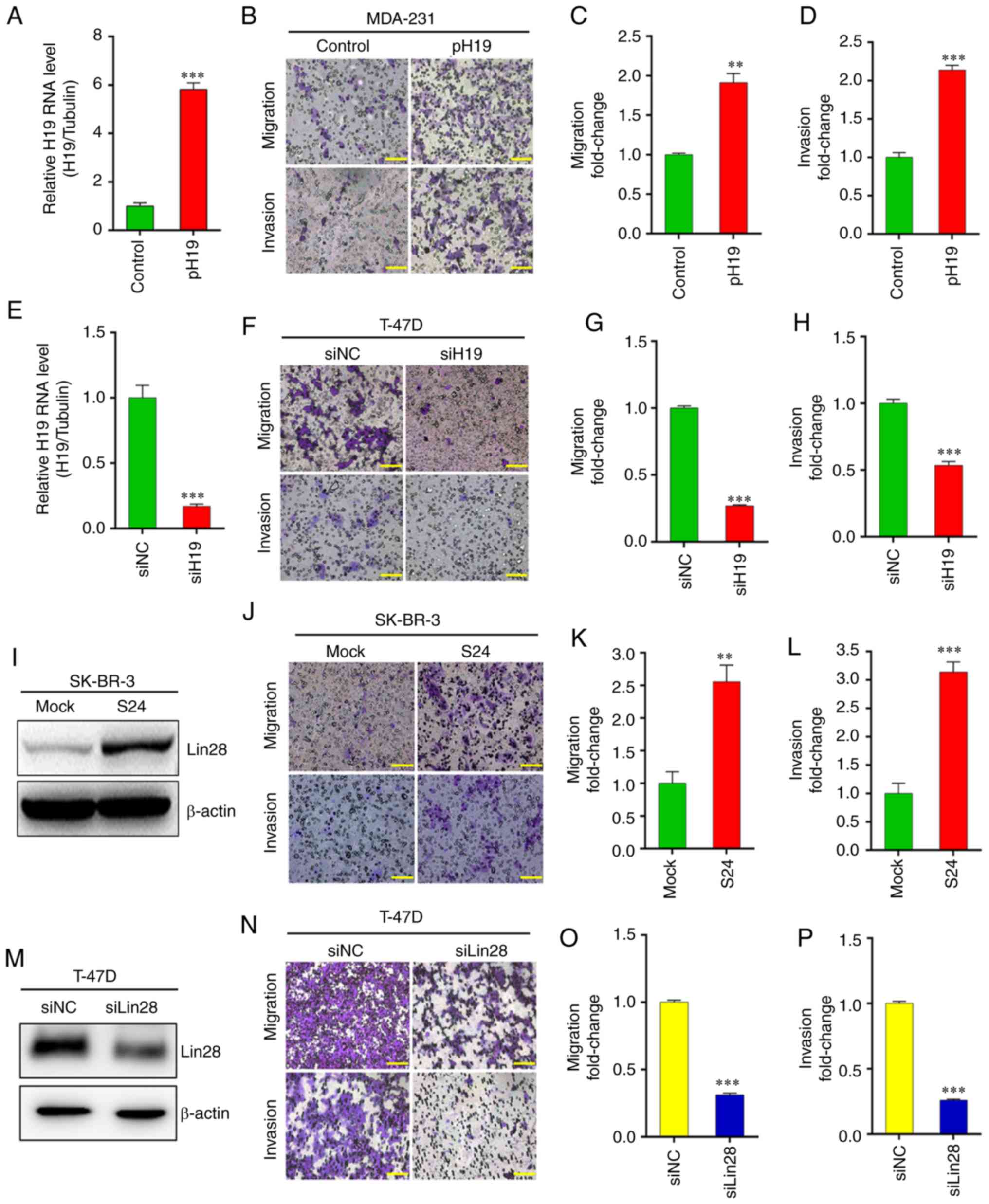
Introduction
Breast cancer (BC) is one of the leading causes of
cancer-associated mortality in females ≤40 years of age globally
(1). Early detection and
comprehensive treatment, which currently consist of surgery,
radiation, chemotherapy, endocrine therapy and targeted therapy,
have substantially improved the prognosis of patients with BC
(2). Nevertheless, a significant
proportion of patients still develop distant metastases, and their
prognoses remain poor (3).
Moreover, the use of conventional therapies has not increased the
overall survival of patients with metastatic BC (4). Therefore, to determine the potential
molecular mechanisms underlying BC metastasis, it is necessary to
further investigate effective biomarkers and molecular targets for
the disease.
The majority of the human genome can be transcribed
into long non-coding RNAs (lncRNAs), which are transcripts >200
nucleotides in length. lncRNAs have attracted increasing attention
for their critical roles in tumor biology, including those in tumor
initiation and progression (5).
First identified in 1991 by Bartolomei et al (6), lncRNA H19 has now been established as
an oncogenic marker in a diverse range of cancer types, including
liver, lung, gastric, bladder, pancreatic and colorectal cancers,
as well as BC (7). Although H19 is
considered an important therapeutic target for various diseases,
the mechanisms and functions of H19 in BC remain poorly understood.
Lin28, an RNA-binding protein and transcription factor, has drawn
considerable attention due to its implicated involved in stem cell
differentiation, normal development, glucose metabolism and cancer
(8). Lin28 is frequently
upregulated in BC, and functions as a oncogene in numerous
cancer-associated processes (9).
Epithelial-mesenchymal transition (EMT) is the
complete trans-differentiation from a functional epithelial cell to
a mesenchymal-like cell, and involves multiple molecular signal
transduction pathways (10).
Numerous observations have indicated that autophagy-related
regulatory pathways, such as the PI3K/AKT/mTOR and beclin-1
pathways, can influence EMT (11,12).
Therefore, further understanding of the specific regulatory
mechanisms between EMT and autophagy is required. Autophagy,
literally defined as self-eating, is a process via which cells
degrade and recycle cytoplasmic material within lysosomes (13). Aside from its role in homeostatic
maintenance, autophagy has also been implicated in a diverse range
of pathologies, including cancer. Autophagic inhibition of EMT has
been identified in pancreatic cancer (14), BC (15) and gastric cancer (16), but the underlying mechanisms of
autophagy in cancer cells remain controversial.
In the present study, the H19/let-7/Lin28 loop was
found to be a requirement for downregulating autophagy in BC cells.
Using Transwell and morphological assays, the H19/let-7/Lin28 loop
was found to promote the EMT of BC cells. Moreover, the
H19/let-7/Lin28 network was found to be involved in the autophagic
inhibition of EMT in BC. Therefore, the results of the present
study indicate that targeting the H19/let-7/Lin28 competitive
endogenous RNA (ceRNA) network may provide putative prevention and
treatment for the metastasis of BC.
Materials and methods
Ethics statement
All patients provided written informed consent for
the use of the remainder of their pathological specimens for
research purposes. The protocols used during the present study, and
the use of human tissue, were approved by the Ethics Committee of
the Sir Run Run Shaw Hospital affiliated with Zhejiang University,
and conducted in full accordance with the ethical principles cited
in the World Medical Association Declaration of Helsinki and local
legislation.
Patient information
From January 2016 to December 2016, a total of 43
patients with BC (42-54 years old) at the Department of Surgical
Oncology, Sir Run Run Shaw Hospital, Zhejiang University were
included in this study. Of these patients, the 23 patients with
lymph node (LN)-positive BC ranged from 42 to 54 years, and the 20
patients with LN-negative BC ranged from 44 to 54 years. Breast
cancer tissues were surgically obtained from patients and frozen at
-80°C.
Cell lines, antibodies and chemical
reagents
The SK-BR-3, MCF-7, MDA-231 and T-47D human BC cell
lines, and the MCF-10A normal human breast cell line were obtained
from the American Type Culture Collection (ATCC). MDA-231 cells
were cultured in Leibovitz's L-15 medium (cat. no. 11415-064) and
the other cell lines were cultured in RPMI-1640 medium (cat. no.
11875-093; both Invitrogen; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (FBS; Gibco; Thermo Fisher
Scientific, Inc.). All cells were incubated at 37°C with 5%
CO2 and 95% humidity. The anti-Lin28 antibody (cat. no.
3533-1) was obtained from Epitomics (Abcam), and the anti-β-Actin
antibody (cat. no. sc-47778) was purchased from Santa Cruz
Biotechnology, Inc. The anti-GAPDH antibody (cat. no. 5174) and
anti-β-Tubulin antibody (cat. no. 2146) were purchased from Cell
Signaling Technology, Inc. Primary antibodies against the EMT
markers E-cadherin (cat. no. 3195) and vimentin (cat. no. 5741)
were purchased from Cell Signaling Technology, Inc. Of the
autophagy markers, anti-p62 (cat. no. PM045) was purchased from MBL
International Co., anti-beclin-1 (cat. no. 3495) was purchased from
Cell Signaling Technology, Inc. and microtubule-associated protein
light chain (LC)3B (cat. no. L7543) was acquired from Sigma-Aldrich
(Merck KGaA). For autophagy induction, cells were treated with 200
nM rapamycin (Rapa; cat. no. 37094; Sigma-Aldrich; Merck KGaA). For
autophagy inhibition, the cells were treated with 50 µM
chloroquine (CQ; cat. no. C6628; Sigma-Aldrich; Merck KGaA). All
cells were incubated at 37°C for 24 h during treatment with these
compounds. For the morphological change assays, the cells were
treated with 2.5 ng/ml transforming growth factor (TGF)-β (R&D
Systems, Inc.) for 24 h at 37°C. Each sample was observed under a
Zeiss LSM 710 confocal microscope (magnification, ×100).
Stable Lin28-expressing SK-BR-3 cell
line
Third-generation Lin28 lentiviral vectors were
generated with the LV-LIN28A plasmid (Addgene, Inc.) and vesicular
stomatitis virus G (Addgene, Inc.), and then transiently
transfected into 2.5×105 293T (ATCC) cells at a ratio of
6:1 using Lipofectamine® 3000 transfection reagent (cat.
no. L3000001; Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol; viral supernatants were
collected 36 h later and filtered through a 0.45-µm filter.
After 5×104 SK-BR-3 cells were inoculated into 48-well
plates for 24 h, viral supernatants (MOI=5) and diethyl-aminoethyl
(DEAE)-dextran (20 µg/ml; Baomanbio, Inc.) were added into
the plate. After culturing for 12 h, the original medium was
replaced with DMEM (Gibco; Thermo Fisher Scientific, Inc.)
containing 10% FBS for culture for another 72 h. After cells were
screened using puromycin (Sigma-Aldrich; Merck KGaA) and cloned,
the SK-BR-3 cell lines exhibiting stable infection were selected.
Overexpression of Lin28 was then verified via western blotting;
clone 1 and 24 (S1 and S24) were found to exhibit stable expression
of Lin28.
RNA/microRNA (miRNA/miR) extraction and
reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from tissue specimens using
an miRNeasy FFPE Kit (cat. no. 217504; Qiagen, Inc.), and was
isolated from cell lines using an E.Z.N.A.® Total RNA
Kit I (cat. no. R6834-02; Omega Bio-Tek, Inc.) according to the
manufacturer's protocols. All samples were eluted in 50 µl
diethyl pyrocarbonate water. RNA was quantified using a NanoDrop™
2000c (NanoDrop Technologies; Thermo Fisher Scientific, Inc.). RT
reactions were performed using random primers from a HiFiScript
cDNA Synthesis Kit (cat. no. CW2569-100; Beijing CoWin Biotech Co.,
Ltd.) according to the manufacturer's protocol. Relative mRNA
expression levels were determined via qPCR (initial denaturation at
95°C for 30 sec, followed by 40 cycles at 95°C for 5 sec and 60°C
for 34 sec) using SYBR® Green Master Mix (cat. no.
CW0957M; Beijing CoWin Biotech Co., Ltd.) and an ABI 7500 Real-Time
PCR System (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Tubulin was used as an internal control for mRNA expression. For
miRNA quantification, RT-qPCR was performed using a Bulge-loop™
miRNA qRT-PCR Starter Kit (cat. no. C10211-2; Guangzhou RiboBio
Co., Ltd.) according to the manufacturer's instructions; the kit
included let-7a and U6 primers (cat. nos. S160726154727 and S1
60325154310; Guangzhou RiboBio Co., Ltd.), the latter of which was
used as an internal control for miRNA expression. All other primers
were purchased from Sangon Biotech Co., Ltd., and their sequences
are listed in Table I. Relative
RNA expression levels were calculated using the 2−ΔΔCq
method (17).
 | Table IPrimers used for quantitative
PCR. |
Table I
Primers used for quantitative
PCR.
| Gene | Primer
sequence |
|---|
| H19 | F:
5′-ACTCAGGAATCGGCTCTGGAA-3′ |
| R:
5′-CTGCTGTTCCGATGGTGTCTT-3′ |
| Lin28 | F:
5′-AGCGCAGATCAAAAGGAGACA-3′ |
| R:
5′-CCTCTCGAAAGTAGGTTGGCT-3′ |
| Tubulin | F:
5′-CGTGTTCGGCCAGAGTGGTGC-3′ |
| R:
5′-GGGTGAGGGCATGACGCTGAA-3′ |
| β-catenin | F:
5′-GATACCTCCCAAGTCCTGTATGAG-3′ |
| R:
5′-GCATCAAACTGTGTAGATGGGATC-3′ |
| ZEB1 | F:
5′-CGCGTCCCTACGGTTTC-3′ |
| R:
5′-CAACCACCACCACATGTTCAG-3′ |
| Twist | F:
5′-TTCAAAGAAACAGGGCGTGG-3′ |
| R:
5′-CCGTCTGGGAATCACTGTCC-3′ |
| Snail | F:
5′-CATCCTTCTCACTGCCATGGA-3′ |
| R:
5′-AGGCAGAGGACACAGAACCAGA-3′ |
| Slug | F:
5′-AGACCCCCATGCCATTGAAG-3′ |
| R:
5′-GGCCAGCCCAGAAAAAGTTG-3′ |
| HMGA2 | F:
5′-CGAAAGGTGCTGGGCAGCTCCGG-3′ |
| R:
5′-CCATTTCCTAGGTCTGCCTCTTG-3′ |
| Beclin-1 | F:
5′-GGCTGAGAGACTGGATCAGG-3′ |
| R:
5′-CTGCGTCTGGGCATAACG-3′ |
| p62 | F:
5′-AAGTCAGCAAACCTGACG-3′ |
| R:
5′-CCATCTGTTCCTCTGGCT-3′ |
Plasmids
pH19 and its corresponding mutant plasmid (pH19mut)
were synthesized as previously described (18), with the 2.6-kb-long amplified H19
sequences inserted into pFLAG-CMV-2 vectors (Sigma-Aldrich; Merck
KGaA). The sequences of wild-type and mutant human H19 are listed
in Table II.
 | Table IISequences of wild-type and mutant
human H19. |
Table II
Sequences of wild-type and mutant
human H19.
| H19 version | Sequence
(5′-3′) |
|---|
| Wild-type |
GTAGGACGAAGCTGGGGGAGGGTCACAGGGATGCCACCCGGGATCCGTAACAGTGTTTATTGATGATGAGTCCAGGGCTCCTGCTGAAGCCCTGGTGGGGAGGGGCACAGAGCGAGATGGGGCGTAATGGAATGCTTGAAGGCTGCTCCGTGATGTCGGTCGGAGCTTCCAGACTAGGCGAGGGCAGGGTGAGGCCTCGGGCACACAGCCGGCGCCCAGTCACCCGGCCCAGATGGAGGGCGGCCGGGCCCTGCACAGGCACTTGCCAAGGTGGCTCACACTCACGCACACTCGTACTGAGACTCAAGGCCGTCTCCACAACTCCAACCAGTGCAAATGACTTAGTGCAAATTAAATTCAGAAGGGACGGGGGAAACAGAGTCGTGGAGGCTTTGAATCTCTCAGAAAAAAGGAAAGACAGGAAAGCTCAGAAACAAAGAGACAGAAGGATGAAAAAGAAGAAGAGGGAGGTGGTGGGGACGGCGTCATCCCGCTGGAGGAGCTCAGCTCTGGGATGATGTGGTGGCTGGTGGTCAACCGTCCGCCGCAGGGGGTGGCCATGAAGATGGAGTCGCCGGTGCGGGGTGGGTGCTGCGGGCGCCGCTGTTCCGATGGTGTCTTTGATGTTGGGCTGATGAGGTCTGGTTCCTCTAGCTTCACCTTCCAGAGCCGATTCCTGAGTCAGGTAGTGCAGTGGTTGTAAAGTGCAGCATATTCATTTCCAAGCTAGAGGGTTTTGTGTCCGGATTCAAAGGCCCAGGCTTGAGCTGGGTAGCACCATTTCTTTCATGTTGTGGGTTCTGGGAGCCCAGAGGGCAGCCATAGTGTGCCGACTCCGTGGAGGAAGTAAAGAAACAGACCCGCTTCTTGCCGCAGCCCCACCAGCCTAAGGTGTTCAGGAAGGCCGGACGCGCCTCCTCTGTCCTCGCCGTCACACCGGACCATGTCATGTCCTGCTTGTCACGTCCACCGGACCTGGCGTCTTGGCCTTCGGCAGCTGGTGGGCACGTCCACCCCAGCTGGAGACCTGGCCTCGTCTCCAGCCCGAACGCTGAGGCACCGATCCCGGCTCCGTCGCCGCCCGCGAGGCCCGCCTTGGCCACGGGCTCTGGAGGCCAGTGCCTCCCGCTCGCCGCCCGCTGCGCTCCTCACCCCTGCCTGCACCATCCTCCCTCCTGAGAGCTCATTCACTCCGCCCCGCCCGCCTCGCCTAGTCTGGTCTCGCCCCATGCCCTTGACTCCCCTGGATGCTGTACTGTCTGCCAAGCCAGCCCCAGGGGCTGAGCGGTGAGGGCATACAGCGTCACCAAGTCCACTGTGGGCCCTCTCCGCACCAGACCCTGGGCCGCAGTGCCTCGTGGGACCGGCGCCCGCAGGCCGAGCCCCTGCAGCCTCCTTGCTGCGCAATGTCCCGGGGCCCCCCTCCCGTGGCCGCTTCGCCCTCCTGGTGACGTCCTGCTGCAACTCCCCGAGCACTGCCTGTCTTCCCGCTCTCCAGCCCTCGAGGCTCCTGTGCCTGCTACTAAATGAATTGCGGTGGGTGAGGTGGCAGCTGGGGACCCCTCTGTCCTGTGTCCCCTGCCATGTCCCTGTCTGACCCAGGCCTGGGGTACCACCCCACGTGCTGGACCCTACGCTAGCCACCCCTGTGCTCCCTCCCTGCCCTCTTGCTCTTTCTGCCTGGAACGGGCCCATAACCCCCCAGCCTTGCGCAGTCTCGGCTCCCCCCGAGAAGATGTCACCTTTGCTAACTCTCCTGCCCCCATCCTGCCCCTCTGCTGGGAGGGTGTCTGCTTCTCCCCGCCAGCCCTCGATCCCCTAAACCTCCTTCTTTCAGAAGGCTGGGGAAGCGAGGCCTGGGGAGGGAAGGGACTCACCTGCCCGGCAGATGGGGTCCTCACCTGCCCGCTGCTGCCAGCTACACCTCCGTTGCCCAGGCCCTGGGATCAAACCCTGCCCACCAGCTCCCCTCGTCCAACCAGCTGCCACGTCCTGTAACCAAAAGTGACCGGGATGAATGCCTGGCTCCCCCTTCTTTCCAGCCCTAGCTCAGGCCCATCGTCCCCAGCTGATGTCGCCCTGTCTGCACGATGCCTGGGCGCCTACTCCACACTCCTCACTGGCCTCAGGCCCCACCAGCCCTGCCTCGAGCTAGCCCCTCCACCCGTCATCACTCCTGCCAGACTCCAGATGTCCAAGGTGCTCCTTGGCTCCCACAAGCTCTCCTCCAGCACCCCATCTTCCCCTGGTTGCCCCTCGGTTCCCCACTTCCCCAGTTTCCCCCGTTACCCCCCACCCATCCCACCCCCTCCCTCACCCTGCTGCGGCCGCAAGCTTGTCGTCATCGTCTTTGTAGTCCATGGTAGATCAATTCTGACGGTTCACTAAACGAGCTCTGCTTATATAGACCTCCCACCGTACACGCCTACCGCCCATTTGCGTCAACGGGGCGGGGTTATTACGACATTTTGGAAAGTCCCGTTGATTTTGGTGCCAAAACAAACTCCCATTGACGTCAATGGGGTGGAGACTTGGAAATCCCCGTGAGTCAAACCGCTATCCACGCCCATTGGTGTACTGCCAAAACCGCATCACCATGGTAATAGCGATGACTAATACGTAGATGTACTGCCAAGTAGGAAAGTCCCGTAAGGTCATGTACTGGGCATAATGCCAGGCGGGCCATTTACCGTCATTGACGTCAATAGGGGGCGGACTTGGCATATGATACACTTGATGTACTGCCAAGTGGGCAGTTTACCGTAAATACTCCACCCATTGACGTCAATGGAAAGTCCCTATTGGCGTTACTATGGGAACATACGTCATTATTGACGTCAATGGGCGGGGGTCGTTGGGCGGTCAGCCAGGCGGGCCATTTACCGTAAGTTATGTAACGCGGAACTCCATATATGGGCTATGAACTAATGACCCCGTAATTGATTACTATTAATAACTAGTCAATAATCAATGTCAACATGGCGGTCATATTGGACATGAGCCAATATAAATGTACATAT |
| Mutant |
GCGGACGAGGAATGGGGAGGGGTCACAGGGATGCCACCCGGGATCCGTAACAGTGTTTATTGATGATGAGTCCAGGGCTCCTGCTGAAGCCCTGGTGGGGAGGGGCACAGAGCGAGATGGGGCGTAATGGAATGCTTGAAGGCTGCTCCGTGATGTCGGTCGGAGCTTCCAGACTAGGCGAGGGCAGGGTGAAAGTGCTATAAGTGCAGGTGCGCCCAGTCACCCGGCCCAGATGGAGGGCGGCCGGGCCCTGCACAGGCACTTGCCAAGGTGGCTCACACTCACGCACACTCGTACTGAGACTCAAGGCCGTCTCCACAACTCCAACCAGTGCAAATGACTTAGTGCAAATTAAATTCAGAAGGGACGGGGGAAACAGAGTCGTGGAGGCTTTGAATCTCTCAGAAAAAAGGAAAGACAGGAAAGCTCAGAAACAAAGAGACAGAAGGATGAAAAAGAAGAAGAGGGAGGTGGTGGGGACGGCGTCATCCCGCTGGAGGAGCTCAGCTCTGGGATGATGTGGTGGCTGGTGGTCAACCGTCCGCCGCAGGGGGTGGCCATGAAGATGGAGTCGCCGGTGCGGGGTGGGTGCTGCGGGCGCCGCTGTTCCGATGGTGTCTTTGATACGACGCTGATGAGGTCTGGTTCCTCTAGCTTCACCTTCCAGAGCCGATTCCTGAGTCAAAGTGCTATAAGTGCAGGTAGTGCAGCATATTCATTTCCAAGCTAGAGGGTTTTGTGTCCGGATTCAAAGGCCCAGGCTTGAGCTGGGTAGCACCATTTCTTTCATGTTGTGGGTTCTGGGAGCCCAGAGGGCAGCCATAGTGTGCCGACTCCGTGGAGGAAGTAAAGAAACAGACCCGCTTCTTGCCGCAGCCCCACCAGCCTAAGGTGTTCAGGAAGGCCGGACGCGCCTCCTCTGTCCTCGCCGTCACACCGGACCATGTCATGTCCTGCTTGTCACGTCCACCGGACCTGGCGTCTTGGCCTTCGGCAGCTGGTGGGCACGTCCACCCCAGCTGGAGACCTGGCCTCGTCTCCAGCCCGAACGCTAAAGTGCTATAAGTGCAGGTGTCGCCGCCCGCGAGGCCCGCCTTGGCCACGGGCTCTGGAGGCCAGTGCCTCCCGCTCGCCGCCCGCTGCGCTCCTCACCCCTGCCTGCACCATCCTCCCTCCTGAGAGCTCATTCACTCCGCCCCGCCCGCCTCGCCTAGTCTGGTCTCGCCCCATGCCCTTGACTCCCCTGGATGCTGTACTGTCTGCCAAGCCAGCCCCAGGGGCTGAGCGGTGAGGGCATACAGCGTCACCAAGTCCACTGTGGGCCCTCTCCGCACCAGACCCTGGGCCGCAGTGCCTCGTGGGACCGGCGCCCGCAGGCCGAGCCCCTGCAGCCTCCTTGCTGCGCAATGTCCCGGGGCCCCCCTCCCGTGGCCGCTTCGCCCTCCTGGTGACGTCCTGCTGCAACTCCCCGAGCACTGCCTGTCTTCCCGCTCTCCAGCCCTCGAGGCTCCTGTGCCTGCTACTAAATGAATTGCGGTGGGTGAAAGTGCTATAAGTGCAGGTTCTGTCCTGTGTCCCCTGCCATGTCCCTGTCTGACCCAGGCCTGGGGTACCACCCCACGTGCTGGACCCTACGCTAGCCACCCCTGTGCTCCCTCCCTGCCCTCTTACTCTTTCTGCCTGGAACGGGCCCATAACCCCCCAGCCTTGCGCAGTCTCGGCTCCCCCCGAGAAGATGTCACCTTTGCTAACTCTCCTGCCCCCATCCTGCCCCTCTGCTGGGAGGGTGTCTGCTTCTCCCCGCCAGCCCTCGATCCCCTAAACCTCCTTCTTTCAGAAGGCTGGGGAAGCGAGGCCTGGGGAGGGAAGGGACTCACCTGCCCGGCAGATGGGGTCCTCACCTGCCCGCTGCTGCCAGCTACACCTCCGTTGCCCAGGCCCTGGGATCAAACCCTGCCCACCAGCTCCCCTCGTCCAACCAGCTGCCACGTCCTGTAACCAAAAGTGACCGGGATGAATGCCTGGCTCCCCCTTCTTTCCAGCCCTAGCTCAGGCCCATCGTCCCCAGCTGATGTCGCCCTGTCTGCACGATGCCTGGGCGCCTACTCCACACTCCTCACTGGCCTCAGGCCCCACCAGCCCTGCCTCGAGCTAGCCCCTCCACCCGTCATCACTCCTGCCAGACTCCAGATGTCCAAGGTGCTCCTTGGCTCCCACAAGCTCTCCTCCAGCACCCCATCTTCCCCTGGTTGCCCCTCGGTTCCCCACTTCCCCAGTTTCCCCCGTTACCCCCCACCCATCCCACCCCCTCCCTCACCCTGCTGCGGCCGCAAGCTTGTCGTCATCGTCTTTGTAGTCCATGGTAGATCAATTCTGACGGTTCACTAAACGAGCTCTGCTTATATAGACCTCCCACCGTACACGCCTACCGCCCATTTGCGTCAACGGGGCGGGGTTATTACGACATTTTGGAAAGTCCCGTTGATTTTGGTGCCAAAACAAACTCCCATTGACGTCAATGGGGTGGAGACTTGGAAATCCCCGTGAGTCAAACCGCTATCCACGCCCATTGGTGTACTGCCAAAACCGCATCACCATGGTAATAGCGATGACTAATACGTAGATGTACTGCCAAGTAGGAAAGTCCCGTAAGGTCATGTACTGGGCATAATGCCAGGCGGGCCATTTACCGTCATTGACGTCAATAGGGGGCGGACTTGGCATATGATACACTTGATGTACTGCCAAGTGGGCAGTTTACCGTAAATACTCCACCCATTGACGTCAATGGAAAGTCCCTATTGGCGTTACTATGGGAACATACGTCATTATTGACGTCAATGGGCGGGGGTCGTTGGGCGGTCAGCCAGGCGGGCCATTTACCGTAAGTTATGTAACGCGGAACTCCATATATGGGCTATGAACTAATGACCCCGTAATTGATTACTATTAATAACTAGTCAATAATCAATGTCAACATGGCGGTCATATTGGACATGAGCCAATATAAATGTACATAT |
Small interfering (si)RNAs, miRNA mimics
and inhibitors, and transfection
H19-siRNA (cat. no. 4390771), let-7 mimics (cat.
nos. 4464066) and the control (cat. nos. AM17110), let-7 inhibitors
(cat. nos. AM17000) and the control (cat. nos. 4464076) were
synthesized by Ambion (Thermo Fisher Scientific, Inc.). Control
siRNA and siLin28 sequences were synthesized by Guangzhou RiboBio
Co., Ltd. (cat. nos. siN05815122147 and siG10118110813). A total of
3-5×105 cells/well were seeded into 24-well plates and
incubated at 37°C (5% CO2) overnight. The cells were
transfected with siRNA (10 µM), miRNA mimics or inhibitors
(5 nM) using siPORT™ NeoFX™ transfection agent (cat. no. AM4511;
Invitrogen; Thermo Fisher Scientific, Inc.); the corresponding
negative controls (siRNA, miRNA mimics or inhibitors) were supplied
by the manufacturers. For plasmid transfection, an equivalent
number of cells were seeded into 24-well plates and incubated
overnight; the cells were transfected with 1-2 µg of each
plasmid using Lipofectamine 3000 transfection reagent, and empty
vector was used as the negative control. The mRNA expression levels
of H19, Lin28 and let-7 were determined by RT-qPCR 48 h
post-transfection, and protein expression was determined by western
blot analysis 72 h post-transfection.
Wound healing, migration and invasion
assays
For the wound-healing assays, 5×105 cells
were seeded into 6-well plates in triplicate. After 24 h (37°C, 5%
CO2), a linear scratch was created across each monolayer
using a sterile pipette tip. The cells were then washed twice with
PBS and incubated in serum-free medium for 24 h, and the wound
widths were measured at 0, 24 and 48 h.
Cell migration was assessed using
Millicell® Hanging Cell Culture Inserts in 24-well
plates (EMD Millipore) according to the manufacturer's
instructions. Briefly, serum-free medium containing
2×105 cells from each subgroup was added to the upper
chamber, and 600 µl medium containing 10% FBS was added to
the lower chamber as a chemoattractant. The cells were incubated
for 24 h at 37°C (5% CO2).
For the invasion assay, the same procedure was used
as for the cell migration assay, but the filters were pre-coated
with 100 µl Matrigel (BD Biosciences) at a 1:3 dilution in
medium; the cells were then incubated for 48 h at 37°C (5%
CO2). Following incubation, the cells on the upper
surface of the membrane were removed using a cotton swab; cells
that had migrated to or invaded the lower membrane were fixed in
methanol for 15 min, and subsequently stained with 0.05% crystal
violet in PBS for 15 min (both at room temperature). Migration and
invasion were assessed by counting the number of stained cells from
10 random fields per filter in each group under a light microscope
(magnification, ×20; Olympus Corporation). The results are depicted
as the mean ± SD, and each experiment was conducted in triplicate
and repeated ≥3 times.
Western blot analysis
Cultured cells were lysed using RIPA buffer (Pierce;
Thermo Fisher Scientific, Inc.) containing a protease inhibitor
cocktail (Roche Diagnostics). The protein concentration of the
lysates was measured using a Protein Assay Kit II (cat. no.
500-0002EDU; Bio-Rad Laboratories, Inc.). Equivalent amounts of
protein (10 µl) were resolved and mixed with 5X Lane Marker
Reducing Sample Buffer (Pierce; Thermo Fisher Scientific, Inc.)
prior to SDS-PAGE (12%). The proteins were transferred to PVDF
membranes (EMD Millipore) and blocked with 5% non-fat milk for 1 h
at 20°C. The membranes were then probed with the aforementioned
primary antibodies (all 1:1,000), washed with TBS-0.1% Tween-20 and
subsequently incubated with corresponding horseradish
peroxidase-conjugated secondary antibodies (1:2,000; cat. nos.
GAM0072 and GAR0072; MultiSciences Biotech Ltd.). The protein bands
were developed using an enhanced chemiluminescent detection system
(Bio-Rad Laboratories, Inc.). All experiments were conducted in
triplicate.
Cell viability assay
Cells were harvested and seeded into 96-well plates
(4×103/well) in triplicate. After culturing for 2 days,
20 µl MTT (Amresco, LLC) was added to each well and the
cells were incubated for a further 4 h at 37°C. The optical density
was determined at least in triplicate against a reagent blank
(wavelength, 490 nm) using a spectrophotometric microplate
reader.
Autophagy flux monitoring
To evaluate the formation of fluorescent LC3B
puncta, p-mCherry-C1-EGFP-hLC3B (LC3B) was used to monitor
autophagy flux; 48 h after LC3B co-transfection with siRNAs, the
cells were washed with 1X PBS and immediately analyzed via confocal
microscopy (magnification, ×100). DAPI Staining Solution (5 mg/ml;
cat. no. C1006; Beyotime Institute of Biotechnology) was used for
immunofluorescence staining at room temperature for 5 min. Each
sample was observed under a Zeiss LSM 710 confocal microscope and
the images were processed using ZEN LE software 2.0 (both Carl
Zeiss AG).
Bioinformatics analysis
The association and prognostic modules of
bc-GenExMiner v4.2 (bcgenex.centregauducheau.fr) were used to
evaluate the associations between the expression of H19 and Lin28,
as well as their prognostic value in human BC.
Statistical analysis
Unless otherwise indicated, all data are presented
as the mean ± SD. Statistical analyses were performed using SPSS
software version 17.0 (SPSS, Inc.). Linear regression analysis and
Pearson's correlation coefficient were used to investigate the
relationship between the expression levels of H19 and Lin28, and
Kaplan-Meier analyses of H19 and Lin28 and log-rank tests were used
to evaluate prognosis. The Mann-Whitney U test was used to compare
the differences between two groups, and data were analyzed using
Kruskal-Wallis with Dunn's multiple comparison test among three or
more groups. All graphs were constructed using Prism 7 version 7.0a
(GraphPad Software, Inc.).
Results
H19/Let-7/Lin28 ceRNA network in BC
The results of the present study showed that the
mRNA expression levels of H19 and Lin28 were higher in BC tissues
with LN metastasis than those without (Fig. 1A and B). Furthermore, linear
regression analysis verified a significant positive relationship
between H19 and Lin28 expression in BC tissue samples collected
from 43 patients (Fig. 1C).
Kaplan-Meier curves were generated using the bc-GenExMiner
database, which revealed that increased levels of H19 and Lin28
expression were both significantly associated with poor survival
time in BC (Fig. 1D and E); this
was validated by a strong positive association between the data
obtained from bc-GenExMiner (Fig.
1F). The expression levels of H19 and Lin28 were also
significantly increased in BC cell lines (MDA-231, MCF-7, SK-BR-3
and T-47D) compared with normal breast cells (MCF-10A; Fig. 1G and H).
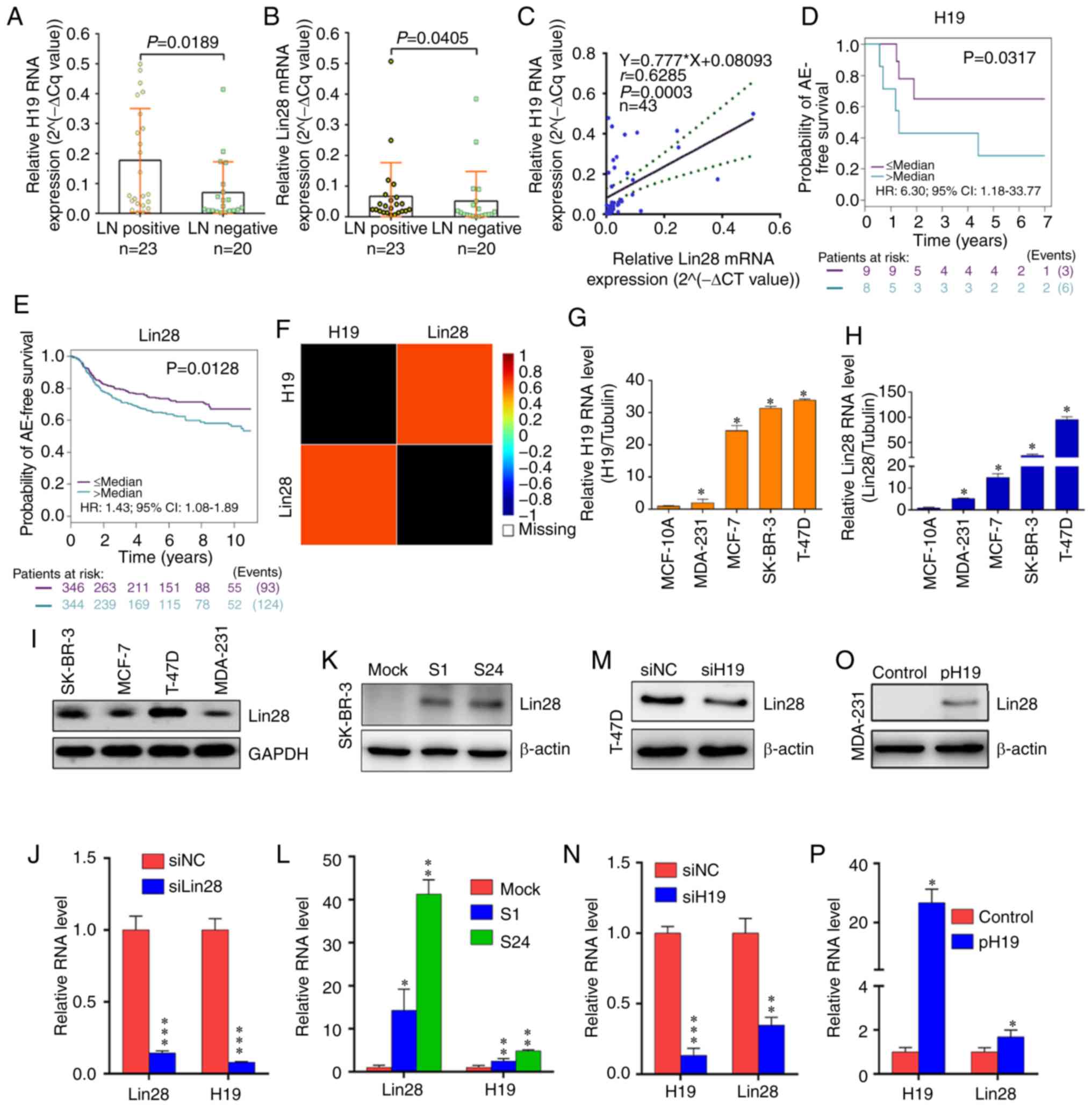 | Figure 1Upregulation of H19 and Lin28
indicates poor survival in BC. BC samples were divided into two
groups based on clinical progression. (A) H19 and (B) Lin28 levels
in the LN metastasis group (n=23) were significantly higher than
those in the non-metastasis group (n=20). (C) Linear regression
analysis indicated an in vivo positive relationship between
the expression levels of H19 and Lin28 in a statistically
significant manner. Pearson's correlation coefficient, P-value and
sample size are indicated in the top left of the plot. Total RNA
was subjected to RT-qPCR analysis; the Cq values were normalized to
Tubulin in each sample. Kaplan-Meier survival curves of (D) H19
(P=0.0317) and (E) Lin28 (P=0.0128) were plotted using log-rank
tests for patients with BC in the bc-GenExMiner v4.2 database. (F)
Correlation map corresponding to the association between H19 and
Lin28 expression was plotted for patients with BC in the
bc-GenExMiner v4.2 database. The numbers represent Pearson's
correlation coefficient. (G) H19 and (H) Lin28 levels were
evaluated via RT-qPCR in four BC cell lines, and normal human
MCF-10A breast cells were used as a control. *P<0.05
vs. MCF-10A. (I) Western blot analysis of Lin28 expression in four
breast cancer cell lines. (J) RT-qPCR analysis of the RNA
expression levels of Lin28 and H19 in T-47D cells transfected with
siLin28. (K) Western blot analysis of Lin28 expression in SK-BR-3
cells S1 and S24. (L) RT-qPCR analysis of the RNA expression levels
of Lin28 and H19 in SK-BR-3 cells S1 and S24; mock was used as a
control. (M) Western blot analysis of Lin28 expression in T-47D
cells transfected with siH19. (N) RT-qPCR analysis of the RNA
expression levels of H19 and Lin28 in T-47D cells transfected with
siH19. (O) Western blot analysis of Lin28 expression in MDA-231
cells transfected with pH19. (P) RT-qPCR analysis of the RNA
expression levels of H19 and Lin28 in MDA-231 cells transfected
with pH19. Results represent the mean ± SD of three independent
experiments. *P<0.05, **P<0.01,
***P<0.001 vs. Control or Mock. BC, breast cancer;
LN, lymph node; RT-qPCR, reverse transcription-quantitative PCR;
Cq, quantification cycle; HR, hazard ratio; CI, confidence
interval; si, small interfering RNA; NC, negative control; S1/S24,
SK-BR-3 clones infected with a Lin28 overexpression lentivirus;
pH19, pFlag-CMV-H19. |
The T-47D cell line was selected to investigate the
association between H19 and Lin28 in BC, as these cells expressed
the highest levels of Lin28 among the cell lines investigated
(Fig. 1I). In T-47D cells
transfected with siLin28, the H19 expression level was
significantly downregulated (Fig.
1J). For the two established SK-BR-3 clones (S1 and S24;
Fig. 1K) (19), RT-qPCR showed that H19 expression
was significantly increased when Lin28 was overexpressed (Fig. 1L). To test the influence of H19 on
Lin28, T-47D cells were transfected with siH19, and a decrease in
the level of Lin28 mRNA and protein expression was observed
(Fig. 1M and N). In addition,
Lin28 expression was examined in MDA-231 cells transfected with
pFlag-CMV-H19, which revealed that the H19 plasmid resulted in the
successful overexpression of Lin28 at both the mRNA and protein
level (Fig. 1O and P).
It is known that a double-negative feedback loop
exists between let-7 and its RNA-binding protein Lin28 (8), and the results of the current study
are consistent with these findings. As presented in Fig. S1A and B, the overexpression of
let-7 in T-47D cells significantly inhibited the expression of
Lin28 at both the mRNA and protein level; furthermore, MDA-231
cells transfected with a let-7 inhibitor exhibited an upregulation
in Lin28 expression (Fig. S1C and
D). Moreover, compared with the mock control, let-7 expression
was significantly downregulated at the mRNA level in S1 and S24
(Fig. S1E). In T-47D cells
transfected with siLin28, let-7 mRNA expression was upregu-lated as
a result of the decreased expression of Lin28 (Fig. S1F).
It has been suggested that H19 may act as a sponge
to antagonize miRNA let-7 (20).
Collectively, lncRNA H19, miRNA let-7 and the transcription factor
Lin28 may potentially form a double-negative ceRNA network in
BC.
H19/let-7/Lin28 loop is required for
autophagy in BC cells
Western blotting was used to determine the role of
H19 in the autophagic regulation of BC, which showed that the
overexpression of H19 decreased the expression of
autophagy-associated molecules (beclin-1 and LC3-II) in MDA-231
cells; conversely, H19 knockdown in T-47D cells led to the
upregulation of beclin-1 and LC3-II, and the downregulation of p62
(Fig. 2A). Autophagy flux
monitoring was then performed to verify the influence of H19 on
autophagy. As shown in Fig. 2B,
autophagy flux was decreased in MDA-231 cells overexpressing H19
and increased in siH19-transfected T-47D cells. A similar trend was
observed for the influence of Lin28 on p62, beclin-1 and LC3-II
(Fig. 2C). RT-qPCR and western
blot analyses revealed that H19 and Lin28 expression was
significantly upregulated by a let-7 inhibitor (Fig. 2D and E); these results further
indicated that the H19/let-7/Lin28 loop may affect autophagy in
T-47D cells (Fig. 2F and G).
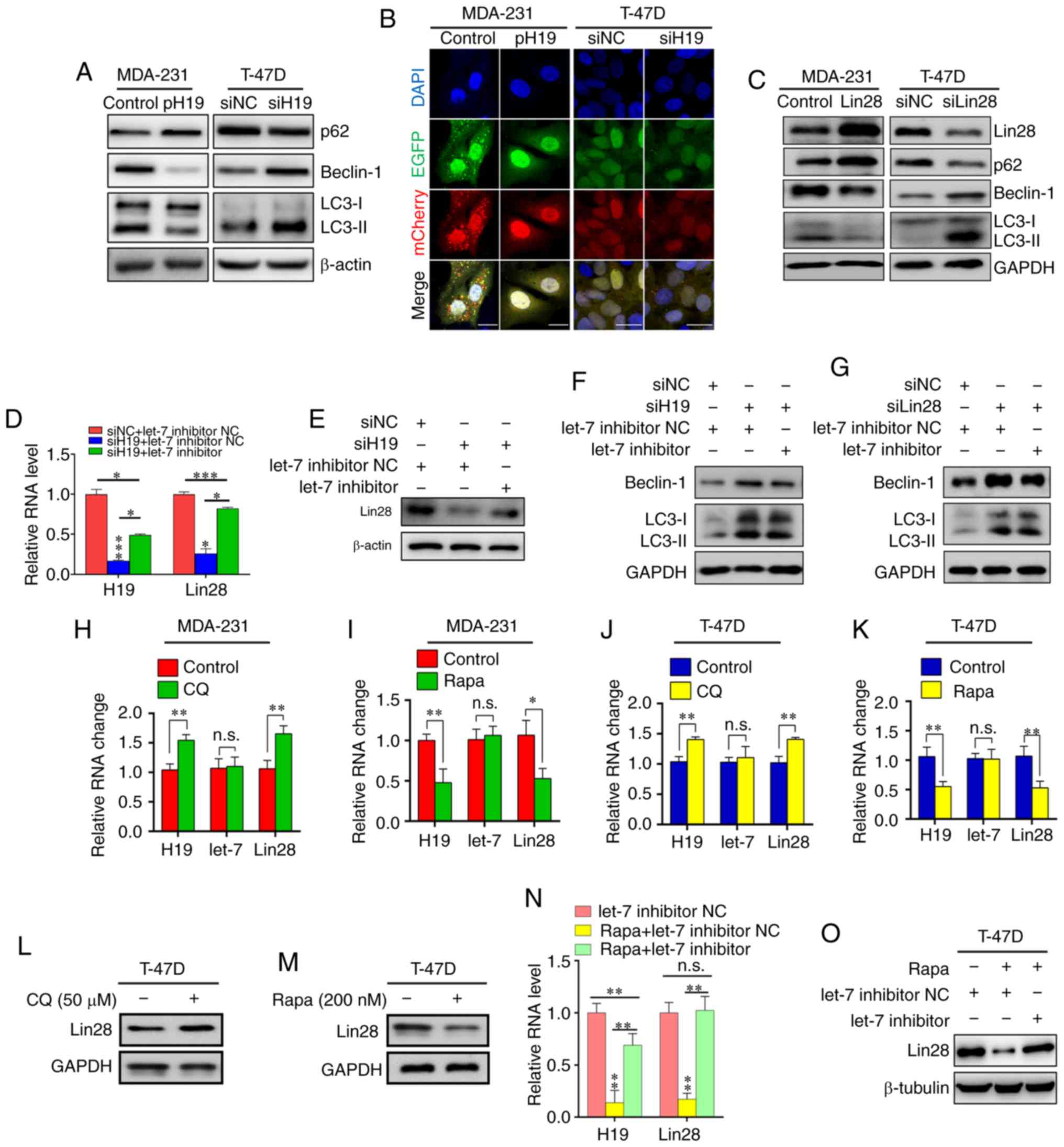 | Figure 2Reciprocal inhibition of the
H19/let-7/Lin28 loop and autophagy in breast cancer cells. (A)
Western blot analysis of autophagy marker expression in MDA-231
cells transfected with pH19 and T-47D cells transfected with siH19.
(B) Measurement of autophagy flux in MDA-231 cells co-transfected
with p-mCherry-C1-EGFP-hLC3B and pH19, and T-47D cells
co-transfected with p-mCherry-C1-EGFP-hLC3B and siH19. Scale bar,
20 µm. (C) Western blot analysis of autophagy marker
expression in MDA-231 cells transfected with Lin28 and T-47D cells
transfected with siLin28. (D) RT-qPCR analysis of the RNA
expression levels of H19 and Lin28 in T-47D cells transfected with
siH19 and let-7 inhibitor. *P<0.05,
***P<0.001 vs. siNC + let-7 inhibitor NC unless
otherwise indicated. (E) Western blot analysis of Lin28 expression
in T-47D cells transfected with siH19 and let-7 inhibitor. (F)
Western blot analysis of autophagy markers expression in T-47D
cells transfected with siH19 and let-7 inhibitor. (G) Western blot
analysis of autophagy markers expression in T-47D cells transfected
with siLin28 and let-7 inhibitor. RT-qPCR analysis of the RNA
expression levels of H19, let-7 and Lin28 in MDA-231 cells
incubated with (H) 50 µM CQ or (I) 200 nM Rapa. RT-qPCR
analysis of the RNA expression levels of H19, let-7 and Lin28 in
T-47D cells incubated with (J) 50 µM CQ or (K) 200 nM Rapa.
Western blot analysis of Lin28 in T-47D cells incubated with (L) 50
µM CQ or (M) 200 nM Rapa. (N) RT-qPCR analysis of the RNA
expression levels of H19 and Lin28 in T-47D cells incubated with
200 nM Rapa and let-7 inhibitor. (O) Western blot analysis of Lin28
in T-47D cells incubated with 200 nM Rapa and let-7 inhibitor.
Results represent the mean ± SD of three independent experiments.
*P<0.05, **P<0.01. si, small
interfering RNA; pH19, pFlag-CMV-H19; RT-qPCR, reverse
transcription-quantitative PCR; LC3, light chain 3; CQ,
chloroquine; Rapa, rapamycin; NC, negative control; n.s., not
significant. |
To further investigate the impact of autophagy on
the H19/let-7/Lin28 loop in BC, MDA-231 and T-47D cells were
treated for 24 h with the autophagic inhibitor CQ and the
autophagic stimulator Rapa. The results showed that CQ upregulated
the expression of both H19 and Lin28 mRNA, but that Rapa had a
negative impact on H19 and Lin28 (Fig.
2H-K), which was also demonstrated by the protein expression
level of Lin28 in T-47D cells (Fig. 2L
and M). Of note, CQ/Rapa could only induce significant changes
in Lin28 mRNA expression in MDA-231 cells, not protein expression
(data not shown). Finally, a rescue experiment was conducted by
co-treating T-47D cells with both Rapa and let-7 inhibitor, the
results of which indicated that the H19/let-7/Lin28 loop may be
involved in Rapa-induced autophagy in BC cells (Fig. 2N and O).
H19/let-7/Lin28 loop promotes EMT in BC
cells
A positive relationship was found between H19 and
Lin28 expression, and LN metastasis (Fig. 1A and B); subsequently, the effects
of H19 and Lin28 overexpression and knockdown on the migration and
invasion of BC cells were explored. Transwell assays showed that in
MDA-231 cells, H19 overexpression significantly promoted migration
and invasion (Fig. 3A-D). H19
knockdown in T-47D cells significantly inhibited migration and
invasion (Fig. 3E-H). Furthermore,
the wound closure time was notably decreased by H19 overexpression
(in MDA-231 cells) and increased by H19 knockdown (in T-47D cells),
compared with the control cells (Fig.
S2A).
The migration and invasion of SK-BR-3 cancer cell
clones was also investigated; the results demonstrated that
upregulation of Lin28 significantly promoted the migration and
invasion of S24 cells compared with the mock control (Fig. 3I-L). Following Lin28 knockdown in
T-47D cells, western blotting demonstrated that Lin28 expression
was significantly reduced compared with negative control (Fig. 3M). As predicted, downregulated
expression of Lin28 significantly attenuated the migration and
invasion of T-47D cells (Fig.
3N-P). These results were supported by wound healing assays
(Fig. S2B), and a rescue
experiment further indicated that H19/let-7/Lin28 enhanced the
migratory and invasive abilities of BC cells (Fig. S3A and B).
Moreover, morphological changes associated with EMT
were observed in SK-BR-3 and T-47D cells; specifically, H19
overexpression and the upregulation of Lin28 by let-7 inhibitor
contributed to EMT (based on the transition from a round cell
morphology to a spindle-shaped morphology with antennae) after 24 h
treatment (Fig. S4). To exclude
the possibility that the aforementioned results were the
consequence of Lin28 and H19 expression on the viability of BC
cells, MTT assays were performed in cells following H19 or Lin28
upregulation/knockdown, which revealed that the metastatic
alterations were not associated with changes in the viability of
the BC cell lines (Fig. S5).
To further confirm that H19 influenced Lin28 by
interacting with let-7 during migration and invasion, pH19mut was
constructed with the let-7 binding site removed (Fig. S6A). Transfection of MDA-231 cells
with pH19mut significantly decreased the mRNA expression level of
Lin28 (Fig. S6B). Western blot
analysis also showed this change in Lin28 expression compared with
the control plasmid group (Fig.
S6C). Finally, Transwell assays showed that pH19mut suppressed
MDA-231 cell migration and invasion (Fig. S6D-F).
Collectively, all above-mentioned results suggested
that H19/let-7/Lin28 promotes migration and invasion via EMT in BC
cells.
H19/let-7/Lin28 loop regulates
EMT-associated genes
EMT is associated with reduced expression of
E-cadherin (21); the present
study indicated that H19 knockdown led to an increase in the
protein expression level of E-cadherin, whereas let-7 inhibitor
restored the effect of H19 on its downstream target (Fig. S3C). Several EMT-inducing
transcription factors, including β-catenin (22), zinc finger E-box-binding homeobox 1
(23), Twist (24), Snail (25), Slug (26) and high mobility group A2 (27), were reported to act in a
gene-specific manner to orchestrate the transcriptional regulation
necessary for EMT in various types of tumor, including BC (28); these factors were also found to be
associated with H19/let-7/Lin28 in the present study. The results
of RT-qPCR verified that H19 knockdown decreased the expression of
EMT-inducing transcription factors, and that let-7 inhibitor
reversed this effect (Fig. S3D).
These observations indicated the involvement of the H19/let-7/Lin28
network in the EMT of BC cells.
H19/let-7/Lin28 loop is involved in the
autophagic inhibition of EMT in BC cells
The present study aimed to investigate the role of
H19 in the interplay between metastasis and autophagy in BC. First,
to determine whether autophagy may inhibit the migration and
invasion of BC cells, T-47D cells were treated with Rapa or CQ for
24 h, and the effect on autophagy was evaluated via western blot
analysis of p62, beclin-1 and LC3-II (Fig. S7). The results showed that
migration and invasion were inhibited by treatment for 24 h with
200 nM Rapa, which was significantly reversed by combined treatment
with 50 µM CQ (Fig. 4A and
B). EMT-associated proteins and EMT-inducing transcription
factors were also investigated; western blotting showed that
vimentin was downregulated and E-cadherin was upregulated following
incubation with Rapa for 24 h, which was again notably reversed by
co-treatment with CQ (Fig. 4C).
These results were further verified at the mRNA level by RT-qPCR
(Fig. 4D). Finally, a rescue
experiment suggested that the overexpression of H19 reversed the
reduced migration and invasion that resulted from autophagic
inhibition (Fig. 4E-H).
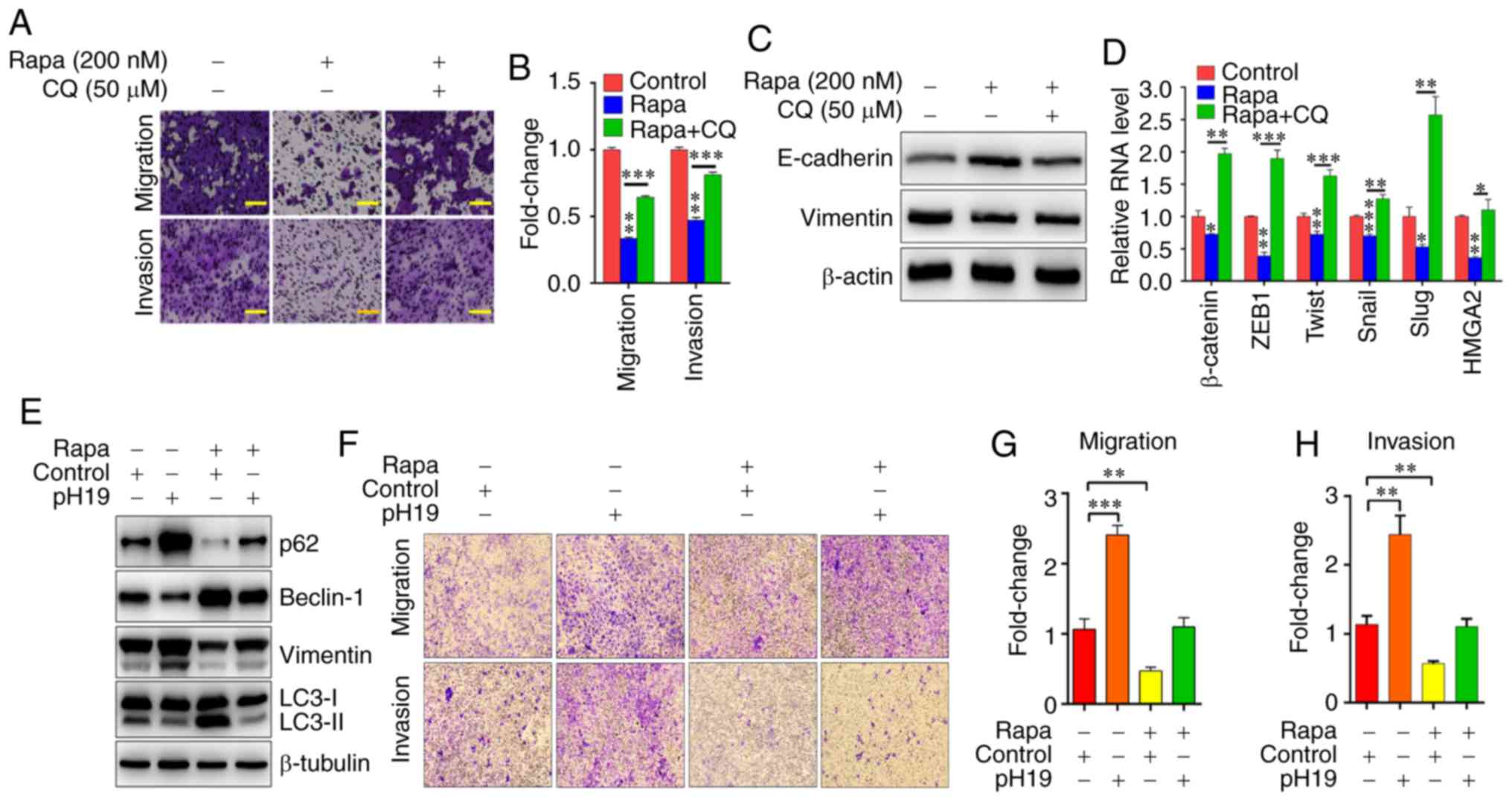 | Figure 4H19/let-7/Lin28 loop regulates the
inhibitory effects of autophagy on the EMT of breast cancer cells.
(A) T-47D cells were incubated with 200 nM Rapa and 50 µM
CQ, and cell migration and invasion were evaluated by Transwell
assays. Scale bar=50 µm. (B) Quantification of migration and
invasion of T-47D cells incubated with 200 nM Rapa and 50 µM
CQ. (C) Western blot analysis of EMT markers. (D) Reverse
transcription-quantitative PCR analysis of EMT-inducing
transcription factors in T-47D cells incubated with 200 nM Rapa and
50 µM CQ. (E) Western blot analysis of autophagy and EMT
markers in T-47D cells treated with 200 nM Rapa and pH19. (F) T-47D
cells were treated with 200 nM Rapa and pH19, and cell migration
and invasion were evaluated by Transwell assays. Scale bar=50
µm. Quantification of (G) migration and (H) invasion rates
of T-47D cells incubated with 200 nM Rapa and pH19. Results
represent the mean ± SD of three independent experiments.
*P<0.05, **P<0.01,
***P<0.001 vs. Con unless otherwise indicated. pH19,
pFlag-CMV-H19; Con, control; CQ, chloroquine; Rapa, rapamycin; EMT,
epithelial-mesenchymal transition; ZEB1, zinc finger E-box binding
homeobox 1; HMGA2, high-mobility group AT-hook 2; LC3, light chain
3. |
Therefore, the results of the present study suggest
that, similar to TGF-β (29),
which induces a notable reduction in autophagy via downregulation
of beclin-1 and LC3-II and upregulation of p62 (Fig. S8A and B) while enhancing EMT
(Fig. S4), H19/let-7/Lin28 may be
involved in the interplay between EMT and autophagy.
Discussion
It is well known that lncRNAs are involved in the
regulation of complex biological processes via various regulatory
mechanisms, which include their interactions with DNA and
chromatin, signaling and regulatory proteins, as well as a variety
of cellular RNA species. Accumulating evidence has demonstrated the
relevance of specific lncRNAs in the initiation and progression of
BC (30). A previous study
suggested that lncRNAs may act as ceRNAs to communicate with other
RNA transcripts via their miRNA response elements (31). For example, lncRNA H19 was found to
bind and antagonize the function of miR-17-5p, leading to the
expression of tyrosine protein kinase YES1 at both the mRNA and
protein level (32).
The lncRNA H19 gene occupies an imprinted region
within chromosome 11 (11p15.5), and has been reported to modulate
the functions of both miRNAs and proteins (33). Although the mechanism and function
of H19 have been studied in various types of cancer, its role in
tumor initiation and progression remains the subject of
controversy. For example, H19 overexpression increased the
metastatic capacity of lung carcinoma (34); however, H19 was also shown to
suppress EMT by upregulating the expression of miR-200 family
members (35). As an oncogenic
regulator in various types of cancer, Lin28 exerts similar
biological functions (36,37). In the present study, the
overexpression of H19 and Lin28 was shown to promote the migration
and invasive ability of BC cells, while H19 and Lin28 knockdown
inhibited these functions. Consistent with these findings, the
expression of H19 was found to be positively associated with that
of Lin28 in BC cell lines. It is therefore proposed that H19 and
Lin28 may play oncogenic roles in the progression of BC
metastasis.
H19 has been reported to regulate autophagy in
numerous types of cancer (38-40).
Furthermore, H19 has been revealed to play important roles in
promoting tumorigenesis through the stimulation of mTOR signaling
(41); this may explain why in the
present study, H19 inhibited the Rapa-induced activation of
autophagy. This supports previous observations that H19
overexpression promoted mTOR phosphorylation and inhibited
autophagic activation in cardiomyocytes (42). As well as mTOR, lncRNAs could also
affect the modulation of autophagy-related genes during the
autophagic process, in a direct or indirect manner (43). These findings may have importance
for the discovery of new mechanisms involved in the interaction of
H19 with autophagy.
As for Lin28 and autophagy, as both Lin28 mRNA and
protein expression were suppressed by autophagic activation, the
same trend was proposed as for H19. There are numerous possible
explanations for this. First, the expression of Lin28 was
demonstrated to be positively associated with that of H19; thus, it
is possible that autophagy could affect the expression of H19 prior
to that of Lin28. Additionally, on account of the negative
association between let-7 and autophagy, it is speculated that
autophagy-related proteins may interact with Lin28 in a direct or
indirect manner outside of the H19/let-7/Lin28 ceRNA network. At
the same time, in the present study, it was noteworthy that
autophagic inhibition or activation could induce significant
changes in Lin28 mRNA expression but not protein expression in
certain cells; there may be further unknowns to be explored
concerning the Lin28 translation process.
There are various possible reasons as to why
inducing autophagy could suppress the expression of H19 and Lin28.
It has been proposed that tumor-suppressive mechanisms of autophagy
contribute to the maintenance of normal bioenergetic functions,
oncogene-induced cell death, degradation of oncogenic proteins,
immune responses that prevent the establishment and proliferation
of malignant cells, anti-inflammatory effects and the maintenance
of normal stem cells (44). As, in
the present study, autophagic activation or inhibition was revealed
to have no significant effect on the expression of let-7, it was
proposed that H19 and Lin28 may be involved in other ceRNA
networks, or influence other cellular mechanisms.
As the results of the present study suggest that
autophagy could inhibit EMT via the H19/let-7/Lin28 network, and as
various studies have previously reported other factors associated
with both EMT and autophagy (45-47),
it is of note that a novel and non-linear relationship links EMT
and autophagy in BC. The interplay between these two biological
processes can be influenced by an intricate web of regulatory
signaling pathways, which may include the H19/let-7/Lin28 ceRNA
network. Other factors may also disrupt the equilibrium between
these two processes, such as the overexpression of TGF-β.
EMT is an important cellular mechanism in embryonic
development and tissue repair, while also contributing to the
progression of various diseases, including cancer (48,49).
In addition to genetic alterations, epigenetic regulation also
plays a significant role in multiple EMT-related processes, such as
the TGF-β, E-cadherin, WNT/β-catenin, Notch, hypoxia and tumor
necrosis factor-α pathways (28).
With regarding to present research hotspots, studies into EMT tend
to focus on lncRNAs and miRNAs. For example, lncRNA metastasis
associated lung adenocarcinoma transcript 1, HOX transcript
antisense RNA and TRE could represent important examples of
lncRNA-mediated regulators of EMT in BC (50). Additionally, let-7 inhibited the
Wnt1/Frizzled/β-catenin pathway in hepa-tocellular carcinoma stem
cells (51). Findings such as
these and the results of the present study further advance present
understanding of the complexity and importance of ceRNA networks
during the progression of tumor metastasis.
The complex relationship between autophagy and EMT,
and their biological significance in the occurrence and development
of cancer, are established. The regulatory crosstalk between
autophagy and EMT has been associated with the
Ras/Raf1/mitogen-activated protein kinase kinase1/2/ERK (52), JAK/STAT (53), integrin (54) and NF-κB signaling pathways
(55).
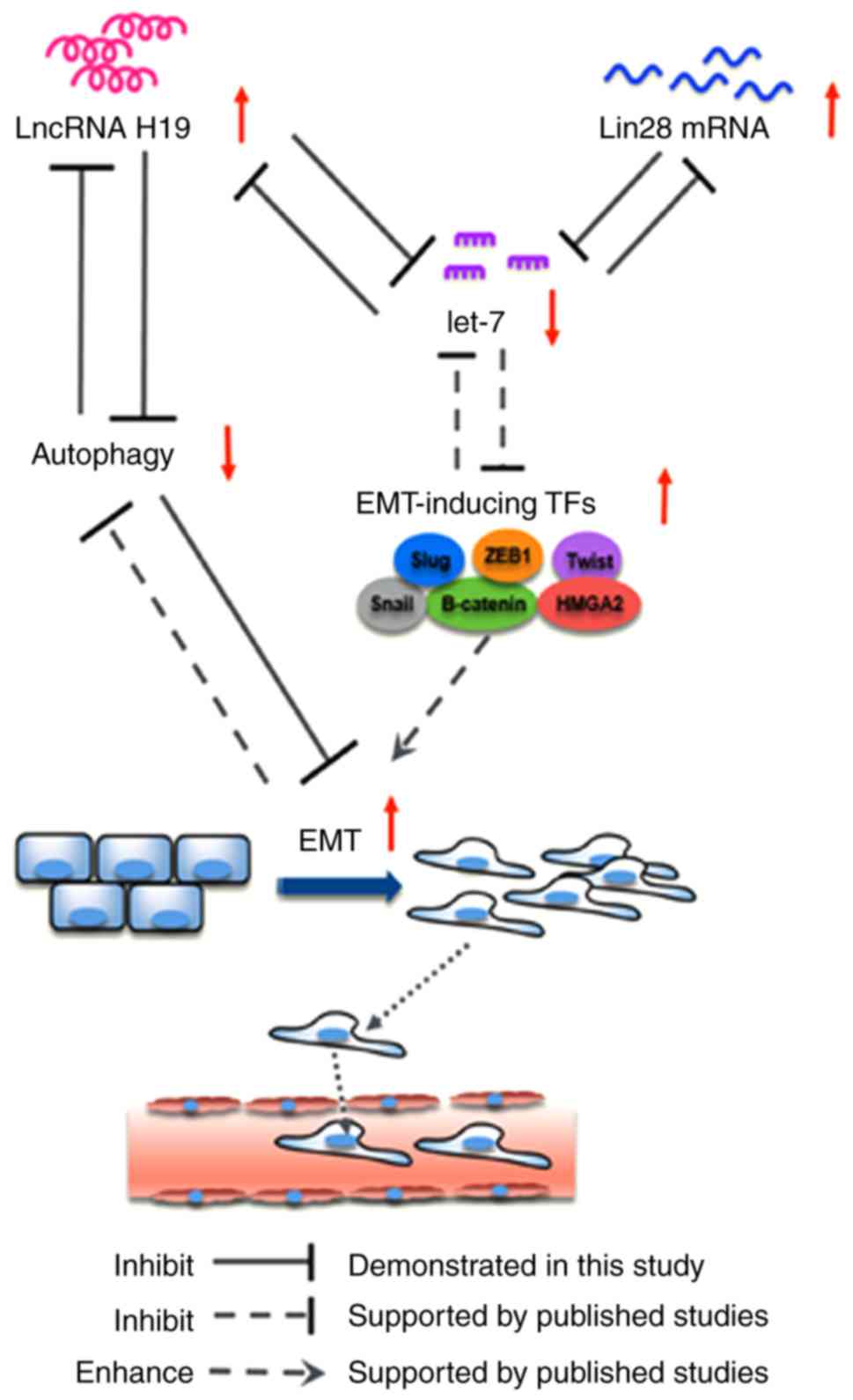
It is of note that the present study demonstrated
interactions among the H19/let-7/Lin28 ceRNA network, EMT and
autophagy, and that this feedback loop is critical for BC migration
and invasion (Fig. 5). Aberrant
molecular events that influence a single element of the loop may
trigger a cascade that enhances the metastatic ability of BC cells
in the absence of the inducing signal. The precise molecular events
involved in the autophagic regulation of H19/let-7/Lin28 require
further investigation. However, the present study is an important
indicator of the potential interplay of ceRNAs, EMT and autophagy
in the diagnosis, prognosis and therapeutic intervention of BC.
Supplementary Data
Funding
The work was supported by the National Natural
Science Foundation of China (grant nos. 81972453, 81972597,
81672729 and 81602471) and the Natural Science Foundation of
Zhejiang Province (grant nos. LY19H160055, LY19H160059,
LY18H160030, LY18H160005 and LY20H160026). It was also sponsored by
the Zheng Shu Medical Elite Scholarship Fund.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HCX, JGS and ZHC performed all experiments, and were
major contributors in writing the manuscript. JJY, BJX and YLJ made
contributions to the conception of the study and collected patient
samples. UJ, JW, WHZ and SDX analyzed data and revised the
manuscript. The study was conceived by LBW and JCZ, who designed
and supervised all research and drafted the manuscript. All authors
reviewed the final version of the manuscript.
Ethics approval and consent to
participate
Informed consent was obtained from all patients. The
present study was approved by the Ethics Committee of the Sir Run
Run Shaw Hospital affiliated with Zhejiang University, and
conducted in full accordance with the ethical principles cited in
the World Medical Association Declaration of Helsinki and local
legislation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
We thank Professor Xiao-Fang Yu (Cancer Institute,
Second Affiliated Hospital, School of Medicine, Zhejiang
University) for his critical and informative advice during the
revision process.
References
|
1
|
Ganz PA and Goodwin PJ: Breast cancer
survivorship: Where are we today? Adv Exp Med Biol. 862:1–8. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Waks AG and Winer EP: Breast cancer
treatment: A review. JAMA. 321:288–300. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang X, Li Y, Zhou Y, Mao F, Lin Y, Guan
J and Sun Q: Diagnostic performance of indocyanine green-guided
sentinel lymph node biopsy in breast cancer: A meta-analysis. PLoS
One. 11:e01555972016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Rosenberg SM and Partridge AH: Management
of breast cancer in very young women. Breast. 24(Suppl 2):
S154–S158. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li CH and Chen Y: Targeting long
non-coding RNAs in cancers: Progress and prospects. Int J Biochem
Cell Biol. 45:1895–1910. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Bartolomei MS, Zemel S and Tilghman SM:
Parental imprinting of the mouse H19 gene. Nature. 351:153–155.
1991. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Li X, Lin Y, Yang X, Wu X and He X: Long
noncoding RNA H19 regulates EZH2 expression by interacting with
miR-630 and promotes cell invasion in nasopharyngeal carcinoma.
Biochem Biophys Res Commun. 473:913–919. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Thornton JE and Gregory RI: How does Lin28
let-7 control development and disease? Trends Cell Biol.
22:474–482. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang H, Zhao Q, Deng K, Guo X and Xia J:
Lin28: An emerging important oncogene connecting several aspects of
cancer. Tumour Biol. 37:2841–2848. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Saitoh M: Involvement of partial EMT in
cancer progression. J Biochem. 164:257–264. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo S, Liang X, Guo M, Zhang X and Li Z:
Migration inhibition of water stress proteins from nostoc commune
vauchvia. activation of autophagy in DLD-1 cells. Int J Biol
Macromol. 119:669–676. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhou WH, Tang F, Xu J, Wu X, Yang SB, Feng
ZY, Ding YG, Wan XB, Guan Z, Li HG, et al: Low expression of beclin
1, associated with high Bcl-xL, predicts a malignant phenotype and
poor prognosis of gastric cancer. Autophagy. 8:389–400. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Klionsky DJ: Autophagy: From phenomenology
to molecular understanding in less than a decade. Nat Rev Mol Cell
Biol. 8:931–937. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liang C, Xu J, Meng Q, Zhang B, Liu J, Hua
J, Zhang Y, Shi S and Yu X: TGFB1-induced autophagy affects the
pattern of pancreatic cancer progression in distinct ways depending
on SMAD4 status. Autophagy. 17:1–15. 2019.
|
|
15
|
Lv Q, Wang W, Xue J, Hua F, Mu R, Lin H,
Yan J, Lv X, Chen X and Hu ZW: DEDD interacts with PI3KC3 to
activate autophagy and attenuate epithelial-mesenchymal transition
in human breast cancer. Cancer Res. 72:3238–3250. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Qin W, Li C, Zheng W, Guo Q, Zhang Y, Kang
M, Zhang B, Yang B, Li B, Yang H and Wu Y: Inhibition of autophagy
promotes metastasis and glycolysis by inducing ROS in gastric
cancer cells. Oncotarget. 6:39839–39854. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Zhou J, Yang L, Zhong T, Mueller M, Men Y,
Zhang N, Xie J, Giang K, Chung H, Sun X, et al: H19 lncRNA alters
DNA meth-ylation genome wide by regulating S-adenosylhomocysteine
hydrolase. Nat Commun. 6:102212015. View Article : Google Scholar
|
|
19
|
Wang L, Yuan C, Lv K, Xie S, Fu P, Liu X,
Chen Y, Qin C, Deng W and Hu W: Lin28 mediates radiation resistance
of breast cancer cells via regulation of caspase, H2A.X and Let-7
signaling. PLoS One. 8:e673732013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kallen AN, Zhou XB, Xu J, Qiao C, Ma J,
Yan L, Lu L, Liu C, Yi JS, Zhang H, et al: The imprinted H19 lncRNA
antagonizes let-7 microRNAs. Mol Cell. 52:101–112. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Gonzalez DM: Medici D. Signaling
mechanisms of the epithelial-mesenchymal transition. Sci Signal.
7:re82014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wu ZQ, Li XY, Hu CY, Ford M, Kleer CG and
Weiss SJ: Canonical Wnt signaling regulates Slug activity and links
epithelial-mesenchymal transition with epigenetic breast cancer 1,
early onset (BRCA1) repression. Proc Natl Acad Sci USA.
109:16654–16659. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Rhodes LV, Tate CR, Segar HC, Burks HE,
Phamduy TB, Hoang V, Elliott S, Gilliam D, Pounder FN, Anbalagan M,
et al: Suppression of triple-negative breast cancer metastasis by
pan-DAC inhibitor panobinostat via inhibition of ZEB family of EMT
master regulators. Breast Cancer Res Treat. 145:593–604. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yang J, Hou Y, Zhou M, Wen S, Zhou J, Xu
L, Tang X, Du YE, Hu P and Liu M: Twist induces
epithelial-mesenchymal transition and cell motility in breast
cancer via ITGB1-FAK/ILK signaling axis and its associated
downstream network. Int J Biochem Cell Biol. 71:62–71. 2016.
View Article : Google Scholar
|
|
25
|
Yook JI, Li XY, Ota I, Hu C, Kim HS, Kim
NH, Cha SY, Ryu JK, Choi YJ, Kim J, et al: A Wnt-Axin2-GSK3beta
cascade regulates snail1 activity in breast cancer cells. Nat Cell
Biol. 8:1398–1406. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Weyemi U, Redon CE, Sethi TK, Burrell AS,
Jailwala P, Kasoji M, Abrams N, Merchant A and Bonner WM: Twist1
and Slug mediate H2AX-regulated epithelial-mesenchymal transition
in breast cells. Cell Cycle. 15:2398–2404. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou Q, Wu H, Fu F, Yi W, Pei L and Zhou M:
RKIP suppresses the proliferation and metastasis of breast cancer
cell lines through up-regulation of miR-185 targeting HMGA2. Arch
Biochem Biophys. 610:25–32. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Felipe Lima J, Nofech-Mozes S, Bayani J
and Bartlett JM: EMT in breast carcinoma-a review. J Clin Med.
5:E652016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lamouille S, Connolly E, Smyth JW, Akhurst
RJ and Derynck R: TGF-β-induced activation of mTOR complex 2 drives
epithelial-mesenchymal transition and cell invasion. J Cell Sci.
125:1259–1273. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang J, Ye C, Xiong H, Shen Y, Lu Y, Zhou
J and Wang L: Dysregulation of long non-coding RNA in breast
cancer: An overview of mechanism and clinical implication.
Oncotarget. 8:5508–5522. 2017.
|
|
31
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Liu L, Yang J, Zhu X, Li D, Lv Z and Zhang
X: Long noncoding RNA H19 competitively binds miR-17-5p to regulate
YES1 expression in thyroid cancer. FEBS J. 283:2326–2339. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Imig J, Brunschweiger A, Brümmer A,
Guennewig B, Mittal N, Kishore S, Tsikrika P, Gerber AP, Zavolan M
and Hall J: miR-CLIP capture of a miRNA targetome uncovers a
lincRNA H19-miR-106a interaction. Nat Chem Biol. 11:107–114. 2015.
View Article : Google Scholar
|
|
34
|
Matouk IJ, Raveh E, Abu-lail R, Mezan S,
Gilon M, Gershtain E, Birman T, Gallula J, Schneider T, Barkali M,
et al: Oncofetal H19 RNA promotes tumor metastasis. Biochim Biophys
Acta. 1843:1414–1426. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang L, Yang F, Yuan JH, Yuan SX, Zhou
WP, Huo XS, Xu D, Bi HS, Wang F and Sun SH: Epigenetic activation
of the MiR-200 family contributes to H19-mediated metastasis
suppression in hepatocellular carcinoma. Carcinogenesis.
34:577–586. 2013. View Article : Google Scholar
|
|
36
|
Balzeau J, Menezes MR, Cao S and Hagan JP:
The LIN28/let-7 pathway in cancer. Front Genet. 8:312017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang S and Baltimore D: RNA-binding
protein Lin28 in cancer and immunity. Cancer Lett. 375:108–113.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Cui C, Li Z and Wu D: The long non-coding
RNA H19 induces hypoxia/reoxygenation injury by up-regulating
autophagy in the hepatoma carcinoma cells. Biol Res. 52:322019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang M, Han D, Yuan Z, Hu H, Zhao Z, Yang
R, Jin Y, Zou C, Chen Y, Wang G, et al: Long non-coding RNA H19
confers 5-Fu resistance in colorectal cancer by promoting
SIRT1-mediated autophagy. Cell Death Dis. 9:11492018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang J, Xie S, Yang J, Xiong H, Jia Y,
Zhou Y, Chen Y, Ying X, Chen C, Ye C, et al: The long noncoding RNA
H19 promotes tamoxifen resistance in breast cancer via autophagy. J
Hematol Oncol. 12:812019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zhang J, Liu CY, Wan Y, Peng L, Li WF and
Qiu JX: Long non-coding RNA H19 promotes the proliferation of
fibroblasts in keloid scarring. Oncol Lett. 12:2835–2839. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhuo C, Jiang R, Lin X and Shao M: LncRNA
H19 inhibits autophagy by epigenetically silencing of DIRAS3 in
diabetic cardiomyopathy. Oncotarget. 8:1429–1437. 2017. View Article : Google Scholar :
|
|
43
|
Xu Z, Yan Y, Qian L and Gong Z: Long
non-coding RNAs act as regulators of cell autophagy in diseases
(review). Oncol Rep. 37:1359–1366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Galluzzi L, Pietrocola F, Bravo-San Pedro
JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J,
Gewirtz DA, Karantza V, et al: Autophagy in malignant
transformation and cancer progression. EMBO J. 34:856–880. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Guo Q, Jing FJ, Xu W, Li X, Li X, Sun JL,
Xing XM, Zhou CK and Jing FB: Ubenimex induces autophagy inhibition
and EMT suppression to overcome cisplatin resistance in GC cells by
perturbing the CD13/EMP3/PI3K/AKT/NF-κB axis. Aging. 11:2019.
|
|
46
|
Liang F, Ren C, Wang J, Wang S, Yang L,
Han X, Chen Y, Tong G and Yang G: The crosstalk between STAT3 and
p53/RAS signaling controls cancer cell metastasis and cisplatin
resistance via the Slug/MAPK/PI3K/AKT-mediated regulation of EMT
and autophagy. Oncogenesis. 8:592019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Han LL, Jia L, Wu F and Huang C: Sirtuin6
(SIRT6) promotes the EMT of hepatocellular carcinoma by stimulating
autophagic degradation of E-cadherin. molecular cancer research:
Mol Cancer Res. 17:2267–2280. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jiang Z, Jones R, Liu JC, Deng T, Robinson
T, Chung PE, Wang S, Herschkowitz JI, Egan SE, Perou CM and
Zacksenhaus E: RB1 and p53 at the crossroad of EMT and
triple-negative breast cancer. Cell Cycle. 10:1563–1570. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Dhamija S and Diederichs S: From junk to
master regulators of invasion: lncRNA functions in migration, EMT
and metastasis. Int J Cancer. 139:269–280. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Jin B, Wang W, Meng XX, Du G, Li J, Zhang
SZ, Zhou BH and Fu ZH: Let-7 inhibits self-renewal of
hepatocellular cancer stem-like cells through regulating the
epithelial-mesenchymal transition and the Wnt signaling pathway.
BMC Cancer. 16:8632016. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Qiu XY, Hu DX, Chen WQ, Chen RQ, Qian SR,
Li CY, Li YJ, Xiong XX, Liu D, Pan F, et al: PD-L1 confers
glioblastoma multi-forme malignancy via Ras binding and Ras/Erk/EMT
activation. Biochim Biophys Acta Mol Basis Dis. 1864:1754–1769.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Hu F, Zhao Y, Yu Y, Fang JM, Cui R, Liu
ZQ, Guo XL and Xu Q: Docetaxel-mediated autophagy promotes
chemoresistance in castration-resistant prostate cancer cells by
inhibiting STAT3. Cancer Lett. 416:24–30. 2018. View Article : Google Scholar
|
|
54
|
Sosa P, Alcalde-Estevez E, Plaza P,
Troyano N, Alonso C, Martínez-Arias L, Evelem de Melo Aroeira A,
Rodriguez-Puyol D, Olmos G, López-Ongil S and Ruíz-Torres MP:
Hyperphosphatemia promotes senescence of myoblasts by impairing
autophagy through Ilk overexpression, a possible mechanism involved
in sarcopenia. Aging Dis. 9:769–784. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Huang M and Xin W: Matrine inhibiting
pancreatic cells epithelial-mesenchymal transition and invasion
through ROS/NF-κB/MMPs pathway. Life Sci. 192:55–61. 2018.
View Article : Google Scholar
|