Introduction
Locally advanced rectal carcinoma (LARC) is the
third most common cancer and the third most common cancer cause of
death globally (1). The prognosis
of patients with LARC has slowly improved during the past decades
in many countries (2), but there
is much room to improve the treatment of patients suffering from
colorectal cancer. At present, the standard procedures to treat
LARC include neoadjuvant radiation therapy (nRT), surgery, and
adjuvant therapy (3). Neoadjuvant
therapy could further downstage tumors, improve the rate of
sphincter preservation, and reduce the rate of local recurrence
(4). For patients with stage II
and III LARC, nRT reduces rates of local recurrence and toxic
effects. Though rectal cancer patients have benefited from
neoadjuvant therapy, the response of patients to neoadjuvant
therapy varies from pathological complete response (pCR) (9–37%) to
complete resistance (5). Patients
who achieved pCR have a prior prognosis compared with those who did
not achieve pCR. The identification of patients at risk of being
radioresistant would avoid the adverse side effects and cost of nRT
and provide the patients alternative options such as surgical
intervention without nRT (6). Even
with nRT to achieve pCR, a substantial number of LARC patients will
experience metastatic progression (7). Currently, it is inconclusive whether
nRT may reduce the risk of metastatic development in rectal cancer
(8). Therefore, identification of
a specific molecular marker could be a target of neoadjuvant
radiotherapeutic interventions. To date, however, no biomarker
predicts tumor response to nRT and prevent metastatic
progression.
Pigment epithelium-derived factor (PEDF; encoded by
SERPINF1 and also known as EPC1), a serpin that has multiple
biological actions, is a differentiation factor for retino-blastoma
cells (9). PEDF is a potent
inhibitor of angiogenesis (10).
Many observations have shown that PEDF is implicated in several
biological processes, such as neurogenesis, neuroprotection,
anti-angiogenesis, retina protection, stem cell renewal, and
inflammation (11). PEDF expresses
in various kinds of cancers, such as lung cancer, pancreatic
cancer, and breast cancer (12–14),
and some research suggests that PEDF could be a new treatment to
benefit cancer patients (15–17).
In colorectal cancer (CRC), PEDF plays a role in inhibiting tumor
growth and metastasis, anti-angiogenesis, decreasing tumor
microvessel density (MVD) and tumor cell apoptosis, and recently,
PEDF has also been identified as a prognostic marker for CRC
(18–20). However, there are scarce data on
the relationship between PEDF and sensitivity to nRT. This study
evaluated the expression of PEDF after nRT and correlated the
expression of PEDF with the clinicopathological parameters and
prognosis of the disease.
Materials and methods
Subjects and samples
This research was approved by the ethics committee
of the Health Science Center of Peking University and the Oncology
Center at Peking University. Written informed consent was obtained
from all the participants prior to the enrollment.
Pre-treatment biopsy tumor samples were obtained at
colonoscopy or rigid sigmoidoscopy from 48 rectal cancer patients.
To ensure consistent quality and tumor presence, both normal and
tumor tissues were evaluated by an experienced pathologist.
We also retrospectively reviewed the data of 197
rectal cancer patients who received 30 Gy/10 fractions (10 f) nRT
at Peking University Cancer Hospital between August 2003 and
October 2009. Each subject conformed to the following inclusion
criteria: i) diagnosis of rectal adenocarcinoma by biopsy; ii)
resectable rectal cancer ≤10 cm from the anal verge; iii) evaluated
by endorectal ultrasound (ERUS) or magnetic resonance imaging (MRI)
before treatment; iv) presence of distant metastases excluded by
imaging examinations; v) neoadjuvant radiotherapy of 30 Gy/10f; vi)
radical surgery following total TME.
Patients with the following characteristics were
excluded: i) cases with familial adenomatous polyposis LARC and
hereditary non-polyposis colorectal carcinoma; ii) previous history
(within 5 years) of malignant tumor; iii) presence of unresectable
cancer; iv) death due to complications or other non-cancer related
causes.
Tumor regression grade
The histology of all surgical specimens was reviewed
and confirmed by an independent element and was classified based on
the Mandard tumor regression grade (TRG) system (21), as follows: i) complete regression
(fibrosis without detectable tissue of tumor); ii) fibrosis with
scattered tumor cells; iii) fibrosis and tumor cells with
preponderance of fibrosis; iv) fibrosis and tumor cells with
preponderance of tumor cells; and v) tumor tissue lacking changes
related to regression.
Tissue microarray and
immunohistochemistry
Formalin-fixed and paraffin-embedded specimens were
microdissected to perform tissue microarrays (TMAs) with a
tissue-arraying instrument (Beecher Instruments, Silver Spring, MD,
USA). The TMAs were analyzed by immunohistochemistry as described
previously (22). Sections were
incubated with monoclonal antibodies against PEDF (1:100, Santa
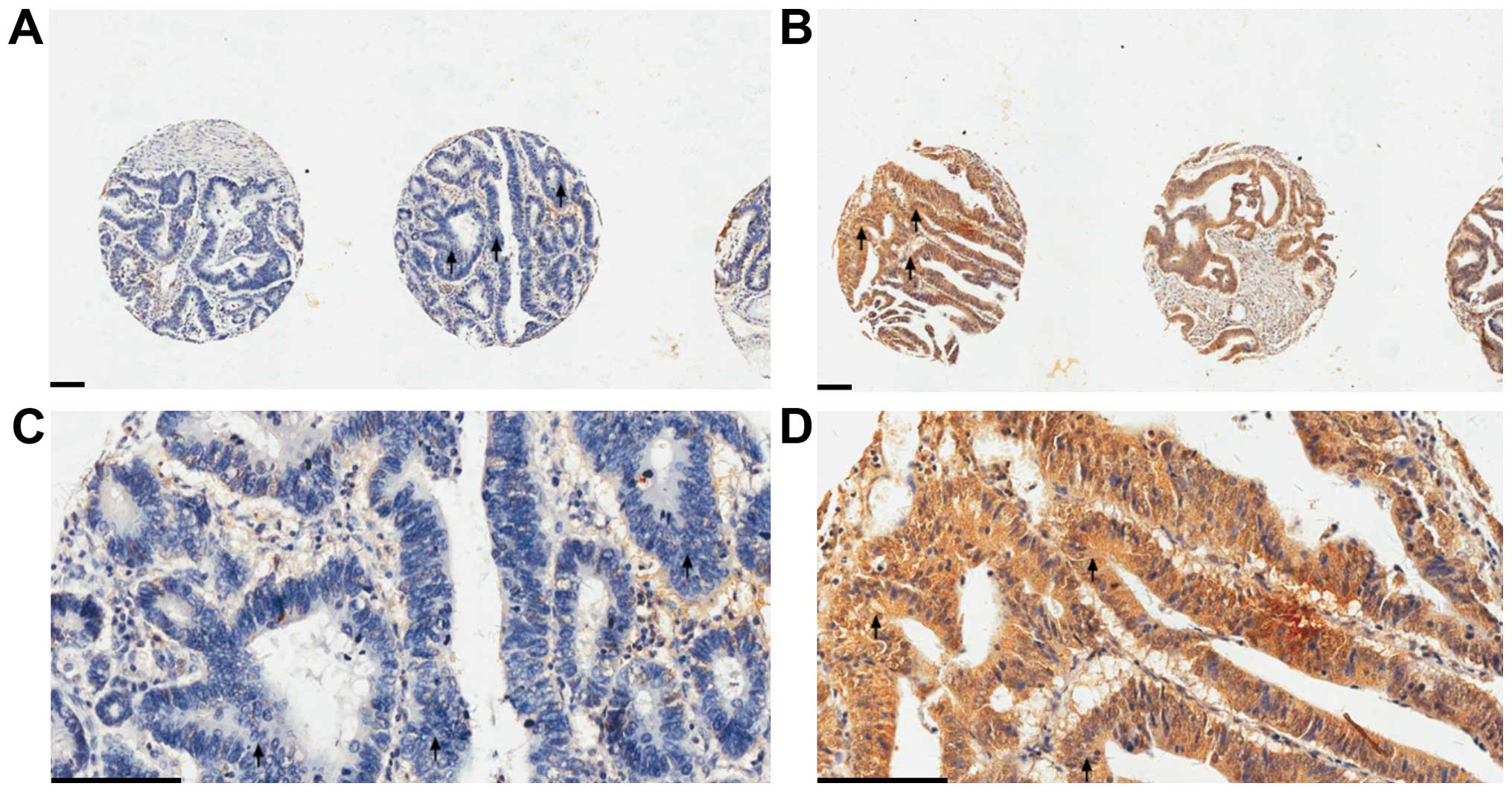
Cruz, Dallas, TX, USA). All images were examined by two experienced
pathologists independently. PEDF immunohistochemical staining of a
tissue sample was graded as either negative (patchy and weak or
negative immunoreactivity) or positive (uniformly intense
immunoreactivity) (Fig. 1).
Subcutaneous tumor xenografts
Ten NOD/SCID mice were randomly divided into
LoVo-PEDF and LoVo-control groups (n=5 per group). Equal amounts of
corresponding cells (2×106) were subcutaneously
implanted on the right buttock of each mouse. After three weeks,
mice were sacrificed and xenografts were harvested and weighed. All
animal experiments were reviewed and approved by the Ethics Review
Committee at the Peking University School of Oncology.
Cell culture and cell lines
Human CRC cell lines (LoVo) from the American Type
Culture Collection were cultured in DMEM medium with 10% fetal
bovine serum (FBS) (Gibco, Grand Island, NY, USA) in a humidified
atmosphere of 5% CO2 at 37°C. The cell lines have been
tested and authenticated by STR profiling.
Animal
Female NOD-SCID mice (4–6-week-old) were provided by
Beijing HFK Bio-technology Co., Ltd. (Beijing, China). Mice were
maintained in a pathogen-free facility and used under the
institutional guidelines for animal care.
Plasmid construction and cell
transfection
PEDF-over-expressing cells were generated by using a
ribozyme transgene system. Ribozyme transgenes specific for PEDF
were synthesised and cloned into pEF6/V5-His TOPO TA expression
plasmids in line with the manufacturer’s protocol (Invitrogen,
Shanghai, China). PEDF ribozyme transgene plasmids and control pEF6
plasmids were subsequently transfected into LoVo cells. Cells were
placed in normal medium overnight to allow recovery and then in
selection medium (normal medium containing blasticidin 5 μg/ml) for
eight days to select for cells containing the plasmid. Following
selection, cells were placed into a maintenance medium (0.5 μg/ml
blasticidin). Following selection, the cells were tested, at the
translational level, for the efficacy of the expression
plasmid.
Irradiation
Irradiation was delivered by linear accelerator at
room temperature. The dose rate was 400 cGy/min. The irradiation
doses were 0, 2, and 4 Gy. After irradiation, the medium with drugs
was adsorbed and replaced with fresh DMEM medium containing 10%
FBS.
Growth curve
LoVo-TR and LoVo-PEDF Cells (3×104
cells/well) were plated in a 96-well plate (Corning Life Science,
MA, USA). After 24 h, the medium was replaced. Growth curves were
determined by placing cells in the IncuCyte ZOOM system (Essen
BioScience), which allows an automated and non-invasive method of
monitoring live cells in culture. A growth curve is constructed
automatically from data points acquired during round-the-clock
kinetic imaging.
Clonogenic survival assay
The LoVo cells were plated at 200 cells per well in
6-well tissue culture plates and allowed to attach for 24 h. The
cells were incubated for 24 h. Immediately after exposure of the
cells to 0, 2, and 4 Gy of radiation, the medium was replaced with
fresh DMEM supplemented with 10% FBS. Cells were incubated under
standard growth conditions for 14 days, and the resultant colonies
were stained with Giemsa. Colonies containing ≥50 were scored
manually.
Wound healing assay
Approximately 5×105 cells per well were
seeded into 6-well culture plates and an incision was made in the
central area of the confluent cells 24 h later. Cell migration near
the wound area in confluent monolayers was monitored under a
microscope (Leica, Germany) at the time of scratching, and after
24, 48, and 72 h. The experiment was performed in triplicate with
three independent repeats.
Transwell migration and invasion
assay
To further evaluate the effect of PEDF on cell
migration and invasion, 2.5×104 cells in 100 μl DMEM
with 1% FBS were plated into the upper chamber of the Transwell
chamber (8-μm pore size; Corning Inc.). A total of 500 μl of DMEM
with 10% FBS was loaded into the lower chamber to serve as a
chemoattractant for the cells. After 24 h, cells migrated to or
invaded the other side of the membrane and were counted and imaged
under a microscope (Leica), after fixing with 2% methanol and
staining with 1% crystal violet solution. The experiments were
repeated three times.
Protein extraction and western
blotting
Cell proteins were extracted using RIPA buffer
containing complete protease. Extracted protein (20 μg) was
separated by 10% SDS-PAGE and blotted onto PVDF membranes
(Millipore, Billerica, MA, USA). Rabbit anti-PEDF (1:1,500
dilution; Abcam, Cambridge, MA, USA), mouse anti-P53 (1:2,000
dilution; Cell Signaling Technology), and mouse anti-GAPDH
(1:10,000 dilution; Bioworld Biotechnology, GA, USA) were used as
primary antibodies. Horseradish-peroxidase-conjugated goat
anti-rabbit or anti-mouse IgG (1:50,000 dilution, cwBiotech,
Beijing, China) was used as secondary antibody. Signals were
detected with a chemiluminescence (ECL) kit (Millipore).
TCGA datasets
One of the largest publicly available cancer
datasets, the Cancer Genome Atlas (TCGA), was used to make the
analysis (23). We downloaded the
normalized gene expression data generated using the Illumina-GA
RNA-seq platform data and Illumina-Hiseq RNA-seqV2 platform data
and the DNA methylation data generated using Illumina Infinium
Human Methylation 450 platform from the cBioPortal through the
cgdsr R package (version 2.7.1). Gene expression values were
transformed as X‘=log2ðX +1Þ, where X represents the normalized
fragments per kilobase transcript per million mapped reads values.
Altogether, we collected 171 rectum adenocarcinoma patients for
analysis from TCGA.
Statistical analysis
Association
Comparison of PEDF expression between the subgroups
of various clinicopathological parameters was evaluated by
χ2 test or Fisher’s exact test. P<0.05 was considered
significant. Pearson correlation coefficient was used to estimate
the strength and significance of the association between two
continuous variables, such as gene expression and DNA
methylation.
Prognosis
Categorical variables were analysed with Pearson’s
χ2 or Fisher’s exact test, and the level of significance
was set at 0.05. The DFS was compared by the log-rank test using a
Kaplan-Meier survival curve. Multivariate Cox proportional hazards
regression was used to determine the independent factors affecting
DFS, with the level of significance set at 0.05.
Gene ontology/pathway analysis
To identify functional clusters of genes
co-expressed with PEDF, we performed Functional Annotation
Clustering using Reactome Pathway database (24) and SRTING online database (25).
Gene set enrichment analysis
To identify the pathways that are significantly
enriched in genes associated with the PEDF, we computed the
association of gene sets with PEDF calls using the following
logistic regression model: C =β0 + β1X described before (26).
Results
The association between PEDF expression
level and clinicopathological features in LARC patients
To determine the relationship between the expression
level of PEDF and clinical pathological variables, we examined PEDF
expression by immunohistology in 197 human LARC tissues received
nRT. As shown in Table I, the
number of positively and negatively stained PEDF was counted.
Consequently, we found 110 patients had positive PEDF expression.
On analysis, low expression of PEDF was significantly correlated
with tumor differentiation (P<0.016), ypT stage (P<0.037),
and ypTNM stage (P<0.033). Of note, the PEDF expression level
was significantly negatively correlated with ypN stage (P=0.006).
Clinicopathological characteristics of all patients are summarized
in Table I.
 | Table IAssociation between
clinicopathological features and pigment epithelium-derived factor
status in the cohort received neoadjuvant radiotherapy. |
Table I
Association between
clinicopathological features and pigment epithelium-derived factor
status in the cohort received neoadjuvant radiotherapy.
| Variates | PEDF−
(%) | PEDF+
(%) | P-value |
|---|
| Gender | | | 0.065 |
| Male | 44 (50.6) | 70 (63.6) | |
| Female | 43 (49.4) | 40 (36.4) | |
| Age | | | 0.926 |
| <65 | 63 (72.4) | 79 (71.8) | |
| >65 | 24 (27.6) | 31 (28.2) | |
| BMI | | | 0.734 |
| <19 | 6 (6.9) | 6 (5.4) | |
| 19–24 | 45 (54.7) | 63 (57.3) | |
| >24 | 36 (41.4) | 41 (37.3) | |
| Surgery | | | 0.67 |
| APR | 26 (29.9) | 36 (32.7) | |
| Non-APR | 61 (70.1) | 74 (67.3) | |
|
Differentiation | | | 0.016 |
| ypCR | 5 (5.7) | 0 (0.0) | |
| G1–2 | 49 (56.3) | 84 (76.4) | |
| G3–4 | 33 (37.9) | 26 (23.6) | |
| TRG | | | 0.064 |
| ypCR | 8 (9.4) | 10 (9.3) | |
| Near ypCR | 57 (67.1) | 83 (76.9) | |
| Minor
regression | 20 (27.5) | 15 (13.9) | |
| ypT | | | 0.037 |
| T0 | 4 (6.4) | 0 (3.3) | |
| T1–2 | 22 (22.3) | 43 (46.2) | |
| T3–4 | 61 (71.3) | 67 (50.5) | |
| ypN | | | 0.006 |
| N0 | 39 (44.8) | 60 (64.5) | |
| N1–2 | 33 (37.9) | 31 (34.5) | |
| N3 | 15 (17.2) | 1 (1.0) | |
| ypTNM stage | | | 0.033 |
| I | 21 (24.1) | 36 (32.7) | |
| II | 19 (21.8) | 25 (31.8) | |
| III | 47 (54.0) | 39 (35.5) | |
| LVI | | | 0.068 |
| Negative | 16 (18.4) | 10 (9.1) | |
| Positive | 71 (81.6) | 100 (90.9) | |
Positive PEDF expression is associated
with prior disease-free survival and overall survival of LARC
patients with nRT
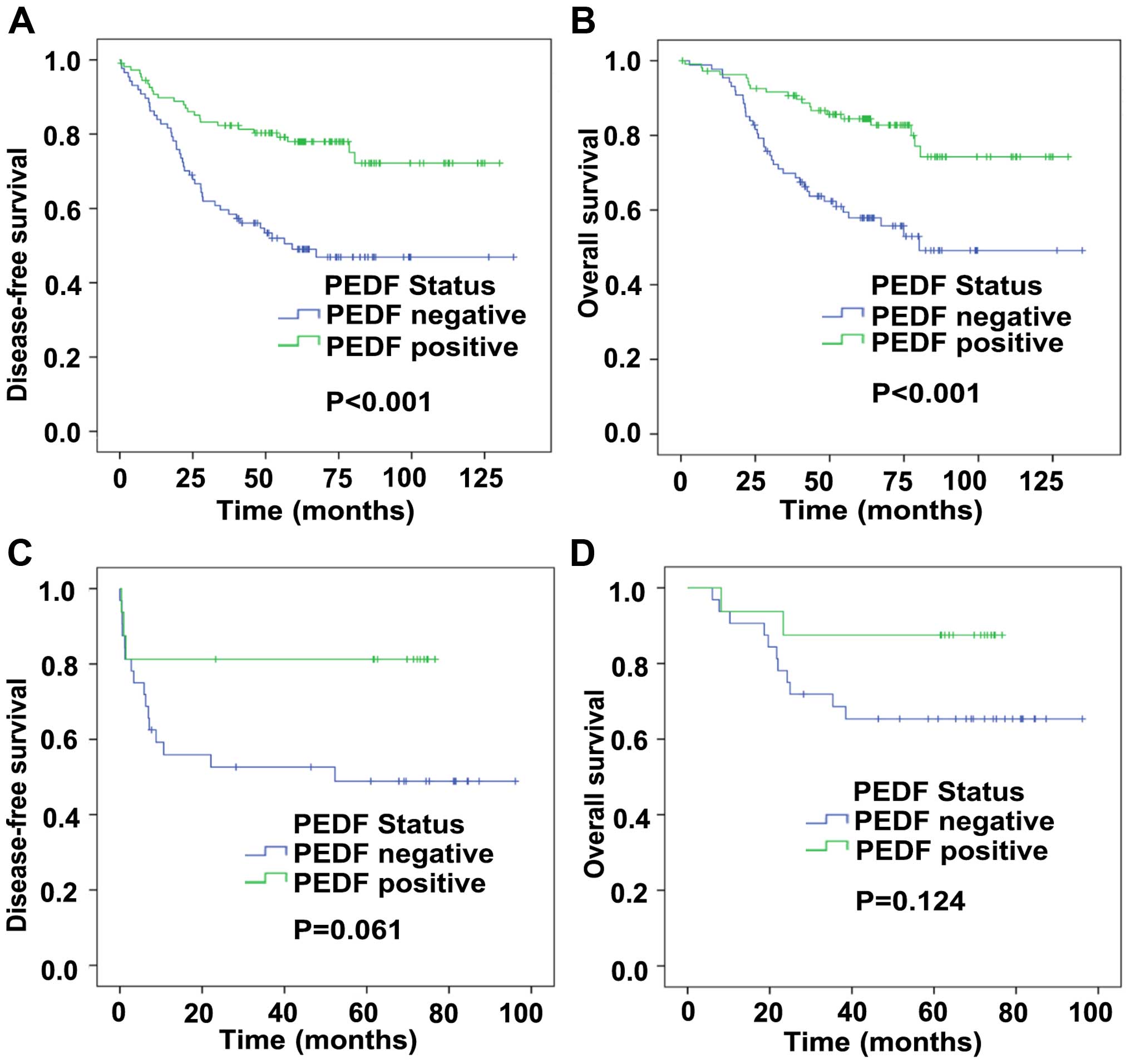
For tumor specimens, Kaplan-Meier curve analysis
revealed that positive expression of PEDF predicted prior patient
disease-free survival (DFS) and over survival (OS). Fig. 2A and B showed that the DFS time
(77.1 vs 49.0%, P<0.001) and OS time (87.1 vs 56.3%, P<0.001)
for patients with positive PEDF expression were significantly
longer than those for patients with negative PEDF expression.
According to a univariate analysis (Table II), several clinicopathological
parameters, including ypN stage, ypTNM stage, and PEDF, were
important prognosistic factors for both DFS and OS. Cox
proportional hazard regression analysis showed only PEDF, was
significant, as an independent factor for DFS [P=0.001; HR, 0.422
(95% CI, 0.249–0.717)] and OS [P=0.003; HR, 0.418 (95% CI,
0.234–0.749)] (Table III). These
data indicated that positive PEDF expression predicted prior
prognosis for LARC patients received nRT.
 | Table IIUnivariate log-rank analysis for
important clinicopathological features and pigment
epithelium-derived factor status in the cohort received neoadjuvant
radiotherapy. |
Table II
Univariate log-rank analysis for
important clinicopathological features and pigment
epithelium-derived factor status in the cohort received neoadjuvant
radiotherapy.
| DFS | OS |
|---|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender | 0.867 | 0.579–1.459 | 0.532 | 0.846 | 0.497–1.438 | 0.536 |
| Age | 1.126 | 0.674–1.881 | 0.650 | 1.299 | 0.751–2.248 | 0.350 |
| BMI | 1.226 | 0.839–1.889 | 0.266 | 1.379 | 0.879–2.162 | 0.162 |
| Surgery | 1.575 | 0.971–2.557 | 0.066 | 1.734 | 1.023–2.940 | 0.041 |
|
Differentiation | 0.968 | 0.479–1.965 | 0.928 | 0.868 | 0.429–1.758 | 0.695 |
| TRG | 0.927 | 0.714–1.205 | 0.571 | 0.799 | 0.491–1.301 | 0.366 |
| ypT | 1.208 | 0.450–3.243 | 0.708 | 1.789 | 1.359–2.356 | 0.000 |
| ypN | 1.599 | 1.252–2.041 | 0.001 | 2.926 | 1.816–4.713 | 0.000 |
| ypTNM | 1.775 | 1.277–2.412 | 0.000 | 1.918 | 1.340–2.745 | 0.000 |
| LVI | 0.577 | 0.365–1.086 | 0.046 | 0.621 | 0.336–1.148 | 0.129 |
| PEDF | 0.381 | 0.233–0.623 | 0.000 | 0.351 | 0.204–0.604 | 0.000 |
 | Table IIIMultivariate analysis for important
cliniopathological features and pigment epithelium-derived factor
status in the cohort received neoadjuvant radiotherapy. |
Table III
Multivariate analysis for important
cliniopathological features and pigment epithelium-derived factor
status in the cohort received neoadjuvant radiotherapy.
| DFS | OS |
|---|
|
|
|
|---|
| Parameter | HR | 95% CI | P-value | HR | 95% CI | P-value |
|---|
| Gender | 0.753 | 0.438–1.293 | 0.303 | 0.744 | 0.409–1.355 | 0.334 |
| Age | 1.27 | 0.729–2.214 | 0.399 | 1.567 | 0.861–2.851 | 0.141 |
| BMI | 1.163 | 0.742–1.832 | 0.509 | 1.285 | 0.781–2.115 | 0.323 |
| Surgery | 1.488 | 0.882–2.509 | 0.137 | 1.716 | 0.971–3.033 | 0.063 |
|
Differentiation | 0.968 | 0.479–1.965 | 0.46 | 0.654 | 0.256–1.670 | 0.375 |
| TRG | 0.912 | 0.529–1.751 | 0.739 | 0.882 | 0.491–1.301 | 0.684 |
| ypT | 1.472 | 0.420–5.156 | 0.545 | 1.319 | 0.373–4.668 | 0.668 |
| ypN | 1.355 | 0.705–2.605 | 0.362 | 1.808 | 0.828–3.950 | 0.137 |
| ypTNM | 1.136 | 0.495–2.610 | 0.763 | 0.9 | 0.341–2.372 | 0.831 |
| LVI | 0.851 | 0.365–1.086 | 0.61 | 0.848 | 0.427–1.686 | 0.639 |
| PEDF | 0.422 | 0.249–0.717 | 0.001 | 0.418 | 0.234–0.749 | 0.003 |
PEDF may predict the radiosensitivity and
prognosis for LARC patients
To determine the relationship between the expression
level of PEDF and radiosensitivity, we examined PEDF expression by
immunohistology in 48 pre-treatment biopsy tissues (Table IV). We found a trend that PEDF had
a negative correlation with TRG, suggesting its role in sensitivity
to nRT (correlation coefficient=−0.290, P=0.046) (Table V). The same as TRG, positive PEDF
expressing patients had a prior DFS time (81.3 vs 50.0%, P=0.061)
than those with negative PEDF expressing (Fig. 2C and D). However, we did not find
the correlation between clinicopathological characteristics and
PEDF status. All these data indicate that PEDF may be a factor to
predict the radiosensitivity and prognosis for LARC patients.
 | Table IVAssociation between
clinicopathological features and PEDF status in the pre-treatment
biopsy specimens. |
Table IV
Association between
clinicopathological features and PEDF status in the pre-treatment
biopsy specimens.
| Variates | PEDF−
(%) | PEDF+
(%) | P-value |
|---|
| Gender | | | 0.112 |
| Male | 23 (71.9) | 7 (43.8) | |
| Female | 9 (28.1) | 9 (56.2) | |
| Age | | | 0.697 |
| <65 | 25 (78.1) | 14 (87.5) | |
| >65 | 7 (21.9) | 2 (12.5) | |
| Surgery | | | 0.750 |
| APR | 22 (68.8) | 10 (62.5) | |
| Non-APR | 10 (31.3) | 6 (37.5) | |
|
Differentiation | | | 0.329 |
| ypCR | 4 (12.5) | 0 (0.0) | |
| G1–2 | 26 (81.3) | 15 (93.8) | |
| G3–4 | 2 (6.3) | 1 (6.3) | |
| TRGa | | | 0.146 |
| ypCR | 1 (4.5) | 2 (18.2) | |
| Near ypCR | 3 (13.6) | 3 (27.3) | |
| Minor
regression | 18 (81.8) | 6 (54.5) | |
| ypT | | | 1.000 |
| T0 | 3 (9.4) | 0 (0.0) | |
| T1–2 | 6 (18.8) | 5 (31.3) | |
| T3–4 | 23 (71.9) | 11 (68.8) | |
| ypN | | | 0.841 |
| N0 | 16 (50.0) | 9 (56.3) | |
| N1–2 | 9 (21.9) | 3 (18.8) | |
| N3 | 7 (17.2) | 4 (25.0) | |
| ypTNM stage | | | 0.802 |
| I | 8 (25.0) | 4 (25.0) | |
| II | 8 (25.0) | 5 (31.3) | |
| III | 16 (50.0) | 7 (43.8) | |
| LVI | | | 0.652 |
| Negative | 28 (87.5) | 15 (93.8) | |
| Positive | 4 (12.5) | 1 (6.2) | |
 | Table VSpearman correlation test between
tumor regression grade and PEDF status in the pre-treatment biopsy
specimens. |
Table V
Spearman correlation test between
tumor regression grade and PEDF status in the pre-treatment biopsy
specimens.
| Variates | PEDF−
(%) | PEDF+
(%) | P-valuea |
|---|
| TRG | | | 0.046 |
| ypCR | 1 (4.5) | 2 (18.2) | |
| Near ypCR | 3 (13.6) | 3 (27.3) | |
| Minor
regression | 18 (81.8) | 6 (54.5) | |
PEDF suppresses LoVo cell proliferation,
spread, migration, and invasion and enhances the radiation
sensitivity
The relationship between increased expression of
PEDF and cell invasion of LARC drove us to explore the possible
biological functions of PEDF in cancer cells; therefore, we
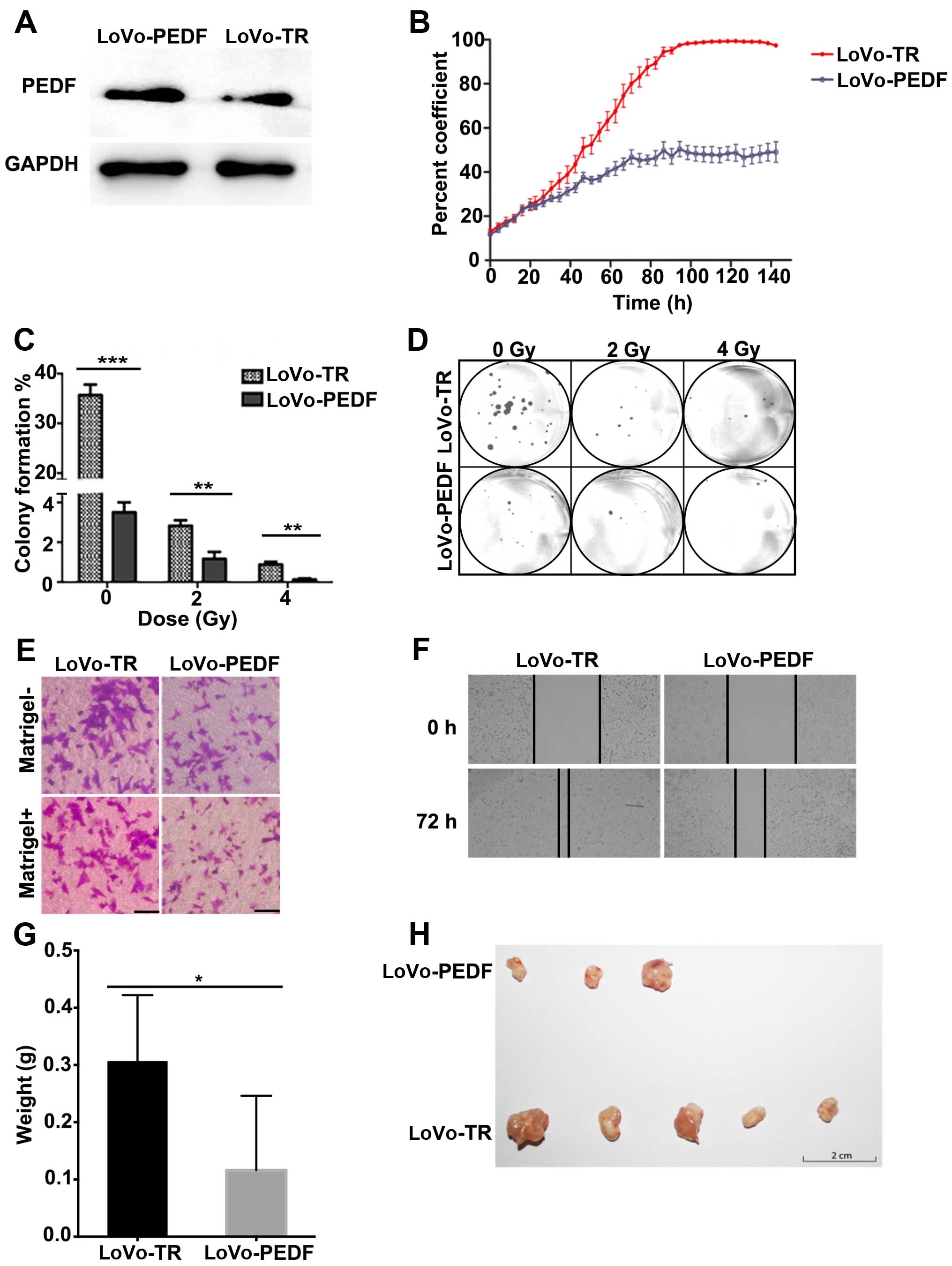
overexpressed PEDF in the highly metastatic LoVo cells. As shown in
Fig. 3A, the expression of PEDF
increased markedly in LoVo-PEDF cells (Fig. 3A). The survival rate and cell
growth curve (Fig. 3B) showed PEDF
played an important role in inhibiting cell proliferation.
Clonogenic survival assay showed PEDF displayed a
radiation-enhancing effect at the irradiation doses of 2 and 4 Gy
in LoVo cells (Fig. 3C and D).
LoVo cells that overexpressed PEDF showed a significant decrease in
cell spreading (Fig. 3F), cell
migration, and invasion (Fig. 3E),
compared with control cells. In vivo, we confirmed PEDF
decreased the ability of LoVo cells to form and grow tumors
(Fig. 3G and H). Collectively,
overexpression of PEDF suppresses cell proliferation, spreading,
migration, and invasion and enhances the radiation effect.
PEDF regulates the double-strand breaks
(DSBs) repair pathway and activates G protein pathway
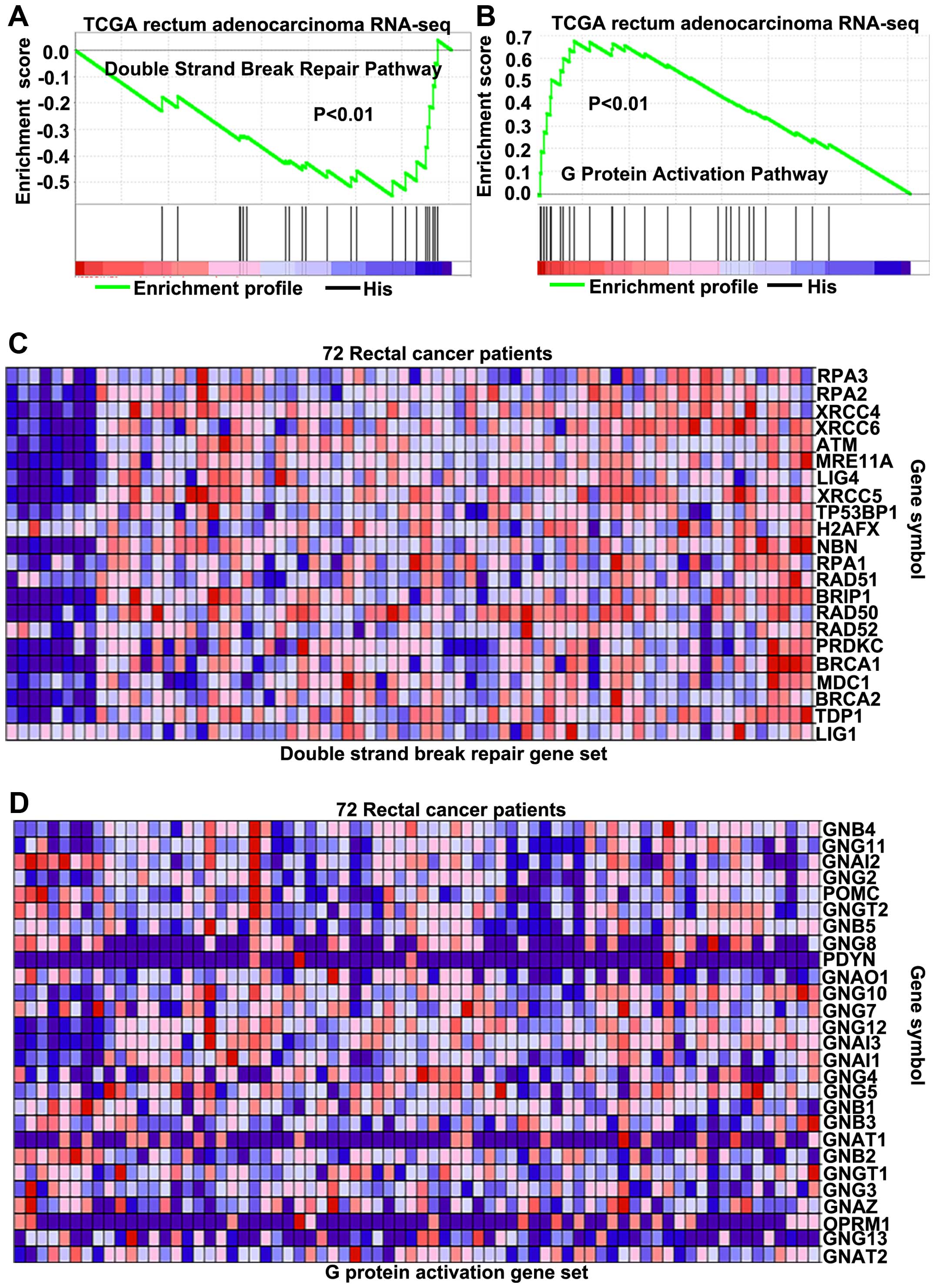
Until now, the exact pathways that PEDF may regulate
in LARC remain unclear. To probe the PEDF-associated pathways on an
unbiased basis, we performed GSEA using the Illumina-GA RNA-seq
platform data of the rectal cancer cohort of The Cancer Genomic
Atlas project (TCGA, 171 patients), GSEA is designed to detect
coordinated differences in expression of predefined sets of
functionally related genes (26).
Among all the 674 predefined ‘Reactome’ gene sets, the DSBs repair
pathway and G protein activation pathways were identified with a
strongly negative and positive association, respectively, with PEDF
expression in the TCGA rectal carcinoma dataset (Fig. 4A and B). By the analysis of LARC
cohort of TCGA, parts of key genes correlated with the DSBs repair
pathway (Fig. 4C) on mRNA levels
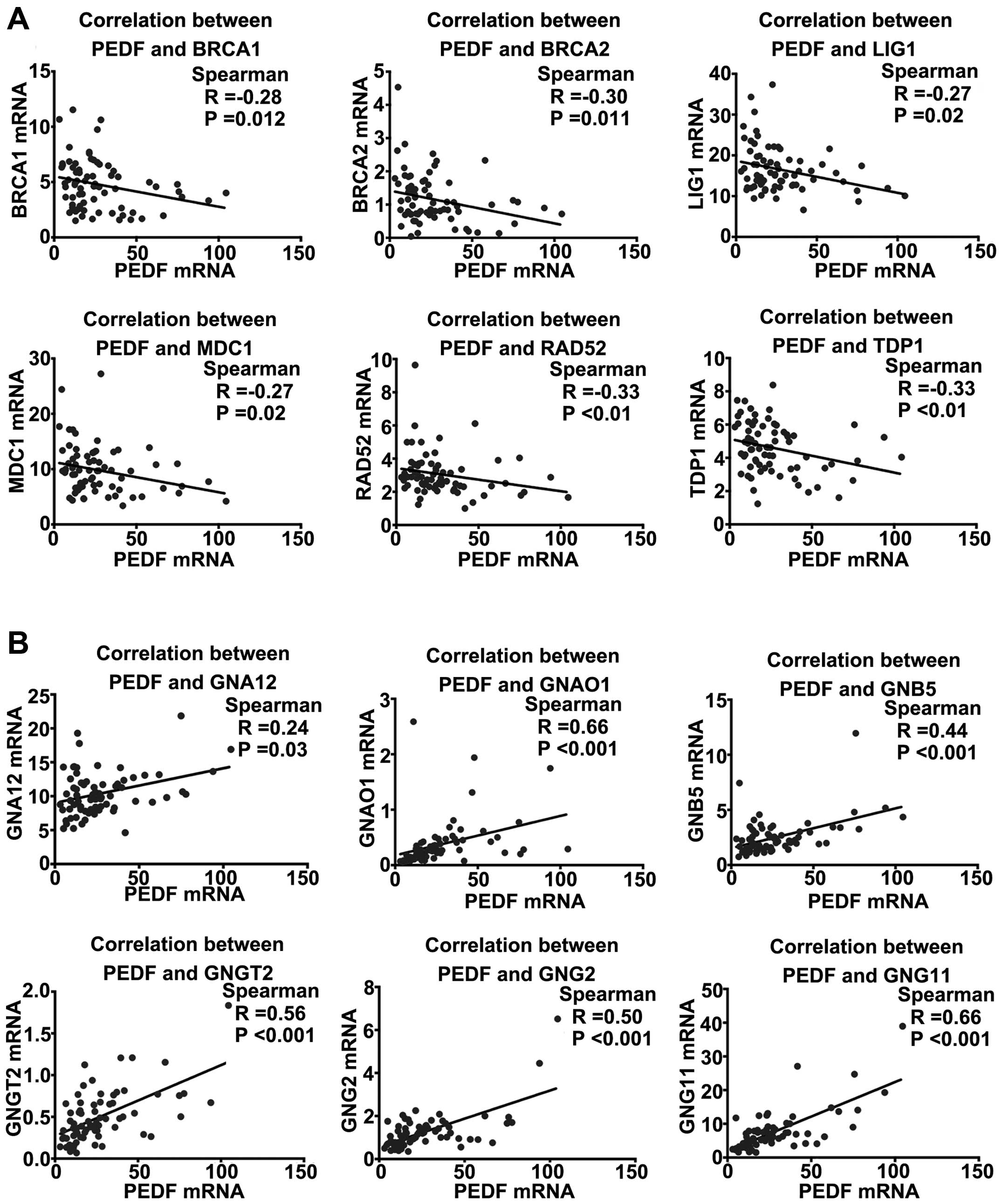
were found to be negatively associated with PEDF (Fig. 5A). For genes correlated with the G
protein activation pathway (Fig.
4D), parts of key genes were found to have positive association
with PEDF (Fig. 5B). These
findings consistently suggest that PEDF may be involved in the
regulation of the DSBs repair pathway and enhancement of G protein
activation pathway.
PEDF activates P53 to perform
functions
To validate how PEDF is involved in the DSBs repair
and G protein activation pathways, the online database STRING
(25) was used to investigate
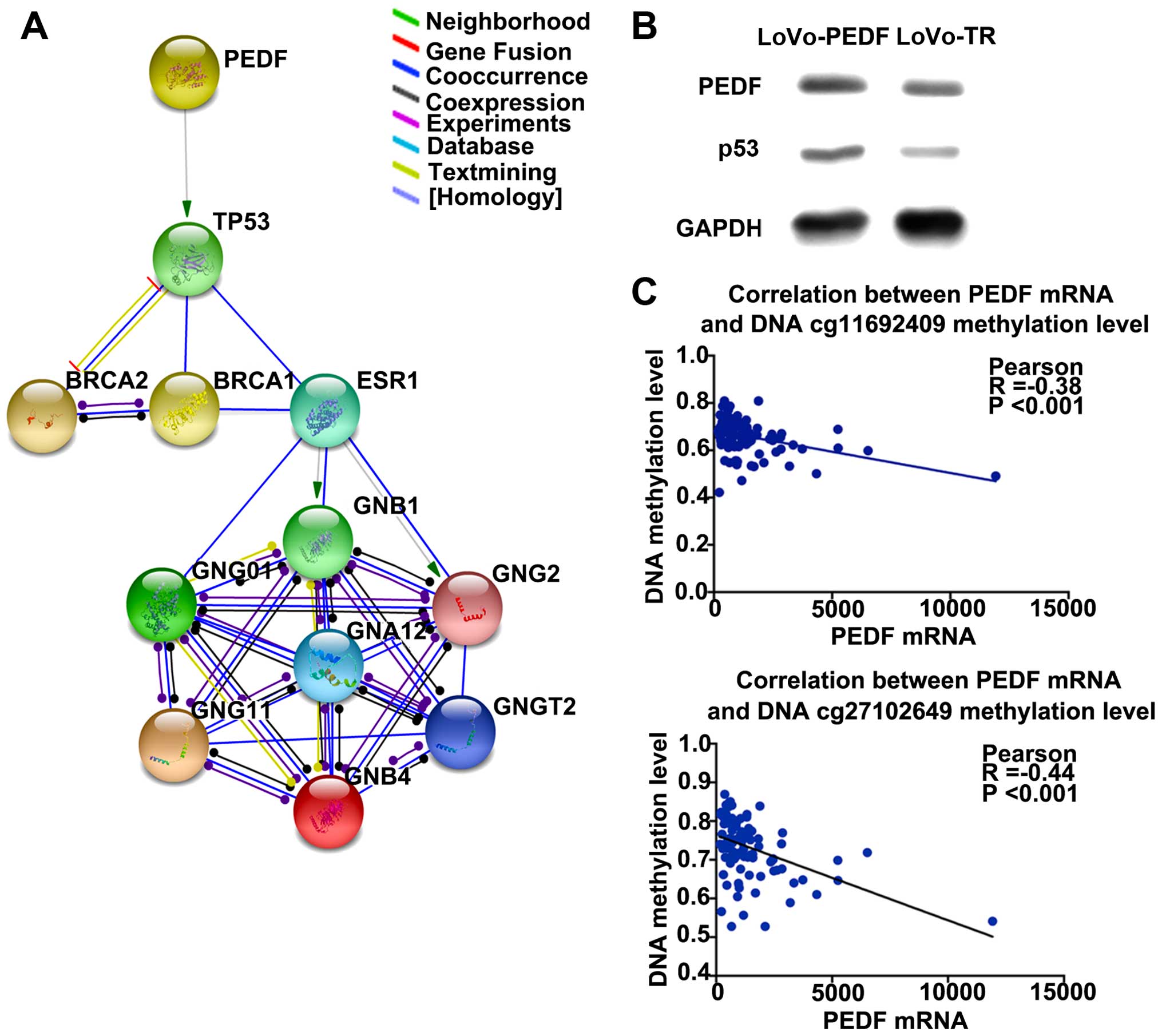
interactions between PEDF and associated genes. By protein
interaction analysis, we found P53 could be the downstream target
gene of PEDF, and interaction network was constructed (Fig. 5A). We then used The Cancer Cell
Line Encyclopedia database (27)
to confirm that P53 is wild-type in LoVo cells and has anticancer
function (data not shown). We further validated that P53 was more
highly expressed in LoVo cells with PEDF overexpression compared
with the control cells (Fig. 5B).
These findings consistently suggest that P53 is a downstream gene
of PEDF.
PEDF expression is controlled by promoter
methylation
The relationship between PEDF expression level and
PEDF promoter methylation status was analyzed using Illumina-Hiseq
RNA-seqV2 platform data and Illumina Infinium Human DNA Methylation
450 beadchip data.
To investigate whether PEDF methylation status
affects the mRNA level in rectal cancer, we used the TCGA date-base
(23) to make the correlation
between methylation of PEDF cg11692409 and PEDF cg27102649 and PEDF
mRNA expression level. The data showed that methylation of the two
genomic coordinates significantly negatively correlated with PEDF
mRNA expression (correlation coefficient=−0.38, P<0.001 and
correlation coefficient=−0.44, P<0.001) (Fig. 6C).
Discussion
PEDF has been found to enhance the antitumor effects
of radiotherapy on nasopharyngeal carcinoma by downregulating VEGF
expression and inhibiting angiogenesis (28), but the prognostic value of PEDF in
response to nRT prognosis for LARC patients is rarely reported. To
the best of our knowledge, this is the first time that PEDF
expression was found to be different between the long-term DFS
group and the short-term DFS group. LARC patients with high levels
of PEDF had prior progression-free survial and tumor regression.
The research demonstrated that PEDF could be a biomarker to predict
the response of the LARC patients to nRT; in vitro and in
vivo data showed that PEDF could be a target for developing
novel therapeutic strategies.
The nRT has become a standard treatment for LARC
patients, and the nRT regimen we adopted in this study was 30 Gy in
10 fractions, which is recommended by the Chinese Anti-Cancer
Association. Our previous data revealed prior response rates and
clinical efficiency compared with traditional and published nRT
regimens (29). However, not all
patients are sensitive to it. Several studies have suggested TRG
can be used to monitor treatment and as a prognostic parameter for
patients who received nRT (30).
The response to nRT varies from sensitivity to resistance, and some
LARC patients suffer side effects and risk of disease progression
during therapy (6). We found a new
marker to predict the TRG before nRT, providing a new therapeutic
strategy for LARC patients. In accordance with our data, PEDF
contributed to radiosensitivity in melanoma therapy. Using
quantitative reverse transcription PCR and immunohistochemical
staining, we previously detected that low PEDF levels were
associated with liver metastasis and disease-free survival in a
large corhort of LARC patients (20).
In this study, we not only revealed that PEDF was an
independent prognostic factor for DFS and OS in Cox regression
analysis but also found that low expression of PEDF in LARC was
associated with a high degree of ypN stage. Many researchers have
found that lymph node metastases are significantly correlated with
decreased survival in several solid tumors (31), and reduced PEDF levels in lung
cancer tissues significantly correlated with lymph node metastasis
(32). In addition to the
anti-angiogenesis effect, PEDF plays a role in apoptosis,
autophagy, and many other antitumor processes in various
malignancies (14,15). All of these suggest that PEDF may
decrease lymph node metastasis in cancers.
Because of the association between PEDF and
clinicopathological factors, we hypothesized that PEDF could
increase the sensitivity of the tumor to nRT. DNA damage and
hypoxia are important biological processes in chemoradiosensitivity
(33,34), and PEDF could induce cancer cell
death by apoptosis and anti-angiogenesis during therapy (18–20).
To confirm this hypothesis, we overexpressed PEDF in LoVo cells.
Compared to the control, LoVo cells that overexpressed PEDF reduced
tumorigenesis, with fewer colonies after exposure to the radiation.
Metastasis and recurrence are the determining factors affecting DFS
time of LARC patients, and angiogenesis has an indispensable
implication on the invasive property of cancer cells (35–37).
Simultaneous treatment with PEDF and radiation has an additive
effect on the downregulation of VEGF expression and on angiogenesis
inhibition (28). Therefore, our
in vitro and in vivo results suggested that the
higher level of PEDF expression promoted the DFS time by
suppressing tumor growth and metastasis in LARC patients.
To further study the mechanism by which PEDF
regulates the radiosensitivity and cell proliferation, we used the
TCGA database to identify the PEDF-associated genes. The GSEA
indicated that PEDF suppresses the DSBs repair pathway and
activates the G protein activation pathway and thus may provide
useful information for targeted therapy. Demonstrating the effect
of PEDF on regulating the pathways by STRING online database, we
provide a more complete network that PEDF activates P53.
Gene expression is regulated by DNA methylation at
CpG island promoter regions (38).
Ha et al found that the methylation status of genes is
associated with radiosensitivity to nRT (39). Our data demonstrated negative
correlation between PEDF expression and its methylation status in
LARC patients. The patients with low PEDF expression usually
possess high-level status of methylation, and our results indicate
that the hypermethylation of PEDF could decrease its mRNA
expression, generating angiogenesis and cancer cell proliferation,
which induces tumor metastasis in LARC patients.
A number of limitations need to be noted regarding
this study. The most important limitation lies in the fact that the
limited pre-treatment sample size could not confirm the trend
between the expression level of PEDF and TRG before nRT, and we
will accumulate more specimens to confirm the relationship. In
addition, there is a variety of CRC cell lines available to us; we
only used LoVo cells to identify the relationship between the level
of PEDF expression and radiosensitivity signatures. The LoVo cell
was used for overexpressed PEDF since it was exclusively lower
expression compared to the other eight LARC cell lines. Researching
ATCC, we determined that the LoVo cell line is derived from
metastatic sites, which is suitable for our purpose. Next, we plan
to validate the direct interaction between P53 and PEDF.
Methylation status is different in CRC cell lines that are derived
from diverse stages of CRC. Epigenetics signatures in LARC patients
are also affected by various factors (38). Much more research is needed to
prove the hypermethylation status of PEDF suppresses its mRNA
transcription.
In concusion, that PEDF has a prognostic value to
LARC patients after nRT. According to the level of PEDF expression,
LARC patients may be stratified into risk subgroups and allow
personalized therapeutic strategies. In addition to prolonging the
DFS of patients, PEDF enhances the sensitivity of a tumor to nRT.
Moreover, PEDF, as an anti-angiogenesis factor, possesses the
potential to be an independent therapeutic agent for LARC
patients.
Acknowledgements
This study was supported by the National Natural
Science Foundation (81372593, 81201965), Beijing Natural Science
Foundation (7132052), and the National High Technology Research and
Development Program of China (863 Program) (no. 2012AA02A506,
2014AA020801). The authors would like to thank Dr Bin Dong, the
Department of Pathology, Peking University Cancer Hospital and
Institute for her technical assistance.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
DeSantis CE, Lin CC, Mariotto AB, Siegel
RL, Stein KD, Kramer JL, Alteri R, Robbins AS and Jemal A: Cancer
treatment and survivorship statistics, 2014. CA Cancer J Clin.
64:252–271. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Brenner H, Kloor M and Pox CP: Colorectal
cancer. Lancet. 383:1490–1502. 2014. View Article : Google Scholar
|
|
4
|
van Gijn W, Marijnen CA, Nagtegaal ID,
Kranenbarg EM, Putter H, Wiggers T, Rutten HJ, Påhlman L, Glimelius
B and van de Velde CJ; Dutch Colorectal Cancer Group. Preoperative
radiotherapy combined with total mesorectal excision for resectable
rectal cancer: 12-year follow-up of the multicentre, randomised
controlled TME trial. Lancet Oncol. 12:575–582. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Huerta S, Gao X and Saha D: Mechanisms of
resistance to ionizing radiation in rectal cancer. Expert Rev Mol
Diagn. 9:469–480. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huerta S, Hrom J, Gao X, Saha D, Anthony
T, Reinhart H and Kapur P: Tissue microarray constructs to predict
a response to chemoradiation in rectal cancer. Dig Liver Dis.
42:679–684. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Gérard JP, Azria D, Gourgou-Bourgade S,
Martel-Lafay I, Hennequin C, Etienne PL, Vendrely V, François E, de
La Roche G, Bouché O, et al: Clinical outcome of the ACCORD 12/0405
PRODIGE 2 randomized trial in rectal cancer. J Clin Oncol.
30:4558–4565. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bosset JF, Calais G, Mineur L, Maingon P,
Stojanovic-Rundic S, Bensadoun RJ, Bardet E, Beny A, Ollier JC,
Bolla M, et al; EORTC Radiation Oncology Group. Fluorouracil-based
adjuvant chemotherapy after preoperative chemoradiotherapy in
rectal cancer: Long-term results of the EORTC 22921 randomised
study. Lancet Oncol. 15:184–190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Steele FR, Chader GJ, Johnson LV and
Tombran-Tink J: Pigment epithelium-derived factor: Neurotrophic
activity and identification as a member of the serine protease
inhibitor gene family. Proc Natl Acad Sci USA. 90:1526–1530. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Dawson DW, Volpert OV, Gillis P, Crawford
SE, Xu H, Benedict W and Bouck NP: Pigment epithelium-derived
factor: A potent inhibitor of angiogenesis. Science. 285:245–248.
1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Becerra SP and Notario V: The effects of
PEDF on cancer biology: Mechanisms of action and therapeutic
potential. Nat Rev Cancer. 13:258–271. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Hong H, Zhou T, Fang S, Jia M, Xu Z, Dai
Z, Li C, Li S, Li L, Zhang T, et al: Pigment epithelium-derived
factor (PEDF) inhibits breast cancer metastasis by down-regulating
fibronectin. Breast Cancer Res Treat. 148:61–72. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guan M, Jiang H, Xu C, Xu R, Chen Z and Lu
Y: Adenovirus-mediated PEDF expression inhibits prostate cancer
cell growth and results in augmented expression of PAI-2. Cancer
Biol Ther. 6:419–425. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kim KJ, Yun JH, Heo JI, Lee EH, Min HS,
Choi TH and Cho CH: Role of pigment epithelium-derived factor in
the involution of hemangioma: Autocrine growth inhibition of
hemangioma-derived endothelial cells. Biochem Biophys Res Commun.
454:282–288. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ek ET, Dass CR and Choong PF: PEDF: A
potential molecular therapeutic target with multiple anti-cancer
activities. Trends Mol Med. 12:497–502. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Manalo KB, Choong PF and Dass CR: Pigment
epithelium-derived factor as an impending therapeutic agent against
vascular epithelial growth factor-driven tumor-angiogenesis. Mol
Carcinog. 50:67–72. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Manalo KB, Choong PF, Becerra SP and Dass
CR: Pigment epithelium-derived factor as an anticancer drug and new
treatment methods following the discovery of its receptors: A
patent perspective. Expert Opin Ther Pat. 21:121–130. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wu QJ, Gong CY, Luo ST, Zhang DM, Zhang S,
Shi HS, Lu L, Yan HX, He SS, Li DD, et al: AAV-mediated human PEDF
inhibits tumor growth and metastasis in murine colorectal
peritoneal carcinomatosis model. BMC Cancer. 12:1292012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cui FY, Song XR, Li ZY, Li SZ, Mu B, Mao
YQ, Wei YQ and Yang L: The pigment epithelial-derived factor gene
loaded in PLGA nanoparticles for therapy of colon carcinoma. Oncol
Rep. 24:661–668. 2010.PubMed/NCBI
|
|
20
|
Ji D, Li M, Zhan T, Yao Y, Shen J, Tian H,
Zhang Z and Gu J: Prognostic role of serum AZGP1, PEDF and PRDX2 in
colorectal cancer patients. Carcinogenesis. 34:1265–1272. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mandard AM, Dalibard F, Mandard JC, Marnay
J, Henry-Amar M, Petiot JF, Roussel A, Jacob JH, Segol P, Samama G,
et al: Pathologic assessment of tumor regression after preoperative
chemoradiotherapy of esophageal carcinoma. Clinicopathologic
correlations. Cancer. 73:2680–2686. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Koumakpayi IH, Le Page C, Mes-Masson AM
and Saad F: Hierarchical clustering of immunohistochemical analysis
of the activated ErbB/PI3K/Akt/NF-kappaB signalling pathway and
prognostic significance in prostate cancer. Br J Cancer.
102:1163–1173. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Cancer Genome Atlas Network. Comprehensive
molecular characterization of human colon and rectal cancer.
Nature. 487:330–337. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Croft D, Mundo AF, Haw R, Milacic M,
Weiser J, Wu G, Caudy M, Garapati P, Gillespie M, Kamdar MR, et al:
The Reactome pathway knowledgebase. Nucleic Acids Res.
42D:D472–D477. 2014. View Article : Google Scholar
|
|
25
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43D:D447–D452. 2015. View Article : Google Scholar
|
|
26
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES, et al: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Barretina J, Caponigro G, Stransky N,
Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV,
Sonkin D, et al: The Cancer Cell Line Encyclopedia enables
predictive modelling of anticancer drug sensitivity. Nature.
483:603–607. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu Z, Fang S, Zuo Y, Zhang Y, Cheng R,
Wang Q, Yang Z, Cai W, Ma J, Yang X, et al: Combination of pigment
epithelium-derived factor with radiotherapy enhances the antitumor
effects on nasopharyngeal carcinoma by downregulating vascular
endothelial growth factor expression and angiogenesis. Cancer Sci.
102:1789–1798. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang L and Gu J: Risk factors for
symptomatic anastomotic leakage after low anterior resection for
rectal cancer with 30 Gy/10 f/2 w preoperative radiotherapy. World
J Surg. 34:1080–1085. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Stipa F, Chessin DB, Shia J, Paty PB,
Weiser M, Temple LK, Minsky BD, Wong WD and Guillem JG: A
pathologic complete response of rectal cancer to preoperative
combined-modality therapy results in improved oncological outcome
compared with those who achieve no downstaging on the basis of
preoperative endorectal ultrasonography. Ann Surg Oncol.
13:1047–1053. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Meimarakis G, Angele MK, Schneider C,
Weidenhagen R, Kalaitzis N, Molki A, Jauch KW, Hatz R and Winter H:
Impact of systematic lymph node dissection in the resection of
pulmonary metastases of solid extrapulmonary tumours. Zentralbl
Chir. 135:556–563. 2010.(In German). View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen J, Ye L, Zhang L and Jiang WG: The
molecular impact of pigment epithelium-derived factor, PEDF, on
lung cancer cells and the clinical significance. Int J Oncol.
35:159–166. 2009.PubMed/NCBI
|
|
33
|
Yamaguchi H, Bhalla K and Wang HG: Bax
plays a pivotal role in thapsigargin-induced apoptosis of human
colon cancer HCT116 cells by controlling Smac/Diablo and Omi/HtrA2
release from mitochondria. Cancer Res. 63:1483–1489.
2003.PubMed/NCBI
|
|
34
|
Dewhirst MW, Cao Y and Moeller B: Cycling
hypoxia and free radicals regulate angiogenesis and radiotherapy
response. Nat Rev Cancer. 8:425–437. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bhadada SV, Goyal BR and Patel MM:
Angiogenic targets for potential disorders. Fundam Clin Pharmacol.
25:29–47. 2011. View Article : Google Scholar
|
|
36
|
Eefsen RL, Engelholm L, Willemoe GL, Van
den Eynden GG, Laerum OD, Christensen IJ, Rolff HC, Høyer-Hansen G,
Osterlind K, Vainer B, et al: Microvessel density and endothelial
cell proliferation levels in colorectal liver metastases from
patients given neo-adjuvant cytotoxic chemotherapy and bevacizumab.
Int J Cancer. 138:1777–1784. 2016. View Article : Google Scholar
|
|
37
|
Yamada Y, Arao T, Matsumoto K, Gupta V,
Tan W, Fedynyshyn J, Nakajima TE, Shimada Y, Hamaguchi T, Kato K,
et al: Plasma concentrations of VCAM-1 and PAI-1: A predictive
biomarker for post-operative recurrence in colorectal cancer.
Cancer Sci. 101:1886–1890. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baylin SB and Ohm JE: Epigenetic gene
silencing in cancer - a mechanism for early oncogenic pathway
addiction? Nat Rev Cancer. 6:107–116. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ha YJ, Kim CW, Roh SA, Cho DH, Park JL,
Kim SY, Kim JH, Choi EK, Kim YS and Kim JC: Epigenetic regulation
of KLHL34 predictive of pathologic response to preoperative
chemoradiation therapy in rectal cancer patients. Int J Radiat
Oncol Biol Phys. 91:650–658. 2015. View Article : Google Scholar : PubMed/NCBI
|