Introduction
Globally, colorectal cancer (CRC) is the third most
common cancer in men (746,000 cases annually, 10.0% of total
cancers) and the second in women (614,000 cases, 9.2% of total
cancers) (1). The early stages of
CRC can be detected by occult blood examination in stool samples or
endoscopic examination. Subsequently, CRC can be treated with
surgery and chemotherapy, which have been shown to increase patient
survival. However, advanced cases exhibiting metastasis to distant
organs are associated with worsened survival rates, indicating the
rationale for further study of the underlying mechanisms of CRC
(1).
Although cancer is a genetic disease, epigenetic
alterations also play a pivotal role in cancer development;
eventually, the hypoxic responses within the tumor microenvironment
characterize the malignant behavior of tumors as a hallmark of
cancer (2,3). Indeed, recent advances in the
high-throughput, next generation sequence technology have aided in
the identification and characterization of long noncoding RNA
(lncRNA), which is a type of noncoding RNA (ncRNA) exceeding 200
nucleotides in length, and epigenetic alterations, which are
defined as mechanisms leading to changes in gene expression related
to cellular biology and tissue homeostasis but not involving
changes in the DNA sequence (4).
HOX transcript antisense RNA (HOTAIR), a human gene located on
chromosome 12, is the first example of lncRNA expressed on one
chromosome and influencing transcription on another chromosome
(5). HOTAIR mediates the
trimethylation of histone H3 at lysine 27 and demethylation of
histone H3 at dimethyl lysine 4 via the recruitment of polycomb
repressive complex 2 and lysine-specific demethylase 1/co-repressor
of RE1-silencing transcription factor (coREST)/REST complex to
target gene promoters. The recruitment of these factors leads to
gene silencing in the nucleus of cancer cells in gastrointestinal
and lung tumors, which is associated with patient prognosis
(6). Subsequently, numerous
studies have indicated that not only mutations or altered
expression of mRNA or protein-coding genes but also mutations and
deregulation of ncRNAs, particularly lncRNA, play major roles in
cancer development (7). It has
been reported that several other lncRNAs, including MALAT1, CCAT2,
SPRY4-IT1, RSU1P2, CCHE1, lncRNA-EBIC and plasmacytoma variant
translocation 1 (PVT1), are also involved in cancer (8). Moreover, PVT1, a newly discovered
lncRNA, is a potential biomarker for clinicopathological
characteristics of different types of cancer (9). Recent studies indicate roles of
lincRNAs in CRC (Table I). In
cancer, lncRNAs are involved in the biological behavior of tumors,
altering cancer cell localization and turnover (10). Although multiple lncRNAs are
involved in the biological processes within the nucleus, and recent
studies have indicated that lncRNAs, such as Xist, nuclear-enriched
abundant transcript 1 (NEAT1), MALAT1 and TERRA, are involved in
various nuclear functions, the differential roles of lncRNAs in the
cytoplasm and nucleus remains poorly understood (11).
 | Table ILncRNAs associated with CRC. |
Table I
LncRNAs associated with CRC.
| LncRNA | Name | Function | Expression | Mechanism | Refs. |
|---|
| LINC-PINT | Long intergenic
non-protein coding RNA, p53 induced transcript | Inhibits cell
proliferation and promotes apoptosis | Downregulated | Interacts with the
polycomb repressive complex 2 (PRC2) to silence gene targets | 26 |
| PR-lncRNA-1 | p53-regulated
lncRNA-1 | Inhibits cell
proliferation, and promotes apoptosis | Downregulated | Enhances p53
transcriptional activation | 27 |
| MALAT1 | Metastasis
associated lung adenocarcinoma transcript 1 | Promotes cell
proliferation and metastasis | Overexpressed | Related to
alternative splicing and active transcription | 28 |
| PTENP1 | Phosphatase and
tensin homolog pseudogene 1 | Inhibits cell
proliferation, migration, invasion and tumor growth | Locus selectively
lost in sporadic colon cancer | Decoy for microRNAs
that target PTEN | 29 |
| CCAT2 | Colon cancer
associated transcript 2 | Promotes tumor
growth, metastasis, chromosomal instability | Overexpressed | Enhancement of WNT
signaling activity | 30 |
| HOTAIR | Hox antisense
intergenic RNA | Cancer progression
by remodeling the chromatin landscape | Overexpressed | Modular scaffold by
interacting with PRC2 | 31 |
| LINC00152 | NA | ceRNA is
suggested | Overexpressed | Hypoxia
pathway | Present |
In contrast to the epigenetic control of lncRNAs,
which bind to histone-modifying enzymes in case of HOTAIR, little
is known regarding the cytoplasmic involvement of lncRNAs. Recent
studies indicated that a small fraction of ncRNAs, such as
microRNAs, functions as competitive endogenous RNA (ceRNA). In the
context of ceRNA, some pseudogene RNAs can act as 'sponges' through
the competitive binding of common miRNA, thereby attenuating
repression via the sequestration of miRNAs from the parental mRNA
(12). Because ceRNAs include all
transcripts, such as mRNA, tRNA, rRNA, lncRNA, pseudogene RNA and
circular RNA, all of which may become targets of miRNA depending on
the spatiotemporal situation, it is suggested that lncRNAs may
function as a 'sponge' and that a competitive balance may exist
between ncRNAs and the target mRNA (12).
In the tumor microenvironment, several processes
within the transcriptional networks of p53 and hypoxia-inducible
factor 1α (HIF1α) are modulated by hypoxia (13), and defines hypoxic glycolysis
metabolism in normoxia, which characterizes cancer cells, as the
so-called Warburg effect, cancer metabolism (14).
In the present study, we analyzed the expression of
lncRNA under normoxic and hypoxic conditions and found that
oncogenic long intergenic noncoding RNA 152 (LINC00152) expression
is increased in the cytoplasm during hypoxia in CRC cells. A
sequence data study indicated that LINC00152 shares the
microRNA-binding sites miR-138 and miR-193, which are located in
the peptide-coding mRNA of the HIF1 gene. A study of clinical
samples further supported the significance of LINC00152.
Materials and methods
Cell lines and cell culture
The human CRC cell line HCT-116 was purchased from
the American Type Culture Collection (Manassas, VA, USA) and
maintained in Dulbecco's modified Eagle's medium (Sigma-Aldrich,
St. Louis, MO, USA) supplemented with 10% fetal bovine serum (FBS)
at 37°C under 5% CO2 in a humidified incubator.
Separation of nuclear and cytoplasmic
fractions
To separate the total cellular fractions into
nuclear and cytoplasmic fractions, we used a PARIS kit (AM1921;
Thermo Fisher Scientific, Yokohama, Japan) according to the
manufacturer's protocol. This kit enables the isolation of both RNA
and protein from the same experimental sample and also permits the
separation of nuclear and cytoplasmic fractions prior to RNA and/or
protein isolation.
Polymerase chain reaction (PCR) array of
lncRNA
We performed an lncRNA PCR array using a Human
Cancer PathwayFinder RT2 lncRNA PCR array (LAHS-002Z;
Qiagen, Hilden, Germany) according to the protocol detailed in the
RT2 Profiler PCR array Handbook. This array profiles the
expression of 84 lncRNAs that are differentially expressed in
tumors compared with normal tissue. Briefly, total RNA was
extracted from cultured cells using TRIzol® RNA
isolation reagents (Thermo Fisher Scientific) as previously
described (15). The RNA from each
fraction was extracted using a PARIS kit. cDNA was synthesized from
500 ng of RNA using an RT2 First Strand kit (Qiagen)
according to the manufacturer's protocol. PCR was performed in an
Applied Biosystems 7900 HT Fast Real-Time PCR system (Applied
Biosystems, CA, USA) using the Thunderbird®
SYBR® qPCR mix (Toyobo Life Science, Osaka, Japan). The
cycling conditions consisted of 95°C for 10 min, 40 cycles of 95°C
for 15 sec, and 60°C for 1 min.
Real-time quantitative reverse
transcriptase-PCR (qRT-PCR)
Total RNA was extracted from the cultured cells
using TRIzol® RNA isolation reagents. cDNA was
synthesized from 1 µg of the total RNA using a High Capacity
RNA-to-cDNA kit (Thermo Fisher Scientific) according to the
manufacturer's protocol. PCR was performed in a Light Cycler™ 2.0
system (Roche Applied Science, Tokyo, Japan) using the
Thunderbird® SYBR® qPCR mix. The expression
level was calculated using the ΔΔCq method (16). The calculated values were then
normalized against the expression of glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) or RPLP0 for mRNA. The PCR reaction mixture
consisted of 5 µl of 1/100 cDNA, 10.0 µl of
Thunderbird™ SYBR® qPCR mix, 4.0 µl of water, and
0.5 µl of each primer. The amplification protocol consisted
of 40 cycles of denaturation at 95°C for 15 sec, annealing at 60°C
for 1 min, and extension at 72°C for 30 sec.
The following primers were used: CA9,
5′-CTAGAGGCTGGATCTTGGAGAA-3′ (forward) and
5′-CTTGGCAGTTAAAAGGAAGTGG-3′ (reverse); LINC00152,
5′-CTCCAGCACCTCTACCTGTTG-3′ (forward) and
5′-GGACAAGGGATTAAGACACACA-3′ (reverse); GAPDH,
5′-AGCCACATCGCTCAGACAC-3′ (forward) and 5′-GCCCAATACGACCAAATCC-3′
(reverse); RPLP0, 5′-AGCCACATCGCTCAGACAC-3′ (forward) and
5′-GCCCAATACGACCAAATCC-3′ (reverse).
Statistical analysis
Statistically significant differences were
determined by a Student's t-test and Fisher's exact probability
test as appropriate. JMP® Pro12 (SAS Institute Inc.,
Cary, NC, USA) was used for all statistical analyses. The data are
reported as the mean ± SEM. Results were considered statistically
significant when P-value of <0.05 was obtained.
Results
Identification of hypoxia-regulated
lncRNA
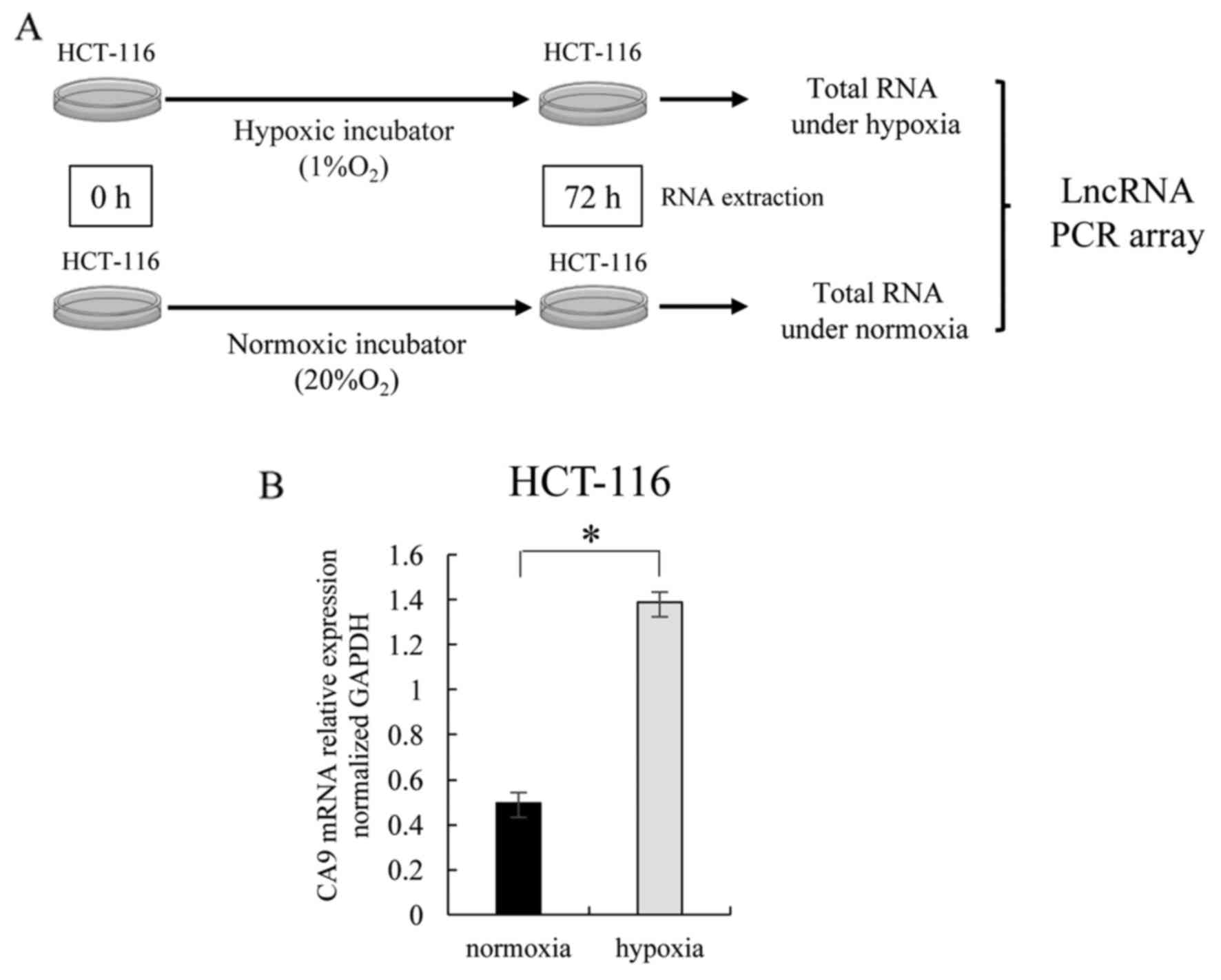
We performed an lncRNA PCR array using the total RNA
of HCT-116 cells incubated for 72 h under conditions of hypoxia (1%
O2) or normoxia (20% O2) (Fig. 1A). As a preliminary experiment, we
assessed the level of carbonic anhydrase 9 (CA9) expression, which
is known to be an indicator of hypoxia (17). CA9 expression was significantly
higher in HCT-116 cells incubated under hypoxic conditions than in
those incubated under normoxic conditions (Fig. 1B). We validated that the hypoxic
environment had been reproduced by the hypoxic incubator. Next we
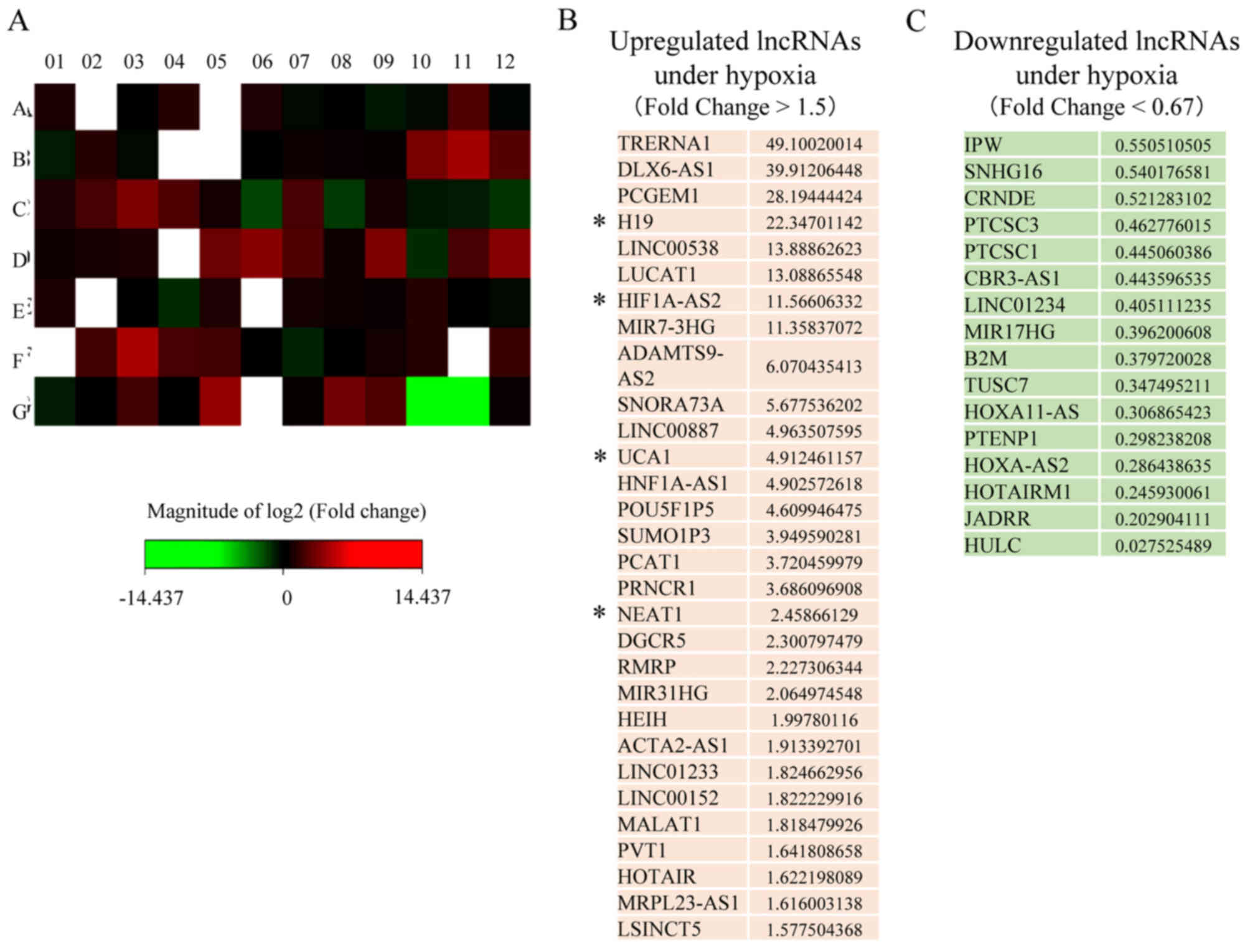
performed study of lncRNA expression using the Human Cancer
PathwayFinder RT2 lncRNA PCR array, which included 84
sets of primers for lncRNAs (Fig.
2A). The lncRNA PCR array revealed that 30 lncRNAs were
upregulated (Fig. 2B) and 16
lncRNAs were downregulated (Fig.
2C) under hypoxic conditions compared with normoxic conditions.
Among the lncRNAs upregulated under hypoxic conditions, some were
previously reported to be upregulated due to hypoxia (18).
Identification of lncRNAs that are
downregulated in the nucleus and upregulated in the cytoplasm under
hypoxic conditions
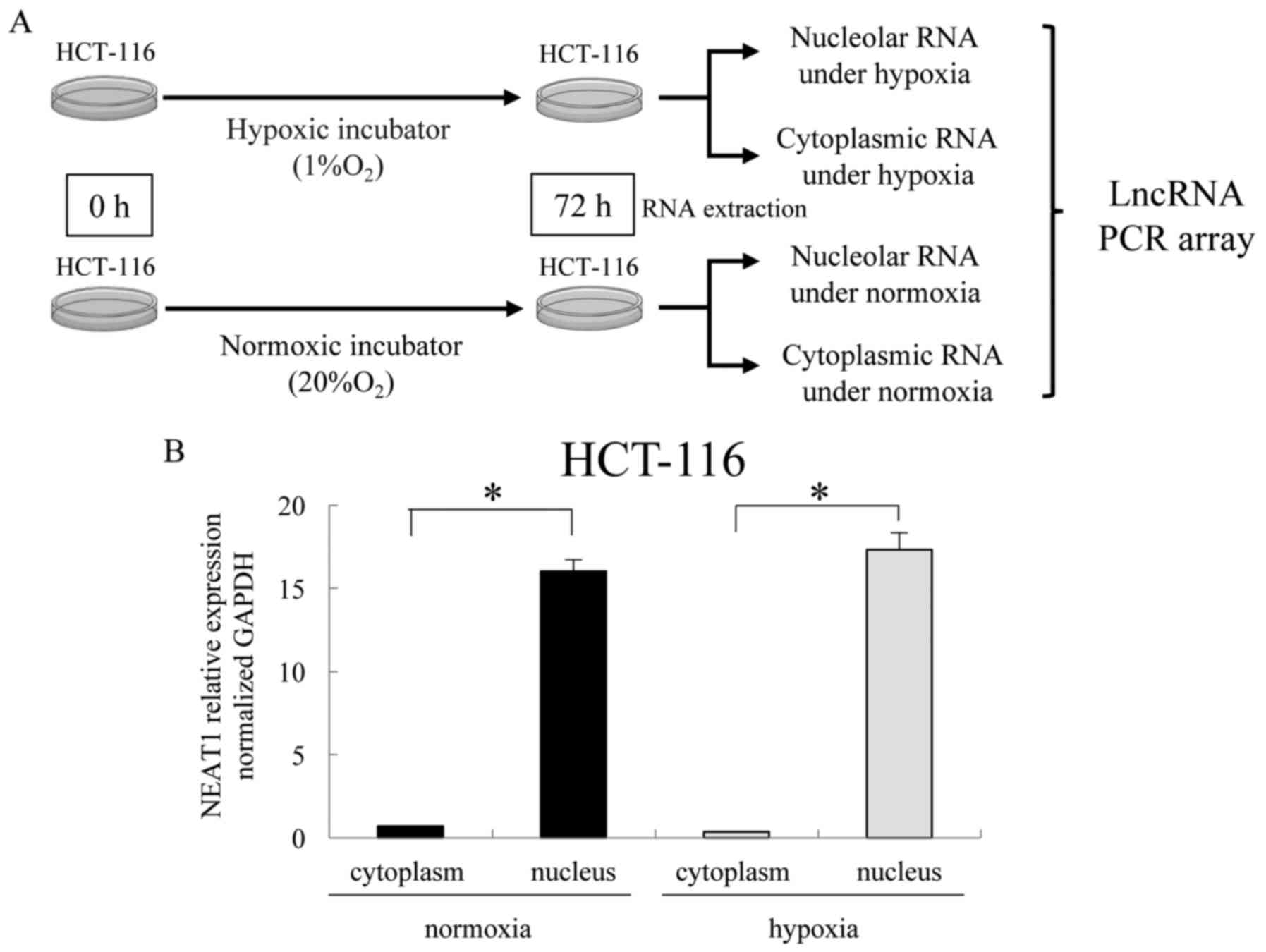
We performed an lncRNA PCR array using nuclear or
cytoplasmic RNA of HCT-116 cells incubated for 72 h under
conditions of hypoxia (1% O2) or normoxia (20%
O2) (Fig. 3A). As a
preliminary experiment, we determined the level of NEAT1
expression, which is known to be a nuclear lncRNA. This preliminary
experiment demonstrated that NEAT1 was mainly expressed in the
nucleus (Fig. 3B), which indicated
adequate separation of the total fraction into nuclear and
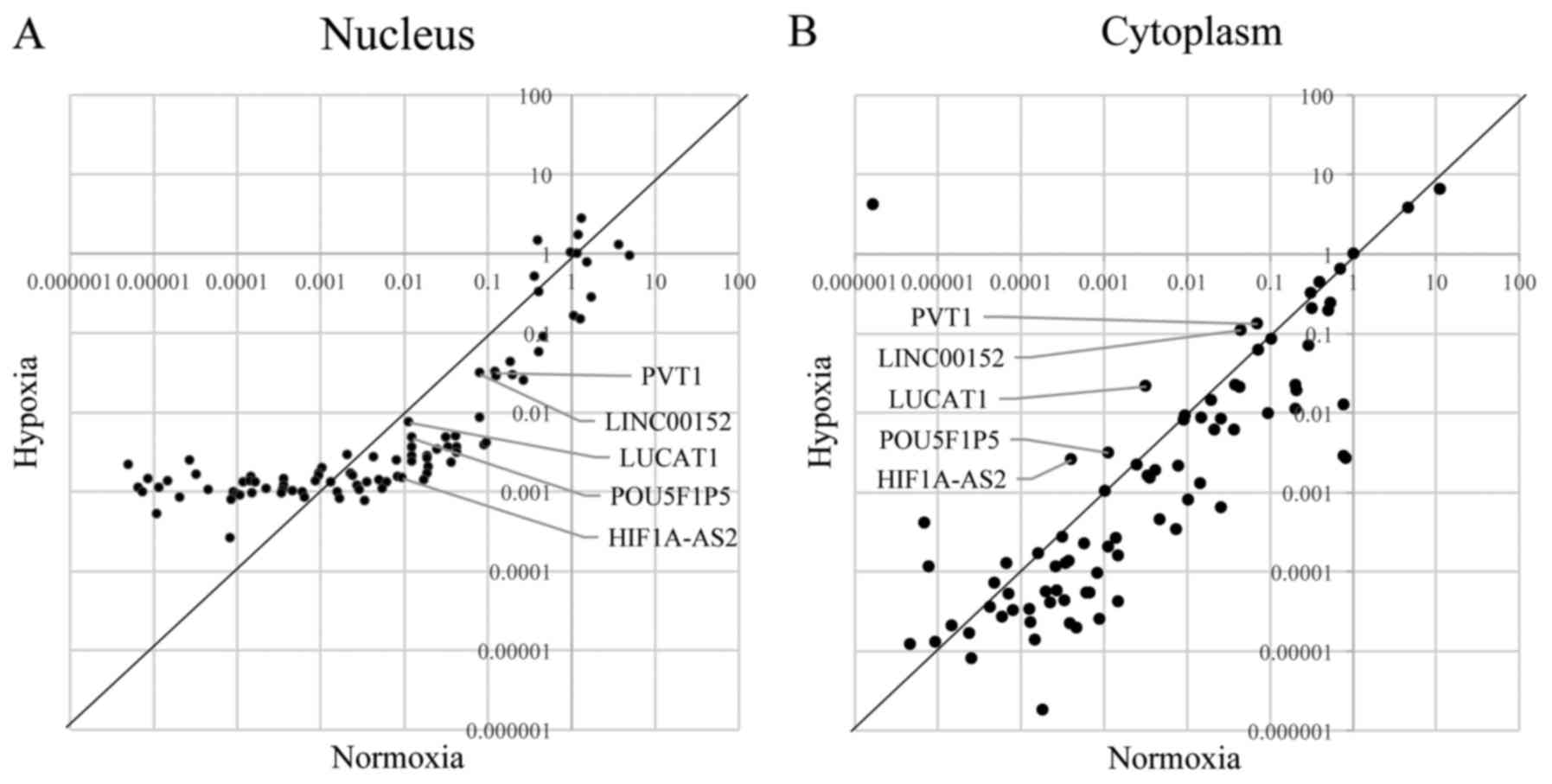
cytoplasmic fractions. Fig. 4A
shows the results of the lncRNA PCR array of the nucleus, and
Fig. 4B shows the results of the
lncRNA PCR array of the cytoplasm. We focused on the lncRNAs that
were downregulated in the nucleus and upregulated in the cytoplasm
under hypoxic conditions because they may function as ceRNAs. The
PCR array revealed that, under hypoxic conditions, nine lncRNAs
were upregulated in the cytoplasm, 46 were downregulated in the
nucleus, and five were downregulated in the nucleus but upregulated
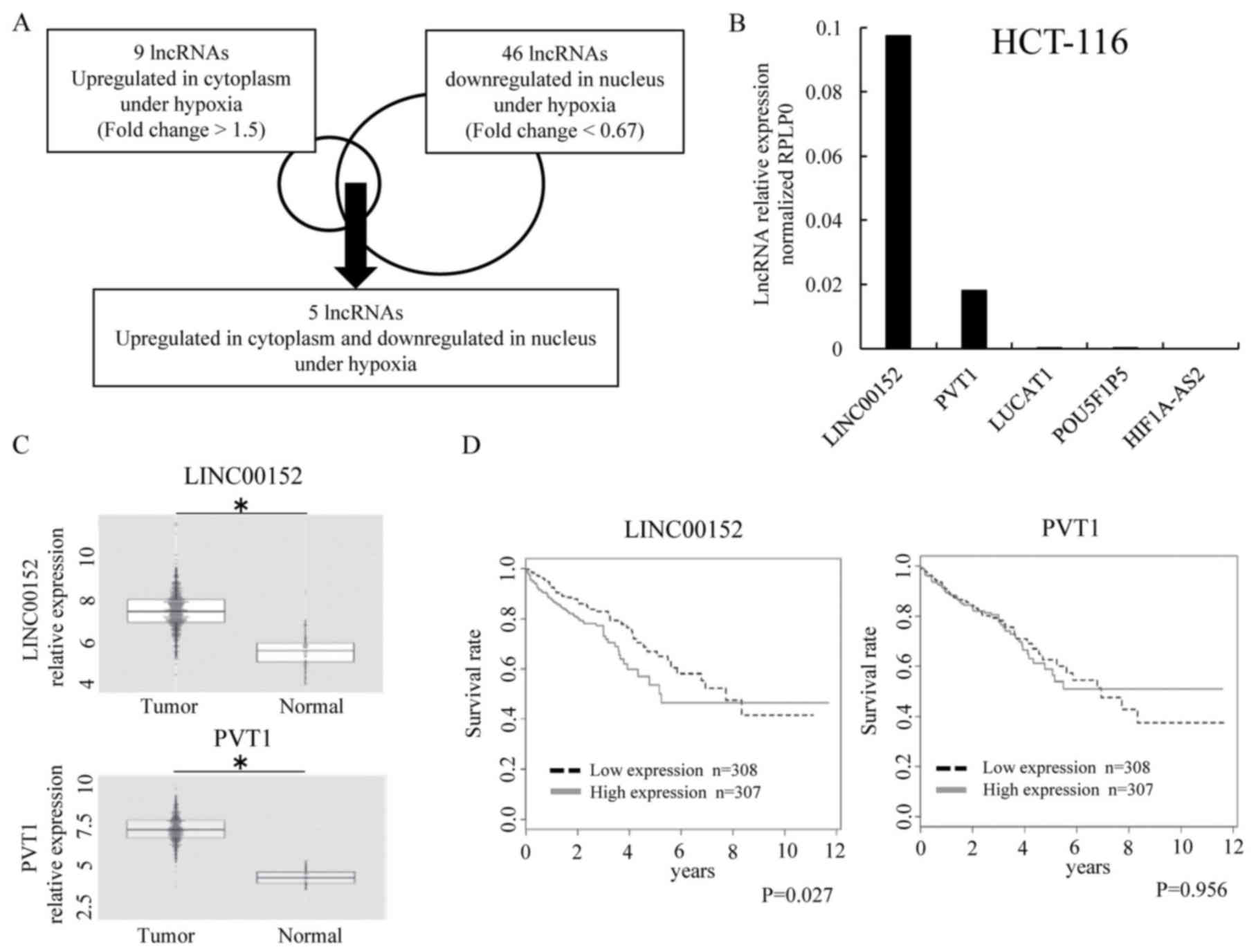
in the cytoplasm (Fig. 5A).
Interestingly, these five lncRNAs were included in the 30 lncRNAs
that were upregulated under hypoxic conditions compared with
normoxic conditions (Fig. 2B).
LINC00152 is associated with poor
prognosis in colorectal cancer
To validate the expression of the five lncRNAs that
were downregulated in the nucleus and upregulated in the cytoplasm,
we performed qRT-PCR using HCT-116 cells. The data showed that the
expression levels of LUCAT1, POU5F1P5 and HIF1A-AS2 were lower than
those of other lncRNAs (Fig. 5B).
Thus, among the five lncRNAs, we focused on LINC00152 and PVT1. To
study the significance of both LINC00152 and PVT1 in CRC patients,
we used the TCGA database of colorectal adenocarcinoma (COADREAD).
Data showed that both LINC00152 and PVT1 exhibited a higher level
of expression in the tumor than in the normal tissue (Fig. 5C), and a high expression of
LINC00152 was associated with a reduced overall survival rate
(P=0.027) (Fig. 5D).
Discussion
In this study, five types of lncRNAs exhibiting
enhanced expression under hypoxic conditions showed decreased
expression in the nucleus and increased expression in the
cytoplasm. Based on the expression data, we focused on LINC00152,
which has relatively high expression among the lncRNAs. LINC00152
is involved in cell growth, cycle arrest, epithelial-mesenchymal
transition (EMT), and invasion in gastric cancers (19). Moreover, the plasma levels of
LINC00152 achieve good diagnostic accuracy for HCC tumorigenesis
and metastasis, and may act as novel biomarkers for HCC (20). Even in the data we examined in the
TCGA database for patients with CRC (COADREAD), the expression of
LINC00152 was more in tumors than in normal tissues, and the
increased expression was associated with poor prognosis, which was
consistent with a previous study (Fig.
5C and D). The present data are compatible with the notion that
LINC00152 exhibits oncogenic function.
It is known that ceRNA binds miRNA in a nucleotide
sequence-dependent manner and regulates miRNA as a decoy. Because
this lncRNA-miRNA interaction occurs in the cytoplasm and most
lncRNA is localized in the nucleus, we supposed that the hypoxia
may induce the transformation of lncRNA function, so that it
changes its localization from the nucleus to the cytoplasm in
response to a hypoxic microenvironment.
Recently, it was reported that LINC00152 competes
with miR-193a-3p as a ceRNA in colorectal cancers and is involved
in the anticancer drug resistance to oxaliplatin (21). In addition, LINC00152 competes with
miR-138 in gall bladder cancers, which suppresses HIF1A downstream
of hypoxia pathway, thereby affecting cancer metastasis and EMT,
which characterizes the metastasis of malignant cancer cells
(22). The database study
[miRcode; http://www.mircode.org; (23)] enabled us to identify the binding
sites of not only previously reported miR-138 and miR-193 but also
others, such as miR-150, miR-155, miR-31 and miR-205, as shared
binding sites for LINC00152 and HIF1A, suggesting their ceRNA
functionality. Thus, it was proposed that LINC00152 expression in
the cytoplasm increases under hypoxic conditions and that it may
act as a decoy for miRNAs involved in malignant cancers.
Previous studies indicated that the hypoxia
inducible system is controlled by the post-translational
modifications, as such involving Hif1 protein degradation (2,3), in
addition to our findings showing that LINC00152 is involved in the
HIF1A mRNA translation by miRNA mechanism. Given that hypoxic
condition induces the stabilization of Hif1 protein, which induces
chemo-resistance and EMT pathway (2,3), the
present study indicates that the LINC00152 pathway collaborates
with the post-translational pathway and enhances the tumor
promoting function of Hif1.
Reportedly the study of a long non-coding antisense
RNA affecting Uchl1 protein translation showed that rapamycin
substantially increased antisense Uchl1 concentration in the
cytoplasmic fraction and decreased antisense Uchl1 concentration in
the nucleolar fraction, which correlates with the expression of
Uchl1 protein, suggesting antisense Uchl1 localization can be
regulated by the mTOR pathway (24). It is supposed that stress-dependent
nucleocytoplasmic shuttling of lncRNAs may be a common strategy to
regulate translation. Although these concepts need further
clarification in future studies, we showed in the present study
that the hypoxic condition induced the translocation of LINC00152
from nucleus to cytoplasm, and may function as ceRNA against miRNA,
which targets HIF1A, and we supposed their relevance with the mTOR
pathway (25).
Previously reported lncRNAs associated with CRC are
shown in Table I. In this study
using colon cancer cells, we identified five types of
hypoxia-induced lncRNA which exhibit decreased expression in the
nucleus and increased expression in the cytoplasm. It is suggested
that LINC00152, which is related to various miRNAs, is associated
with poor prognosis in patients with CRC. Further studies are
necessary to identify useful biomarkers for the development of
innovative therapeutic targets.
Acknowledgments
We thank the members of our laboratories for their
helpful discussions. We thank Drs Tamura and Ikenaga for the
careful arrangement of clinical samples; Dr Yamamoto, Division of
Health Sciences, Osaka University, and Dr Takemasa, Department of
Surgery, Sapporo Medical University, for fruitful discussions. This
study was supported in part by a grant-in-aid for Scientific
Research from the Ministry of Education, Culture, Sports, Science
and Technology and Ministry of Health, Labor and Welfare; and by a
grant from the National Institute of Biomedical Innovation and
Osaka University Drug Discovery Funds. Institutional endowments
were received partially from Taiho Pharmaceutical Co., Ltd.,
Evidence-Based Medical (EBM) Research Center, Idea Consultants,
Inc. (Tokyo, Japan), and Kinshu-kai Medical Corporation (Osaka,
Japan) [to Y.D., M.M. and H.I.]; Chugai Co., Ltd., Yakult Honsha
Co., Ltd., and Merck Co., Ltd. [to Y.D., M.M. and T.S.]. These
funders had no role in the main experimental equipment, supply
expenses, study design, data collection and analysis, decision to
publish, or preparation of the manuscript. This study received
financial support from grants-in-aid for Scientific Research and
P-DIRECT and P-CREATE grants from the Ministry of Education,
Culture, Sports, Science and Technology, MEXT (to M.K., N.N., Y.D.,
M.M. and H.I.); Kobayashi Foundation for Cancer Research (to H.I.);
Kobayashi International Scholarship Foundation (to M.K. and H.I.);
a Grant-in-Aid from the Ministry of Health, Labor, and Welfare (to
M.K., Y.D., M.M. and H.I.); and a Grant-in-Aid for Young Scientists
(B) (Japan Society for the Promotion Science KAKENHI grant no.
17K16549) (to Y.N.).
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen YG, Satpathy AT and Chang HY: Gene
regulation in the immune system by long noncoding RNAs. Nat
Immunol. 18:962–972. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wu L, Zhang L and Zheng S: Role of the
long non-coding RNA HOTAIR in hepatocellular carcinoma. Oncol Lett.
14:1233–1239. 2017.PubMed/NCBI
|
|
7
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A New Paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dong J, Su M, Chang W, Zhang K, Wu S and
Xu T: Long non-coding RNAs on the stage of cervical cancer
(Review). Oncol Rep. 38:1923–1931. 2017.PubMed/NCBI
|
|
9
|
Lu D, Luo P, Wang Q, Ye Y and Wang B:
lncRNA PVT1 in cancer: A review and meta-analysis. Clin Chim Acta.
474:1–7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yoon JH, Kim J and Gorospe M: Long
noncoding RNA turnover. Biochimie. 117:15–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu B and Shan G: Functions of long
noncoding RNAs in the nucleus. Nucleus. 7:155–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
An Y, Furber KL and Ji S: Pseudogenes
regulate parental gene expression via ceRNA network. J Cell Mol
Med. 21:185–192. 2017. View Article : Google Scholar
|
|
13
|
Robertson ED, Semenchenko K and Wasylyk B:
Crosstalk between Mdm2, p53 and HIF1α: Distinct responses to oxygen
stress and implications for tumour hypoxia. Subcell Biochem.
85:199–214. 2014. View Article : Google Scholar
|
|
14
|
Fan C, Tang Y, Wang J, Xiong F, Guo C,
Wang Y, Zhang S, Gong Z, Wei F, Yang L, et al: Role of long
non-coding RNAs in glucose metabolism in cancer. Mol Cancer.
16:1302017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sugimura K, Fujiwara Y, Omori T, Motoori
M, Miyoshi N, Akita H, Gotoh K, Kobayashi S, Takahashi H, Noura S,
et al: Clinical importance of a transcription reverse-transcription
concerted (TRC) diagnosis using peritoneal lavage fluids obtained
pre- and post-lymphadenectomy from gastric cancer patients. Surg
Today. 46:654–660. 2016. View Article : Google Scholar
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Wykoff CC, Beasley NJ, Watson PH, Turner
KJ, Pastorek J, Sibtain A, Wilson GD, Turley H, Talks KL, Maxwell
PH, et al: Hypoxia-inducible expression of tumor-associated
carbonic anhydrases. Cancer Res. 60:7075–7083. 2000.
|
|
18
|
Chang YN, Zhang K, Hu ZM, Qi HX, Shi ZM,
Han XH, Han YW and Hong W: Hypoxia-regulated lncRNAs in cancer.
Gene. 575:1–8. 2016. View Article : Google Scholar
|
|
19
|
Zhao J, Liu Y, Zhang W, Zhou Z, Wu J, Cui
P, Zhang Y and Huang G: Long non-coding RNA Linc00152 is involved
in cell cycle arrest, apoptosis, epithelial to mesenchymal
transition, cell migration and invasion in gastric cancer. Cell
Cycle. 14:3112–3123. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li J, Wang X, Tang J, Jiang R, Zhang W, Ji
J and Sun B: HULC and Linc00152 act as novel biomarkers in
predicting diagnosis of hepatocellular carcinoma. Cell Physiol
Biochem. 37:687–696. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yue B, Cai D, Liu C, Fang C and Yan D:
Linc00152 functions as a competing endogenous RNA to confer
oxaliplatin resistance and holds prognostic values in colon cancer.
Mol Ther. 24:2064–2077. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cai Q, Wang Z, Wang S, Weng M, Zhou D, Li
C, Wang J, Chen E and Quan Z: Long non-coding RNA LINC00152
promotes gallbladder cancer metastasis and epithelial-mesenchymal
transition by regulating HIF-1α via miR-138. Open Biol.
7:1602472017. View Article : Google Scholar
|
|
23
|
Jeggari A, Marks DS and Larsson E:
miRcode: A map of putative microRNA target sites in the long
non-coding transcriptome. Bioinformatics. 28:2062–2063. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Carrieri C, Cimatti L, Biagioli M, Beugnet
A, Zucchelli S, Fedele S, Pesce E, Ferrer I, Collavin L, Santoro C,
et al: Long non-coding antisense RNA controls Uchl1 translation
through an embedded SINEB2 repeat. Nature. 491:454–457. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li L, Zhang GQ, Chen H, Zhao ZJ, Chen HZ,
Liu H, Wang G, Jia YH, Pan SH, Kong R, et al: Plasma and tumor
levels of Linc-pint are diagnostic and prognostic biomarkers for
pancreatic cancer. Oncotarget. 7:71773–71781. 2016.PubMed/NCBI
|
|
27
|
Li N and Richard S: Sam68 functions as a
transcriptional coactivator of the p53 tumor suppressor. Nucleic
Acids Res. 44:8726–8741. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kan JY, Wu DC, Yu FJ, Wu CY, Ho YW, Chiu
YJ, Jian SF, Hung JY, Wang JY and Kuo PL: Chemokine (C-C Motif)
Ligand 5 is Involved in tumor-associated dendritic cell-mediated
colon cancer progression through non-coding RNA MALAT-1. J Cell
Physiol. 230:1883–1894. 2015. View Article : Google Scholar
|
|
29
|
Zhang R, Guo Y, Ma Z, Ma G, Xue Q, Li F
and Liu L: Long non-coding RNA PTENP1 functions as a ceRNA to
modulate PTEN level by decoying miR-106b and miR-93 in gastric
cancer. Oncotarget. 8:26079–26089. 2017.PubMed/NCBI
|
|
30
|
Ling H, Spizzo R, Atlasi Y, Nicoloso M,
Shimizu M, Redis RS, Nishida N, Gafà R, Song J, Guo Z, et al:
CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic
progression and chromosomal instability in colon cancer. Genome
Res. 23:1446–1461. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luo ZF, Zhao D, Li XQ, Cui YX, Ma N, Lu
CX, Liu MY and Zhou Y: Clinical significance of HOTAIR expression
in colon cancer. World J Gastroenterol. 22:5254–5259. 2016.
View Article : Google Scholar : PubMed/NCBI
|