Circular RNA (circRNA) is a common class of
non-coding RNAs produced by back splicing, which is an
unconventional splicing event and are characterized as having no 5′
end cap structures or 3′ end poly-adenylation tails (1). Although Sanger et al (2) discovered this special type of RNA
molecule in 1976, scientists generally believed that it was a
product of incorrect splicing and were generally neglected since.
That was not until 2013, when Hansen et al (3) and Memczak et al (4) reported that circRNA served an
endogenous role as a microRNA (miRNA) sponge, that gradually
changed the perception of researchers. In recent years, with
continuous advancements in sequencing technology, an increasing
number of circRNAs have been discovered and subsequently studied.
Accumulating evidence have suggested that circRNA serves a
significant role in the progression of a number of diseases,
including Alzheimer's disease, cardiovascular disease, diabetes and
cancer (3-6). Since circRNA is highly conserved and
is stably expressed in various tissues and bodily fluids, it can be
applied to predict disease and evaluate the effect of diagnosis and
treatment (7). However, the
emerging role of circRNA as a biomarker in cancer is becoming
particularly prominent.
Digestive system malignancies account for a large
proportion of all cancer cases. According to the 2018 Global Cancer
Statistics Report released by the World Health Organization,
colorectal, gastric, hepatocellular and esophageal cancers were
ranked amongst the top 10 in terms of morbidity (8), whilst colorectal, gastric,
hepatocellular, esophageal and pancreatic cancers were ranked
amongst the top 10 in terms of mortality (8). In addition, colorectal and stomach
cancer are ranked amongst the top five in terms of both morbidity
and mortality (8). These data
suggested that digestive system malignancies have become a
significant threat to global health. In recent decades, diagnosis
and treatment of tumors of the digestive system have been markedly
improved, where several novel strategies have been developed.
However, since patients are frequently diagnosed at an advanced
stage, the survival rate of patients with digestive system
malignancies remain unsatisfactory.
In the present review, the biogenesis, function and
role of circRNA in the development of tumors of the digestive
system were summarized, where the possibility and emerging role of
circRNA as a tumor marker in the digestive system was explored,
providing a referencing point for the study of digestive system
malignancies.
CircRNA is a non-coding RNA that is produced by two
possible models of loop generation previously proposed by Jeck
et al (9): i)
Intron-pairing-driven circularization; and ii) lariat-driven
circularization, both of which are widely accepted.
Intron-pairing-driven circularization occurs as a result of
complementary base pairing between different introns in the
sequence, bringing adjacent exons to close proximity, following
which the spliceosome cut away the adjacent exons and paired
introns to form the circRNA. Lariat-driven circularization relies
on the covalent interaction between the splice acceptor and donor,
resulting in a circRNA containing the exon lariat (9). Intron circRNA is another subtype of
circRNA discovered in recent years (10). The 11-nucleotide C-rich element and
7-nucleotide G-rich element in the parent gene of the intron
circRNA combine to form a circular structure which are then spliced
by the spliceosome (10). The
spliceosome mechanism serves a significant role in the biogenesis
of circRNA, which depends on trans-acting factors and
cis-regulatory elements (11).
Zhang et al (12) used four
thiopurines to label newly generated RNA to reveal that low levels
of circRNA may be a by-product of incomplete pre-RNA splicing,
whilst the transcription of RNA polymerase II (pol II) occurred
simultaneously with the formation of circRNA, suggesting that rapid
extension of the strand may promote the reverse splicing of
complementary paired sequences. In addition, the activity of pol II
is strictly controlled by cis-regulatory elements (12). A number of circRNAs can be detected
after pre-mRNA transcription is complete, suggesting that the
biogenic process of circRNA may be post-transcriptional (13). In addition to pre-RNA and pol II,
the biogenesis of circRNA is also regulated by a variety of
proteins, enzymes, intron sequences and active elements. A previous
study showed that RNA-binding proteins can also serve as regulators
of circRNA production, which is a particularly prevalent mechanism
observed during epithelial-mesenchymal transition (EMT) in tumors
(14). The RNA helicase DExH-box
helicase 9 (DHX9) specifically recognizes the reverse repeat Alu
element, which is involved in the regulation of RNA
post-transcriptional splicing (15). Alu elements are a class of
functional sequences that are widely found in primates and are
closely associated with the biogenesis of circRNAs (15). The expression levels of DHX9
affects the formation of splicing products, the deletion of which
has been found to increase circRNA biogenesis of (15). In addition, exon circularization is
dynamic process that is regulated by neighboring introns, where
individual and flanking introns compete to regulate the formation
of circRNA through base pairing (16). Modifications to the mRNA have been
previously demonstrated to regulate transcription, alternative
splicing, formation of advanced structures, translation and
stability. Tang et al (17)
found that N6-methyladenosine (m6A) modification can
promote the generation of circRNA carrying open reading frames in
mouse male germ cells. In addition, the level of other circRNAs in
tissues can also adversely affect the biogenesis of circRNA
(18).
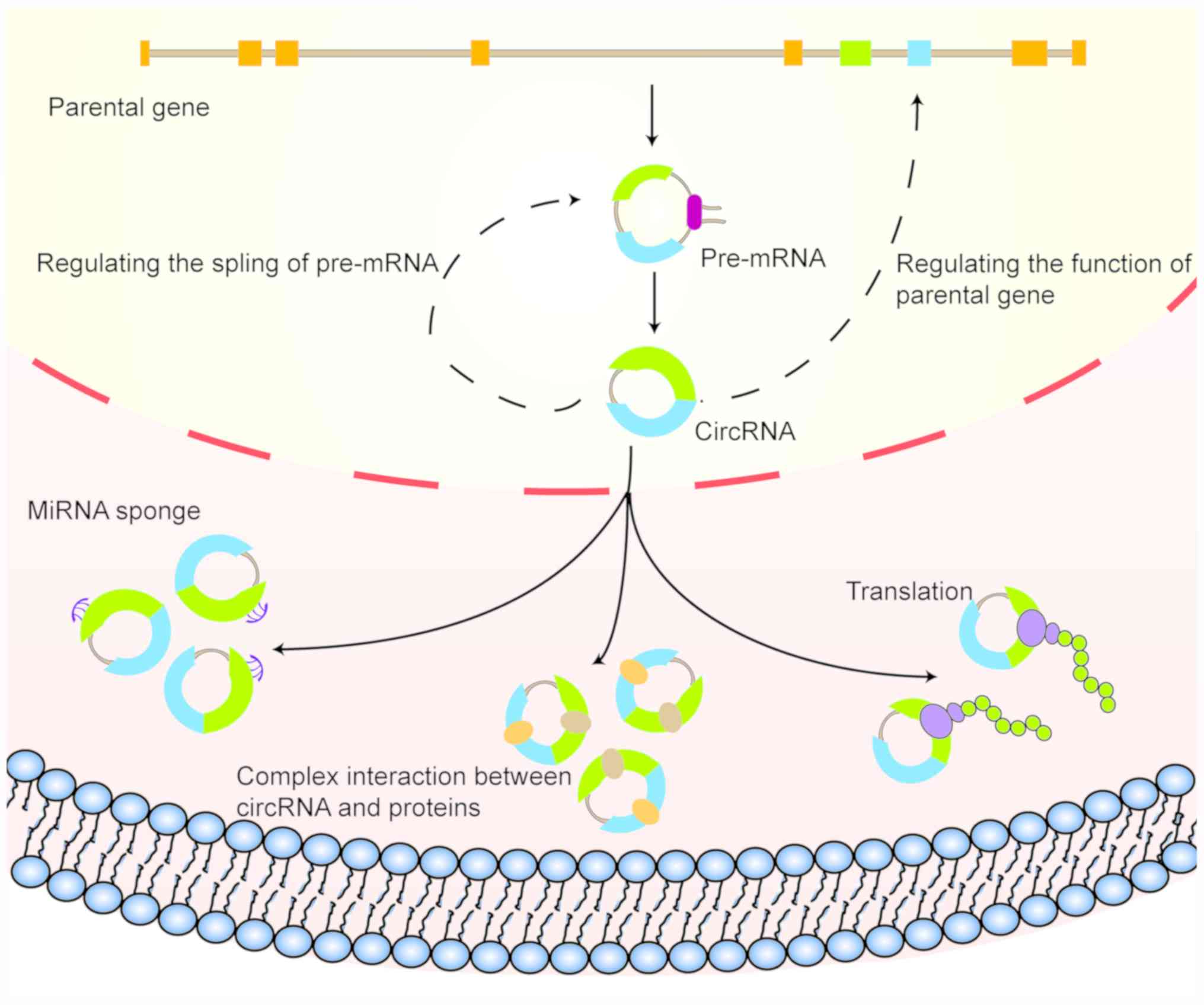
CircRNA can be classified into exon circRNA,
exon-intron circRNA and intron circRNA, according to their origins
(19,20). Exon circRNA is generally more
commonly observed in eukaryotes (19). Compared with corresponding linear
RNA of the same sequence, circRNA is more stable and less
susceptible to degradation by RNase R (21), which is one of the main factors for
circRNA being considered as a potential biomarker.
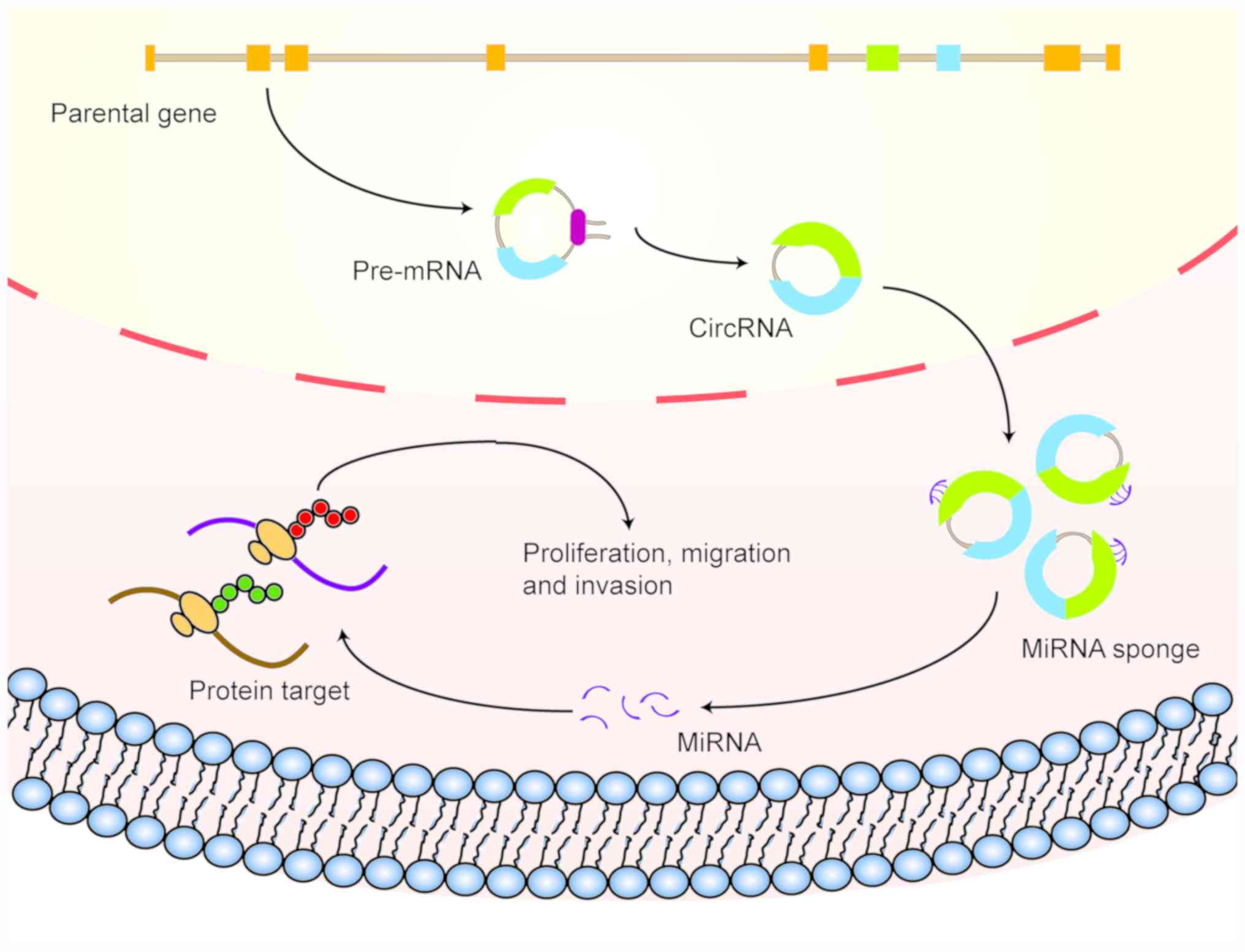
As an important subtype of non-coding RNA, circRNA
has received considerable attention in cancer research in recent
decades, where an increasing number of studies have shown that
circRNA may serve significant roles in a multiple types of
digestive system malignancies. By sponging miRNA, circRNA can
regulate the expression of target proteins, in turn affecting the
pathophysiology of tumors of the digestive system (Fig. 2). This section describes the
differential expression profiles and reported functions of circRNA
in various types of digestive system malignancies.
Esophageal cancer is a tumor of the upper digestive
tract, which ranks 7th and 6th in terms morbidity and mortality
among all cancers, respectively (10). By contrast, gastric cancer ranks
5th and 3rd in terms of morbidity and mortality among all cancers,
respectively (8). Although a
number of advancements have been made in the diagnosis and
treatment of esophageal and gastric cancer in recent years, the
etiology of these two malignancies remain to be fully elucidated
and the survival rates of patients with advanced disease remain
poor. The lack of effective early diagnostic indicators is also an
important cause for the high mortality rates observed in patients
with these two types of malignancies. Accumulating evidence have
shown that circRNA serves a pivotal role in the proliferation,
invasion, migration, cell cycle progression and drug resistance of
esophageal (Table I) and gastric
cancers (Table II).
Since 2016, the number of studies on the
relationship between circRNAs and esophageal cancer has increased
gradually. The elevated expression levels of several circRNAs in
esophageal cancer has been demonstrated to promote the
proliferation of esophageal cancer cells, including circ-discs
large homolog 1, circRNA_100876 and hsa_circ_0067934 (32-34).
In addition to proliferation, aberrant circRNA expression has also
been reported to influence the ability of cancer cells to invade
and migrate, which is a crucial cause of tumor metastasis and
subsequent mortality. Zhong et al (35) found that circ-plasmacytoma variant
translocation (circ-PVT1) upregulated the expression of paired box
proteins and peroxisome proliferator-activated receptors by
sponging miR-4663, resulting in the promotion of esophageal cancer
cell proliferation and migration. The elevated expression levels of
circ-tetratricopeptide repeat domain 17, circ-fibronectin type III
domain containing 3B (circFNDC3B) and hsa_circ_0000337 have also
been revealed to be involved in the proliferation and migration of
esophageal cancer cells (36-38).
Hsa_circ_0006168, circ-protein kinase Cι (circ-PRKCI), c-zinc
finger protein 292 (cZNF292), circRAD23B, circ-ubiquitin associated
protein 2 (circUBAP2) and hsa_circ_0004370 were all found to be
upregulated in esophageal cancer cells and tissues, where they have
the reported function of promoting proliferation, invasion and
migration (39-44). Mechanistically, hsa_circ_0006168,
circ-PRKCI, cZNF292, circRAD23B regulate the PI3K/AKT/mTOR
signaling pathway by sponging miR-100, miR-3680-3p, miR-206 and
miR-5095, respectively; whilst circUBAP2 and hsa_circ_0004370
regulate Rab10 and LIM And SH3 protein 1 by sponging miRNA-422a and
miR-1294, respectively (39-44).
miR-7 is a widely studied miRNA, the expression of which has been
previously found to be reduced as a result of competition from
ciRS7 in esophageal cancer, leading to the activation of NF-κB
signaling, causing changes to the immune micro-environment to
potentiate tumorigenesis (45). In
addition to proliferation, invasion and migration, evidence also
exists demonstrating the role of circRNA in tumor sensitivity to
radiotherapy. He et al (46) previously found that the
down-regulation of circ-vaccinia-related kinase 1 (circVRK1) in
patients with esophageal cancer was associated with a lower
survival rates, whilst the overexpression of circVRK1 could
effectively inhibit EMT and resistance to radiotherapy in
esophageal cancer cells.
Similar to esophageal cancer, research on the
association between circRNA and gastric cancer has been gradually
increasing since 2017. In 2017, studies discovered that
circRNA_100269, circRNA_La ribonucleoprotein 4, hsa_circ_0000705,
hsa_circ_0001895, hsa_circ_00001649, hsa_circ_0000745 and
hsa_circ_0000190 are all significantly downregulated in gastric
cancer, which were found to be closely associated with the
increased cell proliferation, differentiation, subcellular
localization, stage, Borrmann type and invasion (47-53).
By contrast, circ-PVT1 was demonstrated to serve as a proliferative
factor regulating downstream gene expression by sponging miR-125 in
gastric cancer (54). miRNA
sponging remains to be the predominant mechanism through which
circRNA promotes the proliferation, invasion and migration of
gastric cancer cells. By sponging miR-509-3-5p,
circ-myeloid/lymphoid or mixed-lineage leukemia has been found to
be regulated by the expression of GINS4, which binds to and
activates Rac1/cell division cycle to promote cell growth and
metastasis in vivo and in vitro, whilst inhibiting
apoptosis (55). Circ-nuclear
receptor interacting protein 1, circ-cactin, circ-neurofibromatosis
type 1 along with 37 other circRNAs have been previously revealed
to regulate the expression of downstream target genes in gastric
cancer through miRNA sponging to promote the proliferation and
invasion of gastric cancer (Table
II) (56-95). Disorder in fat metabolism is the
main cause of cachexia in patients with advanced gastric cancer and
systemic inflammation (96).
CiRS-133 is a plasma exosomal circRNA that has been found to be
significantly upregulated in gastric cancer (96). Zhang et al (96) found that it can activate PR/SET
domain 16 by inhibiting miR-133, causing the browning of white
adipose tissues in patients with gastric cancer. Other circRNAs can
also affect the invasion and metastasis of gastric cancer by
regulating EMT. The increased expression of circFNDC3B and the
reduced expression of circSMAD7 and circ-hippocampus abundant
transcript 1 (circHIAT1) in gastric cancer has been found to
promote EMT in gastric cancer and induce distant metastasis, which
is the primary cause of poor prognosis in patients with advanced
gastric cancer (97-99). Hsa_circ_0081143 is another circRNA
that is highly expressed in gastric cancer, which is positively
associated with lymph node metastasis and TNM staging in advanced
gastric cancer (100), the
silencing of which can inhibit the development of gastric cancer
and enhance the sensitivity of gastric cancer cells to
chemotherapeutic agents such as cisplatin (100). Similarly, circAKT3 and
circ-fibronectin 1 were found to enhance cisplatin resistance by
sponging miR-198 and miR-182-5p, respectively (101,102).
Pancreatic cancer is a common tumor of the digestive
system. Although the rates of morbidity associated with pancreatic
cancer is not as high as other types of digestive system
malignancies, including gastric, esophageal, colorectal and
hepatocellular cancers, its mortality rate ranks among the highest
(8). Pancreatic cancer is mainly
divided into two subtypes: Pancreatic ductal adenocarcinoma and
pancreatic squamous cell carcinoma, both of which have a poor
prognosis, with a 5-year survival rate of <10% (103). Identifying specific biomarkers
and therapeutic targets is important for improving the survival
rates of patients with pancreatic cancer.
Interestingly, the majority of circRNAs associated
with pancreatic cancer that have been discovered are found to be
upregulated (Table III).
CircRNAs, including hsa_circRNA_0007334, circ-Ras homolog family
member T1, hsa_circ_0006215, circ-zinc finger MYM-type containing
2, circ-disintegrin and metalloproteinase domain-containing protein
(ADAM) 9 and ciRS-7, compete with miRNAs to promote pancreatic
cancer proliferation and inhibit apoptosis (104-109). Other circRNAs can regulate
endothelial monolayer permeability, thereby promoting the invasion
and migration of pancreatic cancer cells. Circ-low density
lipoprotein receptor class a domain containing 3 promotes lymphatic
and venous metastasis by sponging miR-137-3p, whilst
circ-isoleucyl-tRNA synthetase enhances endothelial monolayer
permeability by sponging miR-122 (110,111). Abnormalities in the immune
microenvironment are common changes observed in advanced pancreatic
cancer. Circ_0000977 has been previously found to increase the
expression of hypoxia-inducible factor 1 and ADAM10 by
competitively binding to miR-153, where circ_0000977 silencing can
significantly enhance the cytotoxic effects of natural killer cells
on pancreatic cancer (112). Drug
resistance is another important underlying cause of poor prognosis
in patients with advanced pancreatic cancer. Liu et al
(113) previously found that
knocking down circ-homeodomain interacting protein kinase 3
expression can effectively improve the sensitivity of pancreatic
cancer to gemcitabine. In general, there have been a number of
studies on the relationship between pancreatic cancer and circRNA,
where the role of circRNA in the development of pancreatic cancer
require further exploration.
As of 2018, liver cancer ranks 6th in terms of
prevalence and 4th in terms of mortality rates among other cancers
(8). Liver cancer can be divided
into three main subtypes: Hepatocellular carcinoma, bile duct
carcinoma and mixed carcinoma by cell type, of which hepatocellular
carcinoma is the most frequently observed. Viral hepatitis B (HBV)
is one of the main causes of liver cancer in China whereas
alcoholic fatty liver disease is the main cause in the Western
world (114). Hepatocellular
carcinoma as a result of both HBV infection and excessive alcohol
consumption are associated with high rates of morbidity and
mortality (8). Due to the lack of
symptoms in the early developmental stage of hepatocellular
carcinoma, many are not diagnosed until advanced stages of the
disease (115). Studying the
molecular mechanism of circRNA in hepatocellular carcinoma is of
great importance for the early diagnosis and treatment of this
disease.
The association between circRNA and cancer,
especially that of hepatocellular carcinoma, has become a popular
topic of research over the last 2 years. In early 2020, a total of
56 related research articles reporting the relationship between
circRNA and hepatocellular carcinoma have been published, which is
higher than the number recorded in previous years (Table IV) (137-192). The expression of several circRNAs
has been reported to be dysregulated in hepatocellular carcinoma,
which compete with miRNAs to regulate the expression of downstream
target proteins and affect the proliferation, invasion and
migration of hepatocellular carcinoma. Some of these circRNAs
include hsa_circ_0101432, circ-PVT1, circ-tripartite motif
containing 33-12, circRNA-104718, circ-SET domain containing 3,
actin histidine methyltransferase, has_circ_0078710,
hsa_circ_0064428, circHIAT1, circ-adducin3, hsa_circ_0001649,
circ-ADAM metallopeptidase with thrombospondin type 1 motif 14,
circ_0008450, hsa_circRNA_103809, circ-homer scaffold protein 1,
hsa_circ_0003645, circ-SMG1.72, hsa_circ_0091581, hsa_
circ_0000092, hsa_circ_0056836, circ-TCF4.85, circ-protein arginine
methyltransferase 5 (circPRMT5), circ_0001955, circ-pecanex 1,
circRNA-5692, circ-ATP binding cassette subfamily B member 10 and
circ-ArfGAP with SH3 domain, ankyrin repeat and PH Domain 1
(137,138,140,145,147-150, 155-157,163,166,176-180,183-185,187,189-192). Previous studies have demonstrated
that circRNA in exosomes can regulate the development of
hepatocellular carcinoma by deubiquitylation. Circ-leptin receptor,
circ-matrix metallopeptidase 2 and hsa_circ_0051443 are common
exosomal circRNAs found in hepatocellular carcinoma (142,181,186). Hypoxia is a principal
characteristic of the tumor microenvironment that is considered to
be an important factor affecting sensitivity to radiotherapy and
chemotherapy (146). Yang et
al (146) previously found
that knocking down cZNF292 expression can inhibit angiogenesis,
cell proliferation and resistance to radiotherapy in hepatocellular
carcinoma, which may be due to the hypoxic environment induced by
cZNF292. Similar to cZNF292, the dysregulation of circRNA_101505,
circRNA_104797 and circ_0003418 expression in hepatocellular
carcinoma has been demonstrated to increase the resistance of
hepatocytes to sorafenib and cisplatin, which is considered to be
one of the main underlying causes of poor prognosis in patients
with advanced hepatocellular carcinoma (162,168,174). CircRNA can also directly bind to
protein components of important signaling pathways, thereby
affecting the physiological behavior of hepatocellular carcinoma.
The expression of hsa_circ_0079929 and circ-insulin-like growth
factor 1 receptor in hepatocellular carcinoma was demonstrated to
be significantly reduced, which induced cell cycle arrest and
inhibited apoptosis through the PI3K/AKT/mTOR signal transduction
pathway (139,170). Circ-BTB domain and CNC homolog 1
was found to be highly expressed in hepatocellular carcinoma, which
binds to the embryonic lethal, abnormal vision, drosophila,
homolog-like 1 protein and p27 to regulate cell cycle progression
and promotes the growth of tumor cells (182). Ma et al (161) previously found that circARSP91
enhance innate immune monitoring by enhancing NK cell cytotoxicity
and its downregulation in hepatocellular carcinoma promotes immune
escape of hepatocellular cancer cells.
In conclusion, circRNA serves a significant role in
proliferation, invasion, migration, differentiation, apoptosis,
cell cycle and sensitivity to chemotherapy in hepatocellular
carcinoma. Future research should focus on identifying specific and
sensitive circRNAs that can serve as biomarkers for the early
diagnosis and effective treatment of hepatocellular carcinoma, to
improve the survival rate of patients with hepatocellular
carcinoma.
Compared with hepatocellular carcinoma, although the
rates of morbidity associated with gallbladder cancer is not as
high, the rates of mortality are high where its prognosis poor, due
to the highly invasive properties which is frequently diagnosed
during the advanced stages of the disease (193). Studies have found that circRNA
promotes the proliferation of gallbladder cancer cells through
miRNA sponging and direct binding to proteins. As a miRNA sponge,
circ-forkhead box protein P1 regulates the expression of pyruvate
kinase L/R by sponging miR-370, enhancing the Warburg effect and
promoting the proliferation and invasion of gallbladder cancer
cells (194). In addition,
circ-homeodomain interacting protein kinase 3 has been previously
found to increase the expression of Rho-associated coiled-coil
containing protein kinase 1-CDK6 by sponging miR-124, promoting the
proliferation of gallbladder cancer cells (195). Circ-Erb-B2 binds to the
circErb-B2 receptor tyrosine kinase 2 located in the nucleus, which
regulates proliferation-associated 2G4-dependent ribosomal DNA
transcription and promotes the proliferation of gallbladder cancer
(196). In conclusion, research
on the mechanism of circRNA involvement in the occurrence and
development of gallbladder cancer remains limited (Table V) and several important issues have
yet to be explained. Therefore, there remains room for exploration
in this particular field.
Colorectal cancer ranks 3rd and 2nd worldwide in
terms of morbidity and mortality, respectively, among other types
of cancers. (8). Advancements in
colonoscopy have facilitated the detection and treatment of
precancerous colorectal polyps. However, the mortality and risk
associated with colorectal cancer have changed little since 1997
(197). At present, there is an
urgent need in identifying novel tumor biomarkers and targeted
treatment strategies to improve the early diagnosis and survival
rate of patients with colorectal cancer.
Studies into the relationship between colorectal
cancer and circRNA preceded those of other digestive system
malignancies. In 2015, a study on 31 clinical samples of colorectal
cancer found that the elevated expression of hsa_circ_001988 in
colorectal cancer is closely associated with the differentiation
and infiltration of tumor cells (198). In 2016, circRNA_001569 and
hsa_circ_0000069 were found to be highly expressed in patients with
colorectal cancer (199,200). CircRNA_001569 was demonstrated to
regulate the expression of Bcl-2-associated athanogene 4,
transcription factor E2F5 and formin like 2 by adsorbing miR-145,
though the mechanism mediated by hsa_circ_0000069 remain unknown
(199,200). In 2017, it was found that the
expression of circRNA0003906, circ-BTG3-associated nuclear protein
and hsa_circ_0020397 were dysregulated in colorectal cancer
(201-203). A total of 10 studies reporting
that circRNA regulated the development of colorectal cancer were
published in 2018 (Table VI)
(204-213), which rose to 17 in 2019 and 9
related articles have already been published in the first 2 months
of 2020 (Table VI) (214-239). Over the last 5 years, the number
of studies into the role of circRNA in colorectal cancer has been
increasing annually. Most aberrantly expressed circRNAs in
colorectal cancer exert their function by adsorbing miRNAs,
regulating proteins associated with proliferation, invasion and
metastasis, thereby promoting the progression of colorectal cancer.
Particular circRNAs of interest include hsa_circ_0136666,
circCBL.11, hsa_circRNA_102958, circ-vesicle-associated membrane
protein-associated protein A, circ-formin 2, circ_0021977,
circ_0026344, circ-integrin subunit-α7, has_circ_0055625,
circ-calmodulin regulated spectrin associated protein 1,
hsa_circ_0007142, circ-HECT, UBA and WWE domain containing E3
ubiquitin protein ligase 1, circ-chaperonin containing TCP1 subunit
3, circ_0000218, hsa_circ_0001178, circPRMT5 and hsa_circ_0004277
(215- 220,223,226,227,229-234,236,238). CircRNAs have been reported to
serve either a synergistic or antagonistic role by directly binding
to downstream target proteins to regulate various aspects of
colorectal cancer pathophysiology. Circ-catenin b1 (circ-CTNNB1)
mainly exists in the nucleus and directly binds to DEAD-box
helicase 3/yin and yang 1, resulting in b-catenin activation and
promoting the growth and metastasis of colorectal cancer (225). CircRNA_104916, circ-zinc finger
protein 609 and circ-protein tyrosine kinase 2 have been previously
demonstrated to function in a similar manner to circ-CTNNB1
(221,228,237). Exosome circRNA has been a topic
of particular research interest in recent years, as it is believed
that circRNA may regulate tumor development. Using nanoparticle
tracking analysis and transmission electron microscopy, Pan et
al (222) verified the
exosomes in the peripheral blood of patients with colorectal
cancer. Hsa_circ_0004771, ciRS-122 and circ-ATP binding cassette
subfamily C Member 1 were found to be significantly increased in
these exosomes, suggesting their possible application as a suitable
biomarker for this disease (222,235,239).
A gastrointestinal stromal tumor is a non-epithelial
type of tumor that can occur in the esophagus, stomach, small
intestine, colorectum and abdominal cavity that can cause a variety
of symptoms, including pain, bleeding and abdominal discomfort
(240). Surgical resection is
currently the most common treatment method (240). In a recent study, Jia et
al (241) used a selective
binding complementary competitive endogenous RNA array to analyze
gastrointestinal stromal tumors, which found circ_0084097,
circ_0069765 and circ_0079471 to be aberrantly expressed in
gastrointestinal stromal tumors. Although further study suggested
these circRNAs to serve a regulatory role by sponging mir-144-3p,
mir-142-5-p and mir-485-3p (241), there is no experimental data on
the mechanism of circRNA involved in the development of
gastrointestinal stromal tumors. It remains unclear if circRNA can
be applied as a biomarker and therapeutic target for the diagnosis
of gastrointestinal stromal tumors at present.
Non-coding RNA is comprised of a large family of
different subtypes of RNA molecules. In addition to circRNA, it
also includes miRNA, long non-coding RNA (lncRNA), piwi-interacting
RNA, transfer RNA, ribosomal RNA and small interfering RNA. Over
the past decade, numerous studies have demonstrated that non-coding
RNAs can exert regulatory effects. They serve an irreplaceable role
in transcription, post-transcriptional modification, protein
remodeling and cell signal transduction (242). Supporting this, non-coding RNA
dysfunction has been documented to be associated with the
occurrence and progression of various chronic diseases and tumors
(242).
miRNA is a family of single-stranded, non-coding RNA
that are typically 21-25 nucleotides in length and negatively
regulates >60% of coding genes (243). Most miRNAs contain a 2-7
nucleotide seed sequence at the 3′untranslated region (3′-UTR) end,
which can conservatively bind to a variety of target protein-coding
genes to inhibit target mRNA translation (243). The function of miRNA itself is
under negative regulation by circRNA- or lncRNA-mediated sponging
(243). In a previous study,
miR-7 was found to inhibit osteosar-coma cell proliferation and
migration, but circRNA-CDR1as promotes osteosarcoma growth by
sponging miR-7 (244) Similar
cancer-promoting effects as a result of reduced miR-7 expression
have also been reported in gastric cancer, colorectal cancer,
pancreatic cancer and hepatocellular carcinoma (109,206,245,246). However, it should be noted that
not all miRNAs have a seed sequence at the 3′UTR end. In addition
to directly inhibiting mRNA translation, a number of miRNAs can
also participate in pre-transcriptional regulation by binding to
the argonaute-2 protein in the nucleus (247). However, this phenomenon is yet to
be observed in tumors of digestive system.
LncRNA is a type of non-coding RNA that are >200
nucleotides in length, which has been previously shown to serve a
significant role in maintaining the tumor microenvironment and the
progression of various digestive system malignancies (248). Similar to circRNA, many lncRNAs
also have the ability to sponge miRNAs. Yuan et al (248) found that the lncRNA SWI/SNF
related, matrix associated, actin dependent regulator of chromatin
subfamily C member 2 upregulates transmembrane serine protease 2 by
binding to miR-551b-3p, promoting the proliferation and invasion of
gastric cancer. LncRNA can also directly interact with
double-stranded RNA to regulate the activity of some proteins
(249). A previous study has
found that LINC00665 can activate and enhance the expression of
protein kinase R by protecting it from ubiq-uitin-dependent
degradation, in turn upregulating the NF-κB signaling pathway and
the widespread expression of inflammatory factors, resulting in
changes to the microenvironment and the occurrence of
hepatocellular carcinoma (249).
In short, non-coding RNA forms a complex network of
interactions in the body. where dysfunctions in any one of the
nodes will significantly impact the entire non-coding RNA network,
resulting in the development of diseases. The occurrence and
procession of digestive system malignancies are closely associated
with the aberrant expression of these non-coding RNAs. CircRNA is
an important part of the non-coding RNA family, where its function
in tumors has garnered attention in recent years. The present study
reviewed the research status of circRNA in seven types of tumors in
the digestive system, where the potential mechanism underlying
circRNA function in their respective malignancies were discussed in
detail and the possibility of using circRNA as a diagnostic and
prognostic marker for tumors in the digestive system was
demonstrated. However, this article is associated with limitations,
since it did not discuss in detail the role of circRNAs in cellular
and signal transduction pathways. CircRNA has a complex
intracellular mechanism of action, which is regulated by a variety
of transcriptional regulatory factors and can also affect
downstream biological functions by direct interaction with miRNAs
and proteins. In addition, circRNA can exert biological functions
in the nucleus, cytoplasm or through exosomes secreted into bodily
fluids. The associated in-depth mechanism in digestive system
malignancies remain to be fully elucidated and require further
investigation in future studies.
The present review briefly explored the biogenesis
and function of circRNA and reviewed in detail the current status
of circRNA research in various types of digestive system
malignancies. CircRNA is a class of endogenous non-coding RNA that
is evolutionarily conserved, the expression of which was found to
be dysregulated in a variety of digestive system tumors. In
digestive system malignancies, the majority of circRNAs serve an
endogenous competitive role through miRNA sponging, whilst a
selective number of circRNAs can directly bind to proteins either
as sponges or synergistic factors to regulate tumor growth,
invasion and migration, in turn influencing patient prognosis.
Compared with traditional linear RNA and other families of RNA
molecules, including long-chain non-coding RNA and miRNA, circRNA
is widely expressed in various tissues, cells and bodily fluids,
where its expression is stable, increasing their potential as
biomarkers and targets for therapy. With continuous advancements in
experimental technology and continuing research, it is hoped that
novel functions and modes of action mediated by circRNA will be
discovered and clarified in the future. With the joint efforts of
researchers and clinicians, the pathogenesis of various malignant
tumors of the digestive system will be elucidated, so that patients
will have an increased chance of survival.
The present study was supported by the National
Natural Science Foundation of China (grant nos. 71964021 and
81570783), National Key R&D Program of China (grant nos.
2016YFC1302201 and 2016YFC0107006), Key Research and Development
Program of Gansu Province (grant no. 18YF1FA110), the Key Program
of the Natural Science Foundation of Gansu Province (grant no.
18JR3RA366), the Foundation of The First Hospital of Lanzhou
University (grant no. ldyyyn2018-54), the Open Fund of State Key
Laboratory of Cancer Biology (grant no. CBSKL201718).
All data analyzed during this review are included in
this published article.
HW, YW, and XZ consulted and analyzed the
literature, produced the figures and wrote the manuscript. YZ and
QG were responsible for reviewing and correcting the manuscript. RJ
and YZ proposed and built the theoretical framework of the article.
All authors read and approved the final version of this
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
Not applicable.
|
1
|
Liu CX, Li X, Nan F, Jiang S, Gao X, Guo
SK, Xue W, Cui Y, Dong K, Ding H, et al: Structure and degradation
of circular RNAs regulate PKR activation in innate immunity. Cell.
177:865–880.e21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sanger HL, Klotz G, Riesner D, Gross HJ
and Kleinschmidt AK: Viroids are single-stranded covalently closed
circular RNA molecules existing as highly base-paired rod-like
structures. Proc Natl Acad Sci USA. 73:3852–3856. 1976. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Hansen TB, Kjems J and Damgaard CK:
Circular RNA and miR-7 in cancer. Cancer Res. 73:5609–5612. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Zhu K, Hu X, Chen H, Li F, Yin N, Liu AL,
Shan K, Qin YW, Huang X, Chang Q, et al: Downregulation of circRNA
DMNT3B contributes to diabetic retinal vascular dysfunction through
targeting miR-20b-5p and BAMBI. EbioMedicine. 49:341–353. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen Y, Li Z, Zhang M, Wang B, Ye J, Zhang
Y, Tang D, Ma D, Jin W, Li X and Wang S: Circ-ASH2L promotes tumor
progression by sponging miR-34a to regulate Notch1 in pancreatic
ductal adenocarcinoma. J Exp Clin Cancer Res. 38:4662019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zuo L, Zhang L, Zu J, Wang Z, Han B, Chen
B, Cheng M, Ju M, Li M, Shu G, et al: Circulating circular RNAs as
biomarkers for the diagnosis and prediction of outcomes in acute
ischemic stroke. Stroke. 51:319–323. 2020. View Article : Google Scholar
|
|
8
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar :
|
|
10
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL
and Yang L: Complementary sequence-mediated exon circularization.
Cell. 159:134–147. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Xue W, Li X, Zhang J, Chen S,
Zhang JL, Yang L and Chen LL: The biogenesis of nascent circular
RNAs. Cell Rep. 15:611–624. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liang D and Wilusz JE: Short intronic
repeat sequences facilitate circular RNA production. Genes Dev.
28:2233–2247. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Conn SJ, Pillman KA, Toubia J, Conn VM,
Salmanidis M, Phillips CA, Roslan S, Schreiber AW, Gregory PA and
Goodall GJ: The RNA binding protein quaking regulates formation of
circRNAs. Cell. 160:1125–34. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Aktaş T, Avşar Ilık İ, Maticzka D,
Bhardwaj V, Pessoa Rodrigues C, Mittler G, Manke T, Backofen R and
Akhtar A: DHX9 suppresses RNA processing defects originating from
the Alu invasion of the human genome. Nature. 544:115–119. 2017.
View Article : Google Scholar
|
|
16
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Tang C, Xie Y, Yu T, Liu N, Wang Z,
Woolsey RJ, Tang Y, Zhang X, Qin W, Zhang Y, et al:
m6A-dependent biogenesis of circular RNAs in male germ
cells. Cell Res. 30:211–228. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Panda AC, Grammatikakis I, Munk R, Gorospe
M and Abdelmohsen K: Emerging roles and context of circular RNAs.
Wiley Interdiscip Rev RNA. 8: View Article : Google Scholar : 2017.
|
|
19
|
Aufiero S, van den Hoogenhof MMG, Reckman
YJ, Beqqali A, van der Made I, Kluin J, Khan MAF, Pinto YM and
Creemers EE: Cardiac circRNAs arise mainly from constitutive exons
rather than alternatively spliced exons. RNA. 24:815–827. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Abulizi R, Li B and Zhang CG:
Circ_0071662, a novel tumor biomarker, suppresses bladder cancer
cell proliferation and invasion by sponging miR-146b-3p. Oncol Res.
Nov 18–2019.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang XO, Dong R, Zhang Y, Zhang JL, Luo
Z, Zhang J, Chen LL and Yang L: Diverse alternative back-splicing
and alternative splicing landscape of circular RNAs. Genome Res.
26:1277–1287. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ye T, Yang M, Huang D, Wang X, Xue B, Tian
N, Xu X, Bao L, Hu H, Lv T and Huang Y: MicroRNA-7 as a potential
therapeutic target for aberrant NF-κB-driven distant metastasis of
gastric cancer. J Exp Clin Cancer Res. 38:552019. View Article : Google Scholar
|
|
24
|
Pan M, Li M, You C, Zhao F, Guo M, Xu H,
Li L, Wang L and Dou J: Inhibition of breast cancer growth via
miR-7 suppressing ALDH1A3 activity concomitant with decreasing
breast cancer stem cell subpopulation. J Cell Physiol.
235:1405–1416. 2020. View Article : Google Scholar
|
|
25
|
Liu X, Fu Q, Li S, Liang N, Li F, Li C,
Sui C, Dionigi G and Sun H: LncRNA FOXD2-AS1 functions as a
competing endogenous RNA to regulate TERT expression by sponging
miR-7-5p in thyroid cancer. Front Endocrinol (Lausanne).
10:2072019. View Article : Google Scholar
|
|
26
|
Hollensen AK, Andersen S, Hjorth K, Bak
RO, Hansen TB, Kjems J, Aagaard L, Damgaard CK and Mikkelsen JG:
Enhanced tailored MicroRNA sponge activity of RNA Pol
II-transcribed TuD hairpins relative to ectopically expressed
ciRS7-derived circRNAs. Mol Ther Nucleic Acids. 13:365–375. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang Z, Yang T and Xiao J: Circular RNAs:
Promising biomarkers for human diseases. EBioMedicine. 34:267–274.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Du WW, Yang W, Liu E, Yang Z, Dhaliwal P
and Yang BB: Foxo3 circular RNA retards cell cycle progression via
forming ternary complexes with p21 and CDK2. Nucleic Acids Res.
44:2846–2858. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li X, Liu CX, Xue W, Zhang Y, Jiang S, Yin
QF, Wei J, Yao RW, Yang L and Chen LL: Coordinated circRNA
biogenesis and function with NF90/NF110 in viral infection. Mol
Cell. 67:214–227.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Fan X, Mao M, Song X, Wu P, Zhang
Y, Jin Y, Yang Y, Chen LL, Wang Y, et al: Extensive translation of
circular RNAs driven by N6-methyladenosine. Cell Res.
27:626–641. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Erratum: Circ-DLG 1 promotes the
proliferation of esophageal squamous cell carcinoma [Erratum]. Onco
Targets Ther. 12:2552018. View Article : Google Scholar
|
|
33
|
Cao S, Chen G, Yan L, Li L and Huang X:
Contribution of dysregulated circRNA_100876 to proliferation and
metastasis of esophageal squamous cell carcinoma. Onco Targets
Ther. 11:7385–7394. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xia W, Qiu M, Chen R, Wang S, Leng X, Wang
J, Xu Y, Hu J, Dong G, Xu PL and Yin R: Circular RNA
has_circ_0067934 is upregulated in esophageal squamous cell
carcinoma and promoted proliferation. Sci Rep. 6:355762016.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhong R, Chen Z, Mo T, Li Z and Zhang P:
Potential role of circPVT1 as a proliferative factor and treatment
target in esophageal carcinoma. Cancer Cell Int. 19:2672019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Wang Q, Zhang Q, Sun H, Tang W, Yang L, Xu
Z, Liu Z, Jin H and Cao X: Circ-TTC17 promotes proliferation and
migration of esophageal squamous cell carcinoma. Dig Dis Sci.
64:751–758. 2019. View Article : Google Scholar :
|
|
37
|
Luo G, Li R and Li Z: CircRNA circFNDC3B
promotes esophageal cancer progression via cell proliferation,
apoptosis, and migration regulation. Int J Clin Exp Pathol.
11:4188–4196. 2018.
|
|
38
|
Song H, Xu D, Shi P, He B, Li Z, Ji Y,
Agbeko CK and Wang J: Upregulated circ RNA hsa_circ_0000337
promotes cell proliferation, migration, and invasion of esophageal
squamous cell carcinoma. Cancer Manag Res. 11:1997–2006. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Shi Y, Guo Z, Fang N, Jiang W, Fan Y, He
Y, Ma Z and Chen Y: hsa_circ_0006168 sponges miR-100 and regulates
mTOR to promote the proliferation, migration and invasion of
esophageal squamous cell carcinoma. Biomed Pharmacother.
117:1091512019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shi N, Shan B, Gu B, Song Y, Chu H and
Qian L: Circular RNA circ-PRKCI functions as a competitive
endogenous RNA to regulate AKT3 expression by sponging miR-3680-3p
in esophageal squamous cell carcinoma. J Cell Biochem.
120:10021–10030. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu Z, Hu G, Zhao Y, Xiao Z, Yan M and Ren
M: Silence of cZNF292 suppresses the growth, migration, and
invasion of human esophageal cancer Eca-109 cells via upregulating
miR-206. J Cell Biochem. 121:2354–2362. 2020. View Article : Google Scholar
|
|
42
|
Wu Y, Zhi L, Zhao Y, Yang L and Cai F:
Knockdown of circular RNA UBAP2 inhibits the malignant behaviours
of esophageal squamous cell carcinoma by microRNA-422a/Rab10 axis.
Clin Exp Pharmacol Physiol. Feb 3–2020.Epub ahead of print.
View Article : Google Scholar
|
|
43
|
Lan X, Liu X, Sun J, Yuan Q and Li J:
CircRAD23B facilitates proliferation and invasion of esophageal
cancer cells by sponging miR-5095. Biochem Biophys Res Commun.
516:357–364. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Zhang Z, Lin W, Gao L, Chen K, Yang C,
Zhuang L, Peng S, Kang M and Lin J: Hsa_circ_0004370 promotes
esophageal cancer progression through miR-1294/LASP1 pathway.
Biosci Rep. 39:pii: BSR20182377. 2019.
|
|
45
|
Huang H, Wei L, Qin T, Yang N, Li Z and Xu
Z: Circular RNA ciRS-7 triggers the migration and invasion of
esophageal squamous cell carcinoma via miR-7/KLF4 and NF-κB
signals. Cancer Biol Ther. 20:73–80. 2019. View Article : Google Scholar
|
|
46
|
He Y, Mingyan E, Wang C, Liu G, Shi M and
Liu S: CircVRK1 regulates tumor progression and radioresistance in
esophageal squamous cell carcinoma by regulating
miR-624-3p/PTEN/PI3K/AKT signaling pathway. Int J Biol Macromol.
125:116–123. 2019. View Article : Google Scholar
|
|
47
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1594. 2017. View Article : Google Scholar
|
|
48
|
Zhang J, Liu H, Hou L, Wang G, Zhang R,
Huang Y, Chen X and Zhu J: Circular RNA_LARP4 inhibits cell
proliferation and invasion of gastric cancer by sponging miR-424-5p
and regulating LATS1 expression. Mol Cancer. 16:1512017. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Shao Y, Yang Y, Lu R, Xiao B, Ye G and Guo
J: Identification of tissue-specific circRNA hsa_circ_0000705 as an
indicator for human gastric cancer. Int J Clin Exp Pathol.
10:3151–3156. 2017.
|
|
50
|
Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye
G and Guo J: Decreased expression of hsa_circ_0001895 in human
gastric cancer and its clinical significances. Tumor Biol.
39:10104283176991252017. View Article : Google Scholar
|
|
51
|
Li WH, Song YC, Zhang H, Zhou ZJ, Xie X,
Zeng QN, Guo K, Wang T, Xia P and Chang DM: Decreased expression of
Hsa_ circ_00001649 in gastric cancer and its clinical significance.
Dis Markers. 2017:45876982017. View Article : Google Scholar
|
|
52
|
Huang M, He YR, Liang LC, Huang Q and Zhu
ZQ: Circular RNA hsa_circ_0000745 may serve as a diagnostic marker
for gastric cancer. World J Gastroenterol. 23:6330–6338. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen S, Li T, Zhao Q, Xiao B and Guo J:
Using circular RNA hsa_circ_0000190 as a new biomarker in the
diagnosis of gastric cancer. Clin Chim Acta. 466:167–171. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Chen J, Li Y, Zheng Q, Bao C, He J, Chen
B, Lyu D, Zheng B, Xu Y, Long Z, et al: Circular RNA profile
identifies circPVT1 as a proliferative factor and prognostic marker
in gastric cancer. Cancer Lett. 388:208–219. 2017. View Article : Google Scholar
|
|
55
|
Zhu Z, Yu Z, Rong Z, Luo Z, Zhang J, Qiu Z
and Huang C: The novel GINS4 axis promotes gastric cancer growth
and progression by activating Rac1 and CDC42. Theranostics.
9:8294–8311. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Zhang X, Wang S, Wang H, Cao J, Huang X,
Chen Z, Xu P, Sun G, Xu J, Lv J and Xu Z: Circular RNA circNRIP1
acts as a microRNA-149-5p sponge to promote gastric cancer
progression via the AKT1/mTOR pathway. Mol Cancer. 18:202019.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Zhang L, Song X, Chen X, Wang Q, Zheng X,
Wu C and Jiang J: Circular RNA CircCACTIN promotes gastric cancer
progression by sponging MiR-331-3p and regulating TGFBR1
expression. Int J Biol Sci. 15:1091–1103. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Wang Z, Ma K, Pitts S, Cheng Y, Liu X, Ke
X, Kovaka S, Ashktorab H, Smoot DT, Schatz M, et al: Novel circular
RNA circNF1 acts as a molecular sponge, promoting gastric cancer by
absorbing miR-16. Endocr Relat Cancer. 26:265–277. 2019. View Article : Google Scholar
|
|
59
|
Shen F, Liu P, Xu Z, Li N, Yi Z, Tie X,
Zhang Y and Gao L: CircRNA_001569 promotes cell proliferation
through absorbing miR-145 in gastric cancer. J Biochem. 165:27–36.
2019. View Article : Google Scholar
|
|
60
|
Zhang H, Wang X, Huang H, Wang Y, Zhang F
and Wang S: Hsa_circ_0067997 promotes the progression of gastric
cancer by inhibition of miR-515-5p and activation of X
chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells
Nanomed Biotechnol. 47:308–318. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Wei J, Wang J, Gao X and Qi F:
Identification of differentially expressed circRNAs and a novel
hsa_circ_0000144 that promote tumor growth in gastric cancer.
Cancer Cell Int. 19:2682019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Ouyang Y, Li Y, Huang Y, Li X, Zhu Y, Long
Y, Wang Y, Guo X and Gong K: CircRNA circPDSS1 promotes the gastric
cancer progression by sponging miR-186-5p and modulating NEK2. J
Cell Physiol. 234:10458–10469. 2019. View Article : Google Scholar
|
|
63
|
Liang M, Liu Z, Lin H, Shi B, Li M, Chen
T, Qin L, Niu Q, Yu G, Jiang H and Zhou X: High-throughput
sequencing reveals circular RNA hsa_circ_0000592 as a novel player
in the carcinogenesis of gastric carcinoma. Biosci Rep. 39:pii:
BSR20181900. 2019. View Article : Google Scholar
|
|
64
|
Liang M, Huang G, Liu Z, Wang Q, Yu Z, Liu
Z, Lin H, Li M, Zhou X and Zheng Y: Elevated levels of
hsa_circ_006100 in gastric cancer promote cell growth and
metastasis via miR-195/GPRC5A signalling. Cell Prolif.
52:e126612019. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Huang S, Zhang X, Guan B, Sun P, Hong CT,
Peng J, Tang S and Yang J: A novel circular RNA hsa_circ_0008035
contributes to gastric cancer tumorigenesis through targeting the
miR-375/YBX1 axis. Am J Transl Res. 11:2455–2462. 2019.PubMed/NCBI
|
|
66
|
Wei J, Xu H, Wei W, Wang Z, Zhang Q, De W
and Shu Y: circHIPK3 promotes cell proliferation and migration of
gastric cancer by sponging miR-107 and regulating BDNF expression.
Onco Targets Ther. 13:1613–1624. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Wang N, Lu K, Qu H, Wang H, Chen Y, Shan
T, Ge X, Wei Y, Zhou P and Xia J: CircRBM33 regulates IL-6 to
promote gastric cancer progression through targeting miR-149.
Biomed Pharmacother. 125:1098762020. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
He Y, Wang Y, Liu L, Liu S, Liang L, Chen
Y and Zhu Z: Circular RNA circ_0006282 contributes to the
progression of gastric cancer by sponging miR-155 to upregulate the
expression of FBXO22. Onco Targets Ther. 13:1001–1010. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Pan H, Pan J, Chen P, Gao J, Guo D, Yang
Z, Ji L, Lv H, Guo Y and Xu D: WITHDRAWN: Circular RNA circUBA1
promotes gastric cancer proliferation and metastasis by acting as a
competitive endogenous RNA through sponging miR-375 and regulating
TEAD4. Cancer Lett. Feb 19–2020.Epub ahead of print. View Article : Google Scholar
|
|
70
|
Mo WL, Jiang JT, Zhang L, Lu QC, Li J, Gu
WD, Cheng Y and Wang HT: Circular RNA hsa_circ_0000467 promotes the
development of gastric cancer by competitively binding to MicroRNA
miR-326-3p. Biomed Res Int. 2020:40308262020. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Peng YK, Pu K, Su HX, Zhang J, Zheng Y, Ji
R, Guo QH, Wang YP, Guan QL and Zhou YN: Circular RNA hsa_
circ_0010882 promotes the progression of gastric cancer via
regulation of the PI3K/Akt/mTOR signaling pathway. Eur Rev Med
Pharmacol Sci. 24:1142–1151. 2020.PubMed/NCBI
|
|
72
|
Sun B, Sun H, Wang Q, Wang X, Quan J, Dong
D and Lun Y: Circular RNA circMAN2B2 promotes growth and migration
of gastric cancer cells by down-regulation of miR-145. J Clin Lab
Anal. Feb 5–2020.Epub ahead of print. View Article : Google Scholar
|
|
73
|
Wei W, Mo X, Yan L, Huang M, Yang Y, Jin
Q, Zhong H, Cao W, Wu K, Wu L, et al: Circular RNA profiling
reveals that circRNA_104433 regulates cell growth by targeting
miR-497-5p in gastric cancer. Cancer Manag Res. 12:15–30. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Zhao X, Zhong Q, Cheng X, Wang S, Wu R,
Leng X and Shao L: miR-449c-5p availability is antagonized by
circ-NOTCH1 for MYC-induced NOTCH1 upregulation as well as tumor
metastasis and stemness in gastric cancer. J Cell Biochem. Jan;–14.
2020.Epub ahead of print.
|
|
75
|
Lu J, Zhang PY, Xie JW, Wang JB, Lin JX,
Chen QY, Cao LL, Li P, Zheng CH and Huang CM: Circular RNA
hsa_circ_0006848 Related to ribosomal protein L6 Acts as a novel
biomarker for early gastric cancer. Dis Markers. 2019:38634582019.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Lu J, Zhang PY, Li P, Xie JW, Wang JB, Lin
JX, Chen QY, Cao LL, Huang CM and Zheng CH: Circular RNA hsa_
circ_0001368 suppresses the progression of gastric cancer by
regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun.
512:29–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Liu H, Liu Y, Bian Z, Zhang J, Zhang R,
Chen X, Huang Y, Wang Y and Zhu J: Circular RNA YAP1 inhibits the
proliferation and invasion of gastric cancer cells by regulating
the miR-367-5p/p27 Kip1 axis. Mol Cancer. 17:1512018. View Article : Google Scholar :
|
|
78
|
Li Q, Tang H, Hu F and Qin C: Circular RNA
SMARCA5 inhibits gastric cancer progression through targeting the
miR-346/FBXL2 axis. RSC Adv. 9:18277–18284. 2019. View Article : Google Scholar
|
|
79
|
Fang J, Hong H, Xue X, Zhu X, Jiang L, Qin
M, Liang H and Gao L: A novel circular RNA, circFAT1(e2), inhibits
gastric cancer progression by targeting miR-548g in the cytoplasm
and interacting with YBX1 in the nucleus. Cancer Lett. 442:222–232.
2019. View Article : Google Scholar
|
|
80
|
Wu L, Liu D and Yang Y: Enhanced
expression of circular RNA circ-DCAF6 predicts adverse prognosis
and promotes cell progression via sponging miR-1231 and miR-1256 in
gastric cancer. Exp Mol Pathol. 110:1042732019. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Ding L, Zhao Y, Dang S, Wang Y, Li X, Yu
X, Li Z, Wei J, Liu M and Li G: Circular RNA circ-DONSON
facilitates gastric cancer growth and invasion via NURF complex
dependent activation o transcription factor SOX4. Mol Cancer.
18:452019. View Article : Google Scholar
|
|
82
|
He J, Chen J, Ma B, Jiang L and Zhao G:
CircLMTK2 acts as a novel tumor suppressor in gastric cancer.
Biosci Rep. 39:pii: BSR20190363. 2019. View Article : Google Scholar
|
|
83
|
Lu J, Wang YH, Huang XY, Xie JW, Wang JB,
Lin JX, Chen QY, Cao LL, Huang CM, Zheng CH and Li P: circ-CEP85L
suppresses the proliferation and invasion of gastric cancer by
regulating NFKBIA expression via miR-942-5p. J Cell Physiol. Feb
5–2020.Epub ahead of print.
|
|
84
|
Zhang Z, Wu H, Chen Z, Li G and Liu B:
Circular RNA ATXN7 promotes the development of gastric cancer
through sponging miR-4319 and regulating ENTPD4. Cancer Cell Int.
20:252020. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Deng G, Mou T, He J, Chen D, Lv D, Liu H,
Yu J, Wang S and Li G: Circular RNA circRHOBTB3 acts as a sponge
for miR-654-3p inhibiting gastric cancer growth. J Exp Clin Cancer
Res. 39:12020. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Cai J, Chen Z, Wang J, Wang J, Chen X,
Liang L, Huang M, Zhang Z and Zuo X: circHECTD1 facilitates
glutaminolysis to promote gastric cancer progression by targeting
miR-1256 and activating β-catenin/c-Myc signaling. Cell Death Dis.
10:5762019. View Article : Google Scholar
|
|
87
|
Rong D, Lu C, Zhang B, Fu K, Zhao S, Tang
W and Cao H: CircPSMC3 suppresses the proliferation and metastasis
of gastric cancer by acting as a competitive endogenous RNA through
sponging miR-296-5p. Mol Cancer. 18:252019. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Zhou LH, Yang YC, Zhang RY, Wang P, Pang
MH and Liang LQ: CircRNA_0023642 promotes migration and invasion of
gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci.
22:2297–2303. 2018.PubMed/NCBI
|
|
89
|
Xie Y, Shao Y, Sun W, Ye G, Zhang X, Xiao
B and Guo J: Downregulated expression of hsa_circ_0074362 in
gastric cancer and its potential diagnostic values. Biomark Med.
12:11–20. 2018. View Article : Google Scholar
|
|
90
|
Wang L, Shen J and Jiang Y:
Circ_0027599/PHDLA1 suppresses gastric cancer progression by
sponging miR-101-3p.1. Cell Biosci. 8:582018. View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Tian M, Chen R, Li T and Xiao B: Reduced
expression of circRNA hsa_circ_0003159 in gastric cancer and its
clinical significance. J Clin Lab Anal. 32:2018. View Article : Google Scholar
|
|
92
|
Sun HD, Xu ZP, Sun ZQ, Zhu B, Wang Q, Zhou
J, Jin H, Zhao A, Tang WW and Cao XF: Down-regulation of circPVRL3
promotes the proliferation and migration of gastric cancer cells.
Sci Rep. 8:101112018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 10:5725–5734. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Rong D, Dong C, Fu K, Wang H, Tang W and
Cao H: Upregulation of circ_0066444 promotes the proliferation,
invasion, and migration of gastric cancer cells. Onco Targets Ther.
11:2753–2761. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Liu M, Liu KD, Zhang L, Cai J, Yao HW, Bai
YK and Zhang ZT: Circ_0009910 regulates growth and metastasis and
is associated with poor prognosis in gastric cancer. Eur Rev Med
Pharmacol Sci. 22:8248–8256. 2018.PubMed/NCBI
|
|
96
|
Zhang H, Zhu L, Bai M, Liu Y, Zhan Y, Deng
T, Yang H, Sun W, Wang X, Zhu K, et al: Exosomal circRNA derived
from gastric tumor promotes white adipose browning by targeting the
miR-133/PRDM16 pathway. Int J Cancer. 144:2501–2515. 2019.
View Article : Google Scholar
|
|
97
|
Hong Y, Qin H, Li Y, Zhang Y, Zhuang X,
Liu L, Lu K, Li L, Deng X, Liu F, et al: FNDC3B circular RNA
promotes the migration and invasion of gastric cancer cells via the
regulation of E-cadherin and CD44 expression. J Cell Physiol.
234:19895–19910. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Zhang XY, Xu YY and Chen WY: Upregulation
of circular SMAD7 inhibits tumorigenesis of gastric cancer by
reversing epithelial-to-mesenchymal transition. Eur Rev Med
Pharmacol Sci. 24:1152–1157. 2020.PubMed/NCBI
|
|
99
|
Quan J, Dong D, Lun Y, Sun B, Sun H, Wang
Q and Yuan G: Circular RNA circHIAT1 inhibits proliferation and
epithelial-mesenchymal transition of gastric cancer cell lines
through downregulation of miR-21. J Biochem Mol Toxicol.
34:e224582020. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Xue M, Li G, Fang X, Wang L, Jin Y and
Zhou Q: hsa_ circ_0081143 promotes cisplatin resistance in gastric
cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int.
19:252019. View Article : Google Scholar
|
|
101
|
Huang X, Li Z, Zhang Q, Wang W, Li B, Wang
L, Xu Z, Zeng A, Zhang X, Zhang X, et al: Circular RNA AKT3
upregulates PIK3R1 to enhance cisplatin resistance in gastric
cancer via miR-198 suppression. Mol Cancer. 18:712019. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Huang XX, Zhang Q, Hu H, Jin Y, Zeng AL,
Xia YB and Xu L: A novel circular RNA circFN1 enhances cisplatin
resistance in gastric cancer via sponging miR-182-5p. J Cell
Biochem. Jan 2–2020.Epub ahead of print. View Article : Google Scholar
|
|
103
|
Fang Y, Pu N, Zhang L, Wu W and Lou W:
Chemoradiotherapy is associated with improved survival for resected
pancreatic adenosquamous carcinoma: A retrospective cohort study
from the SEER database. Ann Transl Med. 7:5222019. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Yang J, Cong X, Ren M, Sun H, Liu T, Chen
G, Wang Q, Li Z, Yu S and Yang Q: Circular RNA hsa_circRNA_0007334
is predicted to promote MMP7 and COL1A1 expression by functioning
as a miRNA sponge in pancreatic ductal adenocarcinoma. J Oncol.
2019:76308942019. View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Qu S, Hao X, Song W, Niu K, Yang X, Zhang
X, Shang R, Wang Q, Li H and Liu Z: Circular RNA circRHOT1 is
upregulated and promotes cell proliferation and invasion in
pancreatic cancer. Epigenomics. 11:53–63. 2019. View Article : Google Scholar
|
|
106
|
Zhu P, Ge N, Liu D, Yang F, Zhang K, Guo
J, Liu X, Wang S, Wang G and Sun S: Preliminary investigation of
the function of hsa_circ_0006215 in pancreatic cancer. Oncol Lett.
16:603–611. 2018.PubMed/NCBI
|
|
107
|
An Y, Cai H, Zhang Y, Liu S, Duan Y, Sun
D, Chen X and He X: circZMYM2 competed endogenously with miR-335-5p
to regulate JMJD2C in pancreatic cancer. Cell Physiol Biochem.
51:2224–2236. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Xing C, Ye H, Wang W, Sun M, Zhang J, Zhao
Z and Jiang G: Circular RNA ADAM9 facilitates the malignant
behaviours of pancreatic cancer by sponging miR-217 and
upregulating PRSS3 expression. Artif Cells Nanomed Biotechnol.
47:3920–3928. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Liu L, Liu FB, Huang M, Xie K, Xie QS, Liu
CH, Shen MJ and Huang Q: Circular RNA ciRS-7 promotes the
proliferation and metastasis of pancreatic cancer by regulating
miR-7-mediated EGFR/STAT3 signaling pathway. Hepatobiliary Pancreat
Dis Int. 18:580–586. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yang F, Liu DY, Guo JT, Ge N, Zhu P, Liu
X, Wang S, Wang GX and Sun SY: Circular RNA circ-LDLRAD3 as a
biomarker in diagnosis of pancreatic cancer. World J Gastroenterol.
23:8345–8354. 2017. View Article : Google Scholar
|
|
111
|
Li J, Li Z, Jiang P, Peng M, Zhang X, Chen
K, Liu H, Bi H, Liu X and Li X: Circular RNA IARS (circ-IARS)
secreted by pancreatic cancer cells and located within exosomes
regulates endothelial monolayer permeability to promote tumor
metastasis. J Exp Clin Cancer Res. 37:1772018. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Ou ZL, Luo Z, Wei W, Liang S, Gao TL and
Lu YB: Hypoxia-induced shedding of MICA and HIF1A-mediated immune
escape of pancreatic cancer cells from NK cells: Role of
circ_0000977/miR-153 axis. RNA Biol. 16:1592–1603. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Liu Y, Xia L, Dong L, Wang J, Xiao Q, Yu X
and Zhu H: CircHIPK3 promotes gemcitabine (GEM) resistance in
pancreatic cancer cells by sponging miR-330-5p and targets RASSF1.
Cancer Manag Res. 12:921–929. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Zongyi Y and Xiaowu L: Immunotherapy for
hepatocellular carcinoma. Cancer Lett. 470:8–17. 2020. View Article : Google Scholar
|
|
115
|
Busato D, Mossenta M, Baboci L, Di Cintio
F, Toffoli G and Dal Bo M: Novel immunotherapeutic approaches for
hepato-cellular carcinoma treatment. Expert Rev Clin Pharmacol.
12:453–470. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Yu L, Gong X, Sun L, Zhou Q, Lu B and Zhu
L: The circular RNA Cdr1as act as an oncogene in hepatocellular
carcinoma through targeting miR-7 expression. PLoS One.
11:e01583472016. View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Yao Z, Luo J, Hu K, Lin J, Huang H, Wang
Q, Zhang P, Xiong Z, He C, Huang Z, et al: ZKSCAN1 gene and its
related circular RNA (circZKSCAN1) both inhibit hepatocellular
carcinoma cell growth, migration, and invasion but through
different signaling pathways. Mol Oncol. 11:422–437. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Wang BB, Li JS, Liu YF and Xu Q:
MicroRNA-200b suppresses the invasion and migration of
hepatocellular carcinoma by downregulating RhoA and circRNA_000839.
Tumour Biol. 39:10104283177195772017. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Huang XY, Huang ZL, Xu YH, Zheng Q, Chen
Z, Song W, Zhou J, Tang ZY and Huang XY: Comprehensive circular RNA
profiling reveals the regulatory role of the
circRNA-100338/miR-141-3p pathway in hepatitis B-related
hepatocellular carcinoma. Sci Rep. 7:54282017. View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Fu L, Yao T, Chen Q, Mo X, Hu Y and Guo J:
Screening differential circular RNA expression profiles reveals
hsa_circ_0004018 is associated with hepatocellular carcinoma.
Oncotarget. 8:58405–58416. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Zhu Q, Lu G, Luo Z, Gui F, Wu J, Zhang D
and Ni Y: CircRNA circ_0067934 promotes tumor growth and metastasis
in hepatocellular carcinoma through regulation of
miR-1324/FZD5/Wnt/β-catenin axis. Biochem Biophys Res Commun.
497:626–632. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Zhong L, Wang Y, Cheng Y, Wang W, Lu B,
Zhu L and Ma Y: Circular RNA circC3P1 suppresses hepatocellular
carcinoma growth and metastasis through miR-4641/PCK1 pathway.
Biochem Biophys Res Commun. 499:1044–1049. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Zhang X, Zhou H, Jing W, Luo P, Qiu S, Liu
X, Zhu M, Liang C, Yu M and Tu J: The circular RNA hsa_circ_0001445
regulates the proliferation and migration of hepatocellular
carcinoma and may serve as a diagnostic biomarker. Dis Markers.
2018:30734672018. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Zhang X, Xu Y, Qian Z, Zheng W, Wu Q, Chen
Y, Zhu G, Liu Y, Bian Z, Xu W, et al: circRNA_104075 stimulates
YAP-dependent tumorigenesis through the regulation of HNF4a and may
serve as a diagnostic marker in hepatocellular carcinoma. Cell
Death Dis. 9:10912018. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Zhang X, Luo P, Jing W, Zhou H, Liang C
and Tu J: circSMAD2 inhibits the epithelial-mesenchymal transition
by targeting miR-629 in hepatocellular carcinoma. Onco Targets
Ther. 11:2853–2863. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Zhang J, Liu H, Zhang R and Zhu J:
Circular RNA SLC3A2 promotes hepatocellular carcinoma growth and
invasion by sponging MIR-490-3P and upregulating
PPM1F/AKT/GSK3/β-catenin signaling pathway. Gastroenterology.
154(Suppl 1): S11542018. View Article : Google Scholar
|
|
127
|
Zhang C, Zhang C, Lin J and Wang H:
Circular RNA Hsa_ Circ_0091579 serves as a diagnostic and
prognostic marker for hepatocellular carcinoma. Cell Physiol
Biochem. 51:290–300. 2018. View Article : Google Scholar
|
|
128
|
Yu J, Xu QG, Wang ZG, Yang Y, Zhang L, Ma
JZ, Sun SH, Yang F and Zhou WP: Circular RNA cSMARCA5 inhibits
growth and metastasis in hepatocellular carcinoma. J Hepatol.
68:1214–1227. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Wang B, Chen H, Zhang C, Yang T, Zhao Q,
Yan Y, Zhang Y and Xu F: Effects of hsa_circRBM23 on hepatocellular
carcinoma cell viability and migration as produced by regulating
miR-138 expression. Cancer Biother Radiopharm. 33:194–202. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Meng J, Chen S, Han JX, Qian B, Wang XR,
Zhong WL, Qin Y, Zhang H, Gao WF, Lei YY, et al: Twist1 regulates
vimentin through Cul2 Circular RNA to promote EMT in hepatocellular
carcinoma. Cancer Res. 78:4150–4162. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Luo Z, Mao X and Cui W: Circular RNA
expression and circPTPRM promotes proliferation and migration in
hepatocellular carcinoma. Med Oncol. 36:862019. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Li S, Gu H, Huang Y, Peng Q, Zhou R, Yi P,
Chen R, Huang Z, Hu X, Huang Y and Tang D: Circular RNA
101368/miR-200a axis modulates the migration of hepatocellular
carcinoma through HMGB1/RAGE signaling. Cell Cycle. 17:2349–2359.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Li MF, Li YH, He YH, Wang Q, Zhang Y, Li
XF, Meng XM, Huang C and Li J: Emerging roles of hsa_circ_0005075
targeting miR-431 in the progress of HCC. Biomed Pharmacother.
99:848–858. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Gong Y, Mao J, Wu D, Wang X, Li L, Zhu L
and Song R: Circ-ZEB1.33 promotes the proliferation of human HCC by
sponging miR-200a-3p and upregulating CDK6. Cancer Cell Int.
18:1162018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Chen D, Zhang C, Lin J, Song X and Wang H:
Screening differential circular RNA expression profiles reveal that
hsa_circ_0128298 is a biomarker in the diagnosis and prognosis of
hepatocellular carcinoma. Cancer Manag Res. 10:1275–1283. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Cai H, Hu B, Ji L, Ruan X and Zheng Z:
Hsa_circ_0103809 promotes cell proliferation and inhibits apoptosis
in hepatocellular carcinoma by targeting miR-490-5p/SOX2 signaling
pathway. Am J Transl Res. 10:1690–1702. 2018.PubMed/NCBI
|
|
137
|
Zou H, Xu X, Luo L, Zhang Y, Luo L, Yao Y,
Xiang G, Huang X and Wang G: Hsa_circ_0101432 promotes the
development of hepatocellular carcinoma (HCC) by adsorbing miR-1258
and miR-622. Cell Cycle. 18:2398–2413. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Zhu Y, Liu Y, Xiao B, Cai H, Liu M, Ma L,
Yin H and Wang F: The circular RNA PVT1/miR-203/HOXD3 pathway
promotes the progression of human hepatocellular carcinoma. Biol
Open. 8:pii: bio043687. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Zheng H, Chen T, Li C, Xu C, Ding C, Chen
J, Ju S, Zhang Z, Liang Z, Cui Z and Zhao J: A circular RNA
hsa_circ_0079929 inhibits tumor growth in hepatocellular carcinoma.
Cancer Manag Res. 11:443–454. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Zhang PF, Wei CY, Huang XY, Peng R, Yang
X, Lu JC, Zhang C, Gao C, Cai JB, Gao PT, et al: Circular RNA
circTRIM33-12 acts as the sponge of MicroRNA-191 to suppress
hepatocellular carcinoma progression. Mol Cancer. 18:1052019.
View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Zhang J, Chang Y, Xu L and Qin L: Elevated
expression of circular RNA circ_0008450 predicts dismal prognosis
in hepatocellular carcinoma and regulates cell proliferation,
apoptosis, and invasion via sponging miR-548p. J Cell Biochem.
120:9487–9494. 2019. View Article : Google Scholar
|
|
142
|
Zhang H, Deng T, Ge S, Liu Y, Bai M, Zhu
K, Fan Q, Li J, Ning T, Tian F, et al: Exosome circRNA secreted
from adipocytes promotes the growth of hepatocellular carcinoma by
targeting deubiquitination-related USP7. Oncogene. 38:2844–2859.
2019. View Article : Google Scholar :
|
|
143
|
Zhan W, Liao X, Chen Z, Li L, Tian T, Yu
L, Wang W and Hu Q: Circular RNA hsa_circRNA_103809 promoted
hepatocellular carcinoma development by regulating
miR-377-3p/FGFR1/ERK axis. J Cell Physiol. 235:1733–1745. 2020.
View Article : Google Scholar
|
|
144
|
Zhai Z, Fu Q, Liu C, Zhang X, Jia P, Xia
P, Liu P, Liao S, Qin T and Zhang H: Emerging roles of
hsa-circ-0046600 targeting the miR-640/HIF-1α signalling pathway in
the progression Of HCC. Onco Targets Ther. 12:9291–9302. 2019.
View Article : Google Scholar :
|
|
145
|
Yu J, Yang M, Zhou B, Luo J, Zhang Z,
Zhang W and Yan Z: CircRNA-104718 acts as competing endogenous RNA
and promotes hepatocellular carcinoma progression through
microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci (Lond).
133:1487–1503. 2019. View Article : Google Scholar
|
|
146
|
Yang W, Liu Y, Gao R, Xiu Z and Sun T:
Knockdown of cZNF292 suppressed hypoxic human hepatoma SMMC7721
cell proliferation, vasculogenic mimicry, and radioresistance. Cell
Signal. 60:122–135. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Xu L, Feng X, Hao X, Wang P, Zhang Y,
Zheng X, Li L, Ren S, Zhang M and Xu M: CircSETD3
(Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting
hepatocellular carcinoma growth. J Exp Clin Cancer Res. 38:982019.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Xie B, Zhao Z, Liu Q, Wang X, Ma Z and Li
H: CircRNA has_circ_0078710 acts as the sponge of microRNA-31
involved in hepatocellular carcinoma progression. Gene.
683:253–261. 2019. View Article : Google Scholar
|
|
149
|
Weng Q, Chen M, Li M, Zheng YF, Shao G,
Fan W, Xu XM and Ji J: Global microarray profiling identified
hsa_circ_0064428 as a potential immune-associated prognosis
biomarker for hepatocellular carcinoma. J Med Genet. 56:32–38.
2019. View Article : Google Scholar
|
|
150
|
Wang Z, Zhao Y, Wang Y and Jin C: Circular
RNA circHIAT1 inhibits cell growth in hepatocellular carcinoma by
regulating miR-3171/PTEN axis. Biomed Pharmacother. 116:1089322019.
View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Wang X, Wang X, Li W, Zhang Q, Chen J and
Chen T: Up-regulation of hsa_circ_0000517 predicts adverse
prognosis of hepatocellular carcinoma. Front Oncol. 9:11052019.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Wang L, Long H, Zheng Q, Bo X, Xiao X and
Li B: Circular RNA circRHOT1 promotes hepatocellular carcinoma
progression by initiation of NR2F6 expression. Mol Cancer.
18:1192019. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Wang G, Liu W, Zou Y, Wang G, Deng Y, Luo
J, Zhang Y, Li H, Zhang Q, Yang Y and Chen G: Three isoforms of
exosomal circPTGR1 promote hepatocellular carcinoma metastasis via
the miR449a-MET pathway. EBioMedicine. 40:432–445. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Tan A, Li Q and Chen L: CircZFR promotes
hepatocellular carcinoma progression through regulating
miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch
Biochem Biophys. 661:196–202. 2019. View Article : Google Scholar
|
|
155
|
Sun S, Wang W, Luo X, Li Y, Liu B and Li
X, Zhang B, Han S and Li X: Circular RNA circ-ADD3 inhibits
hepatocellular carcinoma metastasis through facilitating EZH2
degradation via CDK1-mediated ubiquitination. Am J Cancer Res.
9:1695–1707. 2019.PubMed/NCBI
|
|
156
|
Su Y, Xu C, Liu Y, Hu Y and Wu H: Circular
RNA hsa_ circ_0001649 inhibits hepatocellular carcinoma progression
via multiple miRNAs sponge. Aging (Albany NY). 11:3362–3375.
2019.
|
|
157
|
Song C, Li D, Liu H, Sun H, Liu Z, Zhang L
and Hu Y: The competing endogenous circular RNA ADAMTS14 suppressed
hepatocellular carcinoma progression through regulating
microRNA-572/regulator of calcineurin 1. J Cell Physiol.
234:2460–2470. 2019. View Article : Google Scholar
|
|
158
|
Qiu L, Huang Y, Li Z, Dong X, Chen G, Xu
H, Zeng Y, Cai Z, Liu X and Liu J: Circular RNA profiling
identifies circADAMTS13 as a miR-484 sponge which suppresses cell
proliferation in hepatocellular carcinoma. Mol Oncol. 13:441–455.
2019.
|
|
159
|
Pan H, Tang L, Jiang H, Li X, Wang R, Gao
J and Li Q: Enhanced expression of circ_0000267 in hepatocellular
carcinoma indicates poor prognosis and facilitates cell progression
by sponging miR-646. J Cell Biochem. 120:11350–11357. 2019.
View Article : Google Scholar
|
|
160
|
Navarro A: Twist1 activated circRNA-10720
is a new player in hepatocellular carcinoma metastasis. Transl
Cancer Res. 8(S2): S135–S140. 2019. View Article : Google Scholar
|
|
161
|
Ma Y, Zhang C, Zhang B, Yu H and Yu Q:
circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of
natural killer cells against hepatocellular carcinoma via
upregulating UL16 binding protein 1. Oncol Lett. 17:388–397.
2019.PubMed/NCBI
|
|
162
|
Luo Y, Fu Y, Huang R, Gao M, Liu F, Gui R
and Nie X: CircRNA_101505 sensitizes hepatocellular carcinoma cells
to cisplatin by sponging miR-103 and promotes oxidored-nitro
domain-containing protein 1 expression. Cell Death Discov.
5:1212019. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Lin T, Dai Y, Guo X, Chen W, Zhao J, Cao L
and Wu Z: Silencing of hsa_circ_0008450 represses hepatocellular
carcinoma progression through regulation of microRNA-214-3p/EZH2
axis. Cancer Manag Res. 11:9133–9143. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Liang WC, Wong CW, Liang PP, Shi M, Cao Y,
Rao ST, Tsui SK, Waye MM, Zhang Q, Fu WM and Zhang JF: Translation
of the circular RNA circβ-catenin promotes liver cancer cell growth
through activation of the Wnt pathway. Genome Biol. 20:842019.
View Article : Google Scholar
|
|
165
|
Li Z, Hu Y, Zeng Q, Wang H, Yan J, Li H
and Yu Z: Circular RNA MYLK promotes hepatocellular carcinoma
progression by increasing Rab23 expression by sponging miR-362-3p.
Cancer Cell Int. 19:2112019. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Li X and Shen M: Circular RNA
hsa_circ_103809 suppresses hepatocellular carcinoma proliferation
and invasion by sponging miR-620. Eur Rev Med Pharmacol Sci.
23:555–566. 2019.PubMed/NCBI
|
|
167
|
Kou P, Zhang C, Lin J and Wang H: Circular
RNA hsa_ circ_0078602 may have potential as a prognostic biomarker
for patients with hepatocellular carcinoma. Oncol Lett.
17:2091–2098. 2019.PubMed/NCBI
|
|
168
|
Ji L, Xu J, Lin Z, Mao Q, Zhang B and Cai
X: Stabilization of UBQLN1 by circRNA_104797 mediates acquired
sorafenib resistance in hepatocellular carcinoma. J Hepatol.
70:E71–E73. 2019. View Article : Google Scholar
|
|
169
|
Guan Z, Tan J, Gao W, Li X, Yang Y, Li X,
Li Y and Wang Q: Circular RNA hsa_circ_0016788 regulates
hepatocellular carcinoma tumorigenesis through miR-486/CDK4
pathway. J Cell Physiol. 234:500–508. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
170
|
Fu HW, Lin X, Zhu YX, Lan X, Kuang Y, Wang
YZ, Ke ZG, Yuan T and Chen P: Circ-IGF1R has pro-proliferative and
anti-apoptotic effects in HCC by activating the PI3K/AKT pathway.
Gene. 716:1440312019. View Article : Google Scholar : PubMed/NCBI
|
|
171
|
Fang Z, Fan R, Lu Y, Sun Y, Zhao C, Liu L
and Liu X: Circular RNA hsa_circ_0002124 promotes hepatocellular
carcinoma cell proliferation through the MAPK pathway. Transl
Cancer Res. 8:367–378. 2019. View Article : Google Scholar
|
|
172
|
Chen Z, Zuo X, Pu L, Zhang Y, Han G, Zhang
L, Wu J and Wang X: circLARP4 induces cellular senescence through
regulating miR-761/RUNX3/p53/p21 signaling in hepatocellular
carcinoma. Cancer Sci. 110:568–581. 2019. View Article : Google Scholar :
|
|
173
|
Chen Q, Chen Z, Cao S, Guo B, Chen Y, Feng
Z, Wang J, Guo G, Chen X and Huang X: Role of CircRNAs_100395 in
proliferation and metastases of liver cancer. Med Sci Monit.
25:6181–6192. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
174
|
Chen H, Liu S, Li M, Huang P and Li X:
circ_0003418 inhibits tumorigenesis and cisplatin chemoresistance
through Wnt/β-catenin pathway in hepatocellular carcinoma. Onco
Targets Ther. 12:9539–9549. 2019. View Article : Google Scholar :
|
|
175
|
Cao S, Wang G, Wang J, Li C and Zhang L:
Hsa_circ_101280 promotes hepatocellular carcinoma by regulating
miR-375/JAK2. Immunol Cell Biol. 97:218–228. 2019. View Article : Google Scholar
|
|
176
|
Zhao M, Dong G, Meng Q, Lin S and Li X:
Circ-HOMER1 enhances the inhibition of miR-1322 on CXCL6 to
regulate the growth and aggressiveness of hepatocellular carcinoma
cells. J Cell Biochem. Feb 9–2020.Epub ahead of print. View Article : Google Scholar
|
|
177
|
Yu Q, Dai J and Shu M: Hsa_circ_0003645
shows an oncogenic role by sponging microRNA-1299 in hepatocellular
carcinoma cells. J Clin Lab Anal. Feb 28–2020.Epub ahead of print.
View Article : Google Scholar
|
|
178
|
Xiao Y, Liu G, Sun Y, Gao Y, Ouyang X,
Chang C, Gong L and Yeh S: Targeting the estrogen receptor alpha
(ERα)-mediated circ-SMG1.72/miR-141-3p/Gelsolin signaling to better
suppress the HCC cell invasion. Oncogene. 39:2493–2508. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
179
|
Wei X, Zheng W, Tian P, He Y, Liu H, Peng
M, Li X and Liu X: Oncogenic hsa_circ_0091581 promotes the
malignancy of HCC cell through blocking miR-526b from degrading
c-MYC mRNA. Cell Cycle. 19:817–824. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
180
|
Pu J, Wang J, Li W, Lu Y, Wu X, Long X,
Luo C and Wei H: hsa_circ_0000092 promotes hepatocellular carcinoma
progression through up-regulating HN1 expression by binding to
microRNA-338-3p. J Cell Mol Med. Feb 20–2020.Epub ahead of print.
View Article : Google Scholar
|
|
181
|
Liu D, Kang H, Gao M, Jin L, Zhang F, Chen
D, Li M and Xiao L: Exosome-transmitted circ_MMP2 promotes
hepatocellular carcinoma metastasis by upregulating MMP2. Mol
Oncol. Jan 14–2020.Epub ahead of print. View Article : Google Scholar
|
|
182
|
Liu B, Yang G, Wang X, Liu J, Lu Z, Wang
Q, Xu B, Liu Z and Li J: CircBACH1 (hsa_circ_0061395) promotes
hepatocellular carcinoma growth by regulating p27 repression via
HuR. J Cell Physiol. Jan 31–2020.Epub ahead of print.
|
|
183
|
Li Z, Liu Y, Yan J, Zeng Q, Hu Y, Wang H,
Li H, Li J and Yu Z: Circular RNA hsa_circ_0056836 functions an
oncogenic gene in hepatocellular carcinoma through modulating
miR-766-3p/FOSL2 axis. Aging (Albany NY). 12:2485–2497. 2020.
View Article : Google Scholar
|
|
184
|
Gao J, Dai C, Yu X, Yin X-B and Zhou F:
Circ-TCF4.85 silencing inhibits cancer progression through
microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular
carcinoma. Mol Oncol. 14:447–461. 2020. View Article : Google Scholar :
|
|
185
|
Ding Z, Guo L, Deng Z and Li P: Circ-PRMT5
enhances the proliferation, migration and glycolysis of hepatoma
cells by targeting miR-188-5p/HK2 axis. Ann Hepatol. Jan
27–2020.Epub ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
186
|
Chen W, Quan Y, Fan S, Wang H, Liang J,
Huang L, Chen L, Liu Q, He P and Ye Y: Exosome-transmitted circular
RNA hsa_ circ_0051443 suppresses hepatocellular carcinoma
progression. Cancer Lett. 475:119–128. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
187
|
Yao Z, Xu R, Yuan L, Xu M, Zhuang H, Li Y,
Zhang Y and Lin N: Circ_0001955 facilitates hepatocellular
carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release
TRAF6 and MAPK11. Cell Death Dis. 10:945. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
188
|
Chen L, Kong R, Wu C, Wang S, Liu Z and
Liu S, Li S, Chen T, Mao C and Liu S: Circ-MALAT1 functions as Both
an mRNA translation brake and a microRNA sponge to promote
self-renewal of hepatocellular cancer stem cells. Adv Sci (Weinh).
7:19009492019. View Article : Google Scholar
|
|
189
|
Sun P, Fan X, Hu X, Fu X, Wei Q and Zang
Y: circPCNX and pecanex promote hepatocellular carcinoma cell
viability by inhibiting miR-506. Cancer Manag Res. 11:10957–10967.
2019. View Article : Google Scholar
|
|
190
|
Liu Z, Yu Y, Huang Z, Kong Y, Hu X, Xiao
W, Quan J and Fan X: CircRNA-5692 inhibits the progression of
hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP
expression. Cell Death Dis. 10:9002019. View Article : Google Scholar : PubMed/NCBI
|
|
191
|
Fu Y, Cai L, Lei X and Wang D: Circular
RNA ABCB10 promotes hepatocellular carcinoma progression by
increasing HMG20A expression by sponging miR-670-3p. Cancer Cell
Int. 19:3382019. View Article : Google Scholar :
|
|
192
|
Hu ZQ, Zhou SL, Li J, Zhou ZJ, Wang PC,
Xin HY, Mao L, Luo CB, Yu SY, Huang XW, et al: Circular RNA
sequencing identifies CircASAP1 as a key regulator in
hepatocellular carcinoma metastasis. Hepatology. Dec 15–2019.Epub
ahead of print. View Article : Google Scholar : PubMed/NCBI
|
|
193
|
Xu S, Zhan M, Jiang C, He M, Yang L, Shen
H, Huang S, Huang X, Lin R, Shi Y, et al: Genome-wide CRISPR screen
identifies ELP5 as a determinant of gemcitabine sensitivity in
gallbladder cancer. Nat Commun. 10:54922019. View Article : Google Scholar : PubMed/NCBI
|
|
194
|
Wang S, Zhang Y, Cai Q, Ma M, Jin LY, Weng
M, Zhou D, Tang Z, Wang JD and Quan Z: Circular RNA FOXP1 promotes
tumor progression and Warburg effect in gallbladder cancer by
regulating PKLR expression. Mol Cancer. 18:1452019. View Article : Google Scholar : PubMed/NCBI
|
|
195
|
Kai D, Yannian L, Yitian C, Dinghao G, Xin
Z and Wu J: Circular RNA HIPK3 promotes gallbladder cancer cell
growth by sponging microRNA-124. Biochem Biophys Res Commun.
503:863–869. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
196
|
Huang X, He M, Huang S, Lin R, Zhan M,
Yang D, Shen H, Xu S, Cheng W, Yu J, et al: Circular RNA circERBB2
promotes gallbladder cancer progression by regulating
PA2G4-dependent rDNA transcription. Mol Cancer. 18:1662019.
View Article : Google Scholar : PubMed/NCBI
|
|
197
|
Ahnen DJ and Patel SG: Cost-effectiveness
and national effects of initiating colorectal cancer screening for
average-risk persons at age 45 years instead of 50 years.
Gastroenterology. 157:1691–1692. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
198
|
Wang X, Zhang Y, Huang L, Zhang J, Pan F,
Li B, Yan Y, Jia B, Liu H, Li S and Zheng W: Decreased expression
of hsa_ circ_001988 in colorectal cancer and its clinical
significances. Int J Clin Exp Pathol. 8:16020–16025. 2015.
|
|
199
|
Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y,
Yang SS, Zeng ZC, Liao WT, Ding YQ and Liang L: Emerging roles of
circRNA_001569 targeting miR-145 in the proliferation and invasion
of colorectal cancer. Oncotarget. 7:26680–26691. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
200
|
Guo JN, Li J, Zhu CL, Feng WT, Shao JX,
Wan L, Huang MD and He JD: Comprehensive profile of differentially
expressed circular RNAs reveals that hsa_circ_0000069 is
upregulated and promotes cell proliferation, migration, and
invasion in colorectal cancer. Onco Targets Ther. 9:7451–7458.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
201
|
Zhuo F, Lin H, Chen Z, Huang Z and Hu J:
The expression profile and clinical significance of circRNA0003906
in colorectal cancer. Onco Targets Ther. 10:5187–5193. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
202
|
Zhu M, Xu Y, Chen Y and Yan F: Circular
BANP, an upregulated circular RNA that modulates cell proliferation
in colorectal cancer. Biomed Pharmacother. 88:138–144. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
203
|
Zhang XL, Xu LL and Wang F:
Hsa_circ_0020397 regulates colorectal cancer cell viability,
apoptosis and invasion by promoting the expression of the miR-138
targets TERT and PD-L1. Cell Biol Int. 41:1056–1064. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
204
|
Zhang R, Xu J, Zhao J and Wang X:
Silencing of hsa_ circ_0007534 suppresses proliferation and induces
apoptosis in colorectal cancer cells. Eur Rev Med Pharmacol Sci.
22:118–126. 2018.PubMed/NCBI
|
|
205
|
Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T,
Sun H, Pan Y, He B and Wang S: CircHIPK3 promotes colorectal cancer
growth and metastasis by sponging miR-7. Cell Death Dis. 9:4172018.
View Article : Google Scholar : PubMed/NCBI
|
|
206
|
Yuan Y, Liu W, Zhang Y, Zhang Y and Sun S:
CircRNA circ_0026344 as a prognostic biomarker suppresses
colorectal cancer progression via microRNA-21 and microRNA-31.
Biochem Biophys Res Commun. 503:870–875. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
207
|
Wu L, Xia J, Yang J, Shi Y, Xia H, Xiang X
and Yu X: Circ-ZNF609 promotes migration of colorectal cancer by
inhibiting Gli1 expression via microRNA-150. J BUON. 23:1343–1349.
2018.PubMed/NCBI
|
|
208
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Li ZL and
Yang Y: RNA sequencing reveals the expression profiles of circRNA
and indicates that circDDX17 acts as a tumor suppressor in
colorectal cancer. J Exp Clin Cancer Res. 37:3252018. View Article : Google Scholar : PubMed/NCBI
|
|
209
|
Li X, Wang J, Zhang C, Lin C, Zhang J,
Zhang W, Zhang W, Lu Y, Zheng L and Li X: Circular RNA circITGA7
inhibits colorectal cancer growth and metastasis by modulating the
Ras pathway and upregulating transcription of its host gene ITGA7.
J Pathol. 246:166–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
210
|
Jin Y, Yu LL, Zhang B, Liu CF and Chen Y:
Circular RNA hsa_circ_0000523 regulates the proliferation and
apoptosis of colorectal cancer cells as miRNA sponge. Braz J Med
Biol Res. 51:e78112018. View Article : Google Scholar : PubMed/NCBI
|
|
211
|
He JH, Li YG, Han ZP, Zhou JB, Chen WM, Lv
YB, He ML, Zuo JD and Zheng L: The
CircRNA-ACAP2/Hsa-miR-21-5p/Tiam1 regulatory feedback circuit
affects the proliferation, migration, and invasion of colon cancer
SW480 cells. Cell Physiol Biochem. 49:1539–1550. 2018. View Article : Google Scholar
|
|
212
|
Fang G, Ye BL, Hu BR, Ruan XJ and Shi YX:
CircRNA_100290 promotes colorectal cancer progression through
miR-516b-induced downregulation of FZD4 expression and
Wnt/β-catenin signaling. Biochem Biophys Res Commun. 504:184–189.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
213
|
Bian L, Zhi X, Ma L, Zhang J, Chen P, Sun
S, Li J, Sun Y and Qin J: Hsa_circRNA_103809 regulated the cell
proliferation and migration in colorectal cancer via
miR-532-3p/FOXO4 axis. Biochem Biophys Res Commun. 505:346–352.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
214
|
Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W,
Wu C, Zhu D, Wu C and Jiang J: Hsa_circ_0009361 acts as the sponge
of miR-582 to suppress colorectal cancer progression by regulating
APC2 expression. Clin Sci (Lond). 133:1197–1213. 2019. View Article : Google Scholar
|
|
215
|
Jin C, Wang A, Liu L, Wang G and Li G:
Hsa_circ_0136666 promotes the proliferation and invasion of
colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol.
234:7247–7256. 2019. View Article : Google Scholar
|
|
216
|
Li H, Jin X, Liu B, Zhang P, Chen W and Li
Q: CircRNA CBL.11 suppresses cell proliferation by sponging
miR-6778-5p in colorectal cancer. BMC Cancer. 19:8262019.
View Article : Google Scholar : PubMed/NCBI
|
|
217
|
Li R, Wu B, Xia J, Ye L and Yang X:
Circular RNA hsa_ circRNA_102958 promotes tumorigenesis of
colorectal cancer via miR-585/CDC25B axis. Cancer Manag Res.
11:6887–6893. 2019. View Article : Google Scholar :
|
|
218
|
Li XN, Wang ZJ, Ye CX, Zhao BC, Huang XX
and Yang L: Circular RNA circVAPA is up-regulated and exerts
oncogenic properties by sponging miR-101 in colorectal cancer.
Biomed Pharmacother. 112:1086112019. View Article : Google Scholar : PubMed/NCBI
|
|
219
|
Li Y, Li C, Xu R, Wang Y, Li D and Zhang
B: A novel circFMN2 promotes tumor proliferation in CRC by
regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond).
133:2463–2479. 2019. View Article : Google Scholar
|
|
220
|
Lu C, Jiang W, Hui B, Rong D, Fu K, Dong
C, Tang W and Cao H: The circ_0021977/miR-10b-5p/P21 and P53
regulatory axis suppresses proliferation, migration, and invasion
in colorectal cancer. J Cell Physiol. 235:2273–2285. 2020.
View Article : Google Scholar
|
|
221
|
Min L, Wang H and Zeng Y: CircRNA_104916
regulates migration, apoptosis and epithelial-mesenchymal
transition in colon cancer cells. Front Biosci (Landmark Ed).
24:819–832. 2019. View
Article : Google Scholar
|
|
222
|
Pan B, Qin J, Liu X, He B, Wang X, Pan Y,
Sun H, Xu T, Xu M, Chen X, et al: Identification of serum exosomal
hsa-circ-0004771 as a novel diagnostic biomarker of colorectal
cancer. Front Genet. 10:10362019. View Article : Google Scholar
|
|
223
|
Shen T, Cheng X, Liu X, Xia C, Zhang H,
Pan D, Zhang X and Li Y: Circ_0026344 restrains metastasis of human
colorectal cancer cells via miR-183. Artif Cells Nanomed
Biotechnol. 47:4038–4045. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
224
|
Tian J, Xi X, Wang J, Yu J, Huang Q, Ma R,
Zhang X, Li H and Wang L: CircRNA hsa_circ_0004585 as a potential
biomarker for colorectal cancer. Cancer Manag Res. 11:5413–5423.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
225
|
Yang F, Fang E, Mei H, Chen Y, Li H, Li D,
Song H, Wang J, Hong M, Xiao W, et al: Cis-acting circ-CTNNB1
promotes β-catenin signaling and cancer progression via
DDX3-mediated transactivation of YY1. Cancer Res. 79:557–571. 2019.
View Article : Google Scholar
|
|
226
|
Yang G, Zhang T, Ye J, Yang J, Chen C, Cai
S and Ma J: Circ-ITGA7 sponges miR-3187-3p to upregulate ASXL1,
suppressing colorectal cancer proliferation. Cancer Manag Res.
11:6499–6509. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
227
|
Zhang J, Liu H, Zhao P, Zhou H and Mao T:
Has_circ_0055625 from circRNA profile increases colon cancer cell
growth by sponging miR-106b-5p. J Cell Biochem. 120:3027–3037.
2019. View Article : Google Scholar
|
|
228
|
Zhang X, Zhao Y, Kong P, Han M and Li B:
Expression of circZNF609 is down-regulated in colorectal cancer
tissue and promotes apoptosis in colorectal cancer cells by
upregulating p53. Med Sci Monit. 25:5977–5985. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
229
|
Zhou C, Liu HS, Wang FW, Hu T, Liang ZX,
Lan N, He XW, Zheng XB, Wu XJ, Xie D, et al: circCAMSAP1 promotes
tumor growth in colorectal cancer via the miR-328-5p/E2F1 axis. Mol
Ther. 28:914–928. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
230
|
Zhu CL, Sha X, Wang Y, Li J, Zhang MY, Guo
ZY, Sun SA and He JD: Circular RNA hsa_circ_0007142 Is upregulated
and targets miR-103a-2-5p in colorectal cancer. J Oncol.
2019:98368192019. View Article : Google Scholar : PubMed/NCBI
|
|
231
|
Chen HY, Li XN, Ye CX, Chen ZL and Wang
ZJ: Circular RNA circHUWE1 is upregulated and promotes cell
proliferation, migration and invasion in colorectal cancer by
sponging miR-486. Onco Targets Ther. 13:423–434. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
232
|
Li W, Xu Y, Wang X, Cao G, Bu W, Wang X,
Fang Z, Xu Y, Dong M and Tao Q: circCCT3 modulates vascular
endothelial growth factor A and Wnt signaling to enhance colorectal
cancer metastasis through sponging miR-613. DNA Cell Biol.
39:118–125. 2020. View Article : Google Scholar
|
|
233
|
Pei FL, Cao MZ and Li YF: Circ_0000218
plays a carcinogenic role in colorectal cancer progression by
regulating miR-139-3p/RAB1A axis. J Biochem. 167:55–65. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
234
|
Ren C, Zhang Z, Wang S, Zhu W, Zheng P and
Wang W: Circular RNA hsa_circ_0001178 facilitates the invasion and
metastasis of colorectal cancer through upregulating ZEB1 via
sponging multiple miRNAs. Biol Chem. 401:487–496. 2020. View Article : Google Scholar
|
|
235
|
Wang X, Zhang H, Yang H, Bai M, Ning T,
Deng T, Liu R, Fan Q, Zhu K, Li J, et al: Exosome-delivered circRNA
promotes glycolysis to induce chemoresistance through the
miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 14:539–555.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
236
|
Yang B, Du K, Yang C, Xiang L, Xu Y, Cao
C, Zhang J and Liu W: CircPRMT5 circular RNA promotes proliferation
of colorectal cancer through sponging miR-377 to induce E2F3
expression. J Cell Mol Med. 24:3431–3437. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
237
|
Yang H, Li X, Meng Q, Sun H, Wu S, Hu W,
Liu G, Li X, Yang Y and Chen R: CircPTK2 (hsa_circ_0005273) as a
novel therapeutic target for metastatic colorectal cancer. Mol
Cancer. 19:132020. View Article : Google Scholar : PubMed/NCBI
|
|
238
|
Yang L, Sun H, Liu X, Chen J, Tian Z, Xu
J, Xiang B and Qin B: Circular RNA hsa_circ_0004277 contributes to
malignant phenotype of colorectal cancer by sponging miR-512-5p to
upregulate the expression of PTMA. J Cell Physiol. Jan 21–2020.Epub
ahead of print. View Article : Google Scholar
|
|
239
|
Zhao H, Chen S and Fu Q: Exosomes from
CD133+ cells carrying circ-ABCC1 mediate cell stemness
and metastasis in colorectal cancer. J Cell Biochem. 121:3286–3297.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
240
|
Joensuu H, Vehtari A, Riihimäki J, Nishida
T, Steigen SE, Brabec P, Plank L, Nilsson B, Cirilli C, Braconi C,
et al: Risk of recurrence of gastrointestinal stromal tumour after
surgery: An analysis of pooled population-based cohorts. Lancet
Oncol. 13:265–274. 2012. View Article : Google Scholar
|
|
241
|
Jia N, Tong H, Zhang Y, Katayama H, Wang
Y, Lu W, Zhang S and Wang J: CeRNA expression profiling identifies
KIT-Related circRNA-miRNA-mRNA networks in gastrointestinal stromal
tumour. Front Genet. 10:8252019. View Article : Google Scholar : PubMed/NCBI
|
|
242
|
Anastasiadou E, Jacob LS and Slack FJ:
Non-coding RNA networks in cancer. Nat Rev Cancer. 18:5–18. 2018.
View Article : Google Scholar
|
|
243
|
Lewis BP, Burge CB and Bartel DP:
Conserved seed pairing, often flanked by adenosines, indicates that
thousands of human genes are microRNA targets. Cell. 120:15–20.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
244
|
Xu B, Yang T, Wang Z, Zhang Y, Liu S and
Shen M: CircRNA CDR1as/miR-7 signals promote tumor growth of
osteosarcoma with a potential therapeutic and diagnostic value.
Cancer Manag Res. 10:4871–4880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
245
|
Pan H, Li T, Jiang Y, Pan C, Ding Y, Huang
Z, Yu H and Kong D: Overexpression of circular RNA ciRS-7 abrogates
the tumor suppressive effect of miR-7 on gastric cancer via
PTEN/PI3K/AKT signaling pathway. J Cell Biochem. 119:440–446. 2018.
View Article : Google Scholar
|
|
246
|
Kabir TD, Ganda C, Brown RM, Beveridge DJ,
Richardson KL, Chaturvedi V, Candy P, Epis M, Wintle L, Kalinowski
F, et al: A microRNA-7/growth arrest specific 6/TYRO3 axis
regulates the growth and invasiveness of sorafenib-resistant cells
in human hepatocellular carcinoma. Hepatology. 67:216–231. 2018.
View Article : Google Scholar
|
|
247
|
Li H, Fan J, Zhao Y, Zhang X, Dai B, Zhan
J, Yin Z, Nie X, Fu XD, Chen C and Wang DW: Nuclear miR-320
mediates diabetes-induced cardiac dysfunction by activating
transcription of fatty acid metabolic genes to cause lipotoxicity
in the heart. Circ Res. 125:1106–1120. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
248
|
Yuan H, Chen Z, Bai S, Wei H, Wang Y, Ji
R, Guo Q, Li Q, Ye Y, Wu J, et al: Molecular mechanisms of lncRNA
SMARCC2/miR-551b-3p/TMPRSS4 axis in gastric cancer. Cancer Lett.
418:84–96. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
249
|
Ding J, Zhao J, Huan L, Liu Y, Qiao Y,
Wang Z, Chen Z, Huang S, Zhao Y and He X: Inflammation-induced
LINC00665 increases the malignancy through activating PKR/NF-κB
pathway in hepatocellular carcinoma. Hepatology. Feb 21–2020.Epub
ahead of print. View Article : Google Scholar
|