Introduction
Glioma, the most common and malignant brain tumour
in human, originates from neural stromal cells and accounts for 80%
of all malignant brain tumours in adults (1,2).
Glioma can be divided into two groups according to the 2007 World
Health Organisation classification: low grade (grades I and II) and
high grade (grades III and IV) (2). Glioblastoma (GBM), regarded as grade
IV glioma, is the most common histological subtype of glioma in
adults (3). Rapid growth, cellular
heterogeneity, angiogenesis, extensive invasion, hypoxia and
necrosis are the histopathological hallmarks of GBM (4,5).
Currently, surgical resection is the primary treatment method for
GBM, whereas radiotherapy and chemotherapy are common adjuvant
therapeutic approaches (6,7). Despite recent advances in multimodal
therapies, the prognosis of GBM patients remains extremely poor,
with an average 5-year survival rate of only 4–5% (8,9).
Therefore, the elucidation of the molecular mechanisms of GBM and
identification of novel and effective therapeutic strategies for
patients with this disease are urgently needed.
MicroRNAs (miRNAs) are a family of small double-
stranded, non-protein coding and short RNA molecules with lengths
of ~21–25 nucleotides (10).
miRNAs regulate gene expression levels by imperfectly pairing with
complementary sites within the 3′-untranslated regions (3′-UTRs) of
their target messenger RNAs (mRNAs), resulting in either
degradation or translational repression of the target mRNAs
(11,12). A miRNA may regulate numerous target
genes, and one gene may be regulated by multiple miRNAs. Therefore,
>60% of all human genes have been predicted to be regulated by
miRNAs (13). Increasing studies
have shown that miRNAs are involved in regulating various
biological processes, including development, cell proliferation,
differentiation, apoptosis, metabolism and signal transduction
(14–16). Multiple bodies of evidence have
indicated that miRNAs are abnormally expressed in almost all types
of human cancer (17–19). Current studies have acknowledged
that more than half of miRNAs are located in cancer-related genomic
regions; this finding suggests that dysregulation of miRNAs plays
important roles in carcinogenesis and cancer progression (20). Aberrantly overexpressed miRNAs can
act as oncogenes through down-regulation of tumour suppressor
genes, whereas downregulated miRNAs can function as tumour
suppressors through negative regulation of oncogenes (21). Hence, miRNAs may serve as novel
therapeutic targets for anticancer treatments.
Recent accumulating evidence has demonstrated that
miR-485 (also known as miR-485-5p; www.mirbase.org/cgi-bin/mirna_entry.pl?acc=MI0002469)
is involved in the development and progression of several types of
human cancers (22–24). However, the expression level, exact
role and underlying mechanisms of miR-485 in GBM remain unclear.
The present study detected miR-485 in GBM, investigated the exact
roles of miR-485 in GBM progression and elucidated the underlying
molecular mechanism.
Material and methods
Clinical specimens
A total of 27 human GBM tissues were obtained from
patients who were treated with surgical resection in Department of
Neurosurgery, West China Hospital of Sichuan University between
August, 2014 and February, 2016. Twelve normal brain tissues were
obtained from patients with traumatic brain injury and who received
partial resections of normal brain tissue to reduce increased
intracranial pressure. Lineal relative of brain trauma patients
agreed the use of patients normal brain tissues in this study. All
tissue samples were immediately snap-frozen in liquid nitrogen and
stored at −80°C until further use. This study was approved by the
Ethics Committee of West China Hospital of Sichuan University.
Written informed consent was obtained from all patients.
Cell lines
GBM cell lines (U251, U87, LN229 and A172) and
HEK293T cell line were purchased from American Type Culture
Collection (Manassas, VA, USA). The cells were kept in Dulbecco's
modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA, USA)
supplemented with 10% fetal bovine serum (FBS; Invitrogen,
Carlsbad, CA, USA), 100 U/ml penicillin and 100 mg/ml streptomycin
(Invitrogen). Normal human astrocytes (NHAs) were acquired from
ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in
astrocyte medium (ScienCell Research Laboratories). All cells were
grown at 37°C in a humidified chamber containing 5%
CO2.
Oligonucleotides, plasmid and
transfection
miR-485 mimics and miRNA mimics negative control
(miR-NC) were obtained from Shanghai GenePharma (Shanghai, China).
Small interfering RNA targeting p21-activated kinase 4 (si-PAK4)
and its negative control (si-NC) were purchased from Ribobio
(Guangzhou, China). PAK4-overexpressing plasmid (pcDNA3.1-PAK4) and
blank plasmid (pcDNA3.1) were purchased from the Chinese Academy of
Sciences (Changchun, China). All oligonucleotides and plasmids were
transfected into cells using Lipofectamine 2000 (Invitrogen) in
accordance with the manufacturer's instructions. Six hours after
transfection, the culture medium was replaced with fresh DMEM
containing 10% FBS.
RNA isolation and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was isolated from tissue specimens or
cells using the TRIzol reagent (Invitrogen) in accordance with the
manufacturer's protocol. The concentration and quality of total RNA
were determined using a Nanodrop® ND-1000
spectrophotometer (NanoDrop Technologies; Thermo Fisher Scientific,
Inc., Pittsburgh, PA, USA). To quantify miR-485 expression,
complementary DNA (cDNA) was synthesised from total RNA using a
TaqMan miRNA reverse transcription kit (Applied Biosystems,
Carlsbad, CA, USA). Relative miR-485 expression was determined by
TaqMan miRNA PCR kit (Applied Biosystems) using an Applied
Biosystems 7900HT Fast Real-Time PCR system (Applied Biosystems;
Thermo Fisher Scientific, Inc.). For PAK4 mRNA expression, total
RNA was reverse transcribed into cDNA using a PrimeScript RT
Reagent kit (Takara Bio, Inc., Otsu, Japan). A SYBR Premix Ex Taq™
kit (Takara Bio, Inc.) was used to detect PAK4 mRNA expression. U6
and glyceralde-hyde 3-phosphate dehydrogenase (GAPDH) served as
endogenous control for miR-485 and PAK4, respectively. The primers
were designed as follows: miR-485,
5′-CCAAGCTTCACCCATTCCTAACAGGAC-3′ (forward) and
5′-CGGGATCCGTAGGTCAGTTACATGCATC-3′ (reverse); U6,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ (forward) and
5′-CGCTTCACGAATTTGCGTGTCAT-3′ (reverse); PAK4,
5′-AGGGAAGGCGGGAGATGAG-3′ (forward) and 5′-TCAGTTGCTTGTTCGTGC-3′
(reverse); and GAPDH, 5′-GAAGGTGAAGGTCGGAGTC-3′ (forward) and
5′-GAAGATGGTGATGG GAT TTC-3′ (reverse). The relative expression was
analysed by the 2−ΔΔCq method (25).
Cell counting kit 8 (CCK8) assay
The proliferative ability of GBM cells was assessed
using CCK8 assay. Transfected cells were collected at 24 h
post-transfection and seeded into 96-well plates at a density of
3×103 cells/well. Cells were incubated at 37°C in a
humidified 5% CO2 atmosphere for 0, 24, 48 and 72 h. At
each time point, CCK8 assay was performed in accordance with the
manufacturer's instructions. In brief, 10 µl of CCK8
solution (Dojindo, Tokyo, Japan) was added to each well, and then
the cells were incubated at 37°C for another 2 h. The absorbance at
a wavelength of 450 nm was detected with an ELISA reader (Bio-Rad
Laboratories, Inc., Hercules, CA, USA). Each experiment contained
five replicates and repeated at least three times.
Colony formation assay
Transfected cells were harvested and mechanically
dissociated into a single cell suspension. Subsequently, the cells
were seeded into 6-well plates with a density of 1,000 cells/well.
The plates were incubated at 37°C in a humidified atmosphere of 5%
CO2 for 2 weeks. On day 15, the plates were washed with
phosphate-buffered saline (PBS), fixed with 10% formalin and then
stained with Methyl Violet (Beyotime Institute of Biotechnology,
Shanghai, China). The colonies containing at least 50 cells were
counted under a microscope (Olympus IX53; Olympus, Tokyo, Japan).
All experiments were performed at least three times.
Transwell migration and invasion
assay
Transwell migration and invasion assays were
performed using an 8 µm-pore polycarbonate membrane Boyden
chamber insert in a Transwell apparatus (Costar, Cambridge, MA,
USA). For invasion assay, the chamber inserts were pre-coated with
20 µl of Matrigel (1:3 dilution; BD Bioscience, San Jose,
CA, USA). For both assays, transfected cells were harvested and
mechanically dissociated into a single cell suspension.
Subsequently, 5×104 cells in FBS-free medium were added
into the upper chamber, and a 500 µl culture medium
containing 20% FBS was placed as a chemo-attractant in the lower
chamber. The chambers were then incubated for 48 h at 37°C in a 5%
CO2 incubator. Cells on the upper surface were scraped
and washed away, whereas cells on the lower surface were fixed with
100% methanol, stained with 0.5% crystal violet and then subjected
to microscopic inspection (magnification, x200). The values for
migration and invasion abilities were obtained by counting five
fields per membrane.
Flow cytometry analysis
Transfected cells were harvested at 48 h
post-transfection using trypsinisation, washed in ice-cold PBS and
then fixed in 80% ice-cold ethanol in PBS. Cell apoptosis was
determined with the Annexin V-FITC apoptosis detection kit
(Invitrogen) in accordance with the manufacturer's instructions. In
brief, the cells were re-suspended in 300 µl of 1X binding
buffer and then stained with 5 µl of FITC-Annexin V and 5
µl of propidium iodide (PI) for 20 min in the dark at room
temperature. Cell apoptosis was quantified using a flow cytometry
kit (Beckman Coulter Corp., Brea, CA, USA).
Tumour growth assay in nude mice
Eight 4- to 6-week-old BALB/c nude mice were
purchased from Changzhou Cavens Laboratory Animal Center
(Changzhou, China). In brief, transfected U87 cells (26–28)
were harvested and mechanically dissociated into a single cell
suspension. A total of 1×107 U87 cells (29–31)
in 100 µl culture medium were subcutaneously injected into
each nude mouse. The right armpit was injected with U87 cells
transfected with miR-485 mimics. As a control, the left armpit was
injected with U87 cells transfected with miR-NC. Tumour volumes
were measured every 2 days and calculated using the formula 1/2 ×
tumour length × tumour width. At day 30, the nude mice were
sacrificed, and the tumour xenografts were dissected and weighed.
Western blot analysis was also performed to detect protein
expression levels in tumour xenografts.
Bioinformatic analysis and luciferase
reporter assay
Bioinformatic analysis was performed to predict the
putative targets of miR-485 using Pictar (http://www.pictar.mdc-berlin.de/), TargetScan
(http://www.targetscan.org/) and miRanda
(http://www.microrna.org/). 'Human' was selected
as the species, and 'hsa-miR-485' was entered. Putative miRNA: mRNA
interaction was based on the total context score. The more negative
the total context score, the higher the probability of miRNA: mRNA
binding.
Luciferase reporter plasmids, namely,
psiCHECK2-PAK4-3′-UTR wild-type (Wt) and psiCHECK2-PAK4-3′-UTR
mutant (Mut), were synthesised and confirmed by GenePharma
(Shanghai, China). HEK293T cells were seeded into 24-well plates at
a density of 50–60% confluence. One day later, the cells were
co-transfected with wild or mutant-type plasmid together with
miR-485 mimics or miR-NC by using Lipofectamine 2000. After 48 h
incubation, the cells were assayed with the Dual-Luciferase
reporter assay system (Promega, Madison, WI, USA) in accordance
with the manufacturer's instructions. Renilla luciferase was
used for normalisation. Each assay was performed in triplicate and
repeated three times.
Western blot analysis
The total proteins of tissues or cells were
extracted using RIPA lysis buffer (Beyotime Institute of
Biotechnology, Haimen, China) supplemented with 0.1 mg/ml
phenylmethylsulphonyl fluoride, 1 mM sodium orthovanadate and 1
mg/ml aprotinin. A BCA assay kit (Pierce™; Thermo Fisher
Scientific, Inc.) was used to determine protein concentration.
Equal amounts of protein were separated by sodium dodecyl
sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then
transferred onto polyvinylidene difluoride membranes (Millipore,
Billerica, MA, USA). After blocking with 5% skimmed milk at room
temperature for 2 h, the membranes were incubated overnight at 4°C
with primary antibodies. All primary antibodies were purchased form
Santa Cruz Biotechnology, Inc. (Santa Cruz, CA, USA), including
mouse anti-human monoclonal PAK4 (sc-393367; 1:1,000 dilution),
mouse anti-human monoclonal p-AKT (sc-271966; 1:1,000 dilution),
mouse anti-human monoclonal AKT (sc-81434; 1:1,000 dilution), mouse
anti-human monoclonal p-ERK (sc-81492; 1:1,000 dilution), mouse
anti-human monoclonal ERK (sc-514302; 1:1,000 dilution) and mouse
anti-human monoclonal GAPDH antibody (sc-32233; 1:1,000 dilution).
Subsequently, the membranes were washed three times with
Tris-buffered saline containing 0.1% Tween-20 and then probed with
horseradish peroxidase-conjugated secondary immunoglobulin G goat
anti-mouse (sc-2005; 1:5,000 dilution; Santa Cruz Biotechnology,
Inc.) at room temperature for 2 h. Finally, the protein bands were
visualised using enhanced chemiluminescence reagents (Bio-Rad
Laboratories, Inc.) and band densities were analysed using
AlphaEase FC software (version 4.0.1; ProteinSimple, San Jose, CA,
USA).
Statistical analysis
Data are presented as mean ± SD. Data were analysed
using SPSS 17.0 software (SPSS, Inc., Chicago, IL, USA). The
difference between groups was compared with Student's t-test or
one-way ANOVA plus multiple comparisons. The association between
miR-485 and PAK4 mRNA expression was analysed through Spearman's
correlation analysis. P<0.05 was used as the cut off for
statistically significant differences.
Results
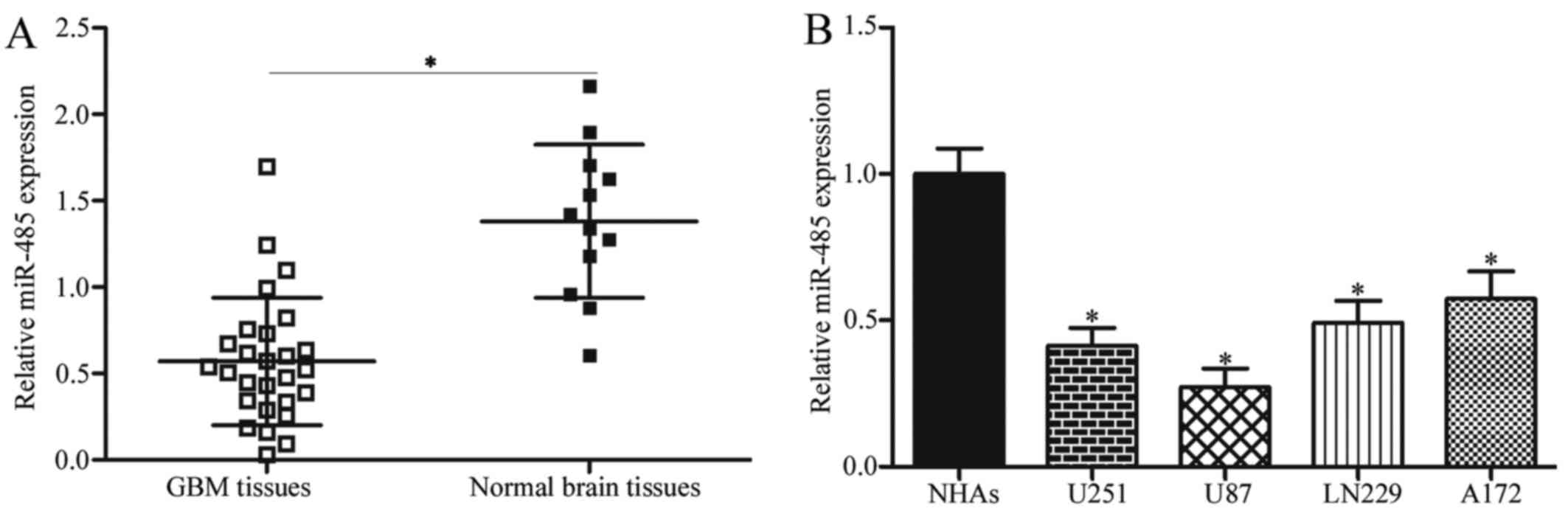
miR-485 is downregulated in GBM tissues
and cell lines
To investigate the potential roles of miR-485 in
GBM, RT-qPCR was first performed to detect miR-485 expression in 27
cases of GBM tissues and 12 cases of normal brain tissues. Results
showed that the expression levels of miR-485 decreased in GBM
tissues in comparison with that in normal brain tissues (P<0.05)
(Fig. 1A). We further examined
miR-485 expression in four GBM cell lines (U251, U87, LN229 and
A172) and NHAs. As shown in Fig.
1B, miR-485 was downregulated in all tested GBM cells compared
with NHAs (P<0.05). These results suggest that miR-485 is
involved in GBM progression.
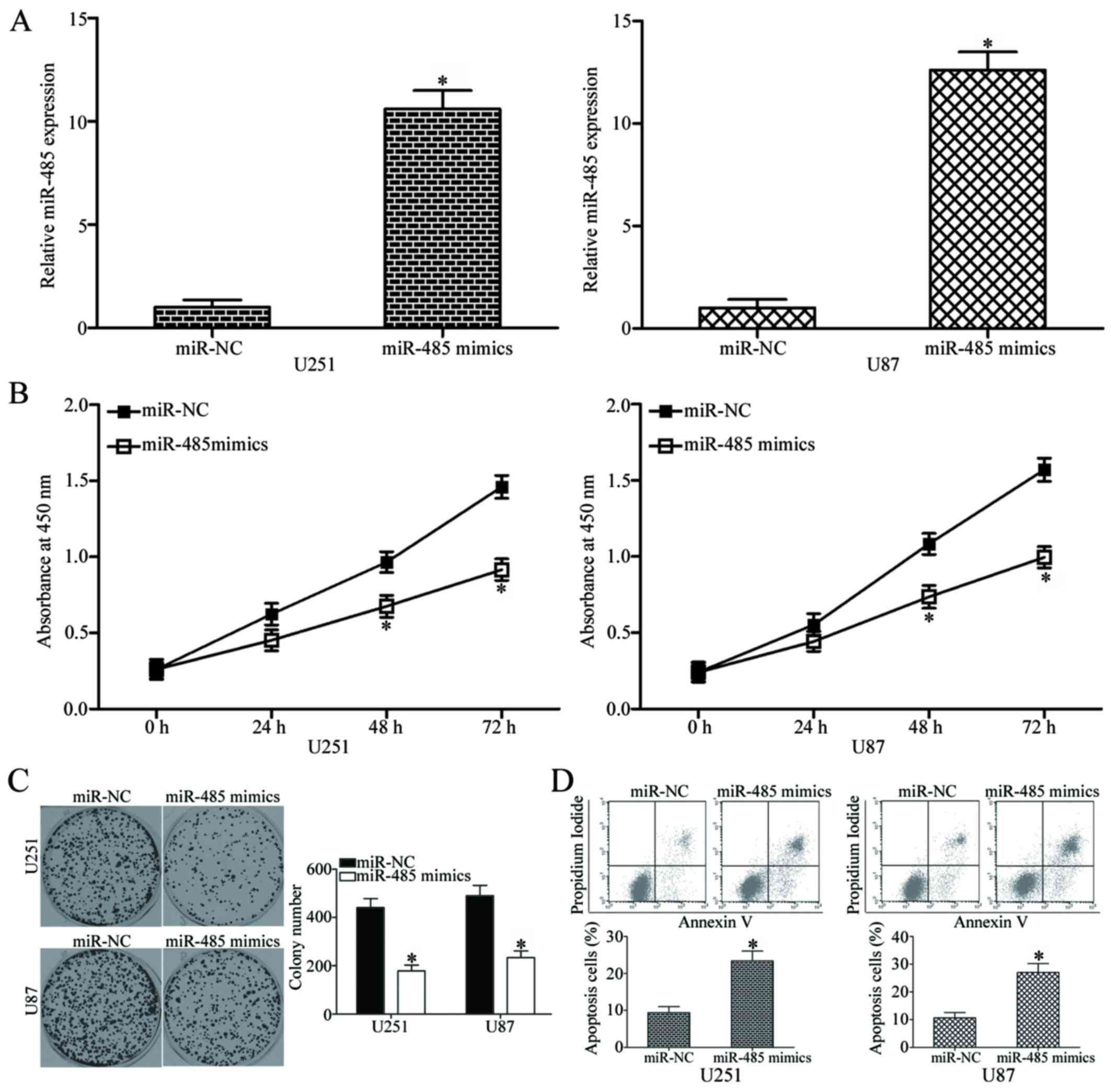
miR-485 overexpression inhibits cell
proliferation and colony formation and increases apoptosis in
GBM
To elucidate the role of miR-485 in GBM progression,
miR-485 mimics or miR-NC was transfected into U251 and U87 cells,
which have relatively low miR-485 expression among the four tested
GBM cell lines. The relative expression of miR-485 after
transfection was evaluated using RT-qPCR, and results showed that
miR-485 was markedly upregulated in the U251 and U87 cells
transfected with miR-485 mimics (P<0.05) (Fig. 2A). We next examined the effect of
miR-485 overexpression on GBM cell proliferation by using CCK8
assay. As shown in Fig. 2B,
upregulation of miR-485 inhibited U251 and U87 cell proliferation
at both 48 and 72 h in comparison with that in the cells
transfected with miR-NC (P<0.05). Furthermore, the results of
the colony formation assay demonstrated that the U251 and U87 cells
transfected with miR-485 mimics exhibited significantly fewer and
smaller colonies in comparison with those transfected with miR-NC
(P<0.05) (Fig. 2C). Flow
cytometry analysis was further conducted to explore the effect of
miR-485 on GBM cell apoptosis. In comparison with the miR-NC group,
the rates of apoptosis in U251 and U87 cells were significantly
increased by miR-485 mimics (P<0.05) (Fig. 2D). Taken together, these results
suggest that miR-485 inhibits GBM cell proliferation and colony
formation and induces apoptosis in vitro.
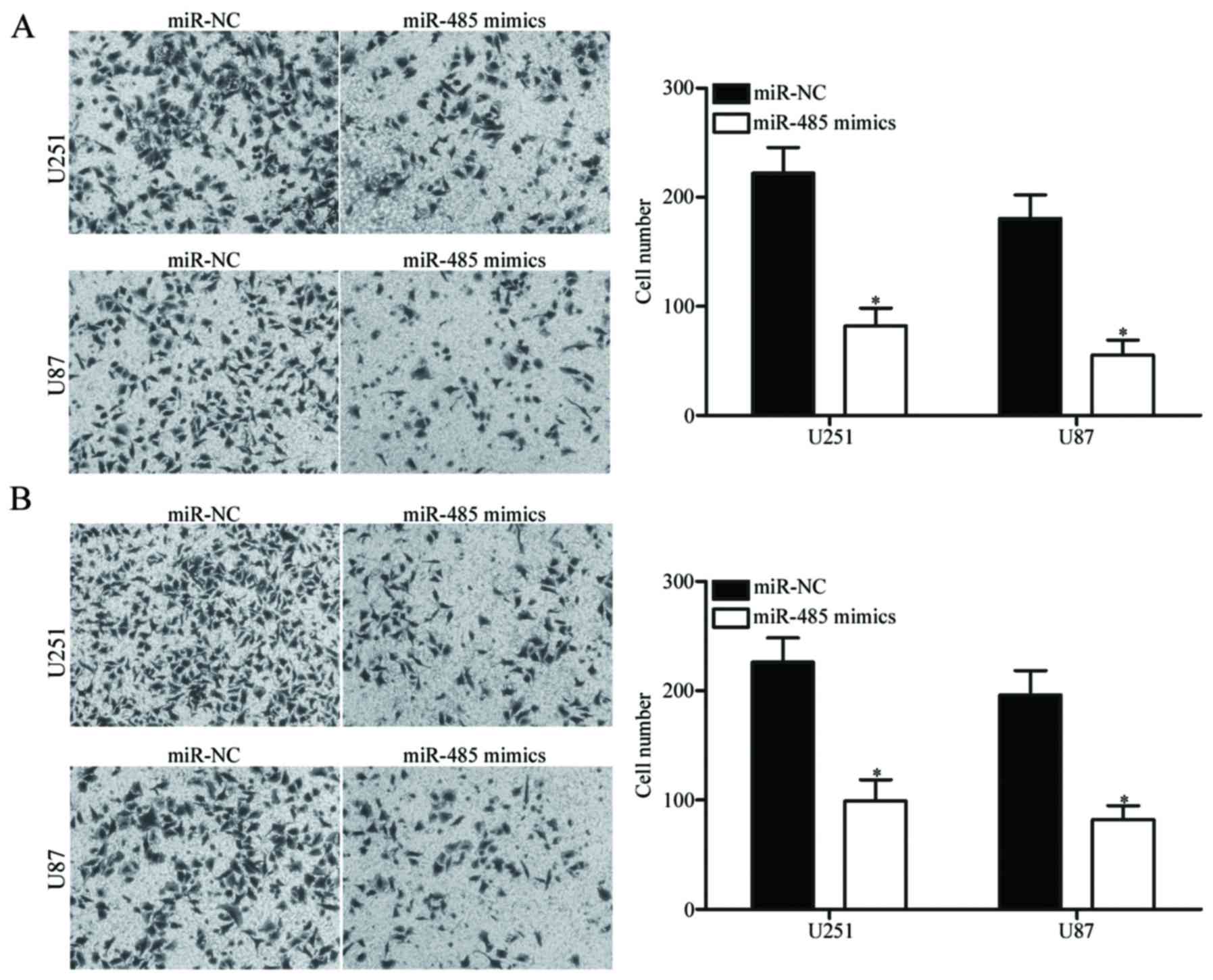
Upregulation of miR-485 decreases GBM
cell migration and invasion
To determine whether the restoration of miR-485
expression affects the migration and invasion of GBM cells,
Transwell migration and invasion assays were carried out in the
U251 and U87 cells transfected with miR-485 mimics or miR-NC. As
shown in Fig. 3A, the migratory
ability of the U251 and U87 cells transfected with miR-485 mimics
was significantly lower than that of the cells transfected with
miR-NC (P<0.05). Similarly, the ectopic expression of miR-485
attenuated the invasion capacities of U251 and U87 cells
(P<0.05) (Fig. 3B). These
findings suggest that miR-485 can inhibit GBM cell metastasis in
vitro.
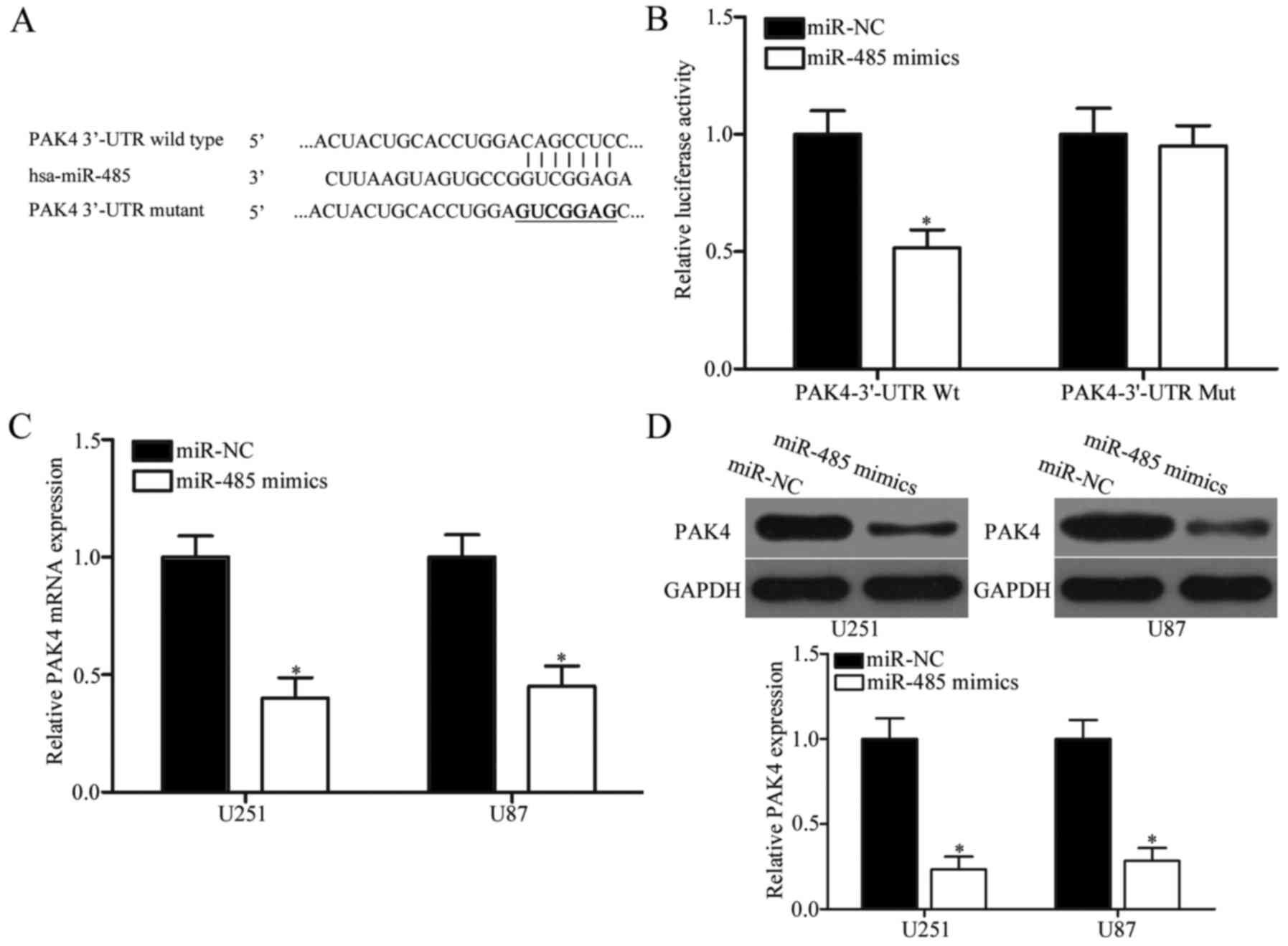
PAK4 is a direct target of miR-485 in
GBM
The potential targets of miR-485 were predicted
through bioinformatic analysis to elucidate the molecular mechanism
underlying the tumour-suppressing roles of miR-485 in GBM. Among
hundreds of candidates, PAK4 was selected as a potential target of
miR-485 (Fig. 4A) because PAK4 is
upregulated in GBM and contributes to GBM progression (32,33).
To examine the hypothesis that miR-485 targets the 3′-UTR of PAK4,
luciferase reporter assay was performed. The wild- or mutant-type
plasmid was transfected into HEK293T cells, together with miR-485
mimics or miR-NC. Fig. 4B shows
that miR-485 upregulation reduced the luciferase activities of the
Wt 3′-UTR of PAK4 (P<0.05). However, no significant difference
was observed in the cells transfected with PAK4 3′-UTR Mut and
miR-485 mimics; this result suggests that miR-485 can directly
target the 3′-UTR of PAK4. RT-qPCR and western blot analyses were
further conducted to explore whether miR-485 regulates endogenous
PAK4 expression. Results indicated that enforced expression of
miR-485 decreased PAK4 expression in U251 and U87 cells at the mRNA
(P<0.05) (Fig. 4C) and protein
(P<0.05) (Fig. 4D) levels.
These results demonstrate that PAK4 is a direct target of miR-485
in GBM.
PAK4 is inversely associated with the
expression of miR-485 in GBM tissues
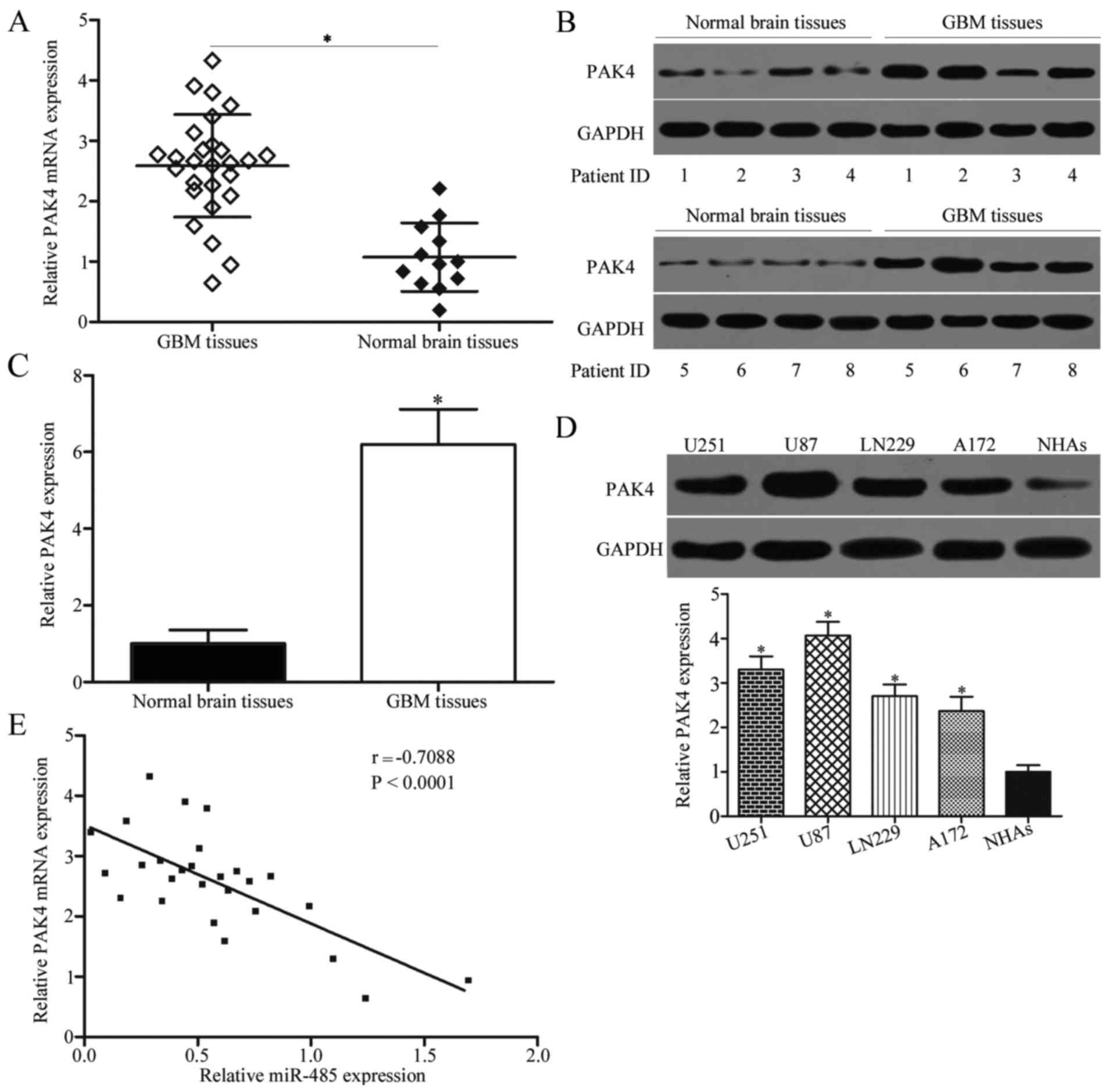
To further characterise the association between PAK4
and miR-485, we detected PAK4 expression in 27 cases of GBM tissues
and 12 cases of normal brain tissues. Results showed that PAK4 mRNA
(P<0.05) (Fig. 5A) and protein
(P<0.05) (Fig. 5B and C) levels
were higher in GBM tissues than in normal brain tissues. We also
measured PAK4 protein expression in GBM cell lines (U251, U87,
LN229 and A172) and NHAs. As shown in Fig. 5D, PAK4 was highly expressed in the
GBM cell lines compared with NHAs (P<0.05). Moreover, Spearman's
correlation analysis revealed a significant and negative
correlation between miR-485 and PAK4 mRNA expression in GBM tissues
(r=−0.7088, P<0.0001) (Fig.
5E).
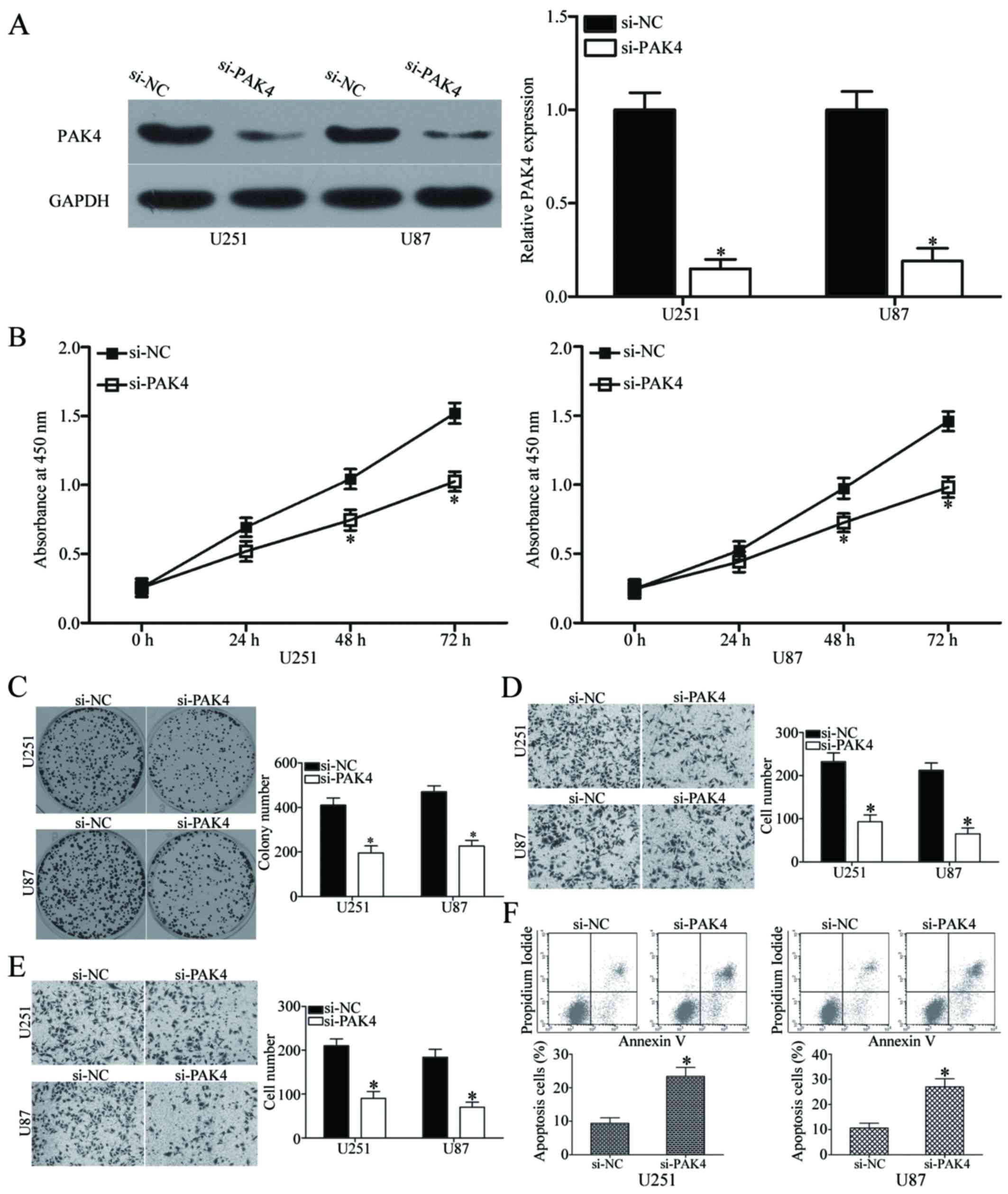
Downregulation of PAK4 exhibits a similar
effect to miR-485 overexpression in GBM cells
Having verified that PAK4 is a direct target of
miR-485 in GBM, we investigated whether PAK4 underexpression exerts
a tumour-suppressing effect similar to miR-485 overexpression in
GBM. We transfected si-PAK4 or si-NC into U251 and U87 cells to
knock down the endogenous expression of PAK4. After transfection,
western blot analysis confirmed that PAK4 was significantly
downregulated in the si-PAK4-transfected U251 and U87 cells
(P<0.05) (Fig. 6A). The effects
of PAK4 knockdown on cell proliferation, colony formation,
apoptosis, migration and invasion were evaluated in U251 and U87
cells using CCK8, colony formation, flow cytometry, Transwell
migration and invasion assays, respectively. Results showed that
PAK4 downregulation dramatically inhibited the proliferation at
both 48 h and 72 h (P<0.05) (Fig.
6B), colony formation (P<0.05) (Fig. 6C), migration (P<0.05) (Fig. 6D), invasion (P<0.05) (Fig. 6E) and promoted apoptosis
(P<0.05) (Fig. 6F) of U251 and
U87 cells. These results are consistent with the effect of miR-485
overexpression on GBM cells and further suggest that PAK4 is a
functional downstream target of miR-485.
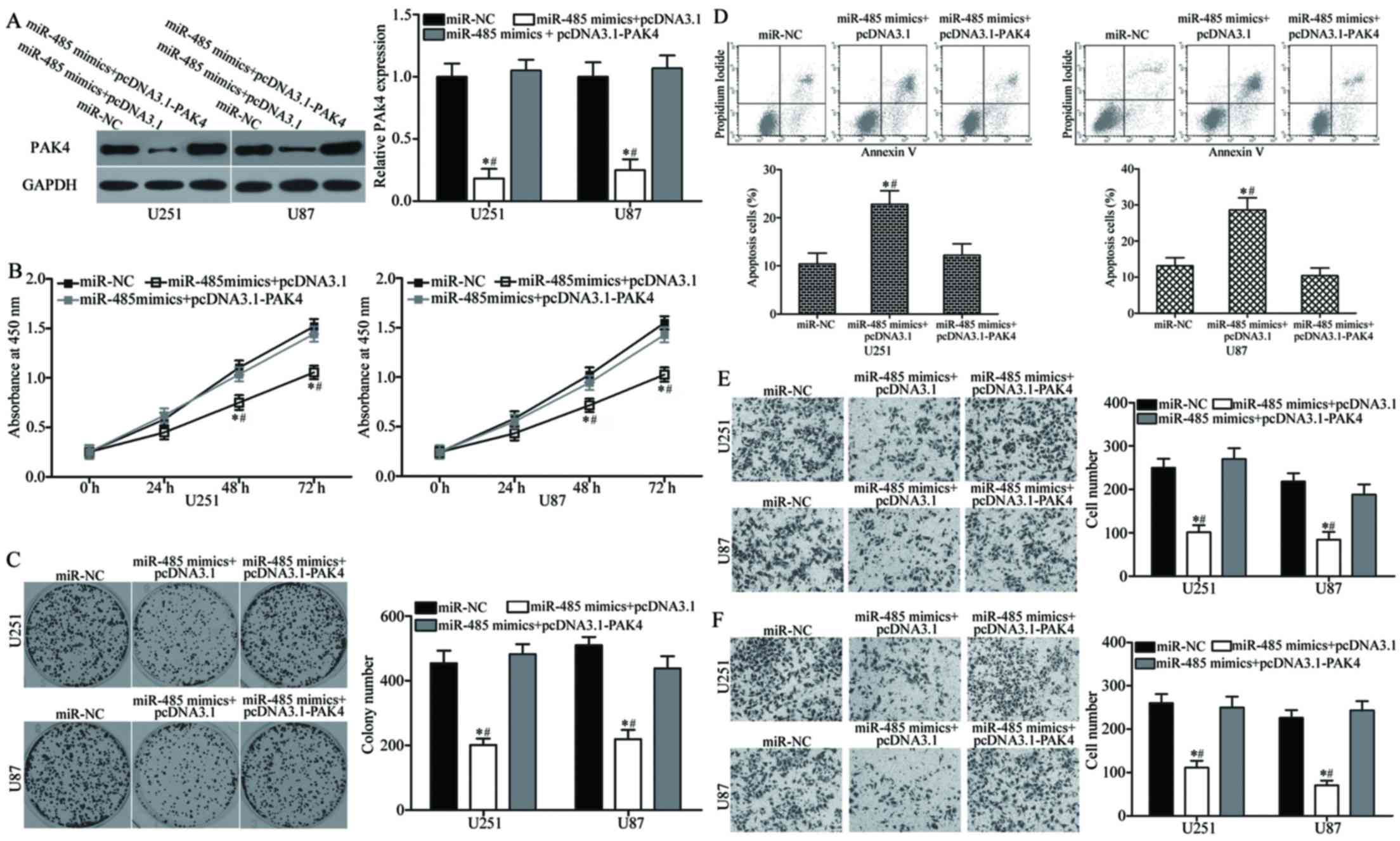
PAK4 overexpression reverses miR-485
tumour-suppressing functions in GBM
We performed a series of rescue experiments to
determine whether PAK4 overexpression can rescue the
tumour-suppressing roles of miR-485 in GBM. U251 and U87 cells were
transfected with miR-485 mimics together with or without
pcDNA3.1-PAK4. Western blot analysis indicated that the decreased
level of PAK4 induced by miR-485 overexpression was rescued by
cotransfection of pcDNA3.1-PAK4 (P<0.05) (Fig. 7A). Functional experiments revealed
that the reintroduction of PAK4 can attenuate the effects of
miR-485 on the proliferation (P<0.05) (Fig. 7B), colony formation (P<0.05)
(Fig. 7C), apoptosis (P<0.05)
(Fig. 7D), migration (P<0.05)
(Fig. 7E) and invasion (P<0.05)
(Fig. 7F) of U251 and U87 cells.
Collectively, these findings suggest that PAK4 acts as a downstream
effector of miR-485 in the regulation of malignant phenotypes of
GBM in vitro.
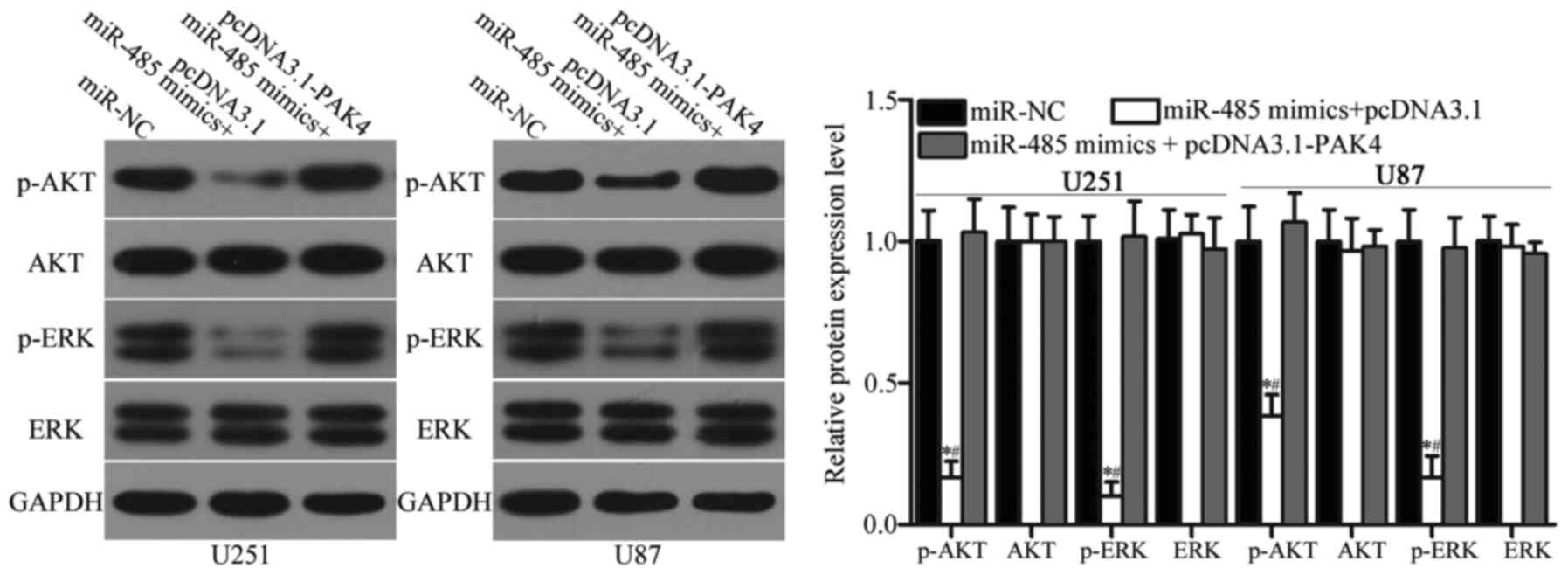
miR-485 inhibits the activation of the
AKT and ERK signalling pathways in GBM
Previous studies reported that PAK4 is involved in
the regulation of the AKT and ERK signalling pathways (34–36).
Considering the regulatory effect of miR-485 on PAK4 in GBM, we
hypothesised that the restoration of miR-485 expression may
inactivate the AKT and ERK signalling pathways. Hence, we detected
the expression levels of AKT, p-AKT, ERK and p-ERK in U251 and U87
cells transfected with miR-NC, miR-485 mimics or miR-485 mimics
cotransfected with pcDNA3.1-PAK4. As shown in Fig. 8, miR-485 overexpression reduced
p-AKT and p-ERK expression in U251 and U87 cells (P<0.05).
However, upregulation of miR-485 did not affect AKT and ERK
expression entirely. In addition, the expression levels of p-AKT
and p-ERK were recovered in the miR-485 mimic-transfected U251 and
U87 cells cotransfected with pcDNA3.1-PAK4. These results suggest
that miR-485 inactivates the AKT and ERK signalling pathways by
regulating PAK4 in GBM.
miR-485 suppresses GBM cell growth in
vivo
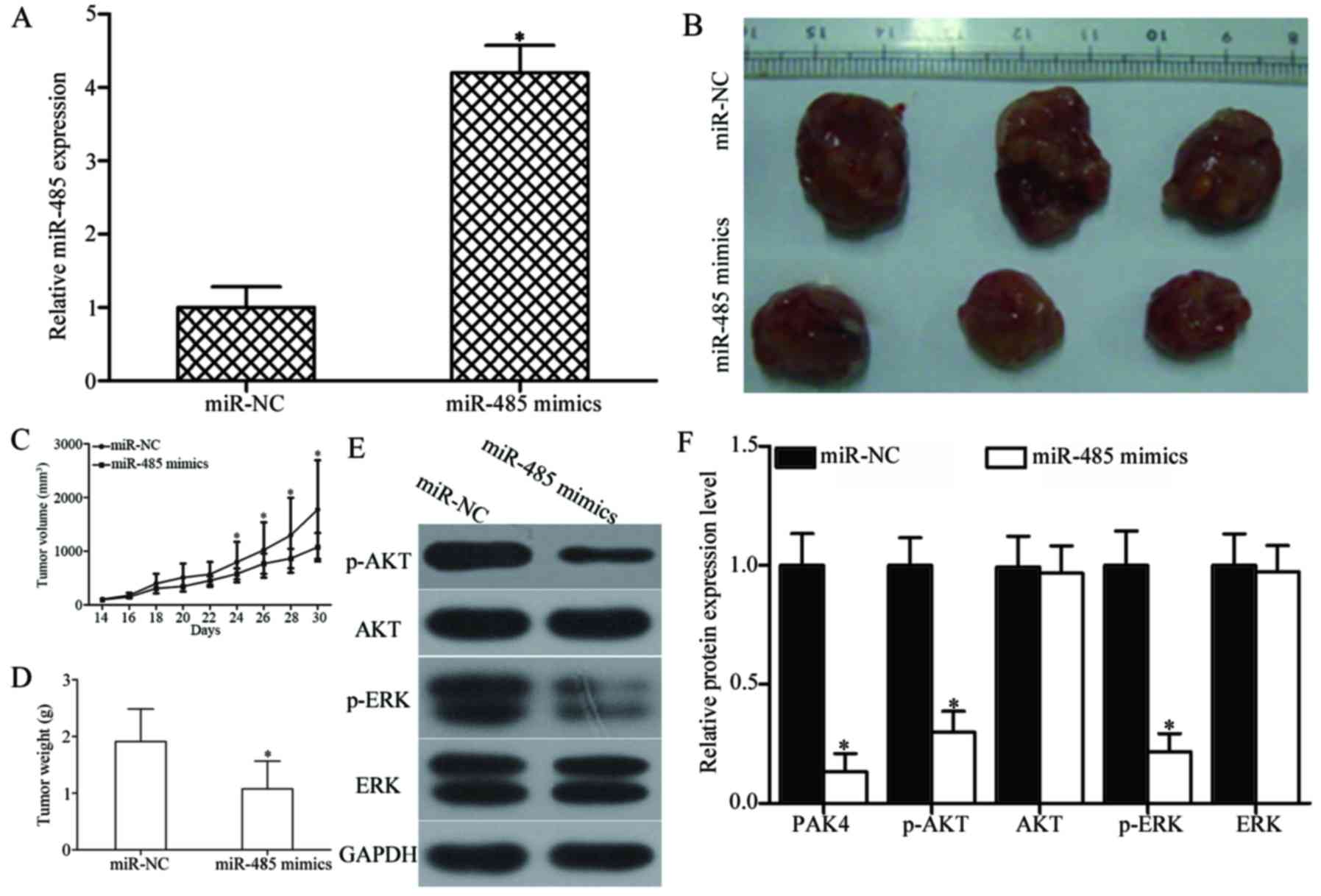
We further explored the effect of miR-485
overexpression on GBM cell growth in vivo. U87 cells
transfected with miR-485 mimics or miR-NC were implanted into the
left and right flanks of nude mice by subcutaneous injection,
respectively. At 30 days, the nude mice were sacrificed. Firstly,
RT-qPCR analysis was performed to detect miR-485 expression in
tumour xenografts. As shown in Fig.
9A, miR-485 expression was significantly upregulated in miR-485
mimics-transfected tumour xenografts (P<0.05). The tumour volume
of the miR-485 mimic groups significantly decreased compared with
that of the miR-NC groups at 30 days (P<0.05) (Fig. 9B and C). Furthermore, the average
tumour weight of the miR-485 mimic groups was significantly lower
than that of the miR-NC group (P<0.05) (Fig. 9D). Western blot analysis also
showed that the expression levels of PAK4, and key components,
including p-AKT and p-ERK, of the AKT and ERK signalling pathways
were significantly reduced in the xenograft tumour tissues with
miR-485 overexpression (P<0.05) (Fig. 9E and F). These results suggest that
miR-485 inhibits GBM tumour growth in vivo by directly
targeting PAK4 and indirectly regulating the AKT and ERK signalling
pathways.
Discussion
miRNA dysregulation is a common feature of human
cancers, including GBM (37,38).
Previous studies have demonstrated that miRNAs are a novel class of
regulatory molecules in various human cancers (39,40).
The abnormal expression of miRNAs is closely related to malignant
biological behaviour, which include proliferation, invasion,
apoptosis, cell cycle and angiogenesis (41–43).
Therefore, miRNAs may be investigated as a novel candidate and
screening tool in the clinical diagnosis, therapy and prognosis of
GBM. The present study initially found that miR-485 was
significantly downregulated in GBM tissues and cell lines.
Upregulation of miR-485 inhibited GBM cell proliferation, migration
and invasion; increased apoptosis in vitro; and reduced
tumour growth in vivo. In addition, PAK4 was validated as a
direct target of miR-485 in GBM. PAK4 was highly expressed in GBM
tissues and negatively correlated with miR-485 expression.
Furthermore, PAK4 downregulation may mimic the functions of miR-485
in GBM cells. PAK4 overexpression can rescue the tumour-suppressing
roles of miR-485 in GBM. Moreover, miR-485 inhibited the activation
of the AKT and ERK signalling pathways in GBM. These findings
suggest that miR-485 can be potentially developed as an effective
therapeutic target for the treatment of patients with GBM.
The abnormal expression of miR-485 has been reported
in numerous types of cancers. For example, the expression level of
miR-485 is decreased in gastric cancer tissues and cell lines. Low
miR-485 expression is correlated with tumour size, invasion depth,
lymph node metastasis and advanced tumour-node-metastasis (TNM)
stage. Gastric cancer patients with low miR-485 expression have a
shorter survival time than those with high miR-485 expression
(44). In breast cancer, miR-485
is downregulated in tumour tissues and negatively correlates with
the development and metastasis potential of breast cancer (45). miR-485 is expressed at low level in
hepatocellular carcinoma tissues and cell lines. Reduced expression
of miR-485 is significantly correlated with tumour size, tumour
number, TNM stage and metastasis in patients with hepatocellular
carcinoma (46,47). The expression level of miR-485 is
reduced in four lung adenocarcinoma cell lines and tissues.
Decreased miR-485 expression is associated with tumour metastasis
in patients with lung adenocarcinoma (22). Downregulation of miR-485 was also
observed in bladder cancer (23)
and melanoma (24). These findings
suggest that miR-485 is a therapeutic marker for diagnosis and
prognosis.
Previous studies demonstrated that miR-485 plays
important roles in tumourigenesis and tumour development. For
instance, Liu et al (43)
reported that miR-485 inhibits migration, invasion and
epithelial-to-mesenchymal transition and increases
cisplatin-induced cell death in oral tongue squamous cell
carcinoma. Kang et al (48)
found that miR-485 upregulation suppresses gastric cancer cell
metastasis, sphere formation in vitro and reduces cell
growth in vitro and in vivo. Lou et al
(45,49) demonstrated that ectopic expression
of miR-485 attenuates the mitochondrial respiration, proliferation
and motility of breast cancer cells. Sun et al (46,47)
revealed that enforced expression of miR-485 represses the
proliferation and metastasis of hepatocellular carcinoma cells
in vitro and decreases tumour growth in vivo. Mou and
Lou (22) indicated that the
restoration of miR-485 expression inhibits cell metastasis and
epithelial-to-mesenchymal transition of lung adenocarcinoma. These
findings suggest that miR-485 can be investigated as a potential
target for the therapeutic treatment of specific types of
cancer.
Several targets of miR-485 have been validated, such
as PAK1 (43) in oral tongue
squamous cell carcinoma, Flot1 (48) in gastric cancer, PGC-1α (45) in breast cancer, stanniocalcin 2
(47) and EMMPRIN (46) in hepatocellular carcinoma, Flot2
(22) in lung adenocarcinoma,
HMGA2 (23) in bladder cancer and
Frizzled7 (24) in melanoma. In
the current study, PAK4 was identified as a direct and functional
downstream target of miR-485 in GBM. PAKs, a family of
serine/threonine protein kinases, are best characterised as
downstream effectors of Rac and Cdc42 (50). PAK4 is a member of the PAK family
and is overexpressed and/or hyperactivated in multiple types of
human cancer, including cervical cancer (51), ovarian cancer (52), anal cancer (53), colorectal cancer (54), breast cancer (55), renal cell carcinoma (56) and head and neck squamous cell
carcinoma (57). PAK4 is
reportedly correlated with tumourigenesis and tumour development by
regulating cell proliferation, apoptosis, migration, invasion,
actin cytoskeletal changes and cytoskeletal organisation (58–60).
In glioma, PAK4 is upregulated and significantly
correlated with pathological grades. PAK4 downregulation represses
glioma cell proliferation, motility and adhesion (32). In addition, PAK4 plays important
roles in GBM cell senescence (33). Considering the importance and role
of PAK4 in GBM, regulating the miR-485/PAK4 axis may offer novel
therapeutic opportunities to target this aggressive cancer.
In conclusion, miR-485 is significantly
downregulated in GBM tissues and cell lines. Upregulation of
miR-485 suppresses GBM cell proliferation, colony formation,
migration and invasion; increases apoptosis in vitro; and
reduces tumour growth in vivo. Furthermore, PAK4 is a direct
and functional target of miR-485 in GBM. Moreover, miR-485
overexpression inhibits the activation of the AKT and ERK
signalling pathways in GBM. Further research exploring the
anticancer role of miR-485 in GBM may contribute to the development
of new therapeutic strategies for patients with this disease.
References
|
1
|
Jungk C, Chatziaslanidou D, Ahmadi R,
Capper D, Bermejo JL, Exner J, von Deimling A, Herold-Mende C and
Unterberg A: Chemotherapy with BCNU in recurrent glioma: Analysis
of clinical outcome and side effects in chemotherapy-naïve
patients. BMC Cancer. 16:812016. View Article : Google Scholar
|
|
2
|
Wu CX, Lin GS, Lin ZX, Zhang JD, Chen L,
Liu SY, Tang WL, Qiu XX and Zhou CF: Peritumoral edema on magnetic
resonance imaging predicts a poor clinical outcome in malignant
glioma. Oncol Lett. 10:2769–2776. 2015.
|
|
3
|
Kovic B and Xie F: Economic evaluation of
bevacizumab for the first-line treatment of newly diagnosed
glioblastoma multiforme. J Clin Oncol. 33:2296–2302. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang R, Luo H, Wang S, Chen W, Chen Z,
Wang HW, Chen Y, Yang J, Zhang X, Wu W, et al: MicroRNA-377
inhibited proliferation and invasion of human glioblastoma cells by
directly targeting specificity protein 1. Neuro Oncol.
16:1510–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lukiw WJ, Cui JG, Li YY and Culicchia F:
Up-regulation of micro-RNA-221 (miRNA-221; chr Xp11.3) and
caspase-3 accompanies down-regulation of the survivin-1 homolog
BIRC1 (NAIP) in glioblastoma multiforme (GBM). J Neurooncol.
91:27–32. 2009. View Article : Google Scholar
|
|
6
|
Nikaki A, Piperi C and Papavassiliou AG:
Role of microRNAs in gliomagenesis: Targeting miRNAs in
glioblastoma multiforme therapy. Expert Opin Investig Drugs.
21:1475–1488. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chaudhry MA, Sachdeva H and Omaruddin RA:
Radiation-induced micro-RNA modulation in glioblastoma cells
differing in DNA-repair pathways. DNA Cell Biol. 29:553–561. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al European Organisation for Research and Treatment of Cancer
Brain Tumor and Radiotherapy Groups; National Cancer Institute of
Canada Clinical Trials Group: Radiotherapy plus concomitant and
adjuvant temozolomide for glioblastoma. N Engl J Med. 352:987–996.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Van Meir EG, Hadjipanayis CG, Norden AD,
Shu HK, Wen PY and Olson JJ: Exciting new advances in
neuro-oncology: The avenue to a cure for malignant glioma. CA
Cancer J Clin. 60:166–193. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Nilsen TW: Mechanisms of microRNA-mediated
gene regulation in animal cells. Trends Genet. 23:243–249. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB: Prediction of mammalian microRNA targets.
Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kushlinskii NE, Fridman MV and Braga EA:
Molecular mechanisms and microRNAs in osteosarcoma pathogenesis.
Biochemistry (Mosc). 81:315–328. 2016. View Article : Google Scholar
|
|
15
|
Dallaire A and Simard MJ: The implication
of microRNAs and endo-siRNAs in animal germline and early
development. Dev Biol. 416:18–25. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu YC, Chang JT, Chan EC, Chao YK, Yeh TS,
Chen JS and Cheng AJ: miR-196, an Emerging Cancer Biomarker for
Digestive Tract Cancers. J Cancer. 7:650–655. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Y, Wang Z, Li Y and Jing R:
MicroRNA-29a functions as a potential tumor suppressor through
directly targeting CDC42 in non-small cell lung cancer. Oncol Lett.
13:3896–3904. 2017.PubMed/NCBI
|
|
18
|
Zhao Y, Li Y, Lou G, Zhao L, Xu Z, Zhang Y
and He F: miR-137 targets estrogen-related receptor alpha and
impairs the proliferative and migratory capacity of breast cancer
cells. PLoS One. 7:e391022012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Guo H, Ingolia NT, Weissman JS and Bartel
DP: Mammalian microRNAs predominantly act to decrease target mRNA
levels. Nature. 466:835–840. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Manikandan J, Aarthi JJ, Kumar SD and
Pushparaj PN: Oncomirs: The potential role of non-coding microRNAs
in understanding cancer. Bioinformation. 2:330–334. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mou X and Liu S: miR-485 inhibits
metastasis and EMT of lung adenocarcinoma by targeting Flot2.
Biochem Biophys Res Commun. 477:521–526. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen Z, Li Q, Wang S and Zhang J: miR 485
5p inhibits bladder cancer metastasis by targeting HMGA2. Int J Mol
Med. 36:1136–1142. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wu J, Li J, Ren J and Zhang D:
MicroRNA-485-5p represses melanoma cell invasion and proliferation
by suppressing Frizzled7. Biomed Pharmacother. 90:303–310. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Wang XP, Zhou J, Han M, Chen CB, Zheng YT,
He XS and Yuan XP: MicroRNA-34a regulates liver regeneration and
the development of liver cancer in rats by targeting Notch
signaling pathway. Oncotarget. 8:13264–13276. 2017.PubMed/NCBI
|
|
27
|
Wang J, Liu H, Tian L, Wang F, Han L,
Zhang W and Bai YA: miR-15b inhibits the progression of
glioblastoma cells through targeting insulin-like growth factor
receptor 1. Horm Cancer. 8:49–57. 2017. View Article : Google Scholar
|
|
28
|
Zhu Q, Gong L, Wang J, Tu Q, Yao L, Zhang
JR, Han XJ, Zhu SJ, Wang SM, Li YH, et al: miR-10b exerts oncogenic
activity in human hepatocellular carcinoma cells by targeting
expression of CUB and sushi multiple domains 1 (CSMD1). BMC Cancer.
16:8062016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Cai WX, Zheng LW, Ma L, Huang HZ, Yu RQ
and Zwahlen RA: Tumorigenicity and validity of fluorescence
labelled mesenchymal and epithelial human oral cancer cell lines in
nude mice. BioMed Res Int. 2016:48979862016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ma L, Zhang X, Wang Z, Chen Y, Wei J and
Hu L: Establishment of a novel myelodysplastic syndrome (MDS)
xenotransplantation model. Clin Lab. 62:1651–1659. 2016. View Article : Google Scholar
|
|
31
|
Wu Z, Cai X, Huang C, Xu J and Liu A:
miR-497 suppresses angiogenesis in breast carcinoma by targeting
HIF-1α. Oncol Rep. 35:1696–1702. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kesanakurti D, Chetty C, Rajasekhar
Maddirela D, Gujrati M and Rao JS: Functional cooperativity by
direct interaction between PAK4 and MMP-2 in the regulation of
anoikis resistance, migration and invasion in glioma. Cell Death
Dis. 3:e4452012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Franovic A, Elliott KC, Seguin L, Camargo
MF, Weis SM and Cheresh DA: Glioblastomas require integrin
αvβ3/PAK4 signaling to escape senescence. Cancer Res. 75:4466–4473.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li SQ, Wang ZH, Mi XG, Liu L and Tan Y:
miR-199a/b-3p suppresses migration and invasion of breast cancer
cells by downregulating PAK4/MEK/ERK signaling pathway. IUBMB Life.
67:768–777. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xue J, Chen LZ, Li ZZ, Hu YY, Yan SP and
Liu LY: MicroRNA-433 inhibits cell proliferation in hepatocellular
carcinoma by targeting p21 activated kinase (PAK4). Mol Cell
Biochem. 399:77–86. 2015. View Article : Google Scholar
|
|
36
|
Fu X, Feng J, Zeng D, Ding Y, Yu C and
Yang B: PAK4 confers cisplatin resistance in gastric cancer cells
via PI3K/Akt- and MEK/ERK-dependent pathways. Biosci Rep.
34:342014. View Article : Google Scholar
|
|
37
|
Huang Q, Zhang XW, Ma YS, Lu GX, Xie RT,
Yang HQ, Lv ZW, Zhong XM, Liu T, Huang SX, et al: Up-regulated
microRNA-299 corrected with poor prognosis of glioblastoma
multiforme patients by targeting ELL2. Jpn J Clin Oncol.
47:590–596. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Yuan Y, Zhang H, Liu X, Lu Z, Li G, Lu M
and Tao X: MicroRNA signatures predict prognosis of patients with
glioblastoma multiforme through the cancer genome atlas.
Oncotarget. 8:58386–58393. 2017.PubMed/NCBI
|
|
39
|
Iorio MV and Croce CM: microRNA
involvement in human cancer. Carcinogenesis. 33:1126–1133. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Garzon R, Marcucci G and Croce CM:
Targeting microRNAs in cancer: Rationale, strategies and
challenges. Nat Rev Drug Discov. 9:775–789. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhi T, Jiang K, Zhang C, Xu X, Wu W, Nie
E, Yu T, Zhou X, Bao Z, Jin X, et al: MicroRNA-1301 inhibits
proliferation of human glioma cells by directly targeting N-Ras. Am
J Cancer Res. 7:982–998. 2017.PubMed/NCBI
|
|
42
|
Zhang P, Kong F, Deng X, Yu Y, Hou C,
Liang T and Zhu L: MicroRNA-326 suppresses the proliferation,
migration and invasion of cervical cancer cells by targeting ELK1.
Oncol Lett. 13:2949–2956. 2017.PubMed/NCBI
|
|
43
|
Liu N, Zhang L, Wang Z, Cheng Y, Zhang P,
Wang X, Wen W, Yang H, Liu H, Jin W, et al: MicroRNA-101 inhibits
proliferation, migration and invasion of human glioblastoma by
targeting SOX9. Oncotarget. 8:19244–19254. 2017.
|
|
44
|
Jing LL and Mo XM: Reduced miR-485-5p
expression predicts poor prognosis in patients with gastric cancer.
Eur Rev Med Pharmacol Sci. 20:1516–1520. 2016.PubMed/NCBI
|
|
45
|
Lou C, Xiao M, Cheng S, Lu X, Jia S, Ren Y
and Li Z: miR-485-3p and miR-485-5p suppress breast cancer cell
metastasis by inhibiting PGC-1α expression. Cell Death Dis.
7:e21592016. View Article : Google Scholar
|
|
46
|
Sun X, Liu Y, Li M, Wang M and Wang Y:
Involvement of miR-485-5p in hepatocellular carcinoma progression
targeting EMMPRIN. Biomed Pharmacother. 72:58–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Guo GX, Li QY, Ma WL, Shi ZH and Ren XQ:
MicroRNA-485-5p suppresses cell proliferation and invasion in
hepatocellular carcinoma by targeting stanniocalcin 2. Int J Clin
Exp Pathol. 8:12292–12299. 2015.
|
|
48
|
Kang M, Ren MP, Zhao L, Li CP and Deng MM:
miR-485-5p acts as a negative regulator in gastric cancer
progression by targeting flotillin-1. Am J Transl Res. 7:2212–2222.
2015.
|
|
49
|
Anaya-Ruiz M, Bandala C and Perez-Santos
JL: miR-485 acts as a tumor suppressor by inhibiting cell growth
and migration in breast carcinoma T47D cells. Asian Pac J Cancer
Prev. 14:3757–3760. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Bokoch GM: Biology of the p21-activated
kinases. Annu Rev Biochem. 72:743–781. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shu XR, Wu J, Sun H, Chi LQ and Wang JH:
PAK4 confers the malignance of cervical cancers and contributes to
the cisplatin-resistance in cervical cancer cells via PI3K/AKT
pathway. Diagn Pathol. 10:1772015. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Helleman J, Jansen MP, Span PN, van
Staveren IL, Massuger LF, Meijer-van Gelder ME, Sweep FC, Ewing PC,
van der Burg ME, Stoter G, et al: Molecular profiling of platinum
resistant ovarian cancer. Int J Cancer. 118:1963–1971. 2006.
View Article : Google Scholar
|
|
53
|
Waalkes S, Atschekzei F, Kramer MW,
Hennenlotter J, Vetter G, Becker JU, Stenzl A, Merseburger AS,
Schrader AJ, Kuczyk MA, et al: Fibronectin 1 mRNA expression
correlates with advanced disease in renal cancer. BMC Cancer.
10:5032010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Song B, Wang W, Zheng Y, Yang J and Xu Z:
p21-activated kinase 1 and 4 were associated with colorectal cancer
metastasis and infiltration. J Surg Res. 196:130–135. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ruiz-Garcia E, Scott V, Machavoine C,
Bidart JM, Lacroix L, Delaloge S and Andre F: Gene expression
profiling identifies fibronectin 1 and CXCL9 as candidate
biomarkers for breast cancer screening. Br J Cancer. 102:462–468.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Liu W, Yang Y, Liu Y, Liu H, Zhang W, Xu
L, Zhu Y and Xu J: p21-activated kinase 4 predicts early recurrence
and poor survival in patients with nonmetastatic clear cell renal
cell carcinoma. Urol Oncol. 33:205.e213–221. 2015.
|
|
57
|
Jerhammar F, Ceder R, Garvin S, Grénman R,
Grafström RC and Roberg K: Fibronectin 1 is a potential biomarker
for radioresistance in head and neck squamous cell carcinoma.
Cancer Biol Ther. 10:1244–1251. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Gnad F, Young A, Zhou W, Lyle K, Ong CC,
Stokes MP, Silva JC, Belvin M, Friedman LS, Koeppen H, et al:
Systems-wide analysis of K-Ras, Cdc42, and PAK4 signaling by
quantitative phosphoproteomics. Mol Cell Proteomics. 12:2070–2080.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Siu MK, Chan HY, Kong DS, Wong ES, Wong
OG, Ngan HY, Tam KF, Zhang H, Li Z, Chan QK, et al: p21-activated
kinase 4 regulates ovarian cancer cell proliferation, migration,
and invasion and contributes to poor prognosis in patients. Proc
Natl Acad Sci USA. 107:18622–18627. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ahmed T, Shea K, Masters JR, Jones GE and
Wells CM: A PAK4-LIMK1 pathway drives prostate cancer cell
migration downstream of HGF. Cell Signal. 20:1320–1328. 2008.
View Article : Google Scholar : PubMed/NCBI
|