Introduction
miRNAs are 20-to-25 mer non-coding RNAs which
incompletely bind to the 3′ untranslated regions (UTR) of multiple
target mRNAs, enhancing their degradation and inhibiting their
translation. MiRNAs participate in the regulation of cell
differentiation, cell cycle progression and apoptosis. Dysregulated
miRNAs play critical roles during carcinogenesis and cancer
progression. The levels of many miRNAs in cancer tissue are lower
than those in normal tissue, a state that contributes to cancer
progression. Thus, abnormally downregulated miRNAs function as
tumor suppressor genes and upregulated miRNAs function as oncogenic
genes (1–3) in tumorigenesis as well as in gastric
cancer. Therefore, it is necessary to identify new aberrant
expression miRNAs in gastric cancers by screening all miRNAs
closely related to tumorigenesis and progression. In recent years,
miRNA microarray including Agilent, Exiqon, Affymetrix, Phalanx
(4–8) or TaqMan miRNA (9,10)
assays have been used to screen candidate miRNA biomarkers from
fresh samples or frozen samples. Because of the differences in
clinical cancer tissue and various methods employed by different
miRNA microarray manufacturers, the screening results are
considerable different, and some screening results of miRNA
microarray are contradictory (11)
also in gastric cancer (9,12–18).
Aberrantly expressed miRNAs between gastric cell and gastric cancer
cells were not fully identified and understood. The profiling
results from this array may identify aberrantly expressed miRNAs as
biomarkers of gastric cancers.
To resolve the issue, we decided to utilize Human
Cancer Pathway Finder miRNA PCR array (MIHS-102Z, Qiagen, Hilgen,
Germany) which profiles the expression of 84 miRNAs differentially
expressed in tumors versus normal tissue. This array provides
cancer researchers with a convenient way to quickly analyze the
miRNAs most relevant to tumorigenesis and screen and identify
aberrantly expressed miRNAs between gastric cell gastric cancer
cells. This array detected expression between gastric cancer and
adjacent non-tumor tissues, and the clinical significance was
analyzed.
Materials and methods
Cell lines
Seven gastric cancer cell lines, AGS, SGC-7901,
MKN-45, MKN-28, MGC-803, BCG-823, HGC-27 and GES-1 were purchased
from the Type Culture Collection of the Chinese Academy of Sciences
(Shanghai, China) and Cancer Institute and Hospital, Chinese
Academy of Medical Sciences (CAMS) (Beijing, China). All the
gastric cancer cell lines were maintained in DMEM supplemented with
10% heat-inactivated fetal bovine serum in a humidified cell
incubator having an atmosphere of 5% CO2 at 37°C.
Exponentially growing cells were used for experiments. All miRNA
PCR primers were purchased from GeneCopoeia™ Inc (Rockville, MD,
USA).
Clinical samples
Fifty-eight gastric cancer samples were obtained
during surgery and used after obtaining informed consent. All
patients underwent curative resection of the primary tumor at WuWei
City Tumor Hospital from the year 2010 to 2012 (WuWei, China, a
high incidene rate area with gastric cancer) (19,20).
All patients had a clear histological diagnosis of gastric cancer,
based on the clinicopathologic criteria. All data, including age,
gender, histological grade, depth, lymph node metastasis, local
invasion, depth of tumor invasion, lymph node metastasis, lymphatic
invasion, venous invasion, Borrmann type and clinical stage were
obtained from clinical and pathologic records. No patient received
neoadjuvant chemotherapy or radiotherapy before surgery and
adjuvant radiotherapy after surgery. Resected cancerous tissues (T)
and paired non-cancerous tissues (N) were immediately cut and
stored in frozen liquid nitrogen, at −80°C until RNA extraction.
Written informed consent was obtained from each patient for his or
her participation in the study. The study protocol conformed to the
ethical guidelines of the 1975 Declaration of Helsinki as reflected
in a priori approval by the ethics committee of the First Hospital
of Lanzhou University.
Total RNA isolation and quality
analysis
Total RNA of gastric cancer cell lines and frozen
tissues of gastric cancer were extracted using RNeasy mini kit
(Qiagen) according to the manufacturer’s instructions.
Concentrations and purity of the RNA samples were assayed by
electrophoresis and spectrophotometric methods.
Human Cancer Pathway Finder miRNA PCR
array expression profiling
Human Cancer Pathway Finder miRNA PCR array were
used to amplify and quantify the expression levels of 84 miRNAs in
GES-1 cell line and AGS, SGC-7901, MKN-45, MKN-28, MGC-803,
BCG-823, HGC-27, 7 gastric cancer cell lines, the differentially
expressed miRNAs were analyzed using the
2-(CTmiRNA-CTRNU6B RNA) method of relative
quantification. qRT-PCR was performed in Bio-Rad CFX96 Real-time
PCR system. Each reaction was performed in a final volume of 25
μl containing 1 μl of cDNA, 0.5 mM of each primer and
1X SYBR Green PCR Master mix (Qiagen). The amplification program
was: denaturation at 95°C for 10 min, followed by 40 cycles of 95°C
for 15 sec, 60°C for 1 min, in which fluorescence was acquired.
Relative changes in gene expression levels were calculated using
ΔΔCt (threshold cycle) method. The housekeeping genes,
such as SNORD61, SNORD68, SNORD72, SNORD95, SNORD96A and RNU6-2,
were used to normalize to the amount of miRNAs. The differentially
expressed miRNAs were analyzed using the formula
2-(CTmiRNA-CTRNU6B RNA) method of relative
quantification. Hierarchical clustering of aberrantly expressed
miRNAs with significantly different expression was performed using
online data analysis tool of Sabiosciences (Qiagen).
miRNA quantification by real-time
qRT-PCR
miRNA quantification by real-time qRT-PCR. SYBR
green qRT-PCR assay was used for miRNA quantification. In brief, 40
ng of total RNA containing miRNA was polyadenylated by poly(A)
polymerase and was reversely transcripted to cDNA using miScript
Reverse Transcription kit according to the manufacturer’s
instructions (GeneCopoeia). miScript SYBR Green PCR kit was used
and miscript Universal primer was provided by the manufacturer
(GeneCopoeia), qRT-PCR was performed in Bio-Rad CFX96 Real-time PCR
system. Each reaction was performed in a final volume of 10
μl containing 2 μl of cDNA, 0.5 mM of each primer and
1X SYBR Green PCR Master mix (GeneCopoeia). The amplification
program was: denaturation at 95°C for 10 min, followed by 40 cycles
of 95°C for 10 sec, 60°C for 30 sec and 72°C for 30 sec, in which
fluorescence was acquired. At the end of the PCR cycles, melting
curve analyses were performed as well as electrophoresis of the
products on 2.5% agarose gels in order to validate the specific
generation of the expected PCR product. Each sample was run in
triplicates for analysis. The expression levels of miRNAs were
normalized to RNU6B. Relative gene expression was calculated as
2-(CTmiRNA-CTRNU6B RNA). Hierarchical
clustering of aberrantly expressed miRNAs with significantly
different expression was performed using the MeV (Multiple
Experiment Viewer) 4.9.1 software and visualized with Treeview
v1.60.
miRNA targeted gene prediction and signal
pathway analyses
We utilized a miRNA target gene prediction database
mirfocus 2.0 (http://mirfocus.org/index.php) to select validated
targets of the differently expressed miRNAs to analyse enriched
KEGG pathways and to annotate the molecular function of the miRNA
targeted genes. The mirfocus 2.0 integrated 5 bioinformatical
Target Prediction Tools: MiRanda, MirTarget2, PicTar, microT and
TargetScanS, and the experimental validated Target Tools include
miRecords, miR2Disease, TarBase and miRTarBase. Enriched KEGG
pathway and Enriched GO term analyses of 4 miRNA-targeted genes
also were performed by mirfocus 2.0. Prediction database support
number was 3, P-value of Fisher Test was P<0.05.
Statistical analysis
Student’s unpaired t-test was used to compare values
of samples of 7 gastric cancer cell lines and samples of GES-1
gastric cell line. Differences between groups were estimated using
the χ2 test. A probability level of 0.05 was chosen for
statistical significance, and all statistical analyses were
performed using the SPSS 11.0 software (SPSS Inc., Chicago, IL,
USA).
Results
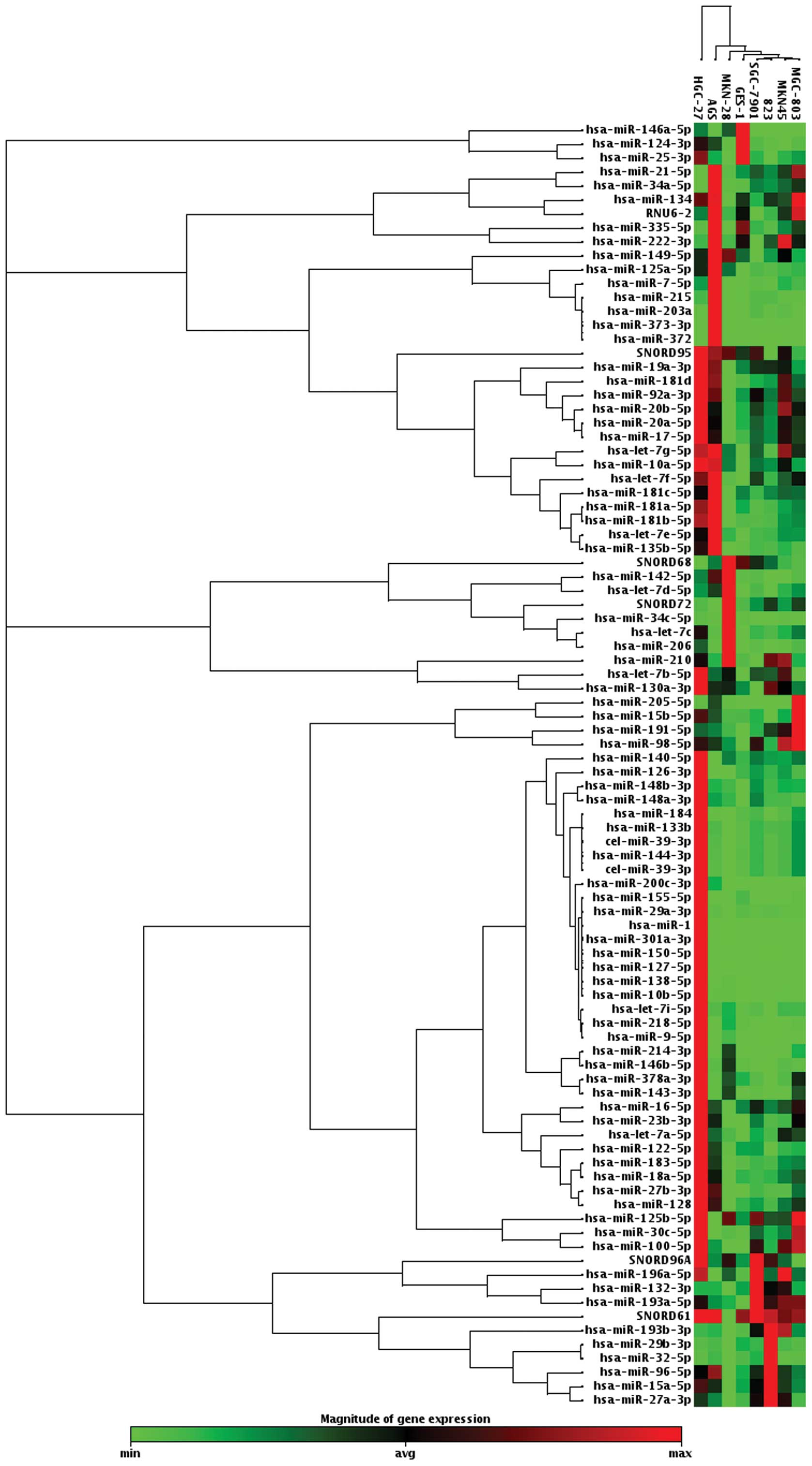
Screening of aberrantly expressed miRNAs
in gastric cancer cell lines
When setting average change >2-fold or <0.5 as
a cut-off level, we found that 37 miRNAs were upregulated, and 5
miRNAs were downregulated in at least 4 of 7 gastric cancer cell
lines compared to GES-1 cell line screened by Human Cancer Pathway
Finder miRNA PCR array among 84 miRNAs related to tumorigenesis
(Fig. 1).
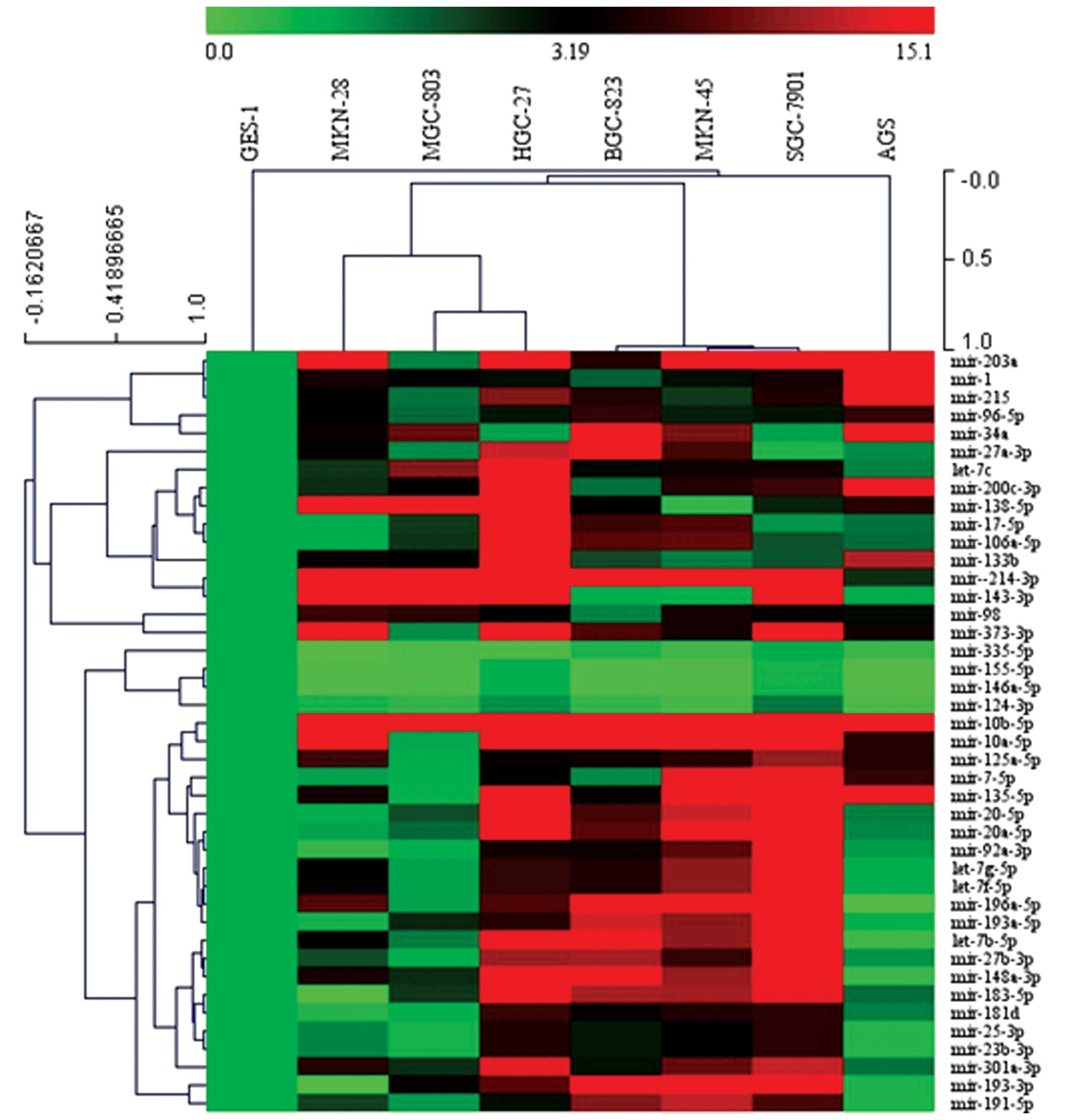
Validation of aberrantly expressed miRNAs
in gastric cancer cell lines
After completing screening, 42 aberrantly expressed
miRNAs were validated by qRT-PCR to verify its expression levels.
Results showed that 38 miRNAs were upregulated in 7 gastric cancer
cell lines compared with the GES-1 cell line, these were mir-1,
mir-7-5p, mir-10a-5p, mir-10b-5p, let-7b-5p, let-7g-5p, let-7c,
let-7f-5p, mir-17-5p, mir-20a-5p, mir-20b-5p, mir-23b-3p,
mir-25-3p, mir-27a-3p, mir-27b-3p, mir-34a, mir-92a-3p, mir-96-5p,
mir-98, mir-106a-5p, mir-125a-5p, mir-133b, mir-135-5p, mir-136-5p,
mir-143-3p, mir-148a-3p, mir-181d, mir-183-5p, mir-191-5p,
mir-193-3p, mir-193a-5p, mir-196a-5p, mir-200c-3p, mir-203a,
mir-215, mir-301a-3p, mir-214-3p, mir-373-3p and 4 miRNAs were
downregulated in 7 gastric cancer cell lines compared with GES-1
cell line, which were mir-124-3p, mir-146a-5p, mir-155-5p and
mir-335-5p. Except mir-25-3p, the expression levels of most miRNAs
were consistent with expression of that by Pathway Finder miRNA PCR
array expression profiles (Figs. 2
and 3).
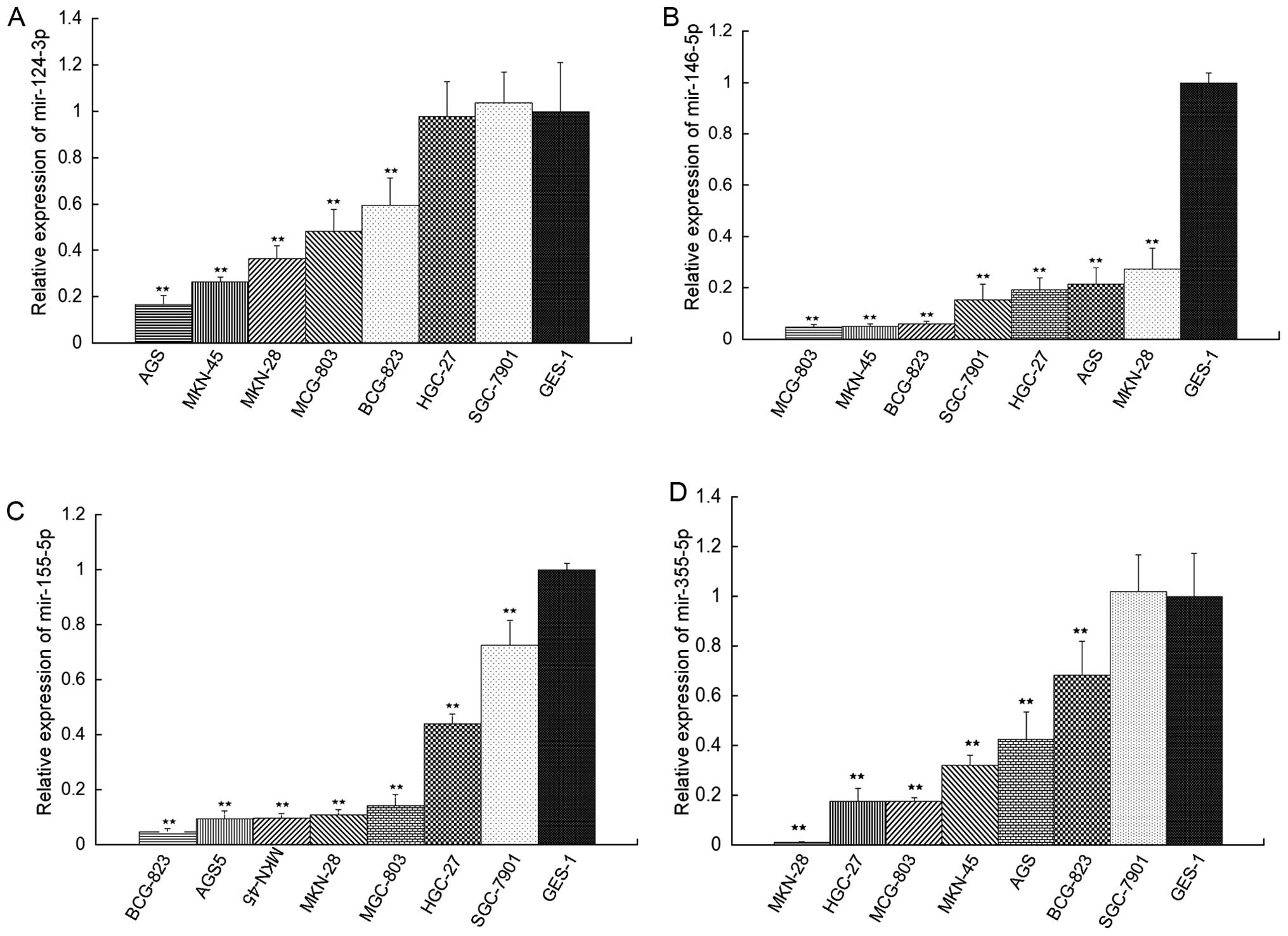
Validation of mir-124-3p, mir-146a-5p,
mir-155-5p and mir-335-5p in gastric cancer tissue compared with
adjacent non-tumor tissues
In light of confusing studies on the expression of
mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p in gastric
tissues, the levels of these 4 miRNAs in 58 cancerous and
corresponding non-cancerous tissues were detected by qRT-PCR
method. Results showed that the number of mir-124-3p in
high-expression group (T/N>2) and low-expression group
(T/N<0.5) amounts to 13 and 45, respectively, according to the
median cancer (T)/non-cancerous (N) tissue ratio of mir-124-3p
expression (Table I). The number
of mir-146a-5p in high-expression group (T/N>2 and
low-expression group (T/N<0.5) was 35 and 20, respectively,
according to the median cancer (T)/non-cancerous (N) tissue ratio
of mir-146a-5p expression (Table
II), the number of mir-155-5p in high-expression group
(T/N>2) and low-expression group (T/ N<0.5) was 26 and 26,
respectively, according to the median cancer (T)/non-cancerous (N)
tissue ratio of mir-155-5p expression (Table III), and the number of mir-335-5p
in high-expression group (T/N>2) and low-expression group (T/
N<0.5) was 30 and 25, respectively, according to the median
cancer (T)/non-cancerous (N) tissue ratio of mir-335-5p expression
(Table IV).
 | Table I.mir-124-3p level and
clinicopathological factors in patients with gastric cancer. |
Table I.
mir-124-3p level and
clinicopathological factors in patients with gastric cancer.
|
Characteristics | mir-124-3p Low
expression (n=45)
| mir-124-3p High
expression (n=13)
| Significance
|
|---|
| n (%) | n (%) | χ2 | P-value |
|---|
| Age | | | 0.481 | 0.488 |
| <59 years | 29 (64.4) | 7 (53.8) | | |
| ≥59 years | 16 (35.6) | 6 (46.2) | | |
| Gender | | | 0.157 | 0.692 |
| Male | 27 (60.0) | 7 (53.8) | | |
| Female | 18 (40.0) | 6 (46.2) | | |
| Degree of tumor
cell differentiation | | | 0.259 | 0.611 |
| Moderate-to-well
differentiated | 16 (35.6) | 3 (23.1) | | |
| Poorly
differentiated | 29 (64.4) | 10 (76.9) | | |
| TNM stage | | | 0.098 | 0.755 |
| I + II | 11 (24.4) | 2 (15.4) | | |
| III + IV | 34 (75.6) | 11 (84.6) | | |
| Local invasion | | | 0.000 | 1.000 |
| T1 + T2 | 10 (22.2) | 3 (23.1) | | |
| T3 | 35 (77.8) | 10 (76.9) | | |
| Lymph node
metastasis | | | 5.784 | 0.016a |
| Positive | 36 (80.0) | 6 (46.2) | | |
| Negative | 9 (20.0) | 7 (53.8) | | |
| Lymphatic
invasion | | | 9.120 | 0.003b |
| Positive | 42 (93.3) | 8 (61.5) | | |
| Negative | 3 (6.7) | 5 (38.5) | | |
| Venous
invasion | | | 1.581 | 0.209 |
| Positive | 5 (11.1) | 0 (0) | | |
| Negative | 40 (88.9) | 13 (100) | | |
| Depth of tumor
invasion | | | 0.646 | 0.421 |
| Mucosa,
submucosa, muscularis propria, subserosa | 22 (48.9) | 8 (61.5) | | |
| Penetration of
serosa, adjacent structures | 23 (51.1) | 5 (38.5) | | |
| Borrmann type | | | 0.625 | 0.429 |
| I + II | 27 (60.0) | 10 (76.9) | | |
| III + IV | 18 (40.0) | 3 (23.1) | | |
 | Table II.mir-146a-5p level and
clinicopathological factors in patients with gastric cancer. |
Table II.
mir-146a-5p level and
clinicopathological factors in patients with gastric cancer.
|
Characteristics | mir-146a-5p Low
expression (n=35)
| mir-146a-5p High
expression (n=20)
| Significance
|
|---|
| n (%) | n (%) | χ2 | P-value |
|---|
| Age | | | 0.000 | 1.000 |
| <59 years | 22 (62.9) | 12 (60.0) | | |
| ≥59 years | 13 (37.1) | 8 (40.0) | | |
| Gender | | | 3.654 | 0.056 |
| Male | 17 (48.6) | 15 (75.0) | | |
| Female | 18 (51.4) | 5 (25.0) | | |
| Degree of tumor
cell differentiation | | | 2.946 | 0.086 |
| Moderate-to-well
differentiated | 11 (31.4) | 11 (55.0) | | |
| Poorly
differentiated | 24 (68.6) | 9 (45.0 | | |
| TNM stage | | | 0.000 | 1.000 |
| I + II | 7 (20.0) | 4 (20.0) | | |
| III + IV | 28 (80.0) | 16 (80.0) | | |
| Local invasion | | | 0.000 | 1.000 |
| T1 + T2 | 21 (60.0) | 12 (60.0) | | |
| T3 | 14 (40.0) | 8 (40.0) | | |
| Lymph node
metastasis | | | 0.074 | 0.786 |
| Positive | 24 (68.6) | 13 (65.0) | | |
| Negative | 11 (31.4) | 7 (35.0) | | |
| Lymphatic
invasion | | | 0.000 | 1.000 |
| Positive | 32 (91.4) | 18 (90.0) | | |
| Negative | 3 (8.6) | 2 (10.0) | | |
| Venous
invasion | | | 3.956 | 0.047a |
| Positive | 12 (34.3) | 2 (10.0) | | |
| Negative | 23 (65.7) | 18 (90.0) | | |
| Depth of tumor
invasion | | | 0.667 | 0.414 |
| Mucosa,
submucosa, muscularis propria, subserosa | 18 (51.4) | 8 (40.0) | | |
| Penetration of
serosa, adjacent strucures | 17 (48.6) | 12 (60.0) | | |
| Borrmann type | | | 0.000 | 1.000 |
| I + II | 21 (60.0) | 12 (60.0) | | |
| III + IV | 14 (40.0) | 8 (40.0) | | |
 | Table III.mir-155-5p level and
clinicopathological factors in patients with gastric cancer. |
Table III.
mir-155-5p level and
clinicopathological factors in patients with gastric cancer.
|
Characteristics | mir-155-5p Low
expression (n=26)
| mir-155-5p High
expression (n=26)
| Significance
|
|---|
| n (%) | n (%) | χ2 | P-value |
|---|
| Age | | | 0.000 | 1.000 |
| <59 years | 18 (69.2) | 15 (57.7) | | |
| ≥59 years | 8 (30.8) | 11 (42.3) | | |
| Gender | | | 0.000 | 1.000 |
| Male | 15 (57.7) | 16 (61.5) | | |
| Female | 11 (42.3) | 10 (38.5) | | |
| Degree of tumor
cell differentiation | | | 0.702 | 0.402 |
| Moderate-to-well
differentiated | 10 (38.5) | 13 (50.0) | | |
| Poorly
differentiated | 16 (61.5) | 13 (50.0) | | |
| TNM stage | | | 0.115 | 0.734 |
| I + II | 5 (19.2) | 6 (23.1) | | |
| III + IV | 21 (80.8) | 20 (76.9) | | |
| Local invasion | | | 0.495 | 0.482 |
| T1 + T2 | 6 (23.1) | 4 (15.4) | | |
| T3 | 20 (76.9) | 22 (84.6) | | |
| Lymph node
metastasis | | | 0.048 | 0.827 |
| Positive | 19 (73.1) | 18 (69.2) | | |
| Negative | 7 (26.9) | 8 (30.8) | | |
| Lymphatic
invasion | | | 0.000 | 1.000 |
| Positive | 24 (92.3) | 25 (96.2) | | |
| Negative | 2 (7.6) | 1 (3.8) | | |
| Venous
invasion | | | 0.885 | 0.347 |
| Positive | 22 (84.6) | 25 (96.2) | | |
| Negative | 4 (15.4) | 1 (3.8) | | |
| Depth of tumor
invasion | | | 1.238 | 0.426 |
| Mucosa,
submucosa, muscularis propria, subserosa | 14 (53.8) | 10 (38.5) | | |
| Penetration of
serosa, adjacent strucures | 12 (46.2) | 16 (61.5) | | |
| Borrmann type | | | 3.914 | 0.048a |
| I + II | 12 (46.2) | 19 (73.1) | | |
| III + IV | 14 (53.8) | 7 (26.9) | | |
 | Table IV.mir-335-5p level and
clinicopathological factors in patients with gastric cancer. |
Table IV.
mir-335-5p level and
clinicopathological factors in patients with gastric cancer.
|
Characteristics | mir-335-5p Low
expression (n=30)
| mir-335-5p High
expression (n=25)
| Significance
|
|---|
| n (%) | n (%) | χ2 | P-value |
|---|
| Age | | | 0.603 | 0.437 |
| <59 years | 21 (70.0) | 15 (60.0) | | |
| ≥59 years | 9 (30.0) | 10 (40.0) | | |
| Gender | | | 0.09 | 0.765 |
| Male | 18 (60.0) | 14 (56.0) | | |
| Female | 12 (40.0) | 11 (44.0) | | |
| Degree of tumor
cell differentiation | | | 6.227 | 0.013a |
| Moderate-to-well
differentiated | 8 (26.7) | 15 (60.0) | | |
| Poorly
differentiated | 22 (73.3) | 10 (40.0) | | |
| TNM stage | | | 0.128 | 0.721 |
| I + II | 6 (20.0) | 6 (24.0) | | |
| III + IV | 24 (80.0) | 19 (76.0) | | |
| Local invasion | | | 0.484 | 0.487 |
| T1 + T2 | 6 (20.0) | 7 (28.0) | | |
| T3 | 24 (80.0) | 18 (72.0) | | |
| Lymph node
metastasis | | | 1.101 | 0.294 |
| Positive | 22 (73.3) | 15 (60.0) | | |
| Negative | 8 (26.7) | 10 (40.0) | | |
| Lymphatic
invasion | | | 4.303 | 0.038a |
| Positive | 28 (93.3) | 17 (68.0) | | |
| Negative | 2 (6.7) | 8 (32.0) | | |
| Venous
invasion | | | 0.110 | 0.740 |
| Positive | 3 (10.0) | 1 (4.0) | | |
| Negative | 27 (90.0) | 24 (96.0) | | |
| Depth of tumor
invasion | | | 0.022 | 0.883 |
| Mucosa,
submucosa, muscularis propria, subserosa | 15 (50.0) | 13 (52.0) | | |
| Penetration of
serosa, adjacent strucures | 15 (50.0) | 12 (48.0) | | |
| Borrmann type | | | 0.638 | 0.425 |
| I + II | 16 (53.3) | 19 (76.0) | | |
| III + IV | 14 (46.7) | 6 (24.0) | | |
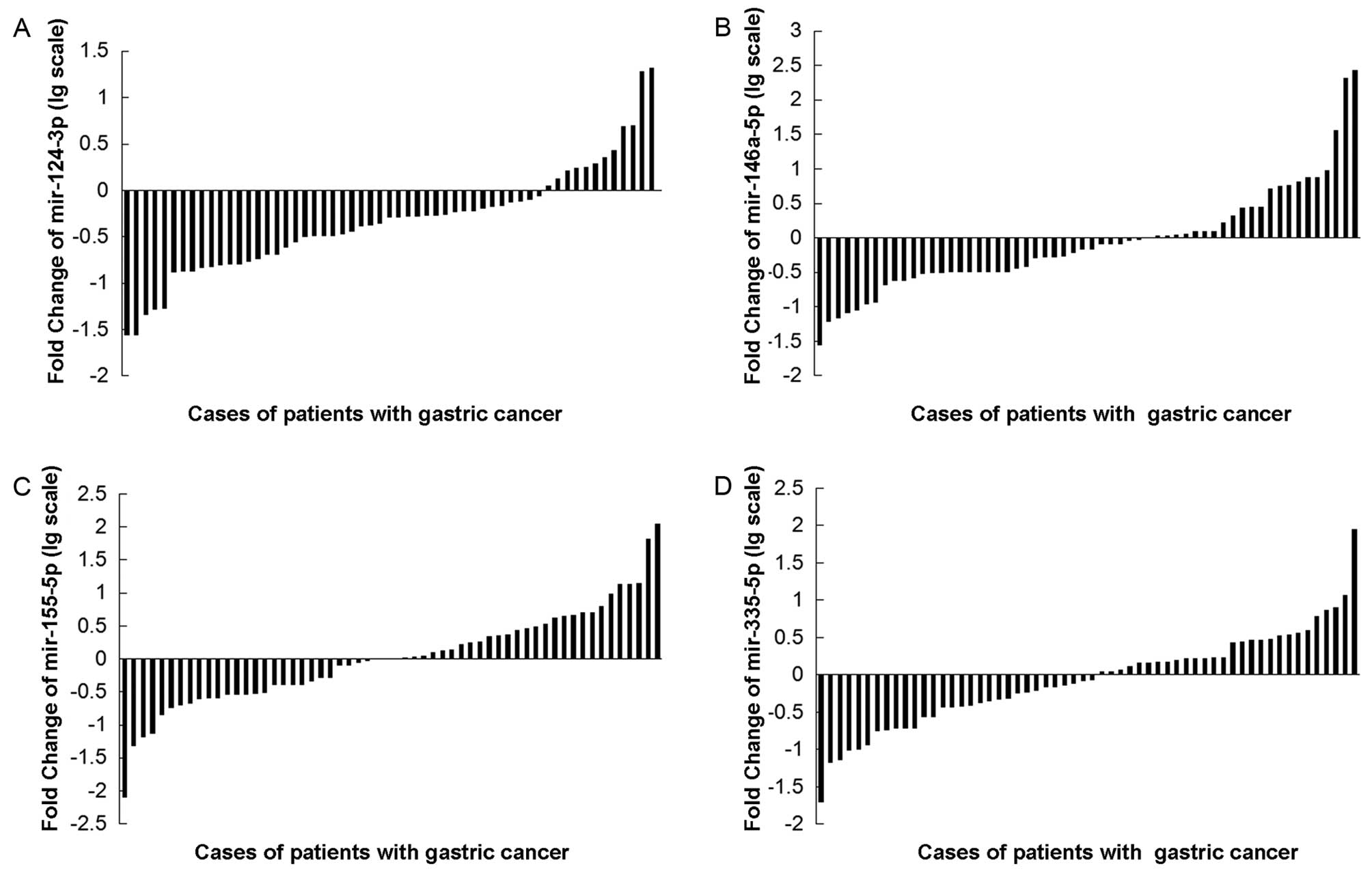
Clinical significance of mir-124-3p,
mir-146a-5p, mir-155-5p and mir-335-5p in gastric cancer tissue
compared with adjacent non-tumor tissues
All clinicopathologic factors were analyzed in
relation to these 4 miRNA levels. The mir-124-3p low-expression
group showed more extensive lymph node metastasis and lymphatic
invasion than the high-expression group (P<0.05; χ2
test). However, no significant differences were observed among age,
gender, degree of tumor cell differentiation, local invasion, depth
of tumor invasion, venous invasion, Borrmann type, and clinical
stage (Table I, Fig. 4A). The mir-146a-5p low-expression
group showed more venous invasion than the high-expression group
(P<0.05; χ2 test). However, no significant
differences were observed in age, local invasion, depth of tumor
invasion, lymph node metastasis, lymphatic invasion, Borrmann type,
and clinical stage (Table II,
Fig. 4B). The mir-155-5p
low-expression group showed higher stage Borrmann type than the
high-expression group (P<0.05; χ2 test). However, no
significant differences were observed in age, local invasion, depth
of tumor invasion, lymph node metastasis, lymphatic invasion,
venous invasion and clinical stage (Table III, Fig. 4C). The mir-335-5p low-expression
group showed more lymphatic invasion and high stage Borrmann type
than the high-expression group (P<0.05; χ2 test).
However, no significant differences were observed in age, gender,
local invasion, depth of tumor invasion, lymph node metastasis,
venous invasion and clinical stage (Table IV, Fig. 4D).
Signaling pathway and gene ontology
analyses of mir-124-3p, mir-146a-5p, mir-155-5p and mir-335-5p
targeted genes
In order to investigate the possible regulation
mechanisms of the mir-124-3p, mir-146a-5p, mir-155-5p and
mir-335-5p in the process of gastric cancer, we utilized a
bioinformatics database mirfocus 2.0 to select plausible targets of
these 4 miRNAs. A total of 1770, 400, 816, 357 target genes were
predicted as the target genes of mir-124-3p, mir-146a-5p,
mir-155-5p and mir-335-5p, respectively. Enriched KEGG pathway
analyses and enriched GO terms showed that most of the targeted
genes, which were regulated by these 4 miRNAs, were involved in the
same pathways.
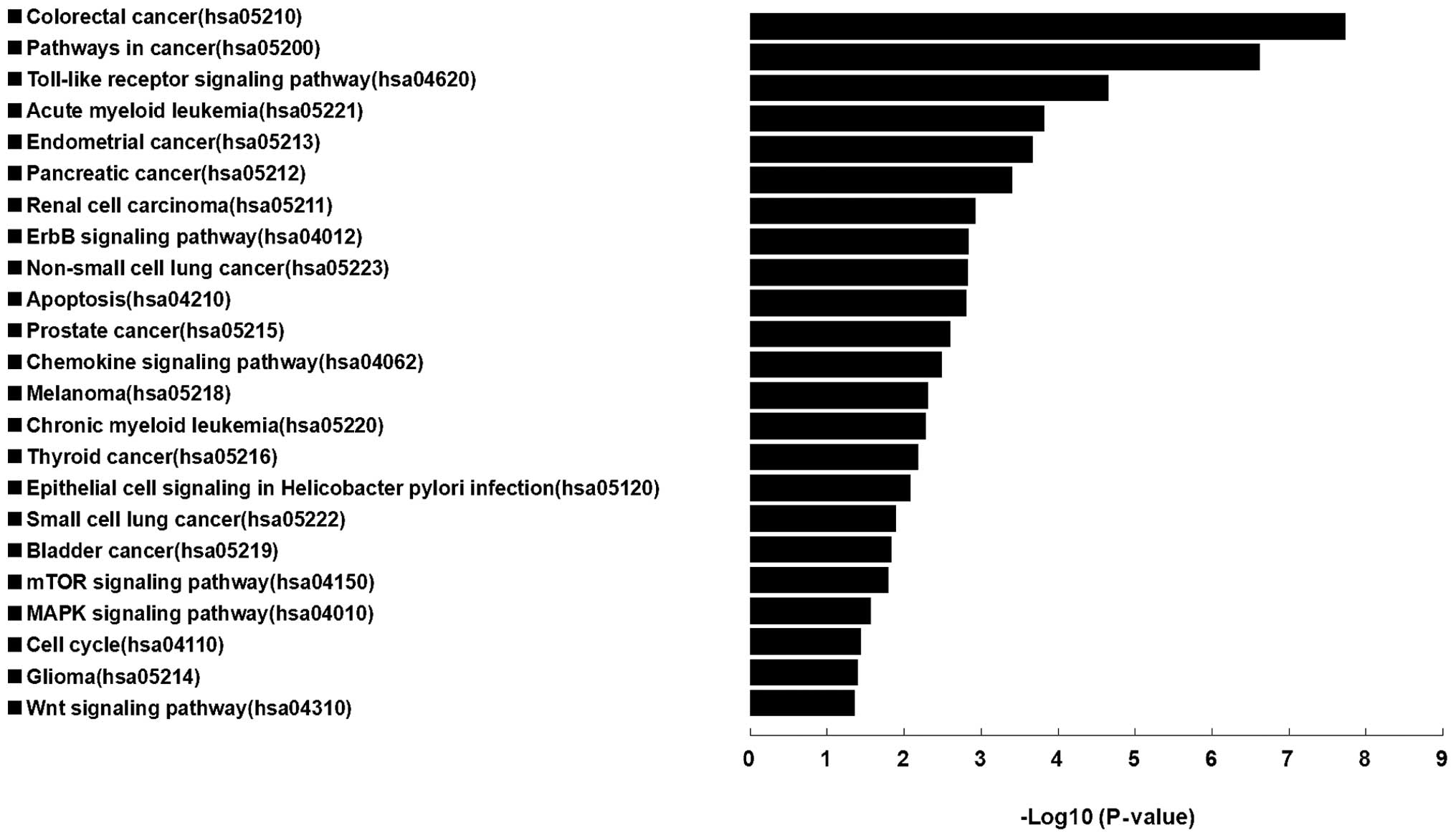
Results of enrichment KEGG pathway indicated that
the target genes of these 4 miRNAs were mainly centralized in
cancer associated terms, which were: colorectal cancer, pathways in
cancer, acute myeloid leukemia, endometrial cancer, adherens
junction, pancreatic cancer, renal cell carcinoma, non-small cell
lung cancer, apoptosis, prostate cancer, melanoma, chronic myeloid
leukemia, thyroid cancer, small cell lung cancer, bladder cancer,
melanogenesis, glioma, and 9 signaling pathways of toll-like
receptor, chemokine, erbb signaling pathway, epithelial cell
signaling in helicobacter pylori infection, cell cycle,
NOD-like receptor, mTOR, MAPK and wnt signaling pathways (Fig. 5).
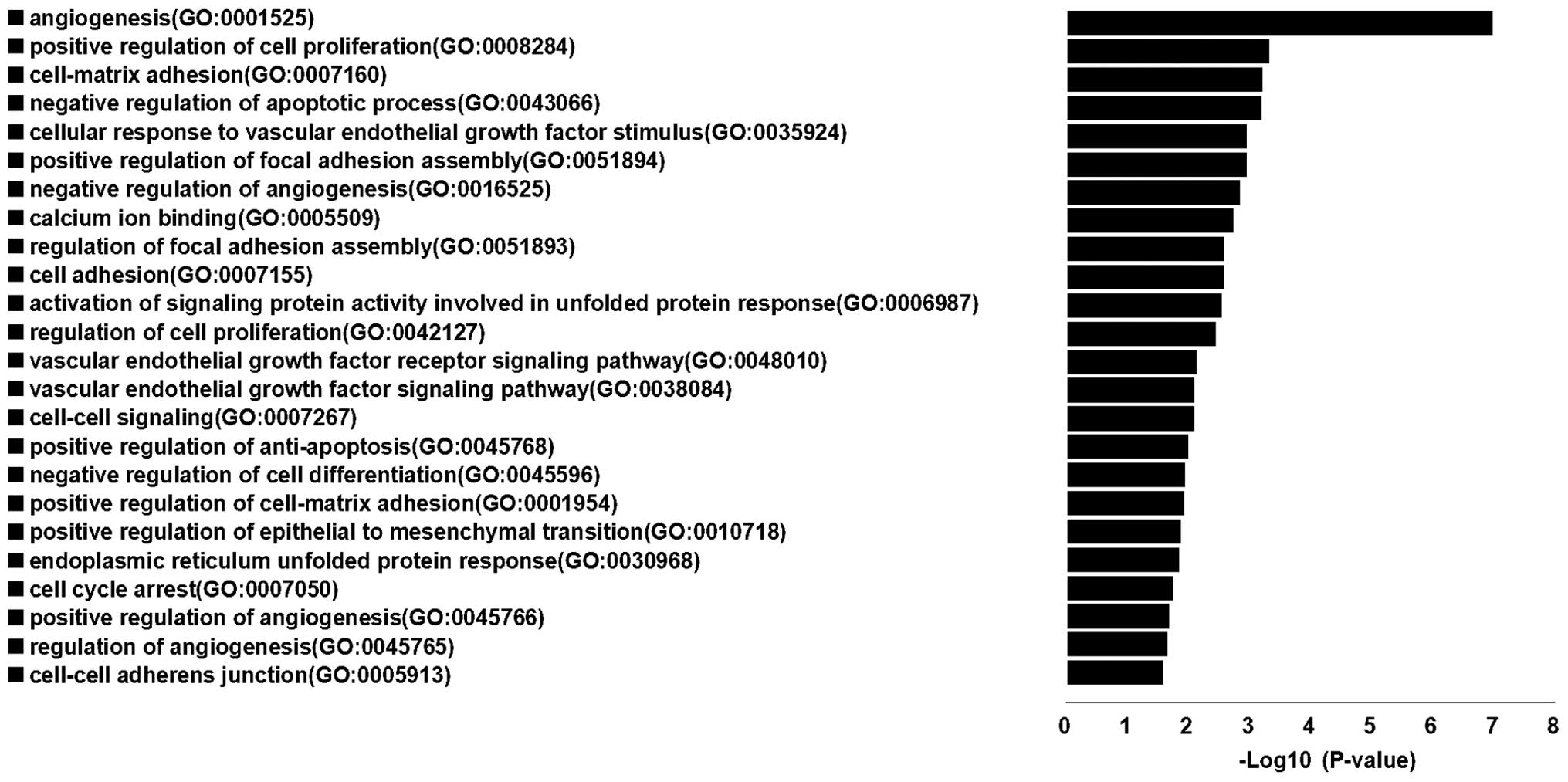
Results of enrichment GO terms indicated that the
target genes of these 4 miRNAs were centralized in cancer
associated terms, which were: angiogenesis, positive regulation of
cell proliferation, cell-matrix adhesion, negative regulation of
apoptotic process, positive regulation of focal adhesion assembly,
cellular response to vascular endothelial growth factor stimulus,
negative regulation of angiogenesis, calcium ion binding, cell
adhesion, regulation of focal adhesion assembly, activation of
signaling protein activity involved in unfolded protein response,
regulation of cell proliferation, vascular endothelial growth
factor receptor signaling pathway, cell-cell signaling, vascular
endothelial growth factor signaling pathway, positive regulation of
anti-apoptosis, negative regulation of cell differentiation,
positive regulation of cell-matrix adhesion, positive regulation of
epithelial to mesenchymal transition, endoplasmic reticulum
unfolded protein response, cell cycle arrest, positive regulation
of angiogenesis, regulation of angiogenesis and cell-cell adherens
junction. The results showed that genes regulated by these 4 miRNAs
participated in most of the important biological process associated
with human cancer (Fig. 6).
Discussion
This study showed that 38 miRNAs were upregulated
and 4 miRNAs downregulated in 7 gastric cancer cell lines compared
to the GES-1 cell line. To our knowledge, this is the first report
on the expression levels of 42 miRNAs in gastric cancer cells which
could facilitate its discovery in gastric cancer tissue and find
new biomarkers of gastric cancer, the 7 miRNAs mir-1, let-7b-5p,
let-7c, let-7g-5p, mir-27b-3p, mir-183-5p, mir-193-3p and mir-203a
were first identified upregulated in gastric cancer cell lines, and
no studies were found in gastric cancer tissues compared with
adjacent normal gastric tissue. In addition, the expression levels
of 4 miRNAs of mir-1, mir-7-5p, mir-133b and mir-143 were
inconsistent with previous studies screened by miRNA microarray
acquired from literature, according to these reports mir-1,
mir-7-5p, mir-133b and mir-143 were downregulated in gastric cancer
tissues and gastric cancer cells (14,21–26).
In previous studies, miRNA microarray has been widely used to
screen aberrant miRNAs between cancer tissue and normal tissue, but
the results indicated that aberrant expression of miRNAs in each
study were different, and the reasons could be associated with
different sample collection from different gastric cancer tissues
which reflect different representations or associate with the miRNA
microarray manufacturing, which employed various methods.
Therefore, Human Cancer Pathway Finder miRNA PCR array was used to
screen and analyse aberrant expression of miRNAs which could be
used to compensate for the disadvantages of miRNA microarray.
Our data indicated that mir-124-3p, mir-146a-5p,
mir-155-5p and mir-335-5p were downregulated in gastric cancer cell
lines and gastric cancer tissues, fully disclosing its expression
levels in gastric cancer lines and gastric cancer tissues. The
downregulated expression of 4 miRNAs showed clinical significance
in gastric cancer tissues comparing with adjacent normal gastric
tissues. Therefore, these 4 downregulated miRNAs are markedly
changed biomarkers in gastric cancer, and new light should be shed
on their target genes and their clinical significance of diagnosis
and prognosis.
In previous studies, upregulation of miR-124
dramatically inhibits the proliferation and tumorigenicity of
gastric cancer cells both in vitro and in vivo
through downregulation of SPHK1 (27), but its clinical significance in
gastric cancer is unknown. Our data found that miR-124-3p was
downregulated in gastric cancers, and low-expression group of
miR-124-3p indicating more extensive lymph node metastasis and
lymphatic invasion than the high-expression group.
Previous studies demonstrate that the level of
miR-146a in cancer tissues was significantly lower than that in the
corresponding non-cancerous tissue in 90 clinical samples of
gastric cancer (28) and
associated with lymph node metastasis and venous invasion.
Overexpression of miR-146a suppressed the migration and invasion of
gastric cancer cells, and the protein level of WASF2 (29). Low expression of miR-146a was
correlated with increased tumor size and poor differentiation in
gastric cancer. Overall survival time of patients with high
miR-146a expression was significantly longer than that of patients
with low expression of miR-146a (30). Moreover, over expression of
miR-146a inhibits the invasion and metastasis of MKN-45 cells in
vitro and in vivo in part due to the downregulation of
L1CAM (31). In addition, some
researchers found that miR-146a expression is upregulated in a
majority of gastric cancers in which it targets CARD10 and COPS8
through inhibiting the activation of NF-κB, thus reducing
expression of NF-κB-regulated tumor-promoting cytokines and growth
factors (32). miR-146a was
upregulated in 20 gastric cancer tissues compared to matched
non-tumor adjacent tissues by directly target SMAD4 (33). Our data confirmed that mir-146a-5p
was downregulated in gastric cancers, and low-expression group of
mir-146a-5p showed more venous invasion than the high-expression
group.
In previous studies, miR-155 was overexpressed in
cancer tissues of 91 patients by formalin-fixed paraffin-embedded
(FFPE) (34), and related to tumor
penetration through serosa and lymph node metastasis (35). However, there is also a study on
miR-155 significantly downregulated in gastric cancer cell lines
compared with GES-1, and overexpression of miR-155 significantly
reduced the protein levels of SMAD2 (36). Our results showed that mir-155-5p
was downregulated in gastric cancers, and low-expression group of
mir-155-5p showed higher stage Borrmann type than the
high-expression group.
Previous reports demonstrate that miR-335 is
down-regulated in gastric cancer cell lines SGC-7901, MGC-803,
BCG-823 and AGS compared with GES-1, and suppresses gastric cancer
invasion in vitro and in vivo, by targeting SP1
directly and indirectly through a Bcl-w-induced signaling pathway
that sequentially involves PI3K, Akt and Sp1 (37). miR-335 has the potential to
recognize the recurrence risk and relate to the prognosis of
gastric cancer patients (38). In
this study, mir-335-5p was also downregulated in gastric cancer
lines and gastric cancer tissues, and low-expression group showed
more lymphatic invasion and high stage Borrmann type than the
high-expression group which was consistent with previous studies
(38).
KEGG pathway enrichment analysis showed that
targeted genes of mir-124-3p, mir-146a-5p, mir-155-5p and
mir-335-5p concentrated on 37 signaling pathways, 24 of 37
signaling pathways were involved in the same pathways relevant to
cancer. Enriched GO terms showed that targeted genes of these 4
miRNAs concentrated on 339 terms, 24 of 339 terms are associated
with same terms relevant to cancer.
In conclusion, Human Cancer Pathway Finder miRNA PCR
array is a new approach to screen and validate aberrantly expressed
miRNAs more effectively and accurately than miRNA microarray. With
this array, we found 38 miRNAs upregulated, and 4 miRNAs
downregulated in 7 gastric cancer cell lines comparing GES-1 cell
line. Among the 38 upregulated miRNAs, 7 miRNAs including mir-1,
let-7b-5p, let-7c, let-7g-5p, mir-27b-3p, mir-183-5p, mir-193-3p
and mir-203a were first identified in gastric cancer cell lines. In
addition, the expression of 4 miRNAs of mir-124-3p, mir-146a-5p,
mir-155-5p and mir-335-5p was also verified in gastric cancer cell
lines and gastric cancer tissues which clarify their confusing,
even paradoxical reports, as their expression levels in gastric
cancer. Clinical significance of these 4 miRNAs in gastric cancer
tissues showed down-regulation relevant to more extensive lymph
node metastasis and lymphatic invasion, more venous invasion, more
high stage Borrmann type, more lymphatic invasion and poor
differentiation. Moreover, bioinformatics analysis indicated
enriched KEGG pathway analysis and gene ontology of mir-124-3p,
mir-146a-5p, mir-155-5p and mir-335-5p were mainly centralized in
pathways and GO terms related to tumorigenesis suggesting 4 miRNAs
could be important biomarkers in gastric cancers.
Acknowledgements
The study was supported by the
Research Fund of Personnel training plan of West Light (no.
201218), Chinese Academy of Sciences; and Open Plan of Key
Laboratory for Gastrointestinal Diseases of Gansu Province (no.
gswcky-2013-004).
References
|
1.
|
Bartels CL and Tsongalis GJ: MicroRNAs:
novel biomarkers for human cancer. Clin Chem. 55:623–631. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Cortes-Sempere M and Ibanez de Caceres I:
microRNAs as novel epigenetic biomarkers for human cancer. Clin
Transl Oncol. 13:357–362. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Shenouda SK and Alahari SK: MicroRNA
function in cancer: oncogene or a tumor suppressor? Cancer
Metastasis Rev. 28:369–378. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Callari M, Dugo M, Musella V, et al:
Comparison of microarray platforms for measuring differential
microRNA expression in paired normal/cancer colon tissues. PLoS
One. 7:e451052012. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
D’Andrade PN and Fulmer-Smentek S: Agilent
microRNA microarray profiling system. Methods Mol Biol. 822:85–102.
2012.PubMed/NCBI
|
|
6.
|
Jang JS, Simon VA, Feddersen RM, et al:
Quantitative miRNA expression analysis using fluidigm microfluidics
dynamic arrays. BMC Genomics. 12:1442011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Ranade AR and Weiss GJ: Methods for
microRNA microarray profiling. Methods Mol Biol. 700:145–152. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Srivastava S, Srivastava AK, Suprasanna P
and D’Souza SF: Identification and profiling of arsenic
stress-induced microRNAs in Brassica juncea. J Exp Bot.
64:303–315. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Katada T, Ishiguro H, Kuwabara Y, et al:
microRNA expression profile in undifferentiated gastric cancer. Int
J Oncol. 34:537–542. 2009.PubMed/NCBI
|
|
10.
|
Li X, Zhang Y, Zhang H, et al: miRNA-223
promotes gastric cancer invasion and metastasis by targeting tumor
suppressor EPB41L3. Mol Cancer Res. 9:824–833. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Gao M, Yin H and Fei ZW: Clinical
application of microRNA in gastric cancer in Eastern Asian area.
World J Gastroenterol. 19:2019–2027. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Carvalho J, van Grieken NC, Pereira PM, et
al: Lack of microRNA-101 causes E-cadherin functional deregulation
through EZH2 up-regulation in intestinal gastric cancer. J Pathol.
228:31–44. 2012.PubMed/NCBI
|
|
13.
|
Guo J, Miao Y, Xiao B, et al: Differential
expression of microRNA species in human gastric cancer versus
non-tumorous tissues. J Gastroenterol Hepatol. 24:652–657. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Li X, Luo F, Li Q, et al: Identification
of new aberrantly expressed miRNAs in intestinal-type gastric
cancer and its clinical significance. Oncol Rep. 26:1431–1439.
2011.PubMed/NCBI
|
|
15.
|
Luo H, Zhang H, Zhang Z, et al:
Down-regulated miR-9 and miR-433 in human gastric carcinoma. J Exp
Clin Cancer Res. 28:822009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Tsukamoto Y, Nakada C, Noguchi T, et al:
MicroRNA-375 is downregulated in gastric carcinomas and regulates
cell survival by targeting PDK1 and 14-3-3zeta. Cancer Res.
70:2339–2349. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Ueda T, Volinia S, Okumura H, et al:
Relation between microRNA expression and progression and prognosis
of gastric cancer: a microRNA expression analysis. Lancet Oncol.
11:136–146. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Yao Y, Suo AL, Li ZF, et al: MicroRNA
profiling of human gastric cancer. Mol Med Rep. 2:963–970.
2009.PubMed/NCBI
|
|
19.
|
Zhang Z-Y, Wu Z-Q and Wang H-J: An
analysis of gastric cancer detection and epidemicity in high
incidental area with gastric cancer, Wuwei City, Gansu Province.
China Cancer. 10:0142009.
|
|
20.
|
Li Y-M, Shi B, Li X, Zhou W-N, Liu H and
Mi D-H: Study of the characteristics of gastric carcinoma in Wuwei
City of Gansu province. Zhonghua pu tong wai ke za zhi. 13:667–669.
2004.(In Chinese).
|
|
21.
|
Kim CH, Kim HK, Rettig RL, et al: miRNA
signature associated with outcome of gastric cancer patients
following chemotherapy. BMC Med Genomics. 4:792011. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Kong D, Piao YS, Yamashita S, et al:
Inflammation-induced repression of tumor suppressor miR-7 in
gastric tumor cells. Oncogene. 31:3949–3960. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Takagi T, Iio A, Nakagawa Y, Naoe T,
Tanigawa N and Akao Y: Decreased expression of microRNA-143 and
-145 in human gastric cancers. Oncology. 77:12–21. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Wen D, Li S, Ji F, et al: miR-133b acts as
a tumor suppressor and negatively regulates FGFR1 in gastric
cancer. Tumour Biol. 34:793–803. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Zhao X, Dou W, He L, et al: MicroRNA-7
functions as an anti-metastatic microRNA in gastric cancer by
targeting insulin-like growth factor-1 receptor. Oncogene.
32:1363–1372. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Zhao Y, Huang J, Zhang L, et al: MiR-133b
is frequently decreased in gastric cancer and its overexpression
reduces the metastatic potential of gastric cancer cells. BMC
Cancer. 14:342014. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Xia J, Wu Z, Yu C, et al: miR-124 inhibits
cell proliferation in gastric cancer through down-regulation of
SPHK1. J Pathol. 227:470–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Kogo R, Mimori K, Tanaka F, Komune S and
Mori M: Clinical significance of miR-146a in gastric cancer cases.
Clin Cancer Res. 17:4277–4284. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Yao Q, Cao Z, Tu C, Zhao Y, Liu H and
Zhang S: MicroRNA-146a acts as a metastasis suppressor in gastric
cancer by targeting WASF2. Cancer Lett. 335:219–224. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Hou Z, Xie L, Yu L, Qian X and Liu B:
MicroRNA-146a is down-regulated in gastric cancer and regulates
cell proliferation and apoptosis. Med Oncol. 29:886–892. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Hou Z, Yin H, Chen C, et al: microRNA-146a
targets the L1 cell adhesion molecule and suppresses the metastatic
potential of gastric cancer. Mol Med Rep. 6:501–506. 2012.
|
|
32.
|
Crone SG, Jacobsen A, Federspiel B, et al:
microRNA-146a inhibits G protein-coupled receptor-mediated
activation of NF-kappaB by targeting CARD10 and COPS8 in gastric
cancer. Mol Cancer. 11:712012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Xiao B, Zhu ED, Li N, et al: Increased
miR-146a in gastric cancer directly targets SMAD4 and is involved
in modulating cell proliferation and apoptosis. Oncol Rep.
27:559–566. 2012.PubMed/NCBI
|
|
34.
|
Kim BH, Hong SW, Kim A, Choi SH and Yoon
SO: Prognostic implications for high expression of oncogenic
microRNAs in advanced gastric carcinoma. J Surg Oncol. 107:505–510.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Liu L, Chen Q, Lai R, et al: Elevated
expression of mature miR-21 and miR-155 in cancerous gastric
tissues from Chinese patients with gastric cancer. J Biomed Res.
24:187–197. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Li CL, Nie H, Wang M, et al: microRNA-155
is downregulated in gastric cancer cells and involved in cell
metastasis. Oncol Rep. 27:1960–1966. 2012.PubMed/NCBI
|
|
37.
|
Xu Y, Zhao F, Wang Z, et al: MicroRNA-335
acts as a metastasis suppressor in gastric cancer by targeting
Bcl-w and specificity protein 1. Oncogene. 31:1398–1407. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Yan Z, Xiong Y, Xu W, et al:
Identification of hsa-miR-335 as a prognostic signature in gastric
cancer. PLoS One. 7:e400372012. View Article : Google Scholar : PubMed/NCBI
|