucidation of multiple
tumor-suppressive miRNAs (12–15).
Our previous studies showed that miR-218 is a frequently
downregulated miRNA and that restoration of this miRNA inhibited
cancer cell migration and invasion in head and neck SCC (HNSCC)
cells (19). We also searched for
downregulated miRNAs in cervical SCC by examining expression
signatures published in public databases (27–31).
These data indicate that miR-218 is frequently downregulated
miRNA in cervical SCC. Thus, we focused on miR-218 and
investigated the functional significance of this miRNA in mediating
cancer pathways.
In the human genome, 2 miR-218 precursor
genes, miR-218-1 and miR-218-2, have identical
sequences in the mature miRNA and map to human chromosomes 4p15.31
and 6q35.1, respectively. Interestingly, the genomic regions of
miR-218-1 and miR-218-2 are located in the introns of
the SLIT2 and SLIT3 genes, respectively.
Downregulation of miR-218 in cancer cells has been shown to
be caused by promoter hypermethylation of SLIT2 and
SLIT3 genes (20).
Silencing of miR-218 by DNA hypermethylation has also been
reported in oral SCC using a function-based screening approach
(21). On the other hand, several
reports have indicated that silencing of miR-218 in cervical
SCC was caused by HPV infection (24,32).
Our expression data showed that miR-218 expression was
significantly reduced in both HPV-positive cells (CaSki, HeLa and
ME180) and HPV-negative cells (Yumoto), in addition to clinical
specimens. Since the molecular mechanisms of miR-218
silencing in cervical SCC are still unclear, further study is
necessary to solve this problem.
In the current study, we found significant
inhibition of cancer cell migration and invasion in cervical SCC
cell lines (CaSki, HeLa, ME180 and Yumoto) by miR-218
restoration, suggesting that miR-218 is a tumor-suppressive
miRNA in cervical SCC. Our previous reports in HNSCC also showed
that miR-218 contributes to cancer cell migration and
invasion (19). The
tumor-suppressive function of miR-218 has also been reported
in several types of cancers, and miR-218 has been shown to
target several oncogenic genes, such as Rictor (oral cancer),
survivin and ROBO1 receptor (nasopharyngeal cancer and gastric
cancer) (20–22). Our recent report also indicated
that restoration of miR-218 inhibited cancer cell
proliferation, migration, and invasion in bladder cancer (23,33).
These data suggested that miR-218 is an important
tumor-suppressive miRNA that is deeply involved in human
cancers.
miRNAs are unique in their ability to regulate many
protein-coding genes. Bioinformatic predictions have indicated that
miRNAs regulate more than 30% of protein-coding genes (34). A single miRNA is capable of
targeting a number of genes to globally regulate biological
processes. The identification of novel cancer pathways and
responsible genes regulated by tumor-suppressive miR-218 in
cervical SCC is the next step for our understanding of cervical SCC
oncogenesis. Thus, we pursued GeneCodis analysis to reveal the
functional significance of these genes potentially regulated by
miR-218 in cervical SCC. The GeneCodis analysis applies many
genes to known pathways in the KEGG Pathway Database, and these
data facilitate our understanding of tumor-suppressive
miRNA-mediated molecular pathways in human cancer. This method of
analysis has previously been used to efficiently identify
tumor-suppressive miRNA-mediated cancer pathways in our laboratory
(19). In the current study, the
GeneCodis analysis revealed 105 signaling pathways, as highlighted
in Table II. In these pathways, we
focused on the focal adhesion pathway and the 48 genes contained
within this pathway.
To search for genes regulated by tumor-suppressive
miR-218 in cervical SCC, we used gene expression profiling
in this study (deposited in the GEO database as accession no.
GSE6791). Among the 48 genes in the focal adhesion pathway, 10 were
upregulated in cervical SCC clinical specimens, indicating that
these genes were candidates for regulation by tumor-suppressive
miR-218 in cervical SCC. From these genes, we focused on
LAMB3, a component of laminin-332, and investigated the
functional significance of this gene.
Laminin-332, a heterotrimer composed of 3 chains
(LAMA3, LAMB3 and LAMC2), is an adhesion substrate for epithelial
cells and regulates epithelial cell migration during epithelial
regeneration and repair processes (35,36).
Several immunohistochemical studies have shown that laminin-332 or
its subunit LAMC2 is expressed in tumor cells at the invasion front
or in budding tumor cells in many types of human cancers, such as
adenocarcinomas of the colon, breast, pancreas and lung, SCC of the
esophagus and melanoma (35). Our
data demonstrated that LAMB3 was directly regulated by
miR-218 and functioned as an oncogene, contributing to
cancer cell migration and invasion in cervical SCC. Many studies
have indicated that laminin-332 binds to several cell-surface
receptors, such as integrins, epidermal growth factor receptor and
syndecan-1 (37–39). Among these binding partners,
integrins are cell surface transmembrane proteins that mediate
extracellular signals and intracellular pathways, leading to
control of the cell cycle, cell migration, and invasion in cancer
cells (40). It will be necessary
to analyze the signal pathways associated with the interactions of
integrins and laminin-332 in cervical SCC in the future.
Acknowledgements
This study was supported by KAKENHI
(C), 24592590.
References
|
1.
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar
|
|
2.
|
Walboomers JM, Jacobs MV, Manos MM, et al:
Human papillomavirus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Muñoz N, Bosch FX, De Sanjosé S, et al:
Epidemiologic classification of human papillomavirus types
associated with cervical cancer. N Engl J Med. 348:518–527.
2003.
|
|
4.
|
Clifford GM, Smith JS, Plummer M, Muñoz N
and Franceschi S: Human papillomavirus types in invasive cervical
cancer worldwide: a meta-analysis. Br J Cancer. 88:63–73. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Band V, Dalal S, Delmolino L and Androphy
EJ: Enhanced degradation of p53 protein in HPV-6 and BPV-1
E6-immortalized human mammary epithelial cells. EMBO J.
12:1847–1852. 1993.PubMed/NCBI
|
|
6.
|
Lechnerl MS, Mackl DH, Finicle AB, Crook
T, Vousden KH and Laiminsl LA: Human papillomavirus E6 proteins
bind p53 in vivo and abrogate p53-mediated repression of
transcription. EMBO J. 11:3045–3052. 1992.
|
|
7.
|
Thomas M, Pim D and Banks L: The role of
the E6-p53 interaction in the molecular pathogenesis of HPV.
Oncogene. 18:7690–7700. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Münger K and Howley PM: Human
papillomavirus immortalization and transformation functions. Virus
Res. 89:213–228. 2002.PubMed/NCBI
|
|
9.
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Nelson KM and WEiss GJ: MicroRNAs and
cancer: past, present, and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar
|
|
12.
|
Kikkawa N, Hanazawa T, Fujimura L, et al:
miR-489 is a tumour-suppressive miRNA target PTPN11 in
hypopharyngeal squamous cell carcinoma (HSCC). Br J Cancer.
103:877–884. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Nohata N, Hanazawa T, Kikkawa N, et al:
Tumour suppressive microRNA-874 regulates novel cancer networks in
maxillary sinus squamous cell carcinoma. Br J Cancer. 105:833–841.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Kano M, Seki N, Kikkawa N, et al: miR-145,
miR-133a and miR-133b: tumor-suppressive miRNAs target FSCN1 in
esophageal squamous cell carcinoma. Int J Cancer. 127:2804–2814.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Moriya Y, Nohata N, Kinoshita T, et al:
Tumor suppressive microRNA-133a regulates novel molecular networks
in lung squamous cell carcinoma. J Hum Genet. 57:38–45. 2012.
View Article : Google Scholar
|
|
16.
|
Hidaka H, Seki N, Yoshino H, Yamasaki T,
et al: Tumor suppressive microRNA-1285 regulates novel molecular
targets: aberrant expression and functional significance in renal
cell carcinoma. Oncotarget. 3:44–57. 2012.PubMed/NCBI
|
|
17.
|
Ichimi T, Enokida H, Okuno Y, et al:
Identification of novel microRNA targets based on microRNA
signatures in bladder cancer. Int J Cancer. 125:345–352. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Yoshino H, Chiyomaru T, Enokida H, et al:
The tumoursuppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Kinoshita T, Hanazawa T, Nohata N, et al:
Tumor suppressive microRNA-218 inhibits cancer cell migration and
invasion through targeting laminin-332 in head and neck squamous
cell carcinoma. Oncotarget. 3:1386–1400. 2012.PubMed/NCBI
|
|
20.
|
Alajez NM, Lenarduzzi M, Ito E, et al:
MiR-218 suppresses nasopharyngeal cancer progression through
downregulation of survivin and the SLIT2-ROBO1 pathway. Cancer Res.
71:2381–2391. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Uesugi A, Kozaki K-I, Tsuruta T, et al:
The tumor suppressive microRNA miR-218 targets the mTOR component
Rictor and inhibits AKT phosphorylation in oral cancer. Cancer Res.
71:5765–5678. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tie J, Pan Y, Zhao L, et al: MiR-218
inhibits invasion and metastasis of gastric cancer by targeting the
Robo1 receptor. PLoS Genet. 6:e10008792010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Tatarano S, Chiyomaru T, Kawakami K, et
al: miR-218 on the genomic loss region of chromosome 4p15.31
functions as a tumor suppressor in bladder cancer. Int J Oncol.
39:13–21. 2011.PubMed/NCBI
|
|
24.
|
Martinez I, Gardiner AS, Board KF, Monzon
FA, Edwards RP and Khan SA: Human papillomavirus type 16 reduces
the expression of microRNA-218 in cervical carcinoma cells.
Oncogene. 27:2575–2582. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Gravitt PE, Peyton CL, Alessi TQ, et al:
Improved amplification of genital human papillomaviruses. J Clin
Microbiol. 38:357–361. 2000.PubMed/NCBI
|
|
26.
|
Mattick JS: RNA regulation: a new
genetics? Nat Rev Genet. 5:316–323. 2004. View Article : Google Scholar
|
|
27.
|
Lee J-W, Choi CH, Choi J-J, et al: Altered
MicroRNA expression in cervical carcinomas. Clin Cancer Res.
14:2535–2542. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Wang X, Tang S, Le S-Y, et al: Aberrant
expression of oncogenic and tumor-suppressive microRNAs in cervical
cancer is required for cancer cell growth. PLoS One. 3:e25572008.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Pereira PM, Marques JP, Soares AR, Carreto
L and Santos MAS: MicroRNA expression variability in human cervical
tissues. PLoS One. 5:e117802010. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Li Y, Wang F, Xu J, et al: Progressive
miRNA expression profiles in cervical carcinogenesis and
identification of HPV-related target genes for miR-29. J Pathol.
224:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Rao Q, Zhou H, Peng Y, Li J and Lin Z:
Aberrant microRNA expression in human cervical carcinomas. Med
Oncol. 29:1242–1248. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Li Y, Liu J, Yuan C, Cui B, Zou X and Qiao
Y: High-risk human papillomavirus reduces the expression of
microRNA-218 in women with cervical intraepithelial neoplasia. J
Int Med Res. 38:1730–1736. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Chiyomaru T, Enokida H, Kawakami K, et al:
Functional role of LASP1 in cell viability and its regulation by
microRNAs in bladder cancer. Urologic Oncol. 30:434–443. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Bartel DP, Lee R and Feinbaum R:
MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell.
116:281–297. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Guess CM and Quaranta V: Defining the role
of laminin-332 in carcinoma. Matrix Biol. 28:445–455. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Marinkovich MP: Laminin 332 in
squamous-cell carcinoma. Nat Rev Cancer. 7:370–380. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Carter WG, Ryan MC and Gahr PJ: Epiligrin,
a new cell adhesion ligand for integrinα3β1 in epithelial basement
membranes. Cell. 65:599–610. 1991.
|
|
38.
|
Schenk S, Hintermann E, Bilban M, et al:
Binding to EGF receptor of a laminin-5 EGF-like fragment liberated
during MMP-dependent mammary gland involution. J Cell Biol.
161:197–209. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Okamoto O, Bachy S, Odenthal U, et al:
Normal human keratinocytes bind to the alpha3LG4/5 domain of
unprocessed laminin-5 through the receptor syndecan-1. J Biol Chem.
278:44168–44177. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Mizejewski GJ: Role of integrins in
cancer: survey of expression patterns. Proc Soc Exp Biol Med.
222:124–138. 1999. View Article : Google Scholar : PubMed/NCBI
|