Introduction
Pancreatic cancer (PC) is a highly malignant tumor
that is characterized by early metastasis. In Western countries, PC
is one of the leading causes of cancer-related mortality (1). Researchers have found that
KRAS oncogene point mutations occur in >95% of patients
with PC (2). Genetic alterations
of oncogenes, such as KRAS and HER-2, and tumor
suppressor genes, such as p53, p16 and SMAD4,
are the main molecular alterations associated with the occurrence
and development of PC (3). Among
these, KRAS oncogene point mutations occur in the early
stages of tumor development, as well as throughout the growth
stages. Oncogenic KRAS promotes pancreatic tumorigenesis
through the activation of multiple downstream pathways, including
phosphatidylinositol-3-kinase, extracellular signal-regulated
kinase and nuclear factor-κB. Since these molecules significantly
affect cell proliferation and differentiation, the timely and
effective treatment of PC through the inhibition of KRAS in
the early stages of tumor development is critical.
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
consisting of 20-22 nucleotides (nt) (4) that play a key role in tumor
development and progression (5).
miRNAs have been found to be differentially expressed at various
stages of PC progression (6).
miRNAs have been previously found to play a key role in the
regulation of KRAS. In particular, miR-193b was verified as
a direct inhibitor of KRAS, resulting in the modulation of
Akt and extracellular signal-regulated kinase pathways and in the
suppression of pancreatic ductal adenocarcinoma (PDAC) cell growth
(7). miR-206 has also been found
to directly target KRAS and to function as a tumor
suppressor in PDAC cells by blocking cell cycle progression,
proliferation, migration and invasion (8), while miR-143 expression has been
found to significantly decrease KRAS mRNA and protein levels
(9). KRAS has also been
identified as a potential direct target of miR-217, which acts as a
tumor suppressor in PDAC cells (10). Furthermore, miR-96 has been shown
to regulate tumorigenesis through the inhibition of cell
proliferative and invasive activities by targeting KRAS
(11).
Long non-coding RNAs (lncRNAs) are an important
class of non-coding RNAs >200 nt in length (12), that rarely participate in protein
coding, but are involved in epigenetic and transcriptional
regulation, as well as in various other levels (13). Recent studies have found that
lncRNAs play a central role in the regulation of a wide range of
biological processes (14,15) and are closely associated with a
number of human diseases, including several types of cancer
(16-18). Furthermore, several differentially
expressed lncRNAs have been identified in primary and metastatic PC
(19); thus, lncRNAs may be
associated with PC metastasis. Kim et al demonstrated that
HOTAIR functions as an oncogene in PC and is associated with
a poor prognosis (20). A previous
microarray analysis revealed that HOTTIP was one of the most
significantly upregulated lncRNAs in PDAC tissues compared with
adjacent normal pancreatic tissues (21), whereas lncRNAs MALAT1 and
HULC have been shown to be associated with a poor prognosis
in PC (22-24). However, there is currently little
information on the association between lncRNAs and KRAS, at
least to the best of our knowledge.
MS2 capsid (coat) protein is the envelope protein
found on bacteriophage MS2 that specifically interacts with MS2
binding sites (MS2bs), which are RNA stem-loop structures composed
of 19 base pairs (25). MS2
protein-MS2bs RNA binding has been used to study intra- and
extracellular RNA-protein interactions (26). Some researchers have even made 12
MS2bs binding sites in series (27). Zhou et al used MS2-MBP to
purify RNA-protein complexes in vitro (28). A pMAL protein expression vector
system was previously used to induce the expression of a target
protein. MBP fusion proteins can enhance the solubility of target
proteins, and MBP can bind with resins. Corcoran et al and
others have utilized an MS2-maltose binding protein (MBP) fusion
protein and resin for affinity chromatography analysis of
RNA-protein complexes and the identification of binding proteins
(29,30). Bardwell et al used MS2
capsid protein and MS2bs structure to purify RNA-protein complexes
(31). SenGupta et al
established an MS2 capsid protein-based yeast RNA 3-hybrid system,
which can identify RNA-protein interactions (32).
The importance of the target genes regulated by
non-coding RNA molecules reflects the importance of these small RNA
molecules. Since KRAS mutations are one of the most
significant genetic alterations in PC, the identification of
non-coding RNAs associated with KRAS regulation is crucial
to the study of the pathogenesis of PC. In this study, we
constructed an expression vector that expresses KRAS
non-coding region coupled with 12 copies of MS2bs in series, and
established a high-throughput library for collecting miRNA- and
lncRNA-omics that regulate KRAS. To the best of our
knowledge, this is the first study to combine RNA-protein
interactions with RNA sequencing to obtain KRAS-associated
non-coding RNAs. We successfully constructed the
pcDNA3.1-KRAS-12xMS2bs plasmid, and used pMAL-c5x-MS2 vectors to
induce the expression of MS2-MBP fusion protein. Subsequently, RNA
that was associated with KRAS was obtained through affinity
chromatography resin experiments. Finally, the miRNA and lncRNA
data of interest were obtained through next-generation RNA
sequencing. We identified a number of miRNA precursors that belong
to the let-7 and miR-34, -30 and -143 families, as well as relevant
lncRNAs and their families (MALAT1, MEG3_2 and TUG1_1-4). Our
databank of non-coding RNAs (mainly miRNAs and lncRNAs) that
regulate KRAS is expected to greatly enhance our
understanding of KRAS regulation-associated tumorigenesis
and may thus aid in the design of novel gene therapies for PC.
Materials and methods
pcDNA3.1-KRAS-12xMS2bs plasmid vector
construction
The MS2 binding site sequence of the 19-nt stem-loop
was ACATGAGGATCACCCATGT. The
sequences were sythesized by Beijing PolePolar Biotechnology Co.,
Ltd. (Beijing, China). The underlined letters in the sequences
indicate MS2 binding sites (MS2bs), which are RNA stem-loop
structures composed of 19 base pairs (26). First, the following sequences were
synthesized: 12xMS2bs-A, 5′-AGCTTGGTACCGAGC TCGGATCCACATGAGGATCACCCATGTACATGAGGATCACCCATGTGAGACCATCCACTAGTCTCGAGTCTAGA
GGGCCCGT-3′; 12 × MS2bs-B, 5′-TAAGCGTCTCACATGAGGATCACCCATGTACATGAGGATCACCCATGTGAGACCCTAGAAGCTCGAGTTA-3′;
12xMS2bs-C, 5′-TAAGCGTCTCCCATGTACATGAGGATCACCCATGTACATGAGGATCACCCATGTCTCGAGTCTAGAGGGCCCGTTTAAACCCGC-3′.
The synthesized sequences were then cloned into a T vector
(Promega, Madison, WI, USA) lacking the BsaI, BsmBI
and XhoI restriction sites and sequenced. 12xMS2bs-B was
digested with BsmBI and XhoI and the fragment was
recovered. The BsaI/XhoI-digested 12xMS2bs-B
fragments were ligated to 12xMS2bs-A, which was digested with
BsaI and XhoI and then sequenced to obtain
12xMS2bs-A4; another BsaI/XhoI-digested the
12xMS2bs-B vector to obtain 12xMS2bs-B4 and the 12xMS2bs-A4 vector
to obtain 12xMS2bs-A6. The 12xMS2bs-C was ligated to 12xMS2bs-B4
and sequenced to obtain 12 × MS2bs-C6, which was ligated to
12xMS2bs-A6 to produce 12xMS2bs-T. Then, 12xMS2bs-T was digested
with BamHI and XhoI to obtain the 12xMS2bs fragment,
which was connected to the pcDNA3.1(+) plasmid (Invitrogen/Thermo
Fisher Scientific, Waltham, MA USA) to obtain the final sequence.
The 12xMS2bs-C6 fragment was also ligated to the 12xMS2bs-B4 vector
and sequenced to produce 12xMS2bs-C10 for future use. The untreated
group was not treated with any transfection reagents and no
transfection plasmid are added. NC was the negative control, a
meaningless control of sequence disruption, and the pcDNA3.1 vector
was used here. MOCK was the blank control, and only transfection
reagents were added, and no transfection plasmid was added.
3′-UTR1-KRAS-12xMS2bs,
3′-UTR2-KRAS-12xMS2bs, 3′-UTR3-KRAS-12xMS2bs, and
3′-UTR4-KRAS-12xMS2bs construction
The KRAS 3′-UTR is very long; thus, to make
it more readily transfectable into PC cells in order to
successfully express RNA, the KRAS 3′-UTR was divided into 4
sections. Subsequently, the abovementioned 12xMS2bs sequences were
cloned into a T vector and sequenced. To obtain the 10xMS2bs
fragment, 12xMS2bs-C10 was digested with BsmBI and
XhoI, and then connected to plasmid B mentioned above to
produce Fragment II. Fragments I and II were amplified by
polymerase chain reaction (PCR) and finally integrated together by
PCR. The integrated fragment was then cloned into a T vector to
obtain the desired target sequence. The target sequence was then
connected to the pcDNA3.1 plasmid to obtain the desired recombinant
plasmid. Human pancreatic adenocarcinoma cells (PANC-1) was
obtained from ATCC (Manassas, VA, USA). The cells were cultured in
appropriate medium (Mediatech, Manassas, VA, USA) containing 10%
fetal bovine serum (Thermo Fisher Scientific) and maintained at
37°C and 5% CO2.
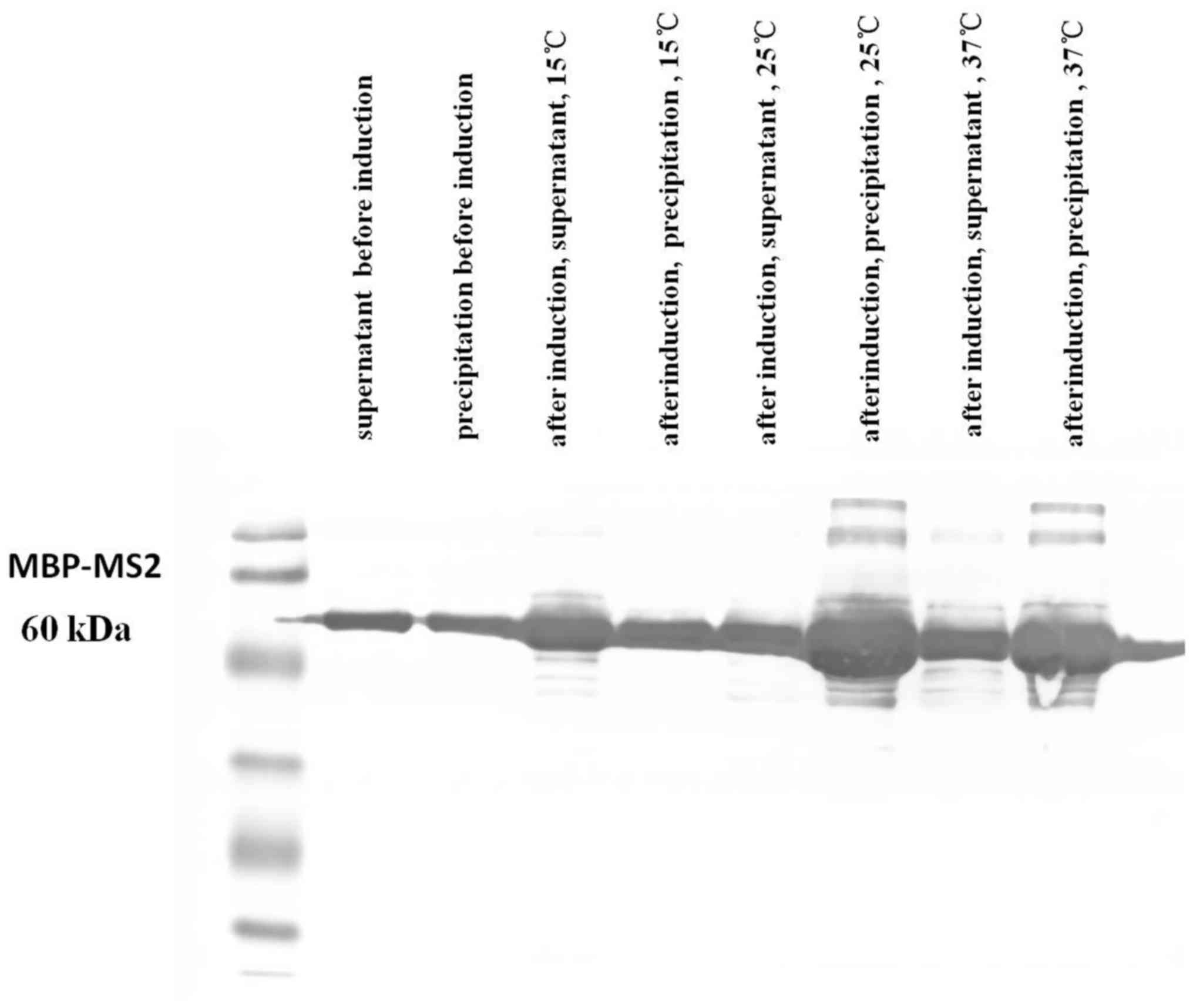
MBP-MS2 fusion protein expression
The PMAL-c5xpMAL vector [New England Biolabs (NEB),
Ipswich, MA, USA) was used for the expression and purification of
fusion proteins. Using the strong promoter ‘Ptac’ and the MBP
translation initiation signal, the target gene (MS2)-malE
(encodes MBP protein) fusion protein was expressed. A small amount
of the recombinant pMAL-c5x plasmid was transformed into
Escherichia coli DE3 (Tiangen Biotech, Beijing, China),
coated on plates and incubated overnight. We selected and collected
a single bacteria which was gently agitated in vitro. The
bacteria was then vigorously shaken. When the optical density (OD)
value was 0.6, isopropyl-β-D-1-thiogalactopyranoside (IPTG) was
added to induce fusion protein expression. The bacterial cells were
collected by centrifugation (15,000 × g, 4°C) and disrupted with
lysis buffer, then centrifuged (15,000 × g, 4°C) again. The
supernatant was collected for further purification with linear
starch and finally rinsed with maltose-containing column eluent to
obtain the MBP-MS2 fusion protein.
Affinity chromatography
We transfected the KRAS-12xMS2bs plasmid into the
PANC-1 cells using Lipofectamine 2000 reagent (Thermo Fisher
Scientific) and total RNA was extracted after 48 h. The column
(#732-6008; Bio-Rad, Hercules, CA, USA) was infused with resin and
washed several times with washing buffer. Subsequently,
KRAS-12xMS2bs RNA and the MBP-MS2 fusion protein were added to the
column followed by incubation on ice for 30 min. The column was
then washed with eluent containing maltose (Bioroyee Biotech,
Beijing, China) and KRAS-12xMS2bs-MS2-MBP complexes were obtained.
Finally, RNA was separated with chloroform and isopropanol to
successfully obtain KRAS-associated RNA.
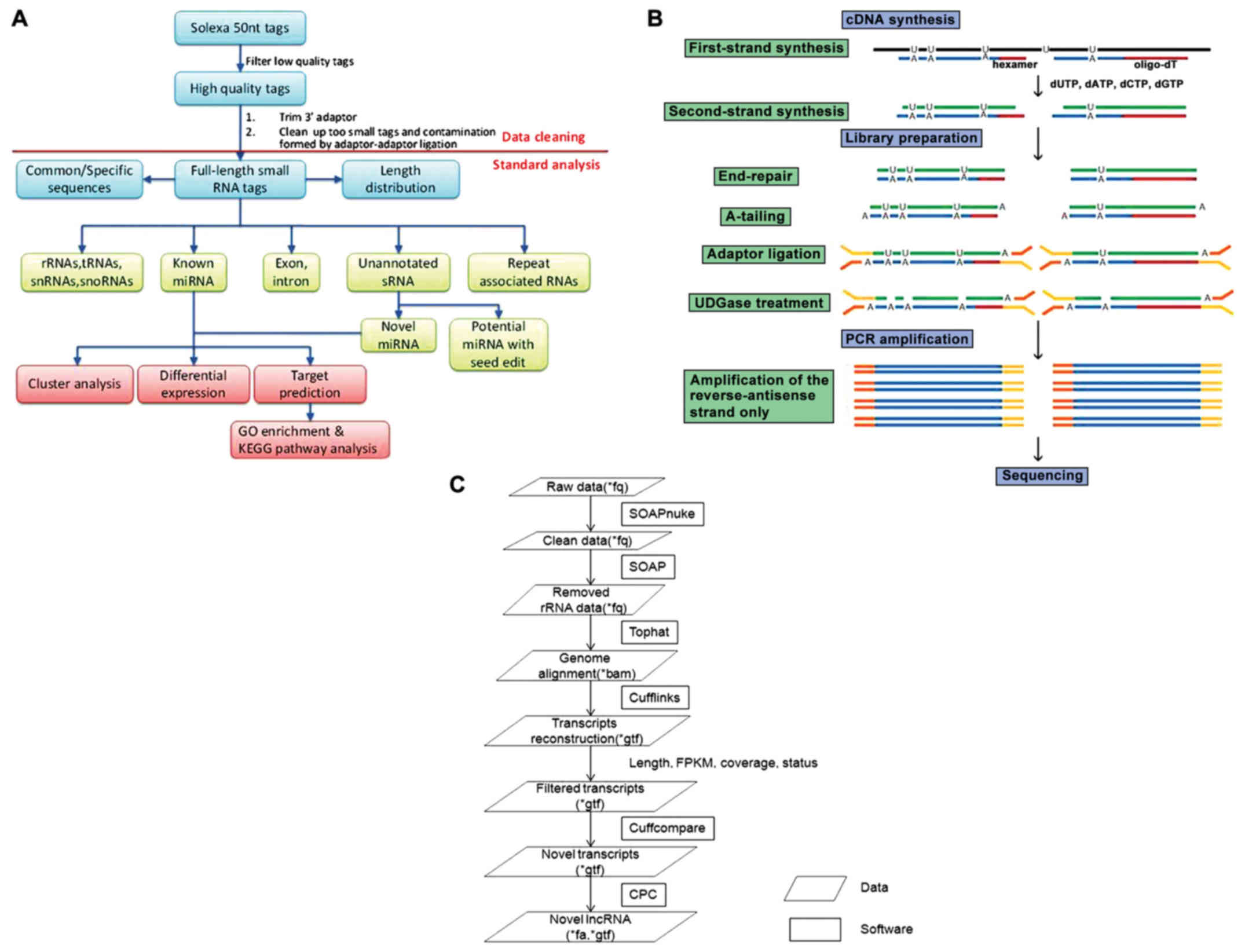
Small RNA sequencing
All sequencing procedures were performed at the
Beijing Genomics Institute (Beijing, China). Following the removal
of adaptor sequences, low-quality tags and contaminants, clean
reads were used for bioinformatics analysis. The small RNA tags
were mapped to the human genome to analyze their expression and
distribution. Subsequently, we screened against the Rfam 10.1 and
GenBank databases to remove the fragments of ribosomal RNA (rRNA),
transfer RNA (tRNA), small nuclear RNA (snRNA) and small nucleolar
RNA (snoRNA). After eliminating repeat-associated small RNA (sRNA),
the degradation fragments of mRNA and identifying conserved miRNAs,
the reads that did not match the above-mentioned databases were
predicted using MIREAP (BGI, Beijing Genomics Institute, Beijing,
China) (Fig. 1A).
lncRNA sequencing
After extracting total RNA from the samples, mRNA
and non-coding RNA were enriched by removing the rRNA with a kit
(BGI, Beijing Genomics Institute); mRNA could also be removed by
removing poly-A RNAs, since lncRNAs also have a poly-A tail. Using
fragmentation buffer (BGI, Beijing Genomics Institute), mRNA and
non-coding RNA were fragmented into short fragments (~200-500 nt),
then first-strand cDNA was synthesized by random hexamer primers
using the fragments as templates, and dTTP was substituted by dUTP
during the synthesis of the second strand. Short fragments were
purified and resolved with Elution Buffer (BGI, Beijing Genomics
Institute) for end reparation and single adenine addition.
Subsequently, the short fragments were connected with adapters and
the second strand degraded using uracil-N-glycosylase. Following
agarose gel electrophoresis, suitable fragments were selected for
PCR amplification as templates. During the quality control (QC)
steps, the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa
Clara, CA, USA) and ABI StepOnePlus Real-Time PCR System (Applied
Biosystems/Thermo Fisher Scientific) were used for the
quantification and qualification of the sample library. Finally,
the library was sequenced using an Illumina HiSeq™ 2000 or another
sequencer when necessary (Fig.
1B). All sequencing procedures were performed at the Beijing
Genomics Institute.
Novel miRNA prediction
miRNA hairpins are mostly located in intergenic
regions, introns, or reverse repeat sequences of exons. The
characteristic hairpin structure of miRNA precursors may be used to
predict novel miRNAs. MIREAP prediction software (http://sourceforge.net/projects/mireap/)
was used to predict novel miRNAs by exploring secondary structure,
the Dicer cleavage site and the minimum free energy of the
unannotated small RNA tags that could be mapped to the genome. Key
search and selection conditions were as follows: i) The tags used
to predict novel miRNA were from unannotated tags, which match the
reference genome, align to intronic regions, or align to antisense
exonic regions; ii) sequences and structures of hairpin miRNA able
to fold into appropriate secondary structures and containing mature
miRNA in one arm of the hairpin precursors were considered as
candidate miRNA genes; iii) both the mature miRNA strand and its
complement had 2-nt 3′-overhangs; iv) hairpin precursors lacking
large internal loops or bulges; v) stable hairpin secondary
structures with a free energy of hybridization ≤−18 kcal/mol; and
vi) alignment results showed at least 5 mature miRNAs with
predicted hairpins.
lncRNA bioinformatics analysis of
single-sample predictions
Primary sequencing data produced by Illumina Hiseq™
2000, referred to as ‘raw reads’, which were filtered into ‘clean
reads’ by removing adaptors, contained and low-quality reads. Due
to unstable rRNA removal efficiency, it was also necessary to
remove the rRNA-containing reads by alignment. We utilized
reference annotation-based assembly to reconstruct transcripts,
while background noise was reduced using fragments per kilobase of
transcript per million mapped reads (FPKM) and coverage threshold.
Novel transcripts were detected through comparison with a
reference, and the coding potential of these transcripts was then
calculated to identify the novel lncRNA. Since lncRNAs may be
processed to yield small RNAs, small RNA precursor prediction was
also performed. lncRNAs were also classified by family, RNA
structure and sequence similarities, all of which helped reveal the
potential function of the lncRNAs (Fig. 1C).
Detecting non-coding RNA molecule
effects
We introduced exogenous miRNA molecules to PC cells
and then isolated RNA to detect the effect (inhibition/activation)
of the non-coding RNA molecules acting on KRAS sequences by
reverse transcription-quantitative PCR (RT-qPCR) and western blot
analyses. miRNA mimics of highly expressed small fragments matching
several miRNA precursors (hsa-let-7a, hsa-let-7i and hsa-miR-30b)
were synthesized (Shanghai GenePharma Co., Ltd., Shanghai, China)
and transfected into the PC cells. For quantitative PCR, total RNA
was isolated from the PC cells using TRIzol reagent
(Invitrogen/Thermo Fisher Scientific), according to the
manufacturer’s instructions. Quantitative PCR assay was performed
with SYBR-Green PCR Master Mix using a StepOnePlus Detection System
(Applied Biosystems/Thermo Fisher Scientific) to detect the KRAS
mRNA expression levels. The KRAS primers used in this study are
listed as follows: forward, 5′-GACTCTGAAGAT GTACCTATGGTCCTA-3′ and
reverse, 5′-CATCATCAACA CCCTGTCTTGTC-3′. For the reaction system, a
96-well plate was used, and each sample was set to 3 replicate
wells with a total volume of 20 µl per well. After the
addition was completed, the samples were centrifuged at 2°C, 800 ×
g for 2 min. The reaction procedure was carried out on a
StepOnePlus Real Time PCR System instrument (Applied Biosystems/
Thermo Fisher Scientific) under the following reaction conditions:
Holding stage, 95°C for 1 min; cycling stage, 95°C for 15 sec, 60°C
for 1 min, 72°C for 30 sec, 40 cycles; melt curve stage, 95°C for
15 sec, 55°C for 1 min, 95°C for 15 sec. Data analysis of the
amplification results was performed using ABI’s StepOne Software
v2.2. Quantification was performed using the 2−ΔΔCq
method (33).
Western blot analysis was performed as
follows: Primary antibodies against the following proteins were
used in this study: KRAS and β-actin
The antibodies against KRAS (ab180772, dilution:
1/200) and β-actin (ab8227, dilution: 1/1,000) were purchased from
Abcam Biotechnology Inc., and the other antibodies were purchased
from Cell Signaling Technology). The cells were digested with 0.25%
trypsin at 48 h prior to transfection. RIPA lysate (pre-added
protease and phosphatase inhibitor) was added to the tube, and
lysed on ice for 1 h, during which the EP tube was inverted every
10 min to promote cell lysis. This was followed by centrifugation
for 30 min at 4°C, 14,000 × g. The supernatant was then carefully
pipetted into a new EP tube, and a small amount was dispensed for
protein concentration determination. The protein concentration was
determined using the BCA Protein Assay kit. The protein
concentration standard curve was prepared based on the protein
standard concentration and the absorbance value. The protein
concentration of the sample was calculated based on the protein
concentration standard curve and the sample dilution factor. A
suitable concentration of separation gel was prepared and added to
the glass interlayer immediately after mixing. Deionized water was
added to the upper layer of the glue and polymerized at room
temperature for 30 min. The ionized water was removed after
condensation. The concentrated glue liquid was then added to the
glass interlayer. The Teflon comb was inserted into the laminated
glue liquid, and polymerization was carried out at room temperature
for 30 min. The protein sample was mixed with the loading buffer
and incubated at 95°C for 5 min. The protein sample was aspirated
and electrophoresis was performed at a loading of 50-100
µg/well. The PVDF membrane was cut to the appropriate size
and the upper left corner was cut as a marker. The PVDF membrane
was equilibrated in methanol for 5 min, and then equilibrated in 1X
transfection buffer. The current intensity and film transfer time
were adjusted according to the molecular weight of the protein, the
thickness of the gel and the pore size of the PVDF membrane. The
parameter settings were: 200 mA ice transfer for 2 h; or 80 mA,
4°C, transfer overnight. The transfer PVDF membrane was then
removed, and we observed whether the pre-dyed Marker was completely
transferred from the gel to the membrane; the PVDF membrane was
then placed in 5% TBST milk at room temperature for 1 h. The
corresponding antibody was diluted in the primary antibody dilution
at an appropriate ratio, the PVDF membrane was removed from the
milk, placed in the diluted primary antibody solution, and
incubated overnight at 4°C with slow shaking. The PVDF membrane was
removed from the primary antibody and washed 5 times with 1X TBST
for 5 min each. The secondary antibody was diluted to a suitable
titer with 5% TBST milk, and the PVDF membrane was placed in a
secondary antibody for 1 h at room temperature on a shaker. The
PVDF membrane was removed from the secondary antibody and washed 4
times with 1X TBST for 5 min each time and 1X TBS for 5 min.
Subsequently, 1 ml of each of the fluorescent luminescent liquids A
and B was mixed (ECL kit, Cat. no. 32106; Pierce
Biotechnology/Thermo Fisher Scientific), dropped on a PVDF
membrane, and incubated for ~1 min. The luminescent liquid on the
surface of the PVDF film was blotted. The PVDF film was placed in a
cassette, and the X film was completed in a dark room. The Thermo
Scientific Pierce ECL Western Blotting Substrate is a highly
sensitive non-radioactive, enhanced luminol-based chemiluminescent
substrate for the detection of horseradish peroxidase (HRP) on
immunoblots. Pierce ECL Western Blotting Substrate enables the
detection of picogram amounts of antigen and allows for easy
detection of HRP using photographic or other imaging methods. Blots
can be repeatedly exposed to X-ray film to obtain optimal results.
The western blots were analyzed using Quantity One software
(Bio-Rad).
Statistical analysis
Statistical analyses were performed using SPSS 17.0
software (SPSS, Chicago, IL, USA). Multigroup comparisons of the
means were carried out by one-way analysis of variance (ANOVA) with
post hoc contrasts by the Student-Newman-Keuls test. Results are
presented as the means ± standard deviation. P-values <0.05 were
considered to indicate statistically significant differences.
Results
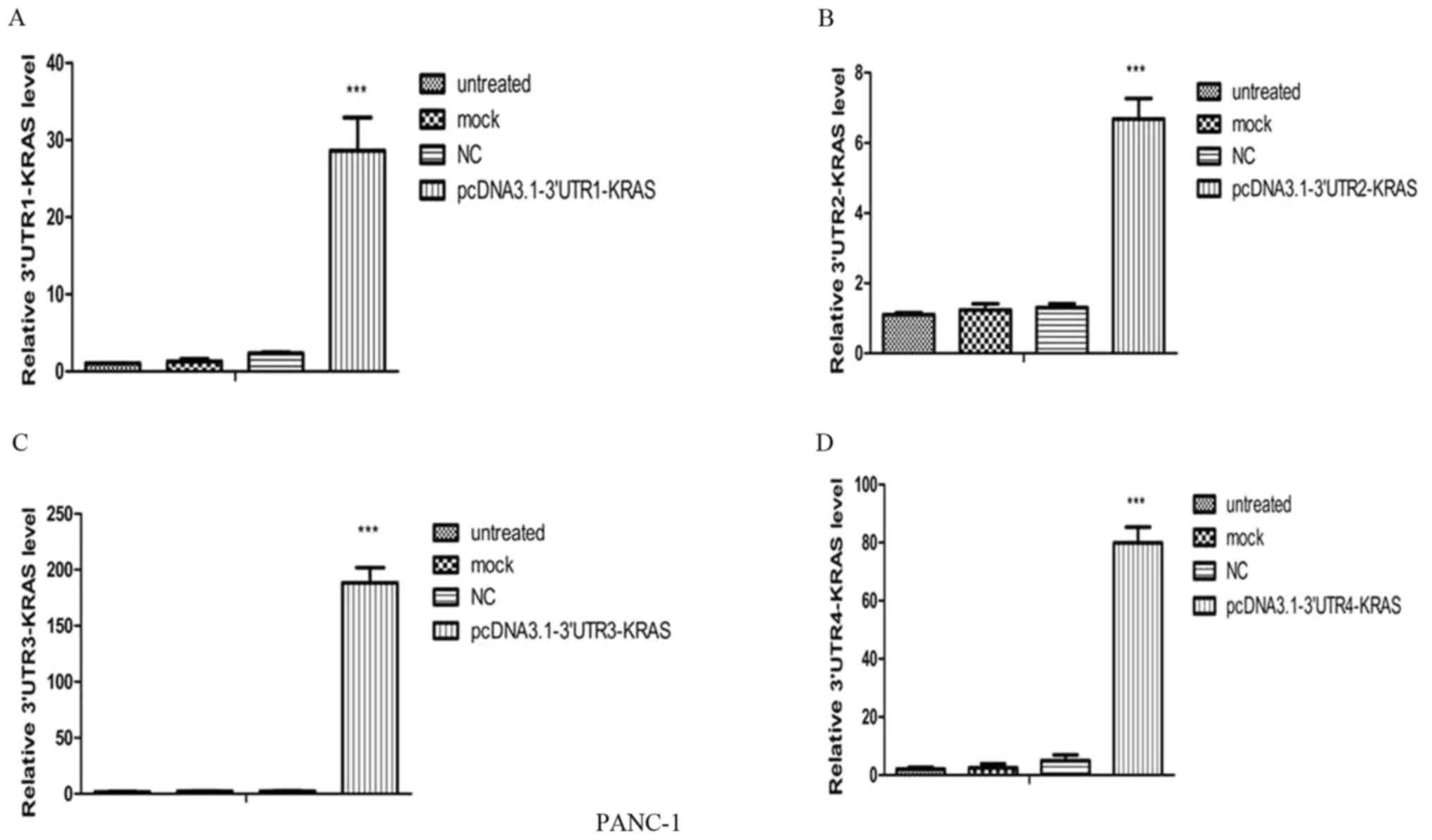
Construction of pcDNA3.1-KRAS-12xMS2bs
expression vector and KRAS-12xMS2bs RNA expression
Following the successful construction of the
pcDNA3.1-KRAS-12xMS2bs plasmid (verified by Sanger sequencing), it
was transfected into the PANC-1 cells using Lipofectamine 2000
reagent, and KRAS-12xMS2bs RNA expression was observed (Fig. 2). Compared with the untreated, mock
and negative control (NC) groups, the pcDNA3.1-3′UTR1-KRAS-12xMS2bs
(Fig. 2A),
pcDNA3.1-3′-UTR2-KRAS-12xMS2bs (Fig.
2B), pcDNA3.1-3′-UTR3-KRAS-12xMS2bs (Fig. 2C) and
pcDNA3.1-3′-UTR4-KRAS-12xMS2bs (Fig.
2D) plasmid groups successfully expressed their corresponding
3′-UTR-KRAS-12xMS2bs RNA (P<0.001). At the same time, the
successful production of the MBP-MS2 fusion protein was observed
(Fig. 3).
miRNA sequencing
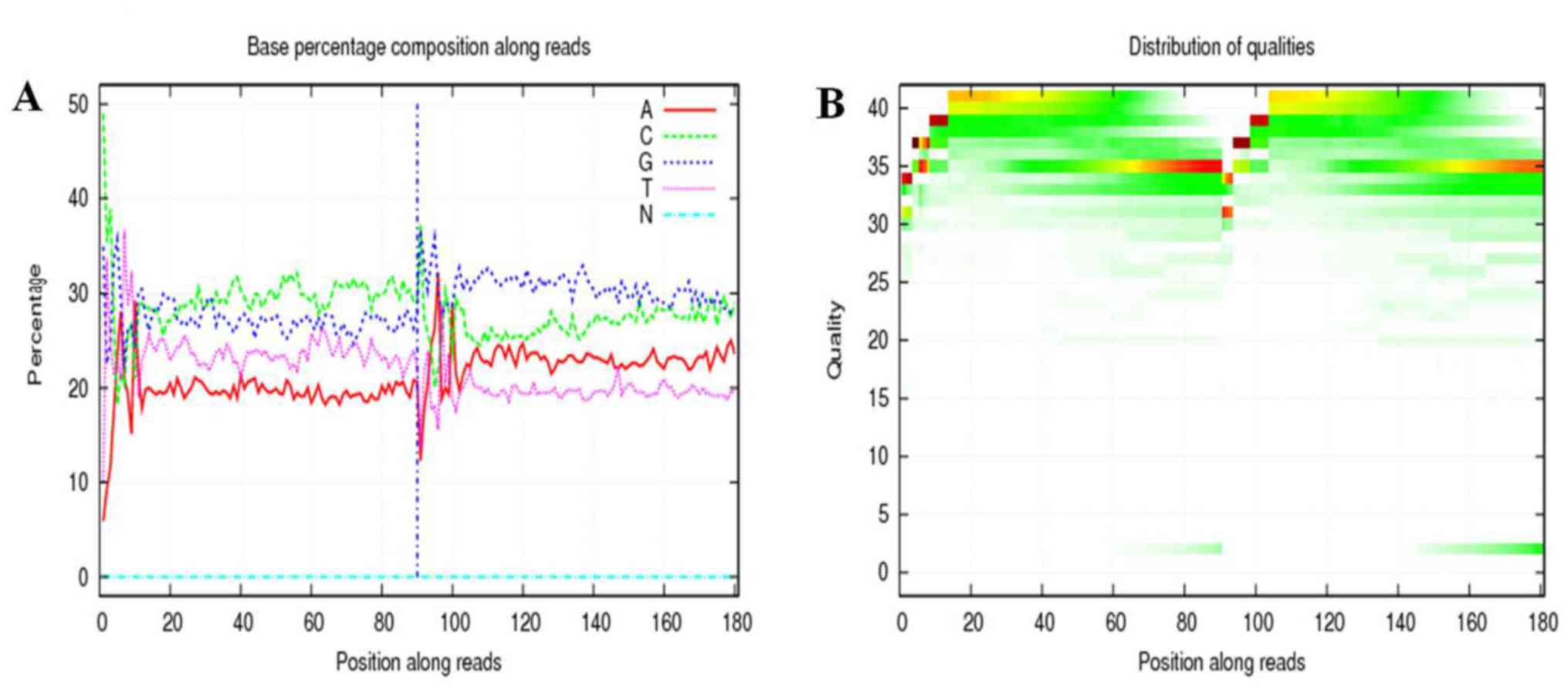
Base composition and quality value distribution maps
of the filtered data indicated the samples with balanced base
compositions. Each point represents the base quality value and
corresponding position of each read (Fig. 4A). The base mass distribution of
reads. The abscissa is the position of the base distributed over
the read, and the ordinate represents the mass of the base. Each
point in the graph represents the base quality value of the
corresponding position in a read. Overall, if the low-mass (<20)
base ratio is low, the sequencing quality of this lane is better
(Fig. 4B).
The distribution of small RNAs among different
categories [such as small cytoplasmic RNAs (scRNAs), snRNAs, rRNAs
and miRNAs] is shown in Fig. 5.
The data on the expression levels of small RNA fragments, their
sequences and precursors are presented in Table I. Family analysis revealed that the
identified miRNAs belonged to the let-7 and miR-34, -30 and -143
families (data not shown). KRAS has been shown to interact
with miRNA from these families, thereby affecting cell
proliferation, apoptosis and migration.
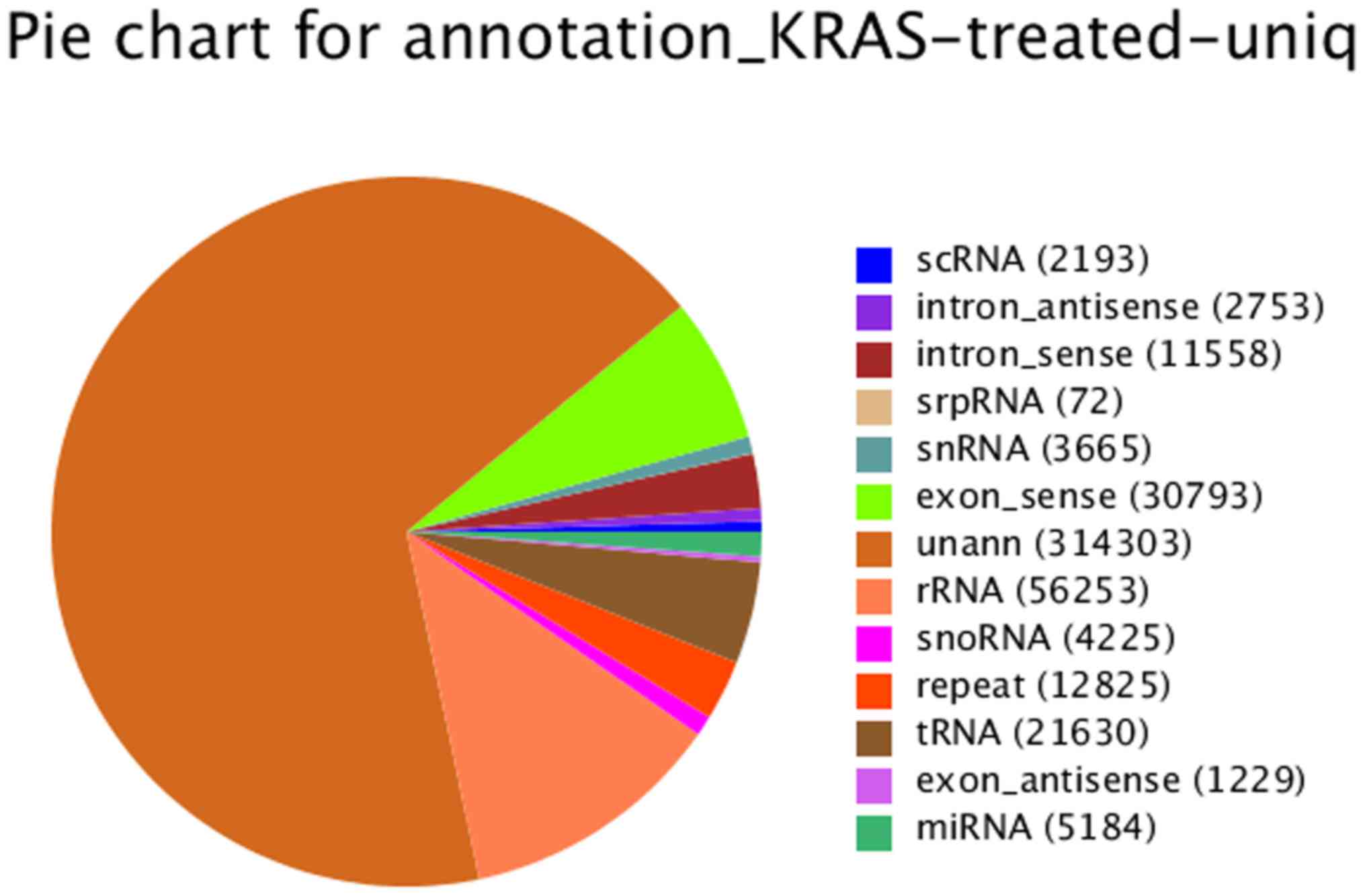 | Figure 5Distribution of small RNAs among
different categories following RNA sequencing [such as small
cytoplasmic RNAs (scRNAs), small nuclear RNAs (snRNAs), ribosomal
RNAs (rRNAs) and microRNAs (miRNAs)]. Small RNAs (sRNAs) from HiSeq
deep sequencing cover almost every kind of RNA, including miRNA,
siRNA, piRNA, rRNA, tRNA, snRNA, snoRNA, repeat associated sRNA and
degraded tags of exon or intron, of which miRNA, siRNA and piRNA
are hot topics in the research field of small RNA. |
 | Table ISmall RNA fragment expression levels,
sequences and precursors. |
Table I
Small RNA fragment expression levels,
sequences and precursors.
| Expression
level | Sequence | Precursors |
|---|
| 686268 |
5′-TCCGCCATGTTGTTGGTGG-3′ | hsa-let-7f |
| 388787 |
5′-TGAGGTAGTAGGTTGTATAGTT-3′ | hsa-let-7a |
| 279651 |
5′-AACATTCAACGCTGTCGGTGAGT-3′ | hsa-mir-181a |
| 176094 |
5′-AAGCTGCCAGTTGAAGAACTGT-3′ | hsa-mir-22 |
| 121080 |
5′-TGTAAACATCCTCGACTGGAAGCT-3′ | hsa-mir-30a |
| 95592 |
5′-TTCACAGTGGCTAAGTTCTGC-3′ | hsa-mir-27b |
| 87762 |
5′-TGAGGTAGTAGTTTGTGCTGTT-3′ | hsa-let-7i |
| 85590 |
5′-AGCTACATCTGGCTACTGGGTCTC-3′ | hsa-mir-222 |
| 60084 |
5′-TGTAAACATCCCCGACTGGAAGCT-3′ | hsa-mir-30d |
| 58675 |
5′-TGTAAACATCCTTGACTGGAAGCT-3′ | hsa-mir-30e |
| 14729 |
5′-TGAGGTAGTAGTTTGTACAGTT-3′ | hsa-let-7g |
| 9400 |
5′-AGAGGTAGTAGGTTGCATAGTT-3′ | hsa-let-7d |
| 7859 |
5′-AACATTCAACGCTGTCGGTGA-3′ | hsa-mir-181a |
| 6654 |
5′-TGTAAACATCCTACACTCAGCT-3′ | hsa-mir-30b |
| 2353 |
5′-TTTGGCACTAGCACATTTTTGCT-3′ | hsa-mir-96 |
miRNA prediction data also included small RNAs that
did not annotate to any database upon classification, but did
annotate to the genomic, intronic and exonic antisense strand
sequences. The 21 newly predicted miRNAs are listed in Table II. Precursor prediction resulted
in 338 unique novel miRNA precursor candidates and 6,311 total
miRNA precursor candidates. Comparison of newly predicted miRNAs to
the miRBase database revealed that a proportion belonged to the
miR-345, -330, -574 and -1243 families (data not shown).
 | Table IINew microRNA precursor statistics in
samples. |
Table II
New microRNA precursor statistics in
samples.
| Name | Nucleotide
sequence |
|---|
| t0004126 |
5′-AGGAACCTTGGAGCTTCGGCAGC-3′ |
| t0016193 |
5′-TCTGACTCCTAGTCCAGGGCT-3′ |
| t0019851 |
5′-ACCAGGCAAGAACTACTGTCT-3′ |
| t0027353 |
5′-AGGAACCTTGGAGCTTCGGCAG-3′ |
| t0031267 |
5′-CAAATGGATAGGATAACACCT-3′ |
| t0037092 |
5′-TGACTGCAGCTACTCTCCCCAAG-3′ |
| t0046425 |
5′-CAAAGCACACGGCCTGCAGAGT-3′ |
| t0050628 |
5′-TGAGTGTGTGTGTGTGAGTGTGA-3′ |
| t0057669 |
5′-TCGAGAATTGCGTTTGGACAAT-3′ |
| t0065832 |
5′-TCACTACCTGACAATACAGCAGT-3′ |
| t0067365 |
5′-CAGCCAGCAGGAGAGAGAGGGAGC-3′ |
| t0068481 |
5′-ATTCCTGTAACTGGGCCAGTTT-3′ |
| t0068961 |
5′-TTCCTGTGATGTTCCTGAGGAAG-3′ |
| t0078411 |
5′-AGGAACCTTGGAGCTTCGGCA-3′ |
| t0084668 |
5′-GGAGGAACCTTGGAGCTTCGGCA-3′ |
| t0137108 |
5′-CGCGGGGCTCCGCGGGCGGCGA-3′ |
| t0140064 |
5′-CGCGGGGCTCCGCGGGCGGCGAG-3′ |
| t0160363 |
5′-TCTGACTCCTAGTCCAGGGC-3′ |
| t0240262 |
5′-CACGATGATGACACTGAGG-3′ |
| t0299819 |
5′-TGAGTGTGTGTGTGTGAGTGTGAAC-3′ |
| t0421760 |
5′-CGGGGAGAGGTCTGGGGAA-3′ |
lncRNA-related miRNA precursor
prediction
Following Dicer and Drosha enzyme
(34,35) shearing, lncRNAs form miRNAs, which
then exert their corresponding effects. Thus, we matched our
lncRNAs to miRBase to identify potential miRNA precursors. lncRNAs
with >90% coverage were selected. MiRPara software (36) was also used to predict miRNA and
their precursors (Table III). A
total of 91 known miRNA precursors and 1,381 predicted new miRNA
precursors were identified (data not shown).
 | Table IIILong non-coding RNA (lncRNA) names
and their corresponding small RNA precursors. |
Table III
Long non-coding RNA (lncRNA) names
and their corresponding small RNA precursors.
| lncRNA | Corresponding small
RNA precursor |
|---|
| TCONS_00062686 | hsa-let-7a-1 |
| n407573 | hsa-let-7a-3 |
| n407573 | hsa-let-7b |
| TCONS_00062686 | hsa-let-7f-1 |
| TCONS_00063039 | hsa-mir-181a-2 |
| TCONS_00014041 | hsa-let-7i |
| n386735 | hsa-mir-18a |
| n386735 | hsa-mir-20a |
| n408292 | hsa-mir-22 |
| TCONS_00019276 | hsa-mir-134 |
| TCONS_00019278 | hsa-mir-154 |
| TCONS_00019274 | hsa-mir-300 |
lncRNA family prediction
lncRNA family class predictions were made based on
structural and sequence features. Rfam is a database that contains
information on various non-coding RNA families, including conserved
regions of RNA secondary structures, cis-acting RNA
elements, as well as other elements. The newly predicted lncRNA
families included MALAT1, MEG3_2, TUG1_1-4, KCNQ1OT1_2, NEAT1_3,
ZEB2_ AS1_3 and DLEU1_1 (Table
IV).
 | Table IVLong non-coding RNA (lncRNA) family
analysis. |
Table IV
Long non-coding RNA (lncRNA) family
analysis.
| Family name | lncRNA |
|---|
| MALAT1 | TCONS_00012377 |
| MEG3_2 | TCONS_00019252 |
| TUG1_1 | TCONS_00040841 |
| TUG1_2 | TCONS_00040841 |
| TUG1_3 | TCONS_00040841 |
| TUG1_4 | TCONS_00040841 |
| KCNQ1OT1_2 | TCONS_00037627 |
| NEAT1_3 | TCONS_00012376 |
| ZEB2_AS1_3 | TCONS_00022706 |
| DLEU1_1 | TCONS_00016977 |
| DLEU1_2 | TCONS_00017846 |
| DLEU2_1 | TCONS_00017847 |
| DLEU2_2 | TCONS_00017851 |
| DLEU2_3 | TCONS_00017854 |
| DLEU2_6 | TCONS_00017854 |
| FAM13A | TCONS_00037223 |
| FTX_1 | TCONS_00066659 |
| JPX_1 | TCONS_00065421 |
| NBR2 | TCONS_00025667 |
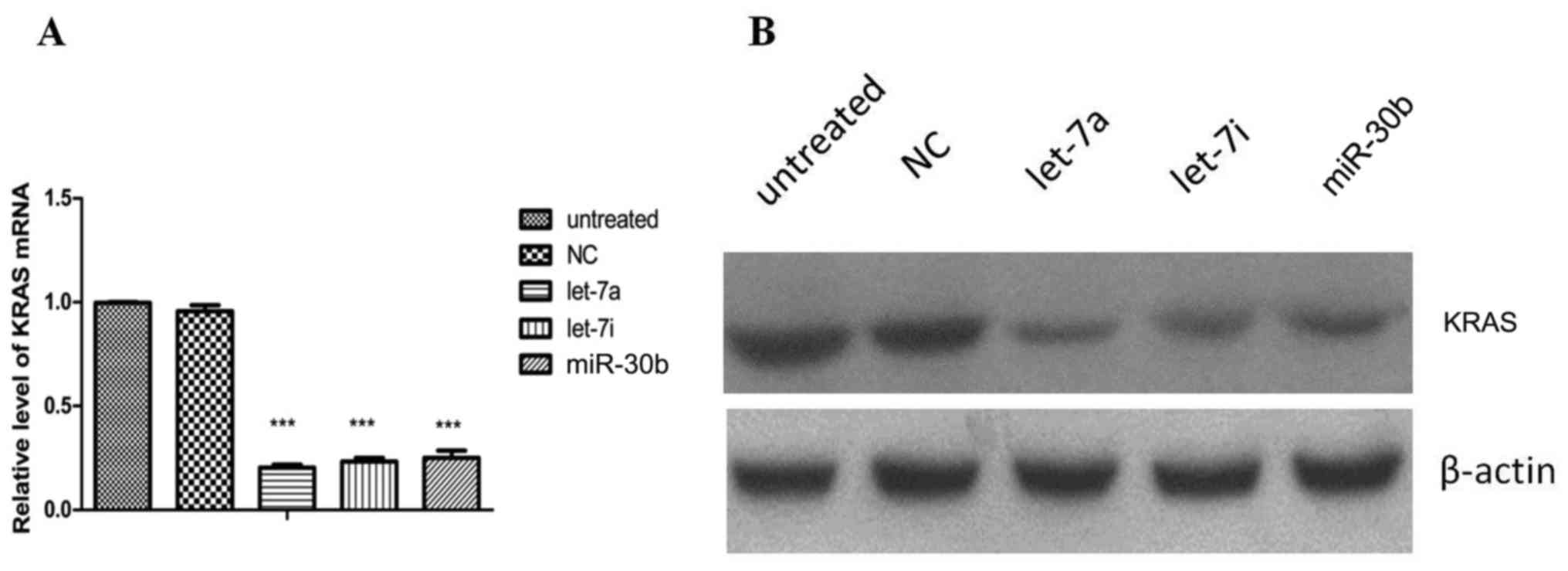
Function verification
miRNA mimics of highly expressed small fragments
matching several miRNA precursors (hsa-let-7a, hsa-let-7i and
hsa-miR-30b) were synthesized (Shanghai GenePharma Co., Ltd.) and
transfected into PC cells. The results demonstrated a decreased
KRAS mRNA and protein expression in the transfected cells (Fig. 6). Compared with the untreated and
negative control (NC) groups, KRAS mRNA (Fig. 6A) and protein (Fig. 6B) levels were both decreased in the
hsa-let-7a, hsa-let-7i and hsa-miR-30b groups.
Discussion
PC is difficult to detect in its early stages, and
metastasis often occurs long before patients develop clinical
symptoms, making surgical resection more difficult. Furthermore, PC
is highly aggressive and resistant to the majority of
chemotherapeutic agents, resulting in a high mortality rate.
KRAS, a member of the RAS gene family, is widely
known to be closely associated with PC. KRAS mutations are
the most common in malignant human cancers (37). Point mutations frequently occur at
codon 12 (GGT to GAT, GTT, or CGT) in PC cells. The mutation
detection rate is <30% in early stages of tumorigenesis,
increasing to ~100% in late-stage PC (38). Generally, KRAS mutations
lead to the abnormal activation of cell proliferation and survival
pathways, thereby playing a key role in PC development.
miR-126 and miR-143 have been shown to act as tumor
suppressors by targeting KRAS (39,40),
which may prove beneficial for patients with PC. Compared with
conventional chemotherapeutic drugs, miRNA molecules are able to
target a certain gene or genes, which would eliminate the
occurrence of serious adverse reactions or side-effects caused by
radiotherapy and chemotherapy. Compared with RNA interference
technology, miRNAs are endogenous small molecules that do not
elicit allergic or other rejection reactions in the body. During
the process of tumor development, some miRNAs are found at
decreased levels or even disappear. If their levels are increased
or restored to normal, it is possible that the malignant behavior
of tumors could be restrained or eliminated. Therefore, gene
therapy strategies involving miRNAs for the treatment of aggressive
diseases, such as PC, should be established. Using miRNAs to
inhibit target genes has many potential advantages and is expected
to become a new method for the study of gene function and a therapy
with broad applications. In the present study, we investigated the
use of miRNAs to inhibit the KRAS oncogene in order to
develop an effective gene therapy for PC.
Overall, we sequenced high levels of several miRNAs
known to target KRAS, such as let-7, miR-30 and miR-34,
demonstrating that our method of specific MS2-MBP and KRAS-MS2bs
affinity chromatography screening of associated non-coding RNAs is
quite feasible. In the present study, the highly expressed small
fragments were found to have miRNA precursors such as hsa-let-7f,
-7i, -7a and -7g, and hsa-miR-30a, -30e, -27a, -27b, -222, -10a,
-10b, -181a and -96. The analysis of known miRNA families revealed
that they belong to the let-7 and miR-34, -30 and -143 families.
The transfection of synthetic miRNA mimics of selected highly
expressed small fragments into PC cells led to a decrease in KRAS
protein levels. Johnson et al first observed that the
expression level of let-7 in lung cancer was decreased, while
RAS expression was increased, confirming the target
relationship between the two (41). Kumar et al found that the
elevated let-7 expression was able to significantly inhibit the
growth of lung cancer cells in humans and mice (42). Let-7 was also found to be
down-regulated in a number of tumors, such as breast, colon,
ovarian and prostate cancers. The loss of let-7 expression in these
tumors plays an important role in promoting the tumorigenic process
(43). Torrisani et al
confirmed that KRAS inhibition by let-7 was weaker in the
pathogenesis of PC (44). Kent
et al confirmed that miR-143/145 was able to inhibit the
expression of KRAS and downstream RREB1 (45). In addition, the effect of miRNA on
KRAS has also been reported in other tumors. Tsang et
al confirmed that miR-18a acts as a tumor suppressor by
inhibiting KRAS in squamous cell carcinoma, colon cancer and
embryonic liver cells (46),
whereas Shin et al found that miR-181 downregulated
KRAS in oral squamous cell carcinoma (47). However, the role of miR-18a and
miR-181 in PC remains to be confirmed by further studies. Several
miRNA molecules (miR-193b, -217 and -96) are downregulated in PC.
These miRNAs inhibit PC cell proliferation, invasion and metastasis
by targeting KRAS (7,10,11).
Based on our current understanding of miRNAs, many more potential
miRNAs remain to be further investigated.
Studies have demonstrated that hsa-let-7a targets
KRAS through its 3′-UTR (48). hsa-let-7g is associated with liver
and breast cancer and tumor metastasis, and regulates cell
proliferation and migration through interacting with KRAS
(49). It has also been
demonstrated that miR-27a acts on the 3′-UTR of KRAS to
inhibit esophageal protein expression (50). Similarly, the present study
detected high expression levels of hsa-miR-27a. In colorectal
cancer, miR-30b and miR-143 target KRAS, acting as tumor
suppressors (51,52), whereas miR-30c was shown to
regulate KRAS expression by combining with its 3′-UTR in
breast cancer (53). Our previous
study demonstrated that miR-96 also targets KRAS (11). As research time was limited, the
experiment of verification of KRAS-3′UTR enrichment and the
effective enrichment of the targets in other cell lines will be
performed in a future study.
In the present study, we also predicted a total of
21 new miRNAs associated with PC, but their expression levels were
low. We also completed secondary structure diagrams of 338 new
pre-miRNAs. The comparison of predicted miRNAs to the miRBase
database revealed they belong to the miR-345, -330, -574 and -1243
families. However, after transfecting synthetic miRNA mimics of the
predicted miRNAs into PC cells, KRAS protein expression was not
significantly affected. A possible explanation may be that these
miRNAs may play an indirect role or act synergistically with other
miRNAs or non-coding RNAs; in this case, KRAS regulation may
not be readily obvious when they are individually transfected into
PC cells.
The role of non-coding lncRNAs in the development of
tumors has become a focus of investigation. lncRNA HEIH is
highly expressed in hepatocellular carcinoma and is an important
proto-oncogene that promotes the progression of this type of cancer
(54). lncRNA PCAT-1 is
PC-specific and known to inhibit BRCA2 expression. It
promotes cancer cell proliferation in vitro and in
vivo and plays an important role in the progression of PC
(55,56). Moreover, Gas5 was found to
be downregulated in breast cancer cells. Gas5 overexpression
can induce apoptosis in breast cancer cells and inhibit their
proliferation (57). lncRNA
MEG3 is a maternally imprinted gene found to downregulate
human gastric, cervical and non-small-cell lung cancer, among
others. MEG3 transfection into HeLa, A549 and other human
tumor cell lines can inhibit tumor cell proliferation and has been
confirmed to act as a tumor suppressor by activation of p53
signaling (58,59).
Unlike protein-coding genes, the role of lncRNAs in
PC carcinogenesis and development remains unclear. As there is a
higher level of temporal, spatial and tissue-specific lncRNA
expression, there is great potential in predicting new lncRNA
involvement in cancers and that may provide valuable downstream
functional data. MALAT1 has been shown to be highly
expressed in PC. MALAT1 expression is significantly
associated with tumor size, stage and depth of invasion, and
patients with high expression of MALAT1 have a worse
prognosis (60). Lu et al,
as well as others, reported that the lncRNA Gas5 negatively
regulates the expression of CDK6, inducing proliferation of
PC cells (61). Kim et al
demonstrated that lncRNA HOTAIR acts as an oncogene in PC
(20). Sun et al, as well
as others, observed low expression of a new lncRNA,
ENST00000480739, in PC; this lncRNA targets HIF-1α to
increase OS-9 expression (62). It has been demonstrated that the
lncRNA H19 increased HMGA2-mediated
epithelial-to-mesenchymal transition through inhibition of let-7,
thereby promoting the invasion and metastasis of PC cells (63). However, the role of lncRNAs in PC
has not been extensively investigated to date. Dicer and
Drosha enzyme shearing of lncRNAs results in the formation
of miRNAs, which can then exert their corresponding cellular
effects. In our experiments, we found the corresponding small
precursor of lncRNA TCONS_00062686 was hsa-let-7a-1, that of
n407573 was hsa-let-7a-3, the precursor of
TCONS_00062686 was hsa-let-7f-1, the precursor of
TCONS_00063039 was hsa-miR-181a-2, and the precursor of
n386735 was hsa-miR-18a. All these lncRNAs and their miRNA
precursors play a key role in tumorigenesis by interacting with
KRAS. In the present study, we obtained the miRNA and lncRNA
data of interest through next-generation RNA sequencing. We
identified several miRNA precursors that belong to the let-7 and
miR-34, -30 and -143 families, as well as relevant lncRNAs and
their families (MALAT1, MEG3_2 and TUG1_1-4).
In future experiments, our aims are to synthesize
and transfect the corresponding small interfering RNA resulting
from lncRNA cleavage into PC cells, and KRAS protein and mRNA
levels to be monitored, along with an extensive analysis of the
mutual regulation between the obtained lncRNA and miRNA. Further
verification of the obtained lncRNA expression in PC
paraffin-embedded tissues will be conducted through RNAscope in
situ hybridization in future experiments, along with an
analysis of miRNA and lncRNA expression levels in blood samples
from PC patient. Our data-bank of non-coding RNAs (mainly miRNAs
and lncRNAs) that regulate KRAS will greatly improve our
understanding of KRAS regulation-associated tumorigenesis
and hopefully lead to the design of gene therapies for PC. The
results should reveal a complex regulatory network that may provide
new insight and novel approaches for more effective PC treatment
strategies.
Funding
This study was supported by the National Nature
Science Foundations of China (grant nos. 8172334, 81341070 and
81472326).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
LZ was involved in the conception and design of the
study, acquisition of data, analysis and interpretation of the
data, and in the drafting of the article; SY, CW and CJ were
involved in the acquisition of data; JC and ZL were involved in the
conception and design of the study; JC gave the final approval of
the article. All authors have reviewed and approved the final
version of the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests to disclose.
Acknowledgments
Not applicable.
References
|
1
|
Ilic M and Ilic I: Epidemiology of
pancreatic cancer. World J Gastroenterol. 22:9694–9705. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Morris JP IV, Wang SC and Hebrok M: KRAS,
Hedgehog, Wnt and the twisted developmental biology of pancreatic
ductal adenocarcinoma. Nat Rev Cancer. 10:683–695. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ying H, Dey P, Yao W, Kimmelman AC,
Draetta GF, Maitra A and DePinho RA: Genetics and biology of
pancreatic ductal adenocarcinoma. Genes Dev. 30:355–385. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Bloomston M, Frankel WL, Petrocca F,
Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C and Croce
CM: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rachagani S, Macha MA, Menning MS, Dey P,
Pai P, Smith LM, Mo YY and Batra SK: Changes in microRNA (miRNA)
expression during pancreatic cancer development and progression in
a genetically engineered KrasG12D;Pdx1-Cre mouse (KC) model.
Oncotarget. 6:40295–40309. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jin X, Sun Y, Yang H, Li J, Yu S, Chang X,
Lu Z and Chen J: Deregulation of the miR-193b-KRAS axis contributes
to impaired cell growth in pancreatic cancer. PLoS One.
10:e01255152015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Keklikoglou I, Hosaka K, Bender C, Bott A,
Koerner C, Mitra D, Will R, Woerner A, Muenstermann E, Wilhelm H,
et al: MicroRNA-206 functions as a pleiotropic modulator of cell
proliferation, invasion and lymphangiogenesis in pancreatic
adenocarcinoma by targeting ANXA2 and KRAS genes. Oncogene.
34:4867–4878. 2015. View Article : Google Scholar :
|
|
9
|
Hu Y, Ou Y, Wu K, Chen Y and Sun W:
miR-143 inhibits the metastasis of pancreatic cancer and an
associated signaling pathway. Tumour Biol. 33:1863–1870. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhao WG, Yu SN, Lu ZH, Ma YH, Gu YM and
Chen J: The miR-217 microRNA functions as a potential tumor
suppressor in pancreatic ductal adenocarcinoma by targeting KRAS.
Carcinogenesis. 31:1726–1733. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yu S, Lu Z, Liu C, Meng Y, Ma Y, Zhao W,
Liu J, Yu J and Chen J: miRNA-96 suppresses KRAS and functions as a
tumor suppressor gene in pancreatic cancer. Cancer Res.
70:6015–6025. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wapinski O and Chang HY: Long noncoding
RNAs and human disease. Trends Cell Biol. 21:354–361. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Novikova IV, Hennelly SP, Tung CS and
Sanbonmatsu KY: Rise of the RNA machines: Exploring the structure
of long non-coding RNAs. J Mol Biol. 425:3731–3746. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tripathi V, Ellis JD, Shen Z, Song DY, Pan
Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al: The
nuclear-retained noncoding RNA MALAT1 regulates alternative
splicing by modulating SR splicing factor phosphorylation. Mol
Cell. 39:925–938. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Muers M: RNA: Genome-wide views of long
non-coding RNAs. Nat Rev Genet. 12:742–743. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen X and Yan GY: Novel human
lncRNA-disease association inference based on lncRNA expression
profiles. Bioinformatics. 29:2617–2624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Harries LW: Long non-coding RNAs and human
disease. Biochem Soc Trans. 40:902–906. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tahira AC, Kubrusly MS, Faria MF, Dazzani
B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC
and Reis EM: Long noncoding intronic RNAs are differentially
expressed in primary and metastatic pancreatic cancer. Mol Cancer.
10:1412011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
21
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar
|
|
23
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kuzmanovic DA, Elashvili I, Wick C,
O’Connell C and Krueger S: The MS2 coat protein shell is likely
assembled under tension: A novel role for the MS2 bacteriophage A
protein as revealed by small-angle neutron scattering. J Mol Biol.
355:1095–1111. 2006. View Article : Google Scholar
|
|
26
|
Batey RT and Kieft JS: Improved native
affinity purification of RNA. RNA. 13:1384–1389. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gong C, Popp MW and Maquat LE: Biochemical
analysis of long non-coding RNA-containing ribonucleoprotein
complexes. Methods. 58:88–93. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhou Z and Reed R: Purification of
functional RNA-protein complexes using MS2-MBP. Curr Protoc Mol
Biol Chapter. 27:32003.
|
|
29
|
Corcoran CP, Rieder R, Podkaminski D,
Hofmann B and Vogel J: Use of aptamer tagging to identify in vivo
protein binding partners of small regulatory RNAs. Methods Mol
Biol. 905:177–200. 2012.PubMed/NCBI
|
|
30
|
Said N, Rieder R, Hurwitz R, Deckert J,
Urlaub H and Vogel J: In vivo expression and purification of
aptamer-tagged small RNA regulators. Nucleic Acids Res.
37:e1332009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bardwell VJ and Wickens M: Purification of
RNA and RNA-protein complexes by an R17 coat protein affinity
method. Nucleic Acids Res. 18:6587–6594. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
SenGupta DJ, Zhang B, Kraemer B, Pochart
P, Fields S and Wickens M: A three-hybrid system to detect
RNA-protein interactions in vivo. Proc Natl Acad Sci USA.
93:8496–8501. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−ΔΔC(T)) Method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
34
|
Cai X, Hagedorn CH and Cullen BR: Human
microRNAs are processed from capped, polyadenylated transcripts
that can also function as mRNAs. RNA. 10:1957–1966. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek
SH and Kim VN: MicroRNA genes are transcribed by RNA polymerase II.
EMBO J. 23:4051–4060. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wu Y, Wei B, Liu H, Li T and Rayner S:
MiRPara: A SVM-based software tool for prediction of most probable
microRNA coding regions in genome scale sequences. BMC
Bioinformatics. 12:1072011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Pylayeva-Gupta Y, Grabocka E and Bar-Sagi
D: RAS oncogenes: Weaving a tumorigenic web. Nat Rev Cancer.
11:761–774. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Rozenblum E, Schutte M, Goggins M, Hahn
SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ, et
al: Tumor-suppressive pathways in pancreatic carcinoma. Cancer Res.
57:1731–1734. 1997.PubMed/NCBI
|
|
39
|
Jiao LR, Frampton AE, Jacob J, Pellegrino
L, Krell J, Giamas G, Tsim N, Vlavianos P, Cohen P, Ahmad R, et al:
MicroRNAs targeting oncogenes are down-regulated in pancreatic
malignant transformation from benign tumors. PLoS One.
7:e320682012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Xu B, Niu X, Zhang X, Tao J, Wu D, Wang Z,
Li P, Zhang W, Wu H, Feng N, et al: miR-143 decreases prostate
cancer cells proliferation and migration and enhances their
sensitivity to docetaxel through suppression of KRAS. Mol Cell
Biochem. 350:207–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kumar MS, Erkeland SJ, Pester RE, Chen CY,
Ebert MS, Sharp PA and Jacks T: Suppression of non-small cell lung
tumor development by the let-7 microRNA family. Proc Natl Acad Sci
USA. 105:3903–3908. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Boyerinas B, Park SM, Hau A, Murmann AE
and Peter ME: The role of let-7 in cell differentiation and cancer.
Endocr Relat Cancer. 17:F19–F36. 2010. View Article : Google Scholar
|
|
44
|
Torrisani J, Bournet B, du Rieu MC,
Bouisson M, Souque A, Escourrou J, Buscail L and Cordelier P: let-7
MicroRNA transfer in pancreatic cancer-derived cells inhibits in
vitro cell proliferation but fails to alter tumor progression. Hum
Gene Ther. 20:831–844. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kent OA, Chivukula RR, Mullendore M,
Wentzel EA, Feldmann G, Lee KH, Liu S, Leach SD, Maitra A and
Mendell JT: Repression of the miR-143/145 cluster by oncogenic Ras
initiates a tumor-promoting feed-forward pathway. Genes Dev.
24:2754–2759. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Tsang WP and Kwok TT: The
miR-18a* microRNA functions as a potential tumor
suppressor by targeting on K-Ras. Carcinogenesis. 30:953–959. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Shin KH, Bae SD, Hong HS, Kim RH, Kang MK
and Park NH: miR-181a shows tumor suppressive effect against oral
squamous cell carcinoma cells by downregulating K-ras. Biochem
Biophys Res Commun. 404:896–902. 2011. View Article : Google Scholar
|
|
48
|
Wang XR, Luo H, Li HL, Cao L, Wang XF, Yan
W, Wang YY, Zhang JX, Jiang T, Kang CS, et al Chinese Glioma
Cooperative Group (CGCG): Overexpressed let-7a inhibits glioma cell
malignancy by directly targeting K-ras, independently of PTEN.
Neuro-oncol. 15:1491–1501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Chen KJ, Hou Y, Wang K, Li J, Xia Y, Yang
XY, Lv G, Xing XL and Shen F: Reexpression of Let-7g microRNA
inhibits the proliferation and migration via K-Ras/HMGA2/snail axis
in hepatocellular carcinoma. Biomed Res Int.
2014:7424172014.PubMed/NCBI
|
|
50
|
Zhu L, Wang Z, Fan Q, Wang R and Sun Y:
microRNA-27a functions as a tumor suppressor in esophageal squamous
cell carcinoma by targeting KRAS. Oncol Rep. 31:280–286. 2014.
View Article : Google Scholar
|
|
51
|
Liao WT, Ye YP, Zhang NJ, Li TT, Wang SY,
Cui YM, Qi L, Wu P, Jiao HL, Xie YJ, et al: MicroRNA-30b functions
as a tumour suppressor in human colorectal cancer by targeting
KRAS, PIK3CD and BCL2. J Pathol. 232:415–427. 2014. View Article : Google Scholar
|
|
52
|
Chen X, Guo X, Zhang H, Xiang Y, Chen J,
Yin Y, Cai X, Wang K, Wang G, Ba Y, et al: Role of miR-143
targeting KRAS in colorectal tumorigenesis. Oncogene. 28:1385–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Tanic M, Yanowsky K, Rodriguez-Antona C,
Andrés R, Márquez-Rodas I, Osorio A, Benitez J and Martinez-Delgado
B: Deregulated miRNAs in hereditary breast cancer revealed a role
for miR-30c in regulating KRAS oncogene. PLoS One. 7:e388472012.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang F, Zhang L, Huo XS, Yuan JH, Xu D,
Yuan SX, Zhu N, Zhou WP, Yang GS, Wang YZ, et al: Long noncoding
RNA high expression in hepatocellular carcinoma facilitates tumor
growth through enhancer of zeste homolog 2 in humans. Hepatology.
54:1679–1689. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Prensner JR, Iyer MK, Balbin OA,
Dhanasekaran SM, Cao Q, Brenner JC, Laxman B, Asangani IA, Grasso
CS, Kominsky HD, et al: Transcriptome sequencing across a prostate
cancer cohort identifies PCAT-1, an unannotated lincRNA implicated
in disease progression. Nat Biotechnol. 29:742–749. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Prensner JR, Chen W, Iyer MK, Cao Q, Ma T,
Han S, Sahu A, Malik R, Wilder-Romans K, Navone N, et al: PCAT-1, a
long noncoding RNA, regulates BRCA2 and controls homologous
recombination in cancer. Cancer Res. 74:1651–1660. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
58
|
Lu KH, Li W, Liu XH, Sun M, Zhang ML, Wu
WQ, Xie WP and Hou YY: Long non-coding RNA MEG3 inhibits NSCLC
cells proliferation and induces apoptosis by affecting p53
expression. BMC Cancer. 13:4612013. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Qin R, Chen Z, Ding Y, Hao J, Hu J and Guo
F: Long non-coding RNA MEG3 inhibits the proliferation of cervical
carcinoma cells through the induction of cell cycle arrest and
apoptosis. Neoplasma. 60:486–492. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiao F, Hu H, Yuan C and Wang L, Jiang W,
Jin Z, Guo Z and Wang L: Elevated expression level of long
noncoding RNA MALAT-1 facilitates cell growth, migration and
invasion in pancreatic cancer. Oncol Rep. 32:2485–2492. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Lu X, Fang Y, Wang Z, Xie J, Zhan Q, Deng
X, Chen H, Jin J, Peng C, Li H, et al: Downregulation of gas5
increases pancreatic cancer cell proliferation by regulating CDK6.
Cell Tissue Res. 354:891–896. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Sun YW, Chen YF, Li J, Huo YM, Liu DJ, Hua
R, Zhang JF, Liu W, Yang JY, Fu XL, et al: A novel long non-coding
RNA ENST00000480739 suppresses tumour cell invasion by regulating
OS-9 and HIF-1α in pancreatic ductal adenocarcinoma. Br J Cancer.
111:2131–2141. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7′s suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|