Introduction
Glioma is the most fatal and common primary
malignant brain tumor of the adult central nervous system with a
3.3% mortality rate and a medical and neurologic morbidity rate of
31.7% (1,2); it is clinically classified into
low-grade glioma (WHO grade I and II) and high-grade glioma (WHO
grade III and IV), according to histopathological malignancy
characteristics (3). Although
combined therapeutic treatments have been improved tremendously in
the last two decades, the 5-year survival rate is still <5%
(4). Therefore, it is necessary to
explore novel therapeutic targets and develop strategies for glioma
treatment.
Long non-coding RNAs (lncRNAs) are key molecules in
the progression of various types of human tumors (5,6),
including glioma pathogenesis (7).
lncRNA LINC00473 is an intergenic lncRNA located on the human 6q27
chromosome (8). Recently,
LINC00473 expression has been found to be elevated in a variety of
human cancers, including non-small cell lung cancer (9), Wilms tumor (10), hepatocellular carcinoma (11), cervical cancer (12), human mucoepidermoid carcinoma
(13), fibrolamellar carcinoma
(14) and gastric cancer (15). However, the clinical importance and
biological functions of LINC00473 in glioma remains unclear.
lncRNAs can function as competing endogenous RNAs
(ceRNAs) to sponge and inhibit the function of specific microRNA
(miRNA) (16). As a ceRNA,
LINC00473 regulates miRNA (miR)-15a-dependent Taxol resistance in
colorectal cancer (17). Moreover,
LINC00473 has been shown to antagonize the tumor suppressor miR-195
to mediate Wilms tumor (10), and
miR-195 serves an antitumoral role in glioma (18-20).
Therefore, we hypothesized that LINC00473 may function as a ceRNA
to link miR-195 and post-transcriptional signaling in glioma
cells.
The targeting of miR-195-5p to YAP1 (Yes-associated
protein 1) has been reported in hepatocellular carcinoma (21), osteosarcoma (22) and colorectal cancer (23). The conserved Hippo signaling
pathway mediates downstream transcriptional co-activators, such as
YAP, which subsequently binds to and activates TEA domain family
members (TEADs) that are responsible for cell proliferation and
apoptosis (24). A previous study
reported that YAP1 was upregulated in glioma cells and induced
glioma growth (25). Ectopic
expression of YAP binds to TEAD and controls the transcription of
pro-proliferative and anti-apoptotic target genes in glioma
(26). TEAD1 was also reported to
be an important regulator of glioma cell migration (27). Therefore, LINC00473 might regulate
glioma progression through YAP1-TEAD1-Hippo signaling pathway.
The present study hypothesized that LINC00473 may
promote glioma development and, as such, investigated the effects
of LINC00473 on tumor proliferation, migration and invasion in
vivo and in vitro. In addition, the downstream targets
of LINC00473 and the underlying mechanisms were also examined. The
results of the present study may contribute to the application of
LINC00473 in glioma therapy.
Materials and methods
Clinical tumor tissues
Paired glioma tissues and para-carcinoma tissues
were obtained from 40 patients (17 male; 23 female; age, 55.8±4.7
years with range of 44-70 years) with primary gliomas between
January 2014 and January 2017, patients with 10-year follow-up
period underwent surgical treatment at Union Hospital Affiliated
with Tongji Medical College of Huazhong University of Science and
Technology (Wuhan, China). Patient glioma statuses were determined
by imaging modalities (MRI scan) and verified though histological
analysis. Tissue specimens were cut into pieces, flash frozen in
liquid nitrogen and stored at −80°C for further experiments. The
present study was approved by the Ethics Committee of Tongji
Medical College, Huazhong University of Science and Technology
(approval no. 2014-S034), and all the patients signed written
informed consent.
Cell culture and transfection
Human glioma cell lines (U251, U118, LN229, U87 and
SHG44) and a normal human astrocyte cell line (NHA) were obtained
from Lonza Group AG and authenticated by short tandem repeat DNA
profiling. Cells were cultured in DMEM supplemented with 10% FBS
(Gibco; Thermo Fisher Scientific, Inc.) in a 37°C constant
temperature incubator with 5% CO2.
Two short hairpin (sh)RNAs against LINC00473
(sh-LINC00473#1, 5′-AAC TGG ATC TTT GCA GAC AGG-3′; sh-LINC00473#2:
5′-AAG AAC CCA AGT CAT ATT CAT-3′) and a scramble negative control
(sh-NC, 5′-CGG AUC AGC UCG CGC UAU CAU CGC A-3′) were inserted into
a pLKO.1 expression vector (BioSettia, Inc.). A total of 40 nM
sh-LINC00473#1, LINC00473#2 or shNC were co-transfected into 293
cells (4×105 cells/well) with psPAX2 and pMD2.G
lentiviral vectors using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) at 37°C for 48 h,
according to the manufacturer’s protocols. Following incubation,
lentiviral particles were harvested by precipitation in
polyethylene glycol followed by centrifugation at 500 × g for 30
min at 4°C, and particles were transduced at 0.5 multiplicity of
infection into U251 or U87 cells (1×106 cells) using
ViraPower™ Packaging Mix (Thermo Fisher Scientific, Inc.) and 8
mg/ml polybrene at 37°C overnight. Cells were treated with 5
µg/ml puromycin (Sigma Aldrich; Merck KGaA) 1 week to
establish stable cell lines.
Full-length of LINC00473 was amplified and cloned
into pcDNA3.1 overexpression vector (Invitrogen; Thermo Fisher
Scientific, Inc.). U251 or U87 cells (4×105 cells/well)
were seeded in 12-well plates and transfected with 300 µg
pcDNA3.1-LINC00473 or the empty vector control (pcDNA3.1) using
Lipofectamine 2000 at 37°C for 48 h, according to the
manufacturer’s protocols, and subsequently used for other
experiments.
miR-195-5p mimics (5′-UAG CAG CAC AGA AAU AUU
GGC-3′), miR-195 inhibitor (inh; 5′-GCC AAU AUU UCU GUG CUG
CUA-3′), miR-NC (5′-UCA CAA CCU CCU AGA AAG AGU AGA-3′ and inh NC
(5′-CAG UAC UUU UGU GUA GUA CAA-3′) were synthesized by Suzhou
GenePharma Co., Ltd. U251 or U87 cells (1×106 cells)
were transfected with miR-195-5p mimics, inhibitors or the
respective controls (40 nM) using Lipofectamine 2000 at 37°C for 48
h, according to the manufacturer’s protocols, and subsequently used
for other experiments. U251 and U87 cells were co-transfected with
i) inh NC and sh-NC, ii) sh-LINC00473 and inh NC, iii) miR-195-5p
inh and sh-NC, and iv) sh-LINC00473 and miR-195-5p inh to determine
the functional roles of LINC00473/miR-195-5p/YAP1-TEAD1-Hippo axis
in regulating glioma progression.
Cell viability assay
Transfected/transduced U251 or U87 cells
(2×103 cells/well) were seeded in 96-well plates and
incubated for 24, 48, 72 and 96 h. Subsequently, 20 µl Cell
Counting Kit-8 (CCK8) solution (Dojindo Molecular Technologies,
Inc.) was added into each well and incubated at 37°C for 2 h,
according to the manufacturer’s protocols. The absorbance was
measured at 450 nm using a Microplate Autoreader (Thermo Fisher
Scientific, Inc.).
Flow cytometry
Transfected/transduced U251 or U87 cells
(1×106 cells) were harvested using trypsin digestion.
For cell cycle analysis, cells were stained with 5 µl
propidium iodide (PI; 100 µg/ml) with 1 U/ml ribonuclease
(Abcam) for 30 min at room temperature, according to the
manufacturer’s protocols. For apoptosis analysis, cells were
resuspended in 100 µl binding buffer (Nanjing KeyGen Biotech
Co., Ltd.) containing 5 µl PI (100 µg/ml) with 1 U/ml
ribonuclease in the dark for 30 min at room temperature, and then
incubated with additional 5 µl of Annexin V-FITC for another
15 min in the dark at room temperature, according to the
manufacturer’s protocols. Cells were subsequently analyzed by FACS
using an Attune flow cytometer (Thermo Fisher Scientific, Inc.)
with software FlowJo 1.1.0 (Tree Star, Inc.) for the determination
of apoptotic rates, which is presented as a combination of early
and late stage apoptosis.
Wound healing assay
Transfected/transduced U251 or U87 cells
(5×105 cells/well) were seeded in six-well plates with
complete medium. Cells ay 90-95% confluence were serum starved for
24 h. Cells with 95% confluence were used for the following
experiment. A 200 µl sterile pipette tip was used to
generate linear scratch wounds, and the plate was washed with PBS
to remove debris and suspended cells. Plates were incubated at 37°C
for 24 h post-wound generation, and images were captured under a
light microscope (magnification, ×100) and the cell migration
distance was measure compared with the 0 h time point.
Transwell invasion assay
Transfected/transduced U251 or U87 cells
(2×104 cells/well) were suspended in 200 µl
serum-free DMEM and plated in the upper Transwell chamber (Corning,
Inc.) containing a pre-coated Matrigel-coated membrane (0.1 ml, 50
µg/ml; BD Biosciences). A total of 400 µl DMEM with
10% FBS was added to the lower chamber. Cells were incubated at
37°C for 24 h, and the invading cells at the bottom of chambers
were stained with 1% crystal violet for 30 min. Cells were imaged
and counted under a light microscope (magnification, ×100) (Olympus
Corp).
Dual-luciferase reporter assay
Potential targets of miR-195-5p and LINC00743 were
predicted using TargetScan release 7.1 (http://www.targetscan.org/vert_71) or starBase v2.0
(http://starbase.sysu.edu.cn),
respectively. Sequences of wildtype (WT) or mutant (MUT)
3’-untranslated regions (UTRs) of LINC00473, YAP1 or TEAD1 were
cloned into pmirGLO luciferase reporter vector (Promega
Corporation). 293 cells (3×104 cells/well) were seeded
in 24-well plates and co-transfected with either miR-195-5p mimics
or NC mimics and pmirGLO-WT-LINC00473, pmirGLO-MUT-LINC00473,
pmirGLO-WT-YAP1, pm i r GLO -MU T-YA P1, pm i r GLO -W T-TEA D1 or
pmirGLO-MUT-TEAD1 using Lipofectamine 2000 at 37°C, according to
the manufacturer’s protocols. At 48 h post-transfection, the
luciferase activities were measured using a Lucifer Reporter Assay
System (Promega Corporation), with firefly luciferase activity
normalized to Renilla luciferase activity.
RNA immunoprecipitation (RIP)
A total of 1×106 trans-fected/transduced
U251 or U87 cells were collected and lysed using Magna RIP Kit (EMD
Millipore, Billerica, MA). Cell lysate was incubated with protein G
Sepharose beads (GE Healthcare) coated with anti-argonaute 2 (Ago2)
antibody (1:50; cat. no. ab186733; Abcam) at 4°C overnight.
Anti-immunoglobulin (Ig)G antibody (1:50; cat. no. ab200699; Abcam)
was used as the negative control, and anti-U1 small nucleoprotein
70 kDa (SNRNP70; 1:50; cat. no. ab83306; Abcam) was used as
positive control. RNA was subsequently isolated for reverse
transcription (RT)-qPCR, described below.
RT-qPCR
Total RNA was isolated from tissues (2 mg) or cell
lines, including cells from RIP, (1×106 cells) using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Ind.),
miRNAs were extracted using miRcute miRNA Isolation kit (Tiangen
Biotech Co., Ltd.). A total of 2 µg RNA was reverse
transcribed into cDNA using the PrimeScript RT Reagent kit (Takara
Bio, Inc.). qPCR was conducted using SYBR Green Master mix (Roche
Diagnostics GmbH) on a ViiA 7 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.), with the following
thermocycling conditions: Initial denaturation at 95°C for 60 sec;
followed by 40 cycles of extension at 95°C for 30 sec and annealing
at 60°C for 40 sec. GAPDH or U6 was used as endogenous controls and
for normalization of mRNA and miRNA expressions, respectively.
Primer sequences are provided in Table
I.
 | Table IPrimer sequences for reverse
transcription-qPCR. |
Table I
Primer sequences for reverse
transcription-qPCR.
| Gene | Sequence
(5′→3′) |
|---|
| GAPDH | F:
ACCACAGTCCATGCCATCAC |
| R:
TCCACCACCCTGTTGCTGTA |
| LINC00473 | F:
TCATTTCCCTACCTGCTCCT |
| R:
CAGTGTCTGCACATCGCTAAT |
|
microRNA-195-5p | F:
GGGGTAGCAGCACAGAAAT |
| R:
TCCAGTGCGTGTCGTGGA |
| YAP1 | F:
CGCTCTTCAACGCCGTCA |
| R:
AGTACTGGCCTGTCGGGAGT |
| TEAD1 | F:
AACATGGAAAGGATGAGCGACT |
| R:
TCAGTCCTTCACAAGCCTGTAGA |
| U6 | F:
CTCGCTTCGGCAGCACA |
| R:
AACGCTTCACGAATTTGCGT |
Western blotting
Total protein was extracted from glioma and
para-cancerous tissues (100 mg) and glioma cell lines
(1×107 cells) using RIPA lysis buffer (Thermo Fisher
Scientific, Inc.). Protein concentrations were quantified by BCA
and a total of 30 µg protein lysate was separated by 10%
SDS-PAGE and subsequently electrotransferred onto PVDF membranes.
After blocking with 5% BSA (Sigma-Aldrich; Merck KGaA) at 37°C for
1 h, the membrane was incubated overnight with primary antibodies
against YAP1 (1:1,500; cat. no. ab56701; Abcam), TEAD1 (1:1,500;
cat. no. ab109080, Abcam), CTGF (1:2,500; cat. no. ab6992; Abcam)
and GAPDH (1:3,000; cat. no. ab9485; Abcam) at 4°C. Following
incubation with a horseradish peroxidase (HRP)-conjugated goat
anti-rabbit (1:5,000; cat. no. ab205718; Abcam) or goat anti-mouse
(1:5,000; cat. no. ab205719; Abcam) secondary antibody at 4°C for 2
h, the immunoreactivities were detected by Enhanced
Chemiluminescence (Nanjing KeyGen Biotech Co., Ltd.). Densitometric
analysis was performed using ImageJ v2.4.1 (National Institutes of
Health) with protein expression levels normalized to GAPDH.
Mouse xenograft assay
All studies involving animals were approved by the
Ethics Committee of Tongji Medical College, Huazhong University of
Science and Technology. Male BALB/c nude mice (n=12; age, 5 weeks;
weight, 20-25 g; Shanghai Experimental Animal Centre) were randomly
separated into two groups, sh-NC and shRNA-LINC00473#2, (n=6
mice/group), and used for tumor formation assay. A total of
1×108 LINC00473#2- or sh-NC-transfected U251 cells in
100 µl PBS were subcutaneously injected in the right flank
of nude mice. Tumors were measured with digital calipers every 7
days; the largest tumor diameter was 4.83 mm, and tumor volume was
calculated. At 5 weeks post-injection, the mice were anaesthetized
with 40 mg/kg sodium pentobarbital (i.p.) and then sacrificed by
10% formalin perfusion fixation of central nervous system; death
was confirmed by completely stopping of the heartbeat and
breathing, as well as disappearance of the foot withdrawal reflex.
The tumor tissues were isolated and weighed, and RNAs and proteins
were extracted for analysis following the aforementioned
protocols.
Hematoxylin and Eosin (H&E)
staining
Glioma tissues from mice were fixed in 4%
paraformaldehyde at 4°C for 24 h, embedded in paraffin and cut into
7 µm sections with an RM2245 microtome (Leica Microsystems
GmbH). The sections were deparaffinized in xylene, rehydrated in
serially diluted ethanol and stained with H&E (Sigma-Aldrich;
Merck KGaA) at 4°C for 10 min. Representative photomicrographs were
captured using a light microscope (Olympus Corporation),
magnification, ×200.
Immunohistochemical (IHC) staining
Mouse glioma tissues were fixed with 4%
paraformaldehyde at 4°C for 24 h, embedded in paraffin and
sectioned (4 µm). After dewaxing with xylene and rehydration
through an ethanol series, sections were incubated in 3%
H2O2 to inhibit endogenous peroxidases. For
antigen retrieval, sections were immersed in Tris-EDTA buffer
containing 0.05% Tween 20 (pH 9.0), in a water bath at 95°C for 30
min. After washing with PBS, the sections were incubated in 4% dry
milk with 0.3% goat serum (Sigma-Aldrich; Merck KGaA) in PBS
solution for 20 min to block non-specific binding, and then
incubated overnight with anti- Ki67 (1:100; cat. no. ab15580;
Abcam) antibody in the presence of 10% rabbit serum (Sigma-Aldrich;
Merck KGaA). After washing with PBS, the sections were incubated
with HRP-conjugated goat anti-rabbit IgG secondary antibody (1:100;
cat. no. ab6721; Abcam). Slides were counterstained with
hematoxylin to stain cell nuclei, dehydrated in an ethanol series
and examined under a light microscope (Olympus Corporation);
magnification, ×200).
Statistical analysis
Experiments were repeated three times and data were
expressed as mean ± SEM. GraphPad Prism software v5.0 (GraphPad
Prism Software, Inc.) was used for statistical analysis by one-way
ANOVA and Tamhane’s T2 post hoc test. Kaplan-Meier method and
log-rank test were used to analyze differences between patients
with high or low levels of LINC00473 expression and overall
survival (OS). χ2 was used to analyze the data in
Table II. Paired Student’s t-test
was used to analyze differences between paired tumor and
para-cancerous tissues. Pearson’s correlation test was used for
correlation analysis. Multivariate and univariate analysis of
prognostic parameters in patients with glioma by Cox regression
analysis. P<0.05 was considered to indicate a statistically
significant difference.
 | Table IIAssociations between LINC00473
expression and clinicopathological features of patients with glioma
were determined by χ2 analysis. |
Table II
Associations between LINC00473
expression and clinicopathological features of patients with glioma
were determined by χ2 analysis.
| Clinicopathological
feature | LINC00473
expression
|
|---|
| Total (n=40) | Low (n=23) | High (n=17) | P-value |
|---|
| Age (years) | | | | |
| <50 | 15 | 7 | 8 | 0.283 |
| ≥50 | 25 | 16 | 9 | |
| Sex | | | | |
| Male | 17 | 7 | 10 | 0.073 |
| Female | 23 | 16 | 7 | |
| Histopathology | | | | |
| Conventional | 22 | 12 | 10 | 0.676 |
| Chordioid | 18 | 11 | 7 | |
| Tumor
recurrence | | | | |
| Yes | 23 | 17 | 6 | 0.015 |
| No | 17 | 6 | 11 | |
| Tumor grade | | | | |
| I-II | 11 | 11 | 0 | 0.001 |
| III | 14 | 9 | 5 | |
| IV | 15 | 3 | 12 | |
Results
lncRNA LINC00473 is highly expressed in
both glioma tissues and cell lines
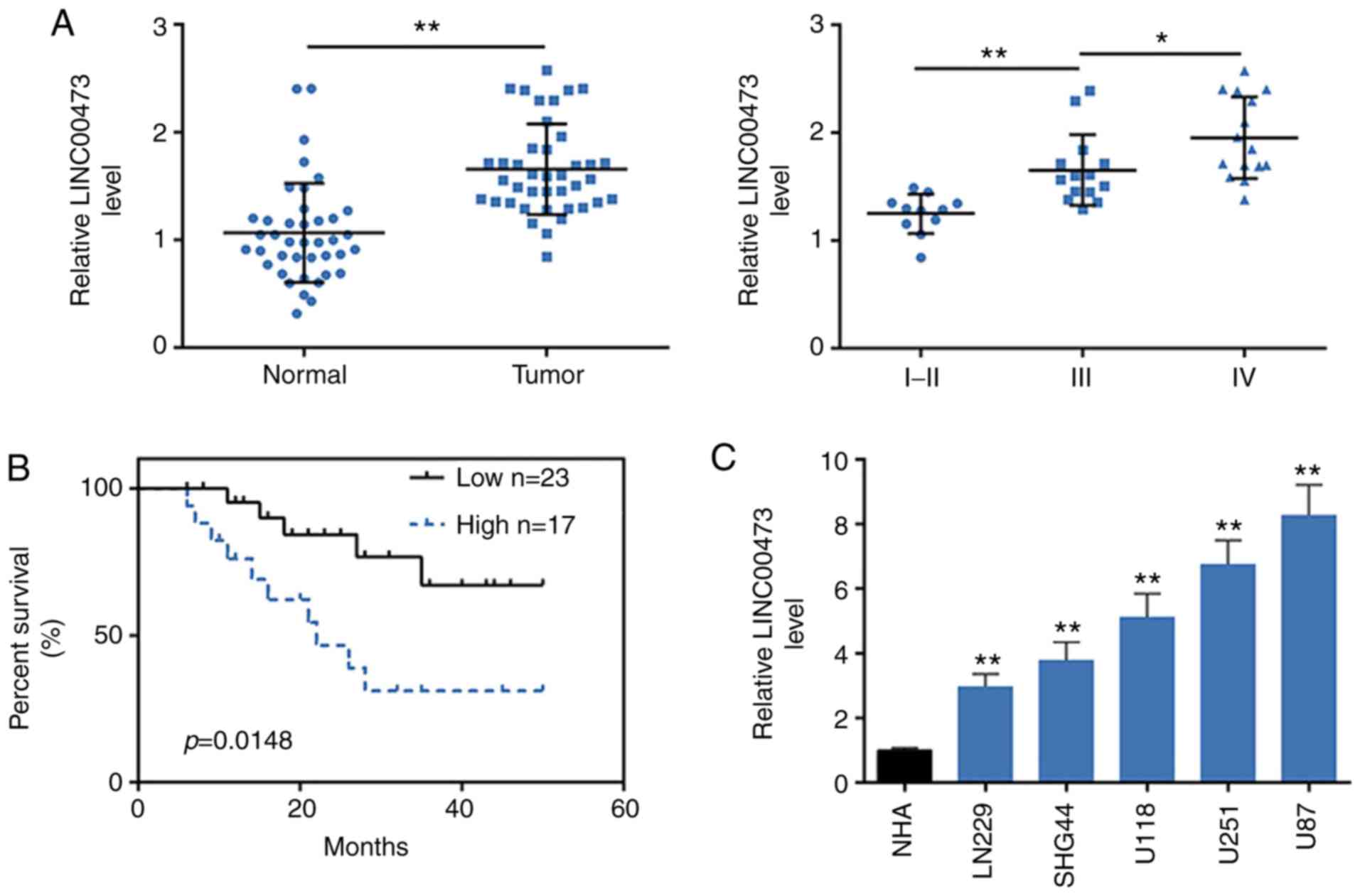
To determine the association between LINC00473 and
glioma tumorigenesis, the expression levels of LINC00473 were first
determined in glioma and para-cancerous tissues from 40 patients.
The results demonstrated that LINC00473 was significantly increased
in tumoral tissues compared with the para-carcinoma tissues
(P<0.01; Fig. 1A). Moreover,
LINC00473 levels were positively related to the WHO grades of
glioma (Fig. 1A), as there was
higher expression level in WHO grade III and IV compared with WHO
grade I and II, which indicated that LINC00473 may be associated
with malignancy of glioma. Median expression level of LINC00473 in
patients was considered as a cut-off, patients were then divided
into two groups: High LINC00473 expression group (fold change ≥2.5;
n=17) and low LINC00473 expression group (n=23). Further
association analysis between LINC00473 expression and
clinicopathological characteristics of patients with glioma
demonstrated that among the 40 patients, high LINC00473 expression
level was significantly associated with tumor recurrence (P=0.015)
and tumor grade (P<0.001) (Table
II). No significant associations were determined between
LINC00473 expression and the other clinicopathological features,
such as the age (P=0.283), histopathology (P=0.676) and sex
(P=0.073) (Table II). The
association between LINC00473 expression and prognosis of patients
with glioma was determined using Kaplan-Meier survival analysis
(Fig. 1B). Patients with high
LINC00473 level exhibited shorter OS compared with patients with
low LINC00473 expression (P=0.0148). Cox multivariate survival
analysis indicated tumor grade (P=0.039) and LINC00473 expression
(P=0.030) were independent risk prognostic factors of glioma
(Table III). However, univariate
analyses identified no clinicopathological features that were
significantly associated with glioma prognosis (Table III). The univariate and
multivariate analysis results suggested that there may be complex
interactions between the independent variables.
 | Table IIIMultivariate and univariate analysis
of prognostic parameters in patients with glioma by Cox regression
analysis. |
Table III
Multivariate and univariate analysis
of prognostic parameters in patients with glioma by Cox regression
analysis.
| Clinicopathological
feature | Multivariate
P-value | Univariate
P-value |
|---|
| Age (years) | | |
| <50 years | 0.288 | 0.181 |
| ≥50 years | | |
| Sex | | |
| Male | 0.11 | 0.275 |
| Female | | |
| Histopathology | | |
| Conventional | 0.233 | 0.733 |
| Chordioid | | |
| Invasion
condition | | |
| Yes | 0.056 | 0.165 |
| No | | |
| Tumor grade | | |
| I-II | 0.039 | 0.244 |
| III | | |
| IV | | |
| LINC00473
expression | | |
| Low | 0.030 | 0.291 |
| High | | |
LINC00473 expression levels were also increased in
human glioma cell lines (U251, U118, LN229, U87 and SHG44) compared
with NHA cells (Fig. 1C). As U251
and U87 cells exhibited the highest expression levels of LINC00473,
they were chosen for the subsequent functional assays.
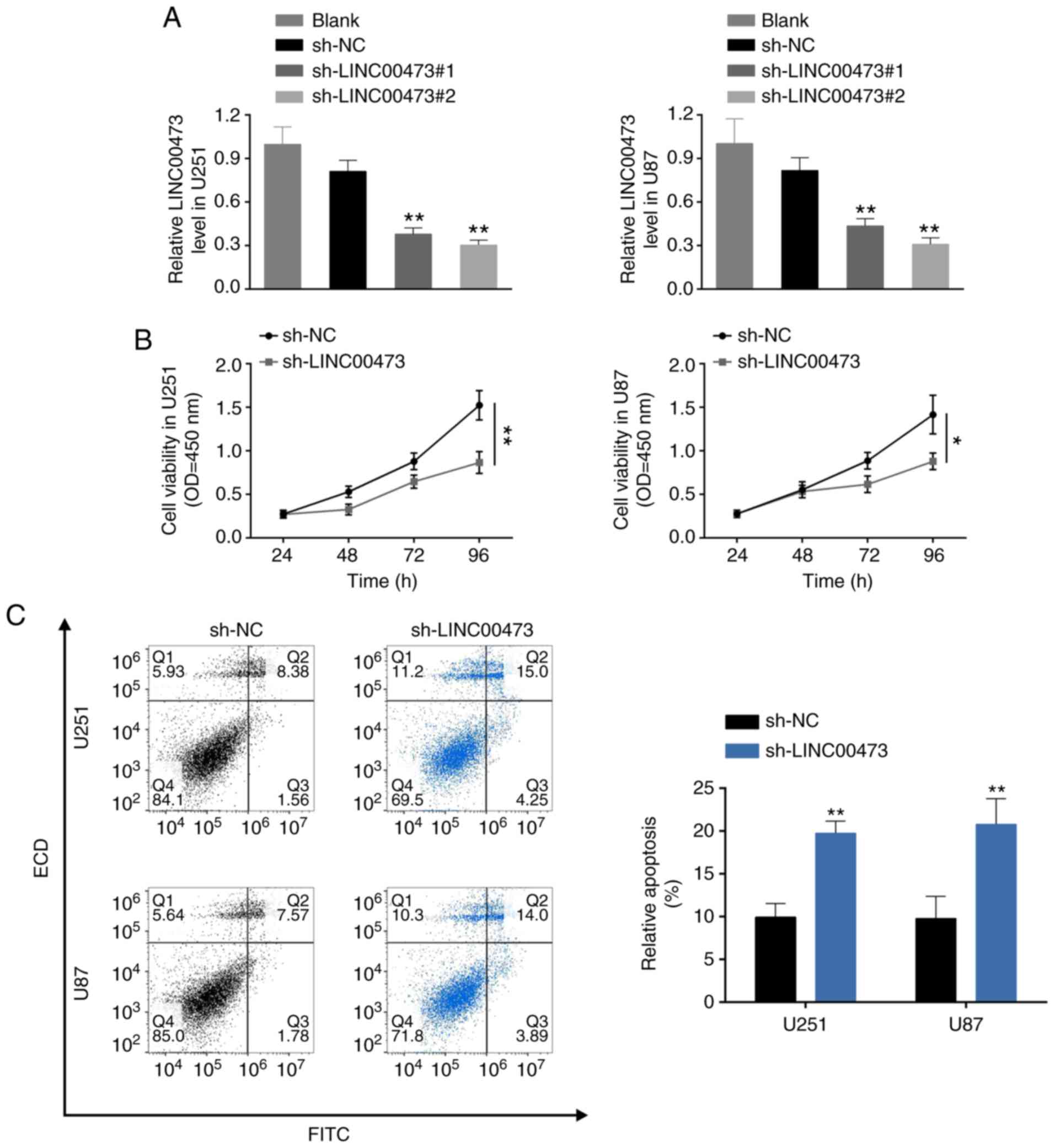
LINC00473 knockdown suppresses glioma
cell proliferation, migration and invasion, and induces
apoptosis
To further investigate the biological role of
LINC00473 in glioma cells, U251 and U87 cells were transfected with
sh-LINC00473#1 and sh-LINC00473#2. RT-qPCR analysis confirmed the
knockdown efficiency of LINC00473 in U251 and U87 cells (Fig. 2A); sh-LINC00473#2 was used for the
subsequent assays, and will be referred to as sh-LINC00473.
Loss-of-function assays demonstrated that LINC00473 knockdown
inhibited cell viability (Fig.
2B), induced apoptosis (Fig.
2C) and blocked cell cycle progression at the G0/G1 phase
(Fig. 2D). Moreover,
down-regulation of LINC00473 also substantially inhibited cell
migration (Fig. 2E) and invasion
(Fig. 2F). In general, LINC00473
knockdown suppressed glioma cell proliferation, migration and
invasion, and induced apoptosis.
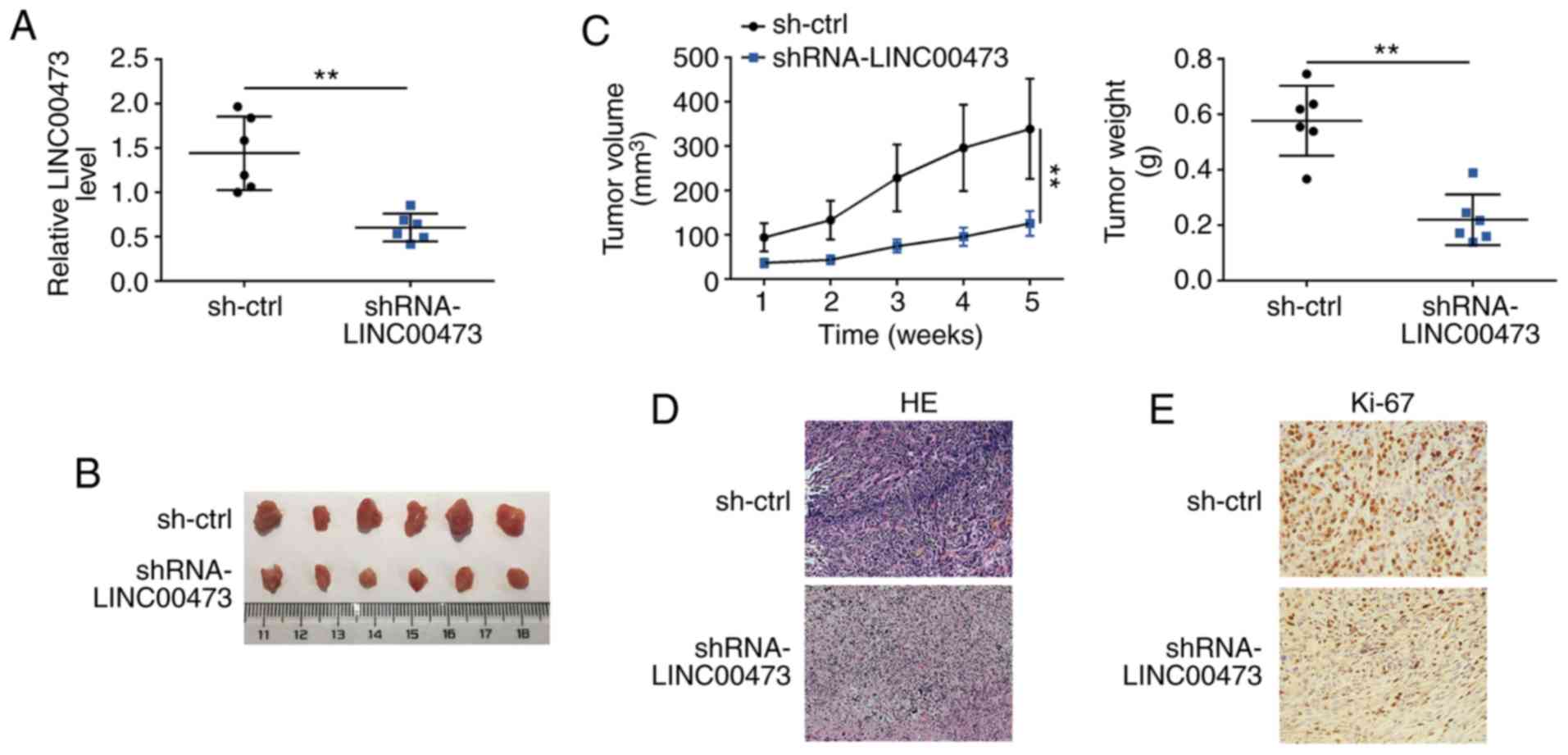
LINC00473 knockdown inhibits glioma
growth in vivo
The effects of LINC00473 on glioma growth were
examined in vivo. At 5 weeks post-transplantation,
subcutaneous tumors were harvested. The knockdown efficiency of
sh-LINC00473 in subcutaneous xenotransplanted tumors was confirmed
by RT-qPCR (Fig. 3A). Mice in the
sh-LINC00473 group exhibited smaller tumor sizes and volumes
compared with those of the sh-NC group (Fig. 3B and C, respectively). Tumor weight
was also significantly decreased in sh-LINC00473 group compared
with the control (Fig. 3C). Tumor
sections were stained with H&E, which revealed that LINC00473
knockdown decreased degrees of malignancy, as tissues in sh-NC
group exhibited dissimilar cells with bigger nucleus (Fig. 3D), and IHC staining demonstrated
that Ki67 expression levels were also notably reduced in the
xenografted tumor tissues of mice injected with
sh-LINC00473-trans-fected cells (Fig.
3E).
LINC00473 directly binds to and decreases
miR-195-5p expression
StarBase was used to determine if LINC00473
contained potential target sites for miR-195-5p (Fig. 4A). Dual luciferase reporter assays
confirmed that miR-195-5p significantly decreased the luciferase
activity of pmirGLO-wt-LINC00473, but did not have an effect on
pmirGLO-mut-LINC00473 (Fig. 4B),
which suggested a potential binding ability between LINC00473 and
miR-195-5p. Results from the RIP assay revealed that LINC00473 and
miR-195-5p were both significantly enriched in Ago2-containing
beads compared with the IgG-containing beads, the same as that of
the SNRNP70 positive control (Fig.
4C). Taken together, these results indicated that there may be
an interaction between LINC00473 and miR-195-5p.
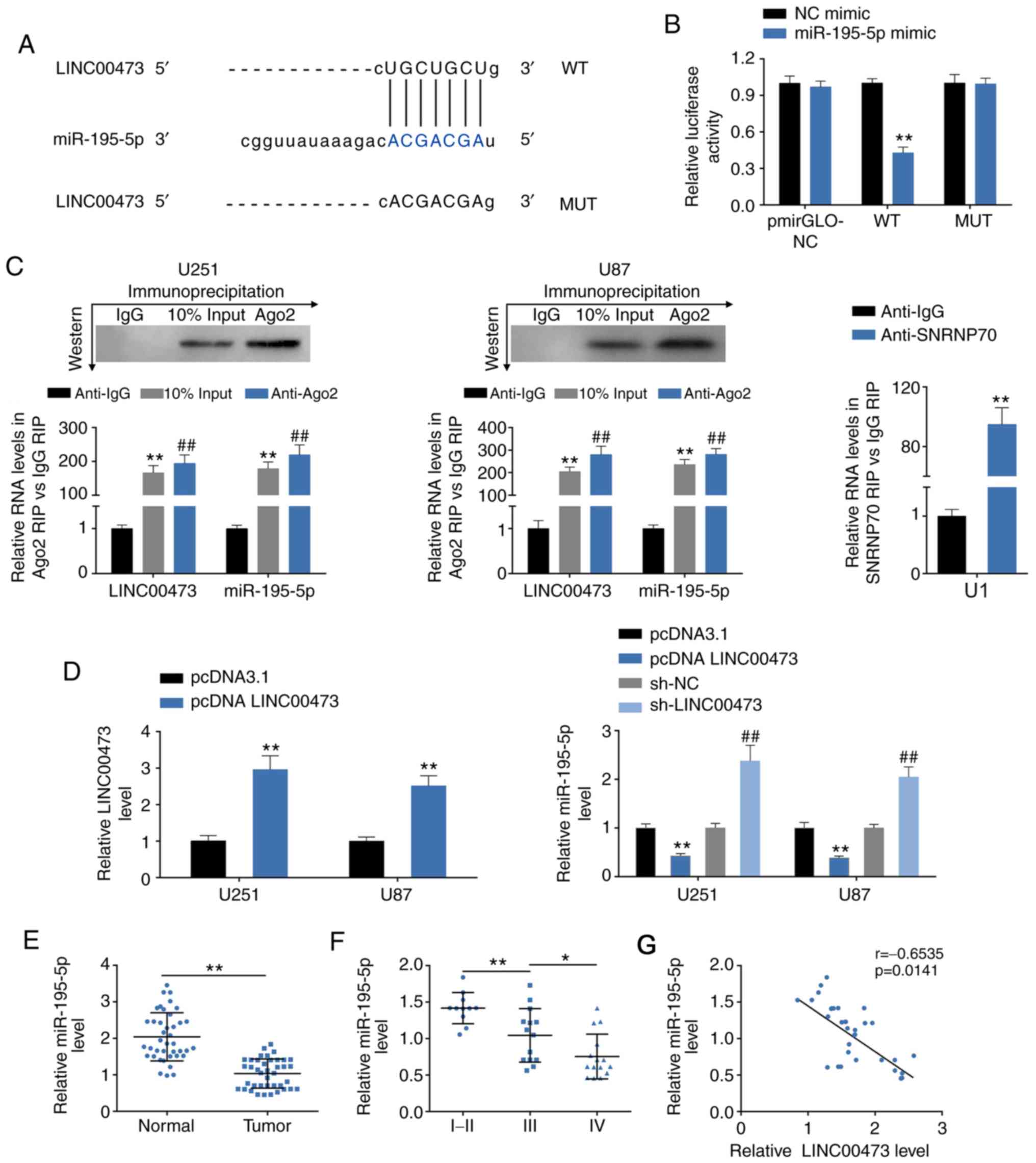 | Figure 4LINC00473 directly binds to and
decreases miR-195-5p expression. (A) Predicted target sites of
miR-195-5p in LINC00473. (B) Effect of miR-195-5p mimics on
luciferase activity of reporter gene with WT or MUT LINC00473
target regions. **P<0.01. (C) RIP assays were
performed using the Ago2 or IgG antibodies, and RT-qPCR was used to
detect the expression levels of miR-195-5p and LINC00473 in U251
and U87 cells. **P<0.01 and ##P<0.01
vs. anti-IgG. (D) Transfection efficiency of pcDNA3.1-LINC00473
overexpression (left) and the effects of pcDNA3.1-LINC00473 and
sh-LINC00473 on miR-195-5p expression in U251 and U87 cells was
detected by RT-qPCR. **P<0.01 vs. pcDNA3.1;
##P<0.01 vs. sh-NC. (E) miR-195-5p expression levels
in glioma tissues and para-carcinoma tissues were determined by
RT-qPCR. n=40; **P<0.01). (F) miR-195-5p expression
levels in high-grade glioma (WHO grade III and IV) and low-grade
glioma (WHO grade I and II) tissues were detect by RT-qPCR.
*P<0.05, **P<0.01. (G) Negative
correlation between LINC00473 and miR-195-5p in glioma tissues.
Ago2, argonaute 2; IgG, immunoglobulin G; miR, microRNA; MUT,
mutant; NC, negative control; RIP, RNA immunoprecipitation;
RT-qPCR, reverse transcription-qPCR; SNRNP70, U1 small
nucleoprotein 70 kDa; WT, wild-type. |
The effects of LINC00473 on miR-195-5p expression
were evaluated by gain- and loss-of-function assays. The
transfection efficiency of pcDNA 3.1-LINC00473 in U251 and U87
cells was confirmed by RT-qPCR (Fig.
4D). miR-195-5p expression levels were significantly increased
upon sh-LINC00473 trans-fection, whereas expression was decreased
upon LINC00473 overexpression, compared with the respective
controls (Fig. 4D). Decreased
miR-195-5p expression levels were also observed in human glioma
tissues (Fig. 4E), and miR-195-5p
expression was determined to be negatively related to the WHO grade
of glioma (Fig. 4E). Bivariate
Pearson’s correlation analysis revealed miR-195-5P expression was
negatively correlated with LINC00473 in glioma tissues (Fig. 4F).
YAP1 and TEAD1 are direct targets of
miR-195-5p
The potential targets for miR-195-5p were predicted
as axin 2, cell division cycle 23, dynactin 5, importin 7, proline
rich 15-like and sortilin 1 using TargetScan. YAP1 and TEAD1 were
also identified as containing miR-195-5p target sites (Fig. 5A) and were selected for the
following experiments. The targeting of miR-195-5p to YAP1 or TEAD1
3′UTRs were confirmed by dual luciferase reporter assay (Fig. 5B). Furthermore, the up- or
downregulation efficiency of miR-195-5p mimics or inh were
determined by RT-qPCR (Fig. 5C).
Similar to the bioinformatics results, miR-195-5p mimics could
decrease both mRNA (Fig. 5C) and
subsequent protein (Fig. 5D)
expression levels of YAP1 and TEAD1, whereas miR-195-5p inh
resulted in significantly increased YAP1 and TEAD1 expressions
compared with the respective controls (Fig. 5C and D). The elevated of YAP1 and
TEAD1 mRNA expression levels were also observed in glioma tissues
(Fig. 5E and F, respectively), and
both of the YAP1 and TEAD1 expressions were positively related to
the WHO grade of glioma (Fig. 5E and
F, respectively). Bivariate Pearson’s correlation analysis
indicated that YAP1 and TEAD1 expression were negatively correlated
with miR-195-5p, but positively correlated with LINC00473 (Fig. 5G and H).
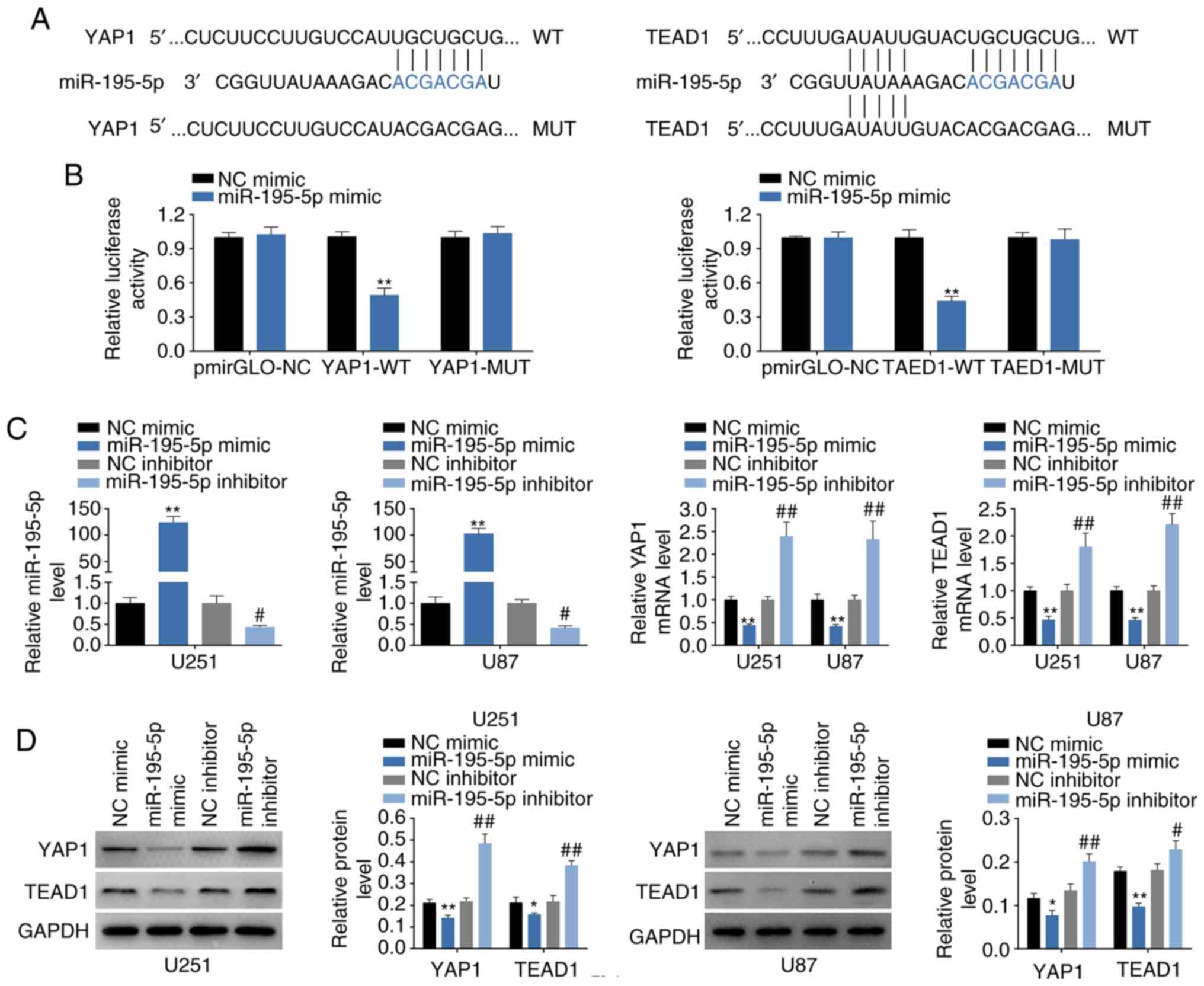 | Figure 5YAP1 and TEAD1 are direct targets of
miR-195-5p. (A) Predicted miR-195-5p target sites in the WT 3′UTR
of in YAP1 and TEAD1; MUT target sites are also shown. (B) Effect
of miR-195-5p on luciferase activity of reporter gene with WT or
MUT 3′UTR of YAP1 and TEAD1. **P<0.01 vs. miR-NC. (C)
Transfection efficiency of miR-195-5p mimics or inh in U251 and U87
cells; and the influence of miR-195-5p mimics or inh on mRNA
expression levels of YAP1 and TEAD1 were detected by RT-qPCR.
**P<0.01 vs. miR-NC; #P<0.05 and
##P<0.01 vs. inh NC. (D) The influence of miR-195-5p
mimics or inh on protein expression levels of YAP1 and TEAD1 was
detected by western blotting. *P<0.05 and
**P<0.01 vs. miR-NC; #P<0.05 and
##P<0.01 vs. inh NC. (E) YAP1 mRNA expression levels
in glioma tissues (Tumor) and para-carcinoma tissues (Normal),
n=40; and in high-grade glioma (WHO grade III and IV) and low-grade
glioma (WHO grade I and II) tissues were detected by RT-qPCR.
*P<0.05 and **P<0.01. (F) TEAD1 mRNA
expression levels in glioma tissues and para-carcinoma tissues
(n=40) and in high-grade glioma (WHO grade III and IV) and
low-grade glioma (WHO grade I and II) tissues were detected by
RT-qPCR. *P<0.05 and **P<0.01. (G)
Negative correlation between miR-195-5p and YAP1 in glioma tissues;
and positive correlation between LINC00473 and YAP1 in glioma
tissues. (H) Negative correlation between miR-195-5p and TEAD1 in
glioma tissues, and positive correlation between LINC00473 and
TEAD1 in glioma tissues. Inh, inhibitor; miR, microRNA; MUT,
mutant; RT-qPCR, reverse transcription-qPCR; TEAD1, TEA domain
family member 1; UTR, untranslated region; WT, wild-type; YAP1,
Yes-associated protein 1. |
LINC00473 promotes glioma progression
through miR-195-5p-mediated YAP1-TEAD1-Hippo signaling
We designed four groups: U251 and U87 cells
transfected with i) inh NC and sh-NC, ii) sh-LINC00473 and inh NC,
iii) miR-195-5p inh and sh-NC, and iv) sh-LINC00473 and miR-195-5p
inh, to determine the functional roles of
LINC00473/miR-195-5p/YAP1-TEAD1-Hippo axis in regulating glioma
progression. CCK-8 assay results revealed that the inhibition of
cell viability by sh-LINC00473 was reversed in cells co-transfected
with miR-195-5p inh (Fig. 6A);
cells trans-fected with miR-195-5p inh exhibited higher viability
compared with cells co-transfected sh-LINC00473 and miR-195-5p inh.
Flow cytometry results revealed that induction of cell apoptosis
(Fig. 6B) and inhibition of cell
cycle (Fig. 6C) by sh-LINC00473
was also reversed in cells co-transfected with miR-195-5p inh;
cells transfected with miR-195-5p inh exhibited lower apoptotic
rates and higher cell cycle ratio (cell number of G1: All the cell
number) compared with cells co-transfected sh-LINC00473 and
miR-195-5p inh. Lastly, the suppressed migratory and invasive
abilities of cells expressing sh-LINC00473 could be reversed by
miR-195-5p inh co-transfection (Fig.
6D and E, respectively); cells transfected miR-195-5p inh
exhibited significant increases in migration and invasion compared
with cells co-transfected sh-LINC00473 and miR-195-5p inh (Fig. 6D and E, respectively). Western blot
analysis demonstrated significantly decreased YAP1, TEAD1 and CTGF
(connective tissue growth factor) levels upon sh-LINC00473
transfection compared with the control group (Fig. 6F), which were reversed in cells
co-transfected with sh-LINC00473 and miR-195-5p inh (Fig. 6F). Cells transfected with
miR-195-5p inh alone exhibited notably higher YAP1 and TEAD1
protein expression levels compared with cells co-expressing
sh-LINC00473 and miR-195-5p inh (Fig.
6F).
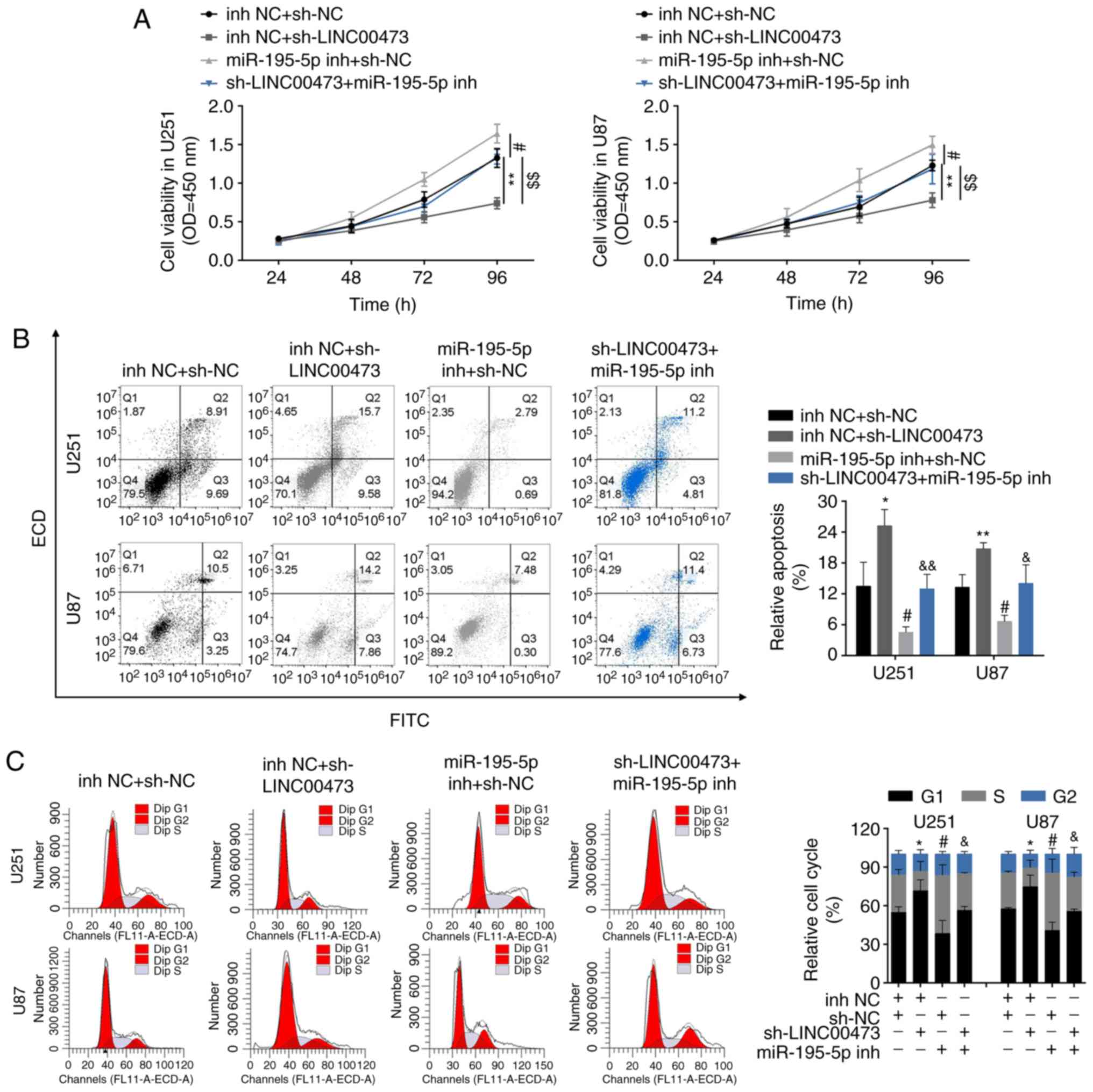 | Figure 6LINC00473 promotes glioma progression
through miR-195-5p-mediated YAP1-TEAD1-Hippo signaling pathway. (A)
The influence of sh-LINC00473 and miR-195-5p inh on cell viability
was detected by Cell Counting Kit-8. (B) The influence of
sh-LINC00473 and miR-195-5p inh on apoptosis was detected by flow
cytometry. (C) The influence of sh-LINC00473 and miR-195-5p inh on
cell cycle progression, specifically G1, was detected by flow
cytometry. LINC00473 promotes glioma progression through
miR-195-5p-mediated YAP1-TEAD1-Hippo signaling pathway. (D) The
influence of sh-LINC00473 and miR-195-5p inh on cell migration as
determined by wound healing assay. (E) The influence of
sh-LINC00473 and miR-195-5p inh on cell invasion was detected by
Transwell assay. (F) The influence of sh-LINC00473 and miR-195-5p
inh on protein expression levels of YAP1, TEAD1 and CTGF were
determined by western blotting. *P<0.05 and
**P<0.01 vs. inh NC + sh-NC; #P<0.05
and ##P<0.01 vs. inh NC + sh-NC;
&P<0.05 and &&P<0.01 vs.
inh NC + sh-LINC00473. CTGF, connective tissue growth factor; inh,
inhibitor; miR, microRNA; NC, negative control; sh, short hairpin
RNA; TEAD1, TEA domain family member 1; YAP1, Yes-associated
protein 1. |
Discussion
Glioma is considered as one of most aggressive
malignancies in central nervous system worldwide; it is clinically
complicated due to various pathogenic genes and proteins (28,29).
lncRNAs are key regulators in glioma pathogenesis (7), and LINC00473 has been reported to be
a novel oncogene of human cancers (10,17).
Therefore, it is important to investigate the regulatory mechanisms
of LINC00473 on prognosis with the aim to treat glioma.
Results from the present study indicated that
LINC00473 was highly expressed in both glioma tissues and cell
lines, which was in line with results from a previous study
(30). Moreover, high LINC00473
expression was not only tightly associated with WHO grades of
glioma, but also related to the OS of the patients, predicting a
poor prognosis in patients with glioma. Poor tumor prognosis is
related to the invasion and metastasis of cancer cells (22). LINC00473 has been correlated with
poor prognosis in non-small cell lung cancer (9), gastric cancer (15), colorectal cancer (17) and head and neck squamous cell
carcinoma (31) by promoting
cancer cell migration and invasion. Therefore, LINC00473 was
speculated to be involved in glioma progression. However, owing to
the small sample size of our current clinical analysis (n=40), the
significant correlation between high LINC00473 expression with
other clinical parameters requires further investigations. A larger
patient cohort is also needed to strengthen the clinical
significance of LINC00473 in patients with glioma.
Consistent with a previous hypothesis (30), our in vitro and in
vivo loss-of-function assays indicated that LINC00473 knockdown
not only inhibited cell proliferation, invasion and migration of
glioma cells, but also blocked cell cycle and induced apoptosis.
In vivo subcutaneous xenotransplanted tumor models revealed
that interference of LINC00473 could suppress tumorigenic ability
of glioma, as shown by the lower expression of Ki67 in sh-LINC00473
group than sh-NC group. Ki67 is a molecular marker that predicts
poor prognosis of glioma patients (32), the reduced expression of Ki67
indicated the potential clinical application of LINC00473 in
treatment of glioma. However, the underlying mechanism remains to
be clarified.
Accumulating evidence suggest that lncRNAs function
as ceRNAs to sponge miRNA, therefore titrating them off the binding
sites on protein-coding mRNAs (33). LINC00473 was previously reported to
sponge miR-15a in colorectal cancer (17), and the binding ability between
LINC00473 and miR-195 was also reported in Wilms tumor (10). In the present study, miR-195-5p was
identified as a target of LINC00473 in glioma cells by the means of
dual luciferase reporter assay and RIP. Moreover, downregulation of
miR-195-5p was found to be associated with poor prognosis of
patients with glioma. In particular, miR-195-5p may inhibit cell
proliferation and induce apoptosis in glioma cells through the
regulation of cell apoptosis-related proteins including Caspase-3,
-8, -9 and Bcl-2 (34). Direct
targets of miR-195 in glioma cells were identified as Sal-like
protein 4 (35), cyclin E1
(36) to affect cell cycle
(37) in the previous studies. The
present study also demonstrated that miR-195-5p expression is
reduced in glioma tissues and cells, and inhibition of miR-195-5p
promoted cell proliferation, migration and invasion of glioma
cells, and inhibited apoptosis.
YAP1 and TEAD1 were identified as targets of
miR-195-5p using online prediction databases and dual luciferase
reporter assay. In vitro loss-of-function assays
demonstrated that LINC00473 knockdown reduced YAP1 and TEAD1
expression, which may suppress glioma progression. In addition,
CTGF, which was reported to promote glioma migration (38), was also decreased upon loss of
LINC00473 expression, which indicated an oncogenic role of
LINC00473. Activation of YAP increased the expression of downstream
target CTGF, leading to stem cell phenotype of glioma (39). Effects of LINC00473 on
epithelial-mesenchymal transition and stemness properties of glioma
cells required further investigation. As LINC00473 is associated
with various lncRNA-miRNA-mRNA regulatory axes and is involved in
signaling pathways in regulation of tumors progression; additional
potential miRNA targets and downstream signaling networks should be
studied in the future. Recently, LINC00473 was reported to promote
glioma progression via regulation of the miR-637/CDK6 axis
(30), confirming various
LINC00473-miRNA-mRNA regulatory axes in regulation of glioma.
In conclusion, the present study demonstrated that
knockdown of lncRNA LINC00473 inhibited cell proliferation,
migration and invasion of glioma cells, as well as blocked cell
cycle and induced cell apoptosis, via regulation of
miR-195-5p/YAP1-TEAD1 axis. These data revealed the relationship
between LINC00473/miR-195-5p/YAP1/TEAD1 regulatory axis and glioma
progression, and may shed light on the potential therapeutic
application of LINC00473 in glioma treatment.
Funding
No funding was received.
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors’ contributions
XW and XBJ conceived and designed the experiments.
XDL and ZYF analyzed and interpreted the results of the
experiments. YZ and XH performed the experiments.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Tongji Medical College, Huazhong University of Science
and Technology (Wuhan, China; approval no. 2014-S034), and all the
patients signed written informed consent. All studies involving
animals were approved by Union Hospital Affiliated with Tongji
Medical College of Huazhong University of Science and Technology
and conducted in accordance with the guidelines set out by Medical
Ethics Committee of Tongji Medical College, Huazhong University of
Science and Technology (approval no. 2014-S034).
Patient consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing
interests.
Acknowledgments
Not applicable.
Reference
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Naderali E, Nikbakht F, Ofogh SN and
Rasoolijazi H: The role of rosemary extract in degeneration of
hippocampal neurons induced by kainic acid in the rat: A behavioral
and histochemical approach. J Integr Neurosci. 17:19–25. 2018.
View Article : Google Scholar
|
|
3
|
Schwartzbaum JA, Fisher JL, Aldape KD and
Wrensch M: Epidemiology and molecular pathology of glioma. Nat Clin
Pract Neurol. 2:494–503; quiz 1 p following 516. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Milano MT, Johnson MD, Sul J, Mohile NA,
Korones DN, Okunieff P and Walter KA: Primary spinal cord glioma: A
surveillance, epidemiology, and end results database study. J
Neurooncol. 98:83–92. 2010. View Article : Google Scholar
|
|
5
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cheetham SW, Gruhl F, Mattick JS and
Dinger ME: Long noncoding RNAs and the genetics of cancer. Br J
Cancer. 108:2419–2425. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Kiang KM, Zhang XQ and Leung GK: Long
non-coding RNAs: The key players in glioma pathogenesis. Cancers
(Basel). 7:1406–1424. 2015. View Article : Google Scholar
|
|
8
|
Pruunsild P, Bengtson CP and Bading H:
Networks of cultured iPSC-derived neurons reveal the human synaptic
activity-regulated adaptive gene program. Cell Rep. 18:122–135.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Chen Z, Li JL, Lin S, Cao C, Gimbrone NT,
Yang R, Fu DA, Carper MB, Haura EB, Schabath MB, et al:
cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer
and mediates tumor growth. J Clin Invest. 126:2267–2279. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhu S, Fu W, Zhang L, Fu K, Hu J, Jia W
and Liu G: LINC00473 antagonizes the tumour suppressor miR-195 to
mediate the pathogenesis of Wilms tumour via IKKα. Cell Prolif.
51:2018. View Article : Google Scholar
|
|
11
|
Chen H, Yang F, Li X, Gong ZJ and Wang LW:
Long noncoding RNA LNC473 inhibits the ubiquitination of survivin
via association with USP9X and enhances cell proliferation and
invasion in hepatocellular carcinoma cells. Biochem Biophys Res
Commun. 499:702–710. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi C, Yang Y, Yu J, Meng F, Zhang T and
Gao Y: The long noncoding RNA LINC00473, a target of microRNA 34a,
promotes tumorigenesis by inhibiting ILF2 degradation in cervical
cancer. Am J Cancer Res. 7:2157–2168. 2017.PubMed/NCBI
|
|
13
|
Chen Z, Lin S, Li JL, Ni W, Guo R, Lu J,
Kaye FJ and Wu L: CRTC1-MAML2 fusion-induced lncRNA LINC00473
expression maintains the growth and survival of human
muco-epidermoid carcinoma cells. Oncogene. 37:1885–1895. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dinh TA, Vitucci EC, Wauthier E, Graham
RP, Pitman WA, Oikawa T, Chen M, Silva GO, Greene KG, Torbenson MS,
et al: Comprehensive analysis of the cancer genome atlas reveals a
unique gene and non-coding RNA signature of fibrolamellar
carcinoma. Sci Rep. 7:446532017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang W and Song Y: LINC00473 predicts
poor prognosis and regulates cell migration and invasion in gastric
cancer. Biomed Pharmacother. 107:1–6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ebert MS and Sharp PA: Emerging roles for
natural microRNA sponges. Curr Biol. 20:R858–R861. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang L, Zhang X, Sheng L, Qiu C and Luo R:
LINC00473 promotes the Taxol resistance via miR-15a in colorectal
cancer. Biosci Rep. 38:pii: BSR20180790. 2018.
|
|
18
|
Hui W, Yuntao L, Lun L, WenSheng L,
ChaoFeng L, HaiYong H and Yueyang B: MicroRNA-195 inhibits the
proliferation of human glioma cells by directly targeting cyclin D1
and cyclin E1. PLoS One. 8:e549322013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhang QQ, Xu H, Huang MB, Ma LM, Huang QJ,
Yao Q, Zhou H and Qu LH: MicroRNA-195 plays a tumor-suppressor role
in human glioblastoma cells by targeting signaling pathways
involved in cellular proliferation and invasion. Neuro Oncol.
14:278–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yilaz Susluer S, Biray Avci C, Dodurga Y,
Ozlem Dogan Sigva Z, Oktar N and Gunduz C: Downregulation of
miR-195 via cyclo-sporin A in human glioblastoma cells. J BUON.
20:1337–1340. 2015.PubMed/NCBI
|
|
21
|
Yu S, Jing L, Yin XR, Wang MC, Chen YM,
Guo Y, Nan KJ and Han LL: MiR-195 suppresses the metastasis and
epithelial-mesenchymal transition of hepatocellular carcinoma by
inhibiting YAP. Oncotarget. 8:99757–99771. 2017.PubMed/NCBI
|
|
22
|
Yang C, Wu K, Wang S and Wei G: Long
non-coding RNA XIST promotes osteosarcoma progression by targeting
YAP via miR-195-5p. J Cell Biochem. 119:5646–5656. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Sun M, Song H, Wang S, Zhang C, Zheng L,
Chen F, Shi D, Chen Y, Yang C, Xiang Z, et al: Integrated analysis
identifies microRNA-195 as a suppressor of Hippo-YAP pathway in
colorectal cancer. J Hematol Oncol. 10:792017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chai J, Xu S and Guo F: TEAD1 mediates the
oncogenic activities of Hippo-YAP1 signaling in osteosarcoma.
Biochem Biophys Res Commun. 488:297–302. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Orr BA, Bai H, Odia Y, Jain D, Anders RA
and Eberhart CG: Yes-associated protein 1 is widely expressed in
human brain tumors and promotes glioblastoma growth. J Neuropathol
Exp Neurol. 70:568–577. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wittig A, Moss RL and Sauerwein WA:
Glioblastoma, brain metastases and soft tissue sarcoma of
extremities: Candidate tumors for BNCT. Appl Radiat Isot. 88:46–49.
2014. View Article : Google Scholar
|
|
27
|
Tome-Garcia J, Erfani P, Nudelman G,
Tsankov AM, Katsyv I, Tejero R, Bin Zhang, Walsh M, Friedel RH,
Zaslavsky E and Tsankova NM: Analysis of chromatin accessibility
uncovers TEAD1 as a regulator of migration in human glioblastoma.
Nat Commun. 9:40202018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Berntsson SG, Merrell RT, Amirian ES,
Armstrong GN, Lachance D, Smits A, Zhou R, Jacobs DI, Wrensch MR,
Olson SH, et al: Glioma-related seizures in relation to
histopathological subtypes: A report from the glioma international
case-control study. J Neurol. 265:1432–1442. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ochiai Y, Sano E, Okamoto Y, Yoshimura S,
Makita K, Yamamuro S, Ohta T, Ogino A, Tadakuma H, Ueda T, et al:
Efficacy of ribavirin against malignant glioma cell lines:
Follow-up study. Oncol Rep. 39:537–544. 2018.
|
|
30
|
Zhang Q, Wang G, Xu L, Yao Z and Song L:
Long non-coding RNA LINC00473 promotes glioma cells proliferation
and invasion by impairing miR-637/CDK6 axis. Artif Cells Nanomed
Biotechnol. 47:3896–3903. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Han PB, Ji XJ, Zhang M and Gao LY:
Upregulation of lncRNA LINC00473 promotes radioresistance of HNSCC
cells through activating Wnt/β-catenin signaling pathway. Eur Rev
Med Pharmacol Sci. 22:7305–7313. 2018.PubMed/NCBI
|
|
32
|
Yuan Y, Xiang W, Yanhui L, Ruofei L,
Shuang L, Yingjun F, Qiao Z, Yanwu Y and Qing M: Ki-67
overexpression in WHO grade II gliomas is associated with poor
postoperative seizure control. Seizure. 22:877–881. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Jia Y, Tian Y, An S and Yang D: Effects of
microRNA-195 on the prognosis of glioma patients and the
proliferation and apoptosis of human glioma cells. Pathol Oncol
Res. Feb 26–2019.Epub ahead of print. View Article : Google Scholar
|
|
35
|
Chen LP, Zhang NN, Ren XQ, He J and Li Y:
miR-103/miR-195/miR-15b regulate SALL4 and inhibit proliferation
and migration in glioma. Molecules. 23:pii: E2938. 2018. View Article : Google Scholar
|
|
36
|
Wang H, Ren S, Xu Y, Miao W, Huang X, Qu
Z, Li J, Liu X and Kong P: MicroRNA-195 reverses the resistance to
temozolomide through targeting cyclin E1 in glioma cells.
Anticancer Drugs. 30:81–88. 2019. View Article : Google Scholar
|
|
37
|
Rizzo A, Donzelli S, Girgenti V, Sacconi
A, Vasco C, Salmaggi A, Blandino G, Maschio M and Ciusani E: In
vitro antineoplastic effects of brivaracetam and lacosamide on
human glioma cells. J Exp Clin Cancer Res. 36:762017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Xie D, Yin D, Wang HJ, Liu GT, Elashoff R,
Black K and Koeffler HP: Levels of expression of CYR61 and CTGF are
prognostic for tumor progression and survival of individuals with
gliomas. Clin Cancer Res. 10:2072–2081. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Zhu G, Wang Y, Mijiti M, Wang Z, Wu PF and
Jiafu D: Upregulation of miR-130b enhances stem cell-like phenotype
in glioblastoma by inactivating the Hippo signaling pathway.
Biochem Biophys Res Commun. 465:194–199. 2015. View Article : Google Scholar : PubMed/NCBI
|