Introduction
Based on the transcript size, non-coding RNAs
(ncRNAs) are divided into two groups: small non-coding RNAs and
long non-coding RNAs (lncRNAs) (1,2).
Biologically, lncRNAs are specifically characterized as RNA
transcripts that are longer than 200 nt in length and do not have
an open reading frame (ORF). To date, several biological functions
of lncRNAs have been described, including i) signal molecules in
gene transcription, ii) decoys to titrate transcription factors,
iii) directors for chromatin, and iv) scaffolds for proteins to
form ribonucleoprotein complexes (1,2).
Differential expression of lncRNAs is a well-known event and
regarded as a hallmark of cancer (3). Many lncRNAs have been characterized
in cancers and they function as tumor suppressors or oncogenes. For
example, imprinted H19 is an lncRNA first reported in mammalian
cells, encoded by a gene located on chromosome 11p15.5; H19 is
involved in fetal and placental growth (4). H19 has been reported as a marker in
breast and cervical cancers (5).
In gastric cancer, the lncRNA H19 promotes carcinogenesis and
metastasis by interacting with miR-675 (4). Other characterized lncRNA oncogenes
and tumor suppressors include ANRIL, HOTTIP, HOTAIR, BCAR4,
lncRNA-p21 and XIST (6).
In cancer, lncRNAs have emerged as critical players
involved in signaling transduction, DNA damage repair, cell cycle
control, epigenetic modifications, and chromosome modeling
(6). For example, the lncRNA
HOTAIR regulates chromatin dynamics and promotes metastasis in
multiple types of cancers, including breast cancer, colorectal
cancer, non-small cell lung carcinoma and hepatocellular carcinoma
(7). The aberrant activation of
cellular signal transduction is well documented in cancer,
promoting unlimited growth and proliferation of cancer cells.
lncRNAs function in cellular signaling pathways and modulate cancer
development and progression. For instance, the lncRNA ATB
stabilizes IL-11 mRNA and activates the STAT3 signaling cascade
which drives cell colonization at distant metastatic sites
(8). BCAR4 (breast cancer
anti-estrogen resistance 4) increases migration and invasion of
breast cancer cells through activation of a non-canonical Hedgehog
signaling pathway (9). CCAT2
(colon cancer-associated transcript 2) and MALAT1
(metastasis-associated lung adenocarcinoma transcript 1) promote
cancer progression and metastasis through activation of the Wnt
signaling pathway (10,11). The present study characterized a
new tumor suppressor lncRNA, named p53-inducible cancer-associated
RNA transcript 1 (PICART1). This PICART1 lncRNA consists of 3
exons, 2533 bp in length (gene ID: 284080) and is encoded by a gene
located at 17q21.33. Our data demonstrated that PICART1 inhibited
cell proliferation and migration in breast and colorectal cancer
cells through suppression of the AKT/GSK3β/β-catenin signaling
pathway, functioning as a tumor suppressor.
Materials and methods
Ethics statement
IRB protocol was approved by Springfield Committee
for Research Involving Human Subjects.
Human sample procurement
Frozen breast (n=50 pairs) and colon (n=50 pairs)
cancer and matched normal adjacent tissues were procured following
approved IRB protocol from the Tissue Bank of Simmons Cancer
Institute at the Southern Illinois University. These frozen
specimens were used for RNA extract and real-time RT-PCR analyses
of PICART1 expression.
Cell culture
Human breast cancer cells (MCF7 and MDA-MB-231) were
cultured in Dulbecco's modified Eagle's medium (DMEM; Thermo Fisher
Scientific, Waltham, MA, USA) supplemented with 10% FBS
(Sigma-Aldrich, St. Louis, MO, USA), 1% penicillin/streptomycin at
37°C, 5% CO2. The basal medium for HEK 293T cells was
the same as that for MCF7 cells, but additional 2.0 mM L-glutamine
was added. Human colorectal cancer cells (HCT116 and HCT8) were
grown in RPMI-1640 medium (Thermo Fisher Scientific) supplemented
with 10% FBS (Sigma-Aldrich), 1% penicillin/streptomycin at 37°C,
and 5% CO2. Human mammary epithelial cells (MCF10A) were
cultured with a commercial MEGM kit (catalog no. CC-3150)
supplemented with 100 ng/ml cholera toxin. All cells were purchased
from the American Type Culture Collection (ATCC) (Manassas, VA,
USA).
RNA preparation and quantitative
RT-PCR
Total RNA was extracted using TRIzol reagent
(Invitrogen, CA, USA). RNA was treated with RNase-free DNase I and
the first strand cDNA was synthesized from 1.0 µg of total
RNA with oligo dT primers and Moloney Murine Leukemia Virus (M-MLV)
reverse transcriptase according to the manufacturer's protocol (New
England Biolab, Beverly, MA, USA). To detect the PICART1
transcript, the SYBR Green method was used with PICART1 primers:
forward 5′-AGGCAGCTACTGTAATAAT-3′ and reverse
5′-GTACCCTGGGCCTTTCTTAC-3′. The internal control was β-actin. ΔΔCt
values were used to determine relative expression levels as fold
changes (12).
Plasmid construction
For ectopic expression of PICART1, its cDNA was
amplified by RT-PCR and then subcloned into the pCDH-CMV-EF1-copGFP
vector (System Biosciences, Mountain View, CA, USA) (13). Conventional sequencing was
conducted to confirm the cDNA sequence. PCR primers were as
follows: F1 forward, 5′-GCTGCTGCTGCTGAAAAGGC-3′, and reverse,
5′-CTGGCAGATTCTGAGCCAGG-3′; F2 forward, 5′-CCTGGCTCAGAATCTGCCAG-3′
and reverse, 5′-GAATTCATGCATAAGGGCCCATGTGC-3′; F3 forward,
5′-ATGCATTCACAGGTGTTTGCCTATGC-3′ and reverse,
5′-ATGCATCCCACTTTGGTATTTTAGTC-3′; shRNA-2 forward,
5′-AGGCAGCTACTGTAATAAT-3′ and reverse, 5′-ATTATTACAGTAGCTGCCT-3′;
shRNA-3 forward, 5′-TCCTGTGGAGAAGCCTCTTTACT-3′ and reverse,
5′-AGTAAAGAGGCTTCTCCACAGGA-3′. High Fidelity Phusion DNA polymerase
(Thermo Fisher Scientific) was used for PCR amplification. PICART1
promoter was amplified by PCR using human genomic DNA as templates
and subcloned into the pGL4 luciferase vector (Promega, Madison,
WI, USA). PCR primers were as follows: −3000-bp forward,
5′-CGAGCCCAGCTGGCTCTCCCAAATG-3′, −1592-bp forward,
5′-ATCTGTGGAATGCAGGGAAGATGTC-3′, −1227-bp forward,
5′-AACAGGCTAGTGCTGGGTACTCG-3′ and −681 bp forward,
5′-TAAGATCCGTGGGTCTATCTTCC TG-3′. The common reverse primer was
5′-CCAGATGCGCCAATGCCAGGTCTCT-3′. Primers for point mutations were
mut1 forward, 5′-CCTCTTCCTCCTGGAACGTCTC CTG-3′ and reverse,
5′-CAGGAGACGTTCCAGGAGGAAGAGG-3′; mut2 forward,
5′-CCTCTTTTTCCTGGATTTTCTCCTG-3 and reverse
5′-GGAGGAAAAATCCCAAAAAAGGCTG-3′.
ChIP (chromatin immunoprecipitation)
assay
MCF7 cells were fixed in 1% formaldehyde for 10 min
and cell lysates were prepared for immunoprecipitation following
protocol according to a previous published study (14). Immune complexes were collected with
50 µl of protein G-agarose beads and DNA was extracted using
proteinase K digestion and phenol/chloroform extraction. PCR
amplification was performed with forward,
5′-CTGAGGCCACGGCAGAGAGA-3′ and reverse, 5′-GGCATGAGGGCGTCTGGTGG-3′
primers at −1763 and −1728 bp of the PICART1 promoter.
Cell transfection and infection
For packaging of lentiviruses, 293T cells were
seeded in 10-cm dishes. Cells were transfected with expression
plasmids pCDH, PICART1, pGreenpuro, shRNA-2 or shRNA-3 with
packaging plasmids REV, VSVG and GAG for 24 h. Then the medium was
changed and cells were continued to culture for 48 h to collect
viral particles (15). For
infection, cells were spread at 60% of confluence and exposed to
lentiviruses (MOI=1) with 1:2000 diluted Polybrene (Millipore,
Billerica, MA, USA).
Western blotting
Cells were lysed in cell lysis buffer (Roche,
Indianapolis, IN, USA) with a protease inhibitor cocktail followed
by centrifugation at 14,000 rpm for 5 min to collect supernatant
proteins. Protein separation, membrane blotting and antibody
probing were conducted as previously described (16). Antibodies used were anti-p53
(dilution 1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA),
anti-pAKT (Thr308) (dilution 1:1000; Cell Signaling Technology,
Danvers, MA, USA), anti-pAKT (Ser473) (dilution 1:1000; Cell
Signaling Technology), anti-AKT (dilution 1:1000; Cell Signaling
Technology), anti-GSK3β (S9) (dilution 1:1000, Cell Signaling
Technology), anti-GSK3β (dilution 1:1000; Cell Signaling
Technology), anti-β-catenin (dilution 1:1000; Cell Signaling
Technology), anti-cyclin D1 (dilution 1:1000; Cell Signaling
Technology), anti-c-Myc (dilution 1:500; Santa Cruz Biotechnology)
and anti-p21Waf/cip1 (dilution 1:1000; Cell Signaling
Technology). β-actin was used as an internal control.
Luciferase activity
The dual luciferase assay kit (Promega, Madison, WI,
USA) was used following the manufacturer's protocol. Cells were
collected and lysed for luciferase assay 48 h after transfection.
Renilla luciferase was used as an internal control for
normalization (17).
Transwell migration and invasion
assays
Transwell migration assays were performed using
8.0-µm pore inserts (BD Biosciences, San Jose, CA, USA). A
total of 2–5×104 cells were suspended in 500 µl
serum-free medium and loaded into upper wells; lower chambers were
filled with 750 µl complete medium with 10% FBS. For the
invasion assays, inserts were coated with 1 mg/ml Mitrigel and
pre-incubated at 37°C for 2 h. A total of 3–7×104 cells
were suspended in 500 µl serum-free medium and loaded into
the coated inserts; lower chambers were filled with 750 µl
complete medium. Migration and invasion chambers were incubated in
a 5% CO2 incubator at 37°C for 36–48 h. Cells were
stained and counted under a microscope for 5 random fields
(18).
Wound healing assays
Cells were seeded in 6-well plates at 90% confluence
and cultured in medium supplemented with 2% FBS. Scratches were
made using a 200-µl tip. Migration distance was estimated
from images (4 fields) taken at each time point (18).
Cell proliferation
For the cell proliferation assays, cells were split
into 96-well plates at 2000–3000 cells/per well. Viable cells were
measured by tetrazolium salt,
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assays over 5 days (19). For cell
counting assays, cells were split into 12-plates. At the indicated
time points, cells were collected and subjected to trypan blue
staining. Viable cells were counted in a Vi-cell counter (Beckman
Coulter, Brea, CA, USA).
Statistical analysis
Data were analyzed by SPSS Statistical software
(SPSS Inc., Chicago, IL, USA). Values are shown as mean ± SD. The
Student's t-test was used for the analyses. A P-value of ≤0.05 was
considered to indicate a statistically significant difference.
Results
Identification of a novel long non-coding
RNA transcript PICART1
p53 is a master transcription factor regulating the
expression of a wide range of coding and non-coding genes. HDM2
(MDM2 in mice) binds to the p53 protein and inhibits its
transactivation activity in target gene expression and triggers
ubiquitination-proteasome degradation (20). A lead compound, RITA (reactivation
of p53 and induction of tumor cell apoptosis), binds to p53 and
thus leads to p53 conformational changes and dissociation from
HDM2, reactivating p53 (21).
Using transcriptome RNA sequencing analysis in MCF7 cells
(p53wt) treated with RITA (100 nM), we recognized 10 new
non-coding RNA species with pivotal expression changes in response
to RITA treatment. Table I
summarizes the basic information of these new RNA transcripts
induced by RITA. A non-coding RNA transcript LOC84080 was
upregulated by RITA/p53 >4.5 fold. This RNA species was 2533 bp
in length, and identified as being a long non-coding RNA (lncRNA).
We named it p53 inducible cancer-associated RNA transcript 1
(PICART1).
 | Table ITop 10 selected LncRNAs in RNA
sequencing. |
Table I
Top 10 selected LncRNAs in RNA
sequencing.
| lncRNA | Regulation | Fold change | Official full
name | Location | Genbank ID | Exon numbers |
|---|
| LOC33161 | Down | −2.65 | Long intergenic
non-protein coding RNA 1001 | Chr11p15.5 | 100133161 | 3 |
| LOC32287 | Down | −1.88 | LOC100132287 | Chr1p36.33 | 100132287 | 3 |
| LOC94362 | Down | −2.86 | LOC100294362 | Chr17 | 100294362 | 2 |
| LOC72216 | Down | −2.60 | LOC100272216 | Chr 5 | 100272216 | 1 |
| LOC30091 | Down | −2.36 | Long intergenic
non-protein coding RNA 886 | Chr3q25.31 | 730091 | 3 |
| LOC28978 | Up | 10.05 | UNC5B antisense RNA
1 | Chr10q22.1 | 728978 | 2 |
| LOC30102 | Up | 1.79 | Quinone
oxidoreductase-like protein 2 pseudogene | Chr1q25.2 | 730102 | 10 |
| LOC00494 | Up | 1.10 | Long intergenic
non-protein coding RNA 494 | Chr20q13.13 | 284749 | 5 |
| LOC88692 | Up | 1.90 | LOC388692 | Chr1q21.2 | 388692 | 2 |
| PICART1 | Up | 4.56 |
LOC284080 |
Chr17q21.33 | 284080 | 3 |
PICART1 is downregulated in breast and
colon cancer cell lines and tissues
As a potential p53 target, PICART1 was first
examined in cancer cell lines and tissues to see whether this
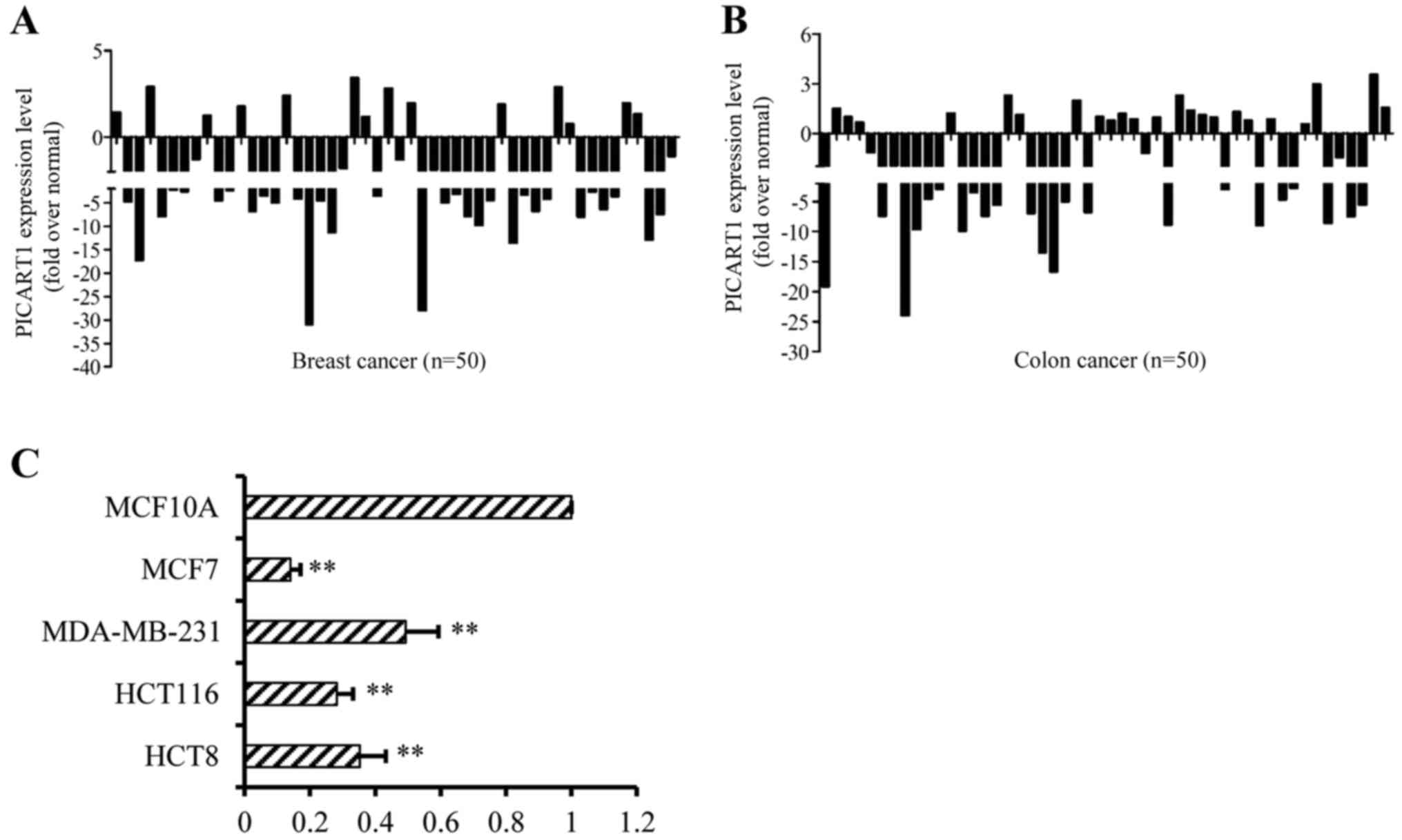
lncRNA species is involved in tumor development and progression. As
shown in Fig. 1A, in 50 paired
breast cancer and adjacent normal tissues, PICART1 expression was
decreased by >2-fold in 32 (64.0%) breast cancer cases compared
to that in the adjacent normal breast tissues, with a maximal
decrease of >30-fold. Similarly, in 50 paired colon cancer and
adjacent normal tissues, PICART1 expression was decreased by
>2-fold in 23 (46%) malignant tissues with a maximal decrease of
>25-fold (Fig. 1B). PICART1
expression was also downregulated in cancer cell lines. As shown in
Fig. 1C, PICART1 expression was
high in non-transformed human mammary epithelial cells MCF10A, but
decreased by ~7.0-, 2.0-, 3.0- and 2.5-fold in the MCF7,
MDA-MB-231, HCT116 and HCT8 cells, respectively, compared to the
MCF10A cells. These data suggest that PICART1 is downregulated in
breast and colon cancer cell lines and tissues, functioning as a
potential tumor suppressor.
PICART1 is upregulated by p53
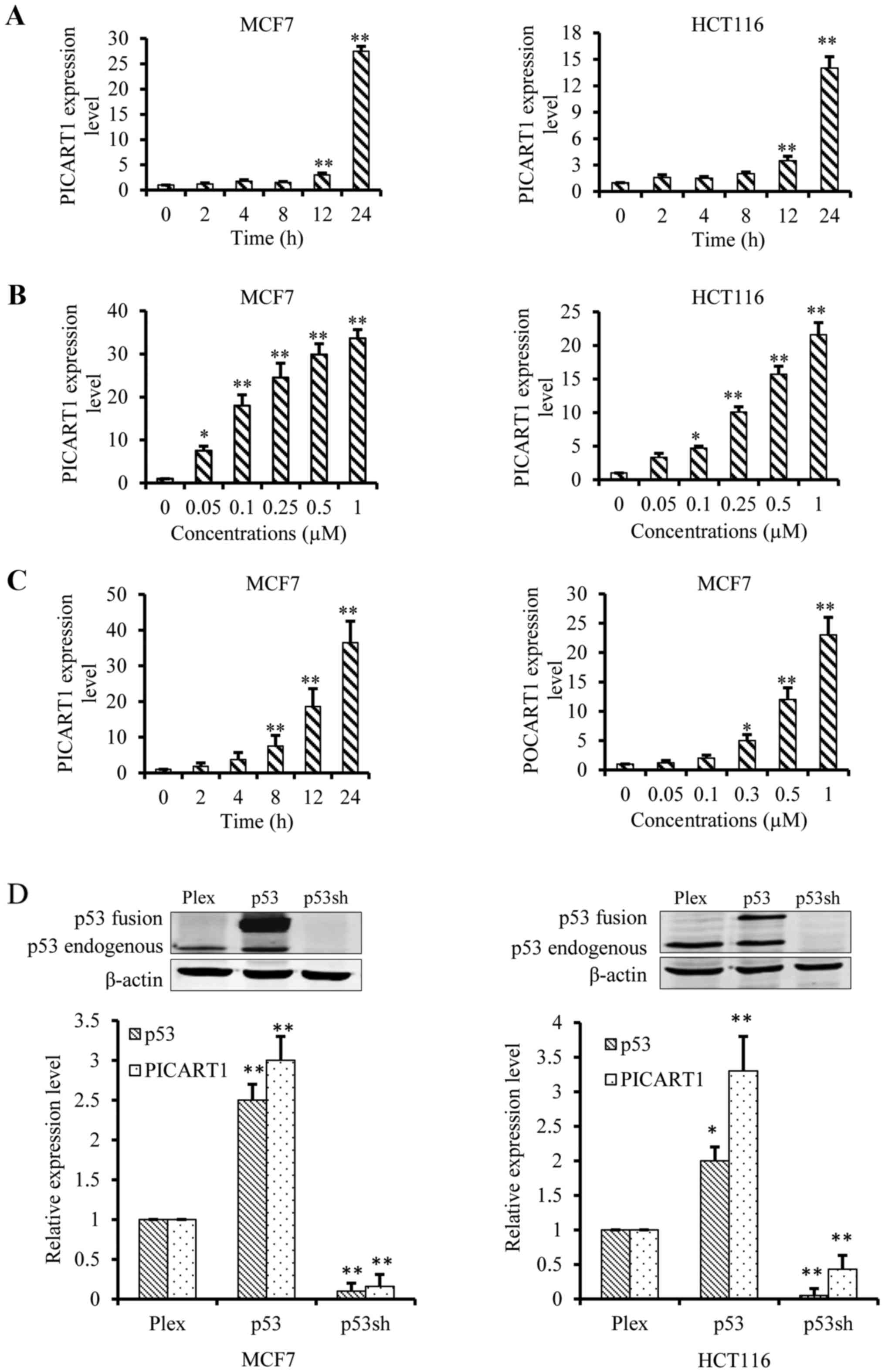
As downregulation of PICART1 was noted in breast and
colon cancers, we aimed to elucidate the regulatory mechanism of
PICART1 expression. We first verified its induction by RITA using
real-time RT-PCR. As shown in Fig.
2A, RITA at 100 nM induced PICART1 expression early at 12 h,
and the induction reached a peak of nearby 15-fold in HCT116 cells
and 30-fold in MCF7 cells at 24 h. Similarly, RITA also induced
PICART1 expression in MCF7 and HCT116 cells in a dose-dependent
manner (Fig. 2B). At 1.0 µM
of RITA, PICART1 was upregulated over 20- and 30-fold in the HCT116
and MCF7 cells, respectively. These data confirmed the induction of
PICART1 by RITA. RITA is a p53 activator (21), and thus we aimed to ascertain
whether RITA induces PICART1 expression through a p53-mediated
mechanism. To test this hypothesis, we determined the effects of
doxorubicin on PICART1 expression. Doxorubicin induces DNA damage
and thus activates p53, which is a different mechanism from that of
RITA. Our results showed that doxorubicin triggered expression of
PICART1 in MCF7 cells in a time- and dose-dependent manner
(Fig. 2C). Furthermore, we
confirmed the regulation of p53 on PICART1 expression by targeted
gene expression or silencing. As shown in Fig. 2D, PICART1 expression in MCF7 cells
was increased ~3-fold by p53 ectopic expression whereas p53
silencing decreased PICART1 expression to 10% compared to the
vector control cells. Similar results were observed in HCT116 cells
(Fig. 2D, right). These data
suggest that p53 induces PICART1 expression.
p53 upregulates PICART1 expression
through a p53 response element in its promoter
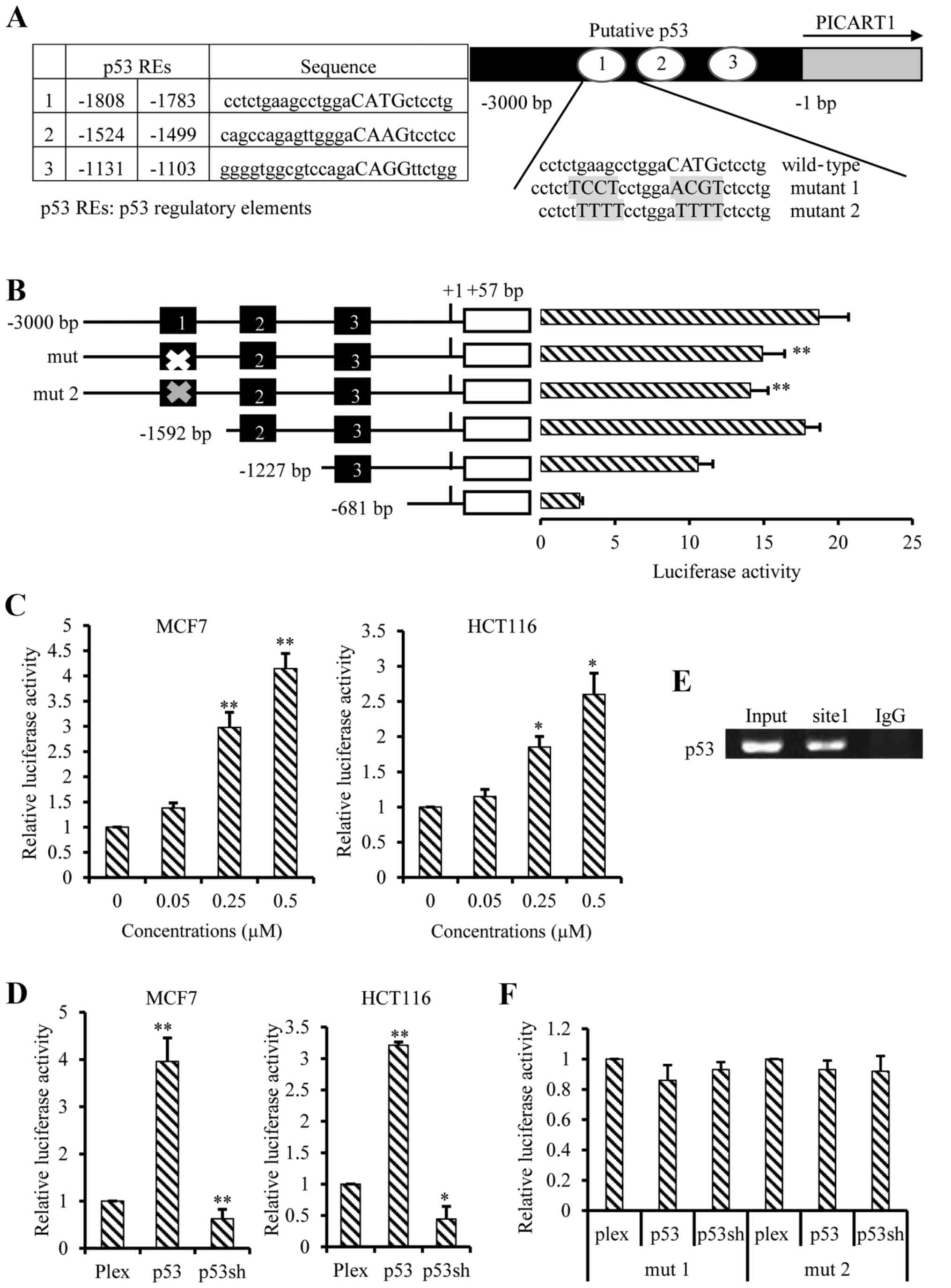
As a transcription factor, p53 may stimulate PICART1
expression through a promoter-mediated mechanism. With a p53 scan
program MatInspector software (https://www.genomatix.de/online), we analyzed the
promoter region of PICART1 and identified 3 putative p53 response
elements ~3000 bp upstream of PICART1 transcripts (Fig. 3A). Specifically, these 3 putative
p53 response elements were located at −1808 to −1783 bp, −1524 to
−1499 bp, and −1131 to −1106 bp. We then cloned this 3000-bp
promoter fragment into a pGL4 basic luciferase reporter vector and
constructed four promoter-luciferase reporter plasmids that
contained −3000, −1592, −1227 and −681 bp, respectively. The −3000
bp hosted all three putative p53 response elements, the −1592 bp
contained putative p53 response elements 2 and 3, and the −1227 bp
had only putative p53 regulatory element 3 (Fig. 3B). These promoter fragments all
demonstrated promoter activity driving luciferase reporter
expression, but the promoter activity of −1227 and −681 bp vectors
was lower compared to the longer −1592 and −3000 bp fragments,
indicating existence of enhancer(s) in the longer region (Fig. 3B).
We further tested the response of these promoter
fragments to RITA. Our results showed that the −3000-bp fragment
was activated by RITA at 0.05 to 0.5 µM in a dose-dependent
manner in both MCF7 and HCT116 cells (Fig. 3C), but the −1592 bp fragment was
not (data not shown). These data suggested that the putative p53
element 1 is functional, but the putative p53 elements 2 and 3 are
not. We then conducted co-transfections of the PICART1 promoter
(−3000 bp) with p53 gene or shRNA in MCF7 and HCT116 cells. Our
results demonstrated that luciferase activity was increased by p53
expression but decreased by p53 silencing compared to the vector
control (Fig. 3D). We examined
binding of p53 to this putative element 1 in the PICART1 promoter
using a ChIP assay. The results demonstrated that p53 binds to the
element 1 (Fig. 3E). We further
characterized this putative p53 element 1 by targeted point
mutations (Fig. 3A, right). As a
result, mutations at the putative p53 responsive element 1 lowered
down basal activity of the PICART1 promoter (Fig. 3B) and eradicated its response to
p53 (Fig. 3F). These data
suggested that p53 upregulates the PICART1 expression through the
p53 responsive element 1 in its promoter.
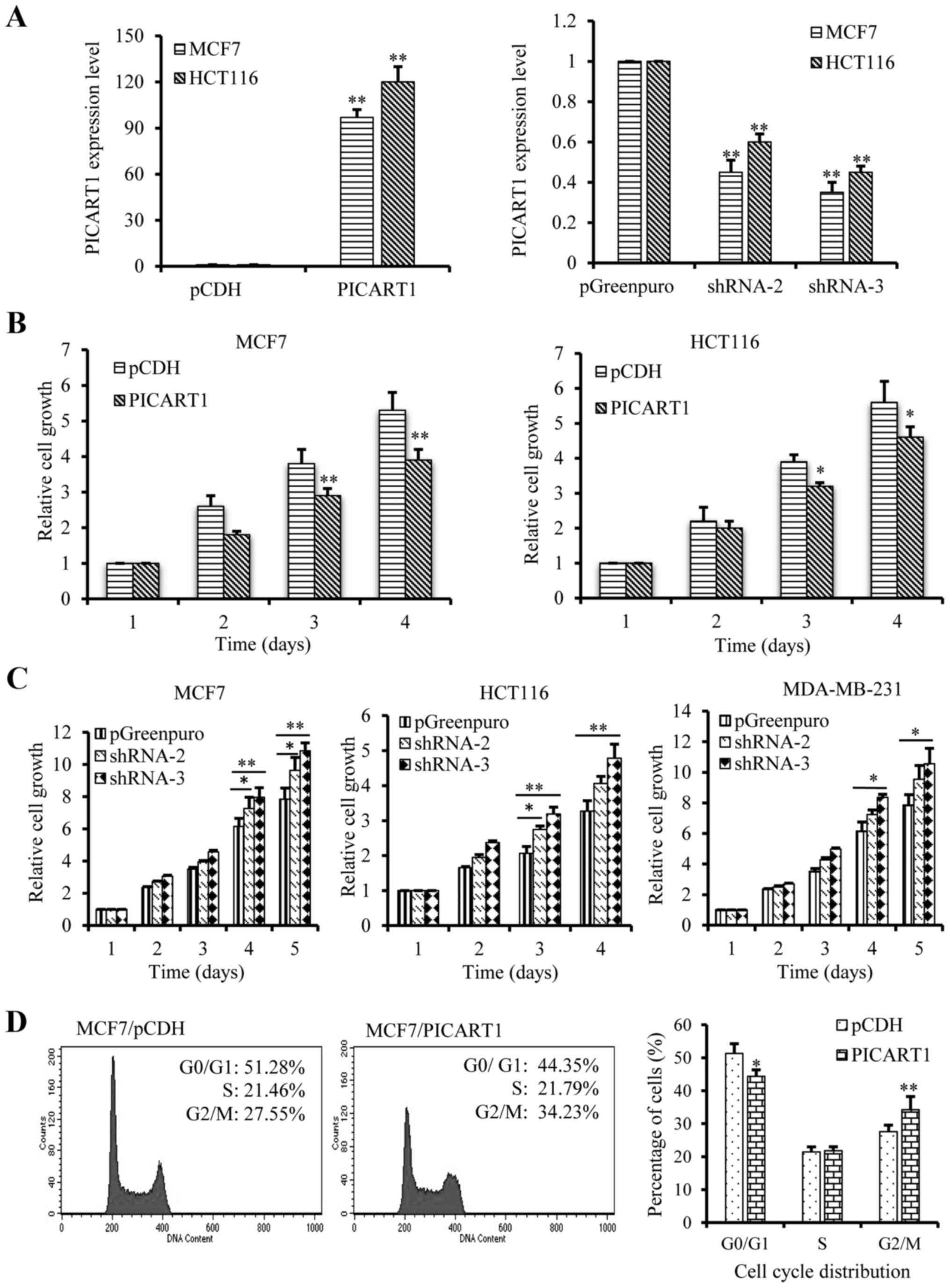
PICART1 inhibits cell proliferation,
migration, and invasion
As a target of p53 tumor suppressor, we further
estimated the biological function of PICART1 in cancer cells, using
the ectopic expression and silencing cell models. Full length
PICART1 cDNA was cloned for ectopic expression, and 3
PICART1-specific shRNA constructs were made for silencing. Fig. 4A shows the targeted ectopic
expression (left) and silencing (right) of PICART1 in MCF7 and
HCT116 cells. The data demonstrated that shRNA-2 and shRNA-3
knocked down PICART1 expression to ~40–60% compared to the vector
control. We estimated the cell growth and proliferation of these
cell lines with targeted expression and silencing of PICART1, and
the results showed that the ectopic expression of PICART1 notably
inhibited the proliferation of MCF7 and HCT116 cells as tested by
MTT assays (Fig. 4B) and viable
cell counting method (data not shown). In contrast, silencing of
PICART1 by shRNAs led to an increase in cell growth and
proliferation in the MCF7, HCT116 and MDA-MB-231 cells (Fig. 4C). Furthermore, FACScan analysis
showed that PICART1 induced cell cycle arrest at the G2/M phase
(Fig. 4D).
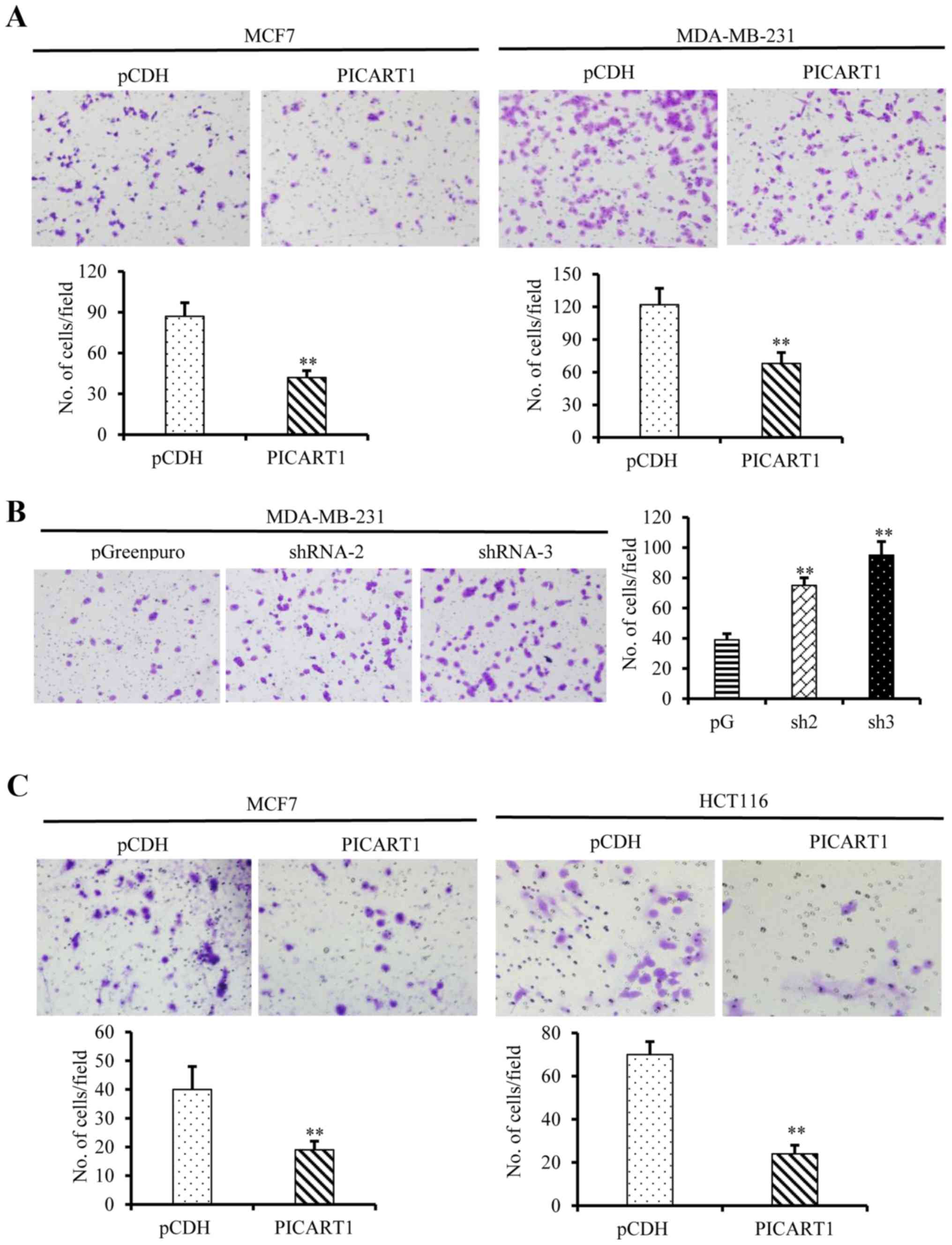
We further investigated the effects of PICART1
expression on migration and invasion of cancer cells. As shown in
Fig. 5A, ectopic expression of
PICART1 in MCF7 and MDA-MB-231 cells led to a notable decrease in
the cell migration through micro-holes in the Transwells. In
contrast, PICART1 silencing enhanced cell migration (Fig. 5B). Similar results were obtained in
the invasion assays, and the ectopic expression of PICART1
suppressed the invasion of MCF7 and HCT116 cells (Fig. 5C). Altogether, these data suggest
that PICART1 inhibits the proliferation, migration, and invasion of
breast and colon cancer cells.
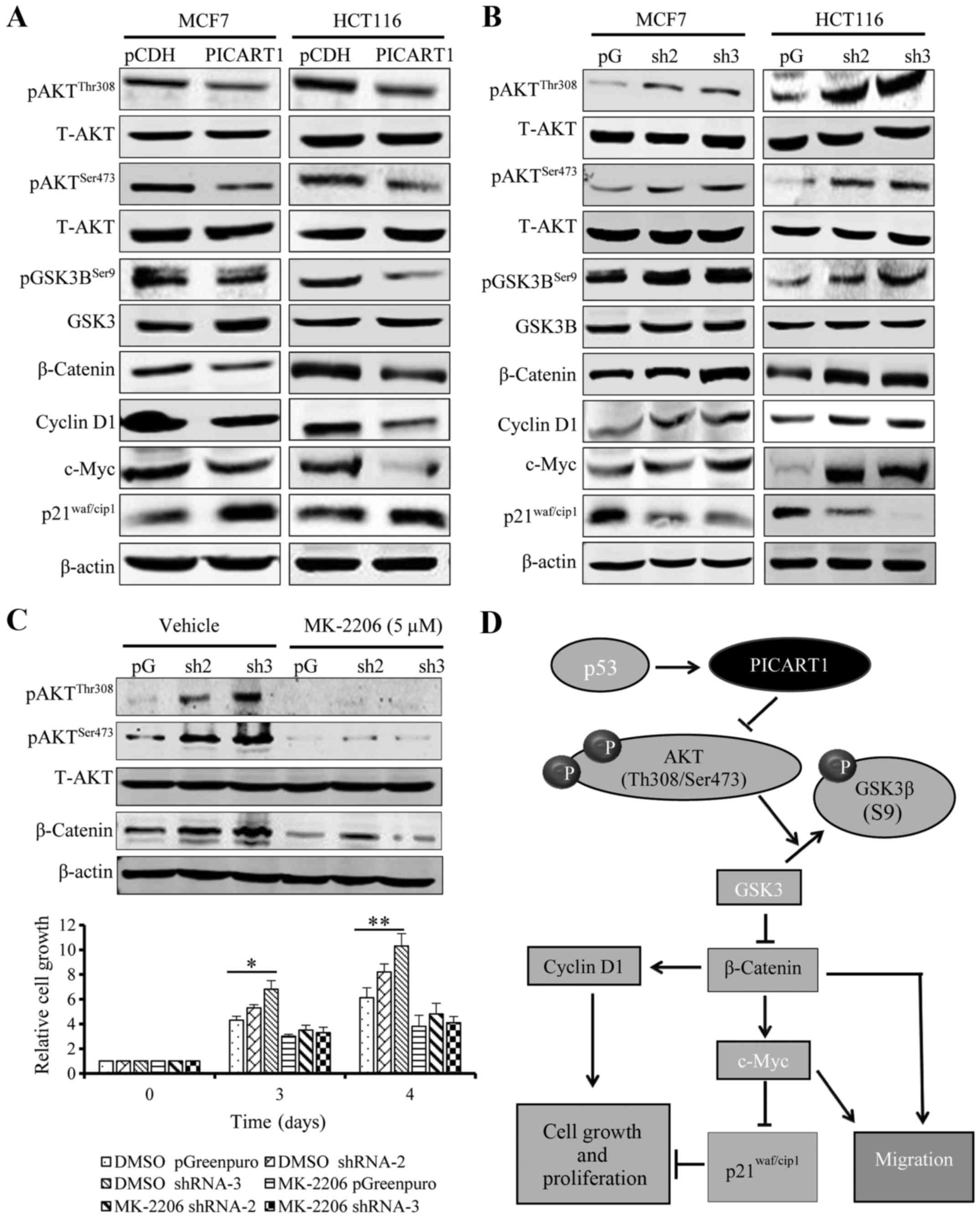
PICART1 regulates cell proliferation
through the AKT/GSK3β/β-catenin signaling pathway
We further investigated the underlying mechanisms
through which PICART1 functions. To address this question, we
explored the effects of PICART1 expression on main oncogenic
signaling pathways, such as AKT and ERK signaling. PICART1 did not
have notable effects on ERK signaling (data not shown). However, as
shown in Fig. 6A, ectopic
expression of PICART1 led to a decrease in pAKT (Thr308 and
Ser473), suggesting that PICART1 may function through the AKT
signaling cascade. We then explored the downstream
targets/effectors of AKT. Our results showed that pGSK3β (Ser9) and
β-catenin expression was decreased by PICART1. β-catenin functions
as a key transcription factor after translocation into the nucleus
(22). As a result, the expression
of cyclin D1 and c-Myc was decreased by PICART1 (Fig. 6A). c-Myc is a suppressor of
p21Waf1/cip1 expression (23). Therefore, in the tested cells, we
observed an increase in p21Waf1/cip1 expression due to
downregulation of c-Myc by PICART1 (Fig. 6A). In contrast, silencing of
PICART1 led to an increase in pAKT (Thr308 and Ser473), pGSK3B
(Ser9), β-catenin, cyclin D1 and c-Myc expression but a decrease in
p21Waf1/cip1 expression (Fig. 6B). To further confirm that PICART1
regulates cell growth and proliferation through the AKT/β-catenin
signaling pathway, we treated PICART1-silenced cells (where AKT
signaling was activated) with 5.0 µM MK-2206, an AKT1/2
inhibitor. Our result showed that exposure to the AKT inhibitor
abolished the stimulation of PICART1 silencing on cell
proliferation (Fig. 6C). Taken
together, our data suggest that PICART1 inhibits proliferation of
MCF7 and HCT116 cells through control of the AKT/GSK3β/β-catenin
pathway (Fig. 6D).
Discussion
Long non-coding RNAs (lncRNAs) are RNA transcripts
in the genome which function as oncogenes or tumor suppressors,
playing a critical role in cancer progression and development
(24). This study characterized a
new lncRNA named PICART1. PICART1 is p53-inducible and functions as
a tumor suppressor via regulating the AKT/GSK3β/β-catenin signaling
pathway. This study also characterized a p53-response element in
the promoter of the PICART1 gene.
p53 is a critical transcription factor that
regulates expression of a wide range of genes, including
protein-coding and non-coding genes. This makes p53 an important
protein in multiple cellular processes, such as DNA repair and
genome stability, cell cycle control, apoptosis, and senescence
(25). As a transcription factor,
p53 functions as a tetramer, binding to a sequence-specific
response element in the promoter of target genes and controlling
gene expression (26). This study
revealed PICART1 as a new lncRNA target of p53, which is supported
by several lines of evidence. The most direct evidence lies in the
effects of targeted expression or silencing of p53 on the
expression of endogenous PICART1 and on its promoter activity, and
in the characterization of an active p53 response element in the
PICART1 promoter. Targeted mutations of this element decreased the
promoter activity at baseline and response to p53 expression or
silencing. In addition, RITA is considered a p53 activator,
restoring the function of p53 from its negative regulator HDM2
(MDM2 in mice); doxorubicin induces DNA damage and thus activates
p53. Our results showed that both RITA and doxorubicin that
activate p53 through different mechanisms triggered PICART1
expression, providing an additional line of evidence that p53
regulates PICART1 expression. However, p53 may not be the sole
signaling pathway that regulates PICART1 expression. It is noted
that in the tested cells, PICART1 expression was not stringently
correlated with the p53 status. The PICART1 level was high in the
non-transformed MCF10A cells that harbors a wild-type p53, but
decreased in MCF7, HCT116, and HCT8 cells that also have a
wild-type p53. It would be interested to further characterize the
other factors that may affect PICART1 expression.
Our data showed that PICART1 expression was higher
in response to RITA exposure than to ectopic p53 expression. This
may be ascribed to the nature of different treatments. Unlike drug
treatment, the efficiency of p53 transfection, i.e., the percentage
of the cells transfected, rarely reaches 100%. Thus, PICART1
expression was affected by p53 delivery only in a portion of cells.
More importantly, p53 is a well-known cell apoptotic tumor
suppressor (27). Gene
transfection of p53 often leads to high expression and resultant
cell death and RNA degradation, thus resulting in lower PICART1
induction observed in p53 delivery than in RITA exposure. In fact,
PICART1 expression was downregulated much more efficiently in the
p53-silenced cells due to the increased survival of these
cells.
Promoter motif analysis recognized 3 putative p53
response elements in the PICART1 promoter, but only one was
functional. The luciferase reporter activity assays showed that the
full length of the promoter fragment (−3000 bp) that contains all
three elements presented a dose-dependent response to RITA
treatment, while the promoter fragment −1592 bp that harbors the
elements 2 and 3 was not responsive to RITA. This excluded the
possibility that the elements 2 and 3 are functional p53 binding
motifs. Therefore, our targeted mutation analyses were focused on
element 1 located at −1808 to −1783 bp, and our study results
confirmed its activity responding to p53, thus strengthening the
conclusion of p53 regulation on the PICART1 expression.
Cancer cells are characterized by unlimited growth
and proliferation and increased migration and invasion, leading to
development of tumors (28).
p53-induced expression, coupled together with the decreased
expression in cancer cells and tissues, suggests that PICART1 may
function as a tumor suppressor. To test this hypothesis, we
investigated the effects of targeted expression or silencing of
PICART1 on proliferation, migration, and invasion of breast and
colon cancer cells. Our result showed that ectopic expression of
PICART1 suppressed cell proliferation and induced cell cycle arrest
in G2/M phase while silencing of PICART1 enhanced cell
proliferation. In addition, PICART1 expression markedly inhibited
cell migration and invasion while silencing of PICART1 promoted
cell migration and invasion. These results, along with the
downregulation in breast and colon cancer, suggest that PICART1
acts as a tumor suppressor.
lncRNAs function as tumor suppressors through
modulating cellular signaling networks (29). PI3K/AKT and Wnt/β-catenin are two
master oncogenic signals regulating cell proliferation, survival,
migration, and invasion (30,31).
AKT is a serine-threonine kinase activated by phosphorylation at
Thr308 and then Ser473 (32). In
the canonical Wnt pathway, Wnt protein binds to Frizzled receptors,
suppressing the activity of glycogen synthase kinase-3β (GSK3β).
Without Wnt signaling, β-catenin associates with Axin, adenomatosis
polyposis (APC), protein phosphatase 2A (PP2A), glycogen synthase
kinase 3β (GSK3β), and casein kinase 1α (CK1α), forming a
destruction complex, degrading β-catenin by targeting for
ubiquitination (33–35). An activation of Wnt signaling
drives the destruction complex to translocation and disruption,
allowing β-catenin accumulation and translocation into the nucleus.
Once in the nucleus, β-catenin associates with Tcf/Lef
transcription factors and drives expression of target genes, such
as cyclin D1 and c-Myc, promoting cell proliferation and migration
(22,36–38).
AKT cross-talks with Wnt/β-catenin signaling through
phosphorylation at Ser9 and inactivation of GSK3β leading to
increase of β-catenin level (39).
In this study, we found that PICART1 suppressed AKT
activity with decreased phosphorylation levels of Thr308 and
Ser473, which in turn lowered the pGSK3β (Ser9) and β-catenin
levels, suppressing cyclin D1 and c-Myc expression. The latter is a
repressive transcription factor of p21Waf1/cip1 by
sequestering SP1 (23). Thus we
observed an increase in p21Waf1/cip1 expression by
PICART1. It is currently unknown how PICART1 suppresses
phosphorylation activation of AKT. Considering the multiple
functional possibilities of lncRNAs, PICART1 may function as an
adaptor to couple AKT with phosphatases, leading to
dephosphorylation. PICART1 may also bind to AKT and block the
access of AKT kinases for phosphorylation. Further study is
warranted. It is also noteworthy that p21Waf1/cip1
regulates both G1/S and G2/M transition (40,41).
In this study, targeted PICART1 expression induced cell cycle
arrest at G2/M phase, but not at G1. It is well known that cell
cycle regulation is a complicated event with involvement of
multiple proteins, which may lead to differential phenotypes
(42).
In summary, this study characterized a new lncRNA,
named PICART1. PICART1 was upregulated by p53 through a
sequence-specific response element in the promoter. PICART1 was
downregulated in breast and colon cancer cells and tissues and
functioned as a tumor suppressor, inhibiting cell proliferation,
migration, and invasion through modulation of the
AKT/GSK3β/β-catenin signaling cascade.
Abbreviations:
|
PICART1
|
p53-inducible cancer-associated RNA
transcript 1
|
|
ANRIL
|
antisense non-coding RNA in the INK4
locus
|
|
HOTTIP
|
HOXA distal transcript antisense
RNA
|
|
HOTAIR
|
HOX transcript antisense RNA
|
|
BCAR4
|
breast cancer anti-estrogen resistance
4
|
|
lincRNA-p21
|
tumor protein p53 pathway corepressor
1
|
|
XIST
|
inactive X specific transcripts
|
|
STAT3
|
signal transducer and activator of
transcription 3
|
|
HDM2
|
human double minute 2 homolog
|
|
RITA
|
reactivation of p53 and induction of
tumor cell apoptosis
|
|
pAKT
|
phosphate protein kinase B (PKB)
|
|
pGSK3β
|
phosphate glycogen synthase kinase
3β
|
Acknowledgments
This research was supported in part by the National
Natural Science Foundation of China (81272918 and 81472465 to
D.C.).
References
|
1
|
Chung SW, Chen YH, Yet SF, Layne MD and
Perrella MA: Endotoxin-induced down-regulation of Elk-3 facilitates
heme oxygenase-1 induction in macrophages. J Immunol.
176:2414–2420. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Da Sacco L, Baldassarre A and Masotti A:
Bioinformatics tools and novel challenges in long non-coding RNAs
(lncRNAs) functional analysis. Int J Mol Sci. 13:97–114. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Schroeder JA, Adriance MC, Thompson MC,
Camenisch TD and Gendler SJ: MUC1 alters beta-catenin-dependent
tumor formation and promotes cellular invasion. Oncogene.
22:1324–1332. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhao Y, Jian W, Gao W, Zheng YX, Wang YK,
Zhou ZQ, Zhang H and Wang CJ: RNAi silencing of c-Myc inhibits cell
migration, invasion, and proliferation in HepG2 human
hepatocellular carcinoma cell line: c-Myc silencing in
hepatocellular carcinoma cell. Cancer Cell Int. 13:232013.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Sahu A, Singhal U and Chinnaiyan AM: Long
noncoding RNAs in cancer: From function to translation. Trends
Cancer. 1:93–109. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bhan A and Mandal SS: lncRNA HOTAIR: A
master regulator of chromatin dynamics and cancer. Biochim Biophys
Acta. 1856:151–164. 2015.PubMed/NCBI
|
|
8
|
Yuan JH, Yang F, Wang F, Ma JZ, Guo YJ,
Tao QF, Liu F, Pan W, Wang TT, Zhou CC, et al: A long noncoding RNA
activated by TGF-β promotes the invasion-metastasis cascade in
hepatocellular carcinoma. Cancer Cell. 25:666–681. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xing Z, Lin A, Li C, Liang K, Wang S, Liu
Y, Park PK, Qin L, Wei Y, Hawke DH, et al: lncRNA directs
cooperative epigenetic regulation downstream of chemokine signals.
Cell. 159:1110–1125. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Gutschner T, Hämmerle M, Eissmann M, Hsu
J, Kim Y, Hung G, Revenko A, Arun G, Stentrup M, Gross M, et al:
The noncoding RNA MALAT1 is a critical regulator of the metastasis
phenotype of lung cancer cells. Cancer Res. 73:1180–1189. 2013.
View Article : Google Scholar
|
|
11
|
Zhao Y, Yang Y, Trovik J, Sun K, Zhou L,
Jiang P, Lau TS, Hoivik EA, Salvesen HB, Sun H, et al: A novel wnt
regulatory axis in endometrioid endometrial cancer. Cancer Res.
74:5103–5117. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shen Y, Ma J, Yan R, Ling H, Li X, Yang W,
Gao J, Huang C, Bu Y, Cao Y, et al: Impaired self-renewal and
increased colitis and dysplastic lesions in colonic mucosa of
AKR1B8-deficient mice. Clin Cancer Res. 21:1466–1476. 2015.
View Article : Google Scholar
|
|
13
|
Cao D, Fan ST and Chung SS: Identification
and characterization of a novel human aldose reductase-like gene. J
Biol Chem. 273:11429–11435. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu Z, Yan R, Al-Salman A, Shen Y, Bu Y,
Ma J, Luo DX, Huang C, Jiang Y, Wilber A, et al: Epidermal growth
factor induces tumour marker AKR1B10 expression through activator
protein-1 signalling in hepatocellular carcinoma cells. Biochem J.
442:273–282. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bu Y, Li X, He Y, Huang C, Shen Y, Cao Y,
Huang D, Cai C, Wang Y, Wang Z, et al: A phosphomimetic mutant of
RelA/p65 at Ser536 induces apoptosis and senescence: An implication
for tumor-suppressive role of Ser536 phosphorylation. Int J Cancer.
138:1186–1198. 2016. View Article : Google Scholar
|
|
16
|
Ma J, Yan R, Zu X, Cheng JM, Rao K, Liao
DF and Cao D: Aldo-keto reductase family 1 B10 affects fatty acid
synthesis by regulating the stability of acetyl-CoA
carboxylase-alpha in breast cancer cells. J Biol Chem.
283:3418–3423. 2008. View Article : Google Scholar
|
|
17
|
Liu Z, Zhong L, Krishack PA, Robbins S,
Cao JX, Zhao Y, Chung S and Cao D: Structure and promoter
characterization of aldo-keto reductase family 1 B10 gene. Gene.
437:39–44. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Huang C, Verhulst S, Shen Y, Bu Y, Cao Y,
He Y, Wang Y, Huang D, Cai C, Rao K, et al: AKR1B10 promotes breast
cancer metastasis through integrin α5/δ-catenin mediated
FAK/Src/Rac1 signaling pathway. Oncotarget. 7:43779–43791.
2016.PubMed/NCBI
|
|
19
|
Yan R, Zu X, Ma J, Liu Z, Adeyanju M and
Cao D: Aldo-keto reductase family 1 B10 gene silencing results in
growth inhibition of colorectal cancer cells: Implication for
cancer intervention. Int J Cancer. 121:2301–2306. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Harris SL and Levine AJ: The p53 pathway:
Positive and negative feedback loops. Oncogene. 24:2899–2908. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Issaeva N, Bozko P, Enge M, Protopopova M,
Verhoef LG, Masucci M, Pramanik A and Selivanova G: Small molecule
RITA binds to p53, blocks p53-HDM-2 interaction and activates p53
function in tumors. Nat Med. 10:1321–1328. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Gartel AL, Ye X, Goufman E, Shianov P, Hay
N, Najmabadi F and Tyner AL: Myc represses the p21(WAF1/CIP1)
promoter and interacts with Sp1/Sp3. Proc Natl Acad Sci USA.
98:4510–4515. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Harris CC: Structure and function of the
p53 tumor suppressor gene: Clues for rational cancer therapeutic
strategies. J Natl Cancer Inst. 88:1442–1455. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Riley T, Sontag E, Chen P and Levine A:
Transcriptional control of human p53-regulated genes. Nat Rev Mol
Cell Biol. 9:402–412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cao Z, Ma J, Chen X, Zhou B, Cai C, Huang
D, Zhang X and Cao D: Uridine homeostatic disorder leads to DNA
damage and tumorigenesis. Cancer Lett. 372:219–225. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schmitz AA, Govek EE, Böttner B and Van
Aelst L: Rho GTPases: Signaling, migration, and invasion. Exp Cell
Res. 261:1–12. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim YG, Kim MJ, Lim JS, Lee MS, Kim JS and
Yoo YD: ICAM-3-induced cancer cell proliferation through the
PI3K/Akt pathway. Cancer Lett. 239:103–110. 2006. View Article : Google Scholar
|
|
31
|
Li X, Xu Y, Chen Y, Chen S, Jia X, Sun T,
Liu Y, Li X, Xiang R and Li N: SOX2 promotes tumor metastasis by
stimulating epithelial-to-mesenchymal transition via regulation of
WNT/β-catenin signal network. Cancer Lett. 336:379–389. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Nakamura T, Hamada F, Ishidate T, Anai K,
Kawahara K, Toyoshima K and Akiyama T: Axin, an inhibitor of the
Wnt signalling pathway, interacts with beta-catenin, GSK-3beta and
APC and reduces the beta-catenin level. Genes Cells. 3:395–403.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Luo J: Glycogen synthase kinase 3beta
(GSK3beta) in tumorigenesis and cancer chemotherapy. Cancer Lett.
273:194–200. 2009. View Article : Google Scholar
|
|
35
|
Minde DP, Anvarian Z, Rüdiger SG and
Maurice MM: Messing up disorder: How do missense mutations in the
tumor suppressor protein APC lead to cancer? Mol Cancer.
10:1012011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kolligs FT, Bommer G and Göke B:
Wnt/beta-catenin/tcf signaling: A critical pathway in
gastrointestinal tumorigenesis. Digestion. 66:131–144. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
MacDonald BT, Tamai K and He X:
Wnt/beta-catenin signaling: Components, mechanisms, and diseases.
Dev Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li YJ, Wei ZM, Meng YX and Ji XR:
Beta-catenin up-regulates the expression of cyclinD1, c-myc and
MMP-7 in human pancreatic cancer: Relationships with carcinogenesis
and metastasis. World J Gastroenterol. 11:2117–2123. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Fang D, Hawke D, Zheng Y, Xia Y,
Meisenhelder J, Nika H, Mills GB, Kobayashi R, Hunter T and Lu Z:
Phosphorylation of beta-catenin by AKT promotes beta-catenin
transcriptional activity. J Biol Chem. 282:11221–11229. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Niculescu AB III, Chen X, Smeets M, Hengst
L, Prives C and Reed SI: Effects of p21(Cip1/Waf1) at both the G1/S
and the G2/M cell cycle transitions: pRb is a critical determinant
in blocking DNA replication and in preventing endoreduplication.
Mol Cell Biol. 18:629–643. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bunz F, Dutriaux A, Lengauer C, Waldman T,
Zhou S, Brown JP, Sedivy JM, Kinzler KW and Vogelstein B:
Requirement for p53 and p21 to sustain G2 arrest after DNA damage.
Science. 282:1497–1501. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chan TA, Hwang PM, Hermeking H, Kinzler KW
and Vogelstein B: Cooperative effects of genes controlling the
G(2)/M checkpoint. Genes Dev. 14:1584–1588. 2000.PubMed/NCBI
|