Introduction
Soft-tissue sarcomas (STSs) are a group of
heterogeneous tumors that account for <1% of all cancers
(1). Therefore, identifying
potential targets in STSs is challenging. The most successful
example is the identification of the activated mutation of
KIT/platelet-derived growth factor receptor A (PDGFRA) in
gastrointestinal stromal tumors (GISTs). Tyrosine kinase inhibitors
against activated KIT/PDGFRA have been successfully developed for
GISTs (2-4). Other examples include rare tumors
such as dermatofibrosarcoma protuberans (DFSPs) (5) or inflammatory myofibroblastic tumor
(6). For most other types of
sarcomas, no satisfactory target therapies are available for
clinical use.
Expression profiling is one of the most commonly
used methods for exploring novel targets and biomarkers.
Nonetheless, identifying treatment targets in sarcoma is difficult
due to its complex genomic background. However, by using genomic
and expression profiling in 183 STSs, Chibon et al (7) established a prognostic gene
expression signature, complexity index in sarcomas (CINSARC),
comprising 67 genes related to mitosis and chromosome management.
CINSARC could predict the metastasis outcome in an independent
validation set of 127 sarcomas (7). By reanalyzing a data set of GISTs,
our group also identified aurora kinase A (AURKA), along with other
genes involving cell cycle and mitosis, as prognostic factors for
recurrence. AURKA could also act as a potential treatment target
(8,9). Therefore, through the use of
expression profiling analysis, identifying potential biomarkers and
targets in sarcoma is possible.
Liposarcoma (LPS) is one of the most frequently
reported cancer of the STSs (10,11).
Chemotherapy is not effective in LPS (12), and at present, no satisfactory
target agents against the disease are available. Expression profile
studies revealed the activation of the cell cycle and checkpoint
pathways in LPS, and provided several possible novel therapeutic
strategies (13). A cyclin
dependent kinase (CDK)4/6 inhibitor was used in the treatment of
LPS (14).
In this study, we reanalyzed two published
microarray data sets of well-differentiated (WD) and
dedifferentiated (DD) LPS to identify potential biomarkers or
targets in LPS. We determined that AURKA could be a potential
biomarker for predicting poor prognosis in LPS. Subsequently, we
identified AURKA as a potential therapeutic target for the
treatment of LPS.
Materials and methods
Bioinformatics analysis
The microarray and limited clinicopathological data
from the Gene Expression Omnibus data set GSE30929 for LPS, a raw
data set from the study of Gobble et al (15), were obtained from the National
Center for Biotechnology Information website (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30929).
Only data of WD LPS and DD LPS were used for analysis. Gene
expression data from GSE30929 were normalized using dChip (16,17)
and filtered with a max/min threshold of ≥5, after which they were
exported for further analysis. For another data set from the study
of Singer et al (13),
including 12 cases of DD LPS and 18 cases of WD LPS, only certain
genes were available from www.cbio.mskcc.org/Public/Liposarcoma. Thus, no
further filtering process was conducted. Both data sets are the
Affymetrix U133A platform (Thermo Fisher Scientific, Inc.). In
order to identify genes showing statistically significant,
concordant differences of expressions between two biological states
(e.g. phenotypes), Gene Set Enrichment Analysis (GSEA), a method
developed by the Broad Institute, was performed using software
downloaded from http://www.broadinstitute.org/gsea/index.jsp (18,19).
The expression levels of individual genes were obtained using
Z-score transformation, and the differences between different
subtypes were then compared using a t-test. In the GSE30929 data
set, data regarding patients' endpoint (distant recurrence) and
follow-up time were obtained (ftp://ftp.ncbi.nlm.nih.gov/geo/series/GSE30nnn/GSE30929/matrix/).
Therefore, distant recurrence-free survival (DRFS) was estimated
via Kaplan-Meier analysis, and a log-rank test was used to compare
the survival curves between groups.
Cell lines and reagents
The LPS cell lines, including LPS853 was kindly
provided by Dr Fletcher JA (Department of Pathology, Brigham and
Women's Hospital), while NDDLS-1 was kindly provided by Dr Ariizumi
(Division of Orthopedic Surgery, Niigata University Graduate School
of Medical and Dental Sciences). LPS853 cells were maintained in
RPMI medium with 15% fetal bovine serum (HyClone; GE Healthcare
Life Sciences), 100 µg/ml penicillin streptomycin (HyClone;
GE Healthcare Life Sciences) and 2 mM L-glutamine solution
(HyClone; GE Healthcare Life Sciences); NDDLS-1 cells were
maintained in RPMI medium with 10% fetal bovine serum (HyClone; GE
Healthcare Life Sciences), 100 µg/ml penicillin streptomycin
(HyClone; GE Healthcare Life Sciences), 2 mM L-Glutamine solution
(HyClone; GE Healthcare Life Sciences) and 60 µg/ml
kanamycin (Thermo Fisher Scientific, Inc.). MLN8237, an
AURKA-selective inhibitor, and chemotherapeutic agents doxorubicin,
topotecan, cisplatin and taxol were purchased from Selleck
Chemicals. For MLN8237, concentrations of 0, 0.02, 0.05, 0.1, 0.5
and 1 µM were used to evaluate mitotic inhibition, the cell
cycle and cytotoxic effects; concentrations of 0, 0.4, 0.8, 1.2,
1.6 and 2 µM were used for the exploring synergistic effects
with chemotherapeutic agents. The concentrations of
chemotherapeutic agents were listed as follows: Cisplatin 0, 4, 8,
12, 16, 20 µM; topotecan 0, 4, 8, 12, 16, 20 µM;
doxorubicin 0, 0.1, 0.2, 0.3, 0.4, 0.5 µM; and taxol 0, 6,
12, 18, 24, 30 µM. Nocodazole was purchased from
Sigma-Aldrich (M1404-2MG; Merck KGaA). 0 µM inhibitor served
as the control. The following antibodies were used for
immunoblotting: β-Actin (cat. no. ab6276-100; 1:10,000; Abcam);
Aurora A (cat. no. 3092; 1:1,000; Cell Signaling Technology, Inc.);
anti-phosphorylated-Aurora A (Thr288; p-Aurora A; cat. no. 3079;
1:1,000; Cell Signaling Technology, Inc.); poly-(ADP-ribose)
polymerase (PARP; cat. no. 9542; 1:1,000; Cell Signaling
Technology, Inc.), Histone H3 (HisH3; cat. no. 9715; 1:1,000; Cell
Signaling Technology, Inc.), anti-phosphorylated (p)HisH3 (Ser10;
pHisH3; cat. no. 9701; 1:2,000; Cell Signaling Technology, Inc.),
p70S6 kinase (cat. no. 9202; 1:1,000; Cell Signaling Technology,
Inc.) and pp70S6 kinase (cat. no. 9205; 1:500; Cell Signaling
Technology, Inc.). The secondary antibodies used were horse
anti-mouse IgG (cat. no. 7076; 1:3,000) and goat anti-rabbit IgG
(cat. no. 7074; 1:3,000), both from Cell Signaling Technology.
Immunoblotting
Immunoblotting was performed as previously described
(20). In brief, cultured
monolayer cells were rinsed in PBS and scraped into lysis buffer
(RIPA Lysis and Extraction Buffer; cat. no. 89901; Thermo Fisher
Scientific, Inc.) containing protease and a phosphatase inhibitor
cocktail (1:100 dilution; Thermo Fisher Scientific, Inc.). Lysates
were first incubated for 30 min at 4°C and then centrifuged for 30
min at 16,000 × g at 4°C. A Pierce BCA Protein Assay Kit (Thermo
Fisher Scientific, Inc.) was employed to determine protein
concentrations. Protein extracts (20-50 µg per lane) were
then electrophoretically separated on SDS-PAGE (8-12% depending on
the molecular weights of proteins), transferred to polyvinylidene
difluoride membranes (PerkinElmer, Inc.), and immunoblotted with
specific antibodies. For primary antibodies, the sample was
incubated at 4°C overnight; for secondary antibody, the sample was
incubated at room temperature for 1 h. Immunoreactive bands were
detected through enhanced chemiluminescence (EMD Millipore) and
exposed to X-ray films. Protein expression was quantified using
ImageJ version 1.51j8 (National Institutes of Health).
Cytotoxicity assay
TACS™ MTT assay (R&D Systems) was used for the
cell cytotoxicity assay. In brief, ~2,000-20,000 cells per 100
µl per well were seeded in 96-well plates at 37°C overnight.
The following day, reagents at different concentrations were added
in triplicate. The plates were subsequently incubated for the 72 h
at 37°C, incubated with 10 µl MTT reagent, and then
incubated for a further 4 h at 37°C. Dimethyl sulfoxide 100
µl/well was added and mixed thoroughly to dissolve the blue
crystals. A Vmax microplate reader (Molecular Devices, LLC) at
wavelengths of 570 nm (test) and 650 nm was used to measure the
absorbance. Cell survival was calculated through the following
equation: % Survival=(mean experimental absorbance/mean control
absorbance) ×100 (21).
The synergistic effect of the applied drug
combination was measured through a combination index (CI), which
was calculated using CalcuSyn software 2.1 (Premier Biosoft
International) (22). CI >1 was
defined as antagonism, CI=1 as additivity, and CI<1 as synergy;
the experiment was performed in triplicate.
A trypan blue exclusion assay (23) was also employed for exploring the
cytotoxic effects of MLN8237. LPS cells were plated at
5×104 cells/well in 24-well plates; then, different
concentrations of MLN8237 was added to each well the next day, and
the plates were incubated at 37°C for 72 h. Subsequently, the cells
were trypsinized and mixed with trypan blue at room temperature and
observed immediately. Viable cells, recognized as cells that did
not absorb trypan blue, were counted using a light microscope
(magnification, ×100; cells were counted in the large, central
gridded square at 1 mm2, then multiplied by
1×104 to estimate the number of cells per ml). All
experiments were performed in triplicate.
Apoptosis assessment through Annexin V
staining
After drug treatment, cells (1×105
cells/well) were washed with 1X PBS once and resuspended in 100
µl staining solution (containing Annexin V fluorescein
isothiocyanate (FITC) and propidium iodide in HEPES buffer, BD
Pharmingen; BD Biosciences). The cells were then incubated at room
temperature for 15 min and then diluted in 1X Annexin V-binding
buffer (BD Biosciences). The percentages of apoptotic cells were
then measured using flow cytometry (BD FACSCanto II, BD
Biosciences; operation software: BD FACSDiva Software v8.0.2;
analysis software: FlowJo 7.6).
Small interfering RNA (siRNA)-mediated
knockdown (KD) of AURKA
For AURKA KD experiment, 5 ×105 of LPS853
cells were transfected with either AURKA siRNA or non-targeting
control pool using ON-TARGETplus system (GE Healthcare Dharmacon,
Inc.) according to manufacturer's protocols (Target Sequence:
UCGAAGAGAGUUAUUCAUA, CGGUAGGCCUGAUUGGGUU, UUCUUAGACUGUAUGGUUA,
AAUAGGAACACGUGCUCUA; 10 µM); DharmaFECT 1 Transfection
Reagent (cat. no. T-2001-03; GE Healthcare Dharmacon, Inc.) was
employed for transfection. The cells were incubated for 48, 72 and
96 h along at 37°C with respective controls, and number of viable
cells was measured using trypan blue exclusion assay, as described
above. The remaining cells then were lysed and the KD efficiency of
AURKA, as well as the protein levels of PARP were confirmed by
western blotting, as aforementioned.
Cell cycle analysis
Cell cycle analysis was performed using flow
cytometry, as previously described (9). After being trypsinized and washed
twice with PBS, cells (1×105 cells/ml) were fixed in 99%
ethanol at -20°C overnight. The following day, the fixed cells were
first washed twice with cold PBS, suspended in 420 µl PBS,
added to 50 µl of 10 mg/ml RNase A (Sigma-Aldrich; Merck
KGaA), and agitated at 37°C for 15 min. The cells were then
maintained at room temperature for 1 h following the addition of 20
µl propidium iodide (0.2 mg/ml). Subsequently, flow
cytometry was conducted using BD FACSCalibur (BD Biosciences) for
relative DNA content based on red fluorescence levels. FACSDiva
Software v8.0.2 was used to calculate the percentages of the cells
in the different phases of the mitotic cell cycle.
Senescence-associated β-galactosidase
(SA-β-gal) assay
SA-β-gal activity was detected using the Cellular
Senescence Assay Kit (EMD Millipore), as described in the
manufacturer's instructions. After being treated with MLN8237 at
37°C for 72 h, adherent LPS853 cells were fixed with 1X fix
solution at room temperature, incubate for 10 min, and stained with
X-gal in a staining solution at pH 6.0 at 37°C for 5 h. The cells
were washed twice with 1X PBS. The percentage of SA-β-gal-positive
cells (the number of positive cells relative to the total number of
cells) was quantified by counting 100 cells in three randomly
selected fields per dish with an OLYMPUS IX51 microscope
(brightfield with phase contrast; magnification, ×200).
Decoy receptor 2 (DcR2) expression
analysis via flow cytometry
The expression of DcR2 was detected using flow
cytometry (1×105 cells/100 µl). LPS853 cells were
first treated with MLN8237 at 37°C for 72 h, washed twice with 1X
PBS, and incubated with Alexa Fluor® 488-labeled
anti-DcR2 (R&D Systems, Inc.) on a shaker at room temperature
for 30 min. Cells were washed twice with 1X PBS and then
resuspended in 1X PBS, the mean of the fluorescence intensity on
the cell surface was measured by flow cytometry using FACSCalibur
(BD Biosciences).
Analysis of interleukin (IL)-1α, IL-6 and
IL-8 expression using reverse transcription-quantitative
transcriptase-polymerase chain reaction (RT-qPCR)
TRIzol® Reagent (Thermo Fisher
Scientific, Inc.) was used for RNA extraction from the cell lines,
in accordance with the manufacturer's protocols. The
SuperScript® III First-Strand Synthesis System
(Invitrogen Thermo Fisher Scientific, Inc.) was used for reverse
transcription with 1 µg RNA (25°C for 10 min, 50°C for 50
min, 85°C for 5 min, hold at 4°C). The copy number for IL-1α, IL-6,
IL-8, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) mRNA was
measured using RT-qPCR with Maxima SYBR® Green/ROX qPCR
Master Mix (Thermo Fisher Scientific, Inc.) and a
LightCycler® 480 System (Roche Diagnostics). The primer
sequences used in the reaction were as follows: IL-1α (forward),
5′-CCG TGA GTT TCC CAG AAG AA-3′; IL-1α (reverse),
5′-ACTGCCCAAGATGAAGACCA-3′; IL-6 (forward), 5′-CATTTGTGGTTGGGTCAG
G-3′; IL-6 (reverse), 5′-AGTGAGGAACAAGCCAGAGC-3′; IL-8 (forward)
5′-CAAGAGCCAGGAAGAAACCA-3′; IL-8 (reverse)
5′-AGCACTCCTTGGCAAAACTG-3′; GAPDH (forward)
5′-GCCAAGGTCATCCATGACAACT-3′; GAPDH (reverse),
5′-GAGGGGCCATCCACAGTCTT-3′ (24).
The qPCR thermocycling conditions were as follows: 95°C for 5 min
and then 45 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C
for 40 sec. The gene expression levels were calculated as described
previously (25).
Statistical and survival analysis
SPSS Statistics 22 (IBM Corp.) was used for
statistical analysis. As aforementioned, survival analysis was
estimated using the Kaplan-Meier method, and the log-rank test was
conducted for survival curve comparison. A Student's t-test was
used for comparing the difference of the expression levels of AURKA
between two subtypes of LPS, and the expression levels of p-Aurora
A in LPS cell lines before and after drug treatment. The difference
in expression levels of pHisH3 (assessed by western blotting), the
percentage of G2/M content (assessed by flow cytometry), percentage
of cell survival (assessed by MTT assay), number of viable cells
(assessed by trypan blue exclusion assay), percentage of apoptotic
cells (assessed by apoptosis assay), and the senescence-related
markers (SA-β-gal staining, flow cytometry of DcR2, and RT-qPCR of
IL-1α, IL-6, and IL-8) between control and treated (all in
triplicate) LPS cell lines were compared using one-way ANOVA, and
Bonferroni test was used as post hoc test. Data was presented as
the mean ± standard deviation; P<0.05 was considered to indicate
a statistically significant difference.
Results
AURKA is significantly upregulated in DD
LPS compared with WD LPS, and DD LPS patients with high AURKA
expression had significantly worse DRFS than those with low
expression
A total of 92 arrays from GSE30929 were obtained,
involving 40 cases of DD LPS and 52 cases of WD LPS (15). Microarray data were first
normalized through dChip, and a total of 17,089 probes were
obtained after filtering with a max/min threshold of ≥5. The
expression values of LPS from GSE30929 and Singer et al (13) were then analyzed using GSEA
software. Subsequently, a heatmap was generated from GESA using the
top 50 genes of each phenotype (DD vs. WD LPS) in each set of
tumors (Fig. S1). By comparing
the top 50 genes that were upregulated in DD LPS in both sets of
data, we identified 12 overlapping genes, namely nuclear and
spindle associated protein 1, maternal embryonic leucine zipper
kinase, AURKA, BAG family molecular chaperone regulator 2, CDC28
protein kinase regulatory subunit 2, kinesin family member 11, DNA
topoisomerase II, Rac GTPase-activating protein 1, abnormal
spindle-like microencephaly-associated protein,
ribonuclo-side-disphosphate reductase subunit M2, retinoic acid
induced 14 and ubiquitin-conjugating enzyme E2 S.
Notably, the top five gene sets enriched in DD LPS
in both sets of data were all involved in cell cycle regulation
(Tables I and II). Among the 12 overlapping genes,
AURKA is a well-known gene involved in cell cycle regulation
(26). Thus, we selected this gene
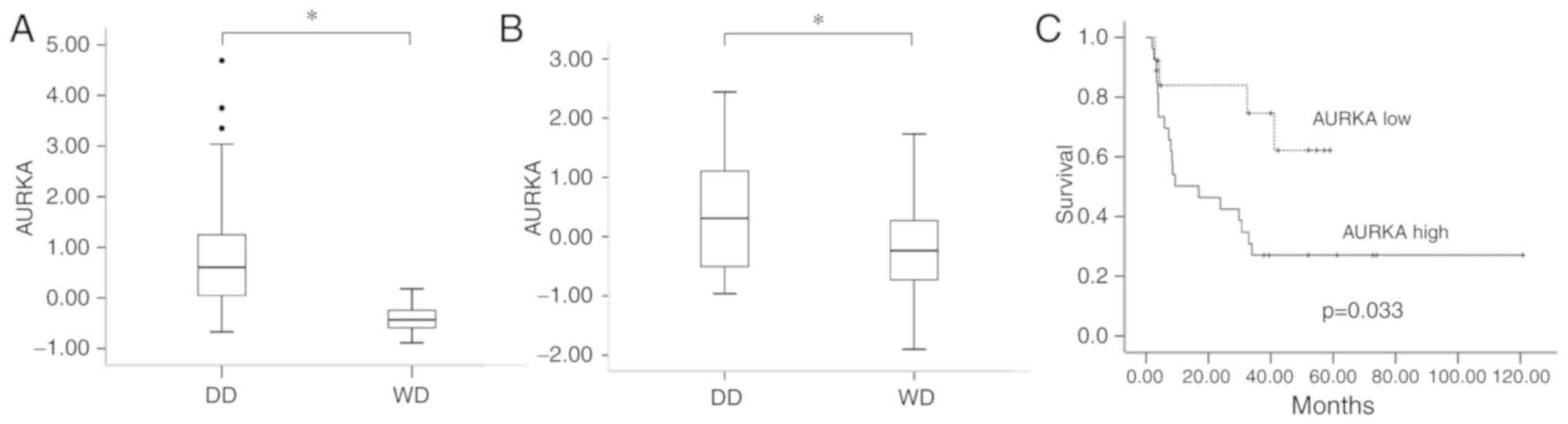
for further analysis. As presented in Fig. 1, in both sets of tumors, AURKA was
significantly upregulated in DD LPS in comparison with WD LPS.
Survival data are available in GSE30929. We then compared the DRFS
data of cases with low AURKA expression (Z-score <0) vs. those
with high AURKA expression in DD LPS. As shown in Fig. 1C, among the 40 cases of DD LPS,
patients with high expression of AURKA in tumors (n=27) had
significantly worse DRFS than those with low expression (n=13).
 | Table ITop 5 gene sets enriched in
dedifferentiated liposarcoma in GSE30929. |
Table I
Top 5 gene sets enriched in
dedifferentiated liposarcoma in GSE30929.
| NAME | NES | Nominal
P-value | FDR q-value |
|---|
| MITOSIS | 2.086395 | 0.002008 | 0.032142 |
|
M_PHASOFITOTIC_CELL_CY CLE | 2.078066 | 0.002004 | 0.019095 |
| SPINDLE | 2.052216 | <0.0001 | 0.019665 |
| M_PHASE | 2.039629 | <0.0001 | 0.016213 |
|
CELL_CYCLE_CHECKPOINT_GO_0000075 | 2.036067 | 0.004124 | 0.013608 |
 | Table IITop 5 gene sets enriched in
dedifferentiated liposarcoma the data of Singer et al
(13). |
Table II
Top 5 gene sets enriched in
dedifferentiated liposarcoma the data of Singer et al
(13).
| NAME | NES | Nominal
P-value | FDR q-value |
|---|
|
MITOTIC_CELL_CYCLE | 1.574733 | 0.016393 | 0.293481 |
| CELL_C YCLE_PROCES
S | 1.567766 | 0.024691 | 0.154675 |
|
CELL_CYCLE_PHASE | 1.547144 | 0.034908 | 0.132447 |
|
CELL_CYCLE_GO_0007049 | 1.533873 | 0.055215 | 0.112444 |
|
REGULATION_OF_CELL_C Y CLE | 1.312828 | 0.179167 | 0.477817 |
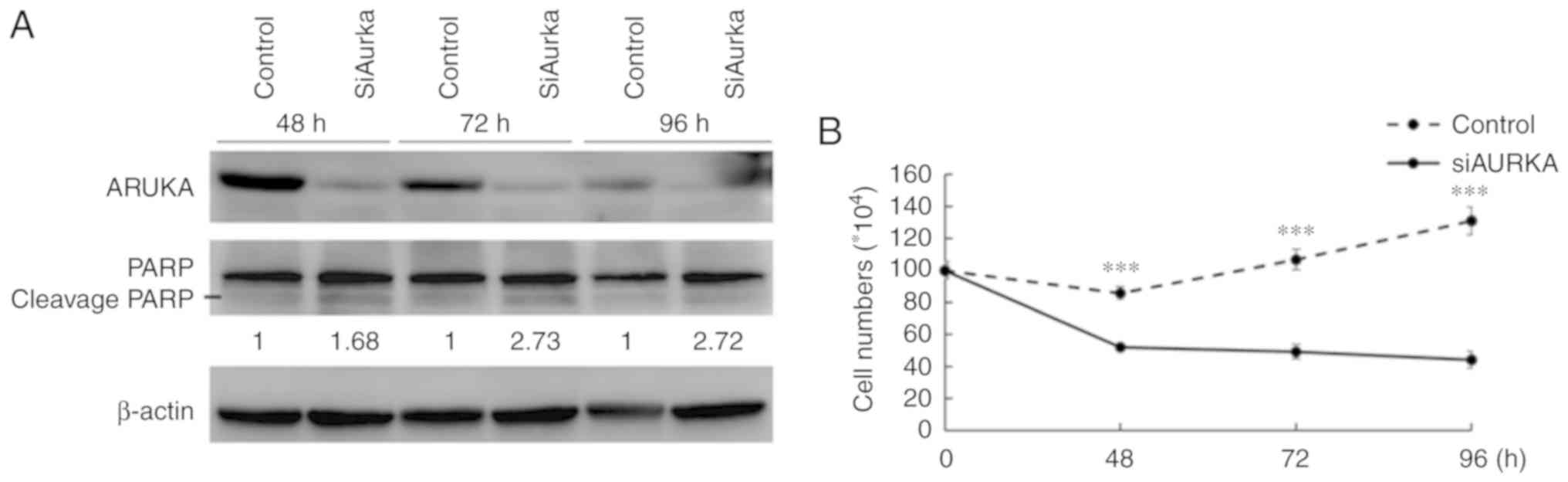
AURKA-KD decreases the number of viable
cells and induces apoptosis in LPS cell lines
AURKA-KD was performed in LPS853 cells using siRNA
against AURKA with appropriate controls. As presented in Fig. 2A, silencing of AURKA was
demonstrated at the protein level using anti-AURKA antibody.
AURKA-KD was also associated with increased cleavage form of PARP,
indicating an apoptosis-inducing effect (Fig. 2A). Furthermore, compared with the
control, AURKA-KD significantly decreased number of viable cells
(as determined by a trypan blue exclusion assay) in LPS853 cells
(Fig. 2B). The results indicated
that AURKA could act as a potential therapeutic target in LPS
cells.
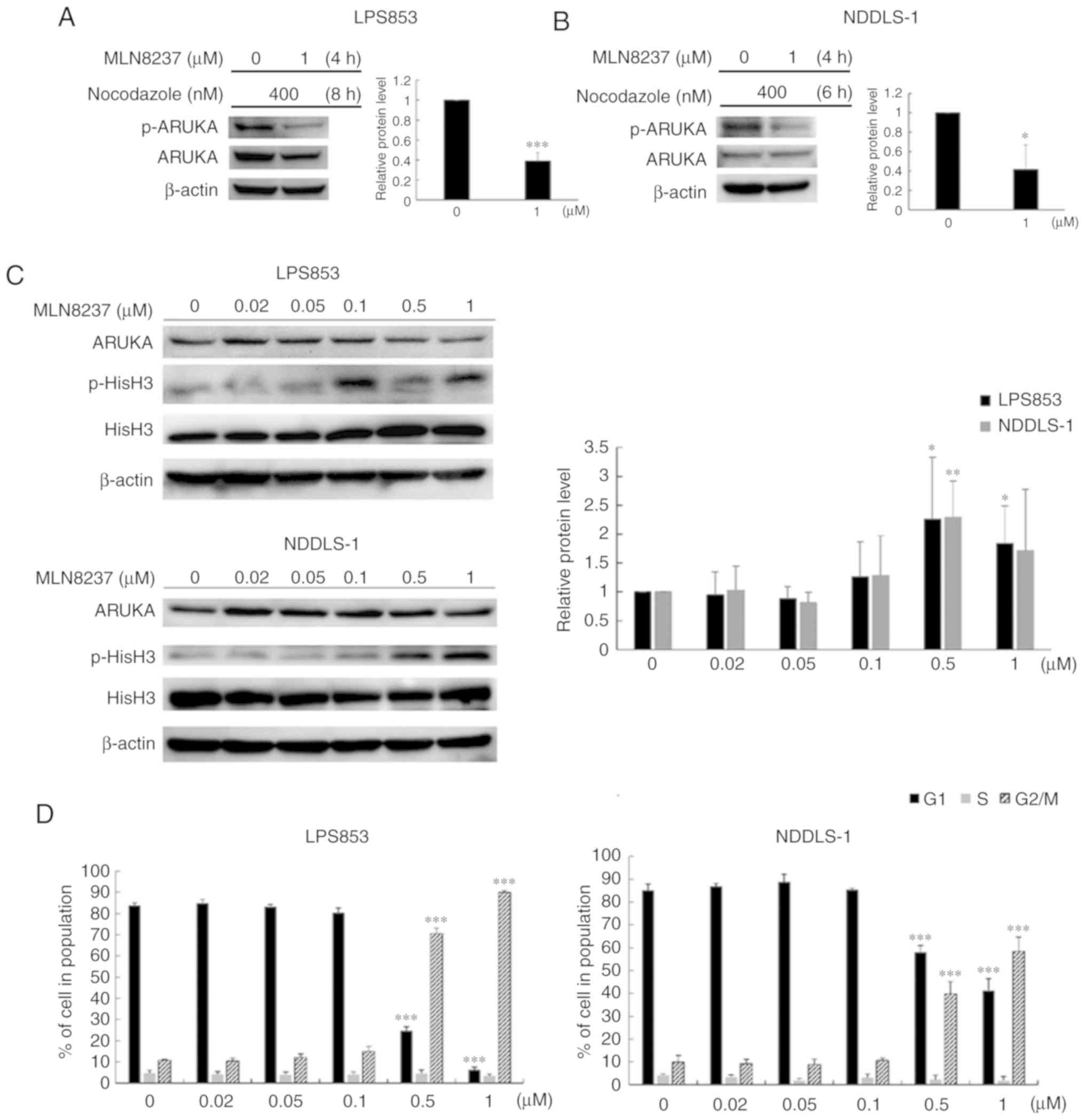
MLN8237 inhibits AURKA and induces G2/M
arrest in LPS cell lines
We used an in vitro model to determine the
potential of AURKA as a therapeutic target in LPS. MLN8237 is a
potent inhibitor of AURKA that reduces the activity of AURKA in a
variety of cancers (9,27,28).
We examined whether MLN8237 inhibits AURKA phosphorylation and
induces mitotic arrest in LPS cells. As shown in Fig. 3, immunoblotting revealed
significantly decreased AURKA phosphorylation levels in
nocodazole-stimulated and MLN8237-treated (treated with nocodazole
400 nM for 8 h then added MLN8237 for 4 h) LPS853 and NDDLS-1
compared with the control (0 µM inhibitor) (Fig. 3A and B). In addition, the levels of
pHisH3, an indicator of mitotic arrest, were significantly
increased in both cell lines, in comparison with control, after
treatment with MLN8237 at 0.5 µM for 8 h; LPS853 cells
exhibited a significant increase in pHis3 expression in response to
1 µM MLN8237 (Fig. 3C).
Flow cytometry analysis of DNA content in LPS853 and NDDLS-1 cells
treated with MLN8237 demonstrated that this compound caused a
significant accumulation of cells at G2/M DNA content, in
comparison with control, in the two LPS cell lines (Figs. 3D and S2). These studies indicated that MLN8237
could target AURKA and induce mitotic arrest in LPS cells.
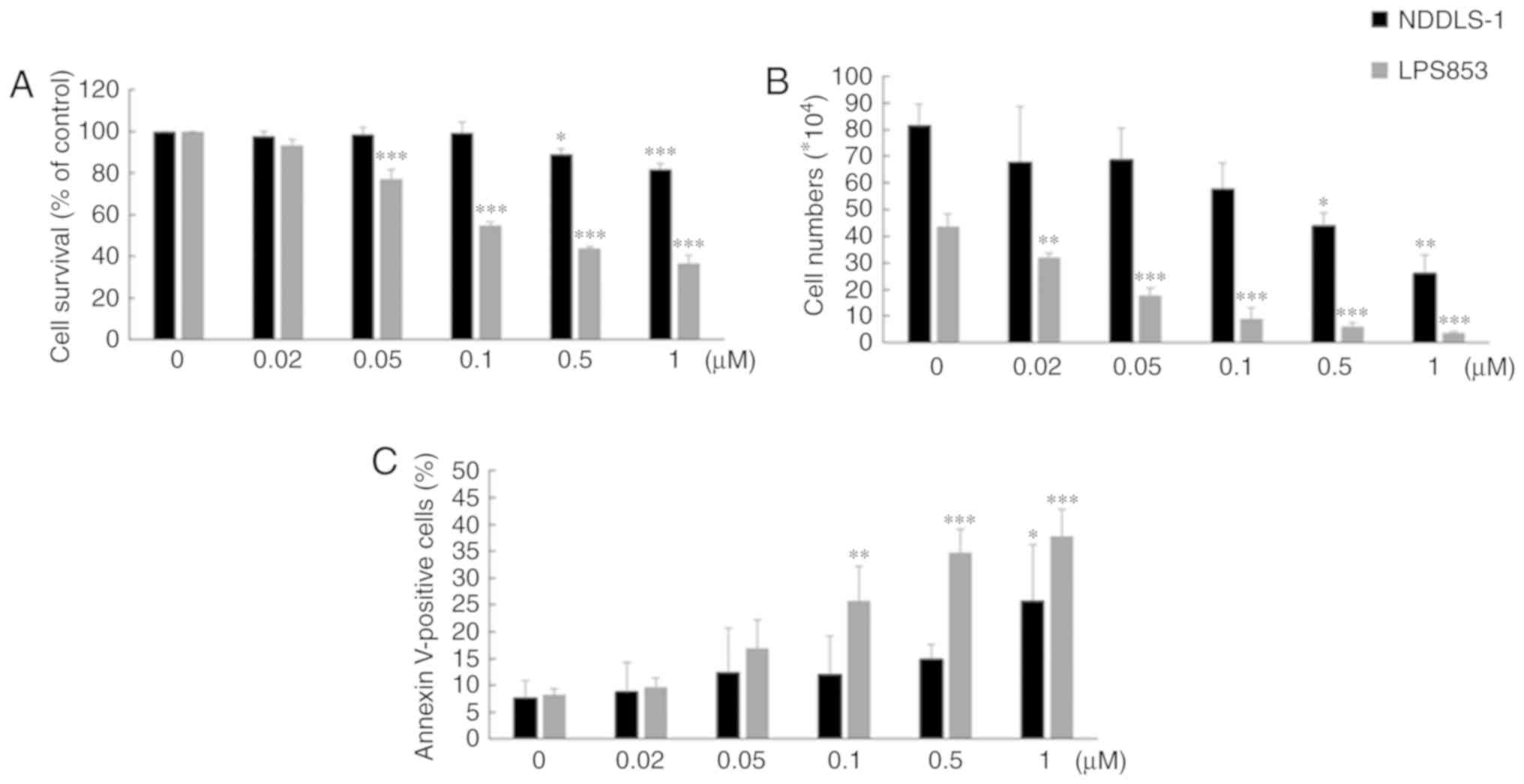
MLN8237 exerts cytotoxic effects by
inducing apoptosis in LPS cell lines
We examined whether MLN8237 could exhibit cytotoxic
activity against LPS cell lines. MLN8237 exerted significant
cytotoxic effects against NDDLS-1 and LPS853 cells as assessed
through an MTT assay (Fig. 4A) or
via a trypan blue exclusion assay (Fig. 4B). A significant induction of
apoptosis in LPS cell lines was also detected by costaining with
propidium iodide and FITC-labeled Annexin V (both early and late
apoptotic cells are included as indicated by total cell number with
positive Annexin V-FITC, Figs. 4C
and S3).
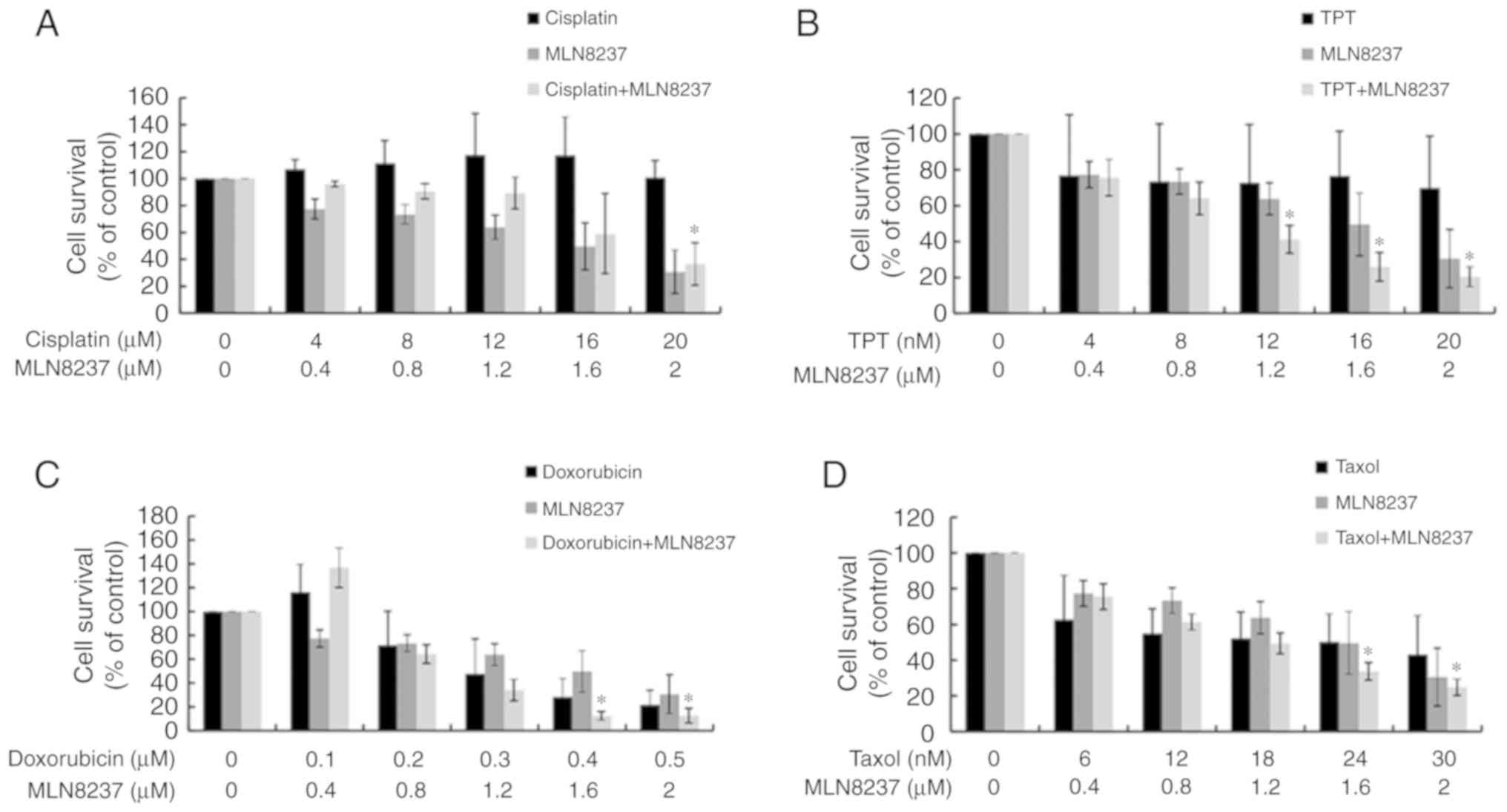
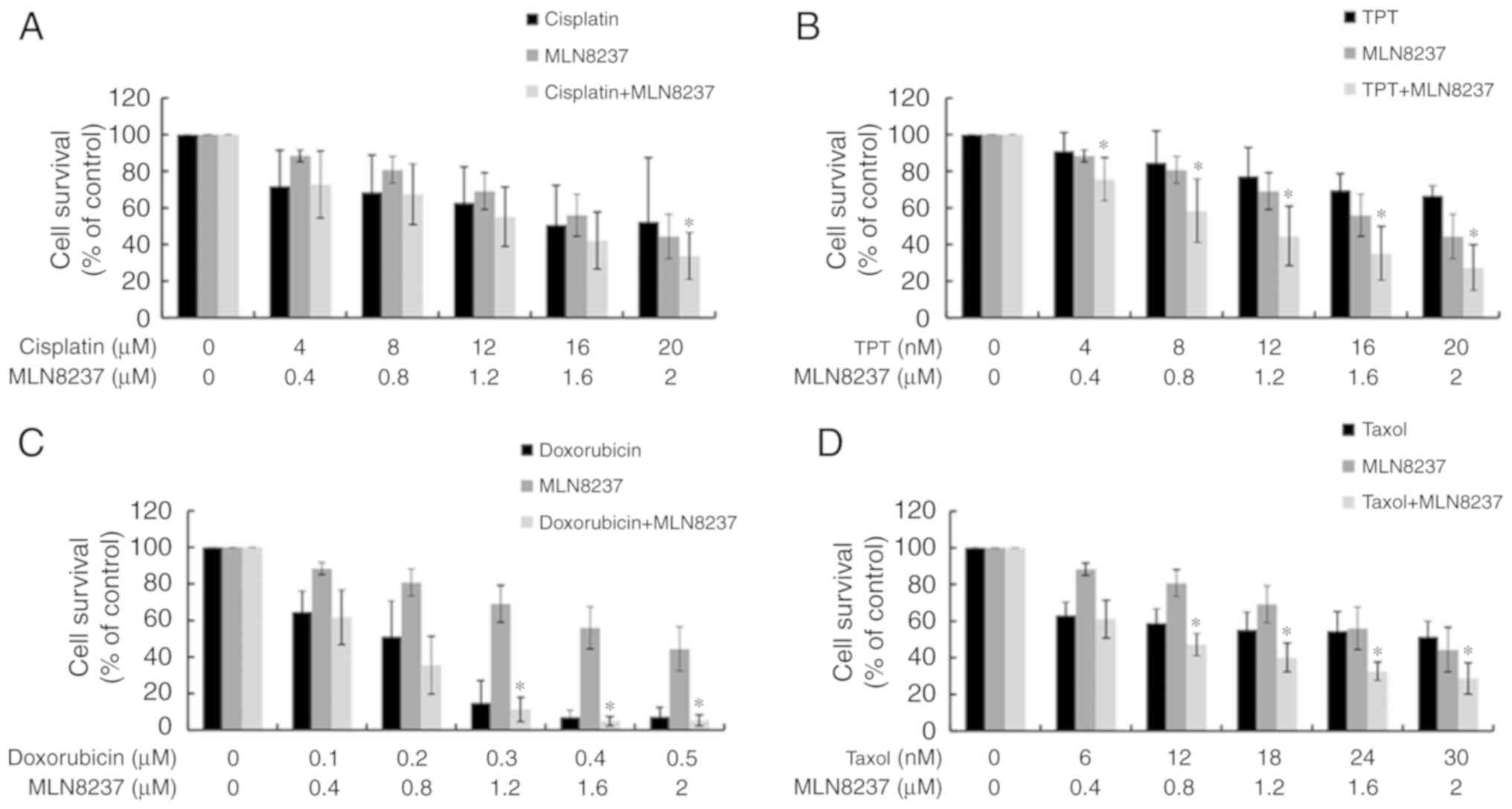
Synergistic effects of combining MLN8237
and chemo-therapeutic agents
As LPS exhibited some sensitivity to chemotherapy,
we next explored the possible synergistic effects of treatment with
both MLN8237 and chemotherapy. As presented in Figs. 5 and 6, except for cisplatin, MLN8237 exhibited
significant synergistic effect (combination index<1.0) with all
other chemotherapeutic agents against LPS cell lines LPS853
(Fig. 5) and NDDLS-1 (Fig. 6) at least at two different
combination dose levels. These results indicate that MLN8237 can
act synergistically with some chemotherapeutic agents, and provide
future chances for combination therapy in LPS.
MLN8237 induces cellular senescence in
LPS cell lines
AURKA inhibition may induce senescence in cells. To
examine whether senescence occurred in LPS cells following
treatment with MLN8237, a SA-β-gal assay was performed in LPS853
cells after being treated with MLN8237 for 72 h. The size of LPS
cells became larger and the shape went flatter after MLN8237
treatment. In addition, MLN8237 treatment significantly increased
SA-β-gal activity in LPS cells compared with the control (Fig. 7A and B). The levels of pp70S6
kinase remained steady, indicating sustained mTOR activity
(Fig. 7C). Furthermore,
administration of MLN8237 significantly increased the expression of
DcR2 (Fig. 7D), a well-known
senescence biomarker, as well as the expression of IL-1α (Fig. 7E), IL-6 (Fig. 7F) and IL-8 (Fig. 7G), cyto-kines associated with the
senescence-associated secretory phenotype (SASP) (29), in LPS853 cells. Of note, the levels
of IL-1α and IL-8 did not significantly increase in response to 0.1
µM MLN8237. Collectively, these results demonstrate that
MLN8237 treatment induces senescence in LPS cells.
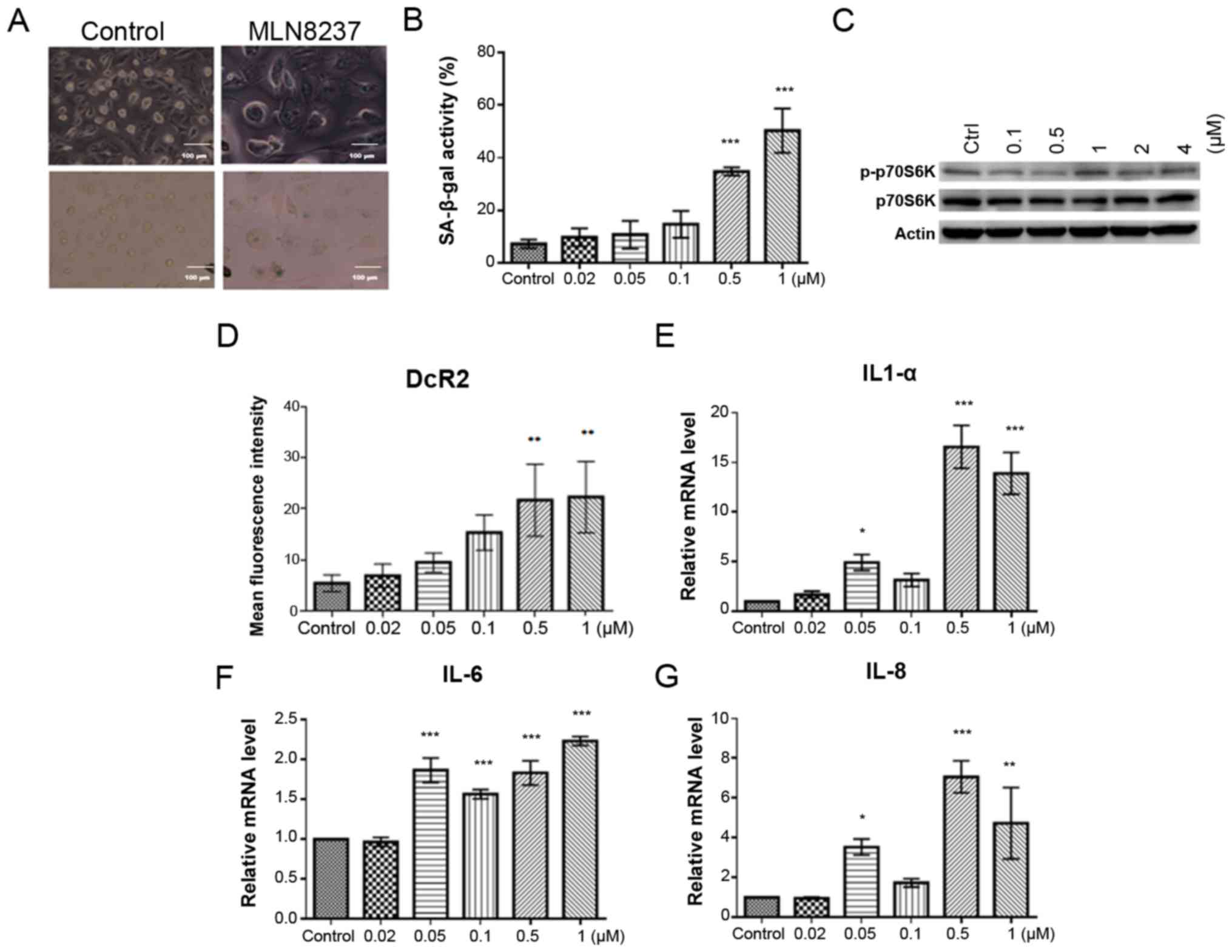 | Figure 7Inhibition of AURKA induces
senescence in LPS853 cells. (A) The size of LPS cells became larger
and the shape went flatter after MLN8237 treatment. The SA-β-gal
activity was also increased (magnification x200, scale bar=100
µm); (B) MLN8237 treatment significantly increased SA-β-gal
activity in LPS cells; (C) the levels of p-p70S6K remained steady,
indicating sustained mTOR activity; MLN8237 treatment increased the
expression of (D) DcR2, a well-known senescence biomarker, and (E)
(IL)-1α, (F) IL-6 and (G) IL-8, cytokines associated with the
senescence-associated secretory phenotype, in LPS853 cells. All
data are presented as the mean ± standard deviation of three
independent experiments. *P<0.05,
**P<0.01, ***P<0.001 vs. control. DcR2,
Decoy receptor 2; IL, interleukin; p, phosphorylated; p70S6K, p70S6
kinase; SA-β-gal, senescence-associated β-galactosidase. |
Discussion
In this study, we identified AURKA as a prognostic
factor as well as potential therapeutic target for the treatment of
LPS. An in vitro assay revealed that the AURKA inhibitor
MLN8237 could inhibit cell growth and induce the apoptosis of LPS
cell lines, and that this inhibitor exhibited a synergistic effect
with several chemotherapeutic agents. MLN8237 was also observed to
induce cellular senescence in LPS cell lines.
LPS has been demonstrated to account for 20-30% of
STS cases (10,11). Chemotherapy has been reported to be
ineffective in LPS; however, recent clinical trials indicated that
eribulin (clinical trial no. NCT01327885) (30,31)
and trabectedin (clinical trial no. NCT01343277) (32) benefited survival in late-line
therapy. Nevertheless, the response rate remains unsatisfactory.
Our previous study reported that Akt1 overexpression could enhance
adipogenesis and lead to lipoma formation in zebrafish (33). However, its role in the
pathogenesis of LPS is still unclear.
WD and DD LPS, the two most common subtypes, shared
common genomic changes with 12q13-15 amplification (34). Mouse double minute 2 and cyclin
dependent kinase (CDK)4 are two candidate genes within this region,
and are routinely used as diagnostic markers for WD and DD LPS
(35). In a phase II study of LPS
(clinical trial no. NCT01209598), palbociclib, a CDK4/6 inhibitor,
achieved a 66% (90% confidence interval, 51-100%) progression-free
rate at 12 weeks, significantly exceeding the primary endpoint of
the trial; nevertheless, there was only one partial response
(14).
In the present study, by reanalyzing two published
microarray data sets of LPS and comparing the top 50 genes that
were overexpressed in DD LPS in both sets of data, we identified 12
overlapping genes. Furthermore, we discovered that the top five
gene sets enriched in DD LPS in both sets of data were all involved
in cell cycle regulation. AURKA, a well-known gene involved in cell
cycle regulation, was then selected among the 12 overlapping genes.
AURKA is one of the top three genes in the CINSARC list (7). Our group (8,9) and
Lagarde et al (36) have
also identified AURKA as an independent prognostic factor in
predicting GIST recurrence. Furthermore, the present study, AURKA
was found to be overexpressed in DD LPS, compared with WD LPS, and
could act as a prognostic factor in LPS.
In an in vitro study, MLN8237, an AURKA
inhibitor, was proposed to inhibit AURKA in LPS cell lines with a
resultant G2/M arrest. MLN8237 was observed to exert a cytotoxic
effect by inducing apoptosis in LPS cell lines. These findings are
consistent with that of our previous study of AURKA in GISTs
(9). In a recent phase II study
using MLN8237 (Alisertib) in treating 72 cases of sarcoma (clinical
trial no. NCT01653028), a 73% 12-week progression-free rate was
achieved in 12 cases of LPS; however, none of them exhibited tumor
response (37). Therefore,
combining cell cycle inhibitors with other agents may be necessary
to achieving improved cytotoxic effects on cancer cells. In the
current study, MLN8237 was exhibited a significant synergistic
effect with several of the chemotherapeutic agents tested against
LPS cell lines. These results indicated a possibility of the
combination of MLN8237 with chemotherapy for treating LPS.
Drugs targeting the cell cycle can cause
therapy-induced senescence (TIS) (38). Senescent cells are permanently
growth arrested in either G1 or G2/M stage of the cell cycle, but
remain viable with sustained survival signaling (39). Of note, these cells can be
visualized through a staining technique that employs a widely
accepted and used marker, SA-b-gal activity (40).
In this study, we demonstrated that MLN8237 could
induce several features of TIS in LPS, including the increased
activity of SA-β-gal in cells with a flattened morphology and
upregulated expression of DcR2, as well as the increased expression
of proinflammatory cytokines IL-1α, IL-6 and IL-8. These
observations comprise called the SASP; prosurvival signaling was
unaffected as suggested by the levels of pp70S6K. Similar findings
have also been reported in our previous studies of the AURKA
inhibitor MLN8237 in treating a GIST cell line (9) and of the polo-like kinase 1 inhibitor
GSK461364 in treating an osteosarcoma cell line (20).
In summary, our study identified AURKA as a
potential biomarker in predicting poor prognosis in LPS. We also
reported the potential of AURKA as a therapeutic target in LPS cell
line models. The novel combination of AURKA inhibitors with
chemotherapy appears to be promising; however, further
investigation is required.
Supplementary Data
Acknowledgments
This manuscript was edited by Wallace Academic
Editing.
Funding
The present study was supported by grants from the
National Science Council (grant nos. NSC 100-2314-B-075-081 a n d
NSC 101-2 314 -B - 0 75 - 0 29), M i n i st r y of Science and
Technology, Taiwan (grant nos. MOST 103-2314-B-075-066 and MOST
105-2314-B-075-059), and Taipei Veterans General Hospital (grant
nos. V102E8-003, V103E8-001, V101C-133, V102C-034, V103C-188,
V104C-099, V104E16-001-MY3-1, V104E16-001-MY3-2, V104D16-001-MY3-3,
V105C-094, V106C-160, V107C-085, V107D32-001-MY2-2 and V108C-108)
and from the Yen Tjing Ling Medical Foundation (grant nos.
CI-100-19, CI-103-6 and CI-105-4) designated to Dr Chueh-Chuan Yen.
This work was also supported by the Taiwan Clinical Oncology
Research Foundation, and the Chong Hin Loon Memorial Cancer and
Biotherapy Research Center of National Yang-Ming University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CCY, SCC, GYH, PKW were responsible for the design
and conception of the study; WYC, YCL, CHY, YCC, JYW were
responsible for data acquisition and interpretation; MHY, YC, MCC
and WMC were responsible for the data analysis and drafting the
work. All authors have given final approval of the version to be
published.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Demetri GD, van Oosterom AT, Garrett CR,
Blackstein ME, Shah MH, Verweij J, McArthur G, Judson IR, Heinrich
MC, Morgan JA, et al: Efficacy and safety of sunitinib in patients
with advanced gastrointestinal stromal tumour after failure of
imatinib: A randomised controlled trial. Lancet. 368:1329–1338.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Blanke CD, Demetri GD, von Mehren M,
Heinrich MC, Eisenberg B, Fletcher JA, Corless CL, Fletcher CD,
Roberts PJ, Heinz D, et al: Long-term results from a randomized
phase II trial of standard-versus higher-dose imatinib mesylate for
patients with unresectable or metastatic gastrointestinal stromal
tumors expressing KIT. J Clin Oncol. 26:620–625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Demetri GD, Reichardt P, Kang YK, Blay JY,
Rutkowski P, Gelderblom H, Hohenberger P, Leahy M, von Mehren M,
Joensuu H, et al: Efficacy and safety of regorafenib for advanced
gastrointestinal stromal tumours after failure of imatinib and
sunitinib (GRID): An international, multicentre, randomised,
placebo-controlled, phase 3 trial. Lancet. 381:295–302. 2013.
View Article : Google Scholar
|
|
5
|
McArthur GA, Demetri GD, van Oosterom A,
Heinrich MC, Debiec-Rychter M, Corless CL, Nikolova Z, Dimitrijevic
S and Fletcher JA: Molecular and clinical analysis of locally
advanced dermatofibrosarcoma protuberans treated with imatinib:
Imatinib Target Exploration Consortium Study B2225. J Clin Oncol.
23:866–873. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Butrynski JE, D'Adamo DR, Hornick JL, Dal
Cin P, Antonescu CR, Jhanwar SC, Ladanyi M, Capelletti M, Rodig SJ,
Ramaiya N, et al: Crizotinib in ALK-rearranged inflammatory
myofibroblastic tumor. N Engl J Med. 363:1727–1733. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chibon F, Lagarde P, Salas S, Pérot G,
Brouste V, Tirode F, Lucchesi C, de Reynies A, Kauffmann A, Bui B,
et al: Validated prediction of clinical outcome in sarcomas and
multiple types of cancer on the basis of a gene expression
signature related to genome complexity. Nat Med. 16:781–787. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yen CC, Yeh CN, Cheng CT, Jung SM, Huang
SC, Chang TW, Jan YY, Tzeng CH, Chao TC, Chen YY, et al:
Integrating bioin-formatics and clinicopathological research of
gastrointestinal stromal tumors: Identification of aurora kinase A
as a poor risk marker. Ann Surg Oncol. 19:3491–3499. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yeh CN, Yen CC, Chen YY, Cheng CT, Huang
SC, Chang TW, Yao FY, Lin YC, Wen YS, Chiang KC, et al:
Identification of aurora kinase A as an unfavorable prognostic
factor and potential treatment target for metastatic
gastrointestinal stromal tumors. Oncotarget. 5:4071–4086. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hung GY, Yen CC, Horng JL, Liu CY, Chen
WM, Chen TH and Liu CL: Incidences of primary soft tissue sarcoma
diagnosed on extremities and trunk wall: A population-based study
in Taiwan. Medicine (Baltimore). 94:e16962015. View Article : Google Scholar
|
|
11
|
Liu CY, Yen CC, Chen WM, Chen TH, Chen PC,
Wu HT, Shiau CY, Wu YC, Liu CL and Tzeng CH: Soft tissue sarcoma of
extremities: The prognostic significance of adequate surgical
margins in primary operation and reoperation after recurrence. Ann
Surg Oncol. 17:2102–2111. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Italiano A, Toulmonde M, Cioffi A, Penel
N, Isambert N, Bompas E, Duffaud F, Patrikidou A, Lortal B, Le
Cesne A, et al: Advanced well-differentiated/dedifferentiated
liposarcomas: Role of chemotherapy and survival. Ann Oncol.
23:1601–1607. 2012. View Article : Google Scholar
|
|
13
|
Singer S, Socci ND, Ambrosini G, Sambol E,
Decarolis P, Wu Y, O'Connor R, Maki R, Viale A, Sander C, et al:
Gene expression profiling of liposarcoma identifies distinct
biological types/subtypes and potential therapeutic targets in
well-differentiated and dedifferentiated liposarcoma. Cancer Res.
67:6626–6636. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dickson MA, Tap WD, Keohan ML, D'Angelo
SP, Gounder MM, Antonescu CR, Landa J, Qin LX, Rathbone DD, Condy
MM, et al: Phase II trial of the CDK4 inhibitor PD0332991 in
patients with advanced CDK4-amplified well-differentiated or
dedifferen-tiated liposarcoma. J Clin Oncol. 31:2024–2028. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gobble RM, Qin LX, Brill ER, Angeles CV,
Ugras S, O'Connor RB, Moraco NH, Decarolis PL, Antonescu C and
Singer S: Expression profiling of liposarcoma yields a multigene
predictor of patient outcome and identifies genes that contribute
to liposarcomagenesis. Cancer Res. 71:2697–2705. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li C and Hung Wong W: Model-based analysis
of oligonucleotide arrays: Model validation, design issues and
standard error application. Genome Biol.
2:RESEARCH00322001.PubMed/NCBI
|
|
17
|
Li C and Wong WH: Model-based analysis of
oligonucleotide arrays: Expression index computation and outlier
detection. Proc Natl Acad Sci USA. 98:31–36. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Subramanian A, Tamayo P, Mootha VK,
Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub
TR, Lander ES and Mesirov JP: Gene set enrichment analysis: A
knowledge-based approach for interpreting genome-wide expression
profiles. Proc Natl Acad Sci USA. 102:15545–15550. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mootha VK, Lindgren CM, Eriksson KF,
Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E,
Ridderstrale M, Laurila E, et al: PGC-1alpha-responsive genes
involved in oxidative phosphorylation are coordinately
downregulated in human diabetes. Nat Genet. 34:267–273. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chou YS, Yen CC, Chen WM, Lin YC, Wen YS,
Ke WT, Wang JY, Liu CY, Yang MH, Chen TH and Liu CL: Cytotoxic
mechanism of PLK1 inhibitor GSK461364 against osteosarcoma: Mitotic
arrest, apoptosis, cellular senescence, and synergistic effect with
paclitaxel. Int J Oncol. 48:1187–1194. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mosmann T: Rapid colorimetric assay for
cellular growth and survival: Application to proliferation and
cytotoxicity assays. J Immunol Methods. 65:55–63. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chou TC and Talalay P: Quantitative
analysis of dose-effect relationships: The combined effects of
multiple drugs or enzyme inhibitors. Adv Enzyme Regul. 22:27–55.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Schmit TL, Nihal M, Ndiaye M, Setaluri V,
Spiegelman VS and Ahmad N: Numb regulates stability and
localization of the mitotic kinase PLK1 and is required for transit
through mitosis. Cancer Res. 72:3864–3872. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhu Y, Xu L, Zhang J, Hu X, Liu Y, Yin H,
Lv T, Zhang H, Liu L, An H, et al: Sunitinib induces cellular
senescence via p53/ Dec1 activation in renal cell carcinoma cells.
Cancer Sci. 104:1052–1061. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Muller-Tidow C, Metzger R, Kügler K,
Diederichs S, Idos G, Thomas M, Dockhorn-Dworniczak B, Schneider
PM, Koeffler HP, Berdel WE and Serve H: Cyclin E is the only
cyclin-dependent kinase 2-associated cyclin that predicts
metastasis and survival in early stage non-small cell lung cancer.
Cancer Res. 61:647–653. 2001.PubMed/NCBI
|
|
26
|
Lens SM, Voest EE and Medema RH: Shared
and separate functions of polo-like kinases and aurora kinases in
cancer. Nat Rev Cancer. 10:825–841. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kelly KR, Ecsedy J, Medina E, Mahalingam
D, Padmanabhan S, Nawrocki ST, Giles FJ and Carew JS: The novel
Aurora A kinase inhibitor MLN8237 is active in resistant chronic
myeloid leukaemia and significantly increases the efficacy of
nilotinib. J Cell Mol Med. 15:2057–2070. 2011. View Article : Google Scholar
|
|
28
|
Qi W, Cooke LS, Liu X, Rimsza L, Roe DJ,
Persky AM, Miller TP and Mahadevan D: Aurora inhibitor MLN8237 in
combination with docetaxel enhances apoptosis and anti-tumor
activity in mantle cell lymphoma. Biochem Pharmacol. 81:881–890.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kuilman T and Peeper DS:
Senescence-messaging secretome: SMS-ing cellular stress. Nat Rev
Cancer. 9:81–94. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Schoffski P, Chawla S, Maki RG, Italiano
A, Gelderblom H, Choy E, Grignani G, Camargo V, Bauer S, Rha SY, et
al: Eribulin versus dacarbazine in previously treated patients with
advanced liposarcoma or leiomyosarcoma: A randomised, open-label,
multicentre, phase 3 trial. Lancet. 387:1629–1637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Demetri GD, Schöffski P, Grignani G, Blay
JY, Maki RG, Van Tine BA, Alcindor T, Jones RL, D'Adamo DR, Guo M
and Chawla S: Activity of eribulin in patients with advanced
lipo-sarcoma demonstrated in a subgroup analysis from a randomized
phase III study of Eribulin versus dacarbazine. J Clin Oncol.
35:3433–3439. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Demetri GD, von Mehren M, Jones RL,
Hensley ML, Schuetze SM, Staddon A, Milhem M, Elias A, Ganjoo K,
Tawbi H, et al: Efficacy and safety of trabectedin or dacarbazine
for metastatic liposarcoma or leiomyosarcoma after failure of
conventional chemotherapy: Results of a phase III randomized
multicenter clinical trial. J Clin Oncol. 34:786–793. 2016.
View Article : Google Scholar :
|
|
33
|
Chu CY, Chen CF, Rajendran RS, Shen CN,
Chen TH, Yen CC, Chuang CK, Lin DS and Hsiao CD: Overexpression of
Akt1 enhances adipogenesis and leads to lipoma formation in
zebrafish. PLoS One. 7:e364742012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Fletcher CD, Akerman M, Dal Cin P, de
Wever I, Mandahl N, Mertens F, Mitelman F, Rosai J, Rydholm A,
Sciot R, et al: Correlation between clinicopathological features
and karyotype in lipomatous tumors. A report of 178 cases from the
chromosomes and morphology (CHAMP) Collaborative study group. Am J
Pathol. 148:623–630. 1996.PubMed/NCBI
|
|
35
|
Dei Tos AP, Doglioni C, Piccinin S, Sciot
R, Furlanetto A, Boiocchi M, Dal Cin P, Maestro R, Fletcher CD and
Tallini G: Coordinated expression and amplification of the MDM2,
CDK4, and HMGI-C genes in atypical lipomatous tumours. J Pathol.
190:531–536. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Lagarde P, Perot G, Kauffmann A, Brulard
C, Dapremont V, Hostein I, Neuville A, Wozniak A, Sciot R,
Schoffski P, et al: Mitotic checkpoints and chromosome instability
are strong predictors of clinical outcome in gastrointestinal
stromal tumors. Clin Cancer Res. 18:826–838. 2012. View Article : Google Scholar
|
|
37
|
Dickson MA, Mahoney MR, Tap WD, D'Angelo
SP, Keohan ML, Van Tine BA, Agulnik M, Horvath LE, Nair JS and
Schwartz GK: Phase II study of MLN8237 (Alisertib) in
advanced/metastatic sarcoma. Ann Oncol. 27:1855–1860. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Huck JJ, Zhang M, McDonald A, Bowman D,
Hoar KM, Stringer B, Ecsedy J, Manfredi MG and Hyer ML: MLN8054, an
inhibitor of Aurora A kinase, induces senescence in human tumor
cells both in vitro and in vivo. Mol Cancer Res. 8:373–384. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Campisi J and d'Adda di Fagagna F:
Cellular senescence: When bad things happen to good cells. Nat Rev
Mol Cell Biol. 8:729–740. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Dimri GP, Lee X, Basile G, Acosta M, Scott
G, Roskelley C, Medrano EE, Linskens M, Rubelj I, Pereira-Smith O,
et al: A biomarker that identifies senescent human cells in culture
and in aging skin in vivo. Proc Natl Acad Sci USA. 92:9363–9367.
1995. View Article : Google Scholar : PubMed/NCBI
|