Introduction
Even though the treatment strategies against
epithelial ovarian cancer have witnessed marked improvements,
epithelial ovarian cancer remains the leading cause of
cancer-related mortality from gynecological cancers (1). As the symptoms are vague and
non-specific, these tumors often present at an advanced stage; in
addition, the molecular mechanisms that govern its dissemination
are not yet fully understood.
MicroRNA (miRNA/miR-126) is an intragenic miRNA. It
is located on human chromosome 9, within the 7th intron of EGFL7
(2). The function of miR-126 in
physiological and pathological processes has been extensively
studied (3-5). miR-126 is found in endothelial cells
and in highly vascularized tissues (6). In vivo studies have
demonstrated miR-126 mutant mice develop leaky blood vessels,
indicating that miR-126 plays an essential role in developmental
angiogenesis and vascular integrity (7). In cancers, the downregulation of
miR-126 is a frequent occurrence. The aberrant expression of
miR-126 is closely related to a variety of cancers, such as
hepatocellular carcinoma, breast cancer, thyroid cancer,
gastrointestinal cancer and lung cancer (2,8).
Research has indicated (5) that
miR-126 can inhibit cell proliferation, invasion and migration by
targeting solute carrier family 7 member 5 (SLC7A5), SRY-box
transcription factor 2 (SOX-2), insulin receptor substrate 1
(IRS-1), homeobox A9 (HOXA9), ADAM metallopeptidase domain 9
(ADAM9), CRK proto-oncogene, adaptor protein (CRK), KRAS, epidermal
growth factor (EGF)-like domain multiple 7 (EGFL7),
phosphoinositide 3-kinase (PI3K) and vascular endothelial growth
factor (VEGF).
The association between miR-126 and epithelial
ovarian cancer has been reported in previous studies (9,10).
However, limited research has been conducted on its expression and
biological function in ovarian cancer (11-13).
In the present study, laser microdissection was used to ensure
specimen homogeneity. The expression of miR-126 was examined using
clinical specimens and in vivo animal and in vitro
cell models. The interaction between miR-126 and VEGF-A and its
role in epithelial ovarian cancer were investigated.
Materials and methods
Tissue samples
Tumor and normal specimens were obtained from the
Gynecologic Tissue Bank of Wuhan Union Hospital with written
informed consent and ethical approval (February, 2004 to June,
2016). The present study was approved by the Ethics Committee of
Union Hospital, Tongji Medical College, Huazhong University of
Science and Technology, China. In total, 10 normal ovarian tissues
(germinal epithelium from patients with adenomyosis or myoma) and
36 malignant epithelial tumors (1 clear cell tumor, 3 endometrioid
carcinomas, 6 mucinous cystadenocarcinomas and 26 serous
cystadeno-carcinomas) were analyzed. No patients had been subjected
to previous chemotherapy. All diagnoses were pathologically
confirmed.
Laser microdissection (LMD)
To ensure the homogeneity of the specimens,
specimens with <90% cancerous content were subjected to LMD. LMD
was performed according to the method described in the study by Cai
et al (14). Briefly,
serial sections (8-µm-thick) and a 4-µm-thick section
were cut from formalin-fixed paraffin-embedded (FFPE) samples for
LMD and H&E staining, respectively. Immediately prior to LMD,
the sections were deparaffinized, stained with 1% cresyl violet
(Sigma-Aldrich; Merck KGaA) for 1 min, dehydrated through 75, 95
and 100% alcohol grades for 30 sec each, and finally immersed in
xylene for 3 min and air-dried for 1 min. The sections were
deparaffinized and dehydrated prior to micro-dissection, which was
carried out using an ArcturusXT LCM instrument (Applied
Biosystems-Life Technologies; Thermo Fisher Scientific, Inc.). The
AutoScan™ XT1.2 analysis software module was used for
visualization. The captured cells (approximately 5,000 per
specimen) were used in subsequent analysis.
Cell lines and culture
SKOV3 and ES2 cell lines were purchased from the
China Center for Type Culture Collection. The former was derived
from adenocarcinoma, and the latter was derived from ovarian clear
cell carcinoma. These two cell lines were propagated in Dulbecco's
modified Eagle's medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.). Cell lines were cultured at 37°C in a humidified
incubator with 5% CO2.
Isolation and culture of HUVECs
Human umbilical cords were collected with written
informed consent and ethical approval provided the the Ethics
Committee of Union Hospital, Tongji Medical College, Huazhong
University of Science and Technology, Wuhan, China. Human umbilical
cords were collected from 24 pregnant women (38-41 weeks of
gestational age, abdominal delivery, singleton, with no
complications) at Wuhan Union Hospital (from March, 2014 to
September, 2014). Endothelial cells (ECs) were isolated from the
umbilical vein vascular wall and cultured as previously described
by Xie et al (15). HUVECs
were identified with two endothelial cell markers, factor VIII
related antigen [von Willebrand factor (vWF)] and CD31 [platelet
endothelial cell adhesion molecule-1 (PECAM-1)]. Prior to
identification, the isolated cells were fixed with 4%
paraformaldehyde and sealed with 1% bovine serum albumin (Gibco;
Thermo Fisher Scientific, Inc.) for 30 min at room temperature
sequentially. For immunofluorescence identification, the isolated
cells were treated with primary antibody to Factor VIII related
antigen (1:100, cat. no. PB0086, Boster Biological Technology) and
CD31 (1:500, cat. no. M01513; Boster Biological Technology) at 4°C
in a humid chamber overnight. Subsequently, the cells were washed
with phosphate-buffered saline (PBS) 3 times at room temperature,
and then incubated with biotinylated secondary antibody from
SABC-Cy3 kit (1:100; cat. no. SA1078; Boster Biological Technology)
for 30 min at at 37°C. Before the cells were counterstained with
SABC-Cy3 from the SABC-Cy3 kit (1:100; cat. no. SA1078; Boster
Biological Technology) in the dark for 30 min at 37°C, they were
washed with PBS 3 times at room temperature. After washing with PBS
5 min for 4 times at room temperature, the cells were visualized
under a fluorescence microscope (IX71; Olympus Corporation).
Transfection of RNA
oligoribonucleotides
Transfection was performed using Lipofectamine 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer's protocol. At 1 day prior to transfection, cells were
plated in 6-well plates (2×105/well). The following day,
or when the cells were 70% confluent, they were transfected with
either 100 pmol of miR-126 mimics (target sequence,
5′-UCGUACCGUGAGUAAUAAUGCG-3′ and 5′-CAUUAU UACUCACG GUACGAU U-3′;
GenePha r ma), miR-126 inhibitors (target sequence, 5′-CGCAUUAUU
ACUCACGGUACGA-3′; GenePharma) or the negative control siRNAs (MNC
and INC) (GenePharma). Subsequent experiments were performed at
24-48 h following transfection.
Lentiviruses packaging and stable cell
lines
Lentiviral constructs with hsa-miR-126 or a
hsa-miR-scrambled control vectors were constructed, packaged and
validated by GenePharma. For LV-miR-126 transfection, SKOV3 and ES2
cells were seeded into 6-well plates at 3.5×105
cells/well. Following propagation for 24 h, viral particles
(3×107) were added.
Recombinant human VEGF
Recombinant human VEGF (cat. no. 100-20; PeproTech)
was added at a concentration of 40 ng/ml to lentivirus-miR-126
(LV-miR-126)-infected cells. Following incubation for 24 h at 37°C,
RNA and protein were collected for RT-qPCR and western blot
analysis, respectively as described below. The infected cells were
used for migration, invasion and cell proliferation
experiments.
Bioinformatics analyses
For miRNA target gene prediction, TargetScan online
software (http://www.targetscan.org/), miRanda
(http://www.microrna.org/microrna/getDownloads.do)
and PicTar (http://pictar.mdc-berlin.de/) were used.
RNA extraction and RT-qPCR
Total RNA was extracted from the FFPE specimens and
cultured cell using the RNeasy FFPE kit (Qiagen) and TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), respectively. cDNA
was synthesized using the RevertAid™ First Strand cDNA Synthesis
kit (MBI Fermentas) according to the supplied protocol. miR-126 was
amplified in triplicate using a Hairpin-it™ miRNAs qPCR
Quantitation kit (GenePharma) on a StepOnePlus system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) following the
manufacturer's instructions. The levels of miR-126 expression were
normalized to U6. The cycling parameters for the reverse
transcription reaction were 16°C for 30 min, 42°C for 30 min, 85°C
for 10 min and a hold at 4°C. The amplification conditions were as
follows: Initial 1 step at 95°C for 3 min, followed by 40 cycles at
95°C for 12 sec and with a final extension step at 62°C for 60 sec.
The primers used were as follows: VEGF-A forward,
5′-GAACTTTCTGCTGTCTTGG-3′ and reverse, 5′-TTTTCTTGTCTTGCTCTATCT-3′;
and β-actin forward, 5′-GCCAACACAGTGCTGTCTGG-3′ and reverse,
5′-GCTCAGGAGGAGCAATGATCTTG-3′. The expression level of each gene
was calculated using the 2−ΔΔCq method (16).
Transwell assay
Cell migration and invasion were measured using
Transwell® chambers (8 µm pore size; Corning
Inc.). For the Transwell assay, at 24 h following transfection with
miR-126 mimics and miRNA-126 inhibitors, 1.0×105 cells
(3 replicates per group) were suspended in serum-free medium and
seeded into the upper chamber with 8 µm pore filters, and
600 µl of DMEM with 10% FBS was added to the lower chamber.
The cells were then incubated for 24 h at 37°C. Migrated cells were
stained with crystal violet (Beyotime Institute of Biotechnology)
for 20 min at room temperature and observed under an optical
microscope (IX71; Olympus Corporation). For the invasion assay, the
protocols were similar to those for the Transwell migration assays,
with the difference being that an aliquot of Matrigel (50
µl) (Corning Inc.) was applied to the upper surface to mimic
the basement membrane. The results were analyzed using Image-Pro
Plus, v6.0 (Media Cyberbetics).
Wound-healing assays
Following 12 h of transfection with miR-126 mimics
and miR-126 inhibitors, the SKOV3 cells were plated at
1×106 per well into 6-well plates and allowed to reach
90% confluence. The monolayer was scratched using a 200 ml pipette
tip after the cells were serum-starved for 12 h and washed with
serum-free medium to remove detached cells. The cells were cultured
with serum-free medium. At 0 and 24 h, an inverted microscope
(IX71; Olympus Corporation) was utilized to visualize the wound
healing and obtain images. The percentage migration was calculated
using the following equation: [Δ area/area (day 0)] ×100.
Cell proliferation assay
Cell proliferation was evaluated using the
Clik-iT® EdU Imaging kit and Click-iT®
reaction cocktail (Invitrogen; Thermo Fisher Scientific, Inc.)
according to the manufacturer's protocol. The results were analyzed
using Image-Pro Plus, v6.0 (Media Cyberbetics). Cell viability was
assessed using trypan blue staining (Invitrogen; Thermo Fisher
Scientific, Inc.). Following transfection, the cells were stained
by 0.4% trypan blue for 3 min at room temperature. The growth rate
was determined by trypsinization and counting the number of viable
cells (trypan blue exclusion) on a hematocytometer in triplicate
every 12 h for 3 days.
F-actin immunofluorescence
Following 48 h of transfection with miR-126 mimics
and miR-126 inhibitors, the SKOV3 cells were cultured at
5×105 per well in 12-well plates and allowed to adhere
for 24 h. SKOV3 cells were grown to 75% confluency on uncoated
glass cover slips in a 12-well plate. The cells were fixed in 3.7%
paraformaldehyde for 10 mins followed 0.1% Triton X-100 for 15 min
at room temperature. The cells were maintained for 60 min at room
temperature in rhodamine-phalloidin solution. Finally, nuclei were
stained with Hoechst 33342 (Invitrogen Life Technologies; Thermo
Fisher Scientific, Inc.) for 30 min at room temperature. Images
were captured using an IX71 digital camera (Olympus
Corporation).
Tube formation assay
Tube formation assay was used to evaluate the
effects of miR-126 on HUVECs according to a previously published
study with a few modifications (17). Briefly, the HUVECs were seeded in a
Matrigel-coated plate at a density of 5×104 cells per
well. Following 1 h of incubation at 37°C, the HUVECs were
incubated with LV-miR-126 supernatant for 24 h at 37°C and 5%
CO2. Tube formation was observed using an inverted
microscope (Leica DMI6000B; Leica Microsystems GmbH).
Enzyme-linked immunosorbent assay (ELISA)
for VEGF
The levels of VEGF-A in conditioned media were
measured using a human ELISA kit (cat. no. K5363-100;
NeoBioscience) as per the manufacturer's instructions.
Protein extraction and western blot
analysis
Cells were washed twice with cold PBS, then lysed
using cell lysis (RIPA) buffer (Beyotime Institute of
Biotechnology). Cell lyses were centrifuged at 13,000 × g for 10
min, and the supernatants were collected and stored in aliquots at
-80°C after the protein concentration was measured using the
bicinchoninic acid (BCA) assay. Total cell extracts were resolved
on a 12% SDS polyacrylamide gels (25 µg of protein were
loaded per lane) and blotted onto a Hybond PVDF membrane (GE
Healthcare Life Sciences). The membranes were blocked with blocking
buffer [5% non-fat dry milk in tris-buffered saline containing 0.1%
(v⁄v) Tween-20 (TBST)] for 1 h at room temperature. The membranes
were then incubated overnight at 4°C with VEGF (1:2,000 dilution;
cat. no. 500-M88; Peprotech), E-cadherin (1:500 dilution; cat. no.
3195; Cell Signaling Technology, Inc.), vimentin (1:500 dilution;
cat. no. 5741; Cell Signaling Technology, Inc.), matrix
metalloproteinase (MMP)2 (1:1,000 dilution; cat. no. 2763-1;
Epitomics; Abcam) and β-actin (1:1,000 dilution; cat. no. sc-8432;
Santa Cruz Biotechnology, Inc.). Following three 10-min washes in
TBST, the membranes were incubated for 2 h at room temperature with
a horseradish peroxidase-conjugated secondary antibody at 1:5,000
in blocking solution (cat. no. sc-2357; Santa Cruz Biotechnology,
Inc.) and visualized using an enhanced chemiluminescence kit
(Pierce; Thermo Fisher Scientific, Inc.).
Histological and immunohistochemical
analysis
Immunohistochemical analyses were performed
according to standard procedures (18). Antibodies and reagents for
immunocytochemistry (IHC) included: Anti-VEGF-A (1:1,00 dilution;
cat. no. BA0407; Wuhan Boster Biological Technology, Ltd.),
anti-MMP2 (1:50 dilution; cat. no. 40994; Cell Signaling
Technology, Inc.), anti-vimentin (1:100 dilution; cat. no. 5741;
Cell Signaling Technology, Inc.), anti-E-cadherin (1:100 dilution;
cat. no. 3195; Cell Signaling Technology, Inc.) and biotinylated
goat anti-rabbit secondary antibody (1:100; cat. no. sc-2004; Santa
Cruz Biotechnology, Inc.). The sections were incubated with the
above-mentioned primary antibodies at 4°C overnight, followed by 1
h of incubation with the secondary antibody. Images were captured
using an IX71 digital camera (Olympus Corporation).
Ovarian tumor xenograft model
Female BALB/c nude mice (n=10; age, 5 weeks; weight,
17-18 g) were randomly separated into two groups, the LV-miR-126
and negative control (NC), (n=5 mice/group) and used for tumor
formation assay. The housing conditions of the animals were as
follows: Temperature, 23±1°C; humidity, 40-70%; 12-h dark/light
cycle; and free access to food and water. LV-miR-126 SKOV3 cells or
negative control cells (5×106 cells /mouse) were
injected subcutaneously into the dorsal flanks of the nude mice.
The tumor sizes were measured every 4 days using micrometer
calipers. Tumor volumes were calculated according to the formula: ½
x length x width2. After 30 days, the mice were
euthanized by cervical dislocation and the tumor tissues were
harvested; death was confirmed by the completely termination of the
heartbeat and breathing, as well as the disappearance of the foot
withdrawal reflex. All animal studies were approved by the
Institutional Animal Care and Use Committee at Tongji Medical
College, Huazhong University of Science and Technology.
Statistical analysis
SPSS software v12.0 (SPSS Inc.) was used to perform
statistical analysis. Data are expressed as the means ± SD.
Multigroup comparisons of the means were carried out by one-way
analysis of variance (ANOVA) test with Tukey's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference. Spearman's correlation analysis was utilized to
determine the correlation between miR-126 and VEGF-A
expression.
Results
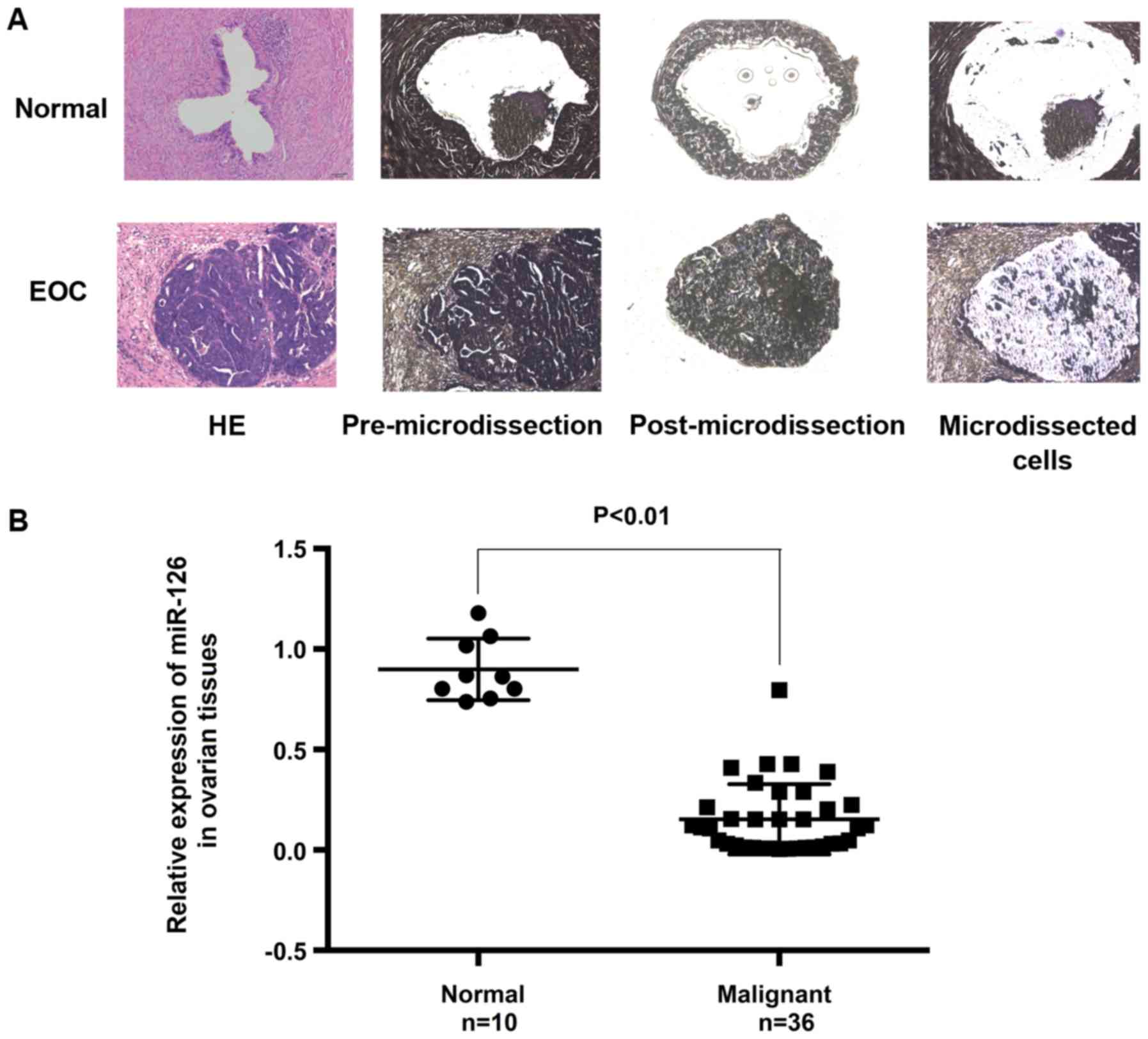
miR-126 expression is downregulated in
epithelial ovarian cancer
Previous reports have associated miR-126 with
epithelial ovarian cancer. In the present study, to clarify whether
miR-126 is downregulated in EOC, the expression of miR-126 was
determined in 46 tissue specimens by RT-qPCR. To ensure the
homogeneity of the specimens, 21 specimens (out of 46 specimens)
with <90% cancerous content were subjected to LMD. The results
revealed that the expression of miR-126 was lower in malignant
tumors than in normal ovarian epithelium (Fig. 1).
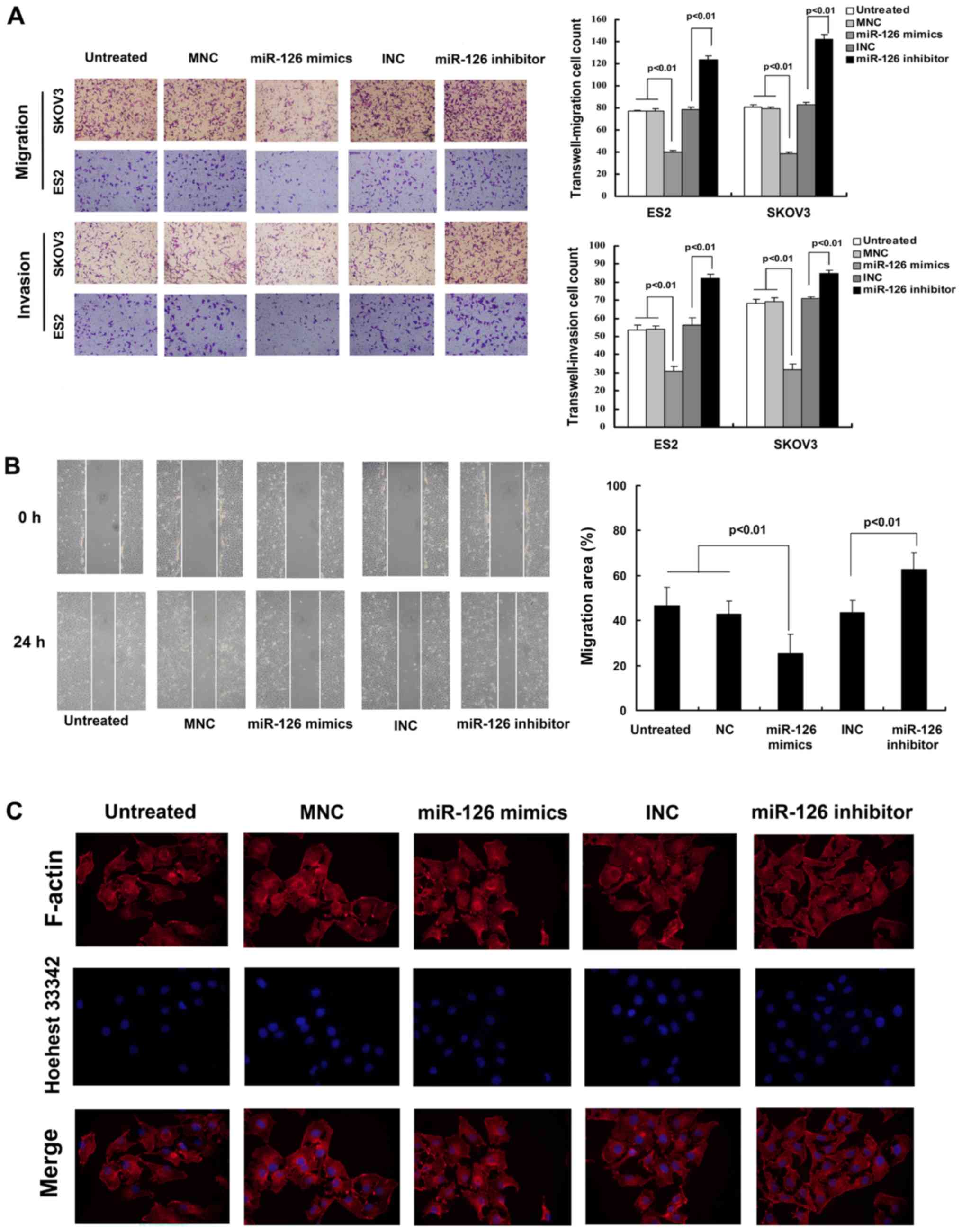
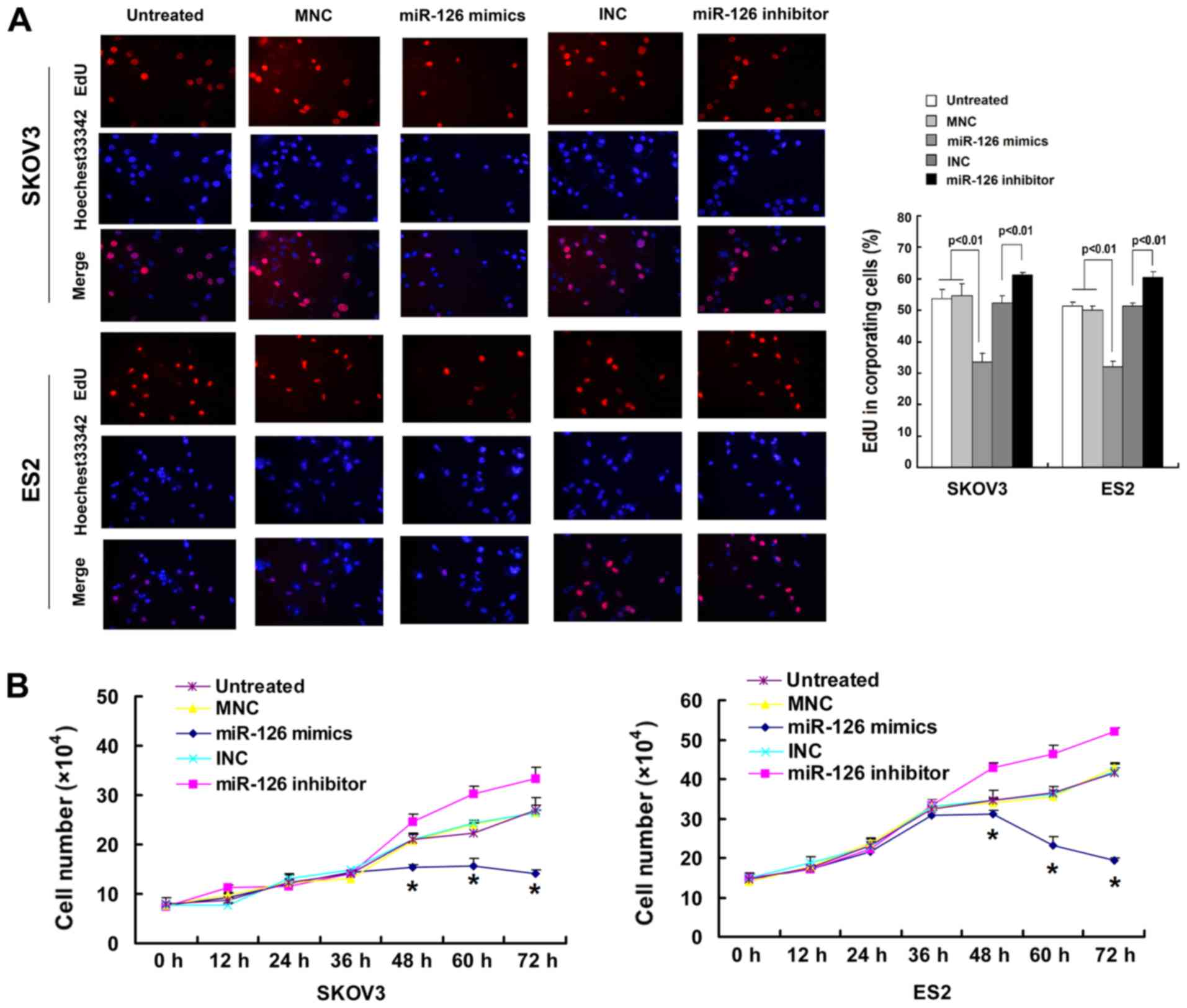
miR-126 suppresses cancer-relevant traits
in vitro
To examine the effects of miR-126 on cell migration
and proliferation, Transwell, wound-healing, EdU incorporation and
trypan blue exclusion assays were used. The SKOV3 and ES2 cells
were transfected with miR-126 mimics or inhibitors. The results
indicated that the ectopic expression of miR-126 decreased the
invasive and migratory ability of the SKOV3 and ES2 cells at 36 h
following transfection (Fig. 2A).
These observations were confirmed by a wound-healing assay
(Fig. 2B). However, the inhibition
of miR-126 reversed these effects. The ectopic expression of
miR-126, however, did not markedly affect the cytoskeleton in
vitro (Fig. 2C). It was also
found that the over-expression of miR-126 decreased the cell
proliferation rates, and the downregulation of miR-126 increased
the cell proliferation rates at 48 h following transfection
(Fig. 3A). These observations were
confirmed by the trypan blue exclusion assay (Fig. 3B). The miR-126 mimic negative
control (MNC) and the miR-126 inhibitor negative control (INC)
failed to influence migration, invasion and proliferation.
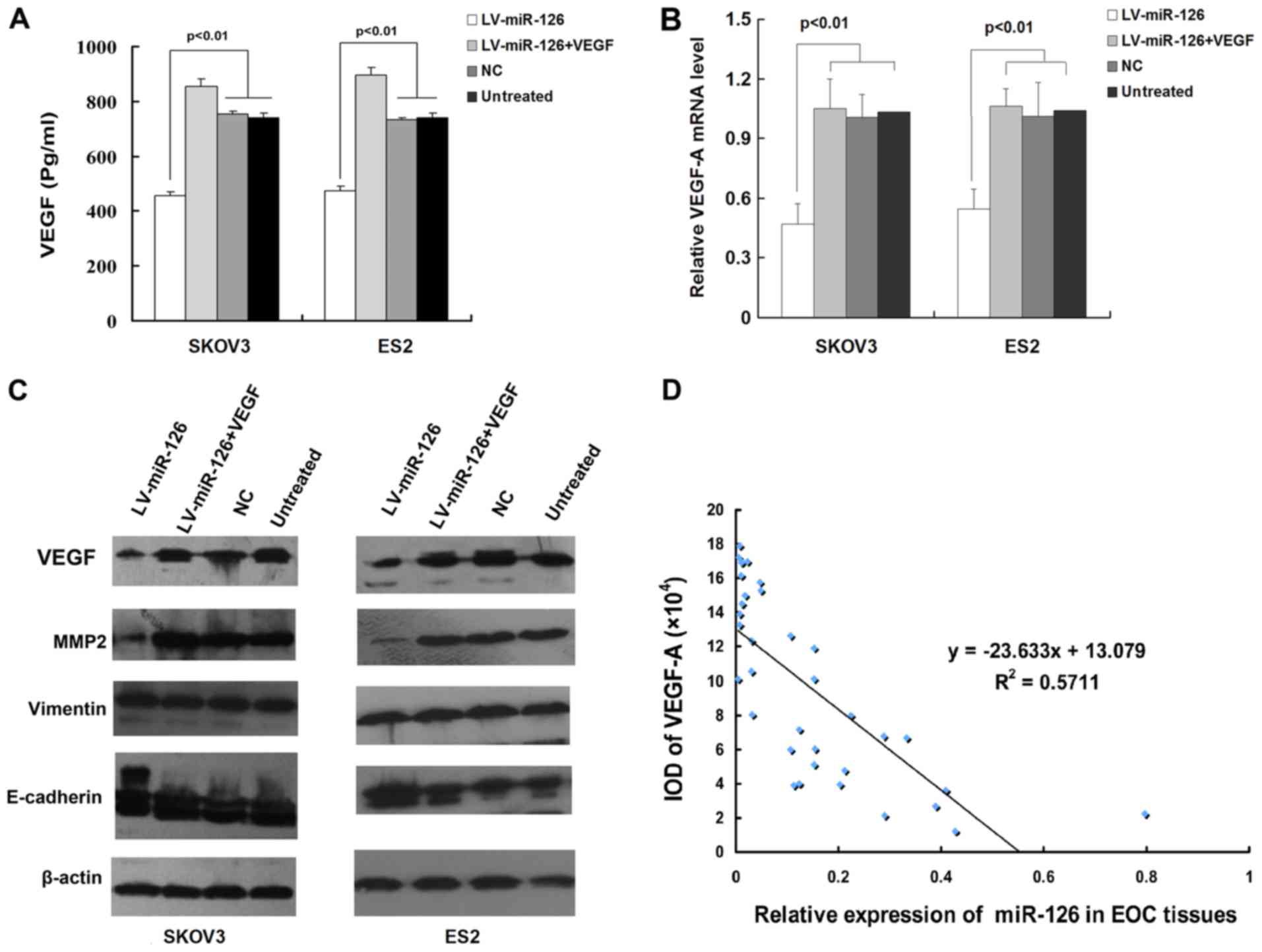
miR-126 regulates the expression of
VEGF-A
It has previously been reported that miR-126 targets
VEGF-A in other tumors (5). In the
present study, to determine whether this is the case in epithelial
ovarian cancer, SKOV3 and ES2 cells were infected with
lentivirus-miR-126 (LV-miR-126), and the expression levels of
VEGF-A were determined by RT-qPCR and ELISA. As shown in Fig. 4A and B, the ectopic expression of
miR-126 decreased the expression of VEGF-A in SKOV3 and ES2 cells
at both the mRNA and protein level. Moreover, infection with
LV-miR-126 downregulated the expression of the VEGF-A effector,
MMP2, and upregulated the expression of the epithelial marker,
E-cadherin, while no difference was observed between the negative
control and untreated cells (Fig.
4C). Despite these findings, no reduction in the mesenchymal
marker, vimentin, was observed when miR-126 expression was
modulated (Fig. 4C). Furthermore,
miR-126 expression negatively correlated with VEGF-A expression in
epithelial ovarian cancer tissues (R2=0.5711; Fig. 4D).
Ectopic VEGF-A expression reverses the
miR-126-mediated effects on the invasion, proliferation defects and
tube formation of HUVECs
To determine whether in vitro phenotypes
associated with miR-126 expression can be reversed by the
restoration of VEGF-A expression, recombinant human VEGF-A was
added to LV-miR-126-infected SKOV3 and ES2 cells and the VEGF-A
levels were assessed. In these cells, the ectopic expression of
VEGF-A reversed the miR-126-imposed invasion and proliferation
defects (Fig. 5A-C). HUVECs were
identified morphologically and by immunofluorescence in terms of
origin and purity. It was found that >95% of the cells presented
typical features of endothelial cells, such as a cobblestone
pattern and the expression of Factor VIII related antigen and CD31
(Fig. 5D and E). Tube formation
assays revealed that the HUVECs treated with LV-miR-126-infected
supernatant formed fewer dendritic and tube-like structures than
what was formed by the control cells. However, the number of these
structures was increased on recombinant human VEGF spike (Fig. 5F).
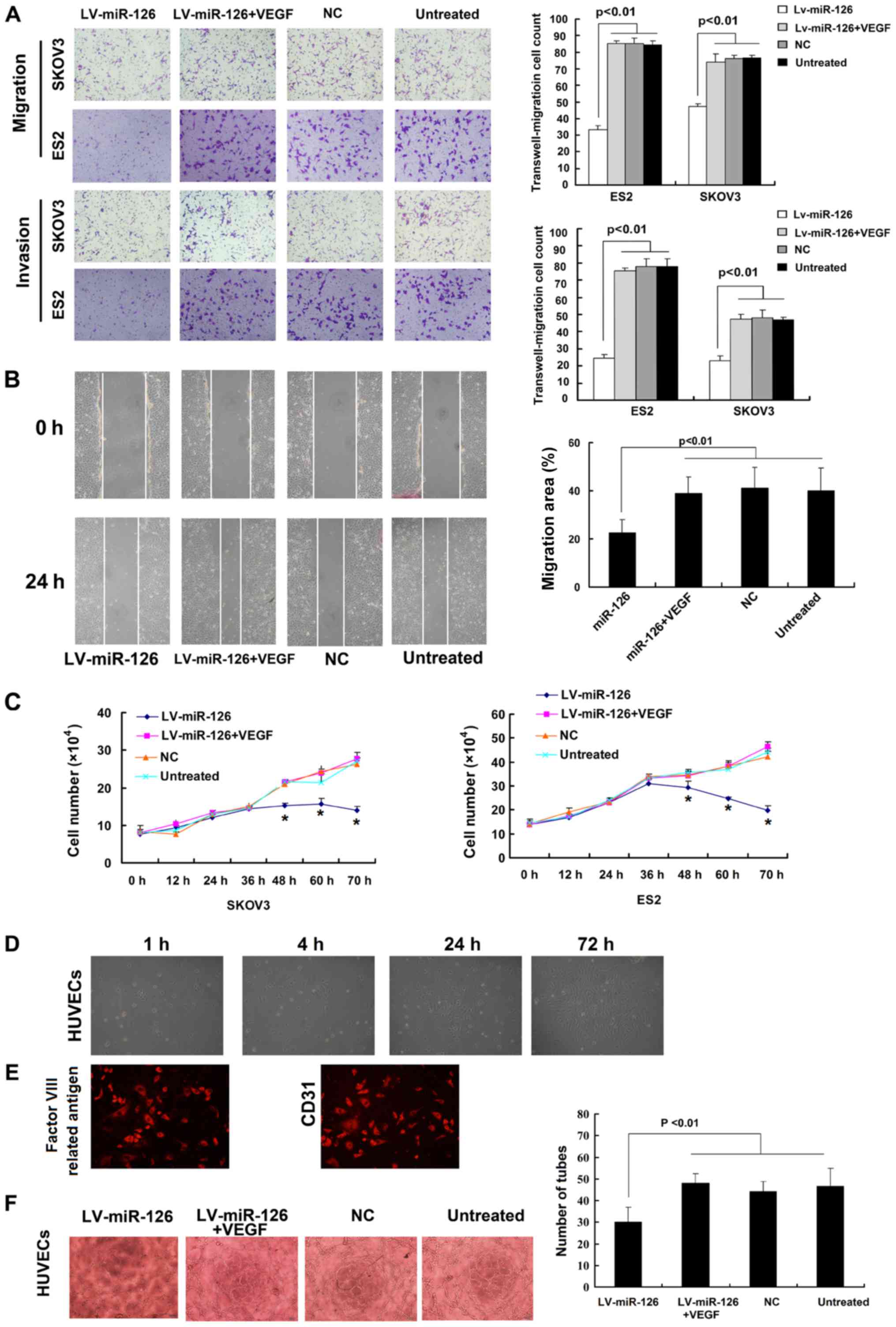 | Figure 5Ectopic expression of VEGF-A
attenuates the miR-126-mediated effects on metastasis and
proliferation. Recombinant human VEGF was added to
lentivirus-miR-126 (LV-miR- 126)-infected cells. (A) Transwell
migration and invasion assays was used to detect cell proliferation
in the LV-miR126, LV-miR-126 + VEGF, NC and untreated groups. (B)
Wound-healing assay to measure the migration of the LV-miR126,
LV-miR-126+VEGF, NC, untreated cells. (C) Trypan blue dye exclusion
assay was conducted to count the number of viable cells at 0, 12,
24, 36, 48, 60 and 72 h in SKOV3 and ES2 cells;
*P<0.05 compared to untreated cells. (D and E)
Identification for isolated HUVECs. (D) These endothelial cells
exhibited a polygonal shape and were arranged in a single layer of
paving stones under an inverted microscope. (E) Identification for
the isolated HUVECs by examining the factor VIII related antigen
and CD31. (F) Observation for branching points of dendritic and
tube-like structures. NC, negative control. |
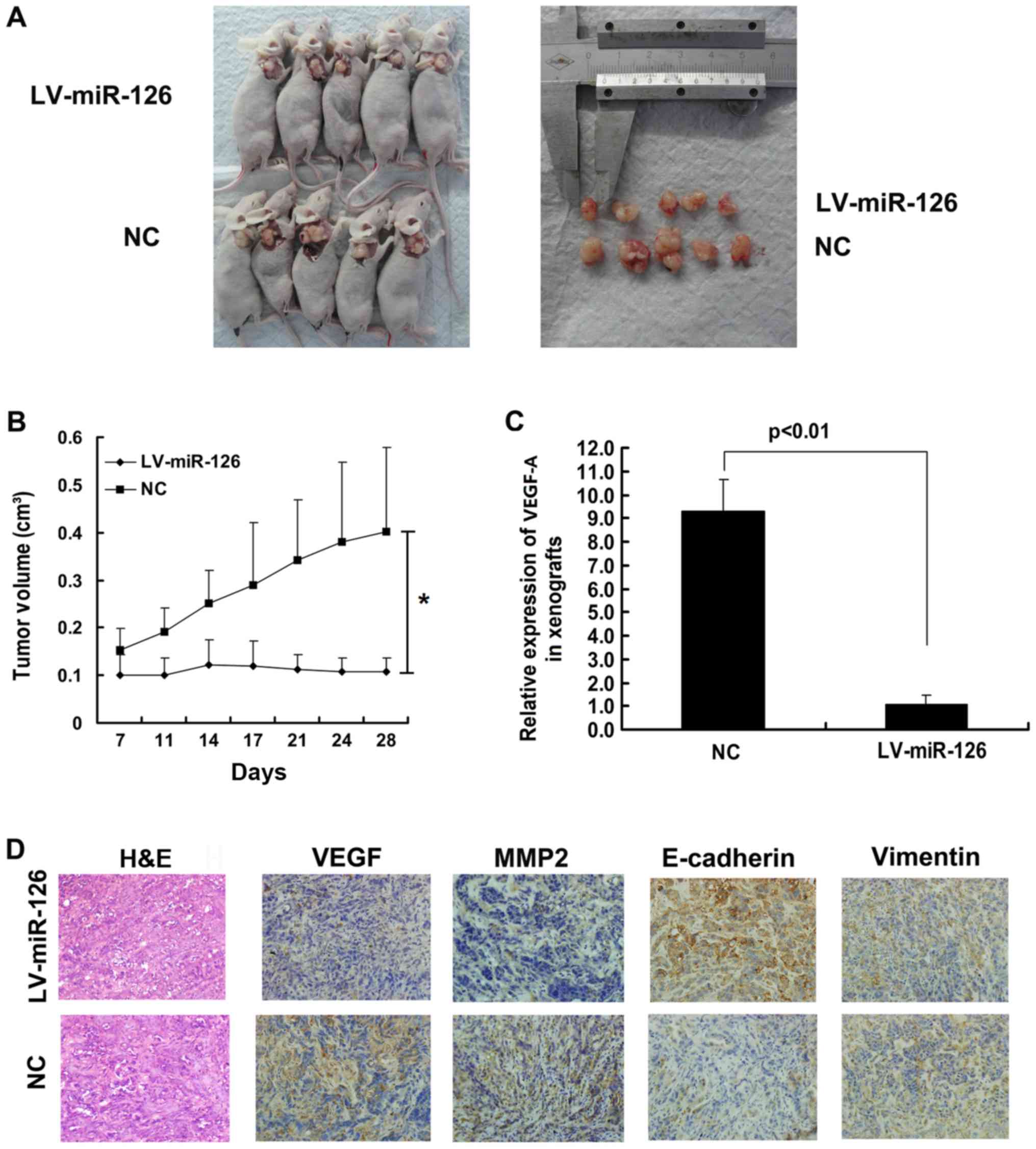
miR-126 inhibits tumor growth in
vivo
To investigate the role of miR-126 in vivo,
LV-miR-126 SKOV3 cells and NC cells were injected subcutaneously
into nude mice. The volume of subcutaneous tumors was measured
every 4 days. The tumor volume was suppressed to a greater extent
in the miR-126 overexpression group than in the NC group (Fig. 6A and B). Moreover, the results of
RT-qPCR revealed that the VEGF-A mRNA level was much lower in the
LV-miR-126 SKOV3 cells than in the NC cells (Fig. 6C). Immunohistochemistry was
performed to investigate the levels of VEGF-A, MMP2, vimentin and
E-cadherin in tumor xenografts. The higher expression of
E-cadherin, and the lower expression of VEGF-A and MMP2 was found
in the LV-miR-126 xenografts compared with the NC xenografts
(Fig. 6D).
Discussion
In ovarian cancer, previous expression profiling of
clinical tumors has provided contradictory findings as regards the
status of miR-126. One study found it to be highly expressed
(19), while it was found to be
downregulated in another (20).
The specific status of miR-126 in ovarian cancer has been far from
conclusive as these profiling studies assessed neither tumor
contents nor purified epithelial cells via microdissection prior to
RNA extraction. Moreover, Chip-based profiling needs to be
validated on a platform with more specificity. Previous studies
have relied on established cell lines (13,21),
which cannot simulate clinical carcinomas in a perfect manner. Cell
lines, for example, accumulate genetic alterations in culture.
Similar to other cancers, epithelial ovarian cancer is also a
heterogeneous population of cells. In the present study, using LMD,
the interference of confounding factors was minimized. The results
revealed that the expression of miR-126 was significantly decreased
in epithelial ovarian cancer compared with normal ovarian
epithelial cells.
VEGF family members, such as VEGF-A, VEGF-B, VEGF-C,
VEGF-D, VEGF-E and PGF are essential modulators of angiogenesis.
Previous studies have demonstrated that VEGF-A is the direct target
of miR-126 in various tumors (22-26).
Phase III clinical trials have provided encouraging results for
anti-VEGF therapy in ovarian cancer (27). It has been reported that
bevacizumab combined with chemotherapy improves progression-free
survival and the objective response rate in patients with
refractory ovarian cancer (28).
Consistent with a previous in vitro study, the results of
the present study demonstrated that miR-126 targeted VEGF-A in ES2
and SKOV3 cells and that miR-126 expression negatively correlated
with VEGF expression levels in epithelial ovarian cancer tissues.
Moreover, the ectopic expression of miR-126 impeded angiogenesis in
the tube formation assay. It is important to note that the addition
of recombinant VEGF-A could not fully mitigate the miR-126-mediated
effects, highlighting the pleiotropic effects of miR-126. It is
necessary to point out that the present study was correlational and
was mainly focused on the association between miR-126 and VEGF-A.
There was no direct evidence that miR-126 directly targeted VEGF-A
in ovarian cancer and investigating the effects of miR-126 on the
other VEGF family members and/or various isoforms of VEGF-A will
better reveal the association between miR-126 and VEGF family
members. Further studies are thus required.
In conclusion, the present study demonstrated that
miR-126 expression was decreased in ovarian cancer and that the
decreased expression of miR-126 promoted ovarian cancer
angiogenesis and invasion by targeting VEGF-A. Moreover, as
metastases and proliferation are responsible for patient mortality
in ovarian cancer, the ability of miR-126 to impede angiogenesis
and invasion may prove to be clinically useful.
Acknowledgments
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81402166).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article or are available from the
corresponding author on reasonable request.
Authors' contributions
The experiments were designed by JCao and SW. LY and
LL performed the experiments (with the assistance of DH, QH and
JCai) and were responsible for data analysis and the drafting of
the article. JCao and SW revised the article critically for
intellectual content. All authors have read and approved the final
manuscript.
Ethics approval and consent to
participate
Tumor and normal specimens were obtained from the
Gynaecologic Tissue Bank of Wuhan Union Hospital with written
informed consent and ethical approval. The present study was
approved by the ethical committee of Union Hospital, Tongji Medical
College, Huazhong University of Science and Technology, China. In
addition, human umbilical cords were collected from pregnant women
with written informed consent and ethical approval by Ethics
Committee of Union Hospital, Tongji Medical College, Huazhong
University of Science and Technology, Wuhan, China. All animal
experiments were approved by the Institutional Animal Care and Use
Committee at Tongji Medical College, Huazhong University of Science
and Technology.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Meister J and Schmidt MHH: miR-126 and
miR-126*: New players in cancer. ScientificWorldJournal.
10:2090–2100. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Fish JE, Santoro MM, Morton SU, Yu S, Yeh
RF, Wythe JD, Ivey KN, Bruneau BG, Stainier DY and Srivastava D:
miR-126 regulates angiogenic signaling and vascular integrity. Dev
Cell. 15:272–284. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang Y, Yang P, Sun T, Li D, Xu X, Rui Y,
Li C, Chong M, Ibrahim T, Mercatali L, et al: miR-126 and
miR-126* repress recruitment of mesenchymal stem cells
and inflammatory monocytes to inhibit breast cancer metastasis. Nat
Cell Biol. 15:284–294. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ebrahimi F, Gopalan V, Smith RA and Lam
AK: miR-126 in human cancers: Clinical roles and current
perspectives. Exp Mol Pathol. 96:98–107. 2014. View Article : Google Scholar
|
|
6
|
Wang S, Aurora AB, Johnson BA, Qi X,
McAnally J, Hill JA, Richardson JA, Bassel-Duby R and Olson EN: The
endothelial-specific microRNA miR-126 governs vascular integrity
and angiogenesis. Dev Cell. 15:261–271. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lagos-Quintana M, Rauhut R, Yalcin A,
Meyer J, Lendeckel W and Tuschl T: Identification of
tissue-specific microRNAs from mouse. Curr Biol. 12:735–739. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roviello G, Bachelot T, Hudis CA,
Curigliano G, Reynolds AR, Petrioli R and Generali D: The role of
bevacizumab in solid tumours: A literature based meta-analysis of
randomised trials. Eur J Cancer. 75:245–258. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Pan C, Stevic I, Müller V, Ni Q,
Oliveira-Ferrer L, Pantel K and Schwarzenbach H: Exosomal microRNAs
as tumor markers in epithelial ovarian cancer. Mol Oncol.
12:1935–1948. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Prahm KP, Høgdall C, Karlsen MA,
Christensen IJ, Novotny GW and Høgdall E: Identification and
validation of potential prognostic and predictive miRNAs of
epithelial ovarian cancer. PLoS One. 13:e02073192018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu G, Cao L, Zhu J, Tan Z, Tang M, Li Z,
Hu Y, Yu R, Zhang S, Song L, et al: Loss of RBMS3 confers platinum
resistance in epithelial ovarian cancer via activation of
miR-126-5p/ β-catenin/CBP signaling. Clin Cancer Res. 25:1022–1035.
2019. View Article : Google Scholar
|
|
12
|
Xiang G and Cheng Y: MiR-126-3p inhibits
ovarian cancer proliferation and invasion via targeting PLXNB2.
Reprod Biol. 18:218–224. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Luo P, Fei J, Zhou J and Zhang W:
microRNA-126 suppresses PAK4 expression in ovarian cancer SKOV3
cells. Oncol Lett. 9:2225–2229. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cai J, Li T, Huang B, Cheng H, Ding H,
Dong W, Xiao M, Liu L and Wang Z: The use of laser microdissection
in the identification of suitable reference genes for normalization
of quantitative real-time PCR in human FFPE epithelial ovarian
tissue samples. PLoS One. 9:e959742014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Xie H, Zou L, Zhu J and Yang Y: Effects of
netrin-1 and netrin-1 knockdown on human umbilical vein endothelial
cells and angio-genesis of rat placenta. Placenta. 32:546–553.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
17
|
Li Q, Cheng K, Wang AY, Xu QG, Fu ZF, He
SY and Xu PX: microRNA-126 inhibits tube formation of HUVECs by
interacting with EGFL7 and down-regulating PI3K/AKT signaling
pathway. Biomed Pharmacother. 116:1090072019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Guo J, Cai J, Yu L, Tang H, Chen C and
Wang Z: EZH2 regulates expression of p57 and contributes to
progression of ovarian cancer in vitro and in vivo. Cancer Sci.
102:530–539. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bu J, Li H, Li XY, Liu LH, Sun W and Xiao
T: Prognostic role of microRNA-126 for survival in malignant
tumors: A systematic review and meta-analysis. Dis Markers.
2015:7394692015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gu L, Li H, Chen L, Ma X, Gao Y, Li X,
Zhang Y, Fan Y and Zhang X: MicroRNAs as prognostic molecular
signatures in renal cell carcinoma: A systematic review and
meta-analysis. Oncotarget. 6:32545–32560. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Luo J, Zhu C, Wang H, Yu L and Zhou J:
MicroRNA-126 affects ovarian cancer cell differentiation and
invasion by modulating expression of vascular endothelial growth
factor. Oncol Lett. 15:5803–5808. 2018.PubMed/NCBI
|
|
22
|
Chen H, Li L, Wang S, Lei Y, Ge Q, Lv N,
Zhou X and Chen C: Reduced miR-126 expression facilitates
angiogenesis of gastric cancer through its regulation on VEGF-A.
Oncotarget. 5:11873–11885. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen CY, Su CM, Hsu CJ, Huang CC, Wang SW,
Liu SC, Chen WC, Fuh LJ and Tang CH: CCN1 promotes VEGF production
in osteoblasts and induces endothelial progenitor cell angiogenesis
by inhibiting miR-126 expression in rheumatoid arthritis. J Bone
Miner Res. 32:34–45. 2017. View Article : Google Scholar
|
|
24
|
Kong D, Ying B, Zhang J and Ying H: The
anti-osteosarcoma property of ailanthone through regulation of
miR-126/VEGF-A axis. Artif Cells Nanomed Biotechnol. 47:3913–3919.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu B, Peng XC, Zheng XL, Wang J and Qin
YW: MiR-126 restoration down-regulate VEGF and inhibit the growth
of lung cancer cell lines in vitro and in vivo. Lung Cancer.
66:169–175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Caporali S, Amaro A, Levati L, Alvino E,
Lacal PM, Mastroeni S, Ruffini F, Bonmassar L, Antonini Cappellini
GC, Felli N, et al: miR-126-3p down-regulation contributes to
dabrafenib acquired resistance in melanoma by up-regulating ADAM9
and VEGF-A. J Exp Clin Cancer Res. 38:2722019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cortez AJ, Tudrej P, Kujawa KA and
Lisowska KM: Advances in ovarian cancer therapy. Cancer Chemother
Pharmacol. 81:17–38. 2018. View Article : Google Scholar :
|
|
28
|
Pujade-Lauraine E, Hilpert F, Weber B,
Reuss A, Poveda A, Kristensen G, Sorio R, Vergote I, Witteveen P,
Bamias A, et al: Bevacizumab combined with chemotherapy for
platinum-resistant recurrent ovarian cancer: The AURELIA open-label
randomized phase III trial. J Clin Oncol. 32:1302–1308. 2014.
View Article : Google Scholar : PubMed/NCBI
|