Introduction
The improvement of the treatment efficacy of lung
cancer is an important issue worldwide. In 2012, 1.8 million
individuals were diagnosed with lung cancer worldwide, resulting in
the death of 1.6 million individuals (1). The majority of lung cancers
(approximately 80%) are classified as non-small cell lung cancer
(NSCLC). NSCLC is subdivided into four histopathological subtypes
as follows: adenocarcinoma (LUAD), squamous cell carcinoma (LUSQ),
large cell carcinoma and neuroendocrine cancer (2). The overall survival rate of patients
with NSCLC is poor as almost half of patients have metastatic
disease at initial diagnosis (3).
In patients with adenocarcinoma, the survival rates are markedly
improved by treatment with epidermal growth factor receptor
(EGFR)-tyrosine kinase inhibitors (TKIs), inhibitor of anaplastic
lymphoma kinase (ALK) and immune check point drugs (4). By contrast, recent targeted molecular
therapies have little benefit to the management of patients with
LUSQ (5–7). Therefore, in order to improve the
prognosis of patients with LUSQ, it is important to analyze the
molecular mechanisms of metastatic pathways using the latest
genomic approaches.
The human genome sequencing project has shown that a
large proportion of non-coding RNAs (ncRNAs) are transcribed
(8). The functions of ncRNAs are
varied and they can act as effective regulatory molecules in a wide
range of biological progresses (9). ncRNAs are categorized into two
classes based on their molecular sizes: long ncRNAs (lncRNAs) and
small ncRNAs (9). MicroRNAs
(miRNAs or miRs) are members of the small ncRNA family. They are
typically 19 to 23 nucleotides length, and they regulate the
expression of protein-coding RNAs or non-coding RNAs (10). miRNAs possess unique properties,
including the ability of a single miRNA species to regulate a vast
number of protein-coding or ncRNAs in human cells (10). Thus, aberrantly expressed miRNAs
can disrupt systematically regulated RNA networks in cancer cells.
In fact, dysregulated miRNAs are deeply involved in the
pathogenesis of human cancers (11).
To elucidate the aggressive nature of LUSQ, we
previously identified miRNAs that play regulatory roles in this
disease (12–16). For example, our previous studies
have revealed that the clustered miRNAs, miR-1 and
miR-133a, markedly inhibit cancer cell aggressiveness by
regulating actin binding protein CORO1C (12). All family members of miR-29
(miR-29a, miR-29b and miR-29c) act as anti-metastatic
miRNAs by targeting lysyl oxidase homolog 2 (LOXL2)
(13,14). The over-expression of LOXL2
has been observed in several types of cancer and the knockdown of
LOXL2 interferes with cancer cell aggressiveness (17,18).
Moreover, miR-206 has been shown to inhibit cancer cell
malignancies by targeting two pivotal tyrosine kinase receptors,
MET and EGFR, in LUSQ (16). These
findings provide new knowledge into the novel molecular mechanisms
underlying the pathogenesis of LUSQ.
The analysis of miRNA expression signatures of head
and neck squamous cell carcinoma (HNSCC) by RNA sequencing revealed
that miR-150-5p was downregulated in cancer tissues
(19). In addition, the
downregulation of miR-150-5p was detected in cancer
signatures derived from prostate cancer and bladder cancer
(20,21). However, the functional significance
of miR-150-5p in LUSQ remains unknown. Thus, in this study,
we focused on miR-150-5p and investigated its functional
significance and the regulatory RNA networks in LUSQ cells.
Materials and methods
Clinical samples and cell lines
In this study, a total of 33 LUSQs and 24
non-cancerous lung specimens apart from the tumors were obtained
from patients who underwent lobectomy at Kagoshima University
Hospital from 2010 to 2013. The clinicopathological data of the
patients with LUSQ (33 LUSQs and 24 non-cancerous lung specimens)
are summarized in Table I. Our
study was approved by the Institutional Review Board for Clinical
Research of Kagoshima University Hospital. Each patient provided
written informed consent and approval prior to obtaining the
samples.
 | Table ICharacteristics of lung cancer and
non-cancerous cases. |
Table I
Characteristics of lung cancer and
non-cancerous cases.
| A, Characteristics
of the lung cancer cases |
|---|
|
|---|
| Lung cancer
patients | n | (%) |
|---|
| Total number | 33 | |
| Median age (range,
years) | 70 (55–88) | |
| Sex | | |
| Male | 31 | 93.9 |
| Female | 2 | 6.1 |
| Pathological
stage | | |
| IA | 5 | 15.2 |
| IB | 9 | 27.3 |
| IIA | 4 | 12.1 |
| IIB | 6 | 18.2 |
| IIIA | 8 | 24.2 |
| IIIB | 1 | 3.0 |
| B, Characteristics
of the non-cancerous cases |
|---|
|
|---|
| Non-cancerous
tissues | n |
|---|
| Total number | 24 |
| Median age (range,
years) | 69 (50–88) |
| Sex | |
| Male | 24 |
| Female | 0 |
Two human LUSQ cell lines (SK-MES-1and EBC-1) were
used in this study as previously described (15,16).
The cell lines, SK-MES-1 and EBC-1, were obtained from the Japanese
Cancer Research Resources Bank (JCRB) and the American Type Culture
Collection (Manassas, VA, USA), respectively.
Mature miRNA and small interfering RNA
transfection into LUSQ cells
The following RNA species were used in this study:
mature miRNAs, Pre-miR™ miRNA Precursors (has-miR-150-5p,
assay ID: PM 10070; Applied Biosystems, Foster City, CA, USA),
negative control miRNA (Applied Biosystems, assay ID: AM 17111),
small interfering RNA (Stealth Select RNAi siRNA, Invitrogen,
si-MMP14 P/N: HSS106639 and HSS106640). RNA species were
incubated with Lipofectamine RNAiMax reagent (Invitrogen, Carlsbad,
CA, USA) and Opti-MEM (Invitrogen) prior to plating. Subsequently,
the complex was added to suspended 1×105 cells per-well
plated in 6-well plates. Mock-transfected cells were transfected
only with Lipofectamine RNAiMax reagent and Opti-MEM at plating.
The transfection procedures were as previously described (15,16).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was isolated using Isogen (Nippon Gene,
Tokyo, Japan) according to the manufacturer's instructions. The
integrity of the RNA was checked using an RNA 6000 Nano assay kit
and a 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA,
USA).
The procedure for PCR quantification was as
previously described (12,15,16).
In brief, we first synthesized cDNA from total RNA of each sample
using the TaqMan Reverse Transcription kit (P/N N8080234 and
4366596, Applied Biosystems). Subsequently, we evaluated the
expression of the gene by TaqMan Real-Time PCR Assays. TaqMan
probes and primers for MMP14 (P/N: Hs1037003_g1; Applied
Biosystems) were assay-on-demand gene expression products.
Stem-loop RT-PCR for miR-150-5p (assay ID: 000473; Applied
Biosystems) was used. Human GUSB (P/N: Hs99999908_m1;
Applied Biosystems) and RNU48 (assay ID: 001006; Applied
Biosystems) were used as normalized controls. All reactions were
performed in triplicate and the ΔΔCt method was employed to
calculate the fold change.
Cell proliferation, migration and
invasion assays
To investigate the functional significance of
miR-150-5p or MMP14 silencing by siRNA knockdown, we
performed cell proliferation, migration and invasion assays using
the SK-MES-1 and EBC-1 cells.
The cells were transfected with 10 nM miRNA or siRNA
by reverse transfection and plated in 96-well plates at
5×104 cells per well. After 72 h, cell proliferation was
determined by XTT assay using Cell Proliferation kit (Biological
Industries, Kibbutz Beit Haemek, Israel).
Cell migration activity was evaluated by wound
healing assay. The cells were seeded in 6-well plates at
1×105 cells per well. At 48 h after transfection, the
cell monolayer was scraped using a P-200 micropipette tip. The
initial gap length and residual gap length 24 h after wounding were
calculated from photomicrographs.
Cell invasion assay was carried out using modified
Boyden chambers, consisting Transwell pre-coated Matrigel membrane
filter inserts with 8 µm pores in 24-well tissue culture plates (BD
Biosciences, Bedford, MA, USA). At 48 h after transfection, the
cells were planted in the inserts at 1×105 per well. At
72 h after transfection, the cells invaded to the lower side were
fixed and stained with Diff-Quick (Sysmex Corp., Kobe, Japan). The
number of cells invaded to the lower surface was determined
microscopically by counting 8 areas of constant size per well. All
experiments were performed in triplicate. The procedures of these
functional assays were as previously described (15,16).
Western blot analysis
At 72 h after transfection, protein lysates (50 µg)
were separated on NuPAGE on 4–12% Bis-Tris gels (Invitrogen) and
transferred onto polyvinylidene fluoride membranes (GE Healthcare
Japan, Tokyo, Japan). Immunoblotting was performed with monoclonal
MMP14 antibody (1:2,000 dilution) (ab51074; Abcam, Cambridge, UK).
GAPDH antibody (1:10,000 dilution) (ab8245; Abcam) was used as an
internal control. The membranes were washed and then incubated with
an anti-rabbit-IgG, HRP-linked anti body (#7074; Cell Signaling
Technology, Danvers, MA, USA). Complexes were visualized with
Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA). The
procedures have been described in our previous studies (15,16).
Identification of miR-150-5p target
oncogenic genes in LUSQ cells
Specific genes regulated by miR-150-5p were
identified by a combination of in silico and comprehensive
gene expression analyses. Our target search strategies were
described as previously (15,16).
First, we selected putative miR-150-5p target genes using
the TargetScan 7.1 database (http://www.targetscan.org/vert_71/). We then performed
comprehensive gene analysis of downregulated genes in
miR-150-5p transfected EBC-1 cells and upregulated genes in
NSCLC from GEO database. The microarray data were deposited into
GEO (http://www.ncbi.nlm.nih.gov/geo/),
with the Accession number: GSE82108. Upregulated genes in NSCLC
clinical specimens were obtained from the GEO database (Accession
number: GSE19188).
Plasmid construction and dual-luciferase
reporter assays
The wild-type or deletion-type sequences of the
3′-untranslated region (UTR) of MMP14 in miR-150-5p
target sites were inserted in the psiCHECK-2 vector (C8021;
Promega, Madison, WI, USA). We used 3 sequences that were putative
miR-150-5p target sites of MMP14 by the TargetScan
database (position 567–573: UUGGGAG; position 804–810: UGGGAGA and
position 1381–1388: UUGGGAGA). The procedure for dual luciferase
reporter assays was as previously described (15,16).
The synthesized DNA was cloned into the psiCHECK-2 vector. EBC-1
cells were transfected with the vector, miRNAs and Lipofectamine
2000 in Opti-MEM (both from Invitrogen). The activities of Firefly
and Renilla luciferases in cell lysates were determined with
a dual-luciferase assay system (E1910; Promega). Normalized data
were calculated as the quotient of Renilla/Firefly
luciferase activities.
Immunohistochemistry staining and
scoring
A tissue microarray containing a total of 30 lung
samples, 20 LUSQ specimens and 10 normal lung samples was obtained
from US Biomax (Derwood, MD, USA; Cat. no. BC04002). The patient
characteristics for the tissue microarray are shown in Table II. The TNM classification of
cancer tissues was according to the 7th edition of the American
Joint Committee on Cancer (22).
 | Table IIImmunohistochemistry status and
characteristics of the lung cancer and non-cancerous cases. |
Table II
Immunohistochemistry status and
characteristics of the lung cancer and non-cancerous cases.
| A,
Immunohistochemical status and characteristics of the LUSQ
cases |
|---|
|
|---|
| Patient no. | Grade | T | N | M | Pathological
stage | Immunohistochemical
staining intensity |
|---|
| 1 | 1 | 3 | 1 | 0 | IIIA | (+) |
| 2 | 1 | 3 | 0 | 0 | IIIA | (++) |
| 3 | 2 | 2 | 1 | 0 | II | (+++) |
| 4 | 2 | 3 | 0 | 0 | IIIA | (++) |
| 5 | 1 | 2 | 0 | 0 | I | (++) |
| 6 | 1 | 2 | 1 | 0 | II | (+++) |
| 7 | 1 | 3 | 1 | 0 | IIIA | (++) |
| 8 | 1 | 2 | 0 | 0 | I | (++) |
| 9 | 1 | 2 | 1 | 0 | II | (++) |
| 10 | 1 | 2 | 0 | 0 | I | (+++) |
| 11 | 2 | 2 | 2 | 0 | IIIA | (++) |
| 12 | 2 | 2 | 0 | 0 | I | (+++) |
| 13 | 2 | 1 | 0 | 0 | I | (+++) |
| 14 | 2 | 1 | 0 | 0 | I | (+++) |
| 15 | 2 | 2 | 1 | 0 | II | (++) |
| 16 | 2 | 3 | 1 | 0 | IIIA | (+++) |
| 17 | 2 | 2 | 0 | 0 | I | (++) |
| 18 | 2 | 2 | 1 | 0 | II | (++) |
| 19 | 2 | 3 | 2 | 0 | IIIA | (+++) |
| 20 | 2 | 2 | 0 | 0 | I | (++) |
| B,
Immunohistochemical status of non-cancerous cases |
|---|
|
|---|
| Patient no. | Immunohistochemical
staining intensity |
|---|
| 91 | (+) |
| 92 | (+) |
| 93 | (+) |
| 94 | (−) |
| 95 | (+) |
| 96 | (+) |
| 97 | (−) |
| 98 | (++) |
| 99 | (+) |
| 100 | (+) |
We confirmed the expression status of MMP14 in the
LUSQ clinical specimens using immunohistochemical staining. The
procedure for immunohistochemistry was as previously described
(16,23,24).
The tissue sections were incubated with the primary rabbit
monoclonal antibody against MMP14 (1:5,000 dilution; ab51074;
Abcam). The slide was treated with biotinylated goat anti-rabbit
antibodies. Diaminobenzidine hydrogen peroxidase was the chromogen
and counterstaining was done with 0.5% hematoxylin. Each tissues
sample was scored on the basis of the intensity and area of
staining. The procedure for score for staining tissues was as
previously described (25).
Statistical analysis
Relationships between two or three variables and
numerical values were analyzed using Mann-Whitney U tests or
Bonferroni-adjusted Mann-Whitney U tests. Expert StatView software
(version 5.0; SAS Institute Inc., Cary, NC, USA) was used for these
analyses. Statistical analysis was carried out as previously
described (19,23).
Results
Expression levels of miR-150-5p in LUSQ
clinical specimens and cell lines
The expression levels of miR-150-5p were
significantly decreased in the cancer tissues compared with
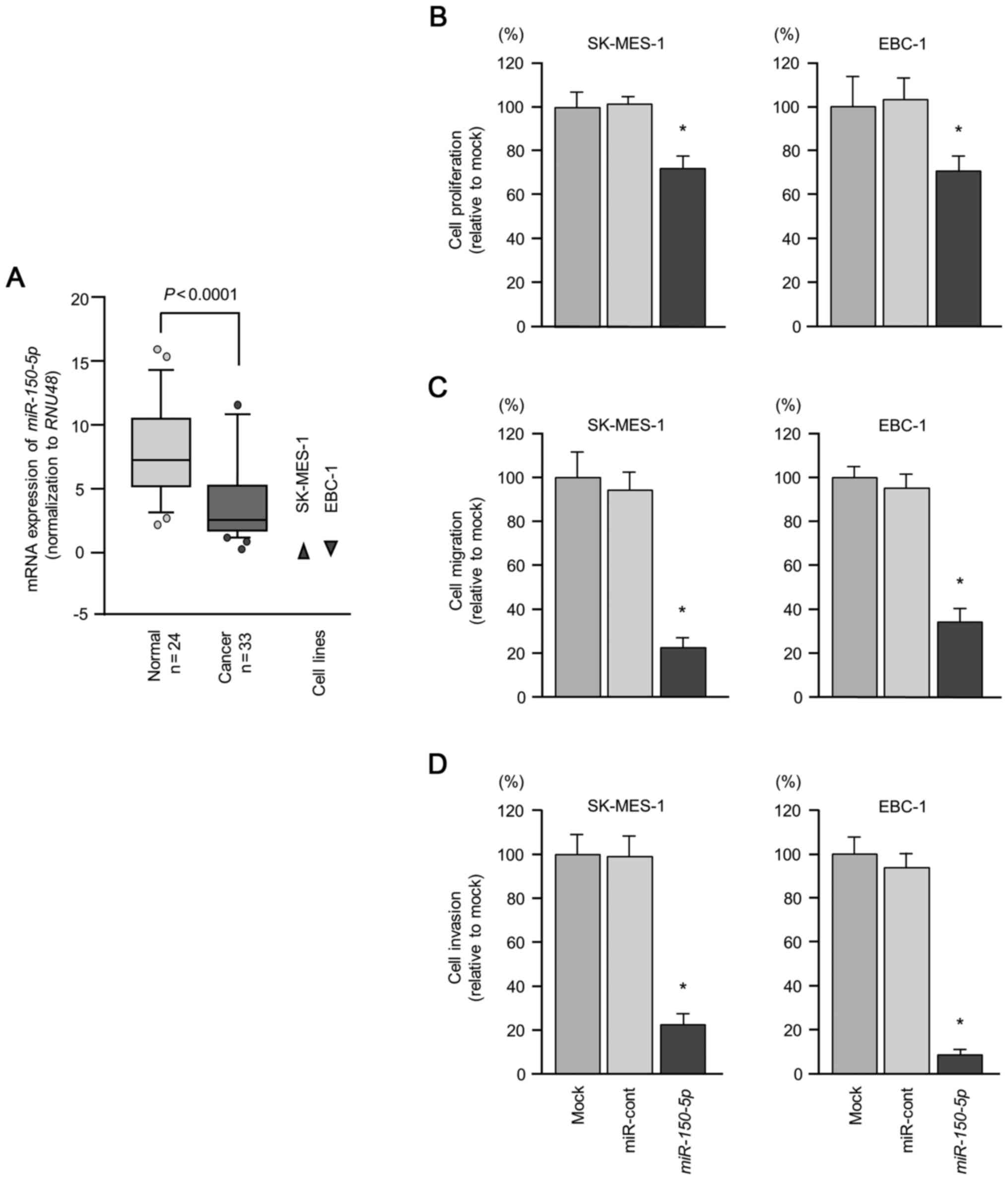
non-cancerous tissues (P<0.0001; Fig. 1A). In addition, the expression
levels of miR-150-5p in the cancer cell lines, SK-MES-1 and
EBC-1, were markedly downregulated (Fig. 1A).
Effects of the ectopic overexpression of
miR-150-5p on cell proliferation, migration and invasion in LUSQ
cell lines
To validate the antitumor effects of
miR-150-5p, we carried out gain-of-function assays by
transfecting miRNA into two LUSQ cell lines (SK-MES-1 and EBC-1).
Cell proliferation was significantly inhibited in the cells
transfected with the mature miR-150-5p in comparison with
the mock (transfection reagent only)- or miR-control-transfected
cells (Fig. 1B). Furthermore, cell
migration and invasion activities were markedly reduced in the
cells transfected with the miR-150-5p expression plasmid
compared to the mock- or miR-control-transfected cells (Fig. 1C and D).
Identification of putative targets of
miR-150-5p regulation in LUSQ cells
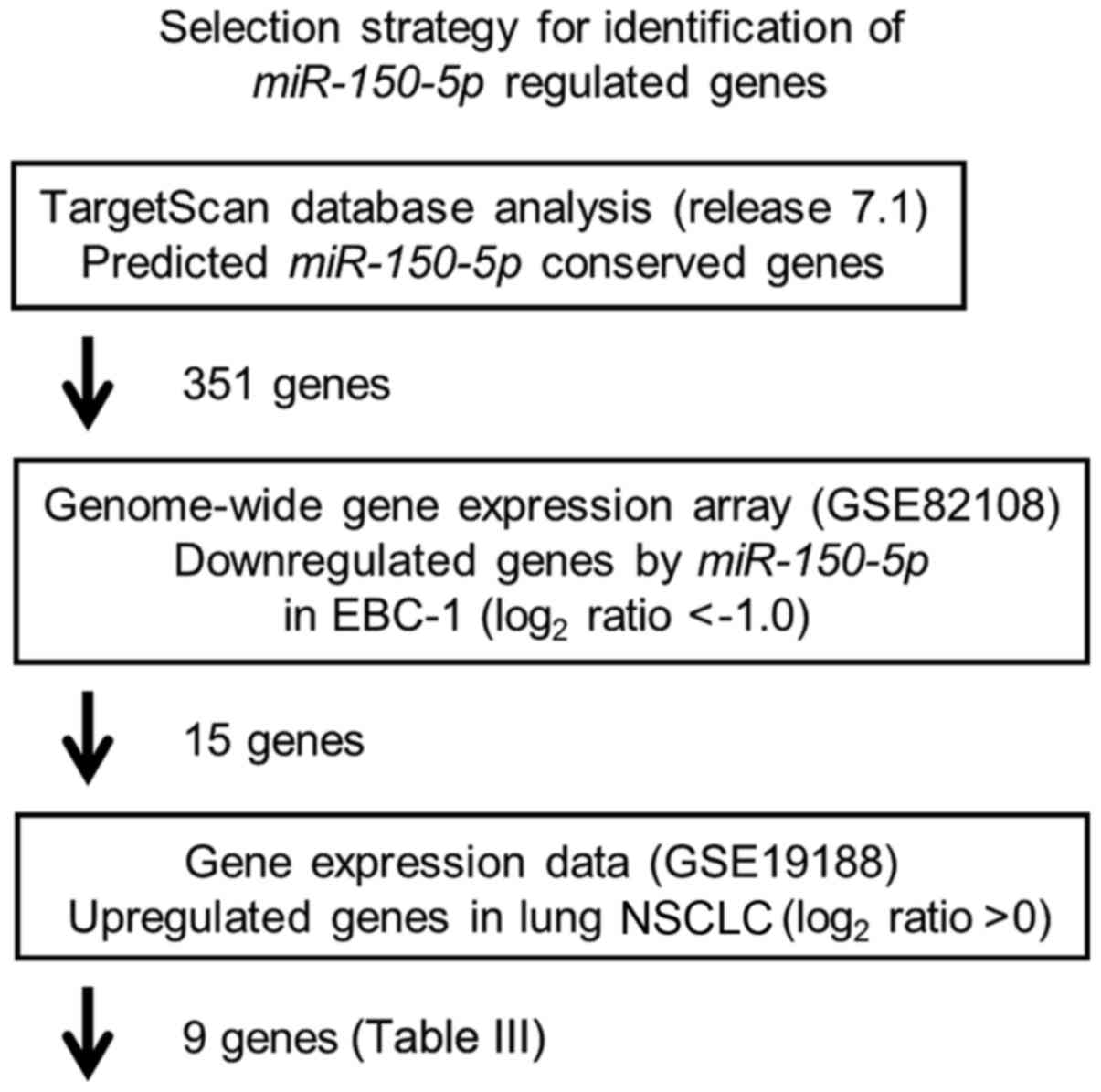
Our strategy for the selection of miR-150-5p
target oncogenic genes is shown in Fig. 2. First, we selected putative
miR-150-5p target genes using the TargetScan 7.1 database
and identified 351 genes. We then performed comprehensive gene
expression analysis using miR-150-5p transfectants of EBC-1
cells, with negative control miRNA transfectants serving as
controls (Accession number: GSE 82108). In this assessment, 15
genes were commonly downregulated (log2 ratio <−1.0).
The gene set was then analyzed with a publicly available gene
expression data set in GEO (Accession number: GSE 19188) and genes
upregulated in LUSQ were selected (fold change >0). A total of 9
genes were identified as candidate targets of miR-150-5p
regulation (Table III).
 | Table IIIPutative target genes regulated by
miR-150-5p in LUSQ cells. |
Table III
Putative target genes regulated by
miR-150-5p in LUSQ cells.
| Gene symbol | Gene name | Conserved | Poorly
conserved | GEO82108
log2 ratio | GEO19188 Fold
change |
|---|
| MMP14 | Matrix
metallopeptidase 14 (membrane-inserted) | 1 | 2 | −1.11 | 1.872 |
| CPD | Carboxypeptidase
D | 1 | 0 | −1.07 | 1.071 |
| LRRC58 | Leucine rich repeat
containing 58 | 1 | 6 | −1.19 | 0.733 |
| CDC73 | Cell division cycle
73 | 1 | 2 | −2.16 | 0.605 |
| ENSA | Endosulfine
alpha | 1 | 5 | −1.79 | 0.501 |
| DSEL | Dermatan sulfate
epimerase-like | 1 | 0 | −1.17 | 0.487 |
| CSNK1A1 | Casein kinase 1,
alpha 1 | 1 | 1 | −1.13 | 0.292 |
| CHD2 | Chromodomain
helicase DNA binding protein 2 | 1 | 2 | −2.14 | 0.260 |
| CAPZB | Capping protein
(actin filament) muscle Z-line, beta | 1 | 4 | −1.18 | 0.201 |
Among these candidate genes, we focused on
MMP14 as the aberrant expression of the matrix
metalloproteinase family enhances cancer cell migration and
invasion (26). The ectopic
overexpression of miR-150-5p significantly inhibited cancer
cell migration and invasion in the LUSQ cells (Fig. 1C and D).
Direct regulation of MMP14 by miR-150-5p
in LUSQ cells
We investigated whether transfection of the LUSQ
cell lines with the miR-150-5p expression plasmid would
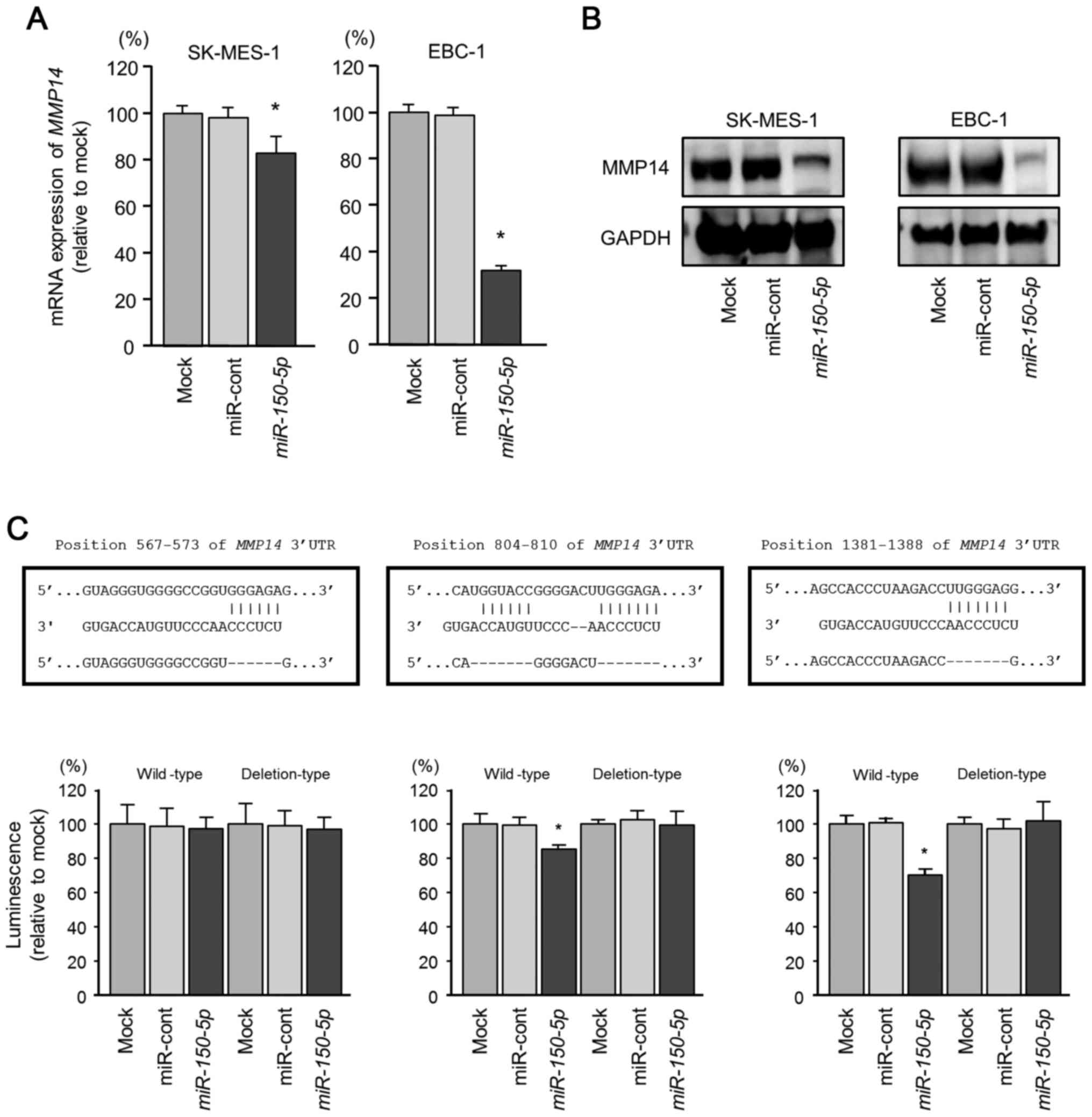
reduce the expression of MMP14/MMP14. The mRNA expression
levels of MMP14 were decreased by transfection with the
miR-150-5p expression plasmid compared with the mock- or
miR-control-transfected cells (Fig.
3A). Furthermore, the protein expression levels of MMP14 were
also decreased by the overexpression of miR-150-5p compared
with the mock- or miR-control-transfected cells (Fig. 3B).
We also performed luciferase reporter assays with a
vector that included the 3′-UTR of MMP14 to confirm that
miR-150-5p directly regulated MMP14 in a
sequence-dependent manner. We used vectors encoding the partial
wild-type or deletion-type sequences of the 3′-UTR of the
MMP14 with miR-150-5p target sites. Three binding
sites for miR-150-5p in the 3′-UTR of MMP14
(positions 567–573, 804–810 and 1381–1388) were predicted by the
TargetScan Human database (Fig.
3C). We observed that the luminescence intensities of the
proteins coded by vectors carrying the wild-type sequences
(positions 804–810 and 1381–1388) were significantly reduced by
co-transfection with miR-150-5p (Fig. 3C). By contrast, transfection with
the deletion-type vector blocked the reduction of luminescence
intensities (Fig. 3C). These
findings indicated that miR-150-5p directly bound to
specific two sites in the 3′-UTR of MMP14.
Effects of knockdown of MMP14 on the
proliferation, migration and invasion of LUSQ cell lines
Loss-of-function assays using siRNA were performed
to examine the function of MMP14 in LUSQ cell lines
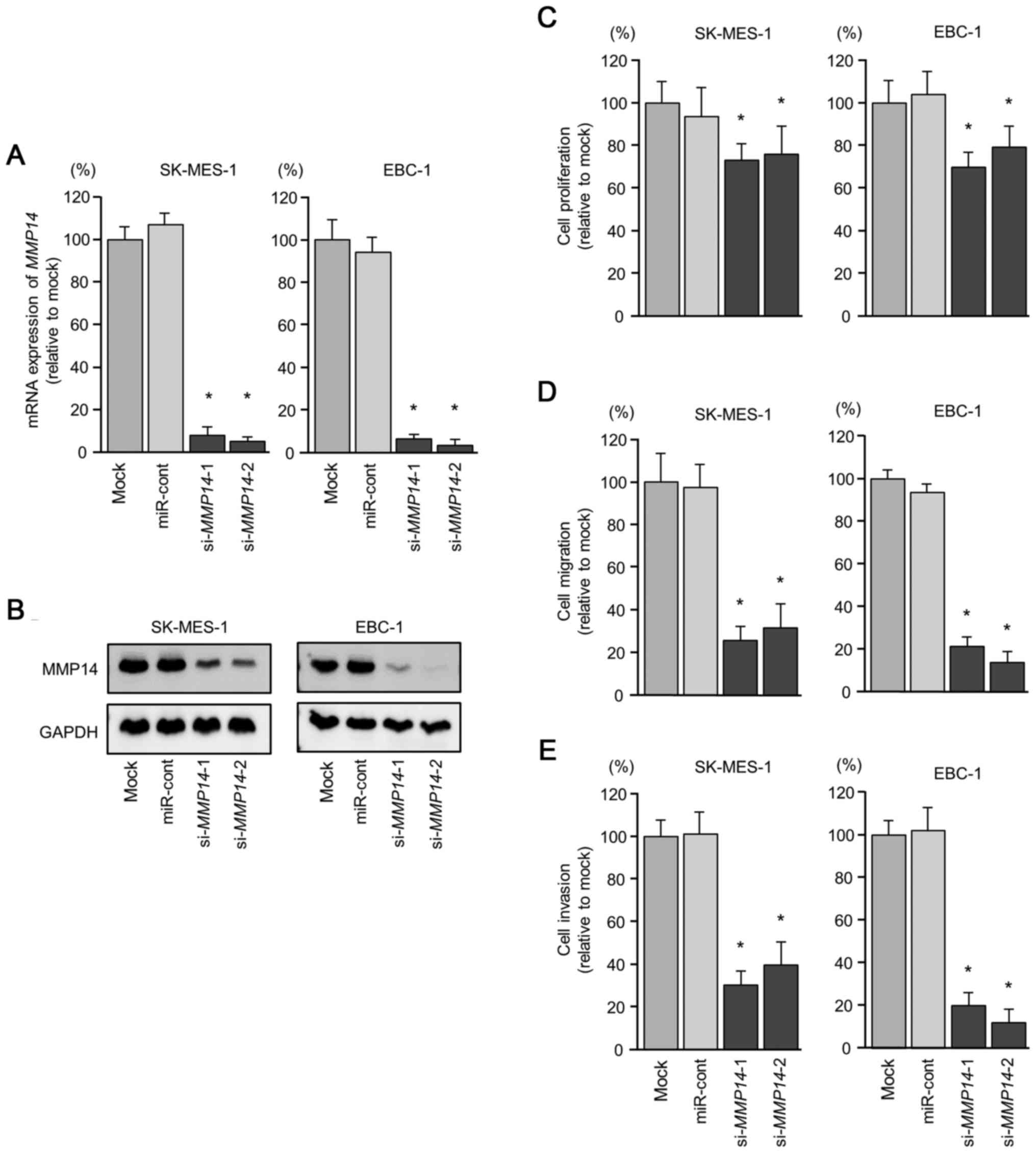
(SK-MES-1 and EBC-1). The mRNA and protein expression levels of
MMP14 were decreased by transfection with si-MMP14 in
the LUSQ cell lines (Fig. 4A and
B).
We then investigated the effects of MMP14
knockdown on the proliferation, migration, and invasion of LUSQ
cell lines. Cancer cell proliferation was significantly reduced in
the si-MMP14-transfected cells (Fig. 4C). In addition, the migration and
invasion activities were significantly attenuated in the
si-MMP14-transfected cells (Fig. 4D and E).
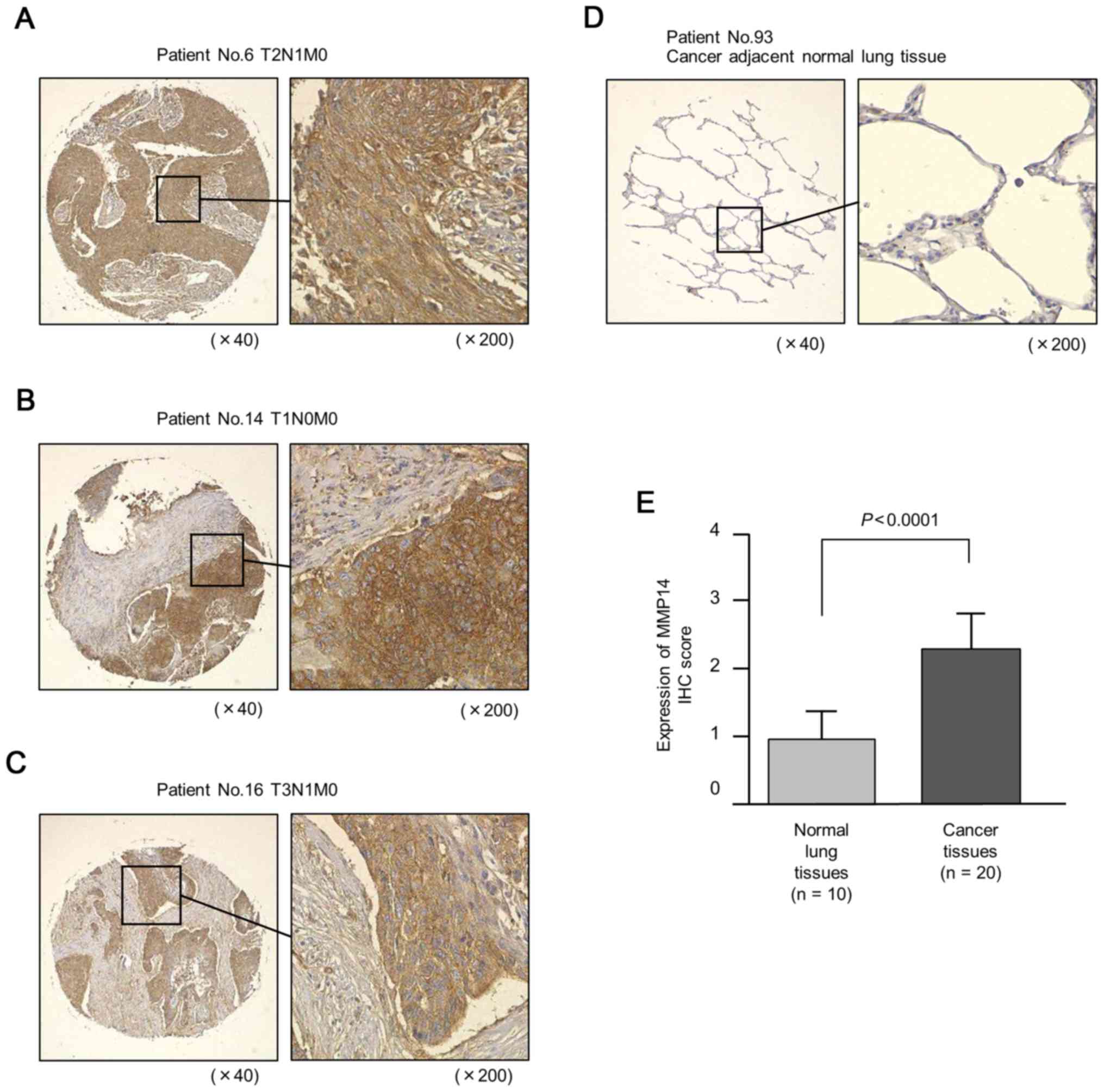
Expression of MMP14 in LUSQ clinical
specimens
We examined the protein expression levels of MMP14
in LUSQ clinical specimens by immunostaining. We confirmed the
overexpression of MMP14 in LUSQ lesions compared with that in
normal tissues (Fig. 5).
Identification of MMP14-regulated genes
in LUSQ cells
To investigate downstream genes regulated by
MMP14, we performed genome-wide gene expression analysis
using si-MMP14 in EBC-1 cells. A total of 92 genes were
identified as MMP14-regulated genes (Table IV).
 | Table IVDownstream genes regulated by
MMP14 in LUSQ cells. |
Table IV
Downstream genes regulated by
MMP14 in LUSQ cells.
| Gene symbol | Gene name | Expression
log2 ratio
|
|---|
|
si-MMP14-1 |
si-MMP14-2 | Average |
|---|
| MMP14 | Matrix
metallopeptidase 14 (membrane-inserted) | −4.19 | −3.72 | −3.96 |
| LMNB1 | Lamin B1 | −3.70 | −2.97 | −3.34 |
| TMEM192 | Transmembrane
protein 192 | −2.61 | −3.48 | −3.05 |
| MKI67 | Marker of
proliferation Ki-67 | −2.17 | −3.85 | −3.01 |
| ENPP1 | Ectonucleotide
pyrophosphatase/phosphodiesterase 1 | −2.77 | −3.11 | −2.94 |
| LPL | Lipoprotein
lipase | −2.81 | −3.05 | −2.93 |
|
SLC7A11-AS1 | SLC7A11 antisense
RNA 1 | −3.90 | −1.90 | −2.90 |
| CDRT1 | CMT1A duplicated
region transcript 1 | −3.92 | −1.77 | −2.85 |
| FAM129A | Family with
sequence similarity 129, member A | −3.68 | −1.97 | −2.83 |
| BZW1 | Basic leucine
zipper and W2 domains 1 | −3.08 | −2.58 | −2.83 |
| LYRM1 | LYR motif
containing 1 | −2.63 | −2.90 | −2.76 |
| CHRNA5 | Cholinergic
receptor, nicotinic, alpha 5 (neuronal) | −3.31 | −2.20 | −2.75 |
| MARCH5 | Membrane-associated
ring finger (C3HC4) 5 | −2.23 | −3.23 | −2.73 |
| DEPDC1 | DEP domain
containing 1 | −3.34 | −2.06 | −2.70 |
| CBX3 | Chromobox homolog
3 | −3.06 | −2.27 | −2.66 |
| ANKRD22 | Ankyrin repeat
domain 22 | −1.50 | −3.76 | −2.63 |
| JKAMP |
JNK1/MAPK8-associated membrane
protein | −1.66 | −3.37 | −2.51 |
| ANTXR1 | Anthrax toxin
receptor 1 | −2.39 | −2.59 | −2.49 |
|
SMAD1-AS1 | SMAD1 antisense RNA
1 | −2.29 | −2.69 | −2.49 |
| ATP5E | ATP
synthase, H+ transporting, mitochondrial F1 complex, epsilon
subunit | −2.31 | −2.64 | −2.47 |
| TOMM20 | Translocase of
outer mitochondrial membrane 20 homolog (yeast) | −2.40 | −2.41 | −2.40 |
| DEFB4A | Defensin, beta
4A | −2.62 | −2.12 | −2.37 |
| ALDH7A1 | Aldehyde
dehydrogenase 7 family, member A1 | −2.48 | −2.25 | −2.37 |
| OSBPL8 | Oxysterol binding
protein-like 8 | −2.70 | −1.99 | −2.34 |
| GLYCTK | Glycerate
kinase | −2.58 | −2.08 | −2.33 |
| SLC16A1 | Solute carrier
family 16 (monocarboxylate transporter), member 1 | −2.82 | −1.82 | −2.32 |
| C5 | Complement
component 5 | −2.18 | −2.42 | −2.30 |
| ASNS | Asparagine
synthetase (glutamine-hydrolyzing) | −3.09 | −1.50 | −2.29 |
| POLE2 | Polymerase (DNA
directed), epsilon 2, accessory subunit | −1.53 | −3.05 | −2.29 |
| SASS6 | Spindle assembly 6
homolog (C. elegans) | −1.94 | −2.62 | −2.28 |
| SMARCA2 | SWI/SNF related,
matrix associated, actin dependent regulator of chromatin,
subfamily a, member 2 | −1.81 | −2.68 | −2.24 |
| FANCD2 | Fanconi anemia,
complementation group D2 | −2.19 | −2.27 | −2.23 |
| TMEM154 | Transmembrane
protein 154 | −2.69 | −1.77 | −2.23 |
| ARHGAP9 | Rho GTPase
activating protein 9 | −2.73 | −1.73 | −2.23 |
| SPATA5 | Spermatogenesis
associated 5 | −2.64 | −1.68 | −2.16 |
|
ARHGAP11A | Rho GTPase
activating protein 11A | −1.71 | −2.59 | −2.15 |
| OSBPL8 | Oxysterol binding
protein-like 8 | −2.12 | −2.16 | −2.14 |
| SUV39H2 | Suppressor of
variegation 3–9 homolog 2 (Drosophila) | −1.77 | −2.50 | −2.13 |
| ACTL8 | Actin-like 8 | −2.36 | −1.85 | −2.10 |
| ESCO2 | Establishment of
sister chromatid cohesion N-acetyltransferase 2 | −2.52 | −1.68 | −2.10 |
| RAB1A | RAB1A, member RAS
oncogene family | −2.07 | −2.13 | −2.10 |
| FBXO4 | F-box protein
4 | −1.76 | −2.42 | −2.09 |
| MAPK6 | Mitogen-activated
protein kinase 6 | −2.32 | −1.86 | −2.09 |
| RNF180 | Ring finger protein
180 | −1.91 | −2.25 | −2.08 |
|
DKFZP434I0714 | Uncharacterized
protein DKFZP434I0714 | −1.77 | −2.38 | −2.08 |
| MTBP | MDM2 binding
protein | −1.93 | −2.21 | −2.07 |
| AIF1L | Allograft
inflammatory factor 1-like | −2.44 | −1.68 | −2.06 |
| MOB3B | MOB kinase
activator 3B | −1.57 | −2.53 | −2.05 |
| CGGBP1 | CGG triplet repeat
binding protein 1 | −1.75 | −2.34 | −2.04 |
|
HIST1H2AI | Histone cluster 1,
H2ai | −2.46 | −1.62 | −2.04 |
| FKBP5 | FK506 binding
protein 5 | −1.79 | −2.27 | −2.03 |
| NUDT21 | Nudix (nucleoside
diphosphate linked moiety X)-type motif 21 | −1.54 | −2.45 | −2.00 |
| NEIL3 | Nei endonuclease
VIII-like 3 (E. coli) | −2.02 | −1.90 − | 1.96 |
| ATG4C | Autophagy-related
4C, cysteine peptidase | −1.64 | −2.26 | −1.95 |
| ECEL1 | Endothelin
converting enzyme-like 1 | −1.89 | −1.95 | −1.92 |
| KIF11 | Kinesin family
member 11 | −1.55 | −2.25 | −1.90 |
| TCF19 | Transcription
factor 19 | −1.64 | −2.16 | −1.90 |
| ZNF681 | Zinc finger protein
681 | −2.03 | −1.75 | −1.89 |
| RRM2 | Ribonucleotide
reductase M2 | −1.66 | −2.10 | −1.88 |
| PLAC8L1 | PLAC8-like 1 | −1.55 | −2.20 | −1.88 |
| IL1B | Interleukin 1,
beta | −1.69 | −2.05 | −1.87 |
| DIS3L | DIS3 like exosome
3′-5′ exoribonuclease | −1.81 | −1.90 | −1.85 |
| CKAP2L | Cytoskeleton
associated protein 2-like | −1.66 | −2.03 | −1.84 |
| RRN3 | RRN3 RNA polymerase
I transcription factor homolog (S. cerevisiae) | −1.54 | −2.15 | −1.84 |
| TMEM180 | Transmembrane
protein 180 | −2.12 | −1.56 | −1.84 |
| HS2ST1 | Heparan sulfate
2-O-sulfotransferase 1 | −1.57 | −2.11 | −1.84 |
| ZNF414 | Zinc finger protein
414 | −1.74 | −1.89 | −1.82 |
|
TMEM194B | Transmembrane
protein 194B | −1.69 | −1.94 | −1.82 |
| STXBP5L | Syntaxin binding
protein 5-like | −1.92 | −1.71 | −1.82 |
| CENPI | Centromere protein
I | −1.98 | −1.64 | −1.81 |
| ERCC6L | excision repair
cross-complementation group 6-like | −1.60 | −2.02 | −1.81 |
| SPC24 | SPC24, NDC80
kinetochore complex component | −1.79 | −1.83 | −1.81 |
|
KRTAP10-3 | Keratin associated
protein 10-3 | −1.85 | −1.76 | −1.80 |
| ID2-AS1 | ID2 antisense RNA 1
(head to head) | −1.55 | −2.02 | −1.78 |
|
GATA2-AS1 | GATA2 antisense RNA
1 | −2.03 | −1.53 | −1.78 |
| FBF1 | Fas (TNFRSF6)
binding factor 1 | −1.55 | −2.01 | −1.78 |
| MED1 | Mediator complex
subunit 1 | −1.64 | −1.91 | −1.78 |
| PLCL2 | Phospholipase
C-like 2 | −2.01 | −1.52 | −1.77 |
| CDKN3 | Cyclin-dependent
kinase inhibitor 3 | −1.56 | −1.92 | −1.74 |
| CSF3 | Colony stimulating
factor 3 (granulocyte) | −1.55 | −1.92 | −1.73 |
| PLAA | Phospholipase
A2-activating protein | −1.53 | −1.88 | −1.70 |
| MCM6 | Minichromosome
maintenance complex component 6 | −1.81 | −1.59 | −1.70 |
| TRIM14 | Tripartite motif
containing 14 | −1.64 | −1.75 | −1.69 |
| TUBA3FP | Tubulin, alpha 3f,
pseudogene | −1.66 | −1.71 | −1.68 |
| OVOS2 | Ovostatin 2 | −1.76 | −1.59 | −1.67 |
| NUCKS1 | Nuclear casein
kinase and cyclin-dependent kinase substrate 1 | −1.58 | −1.75 | −1.67 |
| HOOK1 | Hook
microtubule-tethering protein 1 | −1.62 | −1.66 | −1.64 |
| ZBTB33 | Zinc finger and BTB
domain containing 33 | −1.72 | −1.50 | −1.61 |
| GALNT4 | Polypeptide
N-acetylgalactosaminyltransferase 4 | −1.70 | −1.53 | −1.61 |
| RRM1 | Ribonucleotide
reductase M1 | −1.61 | −1.61 | −1.61 |
| MDM1 | Mdm1 nuclear
protein homolog (mouse) | −1.69 | −1.53 | −1.61 |
| FAM72D | Family with
sequence similarity 72, member D | −1.51 | −1.66 | −1.59 |
Discussion
Recently, several treatment options have been
approved for post-first-line therapy for LUSQ, such as chemotherapy
with or without an angiogenesis inhibitor (27), or immunotherapy (4). However, treatment outcomes do not
appear to have improved. Therefore, clinicians need new treatment
options in their approach to LUSQ. Presumably, they should be based
on the latest molecular analyses of the pathology of this
disease.
In this study, we demonstrated that the restoration
of miR-150-5p significantly attenuated with cancer cell
aggressiveness, suggesting that this miRNA possesses antitumor
activity in LUSQ cells. It has been demonstrated that
miR-150-5p has multiple functions, possessing antitumor
activity or oncogenic functions (28). The overexpression of
miR-150-5p has been reported in several types of cancer
(29,30). In contrast to the overexpression of
miR-150-5p in cancer cells, the antitumor roles of
miR-150-5p have been reported in several types of cancer
through its targeting of oncogenic genes (19,31,32).
Previous studies showed that contradiction resulted as to the
expression pattern of miR-150-5p in lung cancer (28). Our preliminary data demonstrated
that the expression levels of miR-150-5p were reduced in
lung adenocarcinoma clinical specimens (data not shown). One such
study showed that miR-150-5p expression was significantly
reduced in HNSCC tissues and had antitumor roles. A low expression
of miR-150-5p has been shown to be significantly associated
with a poor prognosis of patients with HNSCC (19). Moreover, integrin subunit alpha 3
(ITGA3), integrin subunit alpha 6 (ITGA6) and
tenascin C (TNC) have been identified as oncogenes in HNSCC
that were downregulated by miR-150-5p (19). Previous studies have shown that the
aberrant expression of integrin family genes and the activation of
integrin-mediated oncogenic signaling promotes cancer cell
metastasis and the epithelial-mesenchymal transition (EMT)
phenotype (33,34). Other studies have demonstrated that
SPOCK1 is directly regulated by miR-150-5p in
prostate cancer and esophageal squamous cell carcinoma (31,32).
The aberrant expression of SPOCK1 has been observed in
several types of cancer and has been shown to play pivotal roles in
cancer cell progression, metastasis and drug resistance (35,36).
Of note, the ectopic expression of SPOCK1 induces EMT in
lung cancer (37). These findings
suggest that the downregulation of miR-150-5p induces
several cancer-promoting genes and that its expression is deeply
involved in cancer pathogenesis.
In this study, to identify oncogenic genes targeted
by miR-150-5p, we applied gene expression analysis and in
silico database searches. A total of 9 genes (MMP14, CPD,
LRRC58, CDC73, ENSA, DSEL, CSNK1A1, CHD2 and CAPZB) were
identified as putative targets of miR-150-5p regulation in
LUSQ cells. Among these, we focused on MMP14 as the matrix
metalloproteinase family contributes to cancer cell migration and
invasion. Our luciferase reporter assay revealed that MMP14
was directly regulated by miR-150-5p in LUSQ cells. The
overexpression of MMP14 was observed in LUSQ clinical
specimens and the knockdown of MMP14 by siRNA significantly
interfered in vitro with cancer cell malignancies. These
findings indicate that MMP14 acts as an oncogene regulated
by the antitumor miR-150-5p in LUSQ cells.
MMP14 belongs to the membrane-type matrix
metalloprotease (MT-MMP) family that includes pivotal regulators of
cell invasion, growth and survival in normal cells (26). The upregulation of MMP14 has been
observed in many types of cancer, and the overexpression of MMP14
has been shown to correlate with a poor prognosis in patients with
NSCLC, renal cell carcinoma and breast cancer (38–40).
Activated MMP14 cleaves pro-MMP2 and pro-MMP13 and the activated
form of MMP2 and MMP13 are involved in cancer pathogenesis
(26). A major substrate of MMP14
is type I collagen in ECM components. MMP14 is localized to the
leading edge of invadopodia in migrating cells, achieving spatially
coordinated matrix degradation to support invasion (41). MMP14 influences cancer cells and
cancer niches (ECM and fibroblast cells). MMP14 is mediated through
membrane proteins, e.g., receptor tyrosine kinases (RTK) and
integrins, and these events promote cancer cell aggressiveness
(42,43). For instance, he interaction of
MMP14 and CD44 induces the phosphorylation of the EGF receptor and
enhances downstream activation of the MAPK and PI3K signaling
pathways (44,45).
Proteolytic enzymes, MMPs or MT-MMPs are tightly
controlled by tissue inhibitors of metalloproteases (TIMPs)
(26). The expression and
activation of MMPs and TIMPs are important to biological processes
and essential for tissue homeostasis. Dysregulated MMPs and TIMPs
have been implicated in cancer cell progression and metastasis
(46). Recent studies have
indicated that several downregulated miRNAs caused the aberrant
expression of MMP14 in cancer cells (47,48).
Our recent studies showed the antitumor functions of miR-375
and miR-139-5p/-3p that targeted MMP13 and
MMP11, respectively (49,50).
These MMPs were overexpressed in cancer specimens, and the
restoration of these miRNAs or knockdown of MMP13 or
MMP11 inhibited cancer cell migration and invasion (49,50).
The identification of antitumor miRNAs that regulate novel LUSQ
networks may lead to a better understanding of the aggressiveness
of this disease.
Taken together, this study demonstrates
miR-150-5p acts as an antitumor miRNA by targeting
MMP14 in LUSQ cells. The overexpression of MMP14 may
be enhanced in lung cancer oncogenesis. The exploration of
antitumor miRNA-mediated regulatory networks may lead to the
development of novel treatment strategies for this disease.
Acknowledgments
This study was supported by the Japan Society for
the Promotion of Science (JSPS) Grants-in-Aid for Scientific
Research (KAKENHI), 17K16893, 15K10801, 16K19458, and 17K09660.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Cheng TY, Cramb SM, Baade PD, Youlden DR,
Nwogu C and Reid ME: The international epidemiology of lung cancer:
Latest trends, disparities, and tumor characteristics. J Thorac
Oncol. 11:1653–1671. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Travis WD: Pathology of lung cancer. Clin
Chest Med. 32:669–692. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Reck M, Heigener DF, Mok T, Soria JC and
Rabe KF: Management of non-small-cell lung cancer: Recent
developments. Lancet. 382:709–719. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Reck M, Rodríguez-Abreu D, Robinson AG,
Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe
S, et al KEYNOTE-024 Investigators: Pembrolizumab versus
chemotherapy for PD-L1-positive non-small-cell lung cancer. N Engl
J Med. 375:1823–1833. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sandler AB, Schiller JH, Gray R, Dimery I,
Brahmer J, Samant M, Wang LI and Johnson DH: Retrospective
evaluation of the clinical and radiographic risk factors associated
with severe pulmonary hemorrhage in first-line advanced,
unresectable non-small-cell lung cancer treated with Carboplatin
and Paclitaxel plus bevacizumab. J Clin Oncol. 27:1405–1412. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Scagliotti GV, Parikh P, von Pawel J,
Biesma B, Vansteenkiste J, Manegold C, Serwatowski P, Gatzemeier U,
Digumarti R, Zukin M, et al: Phase III study comparing cisplatin
plus gemcitabine with cisplatin plus pemetrexed in
chemotherapy-naive patients with advanced-stage non-small-cell lung
cancer. J Clin Oncol. 26:3543–3551. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Paz-Ares L, Tan EH, O'Byrne K, Zhang L,
Hirsh V, Boyer M, Yang JC, Mok T, Lee KH, Lu S, et al: Afatinib
versus gefitinib in patients with EGFR mutation-positive advanced
non-small-cell lung cancer: Overall survival data from the phase
IIb LUX-Lung 7 trial. Ann Oncol. 28:270–277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Djebali S, Davis CA, Merkel A, Dobin A,
Lassmann T, Mortazavi A, Tanzer A, Lagarde J, Lin W, Schlesinger F,
et al: Landscape of transcription in human cells. Nature.
489:101–108. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Beermann J, Piccoli MT, Viereck J and Thum
T: Non-coding RNAs in development and disease: Background,
mechanisms, and therapeutic approaches. Physiol Rev. 96:1297–1325.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wiemer EA: The role of microRNAs in
cancer: No small matter. Eur J Cancer. 43:1529–1544. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mataki H, Enokida H, Chiyomaru T, Mizuno
K, Matsushita R, Goto Y, Nishikawa R, Higashimoto I, Samukawa T,
Nakagawa M, et al: Downregulation of the microRNA-1/133a cluster
enhances cancer cell migration and invasion in lung-squamous cell
carcinoma via regulation of Coronin1C. J Hum Genet. 60:53–61. 2015.
View Article : Google Scholar
|
|
13
|
Kamikawaji K, Seki N, Watanabe M, Mataki
H, Kumamoto T, Takagi K, Mizuno K and Inoue H: Regulation of LOXL2
and SERPINH1 by antitumor microRNA-29a in lung cancer with
idiopathic pulmonary fibrosis. J Hum Genet. 61:985–993. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mizuno K, Seki N, Mataki H, Matsushita R,
Kamikawaji K, Kumamoto T, Takagi K, Goto Y, Nishikawa R, Kato M, et
al: Tumor-suppressive microRNA-29 family inhibits cancer cell
migration and invasion directly targeting LOXL2 in lung squamous
cell carcinoma. Int J Oncol. 48:450–460. 2016. View Article : Google Scholar :
|
|
15
|
Mataki H, Seki N, Mizuno K, Nohata N,
Kamikawaji K, Kumamoto T, Koshizuka K, Goto Y and Inoue H:
Dual-strand tumor-suppressor microRNA-145 (miR-145-5p and
miR-145-3p) coordinately targeted MTDH in lung squamous cell
carcinoma. Oncotarget. 7:72084–72098. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mataki H, Seki N, Chiyomaru T, Enokida H,
Goto Y, Kumamoto T, Machida K, Mizuno K, Nakagawa M and Inoue H:
Tumor-suppressive microRNA-206 as a dual inhibitor of MET and EGFR
oncogenic signaling in lung squamous cell carcinoma. Int J Oncol.
46:1039–1050. 2015. View Article : Google Scholar
|
|
17
|
Fukumoto I, Kikkawa N, Matsushita R, Kato
M, Kurozumi A, Nishikawa R, Goto Y, Koshizuka K, Hanazawa T,
Enokida H, et al: Tumor-suppressive microRNAs (miR-26a/b,
miR-29a/b/c and miR-218) concertedly suppressed
metastasis-promoting LOXL2 in head and neck squamous cell
carcinoma. J Hum Genet. 61:109–118. 2016. View Article : Google Scholar
|
|
18
|
Kurozumi A, Kato M, Goto Y, Matsushita R,
Nishikawa R, Okato A, Fukumoto I, Ichikawa T and Seki N: Regulation
of the collagen cross-linking enzymes LOXL2 and PLOD2 by
tumor-suppressive microRNA-26a/b in renal cell carcinoma. Int J
Oncol. 48:1837–1846. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koshizuka K, Nohata N, Hanazawa T, Kikkawa
N, Arai T, Okato A, Fukumoto I, Katada K, Okamoto Y and Seki N:
Deep sequencing-based microRNA expression signatures in head and
neck squamous cell carcinoma: Dual strands of pre-miR-150 as
antitumor miRNAs. Oncotarget. 8:30288–30304. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Goto Y, Kurozumi A, Arai T, Nohata N,
Kojima S, Okato A, Kato M, Yamazaki K, Ishida Y, Naya Y, et al:
Impact of novel miR-145-3p regulatory networks on survival in
patients with castration-resistant prostate cancer. Br J Cancer.
117:409–420. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Itesako T, Seki N, Yoshino H, Chiyomaru T,
Yamasaki T, Hidakaå H, Yonezawa T, Nohata N, Kinoshita T, Nakagawa
M, et al: The microRNA expression signature of bladder cancer by
deep sequencing: The functional significance of the miR-195/497
cluster. PLoS One. 9:e843112014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mirsadraee S, Oswal D, Alizadeh Y, Caulo A
and van Beek E Jr: The 7th lung cancer TNM classification and
staging system: Review of the changes and implications. World J
Radiol. 4:128–134. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Koshizuka K, Hanazawa T, Fukumoto I,
Kikkawa N, Matsushita R, Mataki H, Mizuno K, Okamoto Y and Seki N:
Dual-receptor (EGFR and c-MET) inhibition by tumor-suppressive
miR-1 and miR-206 in head and neck squamous cell carcinoma. J Hum
Genet. 62:113–121. 2017. View Article : Google Scholar
|
|
24
|
Okato A, Goto Y, Kurozumi A, Kato M,
Kojima S, Matsushita R, Yonemori M, Miyamoto K, Ichikawa T and Seki
N: Direct regulation of LAMP1 by tumor-suppressive microRNA-320a in
prostate cancer. Int J Oncol. 49:111–122. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kojima S, Chiyomaru T, Kawakami K, Yoshino
H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, et
al: Tumour suppressors miR-1 and miR-133a target the oncogenic
function of purine nucleoside phosphorylase (PNP) in prostate
cancer. Br J Cancer. 106:405–413. 2012. View Article : Google Scholar :
|
|
26
|
Turunen SP, Tatti-Bugaeva O and Lehti K:
Membrane-type matrix metalloproteases as diverse effectors of
cancer progression. Biochim Biophys Acta. 1864(11 Pt A): 1974–1988.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Maione P, Sgambato A, Casaluce F, Sacco
PC, Santabarbara G, Rossi A and Gridelli C: The role of the
antiangiogenetic ramucirumab in the treatment of advanced non small
cell lung cancer. Curr Med Chem. 24:3–13. 2017. View Article : Google Scholar
|
|
28
|
Wang F, Ren X and Zhang X: Role of
microRNA-150 in solid tumors. Oncol Lett. 10:11–16. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu Q, Jin H, Yang Z, Luo G, Lu Y, Li K,
Ren G, Su T, Pan Y, Feng B, et al: miR-150 promotes gastric cancer
proliferation by negatively regulating the pro-apoptotic gene EGR2.
Biochem Biophys Res Commun. 392:340–345. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Huang S, Chen Y, Wu W, Ouyang N, Chen J,
Li H, Liu X, Su F, Lin L and Yao Y: miR-150 promotes human breast
cancer growth and malignant behavior by targeting the pro-apoptotic
purinergic P2X7 receptor. PLoS One. 8:e807072013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Okato A, Arai T, Kojima S, Koshizuka K,
Osako Y, Idichi T, Kurozumi A, Goto Y, Kato M, Naya Y, et al: Dual
strands of pre-miR-150 (miR-150-5p and miR-150-3p) act as antitumor
miRNAs targeting SPOCK1 in naïve and castration-resistant prostate
cancer. Int J Oncol. 51:245–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Osako Y, Seki N, Koshizuka K, Okato A,
Idichi T, Arai T, Omoto I, Sasaki K, Uchikado Y, Kita Y, et al:
Regulation of SPOCK1 by dual strands of pre-miR-150 inhibit cancer
cell migration and invasion in esophageal squamous cell carcinoma.
J Hum Genet. 62:935–944. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Desgrosellier JS and Cheresh DA: Integrins
in cancer: Biological implications and therapeutic opportunities.
Nat Rev Cancer. 10:9–22. 2010. View Article : Google Scholar
|
|
34
|
Gilcrease MZ: Integrin signaling in
epithelial cells. Cancer Lett. 247:1–25. 2007. View Article : Google Scholar
|
|
35
|
Shu YJ, Weng H, Ye YY, Hu YP, Bao RF, Cao
Y, Wang XA, Zhang F, Xiang SS, Li HF, et al: SPOCK1 as a potential
cancer prognostic marker promotes the proliferation and metastasis
of gallbladder cancer cells by activating the PI3K/AKT pathway. Mol
Cancer. 14:122015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kim HP, Han SW, Song SH, Jeong EG, Lee MY,
Hwang D, Im SA, Bang YJ and Kim TY: Testican-1-mediated
epithelial-mesenchymal transition signaling confers acquired
resistance to lapatinib in HER2-positive gastric cancer. Oncogene.
33:3334–3341. 2014. View Article : Google Scholar
|
|
37
|
Miao L, Wang Y, Xia H, Yao C, Cai H and
Song Y: SPOCK1 is a novel transforming growth factor-β target gene
that regulates lung cancer cell epithelial-mesenchymal transition.
Biochem Biophys Res Commun. 440:792–797. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang YZ, Wu KP, Wu AB, Yang ZC, Li JM, Mo
YL, Xu M, Wu B and Yang ZX: MMP-14 overexpression correlates with
poor prognosis in non-small cell lung cancer. Tumour Biol.
35:9815–9821. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lu H, Hu L, Yu L, Wang X, Urvalek AM, Li
T, Shen C, Mukherjee D, Lahiri SK, Wason MS, et al: KLF8 and FAK
cooperatively enrich the active MMP14 on the cell surface required
for the metastatic progression of breast cancer. Oncogene.
33:2909–2917. 2014. View Article : Google Scholar :
|
|
40
|
Hagemann T, Gunawan B, Schulz M, Füzesi L
and Binder C: mRNA expression of matrix metalloproteases and their
inhibitors differs in subtypes of renal cell carcinomas. Eur J
Cancer. 37:1839–1846. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Gawden-Bone C, Zhou Z, King E, Prescott A,
Watts C and Lucocq J: Dendritic cell podosomes are protrusive and
invade the extracellular matrix using metalloproteinase MMP-14. J
Cell Sci. 123:1427–1437. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gálvez BG, Matías-Román S, Yáñez-Mó M,
Sánchez-Madrid F and Arroyo AG: ECM regulates MT1-MMP localization
with beta1 or alphavbeta3 integrins at distinct cell compartments
modulating its internalization and activity on human endothelial
cells. J Cell Biol. 159:509–521. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Munshi HG and Stack MS: Reciprocal
interactions between adhesion receptor signaling and MMP
regulation. Cancer Metastasis Rev. 25:45–56. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Cho SH, Park YS, Kim HJ, Kim CH, Lim SW,
Huh JW, Lee JH and Kim HR: CD44 enhances the epithelial-mesenchymal
transition in association with colon cancer invasion. Int J Oncol.
41:211–218. 2012.PubMed/NCBI
|
|
45
|
Zarrabi K, Dufour A, Li J, Kuscu C,
Pulkoski-Gross A, Zhi J, Hu Y, Sampson NS, Zucker S and Cao J:
Inhibition of matrix metalloproteinase 14 (MMP-14)-mediated cancer
cell migration. J Biol Chem. 286:33167–33177. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hojilla CV, Mohammed FF and Khokha R:
Matrix metalloproteinases and their tissue inhibitors direct cell
fate during cancer development. Br J Cancer. 89:1817–1821. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Li Y, Kuscu C, Banach A, Zhang Q,
Pulkoski-Gross A, Kim D, Liu J, Roth E, Li E, Shroyer KR, et al:
miR-181a-5p inhibits cancer cell migration and angiogenesis via
downregulation of matrix metalloproteinase-14. Cancer Res.
75:2674–2685. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zuo QF, Cao LY, Yu T, Gong L, Wang LN,
Zhao YL, Xiao B and Zou QM: MicroRNA-22 inhibits tumor growth and
metastasis in gastric cancer by directly targeting MMP14 and Snail.
Cell Death Dis. 6:e20002015. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Osako Y, Seki N, Kita Y, Yonemori K,
Koshizuka K, Kurozumi A, Omoto I, Sasaki K, Uchikado Y, Kurahara H,
et al: Regulation of MMP13 by antitumor microRNA-375 markedly
inhibits cancer cell migration and invasion in esophageal squamous
cell carcinoma. Int J Oncol. 49:2255–2264. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Yonemori M, Seki N, Yoshino H, Matsushita
R, Miyamoto K, Nakagawa M and Enokida H: Dual tumor-suppressors
miR-139-5p and miR-139-3p targeting matrix metalloprotease 11 in
bladder cancer. Cancer Sci. 107:1233–1242. 2016. View Article : Google Scholar : PubMed/NCBI
|