Introduction
Colorectal carcinoma (CRC) is the third most
frequently diagnosed malignancy and the third leading cause of
death among cancer patients in the United States (1). Despite current therapeutic strategies
combining adjuvant chemotherapy, surgery and sometimes
radiotherapy, the prognosis of CRC remains poor, since most CRC
patients have distant metastases at diagnosis or develop recurrent
metastatic CRC following surgical treatment. Although recent
developments in molecular biology have provided insight into the
molecular mechanisms of CRC, the fundamental molecular mechanisms
underlying metastasis in CRC have not been fully elucidated.
Therefore, it is essential to identify metastasis-associated
molecules as effective drug targets and to enhance the
understanding of the mechanisms underlying the metastasis of
CRC.
MicroRNAs (miRNAs) are small non-coding RNAs (~22
nucleotides in length), transcribed from non-protein-coding genes
or introns, which regulate gene expression through repressing
translation and cleaving their mRNAs by binding to complementary
sites in their 3′-untranslated region (3′-UTR) (2). miRNAs regulated the expression of a
wide variety of target genes, and aberrant expression miRNAs cause
them to function as tumor suppressors or oncogenes according to the
role of their target genes (3,4).
Particularly, miRNAs can regulate various biological processes of
tumor cells, including cell proliferation, differentiation,
progression, apoptosis, proliferation, migration and invasion
(5–7). Furthermore, increasing numbers of
miRNAs have been observed in various types of cancer and may be
involved in modulating cancer cell behavior (8–10).
Several aberrantly expressed miRNAs have been proven to be
associated with tumorigenesis, tumor progression and metastasis in
CRC, and taken as prognostic indicators for CRC including miR-150
(11), miR-28-5p/-3p (12), miR-200 (13) and many others (14–16).
miRNA-494 (miR-494), located on chromosome 14q32.31,
has been reported to be highly expressed in retinoblastoma cells
(17) and in the anti-BPDE-induced
human bronchial epithelial cell line 16HBE (18), but the role of miR-494 in
carcinogenesis is still not fully understood. In tumor-expanded
myeloid-derived suppressor cells, it has been reported that miR-494
was able to regulate the expression phosphatase and tensin homolog
deleted on chromosome 10 (PTEN) post-transcriptionally and function
as a micro-oncogene in carcinogenesis (19). In glioma, miR-494 was also proved
to be a target to control invasiveness in cancer therapy (20). These studies suggest that miR-494
may play an important oncogenic role in tumorigenesis and
metastasis. But miR-494 was found downregulated and suppressed cell
proliferation in gastric cancer, A549 lung cancer cells
cholangiocarcinoma (21–23). It is still unclear whether miR-494
was an oncogenic miRNA or a tumor suppressor miRNA in CRC. Thus the
role of miR-494 in carcinogenesis and metastasis of CRC needed
further investigations.
The aim of our study was to investigate the effects
of miR-494 on cell migration and invasion and further discuss the
mechanism of action of miR-494 by identifying its potential target
gene in CRC. We found that miR-494 expression was upregulated in
the majority of CRC tissues, and upregulated miR-494 was
significantly associated with tumor recurrence, metastasis and poor
prognosis in CRC patients. Moreover, in vitro assays showed
that miR-494 could promote tumor migration and invasion of CRC cell
lines by targeting its direct target PTEN. To the best of our
knowledge, this is the first study to examine the expression and
role of miR-494 in prognosis and metastasis of CRC.
Materials and methods
Patients and tissue samples
This study was approved by the Research Ethics
Committee of Xi’an Jiaotong University. Written informed consent
was obtained from all the patients. The specimens were handled and
made anonymous according to the ethical and legal standards. A
total of 247 patients were enrolled in this study. Patients
received curative resection for CRC at the Second Affiliated
Hospital, Xi’an JiaoTong University (Shaanxi, China) between
2001–2008. None of the patients enrolled in this research received
blood transfusion or chemotherapy before surgery. The
clinicopathological information of the patients is shown in
Table I. The follow-up information
of all participants was updated every 3 months by telephone. The
overall survival was defined as the time elapsed from surgery to
death. Information regarding the death of patients was ascertained
from their family. Patients were followed after surgical treatment
until May 2013, with a median follow-up of 83 months (range, 8–139
months). During the follow-up period, 121 patients (49.0%) died of
the disease. The median overall and progression-free survival of
patients was 37 and 34 months, respectively.
 | Table IClinical correlation between miR-494
expression and other clinicopathological features in CRC. |
Table I
Clinical correlation between miR-494
expression and other clinicopathological features in CRC.
| Clinicopathological
features | Total no. of
patients | High miR-494
group | χ2 | P |
|---|
| Age (years) | 247
(57.4±10.7) | 129 | 0.612 | 0.287 |
| ≤60 | 117 | 59 | | |
| >60 | 130 | 70 | | |
| Sex | | | 0.002 | 0.962 |
| Men | 149 | 78 | | |
| Women | 98 | 51 | | |
| Tumor size | | | 2.468 | 0.116 |
| ≤5 cm | 171 | 95 | | |
| >5 cm | 76 | 34 | | |
| Location | | | 0.045 | 0.832 |
| Colon | 167 | 88 | | |
| Rectum | 80 | 41 | | |
| Depth of
invasion | | | 0.118 | 0.732 |
| T1–T2 | 79 | 40 | | |
| T3–T4 | 168 | 89 | | |
| Lymph node
status | | | 7.705 | 0.006a |
| Negative | 145 | 65 | | |
| Positive | 102 | 64 | | |
| Metastasis
status | | | 10.554 | 0.001a |
| Negative | 203 | 101 | | |
| Positive | 44 | 28 | | |
| TNM stage | | | 9.163 | 0.027a |
| I | 42 | 15 | | |
| II | 73 | 39 | | |
| III | 88 | 45 | | |
| IV | 44 | 30 | | |
|
Differentiation | | | 7.650 | 0.022a |
| Well | 119 | 53 | | |
| Moderately | 76 | 41 | | |
| Poorly | 52 | 35 | | |
Quantitative reverse transcriptase PCR
(RT-qPCR) assay
The expression of miR-494 in CRC and corresponding
adjacent tissues were detected by RT-qPCR assay. Briefly, total RNA
was extracted from tissues using TRIzol reagent (Invitrogen Life
Technologies, Carlsbad, CA, USA) according to the manufacturer’s
protocol. Then, miRNA expression levels were quantitated using
TaqMan microRNA real-time RT-PCR kit (Applied Biosystems, Foster
City, CA, USA) according to the manufacturer’s protocol. Data were
analyzed with 7500 Software v.2.0.1 (Applied Biosystems), with the
automatic Ct setting for adapting baseline and threshold for Ct
determination. The universal small nuclear RNA U6 (RNU6B) was used
as an endogenous control for miRNAs. Each sample was examined in
triplicate and the amounts of PCR products produced were
non-neoplasticized to RNU6B.
Cell culture
Human CRC cell lines SW620, SW480, HCT116 and normal
colon epithelium cell line CCD-18Co were obtained from the Cell
Bank of the Chinese Academy of Sciences (Shanghai, China), where
they were characterized by mycoplasma detection, DNA
fingerprinting, isozyme detection and cell vitality detection. They
were cultured in RPMI-1640 (Invitrogen Life Technologies) mediums
supplemented with 10% fetal bovine serum (FBS) (HyClone, Logan, UT,
USA) and cultured in a humidified incubator at 37°C in 5%
CO2.
Oligonucleotide transfection
miR-494 mimics and inhibitors were chemically
synthesized by Shanghai GenePharma (Shanghai, China). Once the
cells were 80% confluent, miR-494 mimics or inhibitor was
transfected into osteosarcoma cells with Lipofectamine 2000
(Invitrogen Life Technologies) according to the manufacturer’s
instructions. Cells were also transfected with scramble
oligonucleotide as negative control (NC). The expression level of
miR-494 in the transfected osteosarcoma cells were identified by
RT-qPCR.
In vitro migration and invasion
assays
Cell migration and invasion capacity were measured
using Transwell migration assays (Millipore, Billerica, MA, USA)
in vitro. The CRC SW620 cells were transfected with miR-494
inhibitors and NCs for 48 h, the CRC SW480 cells were transfected
with miR-494 mimics and NCs for 48 h, and then the cells were
suspended in RPMI-1640 with 10 g/l BSA at a density of
1×106 cells/ml. Then, cell suspensions (150 μl) were
seeded in the upper chamber with aporous membrane coated with (for
the Transwell invasion assay) or without (for the migration assay)
Matrigel (BD Biosciences, San Diego, CA, USA). To attract the
cells, 500 μl of RPMI-1640 with 10% serum was added to the bottom
chamber. After allowing the cells to migrate for 24 h or to invade
for 48 h, the penetrated cells on the filters were fixed in dried
methanol and stained in 4 g/l crystal violet. The numbers of
migrated or invasive cells were determined from five random fields
using a microscope (Olympus, Tokyo, Japan) at ×10
magnification.
Luciferase reporter assay
CRC cells were seeded in a 96-well plate at 60%
confluence. After 24 h, cells were transfected with 120 ng of
wild-type (WT) or mutant (MT) 3′-UTR of PTEN mRNA expression vector
pGL3-target-3′UTR and 30 ng of miR-494 mimics using Lipofectamine
2000. Cells were collected 48 h after transfection, and luciferase
activity was measured using a dual luciferase reporter assay system
according to the manufacturer’s protocol (Promega Corporation,
Madison, WI, USA).
Western blot analysis
Cells were harvested in lysis buffer (50 mM NaCl, 50
mM EDTA, 1% Triton X-100) containing protease inhibitor cocktail
(Roche Diagnostics, Indianapolis, IN, USA). The cell lysates (30
μg) were separated using 10% SDS-PAGE gels and then transferred
onto nitrocellulose membranes (Millipore, Bedford, MA, USA). The
membranes were blocked with 5% non-fat milk diluted in PBS for 2 h
at room temperature before the addition of the appropriate primary
antibody. The antibodies used in this study included anti-PTEN
(1:1,000, no. 9188; Cell Signaling Technology, Inc., Danvers, MA,
USA) and anti-β-actin (1:3,000, A2066; Sigma, St. Louis, MO, USA).
The membranes were then washed with PBS containing 0.05% Tween-20
and incubated with the appropriate HRP-conjugated secondary
antibody (1:5,000; Sigma) for 1 h at room temperature. The bands
were visualized using a chemiluminescence reagent (New England
Nuclear, Boston, MA, USA).
Statistical analysis
Statistical analysis was performed using IBM SPSS
statistical software (version 20.0). Survival curves were estimated
using the Kaplan-Meier method, and distributions were evaluated by
the log-rank test. Cox proportional hazard models of factors
related to survival were used to calculate RRs and identify the
factors that affect survival. The differences in characteristics
between the two groups were examined by the χ2 test and
Fisher’s exact test. All P-values were determined from two-sided
tests, and statistical significance was based on a P-value of
0.05.
Results
Upregulation of miR-494 is associated
with metastasis and recurrence of CRC
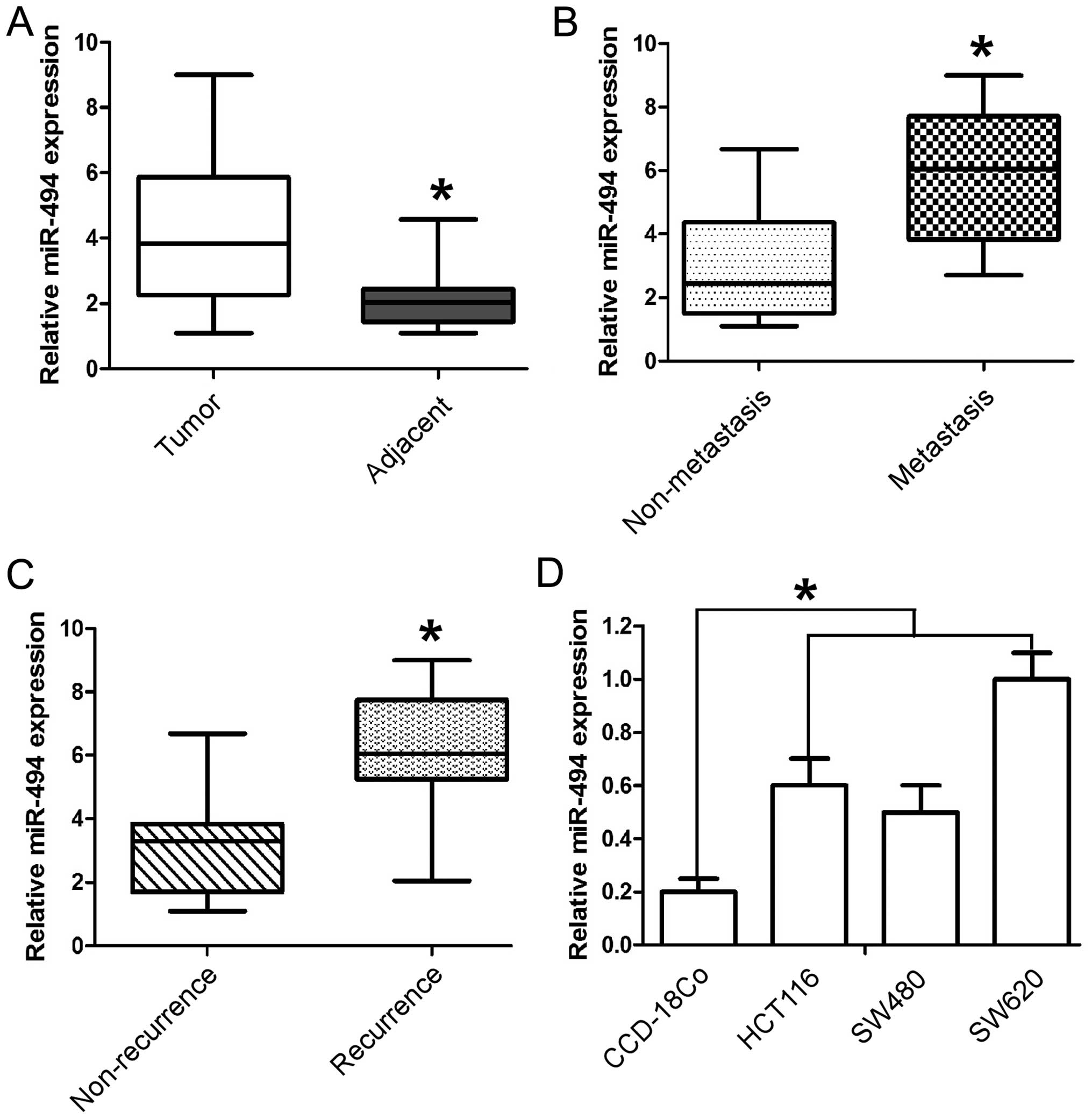
Expression of miR-494 was analyzed in 247 pairs of
CRC and adjacent tissues by RT-qPCR and normalized against an
endogenous control (U6 RNA). As shown in Fig. 1A the expression level of miR-494 in
the CRC tissues was found to be distinctly upregulated when
compared to that in adjacent tissues. Furthermore, in comparison to
the non-metastatic CRC tissues, the miR-494 levels were
significantly higher in metastatic CRC tissues (Fig. 1B). Moreover, miR-494 levels were
upregulated in the tumor tissues obtained from the patients who
suffered CRC recurrence (Fig. 1C).
Collectively, these data indicated that significant upregulation of
miR-494 expression occurred in CRC and was correlated with CRC
relapse and metastasis. To further evaluate the association of
miR-494 with CRC metastasis, we analyzed miR-494 levels in three
CRC cell lines (SW620, SW480, and HCT116) and normal colon
epithelium cell line CCD-18Co. Similarly, we found the expression
of miR-494 was much higher in all three CRC cell lines than that in
the normal colon cell line. It is noteworthy that miR-494 level was
higher in metastatic CRC cells (SW620) than its paired metastatic
cell line (SW480) (Fig. 1D). Taken
together, the above findings suggest that miR-494 levels correlate
with metastatic potential of CRC.
Upregulation of miR-494 is associated
with advanced clinicopathological features of CRC
To determine the clinical significance of miR-494 in
CRC, we analyzed the association of miR-494 expression with various
clinicopathological parameters of CRC tissues. The median miR-494
expression in all 247 patients with CRC was 3.84. The patients were
divided into two groups according to their expression levels of
miR-494, using the median level as a cutoff: the high miR-494
expression group (n=129) and the low miR-494 expression group
(n=118). As shown in Table I,
miR-494 was significantly upregulated in CRC patients with advanced
clinical stage (P=0.027), metastasis status (P=0.001), and positive
lymph node metastasis (P=0.006). We also found a trend of increased
miR-494 level from well to poor differentiation (P=0.022). However,
miR-494 expression was not significantly correlated with gender,
age, tumor size, tumor location and depth of invasion.
Upregulation of miR-494 is associated
with poor prognosis in patients with CRC
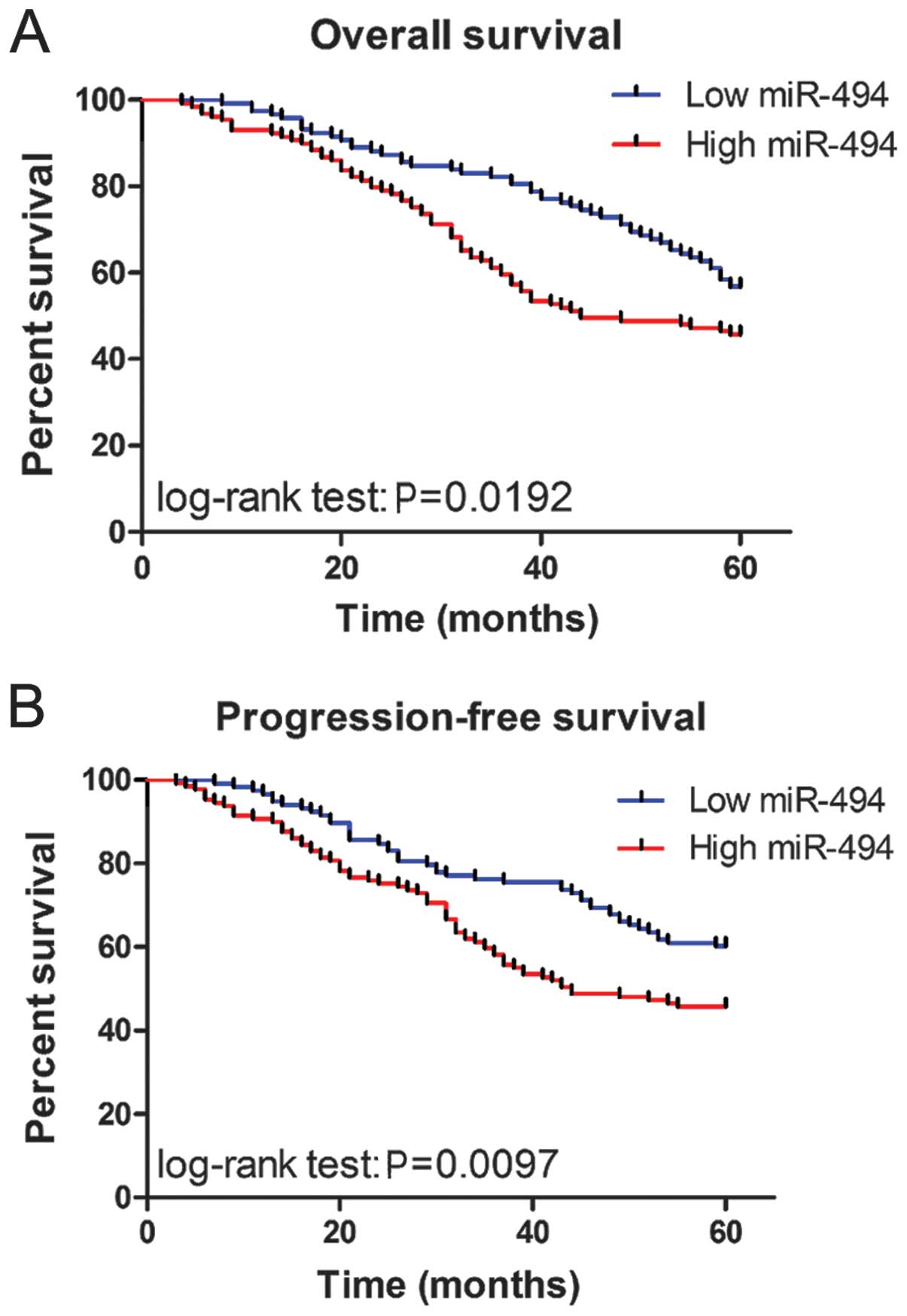
To determine the prognostic value of miR-494
expression in CRC, we analyzed the relationship between the miR-494
and clinical outcome. The relationship between miR-494 expression
overall survival or progression-free survival was investigated
using Kaplan-Meier analysis and log-rank test. Statistically
significant differences in overall survival and progression-free
survival were found between the high miR-494 level group and low
miR-494 level group (Fig. 2A and
B; log-rank test: P=0.0192 and P=0.0097, respectively). The
patients with high miR-494 expression tended to have shorter
overall and progression-free survival time when compared to
patients with low miR-494 expression. Univariate analysis
identified advanced clinical stage, metastasis status, lymph node
status, differentiation status and high expression of miR-494 as
indicators for poor prognosis for overall survival and
progression-free survival (all P<0.05), whereas age, gender and
tumor location were not significantly associated with overall
survival and progression-free survival (Tables II and III). To test whether the prognostic
value of high miR-494 expression was independent of other risk
factors for poor overall and progression-free survival, a
multivariate analysis was performed using a Cox proportional hazard
model. Multivariate analyses including age, gender, tumor location,
tumor stage, miR-494 expression, differentiation status, lymph node
status, depth of invasion and metastasis status demonstrated that
high miR-494 expression was an independent predictor for poor
overall and progression-free survival in CRC patients (HR=2.874,
CI=1.148–3.294, P=0.017 and HR=3.712, CI=1.513–5.272, P=0.009,
respectively). Statistically significant results were also obtained
for advanced tumor stage, and metastasis status, whereas all other
parameters were not significant for overall and progression-free
survival (Tables II and III).
 | Table IICox regression analysis of prognostic
factors for overall survival in colorectal cancer patients
(n=143). |
Table II
Cox regression analysis of prognostic
factors for overall survival in colorectal cancer patients
(n=143).
| Univariate | Multivariate |
|---|
|
|
|
|---|
| HR | 95% CI | P | HR | 95% CI | P |
|---|
| miR-494
expression | 3.617 | (1.208–4.576) | 0.008a | 2.874 | (1.148–3.294) | 0.017a |
| Age | 1.215 | (0.478–1.935) | 0.617 | 1.483 | (0.720–1.816) | 0.764 |
| Sex | 1.415 | (0.787–1.913) | 0.601 | 1.213 | (0.547–1.811) | 0.538 |
| Tumor location | 1.294 | (0.567–2.104) | 0.711 | 0.984 | (0.481–2.506) | 0.762 |
| TNM stage | 2.441 | (1.253–2.872) | 0.021a | 1.716 | (1.319–2.522) | 0.013a |
|
Differentiation | 2.952 | (1.224–2.228) | 0.041a | 3.291 | (0.826–3.773) | 0.077 |
| Tumor size | 1.761 | (0.769–1.927) | 0.319 | 1.629 | (0.813–2.109) | 0.427 |
| Lymph node
metastasis | 2.165 | (1.481–3.251) | 0.014a | 1.957 | (0.957–3.182) | 0.218 |
| Depth of
invasion | 1.291 | (0.521–2.114) | 0.542 | 1.417 | (0.619–2.217) | 0.611 |
| Metastasis
status | 2.761 | (1.274–3.189) | 0.032a | 2.465 | (1.198–2.981) | 0.021a |
 | Table IIICox regression analysis of prognostic
factors for progression-free survival in colorectal cancer patients
(n=143). |
Table III
Cox regression analysis of prognostic
factors for progression-free survival in colorectal cancer patients
(n=143).
| Univariate | Multivariate |
|---|
|
|
|
|---|
| HR | 95% CI | P | HR | 95% CI | P |
|---|
| miR-494
expression | 4.112 | (1.298–5.428) | 0.014a | 3.712 | (1.513–5.272) | 0.009a |
| Age | 1.461 | (0.563–1.892) | 0.702 | 1.287 | (0.641–1.674) | 0.815 |
| Sex | 1.599 | (0.659–2.114) | 0.705 | 1.761 | (0.847–2.832) | 0.642 |
| Tumor location | 1.379 | (0.661–1.907) | 0.743 | 1.211 | (0.575–1.803) | 0.619 |
| TNM stage | 3.115 | (1.418–4.861) | 0.034a | 2.724 | (1.275–3.554) | 0.019a |
|
Differentiation | 2.792 | (1.125–3.219) | 0.029a | 2.491 | (0.751–3.773) | 0.126 |
| Tumor size | 1.551 | (0.659–1.832) | 0.584 | 1.441 | (0.707–1.614) | 0.535 |
| Lymph node
metastasis | 3.144 | (1.524–4.268) | 0.027a | 2.163 | (0.897–3.264) | 0.144 |
| Depth of
invasion | 1.421 | (0.762–2.392) | 0.641 | 1.721 | (0.765–2.319) | 0.597 |
| Metastasis
status | 2.415 | (1.103–2.891) | 0.013a | 2.117 | (1.298–2.771) | 0.011a |
miR-494 promotes the migration and
invasion of CRC cells
As patients with recurrent metastatic CRC present
with poor outcome, and our previous results indicated that miR-494
was significantly associated with progression-free and overall
survival, we aimed to ascertain whether miR-494 affects the cell
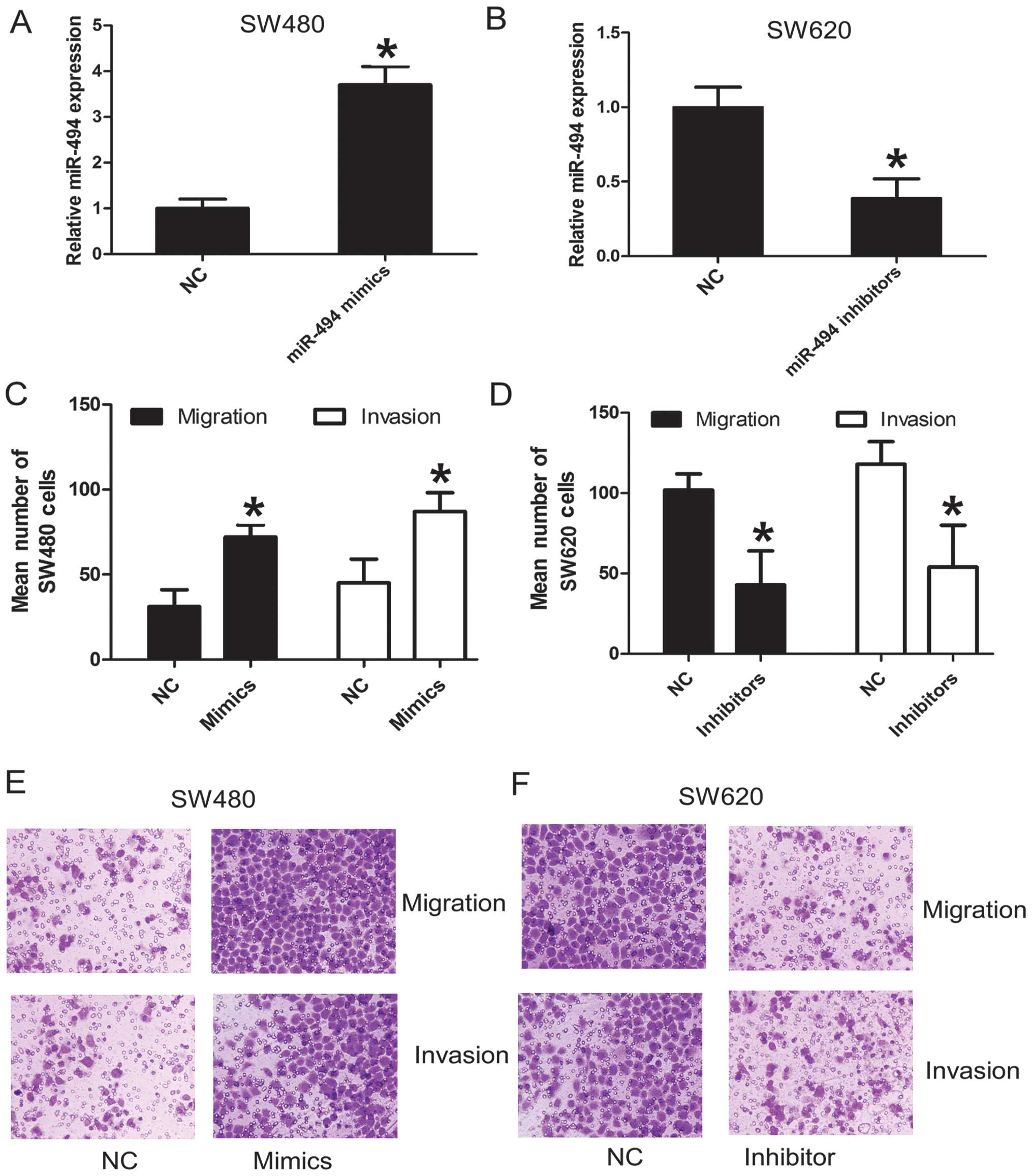
migration and invasion of CRC cells. The SW480 cell line was
derived from the primary adenocarcinoma and SW620 cell line was
derived from metastatic adenocarcinoma of the same patients
(24). Our previous results proved
that SW480 had lower expression of miR-494 than SW620. So they were
used to investigate the role of miR-494 on cell migration and
invasion. SW480 was transiently transfected with miR-494 mimics,
whereas SW620 was transiently transfected with miR-494 inhibitors.
As expected, transfection of miR-494 mimics or miR-494 inhibitors
resulted in an increased or decreased miR-494 expression in SW480
or SW620, respectively (Fig. 3A and
B). Moreover, the migration and invasion assays demonstrated
that miR-494 overexpression significantly increased the migration
and invasion ability of SW480 (Fig. 3C
and E). Conversely, knockdown of miR-494 in SW620 decreased the
migration and invasion ability of SW620 (Fig. 3D and F). These results indicate
that miR-494 functions as oncogenic miRNA and contributes to the
progression and metastasis of CRC cells.
PTEN is a direct target of miR-494 in CRC
cells and inversely correlates with miR-494 in CRC tissues
To characterize the mechanism by which miR-494
promotes CRC metastasis, we searched for potential target genes of
miR-494 using three publicly available databases, TargetScan,
PicTar and miRanda. We were particularly interested in tumor
suppressor gene PTEN, because PTEN was a crucial factor in
various central processes of cancer development and loss of PTEN
was proved to be associated with advanced CRC, liver metastasis,
and poor survival (25,26). It has been proven that PTEN was the
direct target of miR-494 in other cells (19,27).
Considering the tissue-specific and developmental stage-specific
manner of miRNAs, we investigate the relationship of PTEN and
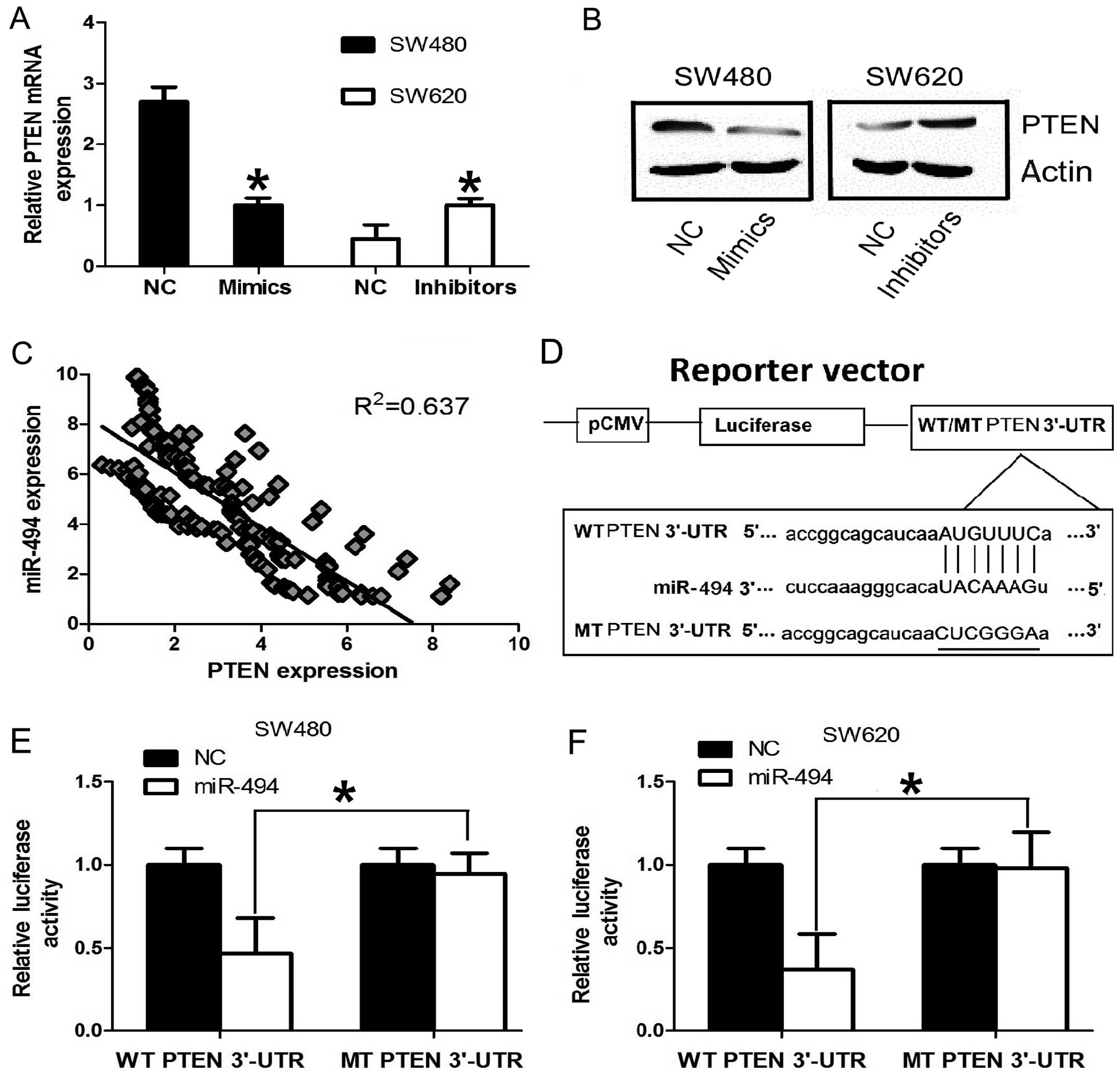
miR-494 in CRC cell lines. In order to confirm PTEN is a
target gene for miR-494 in CRC cells, RT-qPCR was used to detect
the expression of PTEN in CRC cell line SW480 and SW620. The
expression of PTEN at mRNA and protein level was significantly
downregulated after overexpression of miR-494 in SW480 cell line.
Conversely, the expression of PTEN was significantly upregulated
after knockdown miR-494 expression in SW620 cell line (Fig. 4A and B). Furthermore, we assessed
the significance of the miR-494 and PTEN correlation in CRC
tissues. We determined the PTEN mRNA and miR-494 expression in the
same CRC specimens by RT-qPCR. As shown in Fig. 4C, a statistically significant
inverse correlation was revealed by Spearman’s correlation analysis
between mRNA levels of miR-494 and PTEN (r=−0.798; P<0.001).
Taken together, our results suggest that miR-494 negatively
regulates the expression of its potential target gene PTEN
in CRC cell lines and tissues.
We further performed luciferase reporter assay to
verify whether miR-101 directly targeted the 3′-UTR of PTEN in CRC
cells. The target sequence of EZH2 3′-UTR (WT 3′-UTR) or the MT
sequence (MT 3′-UTR) was cloned into a luciferase reporter vector
(Fig. 4D). SW480 and SW620 cells
were then transfected with WT or MT 3′-UTR vector and the miR-494
mimic. As shown in Fig. 4E, a
significant decrease in luciferase activity was noted between the
PTEN WT 3′-UTR group and the NC group in SW480 cells (P<0.05).
The repressive effect was abrogated by point mutation in the core
binding sites of the EZH2 3′-UTR. A similar trend was also found in
the SW620 cells (Fig. 4F). These
results indicate that miR-494 exerts an inhibitory effect on PTEN
expression via interaction with the 3′-UTR of PTEN in CRC
cells.
Discussion
Several miRNAs have been identified as candidate
components of oncogene and tumor suppressor networks in CRC, and
these miRNAs and their targets play critical roles in
carcinogenesis and are important for finding out novel therapeutic
targets. Previous reports showed that miR-494 was downregulated and
functioned as a tumor suppressor in human cholangiocarcinoma
(21), gastric cancer (23) and lung cancer (28). However, other studies demonstrated
that upregulation of miR-494 was associated with several types of
human malignant solid tumors, including hepatocellular carcinoma
(29), non-small cell lung cancer
(30), and carcinoma induced by
anti-BPDE (27). In these types of
cancer, miR-494 seemed to be an oncogene, and promoted tumor cell
proliferation, cell cycle, cell migration and invasion via
regulation of the target genes PTEN, BIM and MCC. However, the
function and clinical relevance of miR-494 in CRC have not yet been
studied. In the current study, our results showed that miR-494 was
upregulated in CRC tissues compared to adjacent tissues, and
miR-494 was also upregulated in the tissues of CRC patients with
lymph node metastasis compared with those without lymph node
metastasis. Accordingly, we confirmed that miR-494 was commonly
overexpressed in CRC cell lines compared with the normal colon
epithelium cell line. These findings suggest that miR-494 may have
vital roles in CRC progression.
We then investigated the clinical relevance of
miR-494 in CRC. It was found that the increased expression of
miR-494 in CRC was significantly associated with the adverse
clinicopathological and histopathological features. Moreover, we
demonstrated that the miR-494 expression level was also predictive
of disease progression and cancer-specific death. Our results
showed that the patients with high miR-494 level are usually at a
significantly higher risk of cancer progression, cancer-specific
death and shorter progression-free and overall survival time.
Multivariate analysis demonstrated that high expression of miR-494
was a statistically significant risk factor affecting both
progression-free and overall survival in CRC patients, which
indicated that miR-494 expression could be an independent predictor
of CRC progression and prognosis. The above results provided the
first evidence supporting that miR-494 was a predictor of poor
prognosis in CRC.
To reveal the role of miR-494 in CRC cells, we
tested the effect of miR-494 on cell migration and invasion. Our
results showed that miR-494 could promote cell migration and
invasion. These results indicate that miR-494 might be a novel
oncogenic miRNA that play important roles in the regulation of
tumor metastasis of CRC. To address the molecular mechanisms
involved in miR-494-mediated changes of biological properties, the
PTEN was selected for further study because it was predicted to be
a target of miR-494 by bioinformatics analysis. We found the
following evidence supporting that miR-494 may be involved the
modulation of PTEN expression. Both the mRNA and protein levels of
PTEN were significantly downregulated after overexpression of
miR-494 in SW480 cell lines. Conversely, the expression of PTEN was
significantly upregulated after knockdown of miR-494 expression in
SW620 cell lines. Furthermore, the mRNA levels of miR-494 inversely
correlated with PTEN levels in CRC tissues. Moreover,
overexpression of miR-494 decreased the luciferase reporter
activity of WT 3′-UTR but not MT 3′-UTR of PTEN. These data confirm
that PTEN is a downstream mediator of miR-494 function in CRC.
The PTEN gene, known also as mutated in
multiple advanced cancer 1 (MMAC1), is a tumor suppressor
gene located at chromosome 10q23.31 (31). The PTEN protein is principally
involved in homeostatic maintenance of PI3K/Akt signaling
originating from EGFR activation. PTEN/PI3K/Akt is highly involved
in carcinogenesis and associated with EMT, cell cycle arrest
(32–34). It has been also confirmed that
PTEN, which counteracts PI3K/Akt activity, is involved in
inhibition of cell proliferation and invasion (35). Thus, loss of PTEN function has an
important impact on multiple aspects of cancer development such as
cell proliferation, apoptosis resistance, angiogenesis, metabolism
regulation, genomic instability, stem cell self-renewal, cell
migration and invasion (36–39).
It was reported that loss of PTEN expression was associated with
liver metastasis, EMT, and enhanced migration and invasion of CRC
(25,26). Our results showed that miR-494
promoted tumor migration and invasion, and miR-494 upregulation was
correlated with PTEN downregulation in CRC. These results indicate
that upregulation of miR-94 in CRC may enhance cell migration and
invasion, at least partially through the downregulation of PTEN
expression.
The role of miR-494 in tumor development proves to
be tissue dependent, causing increased proliferation in H460 lung
cancer cells, hepatocellular carcinoma, breast and transformed
bronchial epithelial cells (19,27,29,30).
However, it induces cell cycle arrest in lung cancer and
cholangiocarcinoma (22,28). Our results demonstrated that
miR-494 increases cell migration and invasion in CRC, which
indicated an oncogenic role of miR-494 in CRC. Moreover, our study
supported the role of PTEN which was the direct target of oncogenic
miR-494 as the tumor suppressor in CRC. In the tissues of CRC
patients, we further confirmed the oncogenic role of miR-494 in CRC
and its inverse relationship with PTEN. These results shed new
light on the role of miR-494 in different types of tumors,
especially in CRC.
Collectively, we showed that miR-494 was upregulated
and its target PTEN was downregulated in CRC tissues. Moreover,
upregulated miR-49 was significantly associated with adverse
clinicopathological and histopathological features of CRC. We found
ectopic expression of miR-494 could significantly promote cell
migration and invasion in CRC cells through directly targeting
PTEN. These results suggested that miR-494 played a role in
inhibiting the development and progression of CRC by targeting PTEN
and may potentially lead to a novel strategy for treatment of
CRC.
Acknowledgements
The authors thank the local doctors and the patients
who participated in our study.
Abbreviations:
|
miR-494
|
miRNA-494
|
|
PTEN
|
phosphatase and tensin homolog deleted
on chromosome 10
|
|
WT
|
wild-type
|
|
MT
|
mutant
|
|
3′-UTR
|
3′-untranslated region
|
|
RT-qPCR
|
quantitative real-time polymerase
chain reaction
|
References
|
1
|
Siegel R, Ma J, Zou Z and Jemal A: Cancer
statistics, 2014. CA Cancer J Clin. 64:9–29. 2014. View Article : Google Scholar
|
|
2
|
van den Berg A, Mols J and Han J:
RISC-target interaction: cleavage and translational suppression.
Biochim Biophys Acta. 1779:668–677. 2008.PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kumar MS, Lu J, Mercer KL, Golub TR and
Jacks T: Impaired microRNA processing enhances cellular
transformation and tumorigenesis. Nat Genet. 39:673–677. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Croce CM and Calin GA: miRNAs, cancer, and
stem cell division. Cell. 122:6–7. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gregory RI and Shiekhattar R: MicroRNA
biogenesis and cancer. Cancer Res. 65:3509–3512. 2005. View Article : Google Scholar
|
|
8
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Volinia S, Calin GA, Liu CG, et al: A
microRNA expression signature of human solid tumors defines cancer
gene targets. Proc Natl Acad Sci USA. 103:2257–2261. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lujambio A, Calin GA, Villanueva A, et al:
A microRNA DNA methylation signature for human cancer metastasis.
Proc Natl Acad Sci USA. 105:13556–13561. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ma Y, Zhang P, Wang F, et al: miR-150 as a
potential biomarker associated with prognosis and therapeutic
outcome in colorectal cancer. Gut. 61:1447–1453. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Almeida MI, Nicoloso MS, Zeng L, et al:
Strand-specific miR-28-5p and miR-28-3p have distinct effects in
colorectal cancer cells. Gastroenterology. 142:886–896. e92012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Pichler M, Ress AL, Winter E, et al:
MiR-200a regulates epithelial to mesenchymal transition-related
gene expression and determines prognosis in colorectal cancer
patients. Br J Cancer. 110:1614–1621. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hwang WL, Jiang JK, Yang SH, et al:
MicroRNA-146a directs the symmetric division of Snail-dominant
colorectal cancer stem cells. Nat Cell Biol. 16:268–280. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Yang X, Zeng Z, Hou Y, et al: MicroRNA-92a
as a potential biomarker in diagnosis of colorectal cancer: a
systematic review and meta-analysis. PLoS One. 9:e887452014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu Y, Zhou Y, Feng X, et al: Low
expression of microRNA-126 is associated with poor prognosis in
colorectal cancer. Genes Chromosomes Cancer. 53:358–365. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao JJ, Yang J, Lin J, et al:
Identification of miRNAs associated with tumorigenesis of
retinoblastoma by miRNA microarray analysis. Childs Nerv Syst.
25:13–20. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen YL, Jiang YG, Greenlee AR, Zhou LL
and Liu LH: MicroRNA expression profiles and miR-10a target in
anti-benzo[a] pyrene-7, 8-diol-9, 10-epoxide-transformed human
16HBE cells. Biomed Environ Sci. 22:14–21. 2009.PubMed/NCBI
|
|
19
|
Liu Y, Lai L, Chen Q, et al: MicroRNA-494
is required for the accumulation and functions of tumor-expanded
myeloid-derived suppressor cells via targeting of PTEN. J Immunol.
188:5500–5510. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kwak SY, Yang JS, Kim BY, Bae IH and Han
YH: Ionizing radiation-inducible miR-494 promotes glioma cell
invasion through EGFR stabilization by targeting p190B rhoGAP.
Biochim Biophys Acta. 1843:508–516. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yamanaka S, Campbell NR, An F, et al:
Coordinated effects of microRNA-494 induce G2/M arrest
in human cholangiocarcinoma. Cell Cycle. 11:2729–2738. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Olaru AV, Ghiaur G, Yamanaka S, et al:
MicroRNA downregulated in human cholangiocarcinoma control cell
cycle through multiple targets involved in the G1/S checkpoint.
Hepatology. 54:2089–2098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li X, Luo F, Li Q, et al: Identification
of new aberrantly expressed miRNAs in intestinal-type gastric
cancer and its clinical significance. Oncol Rep. 26:1431–1439.
2011.PubMed/NCBI
|
|
24
|
Leibovitz A, Stinson JC, McCombs WB 3rd,
McCoy CE, Mazur KC and Mabry ND: Classification of human colorectal
adenocarcinoma cell lines. Cancer Res. 36:4562–4569.
1976.PubMed/NCBI
|
|
25
|
Sawai H, Yasuda A, Ochi N, et al: Loss of
PTEN expression is associated with colorectal cancer liver
metastasis and poor patient survival. BMC Gastroenterol. 8:562008.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bowen KA, Doan HQ, Zhou BP, et al: PTEN
loss induces epithelial-mesenchymal transition in human colon
cancer cells. Anticancer Res. 29:4439–4449. 2009.PubMed/NCBI
|
|
27
|
Liu L, Jiang Y, Zhang H, Greenlee AR and
Han Z: Overexpressed miR-494 down-regulates PTEN gene expression in
cells transformed by anti-benzo(a)pyrene-trans-7,8-dihydrodiol-
9,10-epoxide. Life Sci. 86:192–198. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ohdaira H, Sekiguchi M, Miyata K and
Yoshida K: Micro-RNA-494 suppresses cell proliferation and induces
senescence in A549 lung cancer cells. Cell Prolif. 45:32–38. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lim L, Balakrishnan A, Huskey N, et al:
MicroRNA-494 within an oncogenic microRNA megacluster regulates
G1/S transition in liver tumorigenesis through suppression of
mutated in colorectal cancer. Hepatology. 59:202–215. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Romano G, Acunzo M, Garofalo M, et al:
MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced
apoptosis in non-small-cell lung cancer through BIM
down-regulation. Proc Natl Acad Sci USA. 109:16570–16575. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Molinari F and Frattini M: Functions and
regulation of the PTEN gene in colorectal cancer. Front Oncol.
3:3262014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Sansal I and Sellers WR: The biology and
clinical relevance of the PTEN tumor suppressor pathway. J Clin
Oncol. 22:2954–2963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Castellino RC and Durden DL: Mechanisms of
disease: the PI3K-Akt-PTEN signaling node - an intercept point for
the control of angiogenesis in brain tumors. Nat Clin Pract Neurol.
3:682–693. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Grille SJ, Bellacosa A, Upson J, et al:
The protein kinase Akt induces epithelial mesenchymal transition
and promotes enhanced motility and invasiveness of squamous cell
carcinoma lines. Cancer Res. 63:2172–2178. 2003.
|
|
35
|
Vogt PK, Gymnopoulos M and Hart JR: PI
3-kinase and cancer: changing accents. Curr Opin Genet Dev.
19:12–17. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhang S and Yu D: PI(3)king apart PTEN’s
role in cancer. Clin Cancer Res. 16:4325–4330. 2010.PubMed/NCBI
|
|
37
|
Song MS, Salmena L and Pandolfi PP: The
functions and regulation of the PTEN tumour suppressor. Nat Rev Mol
Cell Biol. 13:283–296. 2012.PubMed/NCBI
|
|
38
|
Stewart AL, Mhashilkar AM, Yang XH, et al:
PI3 kinase blockade by Ad-PTEN inhibits invasion and induces
apoptosis in RGP and metastatic melanoma cells. Mol Med. 8:451–461.
2002.PubMed/NCBI
|
|
39
|
Tamura M, Gu J, Matsumoto K, Aota S,
Parsons R and Yamada KM: Inhibition of cell migration, spreading,
and focal adhesions by tumor suppressor PTEN. Science.
280:1614–1617. 1998. View Article : Google Scholar : PubMed/NCBI
|