Introduction
Cigarette smoking is a well-known risk factor for
the development of certain respiratory diseases, including chronic
obstructive pulmonary diseases (COPD), interstitial pulmonary
fibrosis and lung cancer (1).
Approximately 20–25% of heavy smokers develop clinically
significant COPD while 12–17% of smokers develop lung cancer. Among
these smoking-related pulmonary diseases, lung cancer is the
leading cause of cancer death in Japan and also worldwide (2). The odds ratios of lung cancer for
Japanese smokers are currently 4.5 for men and 4.2 for females,
relative to hospital controls (3).
The most common histological types of smoking-related lung cancer
are lung squamous cell cancer (SCC) and small cell carcinoma. Of
these, the pathogenesis of SCC is the most complex and incompletely
understood. In general, targeted molecular agents developed for
lung adenocarcinoma are largely ineffective against lung SCC and
the current treatment for advanced SCC patients is still cytotoxic
chemotherapy. Unfortunately, modern cytotoxic and targeted
molecular agents, such as pemetrexed and bevacizumab, are not
recommended for patients with SCC (4,5). In
addition to smoking being a well-known risk factor for lung cancer,
previous findings strongly suggest that COPD is also an important,
independent risk factor. For instance, the risk of a lung cancer
diagnosis within six months of a COPD diagnosis in patients was
reported to be 11-fold greater than in patients without COPD
(6). Low-dose helical CT
screenings possess the potential to detect early-stage lung cancer
and have demonstrated 20% lower lung cancer mortalities compared to
chest X-ray screenings (7).
However, it is still difficult to detect lung cancer in a high-risk
population using solely radiographic examinations. Therefore, the
identification of early detection biomarkers for SCC in COPD or
heavy smokers is urgently required as this could have a markedly
beneficial and clinically significant impact on patient
survival.
A number of potential biomarkers for SCC have
already been identified; however, sensitive biomarkers are
presently unavailable to detect early SCC due to their low
sensitivity and specificity. Serum SCC antigen and CYFRA, a
cytokeratin fragment, are tumor markers widely used for the
evaluation of the therapeutic effects of lung cancer treatments,
and in the detection of SCC recurrence, but are not considered
applicable to the early detection of SCC (8,9). In
addition, no single biomarker, with high enough sensitivity and
specificity for the detection of SCC in COPD or heavy smokers, is
likely to be discovered.
In the present study, we compared the serum
proteomic profiles from SCC cases, COPD patients and healthy
smokers to identify any serum biomarkers associated with
smoking-related pulmonary disease, including SCC and COPD. Serum is
a highly complex bodily fluid that contains proteins ranging in
concentrations over at nine orders of magnitude (10). Since high abundance proteins can
interfere in the separation and detection of low abundance
proteins, an immunoaffinity column was used to deplete highly
abundant plasma proteins, and the resulting >2000 protein spots
were separated by two-dimensional difference gel electrophoresis
(2D-DIGE) and mass-spectrometry analysis (MS) (11). After isobaric tagging for relative
and absolute quantification (iTRAQ)-2DLC-MS/MS experiments
(12), we identified candidate
proteins as serum biomarkers of SCC by 2D-DIGE and western blot
analysis. Using enzyme-linked immunosorbent assay (ELISA), we
identified a cancer-associated serum peptide of haptogloblin (HP),
haptoglobin 216 (HP 216), as a novel biomarker for SCC.
Materials and methods
Serum samples
The present study was approved by the institutional
review boards of the Nippon Medical School Hospital and Nippon
Medical School Respiratory Care Clinic. All patients provided
informed consent. Serum samples for 2D-DIGE and MS analysis were
obtained from 11 SCC patients at Nippon Medical School Hospital,
and 7 COPD patients and 7 healthy smokers at Nippon Medical School
Respiratory Care Clinic, all of whom were <60 years of age. The
25 cases were all male patients or healthy volunteers, with a
smoking history of 15 or more pack-years. The first set of serum
samples collected consisted of 12 samples and the second set
consisted of 13 samples. We also used an additional 23 serum
samples, for ELISA, collected from five SCC patients at Nippon
Medical School Hospital, 10 COPD patients and eight healthy smokers
at Nippon Medical School Respiratory Care Clinic, all of whom were
<70 years of age. After blood sampling, each serum was
immediately separated by centrifugation at 1,500 rpm for 15 min at
room temperature. Aliquots of sera were taken and stored at −80ºC
until use.
Removal of high-abundance proteins from
serum samples and 2D-DIGE
Serum samples were processed using an affinity
column (Agilent spin concentrators; Agilent Technologies, Santa
Clara, CA, USA), which was expected to selectively remove abundant
proteins, including albumin, IgG, IgA, transferrin and HP, from
serum samples, according to the manufacturer's instructions.
Protein samples were then labeled with cysteine reactive saturation
dye (GE Healthcare, Buckinghamshire, UK). The labeling reaction was
performed according to the manufacturer's instructions as
previously described (13). An
internal serum control mixture was made by mixing equal portions of
three or four healthy control samples. The internal control sample
was labeled with Cy3 and patient samples were labeled with Cy5 and
2D-DIGE was performed as previously described (13–16).
Each labeled protein sample was loaded onto triplicate IPG DryStrip
gels (24 cm length; pI values ranging from 4 to 7; GE Healthcare).
The equilibrated IPG gels were applied onto 12% polyacrylamide gels
(GE Healthcare), and proteins were then separated using an Ettan
DALT-six electrophoresis system (GE Healthcare). All gel images
were scanned on a Typhoon 9400 Imager (GE Healthcare). Statistics
and the quantitation of protein expression were carried out using
DeCyder 2-D differential analysis software (GE Healthcare). A
paired t-test derived P-value of <0.05 was considered
statistically significant.
Mass spectrometry
To identify proteins corresponding to spots on 2D
gels, mass spectrometry was performed. An automated spot recovery
robot, the Ettan Spot Picker (GE Healthcare), transferred protein
spots from each gel to a 96-well plate. In-gel digestion was then
performed as previously described and tryptic peptides were
subjected to mass spectrometric analysis (13–16).
Peptide map fingerprinting (PMF) analysis was performed with
ESI-LC/Q-TOF-MS (Waters-Micromass) (LabX Media Group Inc., Midland,
ON, Canada). Mass spectra were processed using Analyst QS software
and a Mascot program. We performed a second Swiss-Prot database
search.
Western blot analysis
Protein (10 μg) was separated by gel electrophoresis
on 10% gels, transferred to nitrocellulose membranes and detected
by immunoblotting using specific antibodies and a chemiluminescence
system (GE Healthcare). Antibodies detecting HP, apolipoprotein
(APOA4), α-1-antichymotrypsin, (AACT) and fibronection were
purchased from Abcam (Cambridge, UK).
iTRAQ-2DLC-MS/MS
Protein identification and quantification for
iTRAQ-2DLC-MS/MS experiments was carried out using ProteinPilot 2.0
software according to the manufacturer's protocols (Filgen, Inc.,
Nagoya, Japan) (12). Proteins
were labeled with iTRAQ tags as follows: SCC samples with a 116
isobaric tag; and healthy smoker samples with a 114 isobaric tag. A
protein sequence search was performed using a Swiss-Prot human
database. At least two peptides with 95% confidence were considered
for protein identification and results were then exported into
Excel for manual data interpretation. Protein quantitation for
iTRAQ were carried out using ProteinPilot 2.0 software. One-sample
t-test derived P-value of <0.05 was considered statistically
significant and shown in Table
II.
 | Table IIProtein expression levels, after
iTRAQ analysis, in SCC and COPD cases. |
Table II
Protein expression levels, after
iTRAQ analysis, in SCC and COPD cases.
| Accession no. | Protein name | Peptides (95%) | 116:114 | P-value | EF |
|---|
| IPI00641737.2 | Haptoglobin | 187 | 10.8 | <0.001 | 1.25 |
| IPI00553177.1 | Isoform 1 of
α-1-antitrypsin | 15 | 5.4 | <0.001 | 1.45 |
| IPI00022429.3 | α-1-acid
glycoprotein 1 | 164 | 3.2 | <0.001 | 1.56 |
| IPI00027462.1 | Protein
S100-A9 | 6 | 3.1 | 0.002 | 1.50 |
| IPI00022417.4 | Leucine-rich
α-2-glycoprotein | 56 | 2.9 | <0.001 | 1.20 |
| IPI00876950.1 | Isoform 2 of
inter-α-trypsin inhibitor heavy chain H3 | 51 | 2.9 | <0.001 | 1.18 |
| IPI00478003.3 |
α-2-macroglobulin | 825 | 2.7 | <0.001 | 1.14 |
| IPI01009486.1 | Uncharacterized
protein | 11 | 2.5 | <0.001 | 1.33 |
| IPI00022395.1 | Complement
component C9 | 34 | 2.4 | <0.001 | 1.32 |
| IPI00896380.1 | Isoform 2 of Ig mu
chain C region | 14 | 2.3 | <0.001 | 1.15 |
| IPI01025667.1 |
α-1-antitrypsin | 141 | 2.3 | <0.001 | 1.24 |
| IPI00296608.6 | Complement
component C7 | 34 | 2.0 | <0.001 | 1.32 |
| IPI00064667.5 | β-Ala-His
dipeptidase | 20 | 0.5 | <0.001 | 1.16 |
| IPI00339228.2 | Fibronectin | 126 | 0.4 | <0.001 | 1.23 |
| IPI00657670.1 | Apolipoprotein
C-III variant 1 | 40 | 0.4 | 0.006 | 1.59 |
| IPI00216065.3 | Isoform 2 of
Vitamin K-dependent protein Z | 2 | 0.4 | 0.004 | 1.45 |
Establishment of polyclonal antibodies
for HP peptides and a HP ELISA
Customized, rabbit anti-HP peptide antibodies
against two peptides were raised (PharmaBio, Nagoya, Japan). Serum
samples were eluted from the affinity column, which was expected to
selectively remove HP from serum samples (Agilent spin
concentrators; Agilent Technologies). Duplicates of 28 sera samples
from SCC and COPD patients, and smokers, as well as positive and
negative haptoglobin peptide controls, were coated overnight onto a
96-well plate at 4ºC. Each well was washed twice in PBS and blocked
with 3% BSA/PBS for 1 h at 37ºC. After washing twice, rabbit
anti-HP peptide antibodies (1:1,000 dilution) were added to wells
and incubated for 2 h at 37ºC. Subsequently, FITC-labeled goat
anti-rabbit IgG antibody was added to the wells for 1 h at 37ºC.
Finally, the plate was read at a wavelength of 535 nm on a
fluorescence plate reader (Perkin-Elmer, Inc., Waltham, MA,
USA).
Immunoradiometric assay for CYFRA
Plasma (80 μl) was analyzed using a commercially
available TM-CYFRA 21-1 ELISA kit (DRG Instruments GmbH, Marburg,
Germany) according to the manufacturer's recommendations.
Statistical analysis
The set of spot intensities and prior biological
information were used in statistical analyses. Data from 2D-DIGE
was analyzed by principal component analysis (PCA) using
Impressionist software (Genedata AG, Basel, Switzerland) (13,14).
PCA is a multidimensionality-reduction technique used to visualize
similarities and differences between samples. It belongs to the
class of unsupervised methods.
Results
Identification of proteins in
smoking-related pulmonary disease by 2D-DIGE
We compared serum protein expression profiles of SCC
and COPD patients with those of healthy smokers. Serum samples were
collected from young adult men (<60 years) with a moderate to
heavy smoking habit (>15 pack years) but without comorbidity. An
immunoaffinity column was used to remove abundant serum proteins
and quantitative protein profiles of samples were subsequently
generated by 2D-DIGE. More than 2400 spots were resolved, and
computer-assisted quantitative analysis was performed.
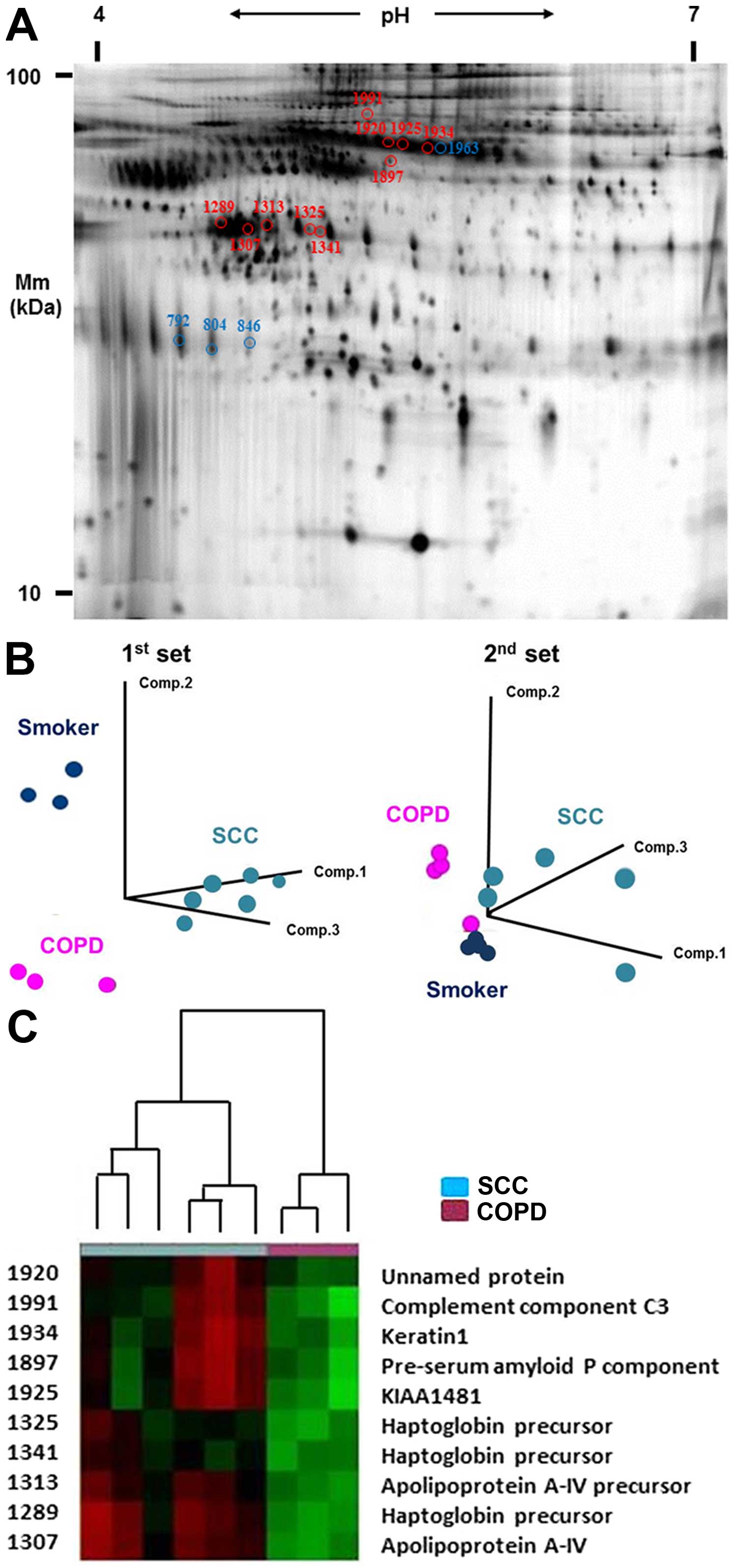
Fig. 1A illustrates
a representative Cy5 image unique to SCC patients. To estimate the
ability of 2D-DIGE profiling to distinguish between patients based
on the types of smoking-related diseases, PCA was performed using a
data set of protein spot profiles. Twelve patients of the first set
of serum samples collected were divided into three groups according
to patients types (Fig. 1B).
Thirteen patients of the second collected set of serum samples were
also divided into three groups according to patient types (Fig. 1B). We tried to identify
differentially expressed protein spots common to both cohorts.
First, we identified 30 serum protein spots that distinguished SCC
from COPD patients using the first set. We validated these protein
expression levels using an independent, second sample set. Of the
30 spots, we identified 10 SCC serum-specific protein spots with
increased intensity when compared with COPD serum in the second set
(Fig. 1A and C). To more
specifically identify those 10 differentially expressed proteins
common to both cohorts, peptide mass fingerprinting of tryptic
digests was performed by mass spectrometry. A database search
successfully identified 10 proteins with expression significantly
higher in SCC than in COPD sera (Table
I and Fig. 1C; P<0.05 for
all). Three spots were identified as HP protein and a further two
were identified as apolipoprotein A-IV (APOA4). We also identified
4 COPD serum-specific protein spots when compared with samples from
smokers in both sets of collected sera (Table I and Fig. 1A). Three of these spots were
α-1-antichymotrypsin (AACT), whose expression was significantly
lower in COPD than in the sera of smokers (P<0.05 for all).
 | Table IThe differentially expressed proteins
in SCC and COPD. |
Table I
The differentially expressed proteins
in SCC and COPD.
| | | 1st set | 2nd set |
|---|
| | |
|
|
|---|
| Spot no. | Protein ID | Protein name | Ratio | P-value | Ratio | P-value |
|---|
| SCC vs. COPD |
| 1289 | gi|306882 | Haptoglobin
precursor | 10.4 | 2.6E-06 | 4.2 | 0.008 |
| 1325 | gi|306882 | Haptoglobin
precursor | 4.9 | 9.1E-06 | 8.1 | 0.009 |
| 1341 | gi|306882 | Haptoglobin
precursor | 4.0 | 0.000053 | 8.7 | 0.003 |
| 1307 | gi|178759 | Apolipoprotein
A-IV | 10.4 | 1.8E-06 | 6.7 | 0.005 |
| 1313 | gi|71773110 | Apolipoprotein A-IV
precursor | 5.9 | 0.000042 | 9.6 | 0.005 |
| 1991 | gi|78101268 | Complement component
C3 | 5.2 | 0.000032 | 2.8 | 0.006 |
| 1897 | gi|337758 | Pre-serum amyloid P
component | 8.7 | 0.000018 | 3.4 | 0.007 |
| 1920 | gi|28317 | Unnamed protein
product | 5.7 | 0.000011 | 1.7 | 0.04 |
| 1925 | gi|7959223 | KIAA1481
protein | 12.0 | 2.1E-07 | 3.8 | 0.02 |
| 1934 | gi|11935049 | Keratin 1 | 9.2 | 1.4E-06 | 2.8 | 0.006 |
| COPD vs.
smoker |
| 1963 | gi|7331218 | Keratin 1 | 4.7 | 7.5E-008 | 1.2 | 0.04 |
| 804 | gi|177809 |
α-1-antichymotrypsin | −5.7 | 8.2E-006 | −1.7 | 0.04 |
| 846 | gi|177809 |
α-1-antichymotrypsin | −4.7 | 1.2E-006 | −2.7 | 0.001 |
| 792 | gi|177809 |
α-1-antichymotrypsin | −3.9 | 1.3E-006 | −1.8 | 0.04 |
To confirm the degree of change in spot intensity
observed by 2D-DIGE, protein samples of the first and second sets,
after affinity column treatment to remove abundant serum proteins,
including HP, were separated by SDS-PAGE, western blotted and
incubated with antibodies against HP. The detection of HP protein
bands had not been expected at the start of this study because of
the expected removal of all high-abundance proteins, including HP,
by the affinity column and this was confirmed in COPD and healthy
smoker sera by western blotting. However, the expression of HP was
detected in all five SCC patient serum samples, in spite of the
removal of high-abundance proteins (Fig. 1D). In addition, high molecular
weight isoforms of APOA4, probably due to post-translational
modifications, were also found in three of five SCC samples
(Fig. 1D). In COPD cases, the
decreased expression of α-1-antichymotrypsin (AACT) was observed by
western blotting (Fig. 1E). These
results suggest that HP and APO-A4 may be involved in the
carcinogenesis of SCC, and the decreased expression of AACT may
contribute to the occurrence of COPD.
Identification, by iTRAQ-2DLC-MS/MS, of
peptides whose expression is affected by the smoking-related
pulmonary disease
To further identify specific peptides or proteins
with post-translational modifications to distinguish SCC patients
from COPD patients, iTRAQ-2DLC-MS/MS was performed. Using pooled
serum samples from SCC and COPD patients. A total of 861 proteins
were filtered and quantified using manually selected filter
exclusion parameters. An additional 2.0-fold change cut-off for all
iTRAQ ratios (ratio >2.0 or <0.5) was selected to classify
proteins as upregulated or downregulated in SCC. Sixteen proteins
were screened from SCC samples, including 12 and 4 proteins
upregulated and downregulated in SCC, respectively (P<0.05 for
all; error factor [EF] <2; Table
II). Notably, HP protein expression was significantly increased
by >10.8-fold in the SCC sample (P<0.001). A total of 187
peptides were used for the identification of HP protein (EF=1.25).
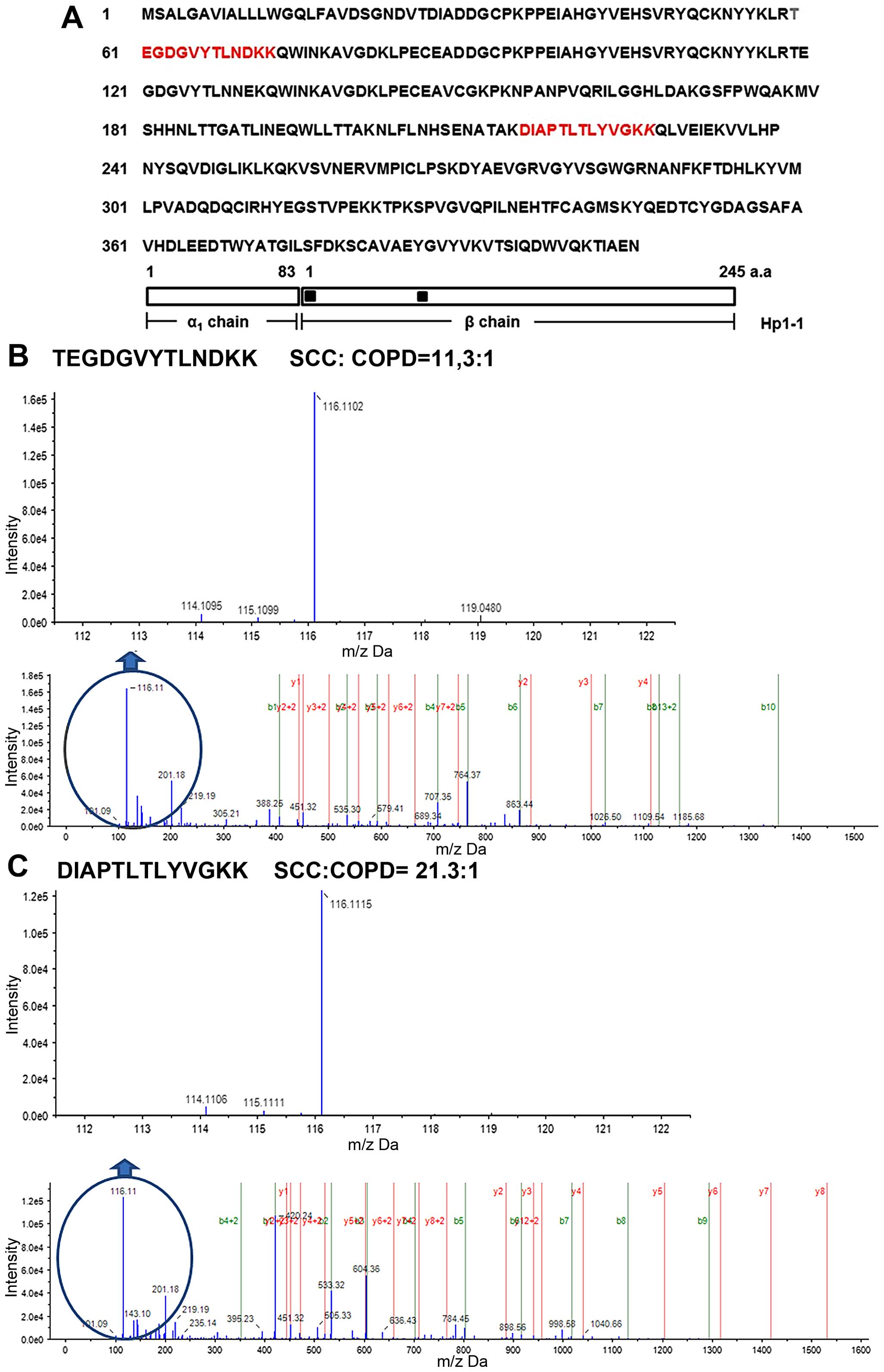
Among these identified HP peptides, two peptides were located in
the N-terminal region of the Hp β-chain, namely, HP60-72 (HP60) and
HP216-228 (HP216) (Fig. 2A), and
showed significantly higher serum levels in SCC patients than in
COPD patients (Fig. 2B and C). A
11.3-fold increase in levels of the HP60 peptide sequence was
observed in SCC compared to COPD samples (Fig. 2B). The HP216 peptide sequence was
also increased by 21.3-fold in SCC compared to COPD samples
(Fig. 2C). As for APOA4, protein
expression levels between SCC and COPD samples remained unchanged
according to the iTRAQ method (results not shown). In addition,
specific post-translational modifications, including
phosphorylation of APOA4, were not found. These results suggest
that two specific HP peptide sequences were candidate serum
biomarkers for SCC.
HP peptide ELISA
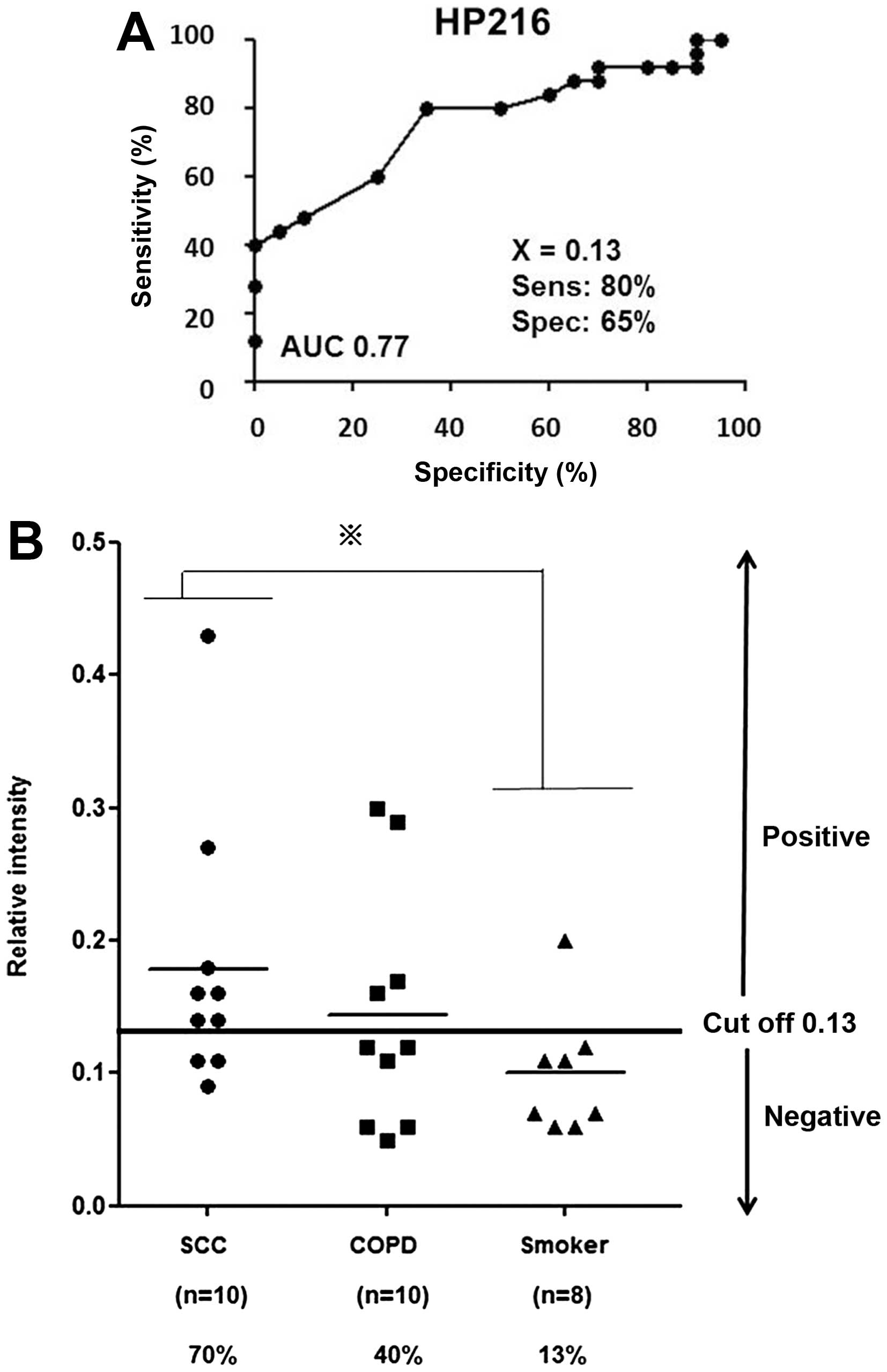
We next assessed the sensitivity and specificity of
these two peptides for SCC diagnosis by receiver operating
characteristic (ROC) curve analysis. The cut-off value was set at
the point whose distance from the (sensitivity, specificity) =
(1,1) reached the minimum (Fig. 3A). To assess the quantitative
reproducibility of these two peptides, we conducted further
validation studies by ELISA using 28 serum samples consisting of
five available SCC samples from the second set of 2D-DIGE and 23
additional serum samples. We first established novel polyclonal
antibodies for HP60 and HP216 and then developed a sandwich ELISA
for the quantification of these specific peptides using samples
from 10 SCC and 10 COPD patients, and eight healthy control
smokers. For HP60, expression levels of the peptide were identical
between SCC and healthy control serum samples (data not shown). In
contrast, significantly high levels of HP216 peptide were found in
sera from SCC patients when compared with those of healthy smoker
controls (Fig. 3B; P<0.05). The
sensitivity of HP216 (cut-off value of 0.13) was 70% (7 of 10) of
SCC patients, 40% (4 of 10) of COPD patients and 13% (1 of 8) of
smoker controls (Fig. 3B). These
results suggest that HP216 is a novel candidate serum biomarker of
SCC patients.
Immunoradiometric assay for CYFRA
We also measured serum CYFRA protein levels,
clinically used as SCC tumor marker, in all 28 cases. The
sensitivity of CYFRA protein level (cut-off value of 3.0) in serum
samples of SCC, and COPD patients, and healthy controls was 70% (7
of 10), 0% (0 of 10) and 0% (0 of 8), respectively (Table III). Interestingly, the detection
of HP216 peptides was positive in three CYFRA false-negative SCC
cases (Table IV). When the
detection of HP216 was combined with that of CYFRA, 100% (10 of 10
patients) of SCC cases were detected (Table IV). These findings suggest that
the SCC-specific peptide HP216 may be used as a diagnostic
biomarker for SCC. A combination of detecting HP216 with CYFRA
protein fragment levels in patient sera may be applicable to
diagnose SCC.
 | Table IIIDetection rates for CYFRA and HP216,
and their combination. |
Table III
Detection rates for CYFRA and HP216,
and their combination.
| No. of cases | CYFRA n, (%) | HP216 n, (%) | Combination n,
(%) |
|---|
| SCC | 10 | 7 (70) | 7 (70) | 10 (100) |
| COPD | 10 | 0 (0) | 4 (40) | |
| Smoker | 8 | 0 (0) | 1 (13) | |
 | Table IVThe results of CYFRA and HP 216, and
their combination. |
Table IV
The results of CYFRA and HP 216, and
their combination.
| No. | Case | CYFRA | HP216 | Combination |
|---|
| 1 | SCC | 14.8 | + | 0.14 | + | + |
| 2 | SCC | 14.4 | + | 0.16 | + | + |
| 3 | SCC | 10.9 | + | 0.14 | + | + |
| 4 | SCC | 9.1 | + | 0.18 | + | + |
| 5 | SCC | 70.1 | + | 0.11 | − | + |
| 6 | SCC | 39.6 | + | 0.09 | − | + |
| 7 | SCC | 6.1 | + | 0.11 | − | + |
| 8 | SCC | 2.3 | − | 0.43 | + | + |
| 9 | SCC | 2.0 | − | 0.16 | + | + |
| 10 | SCC | 1.8 | − | 0.27 | + | + |
Discussion
Smoking history and the existence of COPD are
important risk factors for SCC (1,3).
However, it is not easy to detect SCC in COPD patients and heavy
smokers by the existing method of radiographic examination.
Targeted molecular therapies, such as epidermal growth factor
receptor (EGFR) tyrosine-kinase inhibitor (EGFR-TKI) and anaplastic
lymphoma kinase (ALK) inhibitor, for lung adenocarcinoma have been
developed (17–19); however, SCC shows resistance to
standard chemotherapy and targeted molecular therapy (4,5).
Therefore, serum biomarkers have great potential to facilitate the
early detection of SCC in patients with SCC risk factors such as
COPD and a smoking history, and for the monitoring of cancer
recurrence and treatments in SCC patients, resulting in
improvements in the prognosis for this disease.
Chronic tobacco smoking is a major risk factor for
the development of COPD, but only a relatively small proportion of
smokers actually develop airway obstruction. It is well known that
α-1 antitrypsin deficiency (AATD) is a hereditary disorder
characterized by low circulating levels of the key antiprotease,
α-1 antitrypsin, and is associated with the development of COPD,
often by the third or fourth decade (20). In this study, we found that
decreased serum expression of AACT was observed in COPD patients.
AACT belongs to the class of serine protease inhibitors and is
synthesized in the liver by alveolar macrophages and airway
epithelia (21). Several cases of
COPD were reported to be associated with mutations and
polymorphisms in the AACT gene (22,23).
Although the frequency of AATD is relatively low in Japanese
patients, as is the case with α-1 antitrypsin, decreased serum
expression of AACT in heavy smokers may contribute to the
development of COPD in Japanese.
In the present study, we compared the serum protein
profiles, using 2D-DIGE/MS and iTRAQ/MS analyses, of SCC, COPD, and
healthy smoker cases of identical ages to identify a biomarker for
SCC and COPD patients. In general, a relatively small number of
~200–300 protein spots obtained by 2D-DIGE from serum samples have,
for a long time, been a major concern (10); employing an affinity column was
found to significantly improve the number of protein spots detected
to over 2000 spots. The iTRAQ-based approach covered peptide
profiling, which could not be done by 2D-DIGE (12). Based on two independent proteomics
techniques followed by ELISA, total HP or HP peptide (HP 216), was
identified as a candidate serum biomarker of SCC. Furthermore, SCC
was predicted in all ten SCC patients only when using the detection
of HP216 combined with CYFRA.
We specifically focused on HP as a biomarker of SCC.
HP is classified as an acute phase protein whose serum expression
levels increases with inflammation, infection, and several cancers
in this study (24). The major
site for HP biosynthesis is the liver and its synthesis is
regulated by various inflammatory cytokines, including IL-1, IL-6,
TNF-α and TNF-β. Previous studies have demonstrated increases in
the serum level of total HP protein and HP fragments, as well as
specific HP post-translational modifications, including
fucosylation and glycosylation, in lung and colorectal cancer
patients (25–28). Abnormal HP glycosylation was found
to interfere with tumor metastasis (27), while fucosylated HP was shown to be
a novel biomarker for colorectal cancer (28). In addition, HP has been shown to be
an angiogenic factor in sera from patients with vasculitis and
cancer. It is likely that the causes of these changes may encompass
host-defense and tumor-promoting mechanisms. By processing serum
samples after the removal of high-abundance proteins, we finally
identified HP216 peptide as a candidate SCC biomarker. Both the
origin and function of HP216 are still unknown; however, serum
levels of a two-residue longer peptide, HP216-230, were also
significantly higher in SCC the patient samples in the present
study (data not shown). CYFRA21-1 is a fragment of cytokeratin 19
that has been extensively studied in patients with SCC and has been
demonstrated to be clinically useful (9). These findings suggest the existence
of SCC-associated endo- or exopeptidases responsible for cleavage
at HP amino acid 216 in SCC patients.
No single biomarker will be able to detect all SCC
cases with a high enough specificity and sensitivity. Recent study
demonstrated that combination detection of HP and current tumor
markers could significantly improve the sensitivity and specificity
in diagnosis of lung cancer (29).
Four tumor markers including CEA, NSE, CYFRA as well as HP the
combined together could produce a positive detection rate of 85%,
significantly higher than that of any single test (29). In the present study, all SCC cases
were detected by the measurement of HP216 combined with CYFRA. A
combination of detecting serum HP216 with CYFRA levels may be
applicable to diagnose SCC.
Taken together, the HP peptide HP216, is a promising
cancer biomarker for the early detection of SCC in high-risk lung
cancer populations, including COPD patients and heavy smokers.
Combined with CYFRA determinations, the measurement of serum HP216
may lead to the earlier detection of SCC in at-risk patients and
become a novel diagnostic tool for SCC. A limitation of the study
was the relatively small sample size. Further studies will be
needed to confirm the significance of HP216 using a large number of
subjects and to elucidate the origin and function of the HP
peptide.
Acknowledgements
We thank Dr Tadashi Kondo at Division of Rare Cancer
Research, National Cancer Center Research Institute for technical
advice and helpful discussion. We also thank Ms. Mina Fujishiro for
sample collection. The present study was supported in part by a
Grant-in-Aid from Smorking Research Fundation (to A.G.).
References
|
1
|
JCS Joint Working Group; Japanese Society
for Oral Health; Japanese Society of Oral and Maxillofacial
Surgeons; Japanese Society of Public Health; Japanese Respiratory
Society; Japan Society of Obstetrics and Gynecology; Japanese
Circulation Society; Japan Pediatric Society; Japanese College of
Cardiology; Japan Lung Cancer Society. Guidelines for Smoking
Cessation (JCS 2010) - digest version. Circ J. 76:1024–1043. 2012.
View Article : Google Scholar
|
|
2
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics 2013. CA Cancer J Clin. 63:11–30. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sobue T, Yamamoto S, Hara M, Sasazuki S,
Sasaki S and Tsugane S; JPHC Study Group; Japanese Public Health
Center. Cigarette smoking and subsequent risk of lung cancer by
histologic type in middle-aged Japanese men and women: the JPH
study. Int J Cancer. 99:245–251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Scagliotti GV, Parikh P, von Pawel J,
Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U,
Digumarti R, Zukin M, et al: Phase III study comparing cisplatin
plus gemcitabine with cisplatin plus pemetrexed in
chemotherapy-naive patients with advanced-stage non-small-cell lung
cancer. J Clin Oncol. 26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sandler A, Gray R, Perry MC, Brahmer J,
Schiller JH, Dowlati A, Lilenbaum R and Johnson DH:
Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell
lung cancer. N Engl J Med. 355:2542–2550. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Powell HA, Iyen-Omofoman B, Baldwin DR,
Hubbard RB and Tata LJ: Chronic obstructive pulmonary disease and
risk of lung cancer: The importance of smoking and timing of
diagnosis. J Thorac Oncol. 8:6–11. 2013. View Article : Google Scholar
|
|
7
|
Kovalchik SA, Tammemagi M, Berg CD,
Caporaso NE, Riley TL, Korch M, Silvestri GA, Chaturvedi AK and
Katki HA: Targeting of low-dose CT screening according to the risk
of lung-cancer death. N Engl J Med. 369:245–254. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Torre GC: SCC antigen in malignant and
nonmalignant squamous lesions. Tumour Biol. 19:517–526. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pujol JL, Grenier J, Daurès JP, Daver A,
Pujol H and Michel FB: Serum fragment of cytokeratin subunit 19
measured by CYFRA 21-1 immunoradiometric assay as a marker of lung
cancer. Cancer Res. 53:61–66. 1993.PubMed/NCBI
|
|
10
|
Tirumalai RS, Chan KC, Prieto DA, Issaq
HJ, Conrads TP and Veenstra TD: Characterization of the low
molecular weight human serum proteome. Mol Cell Proteomics.
2:1096–1103. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Okano T, Kondo T, Kakisaka T, Fujii K,
Yamada M, Kato H, Nishimura T, Gemma A, Kudoh S and Hirohashi S:
Plasma proteomics of lung cancer by a linkage of multi-dimensional
liquid chromatography and two-dimensional difference gel
electrophoresis. Proteomics. 6:3938–3948. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Latterich M, Abramovitz M and
Leyland-Jones B: Proteomics: New technologies and clinical
applications. Eur J Cancer. 44:2737–2741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Seike M, Kondo T, Fujii K, Okano T, Yamada
T, Matsuno Y, Gemma A, Kudoh S and Hirohashi S: Proteomic
signatures for histological types of lung cancer. Proteomics.
5:2939–2948. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Seike M, Kondo T, Fujii K, Yamada T, Gemma
A, Kudoh S and Hirohashi S: Proteomic signature of human cancer
cells. Proteomics. 4:2776–2788. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Seike M, Kondo T, Mori Y, Gemma A, Kudoh
S, Sakamoto M, Yamada T and Hirohashi S: Proteomic analysis of
intestinal epithelial cells expressing stabilized beta-catenin.
Cancer Res. 63:4641–4647. 2003.PubMed/NCBI
|
|
16
|
Okano T, Kondo T, Fujii K, Nishimura T,
Takano T, Ohe Y, Tsuta K, Matsuno Y, Gemma A, Kato H, et al:
Proteomic signature corresponding to the response to gefitinib
(Iressa, ZD1839), an epidermal growth factor receptor tyrosine
kinase inhibitor in lung adenocarcinoma. Clin Cancer Res.
13:799–805. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Maemondo M, Inoue A, Kobayashi K, Sugawara
S, Oizumi S, Isobe H, Gemma A, Harada M, Yoshizawa H, Kinoshita I,
et al: Gefitinib or chemotherapy for non-small-cell lung cancer
with mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mitsudomi T, Morita S, Yatabe Y, Negoro S,
Okamoto I, Tsurutani J, Seto T, Satouchi M, Tada H, Hirashima T, et
al; West Japan Oncology Group. Gefitinib versus cisplatin plus
docetaxel in patients with non-small-cell lung cancer harbouring
mutations of the epidermal growth factor receptor (WJTOG3405): An
open label, randomised phase 3 trial. Lancet Oncol. 11:121–128.
2010. View Article : Google Scholar
|
|
19
|
Solomon BJ, Mok T, Kim DW, Wu YL, Nakagawa
K, Mekhail T, Felip E, Cappuzzo F, Paolini J, Usari T, et al:
First-line crizotinib versus chemotherapy in ALK-positive lung
cancer. N Engl J Med. 371:2167–2177. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kelly E, Greene CM, Carroll TP, McElvaney
NG and O'Neill SJ: Alpha-1 antitrypsin deficiency. Respir Med.
104:763–772. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nagareda T, Takeda M, Kojima K, Tanaka A,
Terada N, Yamasaki T, Nagareda T, Ueno H and Kotoh K: Alpha-1
anti-chymotrypsin is increased in human alveolar macrophages by
phorbol myristate acetate or lipopolysaccharide and released from
these activated macrophages by glucocorticoid. J Pathol.
165:319–323. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Poller W, Faber JP, Weidinger S, Tief K,
Scholz S, Fischer M, Olek K, Kirchgesser M and Heidtmann HH: A
leucine-to-proline substitution causes a defective alpha
1-antichymotrypsin allele associated with familial obstructive lung
disease. Genomics. 17:740–743. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ishii T, Matsuse T, Teramoto S, Matsui H,
Hosoi T, Fukuchi Y and Ouchi Y: Association between
alpha-1-antichymotrypsin polymorphism and susceptibility to chronic
obstructive pulmonary disease. Eur J Clin Invest. 30:543–548. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Turner GA: Haptoglobin. A potential
reporter molecule for glycosylation changes in disease. Adv Exp Med
Biol. 376:231–238. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dowling P, O'Driscoll L, Meleady P, Henry
M, Roy S, Ballot J, Moriarty M, Crown J and Clynes M: 2-D
difference gel electrophoresis of the lung squamous cell carcinoma
versus normal sera demonstrates consistent alterations in the
levels of ten specific proteins. Electrophoresis. 28:4302–4310.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bharti A, Ma PC, Maulik G, Singh R, Khan
E, Skarin AT and Salgia R: Haptoglobin alpha-subunit and hepatocyte
growth factor can potentially serve as serum tumor biomarkers in
small cell lung cancer. Anticancer Res. 24:1031–1038.
2004.PubMed/NCBI
|
|
27
|
Hoagland LF IV, Campa MJ, Gottlin EB,
Herndon JE II and Patz EF Jr: Haptoglobin and posttranslational
glycan-modified derivatives as serum biomarkers for the diagnosis
of nonsmall cell lung cancer. Cancer. 110:2260–2268. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Takeda Y, Shinzaki S, Okudo K, Moriwaki K,
Murata K and Miyoshi E: Fucosylated haptoglobin is a novel type of
cancer biomarker linked to the prognosis after an peration in
colorectal cancer. Cancer. 15;118:3036–3043. 2012. View Article : Google Scholar
|
|
29
|
Wang B, He YJ, Tian YX, Yang RN, Zhu YR
and Qiu H: Clinical utility of haptoglobin in combination with CEA,
NSE and CYFRA21-1 for diagnosis of lung cancer. Asian Pac J Cancer
Prev. 15:9611–9614. 2014. View Article : Google Scholar : PubMed/NCBI
|