Introduction
Rhabdomyosarcoma (RMS) is the most common type of
pediatric soft tissue sarcoma worldwide (1) and accounts for 7% of all pediatric
malignant tumors (2). Embryonal
RMS (RME) and alveolar RMS (RMA) are the two main histological
subtypes; RME represents ~70% and RMA ~20% of all RMS cases
(3). RME and gene fusion-negative
RMA are known to be prognostically favorable, whereas paired box
gene 3-fork-head (PAX3-FKHR) or PAX7-FKHR gene fusion-positive RMA
frequently leads to metastatic progression (4-6),
which is present in nearly 20% of RMS cases at time of diagnosis
(7). Even with multimodal
treatment, including systemic chemotherapy, surgery and/or
radiotherapy, the prognosis of advanced stage RMS remains poor
(8). Major treatment issues are
metastatic invasion, local tumor recurrence and drug resistance
(9). Several genes responsible for
metastatic invasion have been identified in different types of
cancer, including prostate, lung, breast and pancreatic cancer
(10,11). One of the genes of the metastatic
cascade encodes the chemokine receptor 4 (CXCR4), a 7-transmembrane
G protein-coupled receptor (12)
that is upregulated in RMS cell lines as well as in lung,
pancreatic and prostate tumors (13).
CXCR4 has been described as a marker of poor
prognosis in acute lymphoblastic leukemia (ALL) and acute myeloid
leukemia (AML) (14). A clinical
study of 40 patients with RMS has demonstrated that high levels of
CXCR4 are associated with unfavorable clinical features, bone
marrow involvement and poor outcomes (15). In addition to its physiological
role, multiple preclinical trials have demonstrated that CXCR4 and
its ligand stromal cell-derived factor 1α (SDF-1α) crucially
influence metastatic invasion, proliferation and angiogenesis in
different tumor types such as breast cancer (16), pancreatic cancer (17), colon cancer (18) and RMS (19-23).
Chemotherapy appears to be involved in drug resistance and may
promote metastasis by the induction of chemokines and chemokine
receptor expression. Such metastasis promotion was observed in
tumor models of melanoma (24,25),
head and neck (26), ovarian and
breast cancer (27,28).
Previous studies have demonstrated that the
combination of chemotherapeutic drugs and CXCR4
inhibitors/antagonists such as AMD3100, AMD11070 and AMD3465 are
successful treatments for various malignancies such as ALL
(29,30), AML (14) and prostate cancer (31). AMD3100 is a member of the bicyclam
family, which was first identified as an antiretroviral small
molecule and was demonstrated to bind selectively as an antagonist
to CXCR4 (32,33). AMD3100 inhibited SDF-1α-induced
HER2/Neu activation in vitro in breast cancer cells and
modestly improved the overall survival of mice with metastatic
ovarian cancer by inhibiting tumor growth in vivo (34-37).
Additionally, AMD3100 has been reported to bind selectively as an
antagonist to CXCR4, thus inhibiting the growth of ovarian cancer
cells and intraperitoneal dissemination (38).
The aim of the present study was to evaluate the
effects of different cytotoxic drugs on CXCR4 expression levels and
to study the impact of a combination of CXCR4 inhibition and
chemotherapy on the treatment of RMS in vitro.
Materials and methods
Cell lines and cell culture
The RMA cell line RH30 (ACC no. 489; DSMZ-German
Collection of Microorganisms and Cell Cultures GmbH) and the RME
cell line RD (ATCC® CCL-136; American Type Culture Collection) were
routinely cultured in DMEM (Biochrom, Ltd.) supplemented with 10%
fetal bovine serum (FBS; Biochrom, Ltd.), 2 mM L-glutamine, 50 U/ml
penicillin and 50 µg/ml streptomycin (Biochrom, Ltd.).
Mycoplasma-negative RMS cell lines were cultured in a humidified
atmosphere incubator (BBD 6220; Heraeus Holding GmbH) with 5%
CO2 at 37°C.
Patient characteristics
The RMS tissues were collected by resection between
January 2015 and January 2019 at the University Hospital of
Tuebingen. Patients (3 female and 3 male) with histologically
proven primary and/or recurrent rhabdomyosarcoma (n=6) were
prospectively recorded in this study. Diagnosis of rhabdomyosarcoma
was histopathologically confirmed by the local Department of
Pathology in Tuebingen as well as by the Reference Pathology
(Institute of Pathology, University of Schleswig-Holstein, Kiel,
Germany) of the Society for Paediatric Oncology and Haematology.
The mean age of the patients was 26 months (range, 13-40 months).
Written informed consent to participate in this study was obtained
from the parents of all patients. This study was approved by the
Ethical Committee of the Medical Faculty of the University Hospital
Tuebingen (approval no. 354/2018A, amendment no. 354/2018B02).
Immunohistochemistry of CXCR4
Formalin-fixed and paraffin-embedded RMS specimens
were cut into 4-µm sections and deparaffinized with xylol
and ethanol (Carl Roth GmbH & Co. KG). Standard and automated
hematoxylin and eosin staining (Sakura Prisma, Sakura Finetek
Germany GmbH) at room temperature was performed for the evaluation
of tumor tissue and areas of necrosis. For demasking the epitopes,
samples were treated with hot citrate buffer (100°C; pH 6.0).
Paraffin sections were then blocked with 3% goat serum (Dako;
Agilent Technologies, Inc.) and incubated overnight at 4°C with
mouse monoclonal anti-CXCR4 antibody (1:50; cat. no. sc-53534;
Santa Cruz Biotechnology, Inc.). The following day, antibody
binding was identified using VECTASTAIN Universal Elite ABC kit
(LINARIS GmbH) and DAB solution (Dako; Agilent Technologies, Inc.).
The slides were then analyzed using a transmitted light Zeiss
Axioskop 40 microscope (Carl Zeiss AG; original magnification,
×100).
Reagents, chemotherapeutics and
antibodies
The CXCR4 antagonist AMD3100, also referred to as
Plerixafor, was purchased from Sigma-Aldrich (Merck KGaA) and was
pre-incubated for 30 min in all experiments before cytotoxic drug
addition. CXCR4 mouse anti-human/CD184 antibody [clone 12G5;
conjugated to allophycocyanin (APC); cat. no. 560936] was obtained
from Becton, Dickinson and Company. Recombinant human SDF-1α was
purchased from ImmunoTools GmbH. Doxorubicin, vincristine and
dactinomycin were obtained from the pharmacy of the University
Hospital of Tuebingen. Paclitaxel was obtained from Enzo Life
Sciences. The doses of cytotoxic agents used in the following
experiments were LD40-80 of doxorubicin (0.2/0.5
µg/ml), vincristine (0.001/0.0025 µg/ml) and
dactinomycin (0.01/0.025 µg/ml). For paclitaxel, 0.01 and 1
µg/ml was used in RH30 cells, and 0.01 and 0.1 µg/ml
was used in RD cells.
Flow cytometry
The percentage of CXCR4-positive cells was measured
by fluorescence‑activated cell sorting (FACS). RMS cells RH30 and
RD were seeded in a 6-well plate at a density of 1×105
cells/well and treated with or without cytotoxic drugs in the
presence or absence of 10 µM AMD3100 for 24 h. After
incubation, the adherent cells were collected by trypsinization,
washed twice with PBS (Biochrom, Ltd.) and stained with CXCR4 mouse
anti-human/CD184 antibody (1:100; cat. no. 560936; Becton,
Dickinson and Company) for 30 min at room temperature in the dark.
Subsequently, the cells were washed again with 200 µl
CellWASH solution (Becton, Dickinson and Company). The surface
expression and the mean fluorescence intensity (MFI) of CXCR4 was
analyzed using a BD FACS Canto II flow cytometer (Becton, Dickinson
and Company) and evaluated with BD FACS Diva software v.8.0
(Becton, Dickinson and Company).
Reverse transcription‑quantitative PCR
(RT‑qPCR)
To evaluate the gene expression level of CXCR4,
total RNA was isolated from RMS cells pretreated for 24 h with or
without cytotoxic drugs using the RNeasy Plus Mini kit (Qiagen
GmbH) according to the manufacturer's instructions. cDNA
synthesis was performed using the High-Capacity cDNA Reverse
Transcription kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. PCR amplification of the
respective genes was performed in a total volume of 20 µl
using 40 ng cDNA, 500 nM forward and reverse primers and 2X GoTaq
qPCR Master Mix (Promega Corporation) according to the
manufacturer's protocol. The following amplification
protocol was applied: 95°C for 5 min, followed by 40 cycles of 60°C
for 30 sec and 72°C for 30 sec. The following primers were used:
CXCR4 forward, 5′-GGTTCCTTCATGGAGTCATAGTC-3′ and
reverse, 5′-CGGTTACCATGGAGGGGATC-3′; and TATA-binding
protein (TBP) forward, 5′-GCCCGAAACGCCGAATAT-3′ and reverse,
5′-CCGTGGTTCGTGGCTCTC-3′. The specificity of the PCR
amplicons was confirmed by analysis of the melting curves. For qPCR
amplification and data analysis, CFX96 Real Time PCR Detection
System (Bio-Rad Laboratories, Inc.) was used. To standardize gene
expression and perform accurate RT-qPCR analysis, normalization
relative to the consistently expressed housekeeping gene TBP was
performed. Relative quantification of gene expression was
calculated using the ΔΔCq method as previously described (39).
Transwell migration assay
Transwell migration assays were performed in 24-well
plates (Corning, Inc.) using Transwell inserts with an 8-µm
pore size (Becton, Dickinson and Company). RMS cells were seeded
into 6-well plates (Corning Inc.) at 5×105 cells/well
and pretreated with or without 0.2 µg/ml doxorubicin, 0.001
µg/ml vincristine and/or 10 µM AMD3100. After 24 h,
750 µl of the culture medium with or without 300 ng/ml human
recombinant SDF1-α was pipetted into the lower chambers of 24-well
plates. Transwell inserts were placed into each well, and
pretreated RH30 and RD cells were seeded into the upper chamber of
the inserts at a density of 5×104 cells/well in culture
medium without FBS. RMS cells were either treated or left untreated
for 24 h and then cultured in a humidified atmosphere at 37°C with
5% CO2. Subsequently, the inserts were moved to 4%
paraformaldehyde (Carl Roth GmbH & Co. KG), incubated for 15
min at room temperature, washed twice with PBS and stained with
Giemsa (Sigma-Aldrich; Merck KGaA). The migrated cells bound to the
lower surface of the membrane were counted with an inverted
Axiovert 135 light microscope at magnification, ×10 (Carl Zeiss AG)
using three different areas of each membrane, and the mean was
calculated with AxioVision Rel v.4.8 software (Carl Zeiss AG).
Evaluation of drug interaction
To evaluate the effect of the combination of AMD3100
with chemotherapeutic agents, the coefficient of drug interaction
(CDI) was calculated as described by Foucquier and Guedj (40). According to the bliss independence
model, which dissects additive from synergistic and antagonistic
effects respectively, CDI was calculated by the formula CDI = [(A +
B) - (A x B)] / AB where AB is the ratio of the absorbance in the
combination of drugs vs. that of the control; and A or B is the
ratio of the absorbance of the single agent group to that of the
control group. CDI values <1, =1 or >1 indicate that the
drugs act in a synergistic, additive or antagonistic manner,
respectively.
Western blotting
To analyze the total protein expression levels of
CXCR4, 5×105 RH30 cells were seeded in 6-well plates and
treated with or without 10 µM AMD3100 for 24 h. RMS cells
were next washed with ice-cold PBS and lysed with cell lysis buffer
(Cell Signaling Technology, Inc.) on ice. The extracts were
centrifuged at 4°C and 14,000 × g for 20 min, and the protein
concentration of the supernatant was determined by a Bradford
assay. Protein samples (30 µg) were subjected to 10%
SDS-PAGE and transferred to a nitrocellulose membrane (VWR
International GmbH). Subsequently, the membranes were blocked for 1
h at room temperature with 10% non-fat dried milk (Carl Roth GmbH
& Co. KG) in TBS (Sigma-Aldrich; Merck KGaA) containing 0.1%
Tween-20 (Carl Roth GmbH & Co. KG). For immunoblotting, the
membranes were incubated overnight at 4°C with an antibody against
CXCR4 (1:100; cat. no. ACR-014; Alomone Labs). Anti-GAPDH antibody
(1:1,000; cat. no. 2118S; Cell Signaling Technology, Inc.) was used
as a loading control. After incubation with a secondary anti-rabbit
IgG antibody conjugated to horseradish peroxidase (1:3,000; cat.
no. 7074S; Cell signaling Technology, Inc.), the proteins were
visualized by the WesternSure® PREMIUM Chemiluminescent
Substrate (LI‑COR Biosciences). Specific bands were quantified with
the Odyssey Fc Imaging System (LI-COR Biosciences), and the levels
of CXCR4 protein were expressed as the ratio of signal intensity of
the target protein relative to that of GAPDH.
Statistics
Data are expressed as the mean ± standard error of
the mean. All experiments were repeated at least three times. A
Shapiro-Wilk test was performed to determine the normal
distribution of the data. All data were tested for significance
using an unpaired Student's t-test with Welch's
correction or one-way ANOVA (with Bonferroni correction) using
GraphPad Prism v.7.0 (GraphPad Software, Inc). P<0.05 was
considered to indicate a statistically significant difference.
Results
Expression of CXCR4 in RMS cell
lines
CXCR4 expression levels were analyzed in two RMS
cell lines RH30 and RD by RT-qPCR and flow cytometry. The results
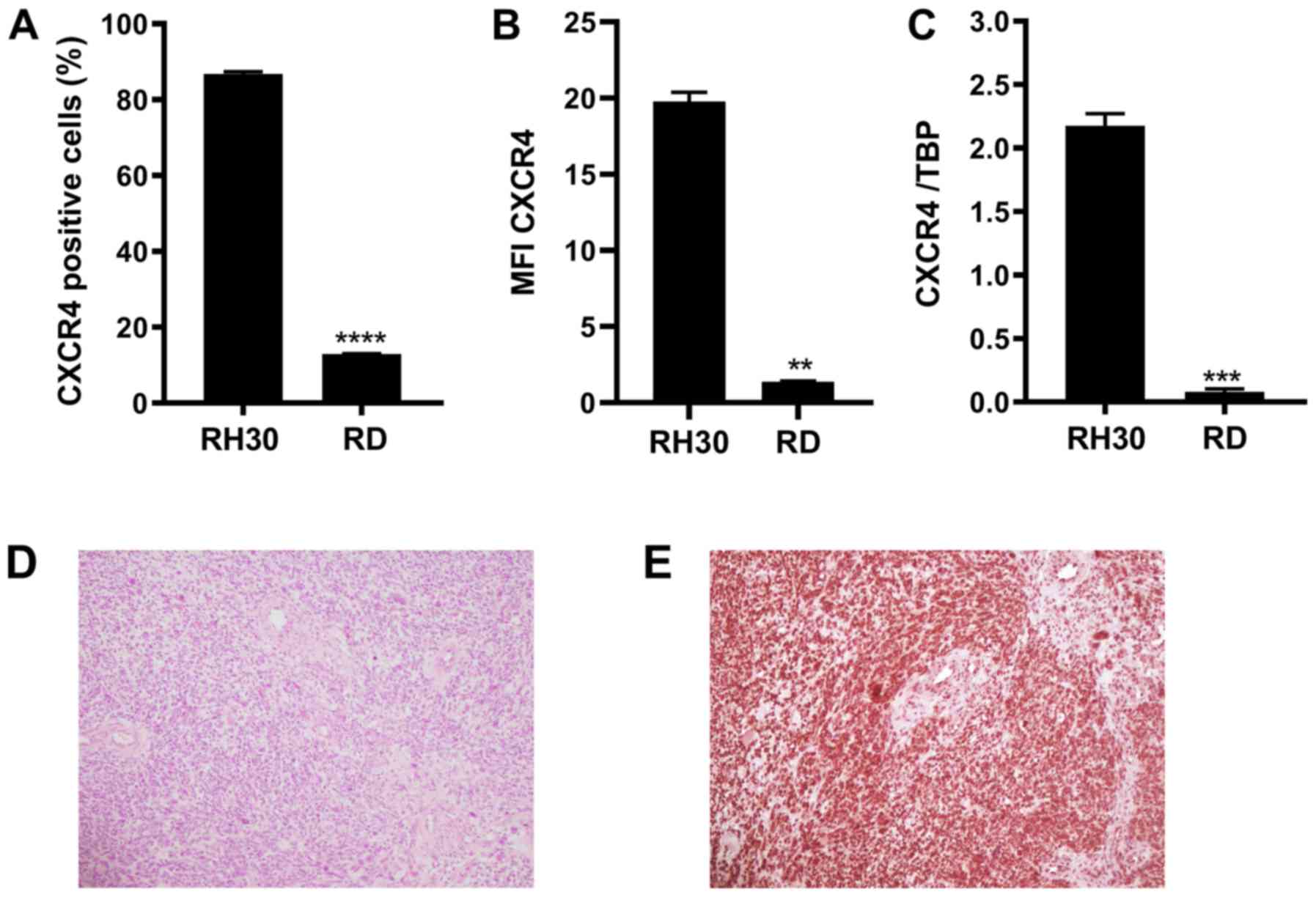
of the flow cytometry assay demonstrated that 86.8% of the alveolar
RMS cells (RH30) and 12.9% of the embryonal RMS cells (RD) were
CXCR4‑positive (Fig. 1A). RH30
cells exhibited significantly higher mean fluorescence intensity
(MFI) values for CXCR4 compared with those in RD cells (Fig. 1B). RT-qPCR was performed to confirm
this expression pattern. As illustrated in Fig. 1C, CXCR4 transcripts were detected
by RT-qPCR in both RH30 and RD RMS cell lines and were present at
significantly higher levels in RH30 cells compared with RD cells.
In addition, immunohistochemical analysis of CXCR4 expression was
performed in paraffin sections obtained from six recently diagnosed
primary RMS cases; intense nuclear staining of CXCR4 was observed
in RMS tissues, whereas lower intensity of CXCR4 was present in
regular adjacent vascular structures (Fig. 1E).
Effects of cytotoxic drugs in combination
with the CXCR4 antagonist AMD3100
MTT assays were performed to obtain the
dose-response curves and the corresponding IC50 values
of chemotherapeutics in RD and RH30 cells (Fig. S1). In addition, further MTT assays
were performed to evaluate the potential effects of the CXCR4
antagonist AMD3100, or the additive or synergistic effects of
chemotherapeutic agents in combination with AMD3100. However, no
significant effects of AMD3100 on RD or RH30 cells were observed,
and no additive or synergistic effects were observed in RD or RH30
cells when combining chemotherapeutic agents with AMD3100 compared
with the effects of the corresponding chemotherapeutic agents alone
(Fig. S2).
CXCR4 expression after treatment with
cytotoxic drugs
The present study next investigated whether the
quantity of CXCR4 mRNA in RMS cells was sensitive to cytotoxic
treatment.
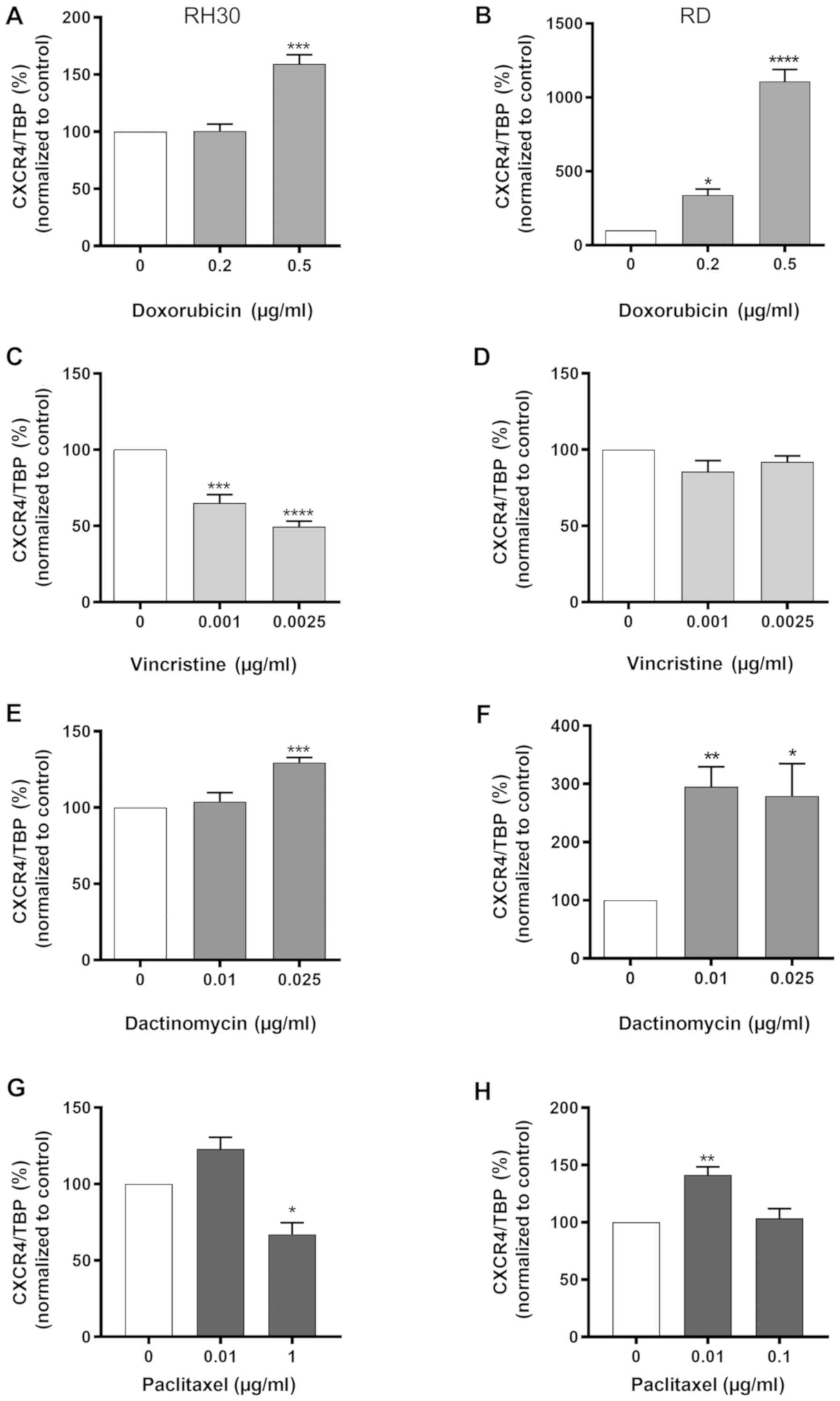
As presented in Fig.
2, CXCR4 mRNA expression was significantly enhanced in the RH30
and RD cells after treatment with 0.5 µg/ml doxorubicin or
0.025 µg/ml dactinomycin compared with that in the untreated
cells. Of note, vincristine and paclitaxel led to a significant
downregulation of CXCR4 mRNA expression in RH30 cells compared with
the untreated cells (Fig. 2C and
G); however, this was not observed in RD cells (Fig. 2D and H).
Flow cytometry analysis was utilized to confirm this
expression pattern. Similar to the RT-qPCR results in Fig. 2, the number of CXCR4-positive cells
and the MFI values in the two RMS cell lines increased
significantly when treated with doxorubicin (Fig. 3A-a and B-a) and dactinomycin
(Fig. 3A-c and B-c) compared with
those in the control group. In addition, vincristine and paclitaxel
also induced a significant reduction of CXCR4-positive cells and
MFI values in RH30 cells compared with the untreated cells
(Fig. 3A-b and A-d), in contrast
to the results observed in RD cells (Fig. 3B-b and B-d).
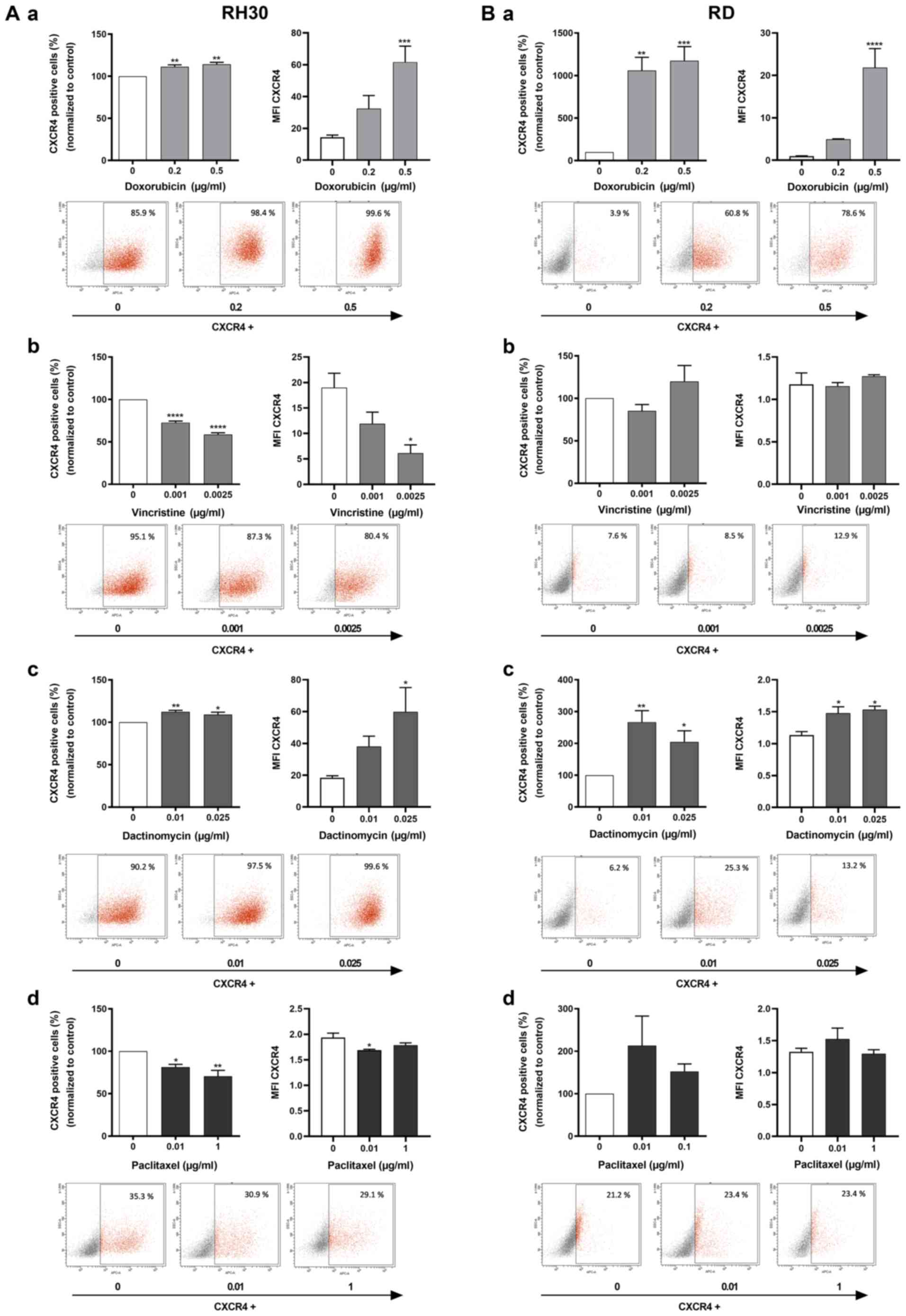 | Figure 3Flow cytometric analysis of CXCR4 in
RMS cell lines after cytostatic treatment. (A and B) CXCR4-positive
tumor cells (%) and respective MFI for CXCR4 expression were
evaluated for RMS cell lines (A) RH30 and (B) RD after a 24 h
treatment with (a) 0, 0.2 and 0.5 µg/ml doxorubicin (n=4),
(b) 0, 0.001 and 0.0025 µg/ml vincristine (n=5), (c) 0, 0.01
and 0.025 µg/ml dactinomycin (n=5) or (d) 0, 0.01 and 0.1
µg/ml paclitaxel in RD cells and 0.01 and 1 µg/ml
paclitaxel in RH30 cells (n=4). (a‑d) Representative plots of CXCR4
expression are presented from one of five experiments under each
graph. Error bars represent SEM. *P<0.05,
**P≤0.01, ***P≤0.001, ****P≤0.0001
vs. the untreated control (100%). CXCR4, CXC chemokine receptor 4;
RMS, rhabdomyosarcoma; MFI, mean fluorescence intensity. |
Effects of AMD3100 treatment on CXCR4
expression
AMD3100 prevents the binding of the anti-CXCR4
antibody 12G5 in Jurkat and SUP-T1 cells (41,42)
as they bind the same epitope. The present study used this
competitive binding to monitor AMD3100 binding to CXCR4 in RMS cell
lines (43).
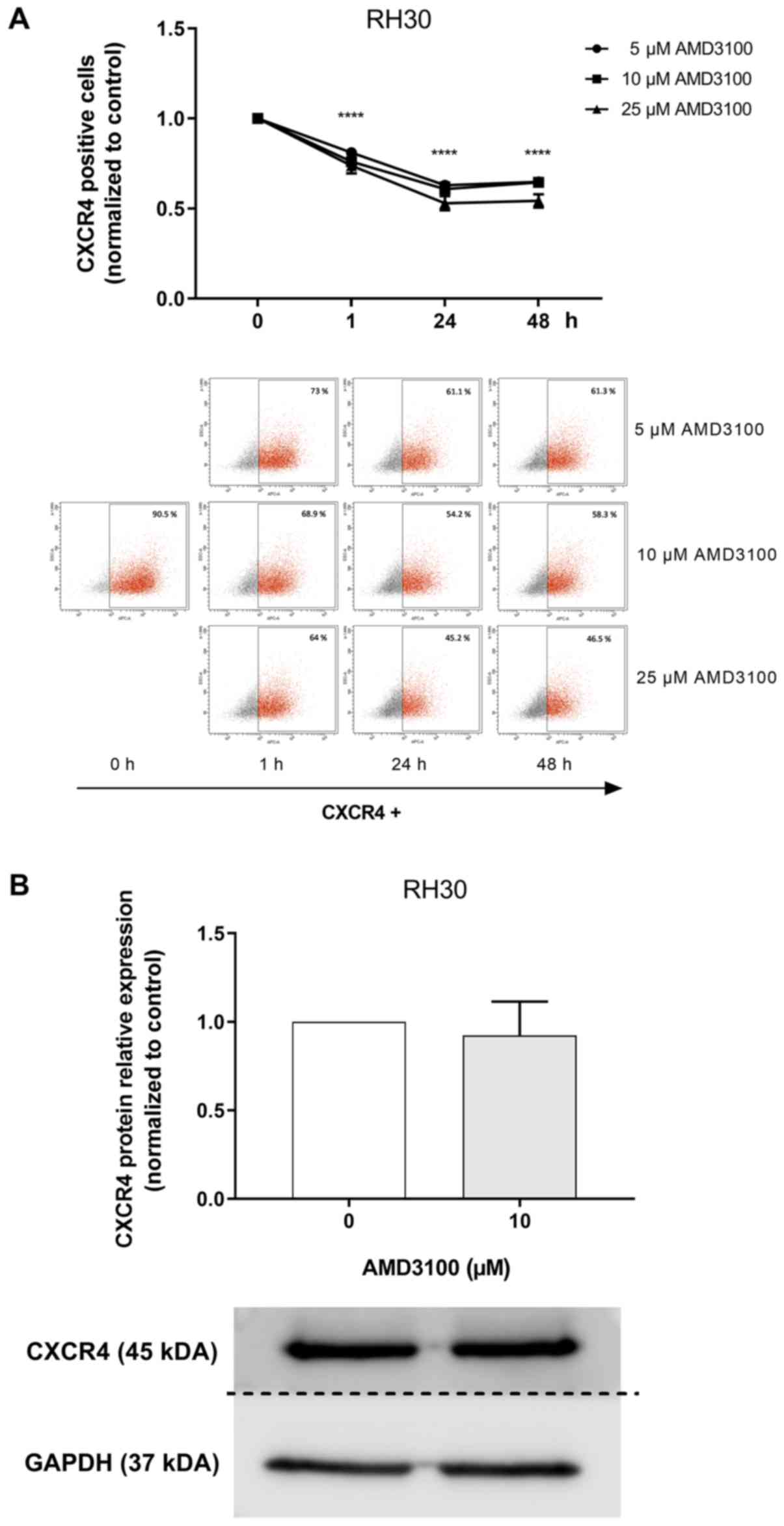
To investigate the optimal time and dose of AMD3100
binding, a competition assay with an anti-CXCR4 antibody (12G5
conjugated to APC) was performed by flow cytometry. The RH30 cell
line was used due to its high levels of CXCR4 expression (Fig. 4). Compared with the untreated
control, treatment with AMD3100 significantly reduced the binding
of the anti-CXCR4 antibody to CXCR4 (Fig. 4A).
The effect of AMD3100 on CXCR4 protein expression
was investigated by western blotting. AMD3100 exerted no effect on
CXCR4 protein levels in RH30 cells (Fig. 4B).
Migratory response of RMS cells to
chemotherapeutic agents in combination with AMD3100
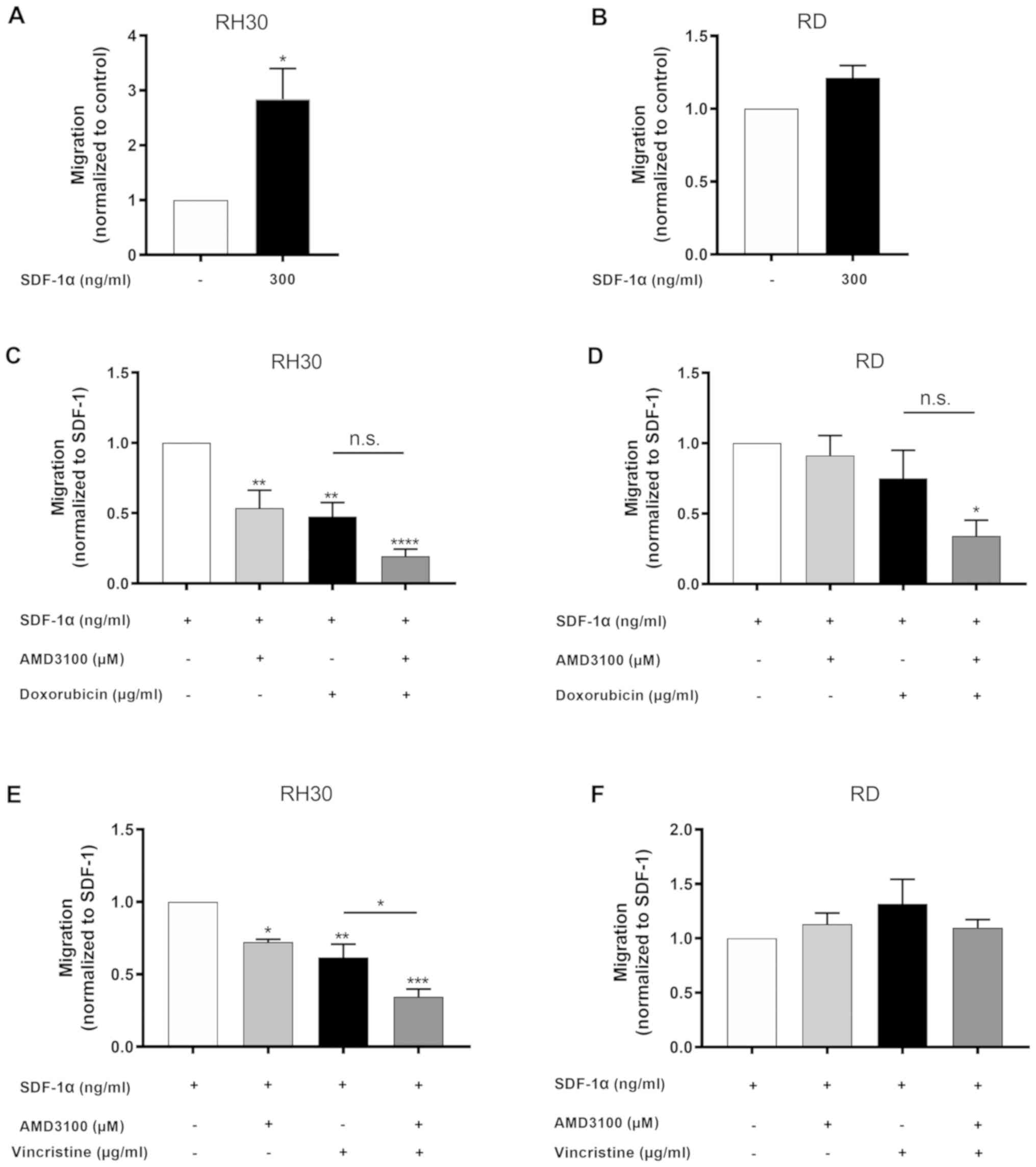
Corroborating the previously reported findings, a
significantly stronger migratory response to SDF-1α (300 ng/ml) was
only observed in RH30 cells compared with that of the untreated
control; by contrast, this was not observed in RD cells (Fig. 5A and B).
The functional impact of CXCR4 inhibition by AMD3100
alone and in combination with doxorubicin or vincristine on the
migration of RMS cell lines was assessed using a Transwell
migration assay. Treatment of RH30 cells, which exhibited high
CXCR4 expression levels, with AMD3100 significantly reduced their
migratory ability (Fig. 5C and E).
This effect was not observed in RD cells, which have low levels of
CXCR4 expression (Fig. 5D and
F).
Doxorubicin significantly reduced the migration of
RH30 cells compared with SDF-1α treatment alone; this effect was
amplified synergistically with the addition of AMD3100 (Fig. 5C). There was no change in the
migration of RD cells with doxorubicin compared with SDF-1α
treatment alone; however, the combination of doxorubicin and
AMD3100 led to a significant synergistic impairment of RD cell
migration compared with SDF-1α treatment alone (Fig. 5D).
The migration of RH30 cells was also significantly
inhibited by vincristine compared with SDF-1α treatment alone, and
this effect was synergistically and significantly stronger with the
addition of AMD3100 (Fig. 5E).
Vincristine alone or in combination with AMD3100 had no effect on
the migration of RD cells (Fig.
5F).
Effects of AMD3100 treatment on clonal
growth
The impact of the CXCR4 antagonist AMD3100 on tumor
cell proliferation was quantified in colony forming assays. As
presented in Fig. S3, the
relative number of clones was significantly decreased in the
presence of 10 and 15 µM AMD3100 in the embryonal RMS cell
line RD, but not in the alveolar RMS cell line RH30.
Discussion
The CXCR4-SDF1α axis is involved in the migration
and metastatic invasion of RMS cells in vitro (21-23).
Emerging evidence suggests that CXCR4 also serves a decisive role
in metastatic disease (44-47).
CXCR4 expression levels are considered to be a prognostic marker
for clinical outcome in soft tissue sarcoma (48). In RMS, high CXCR4 expression is
associated with an aggressive alveolar subtype, unfavorable primary
site, bone marrow infiltration and a poor clinical outcome
(15). To improve this poor
clinical prognosis, it is mandatory to understand the mechanisms
influencing the metastatic behavior of these cells in the presence
of cytotoxic agents and to investigate how chemotherapeutics
regulate the CXCR4 receptor and how this affects RMS cell
migration.
The present study examined how a combination therapy
combining chemotherapy and CXCR4 modulation modulates CXCR4
expression and CXCR4-mediated migration. First, RT-qPCR and flow
cytometry analysis were performed to assess the basal CXCR4
expression levels in RMA (RH30) and RME (RD) cell lines, and the
results demonstrated that CXCR4 receptor expression was
significantly higher in RH30 cells compared with that in RD cells.
This is in agreement with previous studies, which have reported
that CXCR4 expression levels are higher in alveolar compared with
embryonal RMS cell lines (22,49).
Additionally, in a clinical series of 40 RMS cases, significantly
higher expression of CXCR4 was identified in the alveolar subtype
of RMS (15).
The effects of chemotherapeutic agents on the
expression level of CXCR4 in RMS cells were subsequently analyzed.
Both mRNA levels and membrane expression of CXCR4 were
significantly enhanced under treatment with the topoisomerase II
inhibitors doxorubicin and dactinomycin in the RMS cell lines RD
and RH30 compared with the untreated control cells (50). The present study demonstrated for
the first time that chemotherapeutics such as vincristine and
paclitaxel induced a significant downregulation of CXCR4 at the
mRNA and protein level in RMA cells, but not in RME cells. A
possible explanation for this observation may be that
chemotherapeutic drugs such as vincristine exert a destabilizing
effect on micro-tubules and may prevent their depolymerization
(51). Another explanation may be
that vincristine and paclitaxel influence the stabilization of
certain receptors or the membrane trafficking system; this needs to
be clarified in detail in further studies. The upregulating effect
of doxorubicin and dactinomycin is in accordance with previously
published findings that chemotherapeutic agents can upregulate
CXCR4 expression in different tumor models, such as pediatric AML,
head and neck cancer and melanoma (14,25,26,52).
Meta-analyses have reported that high CXCR4
expression levels are associated with a poor outcome/prognosis and
high metastatic rates in breast cancer (46), lung cancer (53) and sarcoma (47,54).
CXCR4 has also been demonstrated to be involved in cell migration
and invasion in soft tissue sarcoma (48). High levels of CXCR4 are associated
with bone marrow involvement and poor outcome in RMS (15). Thus, increased chemokine receptor
CXCR4 expression after cytotoxic treatment with doxorubicin and
dactinomycin may represent a mechanism of therapy resistance and
metastatic behavior in pediatric RMS. To gain an insight into this
mechanism, Transwell assays were performed in the present study,
and the migratory behavior of RH30 and RD cells in response to
SDF1α was studied. SDF-1α, a selective ligand of CXCR4, serves a
pivotal role in the metastatic behavior of tumor cells that express
high levels of CXCR4 (21). In
accordance with the study by Tarnowski et al (55), the results of the present study
confirmed a significant chemotactic response to SDF-1α in RMA
(RH30) cells compared with that of cells in control medium devoid
of SDF-1α. This effect was not observed in RME (RD) cells. CXCR4
expression can be induced by the fused oncoproteins PAX3-FKHR,
which are strongly expressed in RMA but not in RME cells (56), leading to enhanced migration and
adhesion (22). This finding was
supported by the present study. Furthermore, significantly reduced
cell migration following treatment with the CXCR4 antagonist
AMD3100 was observed in RH30 cells, but not in RME cells compared
with the untreated controls. As previously described (22), these results may be attributed to
the high CXCR4 levels of RMA cells and the low CXCR4 expression in
RME cells.
The functional impact of CXCR4 modulation on the
migration of RMS cell lines was evaluated in the present study
using AMD3100 alone and in combination with doxorubicin or
vincristine. The migration of RMA cells that were pre‑treated with
vincristine or doxorubicin was significantly decreased by AMD3100,
whereas the migration of RME cells towards the medium containing
SDF-1α was not significantly influenced by AMD3100. Similar to
these results, the study by Sison et al (14) demonstrated that the
chemotherapeutic agents cytarabine, daunorubicin, etoposide and
methotrexate led to an enhanced expression of CXCR4 in the leukemia
cell lines MOLM-14, MV4-11 and 697. These authors also demonstrated
an enhanced SDF-1α mediated chemotaxis of MOLM-14 cells treated
with cytarabine (14). In the
present study on RMS cells, all the tested chemotherapeutic agents
reduced SDF-1α-induced migration of RMA cells. The reason for this
difference may be that Sison et al (14) only analyzed viable
(annexin-negative) leukemic cells. The present study did not
exhibit increased RMA cell migration induced by chemotherapeutic
drugs possibly due to methodical reasons or due to the fact that
the biological behavior of RMS cells cannot be compared with that
of leukemic cells. Another potential explanation may be the impact
of a different chemokine receptor CXCR7. CXCR7 is expressed in RMS
cells, is encoded on the same chromosome as CXCR4 (23) and has a greater affinity for SDF‑1α
(57). Kalatskaya et al
(32) has revealed that AMD3100
also functions as an agonist for CXCR7. Further studies regarding
the assessment of chemotherapeutic treatment on CXCR7 expression in
conjunction with CXCR4 are needed to reveal new aspects of the role
of the CXCR4-CXCR7-SDF-1α axis.
The results of the present study demonstrated for
the first time that doxorubicin and vincristine in combination with
AMD3100 markedly reduced the migration of RMA cells compared with
the control cells. In the RME cell line RD, this effect could only
be accomplished with doxorubicin. This was in line with a previous
study on an ALL mouse model, which reported that the combination of
AMD11070 and vincristine resulted in lower numbers of leukemic
cells in the bloodstream and higher overall survival rates compared
with the effects of monotherapy with vincristine (29). In small cell lung cancer, only the
combination of chemotherapy and AMD3100 exhibited anti-metastatic
effects (58).
Of note, AMD3100 exerted different effects on the
proliferation and clonogenicity in embryonic RMS cells RD compared
with those in alveolar RMS cells RH30. The reason why these tumor
cell lines exhibited different susceptibility to growth reduction
by a CXCR4 antagonist may be due to their different subtypes.
Although RD cells express significantly lower levels of CXCR4 on
their surface, their growth response is significantly more affected
by AMD3100 compared with that of RH30 cells, which have higher
expression levels of the CXCR4 receptor. On the other hand, CXCR4
and CXCR7 levels are (counter-) balanced with CXCR7, exhibiting a
higher affinity for SDF‑1α compared with CXCR4. Since AMD3100 can
bind to CXCR7, high expression of CXCR7 may account for the reduced
growth rate of RD cells (32,57).
It has previously been demonstrated that AMD3100, a
reversible inhibitor of CXCR4, binds to the SDF-1α binding site of
CXCR4 and thus prevents the binding of the anti-CXCR4 antibody 12G5
(29,59). The present study also observed a
reduced number of CXCR4-positive cells after AMD3100 treatment
compared with untreated cells in the flow cytometry experiments.
However, this was not reflected by the reduced CXCR4 protein levels
in AMD3100-treated cells detected by western blotting. This
suggested that a reduction of CXCR4-positive cells reflected
impaired anti‑CXCR4 antibody binding at the receptor site occupied
by AMD3100 or downregulation of the surface receptor, leading to an
increased concentration of CXCR4 intracellularly while maintaining
stable protein content.
Previous studies have suggested that in RMS, as in
other tumor entities, in addition to the blockade of CXCR4,
inhibition of CXCR7 is also important in the regulation of
angiogenesis, stem cell trafficking and mediating organ‑specific
metastases of cancer (60). The
genes for CXCR7 and CXCR4 are both located on chromosome 2q37.3,
and CXCR7 and CXCR4 share ligand specificity for SDF‑1α, suggesting
that a functional synergy of the two receptors is likely (61-63).
In addition, the CXCR4 antagonist AMD3100 increases CXCR7
expression (32,52). The migration data from the present
study, however, suggested that factors other than SDF-1α may also
modulate/contribute to the migratory behavior of RMS cells, since
doxorubicin increased CXCR4 receptor expression but reduced
migration in RH30 cells, whereas vincristine decreased both CXCR4
expression and migratory behavior in the same tumor cell line. By
contrast, in RD cells, both chemotherapeutic drugs did not change
the migratory behavior.
The sheer expression level of chemokine receptors
may not necessarily be associated with functionality, as
membrane-expressed receptors such as CXCR4 have been identified to
be functionally inactive (64,65).
This functionality has been investigated in detail with regard to
its role in migratory behavior (66). Thus, further investigations are
necessary to identify and illuminate factors that control, regulate
and modulate the CXCR4-CXCR7-SDF-1α axis in (RMS) tumor cells.
As demonstrated by Guo et al (12), not only epigenetic and
transcriptional, but also autocrine mechanisms serve a role in the
regulation of CXCR4/SDF-1α expression. In the present study, all
the investigated cytostatic drugs modulated the expression levels
of CXCR4 in RMS cells. However, a number of other factors are known
to also have a modulatory effect on CXCR4 expression (67); these include nuclear respiratory
factor 1 (68), specificity
protein 1 (69), Serum‑ and
glucocorticoid‑inducible kinase 3 and Spartin activate
atrophin-1-interacting protein 4 (70,71)
as well as hypoxia (56,72,73).
It would therefore be of great interest to investigate in more
detail the chemotherapeutic effects on the factors that control
CXCR4 expression and turnover. In vivo studies may also help
clarify the hierarchy and interplay of the factors that control
CXCR4 expression in sarcomas and may identify targets for
therapeutic options.
Considering the results of the present study and the
published data, intensification of pharmacological therapy by
inhibition of CXCR4 with AMD3100 is a promising approach in the
treatment of pediatric RMS. This treatment may be particularly
advantageous for advanced stages of RMS regarding metastasis and
drug resistance. Thus, low CXCR4 expression levels achieved through
a combination of chemotherapeutic agents with AMD3100 may help
decrease cancer cell migration, which lowers the risk of tumor
recurrence. Further studies will be necessary to translate these
promising results into preclinical trials.
Supplementary Data
Funding
This study was supported by the
Kind-Philipp-Foundation, Donors' Association for German
Science e.V. grant (to SR).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
SR and MJS conducted the experiments. SR, MJS, HB,
SS and ES analyzed the data. SR and ES designed the study. ES, SS,
KS, and GS drafted the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
This study was approved by the Ethical Committee of
the Medical Faculty of the University Hospital Tuebingen (approval
no. 354/2018A, amendment no. 354/2018B02). Written informed consent
to participate in this study was obtained from the parents of all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
The authors would like to thank Mrs. Bettina
Kirchner, Mrs. Melanie Hauth and Mrs. Julia Wenz (Department of
Pediatric Surgery and Pediatric Urology, University Hospital
Tuebingen) for technical support, Mr. Olaf Kupka and Mrs. Stefanie
Saile (IT Department, University Hospital Tuebingen) for IT support
and Mrs. Vanessa Di Cecco (McMaster University) for her support as
a native speaker.
Abbreviations:
|
RMS
|
rhabdomyosarcoma
|
|
CXCR4
|
chemokine receptor 4
|
|
SDF-1
|
stromal cell-derived factor-1
|
|
RME
|
embryonal rhabdomyosarcoma
|
|
RMA
|
alveolar rhabdomyosarcoma
|
|
ALL
|
acute lymphoblastic leukemia
|
|
AML
|
acute myeloid leukemia
|
References
|
1
|
Ognjanovic S, Linabery AM, Charbonneau B
and Ross JA: Trends in childhood rhabdomyosarcoma incidence and
survival in the United States, 1975-2005. Cancer. 115:4218–4226.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Skapek SX, Ferrari A, Gupta AA, Lupo PJ,
Butler E, Shipley J, Barr FG and Hawkins DS: Rhabdomyosarcoma. Nat
Rev Dis Primers. 5:12019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rudzinski ER, Anderson JR, Hawkins DS,
Skapek SX, Parham DM and Teot LA: The World Health Organization
Classification of Skeletal Muscle Tumors in Pediatric
Rhabdomyosarcoma: A report from the children's oncology group. Arch
Pathol Lab Med. 139:1281–1287. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Davicioni E, Anderson MJ, Finckenstein FG,
Lynch JC, Qualman SJ, Shimada H, Schofield DE, Buckley JD, Meyer
WH, Sorensen PH, et al: Molecular classification of
rhabdomyo-sarcoma–genotypic and phenotypic determinants of
diagnosis: A report from the Children's Oncology Group. Am J
Pathol. 174:550–564. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Egas-Bejar D and Huh WW: Rhabdomyosarcoma
in adolescent and young adult patients: Current perspectives.
Adolesc Health Med Ther. 5:115–125. 2014.PubMed/NCBI
|
|
6
|
Dagher R and Helman L: Rhabdomyosarcoma:
An overview. Oncologist. 4:34–44. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Oberlin O, Rey A, Lyden E, Bisogno G,
Stevens MC, Meyer WH, Carli M and Anderson JR: Prognostic factors
in metastatic rhabdo-myosarcomas: Results of a pooled analysis from
United States and European cooperative groups. J Clin Oncol.
26:2384–2389. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Perkins SM, Shinohara ET, DeWees T and
Frangoul H: Outcome for children with metastatic solid tumors over
the last four decades. PLoS One. 9:e1003962014. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Herrmann D, Seitz G, Fuchs J and
Armeanu-Ebinger S: Susceptibility of rhabdomyosarcoma cells to
macrophage-mediated cytotoxicity. OncoImmunology. 1:279–286. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Steeg PS: Tumor metastasis: Mechanistic
insights and clinical challenges. Nat Med. 12:895–904. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Gupta GP and Massagué J: Cancer
metastasis: Building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guo F, Wang Y, Liu J, Mok SC, Xue F and
Zhang W: CXCL12/CXCR4: A symbiotic bridge linking cancer cells and
their stromal neighbors in oncogenic communication networks.
Oncogene. 35:816–826. 2016. View Article : Google Scholar
|
|
13
|
Chatterjee S, Behnam Azad B and Nimmagadda
S: The intricate role of CXCR4 in cancer. Adv Cancer Res.
124:31–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sison EA, McIntyre E, Magoon D and Brown
P: Dynamic chemotherapy-induced upregulation of CXCR4 expression: A
mechanism of therapeutic resistance in pediatric AML. Mol Cancer
Res. 11:1004–1016. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Diomedi-Camassei F, McDowell HP, De Ioris
MA, Uccini S, Altavista P, Raschellà G, Vitali R, Mannarino O, De
Sio L, Cozzi DA, et al: Clinical significance of CXC chemokine
receptor-4 and c-Met in childhood rhabdomyosarcoma. Clin Cancer
Res. 14:4119–4127. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Ma Q, Liu Q, Yu H, Zhao L, Shen S
and Yao J: Blockade of SDF-1/CXCR4 signalling inhibits pancreatic
cancer progression in vitro via inactivation of canonical Wnt
pathway. Br J Cancer. 99:1695–1703. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zeelenberg IS, Ruuls-Van Stalle L and Roos
E: The chemokine receptor CXCR4 is required for outgrowth of colon
carcinoma micrometastases. Cancer Res. 63:3833–3839.
2003.PubMed/NCBI
|
|
19
|
Domanska UM, Kruizinga RC, Nagengast WB,
Timmer-Bosscha H, Huls G, de Vries EG and Walenkamp AM: A review on
CXCR4/CXCL12 axis in oncology: No place to hide. Eur J Cancer.
49:219–230. 2013. View Article : Google Scholar
|
|
20
|
Kucia M, Jankowski K, Reca R, Wysoczynski
M, Bandura L, Allendorf DJ, Zhang J, Ratajczak J and Ratajczak MZ:
CXCR4-SDF-1 signalling, locomotion, chemotaxis and adhesion. J Mol
Histol. 35:233–245. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Strahm B, Durbin AD, Sexsmith E and Malkin
D: The CXCR4-SDF1alpha axis is a critical mediator of
rhabdomyo-sarcoma metastatic signaling induced by bone marrow
stroma. Clin Exp Metastasis. 25:1–10. 2008. View Article : Google Scholar
|
|
22
|
Libura J, Drukala J, Majka M, Tomescu O,
Navenot JM, Kucia M, Marquez L, Peiper SC, Barr FG,
Janowska-Wieczorek A, et al: CXCR4-SDF-1 signaling is active in
rhabdomyosarcoma cells and regulates locomotion, chemotaxis, and
adhesion. Blood. 100:2597–2606. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Grymula K, Tarnowski M, Wysoczynski M,
Drukala J, Barr FG, Ratajczak J, Kucia M and Ratajczak MZ:
Overlapping and distinct role of CXCR7-SDF-1/ITAC and CXCR4-SDF-1
axes in regulating metastatic behavior of human rhabdomyosarcomas.
Int J Cancer. 127:2554–2568. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lev DC, Onn A, Melinkova VO, Miller C,
Stone V, Ruiz M, McGary EC, Ananthaswamy HN, Price JE and Bar-Eli
M: Exposure of melanoma cells to dacarbazine results in enhanced
tumor growth and metastasis in vivo. J Clin Oncol. 22:2092–2100.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kim M, Koh YJ, Kim KE, Koh BI, Nam DH,
Alitalo K, Kim I and Koh GY: CXCR4 signaling regulates metastasis
of chemoresistant melanoma cells by a lymphatic metastatic niche.
Cancer Res. 70:10411–10421. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muller A, Sonkoly E, Eulert C, Gerber PA,
Kubitza R, Schirlau K, Franken-Kunkel P, Poremba C, Snyderman C,
Klotz LO, et al: Chemokine receptors in head and neck cancer:
Association with metastatic spread and regulation during
chemotherapy. Int J Cancer. 118:2147–2157. 2006. View Article : Google Scholar
|
|
27
|
Ratajczak MZ, Jadczyk T, Schneider G,
Kakar SS and Kucia M: Induction of a tumor-metastasis-receptive
microenvironment as an unwanted and underestimated side effect of
treatment by chemotherapy or radiotherapy. J Ovarian Res. 6:952013.
View Article : Google Scholar
|
|
28
|
Shaked Y, Henke E, Roodhart JM, Mancuso P,
Langenberg MH, Colleoni M, Daenen LG, Man S, Xu P, Emmenegger U, et
al: Rapid chemotherapy-induced acute endothelial progenitor cell
mobilization: Implications for antiangiogenic drugs as
chemo-sensitizing agents. Cancer Cell. 14:263–273. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parameswaran R, Yu M, Lim M, Groffen J and
Heisterkamp N: Combination of drug therapy in acute lymphoblastic
leukemia with a CXCR4 antagonist. Leukemia. 25:1314–1323. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Juarez J, Bradstock KF, Gottlieb DJ and
Bendall LJ: Effects of inhibitors of the chemokine receptor CXCR4
on acute lympho-blastic leukemia cells in vitro. Leukemia.
17:1294–1300. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Domanska UM, Timmer-Bosscha H, Nagengast
WB, Oude Munnink TH, Kruizinga RC, Ananias HJ, Kliphuis NM, Huls G,
De Vries EG, de Jong IJ, et al: CXCR4 inhibition with AMD3100
sensitizes prostate cancer to docetaxel chemotherapy. Neoplasia.
14:709–718. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kalatskaya I, Berchiche YA, Gravel S,
Limberg BJ, Rosenbaum JS and Heveker N: AMD3100 is a CXCR7 ligand
with allosteric agonist properties. Mol Pharmacol. 75:1240–1247.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
De Clercq E: Potential clinical
applications of the CXCR4 antagonist bicyclam AMD3100. Mini Rev Med
Chem. 5:805–824. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kajiyama H, Shibata K, Terauchi M, Ino K,
Nawa A and Kikkawa F: Involvement of SDF-1alpha/CXCR4 axis in the
enhanced peritoneal metastasis of epithelial ovarian carcinoma. Int
J Cancer. 122:91–99. 2008. View Article : Google Scholar
|
|
35
|
Smith MC, Luker KE, Garbow JR, Prior JL,
Jackson E, Piwnica-Worms D and Luker GD: CXCR4 regulates growth of
both primary and metastatic breast cancer. Cancer Res.
64:8604–8612. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabioglu N, Summy J, Miller C, Parikh NU,
Sahin AA, Tuzlali S, Pumiglia K, Gallick GE and Price JE:
CXCL-12/stromal cell-derived factor-1alpha transactivates HER2-neu
in breast cancer cells by a novel pathway involving Src kinase
activation. Cancer Res. 65:6493–6497. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li YM, Pan Y, Wei Y, Cheng X, Zhou BP, Tan
M, Zhou X, Xia W, Hortobagyi GN, Yu D, et al: Upregulation of CXCR4
is essential for HER2-mediated tumor metastasis. Cancer Cell.
6:459–469. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ray P, Lewin SA, Mihalko LA, Schmidt BT,
Luker KE and Luker GD: Noninvasive imaging reveals inhibition of
ovarian cancer by targeting CXCL12-CXCR4. Neoplasia. 13:1152–1161.
2011. View Article : Google Scholar
|
|
39
|
Schmid E, Stagno MJ, Yan J, Stournaras C,
Lang F, Fuchs J and Seitz G: Store-operated Ca(2+) entry in
rhabdomyosarcoma cells. Biochem Biophys Res Commun. 477:129–136.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Foucquier J and Guedj M: Analysis of drug
combinations: Current methodological landscape. Pharmacol Res
Perspect. 3:e001492015. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Schols D, Struyf S, Van Damme J, Esté JA,
Henson G and De Clercq E: Inhibition of T-tropic HIV strains by
selective antagonization of the chemokine receptor CXCR4. J Exp
Med. 186:1383–1388. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Poty S, Désogère P, Goze C, Boschetti F,
D'huys T, Schols D, Cawthorne C, Archibald SJ, Maëcke HR and Denat
F: New AMD3100 derivatives for CXCR4 chemokine receptor targeted
molecular imaging studies: Synthesis, anti-HIV-1 evaluation and
binding affinities. Dalton Trans. 44:pp. 5004–5016. 2015,
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Stamatopoulos B, Meuleman N, De Bruyn C,
Pieters K, Mineur P, Le Roy C, Saint-Georges S, Varin-Blank N,
Cymbalista F, Bron D, et al: AMD3100 disrupts the cross-talk
between chronic lymphocytic leukemia cells and a mesenchymal
stromal or nurse-like cell-based microenvironment: Pre-clinical
evidence for its association with chronic lymphocytic leukemia
treatments. Haematologica. 97:608–615. 2012. View Article : Google Scholar :
|
|
44
|
Zlotnik A: New insights on the role of
CXCR4 in cancer metastasis. J Pathol. 215:211–213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang J, Loberg R and Taichman RS: The
pivotal role of CXCL12 (SDF-1)/CXCR4 axis in bone metastasis.
Cancer Metastasis Rev. 25:573–587. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Zhang Z, Ni C, Chen W, Wu P, Wang Z, Yin
J, Huang J and Qiu F: Expression of CXCR4 and breast cancer
prognosis: A systematic review and meta-analysis. BMC Cancer.
14:492014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Laverdiere C, Hoang BH, Yang R, Sowers R,
Qin J, Meyers PA, Huvos AG, Healey JH and Gorlick R: Messenger RNA
expression levels of CXCR4 correlate with metastatic behavior and
outcome in patients with osteosarcoma. Clin Cancer Res.
11:2561–2567. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kim RH, Li BD and Chu QD: The role of
chemokine receptor CXCR4 in the biologic behavior of human soft
tissue sarcoma. Sarcoma. 2011:5937082011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Jankowski K, Kucia M, Wysoczynski M, Reca
R, Zhao D, Trzyna E, Trent J, Peiper S, Zembala M, Ratajczak J, et
al: Both hepatocyte growth factor (HGF) and stromal-derived
factor-1 regulate the metastatic behavior of human rhabdomyosarcoma
cells, but only HGF enhances their resistance to radiochemotherapy.
Cancer Res. 63:7926–7935. 2003.PubMed/NCBI
|
|
50
|
Marinello J, Delcuratolo M and Capranico
G: Anthracyclines as topoisomerase II poisons: From early studies
to new perspectives. Int J Mol Sci. 19:192018. View Article : Google Scholar
|
|
51
|
Mukhtar E, Adhami VM and Mukhtar H:
Targeting microtubules by natural agents for cancer therapy. Mol
Cancer Ther. 13:275–284. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Sison EA, Magoon D, Li L, Annesley CE, Rau
RE, Small D and Brown P: Plerixafor as a chemosensitizing agent in
pediatric acute lymphoblastic leukemia: Efficacy and potential
mechanisms of resistance to CXCR4 inhibition. Oncotarget.
5:8947–8958. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Liang JX, Gao W, Liang Y and Zhou XM:
Chemokine receptor CXCR4 expression and lung cancer prognosis: A
meta-analysis. Int J Clin Exp Med. 8:5163–5174. 2015.PubMed/NCBI
|
|
54
|
Li YJ, Dai YL, Zhang WB, Li SJ and Tu CQ:
Clinicopathological and prognostic significance of chemokine
receptor CXCR4 in patients with bone and soft tissue sarcoma: A
meta-analysis. Clin Exp Med. 17:59–69. 2017. View Article : Google Scholar
|
|
55
|
Tarnowski M, Grymula K, Liu R, Tarnowska
J, Drukala J, Ratajczak J, Mitchell RA, Ratajczak MZ and Kucia M:
Macrophage migration inhibitory factor is secreted by
rhabdomyosarcoma cells, modulates tumor metastasis by binding to
CXCR4 and CXCR7 receptors and inhibits recruitment of
cancer‑associated fibroblasts. Mol Cancer Res. 8:pp. 1328–1343.
2010, View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tarnowski M, Grymula K, Reca R, Jankowski
K, Maksym R, Tarnowska J, Przybylski G, Barr FG, Kucia M and
Ratajczak MZ: Regulation of expression of stromal-derived factor-1
receptors: CXCR4 and CXCR7 in human rhabdomyosarcomas. Mol Cancer
Res. 8:1–14. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cheng X, Wang H, Zhang X, Zhao S, Zhou Z,
Mu X, Zhao C and Teng W: The Role of SDF-1/CXCR4/CXCR7 in Neuronal
Regeneration after Cerebral Ischemia. Front Neurosci. 11:5902017.
View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Taromi S, Kayser G, Catusse J, von
Elverfeldt D, Reichardt W, Braun F, Weber WA, Zeiser R and Burger
M: CXCR4 antagonists suppress small cell lung cancer progression.
Oncotarget. 7:85185–85195. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
De Clercq E: Inhibition of HIV infection
by bicyclams, highly potent and specific CXCR4 antagonists. Mol
Pharmacol. 57:833–839. 2000.PubMed/NCBI
|
|
60
|
Sun X, Cheng G, Hao M, Zheng J, Zhou X,
Zhang J, Taichman RS, Pienta KJ and Wang J: CXCL12 / CXCR4 / CXCR7
chemokine axis and cancer progression. Cancer Metastasis Rev.
29:709–722. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Balabanian K, Lagane B, Infantino S, Chow
KY, Harriague J, Moepps B, Arenzana-Seisdedos F, Thelen M and
Bachelerie F: The chemokine SDF-1/CXCL12 binds to and signals
through the orphan receptor RDC1 in T lymphocytes. J Biol Chem.
280:35760–35766. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Burns JM, Summers BC, Wang Y, Melikian A,
Berahovich R, Miao Z, Penfold ME, Sunshine MJ, Littman DR, Kuo CJ,
et al: A novel chemokine receptor for SDF-1 and I-TAC involved in
cell survival, cell adhesion, and tumor development. J Exp Med.
203:2201–2213. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Heesen M, Berman MA, Charest A, Housman D,
Gerard C and Dorf ME: Cloning and chromosomal mapping of an orphan
chemokine receptor: Mouse RDC1. Immunogenetics. 47:364–370. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Mitra P, De A, Ethier MF, Mimori K, Kodys
K, Shibuta K, Mori M, Madison JM, Miller-Graziano C and Barnard GF:
Loss of chemokine SDF-1alpha-mediated CXCR4 signalling and receptor
internalization in human hepatoma cell line HepG2. Cell Signal.
13:311–319. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Honczarenko M, Douglas RS, Mathias C, Lee
B, Ratajczak MZ and Silberstein LE: SDF-1 responsiveness does not
correlate with CXCR4 expression levels of developing human bone
marrow B cells. Blood. 94:2990–2998. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rabin RL, Park MK, Liao F, Swofford R,
Stephany D and Farber JM: Chemokine receptor responses on T cells
are achieved through regulation of both receptor expression and
signaling. J Immunol. 162:3840–3850. 1999.PubMed/NCBI
|
|
67
|
Busillo JM and Benovic JL: Regulation of
CXCR4 signaling. Biochim Biophys Acta. 1768:952–963. 2007.
View Article : Google Scholar
|
|
68
|
Moriuchi M, Moriuchi H, Turner W and Fauci
AS: Cloning and analysis of the promoter region of CXCR4, a
coreceptor for HIV-1 entry. J Immunol. 159:4322–4329.
1997.PubMed/NCBI
|
|
69
|
Wegner SA, Ehrenberg PK, Chang G, Dayhoff
DE, Sleeker AL and Michael NL: Genomic organization and functional
char-acterization of the chemokine receptor CXCR4, a major entry
co‑receptor for human immunodeficiency virus type 1. J Biol Chem.
273:4754–4760. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Marchese A, Raiborg C, Santini F, Keen JH,
Stenmark H and Benovic JL: The E3 ubiquitin ligase AIP4 mediates
ubiquiti-nation and sorting of the G protein-coupled receptor
CXCR4. Dev Cell. 5:709–722. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Slagsvold T, Marchese A, Brech A and
Stenmark H: CISK attenuates degradation of the chemokine receptor
CXCR4 via the ubiquitin ligase AIP4. EMBO J. 25:3738–3749. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Schioppa T, Uranchimeg B, Saccani A,
Biswas SK, Doni A, Rapisarda A, Bernasconi S, Saccani S, Nebuloni
M, Vago L, et al: Regulation of the chemokine receptor CXCR4 by
hypoxia. J Exp Med. 198:1391–1402. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Wang X, Li C, Chen Y, Hao Y, Zhou W, Chen
C and Yu Z: Hypoxia enhances CXCR4 expression favoring microglia
migration via HIF-1alpha activation. Biochem Biophys Res Commun.
371:283–288. 2008. View Article : Google Scholar : PubMed/NCBI
|