Gastric cancer (GC) is the fourth most common cancer
and the second leading cause of cancer-related mortality (1). Most GC patients present with
advanced-stage disease at the time of diagnosis and have a poor
prognosis (2). Genetic and
epigenetic aberrations have long been thought to be the two main
mechanisms that orchestrate various procedures in the process of GC
tumorigenesis. Recent evidence has shown that epigenetic
dysregulation plays a crucial role in GC development (3). Among epigenetic alternations,
methylation modification is particularly closely associated with GC
oncogenesis and progression. These modifications are catalyzed by a
class of group-transfer enzymes known as methyltransferases.
Although the physiological importance of methylation has been known
for many years and has been described in numerous reports, the
discovery of enzymes that can reverse methylation has shifted the
focus towards the study of methyltransferases, which provides
opportunities for targeted treatment using specific inhibitors. The
present review highlights the current knowledge of functions of
methylation aberrations and may contribute to the understanding of
GC mechanisms. We also discuss the importance of methyltransferases
and the function of newly emerging molecular inhibitors as
anticancer targets in GC.
Epigenetics is formally defined as a heritable and
reversible alterations in gene expression or chromosomal stability
without changes in the underlying DNA sequence (4). Epigenetic factors help regulate many
processes from development to differentiation and are maintained
through multiple cellular cycles. In this way, they play a pivotal
role in normal development. Abnormalities in the epigenetic control
of the normal processes have been increasingly recognized as one of
the mechanisms in cancer initiation and progression (4,5).
In normal cells, DNA methylation occurs
predominantly in repetitive genomic regions, maintaining genomic
integrity while CpG islands, particularly those associated with
promoters, are generally unmethylated. Both hypermethylation and
hypomethylation have been observed in all types of cancer cells.
Global DNA hypomethylation, particularly in repetitive sequences,
plays a vital role across developmental stages and cancer
progression. Genome-wide hypomethylation, characterized by the
large depletion of methylation, takes place mainly on normally
heavily methylated repeat elements such as long interspersed
nuclear element (LINE1) and centromeric satellite repeats (Satα and
Sat2). In general, hypomethylation in cancer cells is associated
with a series of adverse outcomes, including chromosome instability
(CIN), repression of transposable elements, loss of imprinting
(LOI) and activation of oncogenes (6–8).
Local hypermethylation of genes takes place on the cytosines
located in CpG dinucleotides resulting in aberrant gene silencing
including tumor-suppressor, cell cycle regulator and DNA-repair
genes silencing involved in cellular pathways, such as cell cycle,
DNA repair, metabolism, cell adherence, apoptosis and angiogenesis
(9,10). Diverse tumor types have a diverse
region of gene hypermethylation defined as CpG island methylator
phenotype (CIMP). The DNA methylation patterns may be potential
prognostic indicators of cancer patients and used as a biomarker
(11). DNA methylation is mediated
by DNA methyltransferases (DNMTs), which employ
S-adenosyl-methionine (SAM) to add a methyl group to cytosine
residues predominantly in the dinucleotide CpGs (12,13).
There are three active DNMTs in mammals: DNMT1, DNMT3A and DNMT3B.
DNMT1 is primarily a maintenance methyltransferase, and DNMT3A and
DNMT3B are primarily de novo methyltransferases. DNMT1 can
interact with both DNMT3a and DNMT3b to influence transcription. In
addition, DNMT1 and DNMT3a have both been shown to bind to KMT1A
(SUV39H1), a histone methyltransferase (HMT), to mediate repression
via H3K9me (14). The
misregulation of DNA methylation is critical to the initiation and
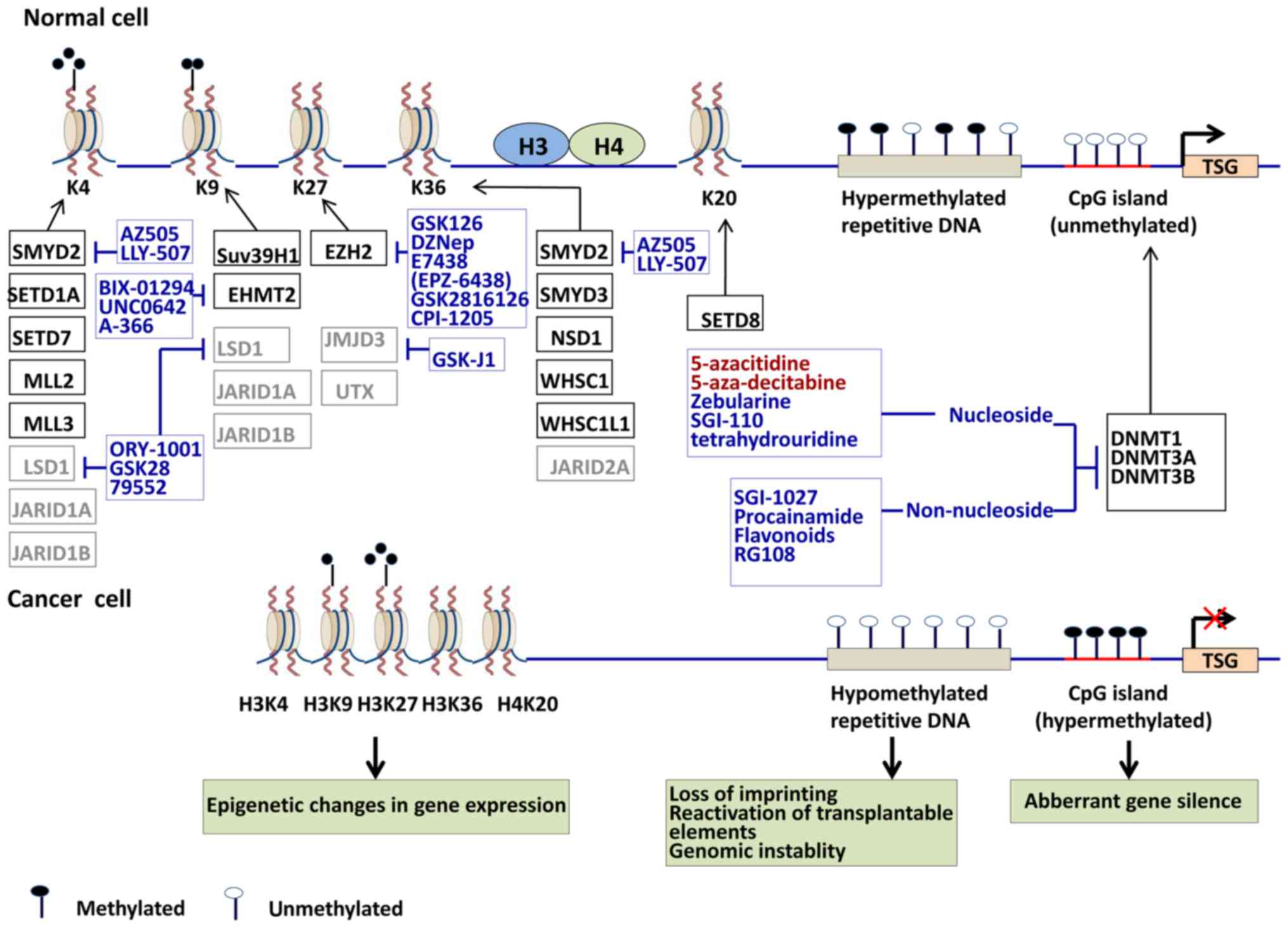
progression of tumorigenesis (Fig.
1) (4,9).
Histone modification is an important epigenetic
regulatory process in cancerous tissues and cells. It involves
transcription regulation through eight different classes of
post-translational modifications, including methylation,
acetylation, phosphorylation, ubiquitynation, sumoylation,
deamination, proline isomerization and ADP-ribosylation of the core
histones (H2A, H2B, H3 and H4) (15,16).
These modifications are thought to contribute to transcription
control through influencing chromatin compactin or signaling to
other protein complexes. In normal cells, the promoters of tumor
suppressor genes are enriched with active transcription markers,
such as H3K4me3 and H4 acetylation, while satellite regions are
enriched with repressive markers, such as H3K27me3, H3K9me2 and
H4K16Ac. In tumor cells, the promoters of tumor suppressor genes
lose nearly all acetylation and acquire repressive markers
including H3K27me3 and H3K9me. Repressive markers such as H4K20me3
and H3K27me3 are lost at repetitive satellite regions leading to
CIN/microsatellite instability (MSI) (Fig. 1). Acetylation and methylation of
histone lysine residues are the most thoroughly studied histone
profiles. Typically, histone acetylation is associated with an
active transcription, whereas methylation may be associated with
either active or repressive states, depending on the target amino
acid residues and the range of methylation (me1, me2 or me3)
(17). A possible explanation is
that histone methylation does not only alter the charge on the
histone tails but also influences the basicity and affinity of
different reader proteins to the methylated sites. Generally, H3K4,
H3K36 and H3K79 methylations are associated with active genes while
H3K9, H3K27 and H4K20 methylations are considered repressive
markers leading to inactive genes.
Epigenetic modifications are not standalone
processes, instead, are subject to considerable crosstalk among the
different types of epigenetic markers. Research in model organisms
has shown that there are extensive links and crosstalk between
histone modifications and DNA methylation. Key to these links are
the readers of histone methylation including plant homeodomains
(PHDs), chromodomains and bromo adjacent homology domains (BAH
domains) and readers of DNA methylation such as the SRA (SET- and
RING-associated), CXXC domain and methyl-CpG-binding domain (MBD).
The unifying molecular feature of GC is a profoundly reshaped
epigenome characterized by global genomic hypomethylation,
gene-specific DNA hyper- or hypo-methylation, aberrant expression
of DNMTs and histone methylation enzymes (18).
Regions of lower-density methylation near CpG
islands (CGIs), defined as 'shores', exhibit great variation in
methylation, including hypomethylation and hypermethylation, across
diverse types of cancers (28).
Currently, a number of hypomethylated tumor-promoting genes and
hypermethylated tumor-suppressor genes (TSG) have also been found
in GC and are associated with oncogene positive transcriptional
regulation during a variety of cellular processes (Table I). The DNA methylation of
chromatin-modifying enzymes (CMEs) that cause changes in chromatin
structure can affect diverse pathways involved in multiple aspects
of GC development and progression. For example, SMARCA5 and MGMT
(O-6-methylguanine-DNA methyltransferase), SWI/SNF related, are
downregulated in GC as a consequence of its promoter methylation
(54,55). The methylation HLTF, encoding a
member of the SWI/SNF family, has been reported in 50% of cases of
GC (56). DNA methylation
biomarkers of certain genes that are abnormally repressed in GC are
appealing in normal gastric epithelium or premalignant lesions as
well as in other body samples (e.g. stool and plasma) early in GC
development. Alterations in DNA methylation can also influence
treatment response in GC. For example, methylation of SULF2 and
BMP4 have been linked to chemotherapeutic responses (57,58).
GC-associated alterations in DNA methylation were
widespread and tend to localize at CGIs (59). Patients with widespread gene
hypermethylation tend to have poor overall survival (59). However, the methylation profile
differs between the intestinal and diffuse types of GC (60). The epithelial cadherin gene CDH1, a
tumor suppressor gene which is downregulated in gastric tumors, is
hypermethylated more frequently in the diffuse type of GC than in
the intestinal type (61,62). Zouridis et al (59) showed that patients with
CIMP-positive GC tend to be younger, with less-differentiated
tumors, and their cases are associated with diffuse histology and
tend to exhibit worse survival outcomes. One potent cause is that
genes methylated by CIMP in GC overlap with those corresponding to
PRC2-targets in embryonic stem cells, suggesting that it has been
present since normal, early development. The strong relationship
between CIMP and H. pylori, EBV, and MSI has been emphasized
in a meta-analysis, but CIMP cannot be used as a prognostic marker
for GC (63).
Notably, the increased expression of DNMT3A in GC is
significantly higher than that of DNMT1 and DNMT3B (64,65).
A recent study demonstrated that the poor overall survival rate of
GC patients is associated with elevated DNMT3A expression, but not
with increased expression of DNMT1 or DNMT3B (66). DNMT3A contributes to the
dysregulation of the cell cycle by repressing p18INK4C
in a DNA methylation-dependent manner (67). DNA methyltransferase genes have
been shown to be mutated in certain cancers. For example, the
DNMT3A gene is mutated in acute myelogenous leukemia,
myeloproliferative disease and myelodysplastic syndrome (68). Zang et al (69) also observed that GC had mutations
in DNMT3A (1/15; 6.7%). In addition, 23.5% allelic loss of DNMT3A
was observed in GC (70).
Increment of functional polymorphism of DNMT1, DNMT3A and DNMT3B
were found in gastric neoplasm and have been found to be associated
with development and progression of GC (65,71–73).
One meta-analysis showed that DNMT1 rs16999593 and DNMT3A rs1550117
could result in GC and that DNMT3B rs1569686 may be a protective
factor against gastric carcinogenesis (73). Furthermore, DNMT1, DNMT3A and
DNMT3B proteins are downregulated through overexpression of
miR-200b and miR-200c, resulting in the global DNA hypomethylation
in GC cell lines (74). In
addition, H. pylori infection could increase DNMT activity
via upregulation of the epidermal growth factor (EGF) and its
receptor or via the release of inflammatory mediators, such as NO
(75,76). In particular, overexpression of
DNMT1 and DNMT3b was found to be associated with EBV infection in
GC (77,78).
Studies conducted on histone methylation of GC have
focused mainly on H3 and H4 (56,79).
Studies have reported that certain histone methylation markers
(H3K4me3, H3K9me3 and H3K27me3) are positively correlated with
clinicopathological characteristics in GC, including tumor stage
and survival (80–83). As a sequence, specific histone
methylation can result in dysregulation of many genes with
important roles in GC (Table II).
For instance, EZH2 mediates H3K27 trimethylation to maintain
transcriptional silencing. EZH2 knockdown represses cell growth,
cell proliferation (RUX3 and ANXA6) (90,91),
invasion and migration (E-cadherin and ArgBP2) (92,93),
and induces cell cycle arrest in GC cells (p53, p21, p14 and p16)
(94,95). In addition, EZH2 promotes the
activation of Wnt and MAPK signaling through downregulation of
CXXC4 expression, figuring an epigenetic mechanism of wnt signaling
activation in GC cells (96,97).
Histone methylation is processed by lysine
methyltransferases. A number of HMTs and histone demethylases
(HDTs) have been found to mediate the addition and removal of
methyl groups from different lysine residues on histones. Indeed,
methylation at different lysine residues on histones has been shown
to display differential functions (69). A current model suggests that
methylated histones are recognized by chromatin effector molecules
(readers), leading to the recruitment of other molecules to alter
the chromatin and/or transcription states (106). In particular, histone methylation
reader proteins such as WD40 repeats, chromo- and bromodomain
proteins, and the PHD finger domain were shown to recruit HMTs to
their target sites. During the past decade, several HMTs and HDTs
have been identified in humans (Fig.
2). All HMTs except KMT4 (DOT1L), contain the SET-domain, which
provides interaction and recognition sites for lysine substrates
and cofactors for general catalysis. SET-domain-containing protein,
a class of HMTs, has been regarded as an important factor in
carcinogenesis (107). HDTs
include amine oxidase-like (AOL) domain-containing demethylases and
Jumonji C (JmjC) domain-containing demethylases (108,109). These HMTs and HDTs have been
shown to methylate histones incorporated in chromatin, free
histones and non-histone proteins (110).
Beyond alteration of the histone code, HMT interacts
with various molecules including DNMT1. For example, EZH2 can
recruit and regulate other epigenetic silencing enzymes, such as
DNMTs, by influencing the binding capacity between DNMTs and the
promoter of target genes (111).
DNMT1-mediated promoter methylation is indispensably maintained by
EZH2-mediated methylation (112,113). LSD family members contain a SWIRM
domain and an AOL domain. The SWIRM domain is present in many
chromatin-interacting enzymes and it can interact with DNA. Several
studies have shown that the expression of several genes in GC can
be regulated by DNA methylation and histone modification
simultaneously (83,114,115). For example, gene expression of
PRDM5 (PR domain containing 5), a member of the kruppel-like zinc
finger family, is downregulated via DNA methylation and H3K27
trimethylation, alleviating the cell-growth-suppressive effect of
PRDM5 (116).
The biological and physiological significance of
non-histone lysine methylation in human tumorigenesis has been
recently explored (117,118). For example, SET and MYND
domain-containing protein 2 (SMYD2) was identified as a lysine
methyltransferase for K370 of p53 (119), K860 and K810 of Rb (120,121) and K528 of PARP1 (122). SMYD2-dependent methylation of Rb
at K810 promotes the cell cycle progression of cancer cells
(121). Moreover, methylation of
K528 on PARP1 enhances its poly(ADP-ribose) activity in cancer
cells (122). Mazur and
colleagues (123) found that
SMYD3 methylated the lysine 260 of MAP3K2 gene, leading to the
activation of the signaling of the Ras/Raf/MEK/ERK in the
development of pancreatic ductal adenocarcinoma and lung
adenocarcinoma.
Inflammation can predispose tissues to cancer, and
it is very likely that DNA methylation is involved in this process
(124). The stomach has been
described as the organ with the highest CpG island hypermethylation
frequency which is age-associated and possibly
inflammation-mediated (29). From
an etiological viewpoint, two pathogens, H. pylori (HP) and
Epstein-Barr virus (EBV), are known to participate in gastric
carcinogenesis. Chronic inflammation in the gastric mucosa due to
HP and EBV infection of gastric epithelial cells has been reported
to cause aberrant promoter methylation, which may contribute to the
tumorigenic mechanisms of these pathogens.
In the multistep development of gastric
carcinogenesis, chronic HP infection progresses over decades,
through stages of chronic gastritis, atrophy, intestinal
metaplasia, adenoma/dysplasia and cancer. Moreover, global DNA
hypomethylation might be implicated in GC associated with HP
infection during early stages (125). According to a study that
investigated the promoters of 48 genes in the gastric mucosae with
or without HP infection, results suggest a possible link between
aberrant DNA methylation and HP infection (126). Furthermore, some of these
HP-related events (such as hypermethylation) are reversed after HP
eradication. However, in patients with preneoplastic lesions,
global DNA methylation decreased over time despite eradication of
HP infection (125).
HP can induce methylation of multiple CpG islands,
especially at sites encoding tumor suppressors such as E-cadherin,
MGMT and the eradication has led to a marked decrease in
methylation levels of these genes (127). Inflammation triggered by HP
infection can induce aberrant DNA methylation, which appears to be
a critical process (128). HP may
induce aberrant DNA methylation through the release of ROS and
nitric oxide (NO) and the activation of DNMT (129). Apart from DNA methylation, HP
also regulates histone modifications in human gastric epithelial
cells through the ROS and histone methyltransferase. For example,
Angrisano et al (130)
reported that HP infection is followed by activation of iNOS gene
expression, chromatin changes at the iNOS promoter (including
decreased H3K9 methylation and increased H3K4 methylation), and
selective release of MBD2 from the iNOS promoter in a GC cell line.
β-catenin directly binds to JMJD2B (KDM4B) promoter and stimulates
JMJD2B expression after HP infection. Increased JMJD2B, together
with NF-κB, binds to COX-2 promoter to enhance its transcription by
demethylating H3K9me3 locally (131).
Taking into account the reversibility of epigenetic
modification, a new field of therapy focusing on key targets, known
as pharmocoepigenomics, has been developed to either block or
reverse the aberrant epigenetic modifications at an early stage
(138). Compounds in preclinical
and clinical stages reported in cancer are summarized in Fig. 1. The present review describes drugs
targeting disordered patterns of DNA and histone methylation and
their potential efficacy in GC (Table III).
As mentioned above, it has been well established
that many genes are hypermethylated in GC (Table I). Two DNA methylation inhibitors,
azacytidine (5-azacitidine) and decitabine (5-aza-deoxycytidine)
have been established for the treatment of myeloid malignancies
(145,146). Recently, more and more DNMT
inhibitors, categorized as nucleoside analogs (decitabine,
zebularine, SGI-110 and tetrahydrouridine) and non-nucleoside
compounds (SGI-1027, procainamide, flavonoids, RG108 and their
derivatives), are actively being explored in the clinical and
preclinical trials as novel treatments for cancer (Fig. 1) (147).
Azacytidine and decitabine are nucleoside analogs of
cytosine that cannot accept a methyl donor at the 5′ position of
the pyrimidine ring and depletes cellular DNMT1. Intracellularly,
azacytidine can convert to decitabine and subsequently be
incorporated into DNA (148).
Results showed that azacitidine inhibited the proliferation and
decreased the level of DNA methylation in GC cell lines (149). Decitabine treatment resulted in
growth suppression and reduced the levels of DNMT3A and DNMT3B
accompanied with the demethylation of P16INK4A gene
(140). CIMP-positive GC lines
appeared to exhibit significant reductions in proliferation after
treatment of DNMT inhibitors such as decitabine compared to
non-CIMP lines (59). The greatest
challenge associated with decitabine is the compound's instability,
which limits its efficacy in treating solid tumors (150).
A recent experimental study showed that the
treatment of DNMT inhibitors alone may be inadequate to reactivate
gene expression (151). Neither
azacytidine nor decitabine has been established as a single-agent
therapy for GC in clinical settings. However, there is evidence
that DNMT inhibitors can increase chemosensitization when combined
with traditional chemotherapy because the reversal of gene
methylation can reactivate the expression of genes involved in
resistance to chemotherapeutic drugs (152). For example, the use of decitabine
and 5-FU in combination showed reactive expression of TFAP2E in GC
by demethylating activity and enhancing the sensitivity of GC cells
to 5-FU (153). Furthermore, a
phase 1 trial of azacytidine combined to EOX (epirubicin,
oxaliplatin and capecitabine) showed that this combination was
well-tolerated with significant clinical and epigenetic responses
(67%) in patients with locally advanced gastric and esophageal
adenocarcinoma (139). More
prospective randomized studies will be needed to further
investigate whether this combination is superior to EOX.
Compared to other DNMTi, zebularine
(2(1H)-pyrimidinone riboside) is a novel DNMT inhibitor with high
oral bioavailability, slight toxicity and high stability (154). Several preclinical studies have
shown that zebularine can form a tight covalent complex with DNMTs
and so reverse the hypermethylation of TSGs in cancer cell lines
(155). Treatment with zebularine
effectively inhibits GC cells proliferation by inducing cell death,
causing apoptosis in a dose-dependent manner. Moreover, zebularine
depletes expression of DNMT protein with re-expression of
epigenetically silenced genes, such as p16 (142). These results indicated that
zebularine is a promising drug for GC therapy, but further
exploration is needed.
In contrast to DNMTi, the agents targeting histone
methylation is still at a primitive level (156). In 2005, the first study
identified the effect of chaetocin, an inhibitor of HMT SUV39H1
(157). Subsequently, more and
more inhibitors of various HMTs and HDTs, such as DOT1L
(EPZ004777), EZH2 (EPZ-6438 and GSK126, EI1) and G9a (BIX-01294),
were investigated as antitumor drugs (Fig. 1). Several inhibitors targeting
DOT1L, EZH2 and LSD1, have entered phase 1/2 human clinical trials
to assess the safety and maximum tolerated dose (Table IV). For example, EPZ-5676, the
most advanced inhibitor for DOT1L that specifically catalyzes the
mono-, di- and tri-methylation of H3K79, is currently in clinical
trials for MLL-rearranged leukemia (NCT02141828 and NCT01684150).
Major clinical studies that have been conducted so far are
summarized in Table IV. Here, we
summarize some common research targets in cancer, especially in
GC.
The first widely used EZH2 inhibitor,
3-deazaneplanocin A (DZNep), is a cyclopentanyl analog of
3-deazaneplanocine that potently interferes with
S-adenosyl-l-homocysteine hydrolase (SAH), which increases cellular
SAH levels, repressing the activity of SAH-dependent HMT (158). It can inhibit PRC2 and remove
H3K27me3 markers in various cancer types (159). Upon exposure to DZNep, EZH2 is
depleted and ubiquitination of wild-type p53 protein is inhibited,
resulting in p53 stabilization and activation of downstream p53
pathways involved in apoptosis, cell cycle arrest and senescence in
GC cell lines (Table III)
(144). However, DZNep has a
short half-life and is both non-specific and toxic in animal models
(160). GSK126 is an
AdoMet-competitive chemical compound targeting Y641-and
A677-mutated EZH2 for the treatment of diffuse large B-cell
lymphoma (DLBCL) (161). A recent
study has shown that GSK126 suppressed tumor migration and
angiogenesis via downregulation of VEGF-A expression in GC cell
lines and rodent animals (162).
The subsequently developed small-molecule EZH2
inhibitor EPZ005687, has shown dose-dependent inhibition of
H3K27me3 in EZH2-wild-type and Y641- and A677-mutant lymphoma cells
as well as in cell lines of other cancer types, including breast
and prostate cancer. In June 2013, a phase 1/2 clinical trial of
EPZ-6438, with better oral bioavailability than EPZ005687 (163), was explored in patients with
advanced solid tumors or B cell lymphomas (NCT01897571). EI1, a
third SAM-competitive inhibitor, inhibits both wild-type and mutant
EZH2 and alters H3K27me2 and H3K27me3 levels in EZH2-mutant DLBCL
cells in a SMARCB1-mutant rhabdoid tumor cell line (164). EZH2 silencing promoted growth
arrest by low concentrations of doxorubicin in p53 mutant gastric
cancer cells (165). The
combination of EZH2 inhibitors and doxorubicin may be a potential
novel approach to GC treatment.
Over the past decade, epigenetic regulation has been
proven to play a pivotal role in cancer pathogenesis. As an
important subgroup, DNA and histone methylation have been reported
to control numerous cancer suppressor genes and proto-oncogenes. In
GC, hyper- and hypo-methylation events are critical to tumor onset.
Of note, histone methylation plays an important role in chromatin
regulators and GC tumorigenesis. The many HMTs and HDMs that have
been identified can mediate the addition and removal of methyl
groups from different lysine residues on histones and display
distinct functions. These methylation enzymes have attracted
considerable attention as potential targets for GC treatment.
Recently, numerous trials have been made to develop and identify
molecules targeting these enzymes. In many studies, the use of
methylation regulators in combination with already existing
chemotherapy results in better outcomes in many studies. However,
these regulators still have risks of off-target toxicity and many
chromatin-modifying enzymes have more diverse functions. Thus,
understanding the underlying epigenetic molecular mechanisms,
discovering new targets and agents will be essential to successful
targeted epigenetic therapy in the future.
We apologize to all groups whose important
contributions to methylation regulation were not mentioned in this
review due to space limitation. The present review was partly
supported by the Innovation Fund of Shanghai Committee of Science
and Technology (no. 14ZR1406600).
|
1
|
Brenner H, Rothenbacher D and Arndt V:
Epidemiology of stomach cancer. Methods Mol Biol. 472:467–477.
2009. View Article : Google Scholar
|
|
2
|
Leung WK, Wu MS, Kakugawa Y, Kim JJ, Yeoh
KG, Goh KL, Wu KC, Wu DC, Sollano J, Kachintorn U, et al Asia
Pacific Working Group on Gastric Cancer: Screening for gastric
cancer in Asia: Current evidence and practice. Lancet Oncol.
9:279–287. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nakamura J, Tanaka T, Kitajima Y, Noshiro
H and Miyazaki K: Methylation-mediated gene silencing as biomarkers
of gastric cancer: A review. World J Gastroenterol. 20:11991–12006.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sharma S, Kelly TK and Jones PA:
Epigenetics in cancer. Carcinogenesis. 31:27–36. 2010. View Article : Google Scholar :
|
|
5
|
Suzuki MM and Bird A: DNA methylation
landscapes: Provocative insights from epigenomics. Nat Rev Genet.
9:465–476. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Eden A, Gaudet F, Waghmare A and Jaenisch
R: Chromosomal instability and tumors promoted by DNA
hypomethylation. Science. 300:4552003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karpf AR and Matsui S: Genetic disruption
of cytosine DNA methyltransferase enzymes induces chromosomal
instability in human cancer cells. Cancer Res. 65:8635–8639. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schulz WA: L1 retrotransposons in human
cancers. J Biomed Biotechnol. 2006:836722006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Esteller M: Epigenetics in cancer. N Engl
J Med. 358:1148–1159. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ehrlich M: DNA methylation in cancer: Too
much, but also too little. Oncogene. 21:5400–5413. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Omenn GS: Strategies for plasma proteomic
profiling of cancers. Proteomics. 6:5662–5673. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Momparler RL and Bovenzi V: DNA
methylation and cancer. J Cell Physiol. 183:145–154. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Robertson DS: Cellular configuration of
DNA and cell division. Med Hypotheses. 57:344–353. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Fuks F, Hurd PJ, Deplus R and Kouzarides
T: The DNA methyltransferases associate with HP1 and the SUV39H1
histone methyltransferase. Nucleic Acids Res. 31:2305–2312. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kouzarides T: Chromatin modifications and
their function. Cell. 128:693–705. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tan M, Luo H, Lee S, Jin F, Yang JS,
Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al:
Identification of 67 histone marks and histone lysine crotonylation
as a new type of histone modification. Cell. 146:1016–1028. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kouzarides T: SnapShot: Histone-modifying
enzymes. Cell. 131:822–822.e1. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gigek CO, Chen ES, Calcagno DQ, Wisnieski
F, Burbano RR and Smith MA: Epigenetic mechanisms in gastric
cancer. Epigenomics. 4:279–294. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ushijima T and Sasako M: Focus on gastric
cancer. Cancer Cell. 5:121–125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Park JH, Park J, Choi JK, Lyu J, Bae MG,
Lee YG, Bae JB, Park DY, Yang HK, Kim TY, et al: Identification of
DNA methylation changes associated with human gastric cancer. BMC
Med Genomics. 4:822011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yoshida T, Yamashita S, Takamura-Enya T,
Niwa T, Ando T, Enomoto S, Maekita T, Nakazawa K, Tatematsu M,
Ichinose M, et al: Alu and Satα hypomethylation in Helicobacter
pylori-infected gastric mucosae. Int J Cancer. 128:33–39. 2011.
View Article : Google Scholar
|
|
22
|
Jang BG and Kim WH: Molecular pathology of
gastric carcinoma. Pathobiology. 78:302–310. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Leodolter A, Alonso S, González B, Ebert
MP, Vieth M, Röcken C, Wex T, Peitz U, Malfertheiner P and Perucho
M: Somatic DNA Hypomethylation in H. pylori-associated high-risk
gastritis and gastric cancer: Enhanced somatic hypomethylation
associates with advanced stage cancer. Clin Transl Gastroenterol.
6:e852015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kosumi K, Baba Y, Ishimoto T, Harada K,
Miyake K, Izumi D, Tokunaga R, Murata A, Eto K, Sugihara H, et al:
Relationship between LINE-1 hypomethylation and Helicobacter pylori
infection in gastric mucosae. Med Oncol. 32:1172015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Shigaki H, Baba Y, Watanabe M, Murata A,
Iwagami S, Miyake K, Ishimoto T, Iwatsuki M and Baba H: LINE-1
hypomethylation in gastric cancer, detected by bisulfite
pyrosequencing, is associated with poor prognosis. Gastric Cancer.
16:480–487. 2013. View Article : Google Scholar :
|
|
26
|
Xiang S, Liu Z, Zhang B, Zhou J, Zhu BD,
Ji J and Deng D: Methylation status of individual CpG sites within
Alu elements in the human genome and Alu hypomethylation in gastric
carcinomas. BMC Cancer. 10:442010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yuasa Y, Nagasaki H, Oze I, Akiyama Y,
Yoshida S, Shitara K, Ito S, Hosono S, Watanabe M, Ito H, et al:
Insulin-like growth factor 2 hypomethylation of blood leukocyte DNA
is associated with gastric cancer risk. Int J Cancer.
131:2596–2603. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hansen KD, Timp W, Bravo HC, Sabunciyan S,
Langmead B, McDonald OG, Wen B, Wu H, Liu Y, Diep D, et al:
Increased methylation variation in epigenetic domains across cancer
types. Nat Genet. 43:768–775. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Song SH, Jong HS, Choi HH, Kang SH, Ryu
MH, Kim NK, Kim WH and Bang YJ: Methylation of specific CpG sites
in the promoter region could significantly down-regulate
p16INK4a expression in gastric adenocarcinoma. Int J
Cancer. 87:236–240. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shu XS, Geng H, Li L, Ying J, Ma C, Wang
Y, Poon FF, Wang X, Ying Y, Yeo W, et al: The epigenetic modifier
PRDM5 functions as a tumor suppressor through modulating
WNT/β-catenin signaling and is frequently silenced in multiple
tumors. PLoS One. 6:e273462011. View Article : Google Scholar
|
|
31
|
Joo JK, Kim SH, Kim HG, Kim DY, Ryu SY,
Lee KH and Lee JH: CpG methylation of transcription factor 4 in
gastric carcinoma. Ann Surg Oncol. 17:3344–3353. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jung HY, Jung KC, Shim YH, Ro JY and Kang
GH: Methylation of the hMLH1 promoter in multiple gastric
carcinomas with microsatellite instability. Pathol Int. 51:445–451.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yousuf A, Bhat MY, Pandith AA, Afroze D,
Khan NP, Alam K, Shah P, Shah MA and Mudassar S: MGMT gene
silencing by promoter hypermethylation in gastric cancer in a high
incidence area. Cell Oncol (Dordr). 37:245–252. 2014. View Article : Google Scholar
|
|
34
|
Cheng YY, Yu J, Wong YP, Man EP, To KF,
Jin VX, Li J, Tao Q, Sung JJ, Chan FK, et al: Frequent epigenetic
inactivation of secreted frizzled-related protein 2 (SFRP2) by
promoter methylation in human gastric cancer. Br J Cancer.
97:895–901. 2007.PubMed/NCBI
|
|
35
|
Nojima M, Suzuki H, Toyota M, Watanabe Y,
Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K,
et al: Frequent epigenetic inactivation of SFRP genes and
constitutive activation of Wnt signaling in gastric cancer.
Oncogene. 26:4699–4713. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ebert MP, Yu J, Hoffmann J, Rocco A,
Röcken C, Kahmann S, Müller O, Korc M, Sung JJ and Malfertheiner P:
Loss of beta-catenin expression in metastatic gastric cancer. J
Clin Oncol. 21:1708–1714. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Oshimo Y, Kuraoka K, Nakayama H, Kitadai
Y, Yoshida K, Chayama K and Yasui W: Epigenetic inactivation of
SOCS-1 by CpG island hypermethylation in human gastric carcinoma.
Int J Cancer. 112:1003–1009. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kim TY, Lee HJ, Hwang KS, Lee M, Kim JW,
Bang YJ and Kang GH: Methylation of RUNX3 in various types of human
cancers and premalignant stages of gastric carcinoma. Lab Invest.
84:479–484. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Byun DS, Lee MG, Chae KS, Ryu BG and Chi
SG: Frequent epigenetic inactivation of RASSF1A by aberrant
promoter hypermethylation in human gastric adenocarcinoma. Cancer
Res. 61:7034–7038. 2001.PubMed/NCBI
|
|
40
|
Hayashi K, Yokozaki H, Goodison S, Oue N,
Suzuki T, Lotan R, Yasui W and Tahara E: Inactivation of retinoic
acid receptor beta by promoter CpG hypermethylation in gastric
cancer. Differentiation. 68:13–21. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yu J, Tao Q, Cheng YY, Lee KY, Ng SS,
Cheung KF, Tian L, Rha SY, Neumann U, Röcken C, et al: Promoter
methylation of the Wnt/beta-catenin signaling antagonist Dkk-3 is
associated with poor survival in gastric cancer. Cancer. 115:49–60.
2009. View Article : Google Scholar
|
|
42
|
Nishigaki M, Aoyagi K, Danjoh I, Fukaya M,
Yanagihara K, Sakamoto H, Yoshida T and Sasaki H: Discovery of
aberrant expression of R-RAS by cancer-linked DNA hypomethylation
in gastric cancer using microarrays. Cancer Res. 65:2115–2124.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mulero-Navarro S and Esteller M:
Epigenetic biomarkers for human cancer: The time is now. Crit Rev
Oncol Hematol. 68:1–11. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Oue N, Matsumura S, Nakayama H, Kitadai Y,
Taniyama K, Matsusaki K and Yasui W: Reduced expression of the TSP1
gene and its association with promoter hypermethylation in gastric
carcinoma. Oncology. 64:423–429. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ganesan K, Ivanova T, Wu Y, Rajasegaran V,
Wu J, Lee MH, Yu K, Rha SY, Chung HC, Ylstra B, et al: Inhibition
of gastric cancer invasion and metastasis by PLA2G2A, a novel
beta-catenin/TCF target gene. Cancer Res. 68:4277–4286. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chan AW, Chan MW, Lee TL, Ng EK, Leung WK,
Lau JY, Tong JH, Chan FK and To KF: Promoter hypermethylation of
Death-associated protein-kinase gene associated with advance stage
gastric cancer. Oncol Rep. 13:937–941. 2005.PubMed/NCBI
|
|
47
|
Yu J, Cheng YY, Tao Q, Cheung KF, Lam CN,
Geng H, Tian LW, Wong YP, Tong JH, Ying JM, et al: Methylation of
protocadherin 10, a novel tumor suppressor, is associated with poor
prognosis in patients with gastric cancer. Gastroenterology.
136:640–51.e1. 2009. View Article : Google Scholar
|
|
48
|
Hu X, Sui X, Li L, Huang X, Rong R, Su X,
Shi Q, Mo L, Shu X, Kuang Y, et al: Protocadherin 17 acts as a
tumour suppressor inducing tumour cell apoptosis and autophagy, and
is frequently methylated in gastric and colorectal cancers. J
Pathol. 229:62–73. 2013. View Article : Google Scholar
|
|
49
|
Haruki S, Imoto I, Kozaki K, Matsui T,
Kawachi H, Komatsu S, Muramatsu T, Shimada Y, Kawano T and Inazawa
J: Frequent silencing of protocadherin 17, a candidate tumour
suppressor for esophageal squamous cell carcinoma. Carcinogenesis.
31:1027–1036. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Hiraki M, Kitajima Y, Koga Y, Tanaka T,
Nakamura J, Hashiguchi K, Noshiro H and Miyazaki K: Aberrant gene
methylation is a biomarker for the detection of cancer cells in
peritoneal wash samples from advanced gastric cancer patients. Ann
Surg Oncol. 18:3013–3019. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Xu L, Li X, Chu ES, Zhao G, Go MY, Tao Q,
Jin H, Zeng Z, Sung JJ and Yu J: Epigenetic inactivation of BCL6B,
a novel functional tumour suppressor for gastric cancer, is
associated with poor survival. Gut. 61:977–985. 2012. View Article : Google Scholar
|
|
52
|
Xie B, Zhou J, Shu G, Liu DC, Zhou J, Chen
J and Yuan L: Restoration of klotho gene expression induces
apoptosis and autophagy in gastric cancer cells: Tumor suppressive
role of klotho in gastric cancer. Cancer Cell Int. 13:182013.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Kwon OH, Park JL, Baek SJ, Noh SM, Song
KS, Kim SY and Kim YS: Aberrant upregulation of ASCL2 by promoter
demethylation promotes the growth and resistance to 5-fluorouracil
of gastric cancer cells. Cancer Sci. 104:391–397. 2013. View Article : Google Scholar
|
|
54
|
Gigek CO, Lisboa LC, Leal MF, Silva PN,
Lima EM, Khayat AS, Assumpção PP, Burbano RR and Smith MA: SMARCA5
methylation and expression in gastric cancer. Cancer Invest.
29:162–166. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Xiong HL, Liu XQ, Sun AH, He Y, Li J and
Xia Y: Aberrant DNA methylation of P16, MGMT, hMLH1 and hMSH2 genes
in combination with the MTHFR C677T genetic polymorphism in gastric
cancer. Asian Pac J Cancer Prev. 14:3139–3142. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hamai Y, Oue N, Mitani Y, Nakayama H, Ito
R, Matsusaki K, Yoshida K, Toge T and Yasui W: DNA hypermethylation
and histone hypoacetylation of the HLTF gene are associated with
reduced expression in gastric carcinoma. Cancer Sci. 94:692–698.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Wang L, Xie L, Wang J, Shen J and Liu B:
Correlation between the methylation of SULF2 and WRN promoter and
the irinotecan chemosensitivity in gastric cancer. BMC
Gastroenterol. 13:1732013. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Ivanova T, Zouridis H, Wu Y, Cheng LL, Tan
IB, Gopalakrishnan V, Ooi CH, Lee J, Qin L, Wu J, et al: Integrated
epigenomics identifies BMP4 as a modulator of cisplatin sensitivity
in gastric cancer. Gut. 62:22–33. 2013. View Article : Google Scholar
|
|
59
|
Zouridis H, Deng N, Ivanova T, Zhu Y, Wong
B, Huang D, Wu YH, Wu Y, Tan IB, Liem N, et al: Methylation
subtypes and large-scale epigenetic alterations in gastric cancer.
Sci Transl Med. 4:156ra1402012. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Selaru FM, David S, Meltzer SJ and
Hamilton JP: Epigenetic events in gastrointestinal cancer. Am J
Gastroenterol. 104:1910–1912. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Yamamoto E, Suzuki H, Takamaru H, Yamamoto
H, Toyota M and Shinomura Y: Role of DNA methylation in the
development of diffuse-type gastric cancer. Digestion. 83:241–249.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Cavallaro U and Christofori G:
Multitasking in tumor progression: Signaling functions of cell
adhesion molecules. Ann NY Acad Sci. 1014:58–66. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zong L and Seto Y: CpG island methylator
phenotype, Helicobacter pylori, Epstein-Barr virus, and
microsatellite instability and prognosis in gastric cancer: A
systematic review and meta-analysis. PLoS One. 9:e860972014.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ding WJ, Fang JY, Chen XY and Peng YS: The
expression and clinical significance of DNA methyltransferase
proteins in human gastric cancer. Dig Dis Sci. 53:2083–2089. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Yang J, Wei X, Wu Q, Xu Z, Gu D, Jin Y,
Shen Y, Huang H, Fan H and Chen J: Clinical significance of the
expression of DNA methyltransferase proteins in gastric cancer. Mol
Med Rep. 4:1139–1143. 2011.PubMed/NCBI
|
|
66
|
Cao XY, Ma HX, Shang YH, Jin MS, Kong F,
Jia ZF, Cao DH, Wang YP, Suo J and Jiang J: DNA methyltransferase3a
expression is an independent poor prognostic indicator in gastric
cancer. World J Gastroenterol. 20:8201–8208. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Cui H, Zhao C, Gong P, Wang L, Wu H, Zhang
K, Zhou R, Wang L, Zhang T, Zhong S, et al: DNA methyltransferase
3A promotes cell proliferation by silencing CDK inhibitor
p18INK4C in gastric carcinogenesis. Sci Rep.
5:137812015. View Article : Google Scholar
|
|
68
|
Ley TJ, Ding L, Walter MJ, McLellan MD,
Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et
al: DNMT3A mutations in acute myeloid leukemia. N Engl J Med.
363:2424–2433. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Zang ZJ, Cutcutache I, Poon SL, Zhang SL,
McPherson JR, Tao J, Rajasegaran V, Heng HL, Deng N, Gan A, et al:
Exome sequencing of gastric adenocarcinoma identifies recurrent
somatic mutations in cell adhesion and chromatin remodeling genes.
Nat Genet. 44:570–574. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kim MS, Kim YR, Yoo NJ and Lee SH:
Mutational analysis of DNMT3A gene in acute leukemias and common
solid cancers. APMIS. 121:85–94. 2013. View Article : Google Scholar
|
|
71
|
Kanai Y, Ushijima S, Kondo Y, Nakanishi Y
and Hirohashi S: DNA methyltransferase expression and DNA
methylation of CPG islands and peri-centromeric satellite regions
in human colorectal and stomach cancers. Int J Cancer. 91:205–212.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Hu J, Fan H, Liu D, Zhang S, Zhang F and
Xu H: DNMT3B promoter polymorphism and risk of gastric cancer. Dig
Dis Sci. 55:1011–1016. 2010. View Article : Google Scholar
|
|
73
|
Li H, Li W, Liu S, Zong S, Wang W, Ren J,
Li Q, Hou F and Shi Q: DNMT1, DNMT3A and DNMT3B polymorphisms
associated with gastric cancer risk: A systematic review and
meta-analysis. EBioMedicine. 13:125–131. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Tang H, Deng M, Tang Y and Xie X, Guo J,
Kong Y, Ye F, Su Q and Xie X: miR-200b and miR-200c as prognostic
factors and mediators of gastric cancer cell progression. Clin
Cancer Res. 19:5602–5612. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Zhang BG, Hu L, Zang MD, Wang HX, Zhao W,
Li JF, Su LP, Shao Z, Zhao X, Zhu ZG, et al: Helicobacter pylori
CagA induces tumor suppressor gene hypermethylation by upregulating
DNMT1 via AKT-NF-κB pathway in gastric cancer development.
Oncotarget. 7:9788–9800. 2016.PubMed/NCBI
|
|
76
|
Wallasch C, Crabtree JE, Bevec D, Robinson
PA, Wagner H and Ullrich A: Helicobacter pylori-stimulated EGF
receptor transactivation requires metalloprotease cleavage of
HB-EGF. Biochem Biophys Res Commun. 295:695–701. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Hino R, Uozaki H, Murakami N, Ushiku T,
Shinozaki A, Ishikawa S, Morikawa T, Nakaya T, Sakatani T, Takada
K, et al: Activation of DNA methyltransferase 1 by EBV latent
membrane protein 2A leads to promoter hypermethylation of PTEN gene
in gastric carcinoma. Cancer Res. 69:2766–2774. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Liang Q, Yao X, Tang S, Zhang J, Yau TO,
Li X, Tang CM, Kang W, Lung RW, Li JW, et al: Integrative
identification of Epstein-Barr virus-associated mutations and
epigenetic alterations in gastric cancer. Gastroenterology.
147:1350–62.e4. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Liu J, Zhu X, Xu X and Dai D: DNA promoter
and histone H3 methylation downregulate NGX6 in gastric cancer
cells. Med Oncol. 31:8172014. View Article : Google Scholar
|
|
80
|
Park YS, Jin MY, Kim YJ, Yook JH, Kim BS
and Jang SJ: The global histone modification pattern correlates
with cancer recurrence and overall survival in gastric
adenocarcinoma. Ann Surg Oncol. 15:1968–1976. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Lee H, Yoon SO, Jeong WY, Kim HK, Kim A
and Kim BH: Immunohistochemical analysis of polycomb group protein
expression in advanced gastric cancer. Hum Pathol. 43:1704–1710.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
He LJ, Cai MY, Xu GL, Li JJ, Weng ZJ, Xu
DZ, Luo GY, Zhu SL and Xie D: Prognostic significance of
overexpression of EZH2 and H3k27me3 proteins in gastric cancer.
Asian Pac J Cancer Prev. 13:3173–3178. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kwon OH, Park JL, Kim M, Kim JH, Lee HC,
Kim HJ, Noh SM, Song KS, Yoo HS, Paik SG, et al: Aberrant
up-regulation of LAMB3 and LAMC2 by promoter demethylation in
gastric cancer. Biochem Biophys Res Commun. 406:539–545. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Komatsu S, Ichikawa D, Hirajima S, Nagata
H, Nishimura Y, Kawaguchi T, Miyamae M, Okajima W, Ohashi T,
Konishi H, et al: Overexpression of SMYD2 contributes to malignant
outcome in gastric cancer. Br J Cancer. 112:357–364. 2015.
View Article : Google Scholar :
|
|
85
|
Liu Y, Deng J, Luo X, Pan Y, Zhang L,
Zhang R and Liang H: Overexpression of SMYD3 was associated with
increased STAT3 activation in gastric cancer. Med Oncol.
32:4042015. View Article : Google Scholar
|
|
86
|
Liu H, Liu Y, Kong F, Xin W, Li X, Liang H
and Jia Y: Elevated levels of SET and MYND domain-containing
protein 3 are correlated with overexpression of transforming growth
factor-β1 in gastric cancer. J Am Coll Surg. 221:579–590. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Liu Y, Liu H, Luo X, Deng J, Pan Y and
Liang H: Overexpression of SMYD3 and matrix metalloproteinase-9 are
associated with poor prognosis of patients with gastric cancer.
Tumour Biol. 36:4377–4386. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Liu Y, Luo X, Deng J, Pan Y, Zhang L and
Liang H: SMYD3 overexpression was a risk factor in the biological
behavior and prognosis of gastric carcinoma. Tumour Biol.
36:2685–2694. 2015. View Article : Google Scholar
|
|
89
|
Cai L, Ma X, Huang Y, Zou Y and Chen X:
Aberrant histone methylation and the effect of Suv39H1 siRNA on
gastric carcinoma. Oncol Rep. 31:2593–2600. 2014.PubMed/NCBI
|
|
90
|
Fujii S, Ito K, Ito Y and Ochiai A:
Enhancer of zeste homologue 2 (EZH2) down-regulates RUNX3 by
increasing histone H3 methylation. J Biol Chem. 283:17324–17332.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Qi Y, Zhang X, Kang Y, Wu J, Chen J, Li H,
Guo Y, Liu B, Shao Z and Zhao X: Genome-wide transcriptional
profiling analysis reveals annexin A6 as a novel EZH2 target gene
involving gastric cellular proliferation. Mol Biosyst.
11:1980–1986. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Fujii S and Ochiai A: Enhancer of zeste
homolog 2 downregulates E-cadherin by mediating histone H3
methylation in gastric cancer cells. Cancer Sci. 99:738–746. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Tong Y, Li Y, Gu H, Wang C, Liu F, Shao Y,
Li J, Cao L and Li F: Microchidia protein 2, MORC2, downregulates
the cytoskeleton adapter protein, ArgBP2, via histone methylation
in gastric cancer cells. Biochem Biophys Res Commun. 467:821–827.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Bai J, Chen J, Ma M, Cai M, Xu F, Wang G,
Tao K and Shuai X: Inhibiting enhancer of zeste homolog 2 promotes
cellular senescence in gastric cancer cells SGC-7901 by activation
of p21 and p16. DNA Cell Biol. 33:337–344. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Jie B, Weilong C, Ming C, Fei X, Xinghua
L, Junhua C, Guobin W, Kaixiong T and Xiaoming S: Enhancer of zeste
homolog 2 depletion induces cellular senescence via histone
demethylation along the INK4/ARF locus. Int J Biochem Cell Biol.
65:104–112. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Lu H, Sun J, Wang F, Feng L, Ma Y, Shen Q,
Jiang Z, Sun X, Wang X and Jin H: Enhancer of zeste homolog 2
activates wnt signaling through downregulating CXXC finger protein
4. Cell Death Dis. 4:e7762013. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Lu H, Jin W, Sun J, Feng L, Lan H, Shen Q,
Ma Y, Li J, Yue Y, Jin H, et al: New tumor suppressor CXXC finger
protein 4 inactivates mitogen activated protein kinase signaling.
FEBS Lett. 588:3322–3326. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Akiyama Y, Koda Y, Byeon SJ, Shimada S,
Nishikawaji T, Sakamoto A, Chen Y, Kojima K, Kawano T, Eishi Y, et
al: Reduced expression of SET7/9, a histone mono-methyltransferase,
is associated with gastric cancer progression. Oncotarget.
7:3966–3983. 2016.
|
|
99
|
Zeng J, Ge Z, Wang L, Li Q, Wang N,
Björkholm M, Jia J and Xu D: The histone demethylase RBP2 is
overexpressed in gastric cancer and its inhibition triggers
senescence of cancer cells. Gastroenterology. 138:981–992. 2010.
View Article : Google Scholar
|
|
100
|
Wang Z, Tang F, Qi G, Yuan S, Zhang G,
Tang B and He S: KDM5B is overexpressed in gastric cancer and is
required for gastric cancer cell proliferation and metastasis. Am J
Cancer Res. 5:87–100. 2014.
|
|
101
|
Li Y, Tian X, Sui CG, Jiang YH, Liu YP and
Meng FD: Interference of lysine-specific demethylase 1 inhibits
cellular invasion and proliferation in vivo in gastric cancer
MKN-28 cells. Biomed Pharmacother. 82:498–508. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Yang H, Liu Z, Yuan C, Zhao Y, Wang L, Hu
J, Xie D, Wang L and Chen D: Elevated JMJD1A is a novel predictor
for prognosis and a potential therapeutic target for gastric
cancer. Int J Clin Exp Pathol. 8:11092–11099. 2015.PubMed/NCBI
|
|
103
|
Hu CE, Liu YC, Zhang HD and Huang GJ:
JMJD2A predicts prognosis and regulates cell growth in human
gastric cancer. Biochem Biophys Res Commun. 449:1–7. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Li W, Zhao L, Zang W, Liu Z, Chen L, Liu
T, Xu D and Jia J: Histone demethylase JMJD2B is required for tumor
cell proliferation and survival and is overexpressed in gastric
cancer. Biochem Biophys Res Commun. 416:372–378. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Zhao L, Li W, Zang W, Liu Z, Xu X, Yu H,
Yang Q and Jia J: JMJD2B promotes epithelial-mesenchymal transition
by cooperating with β-catenin and enhances gastric cancer
metastasis. Clin Cancer Res. 19:6419–6429. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Taverna SD, Li H, Ruthenburg AJ, Allis CD
and Patel DJ: How chromatin-binding modules interpret histone
modifications: Lessons from professional pocket pickers. Nat Struct
Mol Biol. 14:1025–1040. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
He C, Xu J, Zhang J, Xie D, Ye H, Xiao Z,
Cai M, Xu K, Zeng Y, Li H, et al: High expression of trimethylated
histone H3 lysine 4 is associated with poor prognosis in
hepatocellular carcinoma. Hum Pathol. 43:1425–1435. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Zee BM, Levin RS, Xu B, LeRoy G, Wingreen
NS and Garcia BA: In vivo residue-specific histone methylation
dynamics. J Biol Chem. 285:3341–3350. 2010. View Article : Google Scholar :
|
|
109
|
Tsukada Y, Fang J, Erdjument-Bromage H,
Warren ME, Borchers CH, Tempst P and Zhang Y: Histone demethylation
by a family of JmjC domain-containing proteins. Nature.
439:811–816. 2006. View Article : Google Scholar
|
|
110
|
Huang J and Berger SL: The emerging field
of dynamic lysine methylation of non-histone proteins. Curr Opin
Genet Dev. 18:152–158. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Tan JZ, Yan Y, Wang XX, Jiang Y and Xu HE:
EZH2: Biology, disease, and structure-based drug discovery. Acta
Pharmacol Sin. 35:161–174. 2014. View Article : Google Scholar :
|
|
112
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Viré E, Brenner C, Deplus R, Blanchon L,
Fraga M, Didelot C, Morey L, Van Eynde A, Bernard D, Vanderwinden
JM, et al: The Polycomb group protein EZH2 directly controls DNA
methylation. Nature. 439:871–874. 2006. View Article : Google Scholar
|
|
114
|
Ma J, Wang JD, Zhang WJ, Zou B, Chen WJ,
Lam CS, Chen MH, Pang R, Tan VP, Hung IF, et al: Promoter
hypermethylation and histone hypoacetylation contribute to
pancreatic-duodenal homeobox 1 silencing in gastric cancer.
Carcinogenesis. 31:1552–1560. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Meng CF, Zhu XJ, Peng G and Dai DQ:
Promoter histone H3 lysine 9 di-methylation is associated with DNA
methylation and aberrant expression of p16 in gastric cancer cells.
Oncol Rep. 22:1221–1227. 2009.PubMed/NCBI
|
|
116
|
Watanabe Y, Toyota M, Kondo Y, Suzuki H,
Imai T, Ohe-Toyota M, Maruyama R, Nojima M, Sasaki Y, Sekido Y, et
al: PRDM5 identified as a target of epigenetic silencing in
colorectal and gastric cancer. Clin Cancer Res. 13:4786–4794. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Cho HS, Shimazu T, Toyokawa G, Daigo Y,
Maehara Y, Hayami S, Ito A, Masuda K, Ikawa N, Field HI, et al:
Enhanced HSP70 lysine methylation promotes proliferation of cancer
cells through activation of Aurora kinase B. Nat Commun.
3:10722012. View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Hamamoto R, Toyokawa G, Nakakido M, Ueda K
and Nakamura Y: SMYD2-dependent HSP90 methylation promotes cancer
cell proliferation by regulating the chaperone complex formation.
Cancer Lett. 351:126–133. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Huang J, Perez-Burgos L, Placek BJ,
Sengupta R, Richter M, Dorsey JA, Kubicek S, Opravil S, Jenuwein T
and Berger SL: Repression of p53 activity by Smyd2-mediated
methylation. Nature. 444:629–632. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Saddic LA, West LE, Aslanian A, Yates JR
III, Rubin SM, Gozani O and Sage J: Methylation of the
retinoblastoma tumor suppressor by SMYD2. J Biol Chem.
285:37733–37740. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Cho HS, Hayami S, Toyokawa G, Maejima K,
Yamane Y, Suzuki T, Dohmae N, Kogure M, Kang D, Neal DE, et al: RB1
methylation by SMYD2 enhances cell cycle progression through an
increase of RB1 phosphorylation. Neoplasia. 14:476–486. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Piao L, Kang D, Suzuki T, Masuda A, Dohmae
N, Nakamura Y and Hamamoto R: The histone methyltransferase SMYD2
methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in
cancer cells. Neoplasia. 16:257–264.e2. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Mazur PK, Reynoird N, Khatri P, Jansen PW,
Wilkinson AW, Liu S, Barbash O, Van Aller GS, Huddleston M, Dhanak
D, et al: SMYD3 links lysine methylation of MAP3K2 to Ras-driven
cancer. Nature. 510:283–287. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Easwaran H, Tsai HC and Baylin SB: Cancer
epigenetics: Tumor heterogeneity, plasticity of stem-like states,
and drug resistance. Mol Cell. 54:716–727. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Compare D, Rocco A, Liguori E, D'Armiento
FP, Persico G, Masone S, Coppola-Bottazzi E, Suriani R, Romano M
and Nardone G: Global DNA hypomethylation is an early event in
Helicobacter pylori-related gastric carcinogenesis. J Clin Pathol.
64:677–682. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Yamashita S, Tsujino Y, Moriguchi K,
Tatematsu M and Ushijima T: Chemical genomic screening for
methylation-silenced genes in gastric cancer cell lines using
5-aza-2′-deoxycytidine treatment and oligonucleotide microarray.
Cancer Sci. 97:64–71. 2006. View Article : Google Scholar
|
|
127
|
Sepulveda AR, Yao Y, Yan W, Park DI, Kim
JJ, Gooding W, Abudayyeh S and Graham DY: CpG methylation and
reduced expression of O6-methylguanine DNA
methyltransferase is associated with Helicobacter pylori infection.
Gastroenterology. 138:1836–1844. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
128
|
Hur K, Niwa T, Toyoda T, Tsukamoto T,
Tatematsu M, Yang HK and Ushijima T: Insufficient role of cell
proliferation in aberrant DNA methylation induction and involvement
of specific types of inflammation. Carcinogenesis. 32:35–41. 2011.
View Article : Google Scholar
|
|
129
|
Nardone G, Rocco A and Malfertheiner P:
Review article: Helicobacter pylori and molecular events in
precancerous gastric lesions. Aliment Pharmacol Ther. 20:261–270.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Angrisano T, Lembo F, Peluso S, Keller S,
Chiariotti L and Pero R: Helicobacter pylori regulates iNOS
promoter by histone modifications in human gastric epithelial
cells. Med Microbiol Immunol (Berl). 201:249–257. 2012. View Article : Google Scholar
|
|
131
|
Han F, Ren J, Zhang J, Sun Y, Ma F, Liu Z,
Yu H, Jia J and Li W: JMJD2B is required for Helicobacter
pylori-induced gastric carcinogenesis via regulating COX-2
expression. Oncotarget. 7:38626–38637. 2016.PubMed/NCBI
|
|
132
|
Bass AJ, Thorsson V, Shmulevich I,
Reynolds SM, Miller M, Bernard B, Hinoue T, Laird PW, Curtis C,
Shen H, et al Cancer Genome Atlas Research Network: Comprehensive
molecular characterization of gastric adenocarcinoma. Nature.
513:202–209. 2014. View Article : Google Scholar :
|
|
133
|
Kaneda A, Matsusaka K, Aburatani H and
Fukayama M: Epstein-Barr virus infection as an epigenetic driver of
tumorigenesis. Cancer Res. 72:3445–3450. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Sudo M, Chong JM, Sakuma K, Ushiku T,
Uozaki H, Nagai H, Funata N, Matsumoto Y and Fukayama M: Promoter
hyper-methylation of E-cadherin and its abnormal expression in
Epstein-Barr virus-associated gastric carcinoma. Int J Cancer.
109:194–199. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Osawa T, Chong JM, Sudo M, Sakuma K,
Uozaki H, Shibahara J, Nagai H, Funata N and Fukayama M: Reduced
expression and promoter methylation of p16 gene in Epstein-Barr
virus-associated gastric carcinoma. Jpn J Cancer Res. 93:1195–1200.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Ryan JL, Jones RJ, Kenney SC, Rivenbark
AG, Tang W, Knight ER, Coleman WB and Gulley ML: Epstein-Barr
virus-specific methylation of human genes in gastric cancer cells.
Infect Agent Cancer. 5:272010. View Article : Google Scholar
|
|
137
|
Matsusaka K, Kaneda A, Nagae G, Ushiku T,
Kikuchi Y, Hino R, Uozaki H, Seto Y, Takada K, Aburatani H, et al:
Classification of Epstein-Barr virus-positive gastric cancers by
definition of DNA methylation epigenotypes. Cancer Res.
71:7187–7197. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Ballestar E and Esteller M: SnapShot: The
human DNA methylome in health and disease. Cell. 135:1144–1144.e1.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Schneider BJ, Shah MA, Klute K, Ocean A,
Popa E, Altorki NK, Lieberman MD, Schreiner A, Yantiss RK, Christos
PJ, et al: Phase I study of epigenetic priming with azacitidine
prior to standard neoadjuvant chemotherapy for patients with
resectable gastric and esophageal adenocarcinoma. Clin Cancer Res.
View Article : Google Scholar : Epub ahead of
print.
|
|
140
|
Liu J, Xie YS, Wang FL, Zhang LJ, Zhang Y
and Luo HS: Cytotoxicity of 5-Aza-2′-deoxycytidine against gastric
cancer involves DNA damage in an ATM-P53 dependent signaling
pathway and demethylation of P16INK4A. Biomed
Pharmacother. 67:78–87. 2013. View Article : Google Scholar
|
|
141
|
Shin DY, Kim GY, Kim CG, Kim WJ, Kang HS
and Choi YH: Anti-invasive effects of decitabine, a DNA
methyltransferase inhibitor, through tightening of tight junctions
and inhibition of matrix metalloproteinase activities in AGS human
gastric carcinoma cells. Oncol Rep. 28:1043–1050. 2012.PubMed/NCBI
|
|
142
|
Tan W, Zhou W, Yu HG, Luo HS and Shen L:
The DNA methyltransferase inhibitor zebularine induces
mitochondria-mediated apoptosis in gastric cancer cells in vitro
and in vivo. Biochem Biophys Res Commun. 430:250–255. 2013.
View Article : Google Scholar
|
|
143
|
Takeshima H, Wakabayashi M, Hattori N,
Yamashita S and Ushijima T: Identification of coexistence of DNA
methylation and H3K27me3 specifically in cancer cells as a
promising target for epigenetic therapy. Carcinogenesis.
36:192–201. 2015. View Article : Google Scholar
|
|
144
|
Cheng LL, Itahana Y, Lei ZD, Chia NY, Wu
Y, Yu Y, Zhang SL, Thike AA, Pandey A, Rozen S, et al: TP53 genomic
status regulates sensitivity of gastric cancer cells to the histone
methylation inhibitor 3-deazaneplanocin A (DZNep). Clin Cancer Res.
18:4201–4212. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Kaminskas E, Farrell AT, Wang YC, Sridhara
R and Pazdur R: FDA drug approval summary: Azacitidine
(5-azacytidine, Vidaza) for injectable suspension. Oncologist.
10:176–182. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Steensma DP, Baer MR, Slack JL, Buckstein
R, Godley LA, Garcia-Manero G, Albitar M, Larsen JS, Arora S,
Cullen MT, et al: Multicenter study of decitabine administered
daily for 5 days every 4 weeks to adults with myelodysplastic
syndromes: The alternative dosing for outpatient treatment (ADOPT)
trial. J Clin Oncol. 27:3842–3848. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Erdmann A, Halby L, Fahy J and Arimondo
PB: Targeting DNA methylation with small molecules: What's next? J
Med Chem. 58:2569–2583. 2015. View Article : Google Scholar
|
|
148
|
Navada SC, Steinmann J, Lübbert M and
Silverman LR: Clinical development of demethylating agents in
hematology. J Clin Invest. 124:40–46. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Chen XL, Wang FM, Li JJ, He XY, Liu XY and
Ma LB: The effect of two nucleoside antitumor drugs on the
proliferation and DNA methylation of human gastric cancer cells.
Oncol Lett. 10:1919–1923. 2015.PubMed/NCBI
|
|
150
|
Karahoca M and Momparler RL:
Pharmacokinetic and pharmacodynamic analysis of
5-aza-2′-deoxycytidine (decitabine) in the design of its
dose-schedule for cancer therapy. Clin Epigenetics. 5:32013.
View Article : Google Scholar
|
|
151
|
Si J, Boumber YA, Shu J, Qin T, Ahmed S,
He R, Jelinek J and Issa JP: Chromatin remodeling is required for
gene reactivation after decitabine-mediated DNA hypomethylation.
Cancer Res. 70:6968–6977. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Plumb JA, Strathdee G, Sludden J, Kaye SB
and Brown R: Reversal of drug resistance in human tumor xenografts
by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene
promoter. Cancer Res. 60:6039–6044. 2000.PubMed/NCBI
|
|
153
|
Wu FL, Li RT, Yang M, Yue GF, Wang HY, Liu
Q, Cui FB, Wu PY, Ding H, Yu LX, et al: Gelatinases-stimuli
nanoparticles encapsulating 5-fluorouridine and
5-aza-2′-deoxycytidine enhance the sensitivity of gastric cancer
cells to chemical therapeutics. Cancer Lett. 363:7–16. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Chen M, Shabashvili D, Nawab A, Yang SX,
Dyer LM, Brown KD, Hollingshead M, Hunter KW, Kaye FJ, Hochwald SN,
et al: DNA methyltransferase inhibitor, zebularine, delays tumor
growth and induces apoptosis in a genetically engineered mouse
model of breast cancer. Mol Cancer Ther. 11:370–382. 2012.
View Article : Google Scholar
|
|
155
|
Ben-Kasus T, Ben-Zvi Z, Marquez VE, Kelley
JA and Agbaria R: Metabolic activation of zebularine, a novel DNA
methylation inhibitor, in human bladder carcinoma cells. Biochem
Pharmacol. 70:121–133. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Plass C, Pfister SM, Lindroth AM,
Bogatyrova O, Claus R and Lichter P: Mutations in regulators of the
epigenome and their connections to global chromatin patterns in
cancer. Nat Rev Genet. 14:765–780. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Greiner D, Bonaldi T, Eskeland R, Roemer E
and Imhof A: Identification of a specific inhibitor of the histone
methyltransferase SU(VAR)3–9. Nat Chem Biol. 1:143–145. 2005.
View Article : Google Scholar
|
|
158
|
Glazer RI, Hartman KD, Knode MC, Richard
MM, Chiang PK, Tseng CK and Marquez VE: 3-Deazaneplanocin: A new
and potent inhibitor of S-adenosylhomocysteine hydrolase and its
effects on human promyelocytic leukemia cell line HL-60. Biochem
Biophys Res Commun. 135:688–694. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Tan J, Yang X, Zhuang L, Jiang X, Chen W,
Lee PL, Karuturi RK, Tan PB, Liu ET and Yu Q: Pharmacologic
disruption of Polycomb-repressive complex 2-mediated gene
repression selectively induces apoptosis in cancer cells. Genes
Dev. 21:1050–1063. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Sun F, Lee L, Zhang Z, Wang X, Yu Q, Duan
X and Chan E: Preclinical pharmacokinetic studies of
3-deazaneplanocin A, a potent epigenetic anticancer agent, and its
human pharmacokinetic prediction using GastroPlus™. Eur J Pharm
Sci. 77:290–302. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
161
|
McCabe MT, Ott HM, Ganji G, Korenchuk S,
Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A III,
Diaz E, et al: EZH2 inhibition as a therapeutic strategy for
lymphoma with EZH2-activating mutations. Nature. 492:108–112. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
162
|
Chen YT, Zhu F, Lin WR, Ying RB, Yang YP
and Zeng LH: The novel EZH2 inhibitor, GSK126, suppresses cell
migration and angiogenesis via down-regulating VEGF-A. Cancer
Chemother Pharmacol. 77:757–765. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Knutson SK, Warholic NM, Wigle TJ, Klaus
CR, Allain CJ, Raimondi A, Porter Scott M, Chesworth R, Moyer MP,
Copeland RA, et al: Durable tumor regression in genetically altered
malignant rhabdoid tumors by inhibition of methyltransferase EZH2.
Proc Natl Acad Sci USA. 110:7922–7927. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Qi W, Chan H, Teng L, Li L, Chuai S, Zhang
R, Zeng J, Li M, Fan H, Lin Y, et al: Selective inhibition of Ezh2
by a small molecule inhibitor blocks tumor cells proliferation.
Proc Natl Acad Sci USA. 109:21360–21365. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Bai J, Ma M, Cai M, Xu F, Chen J, Wang G,
Shuai X and Tao K: Inhibition enhancer of zeste homologue 2
promotes senescence and apoptosis induced by doxorubicin in p53
mutant gastric cancer cells. Cell Prolif. 47:211–218. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Lee MG, Wynder C, Schmidt DM, McCafferty
DG and Shiekhattar R: Histone H3 lysine 4 demethylation is a target
of nonselective antidepressive medications. Chem Biol. 13:563–567.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Schmidt DM and McCafferty DG:
trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of
the histone demethylase LSD1. Biochemistry. 46:4408–4416. 2007.
View Article : Google Scholar : PubMed/NCBI
|