Introduction
Pancreatic cancer is one of the most lethal
malignancies in the world, with its mortality close to its
incidence (1,2). In recent years, the incidence of
pancreatic cancer keeps rising due to the popularization of the
westernized lifestyle (3).
Approximately 80% of patients with pancreatic cancer are diagnosed
at an advanced stage and miss the chance for curative resection
(2). Pancreatic cancer, as a
highly heterogenous tumor, is a major clinical challenge (2,4,5).
Therefore, identifying subgroups with special biology is urgently
needed for the management of pancreatic cancer.
Carbohydrate antigen 19-9 (CA19-9), also called
sialyl Lewis antigen A, is the most important biomarker for
pancreatic cancer (6-8). The sensitivity in detecting
pancreatic cancer is ~80% for CA19-9 (9). In the population, ~5-10% of
individuals are Lewis antigen negative, with no or low secretion of
CA19-9 (10). In a previous study,
we showed that Lewis-negative patients had poorer outcome than
Lewis-positive patients (11).
Fucosyltransferase 3 (also called Lewis gene), an
α1,3/4-fucosyltransferase, is the key enzyme of CA19-9 biosynthesis
and plays a critical role in protein fucosylation (12). Protein fucosylation has undoubtedly
an important effect on the function of proteins, and affects cancer
development (12). Therefore,
Lewis-negative pancreatic cancer, which is deficient in
fucosylation, may have a special biology, different from
Lewis-positive cancer (11).
However, the characteristics of Lewis-negative pancreatic cancer
are largely unidentified.
In the present study, the characteristics of
Lewis-negative pancreatic cancer in both clinical findings and
basic research were investigated. The clinicopathological
characteristics of 853 patients with pancreatic cancer classified
by Lewis status were examined. Six pancreatic cancer cell lines
were sequenced to determine their Lewis status. Morphological and
molecular features of pancreatic cancer cells classified by Lewis
status were compared. An orthotopic tumor model was
constructed.
Materials and methods
Patients and data collection
Medical data were retrieved from a prospectively
maintained database of the Fudan University Shanghai Cancer Center
(Shanghai, China) from September 2004 to November 2011. Data
including age, sex, tumor location, metastasis, grade, CA19-9,
carbohydrate antigen 125 (CA125, also called MUC16), nerve
invasion, lymphovascular invasion, and lymphatic metastasis were
retrieved. The primary endpoint was overall survival and follow-up
data were updated till October 2019. The study protocol was
authorized by the Ethics Committee of the Fudan University Shanghai
Cancer Center. Written informed consent was acquired from all of
the patients enrolled in the study.
Immunohistochemistry
Tissues were fixed in 10% formalin for 12 h at room
temperature. Formalin-fixed, paraffin-embedded sections (4
µm) of surgically resected pancreatic cancer tissues were
obtained [20 cases of Lewis (-), 19 cases of Lewis (+)]. After
tissue sections were deparaffinized with xylene, the endogenous
peroxidase activity was blocked with 3% H2O2
in methanol at 37°C for 20 min. Sections were incubated with
specific primary antibodies against MUC16 (1:200, cat. no.
60261-1-Ig; ProteinTech Group, Inc.) overnight at 4°C. The antibody
solution was removed, and the sections were washed in wash buffer 3
times for 10 min each. Secondary antibody (GTVision III
immunohistochemical detection kit, GK5005; Gene Tech Co., Ltd.) was
added to each section and the tissues were incubated for 1 h at
room temperature. An avidinbiotin-peroxidase complex solution was
used for the visualization of immunoreactions using
3,3′-diaminobenzidine to detect the protein-antibody complexes.
Protein expression levels were classified as positive and negative
staining using an optical microscope with a magnification of
1:400.
Lewis genotyping
Lewis status was determined by Sanger sequencing
using genomic DNA extracted from blood specimens or pancreatic
cancer cell lines, as previously described (9,11,13).
In order to detect variants in the Lewis gene: T59G, T202C, C314T,
G508A and T1067A, the following primers were used for polymerase
chain reaction amplification: 358F, GGGTGCAGC CAAGCCACAA and 358R,
AGGTGGGAGGCGTGACTT AGG; P1F, ACTTGGAGCCACCCCCTAACTGCCA and 508R,
CGGCCTCTCAGGTGAACCAAGCCGCT).
Cell lines
BxPC-3, SU8686, SW1990, CaPan-1, MiaPaCa-2 and
Panc-1 human pancreatic cancer cells were kindly provided by Stem
Cell Bank, Chinese Academy of Sciences (Shanghai, China). The
CaPan-1, MiaPaCa-2, Panc-1 and SW1990 cells were kept in DMEM
supplemented with 10% FBS and 1% penicillin/streptomycin (Gibco;
Thermo Fisher Scientific, Inc.). The BxPC-3 and SU8686 cells were
kept in RPMI-1640 medium supplemented with 10% FBS and
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.).
All cells were maintained at humidified incubator at a 37°C with 5%
CO2. Only mycoplasma-negative cells were used for the
experiments.
Phase-contrast microscopy and scanning
electron microscopy
Human pancreatic cancer cells were seeded into 10-cm
dishes and images were captured by a phase-contrast microscope
(Leica Microsystems GmbH). For the FEI Quanta 200 scanning electron
microscope (Philips Healthcare), the cells were seeded into 0.8-cm
glass slides treated with polylysine amino acid coating by gold
powder. The cells were fixed in 2.5% glutaraldehyde solution at 4°C
for 5 h. After being washed with 0.1 mol/l phosphoric acid buffer 3
times, the cells were dehydrated by alcohol step by step, replaced
by pure alcohol, dried at the critical point of carbon dioxide, and
then observed and photographed by FEI Quanta 200 scanning electron
microscope after coating. All images were captured by random
fields.
Cell proliferation assay
For cell proliferation, the pancreatic cancer cells
were trypsinized, and 3×103 cells were seeded into
96-well plates (Corning, Inc.). After certain culture periods, 10
µl of Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) were added into the wells and the cells were
incubated at a humidified incubator at 37°C with 5% CO2.
Absorbance was detected on a multifunctional microplate reader at a
wavelength of 450 nm.
Transwell migration assay
Pancreatic cancer cells were trypsinized, and
3×104 cells were seeded into Transwell inserts (8.0 mm
pore; BD Falcon; BD Biosciences) without serum. FBS (10%) and
penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.)
were plated into the lower chamber. After 24 h, the upper side of
the membrane was rubbed with cotton swap and the cells were fixed
in 4% paraformaldehyde and stained by 0.3% crystal violet for 20
min at room temperature. After crystal violet staining, the number
of cells migrating to the basal side insert was counted. Stained
cells were counted in seven randomly selected fields using an
optical microscope with a magnification of 1:400.
Western blot and lectin blot
analyses
Human cells were seeded into 60-mm2
dishes. Cells were harvested and lysed in RIPA buffer, 1 mM PMSF
and 1X Protease Inhibitor Cocktail (Beyotime Institute of
Biotechnology) for 30 min. Protein concentration was determined by
a BCA assay. The protein samples (10 µg) were loaded on a
10% SDS-PAGE and run at 100 V for 80 min in 1X SDS-PAGE running
buffer (25 mM Tris, 192 mM glycine, 1% SDS). Proteins were
transferred onto a 0.45-mm nitrocellulose membrane (EMD Millipore)
using a wet transfer protocol with 1X transfer buffer (25 mM Tris,
192 mM Glycine, 20% methanol) at 300 mA for 110 min at Mini
Trans-Blot Cell Module (Bio-Rad Laboratories, Inc.) in ice box. The
membranes were blocked with PBST (0.02% Tween-20 in PBS) containing
5% skim milk (BD Biosciences) at room temperature. The membranes
were incubated with anti-MUC16 antibody (1:1,000, cat. no.
20077-1-AP; ProteinTech Group, Inc.), anti-EGFR antibody (1:1,000,
cat. no. ab52894; Abcam), anti-STAT3 antibody (1:1,000, cat. no.
ab32143; Abcam) and HRP-conjugated β-actin antibody (1:5,000, cat.
no. HRP-60008; ProteinTech Group, Inc.) in blocking solution on a
shaker at 4°C overnight. Following the primary incubation, the
membranes were incubated with goat-anti-mouse IgG (H+L)-HRP or
goat-anti-rabbit IgG (H+L)-HRP (cat. no. SA00001-2; ProteinTech
Group, Inc.) at 1:5,000 in PBST on a shaker for 1 h at room
temperature. For lectin blot analysis, biotinylated Aleuria
aurantia lentin (AAL, 3 µg/ml) (Vector Laboratories,
Inc.) was incubated with 3% bovine serum albumin on a shaker for 30
min, and then, incubated with 0.1 µg/ml streptavidin-HRP
conjugate (Vector Laboratories, Inc.) in blocking buffer for 20 min
at room temperature. Images were captured after SuperSignal West
Femto ECL (BR11121; Bridgen Co., Ltd.) reaction.
Liquid chromatography-mass spectrometry
(LC-MS) for protein glycosylation
Pancreatic cancer cells (>2×107 cells)
were freshly prepared prior to use. The sample proteins were
extracted using SDT lysis buffer (4% SDS, 100 mM DTT, 100 mM
Tris-HCl pH 8.0). Samples were boiled for 3 min and further
ultrasonicated. Undissolved beads were removed by centrifugation at
16,000 × g for 15 min. The supernatant containing proteins was
collected. Protein digestion was performed with FASP method, as
described by Wiśniewski et al (14). Proteins were subjected to
glycopeptide enrichment and were deglycosylated. Eluted peptides
were collected and dried for further LC-MS analysis (Thermo Fisher
Scientific, Inc.) using a positive or negative ionization mode.
Reverse-phase high-performance liquid chromatography
separation was performed with the EASY-nLC system (Thermo Fisher
Scientific, Inc.) using a self-packed column (75 µm x 150
mm; 3 µm ReproSil-Pur C18 beads, 120 Å; Dr. Maisch GmbH
HPLC) at a flow rate of 300 nl/min. MS data were acquired using a
data-dependent top 20 method dynamically choosing the most abundant
precursor ions from the survey scan (300-1,800 m/z) for HCD
fragmentation. The instrument was run with peptide recognition mode
enabled. A lock mass of 445.120025 Da was used as internal standard
for mass calibration. The full MS scans were acquired at a
resolution of 70,000 at m/z 200, and 17,500 at m/z 200 for MS/MS
scan. MS data were analyzed using MaxQuant software (version
1.6.1.0; Max Planck Institute of Biochemistry) and were searched
against the SwissProt human database (http://www.expasy.ch/sprot/).
Bioinformatics analyses were carried out with
Perseus software (Max Planck Institute of Biochemistry), Microsoft
Excel (Microsoft Corporation) and R statistical computing software
(Free Software Foundation's GNU General Public License; https://www.r-project.org/about.html).
Construction of protein-protein interaction networks was conducted
using the STRING database (https://string-db.org) with Cytoscape software (an
open source software platform provided by the National Resource for
Network Biology). The MS data were analyzed using MaxQuant software
(version 1.6.1.0) and were searched against the SwissProt human
database (20,431 total entries; downloaded, 10/15/2019). The
Motif-X algorithm in the MEME Suite (version 5.1.0) was used for
N-linked glycosylation motif analysis.
Orthotopic animal model
A total of 16 male BALB/c nude (nu/nu) mice (6-8
weeks of age) were purchased from Shanghai SLAC Laboratory Animal
Co., Ltd. and 8 mice were included in each group. Animals were
housed in laminar flow cabinets under specific pathogen-free
conditions. The housing conditions were as follows: temperature
22±1°C, humidity 50%, 12-h dark/light cycle, and ad libitum
access to food and water. Animals were orthotopically injected with
1×106/ml cells into the pancreas (n=8). The mice were
sacrificed at 5-week endpoints to examine tumor weight.
Histological features of tumors were examined by hematoxylin and
eosin (H&E; Beyotime Institute of Biotechnology) staining. All
mouse samples were fixed with 10% buffer formalin at room
temperature (24-36 h) to make formalin-fixed, paraffin-embedded
tissue blocks. H&E staining was performed on 3-mm thick
sections at room temperature for 10 min. The staining was observed
by a light microscope (CKX41; Olympus Corporation), with a
magnification of ×100. All animal procedures were approved by the
Institutional Animal Care Committee of Fudan University (Shanghai,
China).
Statistical analysis
SPSS 19.0 software (IBM Corp.) and Prism statistical
software (version 8; GraphPad Software, Inc.) were used for the
statistical analysis of the data. Unpaired two-tailed Student's
t-tests were used to determine the statistical differences between
two groups. Data were presented as the mean ± standard error of the
mean. Dichotomous variables were analyzed by Chi-square test or
Fisher's exact test. Survival analysis was assessed by the
Kaplan-Meier method and the survival curves were compared by
log-rank tests. P<0.05 was considered to indicate a
statistically significant difference.
Results
Clinicopathological characteristics of
Lewis-negative pancreatic cancer patients
A total of 853 patients with pancreatic cancer were
included to undergo Lewis antigen evaluation and 11.7% of patients
were Lewis negative (Table I). The
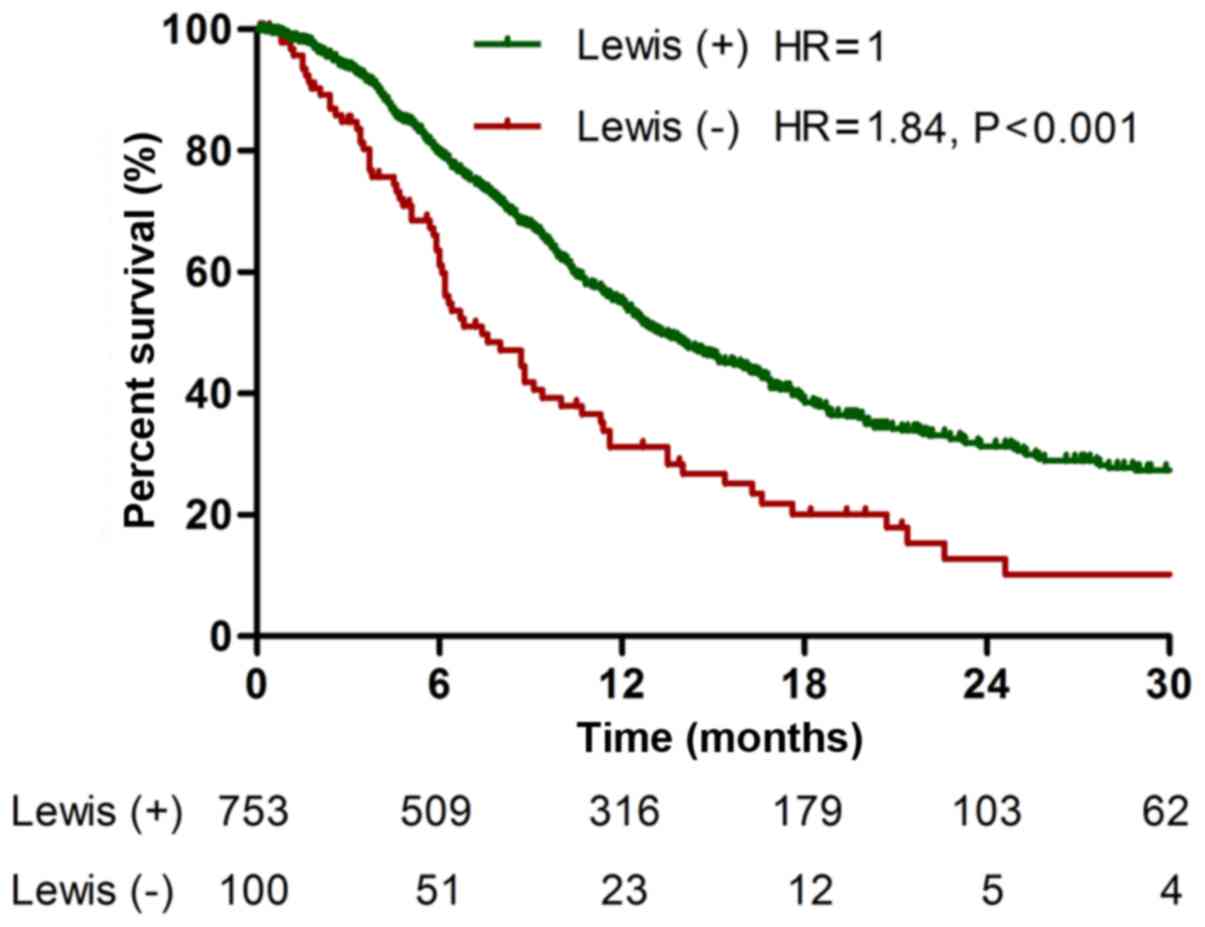
median survival time of Lewis-negative patients was 7.4 months,
which was significantly shorter than that of Lewis-positive
patients (13.3 months, P<0.001; Fig. 1). In addition, Lewis-negative
patients had higher proportion of metastasis (P=0.004) than
Lewis-positive patients. Lewis-negative patients had lower serum
level of CA19-9 (106.0±273.1 U/ml) than Lewis-positive patients
(499.7±635.0 U/ml, P<0.001). However, contrary to CA19-9,
Lewis-negative pancreatic cancer secreted higher level of serum
CA125 (251.9±642.0 U/ml) compared with Lewis-positive cancer
(135.8±401.6 U/ml, P<0.001). These data show that Lewis-negative
pancreatic cancer has aggressive clinicopathological
characteristics with low secretion of CA19-9 and high secretion of
CA125.
 | Table IBaseline characteristics of patients
with pancreatic cancer classified by Lewis status. |
Table I
Baseline characteristics of patients
with pancreatic cancer classified by Lewis status.
|
Characteristics | Total | Lewis positive | Lewis negative | P-value |
|---|
| No. of cases | 853 | 753 | 100 | |
| Median survival
(months) | 12.6 | 13.3 | 7.4 | <0.001 |
| Age (years) | | | | 0.867 |
| ≤60 | 382 | 338 | 44 | |
| >60 | 471 | 415 | 56 | |
| Sex | | | | 0.923 |
| Male | 508 | 448 | 60 | |
| Female | 345 | 305 | 40 | |
| Locationa | | | | 0.582 |
| Head | 431 | 383 | 48 | |
| Others | 421 | 369 | 52 | |
| CA19-9 (U/ml) | 453.5±617.0 | 499.7±635.0 | 106.0±273.1 | <0.001 |
| CA125 (U/ml) | 149.4±437.7 | 135.8±401.6 | 251.9±642.0 | <0.001 |
| Metastasis | | | | 0.004 |
| Yes | 314 | 264 | 50 | |
| No | 539 | 489 | 50 | |
| Gradeb | | | | 0.245 |
| High, medium | 235 | 219 | 16 | |
| Low | 148 | 133 | 15 | |
| Nerve
invasionb | | | | 0.259 |
| Yes | 318 | 290 | 28 | |
| No | 65 | 62 | 3 | |
| Lymphovascular
invasionb | | | | 0.934 |
| Yes | 84 | 77 | 7 | |
| No | 298 | 274 | 24 | |
| Lymph
involvementb | | | | 0.773 |
| Yes | 164 | 150 | 14 | |
| No | 220 | 203 | 17 | |
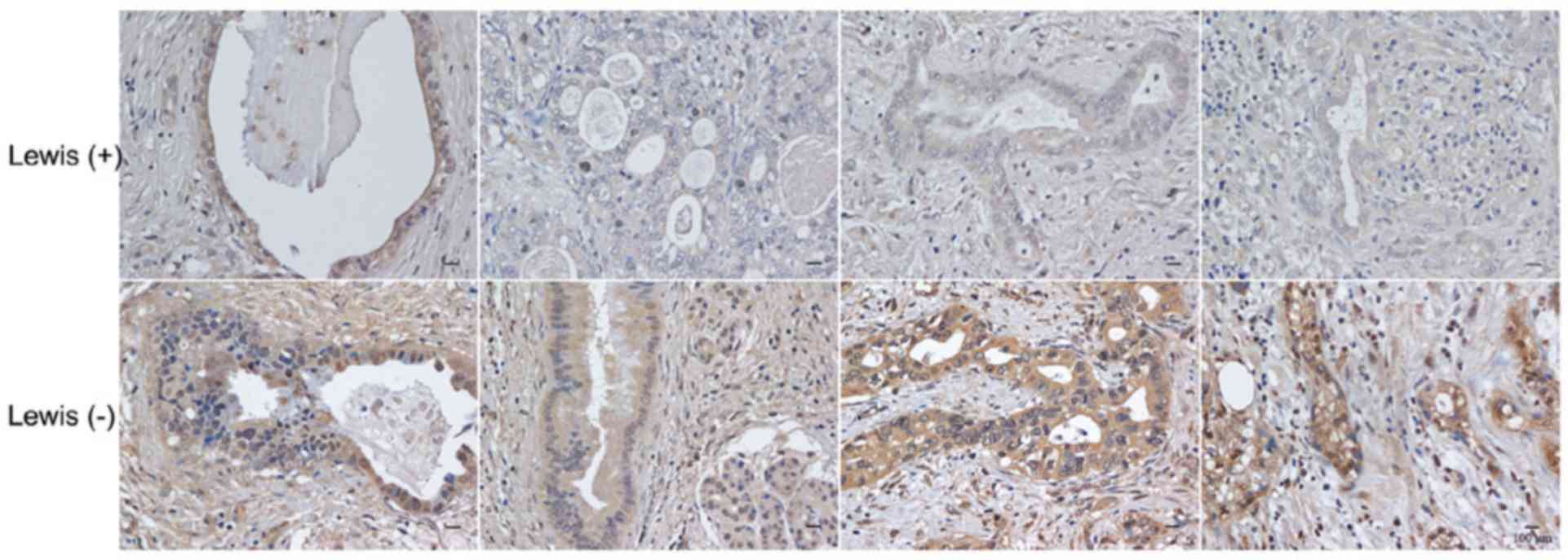
MUC16 expression in pancreatic cancer
tissues
To confirm the association between Lewis status and
CA125 secretion, the expression of MUC16 in pancreatic cancer
tissues was detected by immunohistochemistry. Lewis-negative
pancreatic cancer tissues (16/20) had higher levels of MUC16
expression than Lewis-positive cancer tissues (9/19, P=0.048;
Fig. 2).
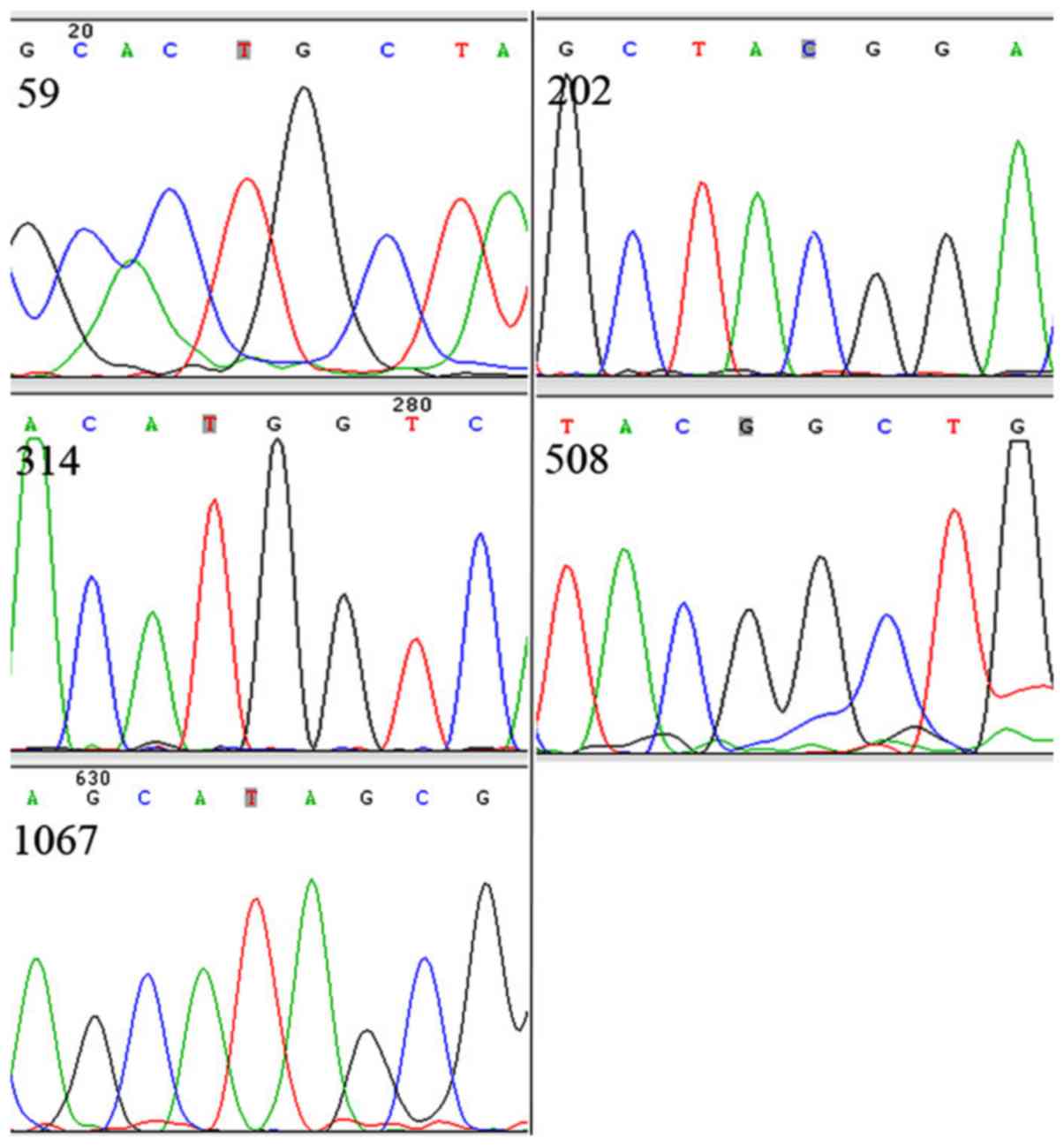
Lewis antigen status of human pancreatic
cancer cell lines
Sanger sequencing of the Lewis gene was carried out
for the determination of the Lewis antigen status of human
pancreatic cancer cell lines (BxPC-3, SU8686, SW1990, CaPan-1,
MiaPaCa-2 and Panc-1). Three cell lines were classified as Lewis
positive (BxPC-3, SU8686 and SW1990) and the other three were
categorized as Lewis negative (CaPan-1, MiaPaCa-2 and Panc-1).
Representative sequencing results are shown in Fig. 3, which demonstrate homozygous
mutations at 202 and 314 alleles of Lewis gene in the Panc-1 cell
line.
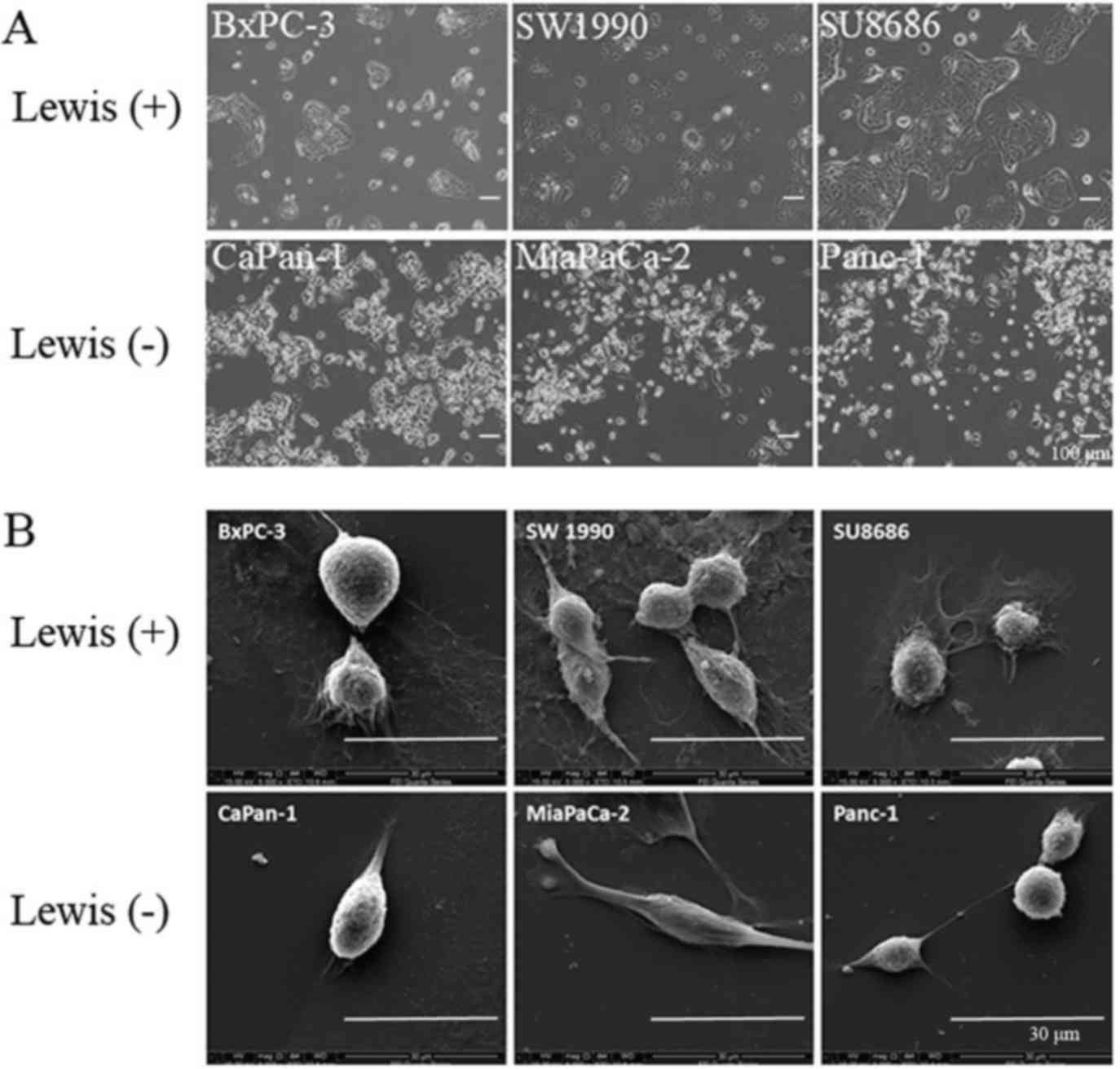
Cell morphology
The difference in morphology between Lewis-positive
and -negative cell lines was examined by phase-contrast microscopy
and scanning electron microscopy. Lewis-positive cell lines
(BxPC-3, SW1990 and SU8686) grew in a cluster pattern, whereas
Lewis-negative cell lines (CaPan-1, MiaPaCa-2 and Panc-1) showed a
shuttle-like morphology by phase-contrast microscopy (Fig. 4A). Scanning electron microscopy
showed that Lewis-positive cells were characterized by abundant
pseudopods closely attached to the culture dish, whereas
Lewis-negative cells were not (Fig.
4B). Hence, these results suggest that there is a difference in
cell morphology between different Lewis phenotype cells.
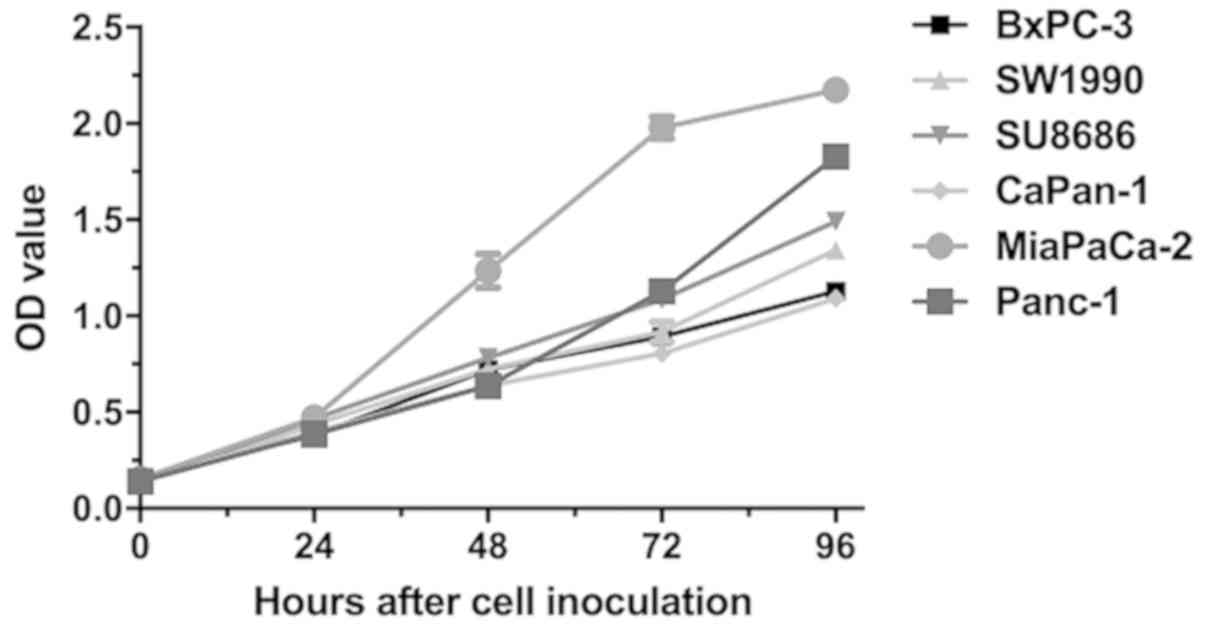
Cell proliferation
The proliferative abilities of Lewis-positive and
-negative cells were evaluated by CCK-8 assay. Overall,
Lewis-negative cells had significantly higher proliferation rate
than Lewis-positive cells at 96 h after seeding (P=0.006; Fig. 5). MiaPaCa-2, a Lewis-negative cell
line, had the highest proliferation rate among all cells.
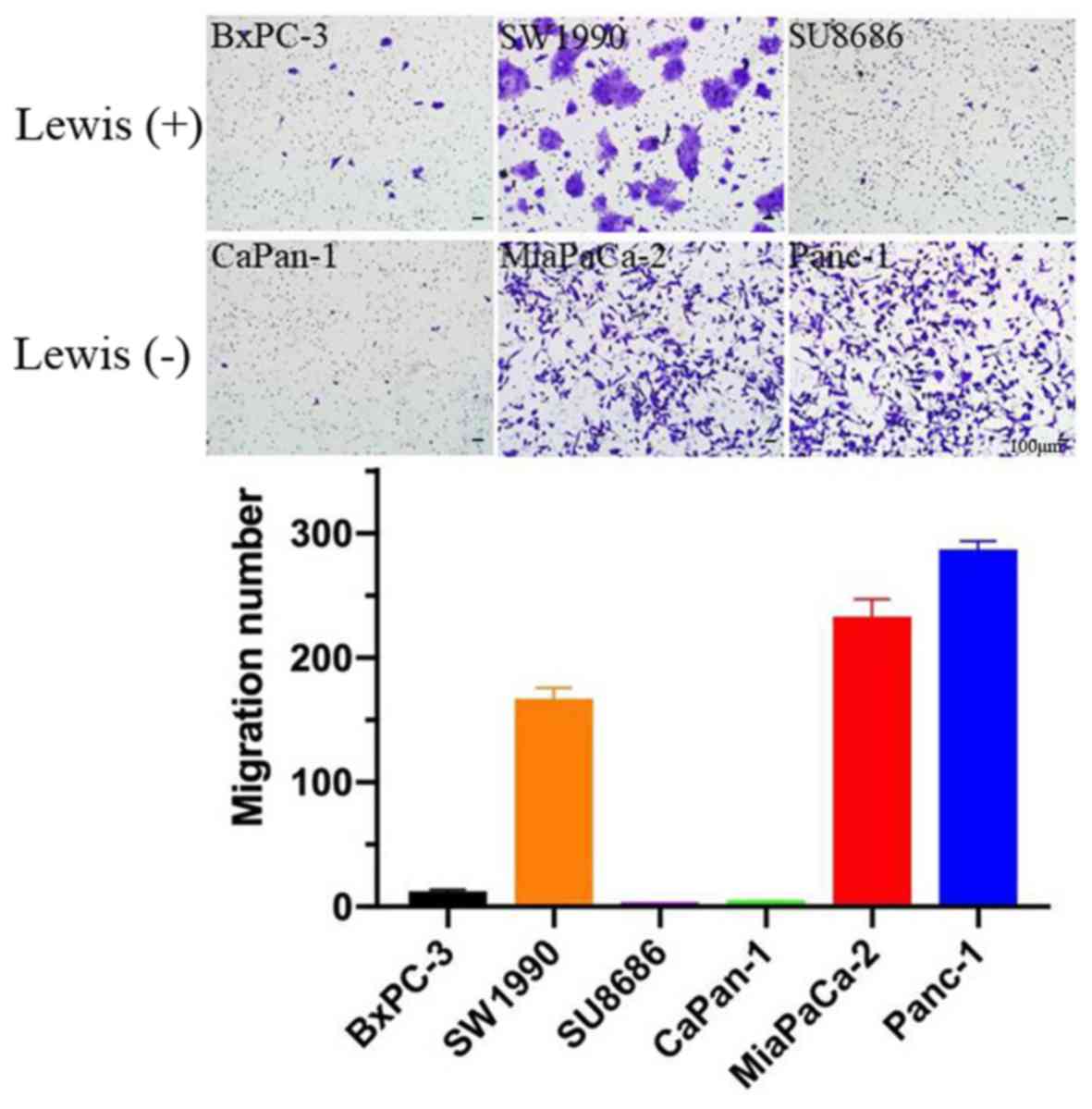
Cell migration
The migration ability of Lewis-positive and
-negative cells was examined by Transwell assay. Approximately
3×104 pancreatic cancer cells were seeded into Transwell
chambers and crystal violet staining was examined after 24 h of
seeding. Overall, Lewis-negative cell lines exhibited higher
migration ability compared with Lewis-positive cell lines (P=0.003;
Fig. 6).
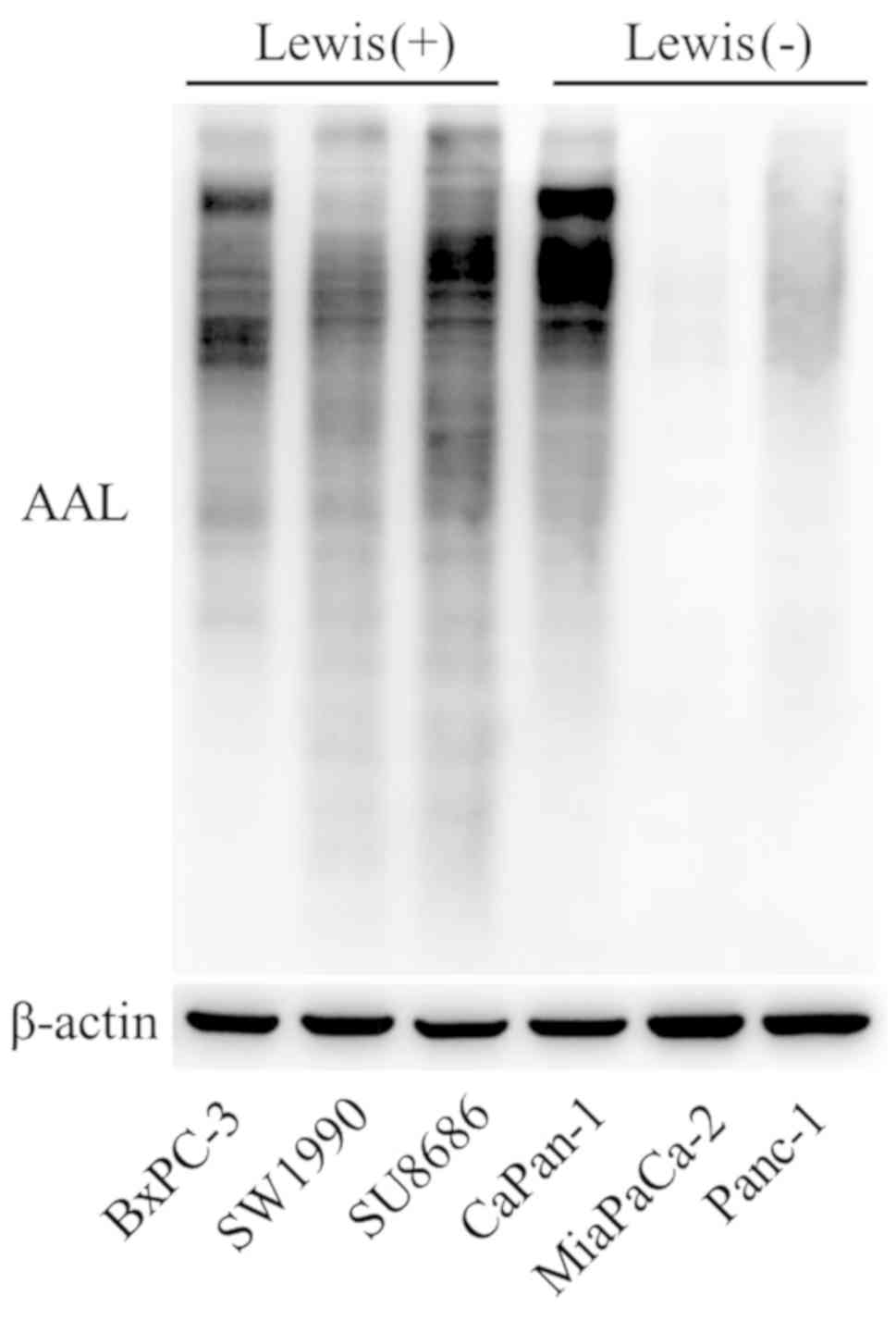
Level of fucosylation
The level of fucosylation in Lewisposi tive and
-negative cells was determined by AAL blotting analysis, which has
been often used as carbohydrate probes for core fucose in
glycoproteins. Lewis-negative cell lines (MiaPaCa-2 and Panc-1)
exhibited lower levels of AAL compared with Lewis-positive cell
lines (Fig. 7). This finding
reveals that the lower fucosylation level may be attributed to the
loss of function of the Lewis gene in Lewis-negative pancreatic
cancer.
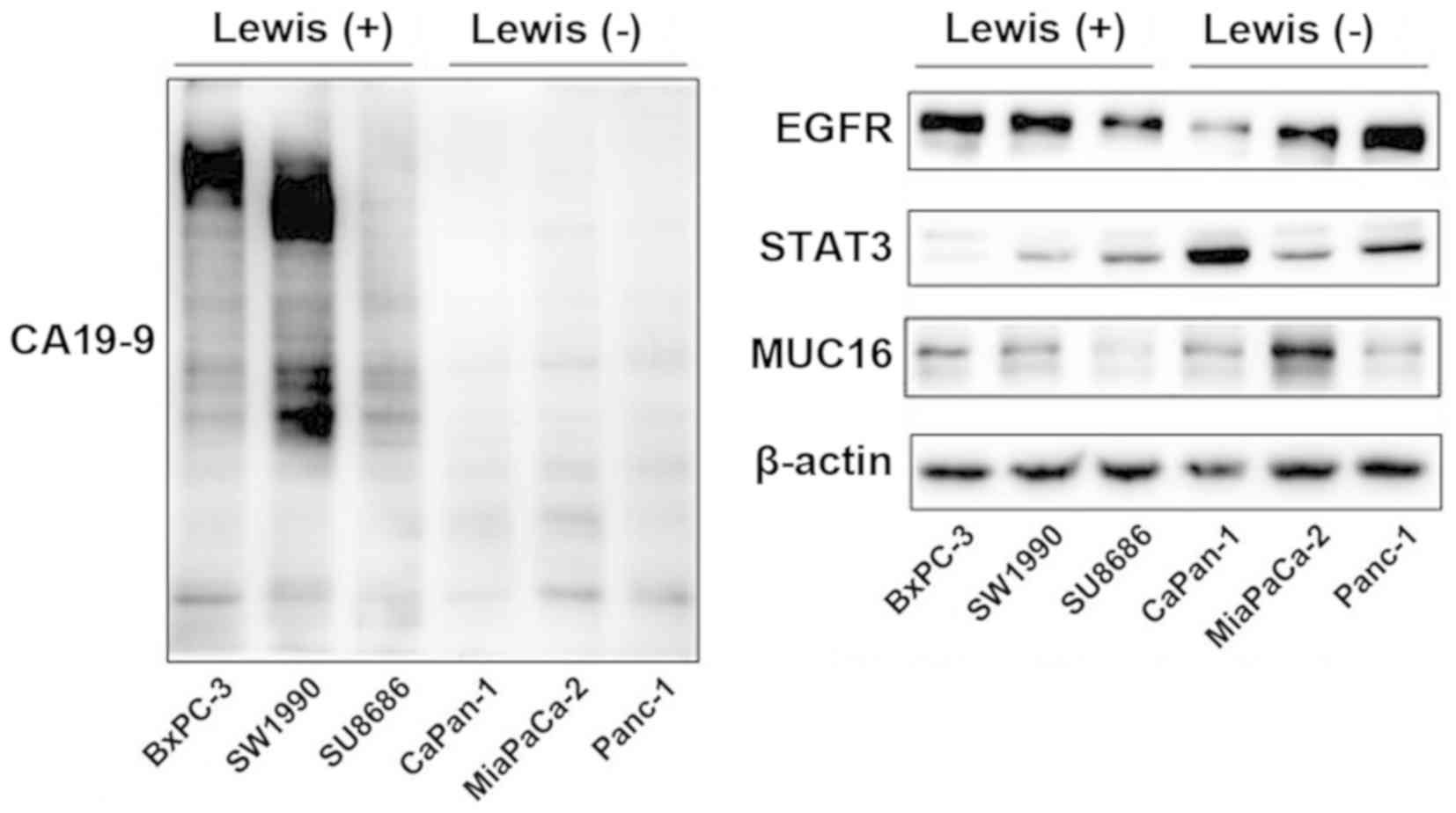
Glycoprotein and protein expression
levels
According to clinical data, Lewis-negative patients
had lower levels of serum CA19-9 than Lewis-positive patients
(Table I). This result was further
verified in pancreatic cancer cell lines. Western blot analysis
revealed that the level of CA19-9 was significantly higher in
Lewis-positive cells than that in Lewis-negative cells (Fig. 8). Lewis-negative cells displayed
higher level of MUC16 compared with Lewis-positive cells. The
association between MUC16 and Lewis status was consistent with the
clinical results of CA125 and Lewis status. Differences in Lewis
genotype had no significant effect on EGFR or STAT3 expression.
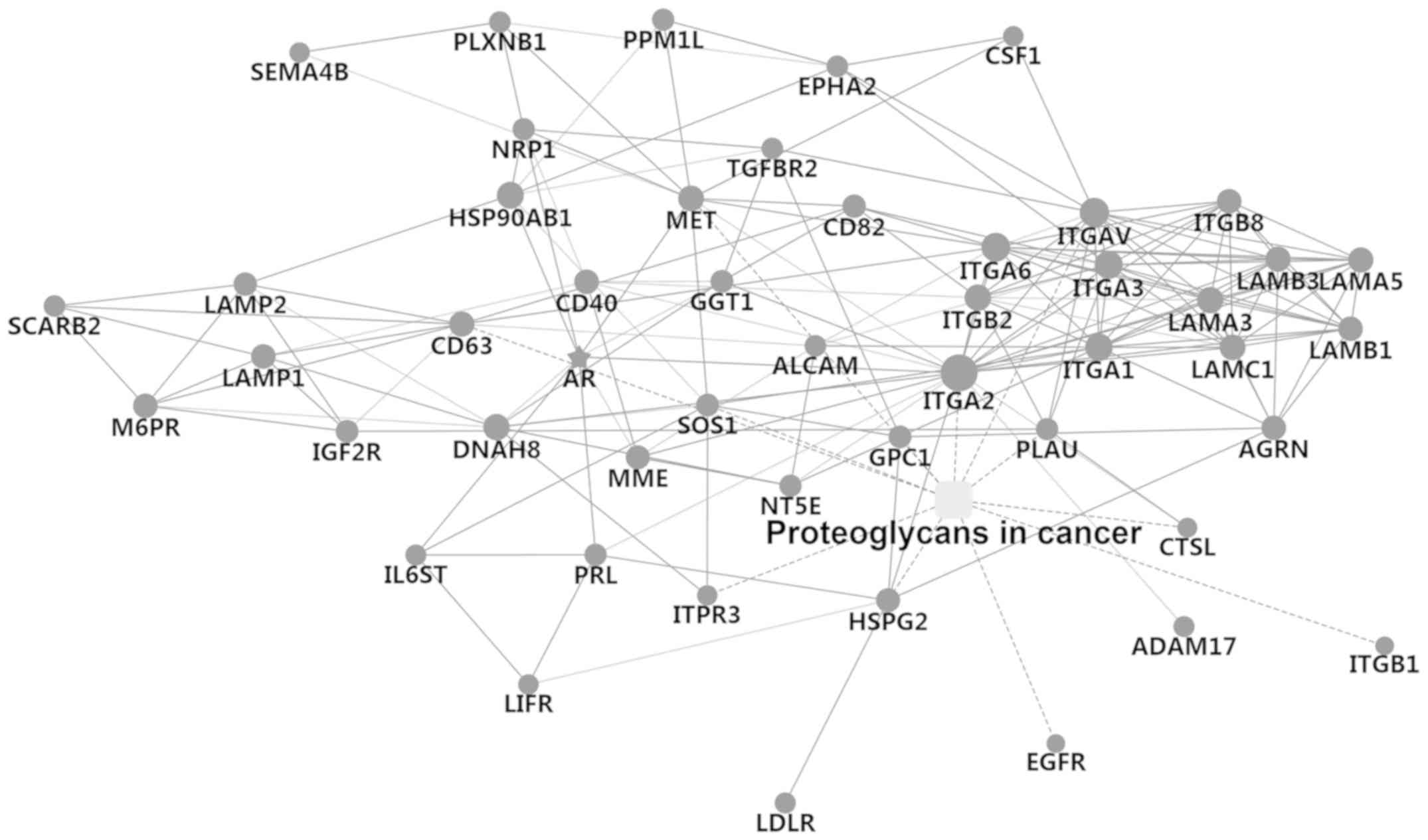
Network of cancer-related
proteoglycans
Lewis gene is a regulator of glycosylation and plays
a key role in fucosylation of proteins. In order to further verify
the role of the Lewis gene on fucosylation, cancer-related
proteoglycans were detected by LC-MS in the Lewis-positive cell
line SU8686 (Fig. 9). Potential
proteoglycan interactions were identified, such as EGFR, HSPG2,
ADAM17, GPC1, ITGA2, CD40, IL6ST and GGT1.
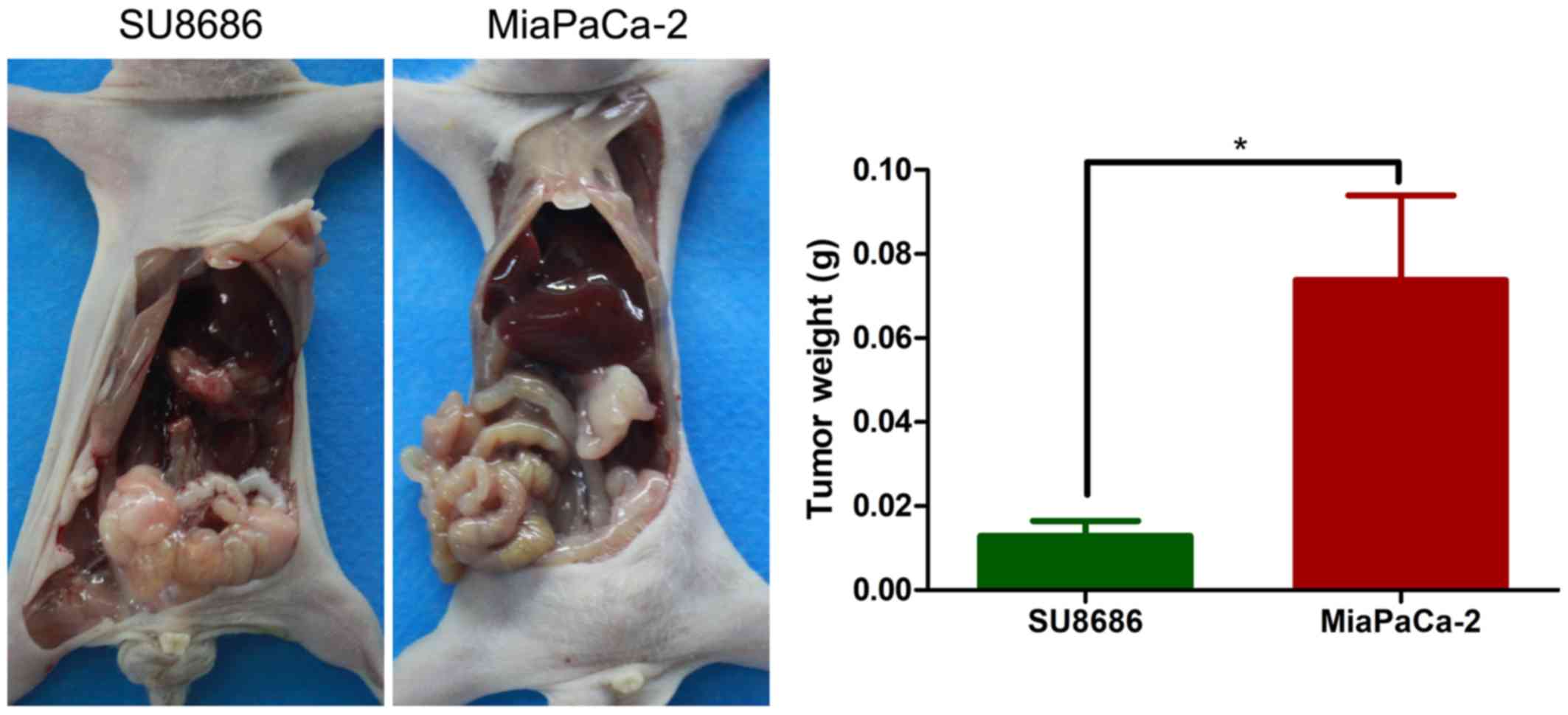
Orthotopic animal model
In order to examine the in vivo growth
ability of pancreatic cancer cell lines classified by Lewis status,
an orthotopic animal model was constructed by injection of tumor
cells into the pancreas. Lewis-negative pancreatic cancer cell line
MiaPaCa-2 corresponded to higher tumor weight than Lewis-positive
pancreatic cell line SU8686 (P=0.008; Fig. 10).
Discussion
Lewis gene is critical for fucosylation and protein
modification (12). In the present
study, a total of 853 patients with pancreatic cancer were included
and 11.7% of patients were Lewis negative. Lewis-negative
pancreatic cancer presented aggressive clinicopathological
characteristics with low secretion of CA19-9 and high secretion of
CA125. Three cell lines were classified as Lewis positive (BxPC-3,
SU8686 and SW1990) and three were classified as Lewis negative
(CaPan-1, MiaPaCa-2 and Panc-1). Lewis-negative pancreatic cancer
cells had a shuttle shape with scarce pseudopods. Overall,
Lewis-negative pancreatic cancer cells demonstrated higher
proliferation rate, higher migration ability, lower fucosylation,
lower expression of CA19-9 and higher expression of MUC16 than
Lewis-positive cells. Potential proteoglycan interactions were
identified by LC-MS, such as EGFR, HSPG2, ADAM17, GPC1, ITGA2,
CD40, IL6ST and GGT1. These findings suggest that Lewis-negative
pancreatic cancer is a unique and aggressive subgroup of pancreatic
cancer with special clinical and molecular features.
CA19-9 is the most widely used biomarker in the
management of pancreatic cancer (6,11,15,16).
Some studies have even reported that CA19-9 is not a bystander but
an effector that could promote pancreatic cancer progression
(17-19). CA19-9 activation could lead to the
modification of fibulin-3, which hyperactivates EGFR signaling and
boosts pancreatic cancer development (18). Approximately 5-10% of the
population are Lewis antigen negative and have no or scarce
secretion of CA19-9 (10).
Therefore, it is reasonable to infer that Lewis-negative pancreatic
cancer is associated with lower levels of CA19-9 secretion. CA19-9
is not recommended as a biomarker for Lewis-negative pancreatic
cancer (11). In the present
study, 11.7% of patients with pancreatic cancer were Lewis
negative. However, 24% of Lewis-negative pancreatic cancer patients
had high secretion of CA19-9 (>37 U/ml), which was also been
reported by previous studies (9,11,20).
Therefore, the potential mechanisms should be explored.
Lewis gene plays an important role in the
fucosylation of proteins, which catalyzes the reaction of adding
fucose to the α1-3,4 position (21,22).
Several studies have shown that the Lewis gene is an oncogene that
could accelerate cancer development (21,23).
Silencing of Lewis by shRNA could reduce the expression of Lewis
antigens and therefore decrease the adhesion abilities of cancer
cells to endothelial cells with E-selectin expression (21). Theoretically, Lewis-negative
pancreatic cancer, which has Lewis gene dysfunction and
fucosylation deficiency, is supposed to be an indolent subgroup for
the role of the Lewis gene in boosting cancer development.
Interestingly, in the present study, Lewis-negative pancreatic
cancer was shown to be an aggressive subgroup of pancreatic cancer
with special clinical and molecular features, which may be
explained by the fact that fucosylation is an important biological
process, and fucosylation deficiency affects both cancer
development and human body physiology.
MUC16, also known as CA125, is a membrane bound
mucin that belongs to the glycoprotein family (24). Fucosylation is an essential process
for MUC16 biosynthesis. MUC16 is an important biomarker for the
diagnosis of various types of cancer, such as ovarian and digestive
cancers (11,15,25).
MUC16 could also be applied in the management of pancreatic cancer,
including diagnosis, predicting resectability, monitoring
therapeutic response and follow-up (11). Importantly, several studies have
reported that MUC16 could promote cancer progression (24,26).
A study has shown that MUC16 could mediate cell-cell adhesion by
affecting the E-cadherin/ β-catenin complex (26). In our previous study, MUC16 was
shown to promote pancreatic cancer progression by Foxp3 expression
and tumor-associated Treg enrichment through the activation of the
IL-6-JAK2/STAT3 pathway (24). In
the present study, Lewis-negative pancreatic cancer was shown to
have higher levels of MUC16 secretion than its counterpart. The
molecular mechanism explaining the association of the Lewis gene
and MUC16 biosynthesis and the effect of MUC16 high secretion on
cancer development undoubtedly deserve further research.
CaPan-1 was confirmed by sequencing to be a Lewis
antigen-negative cell line. However, CaPan-1 presented properties
similar to Lewis antigen-positive cell lines, including low
proliferation rate, low migration ability and high level of
fucosylation. These findings indicate that heterogeneity even
exists in the Lewis-negative subgroup.
The present study is restricted by only presenting
clinicopathological and molecular features of Lewis-negative
pancreatic cancer. The potential mechanisms accounting for the
aggressive properties of Lewis-negative pancreatic cancer should be
investigated. In addition, the clinical value of the identification
of Lewis-negative pancreatic cancer in guiding clinical practice
should be further explored. Efforts should also be paid to the
reasons Lewis-negative pancreatic cancers have Lewis antigen
expression. Finally, the reasons for CaPan-1, a Lewis-negative
pancreatic cancer cell line, having characteristics different from
other Lewis-negative pancreatic cancer cell lines should also be
investigated.
Funding
The study was supported by the National Natural
Science Foundation of China (grant nos. 81625016, 81871940 and
81902417), the Shanghai Natural Science Foundation (grant no.
17ZR1406300), the Shanghai Cancer Center Foundation for
Distinguished Young Scholars (grant no. YJJQ201803), and the Fudan
University Personalized Project for 'Double Top' Original Research
(grant no. XM03190633).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
CL, KJ and SD performed the experiments and the
scientific literature search, and contributed to the figures and
the writing of the manuscript. All authors participated in the data
analysis and reviewed the manuscript. GL and XY conceived and
designed the study and wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study protocol was authorized by the Ethics
Committee of the Fudan University Shanghai Cancer Center (Shanghai,
China). Written informed consent was acquired from all of the
patients enrolled. All animal procedures were approved by the
Institutional Animal Care Committee of Fudan University (Shanghai,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ryan DP, Hong TS and Bardeesy N:
Pancreatic adenocarcinoma. N Engl J Med. 371:2140–2141. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Long J, Luo GP, Xiao ZW, Liu ZQ, Guo M,
Liu L, Liu C, Xu J, Gao YT, Zheng Y, et al: Cancer statistics:
Current diagnosis and treatment of pancreatic cancer in Shanghai,
China. Cancer Lett. 346:273–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wallace DR, Spandidos DA, Tsatsakis A,
Schweitzer A, Djordjevic V and Djordjevic AB: Potential interaction
of cadmium chloride with pancreatic mitochondria: Implications for
pancreatic cancer. Int J Mol Med. 44:145–156. 2019.PubMed/NCBI
|
|
5
|
Suzuki K, Takeuchi O, Suzuki Y and
Kitagawa Y: Mechanisms of metformin's anti tumor activity against
gemcitabine resistant pancreatic adenocarcinoma. Int J Oncol.
54:764–772. 2019.
|
|
6
|
Luo G, Liu C, Guo M, Long J, Liu Z, Xiao
Z, Jin K, Cheng H, Lu Y, Ni Q, et al: CA19-9-Low&Lewis(+)
pancreatic cancer: A unique subtype. Cancer Lett. 385:46–50. 2017.
View Article : Google Scholar
|
|
7
|
Robert M, Jarlier M, Gourgou S, Desseigne
F, Ychou M, Bouché O, Juzyna B, Conroy T and Bennouna J:
Retrospective analysis of CA19-9 decrease in patients with
metastatic pancreatic carcinoma treated with FOLFIRINOX or
gemcitabine in a randomized phase III study (ACCORD11/PRODIGE4).
Oncology. 93:367–376. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Humphris JL, Chang DK, Johns AL, Scarlett
CJ, Pajic M, Jones MD, Colvin EK, Nagrial A, Chin VT, Chantrill LA,
et al: NSW Pancreatic Cancer Network: The prognostic and predictive
value of serum CA19.9 in pancreatic cancer. Ann Oncol.
23:1713–1722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Luo G, Fan Z, Cheng H, Jin K, Guo M, Lu Y,
Yang C, Fan K, Huang Q, Long J, et al: New observations on the
utility of CA19-9 as a biomarker in Lewis negative patients with
pancreatic cancer. Pancreatology. 18:971–976. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo M, Luo G, Lu R, Shi W, Cheng H, Lu Y,
Jin K, Yang C, Wang Z, Long J, et al: Distribution of Lewis and
Secretor polymorphisms and corresponding CA19-9 antigen expression
in a Chinese population. FEBS Open Bio. 7:1660–1671. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo G, Liu C, Guo M, Cheng H, Lu Y, Jin K,
Liu L, Long J, Xu J, Lu R, et al: Potential biomarkers in Lewis
negative patients with pancreatic cancer. Ann Surg. 265:800–805.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gao HF, Wang QY, Zhang K, Chen LY, Cheng
CS, Chen H, Meng ZQ, Zhou SM and Chen Z: Overexpressed
N-fucosylation on the cell surface driven by FUT3, 5, and 6
promotes cell motilities in metastatic pancreatic cancer cell
lines. Biochem Biophys Res Commun. 511:482–489. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo G, Guo M, Jin K, Liu Z, Liu C, Cheng
H, Lu Y, Long J, Liu L, Xu J, et al: Optimize CA19-9 in detecting
pancreatic cancer by Lewis and Secretor genotyping. Pancreatology.
16:1057–1062. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wiśniewski JR, Zougman A, Nagaraj N and
Mann M: Universal sample preparation method for proteome analysis.
Nat Methods. 6:359–362. 2009. View Article : Google Scholar
|
|
15
|
Luo G, Xiao Z, Long J, Liu Z, Liu L, Liu
C, Xu J, Ni Q and Yu X: CA125 is superior to CA19-9 in predicting
the resectability of pancreatic cancer. J Gastrointest Surg.
17:2092–2098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Tsai S, George B, Wittmann D, Ritch PS,
Krepline AN, Aldakkak M, Barnes CA, Christians KK, Dua K, Griffin
M, et al: Importance of normalization of CA19-9 levels following
neoadjuvant therapy in patients with localized pancreatic cancer.
Ann Surg. Oct 11–2018.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Takada A, Ohmori K, Yoneda T, Tsuyuoka K,
Hasegawa A, Kiso M and Kannagi R: Contribution of carbohydrate
antigens sialyl Lewis A and sialyl Lewis X to adhesion of human
cancer cells to vascular endothelium. Cancer Res. 53:354–361.
1993.PubMed/NCBI
|
|
18
|
Engle DD, Tiriac H, Rivera KD, Pommier A,
Whalen S, Oni TE, Alagesan B, Lee EJ, Yao MA, Lucito MS, et al: The
glycan CA19-9 promotes pancreatitis and pancreatic cancer in mice.
Science. 364:1156–1162. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Gebauer F, Wicklein D, Stübke K, Nehmann
N, Schmidt A, Salamon J, Peldschus K, Nentwich MF, Adam G,
Tolstonog G, et al: Selectin binding is essential for peritoneal
carcinomatosis in a xenograft model of human pancreatic
adenocarcinoma in pfp–/rag2– mice. Gut. 62:741–750. 2013.
View Article : Google Scholar
|
|
20
|
Hamada E, Taniguchi T, Baba S and Maekawa
M: Investigation of unexpected serum CA19-9 elevation in
Lewis-negative cancer patients. Ann Clin Biochem. 49:266–272. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Padró M, Cobler L, Garrido M and de Bolós
C: Down-regulation of FUT3 and FUT5 by shRNA alters Lewis antigens
expression and reduces the adhesion capacities of gastric cancer
cells. Biochim Biophys Acta. 1810:1141–1149. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Orntoft TF, Vestergaard EM, Holmes E,
Jakobsen JS, Grunnet N, Mortensen M, Johnson P, Bross P, Gregersen
N, Skorstengaard K, et al: Influence of Lewis
alpha1-3/4-L-fucos-yltransferase (FUT3) gene mutations on enzyme
activity, erythrocyte phenotyping, and circulating tumor marker
sialyl-Lewis a levels. J Biol Chem. 271:32260–32268. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Weston BW, Hiller KM, Mayben JP, Manousos
GA, Bendt KM, Liu R and Cusack JC Jr: Expression of human
alpha(1,3)fucosyltransferase antisense sequences inhibits
selectin-mediated adhesion and liver metastasis of colon carcinoma
cells. Cancer Res. 59:2127–2135. 1999.PubMed/NCBI
|
|
24
|
Fan K, Yang C, Fan Z, Huang Q, Zhang Y,
Cheng H, Jin K, Lu Y, Wang Z, Luo G, et al: MUC16 C
terminal-induced secretion of tumor-derived IL-6 contributes to
tumor-associated Treg enrichment in pancreatic cancer. Cancer Lett.
418:167–175. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fleming ND, Cass I, Walsh CS, Karlan BY
and Li AJ: CA125 surveillance increases optimal resectability at
secondary cytoreductive surgery for recurrent epithelial ovarian
cancer. Gynecol Oncol. 121:249–252. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Akita K, Tanaka M, Tanida S, Mori Y, Toda
M and Nakada H: CA125/MUC16 interacts with Src family kinases, and
over-expression of its C-terminal fragment in human epithelial
cancer cells reduces cell-cell adhesion. Eur J Cell Biol.
92:257–263. 2013. View Article : Google Scholar : PubMed/NCBI
|