Introduction
Endometrial cancer (EnC), originates from the
endometrium and thus, is a gynecological malignancy, commonly
observed in developed nations (1-3),
where post-menopausal women account as the most vulnerable
population. Endometrial adenocarcinoma is the most common
clinicopathological type. Currently, treatment for patients with
EnC relies primarily on tumor stage and histopathological type,
while individualized therapies based on genetic potential biology
are uncommon (4-8). Although a large proportion of
patients with EnC have a good prognosis, even following the
implementation of clinically radical surgical interventions and
advanced adjuvant therapy, some patients still have a poor
prognosis, which is ascribed to the recurrence and metastasis of
tumors following initial treatment (9). Thus, an improved understanding of the
regulatory mechanisms associated with EnC progression is imminently
required in order to improve diagnostics and individualized
treatment regimens for patients with EnC.
Tripartite motif-containing 22 (TRIM22) contains a
RING finger, B-box and coiled-coil domains, and is a member of the
tripartite motif (TRIM) family of proteins. It functions as an E3
ubiquitin ligase, and as a transcriptional regulator involved in
various biological processes (10). Furthermore, a previous clinical
study found that TRIM22 functioned as an oncogenic gene that was
highly expressed in non-small cell lung cancer (NSCLC) tissue,
promoting NSCLC progression and conferring a poor prognosis
(11). Conversely, other studies
have demonstrated that TRIM22 expression is downregulated in tumor
tissue compared to adjacent healthy tissue; this decreased
expression of TRIM22 is associated with high-grade malignancy and a
poor prognosis (12-14). Similarly, TRIM22 regulates
multifarious signaling pathways associated with the expression of
different oncogenic and antioncogenic molecules to subsequently
promote or suppress tumor progression (11-13,15).
Furthermore, TRIM22, as a progestin target gene, has been reported
to improve the prognosis and overall treatment efficacy of patients
with EnC (15). Although
tumor-promoting and tumor-suppressing roles have been described for
TRIM22, its role in EnC remains uncharacterized.
The nuclear factor-KB (NF-κB) pathway is a major
pro-inflammatory signaling pathway. Evidence suggests that it plays
a vital role in carcinogenesis, protecting cells against apoptosis
and promoting resistance to various cancer drugs (16). Following the degeneration of
phosphorylated IκBα, NF-κB, which is located in the cytoplasm, can
readily translocate to the nucleus to regulate the expression of
genes associated with proliferation, migration and invasion
(17). TRIM22 has been described
as a crucial anti-inflammatory cytokine with multi-effect functions
(18-20). Moreover, it has been reported that
TRIM22 can regulate anti-inflammatory responses by disrupting
LTR-driven transcription, which is independent of NF-κB. However,
studies have failed to provide evidence of a role for TRIM22 in the
regulation of tumor growth via the NF-κB signaling pathway.
Nevertheless, as inflammation plays a significant role in
tumorigenesis and malignant progression, the NF-κB signaling
pathway is likely related to TRIM22-mediated tumor control. Hence,
a growing need exists for the elucidation of the specific
underlying molecular mechanisms associated with TRIM22-mediated
malignant tumor regulation, particularly as it pertains to EnC.
Herein, it is demonstrated that TRIM22 expression is
decreased in the tumor tissues of patients with EnC, and to be
associated with tumor stage. Furthermore, the present study
comprehensively analyzed the potential mechanisms of TRIM22
downregulation, as well as the regulatory function of TRIM22 in EnC
cell migration, invasion and proliferation both in vitro and
in vivo. In addition, the data of the present study reveal
that TRIM22 is an important prognostic predictor and a promising
therapeutic target for EnC.
Materials and methods
Patient sample analyses
Ethics committee approval was obtained from the
Reproductive Center of Provincial hospital affiliated to Shandong
University [approval no. (2019) Ethics Approval No. 32]. The
experiments were conducted according to the regulations set out by
the Declaration of Helsinki. All samples were collected from the
Provincial Hospital affiliated to Shandong University (Jinan,
China) from November, 2012 to June, 2016. The clinical specimens
were used in the experiments as follows: Eight pairs of tumor
endometrial tissues and non-tumor endometrial tissues (adjacent
normal tissues and normal endometrial tissues) (matched for each
patient) were used for western blot analysis; 25 normal endometrial
tissues and 74 tumor endometrial tissues were used for
immunohistochemistry (IHC). The patient characteristics presented
in Table I. The specimens were
collected after obtaining informed consent from the patients. All
EnC specimens were diagnosed and assessed in accordance with the
International Federation of Gynecology Oncology (FIGO) criteria
(2009). All the EnC samples must have been diagnosed as endometrial
adenocarcinoma and underwent initial surgery. All the normal
endometrial tissues were collected from patients diagnosed as
having uterine fibroids following hysterectomy. None of the
patients had been administered any pre-operative medical
treatments.
 | Table IDistribution, tissue characteristics
and TRIM22 expression in patients with EnC. |
Table I
Distribution, tissue characteristics
and TRIM22 expression in patients with EnC.
|
Characteristics | Female
| Trim22, means ±
SD | P-value |
|---|
| Case (n=74) | Controls
(n=25) | Totals (n=99) |
|---|
| Age (years) | | | | | 0.0003 |
| 11-20 | - | - | - | - | |
| 21-30 | - | 1 | 1 | - | |
| 31-40 | 1 | 2 | 3 | 43.91±17.20 | |
| 41-50 | 12 | 21 | 33 | 38.88±16.01 | |
| 51-60 | 36 | 1 | 37 | 26.17±11.09 | |
| 61-70 | 22 | - | 22 | 24.22±11.93 | |
| 71-80 | 3 | - | 3 | 22.59±11.00 | |
| Median (years) | 57.2 | 44.88 | 51.04 | | |
| Mini-Maxi
(years) | 38-76 | 22-51 | 22-76 | | |
| Histological
grade | | | | | <0.0001 |
| I | 25 | | 25 | 31.12±7.03 | |
| II | 36 | | 36 | 25.20±7.65 | |
| III | 13 | | 13 | 20.95±5.69 | |
| TNM stage | | | | | <0.0001 |
| I | 26 | | 26 | 32.22±5.60 | |
| II | 26 | | 26 | 29.31±5.16 | |
| III | 19 | | 19 | 18.42±4.96 | |
| IV | 3 | | 3 | 9.84±3.35 | |
| Menstrual cycle
phase | | | | | 0.0056 |
| Proliferative
phase | | 15 | 15 | 42.20±12.45 | |
| Secretory
phase | | 10 | 10 | 56.51±10.92 | |
Cells and cell culture
As described in a previous study (21), KLE (GCC-UT0008RT/GCC-UT0008CS,
http://www.taogene.com/emkt.htm#/PcMerchandises?id=f9c8f996-7ca3-473c-bd2e-8d7525340d03&categoryId=6),
Ishikawa (GCC-UT0004RT/ GCC-UT0004CS, http://www.taogene.com/emkt.htm#/PcMerchandises?id=89692be1-e99d-4a6f-a22d-11441b94b506
&categoryId=6) and RL-952 (GCC-UT0006RT/GCC-UT000 6CS,
http://www.taogene.com/emkt.htm#/PcMerchandises?id=4d36c08c-34cd-4ce4-93a3-1cbdf6c3ce3d&categoryId=6)
EnC cell lines were purchased from GeneChem Co., Ltd. Twenty short
tandem repeat loci plus the gender determining locus, Amelogenin,
were amplified using the PowerPlex® 21 System from
Promega Corp., which tested and authenticated these three EnC cell
lines. All cells were cultured in HyClone™ Dulbecco's modified
Eagle's medium:nutrient mixture F-12 (DMEM/F-12) (HyClone; GE
Healthcare Life Sciences) supplemented with 10% heat-inactivated
fetal bovine serum (FBS) (Biological Industries, Israel), 1%
penicillin and streptomycin (P/S) (HyClone; GE Healthcare Life
Sciences) at 37°C in a humidified 5% CO2 atmosphere.
Immunohistochemical analysis
Paraffin-embedded sections (4-µm-thick) were
dewaxed, hydrated, boiled in citric acid buffer for antigen
retrieval, steeped in 0.3% hydrogen peroxide to interdict the
activity of endogenous peroxidase, blocked with 3% bovine serum
albumin and incubated with primary antibodies at 4°C overnight. The
following day, the sections were incubated with secondary
antibodies (rabbit, PV-9001, ZSGB-Bio) at room temperature for 20
min according to the protocol of the PV-9001 Immunohistochemical
kit (ZSGB-BIO). Primary antibodies were used at the following
dilutions: TRIM22 antibody (1:350, NBP1-81795, Novus Biologicals,
LLC), nucleotide binding oligomerization domain containing 2 (NOD2)
antibody (1:250, NB100-524, Novus Biologicals, LLC), Ki-67 antibody
(1:200, RB-9043-P1, eBioscience; Thermo Fisher Scientific, Inc.).
These sections were then stained with hematoxylin in room
temperature for 3 min, and were then cleared, dehydrated,
hyalinized and mounted. Tissues staining brown in the cytoplasm or
nucleus were regarded as positive. Five fields in each section were
selected for further analysis using a fluorescent microscope
(Olympus Corp.). The quantification of protein expression was
presented with integrated optical density (IOD), using Image-Pro
Plus 6.0 software. The final mean optical density was determined
according to the following equation (equation 1): MOD=(IOD
SUM)/(area SUM), where MOD represents the mean optical density, IOD
SUM is the sum IOD of all selected fields in one image and the area
SUM refers to the sum area of all selected fields.
Immunofluorescence
Paraffin-embedded sections (4-µm-thick) were
dewaxed, hydrated, boiled in citric acid buffer for antigen
retrieval, blocked with 3% bovine serum albumin (Servicebio) and
incubated with primary antibodies TRIM22 (1:200, NBP1-81795, Novus
Biologicals, LLC) and NOD2 (1:100, NB100-524, Novus Biologicals,
LLC) at 4°C overnight. The following day, the sections were
incubated with corresponding secondary fluorescent-conjugated
antibodies (1:300, GB21303, rabbit, Servicebio) in the dark and at
room temperature for 45 min. The sections were counterstained with
4',6'-diamidino-2-phenylindole (DAPI) (Servicebio, China) in the
dark and at room temperature for 10 min, and images were acquired
using a confocal laser scanning microscope (Nikon Copr.). The
staining for TRIM22 was red, that for NOD2 was green, and the
nuclei were stained blue.
Lentivirus infection
Ubi-TRIM22-3FLAG-SV40-EGFP- IRES-puromycin
lentiviral vector (GV358) was constructed by GeneChem Co., Ltd. The
primer sequences were as follows: Forward,
5'-GAGGATCCCCGGGTACCGGTCG CCACCATGGATTTCTCAGTAAAGGTAGACATAG-3' and
reverse, 5'-TCCTTGTAGTCCATACCGGAGCTCGGTGGG CACACAGTCATG-3'. The
TRIM22 gene was used from NCBI (NM_006074). According to the
manufacturer's instructions, the lentiviral vector was transfected
into the Ishikawa and KLE cells using Lipofectamine 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) (these cells were
termed TRIM22 OE). The Ubi-3FLAG-SV40-EGFP-IRES-puromycin
lentiviral vector was used as a control (vector control). The
cellular infection rate and GFP-positive cell number were detected
by fluorescence microscopy at 72 h following infection. Stably
transfected clones of TRIM22 were detected by western blot
analysis.
Western blot analysis
Cells were harvested at predetermined times when the
cells spread out in the dish (approximately 80%) and rinsed twice
with PBS. The tumor tissues were harvested from the mice. Cell
sediments and the tumor tissues were treated with SDS lysis buffer
(Beyotime Institute of Biotechnology), and centrifuged at 14,000 ×
g for 10 min at 4°C. Additionally, extracting the cytoplasmic and
nuclear protein was performed according to the instructions of
NE-PER™ Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher
Scientific, Inc.). Protein samples (20-40µg) were electrophoresed
through 10% polyacrylamide gels and transferred onto 0.45 µm PVDF
membranes (Merck Millipore). The membranes were blocked with 5%
skim milk for 1 h at room temperature, cleared thrice with 1X
Tris-buffered saline and Tween-20 (TBST) (15 min/time), incubated
with primary antibodies overnight at 4°C, washed with 1X TBST
thrice and incubated with secondary antibodies for 1 h at room
temperature, and washed thrice with 1X TBST, developed with
Immobilon Western HRP (ECL; Merck Millipore).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ProteinTech Group,
Inc.) and Lamin B1 (Abcam) were used as reference controls.
Secondary antibodies (ZSGB-BIO) were accompanied with IR dyes.
Blots were detected with the ChemiDoc MP Imaging System (Bio-Rad
Laboratories, Inc.). Primary antibodies were used at the following
dilutions: TRIM22 rabbit polyclonal antibody (1:200, NBP1-81795,
Novus Biologicals, LLC), NOD2 antibody (1:1,000, NB100-524, Novus
Biologicals, LLC), IκBα(E130) antibody (1:1,000, ab32518, Abcam),
p-IκBα (Ser36) [EPR6235 (2)]
antibody (1:10,000, ab133462, Abcam), NF-κb p65 (E379) antibody
(1:50,000, ab32536, Abcam), p-p65(Ser536) (EP2294Y) antibody
(1:10,000, ab76302, Abcam), GAPDH (AG0766) antibody (1:10,000,
60004-1-Ig, Proteintech) and LaminB1 antibody (1:5,000, ab16048,
Abcam) and secondary antibodies (rabbit, mouse, ZB-2301, ZB-2305,
ZSGB-Bio) were used at 1:5,000.
EdU incorporation assay
Cells (1×104 cells/well) were seeded in a
96-well plate in triplicate and incubated at room temperature for
24 h. Subsequently, the cells were incubated with medium containing
50 µM 5-ethynyl-2'-deoxyuridine (EdU) for an additional 2 h. The
absolute ethyl alcohol (95% ethyl alcohol) was used to immobilize
the cells at 4°C. Cell proliferation was assessed using a
Cell-Light™ EdU Cell Proliferation Detection kit according to the
manufacturer's instructions. DNA was stained with Hoechst 33342 at
room temperature for 30 min and observed using an inverted
fluorescence microscope (Olympus Corp.). For each EdU test, 5
fields were randomly selected to image at x50 magnification. The
absolute ethyl alcohol can accelerate the GFP to disappear out of
the cells by enhancing the membrane permeability and, as a result,
GFP has minimal influence on the EdU. The number of EdU-positive
cells was determined according to Hoechst nuclear staining and was
reported as a percentage of the total number of cells in each
field.
Transwell migration and invasion
assay
Cell migration assay was performed using a Transwell
chamber (pore size, 0.8 µm; Merck Millipore), while the invasion
assay was performed using a Transwell chamber coated with Matrigel
(BD Biosciences). Endometrial cells (6×104 cells for
migration assay and 4×104 cells for invasion assay) in
100 µl serum-free DMEM-F12 were placed in the Transwell chamber and
700 µl DMEM-F12 containing 20% FBS was added to the lower chamber.
Following incubation for approximately 24 h at 37°C in a humidified
5% CO2 atmosphere, the migrating or invading cells were
fixed with absolute ethyl alcohol and stained with hematoxylin in
room temperature for 15 min. Cells on the upper surface of the
filter were removed by wiping with a small cotton swab. Five fields
of the fixed cells were imaged using a fluorescent microscope
(Olympus Corp.), and cells were counted.
Cell cycle assay
Cell cycle assay was implemented using the
CycletestTM Plus DNA kit (BD Biosciences). According to the
instructions of the kit, cells (1.5×105) were washed
with the buffered solution and resuspended in A and B solutions to
damage the cell membrane structure, after which they were incubated
with solution C [propidium iodide (PI)] for 10 min in the dark on
ice (4°C). Solution A contained trypsin in a spermine
tetrahydrochloride detergent buffer for the enzymatic
disaggregation of the solid tissue fragments and digestion of cell
membranes and cytoskeletons; solution B contained trypsin inhibitor
and ribonuclease A in citrate-stabilizing buffer with spermine
tetrahydrochloride to inhibit the trypsin activity and to digest
the RNA. Cells were counted with a flow cytometer (Bio-Rad
Laboratories, Inc.) and expressed as the percentage of G1, S and G2
phase cells using ModFit LTV4.1.7 software.
Co-immunoprecipitation (Co-IP)
analysis
Cell lysates (approximately 1.5 mg total protein)
were collected from the TRIM22-overexpressing (TRIM22 OE) Ishikawa
cells using ice-cold non-denaturation lysis buffer. co-IP was
performed according to the manufacturer's protocol of the Thermo
Scientific Pierce Co-IP kit (Thermo Fisher Scientific, Inc.).
TRIM22 and NOD2 antibodies were fixed for 2 h with AminoLink Plus
coupling resin at room temperature. The resin was then washed and
incubated with the cell lysates overnight at 4°C. The following
day, the resin was again washed, and elution buffer was used to
elute the protein combined with the resin. A negative control,
harvested from the vector control-transfected Ishikawa cells, were
treated in the same manner as the Co-IP samples, including
incubation with AminoLink Plus coupling resin combined with TRIM22
and NOD2 antibodies (Novus Biologicals, LLC) overnight at 4°C. This
control allowed us to observe whether the binding protein was
increased in the TRIM22 OE groups. Samples were analyzed by western
blot analysis using rabbit polyclonal anti-NOD2 (Novus Biologicals,
LLC) and mouse monoclonal anti-TRIM22 (Novus Biologicals, LLC)
antibodies.
TRIM22-specific shRNA and
NF-κB-p65-specific shRNA plasmids and transient transfection
Plasmids encoding human TRIM22 shRNA, NF-κB-p65
(RelA) shRNA and scramble shRNA were purchased from Shanghai
Genechem Co., Ltd. A human NF-κB-p65 shRNA plasmid, resistant to
puromycin, was selected to knockdown NF-κB-p65 expression (shR
p65); the TRIM22 shRNA plasmid was then used to knockdown the
expression of TRIM22 (shR TRIM22). TRIM22 OE Ishikawa cells
(2×105) and cells expressing relatively high levels of
TRIM22 (TRIM22 RL-952 cells; 2×105) were seeded into
6-well plates and cultured at 37°C in a humidified 5%
CO2 atmosphere overnight. The following day, the cells
were incubated with the transfection medium containing the shRNA
plasmid and Lipofectamine 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.) for 5-7 h, after which the transfection medium
was replaced with normal growth medium for incubation of the cells
for a further 48 h at 37°C in a humidified 5% CO2
atmosphere. The NF-κB-p65 (RelA) shRNA target sequence was:
5'-CTCCATTGCGGACATGGACTT-3'; shRNA non-target sequence:
5'-TTCTCCGAACGTGTCACGT-3'; TRIM22 shRNA target sequence:
5'-CCAGATATAGACCTCAATA-3'; and TRIM22 shRNA non-target sequence:
5'-TTC TCCGAACGTGTCACGT-3'.
In vivo tumor xenograft measurement
Female nude mice were purchased from Beijing Vital
River Laboratory Animal Technology Co., Ltd., ajoint venture
enterprise of Charles River Laboratories. The use of animals was
approved by the Ethics Committee of Reproductive Hospital
Affiliated to Shandong University. Their care was in accordance
with institutional guidelines. A total of 18 BALB/c female nude
mice which were 4-5-weeks old were kept under specific
pathogen-free (SPF) conditions in the Shandong University
Experimental Animal Room. Ishikawa cells infected with lentivirus
(vector control and TRIM22 OE), were harvested and resuspended in
100 µl Matrigel/PBS (1:1 vol/vol; Corning, Inc.) and subcutaneously
injected into the left flank of each mouse (n=9/group) with
1×107 cells/inoculum(day 0). Tumors of nude mice were
observed every 3 days. Tumor volume was expressed as mm3
and measured using a common ruler with the traditional formula as
follows: (Equation 2): V=½ x length (mm) x width2
(mm).
After 27 days (day 27) (tumor volume ≤1,000 mm3),
the nude mice were anesthetized with a 3-5% (v/v) mixture of
isoflurane (Aerrane; isoflurane, Baxter) in synthetic air (200
ml/min), and were then sacrificed by cervical dislocation and the
tumors were harvested. The tumor tissues were collected for use in
western blot analysis and immobilized in 4% paraformaldehyde for
IHC analysis of TRIM22 and Ki-67 expression, and for hematoxylin
and eosin (H&E)-staining.
Statistical analysis
Data under normal distribution are presented as the
means ± SEM. The Student's unpaired t-test (2-tailed) was used for
comparisons between 2 groups. Comparisons between the tumor and the
adjacent tumor samples were analyzed using a paired t-test
(2-tailed), and one-way ANOVA followed by Tukey's post-hoc test was
applied for comparisons among ≥3 groups. Kaplan-Meier estimation
was performed to investigate the survival probability of the
patients with EnC. GraphPad Prism software version 6.0 was used to
visualize the results of the comparisons, as well as the P-values.
A P-value <0.05 was considered to indicate a statistically
significant difference.
Results
Low expression of TR1M22 in human EnC
indicates malignant transformation
IHC staining was usedd to detect TRIM22
expression in endometrial tissues. IHC analysis revealed
cytoplasmic and nuclear TRIM22 staining in both the normal and
tumorous endometrial tissues (Fig. 1A
and B). However, TRIM22 expression was lower in the EnC
specimens compared to the normal endometrial tissue specimens
(Fig. 1A-C). Moreover, TRIM22
expression was higher in the secretory phase compared to the
proliferative phase in normal endometrial tissues (Fig. 1A and C). An association was
observed between the expression of TRIM22 and the clinical stage.
Specifically, TRIM22 expression decreased with the increasing tumor
stage (Fig. 1B and D, and Table I). Thereafter, western blot
analysis was used to detect TRIM22 expression in 8 paired EnC
samples. The results demonstrated that TRIM22 expression was
decreased in the EnC tissues compared with the normal endometrial
tissues and adjacent normal tissues (Fig. 1E).
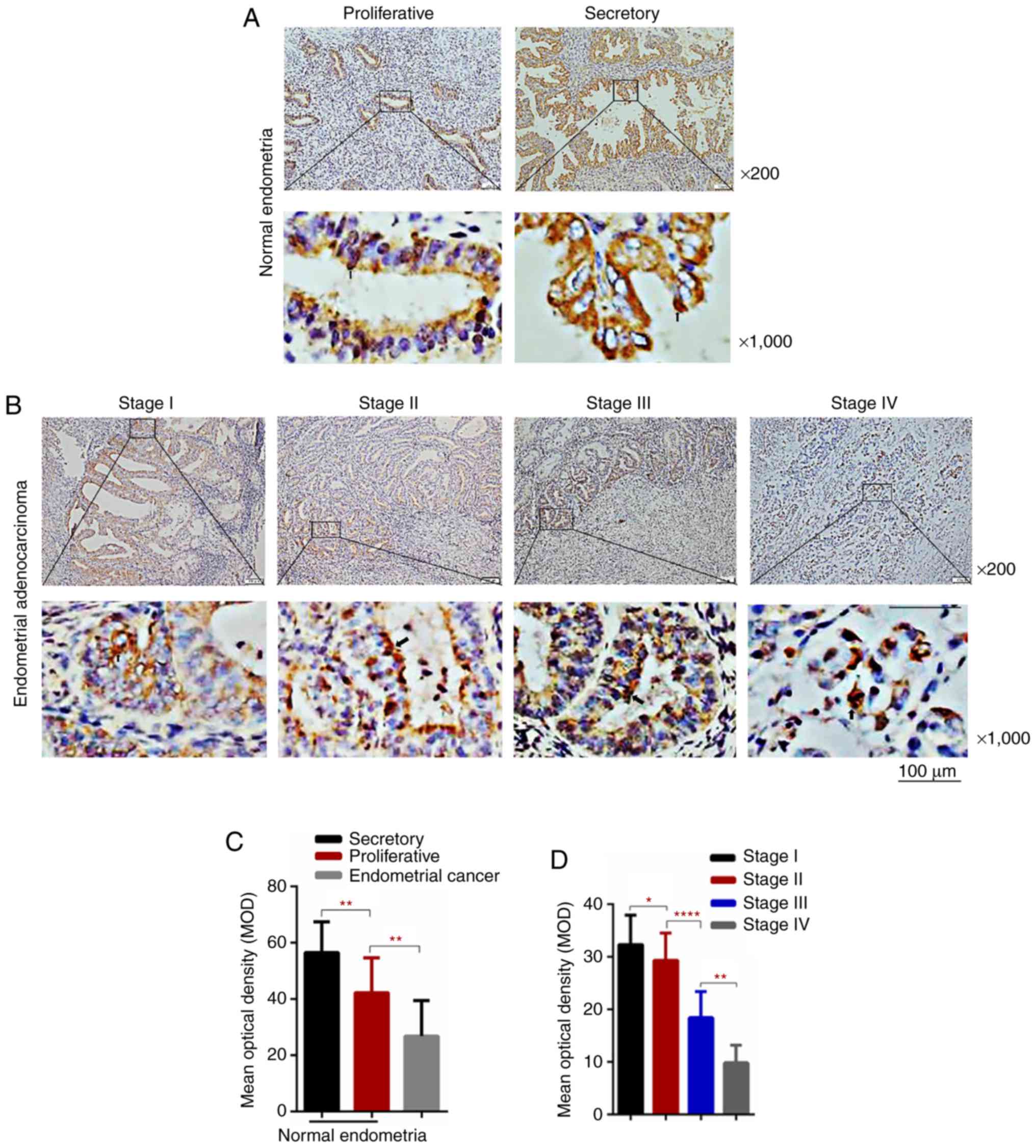 | Figure 1Low TRIM22 levels are associated with
EnC prognosis and a higher overall survival rate. (A and B)
Immunohistochemical assay of TRIM22 expression in normal
endometrial tissues, proliferative and secretory phase,
representative images (magnification, ×200 and ×1,000). TRIM22
expression in EnC tissues (magnification, ×200 and ×1,000). The
cytoplasm and nucleus were stained brown with positive expression.
As observed, TRIM22 levels in EnC were lower than those in normal
endometrial tissue. TRIM22 expression in the secretory phase of
endometrium was higher than that in the proliferative phase. (C)
The IHC MOD of TRIM22 were counted in normal endometrial tissues
and cancerous endometrial tissues. (D) Statistically significant
differences were observed between the different stages of Enc
(stage I, II, III and IV). The figure shows MOD of normal and EnC
tissues stained with TRIM22 for immunohisto- chemistry. Image-pro
Plus and SPSS software were used to measure and analyze each
staining sample. Data are shown as the means ± SD. Scale bar, 100
µm.. Low TRIM22 levels are associated with EnC prognosis and a
higher overall survival rate. (E) TRIM22 protein levels in 8 pairs
of EnC samples were determined by western blot analysis (N, normal
endometrial tissue; A, adjacent normal tissue; T, tumor tissue).
TRIM22 expression and the comparison of TRIM22 expression levels
between EnC specimens and matched adjacent normal tissues in 8
pairs. (F) Kaplan-Meier analysis was used to analyze overall
survival. Overall survival rate of patients with a high TRIM22
expression was significantly higher than that of patients with a
low TRIM22 expression (*P<0.05; **P<0.01; ****P<0.0001).
TRIM22, tripartite motif-containing 22; EnC, endometrial
cancer. |
To further examine the association between the
TRIM22 expression level and EnC malignant conversion, Kaplan-Meier
analysis was applied to analyze the results of IHC MOD and
survival. These results indicated that an increased TRIM22
expression was significantly related to an improved overall
survival of patients with EnC (Fig.
1F). These data support the association between the
downregulated expression of TRIM22 and the malignant transformation
of EnC, while suggesting that TRIM22 may be a promising prognostic
predictor and therapeutic target.
TRIM22 decelerates EnC cell migration,
invasion and proliferation, and inhibits cell cycle
progression
The function of TRIM22 was analyzed in the KLE,
Ishikawa and RL-952 EnC cell lines. Stably transfected TRIM22 OE
Ishikawa and KLE cells were generated which expressed higher levels
of TRIM22; in addition, shR TRIM22 RL-952 cells were generated
which expressed lower levels of TRIM22. Western blot analysis
confirmed that the expression of TRIM22 increased in the TRIM22 OE
Ishikawa and KLE cells, and decreased in the shR TRIM22 RL-952
cells (Fig. S1A and B).
Fluorescence microscopy was used to observe the transfection
efficiency (Fig. S1C and D).
Furthermore, the overexpression of TRIM22 decreased
Ishikawa and KLE cell proliferation, while the opposite effect was
observed in the shRTRIM22 RL-952 cells (Fig. 2A and C). Moreover, the migratory
and invasive abilities of the TRIM22 OE Ishikawa and KLE cells were
markedly restricted; by contrast, shR TRIM22 RL-952 enhanced the
cell migratory and invasive abilities (Fig. 2B and D). Simultaneously, cell cycle
analysis revealed that the number of TRIM22 OE Ishikawa and KLE
cells in the G1 phase increased, while that in the S phase
decreased (Fig. 2E). These results
indicate that TRIM22 suppresses EnC progression.
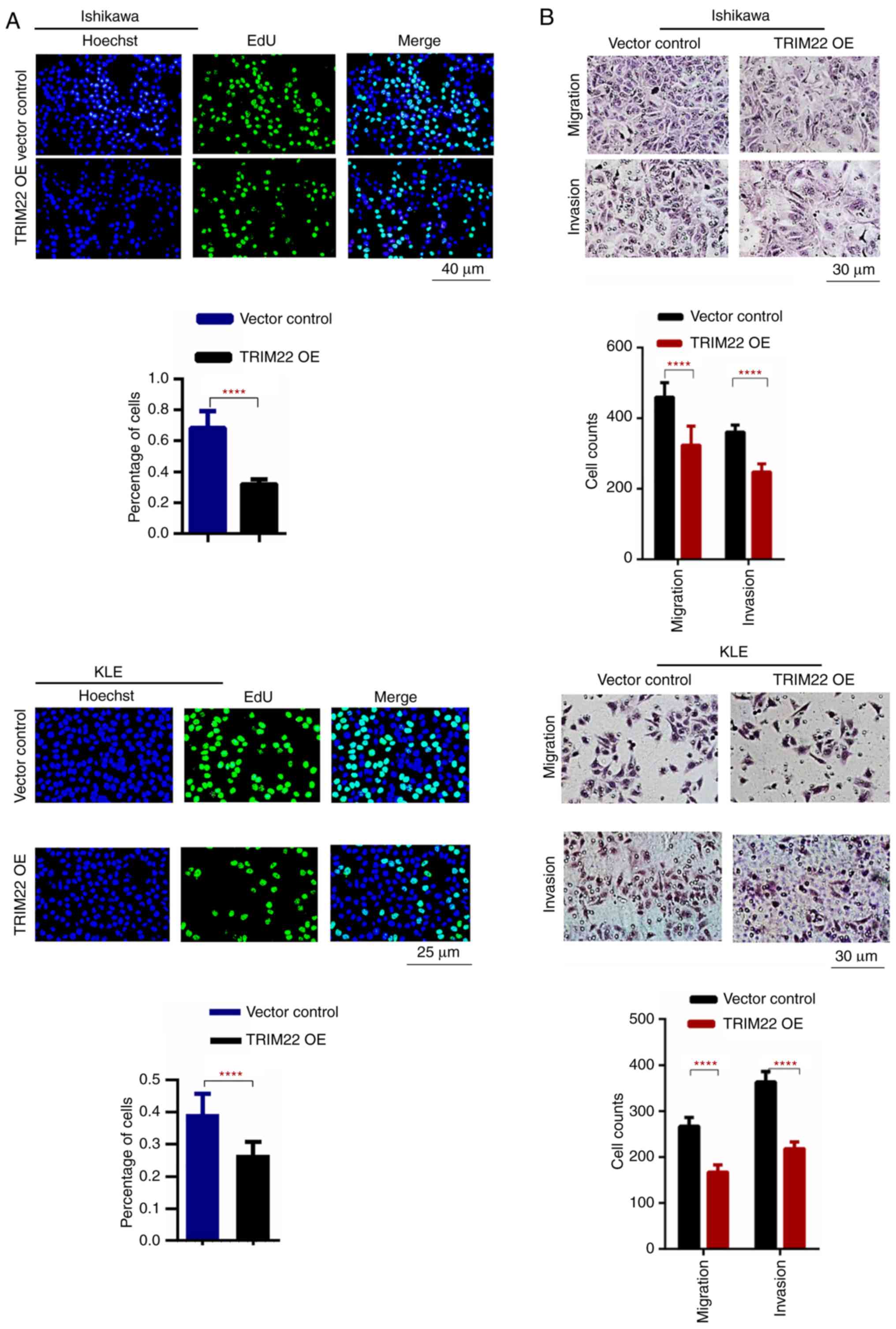 | Figure 2Functional analysis of the effects of
TRIM22 on EnC cell migration, invasion, proliferation and cell
cycle progression. (A) DNA synthesis of Ishikawa and KLE cells was
determined by EdU assay. Green fluorescence indicated that the
cells were EdU-positive. Hoechst staining presenting with blue
fluorescence indicates the total number of cells. Scale bar, 50 µm.
(B) Transwell Matrigel analysis revealed that TRIM22 overexpression
in Ishikawa cells and KLE cells significantly suppressed the
migration and invasion of tumor cells. Scale bar, 100 µm.
Functional analysis of the effects of TRIM22 on EnC cell migration,
invasion, proliferation and cell cycle progression. (C) DNA
synthesis of RL-952 cells was determined by EdU assay. Green
fluorescence indicated that the cells were EdU-positive. Hoechst
staining presenting with blue fluorescence indicates the total
number of cells. Scale bar, 50 µm. (D) Conversely, reducing TRIM22
expression in RL-952 cells improved cell migration and invasion.
Scale bar, 100 µm. Functional analysis of the effects of TRIM22 on
EnC cell migration, invasion, proliferation and cell cycle
progression. (E) Flow cytometry was performed to analyze the PI
staining lentiviral expression vector or lentiviral expression
vector containing TRIM22 cDNA transfected Ishikawa and KLE cells.
Cell cycle analysis indicated that the overexpression of TRIM22 in
Ishikawa and KLE cells led to a higher number of cells in the G1
phase and a lower number in the S phase compared with the control
cells. This confirmed that higher TRIM22 levels decreased EnC cell
growth through G1 cell cycle arrest. Each experiment was conducted
in triplicate (**P<0.01; ***P<0.001; ****P<0.0001).
TRIM22, tripartite motif-containing 22; EnC, endometrial
cancer. |
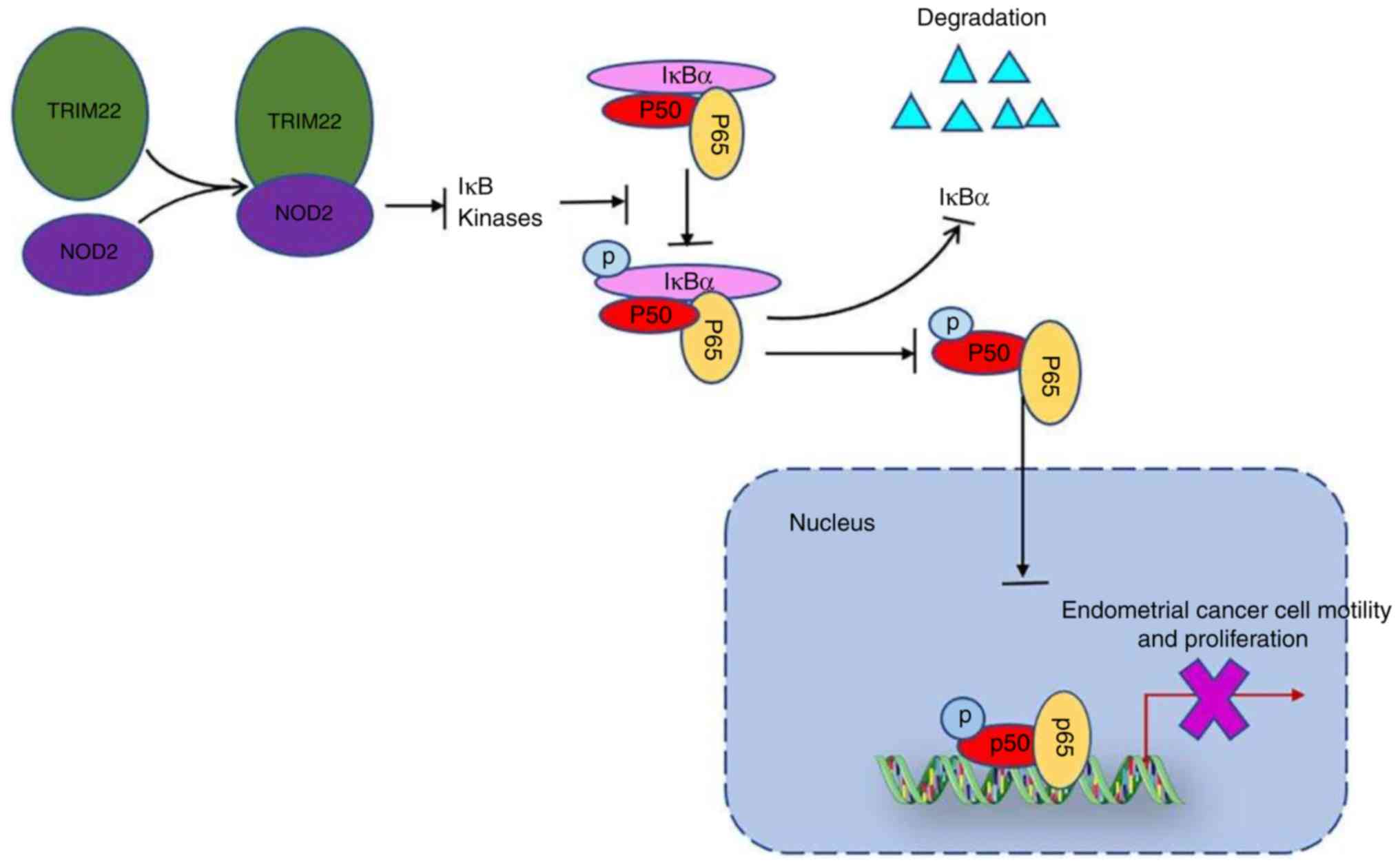
TRIM22-NOD2-NF-κB axis restricts EnC
progression
As an important factor involving multiple aspects of
immunity and inflammation, TRIM22 has been reported to function as
a NOD2-interacting protein, and as a regulator of the NF-κB
signaling pathway, dependent on NOD2 in inflammatory bowel disease
(22). To determine whether the
TRIM22-NOD2 interaction exists in normal endometrial tissues and
endometrial tumor tissues, immunofluorescence and co-IP were
conducted. The results of immunofluorescence assay revealed that
TRIM22 and NOD2 co-localized in both the normal endometrial tissues
and endometrial tumor tissues (Fig.
3A). Furthermore, the expression of NOD2 was decreased in the
tumor tissues, which was consistent with TRIM22 expression
(Fig. 3A). It was also found that
the association between TRIM22 and endogenous NOD2 was weak in the
co-IP experiments in the vector control-transfected Ishikawa cells
and human endometrial tumor cell lines, and this association was
enhanced in the TRIM22 OE Ishikawa cells (Fig. 3B). NOD2 binding to TRIM22 was
further investigated by co-IP. It was confirmed that NOD2 was also
weakly bound to the vector control-transfected Ishikawa cells, and
this association increased in the TRIM22 OE Ishikawa cells
(Fig. 3B). These experiments
demonstrated that TRIM22 directly interacts with NOD2 protein.
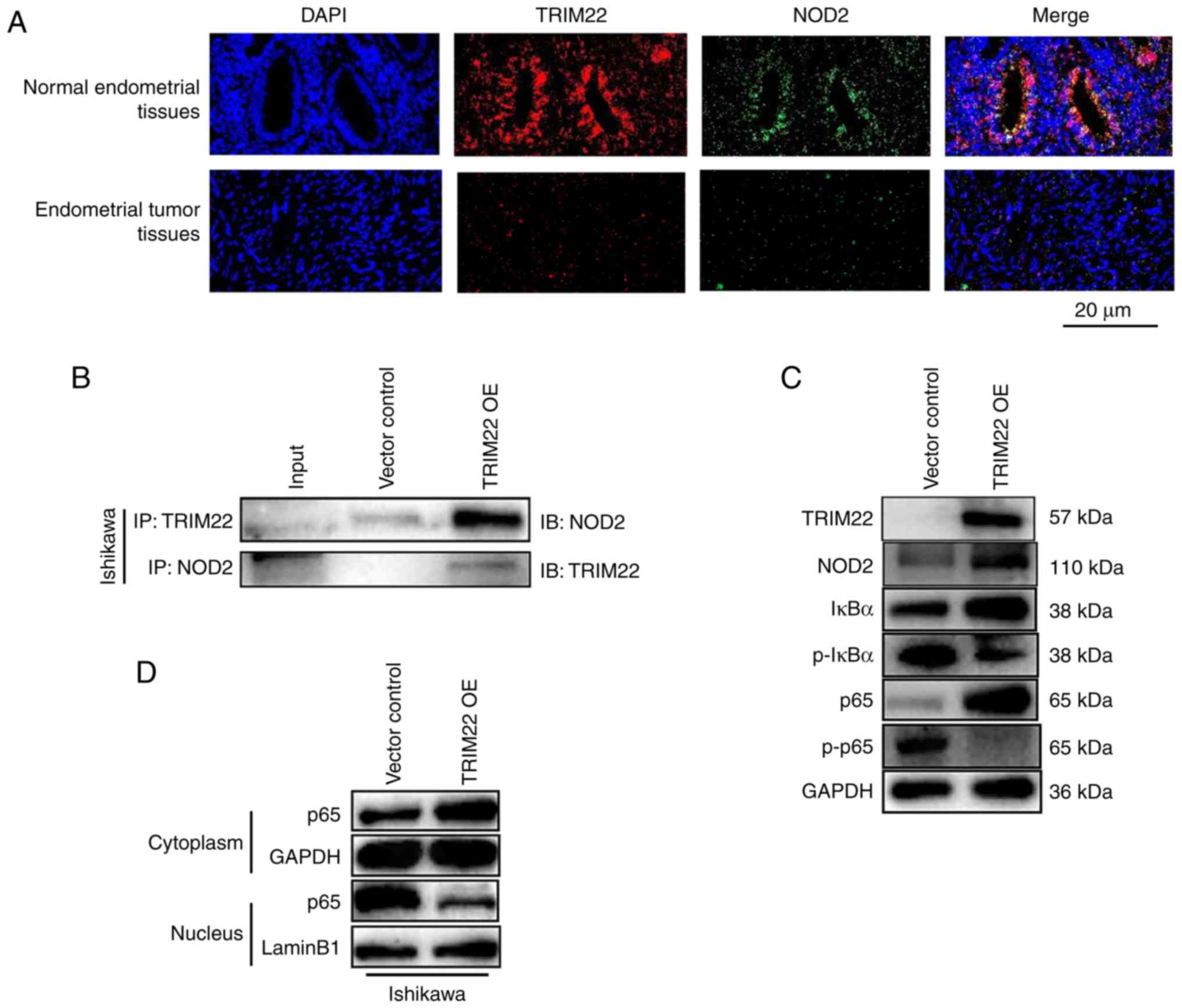 | Figure 3TRIM22 influences EnC cells by
binding NOD2, and suppresses the migration, invasion and
proliferation of EnC cells via the NOD2-NF-κb axis. (A)
Immunofluorescence analysis of TRIM22 and NOD2 in normal
endometrial tissues and endometrial tumor tissues from subjects
undergoing hysterectomy and patients with EnC. TRIM22 and NOD2
colocalization was observed in the cytoplasm of both normal
endometrial tissue and endometrial tumor tissue. Both TRIM22 and
NOD2 colocalization and expression were decreased in EnC tissue.
Scale bar, 20 µm. (B) Ishikawa cells were transfected with
lentiviral construct encoding human TRIM22, and the interaction
between TRIM22 and NOD2 was determined through
co-immunoprecipitation examination. An empty vector construct was
used as IP-negative controls. TRIM22 overexpression construct was
examined. (C) Western blot analysis of IκBα degradation,
phosphorylated Iκbα (Ser 36), p65 and
phosphorylated p65 (Ser 536) in Ishikawa TRIM22 OE cells. (D)
Ishikawa cells were transfected with lentiviral construct encoding
human TRIM22. Western blot analysis was applied to evaluate the
location of p65 in Ishikawa and KLE cells. (E and F) Ishikawa cells
were transfected with lentiviral construct encoding human TRIM22 at
first and followed by transient transfection with
shRNA-NF-κb-p65. Knockdown
NF-κb-p65
attenuated the decrease in the migration, invasion and cell
proliferation which was induced by the overexpression of TRIM22.
The cell proliferative, migratory and invasive abilities of
endometrial tumor cells increased. Scale bars: EdU, 50 µm;
Transwell, 100 µm. ****P<0.0001. TRIM22, tripartite
motif-containing 22; EnC, endometrial cancer; NOD2,
nucleotide-binding oligomerization domain-containing protein 2. |
Since TRIM22 co-localized and co-immunoprecipitated
with NOD2, the question of whether TRIM22 affects the NOD2
signaling pathway was investigated. An alternate study reported
that the role of NOD2 in regulating NF-κB, was bidirectional.
Moreover, NOD2 can activate NF-κB in a number of inflammatory
diseases (23-27) and can inhibit NF-κB in colorectal
tumorigenesis (28-30). The expression of NOD2 and
NF-κB-associated protein was detected in the vector
control-transfected and TRIM22 OE Ishikawa cells, respectively, by
western blot analysis. It was also found that as the expression of
NOD2 increased, that of NF-κB-p65 and IκBα also increased; however,
the phosphorylation of NF-κB-p65 (p-p65) and IκBα (p-IκBα) was
decreased in the TRIM22 OE Ishikawa cells (Fig. 3C). This suggested that TRIM22
induced NOD2 expression and increased the expression of NOD2,
subsequently decreasing NF-κB activation in Ishikawa cells.
Additionally, these results suggest that NOD2 may inhibit NF-κB in
EnC.
Subsequently, the regulatory role of TRIM22 in the
NOD2 signaling pathway was further determined. To identify the
suppressive role of TRIM22 in the translocation of NF-κB-p65,
nuclear and cytoplasmic fractions were prepared from the vector
control-transfected and TRIM22 OE Ishikawa cells. Western blot
analysis revealed that in the cytoplasm, the expression of
NF-κB-p65 in the vector control-transfected cells was lower than
that in the TRIM22 OE cells; however, in the nucleus, the
expression of NF-κB-p65 in the vector control-transfected cells was
higher than that in the TRIM22 OE cells (Fig. 3D). These results suggest that
TRIM22 inhibits NF-κb-p65 from translocating
from the cytoplasm to the nucleus, and thus, it inhibits its
transcriptional regulatory function. Hence, TRIM22 inhibits the
activity of NF-κB-p65. NF-κB-p65 was also knocked down by
transiently transfecting the shRNA-NF-κB-p65 plasmids into TRIM22
OE Ishikawa cells (shR p65). The effect of NF-κB-p65 knockdown was
examined by western blot analysis (Fig. S2). The results of EdU and
Transwell assays revealed that cell proliferation, migration and
invasion of the shR p65 TRIM22 OE Ishikawa cells increased
(Fig. 3E and F). It was thus
suggested that the knockdown of NF-κB-p65 may overcome the reduced
cell proliferation, migration and invasion induced by TRIM22
overexpression. These data indicate that TRIM22 suppresses the
NF-κB signaling pathway by associating with NOD2.
Overexpression of TRIM22 inhibits tumor
growth in vivo
Subsequently, the in vivo effect of TRIM22
was assessed in a BALB/c Ishikawa cell line xenograft model in nude
mice. Nine mice developed palpable tumors at the injection site
following the subcutaneous injection of Ishikawa cells for 7 days.
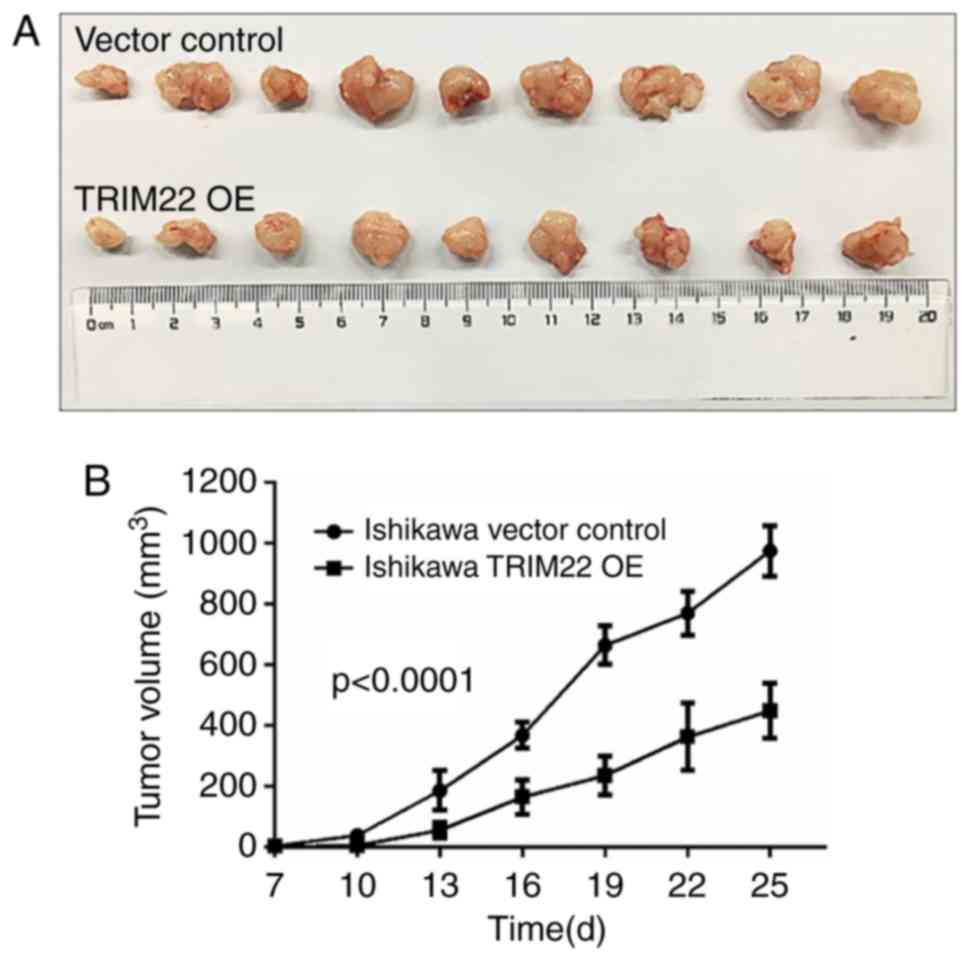
The tumor size was measured twice a week for 4 weeks. TRIM22
overexpression exerted a significant inhibitory effect on tumor
growth by day 25 of the study (Fig.
4). The mean tumor volume of the mice in the TRIM22 OE group
from day 19 was markedly smaller than that of the vector control
group (Fig. 4B).
IHC staining for Ki-67, a cell growth marker, was
used to visualize xenograft tumors size as determined by the tumor
cell proliferation in vivo. The expression of Ki-67 was
significantly decreased in the malignant tumors, which were derived
from the TRIM22 OE Ishikawa cells. The tumor tissues were then
stained with H&E for the observation of the pathological
characteristics in vivo (Fig.
4C).
To validate the regulatory effects of TRIM22 in
vivo, the relevant protein expression in the tumor tissues from
the nude mice were examined. The results demonstrated that the
increased expression of TRIM22 also upregulated IκBα, NF-κB-p65
expression in vivo (Fig.
4D). Taken together, these results indicate that TRIM22
inhibits EnC development and progression by regulating the NF-κB
signaling pathway.
Discussion
The multifactorial nature and complex heterogeneity
of cancer contribute to the high associated global mortality rate
(31). TRIM22 is a member of the
TRIM family of proteins, which have been examined in the context of
a myriad of inflammatory diseases, such as HIV, HBV and HCV
(32-35). However, studies examining the
association between TRIM22 and tumor growth are rare. A previous
study demonstrated that TRIM22 expression was lower in breast
cancer cell lines and tissues compared to in non-malignant mammary
epithelial cell lines and normal breast tissues (12); however, TRIM22 expression has been
found to be increased in NSCLC cell lines (11,36).
Thus, the multi-functional role of TRIM22 in human cancer is rather
complex and may be tissue-specific. Its role in EnC remains
uncharacterized. Herein, it was demonstrated that TRIM22 was
downregulated in both EnC samples (tumor tissues from patients with
EnC) and cancer cell lines. Thereafter, it was demonstrated that a
low expression of TRIM22 in EnC tissues was associated with the
occurrence and a poor prognosis of malignant tumors. Moreover, the
increased expression of TRIM22 inhibited the migration, invasion,
proliferation and cell cycle progression of EnC cells both in
vivo and in vitro. Therefore, it was hypothesized that
TRIM22 overexpression contributes to improved outcomes and
prognoses for patients with EnC, and may thus also be considered as
a promising prognostic factor for EnC.
Tumor formation and development are derived not only
from abnormal gene mutations or disorders in tumor cells, but also
from complex microbial ecosystems that play a vital role in the
formation of systemic innate and inflammatory responses (37). It has been revealed that TRIM
proteins regulate a number of biological processes, including
apoptosis, cell proliferation, innate immunity, autoimmunity,
inflammatory response and tumorigenesis, through different
signaling pathways (20,38). For example, TRIM19 negatively
regulates NF-κB activity by translocating NF-κB to the nucleus
(39). Additionally, TRIM25
regulates RIG-I-mediated antiviral activity (40); TRIM21 has also been described as
playing an important role in regulating specific pro-inflammatory
cytokines by modulating interferon regulatory factors (IRFs)
(41,42); Lastly, TRIM22 has been reported to
accelerate NSCLC progression through the AKT/GSK3β/β-catenin
signaling pathway (11). However,
the regulatory mechanisms employed by TRIM22 in EnC remain unclear.
In the present study, it was demonstrated that TRIM22 inhibits
endometrial cancer progression through the NOD2-NF-κB signaling
pathway. It was also demonstrated that TRIM22 is a NOD2 interacting
protein, and that the overexpression of TRIM22 induces NOD2
expression, subsequently suppressing NF- κB activation.
Mounting evidence suggests that the NF-κB signaling
pathway is involved in the progression of various human tumors,
including those of ovarian cancer (43), prostate cancer (44), cervical cancer (45), as well as head and neck cancer
(46). A recent study demonstrated
that TRIM22 negatively regulates the tumor necrosis factor
receptor-associated factor 6 (TRAF6)-stimulated NF-κB pathway by
binding to the TRIM22 N-terminal RING domain (47). Similarly, it was found that TRIM22
inhibits NF-κB signaling and that the overexpression of TRIM22
inhibits NF-κB-p65
translocation from the cytoplasm to the nucleus. However, the
knockdown of NF-κB-p65 attenuated the effects of
TRIM22 on the cells. However, a previous meta-analysis revealed
that the inflammatory cytokine network is complex, with extensive
interactions. For example, activated NF-κB secretes TNFα, while
TNFα may increase TRIM22 expression, and activate NF-κB (48). A previous study demonstrated that
TRIM22 induced the activation of NF-κB independently of other
transcription factors, such as IRFs, combining with the C-terminal
SPRY domain of TRIM22 (49).
Possibly, the TRIM22 binding site induction of NF-κB activation in
patients with EnC was caused by a deletion mutant. In addition, the
role of TNFα in regulating TRIM22 expression requires further
investigation.
NOD2, the cytosolic NOD-like receptors (NLRs) family
member, functions as a critical player in the regulation of
inflammation (50). NOD2 has been
shown to regulate NF-κB signaling via two distinct pathways. The
first involves NOD2 acting as a sensor of muramyl dipeptide (MDP),
a composition of peptidoglycan present in gram-positive and
gram-negative bacteria; MDP then induces the activation of NF-κB
through NOD2, and thus, NOD2 activates the NF-κB signaling pathway
(51). The second pathway requires
NOD2 to be stimulated by TLR, causing the induction of IRF4, which
subsequently inhibits the activation of NF-κb through interaction with
MyD88 and TRAF6, effectively allowing NOD2 to suppress the NF-κB
signaling pathway (52,53). The present study demonstrated that
the overexpression of TRIM22 induced the expression NOD2, which
subsequently inhibited the activation of NF-κB. However, it remains
unclear as to how NOD2 suppresses NF-κb via TLR and thus, the
specific mechanisms of the TRIM22 and NOD2 interaction warrant
further investigation.
The present study demonstrated that the overall
survival rate of patients with a high TRIM22 expression was
significantly higher compared with that of patients expressing low
levels of TRIM22. TRIM22 expression was associated with the
clinical stage. It was also demonstrated that TRIM22 expression was
higher in the secretory phase than in the proliferative phase,
which suggests that TRIM22 expression may be related to progestin,
and progestin can induce the increased expression of TRIM22, as has
been previously reported (15).
Moreover, progestin is commonly used as an auxiliary treatment for
patients with EnC; TRIM22 has been reported as a future prognostic
predictor in certain types of cancer (11,14).
Although it is commonly considered that the clinical stage has a
minimal association with the biological malignancy of EnC, it was
thus hypothesized that TRIM22 may be a potent prognostic and
diagnostic factor in patients with EnC; however, TRIM22 has been
poorly studied in human tumors and the precise mechanisms between
TRIM22 and progestin remain unclear. Hence, more stringent
selective criteria should be employed to identify the diagnostic
efficiency and true prognostic value of TRIM22. Additionally,
further studies are required to elucidate the mechanisms between
TRIM22 and progestin.
Certain limitations should be noted throughout the
present study. Firstly, the sample size was small. Although the
results of the clinical samples corresponded to TCGA data that has
been previously reported (54),
further studies with larger sample sizes are required in the
future; secondly, the present study only selected three endometrial
cancer cells, Ishikawa, KLE and RL-952 cells, which may limit the
results. Thus, further studies using more EnC cells are warranted
in the future.
In conclusion, the present study demonstrates that
TRIM22 is downregulated in EnC and is associated with clinical
treatment efficacy. Moreover, the present study highlights the
function of TRIM22 in inhibiting EnC cell migration, invasion,
growth and cell cycle progression. It also establishes that TRIM22
directly inhibits NF-κB activity by binding to NOD2 in EnC cells
(Fig. 5). As a result, TRIM22 may
serve as an effective future prognostic indicator, and the
association between progestin and TRIM22 may be a potential basis
for the progestin treatment of patients with EnC.
Supplementary Data
Funding
The present study was supported by the National Key
Research and Development Program of China (grant no.
2018YFC1004801), the Fundamental Research Funds of Shandong
University (grant no. 21520078614063) and the National Natural
Science Foundation of China (grant no. 81571414).
Availability of data and materials
The data associated with the current study are
available from the corresponding author on reasonable request.
Authors' contributions
LipingZ was involved in the investigative aspects of
the study, as well as in the study methodology, data collection,
data analysis, and in the writing of the manuscript. BZ was
involved in data collection and data analysis. MW and ZX designed
part of the study and supervised the study. WK provided resources
and collected the follow-up data of the patients involved. KD was
involved in data collection and providing resources. LinZ was
involved in data analysis. XX and XZ provided resources and
assisted with the animal experiments. LY was involved in project
development, data analysis and in the writing of the manuscript, as
well as in the reviewing and editing of the manuscript. All authors
have read and approved the final manuscript.
Ethics approval and consent to
participate
Ethics Committee approval was obtained from the
Reproductive Center of Provincial Hospital affiliated to Shandong
University. The present study was performed according to the
Declaration of Helsinki. All animal experiments were performed
following the Ethics Committee of Reproductive Center of Provincial
Hospital affiliated to Shandong University. Animal care was in
accordance with institutional guidelines.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgements
The authors would like to express their gratitude to
the Ethics Committee of the Reproductive Center of Provincial
Hospital affiliated to Shandong University for their support.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cancer Genome Atlas Research Network;
Kandoth C, Schultz N, Cherniack AD, AκBani R, Liu Y, Shen H,
Robertson AG, Pashtan I, Shen R, et al: Integrated genomic
characterization of endometrial carcinoma. Nature. 497:67–73. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McAlpine JN, Temkin SM and Mackay HJ:
Endometrial cancer: Not your grandmother's cancer. Cancer.
122:2787–2798. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Weigelt B and Banerjee S: Molecular
targets and targeted therapeutics in endometrial cancer. Curr Opin
Oncol. 24:554–563. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Janzen DM, Rosales MA, Paik DY, Lee DS,
Smith DA, Witte ON, Iruela-Arispe ML and Memarzadeh S: Progesterone
receptor signaling in the microenvironment of endometrial cancer
influences its response to hormonal therapy. Cancer Res.
73:4697–4710. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kim JJ and Chapman-Davis E: Role of
progesterone in endometrial cancer. Semin Reprod Med. 28:81–90.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Banno K, Kisu I, Yanokura M, Tsuji K,
Masuda K, Ueki A, Kobayashi Y, Yamagami W, Nomura H, Susumu N and
Aoki D: Progestin therapy for endometrial cancer: The potential of
fourth-generation progestin (review). Int J Oncol. 40:1755–1762.
2012.PubMed/NCBI
|
|
8
|
Umene K, Banno K, Kisu I, Yanokura M,
Nogami Y, Tsuji K, Masuda K, Ueki A, Kobayashi Y, Yamagami W, et
al: New candidate therapeutic agents for endometrial cancer:
Potential for clinical practice (review). Oncol Rep. 29:855–860.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Morice P, Leary A, Creutzberg C,
Abu-Rustum N and Darai E: Endometrial cancer. Lancet.
387:1094–1108. 2016. View Article : Google Scholar
|
|
10
|
Duan Z, Gao B, Xu W and Xiong S:
Identification of TRIM22 as a RING finger E3 ubiquitin ligase.
Biochem Biophys Res Commun. 374:502–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Liu L, Zhou XM, Yang FF, Miao Y, Yin Y, Hu
XJ, Hou G, Wang QY and Kang J: TRIM22 confers poor prognosis and
promotes epithelial-mesenchymal transition through regulation of
AKT/GSK3β/β-catenin signaling in non-small cell lung cancer.
Oncotarget. 8:62069–62080. 2017.PubMed/NCBI
|
|
12
|
Sun Y, Ho GH, Koong HN, Sivaramakrishnan
G, Ang WT, Koh QM and Lin VC: Down-regulation of tripartite-motif
containing 22 expression in breast cancer is associated with a lack
of p53-mediated induction. Biochem Biophys Res Commun. 441:600–606.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hatakeyama S: TRIM proteins and cancer.
Nat Rev Cancer. 11:792–804. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wittmann S, Wunder C, Zirn B, Furtwangler
R, Wegert J, Graf N and Gessler M: New prognostic markers revealed
by evaluation of genes correlated with clinical parameters in Wilms
tumors. Genes Chromosomes Cancer. 47:386–395. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Saito-Kanatani M, Urano T, Hiroi H,
Momoeda M, Ito M, Fujii T and Inoue S: Identification of TRIM22 as
a progesterone-responsive gene in Ishikawa endometrial cancer
cells. J Steroid Biochem Mol Biol. 154:217–225. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pikarsky E, Porat RM, Stein I, Abramovitch
R, Amit S, Kasem S, Gutkovich-Pyest E, Urieli-Shoval S, Galun E and
Ben-Neriah Y: NF-kappaB functions as a tumour promoter in
inflammation-associated cancer. Nature. 431:461–466. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zandi E, Rothwarf DM, Delhase M, Hayakawa
M and Karin M: The IkappaB kinase complex (IKK) contains two kinase
subunits, IKKalpha and IKκBeta, necessary for IkappaB
phosphorylation and NF-kappaB activation. Cell. 91:243–252. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu H, Pardoll D and Jove R: STATs in
cancer inflammation and immunity: A leading role for STAT3. Nat Rev
Cancer. 9:798–809. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Fan Y, Mao R and Yang J: NF-κb and STAT3
signaling pathways collaboratively link inflammation to cancer.
Protein Cell. 4:176–185. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jefferies C, Wynne C and Higgs R:
Antiviral TRIMs: Friend or foe in autoimmune and autoinflammatory
disease? Nat Rev Immunol. 11:617–625. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong J, Jiao Y, Mu W, Lu B, Wei M, Sun L,
Hu S, Cui B, Liu X, Chen Z and Zhao Y: FκBP51 decreases cell
proliferation and increases progestin sensitivity of human
endometrial adenocarcinomas by inhibiting Akt. Oncotarget.
8:80405–80415. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li Q, Lee CH, Peters LA, Mastropaolo LA,
Thoeni C, Elkadri A, Schwerd T, Zhu J, Zhang B, Zhao Y, et al:
Variants in TRIM22 that affect NOD2 signaling are associated with
very-early-onset inflammatory bowel disease. Gastroenterology.
150:1196–1207. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hugot JP, Chamaillard M, Zouali H, Lesage
S, Cézard JP, Belaiche J, Almer S, Tysk C, O'Morain CA, Gassull M,
et al: Association of NOD2 leucine-rich repeat variants with
susceptibility to Crohn's disease. Nature. 411:599–603. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bonen DK, Ogura Y, Nicolae DL, Inohara N,
Saab L, Tanabe T, Chen FF, Foster SJ, Duerr RH, Brant SR, et al:
Crohn's disease-associated NOD2 variants share a signaling defect
in response to lipopolysaccharide and peptidoglycan.
Gastroenterology. 124:140–146. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Strober W and Watanabe T: NOD2, an
intracellular innate immune sensor involved in host defense and
Crohn's disease. Mucosal Immunol. 4:484–495. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bist P, Cheong WS, Ng A, Dikshit N, Kim
BH, Pulloor NK, Khameneh HJ, Hedl M, Shenoy AR, Balamuralidhar V,
et al: E3 Ubiquitin ligase ZNRF4 negatively regulates NOD2
signalling and induces tolerance to MDP. Nat Commun. 8:158652017.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Pellegrini E, Desfosses A, Wallmann A,
Schulze WM, Rehbein K, Mas P, Signor L, Gaudon S, Zenkeviciute G,
Hons M, et al: RIP2 filament formation is required for NOD2
dependent NF-κb signalling. Nat Commun. 9:40432018. View Article : Google Scholar
|
|
28
|
Watanabe T, Kitani A, Murray PJ, Wakatsuki
Y, Fuss IJ and Strober W: Nucleotide binding oligomerization domain
2 deficiency leads to dysregulated TLR2 signaling and induction of
antigen-specific colitis. Immunity. 25:473–485. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park JH, Kim YG, McDonald C, Kanneganti
TD, Hasegawa M, Body-Malapel M, Inohara N and Nunez G: RICK/RIP2
mediates innate immune responses induced through Nod1 and Nod2 but
not TLRs. J Immunol. 178:2380–2386. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Udden SMN, Peng L, Gan JL, Shelton JM,
Malter JS, Hooper LV and Zaki MH: NOD2 suppresses colorectal
tumorigenesis via downregulation of the tlr pathways. Cell Rep.
19:2756–2770. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Turrini F, Saliu F, Forlani G, Das AT, Van
Lint C, Accolla RS, Berkhout B, Poli G and Vicenzi E:
Interferon-inducible TRIM22 contributes to maintenance of HIV-1
proviral latency in T cell lines. Virus Res. 269:1976312019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Vicenzi E and Poli G: The
interferon-stimulated gene TRIM22: A double-edged sword in HIV-1
infection. Cytokine Growth Factor Rev. 40:40–47. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lim KH, Park ES, Kim DH, Cho KC, Kim KP,
Park YK, Ahn SH, Park SH, Kim KH, Kim CW, et al: Suppression of
interferon-mediated anti-HBV response by single CpG methylation in
the 5'-UTR of TRIM22. Gut. 67:166–178. 2018. View Article : Google Scholar
|
|
35
|
Yang C, Zhao X, Sun D, Yang L, Chong C,
Pan Y, Chi X, Gao Y, Wang M, Shi X, et al: Interferon alpha
(IFNa)-induced TRIM22 interrupts HCV replication by ubiquitinating
NS5A. Cell Mol Immunol. 13:94–102. 2016. View Article : Google Scholar
|
|
36
|
Zhan W, Han T, Zhang C, Xie C, Gan M, Deng
K, Fu M and Wang JB: TRIM59 promotes the proliferation and
migration of non-small cell lung cancer cells by upregulating cell
cycle related proteins. PLoS One. 10:e01425962015. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Clemente JC, Ursell LK, Parfrey LW and
Knight R: The impact of the gut microbiota on human health: An
integrative view. Cell. 148:1258–1270. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Kawai T and Akira S: Regulation of innate
immune signalling pathways by the tripartite motif (TRIM) family
proteins. EMBO Mol Med. 3:513–527. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wu WS, Xu ZX, Hindman WN, Salomoni P,
Pandolfi PP and Chang KS: Promyelocytic leukemia protein sensitizes
tumor necrosis factor alpha-induced apoptosis by inhibiting the
NF-kappaB survival pathway. J Biol Chem. 278:12294–12304. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Gack MU, Shin YC, Joo CH, Urano T, Liang
C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S and Jung JU: TRIM25
RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated
antiviral activity. Nature. 446:916–920. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yang K, Shi HX, Liu XY, Shan YF, Wei B,
Chen S and Wang C: TRIM21 is essential to sustain IFN regulatory
factor 3 activation during antiviral response. J Immunol.
182:3782–3792. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Kong HJ, Anderson DE, Lee CH, Jang MK,
Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC III and
Ozato K: Cutting edge: Autoantigen Ro52 is an interferon inducible
E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression
in macrophages. J Immunol. 179:26–30. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Xu M and Zhang Y: Morin inhibits ovarian
cancer growth through inhibition of NF-κB signaling pathway.
Anticancer Agents Med Chem. 19:2243–2250. 2019. View Article : Google Scholar
|
|
44
|
Lim WK, Chai X, Ghosh S, Ray D, Wang M,
Rasheed SAK and Casey PJ: Ga-13 induces CXC motif chemokine ligand
5 expression in prostate cancer cells by transactivating NF-κB. J
Biol Chem. 294:18192–18206. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Tilborghs S, Corthouts J, Verhoeven Y,
Arias D, Rolfo C, Trinh XB and van Dam PA: The role of nuclear
factor-kappa B signaling in human cervical cancer. Crit Rev Oncol
Hematol. 120:141–150. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Qin X, Yan M, Wang X, Xu Q, Wang X, Zhu X,
Shi J, Li Z, Zhang J and Chen W: Cancer-associated
fibroblast-derived IL-6 promotes head and neck cancer progression
via the osteopontin-NF-kappa B signaling pathway. Theranostics.
8:921–940. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qiu H, Huang F, Xiao H, Sun B and Yang R:
TRIM22 inhibits the TRAF6-stimulated NF-κB pathway by targeting
TAB2 for degradation. Virol Sin. 28:209–215. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Li Q and Verma IM: NF-kappaB regulation in
the immune system. Nat Rev Immunol. 2:725–734. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yu S, Gao B, Duan Z, Xu W and Xiong S:
Identification of tripartite motif-containing 22 (TRIM22) as a
novel NF-κB activator. Biochem Biophys Res Commun. 410:247–251.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Chen GY, Liu M, Wang F, Bertin J and Nunez
G: A functional role for Nlrp6 in intestinal inflammation and
tumorigenesis. J Immunol. 186:7187–7194. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Girardin SE, Boneca IG, Viala J,
Chamaillard M, Labigne A, Thomas G, Philpott DJ and Sansonetti PJ:
Nod2 is a general sensor of peptidoglycan through muramyl dipeptide
(MDP) detection. J Biol Chem. 278:8869–8872. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Watanabe T, Asano N, Meng G, Yamashita K,
Arai Y, Sakurai T, Kudo M, Fuss IJ, Kitani A, Shimosegawa T, et al:
NOD2 down-regulates colonic inflammation by IRF4-mediated
inhibition of K63-linked polyubiquitination of RICK and TRAF6.
Mucosal Immunol. 7:1312–1325. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Watanabe T, Asano N, Murray PJ, Ozato K,
Tailor P, Fuss IJ, Kitani A and Strober W: Muramyl dipeptide
activation of nucleotide-binding oligomerization domain 2 protects
mice from experimental colitis. J Clin Invest. 118:545–559.
2008.PubMed/NCBI
|
|
54
|
Chandrashekar DS, Bashel B, Balasubramanya
SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi BVSK and
Varambally S: UALCAN: A portal for facilitating tumor subgroup gene
expression and survival analyses. Neoplasia. 19:649–658. 2017.
View Article : Google Scholar : PubMed/NCBI
|