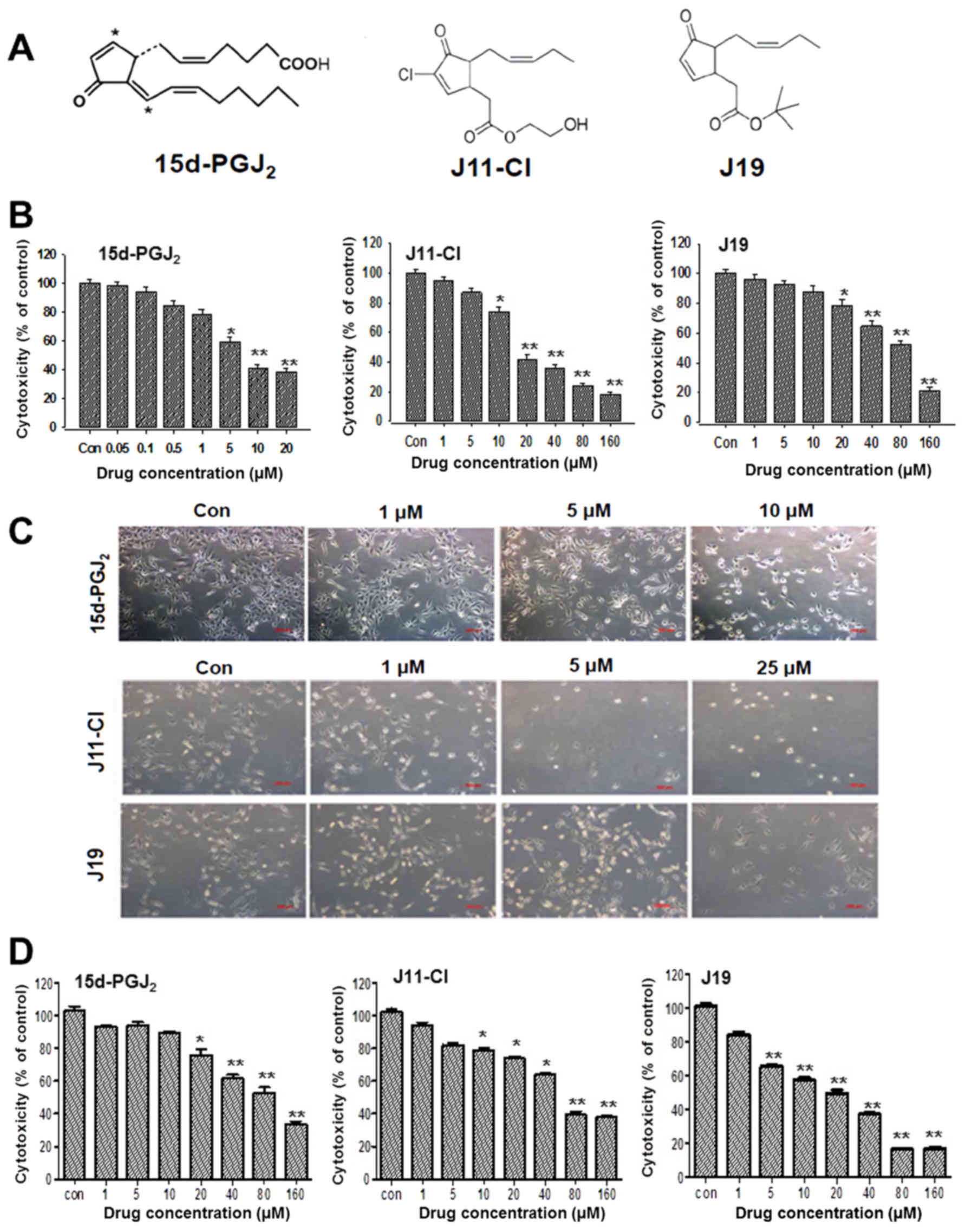
Introduction
Sirtuins (SIRTs) are class III histone deacetylases
(HDACs) that have been identified to serve important biological
functions, including aging, energy mobilization and stress
responses (1,2). Furthermore, SIRTs are involved in the
regulation of cancer cell apoptosis and are potential targets for
novel anticancer drugs that regulate the levels of deacetylated
histone proteins, p53 and several transcriptional factors (3,4).
Several SIRT1 inhibitors, such as Ex527, sirtinol and salermide,
exhibit potent anticancer activity in various cancer cell lines
(5,6). Previously, we demonstrated that a
novel SIRT inhibitor, psammaplin A, increased p53 acetylation and
subsequently induced apoptotic death in MCF-7/Adr cells (7). In addition, Chu et al
(8) demonstrated that patients
with chemoresistant tumors overexpressed SIRT1; furthermore, the
inhibition of SIRT1 expression decreased multidrug resistance 1
(MDR1) expression and increased drug sensitivity.
15-Deoxy-Δ12,14-prostaglandin
J2 (15d-PGJ2) was revealed to exhibit
pharmacological activities, including anti-inflammatory,
anti-fibrotic and apoptotic effects, through peroxisome
proliferator-activated receptor γ-independent signaling pathways
such as the nuclear factor-κB (NF-κB), signal transducer and
activator of transcription 1 (STAT1) and p53-dependent signaling
pathways (9,10). Furthermore, 15d-PGJ2 was
identified to induce apoptosis of various cancer cells through
caspase-dependent signaling pathways (11). A previous study demonstrated that
15d-PGJ2 inhibited the migration of A2780/AD cells,
possibly via NF-κB inhibition resulting from HDAC1 inhibition. The
mechanisms of action underlying these novel effects of
15d-PGJ2 on SIRT1 and HDAC1 gene expression and enzyme
activities were elucidated (12).
In the present study, the effects of novel SIRT1 inhibitors (J11-Cl
and J19), with a 15d-PGJ2 scaffold (11,12),
on ovarian cancer cells were investigated.
Methyl jasmonate is a member of the jasmonate family
of plant stress hormones, the most potent regulator of
defense-associated mechanisms in plants (13). On the basis of its structural
similarity to that of 15d-PGJ2, methyl jasmonate (J-11)
was investigated for SIRT activity, and its functional mechanisms
of regulation of cancer cell death pathways were investigated. A
previous study indicated that an α-haloenone analog, J7, exhibited
enhanced in vitro anti-inflammatory potency (14,15).
Materials and methods
Reagents
15d-PGJ2 (87893-55-8) and 3-methyladenine
(3-MA; 5142-23-4) were purchased from Cayman Chemical Company (Ann
Arbor, MI, USA). J11-Cl and J19 were synthesized in-house. The
chemical structures of the drugs are presented in Fig. 1A. Dulbecco's modified Eagle's
medium (DMEM), fetal bovine serum (FBS) and cell culture
supplements were obtained from Gibco; Thermo Fisher Scientific,
Inc. (Waltham, MA, USA). Primary antibodies against SIRT1 (cat. no.
8469; 1:1,000), SIRT2 (cat. no. 12672; 1:1,000), SIRT4 (cat. no.
sc-135798; 1:500), SIRT5 (cat. no. 8779; 1:1,000), SIRT6 (cat. no.
8771; 1:1,000), B-cell lymphoma-2 (Bcl-2; cat. no. 15071; 1:500),
Bcl-2-associated X protein (Bax; cat. no. 5023; 1:1,000), β-actin
(cat. no. 3700; 1:1,000), light chain 3 (LC3; cat. no. 3868;
1:1,000), beclin-1 (cat. no. 4122; 1:1,000), autophagy-related 3
(Atg3; cat. no. 3415; 1:1,000), Atg5 (cat. no. 12994; 1:1,000),
Atg7 (cat. no. 8558; 1:1,000), α-tubulin (cat. no. 3873; 1:1,000),
cleaved caspase-3 (cat. no. 9661; 1:500), cleaved caspase-9 (cat.
no. 7237; 1:1,000), poly(ADP-ribose) polymerase (PARP; cat. no.
9541; 1:1,000) and acetylated p53 (cat. no. 2570; 1:500) were
purchased from Cell Signaling Technology (Beverly, MA, USA).
Horseradish peroxidase-conjugated secondary antibodies [anti-mouse
immunoglobulin G (IgG); cat. no. sc-516102 or anti-rabbit IgG; cat
no. sc-2357] were purchased from Santa Cruz Biotechnology, Inc.
(Dallas, TX, USA). All other chemicals were purchased from
Sigma-Aldrich; Merck KGaA. All drugs were dissolved in dimethyl
sulfoxide (DMSO) and stored at −20°C until use. Chemical agents
were diluted to appropriate concentrations with culture medium
supplemented with 1% FBS. The final concentration of DMSO was
<0.1% (v/v). DMSO was also present in the corresponding
controls.
SIRT1 enzyme activity
SIRT1 enzymatic activity was assessed using
commercial kits (cat. no. ab156065) from Abcam (Cambridge, UK),
according to the manufacturer's protocol. First, assay buffer [50
mM Tris/HCl, pH 8.0, 137 mM NaCl, 2.7 mM KCl, 1 mM MgCl2
and 1 mg/ml bovine serum albumin (BSA)], SIRT1 enzyme, and either
the solvent dimethylformamide (DMF) or different concentrations of
the drugs (15d-PGJ2, J19 or J11-C1 dissolved in DMF)
were mixed with the substrate (p53) and co-substrate
(NAD+) for 45 min. Deacetylation reactions were
conducted at 37°C for 60 min, and stopped by adding 50 µl
stop solution containing the developer, followed by incubation at
37°C for 30 min. Fluorescence intensity was determined by reading
fluorescence using a SpectraMax M2 microplate reader (Molecular
Devices, LLC, Sunnyvale, CA, USA) with an excitation wavelength of
350 nm and an emission wavelength of 450 nm. Calculations of net
fluorescence were made after subtracting values for a blank
consisting of buffer without NAD.
Docking simulations of sirtinol,
15d-PGJ2, J11-Cl and J19 ligand and target
structure
The X-ray crystal structure of SIRT1 was selected
from the Research Collaboratory for Structural Bioinformatics
Protein Data Bank (PDB; code 4I5I) and prepared using the protein
preparation wizard available in the Glide tool in Maestro (version
10.2; Schrödinger, LLC, New York, NY, USA). During the process, the
missing side and back chains were included (16). The protein preparation wizard
facility has two components: Preparation and refinement. Following
ensuring the chemical accuracy, the preparation component adds
hydrogen and neutralizes a side chain that is neither close to the
binding cavity nor involved in the formation of salt bridges. The
all-atom-optimized potentials for liquid simulations (OPLS-AA)
force field was used for this purpose, and then the active site of
protein was defined. Glide uses the full OPLS-AA force field at an
intermediate docking stage and is considered to be more sensitive
to geometrical detail compared with other docking algorithms. The
water molecule occupying the protein structure was not suitable for
the docking study, therefore it was removed. Finally, the
optimization and minimization processes were performed until the
average root mean square deviation of the non-hydrogen atoms
reached 0.3 Å (17). This was
followed by the generation of energy grids using the Glide
protocol, as previously described (18-20).
The docked ligand was used to define the energy grid boundaries
with the default options. These grids were used to score the ligand
'in place' with the XP scoring function. It should be noted that
considering the computational protocol followed for the generation
of the covalent complexes, the XP Glide Score values can be used
only in a qualitative sense to determine the binding of various
ligands. The Glide XP scoring functions include improvements to the
scoring of hydrogen bonds, the detection of buried polar groups,
and the detection of π-cation and π-π stacking interactions. The
result of the docking calculation is a top-scored predicted complex
that is evaluated using the scoring function. The compounds
sirtinol, 15d-PGJ2, J11-Cl and J19 were prepared using
the LigPrep software (version 3.4; Schrödinger, LLC), which can
generate a number of structures from each input structure with
various ionization states, tautomers, stereochemistries and ring
conformations. This process eliminates molecules using various
criteria including the molecular mass, specified numbers and types
of functional groups present with correct chiralities for each
successfully processed input structure. The OPLS 2005 force field
was used for the optimization, which produced the low-energy isomer
of the ligand (21). Finally, all
ligand molecules formed in the complex structure for input were
docked.
Cell culture
Human ovarian cancer cell lines (SKOV3 and OVCAR3)
and normal kidney epithelial cell lines (HK-2 and NRK-52E were
purchased from the American Type Culture Collection (ATCC;
Manassas, VA, USA). SKOV3, OVCAR3, or NRK-52E cells were maintained
in DMEM supplemented with 10% FBS, 4 mmol L-glutamine, 100 U/ml
penicillin and 100 µg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.). HK-2 cells were maintained in DMEM/Ham's F12
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 5
µg/ml insulin (Gibco; Thermo Fisher Scientific, Inc.), 5
µg/ml transferrin (Gibco; Thermo Fisher Scientific, Inc.),
100 U/ml penicillin, 100 g/ml streptomycin, 0.1 µmol/l
hydrocortisone, 2 nmol/l L-glutamine plus 10% FBS. The cell
cultures were incubated at 37°C in a humidified atmosphere
containing 5% CO2.
Cytotoxicity assay
Cell viability was determined using MTT (5 mg/ml;
Sigma-Aldrich; Thermo Fisher Scientific, Inc.). The cells were
seeded in 96-well plates at a density of 2×103
cells/well. Following incubation at 37°C for 24 h, the cells were
treated with 15d-PGJ2 (0.05-20 µM), J11-C1 (1-160
µM) or J19 (1-160 µM) and cultured for a further 24,
48 or 72 h. Following incubation, 15 µl MTT reagent was
added to each well and plates were incubated at 37°C for 3 h in the
dark. The supernatant was aspirated and formazan crystals were
dissolved in 150 µl DMSO at 37°C for 15 min with gentle
agitation. The absorbance of each well was measured at 540 nm using
a VersaMax microplate reader (Molecular Devices LLC, Sunnyvale, CA,
USA). Three independent experiments were performed for each
condition and normalized to the absorbance of wells containing
medium only (0%) or untreated cells (100%). The half-maximal
inhibitory concentration (IC50) values were calculated
from the sigmoidal concentration-response curves using SigmaPlot
(version 12; Systat Software, Inc., San Jose, CA, USA).
Cell cycle analysis
Cells were treated with 15d-PGJ2 (1, 5 or
10 µM), J11-Cl (1, 5 or 25 µM), or J19 (1, 5 or 25
µM) for 48 h or vincristine sulfate (BML-T117-0005; Enzo
Life Sciences, Inc., Farmingdale, NY, USA) at 5 nM as a positive
control. The total number of cells, including those in suspension
and those adhered to the walls of the wells, were harvested
separately to identify sub-G1 or other cell cycle
stages, and were washed in 1% BSA before fixing in 95% ice-cold
ethanol containing 0.5% Tween-20 for 1 h at −20°C. The cells
(1×106) were washed in a solution containing 1% BSA,
stained with ice-cold propidium iodide (PI) solution (10
µg/ml PI and 100 µg/ml RNase in PBS) and incubated in
the dark for 30 min at room temperature. The data were acquired and
analyzed using a flow cytometer (BD Biosciences, San Jose, CA,
USA).
Detection of apoptosis using Annexin V/PI
staining
Cells were seeded in 6-well plates (Labtek; Nalge
Nunc International, Penfield, NY, USA) at 1×105
cells/well and allowed to attach overnight. After 24 h of
incubation at 37°C, the cells were washed with serum-free medium
and treated with different concentrations of the drugs in 200
µl medium/well for 48 h. Following induction of apoptosis,
the supernatant was collected and the adherent cells
(2×106 cells) were trypsinized from the plates. The
collected cells were washed twice with PBS and centrifuged at 600 ×
g for 5 min. Each pellet was resuspended in 50 µl
4-(2-hydroxyethyl)-1-piperzaine-ethanesulfonic acid (HEPES) buffer
(10 mM HEPES, 135 mM NaCl and 5 mM CaCl2). Cells
(2×106 cells in 100 µl buffer) were transferred
to flow cytometry tubes and 2 µl each of Annexin
V-fluorescein isothiocyanate (FITC) and PI (each at 1 mg/ml) were
added. Following incubation for 5 min at room temperature in the
dark, 400 µl binding buffer was added to each tube. Samples
were analyzed using a flow cytometer (Guava® easyCyte
flow cytometer, EMD Millipore, Billerica, MA, USA).
Western blot analysis
SKOV3 cells were cultured in DMEM at 37°C in a
humidified atmosphere containing 5% CO2. Following
incubation for 24 h, the cells were treated with
15d-PGJ2 (1, 5 or 10 µM), J11-Cl (1, 5 or 25
µM) or J19 (1, 5 or 25 µM), cultured for 48 h,
harvested using trypsin digestion, and then washed twice with
ice-cold PBS. To isolate total proteins, the cells were first
suspended in PRO-PREP™ protein extraction solution (Intron
Biotechnology, Inc., Seongnam, Korea). Protein concentrations were
determined using a Bicinchoninic Acid protein assay kit (Bio-Rad
Laboratories, Inc., Hercules, CA, USA), according to the
manufacturer's protocol. Protein samples (20 µg) of the cell
extracts were separated by SDS-PAGE (6-15% gels) and transferred
onto a polyvinylidene difluoride membrane (EMD Millipore), which
was incubated for 1 h in TNA buffer (25 mM Tris/HCl, pH 8.5, 192 mM
glycine and 20% methanol) and blocked with 5% skimmed milk powder
in PBS. Subsequently, the membrane was incubated with various
primary antibodies against SIRTs, Bax, Bcl-2, β-actin, PARP,
cleaved caspase-3, cleaved caspase-9, p53, acetylated p53, LC3,
beclin-1, Atg3, Atg5 and Atg7 at 4°C overnight. Following washing
for 1 h with TNA buffer, the membrane was incubated with
horseradish peroxidase-conjugated anti-mouse or anti-rabbit
secondary antibodies (1:10,000) for 1 h at room temperature. The
blots were developed using an Enhanced Chemiluminescence Plus kit
(GE Healthcare Life Sciences, Little Chalfont, UK). The data were
acquired and analyzed using an ImageSaver6 (ATTO Corp., Tokyo,
Japan).
Reverse transcription-polymerase chain
reaction (RT-PCR)
Total RNA was isolated using TRIzol®
reagent (Gibco; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. Samples of 2 µg RNA were
reverse-transcribed for 50 min at 42°C in a 20 µl reaction
mixture containing 1 µl oligonucleotide (dT)15
primer (0.5 µg), 10 mM dNTP mixture, 25 mM MgCl2
(4 µl), 0.1 M dithiothreitol (2 µl), RNaseOUT
inhibitor (1 µl; Invitrogen; Thermo Fisher Scientific,
Inc.), Superscript II (50 units) and X10 reverse transcription
buffer (2 µl), followed by denaturation at 68°C for 15 min.
The cDNAs obtained were further amplified using PCR with the
specific primers. The primers sets and PCR conditions are presented
in Table I. The number of PCR
cycles was estimated in a preliminary study and optimized in the
exponential phase of PCR. The PCR products were subjected to
electrophoresis on 2% agarose gels and visualized by ethidium
bromide staining and UV transillumination (WSE-5600 CyanoView; ATTO
Corp.). The molecular sizes of the amplified products were
determined by comparison with a molecular mass marker (100 bp DNA
ladder; Intron Biotechnology, Inc.) that was run in parallel with
the RT-PCR products. Each assay was performed three times.
 | Table IPrimer sequences for the polymerase
chain reaction. |
Table I
Primer sequences for the polymerase
chain reaction.
| Gene name | Primer
sequence | Product length
(bp) | Tm
(°C) | Cycling
conditions |
|---|
| SIRT1 | F:
5′-GACTCCAAGGCCACGGATAG-3′
R: 5′-GTGGAGGTATTGTTTCCGGC-3′ | 110 | 59.89
58.91 | 95°C for 5 min; 34
cycles of 95°C for 15 sec, annealing at 57°C for 15 sec and 72°C
for 15 sec; 72°C for 10 min |
| SIRT2 | F:
5′-GGCAGTTCAAGCCAACCATC-3′
R: 5′-CCACCAAGTCCTCCTGTTCC-3′ | 132 | 59.76
59.96 | 95°C for 5 min; 35
cycles of 95°C for 15 sec, annealing at 57°C for 15 sec and 72°C
for 15 sec; 72°C for 10 min |
| SIRT4 | F:
5′-AGGGTCCTGTGCTTGGATTG-3′
R: 5′-GGTTTCAGATGGCCTCCACA-3′ | 172 | 59.96
59.96 | 95°C for 5 min; 32
cycles of 95°C for 15 sec, annealing at 57°C for 15 sec and 72°C
for 15 sec; 72°C for 10 min |
| GAPDH | F:
5′-GAGTCAACGGATTTGGTCGT-3′
R: 5′-TGTGGTCATGAGTCCTTCCA-3′ | 512 | 58.21
58.27 | 95°C for 5 min; 32
cycles of 95°C for 15 sec, annealing at 56°C for 15 sec and 72°C
for 15 sec; 72°C for 10 min |
Acridine orange staining
SKOV3 cells were seeded in T-25 flasks and treated
with 15d-PGJ2 (1, 5 or 10 µM), J11-Cl (1, 5 or 25
µM) or J19 (1, 5 or 25 µM) for 48 h when the cells
reached 70% confluence. At the appropriate time points, the cells
were treated with 1 µg/ml acridine orange (2.7 µM) in
serum-free medium at 37°C for 15 min at room temperature. Following
washing with PBS, the formation of acidic vesicular organelles
(AVOs) was observed using fluorescence microscopy (FV10i; Olympus
Corp., Tokyo, Japan). The cytoplasm and nuclei of the stained cells
fluoresced bright green, whereas the acidic AVOs fluoresced bright
red. Additionally, green (510-530 nm) and red (650 nm) fluorescence
emission from 1×104 cells exposed to blue (488 nm)
excitation light was determined using a flow cytometer.
Monodansylcadaverine (MDC) incorporation
assay
Autophagic vacuoles were also detected by incubation
with 50 µmol/l MDC in PBS at 37°C for 10 min. Following
incubation, the cells were washed four times with ice-cold PBS and
were fixed with 3.75% paraformaldehyde in PBS. The cells were
immediately analyzed using a confocal microscope (FV10i) equipped
with a filter system (excitation wavelength, 380 nm; emission
filter, 525 nm).
Statistical analysis
Statistical analyses were carried out using GraphPad
Prism software (version 5.03; GraphPad Software, Inc., La Jolla,
CA, USA). All numerical data are presented as the mean ± standard
error of the mean. Statistical analyses were performed using
Student's t-test or Mann-Whitney U test. One-way analysis of
variance with Tukey's post hoc test to assess differences among
specific groups was used for comparison among multiple values.
P<0.05 was considered to indicate a statistically significant
difference.
Results
Effects of 15d-PGJ2, J19 and
J11-C1 on the viability of ovarian cancer cells
Different cancer cell lines were used to screen the
in vitro cytotoxic activity of 15d-PGJ2 and its
derivatives. Cell viability was determined using an MTT assay.
Human ovarian cancer SKOV3 cells were treated with the indicated
concentrations of agents for 48 h. 15d-PGJ2
significantly decreased cell viability in a concentration-dependent
manner with an IC50 of 7.58 µM after 48 h of
treatment. Similarly, J11-C1 and J19 markedly affected SKOV3 cell
viability in a concentration-dependent manner, but the
IC50 values of J11-C1 (17.6 µM) and J19 (83
µM) were much higher compared with that of
15d-PGJ2 (Fig. 1B).
During incubation with 15d-PGJ2, the cell morphology was
altered to an enlarged and elongated form. However, SKOV3 cells
also appeared to be more stretched following J11-Cl treatment, with
a different cell morphology phenotype from that of the control
cells. Following treatment with J19, the cells were initially
rounded and shrunken with larger cytoplasm and distinct cellular
boundaries (Fig. 1C). The
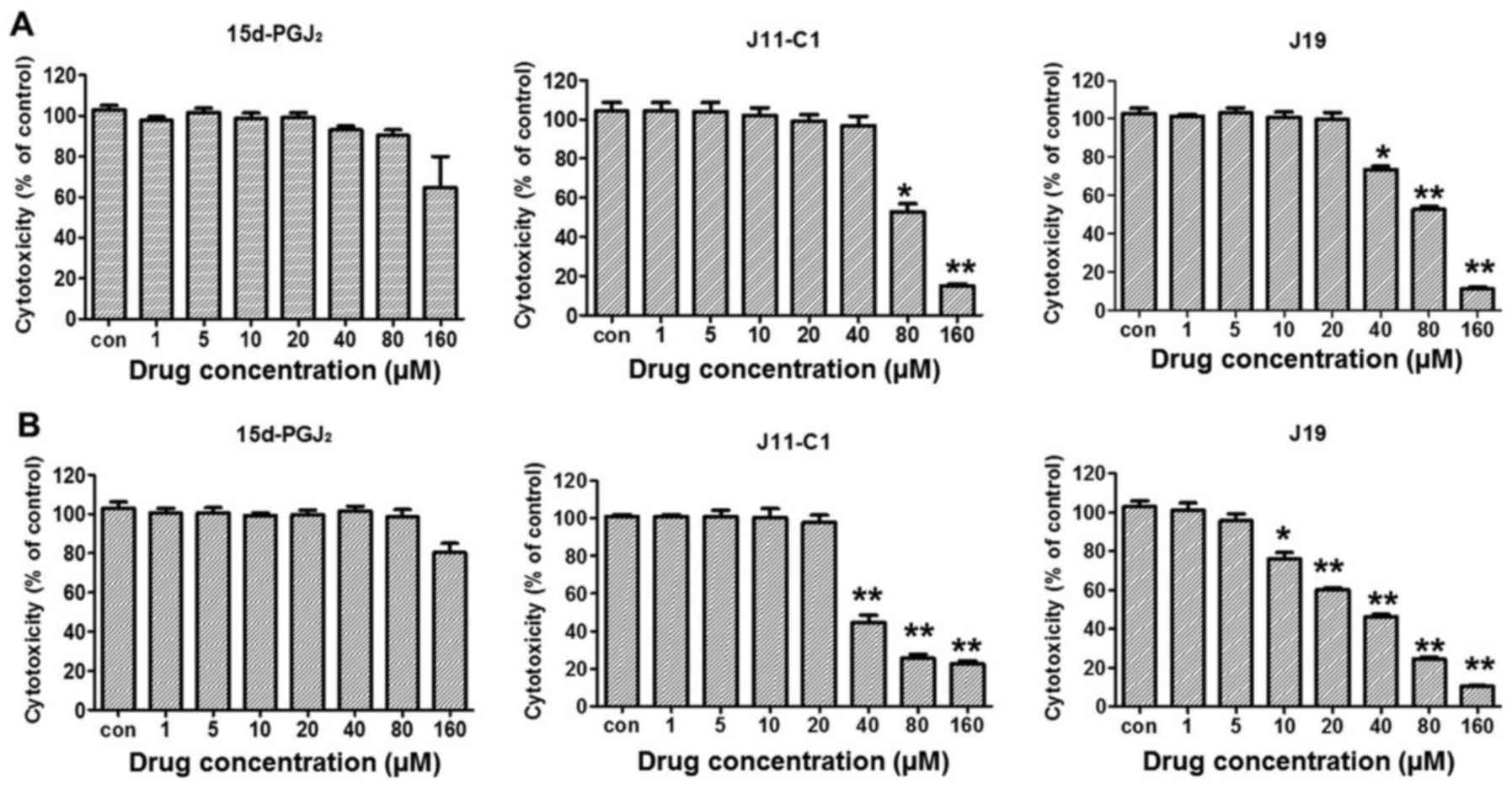
cytotoxicity of 15d-PGJ2 and its derivatives against the
other ovarian cancer cell line used, OVCAR3 cells, were compared.
The results indicated that the cytotoxic effect of
15d-PGJ2, (IC50, 80.7 µM), J11-Cl
(49.2 µM) or J19 (20.3 µM) was lower compared with
that in SKOV3 cells (Fig. 1D),
therefore, SKOV3 cells were selected for our subsequent
experiments. The cytotoxic effect of 15d-PGJ2 and its
derivatives against normal cells was examined using an MTT assay.
Normal kidney proximal tubule epithelial HK-2 or NRK-52E cells were
treated with 15d-PGJ2 or its derivatives for 48 h. No
significant cytotoxicity was observed in HK-2 or NRK-52E cells at
80 µM 15d-PGJ2 (Fig.
2A), which significantly affected SKOV3 cells, suggesting that
the cytotoxic effect of the 15d-PGJ2 was selective
towards cancer cells.
SIRT enzymatic activity and expression in
SKOV3 cells
The effect of 15d-PGJ2 and its
derivatives on SIRT protein expression was examined in SKOV3 cells
using western blotting with specific antibodies against acetylated
SIRT1, SIRT2, SIRT4, SIRT5 and SIRT6. As presented in Fig. 3A, 15d-PGJ2 markedly
decreased the expression of SIRT1, at 1 µM. However, a high
concentration (5 µM) of J11-Cl and J19 markedly decreased
SIRT1 protein levels (Fig. 3A and
B). To confirm the expression of SIRTs in SKOV3 cells, the mRNA
levels of SIRT1, SIRT2 and SIRT4 in SKOV3 cells treated with
15d-PGJ2 (1, 5 or 10 µM), J11-Cl (1, 5 or 25 µM), or
J19 (1, 5 or 25 µM). As presented in Fig. 3C and D, mRNA levels of SIRT1 and
SIRT4 were significantly decreased following 15d-PGJ2
treatment (5 and 10 µM). In addition, the mRNA levels of
SIRT2 were markedly decreased in SKOV3 cells by 25 µM J11-Cl
and J19 treatment (Fig. 3C). The
effects of 15d-PGJ2, J19 and J11-C1 on total SIRT1
enzymatic activity were investigated. Sirtinol was used as a
reference compound for SIRT1 inhibitor. As presented in Fig. 3E, sirtinol significantly inhibited
SIRT1 enzyme activity in a concentration-dependent manner. It was
identified that 15d-PGJ2 and J11-Cl significantly
decreased SIRT1 enzymatic activity at high concentrations. Similar
to the protein levels, J19 only significantly inhibited SIRT1
enzyme activity at high concentrations (Fig. 3E).
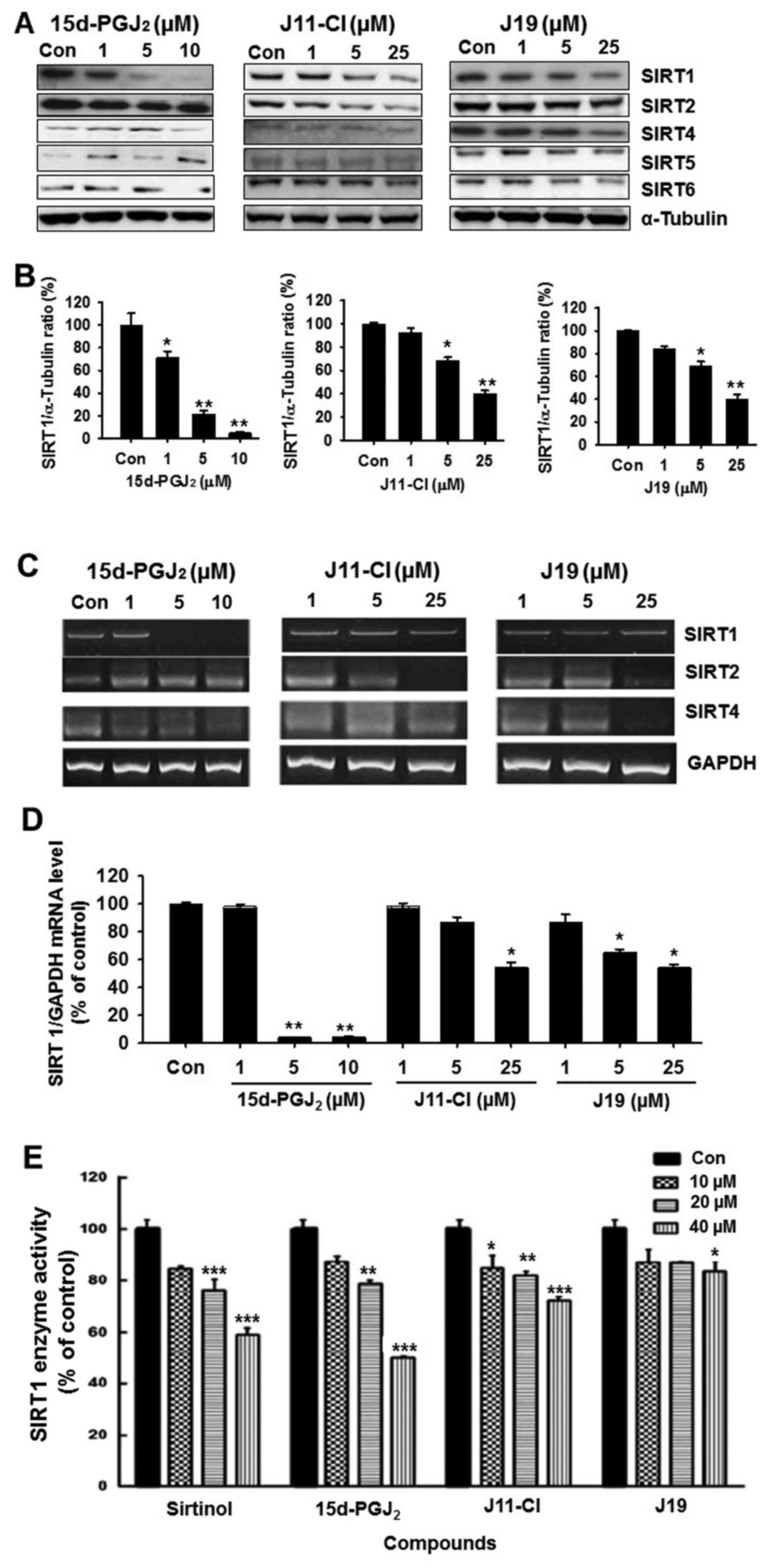 | Figure 3Effects of 15d-PGJ2,
J11-Cl and J19 on SIRT expression and SIRT1 enzyme activity. (A)
SKOV3 cells were treated with various concentrations of
15d-PGJ2, J11-Cl and J19 for 48 h. The proteins were
isolated and western blotting analysis was performed with
antibodies against SIRT1, SIRT2, SIRT4, SIRT5 and SIRT6. Western
blot images are representative of SKOV3 cells from triplicate
experiments. (B) Protein band quantification using densitometry
from three independent experiments. (C) mRNA expression of SIRT1,
SIRT2 and SIRT4 in SKOV3 cells treated with 15d-PGJ2,
J11-Cl and J19. (D) Quantification of mRNA levels from three
independent experiments using densitometry. (E) Effects of
sirtinol, 15d-PGJ2, J11-Cl and J19 on total SIRT1
enzymatic activity using a commercial kit. Results are presented as
the mean ± standard error of the mean of three independent
experiments. *P<0.05, **P<0.01,
***P<0.001 vs. control. 15d-PGJ2,
15-deoxy-Δ12,14-prostaglandin J2; SIRT,
sirtuin; Con, control. |
Molecular docking analysis
The aims of the molecular docking study were to
elucidate whether 15d-PGJ2, J11-Cl or J19 modulate
anticancer targets and identify the actual binding pocket against
the molecular target of SIRT1. The orientations and binding
affinities of the drugs to SIRT1 (PDB code 4I5I) were identified.
The docking results for sirtinol revealed a high docking score of
−9.41 to −8.27 and the formation of a hydrogen bond of 1.94 Å
(–C=O) to the backbone of Gln345 and π-π stacking bond
lengths of 4.7 and 5.2Å with Phe273 and
Phe414. In the docking pose, the chemical natures of the
binding site residues within a radius of 3 Å from the bound
compound are presented in Fig. 4A.
Similarly, the docking results of 15d-PGJ2 revealed a
docking score of −9.43 to −6.82 and formed three hydrogen bonds of
2.37 (O=C–NH–), 2.09 (–C=O) and 1.77 Å (O=C–NH–) to
Asp348 and Ile347 (Fig. 4B). Likewise, the docking results of
J11-C1 revealed a docking score of −7.34 to −3.98 and formed two
hydrogen bonds with lengths of 1.98 Å (–C=O) to PRo271
and of 2.51 Å (O=C–NH–) to Ile347 (Fig. 4C). Lastly, the docking results of
J19 revealed a low docking score of −4.33 to −2.85 and formed two
hydrogen bonds with lengths of 2.18 and 2.24 Å (–NH=C–NH2) to
Arg274 (Fig. 4D). In
the docking pose, the binding site residues were His363,
Ile411, Ala262, Phe413,
Phe273, Phe414, Phe297,
Phe297 and Asn346; thus, the bound drugs
exhibited good binding affinity and marked hydrophobic
interactions, which may lead to greater stability and activity. The
overall interaction of the binding site pocket (within a radius of
3 Å) is summarized in Table
II.
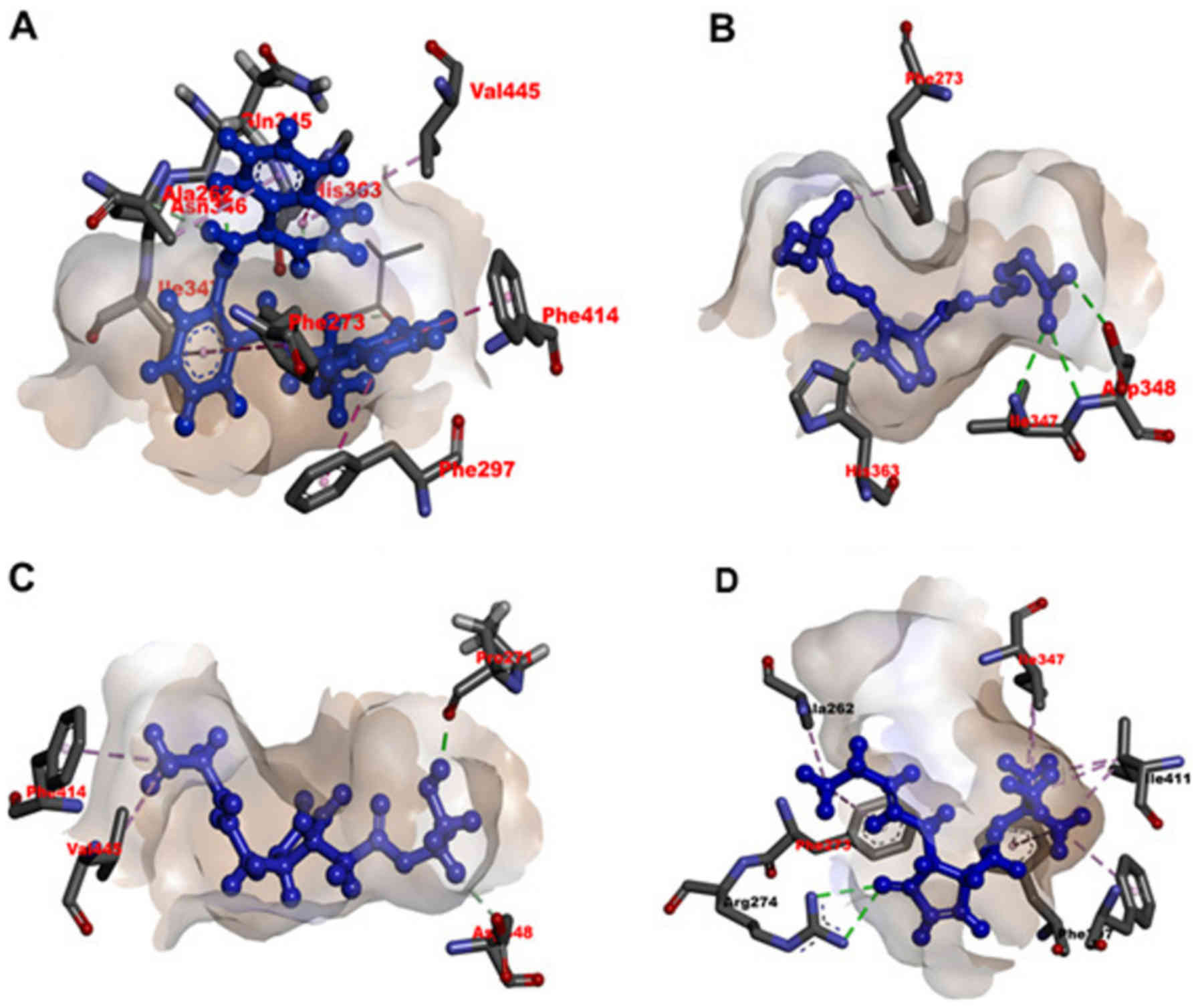 | Figure 4Identification of sirtinol-,
15d-PGJ2-, J11-Cl- and J19-binding sites on SIRT1 using
molecular docking simulations. Effective non-covalent interactions
between the residues of SIRT1 and ligands (A) sirtinol, (B)
15d-PGJ2, (C) J11-Cl and (D) J19 are depicted with
dashed lines. Green, magenta and pink indicate hydrogen bond, π-π
stacking and hydrophobic interaction, respectively. A black dashed
line in a ligand indicates resonance in its functional group.
15d-JG2, 15d-PGJ2,
15-deoxy-Δ12,14-prostaglandin J2. |
 | Table IIIdentification of inhibitor-binding
site in human SIRT1 using molecular docking and dynamics
simulations. |
Table II
Identification of inhibitor-binding
site in human SIRT1 using molecular docking and dynamics
simulations.
| Ligand | Docking score (mean
± standard error of the mean) | Amino acids in the
binding site within 3.0 Å of ligand | Length of hydrogen
bond, Å | No. of hydrogen
bonds |
|---|
| Sirtinol | −8.79±0.52 | Tyr280,
Phe414, Phe413, Val412,
Ile411, Phe297, His363,
Ile316, Phe273, Arg274,
Ile347, Asn346, Gln345,
Ala262, Gly261, Ser441,
Ser442 | 1.94 | 1 |
| 15d-PGJ
2 | −8.10±1.28 | Ala262,
Ser265, Asp348 (a1,
a2, Ile347) (b),
Asn346, Ile270, Phe273,
Arg274, Val445, Tyr280,
Ile411, Phe413, Phe414,
Phe297, Ile316, His363 | a1,
1.77; a2, 2.37; b, 2.09
a, 2.51; b, 1.98 | 3
2 |
| J11-C1 | −5.66±1.68 | Phe297,
Ile316, Ile411, Val412,
Phe413, Phe414, His363,
Gln345, Asn346, Ile347
(a), Asp348, Ser265,
Ala262, Ile270, Pro271
(b), Asp272, Phe273 | | |
| J19 | −3.59±0.74 | Val445,
Tyr280, Phe297, Phe414,
Phe413, Val412, Ile411,
Ile247, His363, Asn246,
Gln345, Gly261, Ala262,
Arg274 (a1, a2),
Phe273 | a1,
2.18; a2, 2.24 | 2 |
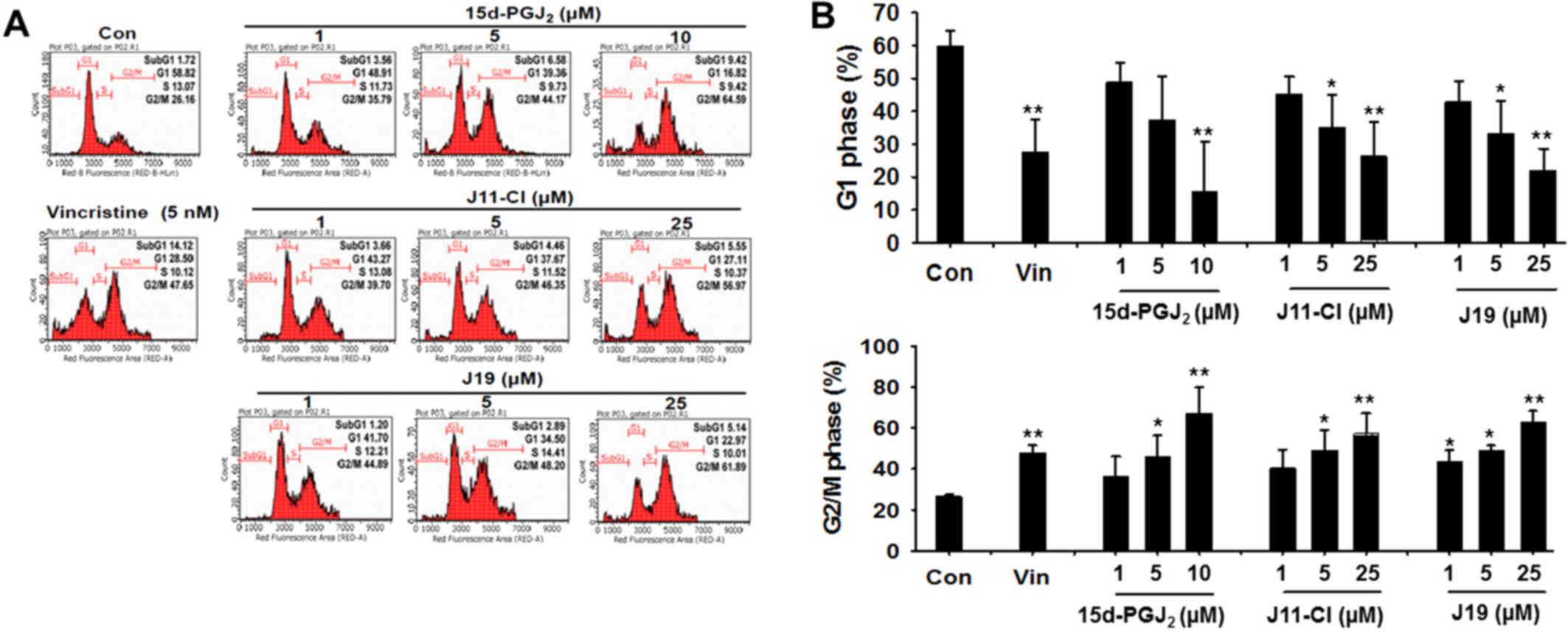
Effects of 15d-PGJ2, J19 and
J11-C1 on cell cycle progression in SKOV3 cells
SIRT inhibitors exhibited moderate cytotoxicity
towards various human cancer cells through the induction of cell
cycle arrest at a specific phase. The effects of
15d-PGJ2 and its derivatives on cell cycle progression
were determined using flow cytometry, and 15d-PGJ2 was
identified to significantly increase the number of cells in
G2/M phase after 48 h of incubation (Fig. 5). Similarly, J11-C1 and J19
significantly increased the population of SKOV3 cells in
G2/M phase following treatment for 48 h. Furthermore,
the proportion of cells in G1 phase was decreased
following treatment of SKOV3 cells with the drugs (Fig. 5).
Effects of 15d-PGJ2, J19 and
J11-C1 on apoptotic death of SKOV3 cells
To evaluate the apoptotic death of SKOV3 cells
following treatment with 15d-PGJ2, J19 or J11-C1,
Annexin V-FITC and PI staining, and western blot analyses were
performed. Concentration-dependent increases in apoptotic cell
death were observed following treatment with 15d-PGJ2
and its derivatives (Fig. 6A and
B). A significant increase in Bax expression and a parallel
decrease in Bcl-2 expression were also observed following treatment
with these drugs (Fig 6C). In
addition, the expression levels of cleaved caspase-3 and -9,
cleaved PARP and acetylated p53 were markedly increased in SKOV3
cells following treatment with high concentration of
15d-PGJ2 and its derivatives (Fig. 6D and E).
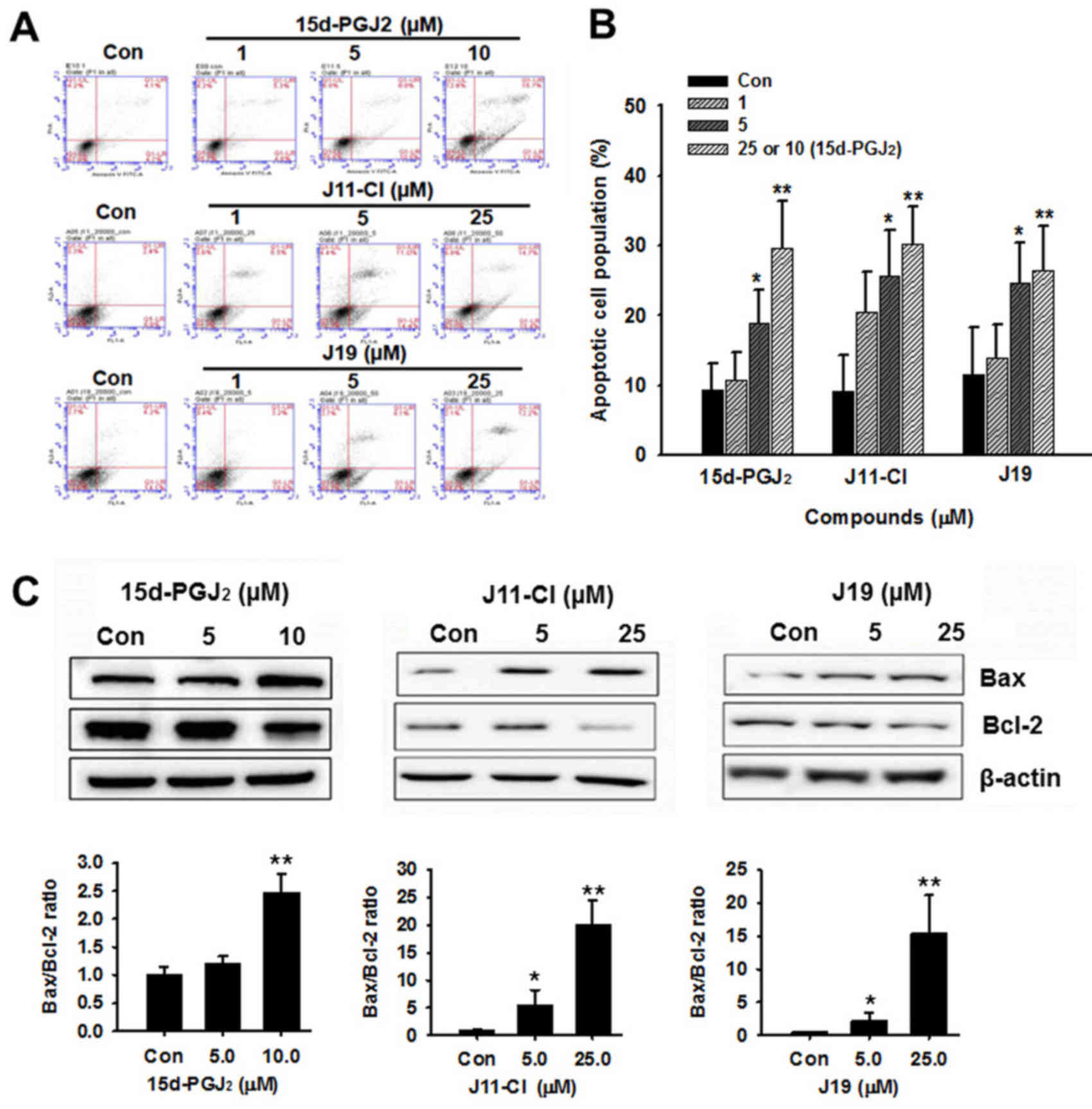 | Figure 6Effects of 15d-PGJ2,
J11-Cl and J19 on apoptotic pathways in SKOV3 cells. (A) SKOV3
cells were treated with 15d-PGJ2, J11-Cl or J19 at the
indicated concentrations for 48 h. The early and late stages of
apoptosis were detected on the basis of Annexin V and propidium
iodide staining using flow cytometry. (B) Quantification of
apoptosis results, presented as the mean ± standard error of the
mean of three independent experiments. (C) Effects of
15d-PGJ2, J11-Cl and J19 on expression of
apoptosis-associated proteins in SKOV3 cells. Changes in Bcl-2 and
Bax expression were determined using western blotting and
quantified from three independent experiments using densitometry.
(D) Effects of 15d-PGJ2, J11-Cl and J19 on the
expression of apoptosis-associated proteins in SKOV3 cells. Changes
in PARP, cleaved caspase-3, cleaved caspase-9, p53 and acetylated
p53 expression levels were determined relative to expression of
β-actin. (E) Quantification of western blot analysis from three
independent experiments using densitometry. *P<0.05,
**P<0.01 vs. control. 15d-PGJ2,
15-deoxy-Δ12,14-prostaglandin J2; Con, control; PARP,
poly(ADP-ribose) polymerase; Ac, acetylated; C-, cleaved; Bcl-2,
B-cell lymphoma 2. |
Effects of 15d-PGJ2, J19 and
J11-C1 on autophagic cell death
To evaluate autophagic cell death induced by
15d-PGJ2, J19 and J11-C1, western blot analysis and
acridine orange staining were performed. The conversion of the
soluble form of LC3-I into the autophagic vesicle-associated form
LC3-II is considered to indicate autophagosome formation (22). As presented in Fig. 7A and B, a high concentration of
15d-PGJ2, J11-Cl and J19 significantly increased the
level of LC3-II, whereas unconjugated LC3-I levels were decreased.
Similar to LC3-II, beclin-1 levels increased following treatment
with 15d-PGJ2, J11-Cl or J19 in a
concentration-dependent manner (Fig.
7A and B). Subsequently, the induction of autophagy was
confirmed by acridine orange staining. Acridine orange is a
lysotropic dye that accumulates in acidic organelles in a
pH-dependent manner (23-26). At neutral pH, acridine orange emits
a green fluorescence, but emits bright red fluorescence within
acidic vesicles when protonated and becomes trapped within the
organelle (2). As presented in
Fig. 7C and D, the control cells
exhibited primarily green fluorescence and minimal red
fluorescence, which indicated a lack of acidic vesicular
organelles. However, drug-treated cells exhibited a more marked red
fluorescence 48 h post-treatment compared with the control cells.
The increased red fluorescence intensity observed in cultured SKOV3
cells following drug treatment indicated enhanced acidification of
vesicular organelles (Fig. 7C).
Fig. 7D presents the mean
fluorescence intensity of the control and drug-treated cells.
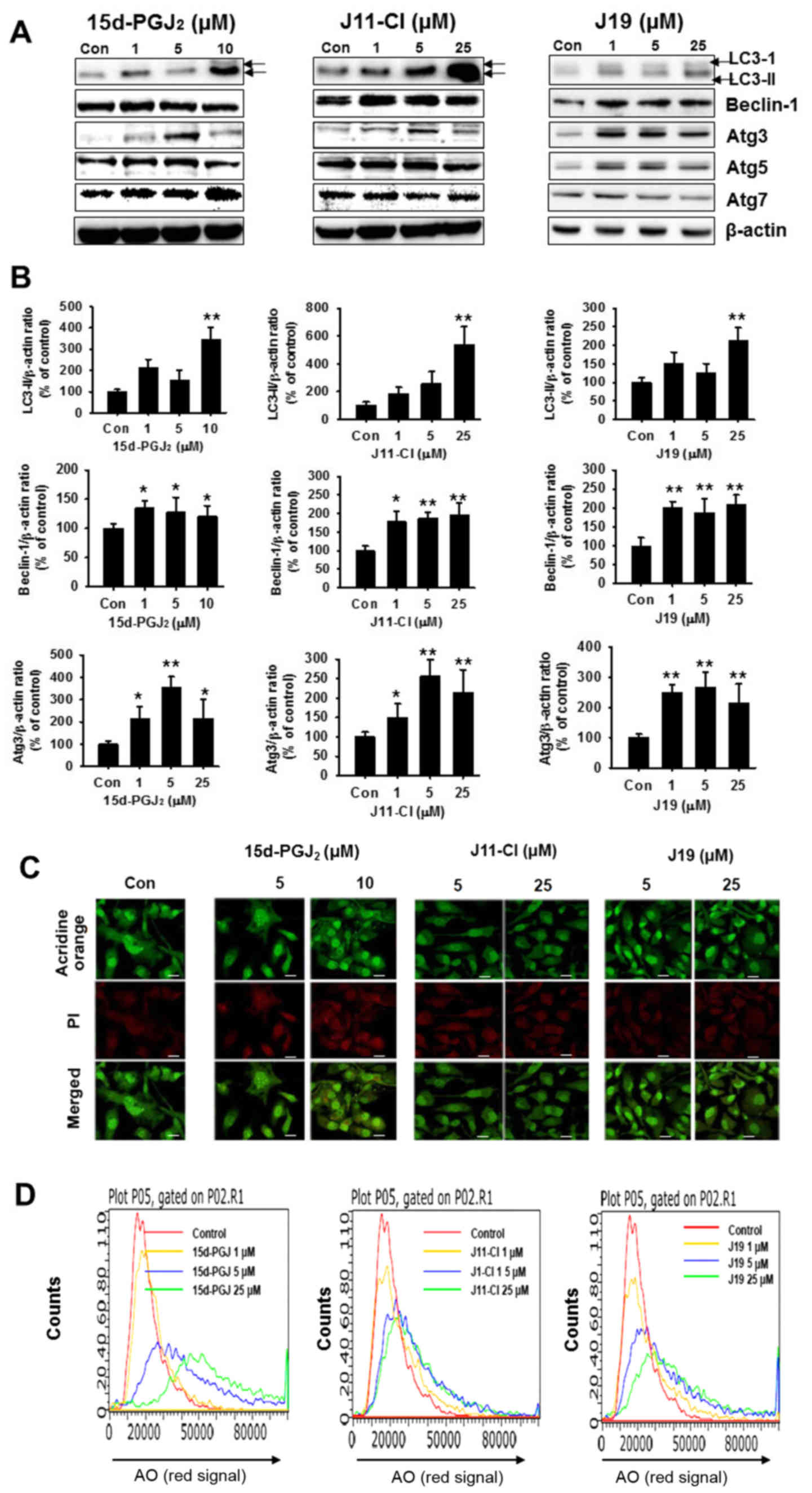 | Figure 7Effects of 15d-PGJ2,
J11-Cl and J19 on the autophagic pathway in SKOV3 cells. (A) SKOV3
cells were treated with 15d-PGJ2, J11-Cl and J19 for 48
h, and expression levels of LC3-II, beclin-1, Atg3, Atg5 and Atg7
were determined using western blotting. (B) Quantification of
western blot assays of three independent experiments using
densitometry. *P<0.05, **P<0.01 vs.
control. (C) Fluorescence of acridine orange-stained SKOV3 cells
treated for 48 h with the indicated drug concentration using
confocal microscopy. Scale bar, 50 µm. (D) Flow cytometric
analysis following acridine orange staining and histogram profiles
of control and drug-treated cells. 15d-PGJ2,
15-deoxy-Δ12,14-prostaglandin J2; LC3, light
chain 3; Atg, autophagy-related; Con, control; AO, acridine
orange. |
Autophagy inhibition ameliorates
cytotoxicity of SKOV3 cells
To investigate the anticancer mechanism of
15d-PGJ2, J19 or J11-C1 against autophagic cell death in
SKOV3 cells, 3-MA (1 mM), an autophagy-specific inhibitor, was
incubated with SKOV3 cells. This agent blocks autophagy by
inhibiting phosphoinositide 3-kinase, an enzyme required for
autophagy (25). As presented in
Fig. 8A, the morphological changes
indicated that 3-MA alone was not cytotoxic to SKOV3 cells.
However, the combination of 3-MA with 15d-PGJ2, J19 or
J11-C1 decreased cell death compared with individual treatment with
15d-PGJ2, J19 or J11-C1 (Fig. 8A). To confirm the molecular
mechanism and whether autophagic cell death was affected by 3-MA
treatment, the expression levels of LC3, beclin-1, Atg5 and Atg7
were determined using western blotting. The expression levels of
LC3-II and beclin-1 in 15d-PGJ2-, J19-, or J11-C1 and
3-MA-treated SKOV3 cells were markedly decreased in SKOV3 cells
(Fig. 8B). Next, the induction of
autophagy was confirmed using MDC staining. The vital dye, which is
commonly used to study autophagy, accumulates in autophagic
vacuoles following a combination of ion trapping and specific
interactions with vacuole membrane lipids (26,27).
As presented in Fig. 8C, the
autophagic cell death induced by 15d-PGJ2 and its
derivatives was decreased by co-treatment of SKOV3 cells with
3-MA.
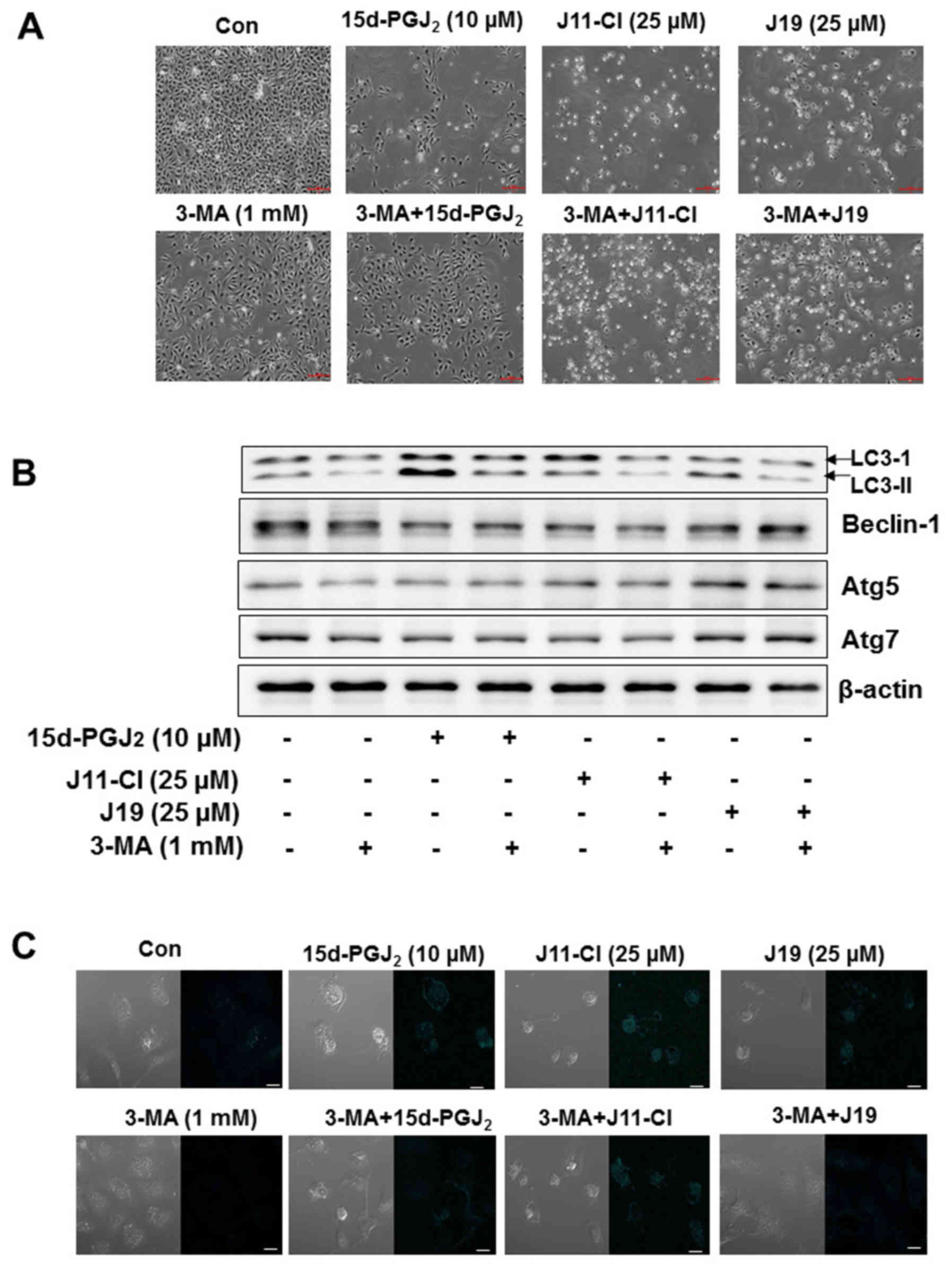 | Figure 8Effects of 3-MA on
15d-PGJ2-, J11-Cl- and J19-induced autophagic death of
SKOV3 cells. SKOV3 cells were cultured for 24 h in DMEM and exposed
to 15d-PGJ2, J11-Cl and J19 in the presence or absence
of 3-MA (1 mM) for 48 h. Autophagy activity was assayed by MDC
incorporation and western blot analysis with anti-LC3,
anti-beclin-1, anti-Atg5 and anti-Atg7 antibodies. (A)
Morphological changes in SKOV3 cells were determined by
co-treatment with 3-MA (1 mM) and drugs. (B) Expression levels of
LC3-II, beclin-1, Atg5 and Atg7 were determined using western blot
analysis. β-actin was used as the internal control. (C) MDC
staining indicated inhibition of autophagy in SKOV3 cells following
co-treatment with 3-MA (1 mM) and drugs. Cells were examined using
confocal microscopy. Scale bar, 10 µm. 3-MA,
3-methyladenine; 15d-PGJ2,
15-deoxy-Δ12,14-prostaglandin J2; LC3, light
chain 3; Con, control; Atg, autophagy-related. |
Discussion
SIRT1 is the most widely studied member of the SIRT
family and is known to modulate cell proliferation,
differentiation, apoptosis, migration and invasion (3,7,28).
SIRT1 controls cellular senescence and is overexpressed in specific
cancer cell types (29,30). In the present study, the effect of
15d-PGJ2 and its derivatives on SIRT1-mediated cell
death pathways were investigated in SKOV3 cells. First, in the
comparison of the cytotoxicity of the tested drugs,
15d-PGJ2 exhibited more potent cytotoxicity compared
with that of J11-Cl and J19, and decreased the expression levels of
SIRT1 protein. Considering the reported electrophilic carbon atom
(C9) of 15d-PGJ2, we hypothesized that the inhibitory
activity of 15d-PGJ2 may be a consequence of the
reactive carbon. The observation that 15d-PGJ2 inhibited
the gene expression of SIRT1 led to the consideration of whether
15d-PGJ2 was able to directly inhibit the activity of
SIRT1. To address this question, an acetylated p53 protein was
selected as a substrate to determine SIRT1 activity. As expected,
SIRT1/2 activity was significantly decreased by
15d-PGJ2. To investigate the contribution of the C9
electrophilic carbon atom to this direct inhibitory activity, the
molecular mechanics of 15d-PGJ2, J11-Cl and J19 on SIRT1
were elucidated using docking simulations. Although the docking
conformation of 15d-PGJ2 with the SIRT1 protein did not
reveal any nucleophile that reacted with C9 within 3 Å of the
chemical, it revealed a higher binding affinity (best docking
score, −9.43) compared with that of the other drugs, with the
formation of three hydrogen bonds with Asp348 and
Ile347. The prior arrangement of the three hydrogen
bonds also appeared to produce greater van der Waals interactions
and electrostatic attractions for this drug. J11-Cl and J19
exhibited moderate (up to −7.34) and low docking scores (up to
−4.33), respectively, to form two hydrogen bonds with
Ile347 and Pro217 and a hydrogen bond with
Arg274; these configurations were less stable and potent
compared with that of 15d-PGJ2. Therefore, the strongly
overlapping conformations of the tested drugs may explain the
difference in binding affinity.
Next, to determine the mechanisms underlying the
action of 15d-PGJ2 and its derivatives, their effects on
apoptotic and autophagic cell death pathways were investigated.
First, cell cycle analysis was performed and it was identified that
15d-PGJ2 and J19 significantly induced G2/M
phase arrest in SKOV3 cells. Previous studies indicated that
sirtinol, a class III HDAC inhibitor, induced senescent-like growth
arrest in breast cancer cells (31). SIRT1 has a more prominent function
in controlling cell growth and survival as it exists in the same
intracellular compartment as most cell cycle and cell death
regulators (7,32). As expected, 15d-PGJ2 and
J19 treatment significantly increased cell cycle arrest in SKOV3
cells. These results are similar to those of a previous study,
which indicated that the inhibition of SIRT1 activates p53 and Bax
gene expression, thereby inducing cell cycle arrest and apoptosis
(33). To investigate the
mechanism underlying the anticancer effects of 15d-PGJ2
and its derivatives, apoptotic cell death was assessed. Flow
cytometric analysis revealed that 15d-PGJ2 and J11-Cl
markedly induced apoptosis and subsequently increased the
sub-G1 phase cell population. The downstream mechanism
of apoptotic cell death was investigated. 15d-PGJ2 and
its derivatives increased the expression of Bax and decreased that
of Bcl-2 in SKOV3 cells.
Autophagy, another cell death pathway that is
important in tumor biology, was also investigated in SKOV3 cells
following treatment with 15d-PGJ2 and its derivatives.
The inhibition of autophagy was suggested to allow the continued
growth of pre-cancerous cells and autophagy may act as a tumor
suppressor (34,35). Generally, cancer cells require
autophagy to survive nutrient-limiting and low-oxygen conditions,
particularly in the poorly vascularized internal region of the
tumor. According to the results of the present study, the
autophagic process led to SKOV3 cell death following treatment with
15d-PGJ2 or its derivatives. Significant increases in
LC3B and Atg7 levels were observed following 15d-PGJ2,
J11-Cl and J19 treatment relative to the control cells, and these
changes were markedly associated with the cytotoxic effects of
these drugs. These results were confirmed by the induction of AVOs
in the cytoplasm that was stained with acridine orange. Most
importantly, novel mechanisms of action of 15d-PGJ2 and
its derivatives were revealed as the inhibition of SIRT1 regulated
multiple mechanisms of tumor cell biology in SKOV3 cells.
In conclusion, the anticancer effects of newly
synthesized 15d-PGJ2 derivatives were investigated and
the underlying molecular mechanisms of these drugs were elucidated.
On the basis of the structural similarity of 15d-PGJ2,
J11-Cl and J19 bind to and inhibit SIRT1 enzyme activity. This
indicated that J11-Cl and J19 may exert anticancer activity by
binding to SIRT1, to enhance apoptotic and autophagic cell death of
ovarian cancer cells. However, limitations to the present study
require attention. Although the results of the present study
clearly indicated that SIRT1 inhibition exhibited potent
anti-cancer activity in ovarian cancer cells, further investigation
to confirm the corresponding effect using in vivo xenograft
models is warranted.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Research Foundation of Korea funded by the Korean government (grant
nos. 2016R1A2B2011071 and 2016R1A4A1011189).
Availability of data and materials
The datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
HSK and MHK conceived and designed the experiments.
IHT, EYP, PD and JYS performed the experiments and statistical
analysis. SS, MHK and SYL performed the docking study. JHJ helped
to analyze and interpret the data, and critically revised the
manuscript. SYL, MHK and HSK drafted the manuscript. All authors
read and approved the final paper.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Nguyen LT, Chen H, Pollock C and Saad S:
SIRT1 reduction is associated with sex-specific dysregulation of
renal lipid metabolism and stress responses in offspring by
maternal high-fat diet. Sci Rep. 7:89822017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chan SH, Hung CH, Shih JY, Chu PM, Cheng
YH, Lin HC and Tsai KL: SIRT1 inhibition causes oxidative stress
and inflammation in patients with coronary artery disease. Redox
Biol. 13:301–309. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wang J, Kim TH, Ahn MY, Lee J, Jung JH,
Choi WS, Lee BM, Yoon KS, Yoon S and Kim HS: Sirtinol, a class III
HDAC inhibitor, induces apoptotic and autophagic cell death in
MCF-7 human breast cancer cells. Int J Oncol. 41:1101–1109. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Park EY, Woo Y, Kim SJ, Kim DH, Lee EK, De
U, Kim KS, Lee J, Jung JH, Ha KT, et al: Anticancer effects of a
new SIRT inhibitor, MHY2256, against human breast cancer MCF-7
cells via regulation of MDM2-p53 binding. Int J Biol Sci.
12:1555–1567. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Peck B, Chen CY, Ho KK, Di Fruscia P,
Myatt SS, Coombes RC, Fuchter MJ, Hsiao CD and Lam EW: SIRT
inhibitors induce cell death and p53 acetylation through targeting
both SIRT1 and SIRT2. Mol Cancer Ther. 9:844–855. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim HB, Lee SH, Um JH, Oh WK, Kim DW, Kang
CD and Kim SH: Sensitization of multidrug-resistant human cancer
cells to Hsp90 inhibitors by downregulation of SIRT1. Oncotarget.
6:36202–36218. 2015.PubMed/NCBI
|
|
7
|
Kim TH and Kim HS, Kang YJ, Yoon S, Lee J,
Choi WS, Jung JH and Kim HS: Psammaplin A induces Sirtuin
1-dependent autophagic cell death in doxorubicin-resistant
MCF-7/adr human breast cancer cells and xenografts. Biochim Biophys
Acta. 1850:401–410. 2015. View Article : Google Scholar
|
|
8
|
Chu F, Chou PM, Zheng X, Mirkin BL and
Rebbaa A: Control of multidrug resistance gene mdr1 and cancer
resistance to chemotherapy by the longevity gene sirt1. Cancer Res.
65:10183–10187. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Smith WL: Prostanoid biosynthesis and
mechanisms of action. Am J Physiol. 263:F181–F191. 1992.PubMed/NCBI
|
|
10
|
Straus DS, Pascual G, Li M, Welch JS,
Ricote M, Hsiang CH, Sengchanthalangsy LL, Ghosh G and Glass CK:
15-deoxy-delta 12,14-prostaglandin J2 inhibits multiple steps in
the NF-kappa B signaling pathway. Proc Natl Acad Sci USA.
97:4844–4849. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cho WH, Choi CH, Park JY, Kang SK and Kim
YK: 15-deoxy-(Delta12,14)-prostaglandin J2 (15d-PGJ2)
induces cell death through caspase-independent mechanism in A172
human glioma cells. Neurochem Res. 31:1247–1254. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
de Jong E, Winkel P, Poelstra K and
Prakash J: Anticancer effects of 15d-prostaglandin-J2 in
wild-type and doxorubicin-resistant ovarian cancer cells: Novel
actions on SIRT1 and HDAC. PLoS One. 6:e251922011. View Article : Google Scholar
|
|
13
|
Ahmad P, Rasool S, Gul A, Sheikh SA, Akram
NA, Ashraf M, Kazi AM and Gucel S: Jasmonates: Multifunctional
roles in stress tolerance. Front Plant Sci. 7:8132016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dang HT, Lee HJ, Yoo ES, Hong J, Bao B,
Choi JS and Jung JH: New jasmonate analogues as potential
anti-inflammatory agents. Bioorg Med Chem. 16:10228–10235. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Choo J, Lee Y, Yan XJ, Noh TH, Kim SJ, Son
S, Pothoulakis C, Moon HR, Jung JH and Im E: A novel peroxisome
proliferator-activated receptor (PPAR)γ agonist 2-hydroxyethyl
5-chloro-4,5-didehydrojasmonate exerts anti-inflammatory effects in
colitis. J Biol Chem. 290:25609–25619. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
LigPrep, version 2.5. Schrödinger, LLC;
New York: 2011
|
|
17
|
Tripathi SK, Muttineni R and Singh SK:
Extra precision docking, free energy calculation and molecular
dynamics simulation studies of CDK2 inhibitors. J Theor Biol.
334:87–100. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Friesner RA, Banks JL, Murphy RB, Halgren
TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK,
et al: Glide: A new approach for rapid, accurate docking and
scoring. 1. Method and assessment of docking accuracy. J Med Chem.
47:1739–1749. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Friesner RA, Murphy RB, Repasky MP, Frye
LL, Greenwood JR, Halgren TA, Sanschagrin PC and Mainz DT: Extra
precision glide: Docking and scoring incorporating a model of
hydro-phobic enclosure for protein-ligand complexes. J Med Chem.
49:6177–6196. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Halgren TA, Murphy RB, Friesner RA, Beard
HS, Frye LL, Pollard WT and Banks JL: Glide: A new approach for
rapid, accurate docking and scoring. 2. Enrichment factors in
database screening. J Med Chem. 47:1750–1759. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Jacobson MP, Pincus DL, Rapp CS, Day TJ,
Honig B, Shaw DE and Friesner RA: A hierarchical approach to
all-atom protein loop prediction. Proteins. 55:351–367. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Murugan S and Amaravadi RK: Methods for
studying autophagy within the tumor microenvironment. Adv Exp Med
Biol. 899:145–166. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chalkiadaki A and Guarente L: The
multifaceted functions of sirtuins in cancer. Nat Rev Cancer.
15:608–624. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pierzyńska-Mach A, Janowski PA and
Dobrucki JW: Evaluation of acridine orange, LysoTracker Red, and
quinacrine as fluorescent probes for long-term tracking of acidic
vesicles. Cytometry A. 85:729–737. 2014. View Article : Google Scholar
|
|
25
|
Shingu T, Fujiwara K, Bögler O, Akiyama Y,
Moritake K, Shinojima N, Tamada Y, Yokoyama T and Kondo S:
Inhibition of autophagy at a late stage enhances imatinib-induced
cytotoxicity in human malignant glioma cells. Int J Cancer.
124:1060–1071. 2009. View Article : Google Scholar
|
|
26
|
Paglin S, Hollister T, Delohery T, Hackett
N, McMahill M, Sphicas E, Domingo D and Yahalom J: A novel response
of cancer cells to radiation involves autophagy and formation of
acidic vesicles. Cancer Res. 61:439–444. 2001.PubMed/NCBI
|
|
27
|
de Duve C, de Barsy T, Poole B, Trouet A,
Tulkens P and Van Hoof F: Commentary. Lysosomotropic agents.
Biochem Pharmacol. 23:2495–2531. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
He S, He C, Yuan H, Xiong S, Xiao Z and
Chen L: The SIRT 3 expression profile is associated with
pathological and clinical outcomes in human breast cancer patients.
Cell Physiol Biochem. 34:2061–2069. 2014. View Article : Google Scholar
|
|
29
|
Wang P, Lv C, Zhang T, Liu J, Yang J, Guan
F and Hong T: FOXQ1 regulates senescence-associated inflammation
via activation of SIRT1 expression. Cell Death Dis. 8:e29462017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen J, Zhang B, Wong N, Lo AW, To KF,
Chan AW, Ng MH, Ho CY, Cheng SH, Lai PB, et al: Sirtuin 1 is
upregulated in a subset of hepatocellular carcinomas where it is
essential for telomere maintenance and tumor cell growth. Cancer
Res. 71:4138–4149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ota H, Tokunaga E, Chang K, Hikasa M,
Iijima K, Eto M, Kozaki K, Akishita M, Ouchi Y and Kaneki M: Sirt1
inhibitor, Sirtinol, induces senescence-like growth arrest with
attenuated Ras-MAPK signaling in human cancer cells. Oncogene.
25:176–185. 2006. View Article : Google Scholar
|
|
32
|
Lin Z and Fang D: The roles of SIRT1 in
cancer. Genes Cancer. 4:97–104. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lee JT and Gu W: SIRT1: Regulator of p53
deacetylation. Genes Cancer. 4:112–117. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yang ZJ, Chee CE, Huang S and Sinicrope
FA: The role of autophagy in cancer: Therapeutic implications. Mol
Cancer Ther. 10:1533–1541. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Sever ON and Demir OG: Autophagy: Cell
death or survive mechanism. J Oncol Sci. 3:37–44. 2017.
|