Introduction
HOXB13 is a transcription factor with a homedomain.
Although the expression of HOXB13 is limited to the normal prostate
and rectum (1), the altered
regulation of HOXB13 expression has been demonstrated in various
cancer tissues originating from the prostate, breast and ovary
(2–4). HOXB13 controls cancer cell
proliferation, but its role varies according to the type of tissue
and the cancer cell environment (5–8). In
addition, HOXB13 has been shown to be overexpressed in
castration-resistant prostate cancer cells (9), suggesting that HOXB13 may play a role
in prostate cancer metastasis. In fact, HOXB13 has recently been
demonstrated to promote prostate cancer invasion by reducing the
level of intracellular zinc (10).
The prostate-derived Ets factor (PDEF) is a new
member of the Ets transcription factor. The PDEF is also known as a
sam-pointed domain containing the Ets transcription factor (SPDEF).
The human Ets family includes 27 members, and the mouse Ets family,
26 members. The DNA-binding domain has 85 amino acids that bind to
DNA sequences with the GGA(A/T) core consensus sequence (11). These Ets transcription factors
regulate target gene expression through cooperation with other
transcription factors and co-factors and play an important role in
a number of cellular processes, including the regulation of cell
differentiation, proliferation, angiogenesis, and apoptosis. In
addition, Ets family members play major roles in tumorigenesis,
particularly in prostate cancer (12–14).
They are aberrantly expressed in specific cases of leukemia and
major solid tumors, including breast and prostate tumors.
The Ets domain of the PDEF preferentially binds to
the GGAT sequence, not to the GGAA sequence preferred by other ETS
proteins (15). Because of the
complexity of binding partners and regulatory mechanisms, the PDEF
has been found to play many roles in various pathophysiological
conditions. Most PDEF target genes are deregulated in the
tumorigenic process and include the prostate-specific antigen
(PSA), survivin, MMP-7, MMP-9, the urokinase-type plasminogen
activator (uPA), maspin, LASP1, VASP, p21 and Slug.
The PDEF has been shown to regulate processes
involved in prostate tumor cell motility and invasion and is lost
during tumor progression and increases in MMP-9 levels, thereby
promoting tumor cell invasion (16). In addition, the siRNA-mediated
reduction of the PDEF induces an EMT-like phenotype such that
mesenchymal markers increase to reduce cell adhesion and increase
cell migration and invasion (17).
The PDEF reduces Slug, a protein required for the EMT (18). By contrast, PDEF overexpression in
PC3 prostate tumor cells influences FAK activity and cell
morphology, reducing cell migration and invasion (16). This clearly suggests that the PDEF
affects multiple pathways, as in the case of many transcription
factors, and that separating the pathways directly regulated by the
PDEF from those indirectly regulated is critical to understanding
the functional role of the PDEF.
This study provides a better understanding of the
role of HOXB13 in prostate cancer metastasis and the mechanisms by
which HOXB13 functions. Because of the highly controversial
function with a very limited understanding of the regulation of
PDEF expression, this study also clarifies the invasive role of the
PDEF in highly malignant PC3 prostate cancer cells. Finally, we
report on the regulatory mechanism underlying PDEF expression.
Materials and methods
Cell culture
LNCaP and PC3 human prostate cancer cell lines were
routinely cultured in RPMI media (Life Technology, Grand Island,
NY, USA) supplemented with 5% FBS at 37°C in an atmosphere
containing 5% CO2. All cultures were fed with fresh
media every 3–4 days.
Plasmids and reagents
pCDNA-HOXB13 was constructed from the pFLAG-HOXB13
backbone to accommodate the neomycin-resistant gene.
pCDNA6-myc-His.c-PDEF was generously provided by Dr C.J. Bieberich
of the University of Maryland, Baltimore, MD. The selection of
HOXB13-positive and PDEF-positive PC3 cells was done with neomycin
(G418 sulfate; Promega Corp., Madison, WI, USA) and blastcidine S
hydrochloride (Sigma-Aldrich Corp., St. Louis, MO, USA),
respectively. Antibodies were purchased as follows: PDEF (Santa
Cruz Biotechnology, Santa Cruz, CA, USA), c-myc (BD Biosciences,
Franklin Lakes, NJ, USA), MMP-9 (Merck Millipore, Billerica, MA,
USA), survivin (Santa Cruz Biotechnology) and β-actin
(Sigma-Aldrich Corp.).
Stable transfection
Cells were grown to 50% confluence in P60 culture
dishes 16 h before transfection. Transfection was carried out using
Lipofectamine 2000 (Life Technology) with either the
pCDNA3.1-HOXB13 or pCDNA6-myc-His.c-PDEF plasmid based on the
manufacturer’s protocol. After a 6 h transfection period, the cells
were washed and fed with a medium containing 5% FBS. At 24 h after
transfection, G418 (200 μg/ml) or blasticidin (2 μg/ml) was added.
The selection process continued with the medium changed every 3
days, and a continuous expansion was made under sufficient
antibiotics (100 μg/ml G418 and 1 μg/ml blasticidine). HOXB13
transfection was also made in PDEF-expressing PC3 cells.
Western blot assay
Cells were grown to 80% confluence in P100 culture
dishes containing 5% FBS-RPMI media. Then the cells were lysed in
protein extraction RIPA buffer (25 mM Tris-HCl pH 7.6, 150 mM NaCl,
1% NP-40, 1% sodium deoxycholate, 0.1% SDS) containing a cocktail
of protease inhibitors (Sigma-Aldrich Corp.). Lysates were
incubated for 30 min on ice, followed by centrifugation for 10 min.
Supernatants were measured for the protein concentration by using
the Bradford assay reagent (Bio-Rad Laboratories, Hercules, CA,
USA). Each cell lysate was loaded onto 10% Bis-Tris gel (Life
Technology) and separated using a Bio-Rad electroporation system.
After the proteins were transferred to the PVDF membrane, primary
antibodies were applied (PDEF; 1–200, HOXB13; 1–100, myc; 1–500,
MMP-9; 1–1,000, survivin; 1–100, β-actin; 1–2,000), followed by
incubation with horse peroxidase-conjugated secondary antibodies.
Enhanced chemiluminescence was used to visualize bands by LAS 3000
(Fujifilm, Tokyo, Japan).
RT-PCR
Cellular RNA from cells using the TRIzol reagent and
its quantity were determined spectrophotometrically. Here 1 μg of
extracted RNA with 100 pM d(T)20 was preheated at 72°C
for 10 min and reverse-transcribed with M-MLV (Promega Corp.) at
42°C for 1 h. PCR was performed in a 20 μl solution containing 4 μl
of 10X PCR buffer (200 mM Tris-HCl pH 8.4, 500 mM KCl), 0.2 mM dNTP
mixture, 1 μl template cDNA and 0.2 μM primers. The primer
sequences were as follows: PDEF fw, 5′-ccgggtctgagcagcgtatc-3′;
PDEF rv, 5′-tgctcaggctcctcaggtgg-3′; HOXB13 fw, 5′-gcctct
gtccttggtgatgaac-3′; HOXB13 rv, 5′-aggccgccatccaggaaaag-3′; β-actin
fw, 5′-gcaccacaccttctacaatgagc-3′; and β-actin rv,
5′-tagcacagcctggatagcaacg-3′. Amplification reactions were
performed using a DNA thermal cycler (Eppendorf, Hamburg, Germany).
PCR product was separated by 1% agarose gel electrophoresis.
Wound-healing assay
For the measurement of cell migration during wound
healing, cells were seeded onto 6-well plates and allowed to grow
to confluence. The confluent cell monolayer was wounded by pressing
a sterile 100 μl pipette tip down onto the plate to cut the cell
sheet and mark the plate with a sharp and visible demarcation at
the wound edge. The medium and debris were aspirated away and
replaced by a fresh serum-free medium, and cells were incubated for
24 h at 37°C. The cells were photographed every 24 h after wounding
by phase contrast microscopy. For the evaluation of the ‘wound
closure’, five randomly selected points along each wound were
marked and the horizontal distance of migrating cells from the
initial wound was measured. This assay was imaged using a
microscope (Leica Microsystems, Wetzlar, Germany). All presented
data are from at least three independent experiments performed in
duplicate.
Matrigel invasion assay
The transwell chamber (Corning Inc., Corning, NY,
USA) was coated with a thin layer of Matrigel (BD Biosciences).
Matrigel was diluted in serum-free media. The 100 μl of gel
suspension was added to the upper chamber and incubated for 1 h at
RT. Then the Matrigel solution was aspirated and washed once with
100 μl of serum-free media. The upper chamber was placed in the
bottom chamber containing 400 μl of media with fibronectin (20
μg/ml). Cells (1×105) were added to the upper chamber in
300 μl of serum-free media. After incubation for 24 h at 37°C,
cells inside the inserts were removed with cotton tips, and the
invaded cells were stained by Diff Quick (Sysmex Corp., Kobe,
Japan) and analyzed using a microscope (Leica Microsystems).
Gelatin zymography
PC3 cells were cultured for 24 h and the supernatant
was collected. The samples were analyzed using a zymographic
technique based on 10% SDS-PAGE with 0.1% (w/v) gelatin
(Sigma-Aldrich Corp.) as the substrate. The samples were loaded
without heating with sample buffer and applied to a non-denaturing
10% polyacrylamide gel containing 1 mg/ml gelatin. After
electrophoresis, the gel was washed twice in 50 mM Tris (pH 7.4)
containing 2.5% (v/v) Triton X-100 for 1 h, followed by two 10 min
rinses in 50 mM Tris (pH 7.4). After SDS removal, the gel was
incubated overnight in 50 mM Tris (pH 7.5) containing 10 mM
CaCl2, 0.15 M NaCl, 0.1% (v/v) Triton X-100, and 0.02%
sodium azide at 37°C under constant and gentle shaking. After
incubation, the gel was stained with 0.25% Coomassie brilliant blue
R-250 and destained in 7.5% acetic acid with 20% methanol.
Gelatinase bands appeared white on a blue background. The activity
of MMP-9 was semi-quantitatively determined by densitometry.
Results
HOXB13 promotes the invasive and
migratory potential of PC3 prostate cancer cells
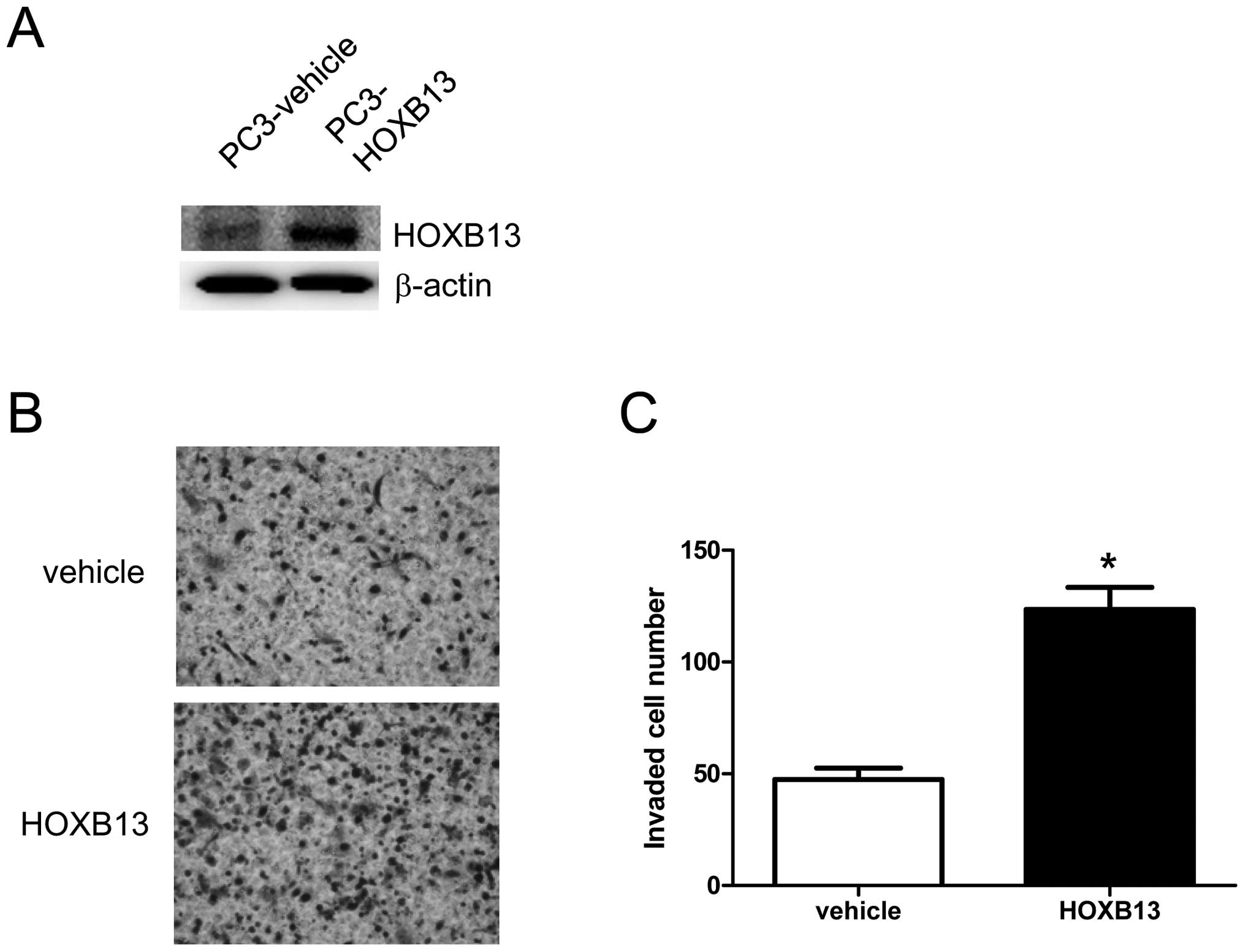
To verify the involvement of HOXB13 in prostate
cancer cell migration and invasion, PC3 cells were stably
transfected with either pCDNA3.1 (vehicle) or pCDNA-HOXB13
(Fig. 1A). Selected cells were
used for Matrigel-coated transwell invasion assays. As shown in
Fig. 1B, there were visibly more
infiltrated cells in HOXB13-transfected cells. Several random
fields were selected, and infiltrated cells were counted (shown as
a bar graph). HOXB13 significantly increased the invasion of PC3
cells through the Martigel-coated transwell (vehicle 47.40±5.13;
HOXB13 123.60±9.84)(Student’s t-test, p<0.05) (Fig. 1C). Then, to determine the
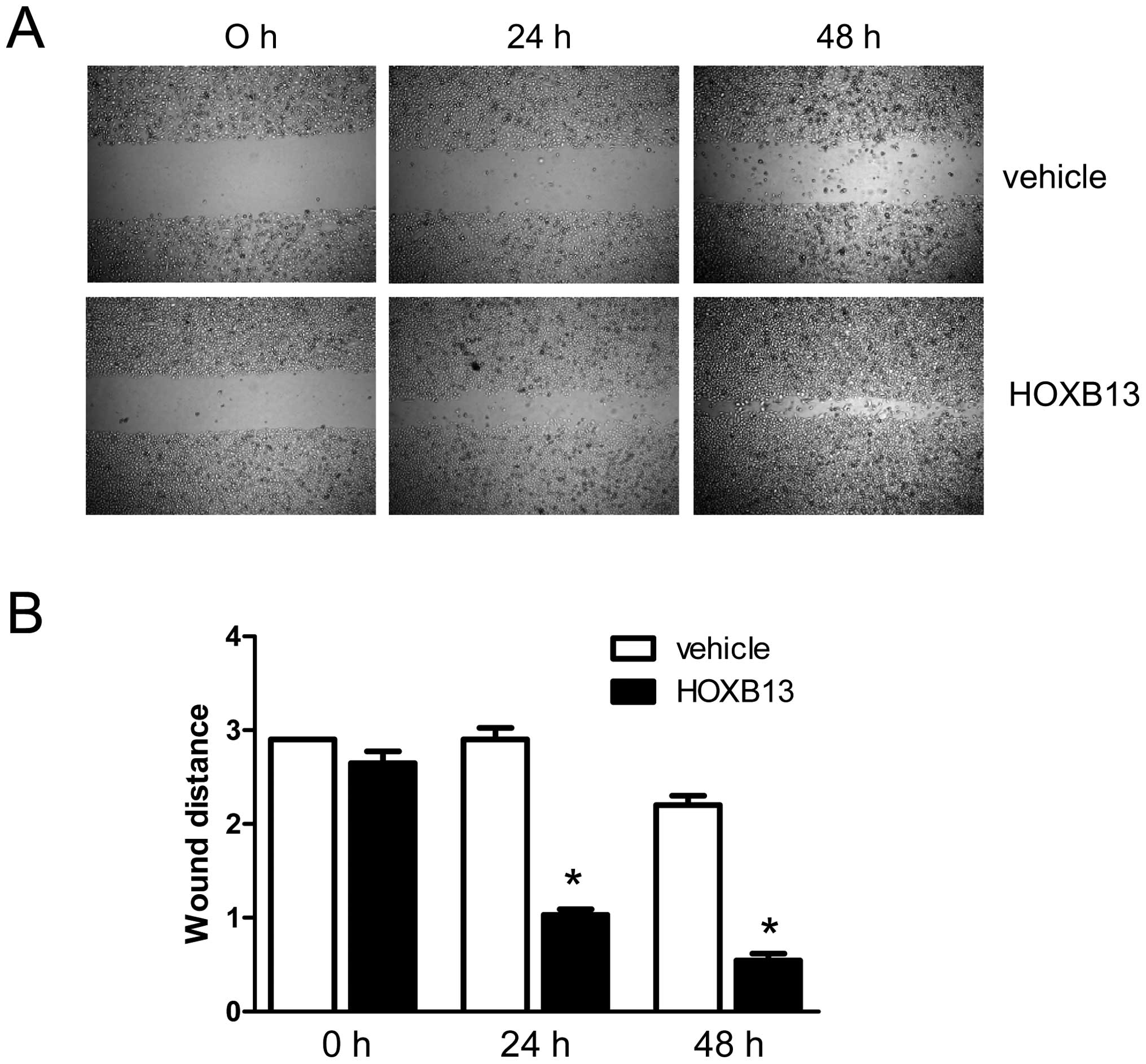
involvement of HOXB13 in the migration of PC3 cells, an in
vitro wound-healing assay was performed using
HOXB13-manipulated PC3 cells. As shown in Fig. 2A, HOXB13 stimulated the migration
of PC3 prostate cancer cells in a time-dependent manner. The bar
graph representation of the migratory property shows the same
result (Fig. 2B). At 24 h, the gap
for the wounded region was 2.90 with control cells, whereas it was
1.03±0.01 with HOXB13-transfected cells (p<0.05). At 48 h, the
gap for the wounded region was 2.20±0.10 with control cells,
whereas it was 0.55±0.07 with HOXB13-transfected cells
(p<0.05).
HOXB13 manipulates the expression of
genes involved in prostate cancer cell migration and adhesion
To profile HOXB13 target genes, LNCaP prostate
cancer cells were infected with either adenoviral HOXB13 or an
empty vehicle, followed by a cDNA microarray analysis, as
demonstrated earlier (8). As shown
in Table I, HOXB13 regulated the
expression of multiple genes involved in cell motility and
adhesion. The most notable gene regulated by HOXB13 was SPDEF, a
SAM-pointed domain containing the Ets transcription factor. The
SPDEF, also known as the PDEF (15), functions as an anti-invasive gene
in prostate or breast cancer (17,19).
 | Table IList of HOXB13 target genes involved
in cancer cell migration and invasion. |
Table I
List of HOXB13 target genes involved
in cancer cell migration and invasion.
| Gene | Fold change | Accession no. | Description |
|---|
| Cell motility |
| CRYAB | −4.88 | NM_001885 | Crystallin, α
B |
| KCNMA1 | −3.28 | NM_002247 | Potassium large
conductance calcium-activated channel, subfamily M, α 1 |
| HMMR | 4.14 | NM_012485 | Hyaluronan-mediated
motility receptor (RHAMM) |
| TNFRSF12A | −3.06 | NM_016639 | Tumor necrosis
factor receptor superfamily, member 12A |
| MYLIP | 3.29 | NM_013262 | Myosin regulatory
light chain interacting protein |
| CKLFSF6 | 3.07 | NM_017801 | Chemokine-like
factor super family 6 |
| LAMA3 | −4.11 | NM_198129 | Laminin, α 3 |
| LAMA1 | −6.07 | NM_005559 | Laminin, α 1 |
| MMP2 | 2.13 | NM_004530 | Matrix
metalloproteinase 2 |
| MMP16 | 7.77 | NM_022564 | Matrix
metalloproteinase 16 |
| MMP27 | 3.78 | NM_022122 | Matrix
metalloproteinase 27 |
| SERPINI1 | 17.76 | NM_005025 | Serine proteinase
inhibitor, clade I, member 1 |
| SERPINC1 | 5.71 | NM_000488 | Serine proteinase
inhibitor, clade C, member 1 |
| SPDEF | −12.38 | NM_012391 | SAM pointed domain
containing ets transcription factor |
| OCLN | 10.84 | NM_002538 | Occludin |
| Cell adhesion |
| STIM2 | 5.28 | NM_020860 | Stromal interaction
molecule 2 |
| DSG4 | 12.72 | NM_177986 | Desmoglein 4 |
| PKD2 | 4.86 | NM_000297 | Polycystic kidney
disease 2 |
| CD47 | 4.05 | NM_198793 | CD47 antigen |
| CDA08 | 3.35 | NM_030790 | T-cell
immunomodulatory protein |
| PPFIA2 | 3.65 | NM_003625 | Protein tyrosine
phosphatase receptor type f (PTPRF), interacting protein, α 2 |
| PPFIBP1 | −4.93 | NM_177444 | PTPRF interacting
protein, binding protein 1 |
| RHOB | −4.51 | NM_004040 | Ras homolog gene
family, member B |
| DST | 3.2 | NM_183380 | Destrin |
| TNFRSF12A | −3.06 | NM_016639 | Tumor necrosis
factor receptor superfamily, member 12A |
| LRIG3 | 3.99 | NM_153377 | Leucine-rich
repeats and immunoglobulin-like domains 3 |
| NRXN1 | 3.33 | NM_138735 | Neurexin 1 |
Establishment of HOXB13 and/or PDEF
stably transfected PC3 prostate cancer cells
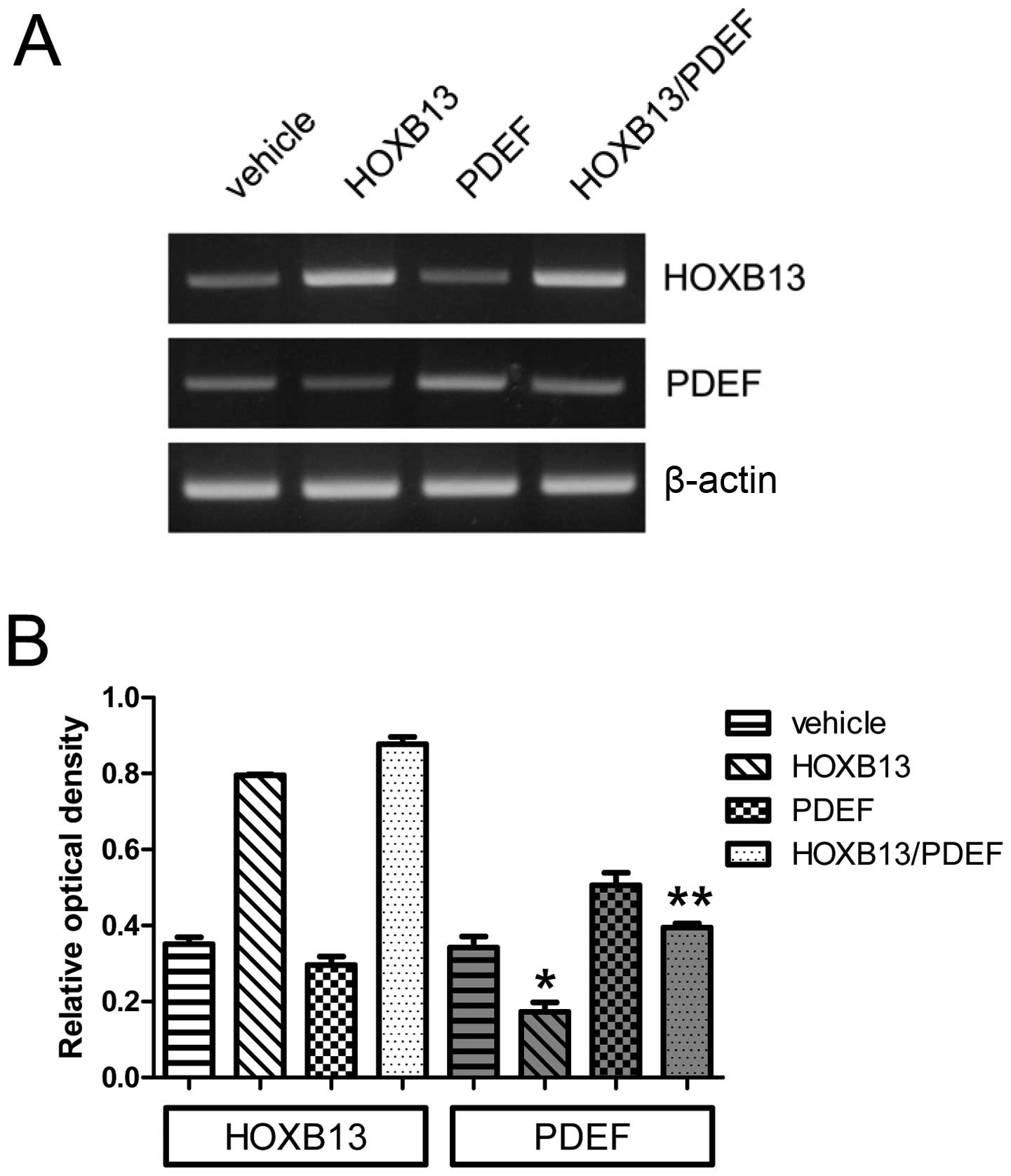
The forced expression of the PDEF was induced in
both wild-type PC3 and PC3-HOXB13 cells. Fig. 3A shows its expression, and Fig. 3B the densitometric evaluation. The
expression of HOXB13 alone significantly reduced PDEF expression
(0.34±0.05 vs 0.17±0.04) (p<0.05), and the expression of both
HOXB13 and PDEF reduced the expression of PDEF (0.40±0.02) in
comparison to the PDEF alone (0.51±0.06) (p<0.05), suggesting
that the reduced expression of the PDEF in HOXB13 and
PDEF-overexpressed PC3 cells derived from the suppression of the
endogenous PDEF by HOXB13.
HOXB13 counteracts the PDEF-mediated
inhibition of prostate cancer cell invasion
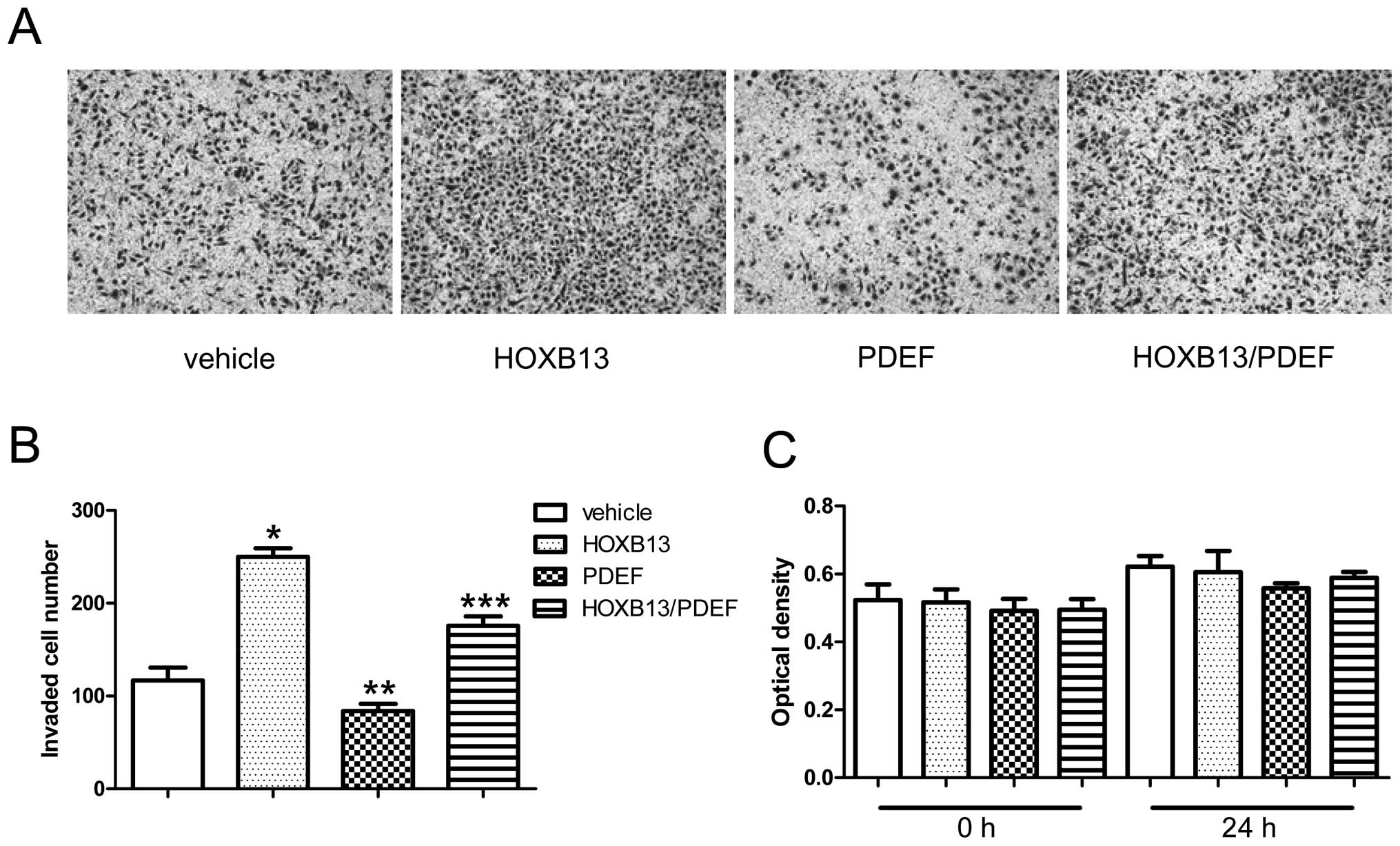
To determine whether the regulation of PDEF
expression would influence the invasive potential of HOXB13,
HOXB13- and/or PDEF-overexpressed PC3 cells were applied to a
transwell assay. Fig. 4A shows the
invaded cells, and Fig. 4B shows
them as a bar graph after the cells were counted on randomly
selected multiple fields. HOXB13 stimulated Matrigel-coated
transwell migration (116.8±13.92 vs 249.8±9.23) (p<0.05),
whereas the PDEF reduced invaded cells (116.8±13.92 vs 83.8±7.89)
(p<0.05) (Fig. 4B). The dual
expression of HOXB13 and the PDEF reduced the number of cells
(175.6±10.36) relative to HOXB13 alone (249.8±9.23) but increased
it relative to the PDEF alone (83.8±7.89) (p<0.05). The
co-overexpression of HOXB13 and the PDEF inhibited cell migration
by ~30% relative to HOXB13 alone and stimulated it by ~50% relative
to the PDEF alone. The MTT in vitro proliferation assay
showed no growth difference up to 36 h of incubation between four
different cell types (Fig. 4C).
These results indicate that HOXB13 manipulated the PDEF-mediated
inhibition of the invasive potential of PC3.
HOXB13 counteracts the expression of PDEF
targets involved in cancer cell invasion
Cancer cells express a high level of matrix
metalloproteinases (MMPs), which are known to facilitate the
initiation, invasion and metastasis of tumors (20). In HOXB13- and PDEF-manipulated
cells, the expression of PDEF target proteins was evaluated,
including MMP-9 (16) and survivin
(19). MMPs represent a family of
enzymes whose function is related mainly to the degradation of
extracellular matrix proteins and are necessary for cell invasion.
In addition, increased MMP activity is associated with tumor
metastasis. Survivin is an anti-apoptotic protein expressed in most
forms of cancer, including prostate cancer. Survivin inhibits
caspase activation, thereby facilitating the negative regulation of
apoptosis, and is a significant contributor to the development of
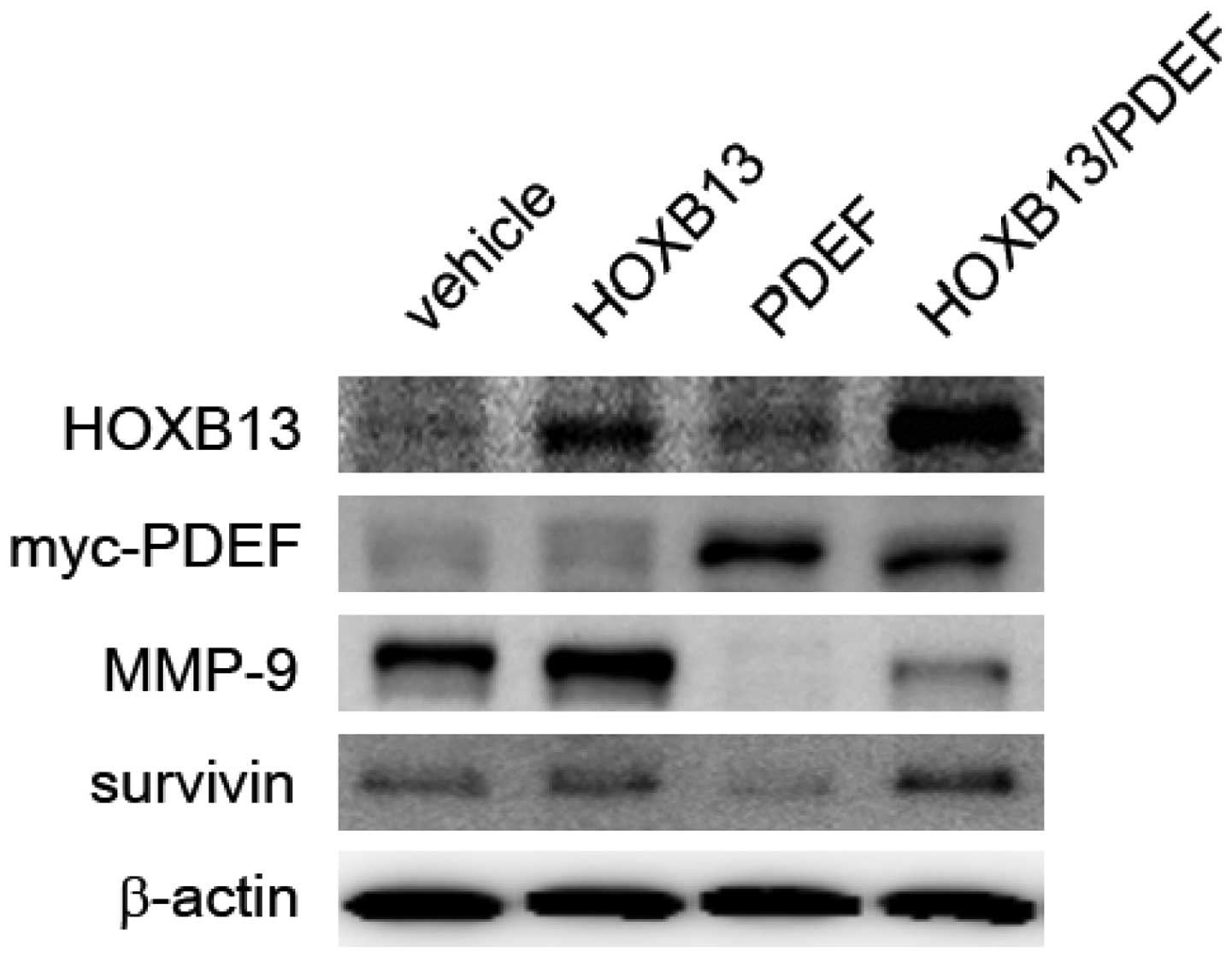
resistance to hormonal therapy in prostate cancer cells (21). As shown in Fig. 5, HOXB13 stimulated the expression
of both MMP-9 and surviving, and their expression was inhibited by
PDEF expression. The expression of HOXB13 partly mitigated the
PDEF-mediated inhibition of MMP-9 and survivin.
HOXB13 restores matrix metalloproteinase
activity suppressed by the PDEF
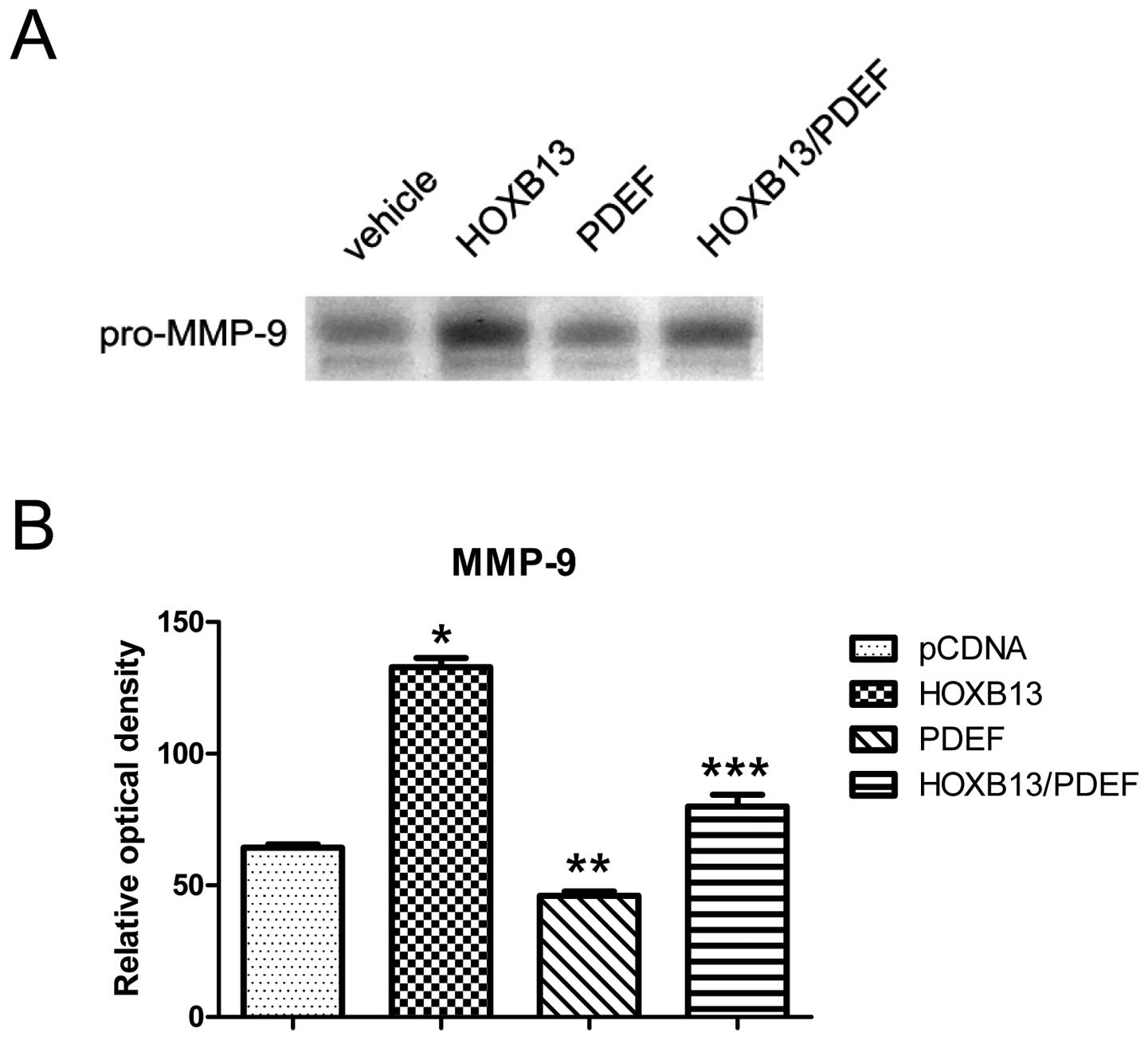
To determine whether the regulation of MMP-9 by
HOXB13-mediated PDEF suppression would show functional activity,
gelatin zymography was performed using conditioned media from each
genetically manipulated PC3 cell line. A major band migrating at
92-kDa molecular mass was observed, and this conceivably
corresponded to latent forms of MMP-9. Gelatin zymography exhibited
the gelatinolytic activity of MMP-9 in conditioned media (Fig. 6A). The conditioned medium from
untransfected PC3 cells was used as a control (lane 1; 64.33±1.28).
MMP-9 was activated by HOXB13 (lane 2; 132.87±3.46), but inhibited
by the PDEF (lane 3; 46.05±1.61). HOXB13 partly mitigated the
reduction in gelanolytic activity by the PDEF (lane 4;79.94±4.54).
A densitometric analysis of the gelatinolytic activity of MMP-9
revealed that HOXB13 significantly increased the PDEF-mediated
repression of MMP-9 activity (p<0.05) (Fig. 6B).
Discussion
The prostate-derived Ets factor (PDEF) is originally
isolated from normal prostate tissue, and its expression has been
demonstrated in other epithelial tissues, including the ovary,
breast, prostate and colon (15,22–24).
The PDEF has been widely investigated for its role in tumor
development and progression, mostly in the case of prostate or
breast cancer. However, this role has been controversial, either
being a positive tumor cell promoter or a tumor suppressor in
various systems under different conditions. Previous studies have
shown that the PDEF generally functions as a negative regulator of
cell growth, migration and invasion (17,19,22,24–26).
The PDEF facilitates breast cancer growth through the negative
regulation of anti-apoptotic survivin or p21 tumor suppressor
expression (19,24,27),
and the loss of the PDEF is associated with increased MMP-9 or SLUG
expression and activity in aggressive prostate cancer for the
promotion of metastatic potential (16,28).
This study is motivated by a research study
demonstrating the role of HOXB13 in promoting prostate cancer cell
invasion (10). A highly invasive
PC3 prostate cancer cell line was employed for the demonstration of
the role of HOXB13 in prostate cancer cell migration and invasion.
The forced expression of HOXB13 inversely regulated the expression
of the PDEF, resulting in the promotion of prostate cancer
migration and invasion. In addition, HOXB13 negatively regulated
the expression of the two most widely known PDEF target proteins,
namely MMP-9 and surviving, which are involved in cancer cell
metastasis and proliferation, respectively. Accordingly, the
expression of HOXB13 and the PDEF in PC3 cells led to partial
restoration of the expression of both PDEF target proteins and
invasive potential, which were inhibited by the PDEF. This implies
that the restoration of invasion by HOXB13 derived from the
suppression of the endogenous PDEF by HOXB13 overexpression. HOXB13
was also involved in cancer cell metastasis. Previous studies have
shown that HOXB13 facilitates the invasion of prostate, breast, and
endometrial cancer cells (3,4,29).
It has been demonstrated that HOXB13 functions as a positive growth
regulator of androgen-independent prostate cancer cell growth by
modulating the RB/E2F1 signaling pathway (9).
The PDEF is also involved in the progression of
prostate, breast and ovary cancer cells (18,22,27,30,31).
PDEF expression is widely observed in benign prostatic tissues and
is downregulated or lost in prostate carcinomas. The PDEF is
inversely correlated with Gleason scores, and patients with
PDEF-positive tumors show significantly longer survival (16,18,31).
In ovarian cancer, positive PDEF expression in patient tumor
lesions is associated with a favorable prognosis with longer
overall survival. The forced expression of the PDEF in
PDEF-negative ovarian tumor cells has been shown to inhibit tumor
cell growth, induce apoptosis, downregulate survivin expression and
reduce its promoter activity (30). The PDEF protein is also reduced in
human invasive breast cancer and is absent in invasive breast
cancer cell lines (22).
The results of this study demonstrate an inverse
correlation between HOXB13 and the PDEF by the mechanism underlying
the targeting of PDEF expression by HOXB13. In normal tissues, the
expression pattern of HOXB13 has been found to be similar to that
of the PDEF, which is exclusively expressed in the prostate and
colon (1). However, HOXB13
expression in the breast has been found to be lower than PDEF
expression (29). Both the PDEF
and HOXB13 are expressed mainly in the nucleus of prostatic
epithelial cells. PDEF expression appears to be inversely
correlated with prostate cancer malignancy, and the alteration of
HOXB13 expression during prostate cancer development is not clearly
observed, presumably because of the multifocal nature of prostate
cancer. Several studies have suggested that HOXB13 expression
changes during prostate cancer development and malignant
progression (5,9,32–34).
HOXB13 expression is correlated with the Gleason score (33) and is overexpressed in
castration-resistant prostate cancer cells relative to
androgen-dependent cancer cells (9). The alteration of HOXB13 expression
has also been observed in tumors from non-prostate cells. The
expression of HOXB13 is downregulated in colon, skin and kidney
tumors (35–39). On the other hand, the
overexpression of HOXB13 has been observed in breast, ovary and
uterine endometrium tumors (2–4).
These mixed results suggest that the role of HOXB13 in cancer cell
growth depends on the tissue and cellular status. For example, the
forced expression of HOXB13 negatively regulates the growth of
prostate cancer cells through the regulation of androgen
receptor-mediated signaling or β-catenin and TCF-mediated signaling
(6–8). In the context of an androgen-free
environment, HOXB13 functions as a growth stimulator, as in the
case of castration-resistant prostate cancer cells. The latter
function appears to be mediated through the regulation of multiple
signaling pathways, including RB/E2F (9) and zinc/NF-κB signals (unpublished
data).
In this study, the results demonstrate that HOXB13
promoted the invasive potential of PC3 prostate cancer cells
through the negative regulation of the PDEF and thus that HOXB13
counteracted the expression of PDEF target genes involved in the
invasion of prostate cancer cells, namely MMP-9. Because of the
lack of information on the PDEF promoter, it remains unclear how
HOXB13 suppresses PDEF expression, although this should broaden
knowledge of the HOXB13-mediated promotion of prostate cancer
malignancy. Given that very little information is available on the
regulation of PDEF expression and on the pattern of mutual HOXB13
and PDEF expression, the results of this study are expected to draw
more research attention to this field.
Acknowledgements
This study was supported by a grant from the
National Research Foundation (NRF) of Korea (MRC, 2011-0030132)
funded by the Korea government, by a Basic Science Research Program
grant (2010-0003838) from the NRF of Korea, and by a grant from the
Chonnam National University Hospital Research Institute of Clinical
Medicine (CRI 120061-31).
References
|
1
|
Sreenath T, Orosz A, Fujita K and
Bieberich CJ: Androgen-independent expression of hoxb-13 in the
mouse prostate. Prostate. 41:203–207. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma XJ, Wang Z, Ryan PD, et al: A two-gene
expression ratio predicts clinical outcome in breast cancer
patients treated with tamoxifen. Cancer Cell. 5:607–616. 2004.
View Article : Google Scholar
|
|
3
|
Miao J, Wang Z, Provencher H, et al:
HOXB13 promotes ovarian cancer progression. Proc Natl Acad Sci USA.
104:17093–17098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhao Y, Yamashita T and Ishikawa M:
Regulation of tumor invasion by HOXB13 gene overexpressed in human
endometrial cancer. Oncol Rep. 13:721–726. 2005.PubMed/NCBI
|
|
5
|
Edwards S, Campbell C, Flohr P, et al:
Expression analysis onto microarrays of randomly selected cDNA
clones highlights HOXB13 as a marker of human prostate cancer. Br J
Cancer. 92:376–381. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jung C, Kim RS, Lee SJ, Wang C and Jeng
MH: HOXB13 homeodomain protein suppresses the growth of prostate
cancer cells by the negative regulation of T-cell factor 4. Cancer
Res. 64:3046–3051. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jung C, Kim RS, Zhang HJ, Lee SJ and Jeng
MH: HOXB13 induces growth suppression of prostate cancer cells as a
repressor of hormone-activated androgen receptor signaling. Cancer
Res. 64:9185–9192. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kim SD, Park RY, Kim YR, et al: HOXB13 is
co-localized with androgen receptor to suppress androgen-stimulated
prostate-specific antigen expression. Anat Cell Biol. 43:284–293.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kim YR, Oh KJ, Park RY, et al: HOXB13
promotes androgen independent growth of LNCaP prostate cancer cells
by the activation of E2F signaling. Mol Cancer. 9:1242010.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim YR, Kim IJ, Kang TW, et al: HOXB13
downregulates intracellular zinc and increases NF-kappaB signaling
to promote prostate cancer metastasis. Oncogene. 7–Oct;2013.(Epub
ahead of print). View Article : Google Scholar : 2013.
|
|
11
|
Sharrocks AD, Brown AL, Ling Y and Yates
PR: The ETS-domain transcription factor family. Int J Biochem Cell
Biol. 29:1371–1387. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Clark JP and Cooper CS: ETS gene fusions
in prostate cancer. Nat Rev Urol. 6:429–439. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kumar-Sinha C, Tomlins SA and Chinnaiyan
AM: Recurrent gene fusions in prostate cancer. Nat Rev Cancer.
8:497–511. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tomlins SA, Bjartell A, Chinnaiyan AM, et
al: ETS gene fusions in prostate cancer: from discovery to daily
clinical practice. Eur Urol. 56:275–286. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Oettgen P, Finger E, Sun Z, et al: PDEF, a
novel prostate epithelium-specific ets transcription factor,
interacts with the androgen receptor and activates
prostate-specific antigen gene expression. J Biol Chem.
275:1216–1225. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Johnson TR, Koul S, Kumar B, et al: Loss
of PDEF, a prostate-derived Ets factor is associated with
aggressive phenotype of prostate cancer: regulation of MMP 9 by
PDEF. Mol Cancer. 9:1482010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gu X, Zerbini LF, Otu HH, et al: Reduced
PDEF expression increases invasion and expression of mesenchymal
genes in prostate cancer cells. Cancer Res. 67:4219–4226. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Turner DP, Findlay VJ, Moussa O, et al:
Mechanisms and functional consequences of PDEF protein expression
loss during prostate cancer progression. Prostate. 71:1723–1735.
2011. View Article : Google Scholar
|
|
19
|
Ghadersohi A, Pan D, Fayazi Z, Hicks DG,
Winston JS and Li F: Prostate-derived Ets transcription factor
(PDEF) downregulates survivin expression and inhibits breast cancer
cell growth in vitro and xenograft tumor formation in vivo. Breast
Cancer Res Treat. 102:19–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Lynch CC and Matrisian LM: Matrix
metalloproteinases in tumor-host cell communication.
Differentiation. 70:561–573. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang M, Latham DE, Delaney MA and
Chakravarti A: Survivin mediates resistance to antiandrogen therapy
in prostate cancer. Oncogene. 24:2474–2482. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Feldman RJ, Sementchenko VI, Gayed M,
Fraig MM and Watson DK: PDEF expression in human breast cancer is
correlated with invasive potential and altered gene expression.
Cancer Res. 63:4626–4631. 2003.PubMed/NCBI
|
|
23
|
Feldman RJ, Sementchenko VI and Watson DK:
The epithelial-specific Ets factors occupy a unique position in
defining epithelial proliferation, differentiation and
carcinogenesis. Anticancer Res. 23:2125–2131. 2003.
|
|
24
|
Moussa O, Turner DP, Feldman RJ, et al:
PDEF is a negative regulator of colon cancer cell growth and
migration. J Cell Biochem. 108:1389–1398. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Turner DP, Findlay VJ, Kirven AD, Moussa O
and Watson DK: Global gene expression analysis identifies PDEF
transcriptional networks regulating cell migration during cancer
progression. Mol Biol Cell. 19:3745–3757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Turner DP, Moussa O, Sauane M, Fisher PB
and Watson DK: Prostate-derived ETS factor is a mediator of
metastatic potential through the inhibition of migration and
invasion in breast cancer. Cancer Res. 67:1618–1625. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schaefer JS, Sabherwal Y, Shi HY, et al:
Transcriptional regulation of p21/CIP1 cell cycle inhibitor by PDEF
controls cell proliferation and mammary tumor progression. J Biol
Chem. 285:11258–11269. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Findlay VJ, Turner DP, Yordy JS, et al:
Prostate-derived ETS factor regulates epithelial-to-mesenchymal
transition through both SLUG-dependent and independent mechanisms.
Genes Cancer. 2:120–129. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ma XJ, Hilsenbeck SG, Wang W, et al: The
HOXB13:IL17BR expression index is a prognostic factor in
early-stage breast cancer. J Clin Oncol. 24:4611–4619. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghadersohi A, Odunsi K, Zhang S, et al:
Prostate-derived Ets transcription factor as a favorable prognostic
marker in ovarian cancer patients. Int J Cancer. 123:1376–1384.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ghadersohi A, Sharma S, Zhang S, et al:
Prostate-derived Ets transcription factor (PDEF) is a potential
prognostic marker in patients with prostate cancer. Prostate.
71:1178–1188. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Fromont G, Chene L, Latil A, et al:
Molecular profiling of benign prostatic hyperplasia using a large
scale real-time reverse transcriptase-polymerase chain reaction
approach. J Urol. 172:1382–1385. 2004. View Article : Google Scholar
|
|
33
|
Jeong TO, Oh KJ, Xuan Nguyen NT, et al:
Evaluation of HOXB13 as a molecular marker of recurrent prostate
cancer. Mol Med Rep. 5:901–904. 2012.PubMed/NCBI
|
|
34
|
Johnson PH, Walker RP, Jones SW, et al:
Multiplex gene expression analysis for high-throughput drug
discovery: screening and analysis of compounds affecting genes
overexpressed in cancer cells. Mol Cancer Ther. 1:1293–1304.
2002.PubMed/NCBI
|
|
35
|
Birkenkamp-Demtroder K, Olesen SH,
Sorensen FB, et al: Differential gene expression in colon cancer of
the caecum versus the sigmoid and rectosigmoid. Gut. 54:374–384.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Hoek K, Rimm DL, Williams KR, et al:
Expression profiling reveals novel pathways in the transformation
of melanocytes to melanomas. Cancer Res. 64:5270–5282. 2004.
View Article : Google Scholar
|
|
37
|
Jung C, Kim RS, Zhang H, et al: HOXB13 is
downregulated in colorectal cancer to confer TCF4-mediated
transactivation. Br J Cancer. 92:2233–2239. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Komuves LG, Ma XK, Stelnicki E, Rozenfeld
S, Oda Y and Largman C: HOXB13 homeodomain protein is cytoplasmic
throughout fetal skin development. Dev Dyn. 227:192–202. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Okuda H, Toyota M, Ishida W, et al:
Epigenetic inactivation of the candidate tumor suppressor gene
HOXB13 in human renal cell carcinoma. Oncogene. 25:1733–1742. 2006.
View Article : Google Scholar
|