Introduction
Breast cancer is one of the most frequent causes of
death among female population worldwide (1). Metastasis is responsible for >90%
of breast cancer mortalities but it is one of the least understood
stages of tumor development. Primary luminal A and B subtypes of
breast cancer represent 60% of the tumors (2). From the two luminal subtypes, luminal
B is the most aggressive tumor (2,3).
Following initial clinical response, 40–50% of these patients
present recurrence with metastases (4).
The crucial role of the tumor microenvironment in
cancer development and metastasis has recently been highlighted
(5). Virchow provided the first
evidence of the interaction between normal tissue and tumor
formation, postulating that cancer originates at sites of chronic
inflammation (6,7).
The tumor microenvironment and chronic inflammation
share several soluble molecules, such as cytokines, growth factors
and metalloproteases, as well as a variety of distinct cell types,
including endothelial cells (ECs) (8). Recruitment of ECs by tumors is
essential in metastasis during tumor vascularization and because
they regulate the intra- and extravasation of tumor cells (9). For a circulating tumor cell to exit
the circulatory system (extravasation), it must first bind to a
blood vessel wall by one of two mechanisms of arrest: physical
occlusion or cell adhesion. The relative prevalence of these
mechanisms depends on the biology of the tumor and the diameter of
the local post-capillary venule (10). The extravasation can vary depending
on the cancer cell type and the extravasation site or target organ,
suggesting that it is determined not only by the metastatic
potential of the tumor cell, but also by the endothelial response
to the unique local endothelial microenvironment (11). In this process soluble factors
derived from both cell types serve in the communication between
tumor and ECs.
During metastasis, ECs function not simply as static
structural cells of perfusing vessels but also as active stromal
regulatory cells with privileged access to the tumors (12). Inflammatory cytokines, including
TNF, IL-1, IL-6 and chemokines as IL-8, may also promote adhesion
and extravasation by increasing vascular permeability and promoting
the survival of tumor cells in the blood circulation. Adherent
tumor cells are dependent on chemokine and cytokine gradients to
direct their migration through EC monolayers. Several inflammatory
cytokines can act at a distance promoting a pro-adhesive phenotype
characterized by an increase of adhesion molecules on the apical
surface of ECs in target organs. Interestingly, a variety of
cytokines can be found in the circulation of cancer patients, and
the expression of chemokines and their receptors correlate with the
aggressiveness of the tumor (13).
The migratory arrest of cancer cells depends on the quality and
quantity of adhesion molecules expressed on ECs, as well as the
adhesion molecule repertoire on the cancer cells (9). In fact, in many cancer types, cell
adhesion molecules (CAMs) are frequently associated with metastatic
progression and adhesion to EC walls in distant organ sites
(14).
Changes in the gene expression of tumor-associated
endothelial cells (TAECs) have been postulated to affect cancer
cell fate (15,16). Analyzing the contribution of tumor
secreted factors to endothelial-recruitment in vivo has
proven to be difficult and conditioned medium (CM) secreted by
different tumor cell lines have been used as model systems,
promoting angiogenesis and a pro-adhesive phenotype in human
umbilical vein endothelial cells (HUVECs) as well as TNF (17–19).
Luminal A and B forms of breast cancer are the most common
presentations of the disease and despite the effective treatment;
the recurrence in the B subtype is associated with metastasis in
most of the patients. Therefore we analyzed the endothelial
transcriptome in response to CM from the human luminal B metastatic
breast cancer cell line ZR75.30. Bioinformatic analysis implicated
NFκB as a key molecular regulator of the vascular pro-adhesive
phenotype activated by CM dominating over other cytokine, chemokine
and growth factor signaling pathways.
Materials and methods
Cell culture
The breast cancer cell lines MCF-7 (luminal A),
ZR75.30 (luminal B) and the monocyte cell line U937 were obtained
from ATCC (Manassas, VA, USA) and maintained in RPMI-1640 medium
supplemented with 10% FBS (both from Gibco-BRL, Grand Island, NY,
USA), penicillin 10,000 U/ml, streptomycin 10 mg/ml and
amphotericin B 25 μg/ml (PAA Laboratories GmbH/GE Healthcare
Bio-Sciences Corp., Piscataway, NJ, USA) in a humidified atmosphere
of 5% CO2 at 37°C.
HUVEC primary culture
HUVECs were isolated and cultured as previously
reported (20) by mixing cells
from at least three human umbilical cords. The resulting cell
cultures were maintained in M199 medium (Gibco-BRL) supplemented
with 10% FBS, 2 mM L-glutamine (Gibco-BRL), 20 μg/ml endothelial
mitogen (Biomedical Technologies, Inc., Stoughton, MA, USA), 5 U/ml
heparin (Laboratorios PISA S.A. de C.V), penicillin 10,000 U/ml,
streptomycin 10 mg/ml and amphotericin B 25 μg/ml, under a
humidified atmosphere of 5% CO2 at 37°C. All HUVEC
cultures used for the experiments were at the third passage. The
local Ethics and Research Committees of the Hospital General Dr
Manuel Gea González, Ministry of Health (Mexico) approved this
protocol (11-62-2014), and all participants signed an informed
written consent form.
Conditioned medium
CM was isolated as previously described (17). Briefly, breast cancer cell lines
were cultured in 100-mm plates until they reached 80% confluence.
The cell layer was first washed 10 times with 10 ml of
PBS/RPMI-1640 (1:1 v/v) without phenol red (Laboratorios Microlab
S.A. de C.V., D.F. Mexico, Mexico) to remove serum components.
Then, cells were maintained in 8 ml of serum-free RPMI without
phenol red, after 48 h the culture medium was collected and
lyophilized. The resulting powder was dissolved in water (1/10 of
the original volume) and dialyzed using a PM-3 Ultrafiltration
Membrane (EMD Millipore, Billerica, MA, USA). The solution was
filtered through a 0.22-μm Millex-GS syringe filter unit, and a
protease inhibitor cocktail was added (cOmplete™ Protease Inhibitor
Cocktail; Roche Applied Science, Indianapolis, IN, USA). The
protein concentration was determined using the Bradford reagent
assay (Bio-Rad, Hercules, CA, USA). The resulting concentrated
preparation was maintained at 4°C until further use.
Bio-Plex assay
CM (50 μl), was analyzed with the Bio-Plex
suspension array system (Bio-Rad) against 26 proteins, following
the manufacturer's instructions.
Sample treatment
For adhesion assay, microarrays and western blots,
confluent HUVECs were treated with 9 μg/ml of the indicated CM or
with 10 ng/ml of human recombinant TNF (R&D Systems, Inc.,
Minneapolis, MN, USA) for the indicated time frames depending on
the experiment. At the end of the treatments, the corresponding
assay was performed as described. Inhibition of IKKs was performed
by pre-incubating HUVECs for 1 h with 10 μM BAY 11-7085
(Calbiochem/Merck KGaA, Darmstadt, Germany). After pre-incubation,
HUVECs were stimulated with 9 μg/ml of ZR75.30-CM. Electrophoretic
mobility shift assay (EMSA) and IκBα western blotting were
performed after 20 min, while CAM western blotting and cell
adhesion were evaluated after 3 h, as described. For time course
assay, HUVECs were starved 4 h previous to stimulation with 9 μg/ml
of ZR75.30-CM, and cell lysates were used for western blot
analysis.
Adhesion assay
This assay was performed as previously described
(17), adherent cells were
visualized in a TMS-F phase-contrast inverted microscope Nikon
Eclipse TS100 (Nikon Instruments, Inc., Melville, NY, USA) and
counted in a β counter (1600TR liquid scintillation analyzer;
Canberra-Packard, Meriden, CT, USA).
RNA isolation and microarrays
TRIzol reagent (Invitrogen Corp., Camarillo, CA,
USA) was used to obtain total RNA from three independent biological
replicates of confluent HUVECs (60-mm plates) treated as indicated.
The preparation of cRNA hybridization to Human Gene 1.0 ST and data
analysis were performed according to Affymetrix™ recommendations.
Differentially expressed genes were determined using the
Partek® Genomics Suite software (Partek, Inc., St.
Louis, MO, USA) with a p<0.05 and a differential fold change of
1.5 on either positive or negative directions. Gene Ontology (GO)
classification was performed through the use of National Cancer
Institute-Database for Annotation, Visualization and Integrated
Discovery (NCI-DAVID) (http://david.abcc.ncifcrf.gov) (21) and the Search Tool for the Retrieval
of Interacting Genes (STRING) software was used to build the
functional gene association networks (string-db. org) (22). Enriched canonical pathways within
the networks of differentially expressed genes were carried out
using the Ingenuity Pathway Analysis (IPA) (Ingenuity®
Systems; www.ingenuity.com) and Protein ANalysis
THrough Evolutionary Relationships (PANTHER) software (www.pantherdb.org) (23).
RT-qPCR
The generation of cDNA was performed using the First
Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Inc., Waltham,
MA, USA) with 2 μg of total RNA as a template. PCR reactions were
performed with Maxima SYBR-Green/ROX qPCR Master Mix (Thermo Fisher
Scientific, Inc.) on a 7300 Real-Time PCR System (Applied
Biosystems, Foster City, CA, USA) using a conventional
amplification protocol. The housekeeping gene ACTIN was used
as an internal control. Primer sequences used for gene expression
analysis are shown in Table I. The
data were analyzed using the 2− Δ ΔCq method (24).
 | Table IPrimer sequences employed for
qPCR. |
Table I
Primer sequences employed for
qPCR.
| Human gene | Forward primer
5′→3′ | Reverse primer
5′→3′ | Accession no. |
|---|
| ACTIN |
TCCCTGGAGAAGAGCTACGA |
AGCACTGTGTTGGCGTACAG | NM_001101 |
| ICAM-1 |
GACCAGAGGTTGAACCCCAC |
GCGCCGGAAAGCTGTAGAT | NM_000201 |
| SELE |
CCGAGCGAGGCTACATGAAT |
GCATCGCATCTCACAGCTTC | NM_000450 |
| VCAM-1 |
TGTTTGCAGCTTCTCAAGCTTTTA |
GTCACCTTCCCATTCAGTGGA | NM_001078 |
| NFKBIA |
CTCCGAGACTTTCGAGGAAATAC |
GCCATTGTAGTTGGTAGCCTTCA | NM_020529 |
| CCL20 |
TGCTGTACCAAGAGTTTGCTC |
CGCACACAGACAACTTTTTCTTT | NM_004591 |
| TNFAIP2 |
GGCCAATGTGAGGGAGTTGAT |
CCCGCTTTATCTGTGAGCCC | NM_006291 |
| TRAF1 |
TCCTGTGGAAGATCACCAATGT |
GCAGGCACAACTTGTAGCC | NM_005658 |
| CXCL2 |
TGCCAGTGCTTGCAGAC |
TCTTAACCATGGGCGATGC | NM_002089 |
| PPP1R3C |
GGTGGCACAGATAGTGATACCT |
ACCATCATTGTTGTCCCAAAAGA | NM_005398 |
| IL-6 |
ACTCACCTCTTCAGAACGAATTG |
CCATCTTTGGAAGGTTCAGGTTG | NM_000600 |
| MAP3K8 |
CTCCCCAAAATGGACGTTACC |
GGATTTCCACATCAGATGGCTTA | NM_005204 |
| CDKN1B |
TAATTGGGGCTCCGGCTAACT |
TGCAGGTCGCTTCCTTATTCC | NM_004064 |
| NFKB2 |
GGGCCGAAAGACCTATCCC |
CAGCTCCGAGCATTGCTTG | NM_002502 |
| TGFB3 |
GGAAAACACCGAGTCGGAATAC |
GCGGAAAACCTTGGAGGTAAT | NM_003239 |
| SORBS1 |
CACAATCGAGAACAGCAAAAACG |
ACCCGCCTACTGTCATCCTTT | NM_001034954 |
Western blotting
Confluent cultures of HUVECs treated as indicated
were lysed using RIPA buffer (150 mM NaCl, 50 mM Tris-HCl pH 7.4,
1% NP-40, 0.25% sodium deoxycholate, 1 mM EDTA pH 8.0, 1 mM PMSF,
1X complete protease inhibitors cocktail, 2 mM
Na3VO4, 10 mM Na2MoO4
and 5 mM NaF) and protein concentration was determined by Bio-Rad
DC protein assay (Bio-Rad). Total cellular proteins (10–40 μg) were
separated via SDS-PAGE, transferred onto PVDF (EMD Millipore),
membranes blocked for 1 h at room temperature in TBS-Tween 0.1%,
with 5% non-fat milk and probed overnight at 4°C with specific
antibodies against anti-VCAM-1, ICAM-1, E-selectin, IκBα, STAT3
(all from Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA),
phospho-STAT3 (Y705), phospho-VEGFR2 (Y1175), and VEGFR2 (all from
Cell Signaling Technology, Inc., Danvers, MA, USA) and β-actin
(Sigma-Aldrich, St. Louis, MO, USA). The membranes were incubated
in the presence of HRP-conjugated secondary antibodies for 1 h at
room temperature (Sigma-Aldrich), and the signals were detected
using enhanced chemiluminescence (SuperSignal West Pico
Chemiluminescent Substrate; Thermo Fisher Scientific, Inc.). The
membranes were stripped as reported by Yeung and Stanley (25) and re-blotted. Optical densitometric
scanning was performed using NIH ImageJ software.
Electrophoretic mobility shift assay
Nuclear protein extracts and cytoplasmic fractions
from HUVECs and NFκB translocation to the nucleus were determined
as previously described (26).
Briefly, 10 μg of the nuclear protein extracts were incubated with
γ-32P-ATP-labeled oligonucleotide containing the
consensus NFκB site (5′-AGTTGAGGGGACTTTCCCAGGC-3′) (Santa Cruz
Biotechnology, Inc.) in the presence of 100-fold excess of
unlabeled specific probe as specific competitor. Samples were
fractionated on a 5% non-denaturing polyacrylamide gel in 1X
Tris-borate-EDTA buffer and DNA-protein complexes visualized on a
Storm PhosphorImager (Molecular Dynamics, San Francisco, CA, USA).
For supershift analysis, 1 μg of anti-p65 antibody (Santa Cruz
Biotechnology, Inc.) was incubated with the nuclear extract for 30
min at room temperature prior to adding the reaction mixtures.
Statistical analysis
The results are expressed as the means ± SE, and
statistical analyses were performed with GraphPad Prism 5.0
software (GraphPad Software, Inc., San Diego, CA, USA). Multiple
comparisons were analyzed using one-way ANOVA with Dunnett's
post-hoc test and two-way ANOVA with Bonferroni correction.
Significance was set at p<0.05.
Results
Differentially expressed genes associated
with the pro-adhesive endothelial phenotype induced by
ZR75.30-CM
Previous study by our group (18) showed that CM secreted from the
breast cancer ZR75.30 cells promotes a pro-adhesive endothelial
phenotype. Interestingly, CM secreted from the breast cancer MCF-7
cells did not promote this phenotype nor induced the expression of
CAMs: ICAM-1, VCAM-1 and E-selectin, therefore we focus on
characterizing the transcriptome of ECs treated with ZR75.30-CM
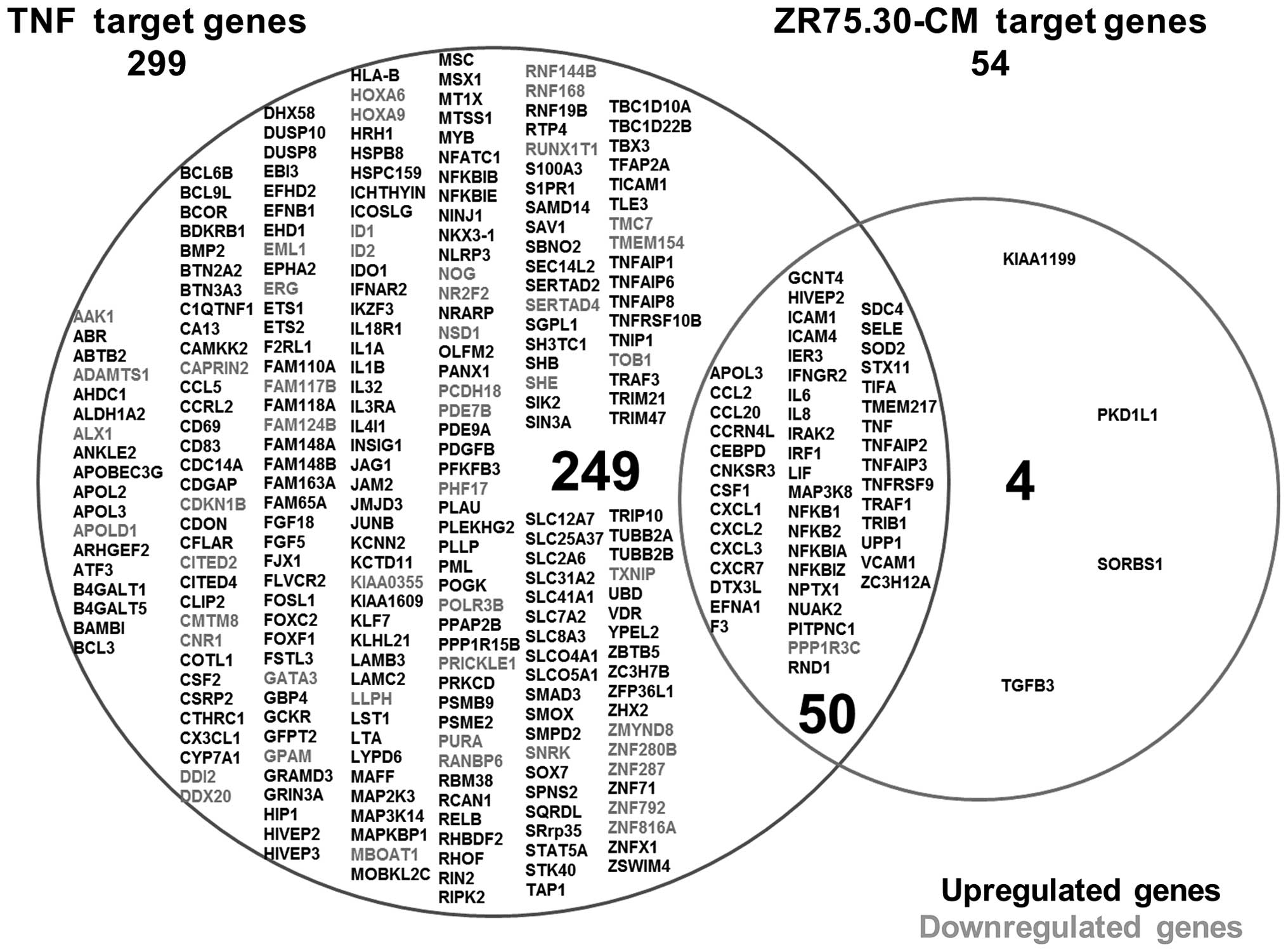
(Fig. 1).
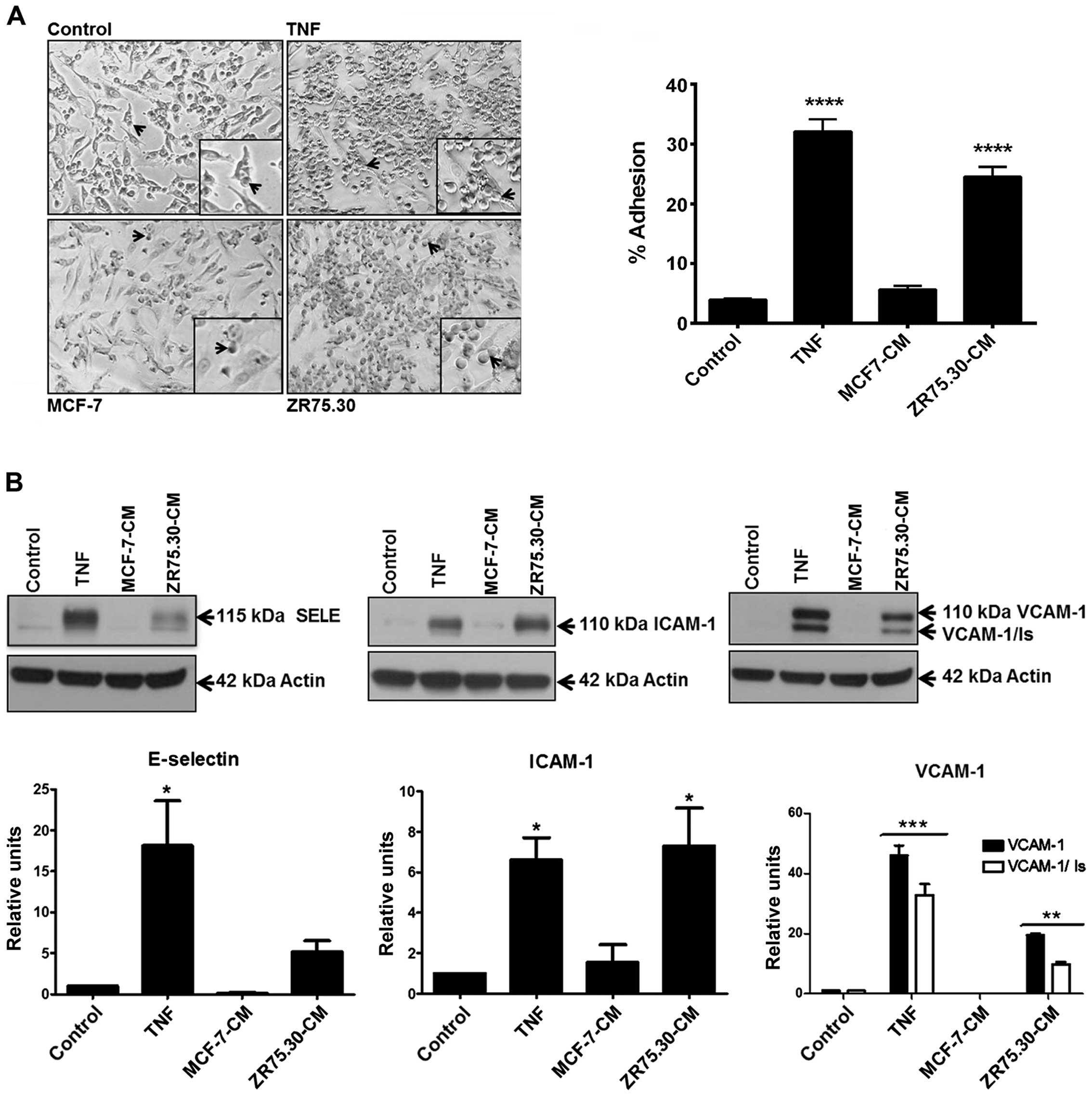 | Figure 1ZR75.30-CM promotes a pro-adhesive
phenotype and expression of CAMs in HUVECs. (A) Micrographs of the
adhesion assay. Control represents basal adhesion of U937 cells to
confluent, untreated HUVECs. HUVECs were pre-treated for 3 h with
TNF (10 ng/ml), ZR75.30-CM (9 μg/ml) or MCF-7-CM (9 μg/ml) prior to
the addition of U937 cells to the HUVEC monolayer. The black arrows
indicate U937 cells adhered to HUVECs. The micrographs were taken
with ×200 magnification. The percentage of U937 cells adhered to
HUVECs was obtained as previously described (14). Data are presented as the means ± SE
of the percentage of the total adherent cells in at least three
independent experiments. ****p<0.0001. (B) A
representative western blotting for ICAM-1, E-selectin and VCAM-1
adhesion molecules employing total HUVEC extracts from cells
treated for 3 h with MCF-7-CM (9 μg/ml), ZR75.30-CM (9 μg/ml) or
TNF (10 ng/ml). Untreated HUVEC extracts were employed as controls.
Actin was used as a loading control. Histograms represent the means
(ratio ICAM-1/actin, E-selectin/actin or VCAM-1/actin) ± SE of
three independent experiments and are expressed as relative units.
*p<0.05, **p<0.01,
***p<0.001. CM, conditioned medium; CAMs, cell
adhesion molecules; HUVECs, human umbilical vein endothelial
cells. |
For the transcriptome analysis a new batch of CM was
tested for induction of the pro-adhesive phenotype in ECs and for
the content of soluble factors using a multiplex assay. We found
enrichment in the inflammatory cytokines TNF, IFN-γ and IL-6, the
hematopoietic cytokine G-CSF, the chemokine IL-8, the growth factor
VEGF and, to a lesser extent, the anti-inflammatory cytokine IL-1Ra
for ZR75.30-CM. MCF-CM contained lower amounts of these factors.
The molecules with the highest differences between ZR75.30-CM and
MCF-7-CM were VEGF (>39,592 vs. 14,231 pg/ml), G-CSF (>28,728
vs. 9,594 pg/ml), IL-8 (>24,800 vs. 8,429 pg/ml), IL-6 (2,891
vs. 1,081 pg/ml), IFN-γ (2,068 vs. 1,315 pg/ml) and TNF (1,274 vs.
771 pg/ml). For VEGF, G-CSF and IL-8 from ZR75.30-CM, the values
were above of detection range. Results from the composition
analysis were similar to those previously reported (18). We used TNF as a positive control
for the induction of a pro-adhesive endothelial phenotype
accompanied by CAM expression. ZR75.30-CM altered the expression of
54 genes in ECs (53 upregulated and 1 down-regulated), whereas TNF
treatment altered the expression of 299 genes (249 upregulated and
50 downregulated). Of the total number of genes affected, 50 were
common to both treatments, 249 were exclusive to the TNF treatment
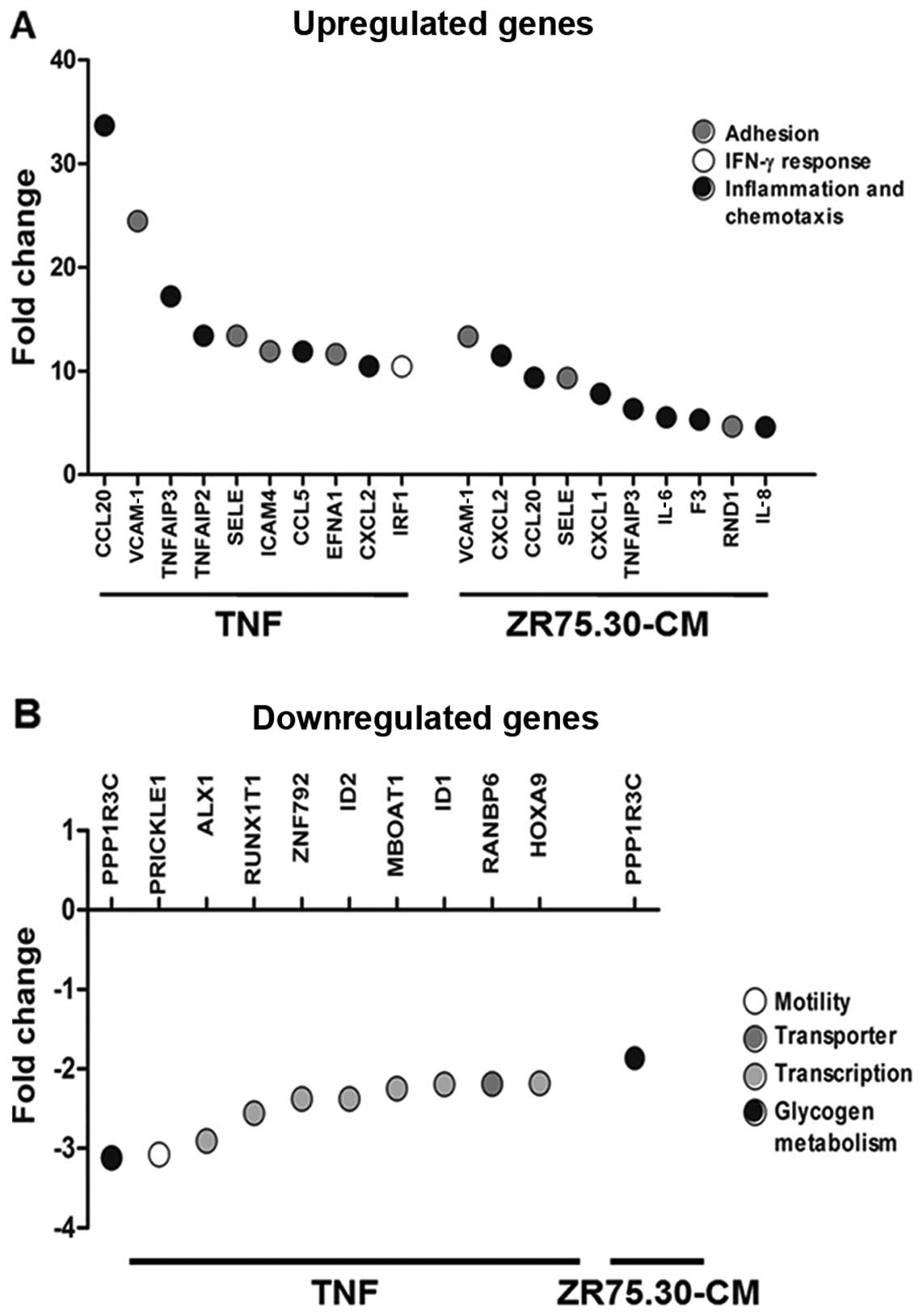
and 4 were exclusive to the ZR75.30-CM treatment (Fig. 2). Among the 10 genes most
upregulated by ZR75.30-CM, are genes related to chemotaxis and
inflammation (CCL20, CXCL2, CXCL1, F3,
IL-6, IL-8 and TNFAIP3) and cell adhesion
(VCAM-1, SELE and RND1). Likewise, the most
upregulated genes after TNF treatment correspond to cell adhesion
process (VCAM-1, SELE, ICAM-4 and
EFNA1), chemotaxis and inflammation (CCL20,
CCL5, CXCL2, TNFAIP3 and TNFAIP2) and
IFN-γ (IRF1) (Fig. 3A). All
genes affected by both treatments showed significantly higher
expression levels with TNF compared to those with ZR75.30-CM with
the exception of CXCL2, for which ZR75.30-CM induced higher
expression (Fig. 3A). The genes
most repressed by TNF were PPP1R3C, PRICKLE1,
ALX1, RUNX1T1, ZNF792, ID2,
MBOAT1, ID1, RANBP6 and HOXA9.
Interestingly, PPP1R3C showed the strongest down-regulation
with TNF and it was the only downregulated gene with ZR75.30-CM
(Fig. 3B).
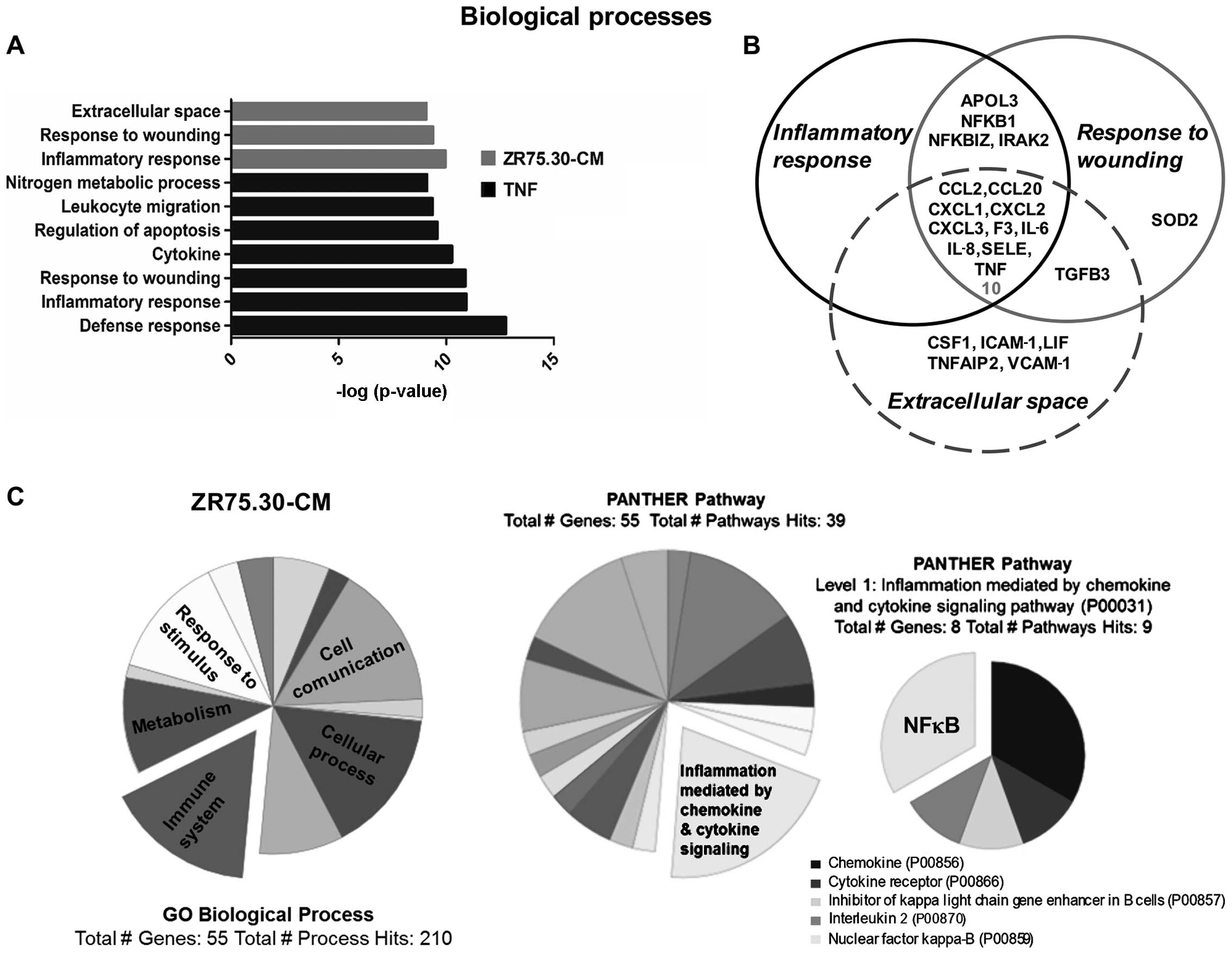
Bioinformatic analysis implicates the
NFκB pathway as a central regulator of the gene expression pattern
in ECs treated with ZR75.30-CM
The induction of a pro-adhesive phenotype directed
the initial approach of this study, however, the bioinformatic
analysis of the microarray by NCI-DAVID software associated the
expression profile with other cellular processes such as:
inflammatory response (p-values of 1.1e–10 for ZR75.30-CM treatment
and 1.2e–11 for TNF), response to wounding (p-value of 4.1e–10 for
ZR75.30-CM treatment and 1.3e–11 for TNF) and extracellular space
(8.5e–10 for ZR75.30-CM) (Fig.
4A). While SELE was present in the three processes identified
by NCI-DAVID, ICAM-1 and VCAM-1 were present only in
the extracellular space process. The genes associated to these
processes also shared chemokines, cytokines and components of the
NFκB pathway known to regulate and promote a pro-adhesive phenotype
(Fig. 4B).
In a similar analysis using PANTHER software the
most representative biological process was the immune system
(Fig. 4C). Among the main pathways
identified by IPA software we found granulocyte adhesion and
diapedesis (p-value of 1.72e–12) as part of ZR75.30-CM treatment
(Table II). NFκB emerged as the
principal regulatory molecule with the highest score (p-value of
8.93e–38) in ZR75.30-CM treatment analyzed by IPA and PANTHER
(Table III and Fig. 4C). Bioinformatic analysis of
TNF-treated ECs also showed NFκB as the main regulator and shared
some biological processes and pathways with ZR75.30-CM treatment
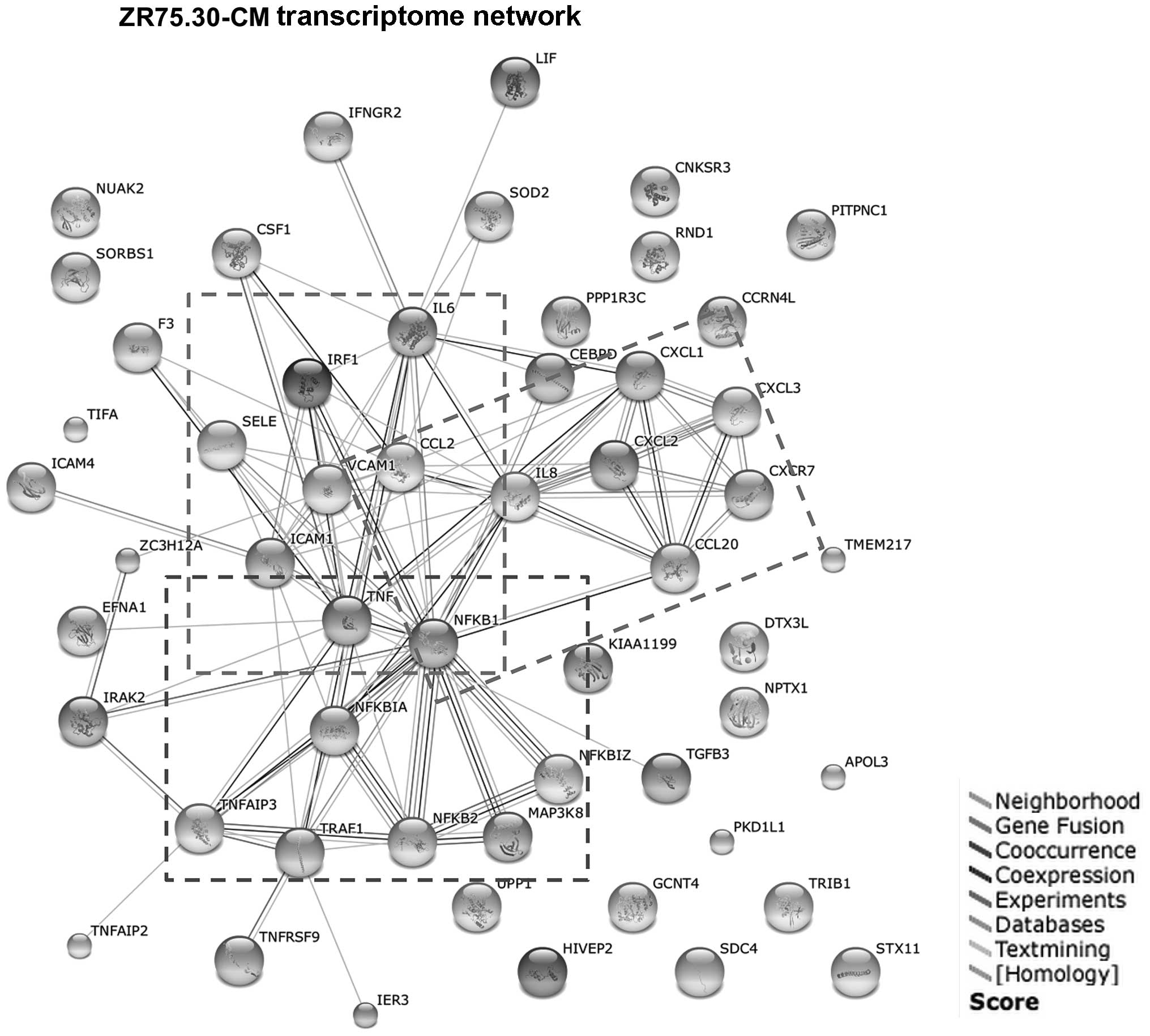
(data not shown). Finally, we generated a functional gene
association network with the STRING software. The resulting network
(Fig. 5) confirmed a cluster of
interactions between TNF, NFKB, and CAMs and
revealed a second cluster of interactions among chemokines and
cytokines. These two clusters are interconnected through
TNF, NFKB, IL-6, IL-8, CCL2 and
CCL20.
 | Table IITop five canonical pathways altered
in HUVEC microarrays with each treatment according to IPA
analysis. |
Table II
Top five canonical pathways altered
in HUVEC microarrays with each treatment according to IPA
analysis.
| IPA analysis |
|---|
|
|---|
| Pathways | P-value | No. of genes |
|---|
| ZR75.30-CM |
| Hepatic
fibrosis/hepatic stellate cell activation | 3.46e–15 | 12 |
| Atherosclerosis
signaling | 3.34e–14 | 11 |
| Granulocyte
adhesion and diapedesis | 1.72e–12 | 11 |
| Role of
macrophages, fibroblasts and | 2.01e–12 | 13 |
| ECs in rheumatoid
arthritis |
| Role of IL-17A in
arthritis | 2.18e–12 | 8 |
| TNF |
| Role of
macrophages, fibroblasts and | 1.5e–15 | 29 |
| ECs in rheumatoid
arthritis |
| Hepatic
fibrosis | 2.03e–12 | 18 |
| TNFR2
signaling | 3.93e–12 | 10 |
| Role of IL-17A in
arthritis | 1.49e–11 | 12 |
| TREM1
signaling | 5.92e–11 | 13 |
 | Table IIITop five molecular regulators altered
in HUVEC microarrays with each treatment when analyzed by IPA or
PANTHER software. |
Table III
Top five molecular regulators altered
in HUVEC microarrays with each treatment when analyzed by IPA or
PANTHER software.
| IPA analysis | PANTHER
analysis |
|---|
|
|
|---|
| Regulators | P-value | No. of genes | Regulators | No. of genes |
|---|
| ZR75.30-CM |
| NFκB complex | 8.93e–38 | 32 | NFκB | 3 |
| TNF | 1.29e–35 | 40 | Chemokines | 2 |
| IL-1B | 1.14e–34 | 33 | Cytokine
receptor | 1 |
| TRADD | 1.98e–33 | 15 | NFKBIA | 1 |
| NFKBIA | 1.79e–32 | 27 | IL-2 | 1 |
| TNF |
| TNF | 1.11e–61 | 119 | NFκB | 7 |
| NFκB complex | 2.42e–56 | 77 | Chemokines | 4 |
| IL-1B | 3.50e–42 | 74 | IL-2 | 4 |
| LPS | 5.49e–41 | 99 | Cytokine
receptor | 2 |
| CD40LG | 1.20e–39 | 54 | NFKBIA | 2 |
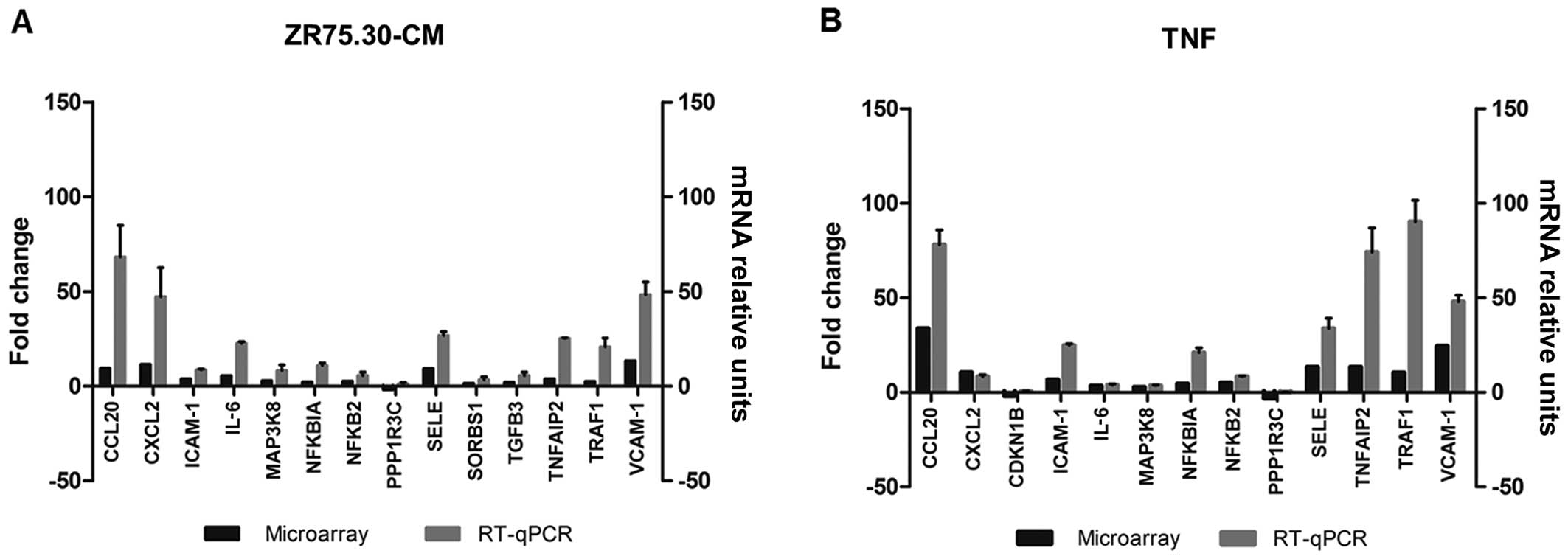
Validation of the transcriptomic response
of ECs to ZR75.30 breast cancer CM
To validate the data obtained in the micro-array and
to compare the expression levels of selected genes between
treatments, we used real-time PCR to quantify the mRNA of 15 genes
involved in processes like cell adhesion (VCAM-1,
SELE, ICAM-1), NFκB pathway (NFKBIA,
NFKB2, MAP3K8, CDKN1B), chemotaxis and
inflammation (CXCL2, CCL20, TGFB3,
IL-6, TNFAIP2, TRAF1) and metabolism
(PPP1R3C, SORBS1). Two of these genes were induced
exclusively with ZR75.30-CM (TGFB3 and SORBS1), one
with TNF (CDKN1B) which was repressed. Real-time PCRs
validated the overexpression of the genes reported in the
microarray. However, downregulation of PPP1R3C and
CDKN1B genes observed in the microarray analysis was not
replicated in the qPCR assays, although their expression level was
lower than control for either TNF or ZR75.30-CM treatment (Fig. 6).
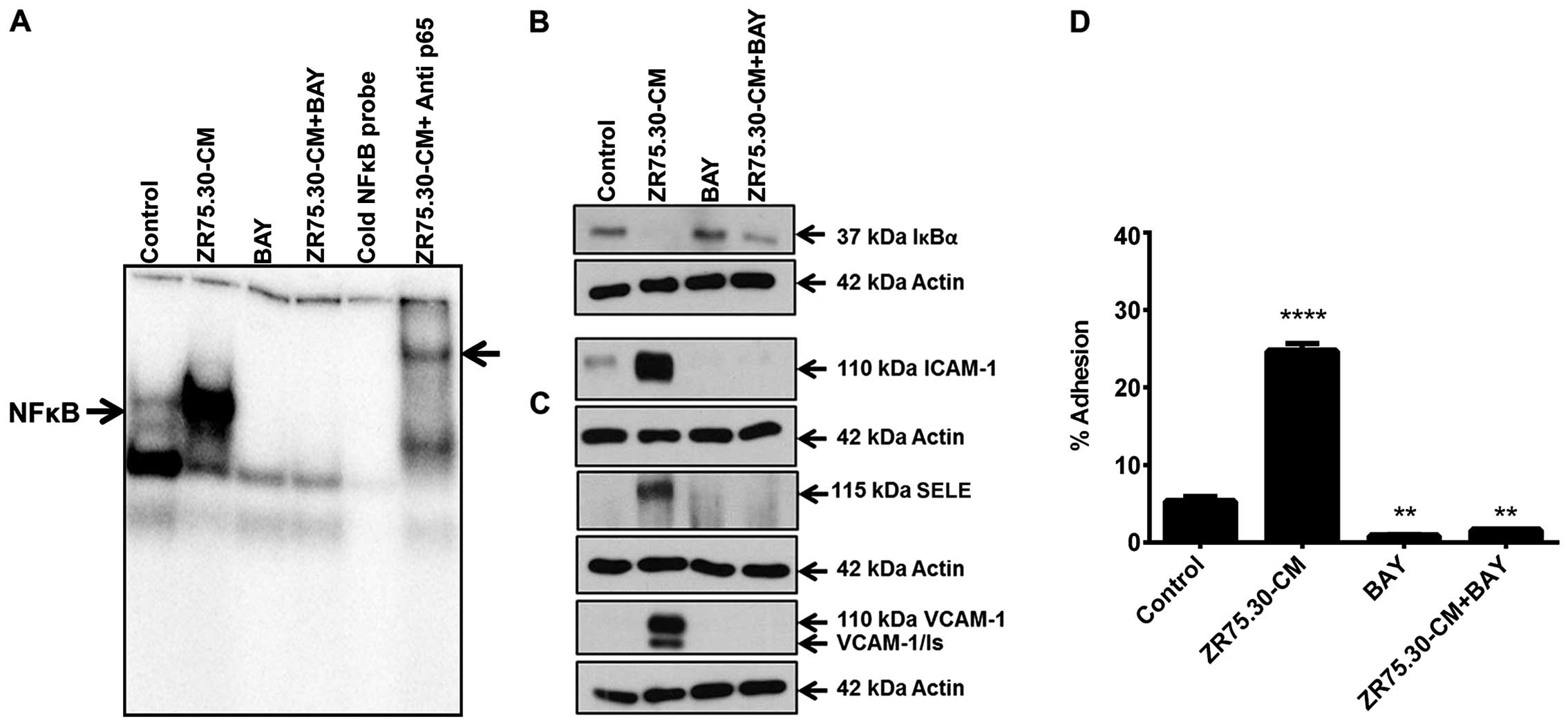
Mechanistic relevance of canonical NFκB
pathway on the pro-adhesive endothelial phenotype in response to
ZR75.30 breast cancer CM
As NFκB emerged as the principal regulator of gene
expression changes after ZR75.30-CM treatment associated to the
pro-adhesive phenotype, and to test the relevance of this pathway
we used a pharmacological inhibition of IκBα phosphorylation by
pre-treating ECs with BAY 11-7085 prior to stimulation with
ZR75.30-CM. EMSA revealed a faint signal from a basal NFκB/DNA
complex present in control cells. The presence of this complex
increased significantly when ECs were treated with ZR75.30-CM
(first and second lanes, respectively). The basal complex
disappeared when ECs were treated with the inhibitor (third lane)
and was barely visible in cells treated with ZR75.30-CM plus
inhibitor (fourth lane). Excess unlabeled NFκB probe completely
eliminated the signal, indicating the specific detection of this
complex (fifth lane) (Fig. 7A).
Supershift assays confirmed that ZR75.30-CM triggered canonical
NFκB activation, evidenced by markedly enhanced supershifted band
in the presence of anti-p65 (sixth lane) (Fig. 7A). Western blotting against IκBα
showed that the basal level of expression in control cells
disappeared when treated with ZR75.30-CM (first and second lane,
respectively). A slight reduction in IκBα expression was observed
in the presence of the inhibitor and CM plus inhibitor (third and
fourth lanes) (Fig. 7B). Western
blotting revealed that the expression of ICAM-1, VCAM-1 and
E-selectin increased with ZR75.30-CM treatment and that the
expression was prevented when the cells were treated with the
inhibitor (third and fourth lanes) (Fig. 7C). Finally, we confirmed the
importance of the NFκB pathway in the adhesion process by showing
that the pro-adhesive phenotype induced by ZR75.30-CM was prevented
when the cells were treated with the inhibitor (Fig. 7D).
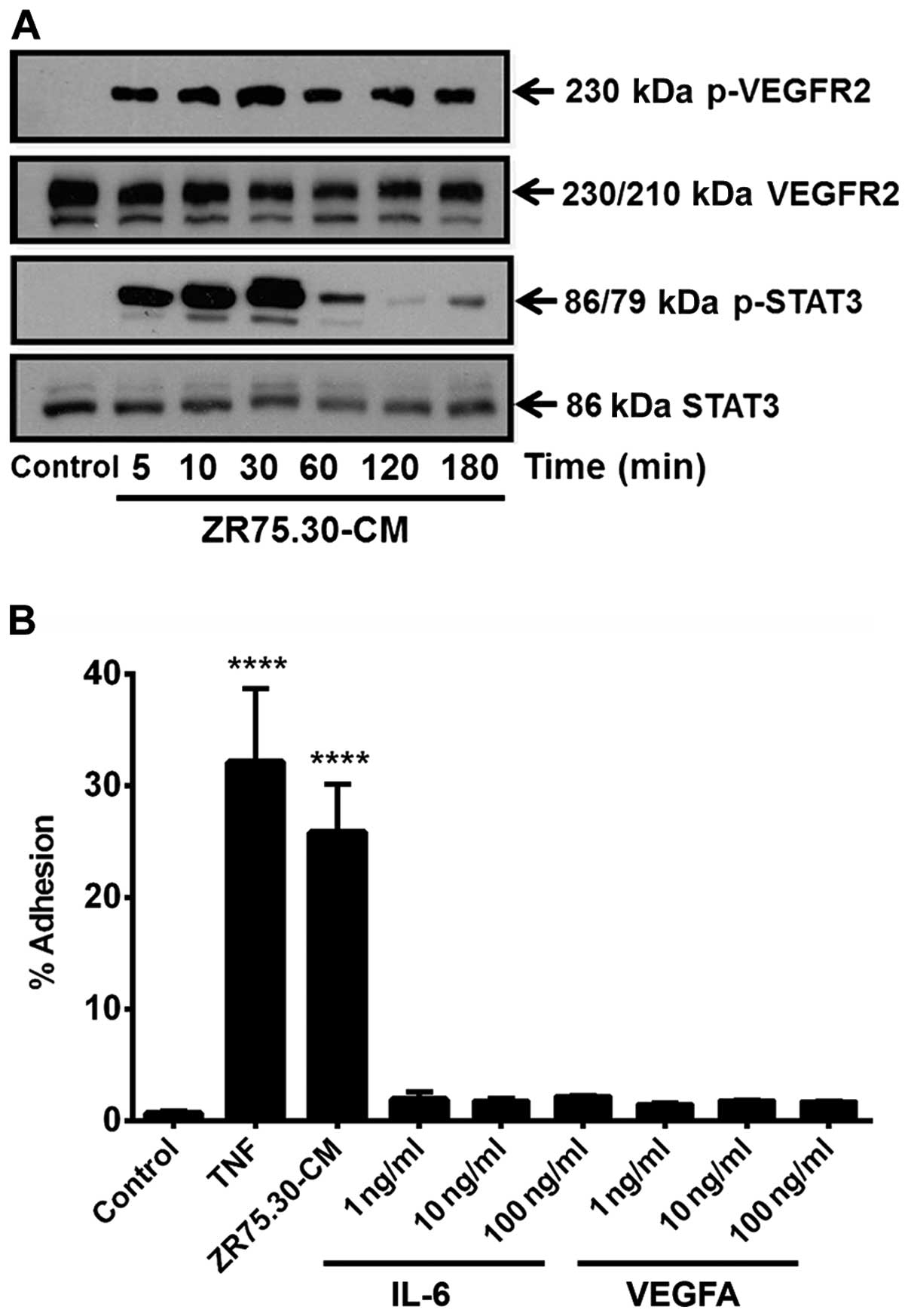
ZR75.30-CM activates early signaling
events related to VEGF and STAT3 pathway
Despite the enrichment in VEGF (>39,592 pg/ml),
G-CSF (>28,728 pg/ml), IL-8 (>24,800 pg/ml) and IL-6 (2,891
pg/ml) in ZR75.30-CM that could affect endothelial gene expression
through STAT3, microarray analysis did not contain classic target
genes related to these pathways. We verified that early signaling
events of these systems were not impaired by analyzing the state of
phosphorylation at residues related to functional activation of
VEGFR2 (Y1175) and STAT3 (Y705) (Fig.
8A). In response to ZR75.30-CM VEGFR2 phosphorylation presents
a biphasic response between 5–180 min. The signal presented a first
peak after 5 min that reached a maximum at 30 min and returned to
the value of the first peak after 1 h. STAT3 phosphorylation also
presented a biphasic response between 5–180 min with a maximum
signal at 30 min that became faint close to 120 min. Taken together
these results indicate that early signaling of these two systems
was not affected. However, when we analyzed the adhesion of HUVECs
in response to recombinant human cytokines IL-6 and VEGF the
adhesion was not significant (Fig.
8B).
Discussion
The tumor cell secretome consists of a complex
mixture of cytokines and growth factors that contribute to the
microenvironment associated with malignancy. These include
paracrine and juxtacrine signals that may be involved at virtually
any stage of tumorigenesis (27).
These tumor soluble factors contribute to recruiting normal stromal
cells into actions that favor tumorigenesis, such as normal ECs for
the intra- and extravasation processes relevant in metastatic
dissemination. Hematogenic dissemination of metastatic cells ends
once the tumor cells attach to ECs in the target organs. Following
adhesion to the apical membrane of the ECs, successful metastasis
requires extravasation followed by metastatic cell proliferation in
the stroma. In cell adhesion and extravasation many ligand-receptor
interactions contribute to these processes, the endothelial
repertoire of CAMs involved includes: selectins, integrins,
cadherins, CD44 and members of the superfamily of CAMs (28). How normal cells integrate and
priorize the information of a mixture of molecules present in the
tumor microenvironment in vivo has been difficult to
approach. However, treatment of normal or cancer cells with CM has
been a useful strategy to perform this kind of studies (29). Considering that pro-inflammatory
cytokines can induce a pro-adhesive endothelial phenotype, we
postulated that the endothelial transcriptome associated to this
phenotype induced by tumor secreted factors from the ZR75.30 breast
cancer cell line could be similar to that induced by
pro-inflammatory cytokines.
Microarray analysis was applied to further
characterize this pro-adhesive endothelial state. Bioinformatic
analysis with NCI-DAVID identified overlapping gene ontology
profiles associated to inflammatory response, wound healing and
extracellular space that was consistent with immune system and cell
communication processes identified by PANTHER analysis. NFκB was
identified as the principal molecular regulator by both IPA and
PANTHER analysis. STRING gene interaction network analysis
confirmed NFκB as a central hub related to CAMs and also revealed
its connection with NFκB through IL-6, IL-8,
CCL2 and CCL20. Interestingly CCL20 was the
third highest gene expressed in response to ZR75.30-CM and the
highest in response to TNF (Fig.
3).
These findings suggest that the EC expression
profile in response to CM includes a group of genes associated to a
transcriptome induced by inflammatory cytokines and moreover, this
response appears to be regulated by NFκB.
TNF is recognized as a classic inducer of a
pro-adhesive endothelial phenotype through NFκB activation
(30–32); however, IL-6, IL-8, IFN-γ and VEGF
can also promote indirect NFκB activation (33–35).
Early STAT3 phosphorylation was confirmed in response to ZR75.30-CM
indicating intact signaling capacity (Fig. 8A). Similarly, the signaling
activity of VEGF present in ZR75.30-CM was confirmed when we
followed the phosphorylation state of VEGFR2 (Fig. 8A). However, these cytokines did not
induce a significant pro-adhesive response compared to ZR75.30-CM
or TNF (Fig. 8B). The
NFκB-dependent transcription appears to be dominant over other
transcription-initiating pathways after 3 h and shows a partial
overlap with VEGF-dependent transcriptome in HUVECs at 0.5–6 h of
exposure to VEGF. The genes shared between VEGF and ZR75.30-CM are:
F3, SELE, CEBPD, CXCL2, IL-8,
NFKBIZ, CXCL1, CXCL3, MAP3K8,
CCL2, VCAM-1, HIVEP2 and CNKSR3
(36). In the case of VEGF
transcriptome related to endothelial proliferation is probably a
later event in time (24 h) and are therefore absent at the time
point analyzed (3 h). In contrast, IL-6 and IL-8 pathways appear in
our bioinformatic analysis (positions 28 and 45, respectively) with
p<e-12. In fact, crosstalk between NFκB/STAT3 and NFκB/VEGF
pathways has been reported (35,37)
suggesting that a signaling crosstalk converged through NFκB gene
expression in our model, perhaps which might priorize gene
expression of this master regulator. A similar transcriptional
dominance was reported in lymphatic ECs treated with MDA-MB231-CM
were, the STAT3 activation prevailed (29).
The transcriptome induced in ECs by ZR75.30-CM
shares 93% of its transcripts with TNF. Only four genes (7%) were
exclusive to the ZR75.30-CM treatment: KIAA1199,
PKD1L1, SORBS1 and TGFB3. In breast cancer,
increase of KIAA1199 expression correlated with
hypomethylation and NFκB binding in the KIAA1199 regulatory
region (38). In contrast,
KIAA1199 repression in colon cancer cells attenuates the Wnt
pathway and reduces proliferation (39). Chimeric products of PKD1L1
with RIF1 have been identified in breast cancer cells
(40). The function of
SORBS1 is related to lipid anabolism and this protein is
also associated with FAK and is a substrate of the c-Abl tyrosine
kinase (41). TGFB3 is
important in cellular differentiation and development, and in ECs,
this isoform participates in homeostasis and maintenance when cells
are subjected to shear stress (42). Hence, SORBS1 and
TGFB3 appear to represent specific genes related to changes
in cellular behavior that promote intercellular interactions.
Although the bioinformatic analysis indicates
TNF/NFκB-activated pathway, only 50 of the 299 genes affected by
recombinant TNF responded to ZR75.30-CM treatment (Fig. 3), suggesting that concomitant
stimulation with the mixture restricted the expression pattern. The
majority of the overlapping genes had the highest fold change with
the TNF treatment. Among the genes repressed by TNF, the
phosphatase PPP1R3C had the highest score, and was the only
gene repressed in response to ZR75.30-CM treatment. IPA analysis
indicates that the repression of this phosphatase is linked to TNF.
Overall, the expression changes induced by ZR75.30-CM have a
smaller magnitude than those induced by TNF.
There are several microarrays of ECs treated with
different cytokines and growth factors. These treatments include
TNF (31,43), IL-1 (44) and VEGF (36,45);
however, expression profiles of ECs treated with IL-6, IL-8 and
IFN-γ are scarce, and transcriptome data for ECs exposed to
components of the tumor microenvironment are even more limited. A
recent study in TAECs identified 49 genes to be associated with
chronic inflammation diseases and cancer; 6 of them constituted an
inflammation-related endothelial-derived gene signature (IREG)
(46). Of these 49 genes
TNFAIP3 was the only one shared with ZR75.30-CM treatment.
We found the gene variant IRF1, which is related to the
IRF7 variant from the IREG. We also identify
inflammation-related diseases (Table
II) as well as TNF and members of the NFκB family to be central
regulators (Table III).
We validated expression changes in 14 of the 54
genes altered by ZR75.30-CM. Several of the validated genes are
important endothelial physiological mediators and have also been
associated with tumorigenesis and malignancy in a variety of cancer
models (Table IV).
 | Table IVFunction of genes validated by
RT-qPCR. |
Table IV
Function of genes validated by
RT-qPCR.
| Gene | Fold change
ZR75.30/TNF | Function | Refs. |
|---|
| CCL20 | 9.35/33.68 | Chemokine involved
in homing during metastasis and is a T-cell chemoattractant | (50) |
| | Actively released
by ECs and epithelial cells, its high expression suggests a
contribution to tumorigenesis | (51,52) |
| CXCL2 | 11.47/10.45 | Chemokine related
to atherosclerosis, angiogenesis and metastasis | (53) |
| | Part of a positive
feedback loop with NFκB in cancer cells, leading to
chemoresistance
Its receptor is expressed by ECs, neutrophils, eosinophils and
macrophages | (54) |
| CDKN1B | (−)/−1.94 | p27, CDK inhibitor.
Binds to cyclin E-CDK2 or cyclin D-CDK4 complexes, controlling the
cell cycle progression | (55) |
| ICAM-1 | 3.81/6.69 | CAM related to
inflammation | (47) |
| | Activates signaling
pathways related to motility. Related to invasion and metastasis of
BC | (48) |
| IL-6 | 5.52/3.46 | Pro-inflammatory
cytokine associated to growth signals, resistance against
apoptosis, vascular inflammatory diseases as atherosclerosis and
cancer progression | (33,56) |
| MAP3K8 | 2.86/2.79 | Serine/threonine
kinase activate in cancer | (57) |
| NFKB2 | 2.58/5.11 | p52 subunit of NFκB
complex | (58) |
| NFKBIA | 2.13/4.46 | Ubiquitin ligase
that inhibits NFκB complex | (58) |
| PPP1R3C | −1.86/−3.11 | Phosphatase
involved in glycogen metabolism. Novel tumor suppressor candidate,
its repression is associated to promoter methylation in MC | (59) |
| | In ECs its
expression has been associated to angiogenesis | (60) |
| SELE | 9.31/13.39 | CAM expressed
exclusively by ECs is related to inflammation process; it has been
related with metastatic dissemination and angiogenesis | (48,61) |
| SORBS1 | 1.52/(−) | Is related to lipid
anabolism. Is also associated with FAK and is a substrate of the
c-Abl tyrosine kinase | (41) |
| TGFB3 | 2.0/(−) | Plays an important
role in cellular differentiation and development. In ECs, this
isoform participates in homeostasis and maintenance to shear
stress | (42) |
| TNFAIP2 | 3.76/13.41 | TNF-inducible
protein 2 recently associated with cancer. Its overexpression is
associated to microvessel density, migration and metastasis | (62) |
| TRAF1 | 2.46/10.36 | Forms a
heterodimeric complex required for TNF-mediated activation of
MAPK8/JNK and NFκB. Interacts with IAPs to mediating the
anti-apoptotic signal from TNF receptors | (63) |
| | TRAF1 has been
associated with rheumatoid arthritis | (64) |
| VCAM-1 | 13.31/24.44 | CAM related to
inflammation, involving in the adhesion and tethering process of
leukocytes | (65) |
| | Expressed by ECs
and tumor cells, associating this expression to metastasis
promotion of lung and bone targets and activation of PI3K/Akt and
NFκB pathway | (49,66) |
Among the validated genes are the CAMs ICAM-1,
E-selectin and VCAM-1 whose expression has been used as marker of
pro-adhesive endothelial phenotype. Expression of these three cell
adhesion molecules has been associated to tumorigenesis and
malignancy (47–49).
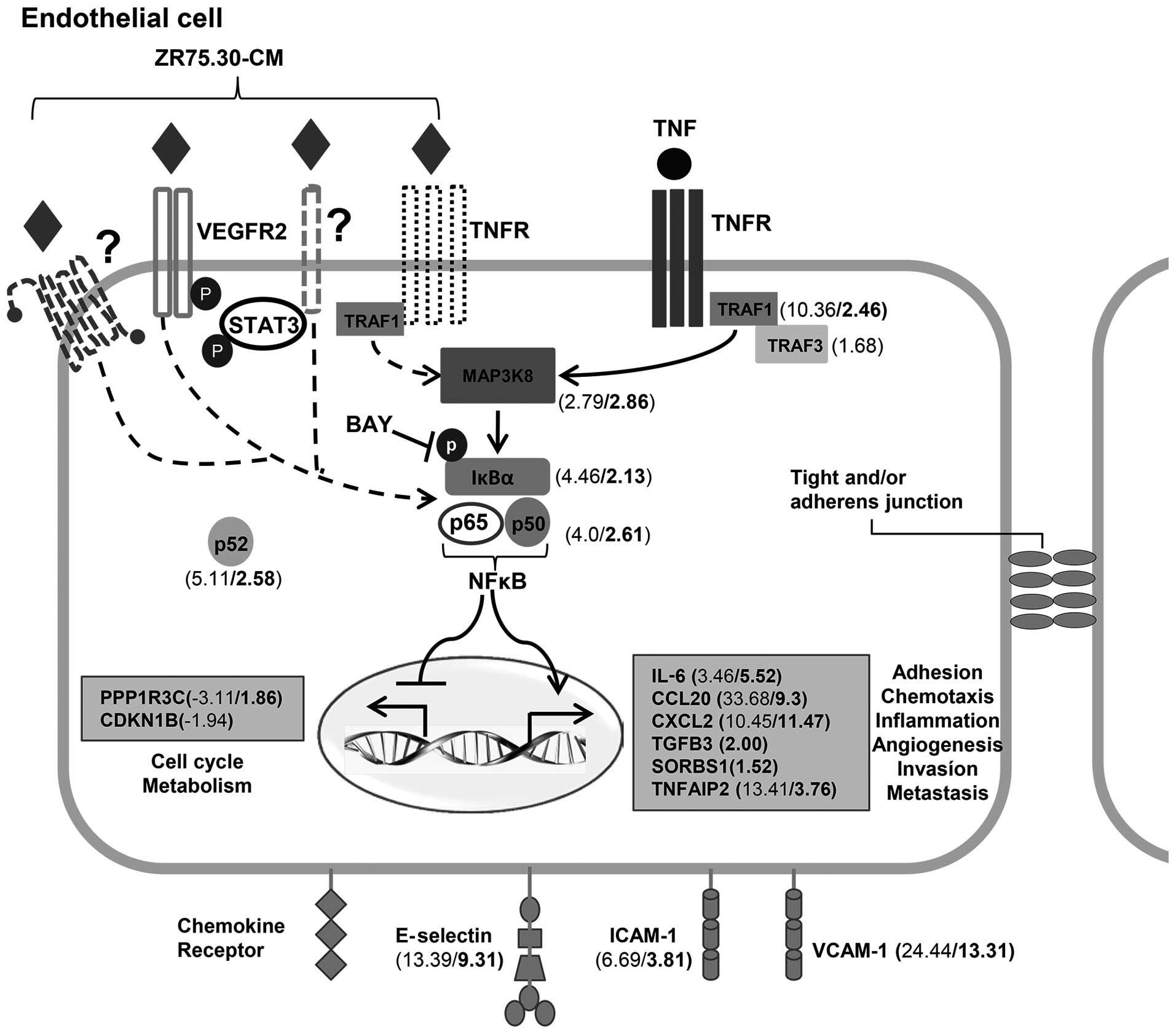
The bioinformatic analysis of microarrays in the
present study indicates that NFκB is a central regulator of the
pro-adhesive phenotype of HUVECs induced by breast cancer secreted
factors (Fig. 9). To further
verify the physiological and molecular relevance of NFκB, we used
BAY 11-7085, a specific inhibitor of IKKs, to interfere with this
pathway. We had previously shown that interference with NFκB
activation in ECs treated with CM from human leukemia prevents NFκB
activation and cell adhesion (17). The fact that we obtained similar
results with CM from a different neoplastic such as breast cancer
suggests that NFκB-dependent activation may be common to epithelial
and hematopoietic neoplastic diseases.
In conclusion, the endothelial transcriptome related
to the pro-adhesive phenotype induced by secreted factors from
ZR75.30 breast cancer cells reveals inflammatory mediators and NFκB
as essential regulators of this phenotype, and pharmacological
inhibition of NFκB validated this prediction. Since all these
changes occur in primary non-transformed human ECs, and this
response is performed by a mixture of cytokines and growth factors
like those secreted by tumor cells, interfering with dominant
transcription pathways could be an alternative therapeutic strategy
to interfere with metastasis.
Acknowledgements
This study was supported by grants from CONACYT to
A.Z.-D. (45519-M and BQO-1104-16/18-2) and P.G. (83597) and DGAPA
to A.Z.-D. (IN226009). J.M.-R. was supported by a CONACYT
pre-doctoral training grant. We thank Dr Jorge Román Audifred
Salomón and Dr Juan Pablo Aragón Hernández from the Hospital
General Dr Manuel Gea González for the facilities in recollection
of umbilical cords.
References
|
1
|
Porter P: ‘Westernizing’ women's risks?
Breast cancer in lower-income countries. N Engl J Med. 358:213–216.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Kennecke H, Yerushalmi R, Woods R, Cheang
MC, Voduc D, Speers CH, Nielsen TO and Gelmon K: Metastatic
behavior of breast cancer subtypes. J Clin Oncol. 28:3271–3277.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakshatri H, Qi G, You J, Kerry B,
Schneider B, Zon R, Buck C, Regnier F and Wang M: Intrinsic
subtype-associated changes in the plasma proteome in breast cancer.
Proteomics Clin Appl. 3:1305–1313. 2009. View Article : Google Scholar
|
|
4
|
Demicheli R, Biganzoli E, Ardoino I,
Boracchi P, Coradini D, Greco M, Moliterni A, Zambetti M, Valagussa
P, Gukas ID, et al: Recurrence and mortality dynamics for breast
cancer patients undergoing mastectomy according to estrogen
receptor status: Different mortality but similar recurrence. Cancer
Sci. 101:826–830. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Balkwill F and Mantovani A: Inflammation
and cancer: Back to Virchow? Lancet. 357:539–545. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karin M: Nuclear factor-kappaB in cancer
development and progression. Nature. 441:431–436. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Coussens LM and Werb Z: Inflammation and
cancer. Nature. 420:860–867. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smith HA and Kang Y: The
metastasis-promoting roles of tumor-associated immune cells. J Mol
Med (Berl). 91:411–429. 2013. View Article : Google Scholar
|
|
10
|
Wirtz D, Konstantopoulos K and Searson PC:
The physics of cancer: The role of physical interactions and
mechanical forces in metastasis. Nat Rev Cancer. 11:512–522. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Stoletov K, Kato H, Zardouzian E, Kelber
J, Yang J, Shattil S and Klemke R: Visualizing extravasation
dynamics of metastatic tumor cells. J Cell Sci. 123:2332–2341.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Franses JW, Baker AB, Chitalia VC and
Edelman ER: Stromal endothelial cells directly influence cancer
progression. Sci Transl Med. 3:66ra52011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Mantovani A, Allavena P, Sica A and
Balkwill F: Cancer-related inflammation. Nature. 454:436–444. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Oppenheimer SB: Cellular basis of cancer
metastasis: A review of fundamentals and new advances. Acta
Histochem. 108:327–334. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
St Croix B, Rago C, Velculescu V, Traverso
G, Romans KE, Montgomery E, Lal A, Riggins GJ, Lengauer C,
Vogelstein B, et al: Genes expressed in human tumor endothelium.
Science. 289:1197–1202. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Seaman S, Stevens J, Yang MY, Logsdon D,
Graff-Cherry C and St Croix B: Genes that distinguish physiological
and pathological angiogenesis. Cancer Cell. 11:539–554. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Estrada-Bernal A, Mendoza-Milla C,
Ventura-Gallegos JL, López-Bojórquez LN, Miranda-Peralta E,
Arechavaleta-Velasco F, Vadillo-Ortega F, Sánchez-Sánchez L and
Zentella-Dehesa A: NF-kappaB dependent activation of human
endothelial cells treated with soluble products derived from human
lymphomas. Cancer Lett. 191:239–248. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Montes-Sánchez D, Ventura JL, Mitre I,
Frías S, Michán L, Espejel-Nuñez A, Vadillo-Ortega F and Zentella
A: Glycosylated VCAM-1 isoforms revealed in 2D western blots of
HUVECs treated with tumoral soluble factors of breast cancer cells.
BMC Chem Biol. 9:72009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Katanasaka Y, Asai T, Naitou H, Ohashi N
and Oku N: Proteomic characterization of angiogenic endothelial
cells stimulated with cancer cell-conditioned medium. Biol Pharm
Bull. 30:2300–2307. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Baudin B, Bruneel A, Bosselut N and
Vaubourdolle M: A protocol for isolation and culture of human
umbilical vein endothelial cells. Nat Protoc. 2:481–485. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar
|
|
22
|
Szklarczyk D, Franceschini A, Wyder S,
Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos
A, Tsafou KP, et al: STRING v10: Protein-protein interaction
networks, integrated over the tree of life. Nucleic Acids Res.
43(D1): D447–D452. 2015. View Article : Google Scholar
|
|
23
|
Mi H, Muruganujan A, Casagrande JT and
Thomas PD: Large-scale gene function analysis with the PANTHER
classification system. Nat Protoc. 8:1551–1566. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
25
|
Yeung YG and Stanley ER: A solution for
stripping antibodies from polyvinylidene fluoride immunoblots for
multiple reprobing. Anal Biochem. 389:89–91. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Machuca C, Mendoza-Milla C, Córdova E,
Mejía S, Covarrubias L, Ventura J and Zentella A: Dexamethasone
protection from TNF-alpha-induced cell death in MCF-7 cells
requires NF-kappaB and is independent from AKT. BMC Cell Biol.
7:92006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hanahan D and Coussens LM: Accessories to
the crime: Functions of cells recruited to the tumor
microenvironment. Cancer Cell. 21:309–322. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Reymond N, d'Água BB and Ridley AJ:
Crossing the endothelial barrier during metastasis. Nat Rev Cancer.
13:858–870. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lee E, Fertig EJ, Jin K, Sukumar S, Pandey
NB and Popel AS: Breast cancer cells condition lymphatic
endothelial cells within pre-metastatic niches to promote
metastasis. Nat Commun. 5:47152014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Perrot-Applanat M, Vacher S, Toullec A,
Pelaez I, Velasco G, Cormier F, Saad HS, Lidereau R, Baud V and
Bièche I: Similar NF-κB gene signatures in TNF-α treated human
endothelial cells and breast tumor biopsies. PLoS One.
6:e215892011. View Article : Google Scholar
|
|
31
|
Viemann D, Goebeler M, Schmid S, Klimmek
K, Sorg C, Ludwig S and Roth J: Transcr iptional profiling of
IKK2/NF-kappa B- and p38 MAP kinase-dependent gene expression in
TNF-alpha-stimulated primary human endothelial cells. Blood.
103:3365–3373. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Das S and Skobe M: Lymphatic vessel
activation in cancer. Ann N Y Acad Sci. 1131:235–241. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brasier AR: The nuclear
factor-kappaB-interleukin-6 signalling pathway mediating vascular
inflammation. Cardiovasc Res. 86:211–218. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Waugh DJ and Wilson C: The interleukin-8
pathway in cancer. Clin Cancer Res. 14:6735–6741. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jośko J and Mazurek M: Transcription
factors having impact on vascular endothelial growth factor (VEGF)
gene expression in angiogenesis. Med Sci Monit. 10:RA89–RA98.
2004.
|
|
36
|
Schweighofer B, Testori J, Sturtzel C,
Sattler S, Mayer H, Wagner O, Bilban M and Hofer E: The
VEGF-induced transcriptional response comprises gene clusters at
the crossroad of angiogenesis and inflammation. Thromb Haemost.
102:544–554. 2009.PubMed/NCBI
|
|
37
|
Grivennikov SI and Karin M: Dangerous
liaisons: STAT3 and NF-kappaB collaboration and crosstalk in
cancer. Cytokine Growth Factor Rev. 21:11–19. 2010. View Article : Google Scholar
|
|
38
|
Kuscu C, Evensen N, Kim D, Hu YJ, Zucker S
and Cao J: Transcriptional and epigenetic regulation of KIAA1199
gene expression in human breast cancer. PLoS One. 7:e446612012.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Birkenkamp-Demtroder K, Maghnouj A,
Mansilla F, Thorsen K, Andersen CL, Øster B, Hahn S and Ørntoft TF:
Repression of KIAA1199 attenuates Wnt-signalling and decreases the
proliferation of colon cancer cells. Br J Cancer. 105:552–561.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Howarth KD, Blood KA, Ng BL, Beavis JC,
Chua Y, Cooke SL, Raby S, Ichimura K, Collins VP, Carter NP, et al:
Array painting reveals a high frequency of balanced translocations
in breast cancer cell lines that break in cancer-relevant genes.
Oncogene. 27:3345–3359. 2008. View Article : Google Scholar :
|
|
41
|
Genua M, Pandini G, Cassarino MF, Messina
RL and Frasca F: c-Abl and insulin receptor signalling. Vitam Horm.
80:77–105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Walshe TE, dela Paz NG and D'Amore PA: The
role of shear-induced transforming growth factor-β signaling in the
endothelium. Arterioscler Thromb Vasc Biol. 33:2608–2617. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou J, Jin Y, Gao Y, Wang H, Hu G, Huang
Y, Chen Q, Feng M and Wu C: Genomic-scale analysis of gene
expression profiles in TNF-alpha treated human umbilical vein
endothelial cells. Inflamm Res. 51:332–341. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mayer H, Bilban M, Kurtev V, Gruber F,
Wagner O, Binder BR and de Martin R: Deciphering regulatory
patterns of inflammatory gene expression from
interleukin-1-stimulated human endothelial cells. Arterioscler
Thromb Vasc Biol. 24:1192–1198. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Rivera CG, Mellberg S, Claesson-Welsh L,
Bader JS and Popel AS: Analysis of VEGF - a regulated gene
expression in endothelial cells to identify genes linked to
angiogenesis. PLoS One. 6:e248872011. View Article : Google Scholar
|
|
46
|
Pitroda SP, Zhou T, Sweis RF, Filippo M,
Labay E, Beckett MA, Mauceri HJ, Liang H, Darga TE, Perakis S, et
al: Tumor endothelial inflammation predicts clinical outcome in
diverse human cancers. PLoS One. 7:e461042012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Konstantopoulos K and Thomas SN: Cancer
cells in transit: The vascular interactions of tumor cells. Annu
Rev Biomed Eng. 11:177–202. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tesarova P, Kalousova M, Zima T, Suchanek
M, Malikova I, Kvasnicka J, Duskova D, Tesar V, Vachek J,
Krupickova-Kasalova Z, et al: Endotelial activation and
flow-mediated vasodilation in young patients with breast cancer.
Neoplasma. 60:690–697. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Lu X, Mu E, Wei Y, Riethdorf S, Yang Q,
Yuan M, Yan J, Hua Y, Tiede BJ, Lu X, et al: VCAM-1 promotes
osteolytic expansion of indolent bone micrometastasis of breast
cancer by engaging α4β1-positive osteoclast progenitors. Cancer
Cell. 20:701–714. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Ghadjar P, Rubie C, Aebersold DM and
Keilholz U: The chemokine CCL20 and its receptor CCR6 in human
malignancy with focus on colorectal cancer. Int J Cancer.
125:741–745. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Marsigliante S, Vetrugno C and Muscella A:
CCL20 induces migration and proliferation on breast epithelial
cells. J Cell Physiol. 228:1873–1883. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bruneau S, Nakayama H, Woda CB, Flynn EA
and Briscoe DM: DEPTOR regulates vascular endothelial cell
activation and proinflammatory and angiogenic responses. Blood.
122:1833–1842. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bendall L: Chemokines and their receptors
in disease. Histol Histopathol. 20:907–926. 2005.PubMed/NCBI
|
|
54
|
Acharyya S, Oskarsson T, Vanharanta S,
Malladi S, Kim J, Morris PG, Manova-Todorova K, Leversha M, Hogg N,
Seshan VE, et al: A CXCL1 paracrine network links cancer
chemoresistance and metastasis. Cell. 150:165–178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Chu IM, Hengst L and Slingerland JM: The
Cdk inhibitor p27 in human cancer: Prognostic potential and
relevance to anticancer therapy. Nat Rev Cancer. 8:253–267. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Naugler WE and Karin M: The wolf in
sheep's clothing: The role of interleukin-6 in immunity,
inflammation and cancer. Trends Mol Med. 14:109–119. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Bièche I, Lerebours F, Tozlu S, Espie M,
Marty M and Lidereau R: Molecular profiling of inflammatory breast
cancer: Identification of a poor-prognosis gene expression
signature. Clin Cancer Res. 10:6789–6795. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Hayden MS and Ghosh S: Shared principles
in NF-kappaB signaling. Cell. 132:344–362. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Bonazzi VF, Irwin D and Hayward NK:
Identification of candidate tumor suppressor genes inactivated by
promoter methylation in melanoma. Genes Chromosomes Cancer.
48:10–21. 2009. View Article : Google Scholar
|
|
60
|
Ferrari N, Pfeffer U, Dell'Eva R,
Ambrosini C, Noonan DM and Albini A: The transforming growth
factor-beta family members bone morphogenetic protein-2 and
macrophage inhibitory cytokine-1 as mediators of the antiangiogenic
activity of N-(4-hydroxyphenyl)retinamide. Clin Cancer Res.
11:4610–4619. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Gout S, Tremblay PL and Huot J: Selectins
and selectin ligands in extravasation of cancer cells and organ
selectivity of metastasis. Clin Exp Metastasis. 25:335–344. 2008.
View Article : Google Scholar
|
|
62
|
Chen LC, Chen CC, Liang Y, Tsang NM, Chang
YS and Hsueh C: A novel role for TNFAIP2: Its correlation with
invasion and metastasis in nasopharyngeal carcinoma. Mod Pathol.
24:175–184. 2011. View Article : Google Scholar
|
|
63
|
Baud V and Karin M: Signal transduction by
tumor necrosis factor and its relatives. Trends Cell Biol.
11:372–377. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Song GG, Bae SC, Kim JH and Lee YH:
Associations between TRAF1-C5 gene polymorphisms and rheumatoid
arthritis: A meta-analysis. Immunol Invest. 43:97–112. 2014.
View Article : Google Scholar
|
|
65
|
Luster AD, Alon R and von Andrian UH:
Immune cell migration in inflammation: Present and future
therapeutic targets. Nat Immunol. 6:1182–1190. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Chen Q, Zhang XH and Massagué J:
Macrophage binding to receptor VCAM-1 transmits survival signals in
breast cancer cells that invade the lungs. Cancer Cell. 20:538–549.
2011. View Article : Google Scholar : PubMed/NCBI
|