Introduction
Osteosarcoma is the most common bone malignancy in
the pediatric age group, with a very high propensity for local
invasion and distant metastasis (1,2). In
recent years, despite the development of new multimodal
therapeutics, the prognosis for patients with this disease is
generally poor (3,4). Therefore, effective therapeutic
methods for osteosarcoma are urgently needed.
Ubiquitin (Ub), a kind of important signaling
protein, is involved in many cellular processes, including cell
cycle regulation, DNA damage response, chromatin remodeling and
proteasome degradation. Similar to phosphorylation, ubiquitin
modification is reversible, and deubiquitinases or DUBs are known
to mediate ubiquitin removal (5).
DUBs can remove Ub moieties from Ub-protein conjugates, resulting
in reduced ubiquitination signaling (6).
The misexpression of DUBs has been demonstrated to
be associated with a number of human diseases, especially cancers
(7). Among them, USP1 is one of
the best-characterized human DUBs, which is involved in the control
of cell differentiation and plays an important role in the
regulation of the cellular response to DNA damage (8).
USP1 has been reported to be overexpressed in
several human malignant tumors (9,10).
Moreover, USP1 has been identified as the DUB responsible for
deubiquitinating Fanconi anemia complementation group I (FANCI),
Fanconi anemia group D2 (FANCD2) and proliferating cell nuclear
antigen (PCNA) in the DNA damage response. PCNA is
monoubiquitinated in response to DNA damage that stalls progression
of the replication fork and then initiates recruitment of
translesion DNA synthesis (TLS) DNA polymerases in the DNA damage
tolerance pathway (11,12). USP1 plays as a regulator of PCNA
ubiquitination in the DNA damage response. It may deubiquitinate
the DNA replication processivity factor, PCNA, thus, contributing
to preventing unscheduled recruitment of error-prone TLS DNA
polymerases, as a safeguard against error-prone TLS of DNA
(13).
USP1-associated factor 1 (UAF1) stabilizes USP1 and
allo-sterically increases its catalytic activity, which is very low
in the absence of the cofactor (14,15).
In addition, UAF1 contributes to targeting USP1 to its nuclear
substrates. USP1 performs its functions in the context of a
heterodimeric complex with UAF1 (16).
RNAi is a powerful approach that silences the
expression of endogenous genes via distinct messenger RNA
degradation pathways. It may be a potential therapeutic agent for
many kinds of diseases, including HIV, cerebrovascular and
cardiovascular diseases, viral hepatitis, metabolic diseases,
neurodegenerative disorders and cancers (17,18).
In the present study, specific-shRNA with lentivirus
vector was employed to knock down USP1 in osteosarcoma U2OS cells,
and the effects of USP1 silencing on cell growth and invasion were
explored.
Materials and methods
Main reagents
Mouse anti-human USP1 polyclonal antibody was
purchased from the ProteinTech Group (Wuhan, China); mouse
anti-human monoclonal antibody to Bcl-2 and mouse anti-human
polyclonal antibodies to MMP-2, GSK-3β, Stat3, cyclin E1, Notch1,
Wnt-1 and cyclin A1 were purchased from ImmunoWay Biotechnology Co.
(Plano, TX, USA); MG132 and mouse anti-human monoclonal antibody to
USP1 were obtained from Merck Millipore (Darmstadt, Germany); mouse
anti-human monoclonal antibody to SIK2 were obtained from BioLegend
(San Diego, CA, USA); TRIzol reagent, Lipofectamine™ 2000, Opti-MEM
and the SuperScript III reverse transcriptase (RT) kit were
obtained from Invitrogen Corp. (Carlsbad, CA, USA); Taq DNA
polymerase was purchased from Fermentas, Inc. (Waltham, MA, USA);
cell culture media and fetal bovine serum (FBS) were purchased from
Gibco-BRL (Grand Island, NY, USA).
Tissue samples, cell lines and cell
culture conditions
Tissue samples from 30 osteosarcomas were obtained
from patients who underwent surgery at the Third Affiliated
Hospital of Soochow University. All participants who had undergone
surgical resection signed an informed consent form for this study.
All osteosarcoma cases had been confirmed both clinically and
pathologically. The experimental protocols for the present study
were approved by the Hospital’s Protection of Human Subjects
Committee.
The human osteosarcoma U2OS cells were purchased
from the Cell Bank of Chinese Academy of Sciences (Shanghai, China)
in 2014 and were identified by short tandem repeat (STR) method in
2015. The cells were cultured in Dulbecco’s modified Eagle’s medium
(DMEM) supplemented with 10% fetal bovine serum (FBS) and
maintained at 37°C in a humidified atmosphere containing 5%
CO2.
Immunohistochemistry
Surgical specimens were fixed in 10% formalin
solution and embedded in paraffin. Four-μm thick sections were
deparaffinized, rehydrated and immersed in 3% hydrogen peroxide
solution for 15 min to block endogenous peroxidase activity. For
antigen retrieval, the sections were boiled in EDTA buffer (pH 9.0)
for 10 min. Histologic sections were pre-treated in 10% normal goat
serum in TBS for 30 min at room temperature in order to block
non-specific binding sites. Then sections were incubated with
polyclonal antibody against USP1 at a dilution of 1:100 in
phosphate-buffered saline (PBS) at 4°C for 1 h. After being rinsed
five times with PBS, sections were incubated with goat anti-mouse
IgG conjugated with horseradish peroxidase at room temperature for
1 h. Sections were developed with diaminobenzidene (DAB) as a
chromogen and counterstained with haematoxylin. The sections were
analyzed under light microscopy.
USP1 gene silence mediated by
lentivirus-delivered shRNA
The human USP1-specific small interfering RNA
(siRNA) sequence is 5′-GAAAGCTCCACATCAATAA-3′, designed with an
online Invitrogen software using the USP1 sequence (GenBank code:
NM_003600) as a reference. The non-silencing sequence
(5′-TTCTCCGAACGTGTCACGT-3′) was used as a scrambled control. The
recombinant lentiviral particles were regenerated in 293T cells by
co-transfection of the modified pGCSIL-GFP viral vector together
with the pHelper 1.0 and pHelper 2.0 plasmids using the
Lipofectamine™ 2000 reagent. The lentiviral particles were referred
to as LV-USP1 siRNA or USP1-siRNA. We generated lentiviruses that
express non-silencing-shRNA as controls and named them scramble
siRNA or scr-siRNA. The osteosarcoma cells transfected with the
USP1-siRNA or scrambled siRNA were designated USP1-siRNA and
scr-siRNA, respectively. For lentiviral transduction, U2OS cells
were cultured in 6-well plates at 70% confluence and infected with
control lentivirus or USP1-specific siRNA lentivirus at MOI of 20.
After 5 days of infection, U2OS cells expressing GFP protein were
observed by fluorescence microscopy to determine the infection
efficiency.
SIK2 siRNA preparation
The siRNA sequences were designed by commercial
software (Applied Biosystems/Ambion, Austin, TX, USA). For SIK2,
the siRNA sense sequence is 5-CUA CCGAGAAGUACAAAUADTDT-3′ and the
antisense sequence is 5′-UAUUUGUACUUCUCGGUAGdTdT-3′. The siRNA
sequences employed as negative controls were 5′-GG
CCUCAGCUGCGCGACGCdTdT-3′ (sense) and 5′-GCGUC GCGCAGCUGGGCCAdTdT-3′
(antisense). The selected sequences were confirmed by NCBI BLAST
(http://www.ncbi.nlm.nih.gov/blast/)
to make sure that the selected genes were specifically targeted.
Chemically synthesized siRNAs were designed and sequenced by
Guangzhou RiboBio, Co., Ltd. (Guangzhou, China).
SIK2 siRNA transfection
U2OS cells were transiently transfected with
synthetic siRNA using Lipofectamine 2000 reagent according to the
manufacturer’s instructions. Briefly, U2OS cells were seeded onto
6-well plates in the maintenance medium at a density of
2×105 cells/well and allowed to grow for 12 h. Then, 5
μl Lipofectamine 2,000 and 200 pmol siRNA were diluted in
serum-free medium to a total volume of 250 μl and incubated for 20
min at room temperature. After the cells were washed with DMEM
medium without FBS, the diluted siRNA-Lipofectamine mix was added
to each well and incubated for 24 h. Then the mix was replaced with
fresh DMEM medium containing 10% FBS. Forty-eight hours after the
transfection, the U2OS cells were collected for protein and RNA
isolation. The osteosarcoma cells transfected with the SIK2-siRNA
or scrambled siRNA were designated SIK2-siRNA and scr-siRNA,
respectively.
Quantitative real-time PCR analysis
Total RNA was extracted from osteosarcoma cells
using TRIzol reagent following the manufacturer’s instructions. RNA
samples (1 μg/reaction) were reverse-transcribed with the RevertAid
First Strand cDNA Synthesis kit according to the manufacturer’s
recommended protocol. The RT reaction was used for amplification
with Taq polymerase and a SYBR-Green RT-PCR kit (Takara, Kyoto,
Japan) was used to detect specific real-time RT-PCR products. The
resulting cDNA was amplified using specific primers, which were
designed by using the GraphPad Prism 5.0 software (GraphPad
Software, Inc., San Diego, CA, USA). The primer sequences were: For
USP1, sense: 5′-AGGTTG CTAGTACAGCGTTTGC-3′ and antisense:
5′-CACTGGATT CCTTGTTTCTATCAGA-3′; for SIK2, sense: 5′-AGTCTGA
AGCCAGGGAAAA-3′ and antisense: 5′-CATGTTGCCAGC AGTTCACC-3′; for
β-actin, sense: 5′-GGCACTCTTCCAGC CTTCC-3′ and antisense:
5′-GAGCCGCCGATCCACAC-3′. Relative gene-expression levels were
calculated using the comparative Ct method, also called the
2−ΔΔCT analysis method.
Cell proliferation assay
U2OS cells were seeded onto 96-well plates and
incubated in corresponding media supplemented with 10% FBS. After
24, 48, 72 and 96 h of incubation, 10 μl of Cell Counting kit-8
(CCK-8; Dojindo Laboratories, Kumamoto, Japan) was added into each
well, followed by 4 h of incubation. The optical density values
were measured at 450 nm using a Bio-Rad microplate reader to
determine cell viability. The results were based on three
independent experiments and are presented as mean ± SD.
Colony-forming assay
Soft agar colony-formation assay was used to
determine the effect of USP1 knockdown on the transformation
capability of U2OS cells. In brief, U2OS cells were counted and
inoculated in 6-well plates at a density of 3,000 cells/well. After
being cultured at 37°C in 5% CO2 in a humidified
incubator for 2 weeks, U2OS cells were immobilized by 4%
paraformaldehyde and stained using Giemsa dye for 20 min. The cells
were rinsed with distilled water. Plates were then scanned and
photographed under an inverted microscope. All experiments were
performed three times.
Flow cytometric analysis
The U2OS cells were harvested and washed with
ice-cold PBS twice. Cells were then resuspended with 100 μl of
Annexin V binding buffer, and incubated with APC-labeled Annexin V
at room temperature for 15 min. Fluorescence-activated cell sorting
(FACS) analysis for Annexin V staining was performed to determine
cell apoptosis. Each experiment was performed in triplicate.
In vitro invasion assay
Twenty-four hours after the infection, the invasion
ability of U2OS cells was determined using a Transwell chamber. A
total of 40 μl Matrigel (10 mg/ml) (BD Biosciences, San Jose, CA,
USA) was coated to 48-well plates and polymerized at 37°C for 30
min. U2OS cells were collected into DMEM medium, supplemented with
1% FBS. In brief, a 100 μl cell suspension containing
2.0×105 cells was added to the upper chamber of each
insert. For each Transwell chamber, 500 μl of DMEM supplemented
with 20% FBS was added in the lower chamber. After 24 h of
incubation at 37°C, cells and Matrigel on the upper surface of the
well were removed. Cells invaded to the lower chamber were fixed
with methanol, stained with 0.5% crystal violet for 20 min and then
were counted under the light microscope. In addition, invaded cells
were segregated, lysed and quantified at 570 nm using
spectrophotometer to evaluate the amount of cells.
Western blot analysis
The U2OS cell pellets were lysed with protein
extraction solution and incubated at 20°C for 20 min. After protein
quantification, the samples were separated by electrophoresis in
SDS-PAGE and then transferred to a PVDF (polyvinylidene fluoride)
membrane. After blocking in Tris-buffered saline Tween-20 (TBST)
containing 5% non-fat milk for 1 h at room temperature, membranes
were subsequently incubated with primary antibodies against USP1,
SIK2, matrix metalloproteinase-2 (MMP-2), glycogen synthase
kinase-3β (GSK-3β), B-cell lymphoma 2 (Bcl-2), signal transducer
and activator of transcription 3 (Stat3), cyclin E1, Notch homolog
1 (Notch1), Wingless-type protein-1 (Wnt-1) and cyclin A1 at 4°C
overnight. After incubation with secondary antibodies (1:1,000;
peroxidase-conjugated anti-mouse IgG) for another 2 h, the
membranes were washed in TBST buffer and protein were detected
using enhanced chemiluminescence. GAPDH and β-actin bands were used
as loading controls.
Effect of MG132 on SIK2 protein
levels
The U2OS cells were cultured in 6-well plates at a
density of 4×104 cells/well. After 24 h of incubation,
cells were treated with various concentrations of MG132 (2, 5 and
10 μmol/l) and cultured for 0, 4, 8 and 16 h. Level of SIK2 protein
was determined using western blot analysis.
Statistical analysis
Data are reported as the mean ± SEM of three
independent experiments. Statistical analysis between comparable
groups was performed using a one-way ANOVA by using GraphPad Prism
5.0 software. In each case, P<0.05 was considered statistically
significant.
Results
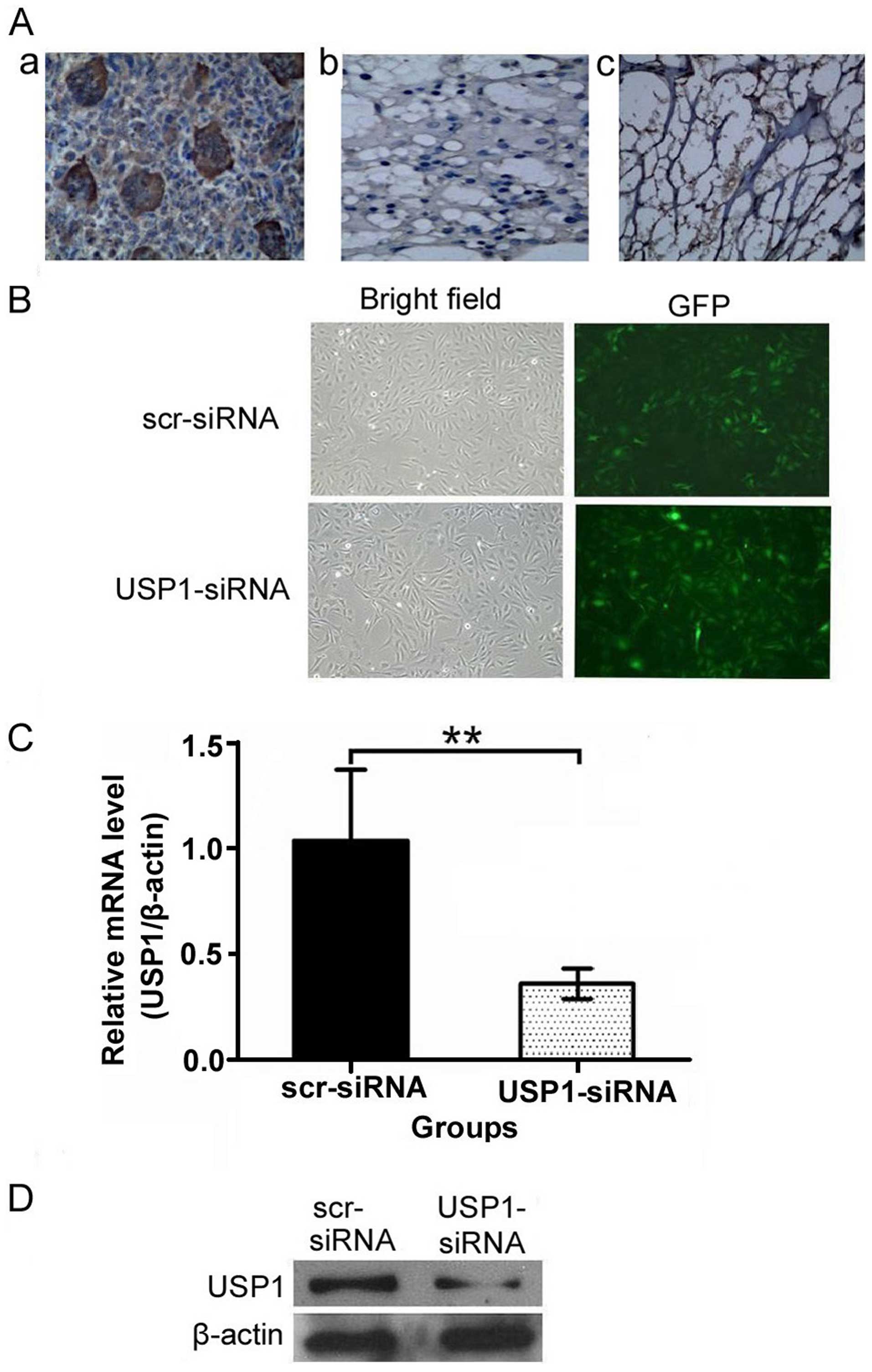
USP1 is overexpressed in osteosarcoma
samples
We examined the expression of USP1 in 30 samples
from patients with osteosarcoma. Of the 30 patients, 26 (86.67%)
were classified as positive expression of USP1. USP1 protein with
positive staining was shown in the nuclei and cytoplasm of
osteosarcoma tissues (Fig. 1A-a),
while rare visible USP1 staining was detected in cartilage tumor
tissues (Fig. 1A-b) and normal
bone tissues (Fig. 1A-c). The USP1
expression in osteosarcoma tissues was significantly higher than
that in cartilage tumor tissues and normal bone tissues.
Construction of a lentiviral vector
mediating RNAi targeting of USP1 (LV-USP1 siRNA) and its effects on
USP1 expression
To further illuminate the relationship between USP1
and osteosarcoma, we constructed a lentivirus-based USP1-specific
siRNA vector and a scramble-siRNA vector. The two vectors were
infected into U2OS cells for 3 days and >90% of the cells showed
a green fluorescence indicating successful infection (Fig. 1B). To confirm whether the
USP1-siRNA had silenced the expression of USP1 mRNA and protein,
real-time PCR and western blot analysis were performed on
lentivirus-infected cells. The results showed that both mRNA
(Fig. 1C) and protein (Fig. 1D) expression levels of USP1 were
significantly reduced by infection of USP1-siRNA, compared to
scr-siRNA. These data suggest that the infection with USP1-siRNA
effectively downregulates USP1 expression in U2OS cells.
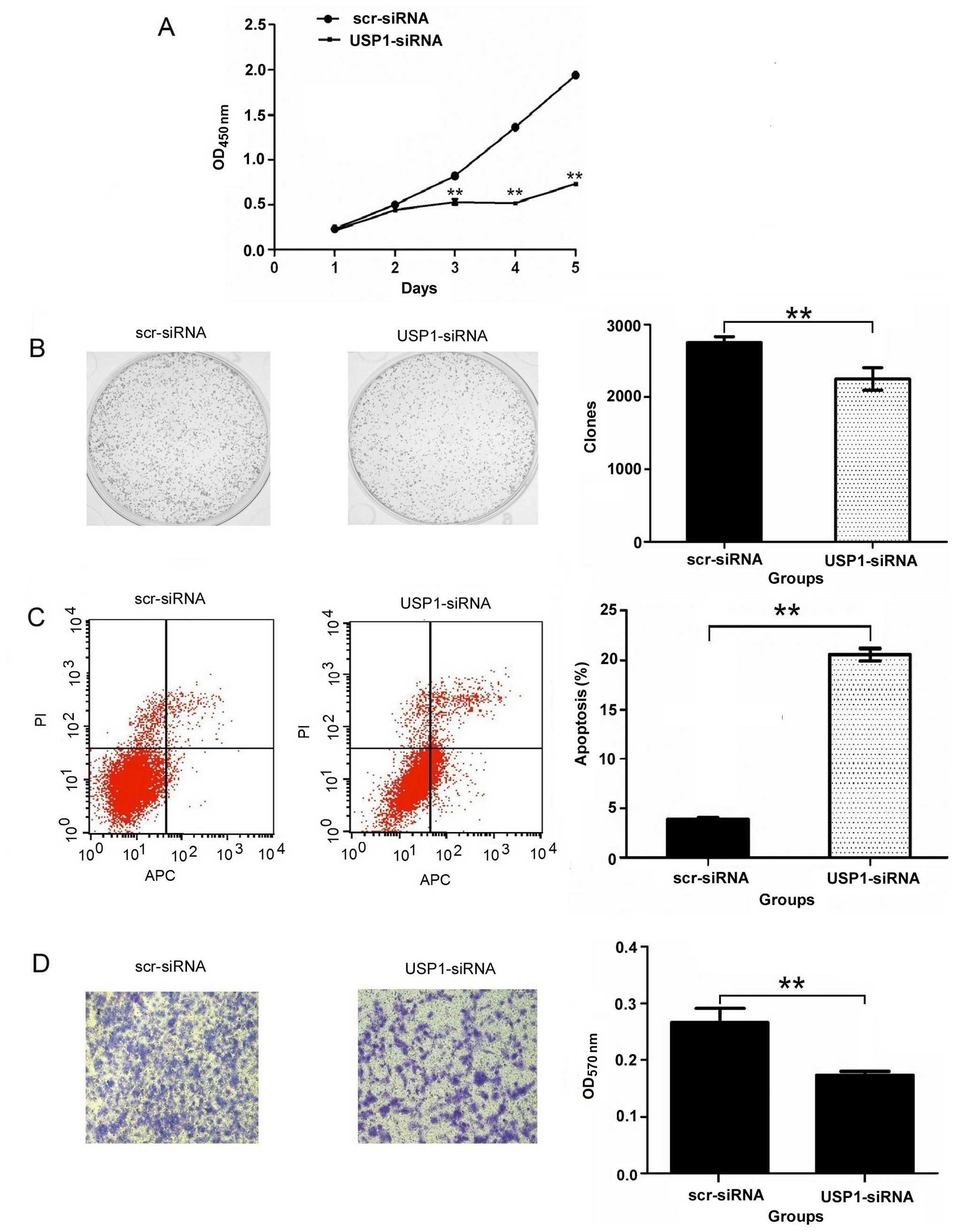
Effects of USP1-siRNA on cell
viability
CCK-8 assay was performed to study the effect of
USP1-siRNA on U2OS cell growth. As shown in Fig. 2A, the cell growth of USP1-siRNA
groups showed a significant (P<0.01) reduction in cell viability
in comparison to the scr-siRNA groups. The result of a colony
formation assay showed that the number of colonies of the
USP1-siRNA group (2249.33±156.31) was significantly less than that
of the scr-siRNA group (2751.00±83.07) in U2OS cells (P<0.01)
(Fig. 2B). These results
demonstrate that the reduction in USP1 expression decreased the
ability of U2OS cells to form colonies. Furthermore, these results
suggest that cell proliferation is reduced due to the silencing of
the USP1 gene.
USP1 silencing induces apoptosis in U2OS
cells
To determine whether USP1 depletion induced cell
apoptosis, flow cytometry was used after the silencing of USP1. The
flow cytometric analysis shows that the percentage of apoptotic
U2OS cells was 3.88±0.22% in the scr-siRNA group cells, and the
percentage of apoptotic cells increased to 20.57±0.64% in the
USP1-siRNA group cells (P<0.01) (Fig. 2C). These data suggest that the
depletion of USP1 specifically induces apoptosis of the U2OS
cells.
Depletion of USP1 inhibits the invasion
of U2OS cells
Next, we performed an in vitro invasion
assay, the results showed that OD value of 570 nm was 0.27±0.04 in
the scr-siRNA group and 0.17±0.01 in the USP1-siRNA group
(P<0.01) (Fig. 2D). These
results demonstrate that USP1-siRNA could reduce the invasion of
U2OS cells.
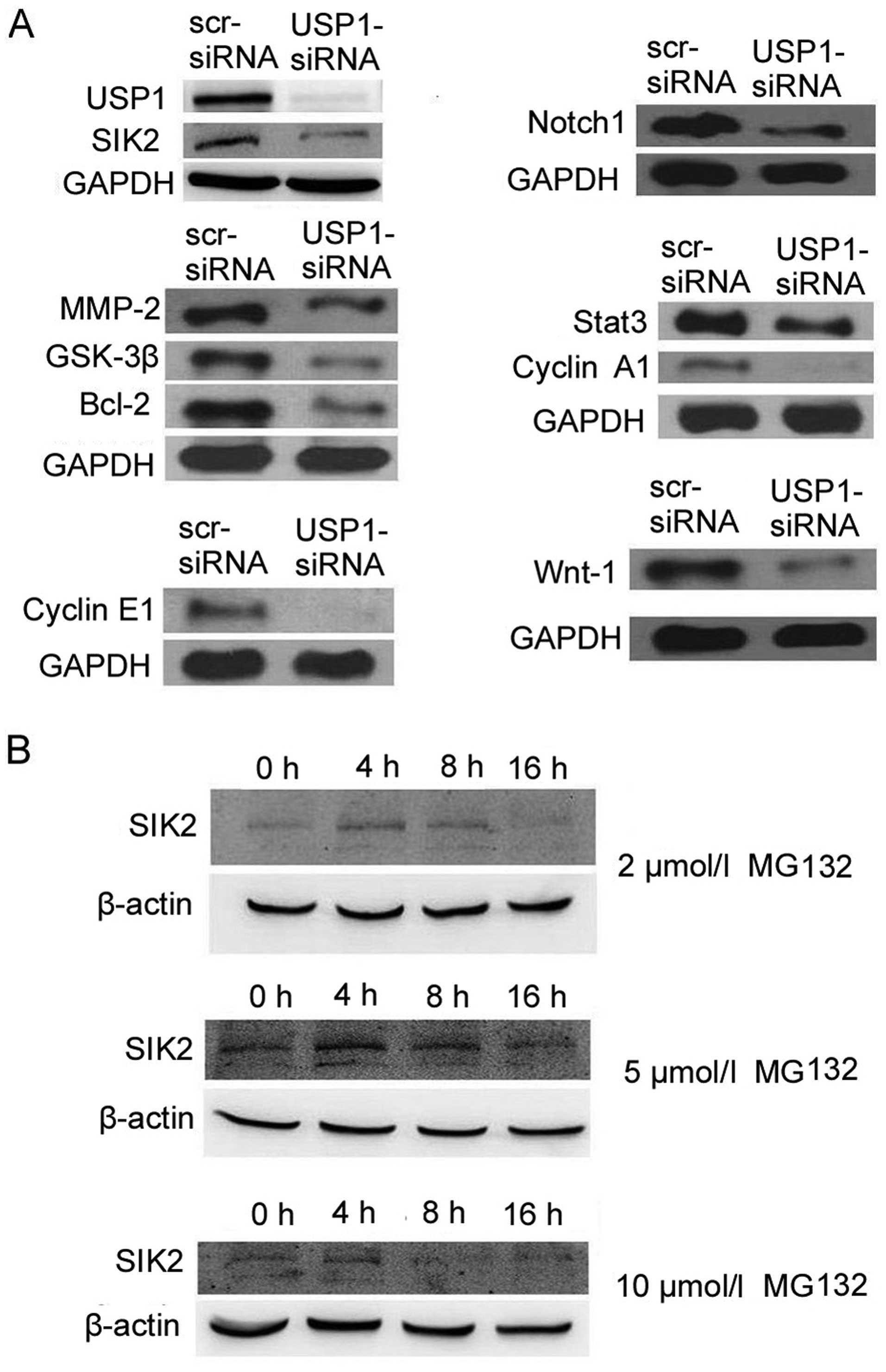
Western blot analysis
The protein levels of related molecules were also
examined. The results showed that USP1 knockdown downregulated
SIK2, MMP-2, GSK-3β, Bcl-2, cyclin E1, Notch1, Stat3, cyclin A1 and
Wnt-1 in U2OS cells (Fig. 3A).
These results suggest that USP1 downregulation inhibits growth and
invasion of U2OS cells through these proteins.
MG132 affects SIK2 protein level
We further examined whether proteasome inhibitor
MG132 can increase the level of SIK2 protein in U2OS cells. The
result showed that cells treated with MG132 (5 μmol/l) for 4 or 8 h
exhibited an elevation of SIK2 protein (Fig. 3B). It suggests that MG132
upregulates SIK2 expression by decreasing its protein degradation,
and therefore, inhibition of proteasome function increases SIK2
protein level in U2OS cells.
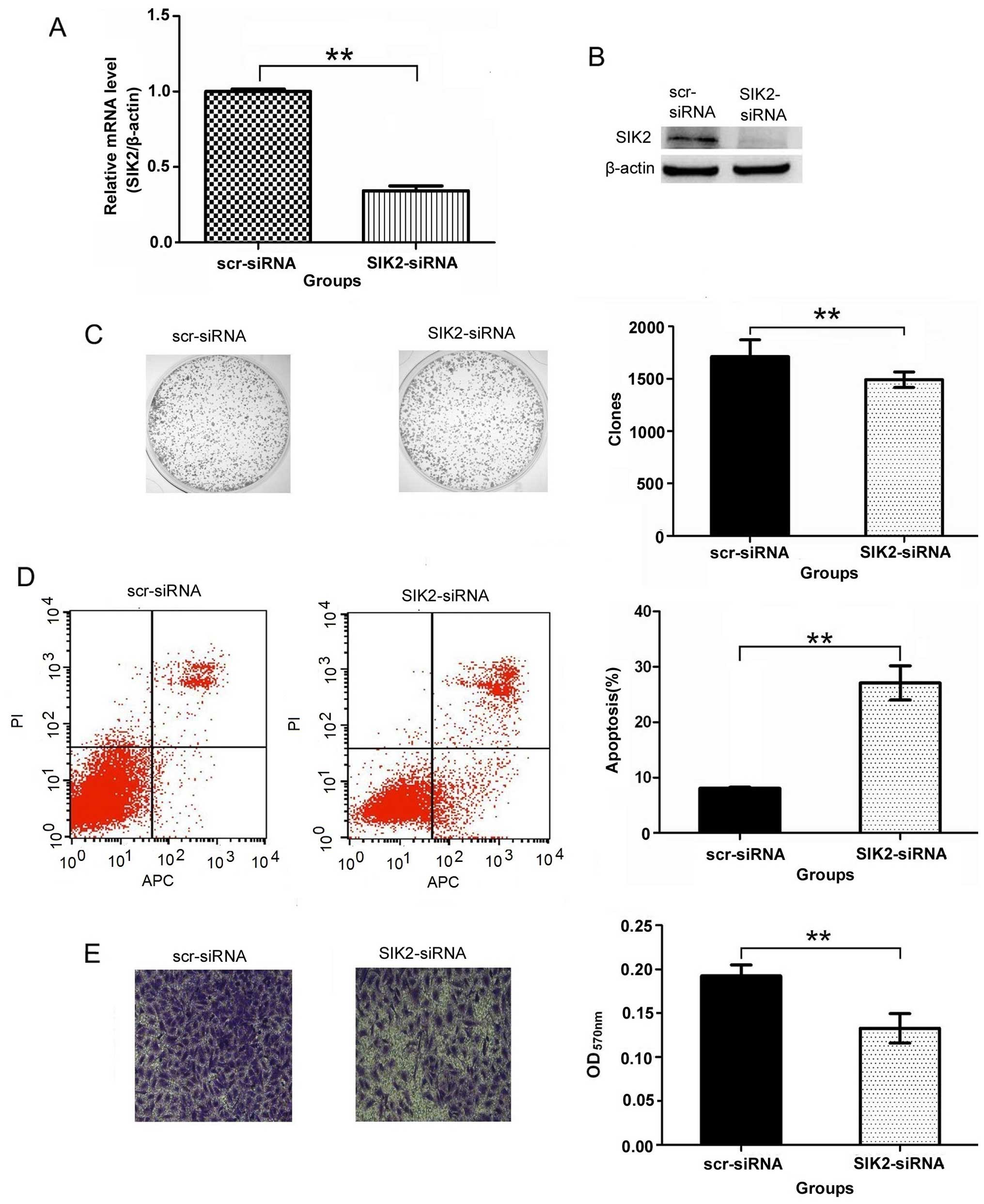
SIK2 siRNA significantly inhibits the
mRNA and protein expression of SIK2
Real-time PCR and western blot analysis were
performed to confirm whether the SIK2 siRNA inhibited the
expression of SIK2 mRNA and protein. The results showed that SIK2
mRNA and protein expression in SIK2-siRNA group were significantly
inhibited compared to scr-siRNA group as shown in Fig. 4A and B. It indicates that the
targeted SIK2 siRNA inhibited significantly the expression of SIK2
gene.
Downregulation of SIK2 suppresses colony
formation in U2OS cells
Colony-forming assays were performed to determine
the effect of SIK2 deletion on colony formation. The results showed
that the number of colonies of the SIK2-siRNA group (1492.00±74.65)
was significantly less than that of the scr-siRNA group
(1713.00±159.46) in U2OS cells (P<0.01) (Fig. 4C). This clearly demonstrates that
the number of the colonies decreased significantly with the SIK2
deletion.
SIK2 silencing induces apoptosis in U2OS
cells
The flow cytometric analysis shows that the
percentage of apoptotic U2OS cells was 8.05±0.20% in the scr-siRNA
group cells, and the percentage of apoptotic cells increased to
27.09±3.07% in the SIK2-siRNA group cells (P<0.01) (Fig. 4D). It indicates that the percentage
of apoptotic cells increased significantly with the SIK2
deletion.
SIK2 is involved in U2OS cell
invasion
The images of cells stained with crystal violet
suggests that knocking down the expression of SIK2 resulted in the
decrease of the amount of invaded cells. Additional observation of
absorption assay also proved that the migration ability of U2OS
cells reduced after knocking down the expression of SIK2. The
results showed that OD value of 570 nm was 0.13±0.02 in the
SIK2-siRNA group and is significantly less than the reading in the
scr-siRNA group (0.19±0.01, P<0.01) (Fig. 4E), all of these data suggest that
SIK2 was involved in the invasion of U2OS cells.
Discussion
Osteosarcoma occurs most frequently at the
metaphysis of long bones during longitudinal growth spurts in
children and young adults (19).
The treatment and prognosis for this disease still need to be
improved.
USP1 belongs to the cysteine protease family, the
USP1 gene encodes a 785 amino acid protein with a predicted
molecular weight of 88.2 kDa (20). USP1 contains the conserved USP
domain that characterizes this DUB family, with an amino-terminal
Cys box motif and a carboxy-terminal, His box motif that contain
the catalytic residues (Cys90, His593 and Asp751) (15,21).
Notably, it has been uncovered that the catalytic activity of USP1
is stimulated through the formation of a tight complex with a WD40
repeat protein UAF1 (22). UAF1
plays a critical role as a cofactor and UAF1 binding induces
conformational changes in USP1 active site increasing the enzyme
activity dramatically by stabilizing it. Importantly, studies show
that several inhibitors of the USP1/UAF1 complex act
synergistically with cisplatin in cancer-derived cell lines,
indicating that this complex may represent a potential therapeutic
target in cancer (23–25).
DUBs play crucial roles in cancer development,
progression and metastasis (26).
Targeting DUBs is a new strategy for anti-tumorigenic therapeutics
(27). USP1-null mice were
hypersensitive to DNA damage, it is likely that targeting USP1
could increase the sensitivity of cancer cells to DNA damaging
agents (28).
In the present study, we found that silencing of
USP1 could inhibit osteosarcoma cell proliferation and invasion,
which provides direct evidence that USP1 may serve as a target for
such tumor treatment. The detailed mechanisms underlying how USP1
functions in osteosarcoma still need to be elucidated in the
future.
Salt-inducible kinase 2 (SIK2) is a multifunctional
serine/threonine kinase of the AMP-activated protein kinase (AMPK)
sub-family, and it plays a role in cAMP response element-binding
protein (CREB)-mediated gene transcription. Previous studies have
demonstrated that increased SIK2 expression is significantly
correlated with poor prognosis in overall survival in patients with
high-grade serous ovarian cancer (HGSC). Recent reports have shown
that SIK2 is required for bipolar mitotic spindle formation and is
a potential target for ovarian cancer therapy. Examination of SIK2
found that it regulates mitotic progression and transcription in
prostate cancer cells. Knockdown of SIK2 expression inhibits growth
and induces cell cycle arrest and apoptosis (29,30).
The results showed that knockdown of USP1 could downregulate
SIK2.
MG132 is a specific proteasome inhibitor and reduces
the degradation of ubiquitin-conjugated proteins in mammalian cells
(31). In order to explore the
relationship between USP1 and SIK2, the U2OS cells were treated
with MG132. The study found that MG132 could upregulate the
expression of SIK2, it suggests that USP1 may stabilize the
expression of SIK2 through protein ubiquitination. In the present
study, we also found that knockdown of SIK2 could inhibit the
ability of forming colonies, induce apoptosis and reduce the
invasiveness of osteosarcoma cells, which indicates the essential
roles of SIK2 in such tumors.
MMP-2, an important member of the MMP family, is
able to degrade extracellular matrix components to promote cancer
cell migration and invasion in multiple tumor types (32–34).
GSK-3β is a ubiquitous serine/threonine kinase that plays different
roles in different types of cancers (35). The GSK-3β gene may function as an
oncogene in various types of human cancer, including colon cancer
and osteosarcoma (36,37). Bcl-2, a kind of classic
proto-oncogene, plays an important regulatory role in many kinds of
malignant tumor cells apoptosis and distant metastasis, such as
liver, lung, kidney, bladder and gastric cancer (38). Experimental and pathological
evidence shows that cyclin E1 deregulation is oncogenic (39,40).
Elevated cyclin E1 is associated with aggressive disease in a
variety of human tumors (41). The
Notch signaling pathway is shown to be an important driver of tumor
growth and metastasis and dysregulated in many cancers (42,43).
Notch1 is required for multiple cell fate determination processes,
the deregulation of the Notch1 signaling cascade plays a crucial
role in some solid tumors (44).
Stat3, a central cytoplasmic transcription factor, is an important
regulator of many biological processes (45). It regulates a number of genes that
are critical to tumor cell proliferation, invasion, metastasis,
angiogenesis and immune evasion (46,47).
Previous studies revealed that cyclin A1 is highly expressed in
many types of cancer, such as cancers of breast, lung and prostate
(48–50). As a member of Wingless family and a
secreted glycoprotein, Wnt-1 binds and activates the frizzled
receptor, triggering a signaling cascade through GSK-3β and
adenomatous polyposis coli protein (APC). Some cancer cells show
upregulation of Wnt-1 signaling and continue to metastasize
(51–53).
Notably, in the present study, all the above factors
were reduced after knowdown of USP1 expression, which indicated
that the inhibition of proliferation and invasion of human
osteosarcoma cells after knockdown of USP1 expression is through
reducing expression of these factors. Further experiments are
required.
It is important to note that USP1 expression is
increased in osteosarcoma tissues compared to cartilage tumor
tissues and normal bone tissues. This suggests an association
between the overexpression of USP1 and osteosarcoma genesis.
In summary, our results suggest that silencing of
USP1 inhibits cell proliferation and invasion in osteosarcoma cells
in vitro, and reduce some tumor-related gene expression,
which in turn provides the potential mechanisms of how UPS1
functions in osteosarcoma cells, and therapeutic targets for such
tumors in the future.
Acknowledgements
The present study was supported by grants from the
Natural Science Foundation of Jiangsu Province (BK20151177), the
National Science Foundation of China (81471263), the Suzhou
Planning Project of Science and Technology (SYS201301), the Natural
Science Foundation for the Youth (no. 81402220) and the Youth
Scientific Research Project of Wuxi Health Bureau (Q201412). The
authors would like to thank Dr Wenxiang Wei (Soochow University,
Suzhou 215123, China) for his sincere help and technical
support.
References
|
1
|
Endo-Munoz L, Evdokiou A and Saunders NA:
The role of osteoclasts and tumour-associated macrophages in
osteosarcoma metastasis. Biochim Biophys Acta. 1826:434–442.
2012.PubMed/NCBI
|
|
2
|
Poletajew S, Fus L and Wasiutyński A:
Current concepts on pathogenesis and biology of metastatic
osteosarcoma tumors. Ortop Traumatol Rehabil. 13:537–545. 2011.
View Article : Google Scholar
|
|
3
|
Allison DC, Carney SC, Ahlmann ER,
Hendifar A, Chawla S, Fedenko A, Angeles C and Menendez LR: A
meta-analysis of osteosarcoma outcomes in the modern medical era.
Sarcoma. 2012:7048722012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Yao C, Wei JJ, Wang ZY, Ding HM, Li D, Yan
SC, Yang YJ and Gu ZP: Perifosine induces cell apoptosis in human
osteosarcoma cells: New implication for osteosarcoma therapy? Cell
Biochem Biophys. 65:217–227. 2013. View Article : Google Scholar
|
|
5
|
Chen ZJ and Sun LJ: Nonproteolytic
functions of ubiquitin in cell signaling. Mol Cell. 33:275–286.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Komander D: The emerging complexity of
protein ubiquitination. Biochem Soc Trans. 37:937–953. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fraile JM, Quesada V, Rodríguez D, Freije
JM and López-Otín C: Deubiquitinases in cancer: New functions and
therapeutic options. Oncogene. 31:2373–2388. 2012. View Article : Google Scholar
|
|
8
|
García-Santisteban I, Peters GJ,
Giovannetti E and Rodríguez JA: USP1 deubiquitinase: Cellular
functions, regulatory mechanisms and emerging potential as target
in cancer therapy. Mol Cancer. 12:912013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Williams SA, Maecker HL, French DM, Liu J,
Gregg A, Silverstein LB, Cao TC, Carano RA and Dixit VM: USP1
deubiquitinates ID proteins to preserve a mesenchymal stem cell
program in osteosarcoma. Cell. 146:918–930. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu Y, Luo X, Hu H, Wang R, Sun Y, Zeng R
and Chen H: Integrative proteomics and tissue microarray profiling
indicate the association between overexpressed serum proteins and
non-small cell lung cancer. PLoS One. 7:e517482012. View Article : Google Scholar
|
|
11
|
Kim H and D’Andrea AD: Regulation of DNA
cross-link repair by the Fanconi anemia/BRCA pathway. Genes Dev.
26:1393–1408. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang TT, Nijman SM, Mirchandani KD,
Galardy PJ, Cohn MA, Haas W, Gygi SP, Ploegh HL, Bernards R and
D’Andrea AD: Regulation of monoubiquitinated PCNA by DUB
autocleavage. Nat Cell Biol. 8:339–347. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fox JT, Lee KY and Myung K: Dynamic
regulation of PCNA ubiquitylation/deubiquitylation. FEBS Lett.
585:2780–2785. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cohn MA, Kowal P, Yang K, Haas W, Huang
TT, Gygi SP and D’Andrea AD: A UAF1-containing multisubunit protein
complex regulates the Fanconi anemia pathway. Mol Cell. 28:786–797.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Villamil MA, Chen J, Liang Q and Zhuang Z:
A noncanonical cysteine protease USP1 is activated through active
site modulation by USP1-associated factor 1. Biochemistry.
51:2829–2839. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yang K, Moldovan GL, Vinciguerra P, Murai
J, Takeda S and D’Andrea AD: Regulation of the Fanconi anemia
pathway by a SUMO-like delivery network. Genes Dev. 25:1847–1858.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Davidson BL and McCray PB Jr: Current
prospects for RNA interference-based therapies. Nat Rev Genet.
12:329–340. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
18
|
Angaji SA, Hedayati SS, Poor RH, Madani S,
Poor SS and Panahi S: Application of RNA interference in treating
human diseases. J Genet. 89:527–537. 2010. View Article : Google Scholar
|
|
19
|
Mirabello L, Troisi RJ and Savage SA:
Osteosarcoma incidence and survival rates from 1973 to 2004: data
from the Surveillance, Epidemiology, and End Results Program.
Cancer. 115:1531–1543. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Fujiwara T, Saito A, Suzuki M, Shinomiya
H, Suzuki T, Takahashi E, Tanigami A, Ichiyama A, Chung CH,
Nakamura Y, et al: Identification and chromosomal assignment of
USP1, a novel gene encoding a human ubiquitin-specific protease.
Genomics. 54:155–158. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Békés M, Okamoto K, Crist SB, Jones MJ,
Chapman JR, Brasher BB, Melandri FD, Ueberheide BM, Denchi EL and
Huang TT: DUB-resistant ubiquitin to survey ubiquitination switches
in mammalian cells. Cell Rep. 5:826–838. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cohn MA, Kee Y, Haas W, Gygi SP and
D’Andrea AD: UAF1 is a subunit of multiple deubiquitinating enzyme
complexes. J Biol Chem. 284:5343–5351. 2009. View Article : Google Scholar :
|
|
23
|
Chen J, Dexheimer TS, Ai Y, Liang Q,
Villamil MA, Inglese J, Maloney DJ, Jadhav A, Simeonov A and Zhuang
Z: Selective and cell-active inhibitors of the USP1/UAF1
deubiquitinase complex reverse cisplatin resistance in non-small
cell lung cancer cells. Chem Biol. 18:1390–1400. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liang Q, Dexheimer TS, Zhang P, Rosenthal
AS, Villamil MA, You C, Zhang Q, Chen J, Ott CA, Sun H, et al: A
selective USP1-UAF1 inhibitor links deubiquitination to DNA damage
responses. Nat Chem Biol. 10:298–304. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wahi D, Jamal S, Goyal S, Singh A, Jain R,
Rana P and Grover A: Cheminformatics models based on machine
learning approaches for design of USP1/UAF1 abrogators as
anticancer agents. Syst Synth Biol. 9:33–43. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
McClurg UL and Robson CN: Deubiquitinating
enzymes as oncotargets. Oncotarget. 6:9657–9668. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
D’Arcy P and Linder S: Molecular pathways:
Translational potential of deubiquitinases as drug targets. Clin
Cancer Res. 20:3908–3914. 2014. View Article : Google Scholar
|
|
28
|
Park E, Kim JM, Primack B, Weinstock DM,
Moreau LA, Parmar K and D’Andrea AD: Inactivation of Uaf1 causes
defective homologous recombination and early embryonic lethality in
mice. Mol Cell Biol. 33:4360–4370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ahmed AA, Lu Z, Jennings NB,
Etemadmoghadam D, Capalbo L, Jacamo RO, Barbosa-Morais N, Le XF,
Vivas-Mejia P, Lopez-Berestein G, et al; Australian Ovarian Cancer
Study Group. SIK2 is a centrosome kinase required for bipolar
mitotic spindle formation that provides a potential target for
therapy in ovarian cancer. Cancer Cell. 18:109–121. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Bon H, Wadhwa K, Schreiner A, Osborne M,
Carroll T, Ramos-Montoya A, Ross-Adams H, Visser M, Hoffmann R,
Ahmed AA, et al: Salt-inducible kinase 2 regulates mitotic
progression and transcription in prostate cancer. Mol Cancer Res.
13:620–635. 2015. View Article : Google Scholar :
|
|
31
|
Jamart C, Raymackers JM, Li An G,
Deldicque L and Francaux M: Prevention of muscle disuse atrophy by
MG132 proteasome inhibitor. Muscle Nerve. 43:708–716. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Aparna M, Rao L, Kunhikatta V and
Radhakrishnan R: The role of MMP-2 and MMP-9 as prognostic markers
in the early stages of tongue squamous cell carcinoma. J Oral
Pathol Med. 44:345–352. 2015. View Article : Google Scholar
|
|
33
|
Yang W, Li Q and Pan Z:
Sphingosine-1-phosphate promotes extravillous trophoblast cell
invasion by activating MEK/ERK/MMP-2 signaling pathways via
S1P/S1PR1 axis activation. PLoS One. 9:e1067252014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen SJ, Yao XD, Peng BO, Xu YF, Wang GC,
Huang J, Liu M and Zheng JH: Epigallocatechin-3-gallate inhibits
migration and invasion of human renal carcinoma cells by
downregulating matrix metalloproteinase-2 and matrix
metalloproteinase-9. Exp Ther Med. 11:1243–1248. 2016.PubMed/NCBI
|
|
35
|
Kotliarova S, Pastorino S, Kovell LC,
Kotliarov Y, Song H, Zhang W, Bailey R, Maric D, Zenklusen JC, Lee
J, et al: Glycogen synthase kinase-3 inhibition induces glioma cell
death through c-MYC, nuclear factor-kappaB, and glucose regulation.
Cancer Res. 68:6643–6651. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Salim T, Sjölander A and Sand-Dejmek J:
Nuclear expression of glycogen synthase kinase-3β and lack of
membranous β-catenin is correlated with poor survival in colon
cancer. Int J Cancer. 133:807–815. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tang QL, Xie XB, Wang J, Chen Q, Han AJ,
Zou CY, Yin JQ, Liu DW, Liang Y, Zhao ZQ, et al: Glycogen synthase
kinase-3β, NF-κB signaling, and tumorigenesis of human
osteosarcoma. J Natl Cancer Inst. 104:749–763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yao K, Xing HC, Wu B, Li Y, Liao AJ, Yang
W and Liu ZG: Effect of TIEG1 on apoptosis and expression of
Bcl-2/Bax and Pten in leukemic cell lines. Genet Mol Res.
14:1968–1974. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hwang HC and Clurman BE: Cyclin E in
normal and neoplastic cell cycles. Oncogene. 24:2776–2786. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Spruck CH, Won KA and Reed SI: Deregulated
cyclin E induces chromosome instability. Nature. 401:297–300. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Hunt KK and Keyomarsi K: Cyclin E as a
prognostic and predictive marker in breast cancer. Semin Cancer
Biol. 15:319–326. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Arcaroli JJ, Quackenbush KS, Purkey A,
Powell RW, Pitts TM, Bagby S, Tan AC, Cross B, McPhillips K, Song
EK, et al: Tumours with elevated levels of the Notch and Wnt
pathways exhibit efficacy to PF-03084014, a γ-secretase inhibitor,
in a preclinical colorectal explant model. Br J Cancer.
109:667–675. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Takebe N, Miele L, Harris PJ, Jeong W,
Bando H, Kahn M, Yang SX and Ivy SP: Targeting Notch, Hedgehog, and
Wnt pathways in cancer stem cells: Clinical update. Nat Rev Clin
Oncol. 12:445–464. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ranganathan P, Weaver KL and Capobianco
AJ: Notch signalling in solid tumours: A little bit of everything
but not all the time. Nat Rev Cancer. 11:338–351. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Darnell JE Jr, Kerr IM and Stark GR:
Jak-STAT pathways and transcriptional activation in response to
IFNs and other extracellular signaling proteins. Science.
264:1415–1421. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hajimoradi M, Mohammad Hassan Z, Ebrahimi
M, Soleimani M, Bakhshi M, Firouzi J and Samani FS: STAT3 is
overactivated in gastric cancer stem-like cells. Cell J.
17:617–628. 2016.PubMed/NCBI
|
|
48
|
Shames DS, Girard L, Gao B, Sato M, Lewis
CM, Shivapurkar N, Jiang A, Perou CM, Kim YH, Pollack JR, et al: A
genome-wide screen for promoter methylation in lung cancer
identifies novel methylation markers for multiple malignancies.
PLoS Med. 3:e4862006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Syed Khaja AS, Dizeyi N, Kopparapu PK,
Anagnostaki L, Härkönen P and Persson JL: Cyclin A1 modulates the
expression of vascular endothelial growth factor and promotes
hormone-dependent growth and angiogenesis of breast cancer. PLoS
One. 8:e722102013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wegiel B, Bjartell A, Ekberg J, Gadaleanu
V, Brunhoff C and Persson JL: A role for cyclin A1 in mediating the
autocrine expression of vascular endothelial growth factor in
prostate cancer. Oncogene. 24:6385–6393. 2005.PubMed/NCBI
|
|
51
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.PubMed/NCBI
|
|
52
|
van Gijn ME, Daemen MJ, Smits JF and
Blankesteijn WM: The wnt-frizzled cascade in cardiovascular
disease. Cardiovasc Res. 55:16–24. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kim YM, Kim IH and Nam TJ: Capsosiphon
fulvescens glycoprotein inhibits AGS gastric cancer cell
proliferation by downregulating Wnt-1 signaling. Int J Oncol.
43:1395–1401. 2013.PubMed/NCBI
|